Abstract
Recent accelerations in multi-modal applications have been made possible with the plethora of image and text data available online. However, the scarcity of analogous data in the medical field, specifically in histopathology, has halted comparable progress. To enable similar representation learning for histopathology, we turn to YouTube, an untapped resource of videos, offering 1,087 hours of valuable educational histopathology videos from expert clinicians. From YouTube, we curate Quilt: a large-scale vision-language dataset consisting of 768,826 image and text pairs. Quilt was automatically curated using a mixture of models, including large language models, handcrafted algorithms, human knowledge databases, and automatic speech recognition. In comparison, the most comprehensive datasets curated for histopathology amass only around 200K samples. We combine Quilt with datasets from other sources, including Twitter, research papers, and the internet in general, to create an even larger dataset: Quilt-1M, with 1M paired image-text samples, marking it as the largest vision-language histopathology dataset to date. We demonstrate the value of Quilt-1M by fine-tuning a pre-trained CLIP model. Our model outperforms state-of-the-art models on both zero-shot and linear probing tasks for classifying new histopathology images across 13 diverse patch-level datasets of 8 different sub-pathologies and cross-modal retrieval tasks2.
1. Introduction
Whole-slide histopathology images are dense in information, and even individual image patches can hold unique, complex patterns critical for tissue characterization. Summarizing this information into a single label is an oversimplification that fails to capture the complexity of the field, which covers thousands of evolving disease sub-types [58]. This highlights the need for more expressive, dense, interconnected representations beyond the reach of a singular categorical label. As such, natural language descriptions can provide this comprehensive signal, linking diverse features of histopathology sub-patch structures [20, 24].
If there were a large-scale vision-language dataset for histopathology, researchers would be able to leverage the significant advancements in self-supervised vision and language pre-training to develop effective histopathology models [48]. Unfortunately, there is a significant scarcity of comprehensive datasets for histopathology. Notable open-source contributions have been made with datasets like ARCH [20] and OpenPath [24]. Yet, these sources are still somewhat limited due to their size, as the former has only ≈ 8K samples and the latter (the largest histopathology vision-language dataset to date) has about 200K samples. Although recent efforts (e.g. PMC-15M [70]) curated 15M image-text pairs across a variety of different biomedical domains from Pubmed [50], whether their samples are specific to histopathology remains ambiguous; worse, their dataset is not openly available.
To address the need for a large-scale vision-language dataset in histopathology, we introduce Quilt: containing 419, 780 images aligned with 768, 826 text pairs. We draw on the insight that publicly available educational YouTube histopathology content represents an untapped potential. We curate Quilt using 1, 087 hours of valuable educational histopathology videos from expert pathologists on YouTube. To extract aligned image and text pairs from the videos, we utilize a mixture of models: large language models, handcrafted algorithms, human knowledge databases, and automatic speech recognition. Quilt does not overlap with any current open-access histopathology data sources. This allows us to merge our dataset with other open-source datasets available. Therefore, to create an even larger and more diverse dataset, we combine Quilt with data from other sources, such as Twitter, research papers, and the Internet, resulting in Quilt-1M. The larger Quilt-1M contains one million image-text pairs, making it the largest public vision-language histopathology dataset to date.
Using Quilt and Quilt-1M, we finetune vision-language models using a contrastive objective between the two modalities. We extensively evaluate it on 13 external histopathology datasets taken across different sub-pathologies. We report zero-shot classification, linear probe, and image-to-text and text-to-image retrieval tasks. Against multiple recently proposed baselines (CLIP [48], PLIP [24], and BiomedCLIP [70]), models trained with Quilt-1M outperform all others. Our ablations identify the importance of Quilt.
Quilt offers three significant advantages: First, Quilt does not overlap with existing data sources; it ensures a unique contribution to the pool of available histopathology knowledge. Second, its rich textual descriptions extracted from experts narrating within educational videos provide more expressive, dense interconnected information. Last, the presence of multiple sentences per image fosters diverse perspectives and a comprehensive understanding of each histopathological image. We hope that both computer scientists and histopathologists will benefit from Quilt’s potential.
2. Related work
We built upon a growing literature applying self-supervised learning and other machine learning methods to medical image understanding.
Machine learning for histopathology.
Early machine learning work in computational pathology primarily relied on weakly-supervised learning, with each whole-slide image (WSI) receiving a single label. The limited nature (single label to many patches) has produced sub-optimal models [12, 26]. Lately, a self-supervised learning approach, which learns useful representations from unlabeled data, has shown some success [26, 13, 12]. Most of this work has been unimodal. They use image augmentations similar to those used for natural images [14], mostly differing by way of consciously injecting domain knowledge. For example, they leverage the compositional nature of H&E stain information of whole-slide images [26], or inject hierarchical morphological information at different magnifications [13], or combine with other modalities like genomic features [12] or with descriptive text [20]. When text data is used, the objectives similarly use augmentations seen in natural language [52]. By contrast, we explore self-supervised mechanisms that learn better histopathology information representations that go beyond a single label, aided by language descriptions.
Medical vision-language datasets.
Learning vision-language representations demands a large dataset of images aligned with descriptive text, a resource that is notably lacking in histopathology. The MIMIC-CXR-JPG v2.0.0 dataset [29], for example, consists of de-identified hospital-sourced chest radiographs and reports. For histopathology, The Cancer Genome Atlas3 provides de-identified PDF-reports for a limited number of WSIs. Despite this resource, the enormous size of this data (reaching up to 120, 0002 pixels) makes processing challenging, limiting its use to a small number of focused studies [41]. A majority of medical vision-language datasets are concentrated in the radiology sub-domain, due to the relatively straightforward process of collecting validated multimodal data [29]. Many models are trained on a subset of PubMed [50] or comparable radiology datasets [71, 23, 18, 45]. PMC-15M [70], a recent subset of PubMed not specific to histopathology, was used to train multiple models. While the models themselves are public, PMC-15M is not, making it hard to determine what portion of it is histopathology-relevant.
Vision-language pairs on histopathology.
One of the first histopathology vision-language datasets, ARCH, contains only 7, 614 accessible image-text pairs [20, 22]. Later on, [24] released OpenPath, a dataset of 200K image-text pairs extracted from Twitter. This was the largest histopathology dataset until Quilt-1M.
Video data for self-supervision.
