Abstract
The intrinsic variability of hepatitis C virus (HCV) envelope proteins E1 and E2 complicates the identification of protective antibodies. In an attempt to identify antibodies to E2 proteins from divergent HCV isolates, we produced HCV E2 recombinant proteins from individuals infected with HCV genotypes 1a, 1b, 2a, and 2b. These proteins were then used to characterize 10 human monoclonal antibodies (HMAbs) produced from peripheral B cells isolated from an individual infected with HCV genotype 1b. Nine of the antibodies recognize conformational epitopes within HCV E2. Six HMAbs identify epitopes shared among HCV genotypes 1a, 1b, 2a, and 2b. Six, including five broadly reactive HMAbs, could inhibit binding of HCV E2 of genotypes 1a, 1b, 2a, and 2b to human CD81 when E2 and the antibody were simultaneously exposed to CD81. Surprisingly, all of the antibodies that inhibited the binding of E2 to CD81 retained the ability to recognize preformed CD81-E2 complexes generated with some of the same recombinant E2 proteins. Two antibodies that did not recognize preformed complexes of HCV 1a E2 and CD81 also inhibited binding of HCV 1a virions to CD81. Thus, HCV-infected individuals can produce antibodies that recognize conserved conformational epitopes and inhibit the binding of HCV to CD81. The inhibition is mediated via antibody binding to epitopes outside of the CD81 binding site in E2, possibly by preventing conformational changes in E2 that are required for CD81 binding.
Hepatitis C virus (HCV), a member of the family Flaviviridae, expresses its proteins from a 9.5-kb positive-sense RNA genome (18). The virus is highly variable, with more than nine distinct genotypes (1, 18). Most patients progress from acute to chronic disease in spite of a robust immune response. Nonetheless, evidence for a humoral immune response providing at least partial protection in clinical and animal model studies is accumulating (6, 9–11, 29, 37) and suggests that neutralizing antibodies have a role in the containment of HCV infection. For a protective immune response, the important viral gene products are the envelope proteins, designated E1 and E2. Both sequence analyses of different isolates and sequential studies of virus isolates in infected patients suggest that the HCV E2 protein is under immune selection leading to selection of variants in the amino-terminal domain of HCV E2, designated hypervariable region 1 (HVR-1) (1, 9, 16–18, 20, 37, 39, 40). Antibodies to HVR-1 appear to mediate virus neutralization in cell culture and chimpanzee protection studies (10, 37). Unfortunately, antibodies to HVR-1 tend to be isolate specific and over time drive the selection of new viral variants that the existing immune response does not recognize (9, 20, 37, 40). Although there has been progress at inducing a broader immune response to HVR-1-related sequences (31), the high mutability of HVR-1 sequences in vivo may allow for the selection of immune escape mutants even against antibodies that recognize the majority of HVR-1 isolates.
Studies using HCV E1 and E2 proteins expressed in mammalian cells showed that infected individuals have an antibody response to HCV E2 composed in part to epitopes that are conformational in nature (15, 23). Studies involving the isolation of human monoclonal or recombinant antibodies to HCV E2 protein showed that a substantial fraction of these antibodies recognize conformational epitopes (2, 3, 13). As to biological function of these domains, investigators have used surrogate assays to provide insights into virus neutralization since the virus cannot be grown in vitro (18). One surrogate assay, referred to as the neutralization of binding (NOB) assay, evaluates the ability of a given antibody or serum to prevent the binding of HCV E2 protein with a human T-cell line (35). The finding that serum antibodies obtained from chimpanzees protected by vaccination were strongly positive in the NOB assay provides support for the relevance of the assay as a measure of virus neutralization activity (35).
The human tetraspanin cell surface protein CD81 (TAPA-1) (for a review, see reference 24) is the target protein bound by HCV E2 in the NOB assay (30). Furthermore, human CD81 binds to free virions and consequently is a possible receptor for HCV (30). At present, however, direct evidence that CD81 is necessary and sufficient for HCV infection has not been obtained (34). Nevertheless, several human monoclonal antibodies (HMAbs) to HCV E2 protein have been reported to inhibit the interaction of HCV 1a E2 with human cells (2, 13). However, it is not known if the epitopes recognized by antibodies that inhibit the interaction of HCV E2 with CD81 are conserved in HCV E2 proteins of different genotypes, nor have the mechanisms underlying the observed inhibition of E2 binding to CD81 been explored. Both breadth of reactivity to multiple HCV genotypes and the ability to interfere with the binding of HCV virions to susceptible cells would be key attributes of a neutralizing antibody if CD81 was a receptor or coreceptor for HCV.
To address these questions, we produced and characterized a panel of HMAbs from peripheral B cells of an individual with asymptomatic HCV infection and having a high serum neutralization of binding titer. These 10 HMAbs to HCV E2 were tested for the ability to bind to HCV E2 proteins of genotypes 1a, 1b, 2a, and 2b and inhibit the interaction of these E2 proteins with human CD81. Additionally, the HMAbs were evaluated for the ability to bind to preformed CD81-E2 complexes. We present evidence indicating that the epitopes recognized by antibodies capable of inhibiting E2 binding to CD81 are conformational and conserved in HCV E2 proteins of multiple genotypes. Also, antibodies that efficiently inhibited the formation of the E2-CD81 complexes retained the ability to recognize preformed E2-CD81 complexes generated by some of the same E2 proteins. Two of the antibodies that inhibited binding of HCV E2 protein to CD81 and did not recognize preformed 1a E2-CD81 complexes also prevented binding of HCV 1a virions to CD81. These antibodies will be useful for testing the hypothesis that antibodies that inhibit the interaction of HCV with CD81 mediate virus neutralization.
MATERIALS AND METHODS
Cell lines and antibodies.
Insect cell line Sf9 was cultured and grown as described elsewhere (22). Mammalian cell lines BSC-1 and HeLa were grown in minimal essential medium (Life Technologies, Bethesda, Md.) supplemented with 10% fetal calf serum and 2 mM glutamine. H. Greenberg (Stanford University) generously provided a mouse monoclonal antibody (E2G) to HCV E2 (8). T. Fuerst (Avant Immunotherapeutics, Needham, Mass.) generously provided the BSC-1 cells, vaccinia virus strains, and pVOTE vector (38).
Generation and identification of HMAbs.
Peripheral B cells were isolated from a single HCV-infected individual. The electrofusion of Epstein-Barr virus (EBV)-activated B cells to heteromyeloma cells produced human hybridomas, using methods as described elsewhere (28). An immunofluorescence assay (IFA) with fixed Sf9 cells expressing recombinant E1 or E2 protein detected HCV-specific antibodies. Sf9 cells infected with recombinant baculovirus Elt (22), Sf9 cells infected with recombinant baculovirus E2t (22), and uninfected cells were combined at a ratio of 1:1:1 and fixed onto HTC Super Cured 24-spot slides (Cel-Line Associates, Newfield, N.J.) with 100% acetone for 10 min at room temperature (RT). Antibody binding to fixed cells was detected by fluorescence microscopy as described previously (14). EBV-activated B cells were selected for electrofusion based on IFA reactivity. Hybridomas of interest from successful electrofusions were cloned by limiting dilution. Antibody purification and biotinylation of HMAb CBH-4G were performed as described elsewhere (14). The heavy- and light-chain subtypes of the antibodies were identified using a commercially available kit (The Binding Site Ltd., Birmingham, United Kingdom).
Expression of HCV E2 proteins.
