Abstract
In addition to highly conserved stem-loop structures located in the 5′- and 3′-nontranslated regions, genome replication of picornaviruses requires cis-acting RNA elements located in the coding region (termed cre) (K. L. McKnight and S. M. Lemon, J. Virol. 70:1941–1952, 1996; P. E. Lobert, N. Escriou, J. Ruelle, and T. Michiels, Proc. Natl. Acad. Sci. USA 96:11560–11565, 1999; I. Goodfellow, Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans, J. Virol. 74:4590–4600, 2000). cre elements appear to be essential for minus-strand RNA synthesis by an as-yet-unknown mechanism. We have discovered that the cre element of poliovirus (mapping to the 2C coding region of poliovirus type 1; nucleotides 4444 to 4505 in 2C), which is homologous to the cre element of poliovirus type 3, is preferentially used as a template for the in vitro uridylylation of VPg catalyzed by 3Dpol in a reaction that is greatly stimulated by 3CDpro (A. V. Paul, E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer, J. Virol. 74:10359–10370, 2000). Here we report a direct correlation between mutations that eliminate, or severely reduce, the in vitro VPg-uridylylation reaction and produce replication phenotypes in vivo. None of the genetic changes significantly influenced translation or polyprotein processing. A substitution mapping to the first A (A4472C) of a conserved AAACA sequence in the loop of PV-cre(2C) eliminated the ability of the cre RNA to serve as template for VPg uridylylation and abolished RNA infectivity. Mutagenesis of the second A (A4473C; AAACA) severely reduced the yield of VPgpUpU and RNA infectivity was restored only after reversion to the wild-type sequence. The effect of substitution of the third A (A4474G; AAACA) was less severe but reduced both VPg uridylylation and virus yield. Disruption of base pairing within the upper stem region of PV-cre(2C) also affected uridylylation of VPg. Virus derived from transcripts containing mutations in the stem was either viable or quasi-infectious.
Poliovirus is a human pathogen belonging to the Picornaviridae that can cause severe neurologic disease. The virus consists of a nonenveloped particle of 60 copies each of four capsid proteins (VP1 to VP4) and a single-stranded RNA genome of positive polarity. The genome (7,441 nucleotides [nt]; Fig. 1) is covalently linked to a small peptide called VPg at the 5′ end (10, 18) via a phosphodiester between the O4-hydroxyl group of a tyrosine and the 5′-terminal uridylic acid (3, 35). A long poly(A) tail follows the heteropolymeric nontranslated region (NTR) at the 3′ end (44). Early in infection the genome of messenger-sense RNA directs the synthesis of a large polyprotein that is processed by virus-encoded proteinases (2Apro, 3Cpro, and 3CDpro) to functional viral proteins. The genomic mRNA must then recruit the RNA-dependent RNA polymerase (3Dpol) to induce synthesis of more plus-stranded RNA via a minus-strand RNA intermediate (11). Newly made plus-strand RNAs can then function as templates in translation and transcription or can be encapsidated to produce progeny virus (for reviews, see references 16, 39, and 42). The entire replication cycle of poliovirus occurs in the cytoplasm (8).
FIG. 1.
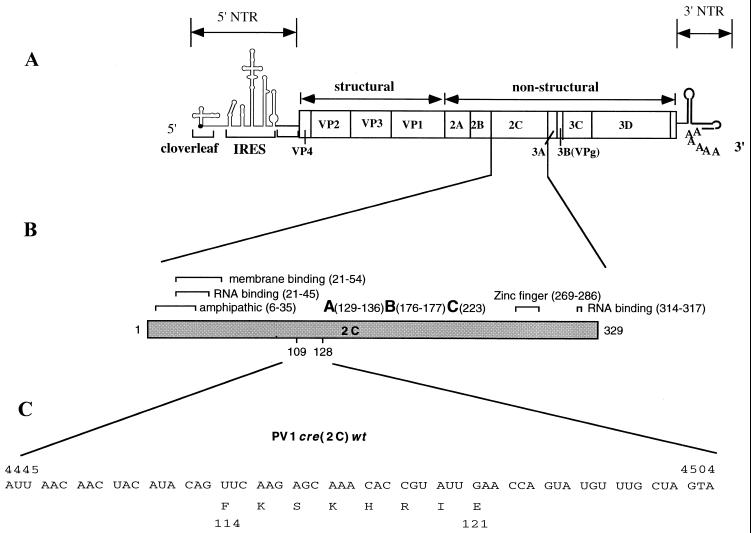
Structure of poliovirus genomic RNA. (A) The single-stranded RNA is covalently linked to VPg at the 5′ end. The 5′NTR consists of two functional domains, the cloverleaf, and IRES elements. The open box depicts the polyprotein with coding regions for different viral proteins. Regions corresponding to structural and nonstructural domains are also indicated. The 3′NTR contains an heteropolymeric region and is polyadenylylated. (B) General features of poliovirus protein 2C with suggested functions as indicated by brackets and amino acid positions. A, B, and C refer to motifs conserved among superfamily 3 helicases and are required for the ATPase activity (adapted from Pfister and Wimmer [30]). Numbers in parentheses indicate amino acid locations within 2C. Poliovirus type 1 cre(2C) is located in the coding region corresponding to amino acids 109 to 127 (16). (C) Nucleotide sequence of PV-cre(2C) and the deduced amino acid sequence. (D) Predicted secondary structure of the PV-cre(2C) nt 4445 to 4504 in 2C and the mutants which were used in this study. Dots indicate the location of nucleotides which are different between type 1 and type 3 PV-cre(2C) (see Fig. 2).
An enigma in poliovirus RNA replication is the mechanism by which the replicating machinery selects its template (1). An overwhelming number of cytoplasmic RNAs are polyadenylylated, just as poliovirus RNA. Since the poly(A) of the poliovirus genome is genetically encoded (9), that is, it is transcribed from a 5′-terminal poly(U) segment on the minus strands, minus-strand synthesis must be initiated at this homopolymeric tail. Yet polioviral RNA polymerase selects only RNA from which it was synthesized, an observation suggesting the presence of specific signals encoded in the genomic RNA in addition to or other than poly(A). The molecular basis for template specificity of RNA-dependent RNA polymerase could be related in part to the recognition of RNA structures at either the 5′ or the 3′ end of the viral genome. Alternatively, the function can be provided by internally located RNA structures.
