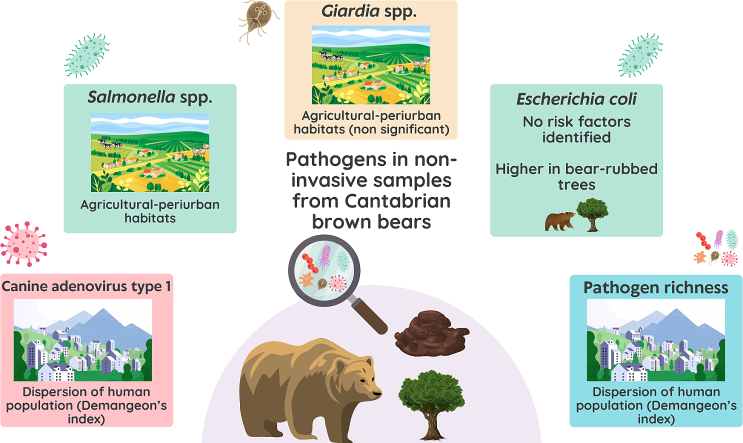
Abstract
Multi-host communities are perfect scenarios for the emergence and spread of pathogens, threatening the recovery of endangered, isolated, or inbred populations, such as the brown bear (Ursus arctos) in northwestern Spain. The population recovery in recent years has forced bears to occupy highly anthropized areas, increasing their interaction with human and domestic animals, with potential consequences for global health. During 2022–2023 a survey of parasites, bacteria and viruses shared between wildlife, domestic animals and humans was performed in this population using non-invasive surveillance, i.e., bear fecal samples (n = 73) and sponge-based sampling of trees (n = 42; 14 rubbed trees and 28 control trees). Pathogen detection rates were defined as the percentage of qPCR or culture-positive samples. Generalized linear models were fitted to assess their relationship with environmental variables including dispersion of the human population, and percentage of agricultural and periurban habitats in a 6 km-buffer around each sample. Canine Adenovirus type 1 (45.2%), Giardia spp. (15.1%), Salmonella spp. (12.3%), and extended-spectrum-beta-lactamases (ESBL) Escherichia coli (1.4%) were identified in fecal samples. In contrast, only five sponges from three rubbed and two control trees resulted positive to E. coli (14.3%). The results suggest that several pathogens are common in the Cantabrian brown bear population and that anthropization of the territory modulates their prevalence and richness.
The effective design of management programs for bear conservation will require a one-health approach, in which genetic analysis of non-invasive samples can be key tools for the sanitary surveillance at the wildlife-livestock-human interface.
Keywords: Brown bear (Ursus arctos), Interface, Canine Adenovirus type 1 (CAdV-1), Giardia spp., Salmonella spp., Non-invasive surveillance, Conservation
Graphical abstract
Highlights
-
•
Shared pathogens are present in Cantabrian brown bear population.
-
•
Level of humanization drives the positivity against pathogens.
-
•
Distribution of human population drives the positivity against pathogens.
-
•
Non-invasive sampling (feces and sponges) is useful for sanitary monitoring.
1. Introduction
Multi-host communities conform perfect scenarios where the pathogens can emerge and spread [1]. This is especially relevant in endangered, isolated, or inbred populations where these pathogens may hinder individual health, threat population recovery, and ultimately promote population declines and/or extinctions at local scales [2]. The endangered Cantabrian brown bear (Ursus arctos) population is located in the Cantabrian Range (northwestern Iberian Peninsula), and constitutes the southwestern distribution limit for this species in Europe [3]. The implementation of conservation strategies have allowed an important recovery of the Cantabrian brown bear distribution and population over the last two decades, currently estimated at 324 individuals, 275 in the western subpopulation and 49 in the eastern subpopulation [4,5]. Consequently, bears have been forced to occupy anthropized areas due to their growing population trends, the development of urban and suburban areas, and the ubiquity of human activities [6,7], which also attract bears looking for food resources of anthropic origin such as crops, livestock, hives, or garbage [6,8]. This spatial overlap may lead to social conflicts [7,9] and consequences for global health [9].
Brown bears are especially susceptible to infectious diseases because of the features of their populations (i.e., small and relatively isolated/fragmented), as well as their behavior and ecology (i.e., apex consumer and facultative scavengers) [10]. Recently, infectious diseases have been identified as an important cause of death in this population (39.7% in the period 1998–2023) [11,12]. The pathogens reported included Clostridium spp., verotoxigenic Escherichia coli, Canine Adenovirus type 1 (CAdV-1), and Canine Distemper Virus (CDV) [[11], [12], [13], [14]]. In addition, parasites such as Dicrocoelium spp., Trichuris spp. and Baylisascaris spp. have been described [9,15,16]. All those pathogens are shared between domestic animals, wildlife and in some cases humans, and in most instances are transmitted through indirect contacts (i.e., shared habitats and resources) at the interface [1,13].
The survey of elusive species, such as the brown bear, has largely been favored by the development of non-invasive genetic material monitoring, such as feces or sponges [[17], [18], [19], [20], [21]], without the need to capture or disturb them [18]. This sampling approach allows to explore ecosystem-level processes such as host-pathogen relationships, making possible the early detection of pathogens and the identification of hotspots for disease transmission [19,20,22].
In this study, we present a survey of shared parasites, bacteria, and viruses – some of them zoonotic – in the Cantabrian brown bear population using non-invasive surveillance. Our aims were to: 1) characterize the richness and detection rate of a set of pathogens in the Cantabrian brown bear population, 2) discuss the effects of land cover variables, livestock numbers and anthropization on the detection rate and richness of microorganisms in the Cantabrian brown bear population, and 3) evaluate the utility of non-invasive sampling in elusive species such as the brown bear.
2. Material and methods
2.1. Study area and sample collection
2.1.1. Study area
The Cantabrian brown bear population inhabits an area of approximately 8600 km2 along the Cantabrian Mountains (northwestern Spain) [23], with an elevation ranging from 100 to 2648 m above sea level (m.a.s.l.). The study area exhibits an oceanic climate with mild temperatures and moderate precipitation throughout the year (900–1400 mm) [23].
2.1.2. Fecal samples
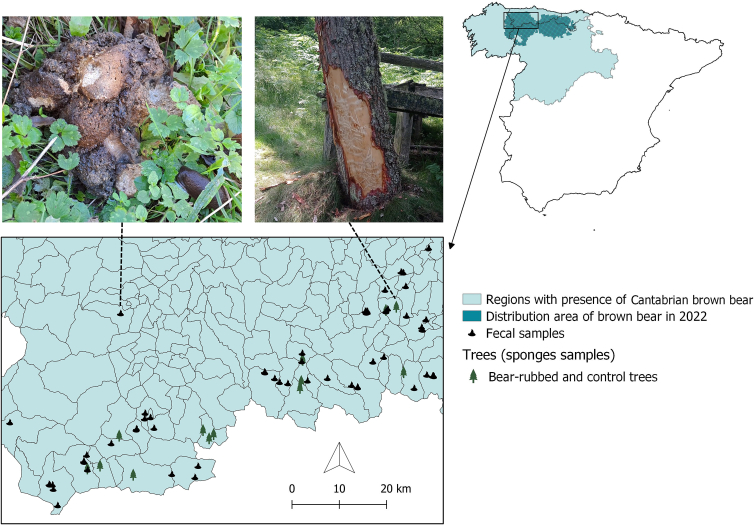
From 21st October to 27th November 2022, 73 fresh fecal samples were collected during routine itineraries and monitoring tasks conducted by rangers working with the species. All the samples were located in the western part of the Cantabrian bear range (Fig. 1), mostly in high-quality areas heavily used by bears during the autumn (i.e., hyperphagia period) [10,24], and regularly visited by rangers during field inspections. Furthermore, sampling during autumn–winter avoids the cubs' period (i.e., April–June; [10]). In order to minimize differences in sampling effort, some areas were reinforced with inspections by researchers and field technicians.
Fig. 1.