Aliquots of plasma positive for HCV RNA were genotyped by the InnoLipa HCV assay (Innogenetics, Leuven, Belgium). RNA used for cloning was prepared with a commercial kit (Purescript RNA kit; Gentra Systems, Minneapolis Minn.) from 125 μl of plasma infected with HCV genotype 1a, 1b, 2a, or 2b. Amplification of RNA was achieved by coupled reverse transcription-PCR with the HCV-specific degenerate primers HCV E2-F1 (5′-CGCATGGCiTGGGAyATGATG-3′) and HCV E2-R1 (5′-CGCGCACrAAGTAsGGyACT-3′. Between 2 and 8 μl of amplified product was then subjected to a second PCR amplification using the forward and reverse primers specific for each genotype (forward primers 1a [CGAAGCTTCATATGATCGCTGGTGCTCACTG G], 1b [CGCATATGGAGCTCGCGGGGGCCCACTGGGGAGT], 2a [CGCTCGAGCCATGGTTGGCGGGGCTCATTGGGGC] and 2b [CGCTCGAGCCATGGTTTTCGGCGGCCATTGGGTG]; reverse primers 1a [GCGGATCCCTGCAGCTACAAACTGGCTTGAAGAATCCA], 1b [GCTCTAGACT GCAGCTATATGCCAGCCTGGAGCACCAT], 2a [TCGAATTCGGATCCTACAAAGCACCTTTTAGGAGATAAGC], and 2b [TCGAATTCGGATCCTACAGAGACGCTTTAAGGAGGTAGGC]). The amplified products were purified and digested with the appropriate restriction enzymes (restriction sites in the primers are underlined). The digested DNAs were then ligated into similarly digested pVOTE-1 or pVOTE-2 vector (38). Expression of HCV E2 protein from vaccinia virus constructs was verified by Western blot analysis of transiently transfected CV-1 cells. The genotypes of the cloned E2 proteins were confirmed by DNA sequencing of the entire insert, using dye terminator methodologies and an automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Plasmids that produced intact HCV E2 were then used to generate recombinant vaccinia virus by homologous recombination into the hemagglutinin locus of vaccinia virus strain VWA (38) as described elsewhere (27). Recombinant vaccinia virus was grown and measured using BSC-1 cells; titers of recombinant virus ranged between 5 × 108 and 10 × 108 PFU/ml.
HCV E2 ELISA.
Monolayers of HeLa cells were grown to 80% confluence and infected at 5 PFU/cell with both VWA and recombinant vaccinia virus or VWA only. HCV recombinants were mixed with wild-type vaccinia virus with an intact hemagglutinin gene to minimize vaccinia virus-induced cytopathic effect observed with hemagglutinin-deficient virus (36). Cells were harvested after 1 day of infection. Extracts were prepared by washing the cells with phosphate-buffered saline (PBS) and then resuspending ∼25 × 106 cells in 1 ml of lysis buffer (150 mM NaCl, 20 mM Tris [pH 7.5], 0.5% deoxycholate, 1.0% Nonidet-P40, 1 mM EDTA, Pefabloc [0.5 mg/ml; Boehringer Mannheim, Indianapolis, Ind.], aprotinin [2 μg/ml], leupeptin [2 μg/ml], pepstatin [1 μg/ml]). Extracts prepared in this manner contained approximately 25 μg of E2 protein per ml. Nuclei were pelleted by centrifugation at 18,000 × g at 4°C for 10 min, and resulting cytoplasmic extracts were stored at 4°C and used for enzyme-linked immunosorbent assay (ELISA) within 24 h of preparation. Microtiter plates were prepared by coating wells with 500 ng of purified Galanthus nivalis lectin (GNA; Sigma, St. Louis, Mo.) in 100 μl of PBS for 1 h at 37°C. Wells were washed with Tris-buffered saline (TBS; 150 mM NaCl, 20 mM Tris-HCl [pH 7.5]) and then blocked with 150 μl of BLOTTO (TBS plus 0.1% Tween 20, 2.5% normal goat serum, and 2.5% nonfat dry milk) by incubation for 1 h at RT. Plates were washed twice with TBS followed by the addition of 15 μl of extract in 100 μl of BLOTTO. After 1.5 h at RT, plates were washed three times with TBS followed by the addition of unlabeled antibodies at various concentrations. Plates were incubated for 1.5 h and washed three times with TBS; then 100 μl of anti-human IgG-alkaline phosphatase conjugate (Promega, Madison, Wis.) diluted 1/5,000 in BLOTTO was added. After 1 h at RT, the plates were washed four times with TBS followed by 30 min of incubation with a 1-mg/ml solution of p-nitrophenyl phosphate (PNPP). Absorbance was measured at 405 nm with a multiwell plate reader (Du Pont Co., Wilmington, Del.).
Assessment of CD81-E2 binding.
Prior to the identification of CD81 as the HCV E2 ligand, the second extracellular domain was termed EC2 (24); however, to prevent confusion between E2 and EC2, we have opted to refer to this region as the large extracellular loop (LEL). The LELs of human and murine CD81 (CD81-LEL) were expressed as fusion proteins with glutathione S-transferase (GST) using the pGEX vector (GST-2T). The proteins were constructed and purified as described previously (12). Five different assays to assess the interaction of recombinant E2 proteins with CD81-LEL were performed. Flow cytometric assessments of E2 binding to CD81 (NOB assays) were performed using methods and HCV E2 proteins previously described (19, 35). Briefly, 1 μg of the HCV E2 1a protein produced in mammalian cells (35) was mixed with serial dilution of antibodies (from 0.1 to 300 μg/ml) and incubated for 30 min at 37°C. Molt-4 cells (105) were added to the mixture and incubated on ice for 1 h. After washing, the amount of HCV-E2 bound to Molt-4 cells was assessed by flow cytometry. The NOB titer is defined as the antibody concentration that results in 50% inhibition of E2 binding.
Antibody capture assays were performed by coating microtiter plate wells with 100 ng of purified CD81-LEL or nonrecombinant GST diluted in PBS. After 1 h at 37°C, wells were washed once with TBS and blocked by incubation with 150 μl of BLOTTO for 1 h at RT. Each well received test antibody and 15 μl of extract from vaccinia virus-infected BSC-1 cells diluted in BLOTTO to a final volume of 100 μl (approximately 300 to 400 ng of E2 for a final concentration of ∼4 μg/ml). Plates were incubated overnight with gentle agitation at 4°C. Wells were then washed three times with TBS followed by addition of appropriate alkaline phosphate-conjugated secondary antibody and PNPP substrate as described above.
Inhibition assays were performed by coating microtiter plate wells with 100 ng of purified human or murine CD81-LEL or 500 ng of GNA. After 1 h at 37°C, wells were washed once with TBS and blocked by incubation with 150 μl of BLOTTO for 1 h at RT. The wells were then washed once with TBS, and various dilutions of test sera or monoclonal antibodies were added in a total volume of 50 μl. Concurrently, extract from BSC-1 cells infected with HCV E2-expressing vaccinia virus was combined with BLOTTO at a ratio of 30% extract to 70% BLOTTO, and biotinylated HMAb CBH-4G was added to a final concentration of 8 μg/ml. The mixture was incubated at RT for 20 min, and 50 μl was added to wells already containing test antibody (resulting in 15 μl of extract/∼400 ng of HCV E2 in each well). The plates were incubated overnight with gentle agitation at 4°C. Wells were then washed three times with TBS followed by the addition of 100 μl of 1/1,000-diluted alkaline phosphatase-conjugated streptavidin (Amersham-Pharmacia Biotech, Piscataway, N.J.). Plates were incubated for 1 h at RT and washed, and alkaline phosphatase activity was quantitated with PNPP substrate as described above. Background signals for binding of E2 to human CD81 were determined from wells coated with murine CD81-LEL; background signals obtained from E2 binding to GNA-coated wells were obtained from extracts of BSC-1 cells infected with wild-type vaccinia virus. Signals obtained with biotinylated CBH-4G and E2 in the presence of competing antibody were compared to signals obtained from CBH-4G and E2 in the absence of competing antibody.
