Abstract
Carnation small viroid-like RNA (CarSV RNA) and its homologous DNA are the two forms of a unique plant retroviroid-like system. CarSV RNA is a 275-nucleotide noninfectious viroid-like RNA, present in certain carnation plants, which can adopt hammerhead structures in both polarity strands and self-cleave accordingly. CarSV DNA is organized as a series of head-to-tail multimers forming part of extrachromosomal elements in which CarSV DNA sequences are fused to sequences of carnation etched ring virus (CERV), a plant pararetrovirus. Analysis of more than 30 CarSV-CERV DNA junctions showed that distinct regions of the viral genome seem able to interact with CarSV DNA. All these junctions were short nucleotide stretches common to both CarSV and CERV DNAs. This suggests a polymerase-driven mechanism for their origin involving an enzyme with low processivity, most likely the viral reverse transcriptase. This view was further supported by the observation that most of CarSV sequences forming part of the junctions correspond either to strong secondary structure motifs in the conformation proposed for CarSV RNA or to its self-cleavage sites, which may have facilitated polymerase jumping. Accompanying the most-abundant CarSV RNA, a series of CarSV RNAs with sequence deletions were previously characterized. Here we have identified some of their corresponding DNA forms, together with other CarSV DNA forms with deletions not found in any CarSV RNA species identified so far. Some of these CarSV DNA forms have also been detected fused to CERV sequences. The existence of these shortened CarSV DNA versions may provide a continuous input of their corresponding transcripts and explain the persistence of CarSV RNAs with defective hammerhead structures for which an RNA-RNA model of amplification seems unlikely.
Carnation small viroid-like RNA (CarSV RNA) and its homologous DNA, have been proposed as the two forms of a unique plant retroviroid-like system (5). CarSV RNA is a 275-nucleotide (nt) circular RNA present in certain carnation plants which can form hammerhead structures in both polarity strands and self-cleave in vitro as predicted by these ribozymes (16). Despite the structural similarities that CarSV RNA shares with viroid and viroid-like satellite RNAs (for reviews on these pathogens, see references 2, 9, 14, 15, and 31), two main differences separate them: CarSV RNA lacks an extracellular infectious phase, i.e., it cannot be transmitted horizontally from plant to plant, and also it exists as a homologous DNA counterpart (5; L. Palkovics, K. Salánki, A. Wittner, E. Tóth, and E. Balázs, 5th Int. Cong. Plant Mol. Biol., abstr. 1019, 1997). CarSV DNA accumulates at low levels in carnation since it is only detectable by PCR amplification, and examination of the patterns of PCR-amplified products indicates that CarSV DNA is organized as a series of head-to-tail multimers (5). The CarSV RNA-DNA element bears a close resemblance to two small linear RNAs from the newt (330 nt) and schistosomes (335 nt), which are also able to adopt hammerhead structures and to self-cleave accordingly, and that are transcribed from tandemly repeated DNA sequences (11, 13). The CarSV RNA-DNA element has also some similarity with the mitochondrial VS RNA from Neurospora, a single-stranded circular molecule of 881 nt able to self-cleave through a nonhammerhead ribozyme, which is transcribed from a low-copy double-stranded circular VS DNA population also organized as a series of head-to-tail multimers (29).
Further analysis showed that CarSV DNA multimers form part of extrachromosomal elements in which the CarSV DNA is directly fused to sequences of Carnation etched ring virus (CERV) (5), a plant pararetrovirus of the family Caulimoviridae which, like the other species of this family, replicates by a mechanism of reverse transcription involving an RNA intermediate (17, 19, 24). Three different CarSV-CERV DNA fusions were initially identified in which the CERV sequences flanking the junctions corresponded to three relatively close positions within the viral genome, mapping specifically within open reading frame (ORF) V, which codes for the reverse transcriptase (5). Moreover, the junctions were characterized by 4 to 6 nt shared by the CarSV and CERV sequences. These observations raise the questions of whether CarSV-CERV DNA fusions are just restricted to some regions of the viral genome or whether they are more widely distributed through the CERV DNA and, if this is so, whether the peculiar structure of the junctions between CarSV and CERV sequences is preserved.
On the other hand, a previous study revealed that the predominant CarSV RNA of 275 nt coexists in vivo with minor amounts of other small circular RNAs. These forms present sequence deletions and/or repetitions with respect to the most-abundant CarSV RNA (CarSV-0) and have been termed CarSV-S (smaller-than-unit) and CarSV-L (longer-than-unit) forms (6). In this context, another unanswered question is whether these minor CarSV RNAs also have DNA counterparts.
In this work, we have addressed these issues in an attempt to get a deeper insight into the structure and possible mechanisms of emergence and maintenance of this singular retroviroid-like element.
MATERIALS AND METHODS
DNA extraction.
Leaves were collected from vegetatively propagated carnation plants (Dianthus caryophyllus L.) of three commercial cultivars ('Indios', 'Carola', and 'Killer') positive for CarSV RNA and DNA as revealed by Northern blot hybridization and PCR amplification, respectively (5). In some control experiments, leaves from a fourth commercial carnation cultivar ('Sarinah') negative for CarSV RNA and DNA were also collected. Total genomic DNA was obtained by a previously reported protocol (7), followed by RNase A treatment, extraction with buffer-saturated phenol, and recovery of nucleic acids by ethanol precipitation. In some specific cases, DNA was further purified by equilibrium sedimentation in a CsCl gradient.
PCR amplification, cloning, and sequencing.
Detection of CarSV DNA forms with sequence deletions with respect to the reference sequence (5) was performed by PCR amplification using two pairs of adjacent CarSV-specific primers of opposite polarities: PI and PII (complementary and identical to nt 257 to 14 and nt 15 to 39 of CarSV-0 RNA, respectively), or PIII and PIV (complementary and identical to nt 174 to 137 and nt 175 to 198 of CarSV-0 RNA, respectively) (Fig. 1). To identify junctions between CarSV and CERV DNAs, a CarSV-specific primer, usually PI or PIII, was used in combination with any of the six CERV-specific primers indicated in Table 1.
