Abstract
The recombinant RNA-dependent RNA polymerase of the bovine viral diarrhea virus specifically requires a cytidylate at the 3′ end for the de novo initiation of RNA synthesis (C. C. Kao, A. M. Del Vecchio, and W. Zhong, Virology 253:1–7, 1999). Using RNAs containing nucleotide analogs, we found that the N3 and C4-amino group at the initiation cytidine were required for RNA synthesis. However, the ribose C2′-hydroxyl of the initiating cytidylate can accept several modifications and retain the ability to direct synthesis. The only unacceptable modification is a protonated C2′-amino group. Quite strikingly, the recognition of the functional groups for the initiation cytidylate and other template nucleotides are different. For example, a C5-methyl group in cytidine can direct RNA synthesis at all template positions except at the initiation cytidylate and C2′-amino modifications are tolerated better after the +11 position. When a 4-thiouracil (4sU) base analog that allows only imperfect base pairing with the nascent RNA is placed at different positions in the template, the efficiency of synthesis is correlated with the calculated stability of the template-nascent RNA duplex adjacent to the position of the 4sU. These results define the requirements for the specific interactions required for the initiation of RNA synthesis and will be compared to the mechanisms of initiation by other RNA-dependent and DNA-dependent RNA polymerases.
De novo (primer-independent) initiation is an important mechanism for viral RNA replication that requires the specific and appropriate interactions of at least four components: (i) an RNA template with a virus-specific initiation nucleotide at or near the 3′ end; (ii) an initiation nucleoside triphosphate (NTP); (iii) a second NTP; and (iv) an RNA-dependent RNA polymerase (RdRp). Improper recognition may reduce or inhibit efficient RNA synthesis. Little is known about specific recognition between RdRp and RNA template moieties for de novo initiation of viral RNA synthesis. Furthermore, the interactions may change as the polymerase undergoes conformational changes during synthesis (11, 32).
Bovine viral diarrhea virus (BVDV) is a member of the genus Pestivirus in the Flaviviridae family, which includes human and animal pathogens, such as Hepatitis C virus (HCV), Dengue virus, and Yellow fever virus (10, 34, 55). BVDV is an enveloped virus containing a single-stranded positive-sense RNA genome (55). In addition to being an important pathogen of livestock (51), BVDV is a model system for HCV infection and for building chimeric viruses with HCV (14, 31, 61, 68).
The BVDV RNA genome is translated as a polyprotein through a cap-independent mechanism after infection. The polyprotein is further processed into individual structural and nonstructural proteins by a combination of cellular and viral proteases (10). The structural proteins are located in the N-terminal portion of the polyprotein, while those associated with replication are present in the C-terminal portion (68). Among the nonstructural proteins, NS5B is an RdRp that can synthesize RNA (3, 69). Double-stranded, RNase-resistant replicative-form RNA and partially double-stranded replication intermediates have been observed early in infection in cells infected by BVDV. When the viral RNA was fully denatured, single-stranded RNAs of genome length were observed (16, 17), indicating that de novo initiation is probably used in vivo. De novo initiation of RNA synthesis of BVDV NS5B has been demonstrated with recombinant BVDV RdRp in a process that requires an initiation cytidylate (+1C) positioned at or near the 3′ end of the template (27). A +1G substitution abolished RNA synthesis, while base substitutions at +2 and +3 had less effect. In addition, a template containing a +1U can interact with BVDV NS5B but initiates RNA synthesis at only 5% of the level found with template containing a +1C. These results demonstrate the highly specific recognition of the moieties in +1C by BVDV NS5B. However, the moieties at and near the template initiation nucleotide required for RNA synthesis have not been systematically examined in any viral RNA template.
We used chemically synthesized RNA containing nucleotide analogs of the +1C to address the features in the RNA required for efficient RNA synthesis in vitro. We also compared the recognition between +1C and other template nucleotides to investigate whether RdRp-template interactions change during RNA synthesis. Initiation of RNA synthesis was found to require several moieties in the cytosine base, while several substitutions at the +1C ribose were acceptable for efficient initiation of RNA synthesis. The riboses at the +2 and +3 positions also contribute to stable interaction with RdRp. Also, the recognition among of base and ribose moieties was found to differ in a position-dependent manner.
MATERIALS AND METHODS
RdRp activity assays and template competition assays.
All RNAs were purified from the denaturing polyacrylamide gels, quantified by spectrophotometry, checked for quality and quantity on a denaturing polyacrylamide gel, and used at 0.125 μM per RdRp reaction. BDVD NS5B was prepared from recombinant Escherichia coli as described previously (62). Standard in vitro RdRp assay mixtures consisted of 5 pmol of the template (unless stated otherwise) and 50 ng of BVDV NS5B in a 40 μl reaction mixture containing 20 mM sodium glutamate (pH 8.2), 12 mM dithiothreitol, 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 1 mM MnCl2, 500 μM GTP, 200 μM ATP, 200 μM UTP, and 250 nM [α-32P]CTP (400 Ci/mmol, 10 mCi/ml; Amersham). MnCl2 was used to increase the level of RNA synthesis. The reaction mixtures were incubated for 60 min at 25°C, and the reactions were stopped by phenol-chloroform (1:1, vol/vol) extraction. The products were precipitated in 6 volumes of ethanol plus 5 μg of glycogen and 0.4 M of ammonium acetate.
Template competition assays were performed with increasing concentrations of various RNA competitors (5, 10, 20, 30, and 40 pmol) and 25 ng of NS5B. The reaction mixture also contained 5 pmol of a template named 3init, which directs a 27-nucleotide (nt) product (27). The amounts of the products generated from the 3init reference template were measured, and the 50% inhibitory concentrations (IC50s) were determined as the concentration of the competitors necessary to reduce synthesis from 3init by 50%.
Analysis of RdRp products.
An 8-μl volume of loading buffer (45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue, and 0.04% [wt/vol] xylene cyanol) was added to each ethanol-precipitated product, and the mixture was denatured by heating at 90°C for 3 min prior to electrophoresis. The products were analyzed on 20% polyacrylamide denaturing gels containing 7 M urea, and the gels were wrapped in plastic and exposed to film at −80°C. The amount of radiolabel incorporated into RdRp products was measured with a PhosphorImager (Molecular Dynamics). The values obtained were expressed relative to those produced from (−)21 to derive the percentage of the activity of modified template. All values represent the means of at least three independent experiments.
RNA templates.
All RNAs were chemically synthesized by Oligos Etc. (Wilsonville, Oreg.) except those in Table 1, which were synthesized using the T7 RNA polymerase. Transcription reactions used cDNA from Operon Inc. (Alameda, Calif.) as described by Kao et al. (27). All the RNAs were purified from denaturing 7 M urea–20% polyacrylamide gels, visually inspected after being stained with toluidine blue, and quantified by using spectrophotometry.
TABLE 1.
Base and ribose substitutions at different positions within (−)21
| Template | % synthesis (mean ± SD)a |
|---|---|
| (−)21 | 100 ± 4 |
| +9 C/U | 127 ± 2 |
| +14 U/ | 128 ± 8 |
| +15 C/U | 109 ± 6 |
| +15 C/A | 105 ± 3 |
| +15 C/G | 150 ± 4 |
| +15 C/dC | 89 ± 10 |
| +16 U/C | 99 ± 21 |
| +20 C/U | 124 ± 6 |
| +20 C/dC | 86.5 ± 3.5 |
All syntheses are normalized to that of (−)21, the prototype template. The percentages are means and SD of at least three independent experiments.
Km and Vmax measurements.
Km and Vmax values of templates for recombinant BVDV NS5B used the same conditions as those described for RdRp activity assay, except that 10 different template concentrations from 0 to 250 nM were used. The Km and Vmax values were obtained by using software created by D. Gilbert (Enzyme Kinetics, Indiana University). Each value is the average of two independent assays.
Thermodynamic calculations of nascent RNA-template interaction.
