Abstract
The first step in the replication of the plus-stranded poliovirus RNA is the synthesis of a complementary minus strand. This process is initiated by the covalent attachment of UMP to the terminal protein VPg, yielding VPgpU and VPgpUpU. We have previously shown that these products can be made in vitro in a reaction that requires only synthetic VPg, UTP, poly(A), purified poliovirus RNA polymerase 3Dpol, and Mg2+ (A. V. Paul, J. H. van Boom, D. Filippov, and E. Wimmer, Nature 393:280–284, 1998). Since such a poly(A)-dependent process cannot confer sufficient specificity to poliovirus RNA replication, we have developed a new assay to search for a viral RNA template in conjunction with viral or cellular factors that could provide this function. We have now discovered a small RNA hairpin in the coding region of protein 2C as the site in PV1(M) RNA that is used as the primary template for the in vitro uridylylation of VPg. This hairpin has recently been described in poliovirus RNA as being an essential structure for the initiation of minus strand RNA synthesis (I. Goodfellow, Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans, J. Virol. 74:4590–4600, 2000). The uridylylation reaction either with transcripts of cre(2C) RNA or with full-length PV1(M) RNA as the template is strongly stimulated by the addition of purified viral protein 3CDpro. Deletion of the cre(2C) RNA sequences from minigenomes eliminates their ability to serve as template in the reaction. A similar signal in the coding region of VP1 in HRV14 RNA (K. L. McKnight and S. M. Lemon, RNA 4:1569–1584, 1998) and the poliovirus cre(2C) can be functionally exchanged in the assay. The mechanism by which the VPgpUpU precursor, made specifically on the cre(2C) template, might be transferred to the site where it serves as primer for poliovirus RNA synthesis, remains to be determined.
The Picornaviridae family of plus-strand RNA viruses includes a large number of pathogens with widely different host range and disease symptoms (38). At the same time, picornaviruses show a strong similarity in their gene organization and in the mechanism by which they replicate their genomes (58). An unusual feature of their genomes is the presence of a small protein VPg, covalently linked to the 5′ end of the RNA. Virus replication in the infected host cell is a two-step process, carried out primarily by the viral RNA polymerase, in conjunction with other viral and possibly also cellular proteins. It takes place in small vesicles that are derived from the host's cellular membranes and with which the nonstructural proteins of the virus are associated. First, the incoming viral RNA is transcribed into complementary minus strands which are then used as templates for the synthesis of the progeny plus strands. Although the basic steps of replication are well known, very little is understood about the details of these processes and in particular about the exact functions of the cis-acting RNA structures contained within picornaviral RNAs (1). One of the important unanswered questions about minus-strand synthesis is how the viral RNA polymerase recognizes and selects only its own RNA as template among the many polyadenylated mRNAs that are present in the host cell (45).
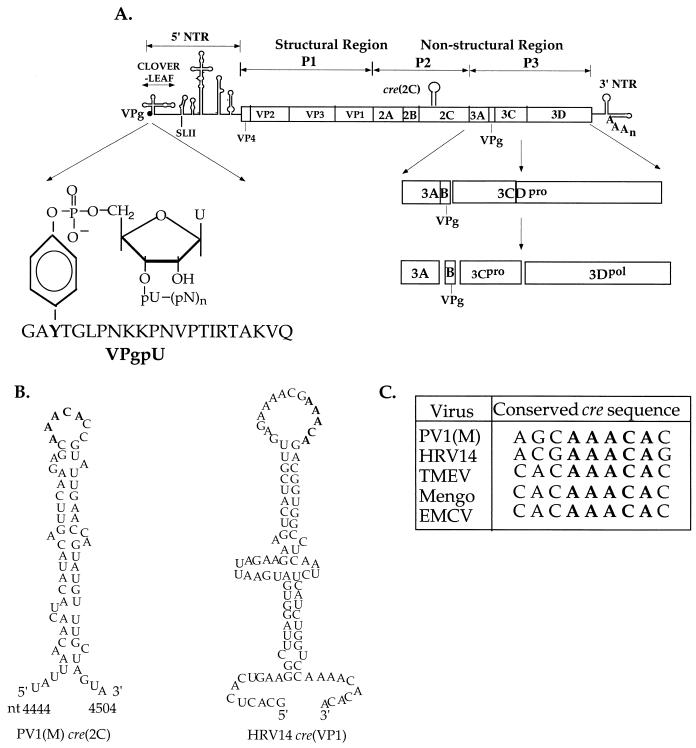
Poliovirus is perhaps the best known member of the Picornaviridae. Its RNA genome of about 7,500 nucleotides (nt) is composed of a long 5′-nontranslated region (NTR), a single open reading frame, a short 3′NTR, and a poly(A) tail (Fig. 1A) (32). The 5′-terminal UMP of the viral RNA is linked to the hydroxyl group of VPg by a phosphodiester bond (Fig. 1A) (2, 56). The 5′NTR consists of two independent domains. The first is a cloverleaf-like structure which is involved both in plus-strand RNA synthesis (3, 4, 24, 70) and in the process of switching from translation to replication (18). The second is a large and complex structure, the internal ribosomal entry site (IRES) (28, 47), which promotes translation of a polyprotein. This polyprotein (Fig. 1A) contains a capsid region (P1) and two nonstructural domains (P2 and P3) (32). The initial cleavage of the polyprotein is carried out by proteinase 2Apro at the P1/P2 site (64). Most other cleavages are mediated by the activities of proteinase 3Cpro and its precursor, 3CDpro (22, 23, 29, 73). The proteins of the P2 domain are predominantly involved in inducing the biochemical and structural changes that occur in the infected cell (8), but 2CATPase is also essential for viral genome replication (49). Those of the P3 region are the ones most directly involved with the process of RNA synthesis. These include the important viral RNA polymerase 3Dpol, which is both primer and template dependent (16). The other members of the P3 domain (Fig. 1A; reviewed in reference 68) are a small membrane-bound protein 3A, the 22-amino-acid terminal protein VPg, proteinase 3Cpro, and two multifunctional precursors 3AB and 3CDpro (3, 4, 24, 69, 70). The binding to the 5′ cloverleaf of 3CDpro, either in a complex with 3AB (24, 70) or with the cellular protein PCBP2 (3, 4, 10, 17, 43), has an important role in plus-strand RNA synthesis. The viral genome is terminated with a poly(A) tail (72) that is attached to a stem-loop structure of the 3′NTR (Fig. 1A). The 3′NTR, whose exact function is not yet known, is important (1, 37, 50–52, 71), but it appears that it is not essential (1, 57, 63) for RNA replication.
FIG. 1.
(A) Structure of poliovirus genomic RNA. The single-stranded RNA genome of poliovirus is shown with the terminal protein VPg (3B) at the 5′ end of the NTR (single line) and the 3′NTR (single line) with the poly(A) tail. The 5′NTR consists of a small cloverleaf and a large IRES element. The location of the cre(2C) element and of SLII of the IRES are indicated. The attachment site of the 5′-terminal UMP of the RNA to the tyrosine of VPg is shown enlarged. The polyprotein contains structural (P1) and nonstructural (P2 and P3) domains. The boxed region containing vertical lines represents the polyprotein with the proteinase cleavage sites. Processing of the P3 domain by proteinases 3C and 3CDpro is shown enlarged. (B) Predicted secondary structures of the HRV14 cre(VP1) (35, 36) and the PV1(M) cre(2C) RNAs (19). Conserved sequences are shown in boldface. (C) Conserved cre sequences in the coding regions of picornaviral RNAs (19, 34–36).
It has been generally believed that the 5′- and 3′NTRs of picornaviral RNAs contain all the cis-acting replication (cre) signals required for RNA replication (1). All natural or synthetic subgenomic picornaviral RNAs that are replication competent contain these terminal structured RNA elements (4, 30, 35, 48). Subgenomic replicons derived from poliovirus or coxsackie B3 virus are fully replication competent even if the entire P1 region is deleted (4, 67). This, however, is not the case with some other members of the Picornaviridae. It was first shown in the case of human rhinovirus 14 (HRV14) that a small RNA hairpin, located in the coding sequences of its capsid protein VP1, is involved in RNA replication (35, 36). Recently the VP2 coding sequences of mengo virus and Theiler's virus (34) were also found to contain such cis-acting RNA structures. In contrast to HRV14 and the cardioviruses, poliovirus contains a cis-replicating element not in the capsid region but in the coding region of protein 2C (19). This RNA hairpin, just like that of HRV14 (36) was found to be position independent for function and required for minus-strand RNA synthesis (19). The internally located cre elements of these different picornaviruses all consist of a relatively simple stem-loop structure but differ in location and in nucleotide sequences (Fig. 1B) (19, 34–36).
