Abstract
Poly ADP-ribose polymerase (PARP) inhibitor monotherapies are selectively effective in patients with pancreatic, breast, prostate, and ovarian cancers with BRCA1 mutations. Cancer patients with more frequent wild-type BRCA show poor responses to PARP inhibitors. Moreover, patients who are initially sensitive to these inhibitors eventually respond poorly to drugs. In the present study, we discover that abrogation of Kruppel-like factor 5 (KLF5) significantly inhibits homologous recombination, which is the main mechanism for DNA double-stranded repair. Furthermore, the downregulation of KLF5 expression promotes the DNA damage induced by olaparib and significantly reduces the IC 50 of the RARP inhibitor in pancreatic cancer cells. Overexpression of BRCA1 reverses the above effects caused by silencing of KLF5. Olaparib combined with a KLF5 inhibitor has an enhanced cytotoxic effect. Mechanistically, we identify BRCA1 as a KLF5 target gene. BRCA1 is positively correlated with KLF5 in PDAC tissue. Our results indicate that inhibition of KLF5 may induce BRCAness in a larger pancreatic cancer subset with proficient BRCA. The combination of KLF5 inhibitors and PARP inhibitors provides a novel treatment strategy to enhance the sensitivity of BRCA1-proficient pancreatic cancer to PARP inhibitors.
Keywords: pancreatic cancer, wild-type BRCA , BRCAness, KLF5, BRCA1
Introduction
There were 466,003 cancer-related deaths and 495,773 new cases of pancreatic cancer (PC) worldwide in 2020 [1]. It is urgent to develop novel treatment strategies beyond surgery and traditional chemotherapy. Strickler et al. [2] demonstrated that patients pretreated with KRAS p. G12C-mutated advanced PC responded robustly to sotorasib and showed good tolerance [2]. Pembrolizumab is recommended for unresectable or metastatic pancreatic ductal adenocarcinomas (PDAC) harboring defective DNA mismatch-repair-deficiency or microsatellite instability-high, which occur in 1%‒2% of PDAC cases. The PARP inhibitor olaparib has been approved for maintenance treatment of pretreated patients with metastatic PDAC with a germline BRCA mutation (4%‒7% of PDAC) [3]. However, PARP inhibitors confer little clinical benefit to most pancreatic cancer patients with wild-type BRCA1/2. Therefore, it is urgent to develop a novel strategy for expanding PARP inhibitors to BRCA-proficient PC.
Krüppel-factor 5 (KLF5) is a member of the KLF family and acts as a transcription factor that is frequently overexpressed in cancers [4]. KLF5 often functions as an oncogenic protein by regulating multiple target genes, such as Slug [5], p27 [6], and Cyclin D1 [7], thus regulating cell migration and proliferation. Hence, the upregulation of KLF5 has been demonstrated to promote the progression of various human malignancies, such as breast cancer [8], bladder cancer [9], colorectal cancer [10], gastric cancer [11], and hepatocellular carcinoma [12]. High expression of KLF5 is significantly correlated with a worse prognosis [12]. Emerging evidence has also revealed that KLF5 drives the progression of pancreatic cancer by inducing proliferation, glycolysis, and immunosuppression [ 13‒ 15]. We previously reported that FBW7 could regulate fibroblast growth factor (FGF)-binding protein-1 in a Myc-dependent way, which mediated the proliferation and migration of PDAC [16]. Emerging evidence has demonstrated that FBW7 acts as a vital tumor suppressor by targeting multiple oncogenic substrates in which the expression of KLF5, another key substrate, is also significantly regulated by FBW7 [17]. Li et al. [18] reported that KLF5 promoted the expression of Rad51, which could elicit DNA repair by homologous recombination. However, the role of KLF5 in PDAC DNA repair and sensitivity to PARP inhibitors remains unclear.
It has been demonstrated that homologous recombination repair (HRR) defects and PARP inhibition are synthetically lethal [19]. The tumor suppressor BRCA1/2 plays a key role in HRR [20]. Therefore, PARP inhibitors could significantly kill cancer cells with BRCA mutations. The inhibitor has been approved to treat patients with prostate, breast, and ovarian cancers with BRCA mutations based on several clinical trials [ 21‒ 23]. However, BRCA wild-type patients, accounting for the majority of all cancer patients, are not recommended for PARP inhibitors. Extensive subsequent studies have focused on expanding the use of these inhibitors to patients. A recent study showed that paclitaxel inhibits CDK1, resulting in weakened BRCA1 phosphorylation, which sensitizes ovarian cancer cells with proficient homologous recombination to PARP inhibitors [24]. Lu et al. [25] reported that inhibition of salt-inducible kinase 2 could produce synthetic lethality with several PARP inhibitors in homologous recombination DNA repair proficiency cancer cells by suppressing PARP enzyme activity and the DNA double-strand break repair pathway. Additionally, Ibrahim et al. [26] reported that suppression of PI3K inhibited BRCA expression and sensitive triple-negative breast cancer with proficient BRCA to PARP inhibition. In general, repression of BRCA expression or protein activity represents a promising strategy to broadly utilize PARP inhibitors in cancers.
In the present study, we explored the potential role of KLF5 in sensitivity to PARP inhibitors in pancreatic cancer cells. We discovered that repression of KLF5 elicited olaparib-induced DNA damage and significantly decreased the IC 50 of olaparib in pancreatic cancer cells. Silencing of KLF5 downregulated the expression of BRCA1 at the transcriptional level, which induced a “BRCAness”. Collectively, our results revealed a promising treatment target by which PARP inhibitors could be utilized to benefit a wider range of patients with pancreatic cancer.
