Abstract
The aim of this study was to produce gene transfer vectors consisting of plasmid DNA packaged into virus-like particles (VLPs) with different cell tropisms. For this purpose, we have fused the N-terminally truncated VP60 capsid protein of the rabbit hemorrhagic disease virus (RHDV) with sequences which are expected to be sufficient to confer DNA packaging and gene transfer properties to the chimeric VLPs. Each of the two putative DNA-binding sequences of major L1 and minor L2 capsid proteins of human papillomavirus type 16 (HPV-16) were fused at the N terminus of the truncated VP60 protein. The two recombinant chimeric proteins expressed in insect cells self-assembled into VLPs similar in size and appearance to authentic RHDV virions. The chimeric proteins had acquired the ability to bind DNA. The two chimeric VLPs were therefore able to package plasmid DNA. However, only the chimeric VLPs containing the DNA packaging signal of the L1 protein were able efficiently to transfer genes into Cos-7 cells at a rate similar to that observed with papillomavirus L1 VLPs. It was possible to transfect only a very limited number of RK13 rabbit cells with the chimeric RHDV capsids containing the L2-binding sequence. The chimeric RHDV capsids containing the L1-binding sequence transfer genes into rabbit and hare cells at a higher rate than do HPV-16 L1 VLPs. However, no gene transfer was observed in human cell lines. The findings of this study demonstrate that the insertion of a DNA packaging sequence into a VLP which is not able to encapsidate DNA transforms this capsid into an artificial virus that could be used as a gene transfer vector. This possibility opens the way to designing new vectors with different cell tropisms by inserting such DNA packaging sequences into the major capsid proteins of other viruses.
One limitation of most viral vectors for gene therapy is the cell tropism of such systems, and it has therefore been reported that viral vectors, such as adenovirus vectors, are poorly transduced into target cells (19, 32). For some applications, the vector should ideally have the capacity to transfect a wide range of cells, and for other applications, it must be restricted to one target cell. To overcome these difficulties, modification of the virus genome by the introduction of novel tropism determinants has been investigated, with or without the deletion of endogenous tropism factors (2, 4). In addition to these viral vectors, the use of artificial virus vectors consisting of DNA packaged in vitro into recombinant virus-like particles (VLPs) was recently described as an alternative method for gene transfer (9, 25). By their nature, such vectors suffer from the same cell tropism limits as recombinant viruses. Modification of the cell tropism determinants of the VLPs is one solution. Another solution is the production of a range of VLP vectors with different cell tropisms. However, we have observed in preliminary studies that not all recombinant VLPs have the capacity to package and transfer DNA plasmids, such as the VP60 capsid of rabbit hemorrhagic disease virus (RHDV) (unpublished data).
RHDV is a member of the Caliciviridae family (14). Its genome is a 7.5-kb positive single-stranded RNA with a viral protein linked to its 5′ terminus (VPg) and a short poly(A) tail linked to its 3′ terminus (31). The nonenveloped icosahedral capsid demonstrated a symmetry of T=3 and is 35 nm in diameter (24). The particle is composed of 90 dimers of the VP60 capsid protein. RHDV cannot be propagated in cell culture and is responsible for a lethal acute viral disease in rabbits characterized by necrotic hepatitis and disseminated intravascular coagulation. Its major capsid protein, VP60, is able to self-assemble into VLPs when expressed in insect cells (11, 17, 22). The VP60 protein is not a DNA-binding protein, and it has been recently shown that the N-terminal 42 amino acids can be deleted without affecting its ability to form VLPs (S. Laurent et al., unpublished data).
In the present study we built synthetic gene vector systems using recombinant pseudoviruses composed of the major capsid protein of RHDV, self-assembled into VLPs in which each of the two putative DNA-binding domains of human papillomavirus type 16 (HPV-16) L1 and HPV-16 L2 proteins have been incorporated (L1BS and L2BS, respectively). The DNA-binding activity of these two domains is sequence independent. Papillomavirions contain two proteins, L1 and L2, which encapsidate a closed, circular, double-stranded DNA of about 8 kbp. The viral capsid of 50 to 55 nm contains 72 pentamers of L1 centered on the vertices of a T=7 icosahedral lattice (1, 28). L2 is present at about 1/30 the abundance of L1 (10). The major capsid protein L1 of HPV can self-assemble into VLPs (10, 20, 21, 26), and capsids consisting of L1 or L1 plus L2 proteins have the ability to transfer genes to a wide range of cell types (25). The two DNA-binding sequences used for the construction of these chimeric VLPs are derived from those identified on the two structural proteins of HPV-16. The first DNA-binding sequence used has been identified at the N-terminal end of the minor capsid protein, L2 (35). The second DNA-binding domain used was identified in a preliminary study at the C terminus of the major capsid protein (L1) of HPV-16 (27). Moreover, it has recently been shown that DNA binding to bovine papillomavirus type 1 (BPV-1) L1 plus L2 VLPs is enhanced by the presence of a 120-bp DNA sequence located in the BPV-1 E1 gene (33).
The two chimeric capsids obtained, VP60Δ-L1BS and VP60Δ-L2BS, were investigated for DNA binding, DNA packaging, and their ability to deliver foreign DNA into a variety of cells with the subsequent expression of the encoded gene.
MATERIALS AND METHODS
Generation of RHDV–HPV-16 L1 or L2 recombinant baculovirus.