Study area and location of non-invasive samples studied in Cantabrian brown bear (Ursus arctos). Territorial divisions in the map refers to administrative units (locally called “parish”). Upper left image: Feces from brown bear. Upper right image: Rubbed tree.
The average distance between the feces collected was 31,713 m (31.7 km) (range: 1.38–171,752.67; SD = 25,935.14).
Around 300 grams (g) from the core of each fresh feces were taken. The outer layer was discarded to avoid potential contamination and the possibility of DNA/RNA being washed out by rain or damaged by sunlight. Sampled bear feces were georeferenced (Fig. 1), introduced into sterile bags, and transported to the laboratory, where they were treated and stored differently depending on their destination: 1) genetic analyses (20 g): dry stored with silica after ethanol soaking following [25]; 2) parasite detection (40 g): stored at −80 °C without any preservative treatment; 3) bacteria detection (40 g): stored at 4 °C and immediately processed; and 4) virus detection (300 mg): stored at 4 °C with RNA later (ThermoFisher Scientific, Madrid, Spain).
To genetically differentiate feces from different individuals, DNA was extracted according to [25] and genotyped using the six microsatellites (MU10, G10L, MU50, MU23, MU59, MU51) most polymorphic in the Cantabrian brown bear western population [25].
2.1.3. Sponges for environmental DNA detection in trees
Environmental DNA samples from 42 trees – 14 bear-rubbed and 28 control trees (i.e., 2 controls/rubbed tree) – were taken during June 2023 using cellulose dry sponges (3 M™ Dry-Sponge; 3 M-España, Madrid, Spain), prehydrated with 15 ml of an isotonic surfactant liquid, which preserves genetic material while rendering microorganisms inactive. June marks the end of the brown bear mating season (April–June) in the study area; therefore, it is more likely to find rubbed trees in the field with recent DNA deposition [26]. First, trees with rubbing signs (i.e., smoothed bark, scratches, bites, lack of vegetation at the base, or bear hair snagged on the bark [26]) were chosen, surveyed and sampled (Fig. 1). Then, two control trees within a 20 m-radius around each rubbed tree were sampled. Control trees were selected, attempting to ensure identical characteristics between control and rubbed trees (i.e., slope, altitude, orientation, tree species, size, etc.), but lacked any evidence of rubbing. All sampled trees were georeferenced (Fig. 1). On each tree, a sponge was scrubbed ten times on its bark and stored at 4 °C.
2.2. Shared pathogen identification
Selected shared pathogens (parasites, bacteria and viruses) were known to be present in the region in both domestic animals and wildlife [[27], [28], [29], [30], [31], [32]] (Table 1). Selected bacteria and parasites were also of zoonotic relevance. Parasites were not analyzed in sponge samples, as they are associated with feces. For each pathogen, the detection rate was calculated as the percentage of PCR or culture-positive samples (separately for feces and trees). For this purpose, fecal samples were treated as individual samples. Detailed information on pathogen detection procedures can be found in Additional file 1.
Table 1.
Shared pathogens studied in non-invasive samples in Cantabrian brown bear (Ursus arctos).
| Type and number (n) of samples | Parasites | Bacteria | Viruses |
|---|---|---|---|
| Feces (n = 73) | Cryptosporidium spp., Giardia spp., Blastocystis sp., Enterocytozoon bieneusi, Encephalitozoon spp. | Salmonella spp., verotoxigenic Escherichia coli and betalactamase (ESBL/AmpC) and/or carbapenemase producing E. coli | Canine Adenovirus type 1 (CAdV-1), Canine Distemper Virus (CDV) |
| Tree-sponges (n = 42) | – | Salmonella spp., verotoxigenic E. coli and colistin resistant E. coli | CAdV-1, CDV |
2.3. Habitat data collection
Land use and vegetation data were obtained from the CORINE land cover map 2018 and were reclassified into four categories: agricultural-periurban settings (including arable land, permanent crops, prairies, heterogeneous agricultural areas, and artificial surfaces), woodlands, scrubland, and natural grassland (i.e., grasslands under no or moderate human influence). A 6 km-radius buffer was considered around each environmental sample (feces or tree sponges) and the percentage of each vegetation type was assigned to each buffer. This radius is similar to the annual home range, i.e., a minimum convex polygon 95% of ∼130 km2, of an adult male, which is usually larger than the one of an adult female [33]. Although all the samples were collected during the hyperphagia period, when brown bears occupy smaller areas, we selected the annual home range because brown bears are flexible dietary generalists that rapidly adapt to seasonal differences in food availability, using a broad variety of habitats in a given year [33].
Mean elevation (m.a.s.l.) of the buffer was calculated from the 2 m resolution DTM PNOA-IGN (National Geographic Institute). The human population densities (individuals/km2) of each administrative unit (locally called “parish”) were obtained from the Spanish National Statistics Institute (INE) for the year 2023. Demangeon's index was selected as a proxy of human spreading across the territory considering a certain population density, i.e., increasing Demangeon's values indicate larger population spreading but not lower population densities. It was calculated for each parish according to Lizana et al. [34]. The density of livestock heads (heads/km2) (including cattle, sheep, goats, and horses) from each parish were calculated from the Asturian Society of Economical and Industrial Studies (SADEI) for the year 2020 (the most updated available data; Additional file 2a and 2b). To extrapolate the information collected at the parish level to each buffer, an average was calculated including the data from those parishes whose centroid was located within each buffer.
The qGIS software version 3.22.5 was used to perform these analyses.
2.4. Statistical analysis
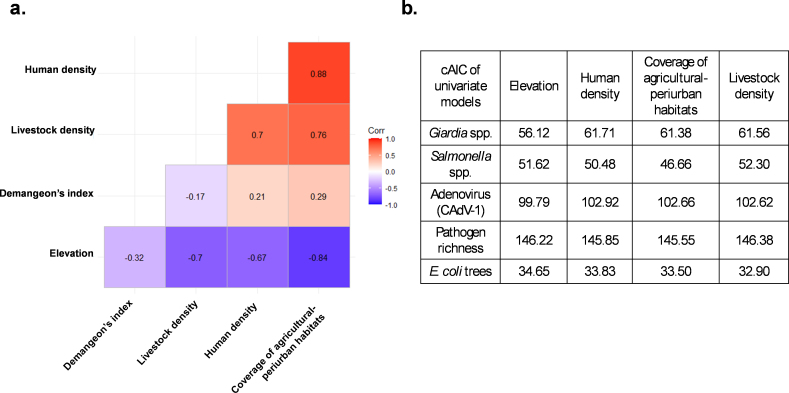
Before multivariate analysis, collinearity among predictors was explored. Numeric variables were scaled, allowing comparisons of their strength in the models. Since correlation > ± 0.60 was detected between several variables, individual models for each predictor were fitted (Additional file 3). Finally, the coverage of agricultural-periurban habitat was selected to be included in the models since it had the highest predictive strength for most pathogens (differences >2 in the corrected Akaike's Information Criterion; AICc) (Additional file 3).
Supplementary Fig. S3.
Additional file 3. Correlations. (a) Correlation matrix for the potential numeric variables to be included in the models. (b) Corrected Akaike's Information Criterion (AICc) for the univariate models including the correlated variables.
Generalized linear models (GLMs) were fitted to assess the association of the positivity to each pathogen (binomial response) with Demangeon's index and coverage of agricultural-periurban settings.
An ordinal multinomial regression model was performed to test the relationship between the pathogen richness (0–2) of each fecal sample (there were no trees with >1 pathogen) and the aforementioned variables.
Selection processes for the final models were performed based on the AICc (Additional files 4–8). When the function yielded the null model as the best model, the next one according to the AICc was selected as the final model, since no >2 ΔAICc values were observed. When multiple models were within 2 ΔAICc threshold, the model with more biological sense was selected.
Normality and the absence of residual pattern in data variation were checked for all the models. The libraries lme4, MASS and MumIn in R software 4.0.2 were used.