ΔG° values were expressed as the sum of the values from each base pair by using the tables provided in reference 57. The results were plotted against the levels of synthesis by using JMP version 3.1.5 (SAS Institute Inc., 1989 to 1994).
RESULTS
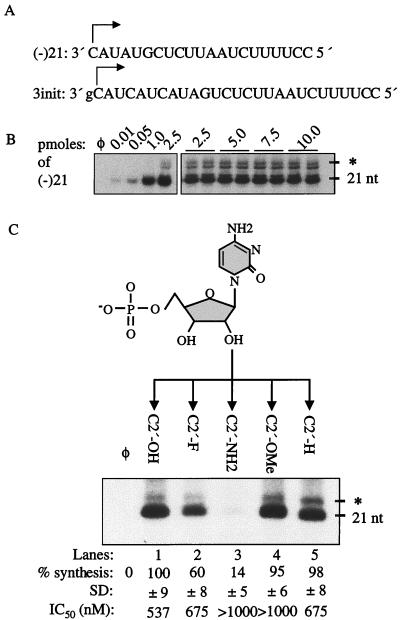
The requirement for a +1C for efficient RNA synthesis indicates that specific nucleotide functional groups are recognized during the initiation of RNA synthesis in vitro. RNA (−)21, containing 21 nt at the 3′ end of the minus-strand BVDV genome, was the prototype used to analyze the nucleotide moieties needed to direct RNA synthesis (Fig. 1A). (−)21 directs the synthesis of a prominent 21-nt RNA, which is correctly initiated and terminated, along with a small amount of 22- and about 24-nt products, presumably due to nontemplated terminal nucleotidyltransferase activity (Fig. 1B). To perform all experiments with NS5B as the limiting component, the synthesis from an increasing amount of (−)21 was determined. Template concentration-dependent synthesis was observed only when (−)21 was present at 60 nM or less (Fig. 1B). Therefore, we routinely use 120 nM template in reactions; thus, changes in the amount of RNA product directed by modified templates will reflect a preference for the polymerase. The results cited below were quantified from three independent RdRp assays whose results were normalized to those from (−)21.
FIG. 1.
(A) RNA sequences used in this study. (−)21, the prototype sequence, is designed by the 21 nt at the 3′ end of the minus-strand BVDV genome and used for analyzing template nucleotides moieties. The 3init template is a reference template for the template competition assays (25). The initiation cytidylates are denoted by arrows. (B) Template titration assay. The (−)21 is present at seven different concentrations, indicated by the numbers above the autoradiograph. φ denotes a control reaction without RNA template. The sizes of the correctly terminated products from (−)21 and products containing a terminally added nucleotide (∗) are indicated to the right of the autoradiogram. (C) Four ribose C2′-hydroxyl modifications are shown below the structure of the +1C ribose, followed by an autoradiogram. The 21-nt RNAs synthesized from (−)21 were separated by denaturing polyacrylamide gel electrophoresis (20% polyacrylamide) and visualized by autoradiography. The quantification, normalized to the control C2′-OH and averaged from at least three independent experiments, is shown below the autoradiogram at each modified template, followed by standard deviation (SD) and IC50.
Ribose modifications.
The role of the +1 ribose in the initiation of RNA synthesis was examined with four templates where the ribose C2′ hydroxyl (C2′-OH) was replaced with fluorine (−F), methoxy (-OCH3), amine (-NH2), and deoxy (-H). These modifications were selected because they affect potential hydrogen bond formation, the size of the C2′ moiety, and/or the equilibrium between the ribose C2′-endo and C3′-endo conformations (58). The RNAs modified with deoxy, OCH3, or F directed RNA synthesis at 98, 95, and 60% relative to (−)21, respectively (Fig. 1C). The results indicate that the C2′-OH does not need to form a hydrogen bond to NS5B during the initiation of RNA synthesis, since F and deoxy cannot accept and donate a hydrogen, respectively. The lower level of RNA synthesis from F compared to deoxy and OCH3 may be due in part to the electronegativity of the F. Ribose with C2′-OH (favoring C3′-endo) and C2′-deoxy (favoring C2′-endo) both directed efficient RNA synthesis, indicating that both sugar conformations at the +1 position were acceptable. The OCH3 substitution directed synthesis at levels comparable to those for (−)21, suggesting that NS5B can tolerate an increase in the size of the modified the ribose C2′ at the +1 position.
RNA C2′-NH2 directed synthesis at only 14% relative to that for (−)21. The pKa of the primary amine is 10.0 ± 0.2 (38), and we speculate that the decreased synthesis from RNA C2′-NH2 could be due to a partial positive charge of the amino group. Since our RdRp reactions were normally carried out at pH 8.2, the protonated NH3+ form should predominate. If a partial positive charge is less strongly preferred, RNA synthesis should increase at higher pH, where the NH2 form is increased. RNA syntheses from C2′-NH2 and (−)21 were tested at several pH values between 5.0 and 9.0 and normalized to those in parallel reaction mixtures containing (−)21 to eliminate possible pH effects on NS5B. At pH 9.0, we observed a reproducible 130% ± 6% increase in RNA synthesis from C2′-NH2 relative to (−)21. In contrast, the relative RNA synthesis was reduced to 33% ± 1% at pH 5.0. These results show that the low level of RNA synthesis of C2′-NH2 was probably due to the existence of an unacceptable NH3+ group at the +1 ribose C2′ position.
To examine whether the efficiency of RNA synthesis reflects the ability of NS5B to interact with the templates, competition assays were carried out using a 3init as a reference template to produce a 27-nt product (Fig. 1A). RNA synthesized from a constant amount of 3init was quantified as a function of competitor concentration, which may produce easily distinguishable 21- and 22-nt RNAs (see Materials and Methods). The IC50 was determined from the mean of three independent experiments. The IC50 of (−)21 was 537 nM. C2′-F and C2′-H had IC50 of 675 nM, slightly increased relative to that of (−)21 (Fig. 1C). In contrast, the IC50 of C2′-OCH3 and C2′-NH2 were greater than 1 μM (Fig. 1C). Despite this, C2′-OCH3 was able to direct synthesis at 95% of that for (−)21 while C2′-NH2 directed synthesis at 14%. These results suggest that a weaker interaction with +1C does not necessarily lead to lower levels of RNA synthesis.
Phosphodiester modifications.
Ionic interaction between phosphodiester backbone of nucleic acids and protein side chains is generally required to stabilize their interaction (45). We tested modifications at the phosphodiester group connecting +1C and +2A to determine (i) the necessity of the negatively charged phosphate in interacting with NS5B, and (ii) whether the polarity of the phosphodiester group affects the interaction between NS5B and RNAs.
RNAs containing specific phosphates (PO43−) replaced with thiolates (SO42−) were made in the context of (−)21 to decrease the localized charge by having one additional electron pair in the oxygens. RNAs with thiolates placed between nt +1 and +2 or between nt +2 and +3 or at both positions directed RNA synthesis at 123% ± 6%, 90% ± 4%, and 113% ± 9%, respectively. In the template competition assay, the RNA with two thiolates at between +1 and +3 had an IC50 of 390 nM, while the singly thiolate-modified template had an IC50 similar to that of (−)21 (M.-J. Kim, unpublished data). These results indicate that an RNA backbone with reduced negative charge near the initiation site allowed more efficient interaction with NS5B and improved RNA synthesis.
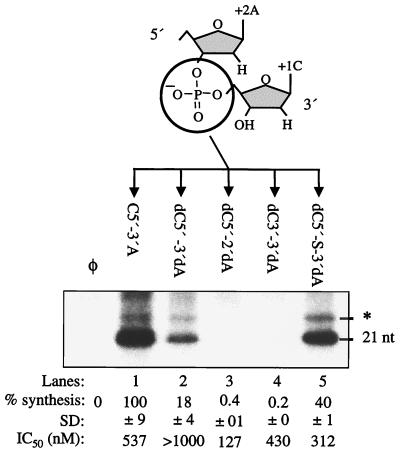
Templates with deoxyriboses at the +1 and +2 positions were used to examine the effects of additional phosphodiester modifications. Some modifications are available only in the deoxyribose form; therefore, we first determined whether deoxyriboses between the +1 and +2 positions in template dC5′-3′dA will affect RNA synthesis. Synthesis from dC5′-3′dA was only 18% of that from (−)21 (Fig. 2, lane 2), in contrast to the results obtained with a deoxy at +1C, which was nearly wild type in RNA synthesis (Fig. 1C, lane 5). Thus, the ribose C2′ moieties at other template positions are recognized differently from those at +1C. These differences will be addressed more thoroughly below.