The VP60 fragment (VP60Δ) was first amplified from a plasmid carrying the VP60 gene (11) with an upper primer containing a 5′ portion encoding the C-terminal end of the L1 (CGCAAAAAACGTAAGCTGACTGCAGAGAACTCATCCGCATCGATT) or L2 (GCAAAACGCACAAAACGTACTGCAGAGAACTCATCCGCATCG) DNA-binding domain and with a lower primer containing a HindIII restriction site (AAGCTTGACATAAGAAAAGCCATTGGCTGTGCCC; the restriction site is in boldface type). In the second PCR, the upper primers are complementary in their 5′ end with the start of the L1 (GGATCCTCTACAACTGCTAAACGCAAAAAACGTAAGCTGACTGCAGAGAACTC) or the L2 (GGATCCATGCGACACAAACGTTCTGCAAAACGCACAAAACGTACTGCAGAGAA) DNA-binding domain and contain a BamHI restriction site. Amplification was performed with 0.7 μM concentrations of each primer and 1.25 IU of Taq polymerase (Life Technologies, Eragny, France). Following amplification, the PCR products were cloned into the pCR2-1 Topo vector (Topo TA cloning; Invitrogen, San Diego, Calif.) and then subcloned into the pFastBacI vector (Life Technologies) after digestion with BamHI and HindIII restriction enzymes. Recombinant baculoviruses encoding VP60Δ-L1BS and VP60Δ-L2BS were generated using the Bac-to-Bac baculovirus expression system according to the manufacturer's instructions (Life Technologies).
Generation of HPV-16 L1 and HPV-16 L1+L2 recombinant baculoviruses.
The HPV-16 L1 gene was first amplified from an HPV-16 DNA-positive biopsy (26) using primers containing BglII sites (upper primer, CCAGATCTATGTCTCTTTGGCTGCCTAGTGAGGC, and lower primer, CCAGATCTTTACAGCTTACGTTTTTTGCGTTTAG). The amplified product was then inserted into pFastBacI to generate HPV-16 L1 VLPs as previously described (27). In addition, the HPV-16 L1 gene was cloned into the pFastBacDual plasmid (Life Technologies) at the BamHI site for expression under the control of the polyhedrin promoter in order to produce HPV-16 L1+L2 VLPs. The HPV-16 L2 gene was then amplified with primers containing XhoI sites (upper primer, TTATTCTCGAGAATATGCGACACAAACGTTCTGCAAAACGCACAAA ACGT, and lower primer, AACCTCTCGAGACTGGGACAGGAGGCAAGTAGACAGTGGCCTCA) and cloned into the pFastBacDual L1 plasmid at the XhoI site for expression under the control of the p10 promoter. Recombinant baculoviruses encoding the pFastBac L1 and the pFastBacDual L1+L2 protein were then generated as described above.
Production and purification of chimeric VLPs.
Sf21 cells, maintained in Grace's insect medium supplemented with 10% fetal calf serum (FCS), were infected with the different recombinant baculoviruses at a multiplicity of infection of 10 and were incubated for 72 h at 27°C. Cells were harvested by centrifugation (500 × g for 5 min), resuspended in phosphate-buffered saline (PBS) containing 0.5% NP-40, and allowed to stand at room temperature for 30 min. Cell lysates were then centrifuged at 14,000 × g for 15 min at 4°C. Supernatants and pellets which represented cytoplasmic and nuclear fractions were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, electrophoresed in a 10% polyacrylamide gel, and either stained with Coomassie blue reagent or blotted onto a nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany) for Western blot analysis. The membrane was probed with a monoclonal anti-RHDV antibody. Specifically bound antibodies were revealed by an anti-mouse immunoglobulin G alkaline phosphatase conjugate (Sigma Aldrich) in the presence of 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT; Sigma Aldrich).
For VLP purification the cytoplasmic fraction was collected as described above and the nuclear fraction was further resuspended in ice-cold PBS and sonicated by three 15-s bursts at 60% maximal power (Vibra Cell; Bioblock Scientific, Strasbourg, France). Fractions were then loaded on the top of a preformed CsCl gradient and centrifuged at equilibrium in a Beckman SW28 rotor (20 h, 27,000 rpm, 4°C). Gradient fractions were analyzed for density by refractometry and were tested for the presence of VP60 protein by enzyme-linked immunosorbent assay using a monoclonal anti-RHDV antibody (12). Immunoreactive fractions were finally pooled and pelleted by ultracentrifugation in a Beckman SW28 rotor (3 h, 27,000 rpm, 4°C). VLPs were resuspended in PBS and protein content was evaluated using the microBCA kit (Pierce, Touzart et Matignon, France).
Determination of DNA-binding activity.
The Southwestern blot assay was based on previously published procedures (3, 15) with some modifications. Briefly, purified chimeric VLPs were denatured in loading buffer and then subjected to SDS-PAGE in a 10% polyacrylamide gel before transfer to a BA 83 nitrocellulose sheet by electroblotting (Hoefer SemiPhor; Pharmacia). After a blocking step with 30 mM HEPES, 100 mM potassium phosphate, and 1% bovine serum albumin, proteins were renatured by overnight incubation at 4°C in a buffer containing 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 200 mM NaCl, 10% glycerol, and 0.1% NP-40. The DNA-binding assay was performed for 30 min at room temperature in a buffer containing 30 mM HEPES, 5 mM MgCl2, and 50 mM NaCl by using a digoxigenin-labeled DNA probe. The probe used is a labeled reference plasmid (Boehringer GmbH, Mannheim, Germany). The membranes were then washed four times with binding buffer and bound DNA was revealed with an antidigoxigenin alkaline phosphatase-conjugated antibody (Boehringer) with NBT and BCIP as substrates.
DNA packaging.
Ten micrograms of purified VLPs was incubated in a 50 mM Tris-HCl buffer (pH 7.5) containing 150 mM NaCl, 1 mM EGTA, and 20 mM dithiothreitol in a final volume of 50 μl at room temperature for 30 min. At this stage, 1 μg (1 μl) of pCMV-β plasmid, 7.2 kbp (Clontech; Ozyme, Montigny le Bretonneux, France) or pBPV-CMV-β plasmid diluted in 49 μl of 50 mM Tris-HCl buffer (pH 7.5)–150 mM NaCl was added to 50 μl of the disrupted VLPs. The preparation was then diluted in 50 mM Tris-HCl buffer (pH 7.5)–150 mM NaCl containing 2 mM CaCl2 and 1% dimethyl sulfoxide (DMSO). CaCl2 molarity was increased stepwise from 2 to 5 mM with an increment of 1 mM/h at 20°C to reach a final volume of 500 μl.