The 5′NTR of the viral genome contains three important structural elements: the terminal peptide VPg, a cis-acting sequence involved in replication (the “cloverleaf”) and the internal ribosomal entry site (IRES), controlling translation (42) (Fig. 1A). The cloverleaf structure, corresponding to the first 100 nt of the viral genome, is an essential component of virus replication (4, 5, 17, 25, 34, 41). This element forms an RNP complex with 3CDpro (a proteinase and RNA binding protein; see references 4, 5, 17, 34, and 45) and with either the viral protein 3AB (17, 40, 41) or with the cellular poly(rC) binding protein (PCBP) (6, 13, 25). Formation of this RNP has been shown to play a role in the initiation of positive strand synthesis (4, 41, 42). More recently, it has been suggested that the interplay between 3CDpro and PCBP with RNA elements within the 5′NTR of poliovirus will lead to the switch from translation to replication (12). This model, however, is difficult to reconcile with the fact that large amounts of 3CDpro are synthesized throughout the replicative cycle that would cause the shutoff of viral translation shortly after the initial rounds of translation. Thus, protein synthesis occurs concomitantly with RNA replication (1). The involvement of the IRES element is controversial but certain mutations in the IRES definitely cause a replication phenotype (see references 1 and 42).
The poliovirus 3′NTR consists of a heteropolymeric sequence, followed by poly(A). Genetic manipulations of the 3′NTR demonstrated the importance of the heteropolymeric region in the replication of enteroviruses (23, 31). On the other hand, the poliovirus 3′NTR can be replaced with that of human rhinovirus type 14 (HRV14), which is entirely different in primary sequence and secondary structure, and yet the resultant chimeric genome replicated with wild-type (wt) kinetics (33). Moreover, deletion of the entire heteropolymeric sequence of the poliovirus 3′NTR severely debilitated but did not abolish viral replication (37). These results suggest a high degree of complexity in the recognition of the poliovirus RNA template involving the 3′-terminal, heteropolymeric region (1, 42).
We have demonstrated previously that the first step in the synthesis of the minus-strand RNA is the uridylylation of the viral protein VPg to form the VPgpUpU, followed by VPg-poly(U) synthesis on a poly(A) template (28). This reaction, catalyzed by 3Dpol, is absolutely dependent upon a template and, under the conditions of the experiment, only poly(A) can fulfill this template function. Since the 5′ terminus of the minus strands is poly(U) (43), this observation met the expectation for the initiation of minus-strand RNA synthesis. However, it did not solve the problem of specificity. We have searched, therefore, for signals bestowing template specificity to the uridylylation reaction (27). However, neither the elements of the 3′-heteropolymeric region nor any structures of the 5′NTR promoted uridylylation (27).
Analyses of HRV14 replicons led to the unexpected discovery of a stem-loop structure in the coding region of the polyprotein whose integrity was essential for replication. McKnight and Lemon (21, 22) demonstrated the existence of a cis-acting replication element (termed cre) in the P1 region of the HRV14 genome that is involved in the initiation of negative-strand RNA synthesis of HRV14. More recently, stem-loop structures of similar function have been discovered in the genome of Theiler's virus (also located in the P1 region) (19) and of poliovirus type 3 (located in the coding region of 2CATPase) (15). A stem-loop structure with nearly identical sequence exists also in the genome of poliovirus type 1 (Mahoney) [PV1(M)]. This element is termed PV-cre(2C) (Fig. 1D and 2).
FIG. 2.
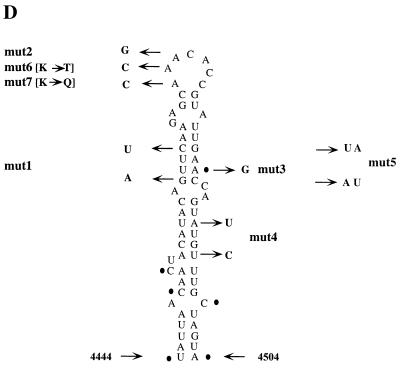
Computer-predicted secondary structures of known picornavirus cre elements. Primary sequences and secondary structures of HRV14 cre (structure 1), poliovirus type 1 cre (structure 2), Theiler's virus cre (structure 3), and EMCV cre (structure 4). The solid line indicates the conserved AAACA motif in the loop of all cre structures. The first nucleotide in the cre sequence, specific for each virus, is indicated. Nucleotide sequences corresponding to poliovirus type 1 PV-cre(2C) which are different from poliovirus type 3 are shown in parentheses.
We have recently made the exciting observation that a highly conserved AAACA motif residing in the loop of PV-cre(2C) can function as template for uridylylation of VPg (27). The efficiency of this reaction, exceeds by far, that of poly(A)-dependent uridylylation with a Mg2+ cofactor. However, it occurs only in the presence of 3CDpro. No other stem-loop structure present within the poliovirus genome can substitute for PV-cre(2C) (27). Here we describe genetic experiments aimed at elucidating the function of PV-cre in genome replication in relation to the uridylylation of VPg. Evidence for the importance of the AAACA motif in the loop and of the secondary structure of the stem has been gathered by analyzing mutant genomes, revertants, and in vitro uridylylation reactions. We have determined a direct correlation between the replication competence of cre(2C) mutants and the ability of the cre element to serve as a template for VPg uridylylation in the in vitro reaction. These results have led us to suggest that two consecutive A residues located on the top of the hairpin (AAACA) are of critical importance for cre function.
MATERIALS AND METHODS
Plasmid construction.
cDNA fragments corresponding to the PV-cre(2C) sequence (nt 4445 to 4504) were obtained by PCR amplification from plasmid pT7PV1 and cloned to pBS to generate the plasmid pBS/cre(2C). This cDNA was amplified using a sense oligonucleotide (P32 N/T7-5′-cre; Table 1) containing an NaeI restriction endonuclease site, the T7 promoter, followed by GG and the 5′-end PV-cre(2C) sequence, and an antisense oligonucleotide (P33 R/N-3′-cre) containing a unique NheI and EcoRI site at the 3′ end of the PV-cre(2C). pBS/cre(2C) mutant plasmids (Fig. 1) were generated using the QuickChange mutagenesis kit (Stratagene) and pBS/cre(2C) as a template as described by the manufacturer. Sense oligonucleotides used for mutagenesis are displayed in Table 1. After mutagenesis, all plasmids were sequenced through the amplified region using Sequenase (U.S. Biochemicals, Cleveland, Ohio). Plasmids digested with NheI and EcoRI were then transcribed with T7 RNA polymerase. To construct full-length mutants, site-directed mutagenesis was carried out using a QuickChange mutagenesis Kit (Stratagene) and pT7PV1 as the template. In this case at least three individual clones for each mutant were analyzed in parallel, and all mutations were verified by dideoxynucleotide sequencing through the 2C region. In addition, mutated 2C fragments were introduced into the poliovirus cDNA replicon pPVM/Luc. The plasmid pPVM/Luc was constructed by X. Li et al. (X. Li, H. Lu, S. Mueller, and E. Wimmer, submitted for publication) and contains a replacement of the poliovirus capsid with the luciferase gene. The 2C-encoding sequences of each mutagenized plasmid (pT7PVM/mut1, pT7PVM/mut2, pT7PVM/mut3, pT7PVM/mut4, pT7PVM/mut6, and pT7PVM/mut7) were excised by digestion with XhoI (nt 4433) and BglII (nt 5600) and were cloned back into PVM/Luc.