Binding of antibodies to preformed E2-CD81 complexes was assessed by coating microtiter plates with 100 ng of purified CD81-LEL or murine CD81-LEL diluted in PBS. After 1 h at 37°C, wells were washed once with TBS and blocked by incubation with 150 μl of BLOTTO for 1 h at RT. Extract from BSC-1 cells infected with HCV E2-expressing vaccinia virus was then diluted in 100 μl of BLOTTO, added to CD81-coated plates, and incubated overnight with gentle agitation at 4°C. Wells were then washed three times with TBS followed by addition of increasing concentrations of test antibodies, also diluted in 100 μl of BLOTTO. After 1.5 h at room temperature, wells were washed and alkaline phosphate-conjugated secondary antibody was added. After incubation for 1 h, wells were washed and bound antibody was detected with PNPP substrate.
The virion-CD81 binding assay was performed as previously described (30). Briefly, 1/4-inch polystyrene beads (Pierce, Rockford, Ill.) were coated overnight with 50 μg of purified recombinant LEL-TRX protein (30) per ml in a citrate buffer (pH 4.0) at room temperature and then blocked for 1 h with 2% bovine serum albumin in 50 mM Tris-Cl (pH 8)–1 mM EDTA–100 mM NaCl (TEN) buffer. Serum containing 5 × 105 HCV RNA genomes was diluted in 200 μl of TEN buffer with 10 μg of purified monoclonal antibodies and incubated for 1 h at 4°C. The diluted serum was then added to the coated beads and incubated at 37°C for 1 to 2 h. After removal of supernatant, each bead was washed five times with 15 ml of TEN buffer, and bound virus was extracted using a commercially available kit (Qiagen, Basel, Switzerland). PCR-mediated evaluation of the RNA copy number was performed using a PE Applied Biosystems 7700 sequence detection system as described elsewhere (30). The copy number of HCV RNA molecules bound to the polystyrene beads was subtracted from values obtained from CD81-coated beads. The percent inhibition of virion binding to CD81-LEL-coated beads was calculated by dividing the number of HCV RNA copies bound in presence of test antibody by the number bound in the presence of an irrelevant antibody.
RESULTS
Identification of HCV HMAbs.
HCV-infected individuals were identified by commercial HCV ELISA. Individuals with a high-titer antibody response to HCV E2 (measured at an optical density [OD] of 405 nm) were identified by IFA with fixed Sf9 cells infected with recombinant baculovirus expressing HCV 1a E2 protein. The individual selected as the B-cell donor was infected with HCV genotype 1b, had a strong antibody response to HCV E2, and had a neutralization of binding assay titer of 1/5,000, which is relatively rare in HCV-infected individuals (19, 35). Alanine aminotransferase values of the donor ranged between 29 and 49 IU (normal is <45) for six blood samples obtained over a period of 27 months. Screening of supernatants from EBV-activated cell cultures was performed by IFA against both HCV E1 and E2. No activated B cells expressing antibodies to HCV E1 were isolated. EBV-activated cells from 30 wells were selected for electrofusions to mouse-human heteromyelomas. Ten of the human hybridomas were cloned by limiting dilution and produced in bulk for subsequent studies (Table 1). All of the HMAbs were IgG1. Sequencing of these IgG1 genes confirmed that each of the 10 hybridomas was derived from an independent B cell and expressed unique CDR3 regions (H. C. Chan, K. G. Hadlock, S. K. H. Foung, and S. Levy, submitted for publication).
TABLE 1.
Isotype and secretion of HCV HMAbs CBH-2 through CBH-17
| Antibody | Target | Fusion partnera | Subtype
|
Antibody secretion (μg/ml)c | |
|---|---|---|---|---|---|
| Heavy | Light | ||||
| CBH-2 | HCV E2 | K6H6/B5 | IgG1 | Kappa | 34–88 |
| CBH-4D | HCV E2 | K6H6/B5 | IgG1 | Lambda | 19–62 |
| CBH-4B | HCV E2 | K6H6/B5 | IgG1 | Kappa | 17–26 |
| CBH-4G | HCV E2 | K6H6/B5 | IgG1 | Kappa | 17–27 |
| CBH-5 | HCV E2 | H73C11 | IgG1 | Kappa | 11–25 |
| CBH-7 | HCV E2 | K6H6/B5 | IgG1 | Kappa | 9–29 |
| CBH-8C | HCV E2 | K6H6/B5 | IgG1 | Kappa | 11–62 |
| CBH-8E | HCV E2 | K6H6/B5 | IgG1 | Kappa | 6 |
| CBH-11 | HCV E2 | K6H6/B5 | IgG1 | Kappa | 23–95 |
| CBH-17 | HCV E2 | K6H6/B5 | IgG1 | Lambda | 27–48 |
| R04b | CMV p64 | K6H6/B5 | IgG1 | Lambda | 119–204 |
Cell line (described in reference 28) used to isolate B cells.
Isotype-matched control antibody.
Obtained from bulk culture of the hybridomas at different times.
Reactivity of HCV HMAbs with HCV E2 of multiple genotypes.
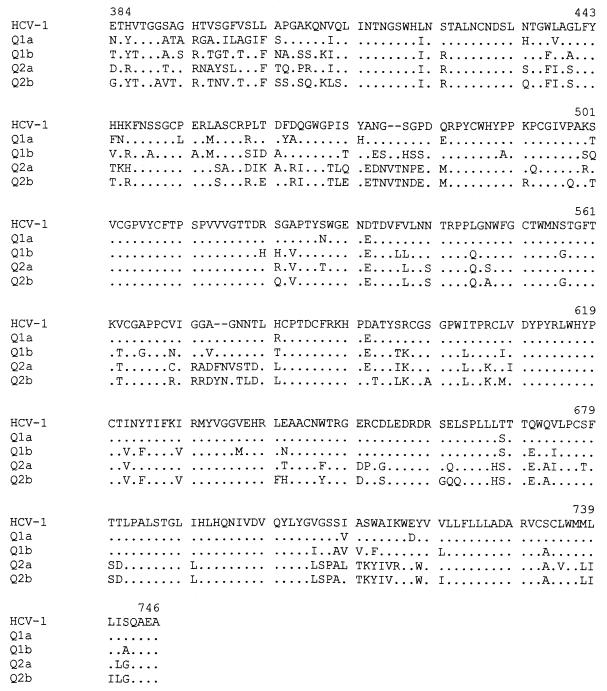
The complete coding sequence of HCV E2 from isolates of HCV genotypes 1a, 1b, 2a, and 2b were PCR amplified from HCV-positive sera and expressed with vaccinia virus using the pVOTE (38) transfer vector (constructs Q1a, Q1b, Q2a, and Q2b for HCV genotypes 1a, 1b, 2a, and 2b, respectively). Genotype selection was based on its divergence and frequency among HCV-infected individuals in the United States (25). The amplified fragments expressed the final 39 amino acids of E1, all of E2, and the amino-terminal 98 amino acids of NS2. Previous studies indicated that similar fragments were correctly processed into full-length E2 when expressed in vaccinia virus (32). Western blot analysis of extracts prepared from infected cells with a murine monoclonal antibody to HCV E2 verified correct expression (data not shown). DNA sequence analysis was performed over the E2 region of the HCV constructs (Fig. 1). The E2 amino acid sequences of Q1a, Q1b, Q2a, and Q2b were approximately 90% homologous with E2 sequences from previously sequenced full-length genomes of genotypes 1a, 1b, 2a, and 2b (reference 1 and references therein). In particular, isolate Q1a was 90.1% homologous and Q1b was 79.9% homologous with the HCV-1 isolate used in the isolation of the HMAbs (Table 2). The most divergent E2 sequences were those of Q1a and Q2b, which were 70% homologous at the amino acid level. As expected, significant diversity was seen in HVR-1 in all of the E2 proteins.