FIG. 2.
Phosphodiester group modifications are shown below the schematic representation of +1C and +2A nucleotides. C5′-3A′, a control in this experiment, represents the phosphodiester between +1C and +2A in (−)21, and all quantifications were normalized to the synthesis from (−)21. The quantification with SD of RNA products and IC50s are shown below the autoradiogram.
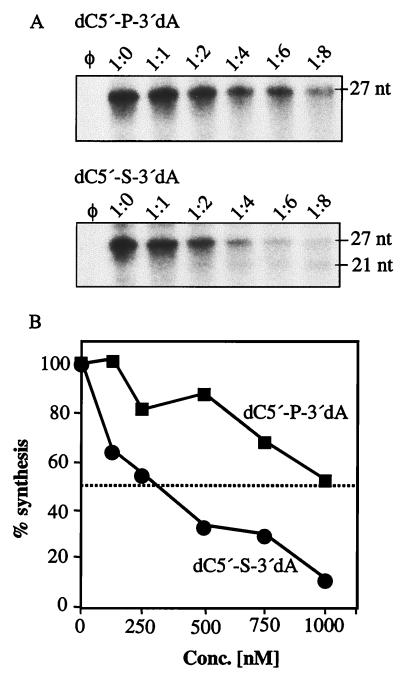
Since a thiolate between the +1 and +2 positions in (−)21 increased RNA synthesis, we placed one between +1dC and +2dA. The resultant RNA directed synthesis at 40%, in comparison to the 18% directed by dC5′-3′dA (Fig. 2, lanes 5 and 2, respectively). This twofold increase is probably due to a more stable interaction with NS5B, as previously seen with thiolate-modified (−)21. In support of this hypothesis, the IC50 of dC5′-P-3′dA was higher than 1 μM while dC5′-S-3′dA had an IC50 of 312 nM (Fig. 3). A decrease in the negative charge in the phosphodiester between the +1 and +2 positions may compensate for the detrimental effect of the deoxyribose modification at +2 position.
FIG. 3.
A thiolate replacement between +1dC and +2dA enhances the interaction with NS5B. (A) Autoradiograms showing products generated from 3init in the presence of competitors, dC-P-dA and dC-S-dA. φ denotes a control reaction without RNA template. The 1:0 reaction mixture contains 5 pmol of 3init without competitor. The amounts of competitor added to the reaction mixtures were 5 pmol (1:1), 10 pmol (1:2), 20 pmol (1:4), 30 pmol (1:6), and 40 pmol (1:8); 27 and 21 nt represent the products from 3init and (−)21, respectively. (B) Reaction products from the template competition assays were quantified, and the mean of at least three independent experiments was plotted, as the percentage of 3init synthesized in the absence of any competitor, against the concentration of competitor.
Next, we examined the effect of the phosphodiester group polarity on RNA synthesis. A 5′-to-2′ polarity between positions +1 and +2 in template dC5′-2′dA and a 3′-to-3′ phosphodiester in template dC3′-3′dA were made and tested. Both RNAs failed to direct RNA synthesis (Fig. 2, lanes 3 and 4), indicating that NS5B requires a 5′-to-3′ polarity between nucleotides for RNA synthesis. In template competition assays, dC3′-3′dA had an IC50 of 430 nM (lane 4). Template dC5′-2′dA had an IC50 of 127 nM despite being unable to direct RNA synthesis (lane 3). The altered polarity may increase nonproductive interaction with NS5B that resulted in no RNA synthesis.
Base modifications.
Templates containing a +1U or a +1G cannot efficiently direct RNA synthesis (27), indicating that specific cytosine moieties are required for efficient initiation of RNA synthesis and/or stable interaction with NS5B. Cytosine C4-amino and N3 are probably important, since they are absent in uridine. The cytosine C4-amino and N3 may be required to form Watson-Crick (W-C) hydrogen bonds with the initiation substrate, GTP, and/or interact directly with NS5B.
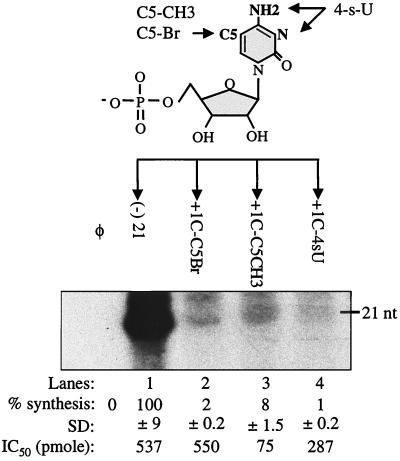
The base analog 4-thiouridine (4sU) was incorporated at the +1 position to change the cytosine N3 and C4-amino to an imino and a C4-thio, respectively. Unlike natural nucleotides, 4sU will make weakened W-C hydrogen bonds with ATP or GTP by being a poor hydrogen bond acceptor (23). RNA synthesis from +1C-4sU was 1% relative to that from (−)21 (Fig. 4, lane 4), demonstrating that the C4-amino and N3 were essential for initiation. In the template competition assay, +1C-4sU had an IC50 of 287 nM (lane 4). This relatively low IC50 suggests that the modification caused an inappropriately tighter interaction with NS5B.
FIG. 4.
The structure of +1C with moieties targeted for modifications in bold. The arrows indicate defined changes in the particular functional groups by incorporating base analogs, 4sU, C5-CH3, and C5-Br. The autoradiogram of the RdRp products is below the schematic of the RNAs containing a modified cytidylate. Quantifications with mean percent synthesis, SD, and IC50 from the template competition assays are shown below the autoradiogram.
Next, we tested whether the non-hydrogen-bonding C5 position of +1C can be modified. RNA +1C-C5Br and +1C-C5CH3, containing a C5 bromyl and a C5 methyl adduct, respectively, failed to direct RNA synthesis (Fig. 4, lanes 2 and 3). The adducts may either sterically affect the proper fit of the initiation complex in the NS5B active site or alter base stacking with the +2A of the template. We used the template competition assay to determine whether interaction with NS5B is affected by the C5 adducts. +1C-C5Br did not appear to be affected in the interaction with NS5B, with an IC50 of 550 nM (lane 2). However, +1C-C5CH3 competed extremely well, with an IC50 of 75 nM (lane 3). This strong interaction may be due to increased hydrophobic interaction with NS5B or the +2A base. Additional experiments are needed to distinguish between these possibilities.
A difference in the recognition of +1C and other nucleotides in the template.
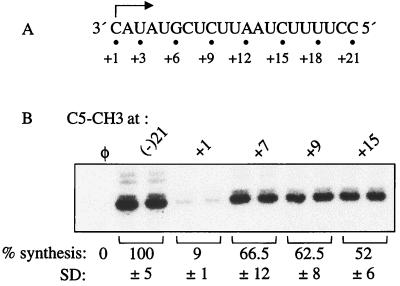
Specific recognition of the +1C led us to ask whether there is a difference in the recognition by NS5B of the initiation cytidylate and other nucleotides in the template. In general, W-C hydrogen bond formation is cited as the main determinant of fidelity for substrate incorporation in nucleic acids synthesis (66). The C5 of cytosine, which does not participate in hydrogen bond formation with GTP or NS5B, was targeted. A C5 methyl group was incorporated in the cytidylate at position +7, +9, or +15 in templates named +7C-C5CH3, +9C-C5CH3, and +15C-C5CH3, respectively. Since all three positions were cytidylates, a direct comparison with a modification at +1C is possible (Fig. 5A).
