Abstract
Apyrase is a class of enzyme that catalyzes the hydrolysis of nucleoside triphosphates/diphosphates (NTP/NDP), which widely involved in regulation of plant growth and stress responses. However, apyrase family genes in maize have not been identified, and their characteristics and functions are largely unknown. In this study, we identified 16 apyrases (named as ZmAPY1-ZmAPY16) in maize genome, and analyzed their phylogenetic relationships, gene structures, chromosomal distribution, upstream regulatory transcription factors and expression patterns. Analysis of the transcriptome database unveiled tissue-specific and abiotic stress-responsive expression of ZmAPY genes in maize. qPCR analysis further confirmed their responsiveness to drought, heat, and cold stresses. Association analyses indicated that variations of ZmAPY5 and ZmAPY16 may regulate maize agronomic traits and drought responses. Our findings shed light on the molecular characteristics and evolutionary history of maize apyrase genes, highlighting their roles in various biological processes and stress responses. This study forms a basis for further exploration of apyrase functions in maize.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-024-01474-9.
Keywords: Maize, Apyrase, Abiotic stress response, Association analysis, Metabolic
Introduction
Apyrase (APY) enzymes, classified as nucleoside triphosphate (NTP) diphosphohydrolases, belong to the superfamily of guanosine diphosphatase 1 (GDA1)-cluster of differentiation 39 (CD39) nucleoside phosphatase. These enzymes have the capability to remove the terminal phosphate from nucleoside triphosphates (NTPs) and nucleoside diphosphates (NDPs) but not from nucleoside monophosphates (NMPs). Based on their subcellular localization, APYs can be broadly categorized into ecto-apyrases and endo-apyrases (Hideaki et al. 1984; Tong et al. 1993; Thomas et al. 1999). ecto-apyrase are located on the cell surface, whereas endo-apyrase are usually located in the endoplasmic reticulum, Golgi and intracellular vesicles (Leal et al. 2005). Some ecto-apyrase possess transmembrane domains at their N- and C-terminals, which are crucial for correct protein folding, membrane targeting, cellular allocation and enzyme activity (Wu et al. 2005; Knowles 2011). The cellular ATP level serves a dual role in providing energy and regulating various cellular processes associated with responses to abiotic stress (Sun et al. 2012). Therefore, the maintenance of cellular ATP homeostasis, regulated in part by apyrases, is essential for preserving normal cell function.
Apyrases are evolutionarily highly conserved (Clark et al. 2014). Their presence in plants was initially identified in potatoes several decades ago (Hideaki et al. 1984). Subsequently, members of the APY family were discovered in the genomes of various plants, including potato (Handa and Guidotti 1996; Riewe et al. 2008), wheat (Liu et al. 2019), soybean (Day et al. 2000), Arabidopsis (Yang et al. 2013), cotton (Clark et al. 2010), rice (Chowdhury et al. 2023), and peanut (Sharif et al. 2023). Transcriptome analysis has revealed specific spatio-temporal expression patterns of APY genes and regulation of these genes by biological and abiotic stresses (Liu et al. 2019; Chowdhury et al. 2023; Clark et al. 2024), indicating potential roles in plant growth, development, and stress responses.
Further studies have elucidated the functions of APY genes in plant growth and development. Arabidopsis APY genes, APY1 and APY2, are predominantly expressed in rapidly growing tissues or those with high auxin levels. Suppression of their expression affects root and shoot growth (Wu et al. 2007). Additionally, APY1/APY2 also participate in the phytochrome-mediated signaling pathway that induces differential growth changes in etiolated seedling tissues (Weeraratne et al. 2022). Another member of the APY gene family, APY7, acts as a negative regulator of cell growth. APY7 modulates the growth-inhibiting effects of RALF1 (Rapid ALkalinization Factor), influencing cell wall architecture, composition, and alters the pH of the extracellular matrix (Gupta et al. 2024). Constitutive expression of pea ectoapyrase, psNTP9, in Arabidopsis and soybeans leads to a more extensive root system architecture (RSA) (Veerappa et al. 2019). Furthermore, the catalytic activity of GS52 ecto-apyrase is crucial for the early infection process of B. japonicum, nodule primordium development initiation, and subsequent nodule organogenesis in soybean (Govindarajulu et al. 2009).
In addition to participating in the regulation of plant growth and development, the APY gene also plays a crucial role in stress response (Clark et al. 2021). Overexpression of the pea APY gene psNTP9 significantly enhances drought resistance and field yield in soybean under both normal watering and drought conditions (Veerappa et al. 2019). Populus euphratica APY genes, PeAPY1 and PeAPY2, enhance drought tolerance by modulating stomatal aperture in Arabidopsis (Zhang et al. 2021b). Furthermore, overexpression of PeAPY2 improves cold resistance by modulating vesicular trafficking and extracellular ATP in Arabidopsis (Deng et al. 2015). Proteome analysis of soybean roots under waterlogging has demonstrated that APY responded to waterlogging stress in soybean (Alam et al. 2010). These findings underscore the multifaceted roles of the APY genes in plant stress responses, highlighting its potential as a key player in enhancing plant resilience to various environmental challenges.
Maize, a primary global food crop, faces significant threats from abiotic stresses. Identifying additional abiotic stress resistance genes and further understanding response system in maize remain crucial. While APY genes has been reported to be involved in various abiotic stresses in Arabidopsis, their presence and biological functions in maize have not been reported. This study aims to characterize APY genes and elucidate their roles. Here, 16 APY family members were identified in maize genome by phylogenetic analysis (Fig. 1; Table S1). APY family expansion patterns and genetic characteristics were analyzed comprehensively (Figs. 1 and 2; Table S2). In addition, potential upstream regulators of ZmAPYs were predicted (Fig. 4). Transcriptome analysis revealed tissue-specific and abiotic stress-responsive expression of ZmAPY genes in maize (Figs. 3 and 5). Association analysis suggested that variations in ZmAPY genes could impact maize agronomic traits and drought responses (Figs. 6 and 7). These findings establish a basis for further investigations into the functions of maize APY genes.
Fig. 1.