FIG. 1.
Amino acid sequences of HCV E2 proteins. The amino acid sequences of the E2 regions of constructs Q1a, Q1b, Q2a, and Q2b are compared to that of the HCV-1 isolate of HCV (5). A dot indicates a conserved amino acid. Amino acids are numbered according to position in the polyprotein of HCV-1.
TABLE 2.
Amino acid homology of HCV E2 proteins
| Protein | % Identity of E2 sequencesa
|
||||
|---|---|---|---|---|---|
| HCV-1 | Q1a | Q1b | Q2a | Q2b | |
| HCV-1 | 100 | ||||
| Q1a | 90.1 | 100 | |||
| Q1b | 79.9 | 78.8 | 100 | ||
| Q2a | 71.9 | 71.4 | 72.5 | 100 | |
| Q2b | 70.8 | 70.0 | 74.1 | 81.7 | 100 |
Comparisons are based on the amino acid sequence of full-length E2 protein (amino acids 384 to 746 of HCV-1 polyprotein). HCV-1 refers to the initial HCV isolate (5).
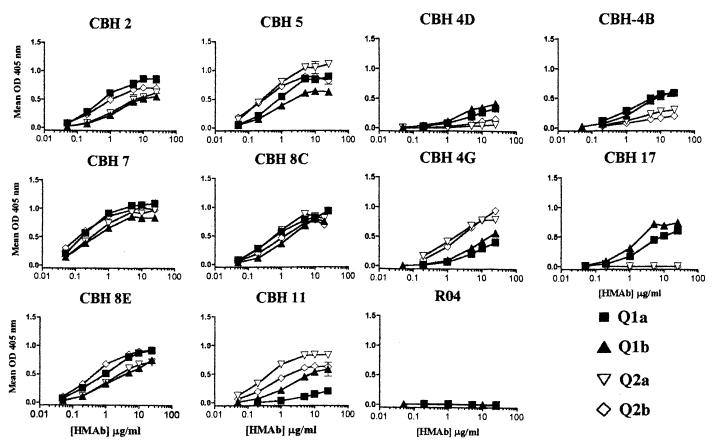
The lectin GNA has been used extensively in the purification of HCV envelope proteins and E2 proteins purified by GNA affinity chromatography are immunogenic in animal models and efficiently recognized by sera from HCV-infected individuals (6, 32). To minimize handling and manipulation, HCV E2 was captured directly from cytoplasmic extracts onto microtiter plates with GNA, using methods similar to those previously described (3). All 10 HCV HMAbs bound to two or more of the HCV E2 constructs (Fig. 2), and no specific signal was obtained with a control HMAb (R04, specific to a cytomegalovirus [CMV] protein). The HMAbs with the highest relative affinities and levels of reactivity to E2 proteins of all four genotypes were CBH-7 and CBH-8C, followed by HMAbs CBH-5, -2, and -8E. HMAb CBH-4G exhibited significantly greater reactivity to HCV E2 proteins of genotypes 2a and 2b, while HMAb CBH-11 was markedly less reactive with Q1a E2 protein. HMAb CBH-17 and to a lesser extent CBH-4D and CBH-4B exhibited preferential binding to E2 proteins of genotype 1a or 1b relative to E2 proteins of genotype 2a or 2b. These variations were not a result of variable efficiencies of capture of the different E2 proteins since the maximum signals obtained with the E2 proteins were very comparable in all experiments. In conclusion, six antibodies, CBH-2, -4G, -5, -7, -8C, and -8E, exhibited significant reactivity with all HCV E2 constructs, indicating that the epitopes recognized by these HMAbs are conserved in E2 proteins of genotypes 1 and 2.
FIG. 2.
HCV antibody reactivity with E2 protein of divergent HCV genotypes. HCV E2 protein expressed by 6 × 105 HeLa cells infected with vaccinia virus Q1a (■), Q1b (▴), Q2a (▿), or Q2b (·⃟) was captured onto wells coated with 500 ng of GNA. Wells were washed and blocked, and bound protein was incubated with the indicated HCV HMAbs and control HMAb (R04) to a CMV protein (14). Values are the mean specific binding (extracts of cells infected with vaccinia virus expressing HCV E2 protein − wild-type vaccinia extracts) of replicate wells. Reactivity of HCV and control HMAbs with proteins from wild-type vaccinia virus-infected cells did not exceed an absorbance of 0.04. Error bars indicate 1 standard deviation from the mean.
These HCV HMAbs were identified by a screening method designed to maximize the selection of antibodies to conformational epitopes. This was verified by comparing the reactions of the HCV HMAbs to both native and denatured HCV 1b E2 proteins (Fig. 3). As shown above, all 10 HCV HMAbs recognized native HCV 1b E2. Treatment of HCV E2 by heating to 56°C in the presence of 0.5% sodium dodecyl sulfate and 5 mM dithiothreitol resulted in complete abrogation of reactivity for 9 of the 10 HCV HMAbs. The exception, HMAb CBH-17, retained approximately 90% of its reactivity with the denatured E2 protein. Western blot analysis of CBH-17 confirmed that it was weakly reactive with HCV envelope proteins as expressed by Q1a or Q1b (data not shown).
FIG. 3.
Nine HCV HMAbs fail to recognize denatured HCV E2. Proteins derived from HeLa cells infected with vaccinia virus Q1b and VWA or VWA alone (gray bars) were either left untreated (white bars) or denatured by incubation with 0.5% sodium dodecyl sulfate and 5 mM dithiothreitol for 15 min at 56°C (black bars). After treatment, proteins were diluted 1:5 in BLOTTO and captured onto wells coated with 500 ng of GNA. Wells were washed and blocked, and bound protein was incubated at 5 μg/ml with HCV HMAbs (x axis) and control HMAb (R04). Bound antibody was detected as described in Materials and Methods. Values for native and denatured HCV 1b are the mean signals obtained from replicate wells. Signals from single wells of native and denatured proteins derived from VWA-infected HeLa cells were indistinguishable and also averaged. Error bars indicate 1 standard deviation from the mean.
Effect of HCV HMAbs on E2 binding to CD81.