FIG. 5.
Recognition of the base C5 moieties. (A) The sequence of (−)21 from 3′ to 5′, with the initiation cytidylate indicated by an arrow and the nucleotide positions indicated by numbers. (B) Autoradiogram showing the RNA products from the templates modified with the base C5 at position +1, +7, +9, or +15. All quantifications were normalized to the synthesis from (−)21, and the mean and SD are shown below the autoradiogram. φ denotes a control reaction performed without template RNA.
RNAs +7C-C5CH3, +9C-C5CH3, and +15C-C5CH3 directed synthesis at 67, 63, and 52% relative to that for (−)21, respectively (Fig. 5B), indicating that C5-methyl adducts at all three positions were acceptable for RNA synthesis. This result is strikingly different from the results obtained with same modification at the +1C position, which abolished the synthesis.
Recognition of template riboses.
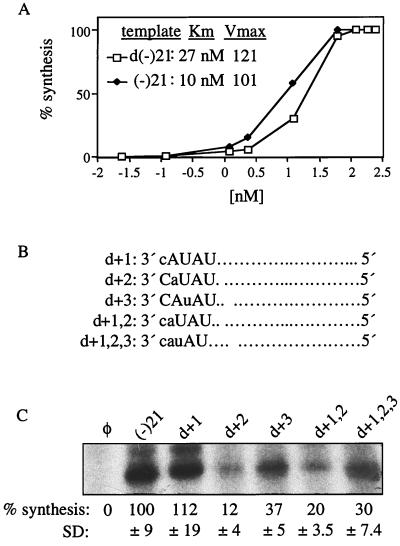
The brome mosaic virus (BMV) RNA-dependent RNA replicase can synthesize RNA from a single-stranded DNA template, but the level of synthesis is eightfold lower than that from RNAs of the identical sequence (46). However, BVDV NS5B has been previously reported to use a DNA template, producing approximately similar amounts of transcripts in comparison to those produced with the RNA template (27). The change of the +1 ribose to a deoxyribose did not observably affect RNA synthesis by the BVDV NS5B (Fig. 1C, lane 5), while deoxyriboses at both the +1C and +2A positions in template d+1,2 reduced RNA synthesis to 18% relative to that for (−)21 (Fig. 2, lane 2). These seemingly contradictory results led us to examine more carefully the use of DNA and RNA templates by the BVDV NS5B. We tested templates (−)21 and d(−)21 to measure Km and Vmax values. The Km for (−)21 was 10 nM, 2.7-fold lower than the Km for d(−)21 (27 nM). However, the Vmax for (−)21 and d(−)21 were apparently similar, at 101 and 121, respectively (Fig. 6A). These results suggest that d(−)21 can direct synthesis as efficiently as (−)21 at template concentrations above 60 nM (Fig. 6A). However, the interaction between NS5B and d(−)21 was less efficient than that between (−)21 and NS5B, indicating that some riboses, perhaps those near the +1C, are needed for proper interaction with NS5B. We do not understand why d+1,2, with only two deoxyriboses at +1C and +2A, was worse at directing synthesis than d(−)21, which is composed entirely of deoxyriboses. However, this result is reproducible and has also been observed with another RdRp (C. C. Kao, unpublished data).
FIG. 6.
Different requirement of the ribose C2′-hydroxyl. (A) Km and Vmax measured for (−)21 and d(−)21. Template concentrations used in the reactions were plotted on a log scale. Each quantification was the mean of two independent experiments. (B) Specific ribonucleotide changed to a deoxyribonucleotide in the context of (−)21. The names of RNAs used are shown to the left, followed by the nucleotides in the affected region. The capital letters represent ribonucleotides, and the lowercase letters denote deoxyribonucleotides. (C) Autoradiogram of the 21-nt RNA products from the templates indicated above. The quantifications, the mean of three independent experiments, are shown below the autoradiogram along with the SD. All values were normalized by the one from (−)21. φ denotes a control reaction lacking template RNA.
To further examine the ribose requirements for RNA synthesis, three templates chimeric for ribose and deoxyribose were made and tested (Fig. 6B). d+2, with a deoxyribose at only the +2 position, directed RNA synthesis at only 12% relative to that for (−)21 (Fig. 6C), indicating that the lower level of synthesis observed with the d+1,2 template was due to the loss of C2′-OH at the +2 position. The IC50 for d+2 was greater than 1 μM, indicating that the C2′-OH of +2A is required to interact with NS5B. Templates d+3 and d+1,2,3 directed synthesis at 37 and 30%, respectively, relative to (−)21 (Fig. 6C), indicating that the C2′-OH at the +3 position is also necessary for proper interaction with NS5B. However, templates with deoxyribose at the +15 or +20 position did not have such severe effects, directing synthesis at 89% ± 10% and 86.5% ± 3.5%, respectively, relative to (−)21 (Table 1).
Kao et al. (27) reported that base substitutions at position +2 and +3 were acceptable for RNA synthesis, suggesting that decreased synthesis from the d+1,2 and d+1,2,3 templates was due to position-dependent recognition of the ribose. To examine further whether base-specific template recognition by NS5B occurs at other positions, the cytidylate at position +9, +15, or +20 was changed to uridylate in RNAs +9C/U, +15C/U, and +20C/U, respectively. In contrast to the severe effect of a uridylate substitution at the +1C position (27), synthesis from all three RNAs was at least 109% relative to that for (−)21 (Table 1). Therefore, the specific preference of base functional groups was observed only with the initiation cytidylate. Taken together, the ribose C2′-OH at specific positions in the RNA helps NS5B to discriminate RNA and DNA molecules but the initiation base is has a specific requirement for recognition. Riboses at specific positions were also required for subgenomic RNA synthesis by the BMV replicase (47).
The BMV replicase transits from initiation to elongation after the synthesis of approximately eight or nine phosphodiester bonds (1, 53, 54). Similar transition points have been reported for other polymerases (for a review, see reference 11). A position-specific recognition of C2′-OHs in the RNA template (Fig. 6C) could correlate with distinct steps of RNA synthesis (i.e., initiation, transition, and elongation) by the BVDV NS5B. We have found that synthesis of template-length products is reduced on a molar basis from a template of 8 nt but not from one of 12 nt. RNAs longer than 8 nt may be necessary for stable interaction with NS5B (M.-J. Kim, unpublished data).
Template ribose C2′-NH2 modifications.
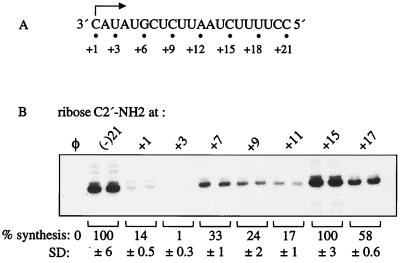
To further examine whether nucleotide recognition is dependent on the template position, we changed the ribose C2′-OH to C2′-NH2 at seven pyrimidines within (−)21 that should span initiation, transition, and elongation: +1, +3, +7, +9, +11, +15, and +17 (Fig. 7A). The ribose C2′-NH2 modifications from positions +1 to +11 all resulted in RNA synthesis of 1 to 33% relative to that for (−)21 (Fig. 7B). However, templates with the ribose C2′-NH2 at +15 or +17 directed synthesis at 100 and 58%, respectively (Fig. 7B). In terms of interaction with NS5B, templates with C2′-NH2 at +1 or +7 were poor competitors (IC50 < 1 μM), but templates with C2′-NH2 at +9, +11, +15, and +17 had similar IC50 to those of (−)21, indicating that the interactions with NS5B were not affected (Kim, unpublished). Unexpectedly, C2′-NH2 at +3 resulted in tighter interaction with NS5B (IC50 = 75 nM). Thus, inefficient RNA synthesis from templates with C2′-NH2 at +1, +3, +7, +9, or +11 was due to several factors, including the inappropriate interaction between NS5B and the ribose C2′ at specific positions in the template and a possible effect on the kinetics of RNA synthesis.
