Abstract
Retroviral populations exhibit a high evolutionary potential, giving rise to extensive genetic variation. Error-prone DNA synthesis catalyzed by reverse transcriptase (RT) generates variation in retroviral populations. Structural features within RTs are likely to contribute to the high rate of errors that occur during reverse transcription. We sought to determine whether amino acids within murine leukemia virus (MLV) RT that contact the deoxyribonucleoside triphosphate (dNTP) substrate are important for in vivo fidelity of reverse transcription. We utilized the previously described ANGIE P encapsidating cell line, which expresses the amphotropic MLV envelope and a retroviral vector (pGA-1). pGA-1 expresses the bacterial β-galactosidase gene (lacZ), which serves as a reporter of mutations. Extensive mutagenesis was performed on residues likely to interact with the dNTP substrate, and the effects of these mutations on the fidelity of reverse transcription were determined. As expected, most substitution mutations of amino acids that directly interact with the dNTP substrate significantly reduced viral titers (>10,000-fold), indicating that these residues played a critical role in catalysis and viral replication. However, the D153A and A154S substitutions, which are predicted to affect the interactions with the triphosphate, resulted in statistically significant increases in the mutation rate. In addition, the conservative substitution F155W, which may affect interactions with the base and the ribose, increased the mutation rate 2.8-fold. Substitutions of residues in the vicinity of the dNTP-binding site also resulted in statistically significant decreases in fidelity (1.3- to 2.4-fold). These results suggest that mutations of residues that contact the substrate dNTP can affect viral replication as well as alter the fidelity of reverse transcription.
Retroviral populations display extensive genetic variability (9, 45). Variation in retroviral populations is generated because of mutations introduced into their genomes during replication by mammalian DNA polymerases, RNA polymerase II, and virally encoded reverse transcriptase (RT). It has been observed in vivo that approximately 32% of the mutations in retroviral replication are generated during the plus-strand DNA synthesis of reverse transcription (31). Therefore, it is likely that error-prone replication by RT is a major contributor to the variation generated in retroviral populations. The innate ability of RT to be less processive than most polymerases and/or the lack of a 3′-5′ exonuclease activity contributes to the error-prone nature of retroviral DNA synthesis (5, 40, 45).
Several amino acid motifs present in different retroviral RTs have been identified that may exert an impact on the fidelity of DNA synthesis during reverse transcription. These include residues of the Tyr-X-Asp-Asp (YXDD) motif, the deoxyribonucleoside triphosphate (dNTP) binding site, the α-helix H of the thumb domain, the conserved Leu-Pro-Gln-Gly (LPQG) motif, the finger domain, and specific amino acids such as E89 and F160 in human immunodeficiency virus type 1 (HIV-1) RT (1, 3, 4, 10, 13, 16–18, 29, 35, 42, 50). The mechanism by which fidelity is maintained may encompass features in RT that involve the local geometry of the active site, the proper positioning of the template-primer complex, or the global effects attributed to different conformational states of the protein (8, 18).
It was previously shown that residues 110 to 116 in HIV-1 RT are located in the active site of RT and have a moderate to high solvent accessibility, suggesting that they may play a role in dNTP binding (22, 44). Furthermore, extensive mutational analysis of residues Y115 and Q151 in HIV-1 RT revealed that substitution mutations at these sites could have drastic effects on substrate dNTP binding (35, 42) as well as resistance to nucleoside analogs (21, 46). Resistance to nucleoside analogs has also been observed with position 116 as well (48). Additional evidence for the importance of this stretch of residues in dNTP binding and catalysis was provided by the ability of the D113 and A114 mutants of HIV-1 RT that exhibit a decrease in the sensitivity to phosphonoformic acid, a pyrophosphate analog (33). It was hypothesized that these residues, in addition to the highly conserved R72, play an important role during pyrophosphate exchange (33, 43). Finally, mutations at K65 have implicated this residue in template-primer complex binding that may ultimately have effects on fidelity (15, 44).
Recently, the structure of a ternary complex of HIV-1 RT catalytically trapped with both template-primer and dTTP substrates was elucidated, which identified the residues that directly contacted the incoming dNTP (20). These included residues K65, R72, D113, A114, Y115, and Q151 of HIV-1 RT. The K65 and R72 side chains as well as the D113 and A114 main chain amide nitrogens (NH) bind to the α, β, and γ phosphates of the incoming dTTP through hydrogen bonding interactions. In addition, it has been illustrated that the side chains of R72, Y115, and Q151 interact with the base of the dTTP. Finally, the main chain NH of residue Y115 was also shown to be in close proximity to the ribose moiety of the dTTP (20).
Since substrate dTTP and the aforementioned amino acids are in close proximity to each other, it is reasonable to hypothesize that substitutions at these positions may alter the binding of the incoming dNTP or the positioning of the template-primer complex or modify the local geometry of the substrate binding pocket (18). This alteration in turn can lead to changes in fidelity. This hypothesis is supported by evidence from mutations introduced at positions K65 and Q151 of recombinant HIV-1 RT, which exhibited decreased mispair extension in vitro as well as resistance to a number of dideoxynucleoside analogs such as 2′,3′-dideoxy-3′-thiacytidine, 3′-azido-3′-deoxythymidine, 2′,3′-dideoxycytidsine, and 2′,3′-dideoxyinosine (14, 18, 47). Finally, a number of mutations introduced at residue Y115 of HIV-1 RT were shown to affect the frequency of misinsertions and mispair extensions (35, 36).
Primary sequence and crystal structure data can be utilized to identify amino acids in murine leukemia virus (MLV) RT that are equivalent to those in HIV-1 RT. Sequence comparisons of several RTs illustrate the presence of highly conserved amino acid motifs (39). The partial structure of the fingers and palm subdomains of MLV RT has been solved (13, 38). Comparison of the fingers and palm subdomains of HIV-1 and MLV RTs reveal similar tertiary structures despite having only ∼25% amino acid sequence identity (13, 19). Furthermore, MLV RT possesses several of the structural motifs present in HIV-1 RT, including the conserved YXDD, Thr-Val-Leu-Asp (TVLD), and LPQG motifs (13, 39). Sequence alignment of common amino acid motifs within the two RTs and a comparison of the crystal structures reveal MLV RT residues that are homologous to those in HIV-1 RT (13, 19). Such comparisons strongly suggest that residues K103, R110, D153, A154, F155, and Q190 of MLV RT are equivalent to the dTTP binding residues K65, R72, D113, A114, Y115, and Q151 in HIV-1 RT, respectively. Furthermore, K103 of MLV RT has been observed to interact with the incoming dNTP, and substitutions at R110 affect processivity (2, 8). Recent studies have provided direct evidence that the F155V mutant of MLV RT is able to incorporate rNTPs during polymerization (12). It has been postulated that F155 of MLV RT may be involved in the selection of dNTPs over ribonucleoside triphosphates (rNTPs) due to steric hindrance between the 2′ OH of rNTP and the F155 side chain (12).
In this study, we sought to determine whether alterations in the structure of MLV RT can affect the fidelity of reverse transcription in vivo. Specifically, we examined whether mutations at residues in the vicinity of the dNTP binding pocket as well as residues that directly contact the substrate dNTP could alter the fidelity of reverse transcription in vivo. We assessed the effects of these mutations on replication, polymerase activity, and fidelity by comparing the mutant MLV RTs to wild-type RT.