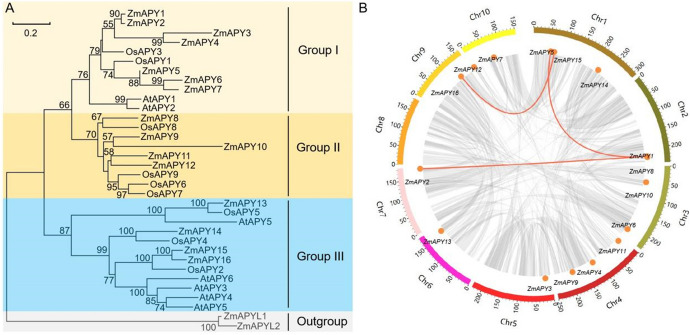
Phylogenetic analysis, chromosomal distribution and collinearity analysis of ZmAPY genes A Phylogenetic analysis of APYs from Z. mays, A. thaliana and O. sativa. A total number of 16 ZmAPYs from maize, 7 AtAPYs from Arabidopsis and 9 OsAPYs from rice were used to construct the phylogenetic tree. All APY transporter members were classified into four groups. Group I-III and outgroup are distinguished by different colors. B Chromosomal distribution and collinearity analysis of ZmAPY genes. The chromosomal location of each ZmAPY gene was mapped according to the maize genome. The chromosome number is indicated next to each chromosome. The syntenic ZmAPY gene pairs are connected by orange lines
Fig. 2.
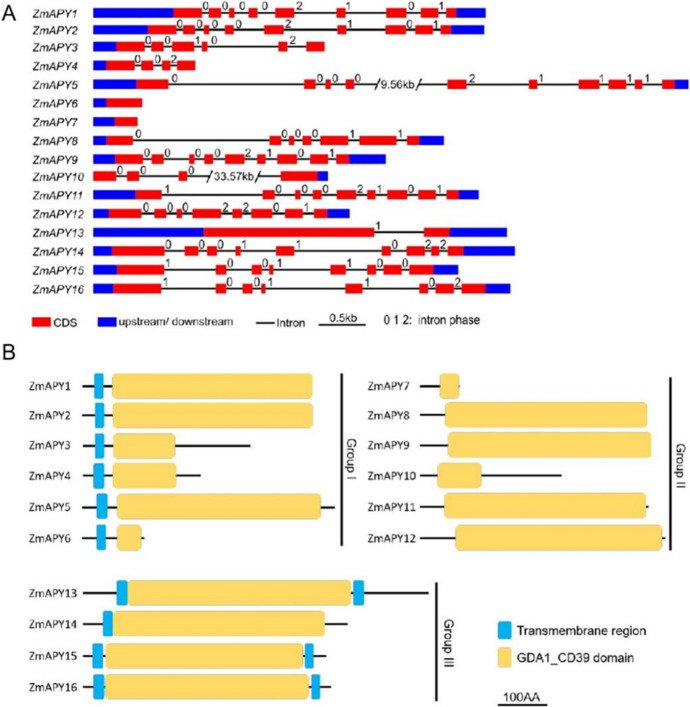
Exon–intron structure and conserved motifs of ZmAPY gene family A Exon–intron structures of ZmAPY genes. Red boxes represent exons (CDS), black lines represent introns, and blue boxes represent 5’ and 3’UTR regions. 0, 1 and 2: Intron phase, defined as the position of the intron within a codon, as introns are located either between codons (phase 0) or within codons (phase 1 and 2). B Predicted conserved protein domains of ZmAPY proteins. The black lines represent the length of each protein sequence, and the conserved protein kinase domains are depicted with colored boxes. blue boxes represent transmembrane region and yellow boxes represent GDA1_CD39 domain
Fig. 4.
Prediction of upstream regulators of APY genes A Upstream regulators of ZmAPYs were predicted by ChIP-seq data of 104 transcription factors and planttfdb software. Orange dots represent ZmAPYs and blue dots represent potential upstream regulators of ZmAPY genes. The red line connects the transcription factors and ZmAPY genes that may have regulatory relationship analyzed by ChIP-seq data, while the blue line connects the transcription factors and ZmAPY genes that may have regulatory relationship predicted by planttfdb software. B Correlation analysis between the expression levels of potential upstream regulators nacttf78, zim36, bzip79, cchh26 and ZmAPYs. The P values were calculated by test of correlation coefficient
Fig. 3.
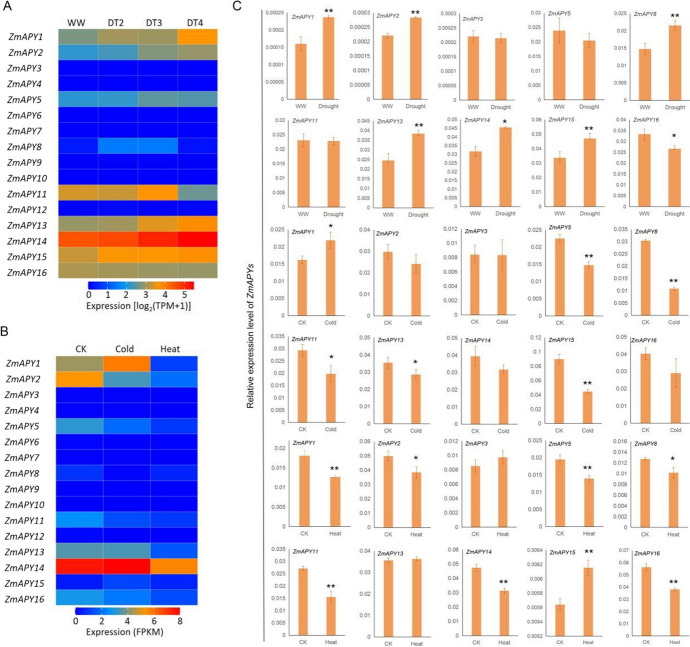
Expression analysis of ZmAPY genes in different tissues The genes were labeled on the left and the tissues were displayed at the top of each column. The gene expression values are reported as square root transformations of the fragments per kilo bases per million mapped reads (FPKM). Different colors in map represent FPKM values, as shown in the bar to the right
Fig. 5.
Expression analysis of ZmAPYs under drought, cold, heat stresses A Expression analysis of ZmAPY genes under three different drought degree stresses. B Expression analysis of ZmAPY genes under cold and heat stresses. The genes were labeled on the left and the abiotic stresses were displayed at the top of each column. The FPKM / TPM values of the ZmAPY genes were used to draw a heat map by using HEMI. C qRT-PCR in analyzing the expression of ZmAPYs under drought, cold and heat stress. Statistical significance was determined by Student’s t test: “*”P < 0.05 and “**”P < 0.01
Fig. 6.
