Abstract
HPV35 has been found in only ~2% of invasive cervical cancers (ICC) worldwide but up to 10% in Sub-Saharan Africa, warranting further investigation and consideration of impact on preventive strategies. We studied HPV35 and ethnicity, in relation to the known steps in cervical carcinogenesis, using multiple large epidemiologic studies in the U.S. and internationally. Combining five U.S. studies, we measured HPV35 positivity and, in Northern California, observed HPV35 type-specific population prevalence and estimated 5-year risk of developing precancer when HPV35-positive. HPV35 genetic variation was examined for differences in carcinogenicity in 1053 HPV35+ cervical specimens from a U.S. cohort and an international collection. African-American women had more HPV35 (12.1% vs 5.1%, P <.001) and more HPV35-associated precancers (7.4% vs 2.1%, P < .001) compared to other ethnicities. Precancer risks after HPV35 infection did not vary by ethnicity (global P = .52). The HPV35 A2 sublineage showed an increased association with precancer/cancer in African-Americans (OR = 5.6 vs A1, 95% CI = 1.3-24.8) and A2 was more prevalent among ICC in Africa than other world regions (41.9% vs 10.4%, P < .01). Our analyses support a strong link between HPV35 and cervical carcinogenesis in women of African ancestry. Current HPV vaccines cover the majority of cervical precancer/cancer across all ethnic groups; additional analyses are required to determine whether the addition of HPV35 to the already highly effective nine-valent HPV vaccine would provide better protection for women in Africa or of African ancestry.
Keywords: African ancestry women, cervical cancer, epidemiology, genetics, HPV35
1 |. INTRODUCTION
Persistent infection with 13 oncogenic types of human papillomavirus (HPV) is the established cause of nearly all invasive cervical cancers (ICC).1,2 While all oncogenic HPV types can cause cervical cancer, they are heterogeneous in prevalence and cancer risk. For example, HPV type 16 (HPV16) is the most common carcinogenic type accounting for ~60% of ICC whereas HPV35, which is genetically the closest related to HPV16, causes ~2% of ICC worldwide,1,3 that is, ~10 000 cases annually.4 Studies have identified variations in the prevalence of oncogenic HPV types among the general population as well as among cervical precancers/cancers across world regions5 and by ethnicity, including the U.S.6–9
Interestingly, HPV35 has increased prevalence in certain countries of Sub-Saharan Africa.10 Specifically, HPV35 is more common among precancerous lesions11,12 and corresponds to a higher fraction of ICC (4%-10%),5,10,12–14 compared to other regions.1,5 Studies in the U.S. suggest that, among women with prevalent infection and cervical precancer, African-American women have decreased HPV16 and increased HPV35, compared to other ethnic groups.6,7,9 This increase of HPV35 has not been thoroughly investigated among invasive cancers in African-American women.6,7,15
Variations in HPV35 carcinogenicity could have implications for cervical cancer prevention among women of African ancestry. The next-generation HPV vaccine covers seven oncogenic HPV types (HPV16, HPV18, HPV31, HPV33, HPV45, HPV52 and HPV58),16 yet, if HPV35 has greater importance among women of African ancestry, adding HPV35 to the vaccine might further improve vaccine effectiveness in African populations.
It is unclear why HPV35 is more prevalent, and perhaps carcinogenic, among women of African ancestry, particularly Sub-Saharan African countries. This variation could reflect the distribution of evolutionary-derived HPV35 variant lineages associated with distinct populations. This has been observed for HPV16.17,18 HPV35 has less known evolutionary genetic variation (ie, lineages) than most other oncogenic HPV types, with only a single main lineage and two sublineages, A1 and A2, as previously reported.19 We know very little about the relationship between HPV35 genetic variation and risk of cervical precancer/cancer, and virtually nothing about whether HPV35 carcinogenicity matches a woman’s ethnicity.20
Here we evaluate ethnic variations in the epidemiology of HPV35 and cervical precancer/cancer in five U.S. studies. In addition, to amassing the largest HPV35 study to date, we whole-genome sequenced 1053 HPV35-positive cervical specimens, collected within the U.S. and internationally, to determine whether HPV35 genetic variation is differentially associated with carcinogenicity among human populations.
2 |. METHODS
2.1 |. Study populations
This manuscript describes a series of analyses performed in six study populations that have been previously described (National Cancer Institute [NCI], Kaiser Permanente Northern California [KPNC], HPV Persistence and Progression [PaP] Cohort, the International Agency for Research on Cancer (IARC) collection and other NCI cohort studies).21–26 For the PaP cohort, the KPNC institutional review board (IRB) approved use of the data, and the National Institutes of Health Office of Human Subjects Research deemed our study exempt from IRB review. For all other NCI studies, local and National Cancer Institute IRBs approved the studies. For the IARC collection, both local and IARC ethical committees approved our study. We examined HPV35 prevalence, risk of cervical precancer given HPV35 infection, pooled HPV35 type attribution in cases, and the HPV35 genome. Details of study populations and sample collection are in the Data Supplement: Study Population and Figure S1.
2.2 |. HPV35 whole-genome sequencing and lineage assignment
We HPV whole-genome sequenced 1053 HPV35-positive specimens from the PaP Cohort (n = 608) and IARC biobank (n = 445; Figure S1) using an in-house designed primer panel and the Ion Torrent platform. Our bioinformatics pipeline was also developed in-house and can be accessed at https://github.com/cgrlab/cgrHPV35. Phylogenetic analyses identified sublineages A1 and A2, as well as a new lineage (named here as B). Mean nucleotide divergence between A1 and A2 was 0.40% (±0.05%) (~30 nucleotide), and the new B lineage differed by 0.78% (±0.08%) and 0.74 (±0.08) from A1 and A2, respectively. To show the relationship between lineages and sublineages, we created a tree with representative HPV35 sequences and used HPV31 as the outgroup to root the tree (Figure 1A).
FIGURE 1.