FIG. 7.
Effects of ribose C2′-NH2 modification. (A) The sequence of (−)21 shown from 3′ to 5′. The initiation cytidylate is indicated by an arrow, and the positions modified are shown by dots and numbers below. (B) Autoradiogram showing the 21-nt RNA products from the templates above, modified with the ribose C2′-NH2 at position +1, +3, +7, +9, +11, +15, or +17. Each modification is shown in duplicate. All quantifications were normalized based on (−)21, and the mean and SD are shown below the autoradiogram. φ denotes a control reaction lacking template RNA.
Template 4sU modifications.
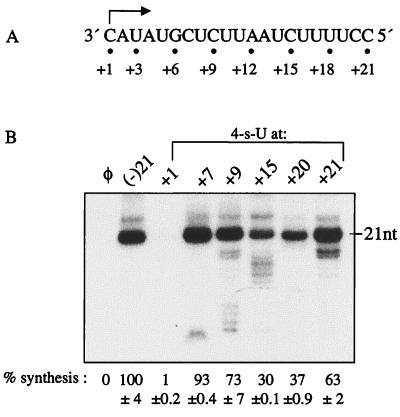
The position-dependent recognition of the ribose C2′ moieties by NS5B led us to determine whether hydrogen bonding between template and substrate nucleotides in RNA synthesis is also modulated a position-dependent manner. To affect hydrogen bonding, 4sU nucleotide analogs were incorporated at position +1, +7, +9, +15, +20, or +21 in (−)21. All positions changed were cytidylates, allowing a direct comparison of the effects of the 4sU modification on synthesis at several positions (Fig. 8).
FIG. 8.
Different tolerances for the 4sU base modifications. (A) Sequence of (−)21, shown from 3′ to 5′. The initiation cytidylate is indicated by an arrow, and the positions modified are shown by dots and numbers below. (B) Autoradiogram showing the 21-nt RNA products synthesized from the various templates denoted above the autoradiogram, modified with the 4sU base analog at position +1, +7, +9, +15, +20, or +21. All quantifications were normalized to (−)21, and the mean and SD are shown below the autoradiogram. φ denotes a control reaction lacking template RNA.
Templates with 4sU modifications at positions +7 to +21 directed RNA synthesis at 30 to 98% of that for (−)21 (Fig. 8). These effects are less severe than those of the same modification at the initiation +1C, where 4sU abolished synthesis (Fig. 4, lane 4). In contrast to the ribose modifications, RNA synthesis does not correlate strictly with the linear position in the template position. For example, 4sU at position +7, +9, or +21 directed synthesis at 93, 73, and 63%, while 4sU at position +15 or +20 directed synthesis at 30 and 38%, respectively (Fig. 8B). These results suggest that mechanisms other than the transition of RNA synthesis from initiation to elongation may be involved in template base recognition. This is logical since base pairing between the template and the substrate NTP is necessary during all stages of synthesis.
The decreased RNA synthesis with 4sU at position +15 or +20 is not due to base-specific recognition by NS5B, because nucleotide substitutions at these positions did not affect efficient RNA synthesis (Table 1). Since 4sU affects hydrogen bonding with the substrate nucleotide, we hypothesize that the decreased RNA syntheses from templates with 4sU may be due to the effects on the template-nascent RNA interaction in the polymerase ternary complex. In support of this hypothesis, we detected less than full-length RdRp products that correlated with the location of the 4sU in the template. Most of the premature RNAs were 1 to 3 nt shorter or longer than the site of the 4sU modification (Fig. 8B). For example, the 4sU at +15 resulted in premature RNAs with lengths ranging from 13 to 18 nt (Fig. 8B, lane 6). Since the amounts and sizes of the prematurely terminated products differed with the position of 4sU, the base pairs flanking the modified site may contribute to the incorporation of the substrate nucleotide.
In ternary complexes of DNA-dependent RNA polymerases, the base pair preceding the one being incorporated in the catalytic site has been proposed to be an important regulating factor for RNA synthesis from a DNA template (19, 37, 40, 63; Kao, unpublished). Thus, we determined whether the neighboring base pairs contributed to the levels of RNA synthesis directed by templates containing 4sU modifications at various positions. A 4sU at position +15 directed the lowest RNA synthesis (Fig. 8B). Thus, the neighboring +14 position was changed from a uridylate to the other three nucleotides while 4sU was retained at the +15 position, resulting in RNAs named A+15S, C+15S, and G+15S. We had previously demonstrated that NS5B does not recognize the +14 position in a sequence-specific manner (Table 1). Therefore, any effect on RNA synthesis will be due to a possible interaction between +14 and the 4sU.
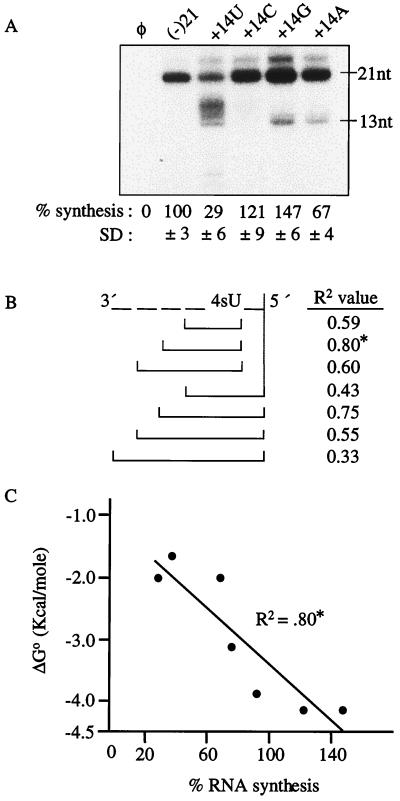
RNAs G+15S, C+15S, and A+15S directed synthesis at 147, 121, and 67%, respectively, relative to (−)21 and at 29% of the synthesis from the template +14U-S (Fig. 9A). Different prematurely terminated products were observed within this set of modified RNAs, indicating that RNA synthesis is affected quantitatively and qualitatively by the template nucleotide immediately 3′ to the 4sU. Higher levels of synthesis from C+15S and G+15S than from A+15S and U+15S suggest that more stable base pairing immediately preceding the 4sU modification site in the RNA duplex helps the substrate nucleotide incorporation opposite the 4sU nucleotide analog.
FIG. 9.
Effect of the neighboring sequence on synthesis in a template with a 4sU base analog. (A) Autoradiogram of 21-nt products and prematurely terminated products from four templates (named above the autoradiogram). All RNAs had the +14 position substituted with a uridylate (U), cytidylate (C), guanylate (G), or adenylate (A), while containing the 4sU base analog at the +15 position. The quantifications (mean and SD) are shown below the autoradiogram. All values were normalized to (−)21, and φ denotes a control reaction lacking template RNA; 21 nt and 13 nt are the relative size of the RNA products. (B) Schematic illustration of the combinations used for the ΔG calculation, with R2 values to the right. “-” Dashes denote a nucleotide next to the position of the 4sU. Each bracket represents a combination for the ΔG calculation. The highest R2 value, indicated by ∗, was used in the graph shown below. (C) Correlation between the level of RNA synthesis and the calculated ΔG, from the combination resulting in the highest R2 value. ΔG calculations were from the values of Turner et al. (57).
Correlation between local RNA duplex stability and synthesis in the presence of 4sU.
Next, we compared RNA synthesis from all eight 4sU-modified templates to the identity of the nearest neighboring nucleotide. Templates containing the 4sU analog and a neighboring 3′ guanylate or cytidylate directed synthesis at 63% (C+21S) to 147% (G+15S) of that for (−)21, while those with a neighboring 3′ adenylate or uridylate directed RNA synthesis at 37 to 73% (Fig. 8 and 9A). Thus, there is a general trend that a more stable upstream base pair will result in better synthesis across from the neighboring 4sU analog. However, the range in synthesis levels suggests that more than one nucleotide near the 4sU modification appears to affect RNA synthesis.