3. Results
3.1. Shared pathogens detected in feces and trees
3.1.1. Feces
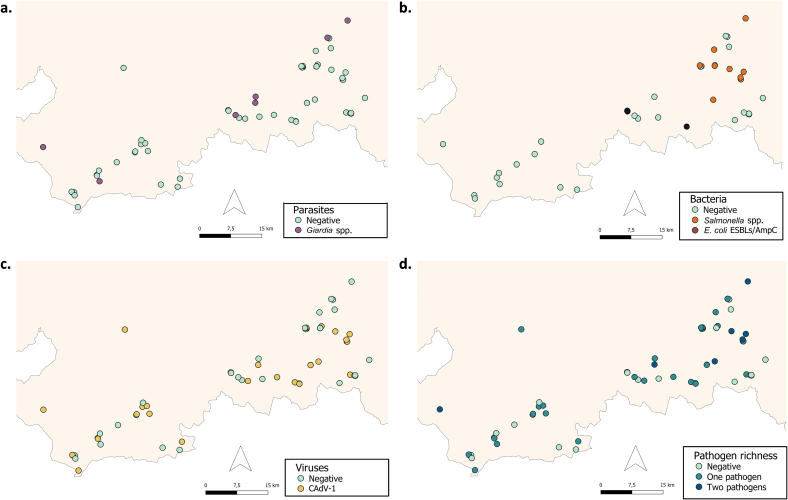
Shared pathogens in fecal samples were found in 60.3% of the samples. Specifically, Giardia spp. (15.07%), Salmonella enterica subsp. enterica (Salmonella spp.) (12.33%), ESBL (1.37%) and CAdV-1 (45.21%) were detected (Additional file 9, Fig. 2). All feces were negative for Cryptosporidium spp., Blastocystis sp., E. bieneusi, Encephalitozoon spp., verotoxigenic E. coli, and CDV. The 22.73% of the fecal samples positive to any pathogen resulted positive for multiple pathogens (10 positive to >1 pathogen/44 positive to ≥1 pathogen). The most frequent co-occurrence was Salmonella spp./CAdV-1 (40%; 4/10) and Giardia spp./CAdV-1 (40%; 4/10), followed by Giardia spp./Salmonella spp. (11.11%; 1/9), and ESBL/CAdV-1 (11.11%; 1/9).
Fig. 2.
Pathogens detected in fecal samples of Cantabrian brown bear (Ursus arctos). (a) Parasites, (b) bacteria, (c) viruses, and (d) number of pathogens detected in 73 feces from brown bear. E. coli ESBLs/AmpC: extended-spectrum beta-lactamases (ESBL), AmpC and carbapenemase producing E. coli. CAdV-1: Canine Adenovirus type 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For Salmonella spp., the most common serovar found was Newport (5.48% of the fecal samples; 4/9 positive samples to Salmonella spp.), followed by Ndolo (2.74% of the fecal samples; 2/9 positive samples to Salmonella spp.), whereas a single sample was positive to Enteritidis, Kottbus and Coeln (Additional file 9). All Salmonella isolates were susceptible (wild type phenotype) to all the antimicrobials included in the EUVSEC3 panel except for tigecycline for two isolates. In the case of the E. coli isolate obtained, it was found to be resistant to cefotaxime, ceftazidime, ampicillin and cefepime.
Genetic analysis to differentiate feces from different bears obtained amplification from 47 fecal samples (64%); however, only the 33 samples (45%) that successfully amplified more than three markers were included in the analysis. Among them, only four couples of samples gave the same genotype (12% of redundant sampling, i.e., feces from the same individuals; sample identifier 23–26, 12–25, 4–42, and 52–59; Additional file 10), identifying a total of 29 different genotypes. In the remaining fecal samples, the extracted DNA was of insufficient quality to perform the genetic analysis.
3.1.2. Trees
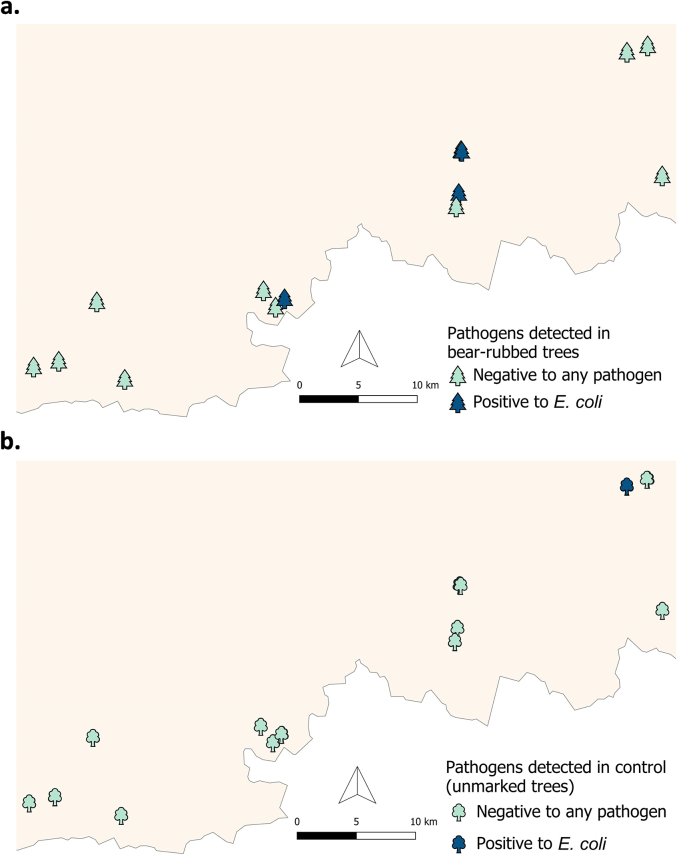
Only five trees were positive to E. coli (11.91%), three of them were bear-rubbed trees (3/14; 21.43%) and two were control trees (2/28; 7.14%) (Fig. 3). All trees were negative to CAdV-1, CDV, and virulence and resistance genes of E. coli.
Fig. 3.
Pathogens detected in trees using sponge-sampling. (a) Bear-rubbed trees (n = 14) and (b) control trees (n = 28). E. coli: Escherichia coli.
3.2. Environmental factors affecting pathogen detection rates
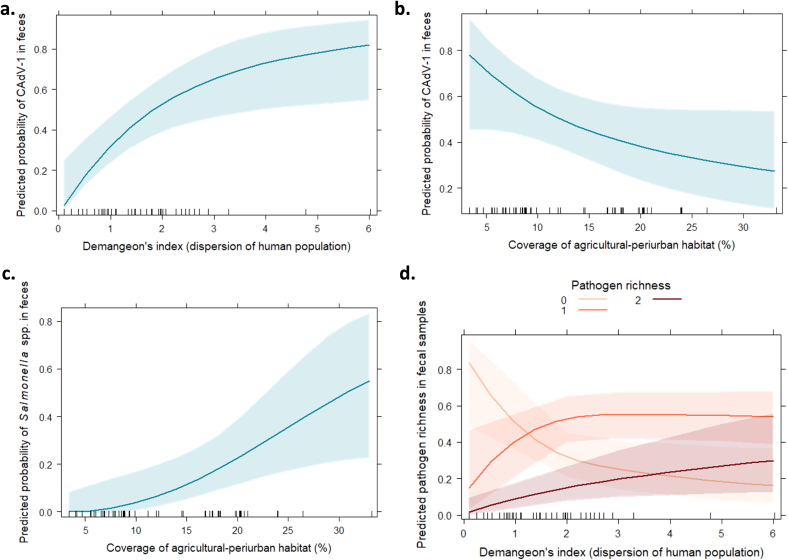
The results revealed that the dispersion of human population (Demangeon's index) and the coverage of agricultural-periurban habitat in a 6 km-buffer explained the positivity of fecal samples to the different pathogens (Table 2, Additional file 2a and 2b). In this regard, the human pressure in the surroundings (percentage of agriculture-periurban habitats in the 6 km-buffer) of each environmental sample (feces and sponges) is displayed in Additional file 2c, and positive fecal samples to any pathogen related to the dispersion of human population (Demangeon's index) is shown in Additional file 2d. According to the models, the probability of being positive to CAdV-1 was associated with the Demangeon's index, being higher in samples collected in areas where human population is dispersed, i.e., most of the human population live spread out from the main foundation nucleus (Table 2 and Fig. 4a). Furthermore, the positivity to CAdV-1 was marginally and negatively related to the coverage of agricultural-periurban habitat (Fig. 4b).