TABLE 1.
Oligonucleotides and templates used for mutagenesis and cloning
| Template | Oligonucleotide | Sequencea |
|---|---|---|
| pT7PVM | mut1 | 5′-CAACTACATACAaTTtAAGAGCAAACACCG-3′ |
| pT7PVM | mut2 | 5′-CAACTACATACAGTTCAAGAGCAAgCACCGTATTGAACCAGTATG-3′ |
| pT7PVM | mut3 | 5′-CAGTTCAAGAGCAAACACCGTATTGAgCCAGTATGTTTGCTAGTACATGGC-3′ |
| pT7PVM | mut4 | 5′-CACCGTATTGAACCAGTtTGcTTGCTAGTACATGGCAGCCC-3′ |
| pPV1cre(2C)mut1 | mut5 | 5′-CAAGAATTCGCTAGCAAACATACTGaTTtAATACGGTG-3′ |
| pT7PVM | mut6 | 5′-CAACTACATACAGTTCAAGAGCAcACACCGTATTGAACCAGTATG-3′ |
| pT7PVM | mut7 | 5′-CAACTACATACAGTTCAAGAGCcAACACCGTATTGAACCAGTATG-3′ |
| pT7PVM | P32 N/T7-5′-cre | 5′-GGGCCGGCTAATACGACTCACTATAGGCTCGAGCATACTATTAAC-3′ |
| pT7PVM | P33 R/N-3′-cre | 5′-CCGAATTCGCTAGCAAACATACTGGTTC-3′ |
Uppercase letters indicate sequences of poliovirus, and lowercase letters indicate mutations from the wt sequence. Restriction sites are in italic.
Computer-based prediction of cre RNA folding.
The secondary structure models and thermodynamic prediction of RNA structures at 37°C were obtained by using the MFOLD 3.0 program with Turner energies and selected constraints on base pairing as indicated. The program was run on a server supporting the Zuker website at the University of Washington (http://www.ibc.wustl.edu/∼zuker/mfold).
Cells and viruses.
HeLa-R19 cell monolayers were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% bovine calf serum. Poliovirus type 1, strain Mahoney, i.e., PV1(M), and its mutant derivatives were amplified in HeLa-R19 cells as described by Lu et al. (20). The titer of viral stocks were determined by standard plaque assay in HeLa R19 monolayers as described elsewhere (20).
Transcription, transfection, and translation.
For the production of RNA transcripts in vitro, 2.0 μg of wt or mutant full-length cDNA of PV1(M) clones were linearized at a unique PvuI restriction site downstream of the viral genome. Conditions for in vitro transcription have been described previously (38). For translation, equal amounts of RNA transcripts were used to program translations in HeLa cell extracts (24). After an overnight incubation at 30°C, aliquots of samples labeled with [35S]translabel (ICN Biochemicals) were analyzed by electrophoresis on sodium dodecyl sulfate–12.5% polyacrylamide gels, followed by autoradiography.
RNA transcripts were transfected into HeLa cell monolayers by the DEAE-dextran method, as described previously (38), and the cells were incubated at 37°C in 1× DMEM containing 0.5% fetal bovine serum.
Viral RNA isolation, reverse transcription-PCR amplification, and sequencing.
RNA was isolated from infected cell lysates or individual viral plaques by using Trizol (Life Technologies). Viral cDNAs were synthesized with Moloney murine leukemia virus reverse transcriptase (Life Technologies) using random hexamers as primers, and cDNA fragments were amplified by using standard PCR and specific oligonucleotides. In some cases, amplified fragments were ligated to plasmid DNA vectors using standard techniques, and resulting plasmid DNAs were sequenced with Sequenase (Amersham, Arlington Heights, Ill.). In other cases, the PCR-amplified cDNAs were sequenced directly using the Sequenase PCR sequencing kit (Amersham).
Characterization of viral growth phenotype.
The plaque phenotypes and titers of wt and mutant PV-cre(2C) viruses were measured by plaque assay on 35-mm dishes of HeLa-R19 cell monolayers. After incubation at 37°C for 48 h or longer, viral plaques were developed by 1% crystal violet staining.
Luciferase assay.
HeLa-R19 cells transfected with RNA transcripts containing wt or mutants PVM/Luc were harvested after 10 h posttransfection by washing them three times with phosphate-buffered saline (PBS) and resuspending the cell pellet in 100 μl of PBS. The cells were then lysed by freeze-thawing, and the process was repeated three times. An aliquot of 20 μl of cell lysate was mixed with 100 μl of luciferin (Promega), and the luciferase activity was measured in an Optocomp I Luminometer (MGM Instruments, Inc.).
Assay for in vitro uridylylation of VPg catalyzed by 3Dpol.
The synthesis of VPgpU and VPgpUpU using wt and mutant PV-cre(2C) RNA templates was measured by an assay described before (27). Briefly, reaction mixtures (total of 20 μl) contained 50 mM HEPES (pH 7.5), 8% glycerol, 3.5 mM magnesium acetate, 0.5 μg of cre(2C), 2 μg of synthetic poliovirus VPg, 1 μg of purified 3D polymerase, 0.75 μCi of [α-32P]UTP (3,000 Ci/mmol; Dupont-NEN), 10 μM unlabeled UTP, and 0.3 to 0.5 μg of 3CDpro [3Cpro(H40A)] with C-terminal His tag. The samples were incubated for 1 h at 34°C and the reactions were then stopped by the addition of 5 μl of gel-loading buffer. Samples were analyzed by Tris-tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad) with 13.5% polyacrylamide. Gels were dried without fixing and autoradiographed. Reaction products present at each time point were quantitated with a PhosphorImager (Molecular Dynamics Storm 860) by measuring the amount of [32P]UMP incorporated into product.
RESULTS
Comparison of cre homologues among members of the Picornaviridae.