An important, but poorly understood, feature of picornaviral RNA replication is that RNA transcription is confined to the viral template and does not involve cellular mRNAs. The conserved RNA structural features in the 3′NTR were predicted to posses all the necessary recognition signals for the RNA polymerase to recognize its specific template and initiate minus-strand synthesis (1, 71). The 3′NTR preceding the poly(A) tract was therefore designated as the origin of replication (oriR) for minus-strand synthesis (51). This proposal was based on numerous genetic studies indicating the importance of the RNA structure that is maintained by “kissing” interactions (37, 50, 51). This, however, has been difficult to reconcile with subsequent genetic and biochemical data. Recently, it was shown that the exchange of the 3′NTR between poliovirus and HRV14 or CBV4 yielded virus growing with nearly wild-type (wt) kinetics (57). Even more unexpected was the finding that the 3′NTR of poliovirus can be deleted without loss of viral viability, although the deleted genome is greatly debilitated with respect to replication (63). In order to explain these conflicting results, Todd et al. (63) have proposed that the primary determinant for template selection might be the localization of viral RNAs on membrane-bound complexes leading to a high local concentration of the replication proteins and the physical proximity of the P2 or P3 gene products to the 3′NTR following translation. Alternatively, the 3′NTRs of poliovirus, CBV4, and HRV14 may all mimic a common structural signal, as yet undefined, promoting the formation of a replication complex (1, 71).
The first step in minus-strand RNA synthesis, the covalent attachment of UMP to the tyrosine residue of VPg, is expected to occur immediately following selection of the polioviral RNA template. We have previously shown that this reaction can be studied in vitro with a simple assay that requires only purified 3Dpol, synthetic VPg, a poly(A) template, UTP, and Mg2+ (45). The products are VPgpU and VPgpUpU, the precursors of VPg-linked poly(U), the 5′ end of minus-strand RNA. This reaction is likely to represent the uridylylation of VPg in vivo, as it is strongly supported by genetic studies with variants of VPg and 3Dpol (A. V. Paul, J. Peters, J. Mugavero, J. H. van Boom, and E. Wimmer, manuscript in preparation). However, although uridylylation in vitro is strictly dependent on poly(A) serving presumably as template, the homopolymeric region can hardly function as the selecting cis-acting signal in vivo. Therefore, we changed the uridylylation assay such that poly(A) was replaced with poliovirus transcript RNA, which offered a direct way of testing the question of template selection. We have also searched for viral and cellular factors that might assist the RNA polymerase either in selecting its template or in the catalysis of the uridylylation reaction. We provide here evidence that in the presence of 3CDpro the primary template in PV1(M) RNA for the in vitro uridylylation of VPg is not the poly(A) tract but the cre(2C) element (19). The specificity of the reaction appears to be provided by the binding activity of 3CDpro to the cre(2C) RNA, either alone or complexed with 3Dpol/VPg. The cre(2C) RNA works in the in vitro reaction only in the context of the plus-strand sequence, suggesting that it is involved in minus-strand RNA synthesis. Mutagenesis of the cre(2C) element reduced or eliminated the ability of that RNA to serve as template in the in vitro reaction and interfered with viral growth but had no effect on the translation and processing of the polyprotein (55). We do not yet know how the VPg-linked precursors are transposed to the poly(A) tail where they might serve as primer for elongation during minus-strand synthesis.
MATERIALS AND METHODS
Construction of plasmids.
Poliovirus cDNA (pT7PVM) (12) nucleotide sequences listed for plasmids or oligonucleotides refer to the full-length (nt 1 to 7525) plus-strand poliovirus cDNA sequence. Mutated nucleotides are underlined in the oligonucleotide sequences.
(i) pET21b/3CD(3Cpro/H40A).
The coding sequences of poliovirus Sabin 2 3CDpro(H40A) (D. W. Kim and E. Wimmer, unpublished) was amplified by PCR using primers 5′-GGGAATTCGGGCCCTGGGTTTGATTAC-3′ (plus-strand sequence) and 5′-CCAAGCTTAAAAGAGTCGAGCCAACG-3′ (minus-strand sequence). The fragment was digested with EcoRI and HindIII and was ligated into the same sites of pET21b (Novagen).
(ii) pET21b/PCBP2.
The coding sequences of PCBP2 were amplified from plasmid pQE30-PCBP2 (gift of B. L. Semler) (43) by PCR using oligonucleotides 5′-CCGGATCCTATGGACACCGGTGTGATT-3′ (plus-strand sequence) and 5′-CGGAAGCTTGTGCTCACAAGAAAAAAGGCA-3′ (minus-strand sequence). The fragment was digested with BamHI and HindIII and was inserted into the BamHI/HindIII-restricted pET21b vector (Novagen).
(iii) pProEX HTb/3CD(3Cpro/H40A).
The coding sequences of poliovirus Sabin 2 3CDpro (3Cpro/H40A) was amplified from plasmid pET21b/3CD(3Cpro/H40A) with oligonucleotides 5′-GGGGAATTCAAGGCCCTGGGTTTGATTACGCAGTGGC-3′ (plus-strand sequence) and 5′-GGGCTGCAGTAAAATGAGTCAAGCCAACGGCGG-3′ (minus-strand sequence). The resulting fragment was cut with PstI and BamHI and was ligated into the same sites of pProEX HTb (Lifetech).
(iv) pPV1(M)cre(2C).
The T7 promoter and the coding sequences of protein 2C (nt 4444 to 4505) from plasmid pT7PVM (12) were inserted into the EcoRI/NaeI sites of plasmid pBR322 (55). The plasmid was digested with NheI or EcoRI and was transcribed with T7 RNA polymerase.
(v) pPV1(M)IRES/SLII.
The SLII (stem loop II) sequences (nt 121 to 165) of the PV1(M) IRES were amplified from plasmid pT7PVM (12) using oligonucleotides 5′-GGGGGATCCTAATACGACTCACTATAGGCAAGTTCAATAGAAGGGG-3′ (plus-strand sequence, also contains T7 promoter sequences) and 5′-CCCGAATTCCTTGTTCGTGGTGGTACTG-3′ (minus-strand sequence). The resulting fragment was cut with EcoRI and BamHI and was inserted into the EcoRI/BamHI sites of pBR322. The plasmid was digested with EcoRI and was transcribed with T7 RNA polymerase.
(vi) pBS/+CL.
The coding sequences of the plus cloverleaf (+CL), nt 1 to 106 of pT7PVM (12), was cloned into the SacI and XhoI sites of Bluescript II KS(+) (Stratagene). Plasmid pBS/+CL (T. Pfister and E. Wimmer, unpublished) was linearized with XhoI and transcribed by T7 RNA polymerase.
(vii) pBS/−CL.
This plasmid (Pfister and Wimmer, unpublished) contains a T7 promoter, and the minus-cloverleaf sequences, nt 1 to 96 of pT7PVM (12) in the SmaI and SacI sites of pUC19 (New England BioLabs). It was linearized with SacI and transcribed with T7 RNA polymerase.
(viii) pT7PV1(M)[no poly(A)].
This construct (K. S. Harris and E. Wimmer, unpublished) contained a SacI site immediately upstream of the poly(A) tail. The plasmid was linearized with SacI prior to transcription with T7 RNA polymerase. The transcript consisted of a full-length poliovirus RNA lacking only the poly(A) tail.
(ix) pT7PV1(M)[no 3′NTR-poly(A)].
This plasmid (Harris and Wimmer, unpublished) contained a SmaI site downstream of nt 7375. It was digested with SmaI and transcribed with T7 RNA polymerase to yield a full-length poliovirus transcript RNA without the 3′NTR and poly(A) sequences.