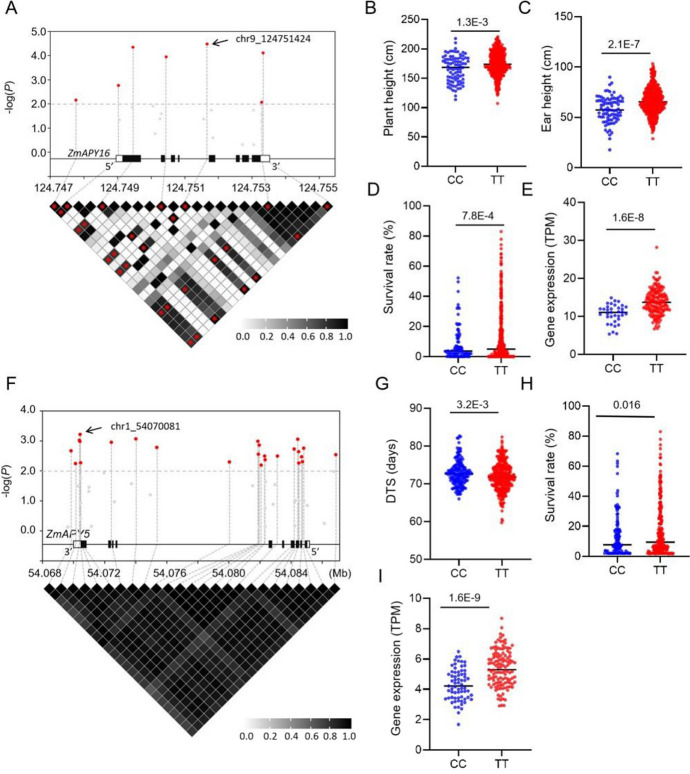
ZmAPYs was associated with agronomic traits and drought resistance of maize A. Association of SNPs from ZmAPY16 with plant height. chr9_124751424 is most significantly associated with plant height compared with other SNPs. SNPs significant associated with plant height (p < 0.01) were marked in red. B-E Genetic variation of ZmAPY16 (chr9_124751424) regulates plant length (B), ear-height (C), drought resistance (D) and ZmAPY5 expression (E) in maize. F Association of SNPs from ZmAPY5 with day to silking (DTS). Chr1_54070081 is most significantly associated with DTS compared with other SNPs. SNPs significant associated with DTS (p < 0.01) were marked in red. G-I Genetic variation of ZmAPY5 (chr1_54070081) regulates silking period (G), drought resistance (H) and ZmAPY5 expression (I) in maize. Statistical significance was determined by Student’s t test
Fig. 7.
Genetic variation in the ZmAPYs regulate the content of drought-induced metabolites A Co-localization analysis of mQTLs with ZmAPY genes. The red lines represent the collinearity relationship of mQTLs under drought stress with ZmAPY genes, while the blue lines represent the collinearity relationship of mQTLs under control conditions with ZmAPY genes. B Genetic variations in the ZmAPY gene region regulate the contents of metabolites induced by drought. Statistical significance was determined by Student’s t test
Materials and methods
Plant materials and growth condition
The maize inbred line B73 was used in this study. Maize seeds are germinated on moist filter paper in a plant growth chamber at 28°C for 2–3 days. Uniformly germinated seeds are then transplanted into soil-filled pots (10 cm × 10 cm × 9 cm) and grown in the chamber until the three-leaf stage for subsequent stress treatments. For drought stress, water is withheld from three-leaf stage maize seedlings for 10 days, while the control group is watered normally. Leaf samples are collected for RNA extraction to analyze ZmAPY genes expression levels. For heat and cold treatments, three-leaf stage maize seedlings were placed in growth chambers at 50°C and 4°C for 4 h and 24 h, respectively, while the control group continues growing at 28°C. Leaf samples are then collected for RNA extraction to evaluate ZmAPY genes expression level.
Identification and Phylogenetic analysis of the ZmAPY genes in maize
Gene ID of all APY genes in Arabidopsis thaliana and Oryza sativa was collected previous study (Chowdhury et al. 2023), protein sequences of these APYs were downloaded from Ensemble Plants (http://plants.ensembl.org/) and aligned for constructing HMM model. Protein sequences for maize (Zea mays) were also collected from Ensemble Plants, and ZmAPY genes were identified using HMMsearch with default parameters. Proteins identified by HMMsearch that lacked conserved domains were removed after submission to SMART (https://smart.embl.de/). Subsequently, MEGA X was used for multiple sequence alignment and phylogenetic tree construction of the identified 18 protein sequences with those from rice and Arabidopsis thaliana's APY proteins. The tree was built using Neighbor-Joining (NJ) method, pair-wise deletion, and bootstrap value set to 1000. As a result, 16 genes were identified as ZmAPY genes based on the evolutionary tree and named as ZmAPY1 -ZmAPY16, respectively. The Synteny analysis of ZmAPY genes was employed by MCScanX by using default parameter.
Prediction of physicochemical properties of ZmAPY proteins
To obtain data such as the number of amino acids, molecular weight, isoelectric point, and average hydrophobicity index of the ZmAPY proteins, a physicochemical property analysis of ZmAPY protein sequences was employed by using an online tool ProtParam on ExPASy (https://web.expasy.org/protparam/). Gene structure analysis was performed by GSDS2.0 (https://gsds.gao-lab.org/). The subcellular localization of ZmAPY proteins were predicted by WoLF PSORT (https://wolfpsort.hgc.jp/) and CELLO (http://cello.life.nctu.edu.tw/).
Expression analysis of ZmAPY genes in different tissues
The expression data for the ZmAPY genes were obtained from various tissues, including seedling, roots, leaves, seeds, shoot apical meristems, internodes, tassel, cob, coleoptite, pericarp and anthers. These data were sourced from the qTeller (https://qteller.maizegdb.org/). The FPKM values of the ZmAPY genes were used to draw a heat map by using HEMI (Deng et al. 2014).
Upstream transcription factor prediction of ZmAPY genes
The 2kb upstream sequences of each ZmAPY gene were obtained from the maize genome for analysis. The PlantTFdb (https://planttfdb.gao-lab.org/) and jbrowse on maizeGDB (https://jbrowse.maizegdb.org/) were used to analyze the retrieved sequences and identify. All predicted upstream transcription factors were visualized by using Gephi (https://gephi.org/). Then expression correlation coefficients between potential upstream transcription factors and ZmAPY genes were calculated by using the expression profiles collected from qTeller.