Replication of picornavirus RNA not only depends upon structural signals residing in the NTRs but also upon cis-acting signals that map to different “internal” regions of the viral genome encoding the polyprotein (1, 42). This was first described for HRV14 by McKnight and Lemon (21, 22), who termed the new genetic element cre (cis acting replication element). cre elements have subsequently been discovered in genomes of cardioviruses (19) and poliovirus type 3 (15). Whereas the cre elements of HRV14 and cardioviruses map to the coding region of the capsid precursor, that of poliovirus type 3 maps to the coding region of the 2CATPase [PV-cre(2C)]. The function of the cre elements is unknown, but the available evidence suggests that they are required for minus-strand RNA synthesis (15, 19, 21, 22). In our search for VPg-uridylylation signals specific for the poliovirus genome, we (27) have discovered that in the presence of 3CDpro the PV-cre(2C) element can replace poly(A) as a specific and highly efficient template for in vitro uridylylation of VPg by 3Dpol.
We have made use of the MFOLD program of Michael Zuker (http://128.252.122.176/∼zuker/rna/) to compare known cre sequences of members of the Picornaviridae family (Fig. 2). Superficially, the stem-loop structures from different genera are quite different in relation to their stems, bulges, or the size of their loops. However, all structures have in common an AAACA motif located in the loop (Fig. 2). The synthesis of VPg-pUpU is expected to occur on a template containing at least two adjacent A's. The conservation of the AAACA motif combined with genetic studies of HRV14 and cardiovirus cre mutants (19, 22) strongly suggested to us a role with a critical function in RNA replication. We speculated that this function may be related to VPg uridylylation (27).
Comparison of wt and mutant PV-cre(2C) in the in vitro uridylylation of VPg.
The discovery that, in the presence of 3CDpro with genomic RNA as template, the 3Dpol-catalyzed uridylylation of VPg occurs primarily on the PV-cre(2C) template and not on poly(A) (27) has prompted us to analyze genetically altered PV-cre(2C) elements. The design of the experiments was based on the assumption that the sequence and structure of the PV-cre(2C) are important for uridylylation. The assays were carried out either with purified cre(2C) RNA or with full-length genomic RNA. The RNAs were incubated with purified 3CDpro and 3Dpol, synthetic VPg, [α-32P]UTP, and Mg2+. The extent of synthesis of VPgpU and VPgpUpU was monitored by polyacrylamide gel electrophoresis as described previously (27, 28).
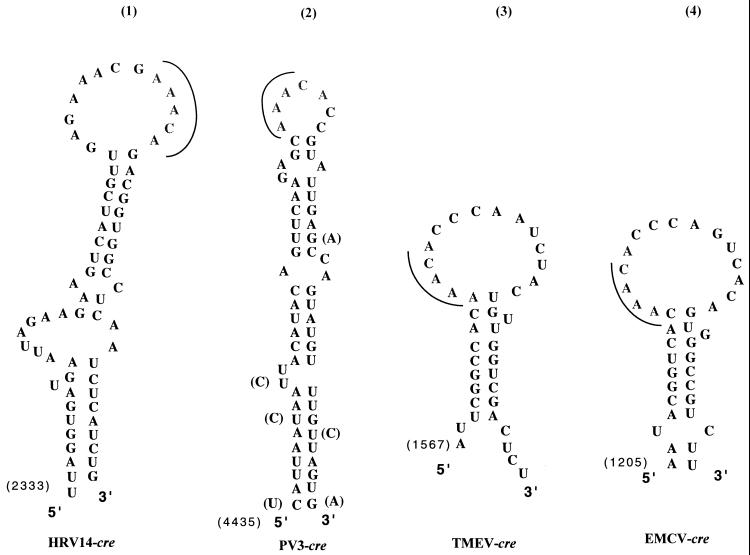
Experiments with purified PV-cre(2C) RNAs are shown in Fig. 3A and B. Of the four mutations in the stem region (mut1, mut3, mut4, and mut5; Fig. 1D), only the mut3 RNA (single base change) supported uridylylation with an efficiency similar to that of wt PV-cre(2C) (Fig. 3A, compare lane 1 with lane 4). This was not unexpected, since the mutation did not significantly alter the structure of the upper stem of PV-cre(2C). However, this mutation is likely to have a profound effect on the structure of the minus-sense cre element (see below). Disruption of the upper stem by the double mutations in mut1 (Fig. 3D) reduced uridylylation of VPg to 2% of the wt reaction (Fig. 3B, lane 2). Neither of the mutations in mut1 resulted in an amino acid change. Interestingly, when the upper stem was restored by introducing two compensatory mutations at nt 4462 and 4487 (AC→AU) and at nt 4465 and 4484 (UG→UA) into mut1 (mut5; Fig. 1D), uridylylation was restored to 23% of the wt reaction (Fig. 3B, lane 2). On the other hand, disruption of the lower part of the stem by the double base change in mut4 resulted in only a slight reduction of uridylylation (Fig. 3B, lane 1). Although limited in scope, the effect of these mutations suggests that the upper stem of PV-cre(2C) serves as an important recognition signal for the RNA to serve as template in uridylylation.
FIG. 3.
Mutant and wt PV-cre(2C) RNAs as templates in VPg uridylylation. The synthesis of VPgpU and VPgpUpU by 3Dpol was assayed as described in Materials and Methods. (A and B) Mutant and wt PV-cre(2C) RNAs as templates for the in vitro reaction. (C) Full-length mutant or wt RNAs as templates in the reaction. The amount of [α-32P]UMP incorporated into the VPgpU and VPgpUpU products was quantitated with a phosphorimager. Autoradiography of the reaction products is shown below.
An interesting pattern emerged from mutations of the AAA triplet of the AAACA motif in the PV-cre(2C) loop. A change of A4474G (AAACA) in mut2 (Fig. 1D) resulted in a fivefold reduction of uridylylation (Fig. 3A, lane 3). The change of A4473C (AAACA) in mut6 decreased the uridylylation even further (Fig. 3B, lane 3). Finally, the function of the PV-cre(2C) element in the in vitro uridylylation reaction was totally eliminated by mutation A4472C (AAACA) in mut7 (Fig. 3B, lane 4). Interestingly, mut6 RNA promoted synthesis of only VPgpU but not of VPgpUpU (Fig. 3B, lane 3).
These experiments were then repeated with in vitro full-length viral RNA transcripts of PV1(M) harboring the mutations described above. Principally, the effect of the mutations was very similar. However, uridylylation was less severely inhibited with mut2 and mut6 (Fig. 3C, lanes 3 and 6, respectively) and more severely inhibited with mut3 (Fig. 3C, lane 4). Remarkably, even in the context of the entire genome carrying poly(A), mut1 and mut7 all but abolished uridylylation (Fig. 3C, lanes 2 and 7, respectively). These results suggest that under the condition of the experiment, the genome does not harbor structures other than PV-cre(2C) that can serve efficiently as a template for uridylylation. This result is in agreement with our recent data (27). We propose that an AA duplet (AAACA) of the PV-cre(2C) loop serves as template for the synthesis of VPgpU and VPgpUpU in vitro by a mechanism further discussed below.