(x) Minigenomes I to III.
Two new XhoI sites were introduced into the full-length pT7PVM (12) clone at nt 750 and 6164 and an MluI site immediately following the poly(A) tail (Kim and Wimmer, unpublished). Following digestion of the mutant plasmid with XhoI the fragment from nt 750 to 6164 was deleted and replaced by the cre(2C) fragment (nt 4444 to 4505) of pT7PVM (12) which contained XhoI linkers at both ends. Insertion of cre (2C) was either in the forward [cre(2C)F] or reverse direction [cre(2C)R]. Minigenome II lacked the cre(2C) sequences. The plasmids were linearized with MluI and were treated with mung bean nuclease (New England BioLabs) before transcription with T7 RNA polymerase.
(xi) pT7PVM (3CproR84S/I86A).
Two mutations were introduced into the 3Cpro domain of pT7PVM using PCR mutagenesis. The first PCR fragment was made with oligonucleotides a (5′-GAGAAATGAAAAGTTCAGCGACGCTAGACCACATATACCTACTCAA ATCAC 3′; plus-strand sequence, nt 5671 to 5721) and b (5′-GGCCCTTTCGTCTTCAAGAATTCCGTT 3′; minus-strand sequence, nt 7520 to 7546). The second fragment was made with oligonucleotides c (5′-GGGTTGGATAGTCAACATCACCAGCC-3′; plus-strand sequence, nt 5230 to 5255) and d (5′-GAGTAGGTATATGTGGTCTAGCGTCGCTGAACTTTTCATTTCTCTTTAGAGTG-3′; minus-strand sequence, nt 5662 to 5714). The two fragments were mixed and used as template in the second PCR step with oligonucleotides b and c. The resulting fragment was digested with EcoRI and BglII and was ligated into similarly cut pT7PVM.
Enzymes.
Poliovirus RNA polymerase was expressed in Escherichia coli BL21(DE3)pLysS from plasmid pT5T-3D which contains the wt 3D coding sequences, preceded by a methionine, and was purified by the method of Pata et al. (44). Poliovirus 3CDpro (3Cpro H40A), containing either an N-terminal or a C-terminal His tag was expressed in E. coli from plasmids pET21b/3CD(3CproH40A) and pProEX HTb/3CD(3Cpro/H40A), respectively. PCBP2 with C-terminal His tag was expressed in E. coli from plasmid pET21b/PCBP2. The proteins were purified by nickel column chromatography (Qiagen). Purified proteins 2C (49) and 2BC (Pfister and Wimmer, unpublished) were a gift of T. Pfister. Purified poly(A) binding protein was a gift of R. Rhoads, and 3AB was purified as described before (33). The plasmid for the expression of His-tagged PCBP2 (43) was a kind gift of Bert Semler.
Assay of RNA polymerase 3Dpol.
The synthesis of VPgpU(pU) was measured by an assay similar to what was described before (45). The reaction mixture (20 μl) contained 50 mM HEPES (pH 7.5), 8% glycerol, 3.5 mM magnesium acetate, 0.5 μg of poly(A) or transcript RNA, 2 μg of synthetic poliovirus VPg (14), 1 μg of purified poliovirus polymerase 3Dpol, 0.75 μCi of [α-32P]UTP (3,000 Ci/mmol; DuPont-NEN), and 10 μM unlabeled UTP. Where indicated, 0.3 to 0.5 μg of purified 3CDpro(3CproH40A), with either N-terminal or C-terminal His tag, was added to the reaction mixtures. After 1 h of incubation at 34°C, the reaction was stopped by the addition of 5 μl of gel loading buffer. The samples were analyzed by Tris-tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad) with 13.5% polyacrylamide. Gels were dried and autoradiographed, and the reaction products were quantitated with a PhosphorImager (Molecular Dynamics Storm 860) by measuring the amount of [32P]UMP incorporated. VPgpU(pU) refers to the sum of VPgpU and VPgpUpU.
In vitro transcription and translation.
Plasmid DNAs were linearized with EcoRI and were transcribed by using T7 RNA polymerase (66). The transcript RNAs were purified by phenol-chloroform extraction and ethanol precipitation. They were translated at 34°C in HeLa cell extracts (40).
RESULTS
We have considered two possible models by which specificity might be provided in vivo to the reaction in which VPg is uridylylated. According to the first model, poly(A) would be the template and the adjacent the 3′NTR would provide the specific viral sequences needed for recognition by the RNA polymerase and/or other replication proteins (45). Alternatively, a poliovirus-specific RNA sequence or structure located somewhere else in the genome could serve both as the site of recognition for the RNA polymerase and the template for the uridylylation reaction. These models were based on the example of other viral polymerases that catalyze the covalent linkage of a nucleotide to a protein (see Fig. 9) (59). In order to distinguish between these two models we have extended our studies to include RNA templates derived from PV1(M) sequences and to search for viral and/or cellular factors that might aid the RNA polymerase either in the template selection or in the VPg-uridylylation reaction.
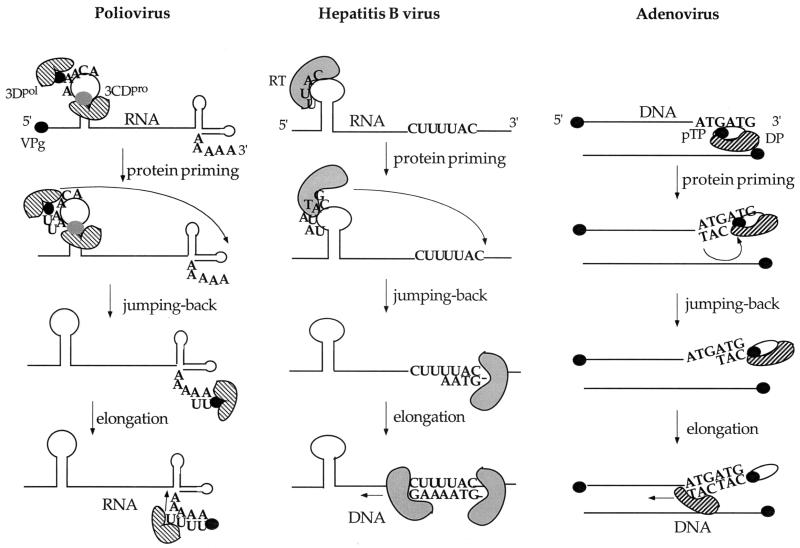
FIG. 9.
Proposed model for poliovirus RNA minus-strand RNA synthesis compared to that of hepatitis B virus and adenovirus. Poliovirus minus-strand synthesis might resemble that used by hepatitis B virus (60) and adenovirus (25). Both DNA and RNA synthesis takes place in three consecutive steps: protein priming, jumping back, and elongation. Poliovirus 3Dpol complexed with VPg and 3CDpro binds to the cre(2C) RNA (19). RNA polymerase 3Dpol catalyzes the covalent linkage of UMP to VPg using the first two A's of the conserved AAACA sequence of the cre(2C) RNA as a template. The complex of 3Dpol and VPgpUpU is released and translocated to the 3′ end of the poly(A) tail. The precursor VPgpUpU then serves as primer for minus-strand RNA synthesis. Abbreviations: RT, reverse transcriptase; pTP, preterminal protein; DP, DNA polymerase.
Uridylylation of VPg on templates derived from PV1(M) RNA.
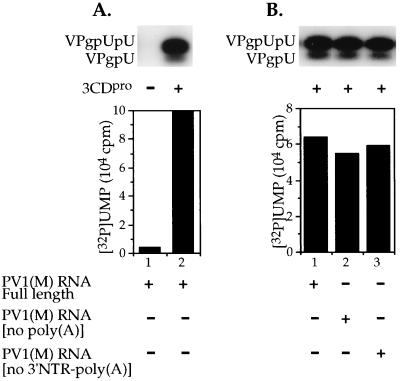
In our preliminary experiments with full-length PV1(M) transcript RNA as template in the 3Dpol-catalyzed VPg-uridylylation reaction we observed a strong stimulation by a preparation of purified 3CDpro (Fig. 2A, compare lane 1 with lane 2). As we will show later, this is in strong contrast to the basic VPg-uridylylation reaction in which synthetic poly(A) serves as the template for 3Dpol (see Fig. 5, compare lane 1 with lane 2). These results suggested that when full-length PV1(M) RNA is the template, a poliovirus-specific RNA sequence or structure is recognized by 3CDpro and/or 3Dpol/VPg.