Recently, the human tetraspanin protein CD81 has been demonstrated to specifically bind to E2, with the involved site localized to CD81-LEL (previously referred to as EC2) (30). The ability of the HMAbs to inhibit binding of HCV 1a E2- to CD81-expressing target cells was assessed via flow cytometry (also referred to as the NOB assay [35]). HMAbs CBH-4D, -4B, -4G, and -17 did not block the binding of E2 to target cells at concentrations of less than 25 μg/ml (Table 3). HMAbs CBH-2, -5, -7, -8C, -8E, and -11 achieved 50% inhibition of E2 binding at concentrations of 1 to 10 μg/ml and can be classified as NOB positive. To confirm results obtained by flow cytometry using E2 proteins of multiple genotypes, we assessed whether the HCV HMAbs could inhibit the interaction of HCV E2 with CD81. Microtiter plates were first coated with purified CD81-LEL–GST fusion protein to which excess HCV E2 was added in the presence of the HCV HMAbs. Because HCV E2 binds specifically to human CD81 (12, 35), the E2 proteins were produced in the green monkey kidney cell line BSC-1 to minimize the effect of endogenous CD81. Neither HCV HMAbs nor control antibodies were captured by purified nonrecombinant GST, nor were the HCV or control antibodies captured by CD81 when combined with extracts of BSC-1 cells infected with wild-type vaccinia virus (data not shown).
TABLE 3.
Inhibition of HCV E2-CD81 binding by HCV HMAbs
| Antibody | NOB 1aa | Mean OD CD81-E2/GST-E2b
|
|||
|---|---|---|---|---|---|
| Q1a | Q1b | Q2a | Q2b | ||
| CBH-2 | 5 | 1.5 | 1.1 | 1.7 | 1.2 |
| CBH-5 | 2 | 1.4 | 1.1 | 2.1 | 1.2 |
| CBH-7 | 7 | 2.6 | 1.2 | 2.6 | 1.2 |
| CBH-8C | 10 | 1.5 | 1.1 | 2.1 | 1.4 |
| CBH-8E | 8 | 1.4 | 1.1 | 1.5 | 1.3 |
| CBH-11 | 3 | 1.2 | 1.1 | 2.0 | 1.3 |
| CBH-4G | — | 18.3 | 16.5 | 10.8 | 15.1 |
| CBH-4B | — | 25.2 | 17.1 | 4.6 | 7.6 |
| CBH-4D | — | 23.3 | 11.5 | 2.0 | 4.0 |
| CBH-17 | — | 8.6 | 10.2 | 1.1 | 1.1 |
| R04 | — | 1.1 | 1.0 | 1.2 | 1.1 |
HMAb reactivity in representative NOB assays, presented as the concentration (micrograms per milliliter) of antibody that results in 50% inhibition of binding of 1 μg of HCV 1a E2 to 105 CD81-expressing Molt-4 cells. Antibodies were tested at concentrations that ranged from 0.1 to 300 μg/ml. —, negative.
Aliquots of BSC-1 cell extracts containing approximately 400 ng of HCV E2 protein were combined with HCV HMAbs (5 μg/ml) in 100 μl of BLOTTO and added to wells coated with 100 ng of GST–CD81-LEL fusion protein or GST as described in Materials and Methods. Values are the mean OD value of antibody captured by CD81 divided by the mean OD value for antibody captured by GST in the presence of the indicated E2 protein. OD values obtained from wells coated with GST ranged between 0.021 and 0.081.
No signal was detected when antibodies CBH-2, -5, -7, -8C, -8E, and -11 were added concurrently with E2 of genotypes 1a, 1b, 2a, and 2b to CD81-coated plates (Table 3). In contrast, positive signals were obtained with HMAbs CBH-4G, CBH-4B, CBH-4D, and CBH-17 in a manner consistent with the reactivity of these HMAbs with GNA-captured E2 (compare binding levels in Fig. 2 and Table 3). In particular, CBH-4G reacted with E2 captured onto CD81-coated plates to equivalent extents with E2 proteins of all four genotypes tested. Incubation of increasing concentrations of HMAbs CBH-4G, -4B, -4D, and -17 with recombinant 1b E2 in CD81-coated plates indicated that these antibodies achieved 50% of maximum binding at concentrations of less than 5 μg/ml (Fig. 4). Binding of E2 and antibody to the CD81-coated plates was not observed with a control HMAb, nor did incubation of the NOB-positive HMAbs CBH-2, CBH-7, and CBH-8C with 1b E2 generate any signal. Thus, HMAbs CBH-4G, -4B, -4D, and -17 react with E2 in E2-CD81 complexes as readily as they react to E2 bound to GNA.
FIG. 4.
Capture of HCV HMAbs and E2 onto CD81-coated plates. Proteins derived from 15 μl of extract derived from BSC-1 cells infected with vaccinia virus Q1b and VWA were combined with the indicated concentrations (x axis) of antibody in a total volume of 100 μl of BLOTTO. The antibody-protein mixture was then added to microtiter plate wells coated with 100 ng of a GST–CD81-LEL fusion protein or nonrecombinant GST and incubated overnight at 4°C. Wells were washed, and bound antibody was detected as described in Materials and Methods. Signals obtained for antibody captured by CD81-coated wells were subtracted from signals obtained from GST-coated wells and averaged. R04 is an isotype matched HMAb that recognizes a CMV protein (14). Values are the average of duplicate wells. Error bars indicate 1 standard deviation from the mean.
The failure of HMAbs CBH-2, -5, -7, -8C, -8E, and -11 to react in the antibody capture experiments could be due to the antibodies failing to bind to CD81-E2 complexes or due to the antibodies inhibiting the formation of CD81-E2 complexes. To discriminate between these possibilities, HMAb CBH-4G was biotinylated and incubated with HCV E2 proteins to label E2 with the biotinylated antibody. The labeled E2 was then combined with increasing concentrations of antibody and added to microtiter plates coated with GNA (which would bind all E2 protein) or CD81-LEL. Results obtained with five of the HMAbs and a control are presented in Fig. 5. None of the antibodies significantly inhibited binding of the labeled 1a or 2a E2 protein to GNA-coated plates. Thus, HMAbs CBH-2, -5, -7, -8C, and -11 did not displace the antibody label on the E2, nor did an irrelevant HMAb, R04, inhibit binding of labeled E2 to either GNA or CD81. In contrast, HMAbs CBH-2, -5, -7, and -8C inhibited binding of labeled 1a or 2a E2 to CD81-LEL. Similar results were obtained with HMAbs CBH-2, -5, -7, and -8C with 1b and 2b E2 proteins and with HMAb CBH-8E and all four E2 proteins (data not shown). CBH-11 inhibited binding of 1b, 2b (data not shown), and 2a E2 but not 1a E2 (Fig. 5) to CD81-LEL, consistent with the reactivity of this HMAb to E2 only. The 50% inhibition values for HMAbs CBH-2, -5, -7, -8C, and -8E ranged from 0.4 to 6.0 μg/ml, consistent with values obtained in the NOB assay (Table 3) or in the E2 binding assays (Fig. 2).
FIG. 5.
Antibody-mediated inhibition of E2 binding to CD81. Extract derived from BSC-1 cells infected with vaccinia virus Q1a and VWA (■, □) or Q2a and VWA (▾, ▿) were diluted in BLOTTO, and biotinylated HMAb CBH-4G was added to a final concentration of 8 μg/ml. The mixture was incubated for 20 min to allow for antibody-mediated labeling of E2. Then aliquots of the labeled E2 proteins were combined with an equal volume of BLOTTO containing increasing amounts (x axis) of competing test antibody (indicated above graph) in wells coated with GNA (closed symbols) or human CD81-LEL (open symbols). R04 is an isotype-matched HMAb that recognizes a CMV protein (14). After overnight incubation at 4°C, wells were washed, strepavidin-conjugated alkaline phosphatase was added, and bound CBH-4G–E2 was detected as described in Materials and Methods. Results are expressed as the percentage signal obtained relative to wells with no competing antibody. Signals are the average values of duplicate wells. Error bars indicate 1 standard deviation from the mean.