The minimum RNA-RNA duplex in an RdRp ternary complex has not been characterized and is still subject to interpretations in better-characterized polymerases (8, 9). We seek to provide a theoretical examination of the ternary complex for NS5B by correlating RNA synthesis from templates with 4sU modifications to the predicted thermodynamic parameters for a number of base pair combinations. This analysis relies on the values for RNA duplexes determined by Turner et al. (57). While speculative, these results can provide a testable model for experimental analysis. Several factors were taken into account: (i) the RNA with 4sU at +1 position was excluded in this analysis, due to the lack of upstream sequences and the clearly different requirements for the initiation nucleotide; (ii) RNA with a 4sU at +21, the 5′ end of the template, was not used since it should have a different thermodynamic constraint due to its being at the end of the duplex (20, 21); (iii) 4sU was treated as a uridylate based on the similarity of functional groups; and (iv) ATP was assumed to be the substrate incorporated opposite the 4sU base analog, since it has the most appropriate W-C geometry. ΔG° values collected from each combination of base pairs were plotted against the percent RNA syntheses to obtain R2 values. A higher R2 value indicates a better correlation between RNA synthesis and the stability of the nascent-template RNA interaction (Fig. 9B).
R2 values ranged from 0.33 to 0.80, indicating that the correlation was significantly influenced by the numbers of neighboring base pairs included in the analysis (Fig. 9B). The highest correlation obtained (R2 = 0.80) was when ΔG° was calculated as a sum of the two base pairs preceding the 4sU site and the base pair between 4sU and ATP (Fig. 9C). Synthesis with 4sU increased along with an increased stability of the template-nascent RNA interaction near the modified site. These results suggest that only a few base pairs near and including the 4sU nucleotide analog are significantly correlated with the ability of the RdRp to complete synthesis on a template with a 4sU modification.
DISCUSSION
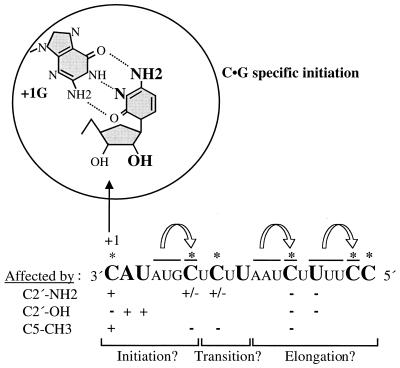
We seek to understand the mechanism of RNA synthesis from RNA templates by the RdRps. Several moieties in the template RNA that are important for de novo initiation of RNA synthesis by the BVDV RdRp were identified. For the initiation cytidylate, specific requirements for the base were found, but several changes at the ribose C2′ were able to direct initiation except for a partially positive charged amino group. Different requirements were found for the noninitiating template nucleotides and the initiation cytidylate. The ribose C2′-OH at the +2 and +3 positions within the template also contributed to RNA synthesis (Fig. 10). The effects of 4sU base modifications were varied in a manner depending on the identity of the neighboring nucleotides (Fig. 10). These observations can be compared and contrasted with the initiation of RNA synthesis by other RdRPs and DNA-dependent RNA polymerases and will provide useful information for structure-function analyses of RdRp ternary complexes.
FIG. 10.
Summary of results presented in this work and a model for RNA synthesis by a viral RdRp. Cytosine moieties and the ribose C2′-hydroxyl of the +1C found to be important for synthesis are shown in bold. The guanine of GTP is shown to identify the hydrogen-bonding and non-hydrogen-bonding faces of the cytosine. Modifications were made in (−)21, and their effects on RNA synthesis are indicated by + and −, where + indicates a severe effect and − indicates tolerance for the modification for RNA synthesis. The effect of neighboring base pairs correlatee with the levels of RNA synthesis in the presence of a 4sU nucleoside analog in the template are indicated by arrows above the (−)21 sequence. Three examples are shown in this diagram. ∗, site of the 4sU modification.
Recognition of the initiation cytidylate.
A cytidylate is the preferred initiation nucleotide for several RdRps, including the BMV replicase, Qβ replicase, BVDV NS5B, and HCV NS5B proteins (6, 24, 30, 49). The preferred position of the initiation cytidylate may be different for each virus. The BVDV NS5B can use either the 3′-most or a penultimate cytidylate but prefers to initiate from the 3′-most cytidylate. HCV RdRp also can direct synthesis from the 3′-terminal cytidylate (70). However, the BMV and turnip yellow mosaic virus replicases require that the initiation cytidylate be at the penultimate position (12, 48, 49, 52).
In contrast to the specific cytidylate requirement for RNA synthesis, pyrimidines, including 4sU, can interact with NS5B in the template competition assay (27) (Fig. 5). This situation is similar to the results obtained with the well-characterized T7 RNA polymerase, where the +1C is not required for the proper positioning of the initiation site in the catalytic pocket of the T7 RNA polymerase (59, 67). Weston et al. (67) demonstrated that the distance between the core promoter elements and the initiation cytidylate could be increased through the addition of flexible linkers, although increased nucleotide misincorporation resulted.
While the initiation nucleotide may not contribute to the T7 promoter recognition, the structure of the T7 polymerase initiation complex provides useful insight into the interactions at the polymerase catalytic active site (9). The distance from the surface of polymerase to the G · C base pairing was within 2.8 Å, suggesting that a hydrogen bond(s) between side chains of the polymerase and the initiation nucleotides can stabilize the ternary complex. The addition of a methyl or a bromo adduct to the non-hydrogen-bonding C5 position of the initiation cytidylate may cause constraints and/or charge requirements at the interface between the non-hydrogen-bonding side of +1C and the pocket of the NS5B RdRp. The nature of the ternary complex of an RdRp remains to be determined.
Polymerases generally have a purine-specific site for the initiation substrate NTP, the i site, and a second nucleotide binding pocket, the i + 1 site (41). The increased stability of the ternary complex has been demonstrated in the presence of a high initiation substrate (GTP) concentration (15). For the BMV replicase, the presence of GTP at the i site resulted in the formation of a more stable initiation complex (53, 54). Our results that the BVDV NS5B requires the N3 and C4-amino groups at the +1C position probably reflect the need to hydrogen bond to the GTP in the i site (Fig. 10). Like other polymerases, the BVDV RdRp requires higher concentrations of initiation substrate nucleotide (25). A higher Km for the initiation nucleotide was reported for the Qβ replicase, the BMV replicase, and the HCV RdRp (6, 27, 29, 52).
Moieties required for stabilization in the template-NS5B interaction.
The ribose C2′-hydroxyl defines the structure of RNAs and contributes to the stability of RNA-protein interactions (7, 13, 35, 64). For the BVDV NS5B protein, the C2′-hydroxyl at the +1 position does not affect RNA synthesis while those at the +2 and +3 positions are more important for efficient initiation (Fig. 10). Deoxyriboses at the +2 and +3 positions may either prevent potential hydrogen bond formation with NS5B or cause a partial structural distortion in the RNA that decreases the efficacy of synthesis and interaction (Fig. 6). The latter scenario is less likely since the (−)21 RNA used in this work does not have a predicted stable structure. In support of this, a similar level of RNA synthesis to that seen with (−)21 was observed with a 12-nt template (Kim et al., unpublished). Rather, our results support the idea that the loss of contact between the +2 and +3 C2′-hydroxyl and the NS5B catalytic site may be critical for stabilizing the initiation complex. The loss of potential hydrogen bonding via the C2′-hydroxyl may account for a decrease in RNA synthesis from d(−)21, d+2, d+3, d+1,2, and d+1,2,3 (Fig. 6C).