FIG. 2.
Uridylylation of VPg in vitro on full-length or truncated PV1(M) transcript RNA templates. The synthesis of VPgpU(pU) was measured as described in Materials and Methods, except that the UTP concentration was 0.1 μM and the poly(A) template was replaced with 1.5 μg of PV1(M) transcript RNA, either full length or truncated. Where indicated, 3CDpro was added to the reaction mixtures. (A) Stimulation by 3CDpro. (B) Full-length and truncated PV1(M) transcript RNAs as templates.
FIG. 5.
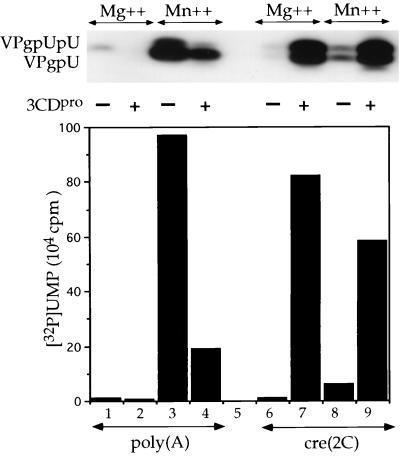
Comparison of poly(A) and cre(2C) RNA as templates in the in vitro uridylylation of VPg. Synthesis of VPgpU(pU) was measured as described in Materials and Methods with either Mg2+ or Mn2+, and the template was varied, as indicated. Some samples contained 3CDpro, as indicated. The top of the figure shows the autoradiography of the reaction products, and the bottom portion displays the quantitation of the data.
The most likely candidate for a function providing specificity appeared to be the 3′NTR, which is not only located adjacent to the poly(A) tail but is also known to bind 3CDpro in the presence of 3AB (24), the precursor of VPg. In this case a deletion of the poly(A) tail and the 3′NTR of PV1(M) RNA would be expected to eliminate the synthesis of VPg-linked precursors. The results of such an experiment, however, have shown that truncation of PV1(M) RNA either by the removal of the poly(A) tract or of the 3′NTR-poly(A) causes only a small reduction in the yield of uridylylated products (Fig. 2B, compare lane 1 with lanes 2 and 3). These data do not support the first model, that the poly(A) tail of PV1(M) RNA is the template for the uridylylation reaction and that the specificity of recognition is provided by the 3′NTR.
Next, we proceeded to test the second model and we searched for a different RNA sequence or structure in PV1(M) RNA that would contain both a specific signal for recognition by 3CDpro and/or 3Dpol/VPg and could also serve as a template in the uridylylation reaction. The only known feature of such a specific RNA template was the presence of at least two adjacent AMP residues on which VPgpU and VPgpUpU could be synthesized. We made transcripts of those RNA structures of PV1(M) RNA which are either known to be, or are likely to be, involved in RNA replication, and these were tested as templates in the reaction in the absence or presence of 3CDpro. As shown on Fig. 3, a stem-loop designated cre(2C) RNA (19) is not only far superior to all of the other RNAs tested but it also requires 3CDpro for optimal activity (compare lane 1 with lane 2) under the conditions of the experiment (see below). The level of uridylylation with 3′NTR-poly(A) (Fig. 3, lanes 3 and 4) with plus (lanes 7 and 8)- or minus (lanes 5 and 6)-strand cloverleaves as templates, is similar to what is obtained with tRNA (lanes 9 and 10), both in the absence and presence of 3CDpro. The same negative results were obtained when stem-loop II (SLII) of the PV1(M) IRES, recently identified as a domain important for RNA replication (27), was tested as template in the reaction (Fig. 3B, lanes 3 and 4).
FIG. 3.
Polioviral cis-replicating RNAs as templates in the in vitro uridylylation of VPg. Synthesis of VPgpU(pU) was measured as described in Materials and Methods, except that poly(A) was replaced by a transcript or tRNA (0.5 μg) template. Where indicated, the reactions contained 3CDpro. The top of the figure shows the autoradiography of the reaction products, and the bottom portion displays the quantitation of the data. (A) Comparison of cre(2C) RNA with 3′NTR-poly(A), −cloverleaf and +cloverleaf RNAs, and tRNA. (B) Comparison of cre(2C) RNA with the IRES SLII RNA.
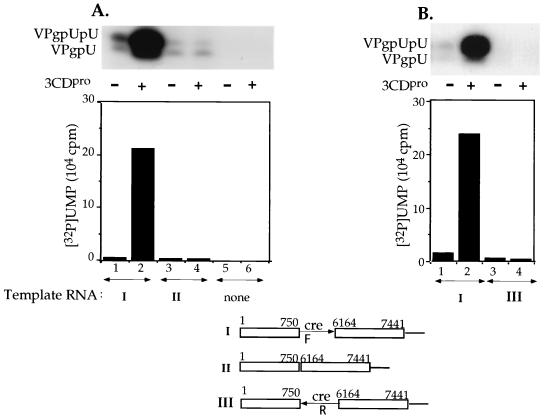
To provide additional proof that in poliovirus RNA the cre(2C) element is the primary template for the uridylylation reaction, we tested some minigenomes. The wt minigenome contained the 5′NTR sequences (nt 1 to 750), the cre(2C) sequences, the 3′-terminal portion of the 3Dpol coding region (nt 6164 to 7369), the 3′NTR, and the poly(A) tail with no additional heteropolymeric sequences (Fig. 4A, construct I). Just as PV1(M) RNA, the wt minigenome RNA contains two potential sites as template for the uridylylation reaction: the cre(2C) element and the poly(A) tail. As shown on Fig. 4A, the wt minigenome is a relatively good template for the reaction (lane 1), and the reaction is strongly stimulated by 3CDpro (lane 2). In comparison, deletion of the cre(2C) element form the wt minigenome (II) greatly reduces the formation of uridylylated products regardless of the presence or absence of 3CDpro (Fig. 4A, lanes 3 and 4). In the absence of any template no product is detectable (Fig. 4A, lanes 5 and 6). In one of the minigenomes (Fig. 4B, construct III) the cre(2C) element was reversed, and this RNA had lost nearly all its ability to function in the reaction (compare lanes 1 and 2 with lanes 3 and 4). These results show not only that the cre(2C) element is essential but also that it is used unidirectionally and only the plus-strand sequence functions in the reaction.
FIG. 4.
Uridylylation of VPg on minigenomic RNA templates. Synthesis of VPgpU(pU) was measured as described in Materials and Methods with minigenomic transcript RNAs (1.0 μg) as templates. Where indicated, 3CDpro was added to the reaction mixtures. Minigenome RNAs (shown below) contained the cre(2C) element either in the forward (F) direction (I) or in the reverse (R) direction (III). The cre(2C) was deleted from RNA II. (A) The cre(2C) element is required for optimal template activity. (B) The reaction requires the plus strand sequence of the cre(2C) element. The top of the figure shows the autoradiography of the reaction products, and below it the quantitation of the data are displayed.
Comparison of poly(A) and cre(2C) RNA as templates for the in vitro uridylylation of VPg by 3Dpol.
The basic assay system, that we have previously described for VPg-uridylylation, used poly(A) as template and Mg2+ as cofactor of 3Dpol (45). The low yield of reaction products obtained under these conditions (45) could be much improved by replacing Mg2+ with Mn2+ (Fig. 5, lanes 1 and 3; A. V. Paul and E. Wimmer, unpublished data). The same effect has also been observed with other DNA polymerases that catalyze nucleotidylylation of proteins, such as the phage Φ29 (15), phage PRD1 (11), and adenovirus (54), or the reverse transcriptase of hepatitis B virus (65). As mentioned above, the reaction with a poly(A) template is not stimulated by 3CDpro, in the presence of either Mg2+ (Fig. 5, lanes 1 and 2) or Mn2+ (lanes 3 and 4). The partial inhibition of the reaction by the 3CDpro preparation (Fig. 5, lanes 1 and 2 and lanes 3 and 4) is most likely due to an inorganic component of the solution since a heated aliquot had the same effect (data not shown). With cre(2C) RNA as the template and Mg2+ as the cofactor of 3Dpol there was a >50-fold stimulation of the reaction by 3CDpro (Fig. 5, compare lane 6 with lane 7). The optimal yield of products with the two different templates was about the same (Fig. 5, lanes 3 and 7). Importantly, Mn2+ did not stimulate uridylylation with cre(2C) to the same extent as Mg2+ (Fig. 5, compare lane 7 with lane 9). Thus, the “manganese effect” all but disappeared when cre(2C) RNA is used as a template in the presence of 3CDpro.