Results from the flow cytometry and in vitro inhibition assays indicated that HMAbs CBH-2, -5, -7, -8C, -8E, and -11 inhibited the binding of E2 to CD81. The inhibitory activity of the HMAbs could be due either to steric hindrance of E2 binding to CD81 or to binding of the HMAbs initiating a conformational change in E2 that prevented E2 from subsequently binding to CD81. These alternative mechanisms were assessed by the ability of HMAbs CBH-2, -5, -7, -8C, -8E, and -11 to bind to preformed complexes of CD81-LEL and recombinant E2 (Fig. 6). Results obtained with these antibodies were compared to results obtained with CBH-4G and a control antibody. No reactivity was observed with any of the E2-CD81 complexes and the control antibody. Consistent with the results of the binding inhibition assay, HMAb CBH-4G efficiently recognized preformed CD81-E2 complexes with E2 proteins of genotypes 1a, 1b, 2a, and 2b. Thus, similar amounts of HCV-E2 complexes were formed using HCV E2 of each genotype tested. HMAbs CBH-2, -5, -7, -8C, -8E, and -11 did not react with preformed 1b E2-CD81 complexes. However, HMAb CBH-7 recognized complexes of HCV 1a E2 with CD81. HMAbs CBH-2, -5, -7, and -8E recognized complexes of HCV 2b E2 with CD81 and HMAbs CBH-7, CBH-5, CBH-8C, and CBH-11 recognized complexes of HCV 2a E2 with CD81. Thus, all six HMAbs that inhibit the interaction of E2 with CD81 can bind to some but not all CD81-E2 complexes.
FIG. 6.
Reactivity of HCV HMAbs to preformed CD81-E2 complexes. Proteins derived from 3 × 105 BSC-1 cells infected with vaccinia virus VWA with Q1a (■, □), Q1b (▴, ▵), Q2a (▾, ▿), and Q2b (⧫, ◊) were added to microtiter plate wells coated with human CD81-LEL (closed symbols and solid lines) or murine CD81-LEL (open symbols and dotted lines) and incubated overnight at 4°C. Wells were washed and then incubated with increasing amounts (x axis) of the indicated HCV HMAb. Bound antibody was detected as described in Materials and Methods. Values for murine CD81 are derived from a single determination. Values for human CD81 are the means of duplicate wells. Error bars indicate 1 standard deviation from the mean.
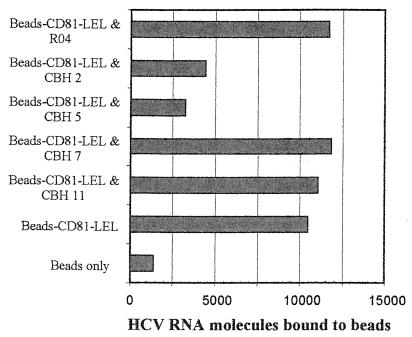
Since recombinant HCV E2 proteins may not mimic the structure of E2 on the surface of a virion, we asked whether antibodies that blocked recombinant E2 binding to CD81 were capable of interfering with virus binding to CD81. Because of the lack of HCV culture assays in vitro, we took advantage of a PCR assay developed to demonstrate binding of envelope-associated HCV RNA to CD81 (30). Briefly, the CD81-LEL was attached to polystyrene beads and incubated with infectious plasma containing a known amount of HCV 1a RNA molecules. After washing, the amount of CD81-associated virus was measured by quantitative RT-PCR. HMAbs CBH-2, CBH-5, CBH-7, and CBH-11 were evaluated at a concentration of 10 μg/ml, which was sufficient to achieve greater than 65% inhibition of E2 binding to CD81 in the NOB and protein inhibition assays (Table 3 and Fig. 5). No inhibition of virus binding was observed with a control antibody or with antibody CBH-7 or CBH-11. In contrast, preincubation of infectious plasma with 10 μg each of HMAbs CBH-2 and CBH-5 per ml inhibited HCV binding to CD81 by 70 and 80% percent, respectively (Fig. 7). These results support the view that HMAbs CBH-2 and CBH-5 can bind HCV virions and have the potential to inhibit binding of virions to CD81 in vivo.
FIG. 7.
HMAbs CBH-2 and CBH-5 inhibit binding of HCV virions to CD81. Number of HCV RNA molecules bound to polystyrene beads (x axis) after HCV 1a chimpanzee serum was combined with 10 μg of the indicated antibodies (y axis) and then allowed to bind to beads coated with CD81-LEL. The virion binding experiment was performed as described in Materials and Methods.
DISCUSSION
This study provides support for the contention that the majority of antibodies capable of inhibiting the interaction of HCV E2 with CD81 are developed to conformational epitopes conserved across genotypes 1 and 2. By producing HMAbs, our approach directly analyzes the human immune response to HCV. We chose an individual with a 2-year history of normal alanine aminotransferase values as the donor for antigen-specific B cells since an asymptomatic infection was more likely to reflect an effective humoral immune response containing HCV-related disease. This individual also had a high NOB titer. In the study by Rosa et al., (35), 60% of 34 HCV-infected individuals exhibited essentially no NOB activity and the remainder exhibited minimal inhibition of E2 binding to CD81. Thus, the antibody response of the B-cell donor used in the production of the HMAbs described herein is exceptional and cannot be assumed to be representative of the antibody response observed in the majority of HCV-infected individuals. We are currently using the HCV HMAbs described in this report to determine the prevalence and titers of similar antibodies in other HCV-infected individuals.
The B-cell donor was infected with genotype 1b. In an effort to obtain cross-reactive antibodies, we used HCV 1a E2 proteins to identify HMAbs to HCV E2. All of the HMAbs also reacted with E2 from a heterologous HCV 1b isolate, Q1b, that was 80% homologous with the HCV 1a isolate used in the selection of HMAbs. We do not, unfortunately, have sequence information on the HCV 1b isolate from the B-cell donor. It is therefore not possible to comment on the relatedness of Q1b to the homologous 1b isolate. All of the isolated hybridomas secreted antibodies with IgG1 heavy chains, which corroborates recent data indicating that the antibody response to HCV E2 is dominated by IgG1 antibodies (4). Nine of the HMAbs recognized conformational epitopes sensitive to denaturation of full-length HCV E2. Future studies with E2 deletion mutants or E2 fragments will be required to determine if the epitopes recognized by the HMAbs can be encompassed in discrete domains of E2. Four HMAbs, CBH-4D, -4B, -4G, and -17, did not inhibit the formation of HCV E2-CD81 complexes. One of these antibodies, CBH-4G, reacted with HCV E2-CD81 complexes of genotypes 1a, 1b, 2a, and 2b, confirming that E2 proteins of genotype 2 are capable of binding to CD81-LEL. The broad reactivity of HMAb CBH-4G with E2-CD81 complexes makes it a useful reagent for quantifying titers of antibody capable of inhibiting binding of E2 with CD81 in HCV sera.