Table 2.
Results of the final generalized linear models (GLMs). GLMs - Canine Adenovirus type 1 (CAdV-1), Giardia spp., Salmonella spp. and Escherichia coli - and ordinal multinomial regression model (pathogen richness) examining the relationship between the positivity to several pathogens and the pathogen richness in non-invasive environmental samples of Cantabrian brown bear (Ursus arctos) and the dispersion of human population (Demangeon's index), coverage of agricultural-periurban habitat in a 6 km-buffer, and type of sampling tree. Statistically significant values are in bold and marginally significant values are marked with an asterisk (*).
| Predictors | Estimates | Confidence interval 95% | p-value |
|---|---|---|---|
| CAdV-1 in fecal samples (n = 72) | |||
| (Intercept) | −0.29 | 0.26 | 0.264 |
| Demangeon's index (dispersion of human population) | 1.05 | 0.37 | 0.005 |
| Coverage of agricultural-periurban habitat (%) | −0.58 | 0.31 | 0.059* |
| Giardia spp. in fecal samples (n = 72) | |||
| (Intercept) | −1.85 | 0.35 | <0.001 |
| Coverage of agricultural-periurban habitat (%) | 0.28 | 0.35 | 0.428 |
| Salmonella spp. in fecal samples (n = 72) | |||
| (Intercept) | −2.78 | 0.67 | <0.001 |
| Coverage of agricultural-periurban habitat (%) | 1.65 | 0.65 | 0.010 |
| Pathogen richness (fecal samples; ordinal) (n = 72) | |||
| 0|1 | −0.41 | 0.25 | 0.010 |
| 1|2 | 2.08 | 0.37 | <0.001 |
| Demangeon's index (dispersion of human population) | 0.65 | 0.26 | 0.013 |
| E. coli in sponges from trees (n = 42) | |||
| (Intercept) | −2.57 | 0.73 | <0.001 |
| Type: bear-rubbed tree | 1.27 | 0.98 | 0.197 |
Fig. 4.
Predicted probability of detecting several pathogens in fecal samples from Cantabrian brown bear (Ursus arctos) population ± confidence interval 95% (error bars or shaded band). (a) Predicted probability of Canine Adenovirus type 1 (CAdV-1) depending on the dispersion of human population (Demangeon's index). (b) Predicted probability of CAdV-1 depending on the coverage of agricultural-periurban habitat (percentage in a 6 km-buffer from each fecal sample). (c) Predicted probability of Salmonella spp. depending on the coverage of agricultural-periurban habitat (percentage in a 6 km-buffer from each fecal sample). (d) Predicted pathogen richness (positive to none −0-, a single pathogen −1- or two pathogens −2-) depending on the dispersion of human population (Demangeon's index). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The risk of Giardia spp. was higher in samples from areas with increased agricultural-periurban coverage, but these results were not statistically significant (Table 2). In this sense, the positivity to Salmonella spp. was significantly associated with higher dominance of agricultural-periurban settings (Table 2 and Fig. 4c).
Finally, the pathogen richness in the fecal samples depended on the dispersion of human populations in the bear range (Table 2 and Fig. 4d). The probability of showing the highest pathogen richness occurred when human populations widely overlap with the bear's habitats, i.e., higher Demangeon's indexes.
For tree sponges, although bear-rubbed trees presented a higher risk of being positive, these results were not statistically significant (Table 2).
Raw data of pathogen detection in feces and tree-sponges can be found in Additional file 10.
4. Discussion
This study has determined the presence of CAdV-1, Giardia spp., Salmonella spp., and ESBL in brown bear feces, as well as E. coli in bear-rubbed trees using non-invasive sampling, which suggests its potential usefulness as a tool to monitor the sanitary status of this species. However, no difference was found in the risk of detecting E. coli in rubbed vs. unmarked (control) trees, thus the use of trees should be further investigated in the future.
The high prevalence of CAdV-1 obtained in fecal samples (45%) reflects the widespread circulation of the virus in the Cantabrian bear population. Since 2014 CAdV-1 has caused the death of three brown bears due to infectious canine hepatitis [12], and it has also been detected in the sympatric Iberian wolf (Canis lupus) [31,32]. CAdV-1 can be transmitted through direct contact with infected animals or contaminated fomites, and is considered stable in the environment, remaining infective for weeks, thus facilitating its detection by non-invasive sampling [35]. The source of CAdV-1 infection for bears might be wolves or domestic dogs, usually not vaccinated in rural areas. In that regard, GLMs related the CAdV-1 positivity to the dispersion of human populations and marginally to lower coverage of agricultural and periurban habitats. The fragmented areas (i.e., those that combine forested areas with small human settlements) constitute engaging environments with anthropogenic food resources for brown bears, which may also favor the interaction between dogs and bears, and thus the potential interspecies transmission of CAdV-1. Therefore, the study of CAdV-1 prevalence in rural dogs, as well as their vaccination, are strongly recommended.
The CDV Morbillivirus, which has caused mortality since 2020 in several carnivore species in the region including brown bear [12,30], was not detected in any fecal nor tree samples. However, it should be noted that RNA viruses are more easily degradable in the environment than DNA ones, suggesting a short life of CDV in the environment and/or the importance of direct transmission routes from infected individuals rather than an absence of circulation of this virus. To the best of our knowledge, there are no previous studies on CDV and CAdV-1 surveillance with non-invasive methods.
Interestingly, a 15% infection rate of Giardia spp. and an apparent absence of Cryptosporidium spp., Blastocystis sp., E. bieneusi, and Encephalitozoon spp. parasites were obtained. Giardia spp. was previously found at a lower infection rate (5%) in sympatric wildlife [29,36]. Unfortunately, lack of amplification of genotyping markers (very likely reflecting the low amount of starting parasite DNA) precluded us from determining the genetic variants involved in these infections. Nevertheless, assemblage A (unknown sub-assemblage) was previously identified in a brown bear fecal sample collected in the same region (GenBank accession number: PP312933), being the first report on the molecular diversity of Giardia infection in wild bears globally. Giardia cysts were identified in resident polar bears (Ursus maritimus) in Alaska [37], but the parasite was not detected in brown bears from Croatia, the United States, and Spain [15,38,39]. Of note, Cryptosporidium spp., E. bieneusi, and Blastocystis sp. have been reported in sympatric wildlife in northern Spain [[27], [28], [29],40,41], and the former also in free-ranging brown bears in Slovakia and Poland [42,43]. In contrast, Blastocystis sp. and E. bieneusi have only been confirmed in captive wild bears from Asia [44,45].
Salmonella spp. and ESBL were detected in fecal samples. Salmonella spp. is a common bacteria isolated from humans and domestic animals [46,47], which has also been occasionally isolated in American black bears (Ursus americanus), e.g., [48]. Five serovars of Salmonella spp. were detected in the Cantabrian bear population, being S. Newport and Ndolo the two most frequently found. Specifically, S. Newport and S. Ndolo were reported in badgers from northern Spain and livestock, respectively, all abundant species in the study area [49,52]. The diversity of Salmonella serovars found in this bear population might be a consequence of its generalist diet and habits, as suggested for other wild carnivores in northern Spain [49]. On the other hand, ESBL was only detected in a fecal sample, and although verotoxigenic E. coli was identified as the cause of death in one Cantabrian brown bear [12], none of the samples tested resulted positive in this study. ESBL has been detected in other wild mammals across the globe, including other bear species, e.g., sloth bear (Melursus ursinus), becoming an increasingly frequent pathogen of concern with potential human and/or animal origin [50].