Phenotypic characterization of PV-cre(2C) mutant genomes.
We then examined the ability of full-length viral RNA transcripts of PV1(M), carrying the same PV-cre(2C) mutations, for their ability to produce virus progeny in transfected HeLa-R19 cells. The analysis faces complications, however, because some of the mutations lead to amino acid changes whose effect on viral replication could conceivably be related to the function of 2CATPase rather than that of the PV-cre(2C). Specifically, mutations in PV-cre(2C) mut6 and mut7 result in amino acid changes K117T and K116Q, respectively. We originally chose the K117T (A4473C) change in mut6 because in enterovirus 71 (ENV71) the loop in the presumptive cre element carries an ACACA instead of an AAACA sequence (7).
Transfection experiments were carried out in HeLa-R19 cell monolayers using similar amounts of wt and mutant RNAs. The growth phenotypes of these constructs are shown in Table 2. Similar to the wt, mut3 transcript RNA showed complete cytopathic effect (CPE) after incubation at 37°C for 24 h posttransfection (p.t.). mut2 and mut4 RNAs produced CPE after 36 h, and mut6 RNA produced CPE after 72 h p.t. Thus, a gradient of replication efficiency (reflected also in a decrease of plaque size) was apparent roughly corresponding to the extent of uridylylation in vitro (Table 2). Significantly, no CPE was detected with mut1 and mut7 transcripts even after 5 days of incubation p.t.
TABLE 2.
Properties of viruses recovered from HeLa-R19 transfected cells
| RNAa | Plaque
|
||||
|---|---|---|---|---|---|
| CPE (h)b | PFU/mlc | Phenotypee | Reversion | Reversionf | |
| PV-cre wt | 24 | 1 × 108 | L, M | ||
| PV-cre mut1 | 0d | Yes | U4465C | ||
| PV-cre mut2 | 36 | 5.8 × 107 | L, M | No | |
| PV-cre mut3 | 24 | 7 × 107 | L, M | No | |
| PV-cre mut4 | 36 | 4 × 107 | L, M | No | |
| PV-cre mut6 | 72 | 4 × 103 | M | Yes | C4473A |
| PV-cre mut7 | 0d | No | |||
cDNA clone used as source of RNA. Two or more clones for each mutant were analyzed.
Hours p.t. to complete CPE.
Amount of infectious virus recovered from transfected cells.
No PFU detected in a 200-μl sample.
Determined by plaque assay. L, large plaques; M, medium-size plaques; L, M, mixture of large and medium-size plaques.
Determined in three individual plaque-purified virus stocks.
We suspected that virus produced in mut6 RNA-transfected cells, showing CPE only after 72 h p.t., might be revertants, that is, mut6 RNA might be quasi-infectious. This was confirmed by sequence analysis of the genomic RNA of three different isolates that revealed the reversion at C4473A, thereby restoring the wt sequence in the AAACA motif. We then also searched for revertants in cells transfected with mut1 and mut7 RNAs after five blind passages in HeLa cells. Indeed, plaques eventually appeared in cells originally transfected with mut1 RNA of which three isolates were sequenced. The genome of all of these variants had regained a C residue at position 4465 but retained the A4462G change (Table 2). Thus, the reversion (U4465C) allowed for partial restoration of the upper stem (see Fig. 1D). A similar result was obtained very recently by Goodfellow et al. (15) with a corresponding mutation in the PV3-cre(2C). These results underline the importance of the integrity of the upper stem region for PV-cre(2C) function. We also conclude that mut1 RNA, just like mut6 RNA, is quasi-infectious.
In contrast to mut1 RNA transfected cells, no revertants of mut7 RNA were detected. Thus, the genetic alteration in mut7 RNA is lethal. This observation covaries with the complete failure of mut7 RNA to serve as template in uridylylation in vitro. It strongly suggests that the lethal phenotype of mut7 RNA relates to its inability to serve as template in the synthesis of VPgpUpU. Alternatively, the K117Q change in the 2CATPase may be lethal for viral proliferation. Currently, we cannot rule out this second possibility.
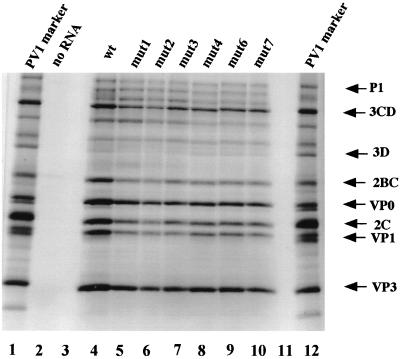
To evaluate whether the defect in replication of mutant RNAs was a consequence of a reduced translation efficiency or aberrant proteolytic processing we translated mut1, mut2, mut3, mut4, mut6, and mut7 RNAs in HeLa cells extracts known to mimic conditions of poliovirus replication (24). RNAs from all the PV-cre(2C) mutants produced all poliovirus-specific polypeptides (Fig. 4, compare lane 4 with lanes 5 to 10). None of the substitutions introduced into the PV cre sequence had a significant effect on protein synthesis or processing in vitro.
FIG. 4.
In vitro protein synthesis and processing directed by the parental PV1(M) RNA and five cre(2C) mutant derivatives. Similar amounts of RNA were translated in HeLa cell extracts at 30°C overnight and then analyzed on 12.5% sodium dodecyl sulfate-polyacrylamide gels. [35S]methionine-labeled PV1(M) proteins extracted from an infected HeLa cell extract were used as markers.
The PV-cre(2C) function is required as positive-sense RNA and cannot be complemented in trans.
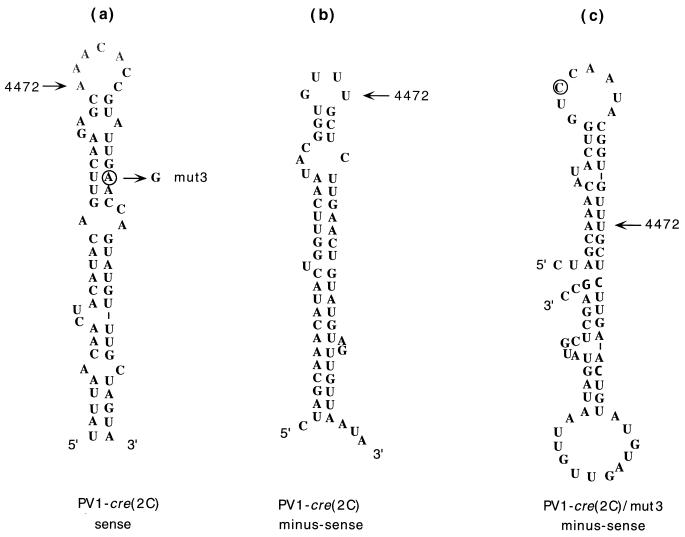
Computer-aided modeling predicts a structure for a putative minus-sense PV-cre(2C) sequence that is shown in Fig. 5b. In the plus sense PV-cre(2C), the mut3 mutation (A4486G) is expected to preserve base pairing of the upper stem (U=A changed to U=G; Fig. 5a), whereas in the putative minus-sense PV-cre(2C) the same mutation would cause major structural alterations (Fig. 5c). Since the A4486G mutation had no effect on viral infectivity, we suggest that only the plus-sense PV-cre(2C) is functional. Support for this conclusion comes from analyses of poliovirus minigenomes containing PV-cre(2C) elements in both the sense and the antisense orientations. In particular, minigenomes containing cre in the sense orientation can be utilized as a template for uridylylation of VPg in vitro. The same minigenomes with the cre(2C) hairpin in the antisense orientation are nonfunctional (27).