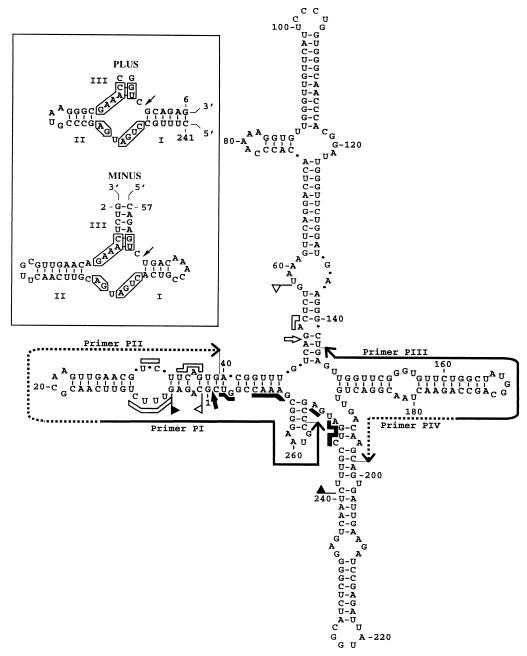
FIG. 1.
Cruciform secondary structure of lowest free energy proposed for the plus polarity CarSV-0 RNA (16). The self-cleavage domains of both polarities are delimited by flags, the 13 residues conserved in most natural hammerhead structures are indicated by bars, and the self-cleavage sites are indicated by arrows. Solid and open symbols refer to the plus and minus polarities, respectively. Continuous and dotted lines with arrowheads denote regions covered by two pairs of adjacent primers with sequences complementary and homologous to CarSV-0 RNA, respectively. (Inset) Hammerhead structures of plus and minus CarSV-0 RNA (16). Arrows denote the predicted self-cleavage sites, helices are labeled I to III, and the 13 conserved residues are boxed. The same numbering is used in the plus and minus polarities.
TABLE 1.
Nucleotide sequence of CERV-specific primers
| Primer | Positiona | Sequenceb |
|---|---|---|
| P1 | 3991–4014 (ORFV) | 5′ TTCTATGGTTGCTGTCATCCATTGC 3′ |
| P2 | 2538–2571 (ORFIV) | 5′ CTCCTAAGAACATGGTATATAgaCCaCTGACTAC 3′ |
| P3 | 1708–1737 (ORFIII) | 5′ GCTGTCTTGTTCCGGAGATGGAAaGATAGC 3′ |
| P4 | 1491–1513 (ORFIII) | 5′ CTGACGCcATGGTGGCCAGATTC 3′ |
| P5 | 1368–1379 (ORFII) | 5′ CTAGAGAGTTCCTTAACCTGAG 3′ |
| P6 | 7866–7888 (S) | 5′ TCTCTGAAAGAGCTGCGAtGGCGG 3′ |
Complementary to the indicated positions of the CERV sequence (accession no. X04658 (19). The ORFs to which the positions belong are shown in parentheses. S corresponds to the intergenic region between ORF I and ORF VI.
Lowercase letters denote nucleotide substitutions with respect to the database CERV sequence.
PCR amplifications were carried out in a volume of 50 μl containing 1 μg of purified total carnation DNA, 500 ng of each primer, 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, a 200 mM concentration each of the four deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase. The PCR cycling profile consisted of a hot start (2 min at 94°C) prior to the addition of 1 U of Taq DNA polymerase (Boehringer Mannheim) followed by 30 rounds of amplification (30 s at 94°C, 30 s at 60°C, 2 min at 72°C) and a final extension step of 10 min at 72°C. In some cases Taq DNA polymerase was replaced by Pfu DNA polymerase and the buffer recommended by the supplier (Stratagene). The PCR-amplified products were separated by electrophoresis in 5% polyacrylamide and/or in 1% agarose gels in 1× TAE (40 mM Tris, 20 mM sodium acetate, 1 mM EDTA [pH 7.0] with acetic acid) that were stained with ethidium bromide. The DNA fragments were eluted from the gel and cloned in the linearized and thymidylated pT7Blue (R) plasmid (Novagen) when amplified with Taq DNA polymerase and into the SmaI-linearized and dephosphorylated pUC18 (Pharmacia) when amplified with Pfu DNA polymerase. Inserts were sequenced manually with dideoxy chain terminators (Pharmacia T7 sequencing kit) or automatically with an ABI PRISM DNA sequencer (Perkin-Elmer).
Southern blot hybridization.
Total DNA (10 μg) from plants of the different carnation cultivars were electrophoresed in 1% agarose gels with 1× TAE. The DNA was blotted to Hybond-N+ membranes (Amersham) and fixed by UV irradiation with a Stratalinker apparatus (Stratagene). The filters were hybridized using a radioactive CERV-specific DNA probe prepared by random priming following standard protocols (28).
Sequence analysis.
Comparisons of the sequences of the PCR-amplified products with those of CarSV and CERV genomes were carried out with the BESTFIT program of the Genetics Computer Group package (8).
RESULTS
Detection of CarSV DNA sequences fused to multiple regions of the CERV genome.
A PCR amplification strategy using different pairs of primers, combining in each case one CarSV- and one CERV-specific primer (Fig. 1 and Table 1), was carried out in order to search for potential CarSV-CERV DNA fusions in different regions of the virus genome. For this purpose, total DNA preparations from carnation plants of the cultivar 'Indios', in which the presence of the CarSV RNA-DNA retroviroid-like element has been previously reported (5), were used as templates. To exclude the possibility that the fused CarSV-CERV sequences could result from intermolecular jumps of the Taq DNA polymerase during the in vitro amplification process, a set of control experiments were performed. To this aim, total DNA preparations from the cultivar 'Indios' were mixed with different amounts of linearized or circular forms of an externally added plasmid, which would mimic the CERV genome, and PCR amplifications were carried out using a CarSV-specific primer and several plasmid-specific primers. Repeated assays following this approach did not produce any fused CarSV-plasmid sequences (the amplification products that appeared occasionally contained sequences of only one of the templates as a result of nonspecific primer annealing), thus validating the strategy to identify CarSV-CERV fusions.