Numerous recent studies have started to tap into video data. For instance, millions of publicly accessible YouTube videos were used to train a vision-language model [68, 69]. Similarly, a causal video model was trained by using sequential gaming videos [6]. Localized narratives [61, 46] provide another example of dense, interconnected supervision for a single image. Despite the untapped potential of video content, video often yields noisier datasets compared to static sources. Recently, the enhanced capabilities of automatic speech recognition models streamlined the curation of large-scale cleaner datasets from videos [68, 6, 70]. Furthermore, the growing versatility of large language models has shown promise as data annotators, information extractors [34, 62, 15, 21], text correctors [66], and as tools for medical information extraction and reasoning [1, 59].
3. Curating Quilt: Overview
Creating a vision-language dataset from videos is a significant undertaking, as not all videos are suitable for our pipeline. Many either lack voiced audio, are not in English, fail to contain medically relevant content, or have insufficient medical relevance—for example, videos that present static images of histopathology content on a slide deck, or those that briefly cover histopathology images in pursuit of a different objective. Conventional automatic speech recognition (ASR) systems also struggle with the specialized requirements of histopathology transcription, necessitating a non-trivial solution. The de-noising of text and image modalities adds further complexity as the videos are typically conversational and, therefore, inherently noisy. Instructors pan and zoom at varying speeds, recording a mix of relevant and irrelevant histopathological visual content in their videos. As such, trivially extracting frames at static intervals fails to capture the data appropriately. To collect Quilt we trained models and handcrafted algorithms that leverage the nuances in the instructors’ textual and visual behavior, ensuring accurate collection and alignment of both modalities.
3.1. Quilt: Collecting medical image and text pairs from YouTube
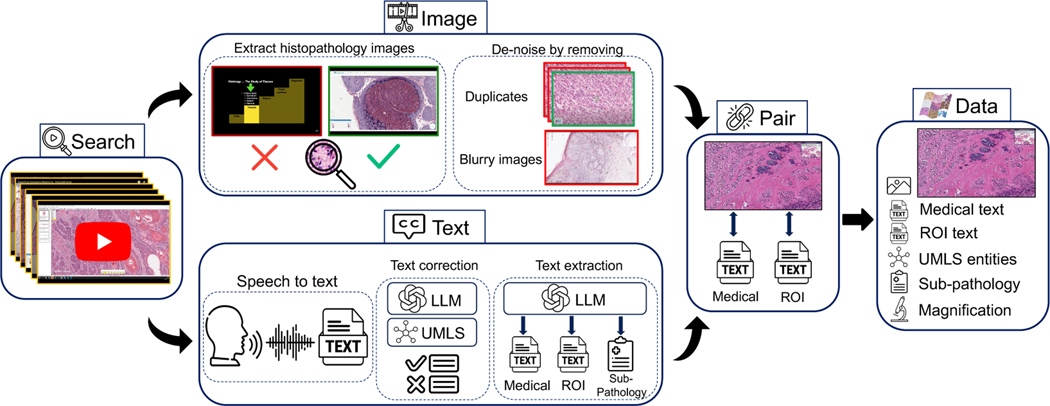
Our proposed dataset curation pipeline involves (1) gathering channel and video data covering the histopathology domain, (2) filtering videos based on a certain “narrative style”, (3) extracting and denoising image and text modalities from videos using various models, tools, and algorithms, (4) postprocessing denoised text by LLMs to extract medical text and finally, (5) splitting and aligning all modalities for curating the final vision-language pre-training (VLP) data. See Figure 1 (and A.1 in the Appendix) for a detailed overview of the pipeline.
Figure 1: Overview of Quilt curation pipeline.
We identify relevant histopathology YouTube videos in Search. For Image extraction, we find and de-noise histopathology frames using trained models. In Text section, we rely on a conventional Automatic Speech Recognition (ASR) model and leverage Unified Medical Language System (UMLS) and large language models (LLMs) for post-processing and ASR error correction. Relevant sub-pathology, medical and region-of-interest (ROI) text are extracted using an LLM. Finally, domain-specific algorithms are used to Pair images and text, eliminating duplicates to yield Quilt, a richly annotated image-text dataset for histopathology.
Collecting representative channels and videos.
Our pipeline begins by searching for relevant channels and video ids on YouTube, focusing on the domain of histopathology. Using keywords spanning 18 sub-pathology fields (see section A.4 in the Appendix), we search among channels before searching for videos to expedite discovery, considering that video searches are time-consuming and the APIs pose limitations on numerous requests [68]. Channels with subscriber count ≥ 300K are excluded to avoid large general science channels, as educational histopathology channels often have fewer subscribers. We then download low-resolution versions of all identified videos.
Filtering for narrative-style medical videos.
For each video within each channel, we exclude videos that are shorter than 1 minute, non-voiced, or have non-English audio. For videos meeting these heuristics, two decisions are made:
Do they have the required medical content, i.e., histopathology image-text pairs?
If so, are they in narrative style – videos wherein the presenter(s) spend a significant time panning and zooming on the WSI, while providing vocal descriptions of image content?
For (A) we automatically identify the relevant videos by extracting keyframes from a video. These keyframes are automatically extracted using FFmpeg 4, marking the beginning or end of a scene (frames containing significant visual changes). The software requires a threshold that determines the minimum amount of visual change required to trigger a keyframe. Through experimentation, we set different thresholds for various video durations, with smaller thresholds for longer videos. Next, we train and use an ensemble of three histopathology image classifiers to identify videos with histopathology images (See section A.3 in the Appendix).
For (B), in which we identify narrative-style videos, we randomly select keyframes predicted to be histopathology. For each such selected frame, we extract the next three histopathology key-frames and compute the cosine similarity between the selected frame and each of the subsequent three frames. If all three have similarity scores ≥ a preset threshold of 0.9, we count it as a narrative streak. A video is identified as narrative style if at least 10% of the selected frames exhibit a narrative streak. Consequently, we download all narrative-style videos at high-resolution. Narrative-style videos typically cover WSIs at various magnifications, hence, we train a tissue-image-magnification classifier to predict the following three scales: {(1 – 10)x, (> 10 – 20)x, (> 20)x}. This provides relevant metadata for downstream objectives.
Text Extraction using ASR and text denoising.
The high costs associated with private medical ASR APIs 5 necessitated the use of a more conventional ASR model: Whisper [49]. As anticipated, this model often misinterprets medical terms, thus requiring the use of post-processing algorithms to minimize its error rates.