Analysis of ZmAPY genes expression in maize under abiotic stresses
Expression profiles data of ZmAPY genes under well-water and drought treatment were collected from qTeller and previous study (Zhang et al. 2019). The FPKM / TPM values of the ZmAPY genes were used to draw a heat map by using HEMI. For qPCR validation, the FastPure Universal Plant Total RNA Isolation kit (Vazyme) was employed for RNA extraction. HiScript III qRT SuperMix for qPCR (Vazyme) was used for reverse transcription to synthesize cDNA. Specifc primers were designed for the 16 ZmAPY genes and utilized for qRT-PCR analysis. The qRT-PCR reactions were performed using the SYBR Green master mix (ChamQ SYBR qPCR Master Mix, Vazyme). Three independent replicates were performed for each treatment. The primers used are described in Table S5.
Association analysis of ZmAPY genes
Association analysis for ZmAPYs was performed by using a maize association mapping population containing 540 inbred lines in a previous study (Liu et al. 2017). Among 1,227,480 high-quality SNP data with minor allele frequency (MAF) ≥ 0.05, 502 SNPs were found in the gene region of all 16 ZmAPYs. maize agronomic traits and drought tolerance trait were also collected from previous studies (Yang et al. 2014). The MLM model were chosen to detect the SNPs significantly associated with maize agronomic traits and drought tolerance by using the TASSEL5.0 program (Bradbury et al. 2007).
Results
Identification of maize APY genes
Totally, we identified 16 ZmAPY family members from the Ensembl Plant database (https://plants.ensembl.org/index.html), named ZmAPY1-ZmAPY16 (Table S1). Their physicochemical properties, including gene ID, protein size, molecular weight (MW), isoelectric point (pI), the grand averages of hydropathicity (GRAVY), instability index and and localization prediction, were characterized and shown in Table S1. The ZmAPY proteins varied in length from 81 to 701 amino acids, with molecular weights ranging from 8.74822 kDa to 76.9779 kDa. Isoelectric points ranged from 4.70 to 11.62 acidic. Most ZmAPY proteins were hydrophilic (GRAVY < 0) except for ZmAPY6 and ZmAPY7, which were hydrophobic (GRAVY > 0). The estimated instability index ranged from 30.91 to 72.05. Subcellular localization predictions indicated that nine ZmAPY proteins were likely localized in the chloroplast, four in the plasma membrane, three in the mitochondria, one in the extracellular space, and one in the nucleus (Table S1), indicating diverse functional roles for these ZmAPY genes.
To explore the phylogenetic relationships of ZmAPYs with other species, a phylogenetic tree was constructed, incorporating 16 ZmAPYs, 7 AtAPYs, and 9 OsAPYs (Fig. 1A; Table S2). The phylogenetic tree topology classified these APY proteins into three Groups: Group I, II, III. In addition, there are two APY-like proteins (ZmAPYL1, ZmAPYL2) in maize that do not belong to Group I-III. Therefore, we classify them as an outgroup (Fig. 1A, Table S2). The expansion of APYs in maize, compared to AtAPYs and OsAPYs, suggests the potential importance of this gene family to regulate biological processes.
Distribution and collinearity analysis of maize APY genes
To investigate features of the ZmAPYs gene family, we analyzed the chromosome distribution of each ZmAPY gene. Our investigation showed that the ZmAPY genes in the maize genome were unevenly distributed across all 9 chromosomes, with the exception of chromosome 8 (Fig. 1B). The number of APY genes varied on each chromosome. Specifically, there is a single APY gene located on chromosomes Chr5, Chr6, Chr7, and Chr9. Chromosomes Chr2, Chr3, and Chr10 each contain two APY genes, while chromosomes Chr1 and Chr4 each have three APY genes. Gene duplications play a crucial role in the expansion of gene families (Konrad et al. 2011). Segmental duplications lead to the presence of large repetitive chromosomal blocks in the genome and are often associated with chromosomal rearrangements and polyploid events (Lallemand et al. 2020). Colinearity analysis indicated the occurrence of three segmental duplication events involving five ZmAPY genes across the maize genome, while no tandem duplications were observed (Fig. 1B).
APY gene structures and predicted protein motifs
Structural differences in exon–intron arrangement serve as sources of gene family variation and species diversity, leading to alterations in gene expression and function. To investigate the conservation and diversity of gene structure within the maize APY gene family, the exons and introns of 16 APY genes were analyzed based on their coding sequences and genomic data. The number of exons varied among the APY genes, ranging from 1 to 10 (Fig. 2A). Most members contained 6–10 exons, with two members in Group I (ZmAPY5 and ZmAPY6) and one member in Group III (ZmAPY13) having only 1 and 2 exons, respectively (Fig. 2A). The number of introns ranged from 0 to 9. Two transposon insertions of 9.56 kb and 33.57 kb were identified in the intron regions of ZmAPY5 and ZmAPY10, respectively. ZmAPY genes with collinearity exhibited similar gene structures. Additionally, members within the same subgroup typically displayed similar motifs and lengths, suggesting functional similarities (Fig. 2A). Analysis of the protein sequences of all ZmAPY gene family members revealed a conserved GDA1_CD39 domain in all proteins, with varying numbers of transmembrane regions. Specifically, members of Group I had one transmembrane region, most members of Group II had two transmembrane regions, and family members of Group III had no transmembrane regions (Fig. 2B). This structural variation may contribute to the functional distinctions observed among different subgroups.
Expression patterns of APY genes in maize
The investigation into tissue-specific gene expression patterns provides valuable insights into the potential biological roles of the ZmAPY genes. Analysis of the expression patterns within the ZmAPY gene family revealed distinct expression profiles among different members (Fig. 3), highlighting their diverse functions. Specifically, ZmAPY8, ZmAPY9, and ZmAPY12 exhibited specific expression in roots, indicating a potential role for these genes in root-related processes. On the other hand, ZmAPY15 and ZmAPY16 showed significantly higher expression levels in anthers compared to other tissues, suggesting their involvement in anther-related functions. Moreover, there was a notable trend of high expression of ZmAPY11 in seeds and endosperm, implying a potential role in seed development and maturation. Conversely, minimal to no detectable expression was observed for ZmAPY3, ZmAPY4, ZmAPY6, and ZmAPY7 across all tissues and organ. Interestingly, similar tissue-specific expression patterns were observed between collinear ZmAPY genes, such as ZmAPY1/ZmAPY2/ZmAPY5 and ZmAPY15/ZmAPY16, suggesting potential functional conservation or shared regulatory mechanisms among these gene clusters. The differential tissue-specific expression patterns observed among the ZmAPY genes indicate their diverse biological roles and potential contributions to various developmental processes and physiological functions in maize.