The pBPV-CMV-β plasmid was obtained by insertion of the 120-bp BPV-1 enhancing packaging sequence (EPS) recently described by Zhao et al. (33) into the EcoRI restriction site of the pCMV-β plasmid. This was achieved by amplification of the DNA extracted from a bovine skin wart containing BPV-1 with specific primers containing an EcoRI site (upper, CCGGAATTCAAGCTTATGCAAAAAAGATCTCATGAAGGAGGA, and lower, CCGGAATTCCTCTTCTCTTACATTTAGCGTGTTTGC).
In order to evaluate the amounts of packaged plasmid DNA, encapsidation experiments were performed using 50 μg of VLPs and 5 μg of plasmids. After refolding, the preparations were treated for 1 h at 20°C with 100 IU of Benzonase (Merck, Darmstadt, Germany). The Benzonase was heat inactivated for 10 min at 65°C in the presence of 25 mM EDTA. The mixture was then incubated in the presence of 3% SDS and 1 mg of proteinase K (Appligene, Illkirch, France) per ml for 2 h at 56°C. Plasmidic DNA was phenol extracted and ethanol precipitated. Recovered DNA was linearized using HindIII restriction enzymes. As a control, the same amount of plasmidic DNA was processed in the same manner with and without Benzonase treatment. Results are expressed as the percentage of Benzonase-protected DNA. Results were assessed using Molecular Analyst software (Bio-Rad, Ivry/Seine, France).
Cell lines.
Five human cell lines (HeLa [cervix, ATCC CCL2], MRC5 [lung, ATCC CCL171], HuH-7 [liver], HepG2 [liver, ATCC HB-8065], and CaCo2 [colon, ATCC HTB37]) and three animal cell lines (Cos-7 [monkey kidney, ATCC CRL1651], CHO [Chinese hamster ovary, ATCC CRL1793], and L929 [mouse muscle, ATCC CCL1]) were used in transfection experiments. Because RHDV infects lagomorphs, rabbit kidney cells (RK13, ATCC CCL37) and hare cells (R17) were also investigated (kind gifts of J. F. Vautherot). Cells grown in monolayers in Dulbecco modified Eagle medium (DMEM)-Glutamax (Life Technologies) supplemented with 10% FCS, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. Cells (5 × 105 cells/well) were seeded in six-well plates (Nunc and Life Technologies) and grown to 80% confluence.
Gene transfer.
The different VLPs were examined for their capacity to mediate the transfer of plasmid DNA to different cell lines. Cells were washed twice with DMEM-Glutamax (Life Technologies) before transfection. After disruption and refolding in the presence of plasmid DNA as described above, 10 μg of VLPs containing 1 μg of plasmid were diluted in 1 ml of culture medium and added to the wells and incubated for 4 h at 37°C. At this stage, the VLPs were removed and 3 ml of DMEM-Glutamax supplemented with 10% FCS was added. The cells were then incubated for 48 h at 37°C. In situ β-galactosidase activity was measured according to a previously described procedure (16). Results are expressed as the number of blue cells per well and are the means of two to three separate experiments.
In some experiments a plasmid coding for luciferase (pCMV-Luc, 7.2 kbp) was used in place of the pCMV-β plasmid. Luciferase gene expression was measured by a luminescence assay (luciferase reporter gene assay with constant light signal; Roche Molecular Biochemicals). The culture medium was discarded and cells were washed three times with PBS. Then 100 μl of PBS and 100 μl of 1× lysis reagent were added. Cell lysate was harvested after 30 min of incubation at room temperature, and luminescence was integrated over 10 s (Victor2; Wallac). Results were expressed as count per second per milligram of cell protein (micro-BCA assay; Pierce).
Cell binding was evaluated by the capacity of the VLPs to inhibit gene transfer obtained with VP60Δ-L1BS pseudovirions composed of the pCMV-Luc plasmid coding for luciferase packaged into VP60Δ-L1BS VLPs. In these competition experiments, the VP60Δ-L1BS pseudovirions were mixed with a 20-fold excess of different VLPs before being added to the cells. Results were expressed as the percent inhibition of the gene transfer activity. Luciferase activity corresponding to incubation of cells with the plasmid alone or with VP60Δ-L1BS pseudovirions and PBS was considered 100 and 0% of inhibition, respectively.
Analysis of transfection efficiency was performed by comparing the means of blue cells observed or the means of luciferase counts per second/per milligram by the F test using EPI-Info 6.04c(US) software.
Transmission electron microscopy.
VLP preparations were applied to carbon-coated grids, negatively stained with 1.5% uranyl acetate, and observed at a nominal magnification of ×50,000 with a JEOL 1010 electron microscope. All the electron micrographs were taken at a magnification of ×30,000 or ×50,000. Subcellular localization of VLPs was analyzed on ultrathin sections. Infected cells were washed in PBS and fixed in 4% formaldehyde and 1% glutaraldehyde in PBS, pH 7.2, for 48 h. Cells were then washed in PBS, harvested, and postfixed with 1% osmium tetroxide for 1 h. After dehydration in graded ethanol, cell pellets were embedded in Epon resin that was allowed to polymerize for 24 h at 60°C. Ultrathin sections were then obtained with a Reichert ultramicrotome, collected onto copper grids, and stained with 1% uranyl acetate and 1% lead citrate in distilled water.