FIG. 5.
Predicted secondary structure of the PV-cre(2C) at 37°C in the sense and antisense strands as modeled by the MFOLD program of Michael Zuker. The position of nucleotide 4472 is indicated for orientation. (a) PV-cre(2C) folding of the sense strand. The circle indicates the location of the substitution corresponding to mut3. (b) Secondary structure of PV-cre(2C) corresponding to the antisense strand. (c) Secondary structure in the antisense strand predicted to be formed by PV-cre(2C) mut3 RNA. A circle highlights the mutation in PV-cre(2C) mut3.
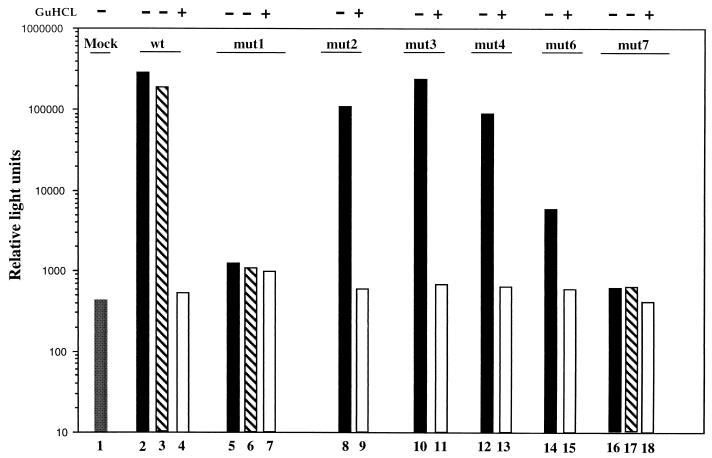
To determine whether the PV-cre(2C) mutants can be complemented in trans, we used poliovirus replicons carrying the luciferase gene instead of the P1 capsid coding region (Li et al., submitted). Relevant cre(2C) substitutions were introduced into full-length plasmids specifying the PVM/Luc replicons (see Materials and Methods). Transcript RNAs corresponding to wt or mutant PV-Luc replicons were then transfected into HeLa cells, and cell extracts were analyzed for luciferase activity at 10 h p.t., a time when the luciferase expression reaches its peak (Li et al., submitted). In parallel experiments we added guanidine hydrochloride (GuHCl) to a final concentration of 2 mM to the cell cultures to estimate the luciferase signal derived from translation of the transfected RNA (Fig. 6, open bars). This is because GuHCl completely inhibits replication of poliovirus RNA (34) without interfering with luciferase activity (2, 32). A high luciferase activity was obtained with wt PVM/Luc and with mut2, mut3, and mut4 RNAs (Fig. 6, lanes 2, 8, 10, and 12). PVM/Luc mut6, however, showed a significantly reduced luciferase signal compared to the wt (Fig. 6, compare lane 14 with lane 2). This was expected from the replication phenotype of this mutant RNA (Table 2). No luciferase activity was detected over time with RNAs corresponding to mut1 and mut7 (Fig. 6, lanes 5 and 16). These results corroborate the data shown before and underline the importance of the cre(2C) element.
FIG. 6.
Transfections of the mutant and the wt PV1/Luc replicon RNAs into HeLa cells. HeLa cell monolayers were transfected with 5 μg of each replicon RNA. The cell lysates were collected at 10 h p.t. The luciferase activity for each cell lysate was determined with a luminometer. In parallel, identical transfections were repeated as described above but with 2 mM GuHCl present. Lanes 3, 6, and 17 correspond to trans-rescue assays with wt transcripts and replication defective PV1/Luc replicons (lanes 3, 6, and 17, respectively). For details, see the text.
HeLa cells were then transfected with PVM/Luc-mut1, -mut7, or -wt RNAs and superinfected 1 h later with full-length PV1 RNA. At 10 h p.t., cell extracts were prepared and assayed for luciferase activity. The results suggested that the replication defect expressed by mut1 and mut7 could not be repaired by providing wt cre(2C) in trans (Fig. 6, compare lane 3 with lanes 6 and 17).
DISCUSSION
cre replication signals mapping to different loci in the open reading frame of the picornavirus genome have been reported for member viruses of the genera Rhinovirus, Cardiovirus, and Enterovirus of Picornaviridae (15, 19, 21, 22). In all cases, these genetic elements may be essential for the initiation of minus-strand RNA synthesis. However, the mechanism by which these cre elements exert their function is still a mystery.
Initiation of poliovirus RNA synthesis is coupled to uridylylation of VPg, whereby the resulting VPgpUpU is serving as primer for the RNA and primer-dependent RNA polymerase 3Dpol (28). In vitro, 3Dpol-catalyzed uridylylation of VPg is stringently dependent on a template. Poly(A), but not any other homopolymer, can serve as a template. However, the efficiency of the reaction is poor with a Mg2+ cofactor (28). Nevertheless, since the 5′ end of minus strands is poly(U), uridylylation at the 3′-terminal poly(A) of genomic RNA, followed by transcription to form poly(U)-terminated minus strands, seemed a sensible mechanism for initiating poliovirus RNA replication. However, cellular mRNAs carrying 3′-terminal poly(A) are abundantly present in the cytoplasm of host cells, a fact calling for signals within viral RNA that would direct uridylylation exclusively to the viral genome. Neither the 3′-heteropolymeric NTR nor any element of the long 5′NTR are able to serve as a specific recognition signal and/or template for uridylylation (27). This led to the discovery that the PV-cre(2C) RNA functions very efficiently to promote uridylylation. Interestingly, the reaction is dependent not only on 3Dpol and PV-cre(2C) but also on the viral polypeptide 3CDpro or the component(s) present in a translation reaction of PVM RNA in HeLa extracts (27).