We propose a four-step text de-noising and quality control pipeline: i) We utilize the Rake keyword extraction algorithm to extract keywords or key-phrases up to four words and refine them by eliminating stopwords [51]. ii) We then cross-check each refined entry against UMLS [7] using the SciSpacy entity linking package [43]. If an entry is not found within UMLS, we check for misspelled words within the entry using a spell-checking algorithm6, instantiated with a specialized list of histopathology terms curated from various histopathology ontology labels and definitions. iii) With this probable list of misspelled keywords, we condition and prompt the LLM with examples to correct the misspelled entry within its context (sentence), and secondly, we task the LLM with identifying additional unconditioned errors/misspelled entries. For both, we leverage a set of manually curated examples to prompt the LLM in-context. For more examples and failure cases, see Table 11 and Figure 9 in the Appendix. iv) Finally, to de-noise the text, we resolve the output mapping of incorrect ↦ correct entries by verifying the corrected words against UMLS and our curated list of histopathology words/phrases. Entries that pass this double-validation process are used to replace the initial noisy transcription. Leveraging domain-specific databases to extract the text and filter out noise allows us to bypass the correction of repetition errors and filler words, such as ’ah’, ’uhm’, ’the’, etc. in tandem, using LLMs allows us to concentrate on correcting medically-relevant misspelled words, rather than correcting non-medically-relevant terms.
From the ASR-corrected text, we extract medical text which describes the image(s) as a whole. Also, when the speaker describes/gestures at visual regions-of-interest through statements like “look here …”, we extract the text entity being described as ROI text. To filter relevant medical text and ROI text from the ASR-corrected text, we utilize LLMs (see Figure 9 in Appendix), a decision rooted in a few compelling reasons: 1) Curating pre-training datasets at a scale that can tolerate higher levels of noise, LLMs are more cost-effective than expert-human (medical) labor. 2) The task does not require LLMs to generate new information but instead they discriminate useful versus irrelevant signals, serving to improve the signal-to-noise ratio of the data. To extract relevant text, we prompt LLMs to filter out all non-medically relevant text, providing context as necessary. See Figure 2 for some example image-text pairs. Lastly, we instruct the LLMs to refrain from introducing any new words beyond the corrected noisy text and set the model’s temperature to zero. Finally, we use LLMs to categorize our videos into one of the 18 identified sub-pathology classes. Similar to the previous tasks, this categorization is done by conditioning with a few examples and prompting the LLM to predict the top three possible classes given the text. More details, prompts, and additional examples are presented in Figure 12 within the Appendix.
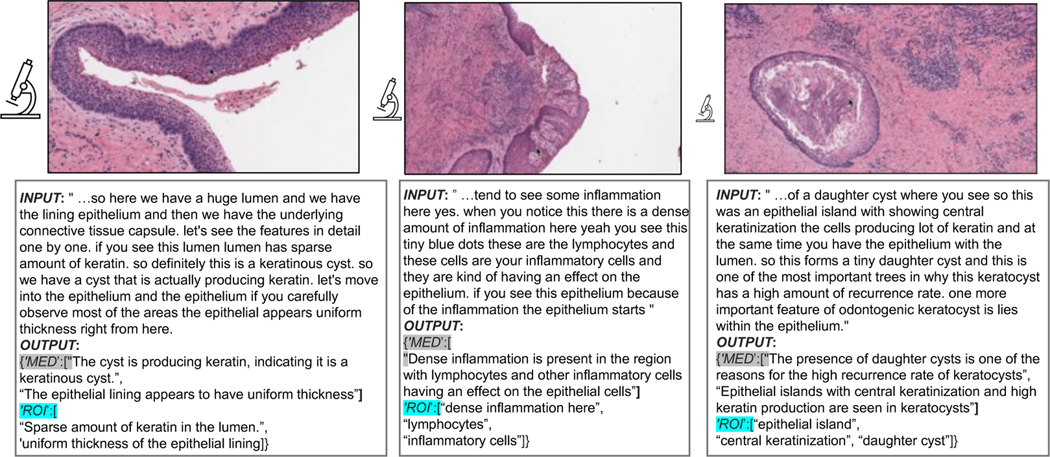
Figure 2: Quilt examples.
Input is the corrected ASR caption for the representative image. Output are the medical and ROI extracted text(s) paired with the image (see Section 3.1). In histopathology, understanding tissue characteristics often involves views from varying magnification levels. Thus, in Quilt we estimate an image’s magnification (indicated by the relative size of the microscope icon).
Image frame extraction and denoising.
For each video, we employ a similar method to that described in Filtering for narrative-style medical videos subsection to extract histopathology key-frames; our method leverages these frames’ times as beacons to break the entire video into time-intervals called chunks from which to extract representative image(s). Next, we extract the median image (pixel-space) of stable (static) frames in each chunk if they exists, else we de-duplicate the histopathology keyframes (beacons of the chunk). In essence, we use the extracted histopathology scene frames as guides for data collection, exploiting the human tendency in educational videos to pause narration during explanation, and we extract the relevant frame(s).
Aligning both modalities.
For each narrative-style video, we perform the following steps to align image and text modalities: First, we compute histopathology time chunks denoted as from keyframes after discriminating histopathology frames using the histopathology ensemble classifier – (scene_chunks). Each scene_chunk is padded with pad_time to its left and right; see Figure 8 and Table 9 in the Appendix for more details.
Text: we use the ASR output to extract the words spoken during each chunk in scene_chunks. Using the method described in Text Extraction using ASR and text denoising subsection, we extract the Medical and ROI caption for this chunk.
Image: we extract representative image(s) for every chunk/time-interval in scene_chunks as described in Filtering for narrative-style medical videos subsection above.
Finally, each chunk in scene_chunks is mapped to texts (both medical and ROI captions) and images. Next we map each medical image to one or more medical text. Using the time interval in which the image occurs, we extract its raw text from ASR and then correct and extract keywords using the Rake method, which we refer to as raw_keywords. We extract keywords from each medical text returned using the LLM, and we refer to these as keywords. Finally, if the raw_keywords occur before or slightly after a selected representative image, and overlap with the keywords in one of the Medical/ROI texts for that chunk, we map the image to the medical/ROI text. Example. keywords: psammoma bodies, match with raw_keyword: psammoma bodies within the ASR-corrected text ‘Meningiomas typically have a meningothelial pattern with lobular-like arrangements and psammoma bodies.’Refer to Figure 7 and Figure 15 in the Appendix for a detailed explanation of the method and examples of aligned image and text.
3.2. Quilt-1M: Combining Quilt with other histopathology data sources
To create Quilt-1M, we expanded Quilt by adding other disparate histopathology image-text open-access sources: LAION, Twitter, and PubMed.
PubMed Open Access Articles.