RESULTS
The short DNA-binding sequences of HPV-16 L1 or L2 were fused by using PCR at the N-terminal extremity of a truncated RHDV-VP60 protein sequence and then cloned into a pFast-BacI vector. Recombinant baculovirus encoding the VP60Δ-L1BS or the VP60Δ-L2BS proteins was generated using the Bac-to-Bac baculovirus expression system. The two chimeric VLPs consisting of the truncated VP60 capsid protein of RHDV with the addition of one of the two DNA-binding domains of the capsid proteins of HPV-16 (Fig. 1) were expressed in Sf21 insect cells by using the recombinant baculoviruses. Three days postinfection, corresponding to the maximal level of expression, the cytoplasmic and nuclear fractions of infected cells were analyzed by SDS-PAGE and immunoblotting using an anti-VP60 monoclonal antibody. A band of around 60 kDa, corresponding to the expected size of the VP60Δ protein, was found, as expected, in the cytoplasmic fraction of infected cells. A band of similar size was identified for both VP60Δ-L1BS and VP60Δ-L2BS, but it was in the nuclear fraction of infected cells (Fig. 2). The DNA-binding capacity of the different proteins was investigated by Southwestern blotting using a digoxigenin-labeled DNA plasmid. The different purified proteins were separated by SDS-PAGE, electrotransferred to nitrocellulose, and hybridized with the digoxigenin-labeled plasmid DNA. As seen in Fig. 3, VP60Δ protein failed to bind plasmid DNA. In contrast, both chimeric proteins VP60Δ-L1BS and VP60Δ-L2BS strongly bound DNA.
FIG. 1.
Schematic maps of VP60-L1BS and VP60-L2BS chimeric proteins.
FIG. 2.
Western blot analysis of VP60Δ-L1BS and VP60Δ-L2BS protein expression. Recombinant capsid proteins were detected in the nuclear fraction of infected sf21 cells with an anti-RHDV monoclonal antibody. Lanes 1 and 2, VP60Δ-L1BS; lanes 4 and 5, VP60Δ-L2BS; lane 3, HPV-16 VLPs.
FIG. 3.
Detection of DNA binding to VP60Δ-L1BS and VP60Δ-L2BS by Southwestern blotting using a digoxigenin-labeled plasmid probe. Lane 1, VP60Δ-L1BS; lane 2, VP60Δ; lane 3, VP60Δ-L2BS.
In order to test VLP subcellular localization, the insect cells were fixed, sectioned, and embedded in Epon resin. Ultrathin sections were then made and stained with uranyl acetate and lead citrate. Analysis of the presence of VLPs by electron microscopy showed that the two VP60 chimeric proteins self-assembled into VLPs (Fig. 4). As expected, the VP60Δ VLPs were found in the cytoplasm of infected cells (Fig. 4a and b). However, VP60Δ-L1BS and VP60Δ-L2BS recombinant VLPs were detected in the nucleus (Fig. 4c and d). In addition, the amount of VLPs was titrated by enzyme-linked immunosorbent assay using anti-VP60 monoclonal antibody, in both the nuclear and cytoplasmic fractions of Sf21-infected cells. For VP60Δ, VP60Δ-L1BS, and VP60Δ-L2BS, the cytoplasm fractions contained 80, 25, and 24% of the total VLP reactivity, respectively. Accordingly, the nuclear fractions contained 20, 75, and 76% of the VLPs, respectively.
FIG. 4.
Subcellular localization of VP60Δ (a and b) and VP60Δ-L1BS (c and d) VLPs by electron microscopy (bars, 200 nm [a and c] and 100 nm [b and d]).
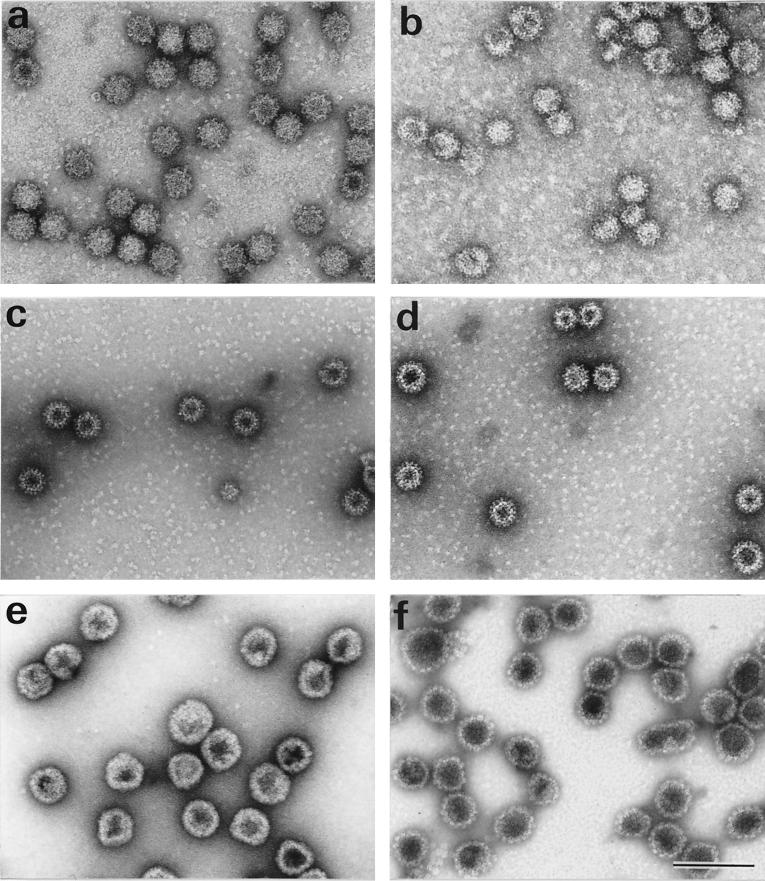
The different VLPs were purified by CsCl gradient ultracentrifugation and observed by electron microscopy after negative staining. These VLPs were similar in size and appearance to authentic RHDV virions (Fig. 5). Some tubular structures and aggregates could also be observed (data not shown). In addition, Fig. 5f shows VLPs obtained by expression of a recombinant baculovirus coding for both HPV-16 L1 and L2 proteins. These VLPs were more regular in size and shape than HPV-16 L1 VLPs (Fig. 5e).
FIG. 5.
Identification of chimeric RHDV and HPV-16 VLPs by transmission electron microscopy. (a) VP60; (b) VP60Δ; (c) VP60Δ-L1BS; (d) VP60Δ-L2BS; (e) HPV-16 L1; (f) HPV-16 L1+L2. Bar, 100 nm.
Packaging and gene transfer capacity of the RHDV–HPV-16 chimeric VLPs.