Biochemical parameters of PV-cre(2C)-dependent uridylylation have been described in detail (27). Here we extend the study of the function of PV-cre(2C) in uridylylation by mutational analyses. The objective of the work is to correlate in vitro data with replication phenotypes. Moreover, experiments have been designed to define structural features of the PV-cre(2C) element important for uridylylation.
McKnight and Lemon (22) carried out an analysis of the HRV14-cre element in replicons and concluded that mutations disrupting base-paired sequences within the helical segment or changing the primary sequence in the loop are lethal for replication. Goodfellow et al. (15) demonstrated the importance of the upper stem region of the PV-cre(2C) for function. Lobert et al. (19) have reported that a CACAAACAC sequence located mainly in the loop of the cardiovirus cre element is essential for RNA replication.
All four cre sequences shown in Fig. 2 (HRV14, poliovirus, Theiler's virus, and encephalomyocarditis virus (EMCV) contain an AAACA motif that may be considered important for cre function. However, even within the genus Enterovirus this motif is not conserved (see Fig. 1 in reference 15). It appears that variations are specific to enterovirus clusters (16). In all C-cluster enteroviruses (polioviruses, coxsackievirus A21, coxsackievirus A24, etc.), the AAACA motif is invariable. On the other hand, in all B-cluster enteroviruses (coxsackievirus A9, coxsackie B viruses, and echoviruses) this motif is AAAUG, assuming that the corresponding RNA segment in the 2C-coding region constitutes a cre element. Exceptions to this pattern are the Sabin strain of PV2 (AAGCA), ENV71 (ACACA; this sequence may have to be updated), and bovine enteroviruses (AAGAA).
Based on our recent studies (27) and the data presented here we argue that the first 2 nt of the AAACA motif in PV-cre(2C) serve as a template in genome-specific uridylylation of picornaviruses (see below). We predict that the first two A residues of the motifs in B-cluster enterovirus (AAAUG) and bovine enteroviruses (AAGAA) in the cre elements serve the same function.
Our analysis targeted the first three A residues of the AAACA motif for mutation. Overall, the effect of these mutations either on uridylylation of VPg or replication phenotype covaried, a result greatly supporting the conclusion that one of the functions of PV-cre(2C) is to serve as template for VPg uridylylation (27). Surprisingly, mutation A4472C of the first A (mut7; CAACA) produced the most severe effect on in vitro uridylylation and replication; it totally abolished VPgpU or VPgpUpU synthesis and conferred a lethal phenotype to the full-length viral RNA. A similar debilitating effect of mutation of the first A residue of the AAACA motif in HRV14-cre on the replication of HRV14 has been reported by McKnight and Lemon (22). Mutation A4473C of the second A (mut6, ACACA) greatly reduced the efficiency of uridylylation and produced quasi-infectious RNA. That is, only RNA that had reverted to the wt sequence was detectable in cells producing progeny virus. Mutation A4474G of the third A (mut2, AAGCA) exerted the smallest negative effect on either uridylylation or the replication phenotype. This ranking of relative importance of the A residues (A ≫ A ≫ A) is difficult to explain. A clue comes from the observation that uridylylation with a PV-cre(2C) RNA harboring the ACACA mutation (mut6) produces predominantly VPgpU (Fig. 3B, lane 3).
We have previously speculated why the in vitro uridylylation at a low UTP concentration with poly(A) as template produces VPgpU, followed by VPgpUpU, but no VPgpUpUpU, etc. (28). We have entertained the possibility that a slide-back mechanism that has been reported for the nucleotidylylation reaction of the terminal proteins of adenovirus or phage Ph29 (36) may be functioning also in poliovirus uridylylation. We could assume, for example, that the first step of uridylylation occurred on A2 of the following poly(A) sequence below: AnA9A8A7A6A5A4A3A2AOH→VPgpU. According to the slide-back mechanism, the second pU to form VPgpUpU would not be encoded from A3 but from A2 again after the uridylylation complex translocated to, and the newly synthesized VPgpU based paired with, AOH. In the case of the AAACA motif, the first step in uridylylation could occur on A4472 (AAACA), followed by translocation of the uridylylation complex to, and base pairing of the newly synthesized VPgpU with, A4473 (AAACA). The second uridylic acid residue would then be transcribed again from A4472. If the base downstream of A4472 is a C residue (ACACA; mut6), uridylylation would be aborted with VPgpU (Fig. 3B, lane 3). If the first base of the “uridylylation motif” is a C residue (CAACA; mut7), no uridylylation is possible (Fig. 3B, lane 4). We note, however, that with full-length poliovirus RNA carrying mut6, both VPgpU and VPgpUpU were synthesized (Fig. 3C, lane 6). It is possible that in the context of the large RNA template the uridylylation complex, after synthesis of VPgpU, transferred to a surrogate AA serving to encode the second pU. Experiments to test the validity of the translocation hypothesis are in progress.
As mentioned before, the lethal phenotype of the A4472C mutation in PV-cre(2C) could also be the result of impaired function(s) of the 2CATPase, rather than related to uridylylation, since the mutation introduced an amino acid change K117Q. We consider this possibility less likely, however, because synthesis and processing of the polyprotein by mut7 RNA is similar to that of wt RNA. Moreover, no significant differences in ATPase activity by recombinant 2C/mut7 could be detected when compared to 2C/wt (T. Pfister, unpublished results). Nevertheless, 2CATPase is a protein with multiple functions (29, 30), and further studies are warranted to clarify this issue.
The AAACA motif is only one of the essential signals recognized by the viral uridylylation machinery. The integrity of the upper stem of PV-cre(2C) is clearly important since the silent double mutation in mut1 abolished uridylylation and replication. However, we were able to uncover revertants from mut1 RNA-transfected cells, an observation suggesting that this mut1 RNA is quasi-infectious and that the mutations in this RNA are not lethal (for a definition of the quasi-infectious phenotype, see references 14 and 16). The single nucleotide revertant restored only partially the upper stem, which apparently was sufficient to affect viral proliferation. However, a detailed analysis of the revertant has yet not been carried out. Compensatory mutations (mut5) of the mut1 double mutation resulted in only partial restoration of the uridylylation reaction (Fig. 3B, lane 2). These observations, together with the replication phenotypes described here and reported by Goodfellow et al. (15), can be interpreted to mean that the recognition of PV-cre(2C) by the uridylylation complex is highly sensitive to structure and/or sequence alterations. Nevertheless, the HRV14-cre has been found to serve efficiently as the template in poliovirus 3Dpol-catalyzed uridylylation in vitro (27). Apparently, the poliovirus uridylylation complex can recognize HRV14-cre just as the poliovirus replication machinery can recognize the HRV14 3′NTR. These observations underline our ignorance of RNA-protein recognition processes related to picornavirus replication (1).