Materials and Methods
Cell culture and small compounds
The human pancreatic duct epithelial cell line hTERT-HPNE and four PDAC cell lines PANC-1, CFPAC-1, BxPC3, and SW1990 were obtained from the American Type Culture Collection (ATCC; Manassas, USA) and cultured based on standard ATCC protocols. In short, hTERT-HPNE and PANC-1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, USA) with 10% fetal bovine serum (FBS; Gibco). BxPC3 cells were cultured in Roswell Park Memorial Institute (RPMI; Gibco) with 10% FBS. CFPAC-1 cells were cultured in Iscove’s modified Dulbecco’s medium containing 10% FBS (Gibco). SW1990 cells were maintained in Leibovitz (L-15; Gibco) with 10% FBS. In addition, 100 U/mL penicillin (Gibco) and 0.1 mg/mL streptomycin (Gibco) were added to all the culture media. SW1990 cells were cultured at 37°C in a humidified incubator without CO 2. The remaining cells were maintained at 37°C in a humidified incubator with 5% CO 2. Olaparib (HY-10162) was purchased from MedChemExpress (Monmouth Junction, USA).
Quantitative real-time PCR (qRT-PCR)
Trizol reagent (Beyotime, Nantong, China) was utilized to isolate and purify total RNA. cDNAs were then obtained after reverse transcription using a PrimeScript RT reagent kit (TaKaRa, Dalian, China). Quantitative real-time PCR was performed as described previously [27]. All reactions were conducted in triplicate. The primers used in this study are presented in Supplementary Table S1.
Western blot analysis
Standard western blot analysis was conducted. Total proteins from hTERT-HPNE and PDAC cells were extracted utilizing RIPA lysis buffer (Beyotime, Shanghai, China). The extracted proteins were electrophoresed in 10% SDS-PAGE gels and then transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, USA). The membranes were incubated with primary antibodies overnight at 4°C and then incubated with the appropriate secondary antibody at room temperature for 1 h. An Enhanced chemiluminescence kit (Beyotime) was used to detect proteins. The antibodies used in this study were as follows: anti-β-actin (1:4000; Proteintech, Chicago, USA), anti-KLF5 (1:1000; Proteintech), anti-BRCA1 (1:1000; Proteintech), anti-Rad51 (1:1000; Proteintech), and HRP-conjugated secondary antibodies (1:3000; Proteintech).
Plasmids
The TRC cloning vector (pLKO.1; Addgene, Cambridge, USA) was utilized to construct shRNA plasmids against KLF5 according to standard protocols [28]. Targets (21 bp) against KLF5 were 5′-CCTATAATTCCAGAGCATAAA-3′ and 5′-GCTGTAATGTATATGGCTTTA-3′. pLKO.1-shKLF5, psPAX2, and pMD2.G were cotransfected into HEK-293T cells at a ratio of 4:3:1 to generate lentiviral particles. The coding sequences of human BRCA1 were cloned and inserted into the lentiviral vector pCDH-CMV-MCS-EF1-puro (SBI, San Francisco, USA) to generate BRCA1 expression plasmids.
Cell viability assay
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Tokyo, Japan) was used to investigate cell viability and cytotoxicity, as previously described [29].
Immunohistochemical staining (IHC)
Clinical tissue samples were obtained from patients diagnosed with pancreatic cancer at Zhongshan Hospital, Fudan University. We obtained the patient’s consent and approval from the Institutional Research Ethics Committee of Zhongshan Hospital, Fudan University. Antibodies against KLF5 and BRCA1 were utilized to perform IHC in paraffin-embedded specimens of tissue according to standard IHC procedures. Anti-KLF5 antibody (Proteintech) and anti-BRCA1 antibody (Proteintech) were used at a dilution factor of 1:400. Positive intensity and proportion were scored as previously described [30].
Colony formation assay
The cells were cultivated in 6-well plates at 500 cells per well for 10 days. The cells were then fixed utilizing 4% paraformaldehyde and stained with 0.1% crystal violet. Finally, the number of colonies was counted under a microscope (Olympus, Tokyo, Japan).
DNA damage detection
To confirm whether DNA is damaged, a DNA Damage Assay kit (Beyotime) was used to detect the DNA damage markers γ-H2AX ( i. e., phosphorylated H2AX) according to the manufacturer’s instructions [31]. The images were taken by a confocal fluorescence microscope (Olympus).
Dual-luciferase assay
The BRCA1 promoter region from ‒2000 to +250 of the transcription start site or its related mutant sequence was cloned and inserted into the pGL3-Basic vector. The firefly and Renilla luciferase activities were measured using the Dual Luciferase Assay kit (Promega, Madison, USA), as previously described [32].
Chromatin immunoprecipitation assay (ChIP)
The ChIP assays were conducted using the EZ-ChIP kit (Millipore), as previously described [33]. Primers to detect BRCA1 promoter occupancy are listed in Supplementary Table S1.
Animal studies
Five-week-old male nude mice were purchased from the Shanghai SLAC Laboratory (Shanghai, China). About 6×10 6 cells were subcutaneously inoculated into the left flank of the mice until the tumor volume reached ~100 mm 3. The mice were randomly divided into four subgroups (5 mice in each group): DMSO, olaparib, ML264, and a combination of olaparib and ML264. Intraperitoneal injections of olaparib (50 mg/kg) or ML264 (20 mg/kg) were administered daily. Next, tumor size was measured every 3 days and the tumor volume was calculated as length×width 2×0.5. At 5 weeks post-implantation, the tumor samples were surgically dissected. The protocol was approved by the Committee on the Ethics of Animal Experiments of Fudan University and conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
Statistical analysis
Experiments were repeated at least three times. All statistical analyses were conducted using SPSS version 19.0 software (IBM) or GraphPad Prism 8. Data are expressed as the mean±SD, and two-tailed unpaired Student’s t tests were used to compare the differences between any two groups. One-way analysis of variance was used for comparisons among different groups. The survival curve was plotted using the Kaplan-Meier method and compared by the log-rank test. Differences were considered significant when P<0.05.