Characterization of the VPg-uridylylation reaction on a cre(2C) template.
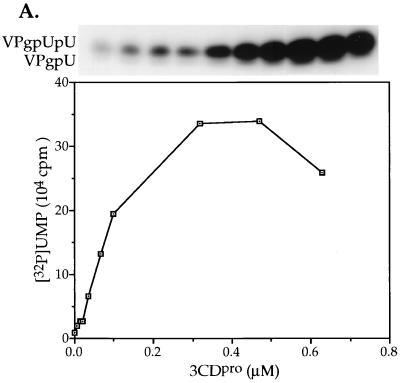
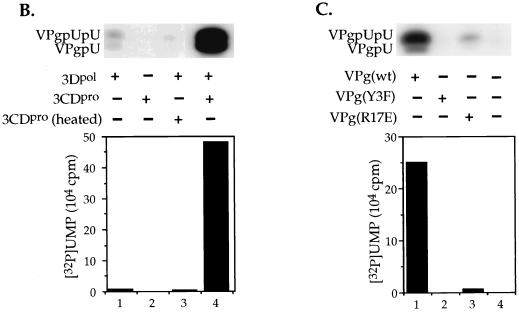
The optimal reaction conditions for the uridylylation of VPg on the cre(2C) RNA (data not shown) are essentially the same as what was found previously with the poly(A) template (45). The exception is the metal requirement and the stimulatory activity of 3CDpro (see above). The concentration of 3CDpro required for optimal stimulation of the reaction on a cre(2C) template was determined to be ca. 0.5 μM (Fig. 6A). This corresponds to a molar ratio of 3CDpro to 3Dpol of ca. 0.5:1. Since 3CDpro is the precursor of 3Dpol, we tested the possibility that this protein also possesses VPg-uridylylating activity. It can be seen from the data in Fig. 6B that there are no products formed when 3Dpol is omitted from the reaction (lane 2), an observation suggesting that 3CDpro only enhances the reaction catalyzed by 3Dpol. A heated aliquot of 3CDpro has no stimulatory activity, demonstrating that the protein itself is the active component of the enzyme preparation (Fig. 6B, lane 3). Other purified viral proteins, GST-linked 2C (49) or 2BC (Pfister and Wimmer, unpublished), 3AB (33), and the cellular proteins His-tagged PCBP2 (43), poly(A) binding protein (31), or uninfected HeLa cell extracts had no stimulatory activity in the reaction (data not shown).
FIG. 6.
Uridylylation of VPg in vitro on a cre(2C) template. The synthesis of VPgpU(pU) was measured as described in Materials and Methods, containing 0.5 μg of cre(2C) transcript RNA. (A) Optimal concentrations of 3CDpro required for stimulation. The amounts of 3CDpro added were varied, as shown in the figure. (B) 3CDpro stimulates the 3Dpol catalyzed reaction. Sample 1 contained no 3CDpro; in sample 2 3Dpol was omitted but 3CDpro was added, and sample 3 contained 3CDpro heated for 3 min at 70°C. (C) VPg specificity of the reaction. Where indicated, wt VPg was either omitted (lane 4) or replaced with a synthetic mutant VPg peptide (2 μg) that contained either the Y3F (lane 2) or the R17E (lane 3) amino acid change. All samples contained 3CDpro. The top of the figure shows the autoradiography of the reaction products and the bottom portion displays the quantitation of the data.
In order to assure that when using a cre(2C) template uridylylation of VPg occurs on the tyrosine of the peptide, we have tested the VPg specificity of the reaction. We have previously shown that in a reaction with a poly(A) template there are no uridylylated products formed when the VPg peptide contains either a Y3F or a R17E mutation or if VPg is omitted (45). The same results are obtained in the presence of the cre(2C) template (Fig. 6C, lanes 1 to 4), confirming the identical nature of the reaction on the two different templates.
Stimulation of VPg uridylylation by translation reactions of PV1(M) RNA.
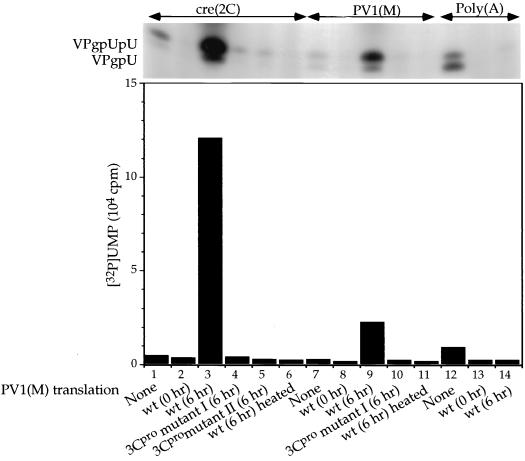
In our search of other viral or cellular proteins that might serve as cofactors and also enhance the uridylylation of VPg, we have tested the effect of translation reactions of PV1(M) RNA in HeLa cell free extracts. As shown on Fig. 7, using either a cre(2C) RNA (compare lane 1 with lane 3) or PV1(M) RNA (compare lane 7 with lane 9) as template, translation reactions (6 h at 34°C) have a strong stimulatory effect. The concentration of the full-length PV1(M) RNA is about 50-fold lower than that of the cre(2C) RNAs, so the yield of products obtained with these templates is not directly comparable. Unincubated translation mixtures (0 h) (Fig. 7, lanes 1 and 2 and lanes 7 and 8) have no effect on the reaction, an observation indicating that the function of a viral protein is involved in the stimulation. When using a poly(A) template, neither the 0-h nor the 6-h translation reactions exhibit a significant effect (Fig. 7, lanes 12 to 14). The similarity of these results to those presented above suggests that 3CDpro is the active component of the translation mixtures. However, we cannot rule out the possibility that other proteins are also involved in the process, in conjunction with 3CDpro.
FIG. 7.
Effect of translation reactions of wt and mutant 3Cpro(R84S/I86A) PV1(M) RNA on the VPg-uridylylation reaction. The synthesis of VPgpU(pU) was measured as described in Materials and Methods, except that the template was varied, as indicated. Translation reaction mixtures (2 μl) were added, as shown in the figure. PV1(M) wt or mutant 3Cpro(R84S/I86A) RNAs, derived from two independently isolated pT7PVM[3Cpro(R84S/I86A)] cDNAs, were translated in HeLa cell extracts (40) for either 0 or 6 h at 34°C, and an aliquot of each translation mixture was heated 3 min at 70°C. The following template RNAs were used: samples 1 to 6, cre(2C) RNA (0.5 μg); samples 7 to 11, PV1(M) transcript RNA (1.5 μg); samples 12 to 14, poly(A) (0.5 μg). The top of the figure shows the autoradiography of the reaction products, and the bottom portion displays the quantitation of the data.