MATERIALS AND METHODS
Plasmids and retroviral vectors.
The construction of the MLV-based retroviral vector pGA-1 was previously described (26). The vector pGA-1 possesses cis-acting elements needed for viral replication. In addition, the vector expresses the bacterial β-galactosidase gene (lacZ) and the neomycin phosphotransferase gene (neo) from a single RNA transcript initiating from the viral promoter. An internal ribosomal entry site of encephalomyocarditis virus is used to ensure the translation of neo (23). Plasmid pLGPS expresses the MLV gag and pol genes from a truncated long terminal repeat (ΔLTR) promoter (37). Plasmid pRMBNB, which was derived from pLGPS, contains three unique restriction sites near the dNTP-binding site of MLV RT. These three restriction sites were generated by the introduction of silent substitutions to facilitate further mutagenesis. The plasmid pSV-A-MLV-env, which expresses the amphotropic MLV envelope gene from the LTR promoter and simian virus 40 enhancer, was obtained from the NIH AIDS Research and Reference Reagent Program (32). Plasmid pBSpac encodes the puromycin N-acetyltransferase gene and therefore confers resistance to puromycin (49). Plasmid pSVα3.6 encodes the α subunit of the murine Na+,K+-ATPase gene and confers resistance to ouabain (28).
Generation of dNTP-binding site MLV RT mutants.
A detailed description of the mutagenic oligonucleotides and strategies used to generate as well as identify mutants is available upon request. Briefly, pRMBNB was generated by two rounds of site-directed mutagenesis using a Chameleon kit (Stratagene) with mutagenic primers and pLGPS as the template. The plasmid pRMBNB was identified by the presence of three unique restriction sites, Bst1107I, NsiI, and BstB1. To ensure the absence of any undesired mutations, the BclI-to-SalI DNA fragment of pRMBNB was subcloned into pLGPS. In addition, the Q190E, Q190H, and Q190N mutants were generated in a similar manner using a Chameleon kit with either pLGPS or pRMBNB as the templates and the appropriate mutagenic primers. The Q190E mutant was generated utilizing pLGPS as the template and screened by the creation of a new Bsu36I site. The BclI-to-SalI DNA fragment of Q190E was subcloned into pLGPS to ensure the absence of any undesired mutations. The Q190E mutant was also generated in pRMBNB and screened by the creation of a new Bsu36I site. Both the Q190H and Q190N mutants were generated using the Chameleon kit with Q190E as the template and screened by the loss of the new Bsu36I site. The NsiI to SalI DNA fragments of Q190H and Q190N were subcloned into pRMBNB and sequenced to ensure the absence of any undesired mutations.
PCR-based mutagenesis with random mutagenic primer sets and pRMBNB as the template was utilized to generate mutants at residues K103 and R110. Amplified DNA fragments of 493 bp were digested with BclI and NsiI and subcloned back into pRMBNB. In addition, the Q190M mutant was generated in a similar manner using PCR-based mutagenesis utilizing mutagenic primers and screened by sequencing. An amplified DNA fragment of 831 bp was digested with NsiI and SalI and subcloned back into pRMBNB.
Mutations introduced at positions 147 to 160 of MLV RT were generated by double-stranded DNA (dsDNA) oligonucleotide-based mutagenesis. These various dsDNA oligonucleotides, which are complementary to the Bst1107I-to-BstBI fragment in pRMBNB and contain the appropriate mutations, were subcloned back into pRMBNB. One or two silent mutations were also introduced into these oligonucleotides to generate new restriction sites to facilitate identification of mutant plasmids. Mutations introduced at positions 147 to 153 of MLV RT were generated by replacing the Bst1107I-to-NsiI fragment of pRMBNB with 25-bp dsDNA oligonucleotides. Mutations introduced at positions 154 to 155 were generated by replacing the Bst1107I-to-BstBI fragment of pRMBNB with 73-bp dsDNA oligonucleotides. Mutations introduced at positions 155 to 160 were generated by replacing the NsiI-to-BstBI fragment of pRMBNB with 48-bp dsDNA oligonucleotides. Mutants were identified by digestion with either XbaI (mutants at positions 147 to 155) or SpeI (mutants at positions 156 to 160).
All mutated plasmids were mapped extensively utilizing various restriction enzymes. Finally, the mutated plasmids were analyzed by DNA sequencing to verify the presence of the desired mutation and the absence of any undesired mutations (ALF Automated Sequencer, Pharmacia).
Cells, transfections, and infections.
D17 dog osteosarcoma cells were transfected with MLV-based vectors or expression constructs and infected with MLV followed by selection for resistance to ouabain or G418 (a neomycin analog) as previously described (25, 26). The D17-based ANGIE P cells were maintained, transfected, and selected for drug resistance in a similar manner (17).
Assay for determining in vivo fidelity.
The ANGIE P cells were plated at a density of 2 × 105 cells per 60-mm-diameter dish and 24 h later were cotransfected with wild-type or mutated pLGPS and pSVα3.6. The transfected cells were selected for resistance to ouabain (10−7 M), and then resistant colonies were pooled, expanded, and plated at a density of 5 × 106 cells per 100-mm-diameter dish. After 48 h, the culture medium containing GA-1 virus was harvested and used to infect D17 target cells plated at a density of 2 × 105 cells per 60-mm-diameter dish. Infected D17 cells were selected for resistance to G418 (400 μg/ml) and stained with 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal) as previously described (17). The observation that the mutant frequency of the wild-type RT was very consistent in 20 or more experiments indicated that any reinfection of the virus producer cells that may have occurred during expansion of the producer cells did not significantly affect the frequencies of lacZ inactivation.
Virus preparation, RT assays, and Western blotting.
Virus isolation, concentration, RT assays, and Western blotting were performed as previously described (17). Briefly, helper cells containing different pLGPS constructs were plated at 5 × 106 cells per 100-mm-diameter dish, viruses were collected 2 days later and centrifuged at 25,000 rpm for 90 min in an SW41 rotor (Beckman) at 4°C. Viral pellets were resuspended in phosphate-buffered saline and virus was stored at −80°C.
Exogenous RT activities were determined as previously described using 50 μg of (20-mer) oligo(dT) (Integrated DNA Technologies) per μl, 100 μg of poly(rA) (Pharmacia) per μl, and 10 μCi of [3H]dTTP (specific activity of 72 Ci/mMol; ICN). The amount of [3H]dTTP incorporated was determined using a scintillation counter (13).
Western blots were used to quantify the amount of protein for the RT assays and performed using standard procedures (41). Briefly, viral proteins were resolved by sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis and transferred to Immobilon membranes (Gelman Sciences, Inc.). Membranes were incubated with primary (monoclonal anti-MLV capsid immunoglobulin G1; 1:10 dilution [American Type Culture Collection]) (7) and secondary (anti-rat immunoglobulin G antibody conjugated to horseradish peroxidase; 1:10,000 dilution [Southern Biotechnology Associates, Inc.]) antibodies, and detection of MLV capsid was performed using an enhanced chemiluminenesence kit (Amersham Pharmacia Biotech). The membranes were then exposed to X-Omat film (Kodak), and the intensity of the p30 band was quantitated using the ImageQuant program (Molecular Dynamics) (17).
RESULTS
Determination of in vivo fidelity.