Prediction of upstream regulators of APY gene
The analysis of variations in expression patterns among different ZmAPY genes has led to the identification of potential upstream regulators that may control APY gene transcription in maize. By utilizing planttfdb software and existing ChIP-seq data of 104 transcription factors (Tu et al. 2020), a total of 251 upstream regulators were predicted (Fig. 4A, Table S3). Subsequently, the correlation between the expression levels of these predicted regulators and ZmAPY gene expression was examined. Significantly, NACTF78, ZIM36, bZIP79, and CCHH26 displayed strong correlations with the transcription levels of ZmAPY2 (R2 = 0.51, P = 5.4 × 10–19), ZmAPY5 (R2 = 0.56, 2.4 × 10–21), ZmAPY9 (R2 = 0.67, P = 1.1 × 10–16), and ZmAPY14 (R2 = 0.60, P = 1.0 × 10–23), respectively (Fig. 4B). Notably, NACTF78 has been previously reported to regulate Fe concentrations in maize kernels, potentially enabling the cultivation of maize varieties with both high yield and high Fe concentrations in their kernels using a molecular marker in the NACTF78 promoter (Yan et al. 2023). Additionally, Vélez-Bermúdez et al. reported that ZML2 (ZIM36) regulates wound-induced lignin genes in maize (Vélez-Bermúdez et al. 2015), while ZmTGA9-1 (bZIP79) has been shown to regulate male sterility in maize (Jiang et al. 2021). The observed correlations between the expression levels of ZmAPY genes and these transcription factors suggest that ZmAPY genes may also play a role in regulating these biological processes, indicating a potential link between APY gene expression and the modulation of Fe concentrations, lignin gene regulation, and male sterility in maize. These findings provide valuable insights into the regulatory network involving ZmAPY genes and their upstream regulators in maize, shedding light on the diverse biological processes influenced by these genes.
Expression analysis of ZmAPYs under drought, cold, heat stresses
To investigate the potential role of APY genes in regulating maize abiotic stress responses, we analyzed the transcription levels of APY genes under drought, cold, and heat stress using RNA-seq data from the maize inbred line B73. Our findings revealed the transcription of the ZmAPY gene is responsive to drought, cold, and heat stress, displaying distinct response profiles (Fig. 5A; 5B). Subsequently, we validated the transcription of ZmAPY genes under drought, cold, and heat stress through qRT-PCR. The results revealed significant upregulation of 6 ZmAPY genes, ZmAPY1, ZmAPY2, ZmAPY8, ZmAPY13, ZmAPY14 and ZmAPY15 under severe drought (DT4), while the transcription of ZmAPY15 was suppressed by drought (Fig. 5C). Cold stress induced the transcription of ZmAPY1 but inhibited the transcription of ZmAPY5, ZmAPY8, ZmAPY11, ZmAPY13 and ZmAPY15 (Fig. 5C). Moreover, heat stress significantly inhibited the transcription of 7 ZmAPY genes, ZmAPY1, ZmAPY2, ZmAPY5, ZmAPY8, ZmAPY11, ZmAPY14, and ZmAPY16, while the transcription of ZmAPY15 was induced by heat (Fig. 5C). It is noteworthy that some of the expression analysis results for the ZmAPY genes are absent from Fig. 5C due to their expression levels falling below the detection threshold of qRT-PCR. The stress-responsive expression patterns of ZmAPY genes suggest their potential regulatory roles in drought, cold, heat, and salt stress responses.
ZmAPYs was associated with agronomic traits and drought resistance of maize
To further investigate the impact of APY on maize agronomic traits and drought resistance, we examined the relationship between SNPs in the ZmAPY gene region and 17 agronomic traits as well as drought phenotypes using the MLM model. Our analysis revealed a significant association between the SNP (chr9_124751424, CC/TT) in ZmAPY16 and maize plant height (Fig. 6A). Subsequent analysis indicated notable differences not only in plant height but also in ear height, drought resistance, and ZmAPY16 expression between the "CC" and "TT" genotypes. Plants with the "TT" allele, showing high ZmAPY6 expression, exhibited greater plant height, ear height, and drought survival rates compared to those with the "CC" allele (Fig. 6B-E), suggesting a positive regulatory role of "TT" allele in maize plant height, ear height, and drought resistance. Additionally, we observed a significant association between the SNP (chr1_54070081, CC/TT) in ZmAPY5 and the spinemaking period in maize (Fig. 6F). Plants with the "TT" allele exhibited lower ZmAPY5 expression levels, delayed flowering, and increased drought survival compared to those with the "CC" allele (Fig. 6G-I), indicating a negative regulation of maize spinemaking and a positive regulation of maize drought resistance by ZmAPY5 "TT" allele.
Genetic variation within ZmAPYs regulate the content of drought-induced metabolites
Metabolites, as small molecules that serve as the end products of metabolic processes and physiological pathways, are known to play crucial roles in plant drought resistance (Kim et al. 2017; Todaka et al. 2017). These compounds can act as osmoprotectants, antioxidants, signaling molecules, and regulators of various stress-responsive pathways in plants (Nakabayashi et al. 2014; Obata et al. 2013; Fàbregas et al. 2018). Analyzing the genome-wide metabolite profiles of 385 maize natural inbred lines grown under well-watered and drought-stressed conditions (Zhang et al. 2021a), we identified metabolite quantitative trait loci (mQTL) for 18 metabolites that co-located with the ZmAPY genes, indicating a potential relationship between ZmAPYs and the levels of these metabolites (Fig. 7A, Table S4). Further investigation into these metabolites revealed that four drought-induced metabolites were influenced by genetic variations within the ZmAPYs gene region. Specifically, genetic variations chr1.S_65348503 and chr1.S_65352471 within the ZmAPY15 gene region were found to regulate the contents of metabolites PN_group_05106 and PN_group_17082, while another genetic variation chr1.S_65352471 within the ZmAPY11 gene region was associated with the regulation of the metabolite PN_group_00505. Additionally, a genetic variation chr10.S_71730551 within the ZmAPY7 gene region was linked to the regulation of the metabolite PN_group_11670 (Fig. 7B). These findings suggest that ZmAPY genes may impact maize drought resistance by modulating the contents of these drought-induced metabolites. This insight highlights the potential role of ZmAPY genes in mediating maize response to drought stress through the regulation of key metabolites involved in stress adaptation and tolerance.