If in vivo the VPgpUpU primer is formed exclusively on the PV-cre(2C) element, as we have proposed (27), the question arises as to how the primer is being translocated to the 3′-terminal poly(A) of plus strands or the 3′-terminal UUUUAAOH of minus strands. Available evidence suggests that transfer must occur in cis but, currently, the mechanism of such translocation in other viral systems is obscure (discussed in reference 27).
Recognition of PV-cre(2C) by the uridylylation complex not only involves polymerase 3Dpol but, surprisingly, also 3CDpro (27). Indeed, as determined by gel shift analysis in the presence of a large excess of competitor RNA, 3Dpol did not reveal binding specificity to PV-cre(2C), whereas 3CDpro was found to form complexes with the RNA probe (E. Rieder, A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer, manuscript in preparation). 3CDpro is not only a potent proteinase but also an RNA binding protein with affinity to the 5′-terminal cloverleaf in the presence of viral (3AB) or cellular (PCBP2) proteins (4, 13, 17, 25, 41, 42). Mutations in 3CDpro that abolish RNA binding of the protein also abolish PV-cre(2C)-dependent uridylylation (27). Thus, we have uncovered a new function for 3CDpro. Surprisingly, 3Dpol failed to supershift the PV-cre(2C)/3CDpro complex in polyacrylamide gel electrophoresis analyses (Rieder et al., in preparation). Perhaps the uridylylation complex PV-cre(2C)/3Dpol/3CDpro can only form in the presence of adequate concentrations of UTP and Mg2+ ions. This possibility is currently being tested.
Apart from 3CDpro, we have observed that a 50-kDa cellular protein has the propensity to bind to PV-cre(2C) RNA in the presence of competitor RNA (Rieder et al., in preparation). The significance of this observation is not known. It should be noted, however, that the uridylylation reaction involving PV-cre(2C), 3Dpol, and 3CDpro is not stimulated by components of the HeLa cell extract, nor can any protein of the HeLa cell extract substitute for 3CDpro (27).
We note that the AAACA motif of the PV-cre(2C) element of the oral Sabin vaccine strain of poliovirus type 2 [PV2(S)] contains the mutation AAGCA. Our data indicate that this mutation (mut2) in the context of PV1(M) RNA reduces the replication efficiency in HeLa cells. It is intriguing to speculate that this mutation may contribute to the attenuation phenotype of PV2(S), just like the uridylylation phenotype of purified 3Dpol that carries mutations of the oral Sabin vaccine strain of poliovirus type 1 (26).
ACKNOWLEDGMENTS
We are indebted to B. Semler and T. Pfister for the gift of 3CDpro and the 2C expression vectors, respectively. We thank Xiaoyu Li for providing the PVM/Luc replicon and M. Shepley for critically reading the manuscript.
This work was supported by NIH NIAID grant 5R37AI15122.
REFERENCES
- 1.Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Lu H H, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–5266. [PubMed] [Google Scholar]
- 4.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 6.Blyn L B, Chen R, Semler B L, Ehrenfeld E. Host cell proteins binding to domain IV of the 5′ noncoding region of poliovirus RNA. J Virol. 1995;69:4381–4389. doi: 10.1128/jvi.69.7.4381-4389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown B A, Pallansch M A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 8.Detzen B M, Lucas J, Wimmer E. Poliovirus single-stranded RNA and double-stranded RNA: differential infectivity in enucleate cells. J Virol. 1978;27:582–586. doi: 10.1128/jvi.27.3.582-586.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsch-Haesler K, Yogo Y, Wimmer E. Replication of picornaviruses. 1. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J Virol. 1975;16:1512–1527. doi: 10.1128/jvi.16.6.1512-1517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanegan J, Pettersson R, Ambros V, Hewlett M, Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci USA. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flanegan J B, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci USA. 1977;74:3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 14.Gmyl A P, Pilipenko E V, Maslova S V, Belov G A, Agol V I. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J Virol. 1993;67:6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond J W, Barclay W, Evans D J. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gromeier M, Wimmer E, Gorbalenya A E. Genetics, pathogenesis, and evolution of picornaviruses. In: Domingo E, Webster R G, Holland J J, editors. Origin and evolution of viruses. New York, N.Y: Academic Press, Inc.; 1999. pp. 287–343. [Google Scholar]
- 17.Harris K S, Xiang W, Alexander L, Paul A V, Lane W S, Wimmer E. Interaction of the polioviral polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors necessary for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 18.Lee Y, Nomoto A, Detjen B, Wimmer E. The genome-linked protein of picornaviruses. I. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobert P E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci USA. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu H-H, Yang C-F, Murdin A D, Klein M H, Harber J J, Kew O M, Wimmer E. Mouse neurovirulence determinants of poliovirus type 1 strain LS-a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J Virol. 1994;68:7507–7515. doi: 10.1128/jvi.68.11.7507-7515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melchers W J, Hoenderop J G, Bruins Slot H J, Pleij C W, Pilipenko E V, Agol V I, Galama J M. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 25.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 26.Paul A V, Mugavero J, Yin J, Hobson S, Schultz S, Van Boom J H, Wimmer E. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology. 2000;272:72–84. doi: 10.1006/viro.2000.0354. [DOI] [PubMed] [Google Scholar]
- 27.Paul A V, Rieder E, Kim D W, van Boom J H, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 29.Pfister T, Jones K W, Wimmer E. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J Virol. 2000;74:334–343. doi: 10.1128/jvi.74.1.334-343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfister T, Wimmer E. Characterization of the nucleotide triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits replication of poliovirus. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 31.Pilipenko E V, Poperechny K V, Maslova S V, Melchers J G, Bruins Slot H J, Agol V I. cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 32.Pincus S E, Wimmer E. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J Virol. 1986;60:793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohll J B, Percy N, Ley R, Evans D J, Almond J W, Barclay W S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothberg P, Harris T, Nomoto A, Wimmer E. The genome-linked protein of picornaviruses. V. O4-(5′-uridylyl)-tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 37.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;78:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wimmer E, Hellen C U, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 40.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang W K, Paul A V, Wimmer E, editors. RNA signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]
- 43.Yogo Y, Teng M, Wimmer E. Poly(U) in poliovirus minus RNA is 5-terminal. Biochem Biophys Res Commun. 1974;61:1101–1109. doi: 10.1016/s0006-291x(74)80397-9. [DOI] [PubMed] [Google Scholar]
- 44.Yogo Y, Wimmer E. Polyadenylic acid at the 3-terminus of poliovirus RNA. Proc Natl Acad Sci USA. 1972;69:1877–1882. doi: 10.1073/pnas.69.7.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ypma-Wong M-F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]