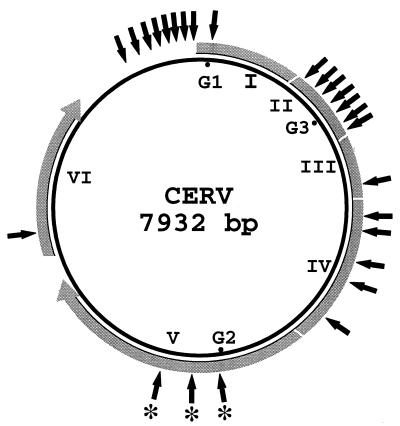
Many different CarSV-CERV junctions were mapped along the CERV genome (Fig. 2). CarSV DNA was observed fused to CERV DNA sequences corresponding to ORF I, in which one junction site was identified, ORF II (seven junction sites), ORF III (one junction site), ORF IV (five junction sites), ORF V (three junction sites), ORF VI (one junction site), and even in the long intergenic region between ORF I and VI (eight junction sites). Therefore, these results indicated that distinct regions of the viral genome are susceptible to sequence recombination with CarSV DNA.
FIG. 2.
Location of CarSV-CERV DNA junctions identified in the carnation cultivar 'Indios' with reference to the CERV circular genome. Viral ORFs are indicated by roman numerals, and G1, G2, and G3 correspond to the presumed gaps in the viral DNA. The outer grey discontinuous lines show the relative length of the coding regions translated in the direction denoted by arrowheads. External solid arrows mark approximate positions in which CarSV-CERV DNA junctions have been identified, and asterisks indicate previously characterized junctions (5).
The common fingerprint of the three CarSV-CERV DNA junctions initially described was a short stretch of nucleotides shared by both DNAs (5). The new CarSV-CERV DNA junctions characterized in the present work displayed the same feature, with the length of the shared sequences varying between 3 and 18 nt (Fig. 3). A perfect match between the sequences forming the junctions and those of CarSV and CERV was generally found with only a few exceptions affecting some nucleotides. It should be pointed out that for comparison purposes we have used the only available complete DNA sequence of a CERV isolate deposited in databases (19), and therefore, it is not surprising that a number of substitutions, or even small deletions and repetitions with respect to this published CERV sequence, were found in the viral DNA (5; data not shown). To establish the precise length of the junctions and to identify those positions that deviate from the match, the CarSV-0 sequence was chosen as a reference. Following this criterion, fusion F6 and 18 shared nt with two mismatches of one nucleotide each; fusion F10, with 9 shared nt, was that with the longest perfect match (Fig. 3). Rearrangements of the CERV DNA itself were also sporadically detected and in some cases involved recombination between sequences of different viral ORFs and between the strands of both polarities. Interestingly, the presence of shared nucleotides was also observed in the CERV-CERV junctions (see example in Fig. 4), suggesting a common underlying mechanism.
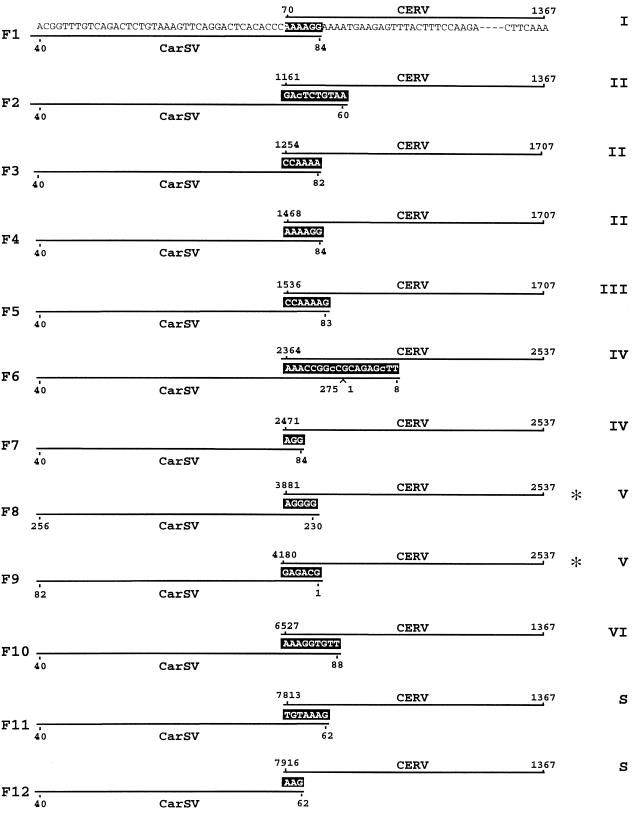
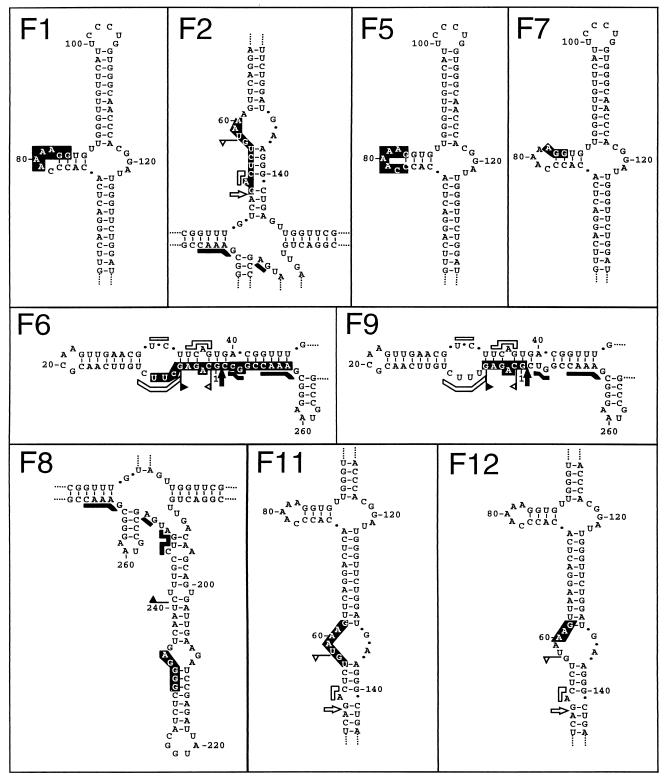
FIG. 3.
Schematic representation of the sequences flanking several CarSV-CERV DNA junctions. CarSV and CERV sequences are shown in F1 and are represented by solid lines in the rest of the sequences shown, and nucleotides common to both sequences are indicated with white letters on a black background. Nucleotide changes in the CarSV sequence are in lowercase type. Numbers refer to nucleotide positions relative to the CarSV-0 RNA and CERV DNA sequences, respectively, excluding those corresponding to primers used for PCR amplification. Asterisks mark junctions identified previously (5). Roman numerals at the right indicate CERV ORFs, and the long viral intergenic region between ORFI and ORFVI is denoted by the letter S.