Discussion
Apyrase (APY) is widely existed in eukaryotes and is highly conserved throughout the evolution of eukaryotes. In plants, apyrase genes have been reported to regulate a variety of biological processes, including root hair development, stomatal movement, and defense responses (Clark et al. 2014; Lim et al. 2014; Wu et al. 2007). However, no apyrase genes have been identified in maize, and their functions remain unclear. In this study, through sequence alignments and phylogenetic analysis, we identified 16 apyrase genes in maize genome and further divided these genes into three subgroups (Fig. 1A). The number of group I ZmAPY genes far exceeds the number of group I apyrase genes in Arabidopsis and rice that identified in a previous study (Clark et al. 2014; Chowdhury et al. 2023). Syntenic analysis revealed that most ZmAPY genes are of the dispersed repeat type, but segmental duplication also played a significant role in the expansion process of group I ZmAPY genes (Fig. 1B). Meanwhile, genes with synteny show higher similarity in expression patterns (Fig. 3). These results suggest that APY genes in maize may have produced functional redundancy through expansion, and since apyrase plays an important regulatory role in plant growth and development and stress responses, the expansion and functional redundancy of gene subgroups generated by segmental and dispersed duplication may enhance the robustness of the maize regulatory network.
The members of the ZmAPY genes are predicted to encode proteins ranging from 81 to 701 amino acid in length, and the PI ranging from 4.70 to 11.62 (Table S1). These results suggest that the differences among ZmAPY genes may be more pronounced than those in rice (in which OsAPY proteins ranging from 451 to 702 amino acid in length, and the PI ranging from 5.44 to 9.34) (Chowdhury et al. 2023). The predicted gene structures of the ZmAPY genes contain one exon to ten exons (Fig. 2A), similar to the gene structures in rice, with two to twelve exons (Chowdhury et al. 2023). Similar to OsAPY proteins, the ZmAPY proteins were predicted to localize to various cellular compartments, including the chloroplast, mitochondrial, plasma membrane, cytoplasm, nuclear and extracellular (Table S1). In summary, ZmAPY genes share similar characteristics with apyrase in rice, but exhibits greater diversity among family members, suggesting that although ZmAPY genes maintain functional conservation, they may also have undergone functional divergence.
Transcription factors are core elements that regulate transcriptional levels during various stages of plant life processes. Analysis of the promoter regions of all ZmAPY genes revealed the presence of diverse upstream transcription factors which may regulate ZmAPY genes expression (Fig. 4). Among the 251 potential upstream transcription factors, several genes have been reported to regulate maize growth and stress response progresses (Table S3). For example, WRKY48 / ZmWRKY40 was predicted to binding the promoter sequences of seven ZmAPY genes, overexpression ZmWRKY40 could enhance drought tolerance in transgenic Arabidopsis by regulating stress-related genes, and the reactive oxygen species (ROS) content in transgenic lines was reduced compared with wild-type plants under drought stress (Wang et al. 2018). ZmBES1/BZR1-5 was predicted to bind the promoter sequences of six ZmAPY genes, ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Meanwhile, ZmBES1/BZR1-5 can also positively regulates kernel size (Sun et al. 2020, 2021). A bZIP transcription factor id1 was predicted to binding the promoter sequences of ten ZmAPY genes, previous study showed that id1 can regulate maize flowering time and floral inductive signals (Colasanti et al. 1998; Muszynski et al. 2006). These results further suggested potential roles of ZmAPY genes in maize growth and stress response progresses.
Maize is a world major food crop, and its yield is seriously threatened by abiotic stresses (Lesk et al. 2016). Although a large number of abiotic stress response genes and loci have been identified in recent years through genetic and molecular biology techniques, due to limited understanding of the plant abiotic stress response system, only a few genes have been successfully applied in commercialization (Zhang et al. 2020). Therefore, it remains essential to further elucidate the abiotic stress response system of maize. Previous studies have shown that the extracellular ATP (eATP) level rises when plants are subjected to stress, leading to the production of reactive oxygen species (ROS) and cell death. Apyrase can regulate the production of ROS by hydrolyzing eATP, and participate in various stress response processes in plants (Deng et al. 2015). By analyzing the transcriptomic data from previous studies, we investigated the expression patterns of ZmAPY genes under different abiotic stresses (Makarevitch et al. 2015; Zhang et al. 2019). The results showed that seven ZmAPY genes responded to drought stress, six to cold stress, and eight to heat stress, and the stress responsive ZmAPY genes were further confirmed by qPCR (Fig. 5C). These results indicated that, like other plants, ZmAPY genes could also be involved in regulating maize's abiotic stress response. Of course, more researches are required to clearly elucidate the function of ZmAPY genes in maize abiotic stress responses.
Natural variations significantly affect maize agronomic traits and stress resistance, and numerous elite alleles have been identified through genetic approaches which significantly accelerated the breeding process (Xiao et al. 2017). To explore the potential application value of ZmAPY genes and their genetic variations in maize breeding, we collected drought tolerance and agronomic traits data of maize association panels published in previous study (Zhang et al. 2019; Yang et al. 2014), and utilized high-density SNP markers to perform association analyses on the ZmAPY gene and its flanking regions. The result indicated that natural variations in ZmAPY16 gene and flanking region were significantly associated with maize plant height (Fig. 6A), further analysis revealed that these variations were also correlated with the ZmAPY16 expression and maize drought tolerance (Fig. 6B-E). Additionally, we found that ZmAPY5 was significantly associated with maize silking time (Fig. 6F), and associated variations were also correlated with the ZmAPY5 expression and maize drought tolerance (Fig. 6G-I). These findings suggest that variations of ZmAPY gene expression may influence maize agronomic traits and drought tolerance. Furthermore, since many metabolic pathways involve energy conversion, while apyrase can affect the levels of ATP/ADP. To investigate the effects of apyrase on maize metabolites, we performed co-localization analysis between ZmAPY variations and previously reported metabolite regulation sites (Zhang et al. 2021a), which revealed that seven ZmAPY genes were co-localized with 18 metabolite regulation sites (Fig. 7A). Some of these metabolites showed significant content change under drought stress (Fig. 7B). Although the functions of these metabolites are not well understood at present, these results suggest that ZmAPY genes and their genetic variations may participate in maize drought response by affecting the levels of drought-responsive metabolites. In summary, we revealed the variations of ZmAPY gene expression among maize populations, which were associated with multiple important maize traits. These results indicate that ZmAPYs and their genetic variations may have potential applications in maize breeding.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
MD, HZ, ZH and XS conceived and designed the research. ZH and JZ performed the experiments and collected the data. HZ and XS supervised the experiments. HZ and XS wrote the manuscript. MD and EN modified manuscript. HJ and SZ polished the manuscript and images. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key Technologies Research and Development Program, China (2022YFE0100500), The National Natural Science Foundation of China (31971954, 32061143031).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
We declare that these experiments comply with the ethical standards in China.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
The original online version of this article was revised due to a retrospective Open Access cancellation.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenghua He and Jie Zhang contributed equally to this work.