We searched the PubMed open-access from 2010–2022, extracting 62,458 histopathology image-text pairs, using our histopathology classifier and multi-plane figure cropping algorithm. The images are categorized into (1) images that are fully histopathology, (2) multi-plane images that contain histopathology sub-figures, and (3) histopathology sub-figures cropped from (1) and (2). See Figure 16, and Section A.2.1 in the Appendix.
Histopathology Image Retrieval from LAION.
The Large-scale Artificial Intelligence Open Network (LAION-5B) [54] curated over 5 billion pairs of images and text from across the Internet, including a substantial volume of histopathology-related data. We tapped into this resource by retrieving 23,240 image and text pairs. See Section A.2.2 in the Appendix.
Twitter Data from OpenPath.
We utilized a list of tweets curated by Huang et al. [24], which totaled up to 55,000 unique tweets and made up 133,526 unique image-text pairs. This exhibits a one-to-many relationship where many images were matched with multiple captions; this differentiated our work from the OpenPath approach. To maintain comparability, we followed their text pre-processing pipeline [24]. See Section A.2.3 in the Appendix.
3.3. Quality
To evaluate our pipeline’s performance, we assess several aspects. First, we calculate the precision of our LLM’s corrections by dividing the number of conditioned misspelled errors replaced (i.e., passed the UMLS check) by the total number of conditioned misspelled words found, yielding an average of 57.9%. We also determined the unconditioned precision of the LLM, similar to the previous step, and found it to be 13.8%. Therefore, we replace our detected incorrect words with the LLM’s correction 57.9% of the time, and 13.8% of the time we replace the LLM’s detected errors with its correction (see Table 11 in the Appendix). To estimate the ASR model’s transcription performance, we compute the total number of errors replaced (both conditioned and unconditioned) and divide it by the total number of words in each video, resulting in an average ASR error rate of 0.79%. To assess the LLM’s sub-pathology classification, we manually annotated top-k sub-pathology types for 100 random videos from our dataset. The LLM’s accuracy for top-3, top-2, and top-1 was 94.9%, 91.9%, and 86.8%, respectively. Also note that, by prompting the LLM to extract only medically relevant text, we further eliminate identifiable information, such as clinic addresses, from our dataset.
3.4. Final dataset statistics
We collected Quilt, from 4504 narrative videos spanning over 1087 hours with over 438K unique images with 768K associated text pairs. The mean length of the text captions is 22.76 words, and 8.68 words for ROI text, with an average of 1.74 medical sentences per image (max=5.33, min=1.0). Our dataset spans a total of 1.469M UMLS entities from those mentioned in the text (with 28.5K unique). The images span varying microscopic magnification scales (0–10x, 10–20x, 20–40x), obtaining (280K, 75K, 107K) images from each scale respectively. Figure 14 (a, c) in the Appendix plots our dataset’s diversity across multiple histopathology sub-domains. This plot shows that the captions cover histopathology-relevant medical subtypes: findings, concepts, organs, neoplastic processes, cells, diseases, and a mix of laboratory and diagnostic procedures. Overall, across all 127 UMLS semantic types, our entities cover 76.2% of medically-related semantic types (e.g., findings, disease, or syndrome) and 23.75% non-medical (e.g., geographic area, governmental or regulatory activity).
4. QuiltNet: Experiments training with Quilt-1M
We use the Contrastive Language-Image Pre-training (CLIP) objective [48] to pretrain QuiltNet using Quilt-1M. CLIP takes a batch of (image, text) pairs and optimizes a contrastive objective to create a joint embedding space. The optimization process involves concurrent training of both image and text encoders to increase the cosine similarity of embeddings from aligned pairs, while decreasing it for unaligned pairs. The objective is minimized via the InfoNCE loss, expressed as:
where and are the embeddings for the aligned -th image and text, respectively. For the image encoder, we use both ViT-B/32 and ViT-B/16 architectures [16]. For the text encoder, we use GPT-2 [47] with a context length of 77, and PubmedBert [70]. We train QuiltNet by finetuning a pre-trained CLIP model on Quilt-1M to enhance its performance in histopathology. Once finetuned, we conduct experiments on two types of downstream tasks: image classification (zero-shot and linear probing) and cross-modal retrieval (zero-shot). We also compare the performance of fine-tuning a pre-trained CLIP model versus training it from scratch.
Downstream histopathology datasets.
We evaluate the utility of QuiltNet on 12 downstream datasets: PatchCamelyon [60] contains histopathology scans of lymph node sections labeled for metastatic tissue presence as a binary label. NCT-CRC-HE-100K [32] consists of colorectal cancer images and is categorized into cancer and normal tissue. For SICAPv2 [56] the images are labeled as non-cancerous, Grade 3–5. Databiox [8] consists of invasive ductal carcinoma cases of Grades I-III. BACH [4] consists of breast tissues labeled as normal, benign, in-situ, and invasive carcinoma. Osteo [5] is a set of tissue patches representing the heterogeneity of osteosarcoma. RenalCell [10] contains tissue images of clear-cell renal cell carcinoma annotated into five tissue texture types. SkinCancer [35] consists of tissue patches from skin biopsies of 12 anatomical compartments and 4 neoplasms that make up the SkinTumor Subset. MHIST [63] contains tissue patches from Formalin-Fixed Paraffin-Embedded WSIs of colorectal polyps. LC25000 [9], which we divide into LC25000 (Lung) and LC25000 (Colon), contains tissue of lung and colon adenocarcinomas. For more details on the datasets refer to C.1 and Table 15 in the Appendix.
Results using zero-shot learning.
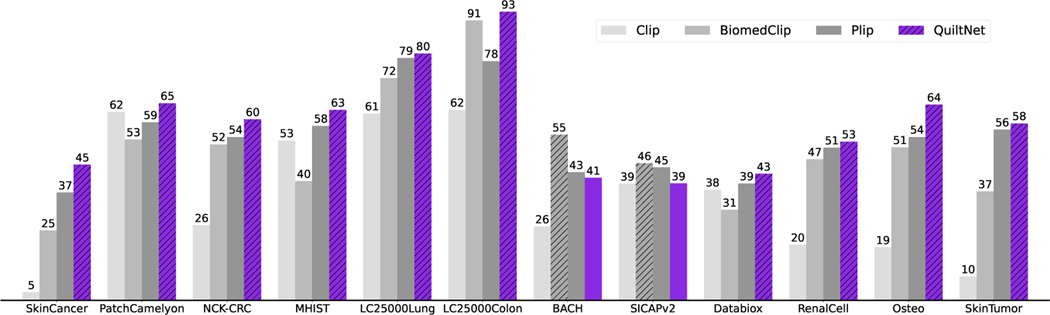
Given the vast diversity of cancer sub-types in histopathology, it is critical that a model maintains comprehensive understanding without requiring specific data for retraining. Thus, we evaluate our model’s zero-shot performance against three state-of-the-art models: CLIP, BiomedCLIP, and PLIP. Our model demonstrates superior performance, as illustrated in Figure 3, where it outperforms the other models in all but two datasets, in which BiomedCLIP performs marginally better. See Table 17 for UMap visualizations and Figure 17 for cross-modal attention visualization comparison in the Appendix. The prompts used for these evaluations are presented in Table 16 in the Appendix. To ensure a fair comparison with BiomedCLIP, which uses a ViT-B/16 and PMB/256 (pre-trained with [70]), we trained three different variants of our model. For detailed insights into the results, please refer to Table 14 in the Appendix.