Results
Knockdown of KLF5 enhances the cytotoxicity of olaparib
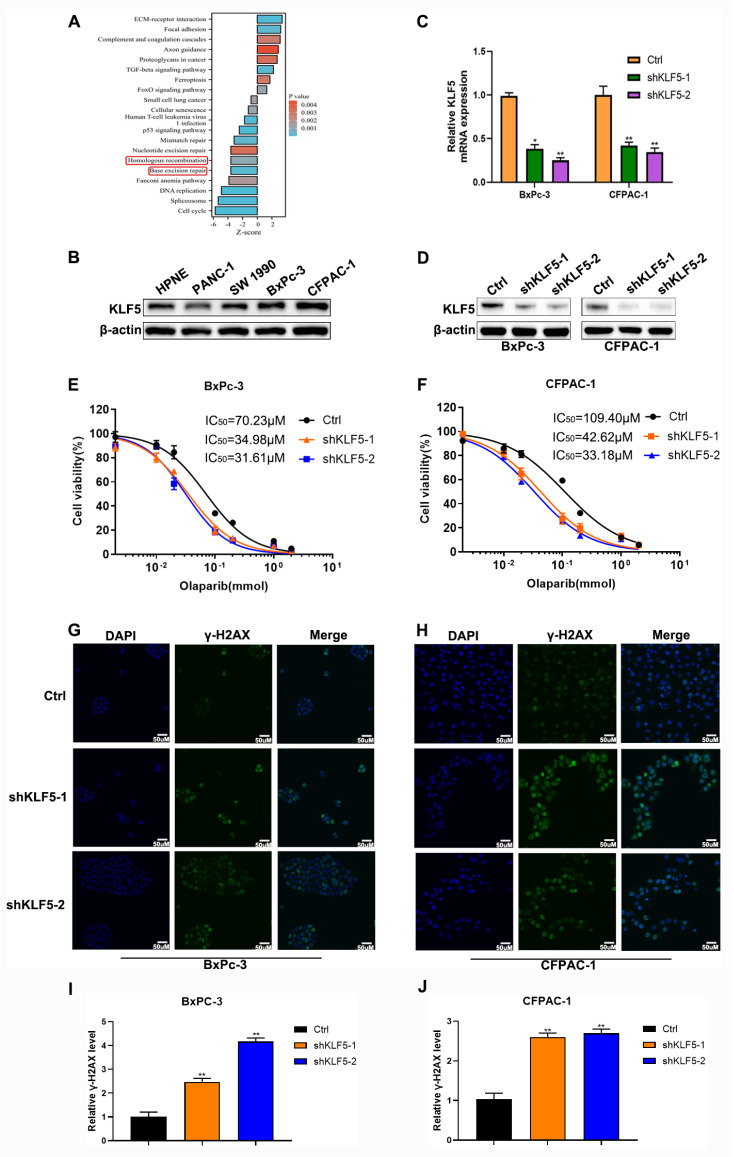
To further investigate the role of KLF5 in the sensitivity of olaparib in PDAC, we reanalyzed the previous transcriptome sequencing in which KLF5 was knocked out in CFPAC-1 cells [34]. KEGG pathway analysis showed that inhibition of KLF5 significantly suppressed homologous recombination ( Figure 1A). We detected KLF5 protein levels in a human pancreatic duct epithelial cell line (hTERT-HPNE) and four pancreatic cancer cell lines to select suitable cell lines for further exploration. Our results indicated that KLF5 expression was higher in the BxPC-3 and CFPAC-1 cell lines ( Figure 1B). Thus, stable KLF5-silenced BxPC-3 and CFPAC-1 cells were established by infection with lentivirus and selected using puromycin. The knockdown efficiency was confirmed by qRT-PCR ( Figure 1C) and western blot analysis ( Figure 1D). To investigate whether KLF5 could decrease the IC 50 of PARP inhibitors in pancreatic cancer cells, we examined the viability of cells cultured in olaparib at gradient concentrations. Abrogation of KLF5 could significantly sensitize BxPC-3 and CFPAC-1 cell lines to olaparib ( Figure 1E,F). Furthermore, we observed that suppression of KLF5 robustly increased the level of the DNA damage marker γ-H2AX induced by olaparib in BxPC-3 cells ( Figure 1G,I) and CFPAC-1 cells ( Figure 1H,J). These results demonstrated that KLF5 could regulate olaparib-induced cytotoxicity in pancreatic cancer cells.
Figure 1 .
Knockdown of KLF5 sensitives PDAC cells to olaparib
(A) KEGG pathway enrichment. (B) Western blot analysis was used to investigate KLF5 expression in several cell lines. (C) qRT-PCR was used to explore the knockdown efficiency of shKLF5. (D) Western blot analysis was used to analyze the knockdown efficiency of shKLF5. (E,F) The curves depict the dose-dependent toxicity of olaparib in BxPC3 and CFPAC-1 cell lines transfected with shKLF5 or shRNA-NC. (G,H) Confocal microscopy suggested γ-H2AX (green) in KLF5-silenced PDAC cells that were pretreated with olaparib (100 μM) or not. (I,J) Relative γ-H2AX level in PDAC cells. *P<0.05, **P<0.01.