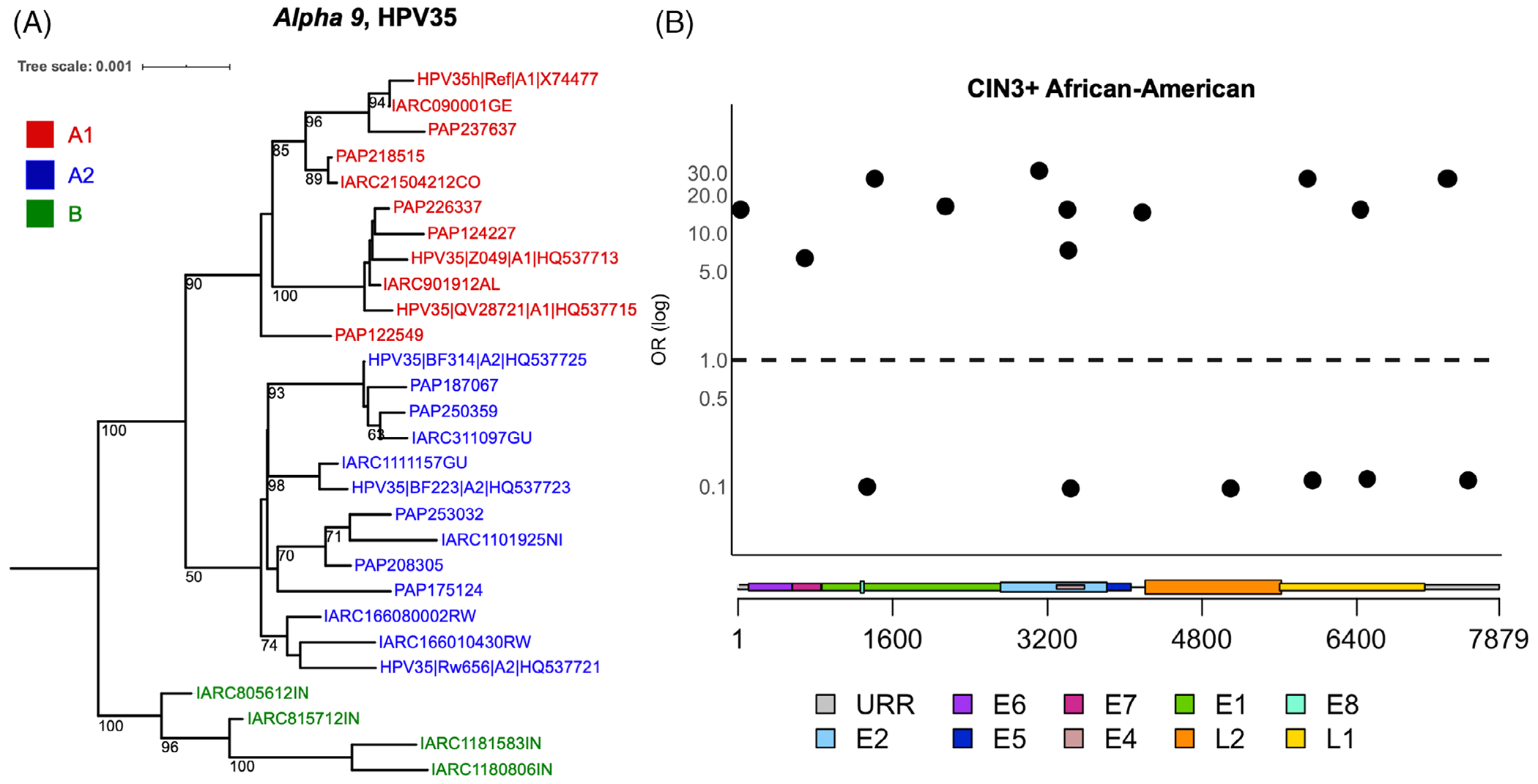
Phylogenetic tree illustrating HPV35 lineages, and individual SNPs across the HPV35 genome associated with CIN3+ in African-American women. A, HPV35 complete genome tree topology. The maximum likelihood (ML) tree was constructed using RAXML from a global alignment of complete HPV35 genome nucleotide sequences from the Persistence and Progression (PaP) Cohort and IARC samples, as well as reference sequences,29 bootstrap support values equal to or higher than 50 are shown on or near branches. HPV31 was used as an outgroup taxon to root the tree (not shown). Distinct variant lineages (ie, termed A and B) and sublineages (eg, termed A1 and A2) were inferred from the tree topology. The bar indicates the nucleotide substitutions per unit change (ie, 0.001) per site. B, SNPs significantly associated with CIN3+ among African-American women in the PaP Cohort. Odds ratios (OR) are shown on the y-axis and HPV35 genome positions by viral gene regions are shown on the x-axis. Circles represent variants significantly (P < .05) associated with CIN3+ compared to other nucleotide at the same genomic position. For details, see Table S4
2.3 |. Statistical analyses
2.3.1 |. Prevalence and risks associated with HPV35 infections in the PaP Cohort
To minimize epidemiologic bias, among 43 205 oncogenic HPV-positive women, we excluded 11 360 women under age 30 and over age 64 at their first cotest, since routine cotesting was not recommended at KPNC at those ages (Figure S1). Women without follow-up (2 or more screening visits or first screening visit that led to histology), prior hysterectomy or high-grade disease were also excluded (n = 3262). Self-assigned ethnicity was obtained from the KPNC cancer registry, mortality files, electronic medical records and previous large study databases. Among the remaining 28 583 women testing HPV-positive, 1728 (6.0%) were excluded because ethnicity information was unavailable; these women did not differ from other women by enrollment cytology or HPV type but they were slightly younger (mean 39.5 vs 41.4 years, P < .001) and less likely to have cervical intraepithelial neoplasia (CIN) grade 3 or worse (CIN3+) histology (3.2% vs 8.6%, P < .001). Ethnicity was reported separately. If women self-reported their ethnicity as Hispanic, they were classified as Hispanic. Women not classified as Hispanic were assigned according to their reported ethnicity: White, Black (includes African-American, and referred to in this article as African-American), Asian (includes Hawaiian/Pacific Islander) or other (includes when multiple etnics were selected). Women categorized as other were excluded from the analyses (n = 305, 1.1%).