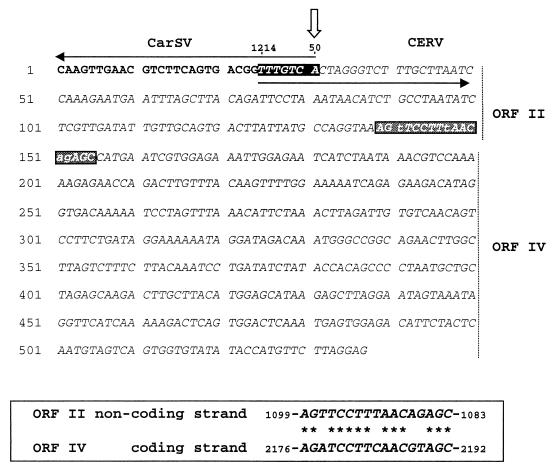
FIG. 4.
CarSV-CERV and CERV-CERV junctions within the same PCR-amplified product. CarSV and CERV sequences appear in boldface and italic type, respectively. White letters on a black background denote the nucleotides common to CarSV and CERV DNAs, and the white perpendicular arrow marks the position of the self-cleavage site of the CarSV-0 RNA minus hammerhead structure. ORFs II and IV to which CERV sequences belong are indicated at the right, and white letters on a grey background show the junction between both CERV sequences. The inset at the bottom shows the sequence similarity between the two CERV sequences at the junction site.
CarSV-CERV DNA junction sites map preferentially to certain structural motifs of CarSV and CERV RNAs.
No obviously preferred sequence motifs could be found in the CarSV-CERV DNA junctions. However, in line with previous results (5), the hammerhead-predicted termini of the linear CarSV RNAs were often involved (Fig. 3 to 5). Indeed, the CarSV boundary was defined by the self-cleavage sites of the plus or minus hammerhead structures in 8 out of the 26 junctions identified in the carnation cultivar 'Indios' (e.g., F2 and F9 in Fig. 3 and 5 and the fusion in Fig. 4). Regarding the CERV sequences flanking the junctions, several mapped relatively close to the presumed gap 1 (e.g., F11 and F12 in Fig. 3) and gaps 2 and 3 (e.g., F8 and F3 in Fig. 3), the regions where synthesis of first- and second-strand viral DNA starts, respectively (19). Analysis of several other regions of the CarSV RNA sequence involved in the formation of CarSV-CERV DNA junctions revealed that certain positions, corresponding to particular loops of the cruciform secondary structure predicted for CarSV-0 RNA, appeared to behave as hot spots for recombination. A good example is the loop around position 80 in the upper arm of CarSV-0 RNA (Fig. 1), because a considerable number of junctions were detected (10 out of 26) in which nucleotides belonging to this loop were implicated; this is the case, among others, for fusions F1, F3, F4, F5, F7, and F10 (Fig. 3 and Fig. 5 and data not shown). Remarkably, fusions F1 and F4 had in common the same CarSV sequence at the junction site (AAAAGG, positions 79 to 84), although they differed in the regions of CERV to which they were fused, which are separated by 1,398 nt and even correspond to different ORFs (Fig. 3). Similar observations were made for junctions involving another loop in the upper arm of CarSV-0 RNA around position 60 (Fig. 3 and 5, fusions F2, F11, and F12).
FIG. 5.
Location on the secondary structure proposed for CarSV-0 RNA of the nucleotides involved in different CarSV-CERV DNA junctions. Fragments of the CarSV-0 RNA structure depicted in Fig. 1 are shown, with the nucleotides common to CarSV and CERV sequences in white against a black background. Notations F1 to F12 on the panels correspond to those in Fig. 3.
Presence of CarSV-CERV DNA elements in CERV-free plants.
CarSV-CERV DNA extrachromosomal elements were initially characterized in plants of the carnation cultivar 'Indios' infected by CERV according to immunological and electron microscopy criteria (5). In order to know whether the presence of the virus was a requisite for detection of such CarSV-CERV elements, PCR amplifications with pairs of CarSV- and CERV-specific primers were undertaken on total DNA preparations from plants of the carnation cultivars 'Carola' and 'Killer', known to be positive for CarSV RNA-DNA but negative for CERV by enzyme-linked immunoassay and electron microscopy. To confirm the CERV status of these plants, Southern blot hybridizations were conducted with total DNA and a radiolabeled probe specific for CERV DNA. As expected, the only samples in which the viral genomic DNA was detected were those from the cultivar 'Indios' known to be infected by the virus (data not shown).
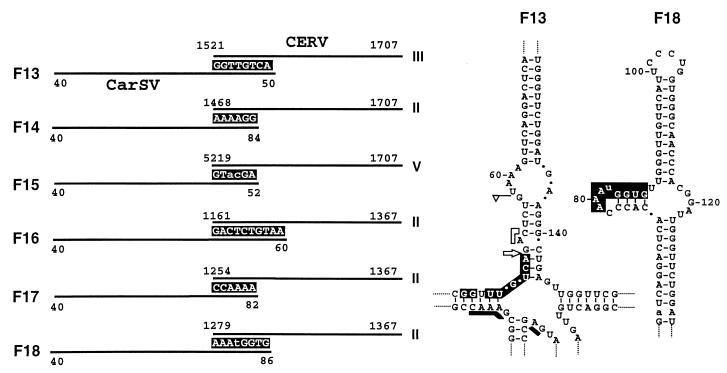
The PCR products obtained from cultivars 'Carola' and 'Killer' exhibited structural features similar to those found previously in the cultivar 'Indios': CarSV-CERV DNA junctions were formed by short stretches of common nucleotides, again preferentially associated to the ends of the linear CarSV-0 RNAs or to some secondary structure motifs thereof as the loop located around position 80 (Fig. 6). Indeed, the self-cleavage site of the minus-polarity CarSV RNA delimited the CarSV sequence in one of the new identified fusions (Fig. 6, F13), and three of these fusions were identical to those previously characterized in the cultivar 'Indios' (compare F14, F16, and F17 in Fig. 6 with F4, F2, and F3 in Fig. 5).
FIG. 6.