Change history
6/10/2024
A Correction to this paper has been published: 10.1007/s11032-024-01482-9
Contributor Information
Hui Zhang, Email: hui_zhang@mail.hzau.edu.cn.
Mingqiu Dai, Email: mingqiudai@mail.hzau.edu.cn.
References
- Alam I, Lee DG, Kim KH, Park CH, Sharmin SA, Lee H, Oh KW, Yun BW, Lee BH. Proteome analysis of soybean roots under waterlogging stress at an early vegetative stage. J Biosci. 2010;35:49–62. doi: 10.1007/s12038-010-0007-5. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Chowdhury AT, Hasan MN, Bhuiyan FH, Islam MQ, Nayon MRW, Rahaman MM, Hoque H, Jewel NA, Ashrafuzzaman M, Prodhan SH. Identification, characterization of Apyrase (APY) gene family in rice (Oryza sativa) and analysis of the expression pattern under various stress conditions. PLoS One. 2023;18:e0273592. doi: 10.1371/journal.pone.0273592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Torres J, Finlayson S, Guan X, Handley C, Lee J, Kays JE, Chen ZJ, Roux SJ. Apyrase (nucleoside triphosphate-diphosphohydrolase) and extracellular nucleotides regulate cotton fiber elongation in cultured ovules. Plant Physiol. 2010;152:1073–1083. doi: 10.1104/pp.109.147637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Morgan RO, Fernandez MP, Salmi ML, Roux SJ. Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development. Plant Sci. 2014;225:107–116. doi: 10.1016/j.plantsci.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Clark G, Brown KA, Tripathy MK, Roux SJ. Recent advances clarifying the structure and function of plant apyrases (Nucleoside Triphosphate Diphosphohydrolases) Int J Mol Sci. 2021;22:3283. doi: 10.3390/ijms22063283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Tripathy MK, Roux SJ. Growth regulation by apyrases: insights from altering their expression level in different organisms. Plant Physiol. 2024;194:1323–1335. doi: 10.1093/plphys/kiad590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/S0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G. Differential expression of two soybean apyrases, one of which is an early nodulin. Mol Plant Microbe Interact. 2000;13:1053–1070. doi: 10.1094/MPMI.2000.13.10.1053. [DOI] [PubMed] [Google Scholar]
- Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Sun J, Zhao R, Ding M, Zhang Y, Sun Y, Wang W, Tan Y, Liu D, Ma X, Hou P, Wang M, Lu C, Shen X, Chen S. Populus euphratica APYRASE2 Enhances Cold Tolerance by Modulating Vesicular Trafficking and Extracellular ATP in Arabidopsis Plants. Plant Physiol. 2015;169:530–548. doi: 10.1104/pp.15.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N, Lozano-Elena F, Blasco-Escámez D, Tohge T, Martínez-Andújar C, Albacete A, Osorio S, Bustamante M, Riechmann JL, Nomura T, Yokota T, Conesa A, Alfocea FP, Fernie AR, Caño-Delgado AI. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat Commun. 2018;9:4680. doi: 10.1038/s41467-018-06861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Kim SY, Libault M, Berg RH, Tanaka K, Stacey G, Taylor CG. GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol. 2009;149:994–1004. doi: 10.1104/pp.108.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Guérin A, Herger A, Hou X, Schaufelberger M, Roulard R, Diet A, Roffler S, Lefebvre V, Wicker T, Pelloux J, Ringli C. Growth-inhibiting effects of the unconventional plant APYRASE 7 of Arabidopsis thaliana influences the LRX/RALF/FER growth regulatory module. PLoS Genet. 2024;20:e1011087. doi: 10.1371/journal.pgen.1011087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa M, Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Hideaki M, Tomoyuki Y, Motokazu T. Activation of ATPase activity in the chromatin fraction of pea nuclei by calcium and calmodulin. Plant Cell Physiol. 1984;25:191–195. [Google Scholar]
- Jiang Y, An X, Li Z, Yan T, Zhu T, Xie K, Liu S, Hou Q, Zhao L, Wu S, Liu X, Zhang S, He W, Li F, Li J, Wan X. CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol J. 2021;19:1769–1784. doi: 10.1111/pbi.13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Matsui A, Tanoi K, Kobayashi NI, Matsuda F, Habu Y, Ogawa D, Sakamoto T, Matsunaga S, Bashir K, Rasheed S, Ando M, Takeda H, Kawaura K, Kusano M, Fukushima A, Endo TA, Kuromori T, Ishida J, Morosawa T, Tanaka M, Torii C, Takebayashi Y, Sakakibara H, Ogihara Y, Saito K, Shinozaki K, Devoto A, Seki M. Acetate-mediated novel survival strategy against drought in plants. Nat Plants. 2017;3:17097. doi: 10.1038/nplants.2017.97. [DOI] [PubMed] [Google Scholar]
- Knowles AF. The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal. 2011;7:21–45. doi: 10.1007/s11302-010-9214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A, Teufel AI, Grahnen JA, Liberles DA. Toward a general model for the evolutionary dynamics of gene duplicates. Genome Biol Evol. 2011;3:1197–1209. doi: 10.1093/gbe/evr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand T, Leduc M, Landès C, Rizzon C, Lerat E. An overview of duplicated gene detection methods: why the duplication mechanism has to be accounted for in their choice. Genes (basel) 2020;11:1046. doi: 10.3390/genes11091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal DB, Streher CA, Neu TN, Bittencourt FP, Leal CA, da Silva JE, Morsch VM, Schetinger MR. Characterization of NTPDase (NTPDase1; ecto-apyrase; ecto-diphosphohydrolase; CD39; EC 3.6.1.5) activity in human lymphocytes. Biochim Biophys Acta. 2005;1721:9–15. doi: 10.1016/j.bbagen.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529:84–87. doi: 10.1038/nature16467. [DOI] [PubMed] [Google Scholar]
- Lim MH, Wu J, Yao J, Gallardo IF, Dugger JW, Webb LJ, Huang J, Salmi ML, Song J, Clark G, Roux SJ. Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 2014;164:2054–2067. doi: 10.1104/pp.113.233429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ni J, Shah FA, Ye K, Hu H, Wang Q, Wang D, Yao Y, Huang S, Hou J, Liu C, Wu L. Genome-wide identification, characterization and expression pattern analysis of APYRASE family members in response to abiotic and biotic stresses in wheat. PeerJ. 2019;7:e7622. doi: 10.7717/peerj.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Luo X, Niu L, Xiao Y, Chen L, Liu J, Wang X, Jin M, Li W, Zhang Q, Yan J (2017) Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol Plant 10(3):414–426 [DOI] [PubMed]
- Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Witt S, Lisec J, Palacios-Rojas N, Florez-Sarasa I, Yousfi S, Araus JL, Cairns JE, Fernie AR. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2013;169:2665–2683. doi: 10.1104/pp.15.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Grosman L, Fernie AR, Wucke C, Geigenberger P. The potato-specific apyrase is apoplastically localized and has influence on gene expression, growth, and development. Plant Physiol. 2008;147(3):1092–1109. doi: 10.1104/pp.108.117564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif Y, Mamadou G, Yang Q, Cai T, Zhuang Y, Chen K, Deng Y, Khan SA, Ali N, Zhang C, Raza A, Chen H, Varshney RK, Zhuang W. Genome-wide investigation of apyrase (APY) genes in peanut (Arachis hypogaea L.) and functional characterization of a pod-abundant expression promoter AhAPY2–1p. Int J Mol Sci. 2023;24:4622. doi: 10.3390/ijms24054622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang C, Zhang X, Deng S, Zhao R, Shen X, Chen S. Extracellular ATP signaling and homeostasis in plant cells. Plant Signal Behav. 2012;7:566–569. doi: 10.4161/psb.19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Yu H, Qu J, Cao Y, Ding L, Feng W, Khalid MHB, Li W, Fu F. Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic arabidopsis. Int J Mol Sci. 2020;21:996. doi: 10.3390/ijms21030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Ding L, Feng W, Cao Y, Lu F, Yang Q, Li W, Lu Y, Shabek N, Fu F, Yu H. Maize transcription factor ZmBES1/BZR1-5 positively regulates kernel size. J Exp Bot. 2021;72:1714–1726. doi: 10.1093/jxb/eraa544. [DOI] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux S. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol. 1999;119:543–552. doi: 10.1104/pp.119.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Zhao Y, Yoshida T, Kudo M, Kidokoro S, Mizoi J, Kodaira KS, Takebayashi Y, Kojima M, Sakakibara H, Toyooka K, Sato M, Fernie AR, Shinozaki K, Yamaguchi-Shinozaki K. Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J. 2017;90:61–78. doi: 10.1111/tpj.13468. [DOI] [PubMed] [Google Scholar]
- Tong CG, Dauwalder M, Clawson GA, Hatem CL, Roux SJ. The major nucleoside triphosphatase in pea (Pisum sativum L.) nuclei and in rat liver nuclei share common epitopes also present in nuclear lamins. Plant Physiol. 1993;101:1005–1011. doi: 10.1104/pp.101.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Mejía-Guerra MK, Valdes Franco JA, Tzeng D, Chu PY, Shen W, Wei Y, Dai X, Li P, Buckler ES, Zhong S. Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat Commun. 2020;11:5089. doi: 10.1038/s41467-020-18832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappa R, Slocum RD, Siegenthaler A, Wang J, Clark G, Roux SJ. Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ. 2019;42:337–353. doi: 10.1111/pce.13425. [DOI] [PubMed] [Google Scholar]
- Vélez-Bermúdez IC, Salazar-Henao JE, Fornalé S, López-Vidriero I, Franco-Zorrilla JM, Grotewold E, Gray J, Solano R, Schmidt W, Pagés M, Riera M, Caparros-Ruiz D. A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell. 2015;27:3245–3259. doi: 10.1105/tpc.15.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Ru JN, Liu YW, Yang JF, Li M, Xu ZS, Fu JD. The Maize WRKYtranscription factor ZmWRKY40 Confers drought resistance in transgenic arabidopsis. Int J Mol Sci. 2018;19:2580. doi: 10.3390/ijms19092580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratne G, Wang H, Weeraratne TP, Sabharwal T, Jiang HW, Cantero A, Clark G, Roux SJ. APYRASE1/2 mediate red light-induced de-etiolation growth in Arabidopsis seedlings. Plant Physiol. 2022;189:1728–1740. doi: 10.1093/plphys/kiac150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Choi LE, Guidotti G. N-linked oligosaccharides affect the enzymatic activity of CD39: diverse interactions between seven N-linked glycosylation sites. Mol Biol Cell. 2005;16:1661–1672. doi: 10.1091/mbc.e04-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ. Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol. 2007;144:961–975. doi: 10.1104/pp.107.097568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu H, Wu L, Warburton M, Yan J. Genome-wide association studies in maize: praise and stargaze. Mol Plant. 2017;10:359–374. doi: 10.1016/j.molp.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Yan P, Du Q, Chen H, Guo Z, Wang Z, Tang J, Li WX. Biofortification of iron content by regulating a NAC transcription factor in maize. Science. 2023;382:1159–1165. doi: 10.1126/science.adf3256. [DOI] [PubMed] [Google Scholar]
- Yang J, Wu J, Romanovicz D, Clark G, Roux SJ. Co-regulation of exine wall patterning, pollen fertility and anther dehiscence by Arabidopsis apyrases 6 and 7. Plant Physiol Biochem. 2013;69:62–73. doi: 10.1016/j.plaphy.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Yang N, Lu Y, Yang X, Huang J, Zhou Y, Ali F, Wen W, Liu J, Li J, Yan J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014;10:e1004573. doi: 10.1371/journal.pgen.1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Fan Y, Sun X, Chen L, Terzaghi W, Bucher E, Li L, Dai M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019;98:697–713. doi: 10.1111/tpj.14267. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao Y, Zhu JK. Thriving under stress: how plants balance growth and the stress response. Dev Cell. 2020;55:529–543. doi: 10.1016/j.devcel.2020.10.012. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wu J, Sade N, Wu S, Egbaria A, Fernie AR, Yan J, Qin F, Chen W, Brotman Y, Dai M. Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol. 2021;22:260. doi: 10.1186/s13059-021-02481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Y, Liu X, Deng J, Yao J, Zhang Y, Deng S, Zhang H, Zhao N, Li J, Zhou X, Zhao R, Chen S. Populus euphratica apyrases increase drought tolerance by modulating stomatal aperture in arabidopsis. Int J Mol Sci. 2021;22:9892. doi: 10.3390/ijms22189892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.