To better represent the entire KPNC Guidelines Cohort testing HPV-positive, weights were applied based upon the sampling probability of a woman being selected for typing (~0.8 for cases vs ~0.3 for controls). The prevalence of HPV35 was compared by ethnicity. Cumulative risks of CIN3+ in the 5 years from time of HPV testing were estimated for each ethnic group among women with HPV35 (regardless of multiple HPV-coinfections). Cumulative 5-year risks of CIN3+ were estimated by jointly fitting a logistic regression model to CIN3+ present at baseline and a proportional hazards model for interval-censored incident CIN3+; joint estimation of the models allowed for the possibility that some CIN3+ diagnosed in follow-up among women not referred to immediate colposcopy were present at the baseline visit.27 Wald-test assessed heterogeneity between estimates. The analyses in this section were performed with R version 3.4.4.
2.3.2 |. HPV35 infections among cervical precancer and cancer cases in NCI studies
Among precancer/cancer, we evaluated the presence of oncogenic HPV types at the visit concurrent with or immediately preceding the diagnostic visit. For women with both single and multiple infections, we stratified cases by study, case subcategory (CIN grade 2 [CIN2], CIN3/AIS and cancer), and ethnic group. Within each stratified analysis of disease and ethnicity, we ranked the frequency of each oncogenic HPV type. For each woman with HPV35, her HPV35 infection was classified using a hierarchical attribution model28 as either: (1) “single HPV35 infection” due to single infection with no other concurrent oncogenic HPV, (2) “HPV35 co-infection, most prevalent” due to positivity with another oncogenic HPV infection that ranked less prevalent within her strata of study, disease and ethnicity (eg, a CIN3 showing HPV35 coinfection with HPV68 or HPV59) or (3) “HPV35 co-infection, less prevalent” due to positivity with another oncogenic HPV infection that ranked more prevalent within her strata of study, disease and ethnicity (ie, HPV35 coinfection with HPV16 or HPV52). The frequency of these three categories of HPV35 infections was compared across ethnic groups. All 95% confidence intervals (CI) and statistical tests in this section were performed with STATA version 14 software.
2.3.3 |. HPV35 genomic variation associated with cervical precancer/cancer
In the PaP Cohort, after HPV-genotyping, a subset of 608 HPV35-positive specimens (all 274 HPV35 CIN2+ cases and 334 of 363 randomly chosen benign HPV35 infections with CIN grade 1 or less [≤CIN1]), were selected for HPV35 whole-genome sequencing, with no restrictions on age or other HPV co-infections (Figure S1). Self-assigned ethnicity was used as a proxy for a women’s genetic background. Odds ratio (OR) and 95% CIs were calculated for CIN2+/CIN3+, using the controls (≤CIN1) as the referent group. Wald-test assessed heterogeneity between ORs. Associations of individual SNPs with CIN2+/CIN3+, by ethnicity, were performed using logistic regression. The significance level of SNP associations was corrected for multiple comparisons using a false discovery rate (FDR), based on the number of polymorphic nucleotides with a minor allele frequency (MAF) >0.03.
In the IARC specimens, the distribution of HPV35 sublineages was compared between Africa and all other regions using Fisher’s exact and Chi-square tests. For this analysis, samples from Sub-Saharan Africa were considered “Africa” and samples from ethnically more diverse North Africa were included with the other world regions. We focused on the case series and did not perform formal case-control analyses because case and control samples were unevenly collected by site/region (Table S1).
Fifty-eight samples (5.5%) were excluded from these analyses due to an uncertain HPV35 sublineage as a result of poor read depth and or poor/spotty coverage across the genome (2.5%) or due to coinfection of >1 HPV35 lineage for which a predominant lineage could not be assigned (3.0%). Because of the predominant etiologic role of HPV16, the summary genetic analyses presented exclude 74 samples from the PaP Cohort and 46 samples from the IARC collection that were concurrently HPV16-positive. For the PaP Cohort, 33 women were excluded since they were self-identified as multiethnic or did not report ethnicity, and one woman with the B lineage was excluded from the association analyses. For the IARC cohort, 17 samples with B lineage and 4 samples with unknown histology were not considered in the analysis. These exclusions restricted our analysis to 478 HPV35-positive specimens from the PaP Cohort and 342 specimens from the IARC collection (Figure S1). Statistical analyses in this last section were performed with R version 3.4.4 and SPSS version 23 software.
3 |. RESULTS
3.1 |. HPV35 prevalence and associated risks in the PaP Cohort
Among 27 402 HPV-positive women, age 30 to 64, of the PaP Cohort, 8230 had HPV35 genotyping information (Table 1; Figure S1). African-American women were more likely to have HPV35 (12.1% vs 5.1% among other women, P < .001). Among women with HPV35, the 5-year CIN3+ risks for African-American women were lower than other women but not significantly different (5.2% vs 10.2% among other women, P = .7; Table 2). Findings were similar for CIN2+ risks and for single HPV35 infections.
TABLE 1.
Prevalence of HPV35 among HPV-positive women ages 30 to 64 in the Persistence and Progression (PaP) Cohort at Kaiser Permanente Northern California by ethnicity
| n | HPV35+ | Weighted percent (95% CI) | Prevalence difference vs other women (95% CI) | P-value | |
|---|---|---|---|---|---|
| Total | 8230 | 478 | 5.8 (5.2-6.3) | – | |
| White | 4142 | 237 | 5.6 (4.8-6.3) | −0.4 (−1.5-0.7) | .3 |
| Hispanic | 1775 | 87 | 5.1 (4.0-6.2) | −0.9 (−2.2-0.4) | .1 |
| Asian | 1609 | 62 | 4.0 (3.0-5.1) | −2.2 (−3.4-0.9) | <.01 |
| African-American | 704 | 92 | 12.1 (9.6-14.6) | +7.0 (4.4-9.5) | <.001 |
Note: Weight based upon sampling probability of the woman being selected for genotyping (higher for cases vs controls). Prevalence difference depicts the absolute higher or lower HPV35 prevalence among women in an ethnic group compared to all other women. Bold indicates Rao-Scott Chi-square test P-value < .0125, threshold used to account for multiple comparisons. In a test for the pairwise comparison using Poisson regression model, the f-statistic is 18.62 and the global P-value is <.0001.