The 3Cpro domain of poliovirus 3CDpro possesses both the proteinase and RNA binding functions of the protein (4, 20). Based on crystal structures recently solved for three picornavirus 3Cpro proteinases, HRV14, HAV, and PV1(M), the RNA binding domain is predicted to reside on the opposite side of the proteolytic active site (reference 41 and references therein). Several amino acids have been identified as being important for either processing or RNA binding activity (9, 20). To determine if the stimulatory effect of 3CDpro in the VPg-uridylylation assay is related to its the RNA binding activity, we tested translations of PV1(M) RNA transcripts that contained mutations affecting RNA binding. We have previously reported that amino acid substitutions R84S or I86A in 3Cpro, which lead to nonviable viral phenotypes, have no deleterious effect on polyprotein processing (20). Amino acid 84 was subsequently shown by both in vivo and in vitro studies to be required for RNA binding (4, 9). Polyprotein processing in translations reactions of PV1(M) transcript RNAs [3Cpro(R84S/I86A)] were found to be essentially the same as in that of wt PV1(M) RNA except for somewhat reduced autoprocessing of 3CDpro (data not shown). Mutant PV1(M) transcript RNAs [3Cpro(R84S/I86A)], made from two independently isolated clones of cDNA (I and II), were translated and were tested in our assay using a cre(2C) RNA or PV1(M) RNA template (Fig. 7). The results clearly indicate that the mutations in 3Cpro that abolish RNA binding also eliminate the ability of the translation reactions to stimulate the VPg-uridylylation reaction (Fig. 7, compare lane 1 with lanes 4 and 5 and lane 7 with lane 10). These results confirm our previous conclusion that 3CDpro is the active component that possesses the stimulatory activity and further show that it is the RNA binding activity of this protein that is important for its function.
The cre(VP1) of HRV14 functions as template for VPg uridylylation by poliovirus 3Dpol.
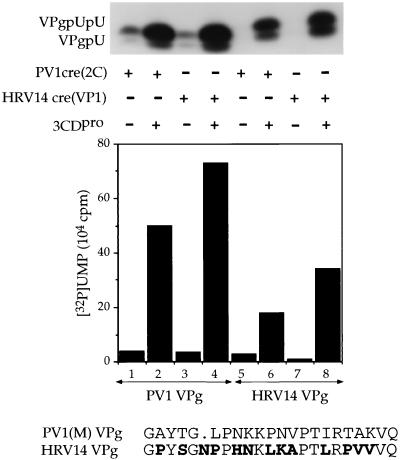
The cre(2C) element of poliovirus (19) is similar in structure to a small stem-loop RNA in the VP1 coding sequence of HRV14 which was previously described by McKnight et al. (35, 36). Although the function of these RNA structures could not be determined, McKnight, Goodfellow, and their coworkers could show a relation to minus-strand RNA synthesis (19, 36). Mutagenesis of the HRV14 structure has shown that the top of the stem and the loop are important for its function (36). We have noted that the loop segment of both the cre(VP1) of HRV14 and the cre(2C) of PV1(M) contain a conserved sequence of 5 nt (AAACA, Fig. 1C). Otherwise the stem portions of the two RNAs are quite different (Fig. 1B). Our results with mutant PV1(M) cre(2C) RNAs in the in vitro assay and also in vivo led to the conclusion that the first 2 A's in the conserved sequence of the loop and the integrity of the top of the stem in the hairpin are important for its function (55). Lobert et al. (34) have shown that for the replication of minigenomic RNAs the cre(VP2) element of Theiler's virus can be functionally replaced by that of Mengo virus [cre(VP2)] but not with that of HRV14 [cre(VP1)]. We were interested to test whether the HRV14 cre(VP1) RNA can replace the PV1(M) cre(2C) RNA in our in vitro assay. Our results show that the cre RNA elements of the two viruses are both good templates for 3Dpol in the uridylylation of PV1(M) VPg (Fig. 8, compare lane 2 with lane 4). Moreover, both reactions are stimulated by poliovirus 3CDpro (Fig. 8, lanes 1 and 2 and lanes 3 and 4). The yield of VPg-linked products obtained with the two cre RNAs is similar (Fig. 8, lanes 2 and 4). These results and our genetic studies (55) suggest that the first two A's of the conserved sequence in both viral cre RNAs are used by poliovirus 3Dpol as template for the synthesis of VPgpUpU. Whether the same sequence of the loop or the structure of the stem of the hairpin is involved in the recognition by 3CDpro and/or 3Dpol/VPg remains to be determined.
FIG. 8.
PV1(M) cre(2C) RNA is functionally exchangeable with the cre(VP1) RNA of HRV14 in the in vitro assay. The synthesis of VPgpU(pU) was measured as described in Materials and Methods, except that the template and VPg were varied, as indicated. The template was either PV1 cre(2C) RNA or HRV14 cre(VP1) RNA. Where indicated, PV1(M) VPg was replaced by HRV14 VPg (2 μg). Samples 2, 4, 6, and 8 contained 3CDpro. The top of the figure shows the autoradiography of the reaction products; below it the quantitation of the data is displayed. The amino acid sequences of PV1(M) and HRV14 VPg are shown below. Amino acid differences between PV1(M) VPg and that of HRV14 are indicated with boldface letters.
The finding that HRV14 VPg is also a good substrate for poliovirus 3Dpol in the uridylylation reaction (Fig. 8, lanes 5 to 8) with either the PV1(M) cre(2C) RNA (lanes 5 and 6) or the HRV14 cre VP1 RNA (lanes 7 and 8) is somewhat unexpected since the VPgs of the two viruses are quite different (Fig. 8). HRV14 VPg, with 23 amino acids, is one residue longer than that of PV1(M) (61). Of these, only 10 amino acids are identical in the two peptides. These include the tyrosine at position 3 and an arginine at position 18 [R17 in PV1(M)]. The in vitro results presented here are in good agreement with our finding that a poliovirus in which the VPg of PV1(M) is replaced by that of HRV14 is viable (Paul et al., in preparation).
DISCUSSION
The experiments reported here were undertaken to obtain further information about the mechanism by which 3Dpol selects its template for transcription. Recognition of specific viral RNA sequences is expected to precede the very first step in minus-strand RNA synthesis which is the uridylylation of VPg by the RNA polymerase. Our simple in vitro assay (45) that measured this biochemical reaction offered a direct way to study the question of specificity. We considered two different models by which specificity could be provided to the reaction in which VPg is uridylylated: first, that the RNA polymerase would be recruited to specific sequences or structures in the 3′NTR, which is spatially close to the poly(A) tail, where uridylylation of VPg would then occur; and second, that specific RNA sequences or structures located somewhere else in PV1(M) RNA could serve both as the recognition site and as the template for the RNA polymerase. Both models were suggested to us by studies of other nucleic acid polymerases that use protein priming for the replication of their genomes. The first model is exemplified by adenovirus, which possesses a linear double-stranded DNA genome and uses sequences near the end of its genome both as a recognition site and as a template to deoxynucleotidylylate the preterminal protein by its DNA polymerase (Fig. 9) (25, 59). Our second model was based on the example of the protein priming reaction carried out by the reverse transcriptase of hepatitis B virus (Fig. 9) (26). This enzyme binds to an internal RNA hairpin and uses it as a template for the synthesis of a short DNA oligonucleotide that is covalently linked to the polymerase molecule. The enzyme then translocates close to the 3′ end of the plus-strand RNA where the oligonucleotide hybridizes to a complementary stretch and is elongated into minus-strand DNA.
In order to distinguish between these possibilities, we have used full-length poliovirus transcript RNA as template for the uridylylation reaction and tested the effect of viral and cellular proteins on these reactions. Purified 3CDpro or translation reactions of poliovirus RNA in HeLa cell extracts strongly stimulated the reaction on full-length poliovirus RNA but not on a poly(A) template, an observation indicating the involvement of a specific viral RNA sequence in the reaction. This lead to the identification of the cre element in the coding region of (2C) (19), both as the primary site of recognition by 3CDpro and/or 3Dpol/VPg and as the template for the in vitro uridylylation of VPg. This conclusion is supported by several lines of evidence. Deletion of the poly(A) tail or of the 3′NTR-poly(A) from PV1(M) RNA resulted only in a small reduction in the ability of such transcripts to serve as templates both in the absence (data not shown) and in the presence of 3CDpro. Similarly, deletion of the cre(2C) sequences from minigenomes that still retain the 3′NTR-poly(A) tail are nearly totally inactive as templates. Finally, transcripts of the small cre(2C) RNA itself are excellent templates in the reaction that requires the RNA binding activity of 3CDpro. In agreement with our data, an essential interaction of 3CDpro with a viral factor of the P2-P3 domain has been recently predicted. These studies involved chimeric poliovirus replicons in which the 3CDpro domain of PV1(M) was replaced by that of coxsackievirus B3 (7).