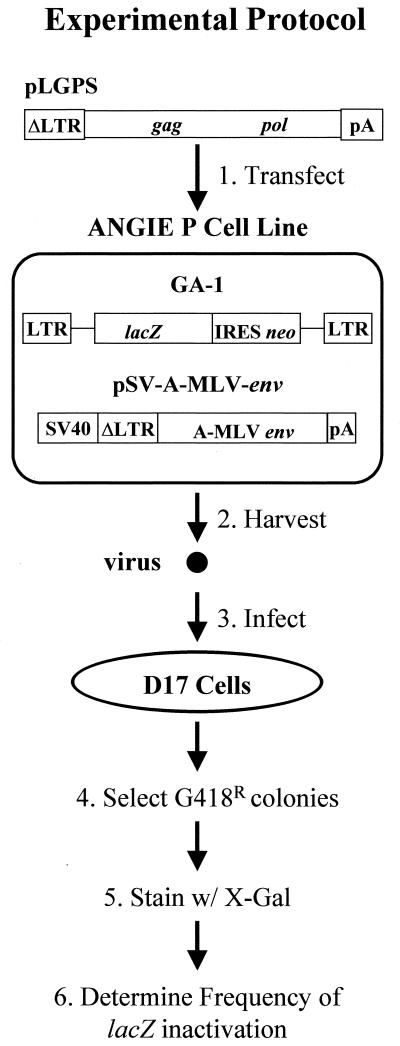
Construction and characterization of the ANGIE P cell line have been previously described (17). Briefly, the ANGIE P cell line expresses the MLV-based retroviral vector pGA-1 and the MLV envelope expression construct pSV-A-MLV-env (Fig. 1). The MLV gag/pol expression construct pLGPS was subjected to site-directed mutagenesis at residues constituting the dNTP-binding site of MLV RT. Subsequently, wild-type or mutated pLGPS was separately introduced into the ANGIE P cells by cotransfection with pSVα3.6. Ouabain-resistant colonies were pooled and expanded; virus was harvested from these pools, serially diluted, and used to infect D17 target cells. Infected D17 cells resistant to G418 were selected and stained with X-Gal, and the frequency of lacZ inactivation was determined by dividing the number of white colonies by the total number of colonies (blue plus white colonies). The frequency of lacZ inactivation provided a measure of the inactivating mutations (white phenotype) introduced into the lacZ gene during one cycle of retroviral replication, and the virus titers were used to determine the effect of mutations on the efficiency of virus replication.
FIG. 1.
Structures of MLV-based constructs and rapid in vivo assay to identify structural determinants in MLV RT that are important for fidelity. The ANGIE P encapsidating cell line is a D17-based cell line expressing both the MLV-based vector pGA-1 and pSV-A-MLV-env. The pGA-1 vector, which contains LTRs and all cis-acting elements of MLV, transcribes the Escherichia coli lacZ and neo genes from the LTR promoter. The internal ribosomal entry site (IRES) of encephalomyocarditis virus is used to express neo. pSV-A-MLV-env expresses the amphotropic MLV envelope from a truncated MLV LTR (ΔLTR) and the simian virus 40 (SV40) promoter-enhancer. Transfection of the wild-type or mutated MLV gag/pol construct pLGPS into the ANGIE P cell line allows for expression of the MLV gag and pol from the ΔLTR (step 1). Infectious viral particles are harvested (step 2) and used to infect D17 cells (step 3). G418r infected colonies are selected (step 4), resistant clones are stained with X-Gal (step 5), and the frequency at which lacZ is inactivated is determined (step 6).
Identification of the dNTP-binding site in MLV RT.
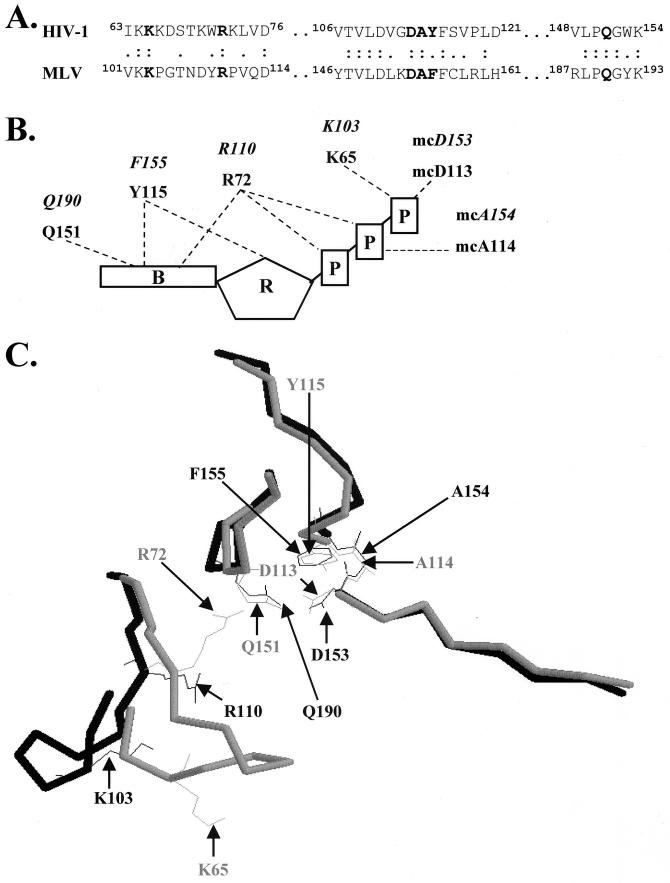
Comparison of both HIV-1 and MLV RT revealed the dNTP-binding site in MLV RT. Amino acid sequence alignment of HIV-1 and MLV RTs revealed the highly conserved TVLD and LPQG motifs (Fig. 2A). This sequence comparison suggested that residues K103, R110, D153, A154, F155, and Q190 of MLV RT are equivalent to residues K65, R72, D113, A114, Y115, and Q151 of HIV-1 RT, respectively. Analysis of the crystal structures of unliganded HIV-1 RT (not bound to DNA or substrate) and MLV RT in the context of the position and orientation of these residues also revealed similarities between the two RTs (Fig. 2B and C). In addition, calculated bond distances between residues constituting the dNTP-binding sites of both RTs in the absence of ligands (13, 19) showed that most distances were within 3 Å, the length of a hydrogen bond (data not shown). The only distances greater than 3 Å involved the R110 position. These lengths were determined by the RasMol program and were characterized as an average bond distance calculated from a number of measurements between the various atoms of the side chain pairs inspected. The equivalence of these MLV RT residues with those in HIV-1 RT is further supported by previous studies (2, 8, 13, 39).
FIG. 2.