Six of the HMAbs recognizing conformational epitopes, CBH-2, -5, -7, -8C, -8E, and -11, inhibited the binding of HCV E2 proteins with CD81-LEL and can be referred to as inhibitory or NOB positive. However, all six antibodies were able to bind to preformed E2–CD81-LEL complexes with some of the same E2 proteins that they efficiently inhibited from binding CD81. Five of the six inhibitory antibodies were reactive with preformed complexes of 2a or 2b E2 proteins with CD81-LEL and not 1a or 1b E2 complexes with CD81. This implies that the tertiary structure of genotype 2 E2 differs from that of genotype 1 E2. However, the ability of an inhibitory antibody to bind to CD81-E2 complexes did not strictly correlate with genotype (i.e., HMAb CBH-7 bound complexes of 1a E2 with CD81-LEL and HMAb CBH-8C did not bind complexes of 2b E2 with CD81-LEL). Thus, the amino acid sequences that underlie the reactivity of inhibitory HMAbs with CD81-E2 complexes may be subject to mutation in vivo. This raises the possibility that sequences in variable regions of E2 affect the reactivity of inhibitory HMAbs with CD81-E2 complexes, even though the actual epitopes recognized by the HMAbs appear to be highly conserved. We also note that the differences in reactivity of the six inhibitory HMAbs with CD81-E2 complexes suggest that the antibodies recognize multiple distinct epitopes. Competition and mutagenesis studies should clarify the total number of unique epitopes recognized by the inhibitory HMAbs.
Previous studies have suggested that the binding site for CD81 in E2 is dependent on E2 conformation (12, 35). The reactivity of the six inhibitory antibodies with preformed E2-CD82 complexes indicates that the epitopes recognized by these antibodies do not directly overlap the CD81 binding site. If the binding sites of the HMAbs and CD81 overlapped, steric hindrance should have prevented simultaneous binding of E2 with the HMAb and CD81. A second possibility is that the epitopes recognized by the inhibitory HMAbs are located nearby to the CD81 binding site, so that a complex of antibody and E2 has a reduced affinity for CD81 due to antibody-mediated steric hindrance. However, the very similar binding patterns of the six inhibitory HMAbs and the noninhibitory HMAb CBH-4G with complexes of genotype 2 E2 and CD81 argue that the epitopes are equivalently accessible. The most likely explanation is that the affinity of the HMAbs for E2 was higher than the affinity of CD81-LEL for E2. Once an antibody-E2 complex was formed, the inhibitory antibodies stabilized E2 protein in a conformation that had a low affinity for CD81. If a complex of E2 and CD81 was already formed, accessibility of the antibody to its epitope may have been affected by the altered conformation of the E2 in the CD81 complex. Alternatively, the epitopes recognized by the NOB-positive antibodies may always be exposed in a CD81-E2 complex, but subsequent binding of the antibody to this epitope may have a variable ability to dissociate E2 from CD81 (i.e., binding of CBH-5 to a complex of 1a E2 and CD81 results in dissociation of 1a E2 from CD81, and binding of CBH-5 to a complex of 2a E2 and CD81 is not sufficient to dissociate 2a E2 from CD81). In the first case, the E2 protein remains associated with CD81; in the second case, the E2 protein would not be present. We are currently exploring the use of biotinylated CBH-4G and other epitope tags to attempt to differentiate between these two possibilities.
Two of the inhibitory antibodies, CBH-2 and CBH-5, were able to prevent the binding of intact HCV virions to CD81. Two other inhibitory antibodies, CBH-7 and CBH-11, did not inhibit the interaction of HCV virions with CD81. The failure of CBH-11 to inhibit binding of HCV virions to CD81 may reflect the poor reactivity of CBH-11 with HCV some 1a isolates (such as isolate Q1a). HMAbs CBH-2 and CBH-5 did not bind preformed 1a E2–CD81-LEL complexes and inhibited binding of virions to CD81. CBH-7 bound to preformed 1a E2–CD81-LEL complexes and did not inhibit binding of virions to CD81. Therefore, failure to bind to preformed complexes of E2 and CD81 may be a better predictor of the ability of an antibody to prevent binding of HCV virions to CD81 in vivo than is inhibition of formation of the CD81-E2 complex. One implication of this proposal would be that of the inhibitory HMAbs, CBH-2 has the greatest potential to effectively inhibit binding of multiple isolates of HCV to CD81 in vivo. The other inhibitory HMAbs would have only limited ability to inhibit binding of HCV 2a/2b virions to CD81. However, testing of CBH-2 and other inhibitory antibodies with HCV virions and recombinant E2 proteins generated from the same sera would be required to confirm this.
The observation that a fraction of the E2 antibodies isolated from this HCV PCR-positive B-cell donor could inhibit the interaction of E2 with CD81 raises an important question. If the B-cell donor had a high titer of potentially neutralizing antibodies, why did this individual continue to exhibit plasma viremia? The most obvious explanation is that CD81 is not the primary receptor for HCV. Antibodies recognizing different epitopes that interfere with the binding of HCV to the putative primary receptor would be the antibodies with neutralization activity. The donor may have had a relatively low titer of this type of antibody. If one assumes that CD81 is involved in HCV infectivity, a second explanation is that antibodies that can inhibit the binding of E2 to CD81 will neutralize the infectivity of the majority of HCV virions but have little effect on cells that are already infected. Studies of the infectivity of HCV innocula in chimpanzee have demonstrated that antibody-coated virions exhibit markedly reduced infectivity compared to free virions (17). Other studies with HCV sera have found that the ability of virions from sera to attach and enter target cells is critically dependent on whether the virions are free or coated with antibodies (21). In addition, studies of E2 expression in mammalian systems indicate that little or no envelope protein is expressed on the cell surface (8, 32). Thus, antibodies would have limited opportunity to bind to infected cells. Clearance of cells that are HCV infected would therefore depend on the action of cytotoxic T lymphocytes, which may or may not be effective (reviewed in reference 33). Assuming that CD81 is a receptor or coreceptor for HCV, individuals with a strong NOB-positive antibody response to E2 may be at a steady state in which minimal de novo infection of susceptible cells occurs while the existing infected cells persist and continue to induce liver damage. Studies in which HCV antisera with high and low NOB activity are assessed for infectivity in naive chimpanzees will be required to more firmly establish a correlation between inhibition of E2-CD81 binding and true virus neutralization.
Overall, multiple HMAbs that recognized conserved epitopes and could inhibit the interaction of HCV E2 with CD81 were obtained from an HCV-infected individual with a high-titer immune response to E2. The antibodies that recognize these epitopes will be useful as reagents to better define the structure of HCV E2. The antibodies will also be very useful reagents in assessing the importance of the interaction between HCV E2 and CD81 in HCV infection. More importantly, if CD81 is confirmed to be a receptor or coreceptor for HCV, the antibodies that inhibited binding of HCV virions to human CD81, CBH-2, and CBH-5 have the potential to mediate virus neutralization. The absence of a true in vitro model for virus neutralization, however, will require that the fundamental proof be obtained by the ability of selected HMAbs to prevent or modify HCV infection in appropriate animal models. If successful, broadly reactive neutralizing antibodies will likely have therapeutic utility. Analogous to the success achieved with hepatitis B immunoglobulin in liver transplantation (7, 26), one possible application is to suppress HCV infection in liver transplant recipients with broadly reactive neutralizing HMAbs.
ACKNOWLEDGMENTS
We thank Ann Warford, Stanford University, for providing HCV genotype-specific sera and genotype testing. Lily Chan, National University of Singapore, generously provided DNA sequencing of the Q1b isolate. Other DNA sequencing was performed by Zhen-yong Keck, whose help and thoughtful comments we gratefully acknowledge. We also thank Sonal Rajyaguru for technical assistance and Wanda Washington for administrative assistance.