The protein priming reactions in which the cre(2C) RNA or poly(A) serve as templates appear to be identical, except that the former reaction is greatly enhanced by the presence of 3CDpro, and Mg2+ is the best cofactor for the polymerase. Our data indicate that both of these in vitro assays can be used to test the reaction in which VPg is uridylylated in vivo. Genetic data with VPg or 3Dpol variants show a strong correlation between viral growth phenotype and yield of uridylylated products obtained in the in vitro reactions (Paul et al., in preparation). It should be noted that poliovirus 3Dpol is not unique among polymerases that are able to use in vitro either a homopolymer or their viral nucleic acid sequences as templates for the protein priming reaction (39). The exact function of 3CDpro in the reaction that uses a cre(2C) RNA template is not yet known. The data strongly suggest that the RNA binding activity of this viral proteinase is involved. A mutation in the 3Cpro(R84S) domain of 3CDpro was previously shown to eliminate the ability of the protein to bind to the 5′ cloverleaf (4, 9) but not affect the processing of the polyprotein (9, 20). Translation reactions of a PV1(M) RNA [3Cpro(R84S/I86A)] exhibited no stimulatory activity of the in vitro uridylylation reaction. The most likely function of 3CDpro is to enhance the binding of 3Dpol/VPg to the template either by complexing with the other two proteins or by stabilizing the structure of the cre RNA or both. In this respect the function of 3CDpro would resemble that of Mn2+ when poly(A) is used as template. The enhanced cofactor activity of Mn2+ over that of Mg2+ during elongation of oligonucleotide primers is known to be due to a reduction in the Km value for 3Dpol binding to primer/template (5). In any case, we describe a new function of 3CDpro: the selection of a specific RNA structure in the viral genome for uridylylation of VPg by 3Dpol to occur, thereby providing all-important specificity to this event.
The cre(VP1) of HRV14 (35, 36) is functionally exchangeable in the in vitro assay with the PV1(M) cre(2C) element, an observation suggesting that although these structures are located in different regions of the viral RNA, they do have identical in vivo functions. Both of these RNAs were shown to function only in the plus-strand sequence and to be required for minus-strand RNA synthesis (19, 36). Whether or not the VPgpUpU made on the cre(2C) template is also used for plus-strand synthesis remains to be determined. It is interesting that PV1(M) transcript RNAs lacking the two initial 5′UMP residues are not only infectious but, remarkably, the virus produced during infection has regained the authentic pUpU end (21). This implies that initiation of plus-strand synthesis is independent of the presence of the two terminal AMPs of the minus strand.
It should be emphasized that we cannot rule out the possibility that at some stage during the viral life cycle poly(A) itself also serves as template in the reaction, albeit only with low efficiency compared to the cre(2C) RNA. However, although in the presence of Mn2+ poly(A) is an efficient template for the uridylylation of VPg in vitro, the cre(2C) element would be superior to it in vivo for several reasons. First, by using a specific and internal viral RNA sequence/structure both as the site of recognition and as the template for VPgpU(pU) synthesis the possibility that the wrong poly(A)-tailed mRNA would be selected by 3Dpol for transcription would be reduced. Second, the presence of another viral protein, 3CDpro, during VPg-uridylylation would provide additional specificity to the reaction. Finally, the use of Mg2+, instead of Mn2+, as cofactor of 3Dpol in this reaction would enhance the overall specificity of minus RNA synthesis. In general Mn2+ is known to relax the specificity of nucleic acid polymerases toward their substrates and templates (5, 62; Paul and Wimmer, unpublished).
The internally located cre elements of picornaviruses thus far identified all consist of a small and relatively simple hairpin structure (19, 34–36). The only conserved sequence in these RNAs (19, 34–36) is a stretch of 5 nt (AAACA; Fig. 1C) found in the loop or bulge. Studies by McKnight et al. (35, 36), Lobert et al. (34), and us (55) have shown that the first A in the sequence is essential for viral viability. Consistent with the viral viability data is our observation that in the context of either the cre(2C) RNA or the PV1(M) RNA, the first A of the AAACA sequence is indispensable for the in vitro VPg-uridylylation reaction (55). Mutagenesis of the second A reduces the yield of uridylylated products, while a change in the third A has no significant effect (55).
The loop portion of SLII of the PV1(M) IRES contains a sequence (AAACCA) (27) that is very similar to the conserved sequence in the internal cre RNAs discussed above. Deletion of these nucleotides from SLII were found to result in a defective replication phenotype (27). Despite its strong similarity to the essential sequences in cre(2C) RNA, the SLII RNA is nearly inactive as template in the VPg-uridylylation reaction either in the absence or in the presence of 3CDpro. These results provide further proof that in the presence of 3CDpro the cre(2C) RNA is a unique structure in poliovirus RNA in that it serves as an efficient and specific template for the reaction.
Unfortunately, we have not yet been able to achieve efficient elongation reaction of the precursors either on a full-length PV1(M) template or on the minigenomes. The addition of purified viral proteins 2C, 2BC, and 3AB or cellular poly(A) binding protein had no effect on this process (data not shown). The same results were obtained when the reaction was supplemented with uninfected or poliovirus-infected HeLa cell extracts (data not shown). However, the small amount of polymeric product is difficult to detect under the conditions of our assay because there is always a relatively high background of such product made in a VPg-independent reaction. This is due to the terminal uridylyl transferase activity of the polymerase (53), to oligonucleotide priming by traces of degraded RNA, or to hairpin priming of nicked high-molecular-weight RNA. Our inability to obtain elongation in the in vitro reaction might be related to the fact that the mutant 3CDpro used in the assay is unable to undergo autoprocessing, possibly a prerequisite to the release of the 3Dpol-VPgpUpU complex from the cre(2C) RNA. Alternatively, this RNA element might have to be located on the same genome from which 3Dpol and/or 3CDpro were translated. This possibility is suggested by the finding that a mutation in the cre(2C) RNA cannot be complemented in trans (19, 55). It has been known for many years that in poliovirus translation and replication are coupled (1, 42, 68). Finally, our failure to achieve efficient elongation with VPgpUpU primer might be due to the lack of, or the insufficiency of, one or more cellular or viral factors that are required for complete minus-strand RNA synthesis. Barton and Flanegan (6) have concluded that in the in vitro translation-transcription system that synthesizes viable virus (40), the guanidine-sensitive ATPase activity of 2C (49), and a soluble HeLa cell factor are required for minus-strand RNA synthesis. The function of this putative cellular factor in RNA replication remains unknown.
It should be noted that free VPgpUpU has been found in the cytoplasm of poliovirus-infected cells (13). Thus, the precursor to elongation is clearly synthesized preceding and most likely independently of the elongation reaction. We do not know, however, what happens to VPgpUpU after it has been synthesized on the cre(2C) RNA. It is most likely that the precursors are translocated to the 3′ end of the poly(A) tail and then elongated into full-length minus-strand RNA (Fig. 9) in a mechanism similar to what is used by the reverse transcriptase of hepatitis B virus (26, 60) (Fig. 9) or the DNA polymerase of adenovirus (25, 59) (Fig. 9). All three of these protein-primed nucleic acid synthesis systems are characterized first by the abortive synthesis of a short oligonucleotide attached to the hydroxyl group of a tyrosine or serine of a viral protein (terminal or preterminal protein, or polymerase). The template for this reaction is located either on an internal hairpin structure (poliovirus and hepatitis B virus) or on a partially opened double-stranded structure near the end of the genome (adenovirus). The second step involves the translocation of the nucleotidylylated protein-polymerase complex back to or near the 3′ end of the template strand. This is followed by the elongation of the protein-linked oligonucleotide primer into minus strands by the polymerase. The last two steps take place only with low efficiency (reference 26 and see above) and probably involve a conformational change in the structure of the polymerase. The affinity of this enzyme for its template is expected to be relatively high for the initiation step but subsequently low so that it can be released and translocated and then move along its template during elongation of the primer. For optimal minus-strand synthesis adenovirus recruits three cellular proteins (NFI, NFII, and Oct I), and the viral DNA binding protein (25) and hepatitis B virus uses a chaperone complex of at least two proteins (hsp90 and p23) (26, 60). At this time one can only speculate as to how poliovirus achieves the translocation and elongation steps. One possibility is that the polymerase, with the help of additional protein(s), undergoes a structural change such as dissociation of oligomers (44) into monomers. Both the RNA binding (44) and uridylylating activity of 3Dpol (45) is enhanced under conditions that favor oligomerization of the protein, while the elongation step has no such requirement (46). Our in vitro assay that uses purified proteins to study the steps of minus-strand RNA synthesis offers a useful system for the identification of all the required components of this very complex process.