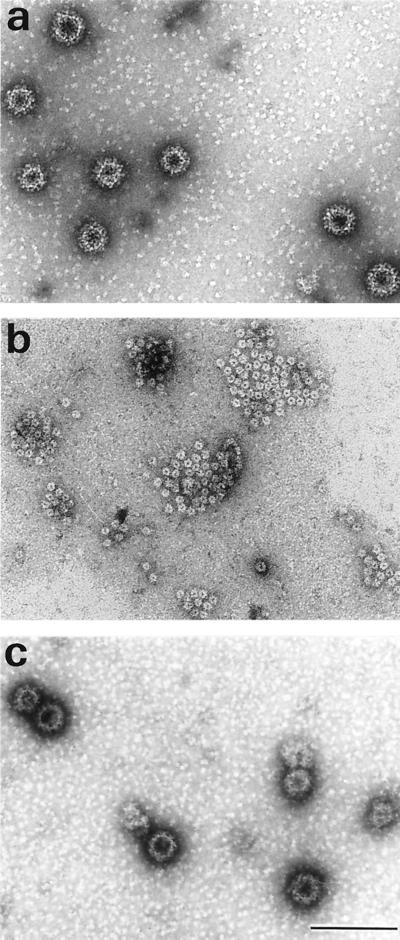
To explore the potential of the VLPs for the packaging of exogenous DNA we investigated whether the VLPs were dissociable and whether reassociation could be initiated to reconstitute intact VLPs. The principle of dissociation is the removal of Ca2+ ions and reduction of disulfide bonds. As shown previously for HPV-16 L1 VLPs (25), it was possible to disassemble RHDV capsids as the two chimeric RHDV-HPV VLPs. Figure 6 shows that under the conditions used, VLPs (Fig. 6a) were completely disassembled into structures resembling capsomers (Fig. 6b). Intact VLPs could then be reconstituted during a multistep reaction by increasing the concentration of calcium and decreasing the concentration of the reducing agent. To investigate whether foreign DNA could be packaged into VLPs, plasmid DNA was added to capsomers obtained after dissociation of the VLPs, and the preparation was then diluted in a buffer containing 5 mM CaCl2 and 1% DMSO in order to refold the VLPs (Fig. 6c). About 30 to 50% of the capsomers seemed to reassemble into VLPs.
FIG. 6.
Purified VP60Δ-L2BS VLPs (a), capsomer-like structures obtained after treatment of VLPs with EGTA and dithiothreitol (b), VP60Δ-L2BS VLPs refolded after reassembly of the capsomers in the presence of CaCl2, DMSO and β-galactosidase plasmid (c). Bar, 100 nm.
To demonstrate that plasmid DNA was packaged into VLPs, reconstituted VLPs were treated with DNase. After disassembly of the VLPs and reassembly in the presence of DNA, the mixture was treated with Benzonase. The detection of DNA after treatment is indicative of its uptake by VLPs. As shown in Fig. 7, VP60Δ-L1BS and VP60Δ-L2BS VLPs, but not VP60Δ VLPs, are able to protect pCMV-β plasmid DNA from digestion. DNA protection was estimated to be 12% for DNA introduced during the reassociation of the VP60Δ-L1BS VLPs (Table 1), compared to 18% observed when DNA was packaged in HPV-16 L1 VLPs. A lower level of protection from Benzonase degradation (9.5%) was observed with the VP60Δ-L2BS VLPs.
FIG. 7.
DNA protection assay using VP60Δ VLPs (lane 1), VP60Δ-L1BS VLPs (lane 2), VP60Δ-L2BS (lanes 3 and 4), and HPV-16 L1 VLPs (lane 5). M, molecular weight markers (λ phage digested with EcoRI and HindIII).
TABLE 1.
Nuclear localization, DNA binding, and DNA protection with HPV-16 L1 VLPs and HPV-16–RHDV chimeric VLPs
| VLP | Nuclear localization | DNA binding | DNA protection (%) |
|---|---|---|---|
| RHDV VP60 | − | − | 0 |
| HPV-16 L1 | + | + | 18 |
| VP60Δ-L1BS | + | + | 12 |
| VP60Δ-L2BS | + | + | 9.5 |
Transfer efficiency of the pCMV-β plasmid packaged into the different VLPs was initially investigated in Cos-7 cells. As shown in Fig. 8, the VP60Δ-L1BS VLPs and HPV-16 L1 VLPs can efficiently transduce packaged plasmid DNA into cells, resulting in the expression of a functional β-galactosidase as demonstrated by the presence of blue cells after addition of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The results indicate that VP60Δ and VP60Δ-L2BS VLPs could not transfer genes into Cos-7 cells. As we had previously demonstrated that many cell lines could be transfected with HPV-16 VLPs, we investigated 10 cell lines for their capacity to be transfected by the pCMV-β plasmid packaged in HPV-16 L1, VP60Δ-L1BS, and VP60Δ-L2BS VLPs. The results (Table 2) indicate that all cell lines investigated could be transfected by the HPV-16 L1 VLP vector. VP60Δ-L1BS VLPs were not able to transfect human cell lines or L929 rodent cells. However, it must be noted that rabbit RK13 and hare R17 cell lines are efficiently transfected with VP60Δ-L1BS VLPs. The level of transfection in these two cell lines was higher than that observed with HPV-16 L1 VLPs, as demonstrated by the observation of a higher number of blue cells (Table 2). Transfection with the VP60Δ-L2BS vector gave no transfer in the different cell lines tested, with the exception of a very limited level of gene transfer into the rabbit RK13 cell line.
FIG. 8.
Demonstration of gene expression of β-galactosidase in Cos-7 (a and b), R17 (c), HuH-7 (d), and CaCo2 (e and f) cells after gene transfer using VP60Δ-L1BS (a, c, d, and e) or HPV-16 L1 (b and f) capsids. Transfected cells expressing β-galactosidase were stained with X-Gal.
TABLE 2.