The dNTP-binding sites of MLV and HIV-1 RTs. (A) Primary sequence homology of the MLV and HIV-1 RT dNTP-binding sites. HIV-1 and MLV RTs were aligned through their TVLD, LPQG, and YXDD amino acid motifs. Residues of the dNTP-binding site of MLV RT were determined by further alignment with residues proposed to constitute the dNTP-binding site of HIV-1 RT. Two dots represent residues that are identical, whereas one dot represents conservative changes between the two RTs. Boldface lettering represents residues constituting the dNTP-binding sites in both HIV-1 and MLV RTs. (B) Structure of HIV-1 RT dNTP-binding site (modified from Huang et al. [20]). The structure shown contains amino acid positions K65, R72, D113, A114, Y115, and Q151 interacting with the dTTP substrate. Italic lettering represents the equivalent residues in MLV RT constituting the dNTP-binding site. Components of the dTTP substrate are denoted as B (base), R (ribose sugar), and P (phosphate). Dashed lines represent interactions between the amino acid residues of RT and the dTTP substrate. mc, main chain interactions between the substrate and the RT. (C) Comparison of structures of MLV and HIV-1 RTs constituting the dNTP-binding sites (13, 19). The backbone of HIV-1 RT (amino acid sequences 63 to 76, 106 to 121, and 148 to 154) was superimposed on the homologous structure in MLV RT (amino acid sequences 101 to 114, 146 to 161, and 187 to 193). Free HIV-1 RT (not bound to DNA or substrate dNTP) is represented in gray; MLV RT is represented in black. Side chains of residues creating the dNTP-binding sites of MLV and HIV-1 RTs are illustrated as wire frames. The structures were superimposed using the RasMol Program to maximize the overlaps between the peptide backbones of the HIV-1 and MLV RTs.
Mutational analysis of the MLV RT dNTP-binding site and effects on fidelity.
MLV RT was mutated at six different residues hypothesized to contact the incoming dNTP. Several conserved and nonconserved substitutions were introduced at residues K103, R110, D153, A154, F155, and Q190 of MLV RT. The effects of these various mutations on the frequency of lacZ inactivation were determined and compared with the frequency of inactivation obtained with wild-type MLV RT in parallel experiments (Table 1). Utilization of the wild-type pLGPS construct resulted in the inactivation of lacZ with a frequency of 5.4% ± 0.23% (mean ± standard error [13 independent experiments]) during one cycle of retroviral replication. These results were comparable to results obtained previously (17).
TABLE 1.
Effects of mutations in dNTP-binding site residues of MLV RT on the frequency of lacZ inactivation
| MLV RT genotype | No. of expts. | No. of mutant colonies/ total no. of coloniesa | Frequency (%) of lacZ inactivation (mean ± SE)b | Relative change in inactivation of lacZc | Relative viral titerd |
|---|---|---|---|---|---|
| Wild type | 13 | 522/10,300 | 5.4 ± 0.23 | 1.0 | 1.0 |
| Mutant | |||||
| K103R | 2 | 154/2,740 | 5.6 ± 0.35 | No change | 0.04 |
| K103L | 2 | NCe | NC | NAf | |
| K103S | 2 | NC | NC | NA | <0.0001 |
| K103W | 2 | NC | NC | NA | |
| R110C | 2 | NC | NC | NA | |
| R110H | 2 | NC | NC | NA | |
| R110I | 2 | NC | NC | NA | |
| R110L | 2 | NC | NC | NA | <0.0001 |
| R110M | 2 | NC | NC | NA | |
| R110S | 2 | NC | NC | NA | |
| R110T | 2 | NC | NC | NA | |
| D153A | 4 | 154/1,816 | 8.4 ± 0.62 | 1.6 | 0.0007 |
| D153C | 2 | 65/1,154 | 5.7 ± 0.23 | No change | 0.0007 |
| D153Q | 2 | 58/1,150 | 5.2 ± 0.15 | No change | 0.001 |
| D153S | 2 | 111/2,241 | 5.1 ± 0.15 | No change | 0.03 |
| D153F | 2 | NC | NC | NA | <0.0001 |
| D153R | 2 | NC | NC | NA | |
| A154S | 2 | 208/2,963 | 7.0 ± 0.10 | 1.3 | 0.015 |
| A154E | 2 | NC | NC | NA | |
| A154F | 2 | NC | NC | NA | |
| A154L | 2 | NC | NC | NA | <0.0001 |
| A154N | 2 | NC | NC | NA | |
| A154P | 2 | NC | NC | NA | |
| A154R | 2 | NC | NC | NA | |
| F155W | 2 | 27/182 | 15.1 ± 1.44 | 2.8 | 0.0004 |
| F155Y | 2 | 131/2,474 | 5.3 ± 0.55 | No change | 0.31 |
| F155D | 2 | NC | NC | NA | |
| F155E | 2 | NC | NC | NA | |
| F155I | 2 | NC | NC | NA | <0.0001 |
| F155M | 2 | NC | NC | NA | |
| F155V | 2 | NC | NC | NA | |
| Q190M | 2 | 44/836 | 5.3 ± 0.40 | No change | 0.6 |
| Q190E | 2 | NC | NC | NA | |
| Q190H | 2 | NC | NC | NA | <0.0001 |
| Q190N | 2 | NC | NC | NA |
The number of mutant colonies that displayed a white colony phenotype and total number of colonies that were observed in 2 to 13 independent experiments.
The frequency of lacZ inactivation was calculated as follows: (number of mutant colonies in each experiment ÷ by total number of colonies) × 100. The standard errors were determined by using the two-sample t test.
The relative change in the frequency at which the lacZ gene was inactivated was calculated as follows: frequency of lacZ inactivation observed with mutant MLV RT ÷ by frequency of lacZ inactivation observed with the wild-type MLV RT. Statistical analysis using a two-sample t test showed that the D153A, A154S, and F155W mutants of MLV RT displayed mutant frequencies significantly higher than that for the wild-type MLV RT (P < 0.03). The mutant frequencies obtained with the K103R, D153C, D153Q, D153S, F155Y, and Q190M mutants of MLV RT were not significantly different from that of the wild-type MLV RT (P > 0.05).
The virus titer for each experimental group was determined by serial dilutions and infections. The average virus titer obtained with the wild-type MLV RT for this set of experiments was 9.8 × 104 CFU/ml. The relative virus titer represents a ratio of the virus titer of mutant MLV RT obtained in each experiment divided by the virus titer obtained with the wild-type MLV RT in parallel experiments.
NC, no colonies present after G418 selection, signifying reductions in viral titers of >10,000-fold.
NA, not applicable, since the relative change in the inactivation of the lacZ gene could not be determined, due to the absence of colonies after G418 selection.
As illustrated in Table 1, the majority of mutations introduced at these positions dramatically reduced viral titers by >10,000-fold in comparison to the wild-type RT (9.8 × 104 CFU/ml). Relative changes in the inactivation of lacZ could therefore not be determined. Mutants with severe reductions in titer (>10,000-fold) were observed for all dNTP-binding site positions analyzed. The results suggested that these amino acid positions are critical for the proper function of the polymerase, consistent with previous observations showing that they are highly conserved among retroviral species (39). However, nine mutants of residues constituting the dNTP-binding site in MLV RT did permit viral replication to the extent that relative changes in fidelity could be measured. Three of the nine mutants (A154S, D153A, and F155W) exhibited increases in the frequency with which lacZ was inactivated by 1.3- to 2.8-fold relative to the wild-type RT (5.4% ± 0.23%). These changes were shown to be statistically different from the frequency of lacZ inactivation by the wild-type RT (P values ranging from 0.03 to 0.00003 in two-sample t tests). In addition, these mutations also reduced viral titers 66- to 2,500-fold in comparison to wild-type RT. Conversely, six of the nine mutants (K103R, D153C, D153Q, D153S, F155Y, and Q190M) did not display a significant change in the frequency of lacZ inactivation (5.1% ± 0.15% to 5.7% ± 0.23%) and were shown to be similar to wild-type RT (P > 0.5). The K103R, D153C, D153Q, and D153S mutants also exhibited decreases in viral titers ranging from 25- to 1,429-fold in comparison to the wild-type RT (Table 1). In contrast, the F155Y and Q190M mutants exhibited virus titers similar to wild-type MLV RT.