BRCA1 is positively correlated with KLF5 expression
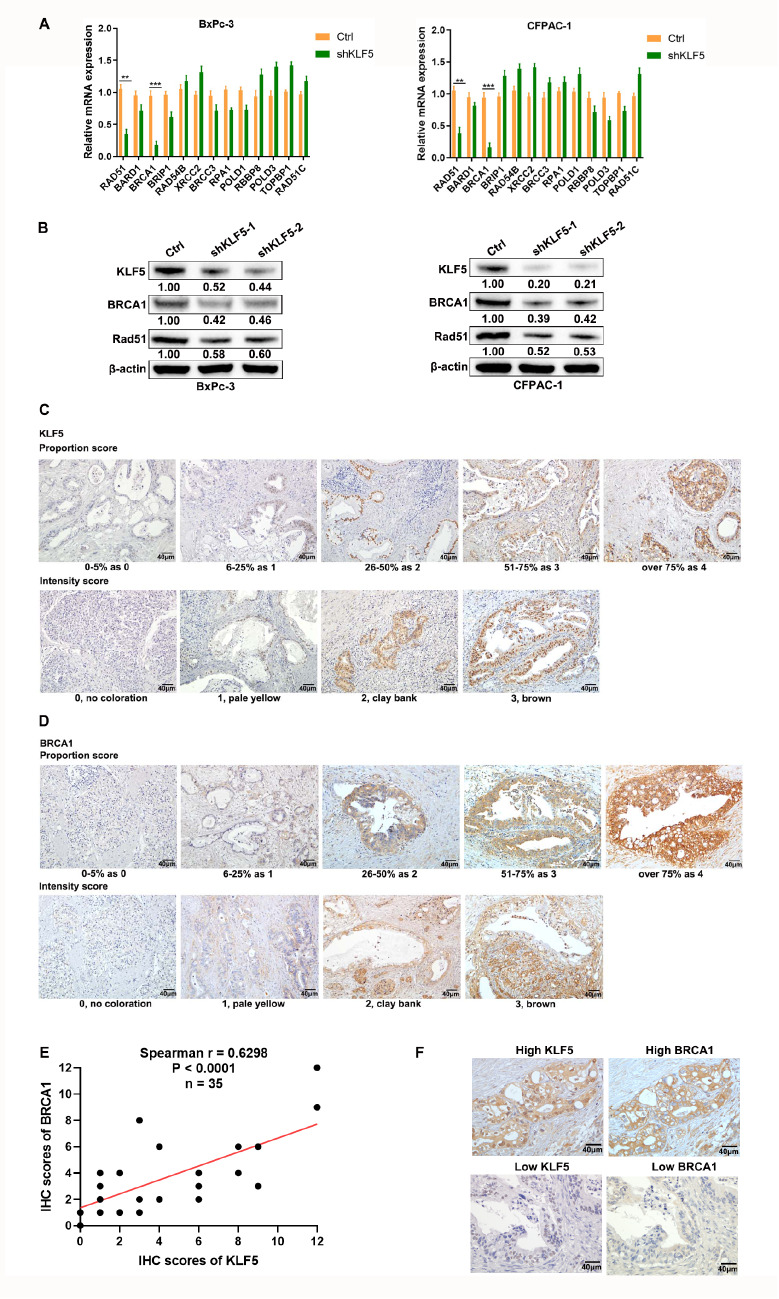
There were 13 differentially expressed genes in the above homologous recombination pathway ( Figure 1A). qRT-PCR results suggested the most significant difference in BRCA1 expression, followed by Rad51 in BxPC-3 and CFPAC-1 cells ( Figure 2A), which was confirmed by western blot analysis ( Figure 2B). These results indicated that BRCA1 may be the key for KLF5 to regulate olaparib sensitivity. To further verify the results of the cell experiments, we investigated the correlation between BRCA1 and KLF5 in PDAC patients. The semiquantitative IHC scores of BRCA1 and KLF5 were obtained by multiplying the intensity scores and proportion scores. The representative expressions of BRCA1 and KLF5 are shown in Figure 2C,D. Then, we performed a statistical analysis of the relationship between BRCA1 and KLF5, and the results suggested that there was a robust positive correlation between BRCA1 and KLF5 in PDAC patients ( Figure 2E). Two typically positive examples of BRCA1 and KLF5 expression are shown in Figure 2F.
Figure 2 .
Correlation between the expression of KLF5 and homologous recombination-related genes
(A) The mRNA levels of KLF5 and genes enriched in homologous recombination pathways in PDAC cells were measured by qPCR. (B) The protein levels of KLF5, BRCA1, and Rad51 were analyzed by western blot analysis. (C,D) Representative micrographs suggesting the intensity score and proportion score of KLF5 and BRCA1. (E) KLF5 was positively correlated with BRCA1 expression in pancreatic cancer patients, as indicated by IHC and scoring. (F) Patients with higher level of KLF5 showed higher BRCA1 expression. **P<0.01, ***P<0.001, ****P<0.0001.
BRCA1 is a downstream target of KLF5
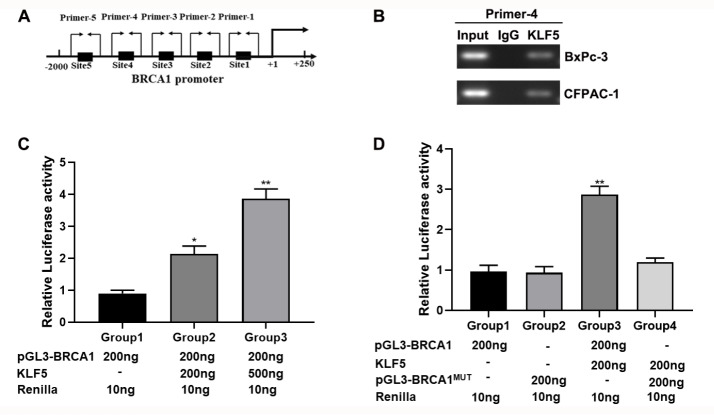
Based on the above results, we hypothesized that BRCA1 may be a target of KLF5. We observed that KLF5 had several possible binding sites in the BRCA1 promoter region ( Figure 3A). We then performed a ChIP assay with a KLF5 antibody to validate that KLF5 binds to the BRCA1 promoter. The results suggested that KLF5 bound to the BRCA1 promoter at the site of primer 4 ( Figure 3B) instead of the other four sites ( Supplementary Figure S1A). The subsequent luciferase assays indicated that regulation of KLF5 expression increased BRCA1 promoter activity in a dose-dependent manner ( Figure 3C). These results were further invalidated by the mutation of BRCA1 binding sites in which the sequence was mutated from TCCCCTTCCC into GAAAAGGAAA ( Figure 3D).