According to our best model, Giardia spp. and Salmonella spp. positivity tended to be associated with higher coverages of agricultural-periurban habitat, reflecting that bears may acquire these pathogens through consumption of contaminated human waste and water, i.e., water courses which received run-off from farm buildings, slurry, human sewage, and feces of wildlife linked to peri-urban habitats [1]. In this line, an elevated pathogen richness was evidenced in areas with higher Demangeon's index.
Sponge sampling gave only E. coli detection on trees. The potential detection of pathogens on trees might come from urine, saliva, nasal discharges, scratches, or hair snagged on the tree bark. However, the detection capacity of sponges in trees may be more susceptible to variables such as the sampling time, the environmental resistance of pathogens, the humidity, and other climatic or habitat factors compared to feces where pathogens are less exposed to environmental damages (e.g., sunlight). Furthermore, finding pathogens in sponge samples or even feces does not imply viable organisms at sufficient concentration for being infective, but pathogen presence in the bear ecosystem [20], confirming its utility for passive surveillance [[20], [21], [22]].
Overall, in our study environmental non-invasive sampling allowed detection of pathogens overcoming the main constraints derived from direct wildlife sampling (i.e., induced stress, increased effort, elusive species, etc.). It also enhanced the epidemiological understanding of viral, parasite and bacterial infections shared at the bear-human interface. This is an important finding since it opens an avenue for a non-invasive sampling of multiple pathogen markers in studies of the ecosystem sanitary status. Specifically, sponge samples may gather genetic material from a significant portion of animals, whereas feces represent the contribution of each individual animal. In addition, sponges used in this study are temperature-resistant and biosafe since pathogens are fixed in preserving liquid [20,21].
Limitations of this study may have been (i) the potential degradation of pathogen DNA/RNA in the environment, especially in trees, which might have prevented its correct detection by diagnostic techniques; and (ii) the redundant sampling of 12% of feces from the same four animals. In that regard, feces and tree sponges were studied to deepen in the knowledge of the presence of pathogens at the interface, not for prevalence studies and diagnostic purposes, which can be reached by necropsies and subsequent analysis, among others.
5. Conclusions
This study reveals that land disturbance and how human population is distributed throughout the territory drives the positivity against several pathogens and higher pathogen richness, with varying results depending on the pathogen. None of the pathogens studied except CAdV-1 seem to be a health issue for bears; however, the ongoing increase in brown bear population together with its territorial expansion in human-modified landscapes will undoubtedly bring more bear exposure to shared pathogens at the interface, some of which might be potentially pathogenic to bears. The study also highlights the usefulness of non-invasive sampling as a tool to monitor and survey the sanitary status of elusive species such as the brown bear, offering new insights into epidemiological research which can help in conservation programs of this and other large carnivore populations. For this reason, including these non-invasive tools in the current integrated wildlife monitoring official programs is recommended.
The following are the supplementary data related to this article.
Additional file 1 Shared pathogen detection procedures.
Supplementary Fig. S2.
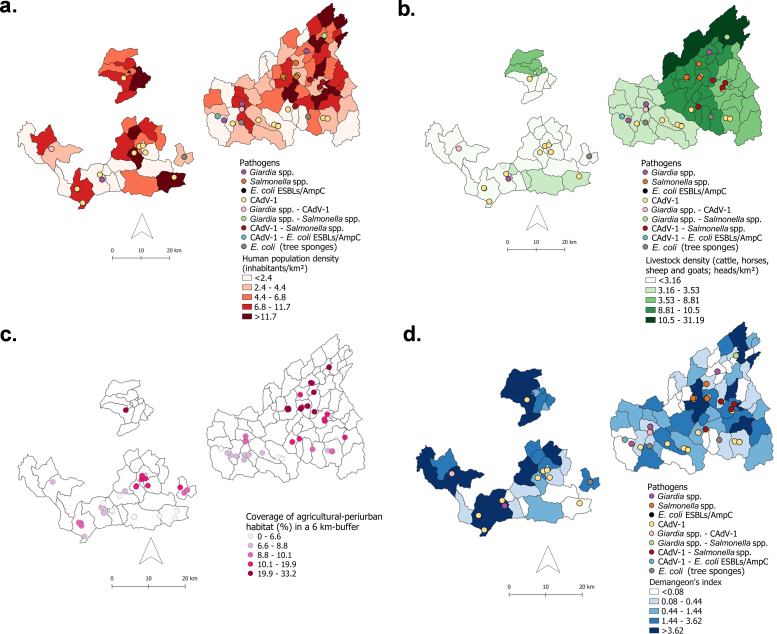
Additional file 2. Pathogens related to human, livestock and vegetation variables. Parasites, bacteria, and viruses detected in fecal samples of Cantabrian brown bear (Ursus arctos), in relation to (a) human population density (inhabitants/km2), (b) number of livestock heads in the study area, (c) fecal and sponge samples depending on the coverage of agricultural-periurban habitat (%) in a 6 km-buffer and, (d) the dispersion of human population (Demangeon's index). E. coli ESBLs/AmpC: extended-spectrum beta-lactamases (ESBL), AmpC and carbapenemase producing E. coli. CAdV-1: Canine Adenovirus type 1.
Additional file 4. Model selection for Canine Adenovirus type 1 (CAdV-1). Details of model selection for the predicted positivity to CAdV-1 in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 5. Model selection for Giardia spp. Details of model selection for the predicted positivity to Giardia spp. in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold and final model is highlighted in red. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 6. Model selection for Salmonella spp. Details of model selection for the predicted positivity to Salmonella spp. in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 7. Model selection for pathogen richness. Details of model selection for the pathogen richness detected in fecal samples of Cantabrian brown bear and the elevation, dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold font. K means the number of variables in the model; cAIC corrected Akaike's Information Criterion; Δi difference of cAIC with respect to the best model; and wi Akaike weight.
Additional file 8. Model selection for Escherichia coli. Details of model selection for the predicted positivity to E. coli in sponge samples from trees based on dispersion of human population (Demangeon's index), coverage of agricultural-periurban habitat (%) in a 6 km-buffer, and type of tree (marked – bear rubbed- or unmarked -control-). The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 9. Detection rate (%) of different pathogens from 73 fecal samples of Cantabrian brown bears (Ursus arctos).
Additional file 10. Raw data of feces and tree-sponge sampling in Cantabrian brown bear (Ursus arctos).
Ethics approval and consent to participate
The International Union for Conservation of Nature (IUCN) Policy Statement on Research Involving Species at Risk of Extinction and the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) were meticulously followed. The national and regional legislation on Cantabrian brown bear sampling (Consejería de Fomento, Cooperación Local y Prevención de Incendios del Principado de Asturias) were also compiled. Ethics approval was unnecessary according to Spanish national regulations (Real Decreto 53/2013).
Funding sources
This work is a contribution to the I+D+i research projects: 1) PID2022-141906OB-C21 and PID2022-141906OB-C22 funded by MCIN/AEI/10.13039/501100011033/FEDER, UE; B) PCTI 2021–2023 (GRUPIN: IDI2021–000102) funded by Principado de Asturias and FEDER; C) PLEC2021-008113 funded by MCIN/AEI/10.13039/501100011033/ and the European Union NextGeneration EU/PRTR. GHG was funded by Junta de Castilla y León and FSE (LE036–20); PB by Juan de la Cierva post-doc fellowship DC2022–049103-I; and AGR by the EU-NextGenerationEU funds through the 2021–2023 Margarita Salas call for the requalification of the Spanish university system, convened by the Universidad de León. DGB is the recipient of a Sara Borrell Research Contract (CD19CIII/00011) funded by the Spanish Ministry of Science, Innovation, and Universities. Additional funding was obtained from the Health Institute Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness, under project PI19CIII/00029.