Effects of mutations at the F156 position of MLV RT on fidelity.
We hypothesized that the F156 residue of MLV RT played an indirect role in substrate binding because of its proximity to other residues that constitute the dNTP-binding site. Specifically, analysis of the MLV RT crystal structure suggested that the bulky hydrophobic nature of F156 might play a role in the proper positioning of Q190, which is important for dNTP binding. Therefore, we investigated the effects of seven mutations (F156I, F156L, F156M, F156Q, F156V, F156W, and F156Y) of this residue in MLV RT on the frequency of lacZ inactivation and viral replication (Table 2). As expected, substitutions at the F156 position were tolerated to a greater extent than residues that directly contacted the incoming dNTP and the effects on fidelity could be determined for four of the seven F156 mutants. The frequency with which lacZ was inactivated by mutants F156L and F156M increased by 1.6- and 1.7-fold relative to the wild-type RT (5.4% ± 0.25% [obtained from eight independent experiments]). These changes were shown to be statistically different from the frequency of lacZ inactivation by the wild-type RT (P < 0.001). Both the F156Y and F156W mutants of MLV RT displayed a slight increase in fidelity, as evidenced by the frequencies of lacZ inactivation, of 4.3% ± 0.27% and 4.3% ± 0.15%, respectively, compared to wild-type MLV RT (P < 0.05). All of these mutants displayed reductions in viral titers relative to the wild-type RT (8.1 × 104 CFU/ml) that ranged from 22- to 500-fold. Finally, the F156I, F156Q, and F156V mutants displayed reductions in virus titers greater than 10,000-fold, and relative changes in fidelity for these mutants could not be determined (Table 2).
TABLE 2.
Effects of F156 mutations in MLV RT on the frequency of lacZ inactivation
| MLV RT genotype | No. of expts. | No. of mutant colonies/total no. of coloniesa | Frequency (%) of lacZ inactivation (mean ± SE)b | Relative change in inactivation of lacZc | Relative viral titerd |
|---|---|---|---|---|---|
| Wild type | 8 | 283/5,524 | 5.5 ± 0.24 | 1.0 | 1.0 |
| Mutant | |||||
| F156L | 2 | 85/955 | 8.9 ± 0.20 | 1.6 | 0.006 |
| F156M | 1 | 20/208 | 9.6 ± 0.00 | 1.7 | 0.006 |
| F156W | 2 | 57/1,340 | 4.3 ± 0.15 | 0.8 | 0.016 |
| F156Y | 4 | 268/6,391 | 4.3 ± 0.27 | 0.8 | 0.046 |
| F156I | 2 | NCe | NC | NAf | |
| F156Q | 2 | NC | NC | NA | <0.0001 |
| F156V | 2 | NC | NC | NA |
The number of mutant colonies that displayed a white colony phenotype and the total number of colonies that were observed in one to eight independent experiments.
The frequency of lacZ inactivation was calculated as described in footnote b of Table 1.
The relative change in the frequency at which the lacZ gene was inactivated was calculated as described in footnote c of Table 1. Statistical analysis using a two-sample t test showed that the F156L and F156M mutants of MLV RT displayed mutant frequencies significantly higher than that for the wild-type MLV RT (P < 0.001). The F156Y and F156W mutants exhibited mutant frequencies less than and statistically different from that of the wild-type MLV RT (P < 0.05).
The virus titer for each experimental group was determined by serial dilutions and infections. The average virus titer obtained with wild-type MLV RT in this set of experiments was 8.1 × 104 CFU/ml. The relative virus titer represents a ratio of the virus titer of mutant MLV RT obtained in each experiment divided by the virus titer obtained with the wild-type MLV RT in parallel experiments.
NC, no colonies present after G418 selection, signifying reductions in viral titers of >10,000-fold.
NA, not applicable, since the relative change in the inactivation of the lacZ gene could not be determined, due to the absence of colonies after G418 selection.
Effects of mutations at residues flanking the MLV RT dNTP-binding site on fidelity.
Regions of MLV RT (positions 147 to 152 and 157 to 161) flanking the residues of the dNTP-binding site were also subjected to mutagenesis, and the effects of these mutations on the fidelity of reverse transcription and viral replication were determined. The data obtained are summarized in Table 3. The frequencies of lacZ inactivation by wild-type MLV RT obtained from six independent experiments were again highly reproducible (5.3% ± 0.26%). The T147A, L151F, K152A, C157A, and H161A mutants exhibited frequencies of lacZ inactivation that were 1.2- to 2.4-fold higher than the wild-type RT (P < 0.04). On the other hand, the R159A mutant was found to be statistically similar to wild-type RT (P > 0.05). The L151F and the T147A mutants displayed an 8- and 50-fold reduction in viral titers, respectively. In contrast, the K152A, C157A, and H161A mutants did not exhibit substantial changes in viral titers. Finally, viral replication of the V148D, L149W, and D150E mutants was reduced by >10,000-fold, and the frequency of lacZ inactivation for these mutants could not be determined (Table 3).
TABLE 3.
Effects of mutations in residues proximal to residues of the dNTP-binding site of MLV RT on the frequency of lacZ inactivation
| MLV RT genotype | No. of expts. | No. of mutant colonies/ total no. of coloniesa | Frequency (%) of lacZ inactivation (mean ± SE)b | Relative change in inactivation of lacZc | Relative viral titerd |
|---|---|---|---|---|---|
| Wild type | 6 | 271/5,220 | 5.3 ± 0.26 | 1.0 | 1.0 |
| Mutant | |||||
| T147A | 2 | 66/969 | 6.8 ± 0.10 | 1.3 | 0.02 |
| V148D | 2 | NCe | NC | NAf | <0.0001 |
| L149W | 2 | NC | NC | NA | <0.0001 |
| D150E | 2 | NC | NC | NA | <0.0001 |
| L151F | 2 | 413/3,304 | 12.5 ± 0.50 | 2.4 | 0.12 |
| K152A | 2 | 109/1,438 | 7.6 ± 0.65 | 1.4 | 0.71 |
| C157A | 2 | 69/1,031 | 6.8 ± 0.20 | 1.3 | 1.10 |
| R159A | 2 | 51/797 | 6.3 ± 0.35 | No change | 0.51 |
| H161A | 2 | 139/2,202 | 6.3 ± 0.20 | 1.2 | 0.45 |
The number of mutant colonies that displayed a white colony phenotype and total number of colonies that were observed in two to six independent experiments.
The frequency of lacZ inactivation was calculated as described in footnote b of Table 1.
The relative change in the frequency at which the lacZ gene was inactivated was calculated as described in footnote c of Table 1. Statistical analysis using a two-sample t test showed that the T147A, L151F, K152A, C157A, and H161A mutants of MLV RT displayed mutant frequencies significantly higher than that of the wild-type MLV RT (P < 0.04). The mutant frequency obtained with the R159A mutant of MLV RT was not significantly different from that of the wild-type MLV RT (P > 0.09).
The virus titer for each experimental group was determined by serial dilutions and infections. The average virus titer obtained with the wild-type MLV RT in this set of experiments was 3.6 × 104 CFU/ml. The relative virus titer represents a ratio of the virus titer of mutant MLV RT obtained in each experiment divided by the virus titer obtained with the wild-type MLV RT in parallel experiments.