Figure 3 .
KLF5 is involved in BRCA1 transcription in PDAC
(A) The position of the KLF5 binding sites in the BRAC1 promoter. (B) KLF5 binds to the sites of the BRAC1 promoter region in BxPC-3 and CFPAC-1 cells, as detected by ChIP assay. (C) KLF5 mediated BRAC1 promoter activity in HEK-293T cells. (D) KLF5 did not regulate mutated BRAC1 promoter activity in HEK-293T cells. *P<0.05, **P<0.01.
Overexpression of BRCA1 reverses enhanced cytotoxicity of olaparib induced by silencing of KLF5
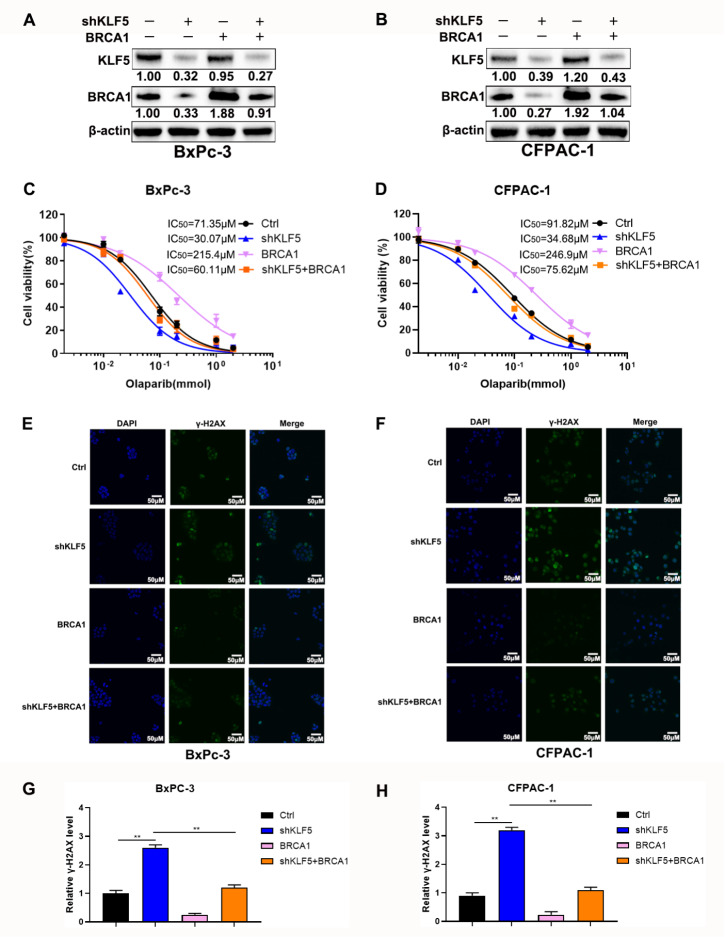
Our results suggested that KLF5 could mediate DNA damage and cytotoxicity induced by olaparib by regulating the expression of BRCA1. Subsequent western blot analysis suggested that overexpression of BRCA1 significantly reversed its decreased level caused by KLF5 silencing in BxPC-3 and CFPAC-1 cells ( Figure 4A,B). Further results showed that increased BRCA1 significantly attenuated the enhanced cytotoxicity of olaparib induced by silencing of KLF5 ( Figure 4C,D). However, overexpression of Rad51 could only slightly weaken the effect of inhibition of KLF5 ( Supplementary Figure S1B,C). The above results indicated that BRCA1 rather than Rad51 plays a critical role in KLF5-mediated sensitivity of pancreatic cancer cells to olaparib. Additionally, upregulation of BRCA1 impaired the increased DNA damage induced by the inhibition of KLF5 in BxPC-3 cells ( Figure 4E,G) and CFPAC-1 cells ( Figure 4F,H).
Figure 4 .
Overexpression of BRCA1 reverses increased sensitivity to olaparib induced by silenced KLF5
(A,B) Western blot analysis was utilized to measure the protein levels of KLF5 and BRCA1 in BxPC-3 and CFPAC-1 cells. (C,D) The CCK-8 assay depicted the dose-dependent toxicity of olaparib in BxPC3 and CFPAC-1 cell lines transfected with shKLF5 and BRCA1. (E,F) Confocal microscopy showed γ-H2AX (green) in BxPC-3 and CFPAC-1 cells that were pretreated with olaparib (100 μM) or not. (G,H) Relative γ-H2AX level in PDAC cells. **P<0.01
KLF5 inhibition synergistically enhances PARP inhibitor activity
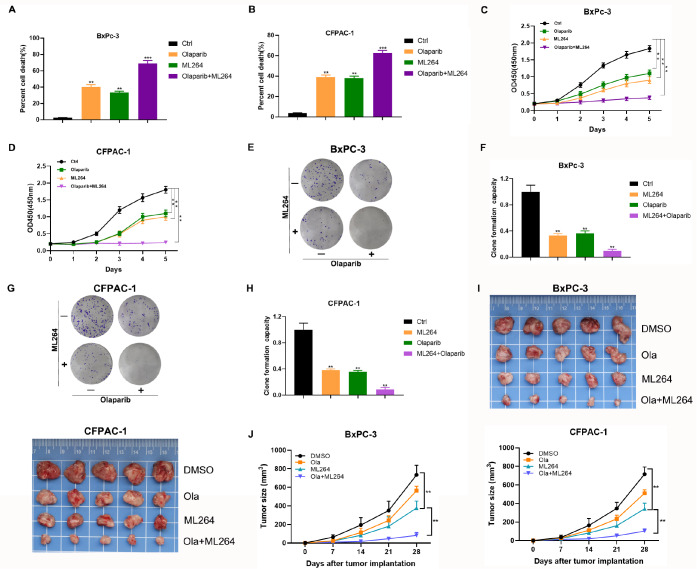
The above results indicated that inhibition of KLF5 may sensitize PDACs to PARP inhibitors. Therefore, we sought to explore the pharmacological significance of inhibiting KLF5. As expected, olaparib or ML264 inhibited cell viability, and the lethal effect of olaparib combined with ML264 was much better than that of monotherapy ( Figure 5A,B). CCK-8 assay results indicated that olaparib or ML264 alone repressed cell growth, and the combination of the two inhibitors further decreased cell viability ( Figure 5C,D). Olaparib or ML264 alone suppressed colony formation in BxPC-3 and CFPAC-1 cells, and the addition of ML264 significantly enhanced the inhibitory effect of olaparib ( Figure 5E‒H).