Figure 3:
QuiltNet, outperforms out-of-domain CLIP baseline and state-of-the-art histopathology models across 12 zero-shot tasks, covering 8 different sub-pathologies (accuracy percentage provided).
Results using linear probing.
We assess the few-shot and full-shot performance of our model by conducting linear probing with 1%, 10%, and 100% of the training data, sampled with three different seeds; we report the average accuracy and their standard deviation in Table 1. We deploy our evaluation across four distinct datasets, specifically those with dedicated training and testing sets among our external datasets. Remarkably, our model, utilizing the ViT-B/32 architecture with GPT/77, outperforms its counterparts, BiomedCLIP, PLIP, and CLIP, in most datasets. On the NCT-CRC and SICAPv2 datasets, our model surpasses even the fully supervised performance using only 1% of the labels. Also, note that for some results 10% does better than 100%; this is because we are sampling from each class equally, and thus the 10% subset contains a more balanced training set than 100%, for datasets that are very imbalanced, resulting in sub-optimal performance at 100%.
Table 1: Linear probing.
Classification results, denoted as accuracy % (standard deviation). Camelyon denotes the PatchCamelyon dataset. Supervised results are from each dataset’s SOTA models.
| Dataset | %shot | ViT-B/32 | ViT-B/16 | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| CLIP | PLIP | QuiltNet | CLIP | QuiltNet | BiomedCLIP | QuiltNet | ||
| Supervised(%) | GPT/77 | GPT/77 | GPT/77 | GPT/77 | GPT/77 | PMB/256 | PMB/256 | |
| NCT-CRC [32] | 1 | 91.0 (0.10) | 93.75 (0.09) | 94.64(0.22) | 90.96 (0.10) | 93.36 (0.23) | 92.14(0.12) | 93.55 (0.25) |
| 10 | 92.02(1.30) | 93.83 (0.06) | 95.30 (0.03) | 92.58 (0.12) | 93.85 (0.04) | 92.90 (0.07) | 93.72 (0.08) | |
| (94.0) | 100 | 91.83 (0.01) | 94.16 (0.01) | 95.22 (0.01) | 92.26 (0.09) | 93.76 (0.02) | 92.97 (0.05) | 93.60 (0.01) |
|
| ||||||||
| Camelyon [60] | 1 | 80.38 (0.16) | 87.26 (0.23) | 87.62 (0.35) | 80.28 (0.20) | 84.78 (0.14) | 83.63 (0.44) | 83.48 (0.18) |
| 10 | 82.67 (0.19) | 87.48 (0.08) | 87.55 (0.03) | 82.20 (0.04) | 86.77 (0.09) | 84.18 (0.15) | 84.42 (0.10) | |
| (97.5) | 100 | 82.80 (0.01) | 87.34 (0.01) | 87.48 (0.01) | 82.55 (0.02) | 86.81 (0.04) | 84.23 (0.01) | 84.44 (0.02) |
|
| ||||||||
| SkinCancer [35] | 1 | 84.27 (0.22) | 91.07 (0.25) | 90.93 (0.25) | 85.62 (0.16) | 88.29 (0.15) | 87.53 (0.21) | 88.06 (0.20) |
| 10 | 89.0 (0.02) | 93.39 (0.05) | 92.99 (0.02) | 90.28 (0.01) | 91.20 (0.0) | 89.23 (0.03) | 90.03 (0.02) | |
| (93.3) | 100 | 89.02 (0.02) | 93.29 (0.01) | 93.03 (0.02) | 90.29 (0.03) | 91.20 (0.0) | 89.16 (0.02) | 89.91 (0.01) |
|
| ||||||||
| SICAPv2 [56] | 1 | 52.45 (2.41) | 65.76 (2.65) | 69.92 (1.02) | 56.01 (0.66) | 66.86 (1.16) | 69.43 (1.03) | 68.49 (1.06) |
| 10 | 62.24 (0.65) | 69.23 (0.43) | 74.14 (0.38) | 63.70 (0.69) | 72.37 (0.65) | 71.61 (0.31) | 72.48 (0.42) | |
| (67.0) | 100 | 65.75 (0.16) | 73.0 (0.14) | 75.48 (0.12) | 68.74 (0.10) | 74.14(0.16) | 74.57 (0.04) | 74.60 (0.17) |
Results using cross-modal retrieval.
In our study, we evaluate cross-modal retrieval efficacy by examining both zero-shot text-to-image and image-to-text retrieval capabilities. We accomplish this by identifying the nearest neighbors for each modality and then determining whether the corresponding pair is within the top nearest neighbors, where . Our experiments are conducted on two datasets: our holdout dataset from Quilt-1M and the ARCH dataset. Results are in Table 2.
Table 2:
Cross-modal retrieval results on the Quilt-1M holdout set and ARCH dataset. In each cell, the results are displayed in the format (%/%), with Quilt-1M holdout results on the left and ARCH results on the right. The best-performing results are highlighted in bold text.
| model | config | Text-to-Image (%) | Image-to-Text (%) | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| R@1 | R@50 | R@200 | R@1 | R@50 | R@200 | ||
| CLIP | ViT-B/32|GPT/77 | 0.49/0.07 | 4.73/2.42 | 10.15/7.21 | 0.39/0.05 | 3.99/2.52 | 8.80/7.22 |
| PLIP | ViT-B/32|GPT/77 | 1.05/0.56 | 10.79/13.10 | 21.80/29.85 | 0.87/0.74 | 11.04/13.75 | 21.63/29.46 |
| QuiltNet | ViT-B/32|GPT/77 | 1.17/1.41 | 16.31/19.87 | 31.99/39.13 | 1.24/1.35 | 14.89/19.20 | 28.97/38.57 |
|
| |||||||
| CLIP | ViT-B/16|GPT/77 | 0.83/0.09 | 5.63/2.73 | 11.26/8.72 | 0.66/0.13 | 5.02/3.09 | 10.82/9.04 |
| QuiltNet | ViT-B/16|GPT/77 | 2.42/1.29 | 22.38/20.30 | 41.05/40.89 | 2.00/1.01 | 21.66/16.18 | 39.29/34.15 |
| BiomedCLIP | ViT-B/16(224)|PMB/256 | 4.34/8.89 | 14.99/53.24 | 25.62/71.43 | 3.88/9.97 | 13.93/52.13 | 23.53/68.47 |
| QuiltNet | ViT-B/16(224)|PMB/256 | 6.20/8.77 | 30.28/55.14 | 50.60/77.64 | 6.27/9.85 | 31.06/53.06 | 50.86/73.43 |
5. Discussion
Limitations.