TABLE 2.
Five-year cumulative risk of CIN2+ and CIN3+ given HPV35 infection vs all remaining women, among women ages 30 to 64 in the Persistence and Progression (PaP) Cohort at Kaiser Permanente Northern California by ethnicity
| Type ethnicity | CIN2+ | CIN3+ | ||||
|---|---|---|---|---|---|---|
| 5-year cumulative risk | Risk of ethnic group vs other women combined (absolute difference) | 5-year cumulative risk | Risk of ethnic group vs other women combined (absolute difference) | |||
| % | 95% CI | % | 95% CI | |||
| HPV35+ | ||||||
| White | 27.3 | 22.2-32.5 | +4.6 | 9.7 | 6.4-12.9 | +0.8 |
| Hispanic | 21.5 | 14.3-28.7 | −4.3 | 10.9 | 5.6-16.2 | +2.1 |
| Asian | 21.5 | 12.7-30.3 | −4.0 | 11.0 | 4.7-17.3 | +2.0 |
| African-American | 25.1 | 17.3-32.9 | +0.1 | 5.2 | 1.5-8.8 | −5.0 |
| P-value (Wald-test) | .52 | .16 | ||||
Note: Weight based upon sampling probability of the woman being selected for genotyping (higher for cases vs controls). Risk difference depicts the absolute higher or lower risk of CIN2+ and CIN3+ among women in an ethnic group compared to all other women. P-value = Wald test for heterogeneity.
Abbreviations: CIN2+, cervical intraepithelial neoplasia (CIN) grade 2, CIN grade 3 and cancer; CIN3+, CIN grade 3 and cancer.
3.2 |. HPV35 among precancer and cancer cases from NCI studies
The HPV35 prevalence in CIN2+ cases from five pooled NCI studies varied by ethnicity (Figure 2; Table S2). Among CIN3 cases, African-American women were more likely to have a “single HPV35” infection (5.5% vs 1.9%, P < .001). The combined prevalence of both “single HPV35 infections” and “HPV35 co-infections, less prevalent” was also higher among African-American women (7.4% vs 2.1%, P < .001). Findings were similar for CIN2 cases. Few ICC among African-American women (n = 32) resulted in limited statistical power to compare HPV35 infections.
FIGURE 2.
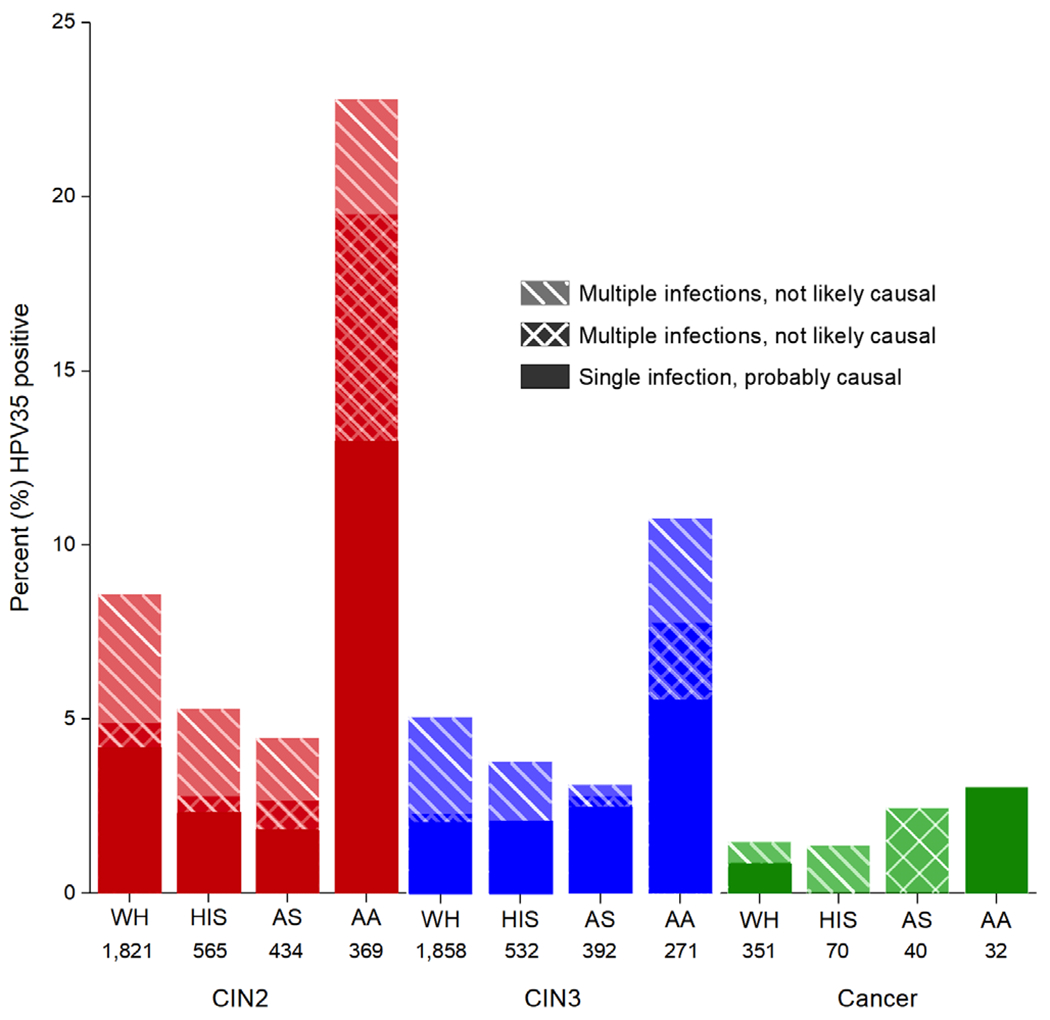
Prevalence of HPV35 among CIN2, CIN3 and cancer cases testing positive for carcinogenic HPV from 5 studies. CIN2, cervical intraepithelial neoplasia (CIN) grade 2; CIN3, CIN grade 3; WH, White; HIS, Hispanic; AS, Asian; AA, African-American. n, number of women with tested specimens. Among CIN2 and CIN3 cases, African-American women were more likely to have a “single HPV35 infection” (13.0% vs 3.5%, P < .001 for CIN2 and 5.5% vs 1.9%, P < .001 for CIN3) and both a “single HPV35 infection” and “HPV35 co-infection, most prevalent” (18.7% vs 4.2%, P < .001 for CIN2 and 7.4% vs 2.1%, P < .001 for CIN3). Differences were not statistically significant for cancers (P = .1). The number of cases from the studies presented in Table S5 and percent with 95% confidence intervals are presented in Table S2
3.3 |. Genomics of HPV35 in PaP Cohort nested case-control study
Based on the elevated HPV35 prevalence and positivity in precancers diagnosed in African-American women, we designed a nested case-control analysis within the PaP Cohort to assess whether HPV35 genetic variation (ie, lineages/sublineages and individual SNPs) is associated with cervical precancer/cancer and if a woman’s ethnicity modify these associations. We observed that finer classifications of HPV35 based on genetic variation across the viral whole-genome, defined here as sublineages (ie, A1 and A2) were differentially distributed by ethnicity for precancer cases (P < .001 for CIN2+ and CIN3+) but not for controls (P = .7; Table 3).