Figure 5 .
KLF5 inhibitors increase the sensitivity of PDAC cells to olaparib
(A,B) Cell death was determined in BxPC-3 and CFPAC-1 cells treated with olaparib (100 μM) and ML264 (10 μM) using a CCK-8 assay. (C,D) The proliferation of BxPC-3 cells was evaluated in BxPC-3 and CFPAC-1 cells treated with olaparib and ML264 by CCK-8 assay. (E,F) The influence of olaparib and ML264 on BxPC-3 cells was determined by colony formation assay. (G,H) The influence of olaparib and ML264 on CFPAC-1 cells was measured by colony formation assay. (I) The mice were randomly divided into DMSO, olaparib, ML264, and olaparib+ML264 groups and treated as described in the Methods. (J) The tumor sizes were measured using Vernier calipers. Tumor growth curves were constructed based on the tumor volumes measured in the mice. **P<0.01, ***P<0.001.
We then established pancreatic xenograft tumor models to further determine whether inhibition of KLF5 sensitizes PDACs to PARP inhibitors in vivo. The combination of olaparib and ML264 significantly decreased the tumor growth rate and size when compared to monotherapy ( Figure 5I,J).
Discussion
Considering the vital role of olaparib in maintenance treatment in patients with unresectable PDAC, developing a novel strategy inducing “BRCAness” could enable more patients to benefit clinically from PARP inhibitors. Our present results suggested that repression of KLF5 promoted olaparib-induced DNA damage in pancreatic cancer cells and increased olaparib sensitivity by suppressing the transcription of BRCA1, which could be reversed by upregulation of BRCA1. Furthermore, olaparib combined with an inhibitor of KLF5 showed stronger cytotoxicity to pancreatic cancer cells.
BRCAness refers to a phenotype of BRCA1/2 mutation. It depicts the state in which an HRR defect occurs in a tumor without a germline BRCA1/2 mutation [35]. The loss or mutation of genes such as ATM, ATR, CDK12, CHEK2, and FANCA involved in HR is also considered to induce BRCAness and may alter the therapeutic efficacy of PARP inhibitors and platinum [36]. Additionally, cancer cells inevitably develop resistance to PARP inhibitors. The most recognized explanation is the restoration of BRCA1/2 function by losing the frameshift caused by the original mutation and restoring the open reading frame [37]. Genetic reversal of genetic mutations could also result in the expression of full-length wild-type proteins [38].
Cancer cells can quickly obtain resistance to PARP inhibitors by losing hypermethylation of the BRCA1 or RAD51C promoter and restoring mRNA and functional protein expression [ 39, 40]. Inhibition of BRCA1 may induce a BRCAness-like situation, which could significantly expand the clinical application and overcome potential resistance to PARP inhibitors.
Krüppel-like factors widely regulate cancer cell metastasis and proliferation, the tumor microenvironment, and cancer stem cells [41]. Based on the significant correlation between high expression of KLF5 and poor prognosis of pancreatic cancer [18], the vital role of KLF5 as a substrate of FBW7 was indicated by our previous study [16]. We explored whether KLF5 regulates the sensitivity to PARPis in pancreatic cancer. Our results indicated that abrogation of KLF5 facilitated DNA damage induced by olaparib and increased sensitivity to PARP inhibitors, which could be reversed by overexpression of BRCA1. Our study also revealed that the expression of Rad51 could be regulated modestly by KLF5, which is consistent with Sun’s research [18]. The above results suggested that inhibition of KLF5 may also induce BRCAness by downregulating Rad51.
Our investigation showed that BRCA1 could be regulated by KLF5 at the transcriptional level. As a key transcription factor, KLF5 mainly mediates the transcription of target genes by binding to specific regions of the promoter. BRCA1 has multiple transcripts, and the most commonly used transcript sequence could contain over 5500 bp [42]. BRCA1/2 mutations include point mutations, small fragment insertions/deletions, and large fragment rearrangements [ 43, 44]. There are no mutation hotspots, and mutations could occur in the entire length of the BRCA1/ 2 gene compared with EGFR, BRAF, Kras, etc. [ 43, 44]. Additionally, its expression level and protein activity are regulated by multiple mechanisms. Chen et al [45] discovered that miR-9 mediates the suppression of BRCA1 and hinders DNA damage repair in ovarian cancer cells. It has been reported that phosphorylated ETS transcription factors induced by the ERK pathway could repress the BRCA1 promoter [ 46, 47]. Moreover, phosphorylated ETS1 could downregulate the expression of BRCA1/2 [26]. It has been well established that the translated BRCA1 protein must be phosphorylated to achieve DNA damage repair function [ 48, 49]. BRCA1 can be phosphorylated by many kinases, such as CDK1, 2, casein kinase 2, and DNA damage-responsive kinases, including ATM, ATR, hCds1, and AKT [ 48‒ 53]. Johnson et al. [54] reported that suppression of CDK1 impairs the ability of cells to repair DNA. In addition, the prolyl isomerase Pin1 could maintain BRCA1 by preventing the ubiquitination of BRCA1 [55].