Despite the promising results, Quilt was curated using several handcrafted algorithms and LLMs. Such curation methods, while effective, introduce their own biases and errors. For instance, our histopathology classifier had occasional false positives (≈ 5%) confirmed by human evaluation. Occasionally, ASR can misinterpret a medical term and transcribe it as a different existing term, such as transcribing ’serous carcinoma’ as ’serious carcinoma’. Unfortunately, such errors are not rectifiable using our current pipeline (see Table 11 in the Appendix). While not directly a limitation of our dataset, training a CLIP model trained from scratch underperformed compared to fine-tuning a pre-trained CLIP (see Table 14 in the Appendix). This suggests that a million image-text pairs may still not be sufficient.
Data Collection and Societal Biases
Aligning in strategies with [68], we release Quilt derived from public videos, taking structured steps to limit privacy and consent harms (see A.5 in the Appendix). Complying with YouTube’s privacy policy, we only provide video IDs, allowing users to opt-out of our dataset. Researchers can employ our pipeline to create Quilt. Regarding societal biases, a significant portion of our narrators originate from western institutions, a situation that is further amplified by our focus on English-only videos. Consequently, QuiltNet may exhibit inherent biases, potentially performing better on data associated with these demographics, while possibly underperforming when applied to other cultural or linguistic groups.
Conclusion.
We introduced Quilt-1M, the largest open-sourced histopathology dataset to date. Empirical results validate that pre-training using Quilt is valuable, outperforming larger state-of-the-art models like BiomedCLIP across various sub-pathology types and tasks including zero-shot, few-shot, full-shot, and cross-modal retrieval. We established a new state-of-the-art in zero-shot, linear probing, and cross-modal retrieval tasks in the field of Histopathology.
Supplementary Material
Acknowledgments
Research reported in this study was supported by the National Cancer Institute under Awards No. R01 CA151306, R01CA225585, R01 CA201376, and R01 CA200690 and the Office of the Assistant Secretary of Defense for Health Affairs through the Melanoma Research Program under Awards No. W81XWH-20-1-0797 and W81XWH-20-1-0798. Opinions, conclusions, and recommendations are those of the authors.
Footnotes
The data and code will be available at Quilt-1M
References
- [1].Agrawal M, Hegselmann S, Lang H, Kim Y, and Sontag D. Large language models are zero-shot clinical information extractors. arXiv preprint arXiv:2205.12689, 2022. [Google Scholar]
- [2].Amith M, Cui L, Roberts K, Xu H, and Tao C. Ontology of consumer health vocabulary: providing a formal and interoperable semantic resource for linking lay language and medical terminology. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1177–1178. IEEE, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Araujo A, Chaves J, Lakshman H, Angst R, and Girod B. Large-scale query-by-image video retrieval using bloom filters. arXiv preprint arXiv:1604.07939, 2016. [Google Scholar]
- [4].Aresta G, Araújo T, Kwok S, Chennamsetty SS, Safwan M, Alex V, Marami B, Prastawa M, Chan M, Donovan M, et al. Bach: Grand challenge on breast cancer histology images. Medical image analysis, 56:122–139, 2019. [DOI] [PubMed] [Google Scholar]
- [5].Arunachalam HB, Mishra R, Daescu O, Cederberg K, Rakheja D, Sengupta A, Leonard D, Hallac R, and Leavey P. Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PloS one, 14(4):e0210706, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baker B, Akkaya I, Zhokov P, Huizinga J, Tang J, Ecoffet A, Houghton B, Sampedro R, and Clune J. Video pretraining (vpt): Learning to act by watching unlabeled online videos. Advances in Neural Information Processing Systems, 35:24639–24654, 2022. [Google Scholar]
- [7].Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res, 32(Database-Issue):267–270, 2004. URL http://dblp.uni-trier.de/db/journals/nar/nar32.html#Bodenreider04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bolhasani H, Amjadi E, Tabatabaeian M, and Jassbi SJ. A histopathological image dataset for grading breast invasive ductal carcinomas. Informatics in Medicine Unlocked, 19:100341, 2020. [Google Scholar]
- [9].Borkowski AA, Bui MM, Thomas LB, Wilson CP, DeLand LA, and Mastorides SM. Lung and colon cancer histopathological image dataset (lc25000). arXiv preprint arXiv:1912.12142, 2019. [Google Scholar]
- [10].Brummer O, Polonen P, Mustjoki S, and Bruck O. Integrative analysis of histological textures and lymphocyte infiltration in renal cell carcinoma using deep learning. bioRxiv, pages 2022–08, 2022. [Google Scholar]
- [11].Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, and Joulin A. Emerging properties in self-supervised vision transformers. In Proceedings of the IEEE/CVF international conference on computer vision, pages 9650–9660, 2021. [Google Scholar]
- [12].Chen RJ, Lu MY, Weng W-H, Chen TY, Williamson DF, Manz T, Shady M, and Mahmood F. Multimodal co-attention transformer for survival prediction in gigapixel whole slide images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 4015–4025, 2021. [Google Scholar]
- [13].Chen RJ, Chen C, Li Y, Chen TY, Trister AD, Krishnan RG, and Mahmood F. Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16144–16155, 2022. [Google Scholar]
- [14].Chen T, Kornblith S, Norouzi M, and Hinton G. A simple framework for contrastive learning of visual representations. In International conference on machine learning, pages 1597–1607. PMLR, 2020. [Google Scholar]
- [15].Ding B, Qin C, Liu L, Bing L, Joty S, and Li B. Is gpt-3 a good data annotator? arXiv preprint arXiv:2212.10450, 2022. [Google Scholar]
- [16].Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929, 2020. [Google Scholar]
- [17].Doukhan D, Carrive J, Vallet F, Larcher A, and Meignier S. An open-source speaker gender detection framework for monitoring gender equality. In Acoustics Speech and Signal Processing (ICASSP), 2018 IEEE International Conference on. IEEE, 2018. [Google Scholar]