NC, no colonies present after G418 selection, signifying reductions in viral titers of >10,000-fold.
NA, not applicable, since the relative change in the inactivation of the lacZ gene could not be determined, due to the absence of colonies after G418 selection.
RT activities of the dNTP-binding site mutants.
Viruses generated from either wild-type or mutated pLGPS were harvested, concentrated by ultracentrifugation, and analyzed by Western blotting (data not shown). Western blots using an anti-MLV capsid antibody were quantified to estimate the amount of capsid protein present in the viral preparations and to ensure that equivalent amounts of viral proteins were used for determination of RT activities (summarized in Table 4).
TABLE 4.
RT activities of dNTP-binding site mutants of MLV RT
| MLV RT genotype | % RT activity (mean ± SE)a | Viral titer (%)b |
|---|---|---|
| Wild type | 100 | 100 |
| Mutant | ||
| T147A | 15.4 ± 0.9 | 2.0 |
| V148D | 3.0 ± 0.9 | <0.01 |
| L149W | 1.0 ± 0.3 | <0.01 |
| D150E | 1.3 ± 0.4 | <0.01 |
| L151F | 86.7 ± 0.4 | 12 |
| K152A | 37.9 ± 15 | 71 |
| K103L | 0.9 ± 0.5 | <0.01 |
| K103R | 15.6 ± 4.5 | 4.0 |
| K103W | 0.5 ± 0.2 | <0.01 |
| R110C | 0.8 ± 0.6 | <0.01 |
| R110H | 0.9 ± 0.6 | <0.01 |
| R110I | 0.5 ± 0.1 | <0.01 |
| R110L | 0.2 ± 0.1 | <0.01 |
| R110M | 0.3 ± 0.0 | <0.01 |
| R110S | 0.3 ± 0.1 | <0.01 |
| R110T | 0.4 ± 0.1 | <0.01 |
| D153A | 6.7 ± 1.0 | 0.07 |
| D153C | 9.9 ± 2.8 | 0.07 |
| D153F | 2.4 ± 0.3 | <0.01 |
| D153Q | 8.0 ± 0.6 | 0.1 |
| D153R | 1.3 ± 0.7 | <0.01 |
| D153S | 23. ± 4.3 | 3.0 |
| A154E | 0.3 ± 0.1 | <0.01 |
| A154F | 0.2 ± 0.0 | <0.01 |
| A154L | 0.4 ± 0.0 | <0.01 |
| A154N | 0.2 ± 0.1 | <0.01 |
| A154P | 0.3 ± 0.1 | <0.01 |
| A154R | 0.4 ± 0.2 | <0.01 |
| A154S | 5.5 ± 0.9 | 1.5 |
| F155D | 0.2 ± 0.2 | <0.01 |
| F155E | 0.9 ± 0.1 | <0.01 |
| F155I | 4.9 ± 3.6 | <0.01 |
| F155M | 2.0 ± 0.4 | <0.01 |
| F155V | 5.5 ± 2.6 | <0.01 |
| F155W | 8.0 ± 3.5 | 0.04 |
| F155Y | 161.4 ± 36.9 | 31 |
| F156I | 0.8 ± 0.4 | <0.01 |
| F156L | 2.4 ± 1.3 | 0.6 |
| F156M | 8.5 ± 3.1 | 0.6 |
| F156Q | 1.1 ± 0.5 | <0.01 |
| F156V | 1.5 ± 0.3 | <0.01 |
| F156W | 11.1 ± 0.9 | 1.6 |
| F156Y | 14.8 ± 1.2 | 3.0 |
| C157A | 56.1 ± 6.0 | 110 |
| R159A | 30.6 ± 6.5 | 51 |
| H161A | 43.6 ± 19.1 | 45 |
| Q190E | 1.0 ± 0.4 | <0.01 |
| Q190H | 0.3 ± 0.1 | <0.01 |
| Q190M | 51.6 ± 2.2 | 60 |
| Q190N | 0.5 ± 0.1 | <0.01 |
Average RT activities determined using virion-associated RT. The activities shown are relative to the wild-type MLV RT (set to 100%) using the poly(rA)-oligo(dT) template-primer complex. The results represent averages of two to three experiments. RT activities of all mutants seem to be statistically different from that of the wild-type MLV RT (P < 0.002), except for those of the L151F, K152A, F155Y, C157A, and H161A mutants (P > 0.05).
Viral titers of mutants as a percentage of the wild-type titer (set at 100%). Viral titers represent the ratio of the virus titer generated from mutant MLV RT obtained in each experiment divided by the virus titer generated from the wild-type MLV RT in parallel experiments and multiplied by 100.
In general, mutants (31 out of 50) possessing RT activities of less than 3% of the wild-type RT activity also had drastic reductions in viral titers (>10,000-fold or 0.01%, relative to the wild type). Only the F156L mutant displayed an RT activity of 2.4% relative to the wild type yet produced a low but detectable viral titer (0.6% in comparison to the wild type). In addition, the F155I and F155V mutants displayed slightly higher RT activities (4.9 and 5.5%, respectively) but did not produce a detectable viral titer. The RT activities of 18 of the remaining mutants ranged from 5.5 to 161.4% relative to the wild-type MLV RT and exhibited detectable viral titers. With the exception of those of the L151F, K152A, F155Y, C157A, and H161A mutants, the RT activities of all mutants were statistically different from the wild-type RT activity (P < 0.002). The RT activity of the K103S mutant could not be compared with the wild-type RT activity, because the mutation apparently generated a defect in Gag processing and the processed capsid protein could not be detected upon Western blotting analysis (data not shown). Therefore, the RT activity of the K103S mutant could not be normalized to the wild-type RT. For all but the Q190M mutant, the virus titers were reduced to a greater extent than RT activities, suggesting that other steps in reverse transcription were also affected by these mutations.
DISCUSSION
In this study, we compared the MLV and HIV-1 RT structures to identify residues of MLV RT that contact the dNTP substrate. Mutational analysis of these residues in MLV RT strongly suggests that they are critical for proper polymerase function and viral replication. The location and orientation of MLV RT residues D153, A154, F155, and Q190 are essentially identical to those of HIV-1 RT residues D113, A114, Y115, and Q151, respectively (Fig. 2A and C) (13, 19). In addition, the distances between the side chains of these residues, as well as the MLV RT residue K103, are very similar to those of the equivalent residues of HIV-1 RT (within 3 Å). However, the MLV RT residue R110 appears in some cases to be as much as 4.0 Å farther away and as much as 6.0 Å closer to the other dNTP-binding site residues in comparison to the homologous distances in HIV-1 RT. It is possible that the MLV RT finger domain that contains the K103 and R110 residues undergoes a larger movement upon binding the dNTP substrate than does the HIV-1 RT. It has been proposed that the R110 residue is involved in the conformational changes that occur during polymerization (8). Alternatively, their location may be altered in the MLV RT crystal structure, since crystallized protein lacks the thumb, connection, and RNase H domains of MLV RT. It is to be noted that all mutations at the R110 position also resulted in very low RT activities, which was consistent with previous observations (8).