Authors' contributions
GHG, JN, AFG, AdM and AGR performed the field work. GHG, AD, DGB, MUR, MPS, LJR, DC and AB performed the laboratory analysis. PB performed the statistical analysis. AB, JN, CG and LD conceptualized the study. AB and CG obtained the funding. PB and AB wrote the first draft of the manuscript. All authors contributed critically to the final draft and gave approval for publication.
CRediT authorship contribution statement
Gloria Herrero-García: Investigation, Methodology, Writing – review & editing. Patricia Barroso: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Alejandro Dashti: Investigation, Methodology, Writing – review & editing. David González-Barrio: Investigation, Methodology, Writing – review & editing. Javier Naves: Conceptualization, Data curation, Methodology, Validation, Writing – review & editing. Alberto Fernández-Gil: Investigation, Methodology, Writing – review & editing. María Ugarte-Ruiz: Investigation, Methodology, Writing – review & editing. Marta Pérez-Sancho: Investigation, Methodology, Writing – review & editing. Luis José Royo: Investigation, Methodology, Writing – review & editing. David Carmena: Data curation, Investigation, Methodology, Writing – review & editing. Arturo de Miguel: Methodology, Writing – review & editing. Alberto García-Rodríguez: Investigation, Methodology, Writing – review & editing. Christian Gortázar: Conceptualization, Data curation, Funding acquisition, Validation, Writing – review & editing. Lucas Domínguez: Conceptualization, Data curation, Supervision, Validation, Writing – review & editing. Ana Balseiro: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
The authors thank colleagues from University of León (especially B. Rabanal from Laboratorio de Técnicas Instrumentales), VISAVET-Universidad Complutense de Madrid (especially M. García, N. Maassoumi Nouha and E. Rivero), University of Oviedo, Fundación Oso de Asturias (especially J. Tuñón) and Consejería de Fomento, Cooperación Local y Prevención de Incendios del Principado de Asturias, for their invaluable support. Authors also thank the rangers and technicians F. Somoano, J. Díaz, D. Ramos, S. Tejerio, J.J. Congregado, E. García, A. Fernández and M. de Gabriel for their collaboration in the field work.
Contributor Information
Gloria Herrero-García, Email: gherrg01@estudiantes.unileon.es.
Patricia Barroso, Email: pbars@unileon.es.
Javier Naves, Email: jnaves@ebd.csic.es.
Alberto Fernández-Gil, Email: albertofg@ebd.csic.es.
María Ugarte-Ruiz, Email: maria.ugarte@ucm.es, maria.ugarte@ucm.es.
Marta Pérez-Sancho, Email: maperezs@ucm.es.
Luis José Royo, Email: royoluis@uniovi.es.
David Carmena, Email: dacarmena@isciii.es.
Christian Gortázar, Email: christian.gortazar@uclm.es.
Lucas Domínguez, Email: lucasdo@visavet.ucm.es.
Ana Balseiro, Email: abalm@unileon.es.
Data availability
Data will be made available on request.
References
- 1.Daszak P., Cunningham A.A., Hyatt A.D. Emerging infectious diseases of wildlife-threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen A.B., Jones K.E., Nunn C.L., Altizer S. Infectious diseases and extinction risk in wild mammals. Conserv. Biol. 2007;21:1269–1279. doi: 10.1111/j.1523-1739.2007.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nores C., Naves J. In: El Oso Pardo (Ursus arctos) En España. Naves J., Palomero G., editors. ICONA; Madrid, Spain: 1993. Distribución histórica del oso pardo en la península Ibérica; pp. 13–33. [Google Scholar]
- 4.Pérez T., Naves J., Vázquez J.F., Seijas J., Corao A., Albornoz J., Domínguez A. Evidence for improved connectivity between Cantabrian brown bear subpopulations. Ursus. 2010;21:104–108. doi: 10.2192/09SC018.1. [DOI] [Google Scholar]
- 5.López-Bao J.V., Godinho R., Palomero G., Ballesteros F., Blanco J.C., Jiménez J. In: Osos Cantábricos. Palomero G., Ballesteros F., Blanco J.C., López-Bao J.V., editors. Demogr. Coexistencia y Retos Conserv; Lynx Edicions, Madrid, Spain: 2021. Seguimiento de la población de oso cantábrico en un escenario de expansión; pp. 23–38. [Google Scholar]
- 6.Zarzo-Arias A., Penteriani V., del M Delgado M., Torre P. Peón, García-González R., Mateo-Sánchez M.C., García P. Vázquez, Dalerum F. Identifying potential areas of expansion for the endangered brown bear (Ursus arctos) population in the Cantabrian Mountains (NW Spain) PLoS One. 2019;14 doi: 10.1371/journal.pone.0209972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naves J., Ordiz A., Fernández-Gil A., Penteriani V., del M Delgado M., López-Bao J.V., Revilla E., Delibes M. Patterns of brown bear damages on apiaries and management recommendations in the Cantabrian Mountains, Spain. PLoS One. 2018;13 doi: 10.1371/journal.pone.0206733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penteriani V., Zarzo-Arias A., Novo-Fernández A., Bombieri G., López-Sánchez C.A. Responses of an endangered brown bear population to climate change based on predictable food resource and shelter alterations. Glob. Chang. Biol. 2019;25:1133–1151. doi: 10.1111/gcb.14564. [DOI] [PubMed] [Google Scholar]
- 9.Costa H., Hartasánchez R., Santos A.R., Camarão A., Cruz L., Nascimento M., Gomes L., Madeira de Carvalho L.M. Preliminary findings on the gastrointestinal parasites of the brown bear (Ursus arctos) in the Cantabrian mountains, Spain. Vet. Parasitol. Reg. Stud. Rep. 2022;28 doi: 10.1016/j.vprsr.2021.100681. [DOI] [PubMed] [Google Scholar]
- 10.Purroy F.J. In: Encicl. Virtual Los Vertebr. Españoles. Salvador A., Barja I., editors. Museo Nacional de Ciencias Naturales, CSIC; Madrid: 2017. Oso pardo – Ursus arctos; pp. 1–36.https://digital.csic.es/bitstream/10261/112114/8/ursarc_v3.pdf [Google Scholar]
- 11.Balseiro A., Royo L.J., Gayo E., Balsera R., Alarcia O., García Marín J.F. Mortality causes in free-ranging Eurasian Brown bears (Ursus arctos arctos) in Spain 1998–2018. Animals. 2020;10:1538. doi: 10.3390/ani10091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balseiro A., Herrero-García G., García-Marín J.F., Cubero D., de Pedro G., Oleaga A., García-Rodríguez A., Espinoza I., Rabanal B., Aduriz G., Tuñon J., Gortázar C., Royo L.J. New threats in the recovery of large carnivores inhabiting human-modified landscapes: the case of the Cantabrian brown bear (Ursus arctos) Vet. Res. 2024;55:24. doi: 10.1186/s13567-024-01279-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García Marín J.F., Royo L.J., Oleaga A., Gayo E., Alarcia O., Pinto D., Martínez I.Z., González P., Balsera R., Marcos J.L., Balseiro A. Canine adenovirus type 1 (CAdV-1) in free-ranging European brown bear (Ursus arctos arctos): a threat for Cantabrian population?, Transbound. Emerg. Dis. 2018;65:2049–2056. doi: 10.1111/tbed.13013. [DOI] [PubMed] [Google Scholar]
- 14.Balseiro A., Oleaga Á., Polledo L., Aduriz G., Atxaerandio R., Kortabarria N., Marín J.F.G. Clostridium sordellii in a Brown bear (Ursus arctos) from Spain. J. Wildl. Dis. 2013;49:1047–1051. doi: 10.7589/2013-03-065. [DOI] [PubMed] [Google Scholar]