(Left) Schematic representation of the sequences flanking several CarSV-CERV DNA junctions from virus-free plants around the junction sites. CarSV and CERV sequences are indicated by solid lines, and nucleotides common to both sequences are shown as white letters on a black background. Nucleotide changes in the CarSV sequence are in lowercase type. Other details are as described in the legend to Fig. 3. (Right) Portion of the secondary structure proposed for CarSV-0 RNA (Fig. 1), with the nucleotides involved in the junction corresponding to fusions F13 and F18 depicted as white letters on a black background. Other symbols used are as described in previous figure legends.
CarSV DNA forms with sequence deletions.
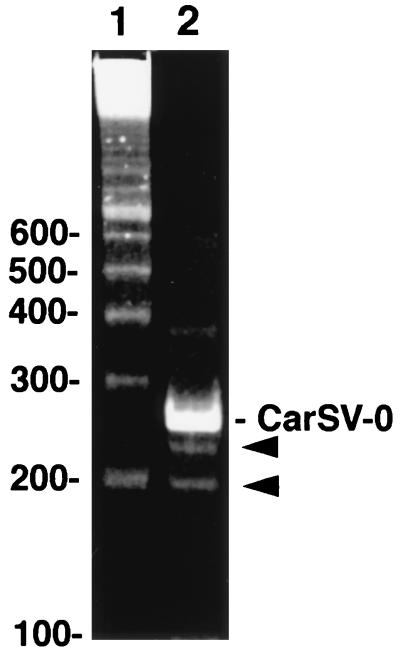
As reported previously, PCR amplification of CarSV DNA with adjacent primers of opposite polarities (Fig. 1, PI and PII or PIII and PIV) yielded a series of major bands upon polyacrylamide gel electrophoresis, corresponding to the monomer and to multimers, reflecting the tandem repeat structure of the CarSV DNA moiety in the extrachromosomal CarSV-CERV elements (5). However, we eventually observed in gels stained with ethidium bromide some additional minor bands migrating faster than the monomeric CarSV DNA (see an example of two of these bands in Fig. 7). Southern blot hybridization with a CarSV specific probe revealed that they had CarSV-derived sequences (data not shown). Cloning and sequencing of some of these minor PCR amplification products showed that they contained deletions from the complete CarSV sequence.
FIG. 7.
Analysis by PAGE and ethidium bromide staining of PCR-amplified products from carnation DNA. Lane 1, DNA ladder of 100-bp multimers (the sites of the smaller markers are shown at left); lane 2, PCR-amplified fragments from the cultivar 'Indios' using the CarSV-specific primers PI and PII (Fig. 1). The position of the monomeric CarSV-0 DNA is indicated, and bands corresponding to smaller CarSV-specific products are marked with arrowheads.
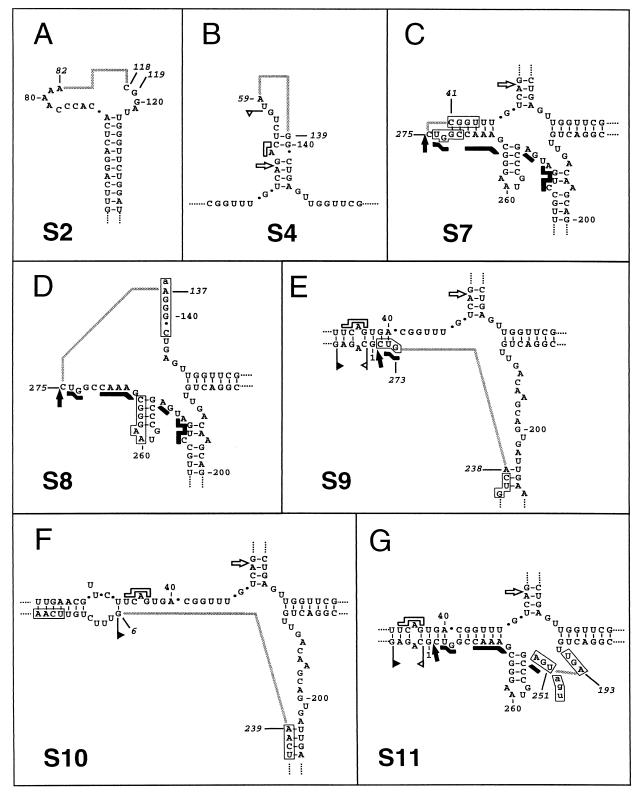
The primary structure of two of these CarSV DNA deletion forms corresponded to the CarSV-S2 and -S4 RNA species characterized before (6), which lack regions encompassing the terminal hairpin of the upper arm of the CarSV-0 RNA cruciform structure and almost the entire upper arm, respectively (Fig. 8A and B). A certain degree of sequence heterogeneity was observed in the boundaries of the deleted region among the different variants constituting a particular CarSV-S RNA group (6). In line with this, two variants of CarSV-S2 DNA differing in the 3′ terminus of the deletion were found (Fig. 8A), and minor differences between the characterized CarSV-S2 DNA sequences and their RNA counterparts were also detected. Regarding the CarSV-S4 DNA form (Fig. 8B), its sequence was identical to that of class I variants of the CarSV-S4 RNA group (6).
FIG. 8.
Schematic representation of CarSV DNA forms with deletions. Deletions found in CarSV DNA are represented on the secondary structure proposed for CarSV-0 RNA. Numbers in italics mark nucleotides at the deletion boundaries, and grey connecting lines represent deleted regions. In panel A two nucleotides are indicated in the 3′ deletion boundary as a consequence of sequence heterogeneity in different clones. Nucleotides forming part of direct repeats found close to the deletion boundaries are boxed and in capital letters when not included in the missing region and are in lowercase letters when included. Other symbols used are as described in previous figure legends.