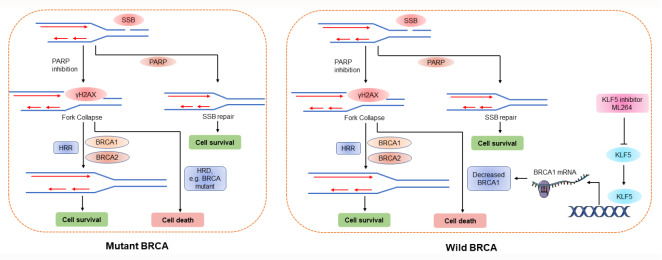
In conclusion, our study indicated that repression of KLF5 could render BRCA1-proficient pancreatic cancer cells BRCA1 deficiency and thus sensitized them to PARP inhibition. We also preliminarily investigated the underlying mechanism ( Figure 6).
Figure 6 .
Schematic representation of the role of KLF5 in DNA damage repair and its underlying mechanism
Downregulation of KLF5 significantly inhibits the expression of BRCA1 at the transcriptional level. Abrogation of KLF5 drives “BRCAness” and empowers PDAC cells with sensitivity to olaparib in BRCA1-proficient pancreatic cancer cells.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 82273382, 81827807, 81972218, 81972257, 82103409, 82272929, and 82173116), Shanghai ShenKang Hospital Development Centre Project (No. SHDC2020CR2017B), the Program of Shanghai Academic/Technology Research Leader (No. 23XD1400600), China Postdoctoral Science Foundation (No. 2021M690037), Shanghai Sailing Program (No. 21YF1407100), Science and Technology Planning Project of Yunnan Province (No. 202305AF150148), and Shanghai Municipal Health Commission (No. 20224Y0307).
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. . 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Strickler JH, Satake H, George TJ, Yaeger R, Hollebecque A, Garrido-Laguna I, Schuler M, et al. Sotorasib in KRAS p.G12C-mutated advanced pancreatic cancer . N Engl J Med. . 2023;388:33–43. doi: 10.1056/NEJMoa2208470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park JO, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer . N Engl J Med. . 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. . 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R, Shi P, Zhou Z, Zhang H, Li W, Zhang H, Chen C. Krüpple‐like factor 5 is essential for mammary gland development and tumorigenesis. J Pathol. . 2018;246:497–507. doi: 10.1002/path.5153. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, Nie Z, Zhou Z, Zhang H, Liu R, Wu J, Qin J, et al. The interplay between TEAD4 and KLF5 promotes breast cancer partially through inhibiting the transcription of p27 Kip1 . Oncotarget. . 2015;6:17685–17697. doi: 10.18632/oncotarget.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulationin the TSU-Pr1 human bladder cancer cell line. Int J Cancer. . 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 8.Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F, Wang Z, et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene. . 2016;35:2040–2051. doi: 10.1038/onc.2015.263. [DOI] [PubMed] [Google Scholar]
- 9.Pattison JM, Posternak V, Cole MD. Transcription factor KLF5 binds a cyclin E1 polymorphic intronic enhancer to confer increased bladder cancer risk. Mol Cancer Res. . 2016;14:1078–1086. doi: 10.1158/1541-7786.MCR-16-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz de Sabando A, Wang C, He Y, García-Barros M, Kim J, Shroyer KR, Bannister TD, et al. ML264, a novel small-molecule compound that potently inhibits growth of colorectal cancer. Mol Cancer Ther. . 2016;15:72–83. doi: 10.1158/1535-7163.MCT-15-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu T, Ma P, Wang W, Shuai Y, Wang Y, Yu T, Xia R, et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. . 2019;26:2179–2193. doi: 10.1038/s41418-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An T, Dong T, Zhou H, Chen Y, Zhang J, Zhang Y, Li Z, et al. The transcription factor Krüppel-like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Commun. . 2019;508:159–168. doi: 10.1016/j.bbrc.2018.11.084. [DOI] [PubMed] [Google Scholar]
- 13.He P, Yang JW, Yang VW, Bialkowska AB. Krüppel-like factor 5, increased in pancreatic ductal adenocarcinoma, promotes proliferation, acinar-to-ductal metaplasia, pancreatic intraepithelial neoplasia, and tumor growth in mice. Gastroenterology. . 2018;154:1494–1508.e13. doi: 10.1053/j.gastro.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia W, Bai H, Deng Y, Yang Y. PLA2G16 is a mutant p53/KLF5 transcriptional target and promotes glycolysis of pancreatic cancer . J Cell Mol Medi. . 2020;24:12642–12655. doi: 10.1111/jcmm.15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Yuan S, Norgard RJ, Yan F, Sun YH, Kim IK, Merrell AJ, et al. Epigenetic and transcriptional control of the epidermal growth factor receptor regulates the tumor immune microenvironment in pancreatic cancer. Cancer Discov. . 2021;11:736–753. doi: 10.1158/2159-8290.CD-20-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Liu M, Hu Q, Xu W, Liu W, Sun Q, Ye Z, et al. FGFBP1, a downstream target of the FBW7/c-Myc axis, promotes cell proliferation and migration in pancreatic cancer. Am J Cancer Res. 2019, 9: 2650–2664 . [PMC free article] [PubMed]
- 17.Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. . 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Kong R, Chen H, Zhao Z, Li L, Li J, Hu J, et al. Overexpression of KLF5 is associated with poor survival and G1/S progression in pancreatic cancer. Aging. . 2019;11:5035–5057. doi: 10.18632/aging.102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. . 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci. . 2001;114:3591–3598. doi: 10.1242/jcs.114.20.3591. [DOI] [PubMed] [Google Scholar]
- 21.de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, Chi KN, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. . 2020;382:2091–2102. doi: 10.1056/NEJMoa1911440. [DOI] [PubMed] [Google Scholar]