ACKNOWLEDGMENTS
We thank J. Mugavero and F. Maggiore for excellent technical assistance. We thank Karla Kirkegaard for the generous gift of plasmid pT5T-3D, Bert Semler for plasmid pQUE30-PCBP2, Thomas Pfister for plasmid pBS+CL and pBS−CL, and Kevin Harris for plasmid pT7PV1(M)[no poly(A)] and pT7PV1(M)[no 3′ NTR-poly(A)]. Purified poly(A) binding protein was a gift from R. Rhoads; purified 2C and 2BC were gifts from Thomas Pfister.
This work was supported by the National Institute of Allergy and Infectious Diseases of the NIH (5R37AI15122).
REFERENCES
- 1.Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–5266. [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′ end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 6.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell Y C, Semler B L, Ehrenfeld E. Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase. J Virol. 1999;73:9413–9421. doi: 10.1128/jvi.73.11.9413-9421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair W S, Parsley T B, Bogerd H P, Towner J S, Semler B L, Cullen B R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 10.Blyn L B, Swiderek K M, Richards O, Stahl D C, Semler B L, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: Identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA. 1996;93:1115–1120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldentey J, Blanco L, Savilahti H, Bamford D H, Salas M. In vitro replication of bacteriophage PRD1 DNA. Metal activation of protein-primed initiation and DNA elongation. Nucleic Acids Res. 1992;20:3971–3976. doi: 10.1093/nar/20.15.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X, Kuhn R J, Wimmer E. Replication of poliovirus RNA containing two VPg coding sequences leads to a specific deletion event. J Virol. 1993;67:5572–5578. doi: 10.1128/jvi.67.9.5572-5578.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford N M, Baltimore D. Genome-linked protein VPg of poliovirus is present as free VPg and VPgpUpU in poliovirus-infected cells. Proc Natl Acad Sci USA. 1983;80:7452–7455. doi: 10.1073/pnas.80.24.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreef-Tromp C M, van den Elst H, van den Boogaart J E, van der Marel G A, van Boom J H. Solid-phase synthesis of an RNA nucleopeptide fragment from the nucleoprotein of poliovirus. Nucleic Acids Res. 1992;20:2435–2439. doi: 10.1093/nar/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteban J A, Bernad A, Salas M, Blanco L. Metal activation of synthetic and degradative activities of Φ29 DNA polymerase, a model enzyme for protein-primed DNA replication. Biochemistry. 1992;31:350–359. doi: 10.1021/bi00117a006. [DOI] [PubMed] [Google Scholar]
- 16.Flanegan J B, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci USA. 1977;74:3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 18.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond J W, Barclay W, Evans D J. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerle T, Molla A, Wimmer E. Mutational analysis of the proposed FG loop of poliovirus proteinase 3C identifies amino acids that are necessary for 3CD cleavage and might be determinants of a function distinct from proteolytic activity. J Virol. 1992;66:6028–6034. doi: 10.1128/jvi.66.10.6028-6034.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmon S A, Richards O C, Summers D F, Ehrenfeld E. The 5′-terminal nucleotides of hepatitis A virus RNA, but not poliovirus RNA, are required for infectivity. J Virol. 1991;65:2757–2760. doi: 10.1128/jvi.65.5.2757-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris K S, Hellen C U T, Wimmer E. Proteolytic processing in the replication of picornaviruses. Semin Virol. 1990;1:323–333. [Google Scholar]
- 23.Harris K S, Reddigari S R, Nicklin M J H, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of the poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 25.Hay R T. Adenovirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 699–719. [Google Scholar]
- 26.Hu J, Seeger C. RNA signals that control DNA replication in hepadnaviruses. Semin Virol. 1997;8:205–211. [Google Scholar]
- 27.Ishii T, Shiroki K, Iwai A, Nomoto A. Identification of a new element for RNA replication within the internal ribosomal entry site of poliovirus RNA. J Gen Virol. 1999;80:917–920. doi: 10.1099/0022-1317-80-4-917. [DOI] [PubMed] [Google Scholar]
- 28.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jore J, De Geus B, Jackson R J, Pouwels P H, Enger-Valk B E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988;69:1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan G, Racaniello V R. Construction and characterization of poliovirus subgenomic replicons. J Virol. 1988;62:1687–1696. doi: 10.1128/jvi.62.5.1687-1696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Clevage of poly(A)-binding protein by coxackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 33.Lama J, Paul A V, Harris K S, Wimmer E. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- 34.Lobert P-E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci USA. 1999;96:11560–1165. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melchers W J G, Hoenderop J G J, Bruins Slot H J, Pleij C W A, Pilipenko E V, Agol V I. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 665–712. [Google Scholar]
- 39.Mendez J, Blanco L, Esteban J A, Bernad A, Salas M. Initiation of Φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding back mechanism for protein primed DNA replication. Proc Natl Acad Sci USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molla A, Paul A V, Wimmer E. Cell-free de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 41.Mosimann S C, Cherney M M, Sia S, Plotch S, James M N G. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 42.Novak J E, Kirkegaard K. Coupling between genome translation and replication in a RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 43.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 44.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 45.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 46.Paul A V, Mugavero J, Yin J, Hobson S, Schultz S, van Boom J H, Wimmer E. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology. 2000;272:72–84. doi: 10.1006/viro.2000.0354. [DOI] [PubMed] [Google Scholar]
- 47.Pelletier J, Sonnenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 48.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 50.Pilipenko E V, Maslova S V, Sinyakov A N, Agol V I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilipenko E V, Poperechny K V, Maslova S V, Melchers W J G, Bruins Slot H J, Agol V I. cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (kissing) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 52.Pierangeli A, Bucci M, Pagnotti P, Degener A M, Perez Bercoff R. Mutational analysis of the 3′-terminal extra-cistronic region of poliovirus RNA: secondary structure is not the only requirement for minus strand RNA replication. FEBS Lett. 1995;374:327–332. doi: 10.1016/0014-5793(95)01127-z. [DOI] [PubMed] [Google Scholar]
- 53.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pronk R, van Driel W, van der Vliet P C. Replication of adenovirus DNA in vitro is ATP-independent. FEBS Lett. 1994;337:33–38. doi: 10.1016/0014-5793(94)80624-1. [DOI] [PubMed] [Google Scholar]
- 55.Rieder E, Paul A V, Kim D W, van Boom J H, Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothberg P G, Harris T J R, Nomoto A, Wimmer E. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rueckert R R. Picornaviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 59.Salas M, Miller J T, Leis J, DePamhilis M L. Mechanisms of priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 60.Seeger C, Mason W S. Replication of the hepatitis virus genome, 815–831. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 61.Stanway G, Hughes P J, Mountford R C, Minor P D, Almond J W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabor S, Richardson C C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toyoda H, Nicklin M J H, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus proteins. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 65.Urban M, McMillan D J, Canning G, Newell A, Brown E, Mills J S, Jupp R. In vitro activity of hepatitis B virus polymerase: requirement for distinct metal ions and the viral epsilon stem loop. J Gen Virol. 1998;79:1121–1131. doi: 10.1099/0022-1317-79-5-1121. [DOI] [PubMed] [Google Scholar]
- 66.van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Kuppeveld F J M, Galama J M D, Zoll J, Melchers W J G. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B: a moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J Virol. 1995;69:7782–7790. doi: 10.1128/jvi.69.12.7782-7790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 69.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 70.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiang W, Paul A V, Wimmer E. RNA signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]
- 72.Yogo Y, Wimmer E. Polyadenylic acid at the 3′ terminus of poliovirus RNA. Proc Natl Acad Sci USA. 1972;69:1877–1882. doi: 10.1073/pnas.69.7.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]