- [18].Eslami S, de Melo G, and Meinel C. Does clip benefit visual question answering in the medical domain as much as it does in the general domain? arXiv preprint arXiv:2112.13906, 2021. [Google Scholar]
- [19].Fragoso G, de Coronado S, Haber M, Hartel F, and Wright L. Overview and utilization of the nci thesaurus. Comparative and functional genomics, 5(8):648–654, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gamper J and Rajpoot N. Multiple instance captioning: Learning representations from histopathology textbooks and articles. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16549–16559, 2021. [Google Scholar]
- [21].Gilardi F, Alizadeh M, and Kubli M. Chatgpt outperforms crowd-workers for text-annotation tasks. arXiv preprint arXiv:2303.15056, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].He X, Zhang Y, Mou L, Xing E, and Xie P. Pathvqa: 30000+ questions for medical visual question answering. arXiv preprint arXiv:2003.10286, 2020. [Google Scholar]
- [23].Huang S-C, Shen L, Lungren MP, and Yeung S. Gloria: A multimodal global-local representation learning framework for label-efficient medical image recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision, pages 3942–3951, 2021. [Google Scholar]
- [24].Huang Z, Bianchi F, Yuksekgonul M, Montine T, and Zou J. Leveraging medical twitter to build a visual–language foundation model for pathology ai. bioRxiv, pages 2023–03, 2023. [DOI] [PubMed] [Google Scholar]
- [25].Hussain E, Mahanta LB, Borah H, and Das CR. Liquid based-cytology pap smear dataset for automated multi-class diagnosis of pre-cancerous and cervical cancer lesions. Data in brief, 30:105589, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ikezogwo WO, Seyfioglu MS, and Shapiro L. Multi-modal masked autoencoders learn compositional histopathological representations. arXiv preprint arXiv:2209.01534, 2022. [Google Scholar]
- [27].Ilharco G, Wortsman M, Wightman R, Gordon C, Carlini N, Taori R, Dave A, Shankar V, Namkoong H, Miller J, Hajishirzi H, Farhadi A, and Schmidt L. Openclip, July 2021. URL 10.5281/zenodo.5143773. [DOI] [Google Scholar]
- [28].Jobin K, Mondal A, and Jawahar C. Docfigure: A dataset for scientific document figure classification. In 2019 International Conference on Document Analysis and Recognition Workshops (ICDARW), volume 1, pages 74–79. IEEE, 2019. [Google Scholar]
- [29].Johnson AE, Pollard TJ, Greenbaum NR, Lungren MP, Deng C.-y., Peng Y, Lu Z, Mark RG, Berkowitz SJ, and Horng S. Mimic-cxr-jpg, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jupp S, Malone J, Burdett T, Heriche J-K, Williams E, Ellenberg J, Parkinson H, and Rustici G. The cellular microscopy phenotype ontology. Journal of biomedical semantics, 7: 1–8, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Karishma Z. Scientific document figure extraction, clustering and classification. 2021. [Google Scholar]
- [32].Kather JN, Halama N, and Marx A. 100,000 histological images of human colorectal cancer and healthy tissue. Zenodo 10, 5281, 2018. [Google Scholar]
- [33].Kembhavi A, Salvato M, Kolve E, Seo M, Hajishirzi H, and Farhadi A. A diagram is worth a dozen images. In Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part IV 14, pages 235–251. Springer, 2016. [Google Scholar]
- [34].Kojima T, Gu SS, Reid M, Matsuo Y, and Iwasawa Y. Large language models are zero-shot reasoners. arXiv preprint arXiv:2205.11916, 2022. [Google Scholar]
- [35].Kriegsmann K, Lobers F, Zgorzelski C, Kriegsmann J, Janßen C, Meliß RR, Muley T, Sack U, Steinbuss G, and Kriegsmann M. Deep learning for the detection of anatomical tissue structures and neoplasms of the skin on scanned histopathological tissue sections. Frontiers in Oncology, 12, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu J, Wang Q, Fan H, Wang S, Li W, Tang Y, Wang D, Zhou M, and Chen L. Automatic label correction for the accurate edge detection of overlapping cervical cells. arXiv preprint arXiv:2010.01919, 2020. [Google Scholar]
- [37].Liu S, Zhu C, Xu F, Jia X, Shi Z, and Jin M. Bci: Breast cancer immunohistochemical image generation through pyramid pix2pix. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 1815–1824, 2022. [Google Scholar]
- [38].Liu Z, Luo P, Wang X, and Tang X. Deep learning face attributes in the wild. In Proceedings of International Conference on Computer Vision (ICCV), December 2015. [Google Scholar]
- [39].Liu Z, Mao H, Wu C-Y, Feichtenhofer C, Darrell T, and Xie S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 11976–11986, 2022. [Google Scholar]
- [40].Loshchilov I and Hutter F. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101, 2017. [Google Scholar]
- [41].Marini N, Marchesin S, Otálora S, Wodzinski M, Caputo A, Van Rijthoven M, Aswolinskiy W, Bokhorst J-M, Podareanu D, Petters E, et al. Unleashing the potential of digital pathology data by training computer-aided diagnosis models without human annotations. NPJ digital medicine, 5(1):102, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morris D, Müller-Budack E, and Ewerth R. Slideimages: a dataset for educational image classification. In Advances in Information Retrieval: 42nd European Conference on IR Research, ECIR 2020, Lisbon, Portugal, April 14–17, 2020, Proceedings, Part II 42, pages 289–296. Springer, 2020. [Google Scholar]
- [43].Neumann M, King D, Beltagy I, and Ammar W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. In Proceedings of the 18th BioNLP Workshop and Shared Task, pages 319–327, Florence, Italy, Aug. 2019. Association for Computational Linguistics. doi: 10.18653/v1/W19-5034. URL https://www.aclweb.org/anthology/W19-5034. [DOI] [Google Scholar]
- [44].Noy NF, Musen MA, Mejino JL Jr, and Rosse C. Pushing the envelope: challenges in a frame-based representation of human anatomy. Data & Knowledge Engineering, 48(3): 335–359, 2004. [Google Scholar]
- [45].Pelka O, Koitka S, Rückert J, Nensa F, and Friedrich CM. Radiology objects in context (roco): a multimodal image dataset. In Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis: 7th Joint International Workshop, CVII-STENT 2018 and Third International Workshop, LABELS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Proceedings 3, pages 180–189. Springer, 2018. [Google Scholar]