This work was supported in part by PHS grants DA-06596 and HL-33811 to S.K.H.F., AI40035 and NIH P51 R13986 to R.E.L., and CA-34233 to S.L.
REFERENCES
- 1.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–62. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 2.Burioni R, Plaisant P, Manin A, Rosa D, Delli Carri V, Bugli F, Solforosi L, Abrignani S, Varaldo P E, Fadda G, Clementi M. Dissection of human humoral immune response against hepatitis C virus E2 glycoprotein by repertoire cloning and generation of recombinant Fab fragments. Hepatology. 1998;28:810–814. doi: 10.1002/hep.510280331. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso M S, Siemoneit K, Sturm D, Krone C, Moradpour D, Kubanek B. Isolation and characterization of human monoclonal antibodies against hepatitis C virus envelope glycoproteins. J Med Virol. 1998;55:28–34. [PubMed] [Google Scholar]
- 4.Chen C, Sallberg M, Sonnerborg A, Weiland O, Mattsson L, Jin L, Birkett A, Peterson D, Milich D R. Limited humoral immunity in hepatitis C infection. Gastroenterology. 1999;116:153. doi: 10.1016/s0016-5085(99)70237-4. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghten M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo Q-L, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghten M, Muchmore E. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson R C. Management of posttransplantation viral hepatitis—hepatitis B. Liver Transplant Surg. 1998;4(5 Suppl. 1):S73–S78. [PubMed] [Google Scholar]
- 8.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feray C, Gigou M, Samuel D, Ducot B, Maisonneuve P, Reynes M, Bismuth A, Bismuth H. Incidence of hepatitis C in patients receiving different preparations of hepatitis B immunoglobulins after liver transplantation. Ann Intern Med. 1998;128:810–816. doi: 10.7326/0003-4819-128-10-199805150-00003. [DOI] [PubMed] [Google Scholar]
- 12.Flint M, Maidens C, Loomis-Price L D, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habersetzer F, Fournillier A, Dubuisson J, Rosa D, Abrignani S, Wychowski C, Nakano I, Treppo C, Desgranges C, Inchauspe G. Characterization of human monoclonal antibodies specific to the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology. 1998;249:32–41. doi: 10.1006/viro.1998.9202. [DOI] [PubMed] [Google Scholar]
- 14.Hadlock K G, Rowe J, Perkins S, Bradshaw P, Song G-Y, Cheng C, Yang J, Gascon R, Halmos J, Rehman S-M, McGrath M, Foung S K H. Neutralizing human monoclonal antibodies to conformational epitopes of human T-cell lymphotropic virus type 1 and 2 gp46. J Virol. 1997;71:5828–5840. doi: 10.1128/jvi.71.8.5828-5840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada S, Suzuki R, Ando A, Watanabe Y, Yagi S, Miyamura T, Saito I. Establishment of a cell line constitutively expressing E2 glycoprotein of hepatitis C virus and humoral response of hepatitis C patients to the expressed protein. J Gen Virol. 1994;76:1223–1231. doi: 10.1099/0022-1317-76-5-1223. [DOI] [PubMed] [Google Scholar]
- 16.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 17.Hijikata M, Shimizu Y K, Kato H, Iwamoto A, Shih J W, Alter H J, Purcell R H, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 19.Ishii K, Rosa D, Watanabe Y, Katayama T, Harada H, Wyatt C, Kiyosawa K, Aizaki H, Matsuura Y, Houghton M, Abrignani S, Miyamura T. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology. 1998;28:1117–1120. doi: 10.1002/hep.510280429. [DOI] [PubMed] [Google Scholar]
- 20.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.68.8.4776-4784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura Y, Hayashida K, Ishibashi H, Niho Y, Akazawa K, Yanagi Y. Attachment of Hepatitis C virus to cultured cells: a novel predictive factor for successful interferon therapy. J Med Virol. 1998;56:25–32. [PubMed] [Google Scholar]
- 22.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 23.Lesniewski R, Okasinski G, Carrick R, Van Sant C, Desai S, Johnson R, Scheffel J M, Moore B, Mushahwar I. Antibody to hepatitis C virus second envelope (HCV-E2) glycoprotein: a new marker of HCV infection closely associated with viremia. J Med Virol. 1995;45:415–422. doi: 10.1002/jmv.1890450411. [DOI] [PubMed] [Google Scholar]
- 24.Levy S, Todd S C, Maecker H T. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 25.Mahaney K, Tedeschi V, Maertens G, Di Bisceglie A M, Veergalla J, Hoffnagle J H, Sallie R. Genotypic analysis of hepatitis C virus in American patients. Hepatology. 1994;20:1405–1411. doi: 10.1002/hep.1840200605. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz J S, Martin P, Conrad A J, Markmann J F, Seu P, Yersiz H, Goss J A, Schmidt P, Pakrasi A, Artinian L, Murray N G, Imagawa D K, Holt C, Goldstein L I, Stribling R, Busuttil R W. Prophylaxis against hepatitis B recurrence following liver transplantation using combination lamivudine and hepatitis B immune globulin. Hepatology. 1998;28:585–589. doi: 10.1002/hep.510280241. [DOI] [PubMed] [Google Scholar]
- 27.Moss B, Earl P. Expression of proteins in mammalian cells using vaccinia viral vectors. In: Ausubel F, Brent R, Kingston R, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1994. pp. 16.15.1–16.18.10. [Google Scholar]
- 28.Perkins S, Foung S. Stabilizing antibody secretion of human Epstein-Barr virus-activated B-lymphocytes with hybridoma formation by electrofusion. Methods Mol Biol. 1995;48:295–307. doi: 10.1385/0-89603-304-X:295. [DOI] [PubMed] [Google Scholar]
- 29.Piazza M, Sagliocca L, Tosone G, Guadagnino V, Stazi M A, Orlando R, Guglielmo B, Domenico R, Abrignani S, Palumbo F, Manzin A, Clementi M. Sexual transmission of the hepatitis C virus and efficacy of prophylaxis with intramuscular immune serum globulin. Arch Intern Med. 1997;157:1537–1544. [PubMed] [Google Scholar]
- 30.Pileri O, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 31.Puntoriero G, Meola A, Lahm A, Zucchelli S, Ercole B B, Tafi R, Pezzanera M, Mondelli M U, Cortese R, Tramontano A, Galfre G, Nicosia A. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 1998;17:3521–3533. doi: 10.1093/emboj/17.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehermann B, Chisari F V. Cell mediated immune response to the hepatitis C virus. Curr Top Microbiol Immunol. 2000;242:299–325. doi: 10.1007/978-3-642-59605-6_14. [DOI] [PubMed] [Google Scholar]
- 34.Rice C M. Is CD81 the key to hepatitis C virus entry? Hepatology. 1999;29:990–992. doi: 10.1002/hep.510290356. [DOI] [PubMed] [Google Scholar]
- 35.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q-L, Chien D, Pileri P, Houghten M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seki M, Oie M, Ichihashi Y, Shida H. Hemadsorption and fusion inhibition activities of hemagglutinin analyzed by vaccinia virus mutants. Virology. 1990;175:372–384. doi: 10.1016/0042-6822(90)90422-n. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q-L, Houghton M, Han J H. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 40.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]