The rest of CarSV DNA deletion forms characterized here, named CarSV-S7 to -S11 DNA (Fig. 8), did not correlate with formerly identified RNA species although they generally shared some structural characteristics with them. Thus, the self-cleavage site of the plus hammerhead structure defined the 5′ boundary of the deleted region in CarSV-S7 and -S8 DNAs (Fig. 8C and D), resembling the situation found for some CarSV-S1 and -S3 RNA variants (6). In CarSV-S7 DNA, a considerable part of the left arm of the CarSV-0 RNA cruciform structure was absent, including nucleotides required to form the plus and minus hammerhead structures (Fig. 8C). The almost-complete removal of the left arm of the CarSV-0 RNA cruciform structure, just leaving a residual helix, has no precedent among the deleted forms of CarSV RNA identified so far, although several short deletions or repetitions map at this same region of the CarSV-0 RNA (6). CarSV-S8 DNA had a deletion spanning from the plus self-cleavage site through both the left and upper arms of the CarSV-0 RNA cruciform structure, with a few nucleotides remaining in each case, being the 3′ boundary of the deletion similar to that of CarSV-S4 RNA (6). The whole and part of the regions involved in forming the minus and plus CarSV-0 RNA hammerhead structures, respectively, were absent. CarSV-S8 DNA, with only 139 nt (Fig. 8D), together with CarSV-S6 RNA of 135–136 nt (6), represent the most extensively deleted forms among the CarSV DNA and RNA variants characterized so far. Some parallels can be drawn between these two forms. In CarSV-S6 RNA, the site in the plus-polarity strand corresponding to the self-cleavage site of the minus hammerhead structure defined one of the deletion boundaries, giving rise to a circular molecule with just two arms, the lower and the left arms of the CarSV-0 RNA cruciform structure. In CarSV-S8 DNA, the self-cleavage site of the plus polarity hammerhead structure delimited one of the deletion boundaries, retaining the sequences of the lower and right arms of the CarSV-0 RNA cruciform structure (Fig. 8D).
The deletions characterized previously at the RNA level map at three arms of the CarSV-0 RNA cruciform structure, the lower arm being unaffected (6). By contrast, in three of the identified CarSV DNA deletion forms, S9, S10, and S11, the deleted region comprised portions of this lower arm (Fig. 8E to G). CarSV-S9 and -S10 DNAs had deletions with almost identical 5′ boundaries at positions 238 and 239, respectively (Fig. 8E and F) which, in turn, almost coincided with the 5′ boundary of the repetition found in CarSV-L1 RNA (position 241 of CarSV-0 RNA [6]). Indeed, CarSV-S9 DNA lacked basically the same repeated sequence of class I variants of CarSV-L1 RNA, whose 3′ limit is defined by the self-cleavage site of the plus hammerhead structure (6). CarSV-S10 DNA differed from CarSV-S9 DNA in the size of the deletion, which entirely covered the sequences forming the plus-polarity hammerhead structure (Fig. 8F). Concerning CarSV-S11 DNA, its most remarkable feature was the lack of the lower arm of CarSV-0 RNA cruciform structure including a small portion of the sequences forming the plus hammerhead structure (Fig. 8G).
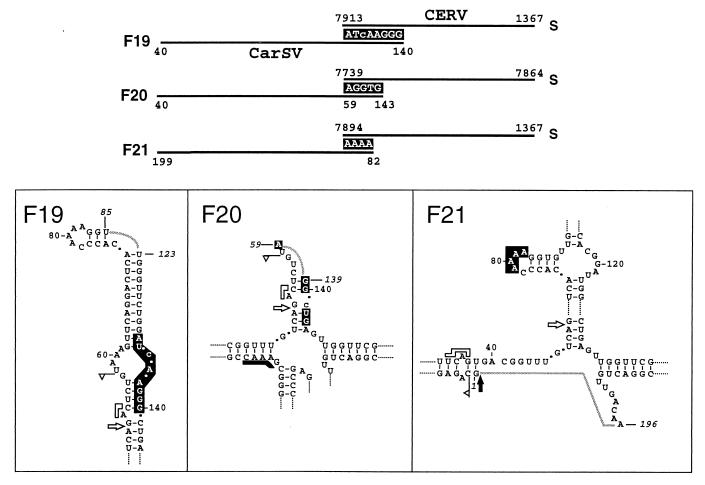
Fusions between CarSV DNA deletion forms and CERV DNA.
The characterization of several PCR amplification products, obtained with pairs of CarSV- and CERV-specific primers, showed the existence of CarSV DNA deletion forms directly fused to CERV DNA. This is the case of fusions F19, F20, and F21 in which the CERV sequences correspond to the long intergenic region between ORFs I and VI (Fig. 9). In fusion F19, the CarSV DNA sequence showed the same deletion as class II variants of the CarSV-S2 RNA group (6). Here again, the junction between both DNAs presented a shared stretch of 8 nt (Fig. 9). In fusion F20, the CarSV DNA sequence showed the same deletion as class I variants of the CarSV-S4 RNA group, and it was found in a peculiar context, since the junction (5 nt) with the CERV sequence included nucleotides at the two boundaries of the deleted region. Finally, in fusion F21, another CarSV DNA deletion form lacking most of the nucleotides corresponding to the lower arm of the CarSV-0 RNA cruciform structure, was found. In this case, the deletion was identical to the repetition observed in RNA variants belonging to the CarSV-L2 (class I) and CarSV-L3 (class III) groups (6), with the 3′ boundary delimited once again by the self-cleavage site of the plus hammerhead structure. Moreover, the junction between this CarSV DNA deletion form and the CERV sequence mapped to the loop around position 80 of the CarSV-0 RNA, further confirming this region of the CarSV sequence as a hot spot for recombination.
FIG. 9.
Schematic representation of the CarSV DNA deletion forms found directly fused to CERV DNA. In the upper part of the figure, CarSV and CERV sequences are indicated by solid lines and nucleotides common to both sequences are shown as white letters on a black background. In the lower panels deletions found in CarSV DNA are represented on the secondary structure proposed for CarSV-0 RNA. Numbers in italics mark the nucleotides at the boundaries, and grey connecting lines represent the deleted regions. Other symbols used are as described in previous figure legends.
DISCUSSION
The singular properties of the CarSV RNA-DNA retroviroid-like system, and particularly the finding that CarSV DNA forms part of extrachromosomal elements in which it is fused to DNA sequences of a plant pararetrovirus, have prompted the present study aimed at gaining a deeper insight into this unique interaction between viral and viroid-like sequences. More than 30 CarSV-CERV DNA fusions from different carnation cultivars were characterized. The primary structure of the junctions between both types of nonhomologous sequences provided sound support for the previously proposed origin of such elements, assumed to result from a template switching mechanism whose hallmark would be the common sequences found in the junction between the two DNAs involved (5). Homologous and nonhomologous template switchings of the reverse transcriptase, a polymerase endowed with an intrinsic low processivity, seem to occur frequently during replication of members of the family Caulimoviridae, to which CERV belongs, leading to the appearance of rearranged molecules (17, 21, 32, 33). The consistent presence in CarSV-CERV DNA junctions of stretches of nucleotides common to both sequences suggests their emergence through a replicase-driven template switching mechanism in which the reverse transcriptase with the nascent strand would dissociate from one template and, after annealing at the second template, would resume DNA elongation. A similar replicase-driven mechanism has been advanced for recombination in RNA viruses (for reviews on this topic, see references 23, 25, and 30).