- 22.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation . N Engl J Med. . 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 23.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. . 2017;18:1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 24.Yanaihara N, Yoshino Y, Noguchi D, Tabata J, Takenaka M, Iida Y, Saito M, et al. Paclitaxel sensitizes homologous recombination-proficient ovarian cancer cells to PARP inhibitor via the CDK1/BRCA1 pathway. Gynecol Oncol. . 2023;168:83–91. doi: 10.1016/j.ygyno.2022.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z, Mao W, Yang H, Santiago-O’Farrill JM, Rask PJ, Mondal J, Chen H, et al. SIK2 inhibition enhances PARP inhibitor activity synergistically in ovarian and triple-negative breast cancers. J Clin Invest. . 2022;132:e146471. doi: 10.1172/JCI146471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibrahim YH, García-García C, Serra V, He L, Torres-Lockhart K, Prat A, Anton P, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-Proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. . 2012;2:1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Hu Q, Ye S, Xiang L. Inhibition of the PIN1-NRF2/GPX4 axis imparts sensitivity to cisplatin in cervical cancer cells. Acta Biochim Biophys Sin. . 2022;54:1325–1335. doi: 10.3724/abbs.2022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. . 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 29.Ye Z, Zhuo Q, Hu Q, Xu X, Mengqi liu X, Zhang Z, Xu W, et al. FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. . 2021;38:101807. doi: 10.1016/j.redox.2020.101807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Q, Qin Y, Ji S, Xu W, Liu W, Sun Q, Zhang Z, et al. UHRF1 promotes aerobic glycolysis and proliferation via suppression of SIRT4 in pancreatic cancer. Cancer Lett. . 2019;452:226–236. doi: 10.1016/j.canlet.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Shi C, Wu X, Zhang Y, Liu H, Wang X, Huang C, et al. Light activation of iridium (III) complexes driving ROS production and DNA damage enhances anticancer activity in A549 cells. J InOrg Biochem. . 2022;236:111977. doi: 10.1016/j.jinorgbio.2022.111977. [DOI] [PubMed] [Google Scholar]
- 32.Hu Q, Dai J, Zhang Z, Yu H, Zhang J, Zhu X, Qin Y, et al. ASS1-mediated reductive carboxylation of cytosolic glutamine confers ferroptosis resistance in cancer cells. Cancer Res. . 2023;83:1646–1665. doi: 10.1158/0008-5472.CAN-22-1999. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Qin Y, Hu Q, Liu W, Ji S, Xu W, Fan G, et al. SETD8 potentiates constitutive ERK1/2 activation via epigenetically silencing DUSP10 expression in pancreatic cancer. Cancer Lett. . 2021;499:265–278. doi: 10.1016/j.canlet.2020.11.023. [DOI] [PubMed] [Google Scholar]
- 34.Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, Natoli G. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer . EMBO J. . 2016;35:595–617. doi: 10.15252/embj.201592404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. . 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA, et al. Gene expression profile of BRCA ness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer . J Clin Oncol. . 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, Wurz K, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma . Cancer Res. . 2009;69:6381–6386. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murthy P, Muggia F. PARP inhibitors: clinical development, emerging differences, and the current therapeutic issues. Cancer Drug Resist. . 2019;2:665–679. doi: 10.20517/cdr.2019.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernards SS, Pennington KP, Harrell MI, Agnew KJ, Garcia RL, Norquist BM, Swisher EM. Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated ovarian carcinoma. Gynecol Oncol. . 2018;148:281–285. doi: 10.1016/j.ygyno.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Abdallah R, Zhao S, Garinet S, Hormigos K, Le Corre D, Cros J, Perez Toralla K, et al. BRCA1 and RAD51C promotor methylation in human resectable pancreatic adenocarcinoma. Clin Res Hepatol Gastroenterol. . 2022;46:101880. doi: 10.1016/j.clinre.2022.101880. [DOI] [PubMed] [Google Scholar]
- 41.Tetreault MP, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer. . 2013;13:701–713. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- 42.Pettigrew CA, French JD, Saunus JM, Edwards SL, Sauer AV, Smart CE, Lundström T, et al. Identification and functional analysis of novel BRCA1 transcripts, including mouse Brca1-Iris and human pseudo-BRCA1. Breast Cancer Res Treat. . 2010;119:239–247. doi: 10.1007/s10549-008-0256-2. [DOI] [PubMed] [Google Scholar]
- 43.Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, Olah E, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations . Hum Mutat. . 2018;39:593–620. doi: 10.1002/humu.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, Guo X, Tang L, Chen M, Luo X, Peng L, Xu X, et al. Analysis of BRCA1/2 mutation spectrum and prevalence in unselected Chinese breast cancer patients by next-generation sequencing. J Cancer Res Clin Oncol. . 2017;143:2011–2024. doi: 10.1007/s00432-017-2465-8. [DOI] [PubMed] [Google Scholar]
- 45.Sun C, Li N, Yang Z, Zhou B, He Y, Weng D, Fang Y, et al. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J Natl Cancer Institute. . 2013;105:1750–1758. doi: 10.1093/jnci/djt302. [DOI] [PubMed] [Google Scholar]
- 46.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. . 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 47.Baker KM, Wei G, Schaffner AE, Ostrowski MC. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1promoter. J Biol Chem. . 2003;278:17876–17884. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- 48.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-Dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. . 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 49.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. . 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 50.Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, Khanna KK. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000, 60: 3299–304 . [PubMed]
- 51.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. . 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J. Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res. 2000, 60: 5037–5039 . [PubMed]
- 53.Johnson N, Cai D, Kennedy RD, Pathania S, Arora M, Li YC, D′Andrea AD, et al. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell. . 2009;35:327–339. doi: 10.1016/j.molcel.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson N, Li YC, Walton ZE, Cheng KA, Li D, Rodig SJ, Moreau LA, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med. . 2011;17:875–882. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo ML, Zheng F, Chen W, Liang ZM, Chandramouly G, Tan J, Willis NA, et al. Inactivation of the prolyl isomerase pin1 sensitizes BRCA1-proficient breast cancer to PARP inhibition. Cancer Res. . 2020;80:3033–3045. doi: 10.1158/0008-5472.CAN-19-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.