TABLE 3.
HPV35 A1 and A2 sublineage associations with CIN2+ and CIN3+ from women in the Persistence and Progression (PaP) Cohort at Kaiser Permanente Northern California by a women’s ethnicity
| Ethnicity/sublineage | Controls (%) | CIN2+ (%) | OR (95% CI) | CIN3+ (%) | OR (95% CI) |
|---|---|---|---|---|---|
| All ethnic/multiethnic groups | |||||
| A1 | 250 (84.2) | 192 (89.7) | ref | 59 (85.5) | ref |
| A2 | 47 (15.8) | 22 (10.3) | 0.6 (0.3-1.0) | 10 (14.5) | 0.9 (0.4-1.9) |
|
| |||||
| White | |||||
| A1 | 101 (83.5) | 111 (97.4) | ref | 33 (97.1) | ref |
| A2 | 20 (16.5) | 3 (2.6) | 0.1 (0.0-0.5) | 1 (2.9) | 0.2 (0.0-1.2) |
|
| |||||
| Hispanic | |||||
| A1 | 50 (83.3) | 24 (82.8) | ref | 10 (76.9) | ref |
| A2 | 10 (16.7) | 5 (17.2) | 1.0 (0.3-3.4) | 3 (23.1) | 1.5 (0.4-6.5) |
|
| |||||
| Asian | |||||
| A1 | 31 (91.2) | 18 (90.0) | ref | 9 (90.0) | ref |
| A2 | 3 (8.8) | 2 (10.0) | 1.2 (0.2-7.5) | 1 (10.0) | 1.2 (0.1-12.4) |
|
| |||||
| African-American | |||||
| A1 | 45 (81.8) | 34 (75.6) | ref | 4 (44.4) | ref |
| A2 | 10 (18.2) | 11 (24.4) | 1.5 (0.5-3.8) | 5 (55.6) | 5.6 (1.3-24.8) |
|
| |||||
| P-value (Wald-test) | 0.02 | 0.05 | |||
Note: For these analyses, there were no age restrictions. The only restriction was HPV16 co-infections. Among CIN2+ and CIN3+ cases, the distribution of A1/A2 significantly varied by ethnicity (Chi-square tests, P < .001 and P < .01, respectively). Among controls, the distribution of A1/A2 did not significantly differ by ethnicity (Chi-square tests, P = .7). Bold indicates P-value < .05 for logistic regression and Wald-test for heterogeneity.
Abbreviations: CIN2+, cervical intraepithelial neoplasia (CIN) grade 2, CIN grade 3 and cancer; CIN3+, CIN grade 3 and cancer; OR, odds ratio; 95% CI, 95% confidence intervals from logistic regression.
The A2 sublineage was significantly associated with CIN3+ among African-American women (OR = 5.6, 95% CI = 1.3-24.8; Table 3), but not among all other ethnic groups combined (OR = 0.5, 95% CI = 0.2-1.4; Figure S2). For CIN2+, the A2 association was weaker and not statistically significant among African-American women (OR = 1.5, 95% CI = 0.5-3.8). Wald-test confirmed differences between ORs. Interestingly, among women with A2, African-American ethnicity was associated with CIN2+ (OR = 3.6 vs among other women, 95% CI = 1.2-11.0) and suggestively associated with CIN3+ (OR = 3.3 vs among other women, 95% CI = 0.8-13.8; Table S3). Conversely, among women with the A1 sublineage, African-American ethnicity was inversely associated with CIN3+ (OR = 0.3 vs among other women, 95% CI = 0.11-0.91; Table S3).
Despite the low genetic diversity of HPV35, we were able to pinpoint SNPs associated with CIN3+ among African-American women from 490 polymorphic sites across the HPV35 genome. Twelve individual SNPs were associated with CIN3+ (ORs ranged from 15 to 31) only in African-American women, compared to other nucleotides at the same genomic position, including one SNP mapped to the E7 oncogene (position 748, OR = 6.4, 95% CI = 1.1-35.6) (Figure 1B; Table S4). In contrast, six individual SNPs were inversely associated with CIN3+ in African-Americans. Of note, these 18 SNPs are not the A1/A2 lineage defining SNPs previously reported.29
Interestingly, when considering the 12 individual SNPs associated with CIN3+ among African-Americans, women usually had a combination of these SNPs (ie, they had 2-11 of these SNPs together, and one was independent [Table S4]). The African-American women with two or more of these 12 individual SNPs were always assigned to the A2 sublineage, and the African-American women with two or more of these individual SNPs were strongly associated with increased CIN3+ (OR = 68.8, 95% CI = 6.4-739.0, data not shown).