Efficacy of gene transfer in 10 cell lines using chimeric HPV-16–RHDV VLPs
| Cell lines | Gene transfer (no. of blue cells/well) obtained with:
|
||
|---|---|---|---|
| VP60Δ-L1BS | VP60Δ-L2BS | HPV-16 L1 | |
| RK13 | 416 | 9 | 208 |
| R17 | 757 | 0 | 500 |
| L929 | 0 | 0 | 808 |
| CHO | 0 | 0 | 90 |
| Cos-7 | 3,200 | 0 | 5,000 |
| HeLa | 0 | 0 | 60 |
| HepG2 | 0 | 0 | 56 |
| CaCo2 | 2 | 0 | 3,680 |
| HuH-7 | 0 | 0 | 224 |
| MRC5 | 0 | 0 | 15 |
It has been shown that the presence of L2 in HPV VLPs dramatically increased their gene transfer efficiency (29) and that packaging with BPV-1 L1+L2 VLPs is increased in the presence of a papillomavirus DNA sequence (33), suggesting that specific DNA binding to L2 is necessary to obtain efficient packaging. We therefore investigated gene transfer into RK13 cells using VLPs consisting of HPV-16 L1, HPV-16 L1+L2, VP60Δ-L1BS, or VP60Δ-L2BS proteins in which the pCMV-β plasmid or the same plasmid containing the BPV EPS was packaged (Table 3). No transfection of RK13 cells was observed with VP60Δ-L2BS VLP vectors, whatever the plasmid used. However, when L1 VLPs, L1+L2 VLPs, or VP60Δ-L1BS VLPs were used, a two- to threefold increase in the level of gene transfer was observed when the packaged plasmid contained the BPV EPS in comparison to the level obtained with a similar plasmid without this DNA sequence (P = 0.0005, 0.0015, and 0.04, respectively).
TABLE 3.
β-Galactosidase gene transfer into RK13 cells using plasmids with or without the BPV-1 EPS
| VLP | Gene transfer (no. of blue cells/well) obtained with:
|
|
|---|---|---|
| pCMV-β | pCMV-β–BPV-1-EPS | |
| L1 | 380 ± 42 | 915 ± 148 |
| L1+L2 | 1,130 ± 155 | 2,875 ± 1,025 |
| VP60Δ-L1BS | 680 ± 183 | 1,329 ± 451 |
| VP60Δ-L2BS | 0 | 0 |
In addition, the number of transfected cells increased from 380 with L1 VLPs to 1,130 with L1+L2 VLPs (P = 0.0001) when the pCMVβ plasmid was used and from 915 to 2,875 (P < 0.01) when the pCMVβ/BPV-1-EPS plasmid was used.
These results were confirmed using the pCMV-Luc plasmid. In experiments using RK13 cells, luciferase activity of (1.2 ± 0.3) × 104 and (1.5 ± 0.1) × 104 cps/mg was observed with VP60Δ-L2BS pseudovirions and plasmid DNA alone, respectively. With L1 VLPs, L1+L2 VLPs, and VP60Δ-L1BS VLPs, the results were (7 ± 1.3) × 106, (2 ± 0.8) × 107, and (1.2 ± 0.2) × 107 cps/mg, respectively.
In order to investigate the capacity of VP60Δ-L2BS VLPs to bind to RK13 cells, VP60Δ-L1BS pseudovirions were incubated with a 20-fold excess of L1, VP60Δ-L1BS, and VP60Δ-L2BS VLPs and then added to the cells (Table 4). Levels of inhibition of the luciferase gene transfer were 95 and 98% when VP60Δ-L1BS pseudovirions were mixed with VP60Δ-L1BS and L1 VLPs, respectively. However, only 2% inhibition was detected when VP60Δ-L2BS VLPs were used as competitors.
TABLE 4.
Inhibition of luciferase gene transfer in RK13 cells mediated by VP60Δ-L1BS pseudovirions
| Inhibitor | Luciferase activity (cps/mg) | Inhibition (%) |
|---|---|---|
| PBS | 1.0 × 107 | 0 |
| VP60Δ-L1BS | 5.0 × 105 | 95 |
| L1 16 | 1.7 × 105 | 98 |
| VP60Δ-L2BS | 0.98 × 107 | 2 |
DISCUSSION
We have shown in this study that the insertion of one of the DNA-binding sequences present in the two structural proteins of HPV-16 at the N-terminal extremity of the RHDV VP60 gene is able to transform this protein into a DNA-binding protein. Moreover, the chimeric VP60 proteins conserved their ability to form VLPs that are similar in size and morphology to authentic virions and VP60Δ VLPs. The VP60Δ protein with a deletion of 45 N-terminal amino acids localized in the cytoplasm of infected cells, as observed for the full-length VP60 protein (18). The chimeric VP60Δ-L1BS and VP60Δ-L2BS VLPs still formed VLPs but were found in the nuclei of infected cells. This confirms the observation of Zhou et al. (34) that the 11 C-terminal amino acids of the HPV-16 L1 protein contained a strong nuclear localization signal. As VP60Δ-L2BS VLPs also localized in the nuclei of infected cells, the 12 N-terminal amino acids of the HPV-16 L2 protein also contained a nuclear localization signal, in agreement with results observed for HPV-6 L2 (23).