Mutations of residues constituting the dNTP-binding site of MLV RT resulted in statistically significant decreases in fidelity of up to 2.8-fold relative to the wild-type MLV RT. It is remarkable that in some cases, reductions in viral titers of >1,000-fold resulted in only minor alterations in fidelity. One possible explanation is that these mutations severely affected the efficiency of specific steps in reverse transcription. For example, mutations that reduced the efficiency of minus-strand DNA synthesis initiation would be expected to have a strong effect on viral titer but not necessarily have a great effect on RT fidelity. Another possible explanation is that the mutations greatly increased the frequency of specific types of mutations. A 10-fold increase in the frequency of a transition substitution is expected to increase the overall frequency of lacZ inactivation by only about 2.5-fold (it is estimated that approximately 16% of all mutations are one transition type because 80% of the mutations are substitutions, 80% of the substitutions are transitions, and there are four different transition types). This hypothesis could be verified by characterizing the nature of mutations generated by mutant RTs. Finally, it is possible that RTs have evolved to minimize the effects of mutations on their fidelity and that it is not possible to drastically alter the accuracy of DNA synthesis during viral replication by introducing single amino acid substitutions. However, a more extensive mutational analysis of RT is needed to verify this hypothesis. Depending on the nature and frequency of the mutations generated by mutant RTs, it is possible that some of the RT mutants will alter the spectrum of viral variants generated in viral populations. However, it is unlikely that a 2.8-fold decrease in the fidelity of RT will have a large effect on the evolution of large HIV-1 populations that rapidly undergo multiple rounds of replication in infected patients (9). Under these conditions, the selective forces are likely to have a greater impact on the viral population than small changes in the mutation rate of the virus.
Most of the substitutions introduced at the F155 position of MLV RT resulted in severe reductions in viral titers (>10,000-fold). The results obtained with the F155M, F155I, and F155V mutants were consistent with previous observations (11). In addition, the result that the F155Y mutant did not significantly affect viral titer was also consistent with previous observation (11). The same study previously reported that the F155W mutant was not viable and the RT activity could not be detected 10 days after transfection of viral DNA. In our study, the F155W mutant did produce a low viral titer (2,500-fold less than the wild type) and displayed very low RT activity. These slightly different results are likely to be due to the fact that we used a single cycle assay in which extremely low levels of viral replication could be detected.
The D153A, A154S, and F155W substitutions of MLV RT increased the frequencies of lacZ inactivation by 1.6-, 1.3-, and 2.8-fold, respectively (Table 1). All three homologous residues in HIV-1 RT have been observed to form hydrogen bonds (D113 and A114) or other interactions (Y115) with the incoming dTTP substrate (20), and mutations in at least the A114 and Y115 residues have been implicated in alterations of the affinity for the dNTP substrate (34–36). Interestingly, coordination of the triphosphate moiety of the dTTP substrate by the D113 and A114 residues of HIV-1 RT is believed to be mediated by the main chain amino (NH) groups of these residues, and the side chains may not have a direct effect on dNTP binding (20). This lack of side chain involvement might account for the fact that several substitutions of the D153 position of MLV RT could complete replication (equivalent to D113 in HIV-1 RT). The D153F and D153R substitutions of MLV RT with amino acids containing large side chains displayed drastic reductions in titers (>10,000-fold) and RT activities (30- to 50-fold), whereas the D153A, D153C, D153Q, and D153S substitutions of MLV RT with smaller side chains had less severe effects on viral replication and RT activity (Table 1). The data suggest that amino acid side chains containing more than two carbons alter the active site in the polymerase in a way that is incompatible with efficient catalysis.
In this study, the F155W mutant of MLV RT exhibited the largest increase in the frequency of lacZ inactivation (2.8-fold). The effect on fidelity is in agreement with previous observations that mutations of the equivalent Y115 residue of HIV-1 RT resulted in alterations in in vitro processivity, frequency of misinsertions, and mispair extensions (18, 35, 36). In addition, the reduction of RT activity associated with the F155 mutants, including F155W (Table 4), may be due to a decrease in the affinity for substrate dNTPs that may have drastically impeded catalysis as previously proposed for the F115 position of HIV-1 RT (36). The aromatic side chains of the HIV-1 RT F115 and MLV RT F155 were shown to play a role in the selection of dNTPs over rNTPs (6, 12). Our results also support the importance of a bulky side chain at this position, since only the F155W and F155Y mutants generated detectable virus titers (Table 1). The F155V mutant of MLV RT was observed to incorporate UTP (12). The F155V mutant in our study exhibited an 18-fold reduction in RT activity, and the incorporation of rNTPs into the proviral DNA in vivo may account for the >10,000-fold reductions in titer (Table 4).
The majority of substitutions introduced at the K103, R110, and Q190 positions in MLV RT were characterized by defects in viral replication (Tables 1 and 4). Several mutants of the R72 and Q151 positions of HIV-1 RT that are equivalent to the R110 and Q190 of MLV RT were previously shown to result in drastic reductions in processivity as well as nucleotidyltransferase activity (27, 42, 43). Furthermore, similar observations were made for several substitutions of the R110 residue and the Q190N substitution of MLV RT (8, 24). Therefore, our results support previous studies indicating that these residues are critical for catalysis (8, 18, 30, 42–44). The K65A and Q151N mutants of HIV-1 RT were previously observed to be more accurate than the wild-type HIV-1 RT in vitro (18). However, the equivalent Q190N mutant of MLV RT reduced viral titers by >10,000-fold, and its effect on in vivo fidelity could not be determined. The Q190M mutant of MLV RT exhibited no change in fidelity, viral replication, or RT activity (Tables 1 and 4), which was in agreement with previously published in vitro results obtained with the equivalent Q151M mutant of HIV-1 RT (18).
Our results indicate that the size of the substituted amino acid side chain at the F156 position of MLV RT can affect fidelity, viral replication, and RT activity (Tables 2 and 4). The side chain of the equivalent F116 position of HIV-1 RT is believed to form a part of the pocket that accommodates the 3′ OH of the dNTP substrate (20). In addition, analysis of MLV RT crystal structure suggests that the F156 side chain might also be important for correct positioning of the Q190 side chain so that it can properly contact the dNTP substrate (13). Small side chains (F156I, F156Q, and F156V) exhibited no detectable levels of viral titer, whereas mutants possessing larger side chains (F156L, F156M, F156W, and F156Y) permitted viral replication (Table 2). It is possible that substitutions with amino acids possessing smaller side chains disrupt interactions between the F156 and Q190 residues in MLV RT, leading to a defect in reverse transcription.
Finally, as expected from previous analyses of the D110 position of HIV-1 RT, which is involved in the coordination of the metal cation, the equivalent D150 residue of MLV RT was highly conserved and did not tolerate the D-to-E substitution (Table 3) (13, 33, 34). Substitutions of the V148 and L149 positions also resulted in undetectable viral titers, perhaps because of their proximity to D150. It is interesting to note that the L151F substitution C terminal to D150 was tolerated and resulted in a 2.4-fold decrease in fidelity (Table 3).
The results of these studies suggest that amino acids that directly contact the dNTP substrate can have an affect on the efficiency of viral replication as well as fidelity of reverse transcription. In general, the substitution mutations of residues that directly contacted the substrate dNTP resulted in drastic reductions in viral titers. Therefore, future studies will be targeted to residues that are in close proximity to the residues that contact the substrate. These changes may be more likely to alter the dNTP-binding site and affect processivity and fidelity without affecting the essential residues that contact the dNTP. In addition, experiments to determine the spectrum of mutations generated by specific mutants of RT that led to the largest increases in the overall mutation rate will be performed.