3.4 |. Worldwide distribution of HPV35 variation
To assess if HPV35 genetic variation (ie, lineages/sublineages) also varied in prevalence across the globe, we investigated HPV35-positive IARC specimens collected from different worldwide regions. The distribution of A1 and A2 were similar for cases and controls for women outside of Africa as well as women in Africa (Table 4). When comparing Africa to all other geographic regions, the A2 sublineage was responsible for a higher proportion of ICC in Africa (41.9% vs 10.4% from all other regions, Chi-square P < .01) and high-grade lesions (42.9% vs 6.3% from all other regions, Fisher’s exact P = .03; Table 4). Similar results were observed among benign infections from Africa (54.1% vs 5.1% from all other regions, Chi-square P < .0001). The A1 sublineage was significantly more frequent than A2 in non-African world regions for ICC (89.6% vs 10.4%), high-grade lesions (93.8% vs 6.3%) and benign infection (94.9% vs 5.1%, respectively). The new B lineage was uniquely found in Asia, particularly in Bhutan and India (Table S1).
TABLE 4.
HPV35 A1 and A2 sublineage distribution among ICC, abnormal lesions and benign infections by world region using IARC specimens
| Sublineages Regions | Benign infections |
HSIL/CIN2/CIN3 |
ICC |
P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | 95% CI | n | % | 95% CI | n | % | 95% CI | ||
| Non-Africa | ||||||||||
| A1 | 93 | 94.9 | 88.6-97.8 | 15 | 93.8 | 71.7-99.7 | 43 | 89.6 | 77.8-95.5 | |
| A2 | 5 | 5.1 | 2.2-11.4 | 1 | 6.3 | 0.3-28.3 | 5 | 10.4 | 4.5-22.2 | .46a |
|
| ||||||||||
| Africa | ||||||||||
| A1 | 62 | 45.9 | 37.7-54.3 | 8 | 57.1 | 32.6-78.6 | 18 | 58.1 | 40.8-73.6 | |
| A2 | 73 | 54.1 | 45.7-62.3 | 6 | 42.9 | 21.4-67.4 | 13 | 41.9 | 26.4-59.2 | .39a |
|
| ||||||||||
| P-value | <.0001 b | .03 a | <.01 b | |||||||
Note: For this analysis, samples from Sub-Saharan Africa were considered “Africa” and samples from North Africa were included with the other world regions as “Non-Africa”. Bold indicates P-value < .05 for
Fisher’s exact test and
Chi-square test.
Abbreviations: CIN2, Cervical intraepithelial neoplasia (CIN) grade 2; CIN3, CIN grade 3; HSIL, high-grade squamous intraepithelial lesion; ICC, invasive cervical cancers.
4 |. DISCUSSION
Through a series of cross-sectional and longitudinal studies and a comprehensive HPV35 genomic approach, we detected a higher prevalence of HPV35 among precancer and elevated etiologic importance of the HPV35 A2 sublineage in women of African ancestry in the U.S.
First, we showed that as a result of higher HPV35 prevalence, and not risk (the 5-year CIN3+ risk associated with an HPV35 was similar by ethnicity), HPV35 (including single HPV35 infections) was more common in cervical precancer cases among African-American women (5.0% vs 1.9% in other ethnic groups). Similar to other U.S. studies,6 the proportion of HPV35 in cervical cancers did not differ by ethnicity, perhaps due to few cases. Our findings coincide with the global observation that HPV35 is of greater importance for cervical cancer among women of African ancestry.5,12,13,29 The observed ethnic variations of HPV35 epidemiology should be placed in a larger context of other oncogenic HPV types. For all ethnic groups, HPV16 and HPV18 are associated with the majority of cervical precancers and cancers and the current 9-valent HPV vaccine has been reported to prevent 80% of HPV-positive cervical precancers and cancers in the U.S.6,30 We observed that for women with CIN2 or CIN3 and HPV35 infection, African-American women had a greater percentage of coinfections compared with other ethnic groups, following a similar pattern observed in a global comparison of cervical cancers from Africa vs other world regions.31 We could not confirm this trend for cervical cancers.
Second, we refined our investigation of this association between HPV35 and African ancestry by whole genome sequencing 1053 HPV35-positive specimens from the U.S. and around the world and found that the genetic variation of HPV35, in particular the A2 sublineage and related SNPs, was associated with precancer/cancer in African-American women and corresponded to a large proportion of ICC in Africa.
In the U.S., the A2 sublineage was strongly associated with cervical precancers among African-American women, whereas the A1 sublineage was associated with cervical precancer among White women. African-American women had a 10-fold significant increase in CIN3+ if infected with A2 compared to White women (OR = 10.0, 95% CI = 1.0-97.5; data not shown). Of note, in the PaP Cohort, the absolute prevalence of A2 among all CIN2+ cases was 0.6%, and 3.7% among CIN2+ cases in African-American women. This is an example of an epidemiologic observation (eg, higher prevalence of HPV35 among precancers in African-American compared to other ethnic groups) being strengthened by specificity of association.32 The relationship between the A2 sublineage and women of African ancestry shown here (ie, increased CIN3+ associations) could be a consequence of thousands of years of virus-host interactions where certain lineages of HPV35 became better adapted and able to persist in these women. However, a likely explanation is that a genetic bottleneck due to the different out-of-Africa events and/or gene introgression events from archaic human populations may have rendered modern humans of non-African ancestry less susceptible to infection and/or progression with HPV35, and specifically the A2 sublineage. HPV16 and HPV18 sublineages have also been associated with cervical precancer/cancer in certain ethnic groups.17,33 A viral-host interaction has been posited for HPV16, where both modern-human migration and reproductive events, with introgression of immune related alleles, confer a host niche adaptation potentially influencing phenotype such as cervical cancer.34,35
Globally, the A2 sublineage was more prevalent in ICC, as well as in benign infections, in Africa compared with other world regions where its prevalence was 2.6 times lower. One potential explanation is that A2 is better adapted to persist in women of African populations and the increased prevalence is due to ancient geographic isolation. In contrast, in the U.S., A2 was equally prevalent among controls for all ethnic groups probably related to recent ethnic admixture in the U.S., that could allow A2 to circulate easier among women from various ethnic groups. Nevertheless, A2 was more prevalent among precancers/cancers in African-American women, resulting in increased risk associated with A2 in this population. It is also possible that the increased A2 among benign infections in Sub-Saharan Africa could be linked to the high prevalence of HIV infection in this region. Two studies in Africa showed that HPV35 was more prevalent among HIV + compared to HIV-negative women in both cases and controls.10,36 Further investigation into HPV35 genetic variation, geographic dispersion is needed.