BVDV NS5B does not recognize the template in a base-specific manner except at the +1C. No difference in RNA synthesis was observed with several nucleotide substitutions within the template (Table 1) (27). However, the template sequence may contribute to the mechanism of RNA synthesis. For the alphavirus-like viral RNAs, Sivakumaran et al. (50) found specific preferences for adenylates and uridylates near the initiation nucleotide. Also, transcription studies using the DNA template have revealed that the template sequences could affect the successful completion of RNA synthesis (28, 37, 63). It is also possible that template specificity for the BVDV replication may be determined by RNA sequences or protein subunits that are not included in this study.
Different recognition between the initiation nucleotide and others in the template.
Conformational changes of polymerases and templates are known to accompany the steps involved in transcription, template binding, initiation complex formation, and transition from the initiation to elongation (11). We have found that ribose C2′-NH2 modifications had less detrimental effects on RNA synthesis when the modifications were beyond position +11 (Fig. 10). This result is consistent with a change in the interaction of the polymerase and the template that results from the transition to elongation (Fig. 10). Furthermore, results of the template competition assays indicate that the interaction between NS5B and the template was inappropriate when the ribose at position +1, +3, or +7 had an amino group. Interestingly, while amino modifications at positions +9 and +11 resulted in similar IC50s to that for wild-type RNA, synthesis was less than 24% of that for (−)21. The BMV replicase is known to have a more stable interaction with the template after RNA synthesis has proceeded for 8 to 10 nt (26, 53, 54). This length is in good agreement with the minimal length of the RNA template that can bind to the poliovirus 3Dpol and is consistent with the poliovirus 3Dpol structure (2, 22). In addition, the recently solved T7 polymerase structure has revealed the pocket accommodating the DNA template up to position +7 until the transition occurs from initiation to elongation (9). Thus, we speculate that positions +9 through +11 may represent a site for the NS5B protein to make a transition to elongation (Fig. 10).
Nucleotide polymerization and the ternary complex.
The placements of the 4sU along the template, weakening W-C hydrogen bonding and/or increasing nucleotide misincoporation, resulted in a wide range of synthesis that cannot be correlated with the possible transition from initiation to elongation (Fig. 8). Our results suggest that the local sequence(s), rather than the position of the ternary complex, may regulate the interaction between the template and substrate NTPs.
The efficiency of nucleotide incorporation over a templated 4sU analog correlated best with the identity of the nucleotide(s) 3′ of the analog (Fig. 9). This observation suggests that the thermodynamics of the template-nascent RNA interaction contribute to the incorporation of the incoming nucleotide and the completion of RNA synthesis. While the effects of nascent RNA-template interaction have not been examined for any RdRp system, transcription studies using the DNA template have revealed that the template sequences influence the successful completion of RNA synthesis (28, 37, 63). Also, the local duplex stability and the base stacking affecting product synthesis have been demonstrated in polymerases including the T4 polymerase and L414 DNA polymerase (4, 5, 39, 43).
We obtained the best correlation between the level of RNA synthesis and the calculated ΔG° of a 3- to 4-nt duplex that includes two adjacent base pairs and the presumed ATP-4sU base pair (Fig. 9C). The correlation was clearly dependent on the assumption of the duplex length and is intended to provide a model for additional analysis. The assumption of duplex length was a function of models of the transcriptional complex (TC). The monotonic transcription model has postulated a RNA-DNA hybrid of 9 to 12 bp (63). This duplex then provides the thermodynamic barrier to TC dissociation. The inchworm model of elongation, proposed by Chamberlin and colleagues (8), has its basis in the difference in sizes of RNA and DNA footprints in the halted TC structure (36, 60, 65). Chamberlin (8) argues that perhaps the only 2 or 3 bp of the RNA-DNA hybrid provide the thermodynamic stability in TC complex. A similar hypothesis based on the ternary complex of the T7 RNA polymerase crystallized with a 4-nt template and a 3-nt nascent RNA indicates that there is only space for a duplex shorter than 4 bp, including the one at the catalytic site (9). Recent findings with the E. coli RNA polymerase suggest synthesis by a sliding-clamp model of RNA polymerase translocation that combines the inchworm and monotonic models in one (29, 37, 42). Although the exact mechanism has not been addressed in this work, our observations most closely resemble the sliding-clamp model because of the patterns of prematurely terminated RNAs, which are also affected by the stability of most nearest-neighbor W-C base pairs (Fig. 8 and 9). A similar observation has been demonstrated with the E. coli RNA polymerase (56). Lastly, the correlation of 3 to 4 bp having the most significant effect on synthesis over the weaken hydrogen bonding 4sU does not imply that the duplex is limited to 3 to 4 nt. Additional base pairing may exist but may not contribute significantly to synthesis over the 4sU-modified position.
Several rules appear to govern the incorporation of the incoming NTP in addition to hydrogen bonding between the nucleotides in template and nascent RNA. In general, hydrogen bond formation is considered informational whereas base stacking is considered noninformational (for a review, see reference 18). Consequently, the hydrogen bonding for A · T and G · C is usually emphasized as primarily responsible for the polymerase fidelity. However, through the use of nonpolar isosteric thymidine analogs, Moran et al. (33) demonstrated that the shape of the nucleotides is also a determining factor for the specificity of the NTP-template interaction. Goodman (18) also suggested that the geometrical selection via an induced fit mechanism played an important role in the fidelity of polymerases. Our results suggest that the neighboring base pairs in the RNA duplex contribute to the ability to overcome a 4sU-nucleotide analog and allow polymerase to complete the synthesis.
Concluding remarks.
Successful viral RNA replication is a multistep process that includes (i) interaction between the promoter and the replicase; (ii) initiation, consisting of the binding of the initiation nucleotide and abortive cycling; (iii) transition of the polymerase to a high-affinity interaction with the RNA template; and (iv) termination of RNA synthesis and release of the nascent RNA from the ternary complex (for reviews, see references 1 and 32). Our work qualitatively examines the nucleotide moieties needed for successful initiation of RNA synthesis by a recombinant viral RdRp. The moieties identified may affect NS5B-RNA interaction and/or nascent RNA-template RNA interaction. Moreover, we observed significant differences in the moieties required for efficient initiation and for elongation. This work should establish the framework for a detailed kinetic analysis of the mechanism of RNA synthesis by an RdRp.
ACKNOWLEDGMENTS
We thank the IU cereal killers for helpful discussion during this work.
The Kao lab acknowledges support from the NSF (MCB9807800) and USDA (9902503). M.-J. Kim acknowledges a fellowship from the Samuel Nobel Foundation, and C. C. Kao is the recipient of a Linda and Jack Gill fellowship.