ACKNOWLEDGMENTS
We especially thank Wei-Shau Hu for valuable intellectual input and discussions throughout this project and Steve Hughes for his intellectual input and discussion of the manuscript. We thank Benjamin Beasley, Sara Cheslock, Que Dang, Krista Delviks, Wei-Shau Hu, Carey Hwang, Timur Kabdulov, Terence Rhodes, Yegor Voronin, and Wen-Hui Zhang for their critical reading of the manuscript and discussion of results. We also thank Erin White and Ronald Mudry, Jr., for their technical support in generating pRMBNB and several of the dNTP-binding site mutants. Finally, we extend our thanks to Anne Arthur for her editorial expertise and revisions.
This work was supported by the Public Health Service grant CA58875 from the National Institutes of Health and by the HIV Drug Resistance Program, National Cancer Institute.
REFERENCES
- 1.Bakhanashvili M, Avidan O, Hizi A. Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett. 1996;391:257–262. doi: 10.1016/0014-5793(96)00747-8. [DOI] [PubMed] [Google Scholar]
- 2.Basu A, Nanduri V B, Gerard G F, Modak M J. Substrate binding domain of murine leukemia virus reverse transcriptase. Identification of lysine 103 and lysine 421 as binding site residues. J Biol Chem. 1988;263:1648–1653. [PubMed] [Google Scholar]
- 3.Beard W A, Bebenek K, Darden T A, Li L, Prasad R, Kunkel T A, Wilson S H. Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase. Molecular nature of polymerase-nucleic acid interactions. J Biol Chem. 1998;273:30435–30442. doi: 10.1074/jbc.273.46.30435. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek K, Beard W A, Casas-Finet J R, Kim H R, Darden T A, Wilson S H, Kunkel T A. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J Biol Chem. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- 5.Borman A M, Quillent C, Charneau P, Kean K M, Clavel F. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G→A hypermutation during negative-strand viral DNA synthesis. Virology. 1995;208:601–609. doi: 10.1006/viro.1995.1191. [DOI] [PubMed] [Google Scholar]
- 6.Boyer P L, Sarafianos S G, Arnold E, Hughes S H. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 2000;97:3056–3061. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury K, Kaushik N, Pandey V N, Modak M J. Elucidation of the role of Arg 110 of murine leukemia virus reverse transcriptase in the catalytic mechanism: biochemical characterization of its mutant enzymes. Biochemistry. 1996;35:16610–16620. doi: 10.1021/bi961462l. [DOI] [PubMed] [Google Scholar]
- 9.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 10.Drosopoulos W C, Prasad V R. Increased polymerase fidelity of E89G, a nucleoside analog-resistant variant of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1996;70:4834–4838. doi: 10.1128/jvi.70.7.4834-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G, Goff S P. Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 14.Gu Z, Arts E J, Parniak M A, Wainberg M A. Mutated K65R recombinant reverse transcriptase of human immunodeficiency virus type 1 shows diminished chain termination in the presence of 2′,3′-dideoxycytidine 5′-triphosphate and other drugs. Proc Natl Acad Sci USA. 1995;92:2760–2764. doi: 10.1073/pnas.92.7.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez-Rivas M, Ibanez A, Martinez M A, Domingo E, Menendez-Arias L. Mutational analysis of Phe160 within the “palm” subdomain of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1999;290:615–625. doi: 10.1006/jmbi.1999.2880. [DOI] [PubMed] [Google Scholar]
- 17.Halvas E K, Svarovskaia E S, Pathak V K. Development of an in vivo assay to identify structural determinants in murine leukemia virus reverse transcriptase important for fidelity. J Virol. 2000;74:312–319. [PMC free article] [PubMed] [Google Scholar]
- 18.Harris D, Kaushik N, Pandey P K, Yadav P N, Pandey V N. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 19.Hsiou Y, Ding J, Das K, Clark A D, Jr, Hughes S H, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 21.Iversen A K, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin J, Kaushik N, Singh K, Modak M J. Analysis of the role of glutamine 190 in the catalytic mechanism of murine leukemia virus reverse transcriptase. J Biol Chem. 1999;274:20861–20868. doi: 10.1074/jbc.274.30.20861. [DOI] [PubMed] [Google Scholar]
- 25.Julias J G, Hash D, Pathak V K. E-vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julias J G, Kim T, Arnold G, Pathak V K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J Virol. 1997;71:4254–4263. doi: 10.1128/jvi.71.6.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaushik N, Harris D, Rege N, Modak M J, Yadav P N, Pandey V N. Role of glutamine-151 of human immunodeficiency virus type-1 reverse transcriptase in RNA-directed DNA synthesis. Biochemistry. 1997;36:14430–14438. doi: 10.1021/bi970645k. [DOI] [PubMed] [Google Scholar]
- 28.Kent R B, Emanuel J R, Ben Neriah Y, Levenson R, Housman D E. Ouabain resistance conferred by expression of the cDNA for a murine Na+,K+-ATPase alpha subunit. Science. 1987;237:901–903. doi: 10.1126/science.3039660. [DOI] [PubMed] [Google Scholar]
- 29.Kim B, Hathaway T R, Loeb L A. Fidelity of mutant HIV-1 reverse transcriptases: interaction with the single-stranded template influences the accuracy of DNA synthesis. Biochemistry. 1998;37:5831–5839. doi: 10.1021/bi972672g. [DOI] [PubMed] [Google Scholar]
- 30.Kim B, Hathaway T R, Loeb L A. Human immunodeficiency virus reverse transcriptase. Functional mutants obtained by random mutagenesis coupled with genetic selection in Escherichia coli. J Biol Chem. 1996;271:4872–4878. doi: 10.1074/jbc.271.9.4872. [DOI] [PubMed] [Google Scholar]
- 31.Kim T, Mudry R A, Jr, Rexrode II C A, Pathak V K. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J Virol. 1996;70:7594–7602. doi: 10.1128/jvi.70.11.7594-7602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larder B A, Purifoy D J, Powell K L, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 34.Lowe D M, Parmar V, Kemp S D, Larder B A. Mutational analysis of two conserved sequence motifs in HIV-1 reverse transcriptase. FEBS Lett. 1991;282:231–234. doi: 10.1016/0014-5793(91)80484-k. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Hernandez A M, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Hernandez A M, Gutierrez-Rivas M, Domingo E, Menendez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Najmudin S, Cote M L, Sun D, Yohannan S, Montano S P, Gu J, Georgiadis M M. Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain. J Mol Biol. 2000;296:613–632. doi: 10.1006/jmbi.1999.3477. [DOI] [PubMed] [Google Scholar]
- 39.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sarafianos S G, Pandey V N, Kaushik N, Modak M J. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 43.Sarafianos S G, Pandey V N, Kaushik N, Modak M J. Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. J Biol Chem. 1995;270:19729–19735. doi: 10.1074/jbc.270.34.19729. [DOI] [PubMed] [Google Scholar]
- 44.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 45.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tisdale M, Alnadaf T, Cousens D. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob Agents Chemother. 1997;41:1094–1098. doi: 10.1128/aac.41.5.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueno T, Shirasaka T, Mitsuya H. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J Biol Chem. 1995;270:23605–23611. doi: 10.1074/jbc.270.40.23605. [DOI] [PubMed] [Google Scholar]
- 48.Vandamme A M, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antivir Chem Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]
- 49.Vara J A, Portela A, Ortin J, Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14:4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]