Our HPV whole-genome sequencing approach identified a new HPV35 B lineage, highly prevalent in India and attributed to 43.0% of ICC in South Asia. The HPV35 B lineage warrants further study to evaluate its potential regional importance among women of South Asian ancestry.
We pinpointed SNPs associated with precancer/cancer among African-American women. One mapped to the E7 ORF at position 748 and was associated with CIN3+ only in African-American women. This DNA change (C>T) and trinucleotide motif are consistent with potentially being induced by the antiviral enzymatic activity of APOBEC3 (ie, the 3-bp motif 5′TCW3]′), a host’s intracellular immune response activated upon HPV infection.37 Due to the importance of E7 as an oncogene, future studies are needed to clarify the potential function of this variant.
We could not assess directly the association of HPV35 and invasive cancers in the U.S. due to small numbers; therefore, we studied precancerous lesions as the surrogate endpoint. Future studies of HPV35 genetic variation (including lineages/sublineages and finer SNPs) from diverse populations, using a molecular approach based on human genetic markers to more precisely define ancestry, are needed to understand the geographic dispersion of these sublineages with important risk differences in African ancestry populations.
In conclusion, our large epidemiologic analyses confirmed the importance of HPV35 in case attribution for cervical precancer among African-American women. Our comprehensive evaluation of the HPV35 genome allowed us to resolve sublineages and linked SNPs, providing striking evidence that the A2 sublineage is strongly associated with precancer/cancer in women of African ancestry, possibly a sign of increased evolutionary fitness in this host group. Significant differences in the proportion of cervical cancers with HPV35 were not observed, perhaps due to few cancers, but also because other more oncogenic types, namely HPV16 and HPV18, are associated with the majority of cervical cancers for all ethnic groups. Considering that vaccination and screening approaches that cover all of the highest-risk types are needed for optimal global prevention coverage,1 further research is required to consider whether the addition of HPV35 to the next generation of vaccines would result in a meaningfully broader coverage for women of African ancestry. Similarly, type-specific HPV screening tests might be improved by recognizing the increased importance of HPV35 in women of African ancestry. If the growing body of evidence remains consistent, prevention programs, especially those in populations without quality screening, should consider how the addition of HPV35 to HPV vaccines would result in improved cervical cancer prevention, particularly in Africa, where the burden of cervical cancer is the highest in the world.
Supplementary Material
What’s new?
HPV35 accounts for 2% of invasive cervical cancers worldwide, but possibly as much as 10% in sub-Saharan Africa. These authors found that African-American women had higher rates of HPV35 infection, and more precancers associated with HPV35, than women of other ethnicities. However, precancer risk by HPV35 status did not vary by ethnicity. Genetic testing uncovered an association between precancer and the A2 sublineage of HPV35 within Africa and among African-American women in the US. Additionally, particular HPV35 SNPs, including one in the E7 oncogene, were associated with precancer or cancer. Recognizing the importance of HPV35 in women of African ancestry could help improve HPV screening tests or vaccines.
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization. Our study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH. This project has been funded in whole or in part with federal funds from the National Cancer Institute, NIH (HHSN261200800001E); and, the National Cancer Institute (CA78527) and the Einstein Cancer Research Center (P30CA013330) from the National Cancer Institute (to R.D.B.). Work at IARC was supported by a grant from the Institut National du Cancer (INCa), France (SHSESP 16-006).
Abbreviations:
- AIS
adenocarcinoma in situ
- CI
confidence intervals
- CIN
cervical intraepithelial neoplasia
- CIN1
cervical intraepithelial neoplasia grade 1
- CIN2
cervical intraepithelial neoplasia grade 2
- CIN3
cervical intraepithelial neoplasia grade 3
- FDR
false discovery rate
- HIV
human immunodeficiency virus
- HPV
human papillomavirus
- HPV35
human papillomavirus type 35
- IARC
International Agency for Research on Cancer
- ICC
invasive cervical cancer
- KPNC
Kaiser Permanente Northern California
- MAF
minor allele frequency
- NCI
National Cancer Institute
- OR
odds ratio
- PaP
persistence and progression
- SNP
single nucleotide polymorphism
Footnotes
CONFLICT OF INTEREST
Phillip Castle has received HPV tests and assays at a reduced or no cost for research from Roche, Becton Dickinson, Cepheid and Arbor Vita Corporation. Maria Demarco has received masked HPV and cytology test results at no cost from Roche Molecular Systems, Becton Dickinson Diagnostics and Qiagen for independent evaluations of these technologies. Nicolas Wentzensen has received cervical cancer screening assays in-kind or at a reduced cost from Roche and Becton Dickinson for study purposes. All other authors declare no competing interests.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
ETHICS STATEMENT
For the PaP cohort, the Kaiser Permanente Northern California institutional review board (IRB) approved use of the data, and the National Institutes of Health Office of Human Subjects Research deemed our study exempt from IRB review. For all other National Cancer Institute (NCI) studies, local and NCI IRBs approved the studies. For the International Agency for Research on Cancer (IARC) collection, both local and IARC ethical committees approved our study.
DATA ACCESSIBILITY
The data that support the findings of this manuscript have been submitted to the GenBank (accession numbers MT217148-MT218011).