We observed that VP60Δ-L1BS and VP60Δ-L2BS VLPs could be disassembled and reassembled under the same conditions as those used previously for HPV-16 L1 VLPs and to package a 7.2-kbp DNA plasmid. However, the DNA protection from enzymatic degradation was lower with VP60Δ-L2BS VLPs than with VP60Δ-L1BS VLPs. The results obtained are in agreement with the fact that truncation of the DNA-binding nuclear localization sequence of the HPV-16 L1 protein results in the loss of its DNA packaging capacities in VLP disassembly-reassembly experiments (27). A similar result was also observed for polyomavirus VLPs (7). These findings suggest that the addition of a short amino acid sequence containing a DNA-binding sequence is sufficient to confer on a VLP the capacity to encapsidate DNA. However, only the VP60Δ-L1BS VLPs were able to efficiently transfer the gene into Cos-7 cells. It has recently been shown that DNA encapsidation by BPV-1 VLPs is enhanced by a specific DNA sequence of the papillomavirus genome (33). This 120-bp sequence, named EPS, was also recognized by HPV-6b VLPs despite the phylogenic difference between these two papillomavirus types. Zhao et al. (33) have therefore suggested that other papillomaviruses may use the same packaging sequence. In order to test whether the low level of gene transfer observed with VP60Δ-L2BS VLPs was due to the absence of a papillomavirus DNA sequence recognized by L2BS, we investigated gene transfer using plasmids with and without the EPS packaged into VLPs containing L1 or L2 binding sequences. Our results indicate that gene transfer was enhanced in the presence of the BPV EPS with capsids containing the L1BS protein. If no gene transfer was observed with VP60Δ-L2BS VLPs whatever the plasmid used, an increase in DNA packaging was observed with the plasmid containing the BPV EPS (data not shown). This confirmed that the EPS is not papillomavirus type specific as suggested by Zhao et al. (33) and suggested that the BPV EPS is recognized by both L1BS and L2BS. In addition, the results suggested that the low level of gene transfer observed with VP60Δ-L2BS capsids is not a consequence of specificity of the DNA binding. The difference between the gene transfer capacities of VP60Δ-L1BS and VP60Δ-L2BS VLPs could be related to the presence of a heparin-binding sequence in the L1BS inserted at the N terminus of the RHDV VP60 protein (8). Its presence in VP60Δ-L1BS might allow the interaction between VLPs and cells, thus favoring attachment to a cell receptor and internalization of the capsid. Inhibition experiments are in agreement with such hypotheses, since an absence of gene transfer inhibition was observed between VP60Δ-L1BS pseudovirions and VP60Δ-L2BS VLPs.
To investigate the cell tropism of the VP60Δ-L1BS VLPs, we compared gene transfer in 10 cell lines to the transfer observed with HPV-16 VLPs. All cell lines investigated could be transfected with HPV-16 L1 VLPs. In addition to previously published findings (25), we transfected rabbit and hare kidney cells, HepG2 and HuH-7 human liver cells, and L929 mouse cells, confirming the presence of HPV-16 receptors on numerous cells. In contrast, no gene transfer was observed in human cell lines with VP60Δ-L1BS VLPs. However, the results were better than those observed with HPV-16 VLPs in rabbit and hare kidney cells. This suggests that RHDV and HPV-16 cell receptors are different. Alpha-6 integrin has been identified as a cell receptor candidate for HPV-16 (5, 13), but no cell receptor has so far been identified for RHDV. Glycosaminoglycans (GAGs) have also been proposed as cell receptors for HPV-16, and virus sequences interacting with these cell components have been identified at the C terminus of the L1 protein (8). As this HPV-16 viral sequence was introduced into the VP60-L1BS, this could explain why gene transfer was much more efficient with VP60Δ-L1BS VLPs than with VP60Δ-L2BS VLPs. However, if GAGs act as the only cell receptor for HPV-16 VLPs, similar cell tropism would be observed with VP60Δ-L1BS and L1 VLPs. Our results thus support the hypothesis that proposes the presence of two cell receptors in HPV-16 VLPs, explaining a difference in cell tropism for HPV-16 VLPs and chimeric L1-RHDV capsids. This could also suggest that virus binding to cells is mediated by interaction between the virus and the cell GAGs followed by internalization of the capsid by a cell receptor, which is the alpha-6 integrin in the case of HPV-16 and is an unknown receptor for RHDV.
In conclusion, we demonstrated the possibility of producing recombinant chimeric VLPs by the addition of a DNA-binding sequence to the capsid protein of RHDV, a protein unable to bind DNA and transfer plasmid DNA by itself. These VLPs have a restricted cell tropism compared to that observed previously for HPV-16 L1 (25) or polyomavirus VP1 VLPs (6). This suggests the possibility of developing new vectors by producing chimeric VLPs based on different capsids with different DNA packaging capacities and different cell tropisms.
ACKNOWLEDGMENTS
This study was supported by grants from the Association pour la Recherche sur le Cancer (no. 9977) and Biotechnocentre. Slimane El Mehdahoui was supported by a fellowship from the Fondation de France.
We thank J. F. Vautherot, P. Velge (INRA, Nouzilly, France), and P. Roingeard (Faculté de Medécine, Tours, France) for the generous gift of cell lines.