- [46].Pont-Tuset J, Uijlings J, Changpinyo S, Soricut R, and Ferrari V. Connecting vision and language with localized narratives. In ECCV, 2020. [Google Scholar]
- [47].Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I, et al. Language models are unsupervised multitask learners. OpenAI blog, 1(8):9, 2019. [Google Scholar]
- [48].Radford A, Kim JW, Hallacy C, Ramesh A, Goh G, Agarwal S, Sastry G, Askell A, Mishkin P, Clark J, et al. Learning transferable visual models from natural language supervision. In International conference on machine learning, pages 8748–8763. PMLR, 2021. [Google Scholar]
- [49].Radford A, Kim JW, Xu T, Brockman G, McLeavey C, and Sutskever I. Robust speech recognition via large-scale weak supervision. arXiv preprint arXiv:2212.04356, 2022. [Google Scholar]
- [50].Roberts RJ. Pubmed central: The genbank of the published literature, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rose S, Engel D, Cramer N, and Cowley W. Automatic keyword extraction from individual documents. Text mining: applications and theory, pages 1–20, 2010. [Google Scholar]
- [52].Santos T, Tariq A, Das S, Vayalpati K, Smith GH, Trivedi H, and Banerjee I. Pathologybert–pre-trained vs. a new transformer language model for pathology domain. arXiv preprint arXiv:2205.06885, 2022. [PMC free article] [PubMed] [Google Scholar]
- [53].Schofield PN, Sundberg JP, Sundberg BA, McKerlie C, and Gkoutos GV. The mouse pathology ontology, mpath; structure and applications. Journal of biomedical semantics, 4(1): 1–8, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schuhmann C, Beaumont R, Vencu R, Gordon C, Wightman R, Cherti M, Coombes T, Katta A, Mullis C, Wortsman M, et al. Laion-5b: An open large-scale dataset for training next generation image-text models. arXiv preprint arXiv:2210.08402, 2022. [Google Scholar]
- [55].Selvaraju R, Cogswell M, Das A, Vedantam R, Parikh D, and Batra D. Grad-cam: Visual explanations from deep networks via gradient-based localization. arxiv 2016. arXiv preprint arXiv:1610.02391. [Google Scholar]
- [56].Silva-Rodríguez J, Colomer A, Sales MA, Molina R, and Naranjo V. Going deeper through the gleason scoring scale: An automatic end-to-end system for histology prostate grading and cribriform pattern detection. Computer methods and programs in biomedicine, 195:105637, 2020. [DOI] [PubMed] [Google Scholar]
- [57].Singh A, Natarajan V, Shah M, Jiang Y, Chen X, Batra D, Parikh D, and Rohrbach M. Towards vqa models that can read. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pages 8317–8326, 2019. [Google Scholar]
- [58].Singh H and Graber ML. Improving diagnosis in health care–the next imperative for patient safety. The New England journal of medicine, 373(26):2493–2495, 2015. [DOI] [PubMed] [Google Scholar]
- [59].Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, Scales N, Tanwani A, Cole-Lewis H, Pfohl S, et al. Large language models encode clinical knowledge. arXiv preprint arXiv:2212.13138, 2022. [Google Scholar]
- [60].Veeling BS, Linmans J, Winkens J, Cohen T, and Welling M. Rotation equivariant cnns for digital pathology. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, 2018, Proceedings, Part II 11, pages 210–218. Springer, 2018. [Google Scholar]
- [61].Voigtlaender P, Changpinyo S, Pont-Tuset J, Soricut R, and Ferrari V. Connecting Vision and Language with Video Localized Narratives. In IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2023. [Google Scholar]
- [62].Wang S, Liu Y, Xu Y, Zhu C, and Zeng M. Want to reduce labeling cost? gpt-3 can help. arXiv preprint arXiv:2108.13487, 2021. [Google Scholar]
- [63].Wei J, Suriawinata A, Ren B, Liu X, Lisovsky M, Vaickus L, Brown C, Baker M, Tomita N, Torresani L, et al. A petri dish for histopathology image analysis. In Artificial Intelligence in Medicine: 19th International Conference on Artificial Intelligence in Medicine, AIME 2021, Virtual Event, June 15–18, 2021, Proceedings, pages 11–24. Springer, 2021. [Google Scholar]
- [64].Weitz P, Valkonen M, Solorzano L, Carr C, Kartasalo K, Boissin C, Koivukoski S, Kuusela A, Rasic D, Feng Y, et al. Acrobat–a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology. arXiv preprint arXiv:2211.13621, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wright PS, Briggs KA, Thomas R, Smith GF, Maglennon G, Mikulskis P, Chapman M, Greene N, Phillips BU, and Bender A. Statistical analysis of preclinical inter-species concordance of histopathological findings in the etox database. Regulatory Toxicology and Pharmacology, 138:105308, 2023. [DOI] [PubMed] [Google Scholar]
- [66].Wu H, Wang W, Wan Y, Jiao W, and Lyu M. Chatgpt or grammarly? evaluating chatgpt on grammatical error correction benchmark. arXiv preprint arXiv:2303.13648, 2023. [Google Scholar]
- [67].Wu W, Mehta S, Nofallah S, Knezevich S, May CJ, Chang OH, Elmore JG, and Shapiro LG. Scale-aware transformers for diagnosing melanocytic lesions. IEEE Access, 9: 163526–163541, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zellers R, Lu X, Hessel J, Yu Y, Park JS, Cao J, Farhadi A, and Choi Y. Merlot: Multimodal neural script knowledge models. Advances in Neural Information Processing Systems, 34:23634–23651, 2021. [Google Scholar]
- [69].Zellers R, Lu J, Lu X, Yu Y, Zhao Y, Salehi M, Kusupati A, Hessel J, Farhadi A, and Choi Y. Merlot reserve: Neural script knowledge through vision and language and sound. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16375–16387, 2022. [Google Scholar]
- [70].Zhang S, Xu Y, Usuyama N, Bagga J, Tinn R, Preston S, Rao R, Wei M, Valluri N, Wong C, et al. Large-scale domain-specific pretraining for biomedical vision-language processing. arXiv preprint arXiv:2303.00915, 2023. [Google Scholar]
- [71].Zhang Y, Jiang H, Miura Y, Manning CD, and Langlotz CP. Contrastive learning of medical visual representations from paired images and text. In Machine Learning for Healthcare Conference, pages 2–25. PMLR, 2022. [Google Scholar]
- [72].Zhou B, Lapedriza A, Khosla A, Oliva A, and Torralba A. Places: A 10 million image database for scene recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.