REFERENCES
- 1.de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. [DOI] [PubMed] [Google Scholar]
- 2.Bouvard V, Baan R, Straif K, et al. A review of human carcinogens—part B: biological agents. Lancet Oncol. 2009;10:321–322. [DOI] [PubMed] [Google Scholar]
- 3.Arbyn M, Castellsague X, de Sanjose S, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. [DOI] [PubMed] [Google Scholar]
- 4.Global Burden of Disease Cancer Consortium, Fitzmaurice C, Allen C, et al. Global, regional, and National Cancer Incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131:2349–2359. [DOI] [PubMed] [Google Scholar]
- 6.Saraiya M, Unger ER, Thompson TD, et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 2015;107:djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hariri S, Steinau M, Rinas A, et al. HPV genotypes in high grade cervical lesions and invasive cervical carcinoma as detected by two commercial DNA assays, North Carolina, 2001-2006. PLoS One. 2012;7:e34044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Unger ER, Hariri S, Steinau M, Markowitz LE. Prevalence of 9-Valent human papillomavirus types by race/ethnicity in the Prevaccine era, United States, 2003–2006. Sex Transm Dis. 2016;43: 633–636. [DOI] [PubMed] [Google Scholar]
- 9.Vidal AC, Smith JS, Valea F, et al. HPV genotypes and cervical intraepithelial neoplasia in a multiethnic cohort in the southeastern USA. Cancer Causes Control. 2014;25:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford GM, de Vuyst H, Tenet V, Plummer M, Tully S, Franceschi S. Effect of HIV infection on human papillomavirus types causing invasive cervical cancer in Africa. J Acquir Immune Defic Syndr. 2016;73:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali-Risasi C, Verdonck K, Padalko E, vanden Broeck D, Praet M. Prevalence and risk factors for cancer of the uterine cervix among women living in Kinshasa, The Democratic Republic of the Congo: a cross-sectional study. Infect Agent Cancer. 2015;10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellsague X, Klaustermeier J, Carrilho C, et al. Vaccine-related HPV genotypes in women with and without cervical cancer in Mozambique: burden and potential for prevention. Int J Cancer. 2008; 122:1901–1904. [DOI] [PubMed] [Google Scholar]
- 13.Okolo C, Franceschi S, Adewole I, et al. Human papillomavirus infection in women with and without cervical cancer in Ibadan, Nigeria. Infect Agent Cancer. 2010;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny L, Adewole I, Anorlu R, et al. Human papillomavirus prevalence and type distribution in invasive cervical cancer in sub-Saharan Africa. Int J Cancer. 2014;134:1389–1398. [DOI] [PubMed] [Google Scholar]
- 15.Hopenhayn C, Christian A, Christian WJ, et al. Prevalence of human papillomavirus types in invasive cervical cancers from 7 US cancer registries before vaccine introduction. J Low Genit Tract Dis. 2014;18:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Food and Drug Administration. Highlights of Prescribing Information. Gardasil 9 (Human Papillomavirus 9-Valent Vaccine, Recombinant). Suspension for Intramuscular Injection Initial U.S. Approval. Silver Spring, MD: FDA; 2014. [Google Scholar]
- 17.Mirabello L, Yeager M, Cullen M, et al. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J Natl Cancer Inst. 2016;108:djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi LF, Kiviat NB, Hildesheim A, et al. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst. 2006;98:1045–1052. [DOI] [PubMed] [Google Scholar]
- 19.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiffman M, Rodriguez AC, Chen Z, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaMere BJ, Howell R, Fetterman B, Shieh J, Castle PE. Impact of 6-month frozen storage of cervical specimens in alkaline buffer conditions on human papillomavirus genotyping. J Virol Methods. 2008;151:298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188:1383–1392. [DOI] [PubMed] [Google Scholar]
- 23.A randomized trial on the management of low-grade squamous intra-epithelial lesion cytology interpretations. Am J Obstet Gynecol. 2003;188:1393–1400. [DOI] [PubMed] [Google Scholar]
- 24.Wang SS, Zuna RE, Wentzensen N, et al. Human papillomavirus cofactors by disease progression and human papillomavirus types in the study to understand cervical cancer early endpoints and determinants. Cancer Epidemiol Biomarkers Prev. 2009;18:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wentzensen N, Walker JL, Gold MA, et al. Multiple biopsies and detection of cervical cancer precursors at colposcopy. J Clin Oncol. 2015;33:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network, Albert Einstein College of Medicine, Analytical Biological Services, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun N, Cheung LC, Pan Q, Schiffman M, Katki HA. Flexible risk prediction models for left or interval-censored data from electronic health records. Ann Appl Stat. 2017;11:1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wentzensen N, Schiffman M, Dunn T, et al. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer. 2009;125:2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gage JC, Ajenifuja KO, Wentzensen NA, et al. The age-specific prevalence of human papillomavirus and risk of cytologic abnormalities in rural Nigeria: implications for screen-and-treat strategies. Int J Cancer. 2011;130(9):2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hariri S, Unger ER, Schafer S, et al. HPV type attribution in high-grade cervical lesions: assessing the potential benefits of vaccines in a population-based evaluation in the United States. Cancer Epidemiol Biomarkers Prev. 2015;24:393–399. [DOI] [PubMed] [Google Scholar]
- 31.Pimenoff VN, Tous S, Benavente Y, et al. Distinct geographic clustering of oncogenic human papillomaviruses multiple infections in cervical cancers: results from a worldwide cross-sectional study. Int J Cancer. 2019;144:2478–2488. [DOI] [PubMed] [Google Scholar]
- 32.Hill AB. The environment and disease: association or causation? Proc R Soc Med. 1965;58:295–300. [PMC free article] [PubMed] [Google Scholar]
- 33.Xi LF, Koutsky LA, Hildesheim A, et al. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol Biomarkers Prev. 2007;16:4–10. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, DeSalle R, Schiffman M, et al. Niche adaptation and viral transmission of human papillomaviruses from archaic hominins to modern humans. PLoS Pathog. 2018;14:e1007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pimenoff VN, de Oliveira CM, Bravo IG. Transmission between archaic and modern human ancestors during the evolution of the oncogenic human papillomavirus 16. Mol Biol Evol. 2017;34:4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy D, Njala J, Stocker P, et al. High-risk human papillomavirus in HIV-infected women undergoing cervical cancer screening in Lilongwe, Malawi: a pilot study. Int J STD AIDS. 2015;26:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren CJ, Westrich JA, Doorslaer KV, Pyeon D. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses. 2017;9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this manuscript have been submitted to the GenBank (accession numbers MT217148-MT218011).