- 15.Remesar S., Busto C., Díaz P., Rivas Ó., López-Bao J.V., Ballesteros F., García-Dios D. Presence of gastrointestinal and bronchopulmonary parasites in Cantabrian brown bears. Eur. J. Wildl. Res. 2024;70:23. doi: 10.1007/s10344-024-01779-2. [DOI] [Google Scholar]
- 16.Valderrábano Cano E., Penteriani V., Vega I., del M Delgado M., González-Bernardo E., Bombieri G., Zarzo-Arias A., Fernández R. Sánchez-Andrade, Paz-Silva A. Influence of seasonality and biological activity on infection by helminths in Cantabrian bear. Int. J. Parasitol. Parasites Wildl. 2024;23 doi: 10.1016/j.ijppaw.2024.100916. 100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Bao J.V., Godinho R., Rocha R.G., Palomero G., Blanco J.C., Ballesteros F., Jiménez J. Consistent bear population DNA-based estimates regardless molecular markers type. Biol. Conserv. 2020;248 doi: 10.1016/j.biocon.2020.108651. [DOI] [Google Scholar]
- 18.Bohmann K., Evans A., Gilbert M.T.P., Carvalho G.R., Creer S., Knapp M., Yu D.W., de Bruyn M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014;29:358–367. doi: 10.1016/j.tree.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Alves P.C., Gavier-Widen D., Ferroglio E., Queirós J., Rafael M., Santos N., Silva T., Gonçalves C., Vada R., Zanet S., Smith G., Gethöffer F., Keuling O., Staubach C., Sauter-Louis C., Blanco J., Podgorski T., Larska M., Richomme C., Knauf S., Rijks J.M., Pasetto C., Benatti F., Poncina M., Gómez A., Dups-Bergmann J., Neimanis A., Vicente J. Literature review on the main existing structures and systematic/academic initiatives for surveillance in the EU for zoonoses in the environment and the methods for surveillance of pathogens in the environment. EFSA Support. Publ. 2022;19 doi: 10.2903/sp.efsa.2022.EN-7792. [DOI] [Google Scholar]
- 20.Martínez-Guijosa J., Romero B., Infantes-Lorenzo J.A., Díez E., Boadella M., Balseiro A., Veiga M., Navarro D., Moreno I., Ferreres J., Domínguez M., Fernández C., Domínguez L., Gortázar C. Environmental DNA: a promising factor for tuberculosis risk assessment in multi-host settings. PLoS One. 2020;15 doi: 10.1371/journal.pone.0233837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-de-Mera I.G., Del-Río F.J. Rodríguez, Fuente J., Pérez-Sancho M., Hervás D., Moreno I., Domínguez M., Domínguez L., Gortázar C. Detection of environmental SARS-CoV-2 RNA in a high prevalence setting in Spain, Transbound. Emerg. Dis. 2021;68:1487–1492. doi: 10.1111/tbed.13817. [DOI] [PubMed] [Google Scholar]
- 22.Reza-Varzandi A., Zanet S., Barroso P., Occhibove F., Vada R., Benatti F., Palencia P., Ferroglio E. Detection of African swine fever virus and wild boar eDNA in soil and turbid water samples: towards environmental surveillance. Eur. J. Wildl. Res. 2024;70:4. doi: 10.1007/s10344-023-01758-z. [DOI] [Google Scholar]
- 23.Naves J., Fernández-Gil A., Rodríguez C., Delibes M. Brown bear food habits at the border of its range: a long-term study. J. Mammal. 2006;87:899–908. doi: 10.1644/05-MAMM-A-318R2.1. [DOI] [Google Scholar]
- 24.Ballesteros F., López-Bao J.V., Blanco J.C., Palomero G., Planella A. In: 26th Int. Conf. Bear Res. Manag., Lujbliana, Slovenia. 2018. Exceptional aggregation of Cantabrian brown bears during hyperphagia; p. 108. [Google Scholar]
- 25.Pérez T., Vázquez F., Naves J., Fernández A., Corao A., Albornoz J., Domínguez A. Non-invasive genetic study of the endangered Cantabrian brown bear (Ursus arctos) Conserv. Genet. 2009;10:291–301. doi: 10.1007/s10592-008-9578-1. [DOI] [Google Scholar]
- 26.González-Bernardo E., Bagnasco C., Bombieri G., Zarzo-Arias A., Ruiz-Villar H., Morales-González A., Lamamy C., Ordiz A., Cañedo D., Díaz J., Chamberlain D.E., Penteriani V. Rubbing behavior of European brown bears: factors affecting rub tree selectivity and density. J. Mammal. 2021;102:468–480. doi: 10.1093/jmammal/gyaa170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dashti A., Santín M., Köster P.C., Bailo B., Ortega S., Imaña E., Habela M.Á., Rivero-Juarez A., Vicente J., Conejero C., González-Crespo C., Garrido C., Gassó D., Andrea Murillo D., Serrano E., Mentaberre G., Torres-Blas I., Estruch J., Pastor J., Ramón López-Olvera J., Escobar-González M., Valldeperes M., Mesalles M., López O., Álvarez R., Cuenca R., Velarde R., Lavín S., Arnal M.C., de Luco D.F., Morrondo P., Armenteros J.A., Balseiro A., Cardona G.A., Martínez-Carrasco C., Ortiz J.A., Calero-Bernal R., Carmena D., González-Barrio D. Zoonotic Enterocytozoon bieneusi genotypes in free-ranging and farmed wild ungulates in Spain. Med. Mycol. 2022;60 doi: 10.1093/mmy/myac070. [DOI] [PubMed] [Google Scholar]
- 28.Vioque F., Dashti A., Santín M., Ruiz-Fons F., Köster P.C., Hernández-Castro C., García J.T., Bailo B., Ortega S., Olea P.P., Arce F., Chicharro C., Nieto J., González F., Viñuela J., Carmena D., González-Barrio D. Wild micromammal host spectrum of zoonotic eukaryotic parasites in Spain. Occurrence and genetic characterisation. Transbound. Emerg. Dis. 2022;69 doi: 10.1111/tbed.14643. [DOI] [PubMed] [Google Scholar]
- 29.Dashti A., Köster P.C., Bailo B., de Las Matas A.S., Habela M.Á., Rivero-Juarez A., Vicente J., Serrano E., Arnal M.C., de Luco D.F., Morrondo P., Armenteros J.A., Balseiro A., Cardona G.A., Martínez-Carrasco C., Ortiz J.A., Carpio A.J., Calero-Bernal R., González-Barrio D., Carmena D. Occurrence and limited zoonotic potential of Cryptosporidium spp., Giardia duodenalis, and Balantioides coli infections in free-ranging and farmed wild ungulates in Spain. Res. Vet. Sci. 2023;159:189–197. doi: 10.1016/j.rvsc.2023.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Oleaga A., Vázquez C.B., Royo L.J., Barral T.D., Bonnaire D., Armenteros J.Á., Rabanal B., Gortázar C., Balseiro A. Canine distemper virus in wildlife in South-Western Europe. Transbound. Emerg. Dis. 2022;69 doi: 10.1111/tbed.14323. [DOI] [PubMed] [Google Scholar]
- 31.Oleaga A., Balseiro A., Espí A., Royo L.J. Wolf (Canis lupus) as canine adenovirus type 1 (CAdV-1) sentinel for the endangered Cantabrian brown bear (Ursus arctos arctos) Transbound. Emerg. Dis. 2022;69:516–523. doi: 10.1111/tbed.14010. [DOI] [PubMed] [Google Scholar]
- 32.Millán J., López-Bao J.V., García E.J., Oleaga Á., Llaneza L., Palacios V., de la Torre A., Rodríguez A., Dubovi E.J., Esperón F. Patterns of exposure of Iberian wolves (Canis lupus) to canine viruses in human-dominated landscapes. Ecohealth. 2016;13:123–134. doi: 10.1007/s10393-015-1074-8. [DOI] [PubMed] [Google Scholar]
- 33.de Gabriel Hernando M., Karamanlidis A.A., Grivas K., Krambokoukis L., Papakostas G., Beecham J. Reduced movement of wildlife in Mediterranean landscapes: a case study of brown bears in Greece. J. Zool. 2020;311:126–136. doi: 10.1111/jzo.12768. [DOI] [Google Scholar]
- 34.Lizana V., Gortázar C., Muniesa A., Cabezón Ó., Martí-Marco A., López-Ramon J., Cardells J. Human and environmental factors driving toxoplasma gondii prevalence in wild boar (Sus scrofa) Res. Vet. Sci. 2021;141:56–62. doi: 10.1016/j.rvsc.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Woods L.W. In: Infect. Dis. Wild Mamm., Third Edit. Williams E.S., Barker I.K., editors. Iowa State University Press/Ames; 2001. Adenoviral diseases; pp. 202–213. [Google Scholar]