Several hot spots have been proposed as being involved in replicase-driven recombination in caulimoviruses, including the 5′ end of the linear molecules and the transcription and reverse transcription initiation sites (10, 17). Some of these regions of the CERV genome were often involved in CarSV-CERV junctions. Moreover, the termini of the linear plus and minus CarSV RNAs, resulting from the hammerhead-mediated self-cleavage, were also frequently found in the junctions. Pertinent here is the observation that the most-abundant 5′ terminus of the minus monomeric linear CarSV-0 RNAs isolated from carnation tissue is that predicted by its hammerhead structure (J.-A. Daròs, unpublished data), indicating that this ribozyme is active not only in vitro but also in vivo. Therefore, the structure of some of the CarSV-CERV junctions supports a forced copy choice mechanism resembling that advanced for retroviruses (18), which is similar to the breakage-induced template switching mechanism proposed in RNA virus recombination (25).
On the other hand, it has been suggested that pausing of the reverse transcriptase, usually caused by secondary structure elements, facilitates template switching (27). In this context it is difficult to predict the folding of long RNAs resulting from transcription of the complete CERV genome. However, the CarSV DNA sequences at the crossover sites corresponded very often to regions of strong secondary structure, like the hairpin loops around positions 60 and 80, in the most-stable cruciform conformation predicted for CarSV-0 RNA (Fig. 1). In summary, similar discontinuous transcription mechanisms may account for the intermolecular fusions between CarSV and CERV DNAs, as well as for the intramolecular rearrangements observed between different regions within CERV DNA (Fig. 4), because the same blueprint—sequence similarity between the parental molecules—is found at the junction sites.
The finding of CarSV DNA sequences fused to CERV sequences in the carnation cultivars 'Carola' and 'Killer' extends the observations made initially with the cultivar 'Indios' and suggests that this is probably the general situation, further supporting the hypothesis of the involvement of the viral reverse transcriptase in the generation of the CarSV DNA forms. Moreover, the absence of the virus in the plants of cultivars 'Carola' and 'Killer' indicates that CERV infection is not a prerequisite for the presence of CarSV-CERV elements. However, PCR amplifications suggest that these elements are more abundant in CERV-infected plants, a situation which could result from higher levels of reverse transcriptase activity derived from the complete viral genomes. In any case the extrachromosomal CarSV-CERV elements, once generated, appear to replicate autonomously without the assistance of the virus, a view corroborated by the observation that these elements, but not the virus, are seed transmissible (5).
The possibility that CarSV RNA may replicate through a rolling-circle mechanism such as that proposed for viroids and viroid-like satellite RNAs (1, 3, 4, 12, 20, 26) has been raised on the basis of their structural resemblances that include small size, circularity, limited sequence similarity, and, particularly, ability of both polarity strands to self-cleave through hammerhead ribozymes (16). Moreover, longer-than-unit plus and minus CarSV RNAs have been found in carnation, providing additional support for a rolling circle model of replication (5). However, the characterization of some CarSV-S RNA forms with deletions affecting sequences of the hammerhead structures posed the questions of how these minor RNA species could accumulate following RNA-RNA replication through a rolling circle with ribozymatic processing (6). Our present finding that, like the predominant CarSV-0 RNA, other minor accompanying CarSV-S RNA forms also have a DNA counterpart might explain why some of them persist in plants, particularly those lacking sequences of the plus hammerhead structure, such as CarSV-S1 and -S3 RNAs, which are proposed to have emerged in some replicative rounds as a consequence of polymerase jumps promoted by template regions with a stable folding (6). These deletion forms were considered end products because they would lead to replicative intermediates with impaired self-cleavage and were assumed to accumulate in vivo because their high content in secondary structure would provide them with some protection against RNases. However, an alternative situation can now be envisaged in which CarSV RNA minus-oligomers containing intact and defective monomeric units, the latter resulting from polymerase jumps in some of the transcription rounds, would serve as templates for the CERV reverse transcriptase. Subsequent transcription from these DNA templates would lead to complete and defective linear monomers which, after ligation, would produce the circular CarSV RNAs detected in vivo.
As stated above, RNAs homologous to some of the CarSV DNA deletion forms, CarSV-S7 to -S11 (Fig. 8), have not been detected in carnation. Perhaps they form part of transcriptionally inactive units. Alternatively, CarSV-S7 to -S11 RNAs may be transcribed from their DNA counterparts but do not accumulate to detectable levels because they may be especially unstable. The possibility that an input of CarSV-S7 to -S11 transcripts initiates an RNA-based replication can be discarded, because these transcripts have serious functional defects: CarSV-S7 and -S8 RNAs lack sequences of both plus and minus hammerhead structures, and CarSV-S9 to -S11 RNAs lack sequences of the plus hammerhead structure.
It is worth noting that short nucleotide repeats seem to have influenced the generation of the CarSV-S7 to -S11 DNAs or, alternatively, of their corresponding putative RNAs from which these DNAs may have been reverse transcribed. Thus, in the S7 form, the same sequence (CGGU) flanks the deletion (Fig. 8C), and a similar situation was observed in the S8 and S9 forms with the AAGGGC and GUC sequences in the close vicinity and flanking the deletions, respectively (Fig. 8D and E). In the S10 form, the tetranucleotide UCAA is adjacent and close to the 5′ and 3′ deletion borders, respectively (Fig. 8F). Finally, in the S11 form, a triplet (UGA) flanks both sides of the deletion and, moreover, the same UGA triplet (positions 248 to 250 in CarSV-0 RNA) is also adjacent to the 3′ deletion boundary and forms part of the deleted region itself (Fig. 8G). Whether these CarSV DNAs originated from their hypothetical RNA counterparts or whether the deletions occurred during DNA synthesis on nondefective RNA templates is an open question. However, the observation that the crossover sites do not present the shared nucleotides that are the hallmark of the CarSV-CERV DNA junctions, suggests that these deletions have probably occurred at RNA level by mechanisms similar to those proposed for the generation of CarSV-S1 to -S6 RNAs (6).