REFERENCES
- 1.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dmitriev I, Krasnykh V, Miller C R, Wang M, Kashentseva E, Mikheeva G, Belousova N, Curiel D T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooley S, Welter C, Blin N. Nonradioactive Southwestern analysis using chemiluminescent detection. Biotechniques. 1992;13:540–543. [PubMed] [Google Scholar]
- 4.Douglas J T, Miller C R, Kim M, Dmitriev I, Mikheeva G, Krasnykh V, Curiel D T. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat Biotechnol. 1999;17:470–475. doi: 10.1038/8647. [DOI] [PubMed] [Google Scholar]
- 5.Evander M, Frazer I H, Payne E, Qi Y M, Hengst K, McMillan N A J. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J Virol. 1997;71:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forstova J, Krauzewicz N, Sandig V, Elliott J, Palkova Z, Strauss M, Griffin B E. Polyomavirus pseudocapsids as efficient carriers of heterologous DNA into mammalian cells. Hum Gene Ther. 1995;6:297–306. doi: 10.1089/hum.1995.6.3-297. [DOI] [PubMed] [Google Scholar]
- 7.Gillock E T, An K, Consigli R A. Truncation of the nuclear localization signal of polyomavirus VP1 results in a loss of DNA packaging when expressed in the baculovirus system. Virus Res. 1998;58:149–160. doi: 10.1016/s0168-1702(98)00113-0. [DOI] [PubMed] [Google Scholar]
- 8.Joyce J G, Tung J S, Przysiecki C T, Cook J C, Lehman E D, Sands J A, Jansen K U, Keller P M. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem. 1999;274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 9.Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. In vitro construction of pseudovirions of human papillomavirus type 16: incorporation of plasmid DNA into reassembled L1/L2 capsids. J Virol. 1998;72:10298–10300. doi: 10.1128/jvi.72.12.10298-10300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirnbauer R, Taub J, Greenstone H, Roden R, Dürst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurent S, Vautherot J-F, Madelaine M-F, Le Gall G, Rasschaert D. Recombinant rabbit hemorrhagic disease virus capsid protein expressed in baculovirus self-assembles into viruslike particles and induces protection. J Virol. 1994;68:6794–6798. doi: 10.1128/jvi.68.10.6794-6798.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent S, Vautherot J F, Le Gall G, Madelaine M F, Rasschaert D. Structural, antigenic and immunogenic relationships between European brown hare syndrome virus and rabbit haemorrhagic disease virus. J Gen Virol. 1997;78:2803–2811. doi: 10.1099/0022-1317-78-11-2803. [DOI] [PubMed] [Google Scholar]
- 13.McMillan N A, Payne E, Frazer I H, Evander M. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology. 1999;261:271–279. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- 14.Meyers G, Wirblich C, Thiel H J. Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991;184:664–676. doi: 10.1016/0042-6822(91)90436-f. [DOI] [PubMed] [Google Scholar]
- 15.Moreland R B, Montross L, Garcea R L. Characterization of the DNA-binding properties of the polyomavirus capsid protein VP1. J Virol. 1991;65:1168–1176. doi: 10.1128/jvi.65.3.1168-1176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller M, Gissmann L, Cristiano R J, Sun X-Y, Frazer I H, Jenson A B, Alonso A, Zentgraf H, Zhou J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J Virol. 1995;69:948–954. doi: 10.1128/jvi.69.2.948-954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagesha H S, Wang L F, Hyatt A D, Morrissy C J, Lenghans C, Westbury H A. Self assembly, antigenicity, and immunogenicity of the rabbit haemorrhagic disease virus. Arch Virol. 1995;140:1095–1108. doi: 10.1007/BF01315418. [DOI] [PubMed] [Google Scholar]
- 18.Park J H, Ochiai K, Itakura C. Detection of rabbit haemorrhagic disease virus particles in the rabbit liver tissues. J Comp Pathol. 1992;107:329–340. doi: 10.1016/0021-9975(92)90008-i. [DOI] [PubMed] [Google Scholar]
- 19.Pickles R J, McCarty D, Matsui H, Hart P J, Randell S H, Boucher R C. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol. 1998;72:6014–6023. doi: 10.1128/jvi.72.7.6014-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose R C, Bonnez W, Reichman R C, Garcea R L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasagawa T, Pushko P, Steers G, Gscheimeissner S E, Hajibagheri M, Finch J, Crawford L, Tommasino M. Synthesis and assembly of virus like particles of human papillomavirus type 6 and type 16 in fission yeast schizosaccharomyces pombe. Virology. 1995;206:403–410. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 22.Sibilia M, Boniotti M B, Angoscini P, Capucci L, Rossi C. Two independent pathways of expression lead to self-assembly of the rabbit hemorrhagic disease virus capsid protein. J Virol. 1995;69:5812–5815. doi: 10.1128/jvi.69.9.5812-5815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X Y, Frazer I H, Müller M, Gissmann L, Zhou J. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology. 1995;213:321–327. doi: 10.1006/viro.1995.0005. [DOI] [PubMed] [Google Scholar]
- 24.Thouvenin E, Laurent S, Madelaine M F, Rasschaert D, Vautherot J F, Hewat E A. Bivalent binding of a neutralising antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J Mol Biol. 1997;270:238–246. doi: 10.1006/jmbi.1997.1095. [DOI] [PubMed] [Google Scholar]
- 25.Touzé A, Coursaget P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 1998;26:1317–1324. doi: 10.1093/nar/26.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touzé A, El Mehdaoui S, Sizaret P-Y, Mougin C, Muñoz N, Coursaget P. The L1 major capsid protein of human papillomavirus type 16 variants affects yield of virus-like particles produced in an insect cell expression system. J Clin Microbiol. 1998;36:2046–2051. doi: 10.1128/jcm.36.7.2046-2051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Touzé A, Mahé D, El Mehdaoui S, Combita A L, Bousarghin L, Coursaget P. The 9 C-terminal amino acids of the major capsid protein of the human papillomavirus type 16 are essential for DNA binding and gene transfer capacity. FEMS Microbiol Lett. 2000;189:121–127. doi: 10.1111/j.1574-6968.2000.tb09217.x. [DOI] [PubMed] [Google Scholar]
- 28.Trus B L, Roden R B, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- 29.Unckell F, Streeck R E, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol. 1997;71:2934–2939. doi: 10.1128/jvi.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpers C, Unckell F, Schirmacher P, Streeck R E, Sapp M. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J Virol. 1995;69:3258–3264. doi: 10.1128/jvi.69.6.3258-3264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirblich C, Meyers G, Ohlinger V F, Capucci L, Eskens U, Haas B, Thiel H-J. European brown hare syndrome virus: relationship to rabbit hemorrhagic disease virus and other calicivirus. J Virol. 1994;68:5164–5173. doi: 10.1128/jvi.68.8.5164-5173.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zabner J, Freimuth P, Puga A, Fabrega A, Welsh M J. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J Clin Investig. 1997;100:1144–1149. doi: 10.1172/JCI119625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K N, Frazer I H, Jun Liu W, Williams M, Zhou J. Nucleotides 1506–1625 of bovine papillomavirus type 1 genome can enhance DNA packaging by L1/L2 capsids. Virology. 1999;259:211–218. doi: 10.1006/viro.1999.9714. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Doorbar J, Sun X Y, Crawford L V, McLean C S, Frazer I H. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology. 1991;185:625–632. doi: 10.1016/0042-6822(91)90533-h. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J, Sun X-Y, Louis K, Frazer I H. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J Virol. 1994;68:619–625. doi: 10.1128/jvi.68.2.619-625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]