REFERENCES
- 1.Adkins S, Stawicki S S, Faurote G, Siegel R W, Kao C C. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman M T, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- 3.Behrens S E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA-polymerase of hepatitis C virus. EMBO J. 1997;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 4.Bessman M J, Reha-Krantz L J. Studies on the biochemical basis of spontaneous mutation. V. Effects of temperature on mutation frequency. J Mol Biol. 1977;116:115–123. doi: 10.1016/0022-2836(77)90122-x. [DOI] [PubMed] [Google Scholar]
- 5.Bloom L B, Otto M R, Beechem J M, Goodman M F. Influence of 5′-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminoputine by Klenow fragment. Biochemistry. 1993;32:11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal T. Qβ replicase template specificity: different templates require different GTP concentrations for initiation. Proc Natl Acad Sci USA. 1980;77:2601–2605. doi: 10.1073/pnas.77.5.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridya N, Uhlenbeck O C. The role of 2′-hydroxyl groups in an RNA-protein interaction. Biochemistry. 1995;34:12363–12368. doi: 10.1021/bi00038a033. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlin M J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1995;88:1–21. [PubMed] [Google Scholar]
- 9.Cheetham G M T, Steitz T A. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 10.Collett M S, Larson R, Belzer S, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 11.Dehaseth P L, Zupancic M L, Record M T., Jr RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deiman B A L M, Koenen A K, Verlann P W G, Pleij P W G. Minimal template requirement for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998;72:3065–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dock-Bregeon A C, Chevrier B, Podjarny A, Moras D, de Bear J, Gough G R, Gilham P T, Johnson J E. High resolution structure of the RNA complex [U(U-A)6A]2. Nature. 1988;335:375–378. doi: 10.1038/335375a0. [DOI] [PubMed] [Google Scholar]
- 14.Frolov I, McBride M S, Rice C M. Cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaal T, Bartlett M S, Ross W, Turnbough C L, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 16.Gong Y, Shannon A, Westaway E G, Gowans E J. The replicative intermediate molecule of bovine viral diarrhoea virus contains multiple nascent strands. Arch Virol. 1998;143:399–404. doi: 10.1007/s007050050296. [DOI] [PubMed] [Google Scholar]
- 17.Gong Y, Trowbridge R, Macnaughton T B, Westaway E G, Shannon A D, Gowans E J. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J Gen Virol. 1998;77:2729–2736. doi: 10.1099/0022-1317-77-11-2729. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M F. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman M F, Gore W C, Muzyczka N, Bessman M J. Studies on the biochemical basis of spontaneous mutation. III. Rate model for DNA polymerase-effected nucleotide misincorporation. J Mol Biol. 1974;88:423–435. doi: 10.1016/0022-2836(74)90492-6. [DOI] [PubMed] [Google Scholar]
- 20.Gray D M. Derivation of nearest neighbor properties from data on nucleic acid oligomers. I. Simple sets of independent sequences and the influence of absent nearest neighbor. Biopolymer. 1997;42:783–793. doi: 10.1002/(sici)1097-0282(199712)42:7<783::aid-bip4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 21.Gray D M. Derivation of nearest neighbor properties from data on nucleic acid oligomers. II. Thermodynamic parameters of DNA · RNA hybrids and DNA duplex. Biopolymer. 1997;42:795–810. doi: 10.1002/(sici)1097-0282(199712)42:7<795::aid-bip5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 22.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey G A, Saenger W. Hydrogen bonding in biological structures. New York, N.Y: Springer-Verlag; 1991. [Google Scholar]
- 24.Jorgensen S E, Buch L B, Nierlich D P. Nucleoside triphosphate termini from RNA synthesized in vivo by Escherichia coli. Science. 1969;164:1067–1070. doi: 10.1126/science.164.3883.1067. [DOI] [PubMed] [Google Scholar]
- 25.Joyce C M. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao C C, Sun J. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase. Virology. 1996;253:1–7. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao C C, Del Vecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 28.Keene R, Luse D. Initially transcribed sequences strongly affect the extent of abortive initiation by RNA polymerase II. J Biol Chem. 1999;274:11526–11534. doi: 10.1074/jbc.274.17.11526. [DOI] [PubMed] [Google Scholar]
- 29.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backword, leaving the 3′ end of the RNA intact and extruded. Proc Acad Natl Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo G, Hamatake R K, Mathis D M, Racela J, Rigat K L, Lemm J, Colonno R J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from cDNA clones: a cellular insert determined NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mooney R A, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran S, Ren R X-F, Kool E T. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy F. Virus taxonomy. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 15–57. [Google Scholar]
- 35.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 36.Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- 37.Nudler E, Mustaev A, Lukhatanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 38.Perrin D D. pKa prediction for organic acids and bases. London, United Kingdom: Champman & Hall; 1992. [Google Scholar]
- 39.Petruska J, Goodman M F. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. J Biol Chem. 1985;260:7533–7539. [PubMed] [Google Scholar]
- 40.Press R C, Bessman M J. Influence of local nucleotide sequence on substitution of 2-aminoputine for adenine during deoxyribonucleic acid synthesis in vitro. Biochemistry. 1983;22:4905–4915. doi: 10.1021/bi00290a006. [DOI] [PubMed] [Google Scholar]
- 41.Reddy P S, Chatterji D. Evidence for a pyrimidine-nucleotide-specific initiation site (the i site) on Escherichia coli RNA polymerase: proximity relationship with the inhibitor binding domain. Eur J Biochem. 1994;225:737–745. doi: 10.1111/j.1432-1033.1994.00737.x. [DOI] [PubMed] [Google Scholar]
- 42.Reeder T C, Hawley D K. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 43.Ronen A, Rahat A. Mutagen specificity and position effects on mutation in T4 rII nonsense sites. Mutat Res. 1976;34:21–34. doi: 10.1016/0027-5107(76)90258-x. [DOI] [PubMed] [Google Scholar]
- 44.Saenger W. Principles of nucleic acid structure. New York, N.Y: Springer-Verlag; 1984. [Google Scholar]
- 45.Saenger W, Heinemann U. Protein-nucleic acid interaction. Boca Raton, Fla: CRC Press, Inc.; 1989. [Google Scholar]
- 46.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel R W, Bellon L, Beigelman L, Kao C C. Use of DNA, RNA and chimeric templates by a viral RNA-dependent RNA polymerase: evolutionary implications for the transition from the RNA to the DNA world. J Virol. 1999;73:6424–6429. doi: 10.1128/jvi.73.8.6424-6429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh R N, Dreher T W. Specific site selection in RNA resulting from a combination of non-specific secondary structure and –CCR- box: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–1095. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivakumaran K, Kao C C. Initiation of genomic positive strand synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J Virol. 1999;73:6415–6423. doi: 10.1128/jvi.73.8.6415-6423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivakumaran K, Kim C-H, Tayon R, Jr, Kao C C. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J Mol Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorenson J T, Enevoldsenm C, Houe H. A stochastic model for simulation of the economic consequences of bovine virus diarrhea infection in a dairy herd. Prev Vet Med. 1995;23:215–227. [Google Scholar]
- 52.Sun J, Adkins S, Faurote G, Kao C C. Initiation of (−)-strand RNA synthesis catalyzed by the brome mosaic virus RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 53.Sun J, Kao C C. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology. 1997;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 54.Sun J, Kao C C. Characterization of RNA products associated with or aborted by a viral RNA-dependent RNA polymerase. Virology. 1997;236:348–353. doi: 10.1006/viro.1997.8742. [DOI] [PubMed] [Google Scholar]
- 55.Thiel H J, Plugemann G W, Moenning V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippencott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 56.Toulmé F, Guérin M, Robichon N, Leng M, Rahmouni A R. In vivo evidence for back and forth oscillations of the transcription elongation complex. EMBO J. 1999;18:5052–5060. doi: 10.1093/emboj/18.18.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner D H, Sugimoto N, Freier S M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 58.Uesugi S, Miki H, Ikehara M, Iwahashi H, Kyogoku Y. A linear relationship between electronegativity of 2′-substituents and conformation of adenine nucleotides. Tetrahedron Lett. 1979;42:4073–4076. [Google Scholar]
- 59.Ujvári A, Martin C T. Identification of a minimal binding element within the T7 RNA polymerase promoter. J Mol Biol. 1997;273:775–781. doi: 10.1006/jmbi.1997.1350. [DOI] [PubMed] [Google Scholar]
- 60.Uptain S M, Kane C M, Chamberin M J. Basic mechanisms of transcription elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 61.Vassilev V B, Collette M C, Donis R O. Authenic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vicky C H, Lai C, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y N. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Von Hippel P H, Yager T D. Transcription elongation and termination are competitive kinetic processes. Proc Natl Acad Sci USA. 1991;88:2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang A H-J, Fujii S, van Boom J H, van der Marel G A, van Boeckel S A A A, Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA-RNA hybrid helix joined to double helical DNA. Nature. 1982;299:601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Meier I, Chan C L, Feng G, Lee D N, Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 66.Watson J D, Crick F H C. Genetic implication of the structure of deoxyribonucleic acid. Nature (London) 1953;171:864–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 67.Weston B F, Kuzmine L, Martin C T. Positioning of the start site in the initiation of transcription by bacteriophage T7 RNA polymerase. J Mol Biol. 1997;272:21–30. doi: 10.1006/jmbi.1997.1199. [DOI] [PubMed] [Google Scholar]
- 68.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polypeptide cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong W, Uss A S, Ferrari E, Lau J Y N, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]