Interestingly, the CarSV-CERV association found in carnation shows a close parallel with a situation reported recently in an animal system, in which the presence of a DNA form from a nonretroviral RNA virus (lymphocytic choriomeningitis virus) was detected by PCR in cells after the disappearance of the replicating virus (22). The generation of such a DNA, which was postulated to exist extrachromosomally, seems to be the result of the interaction of the viral RNA with an endogenous reverse transcriptase activity, a mechanism similar to that proposed for the origin of the carnation retroviroid-like system.
ACKNOWLEDGMENTS
We are grateful to A. Ahuir for taking care of the plants and for technical assistance, V. Pallás for critical reading of the manuscript and suggestions, and Barraclough-Donnellan for revision of the English version of the manuscript.
This work was partially supported by grants PB95-0139 and PB98-0500 from the Comisión Interministerial de Ciencia y Tecnología de España to R.F. A.V., J.-A.D., and C.H. were the recipients of postdoctoral contracts from the Ministerio de Educación y Cultura de España.
REFERENCES
- 1.Branch A D, Robertson H D. A replication cycle for viroids and other small infectious RNAs. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 2.Bruening G, Feldstein P A, Buzayan J M, van Tol H, Passmore B K, deBear J, Gough G R, Gilham P T, Eckstein F. Satellite tobacco ringspot virus RNA: self-cleavage and ligation reactions in replication. In: Maramorosch K, editor. viroids and satellites: molecular parasites at the frontier of life. Boca Raton, Fla: CRC Press; 1991. pp. 141–158. [Google Scholar]
- 3.Bruening G, Passmore B K, van Tol H, Buzayan J M, Feldstein P A. Replication of a plant virus satellite RNA: evidence favors transcription of circular templates of both polarities. Mol Plant-Microbe Interact. 1991;4:219–225. doi: 10.1094/mpmi-4-219. [DOI] [PubMed] [Google Scholar]
- 4.Daròs J A, Marcos J F, Hernández C, Flores R. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daròs J A, Flores R. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci USA. 1995;492:6856–6860. doi: 10.1073/pnas.92.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daròs J A, Flores R. Characterization of multiple circular RNAs derived from a plant viroid-like RNA by sequence deletions and duplications. RNA. 1995;1:734–744. [PMC free article] [PubMed] [Google Scholar]
- 7.Dellaporta S L, Wood J, Hicks J B. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programmes for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener T O. Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J. 1991;5:2808–2813. doi: 10.1096/fasebj.5.13.1717335. [DOI] [PubMed] [Google Scholar]
- 10.Dixon L K, Nyffenegger T, Delley G, Martínez-Izquierdo J, Hohn T. Evidence for replicative recombination in CaMV. Virology. 1986;150:463–468. doi: 10.1016/0042-6822(86)90310-7. [DOI] [PubMed] [Google Scholar]
- 11.Epstein L M, Gall J G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 12.Feldstein P A, Hu Y, Owens R A. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc Natl Acad Sci USA. 1998;95:6560–6565. doi: 10.1073/pnas.95.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferbeyre G, Smith J M, Cedergren R. Schistosome satellite DNA encodes active hammerhead structures. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores R, Di Serio F, Hernández C. Viroids: the non-coding genomes. Semin Virol. 1997;8:65–73. [Google Scholar]
- 15.Flores R, Randles J W, Bar-Joseph M, Diener T O. Viroids. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, McGeoch D J, Maniloff J, Mayo M A, Pringle C R, Wickner R B, editors. Virus taxonomy, 7th report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. pp. 1009–1024. [Google Scholar]
- 16.Hernández C, Daròs J A, Elena S F, Moya A, Flores R. The strands of both polarities of a small circular RNA from carnation self-cleave in vitro through alternative double- and single-hammerhead structures. Nucleic Acids Res. 1992;20:6323–6329. doi: 10.1093/nar/20.23.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohn T. Recombination of a plant pararetrovirus: cauliflower mosaic virus. In: Paszkowski J, editor. Homologous recombination and gene silencing in plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 25–38. [Google Scholar]
- 18.Hu W-S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 19.Hull R, Sadler J, Longstaff M. The sequence of carnation etched ring virus DNA: comparison with cauliflower mosaic virus and retrovirus. EMBO J. 1986;5:3083–3090. doi: 10.1002/j.1460-2075.1986.tb04614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchins C J, Keese P, Visvader J E, Rathjen P D, McInnes J L, Symons R H. Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol Biol. 1985;4:293–304. doi: 10.1007/BF02418248. [DOI] [PubMed] [Google Scholar]
- 21.Király L, Bourque J E, Schoelz J E. Temporal and spatial appearance of recombinant viruses formed between cauliflower mosaic virus (CaMV) and CaMV sequences present in transgenic Nicotiana bigelovii. Mol Plant-Microbe Interact. 1998;11:309–316. [Google Scholar]
- 22.Klenerman P, Hengartner H, Zinkernagel R M. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301. doi: 10.1038/36876. [DOI] [PubMed] [Google Scholar]
- 23.Lai M M C. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews R E F. Plant virology. 3rd ed. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 25.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 26.Navarro J A, Daròs J A, Flores R. Complexes containing both polarity strands of avocado sunblotch viroid: identification in chloroplasts and characterization. Virology. 1999;253:77–85. doi: 10.1006/viro.1998.9497. [DOI] [PubMed] [Google Scholar]
- 27.Pennington R E, Melcher U. In planta deletion of DNA inserts from the large intergenic region of cauliflower mosaic virus DNA. Virology. 1993;192:188–196. doi: 10.1006/viro.1993.1021. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fristch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Saville B J, Collins R A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 30.Simon A, Bujarski J J. RNA-RNA recombination and evolution in virus infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 31.Symons R H. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaden V R, Melcher U. Recombination sites in cauliflower mosaic virus DNAs: implications for mechanisms of recombination. Virology. 1990;177:717–726. doi: 10.1016/0042-6822(90)90538-3. [DOI] [PubMed] [Google Scholar]
- 33.Wintermantel W M, Schoelz J E. Isolation of recombinant viruses between cauliflower mosaic virus and a viral gene in transgenic plants under conditions of moderate selection pressure. Virology. 1996;223:156–164. doi: 10.1006/viro.1996.0464. [DOI] [PubMed] [Google Scholar]