- 36.Mateo M., de Mingo M.H., de Lucio A., Morales L., Balseiro A., Espí A., Barral M., Lima Barbero J.F., Habela M.Á., Fernández-García J.L., Bernal R.C., Köster P.C., Cardona G.A., Carmena D. Occurrence and molecular genotyping of Giardia duodenalis and Cryptosporidium spp. in wild mesocarnivores in Spain. Vet. Parasitol. 2017;235:86–93. doi: 10.1016/j.vetpar.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Van Hemert C., Ballweber L.R., Sinnett D.R., Atwood T.C., Fischbach A., Gustine D.D., Pabilonia K.L. Giardia and Cryptosporidium in resident wildlife species in Arctic Alaska. Food Waterborne Parasitol. 2023;32 doi: 10.1016/j.fawpar.2023.e00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacha R.E., Clark G.W., Williams E.A., Carter A.M., Scheffelmaier J.J., Debusschere P. Small rodents and other mammals associated with mountain meadows as reservoirs of Giardia spp. and Campylobacter spp. Appl. Environ. Microbiol. 1987;53:1574–1579. doi: 10.1128/aem.53.7.1574-1579.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck R., Sprong H., Lucinger S., Pozio E., Cacciò S.M. A large survey of Croatian wild mammals for Giardia duodenalis reveals a low prevalence and limited zoonotic potential. Vector-Borne Zoonotic Dis. 2011;11:1049–1055. doi: 10.1089/vbz.2010.0113. [DOI] [PubMed] [Google Scholar]
- 40.Calero-Bernal R., Santín M., Maloney J.G., Martín-Pérez M., Habela M.A., Fernández-García J.L., Figueiredo A., Nájera F., Palacios M.J., Mateo M., Balseiro A., Barral M., Lima-Barberoi J.F., Köster P.C., Carmena D. Blastocystis sp. subtype diversity in wild carnivore species from Spain. J. Eukaryot. Microbiol. 2020;67:273–278. doi: 10.1111/jeu.12772. [DOI] [PubMed] [Google Scholar]
- 41.M. Santín, R. Calero-Bernal, D. Carmena, M. Mateo, A. Balseiro, M. Barral, J.F. Lima Barbero, M.A. Habela. Molecular characterization of Enterocytozoon bieneusi in wild carnivores in Spain. J. Eukaryot. Microbiol. 65 (2018) 468-474, doi: 10.1111/jeu.12492. [DOI] [PubMed]
- 42.Ravaszova P., Halanova M., Goldova M., Valencakova A., Malcekova B., Hurníková Z., Halan M. Occurrence of Cryptosporidium spp. in red foxes and brown bear in the Slovak Republic. Parasitol. Res. 2012;110:469–471. doi: 10.1007/s00436-011-2523-0. [DOI] [PubMed] [Google Scholar]
- 43.Kvac M., Myskova E., Holubova N., Kellnerova K., Kicia M., Rajsky D., McEvoy J., Feng Y., Hanzal V., Sak B. Occurrence and genetic diversity of Cryptosporidium spp. in wild foxes, wolves, jackals, and bears in Central Europe. Folia Parasitol. 2021;68 doi: 10.14411/fp.2021.002. [DOI] [PubMed] [Google Scholar]
- 44.Wang S.-N., Sun Y., Zhou H.-H., Lu G., Qi M., Liu W.-S., Zhao W. Prevalence and genotypic identification of Cryptosporidium spp. and Enterocytozoon bieneusi in captive Asiatic black bears (Ursus thibetanus) in Heilongjiang and Fujian provinces of China. BMC Vet. Res. 2020;16:84. doi: 10.1186/s12917-020-02292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y.-L., Zhao N., Yang Y., Li Y., Zhang X., Chen J., Peng X., Zhao W. Molecular identification and subtype analysis of Blastocystis in captive Asiatic black bears (Ursus thibetanus) in China’s Heilongjiang and Fujian provinces. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.993312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mentaberre G., Porrero M.C., Navarro-Gonzalez N., Serrano E., Domínguez L., Lavín S. Cattle drive Salmonella infection in the wildlife-livestock interface. Zoonoses Public Health. 2013;60:510–518. doi: 10.1111/zph.12028. [DOI] [PubMed] [Google Scholar]
- 47.Ramos C.P., Xavier R.G.C., Leal C.A.G., Facury Filho E.J., de Carvalho A.U., Viegas F.M., Pires I.H., Lopes E.O., Lobato F.C.F., Silva R.O.S. Antimicrobial susceptibility and molecular characterization of Salmonella serovar Ndolo isolated from outbreaks in cattle and horses. Ciência Rural. 2018;48 doi: 10.1590/0103-8478cr20180688. [DOI] [Google Scholar]
- 48.Dunbar M., Wooding J.B., Thomas L.A. Salmonella Hartford infection in a Florida black bear (Ursus americanus floridanus) Florida Sci. 1995;58:252–254. https://www.jstor.org/stable/24320783 [Google Scholar]
- 49.Millan J., Aduriz G., Moreno B., Juste R.A., Barral M. Salmonella isolates from wild birds and mammals in the Basque Country (Spain) OIE Rev. Sci. Tech. 2004;23:905–911. doi: 10.20506/rst.23.3.1529. [DOI] [PubMed] [Google Scholar]
- 50.VinodhKumar O.R., Karikalan M., Ilayaraja S., Sha A.A., Singh B.R., Sinha D.K., Chandra Mohan S., Pruthvishree B.S., Pawde A.M., Sharma A.K. Multi-drug resistant (MDR), extended spectrum beta-lactamase (ESBL) producing and carbapenem resistant Escherichia coli in rescued sloth bears (Melursus ursinus), India. Vet. Res. Commun. 2021;45:163–170. doi: 10.1007/s11259-021-09794-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Shared pathogen detection procedures.
Additional file 4. Model selection for Canine Adenovirus type 1 (CAdV-1). Details of model selection for the predicted positivity to CAdV-1 in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 5. Model selection for Giardia spp. Details of model selection for the predicted positivity to Giardia spp. in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold and final model is highlighted in red. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 6. Model selection for Salmonella spp. Details of model selection for the predicted positivity to Salmonella spp. in fecal samples of Cantabrian brown bears based on dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 7. Model selection for pathogen richness. Details of model selection for the pathogen richness detected in fecal samples of Cantabrian brown bear and the elevation, dispersion of human population (Demangeon's index) and coverage of agricultural-periurban habitat (%) in a 6 km-buffer. The estimate of each variable in the model is included. Best model is highlighted in bold font. K means the number of variables in the model; cAIC corrected Akaike's Information Criterion; Δi difference of cAIC with respect to the best model; and wi Akaike weight.
Additional file 8. Model selection for Escherichia coli. Details of model selection for the predicted positivity to E. coli in sponge samples from trees based on dispersion of human population (Demangeon's index), coverage of agricultural-periurban habitat (%) in a 6 km-buffer, and type of tree (marked – bear rubbed- or unmarked -control-). The estimate of each variable in the model is included. Best model is highlighted in bold. K indicates the number of variables in the model; AICc corrected Akaike's Information Criterion; Δi difference of AICc with respect to the best model; and wi Akaike weight.
Additional file 9. Detection rate (%) of different pathogens from 73 fecal samples of Cantabrian brown bears (Ursus arctos).
Additional file 10. Raw data of feces and tree-sponge sampling in Cantabrian brown bear (Ursus arctos).
Data Availability Statement
Data will be made available on request.