Abstract
We used a herpes simplex virus type 2 (HSV-2) mutant with a deletion in the RR1 (ICP10) PK domain (ICP10ΔPK) and an MEK inhibitor (PD98059) to examine the role of ICP10 PK in virus growth. In HSV-2-infected cells, ICP10 PK binds and phosphorylates the GTPase activating protein Ras-GAP. In vitro binding and peptide competition assays indicated that Ras-GAP N-SH2 and PH domains, respectively, bind ICP10 at phosphothreonines 117 and 141 and a WD40-like motif at positions 160 to 173. Binding and phosphorylation did not occur in cells infected with ICP10ΔPK. GTPase activity was significantly lower in HSV-2- than in ICP10ΔPK-infected cells. Conversely, the levels of activated Ras and mitogen-activated protein kinase (MAPK), and the expression and stabilization of the transcription factor c-Fos were significantly increased in cells infected with HSV-2 or a revertant virus [HSV-2(R)] but not with ICP10ΔPK. PD98059 inhibited MAPK activation and induction-stabilization of c-Fos. Expression from the ICP10 promoter was increased in cells infected with HSV-2 but not with ICP10ΔPK, and increased expression was ablated by PD98059. ICP10 DNA formed a complex with nuclear extracts from HSV-2-infected cells which was supershifted by c-Fos antibody and was not seen with extracts from ICP10ΔPK-infected cells. Complex formation was abrogated by PD98059. Onset of HSV-2 replication was significantly delayed by PD98059 (14 h versus 2 h in untreated cells), a delay similar to that seen for ICP10ΔPK. The data indicate that Ras-GAP phosphorylation by ICP10 PK is involved in the activation of the Ras/MEK/MAPK mitogenic pathway and c-Fos induction and stabilization. This results in increased ICP10 expression and the timely onset of HSV-2 growth.
Signaling pathways, the ultimate targets of which are nuclear transcription factors, determine the cell's ability to respond to external stimuli. Transduced signals can be interpreted as mitogenic-proliferative, differentiating, or apoptotic, depending on the cell type and the nature and duration of the stimulus. The mitogenic Ras/MEK/MAPK pathway is initiated by growth factor mediated activation of cognate receptors on the cell surface causing the membrane bound G protein Ras to adopt an active, GTP-bound state. Ras coordinates the activation of a cascade of serine (Ser)-threonine (Thr) kinases that begins with Raf and is followed by MAP kinase kinase 1 and 2 (MEK1/2) and mitogen-activated protein kinase (MAPK1/2) and culminates in the expression of the transcription factor c-Fos (41, 46, 48, 67) which is important for promoting cell cycle progression into S phase (14, 39, 44). The GTPase-activating protein Ras-GAP, a major negative regulator of Ras activity, acts to enhance the weak intrinsic Ras GTPase activity by accelerating the hydrolysis rate of bound GTP to GDP (7, 27, 31, 48, 57). Ras-GAP inactivation, for example by phosphorylation on Ser-Thr residues, has been implicated in Ras activation (11, 24, 66, 89). The specificity of the signal transduction is determined by protein domains such as SH2, SH3, and PH that bind unique motifs in target proteins for recruitment into signaling complexes (55, 63).
Viruses depend on cells for their replication. They take advantage of preexisting signaling pathways or activate them through various strategies, including activation of the RAS/MEK/MAPK pathway by Ras-GAP inactivation (32). Vaccinia virus (42), simian virus 40 (SV40) (77), human immunodeficiency virus (HIV) type 1 (35), herpesvirus saimiri (38), and coxsackievirus B3 (32) depend upon the activated RAS/MEK/MAPK pathway for growth. Herpes simplex virus type 1 (HSV-1) increases the levels of transcription factors c-Jun (37) and NF-κB (22, 23), and its replication is enhanced by activation of the c-Jun N-terminal kinase (JNK) and p38 of the stress-activated signal pathway (58, 88). By contrast, HSV-2 increases c-Fos transcription (26), suggesting that the two HSV serotypes use different strategies to take advantage of signaling pathways. However, the mechanism responsible for c-Fos induction in HSV-2-infected cells, the contribution of the Ras/MEK/MAPK pathway, and their relationship to virus replication are still unknown.
The large subunits of HSV-1 and HSV-2 ribonucleotide reductase (RR1) differ from their counterparts in eukaryotic and prokaryotic cells and in other viruses in that they have an intrinsic PK activity (2, 5, 12, 15, 17, 49, 50, 60, 65). The RR1 promoter has an octamer-TAATGARAT sequence that responds to the VP16-Oct1 complex (18, 78, 86, 87). RR1 is expressed with apparently biphasic kinetics that consist of immediate-early (IE; also known as α) and early components (3, 30, 84, 86, 87, 90). Expression is independent of the regulatory IE protein ICP4 (18, 78, 86, 87). AP-1 cognate sites in the HSV-2 RR1 (also known as ICP10) promoter are required for basal expression (86, 87, 90). ICP10 PK is required HSV-2 growth. A temperature-sensitive HSV-2 mutant that is negative for ICP10 PK activity at the nonpermissive temperature failed to grow under these conditions (69). A mutant deleted in the ICP10 PK domain (ICP10ΔPK) had two apparently distinct replication defects, including a significant delay in growth onset in dividing (10% serum) and nondividing (1% serum) cells, and impaired growth in nondividing cells (73). However, the mechanism(s) whereby ICP10 PK contributes to HSV-2 growth is still unclear. The studies described in this report were designed to address this question.
MATERIALS AND METHODS
Viruses and plasmids.
HSV-2 (G strain), the PK-deleted mutant ICP10ΔPK, and the revertant virus HSV-2(R) were as described earlier (73). ICP10 and mutant proteins used in binding assays are listed in Table 1. The construction of eukaryotic expression vectors for these proteins and the establishment of constitutively expressing cell lines were described (12, 50, 60). They include kinase negative mutants p139TM (deleted in the transmembrane domain), p140lys (mutated in both ATP-binding sites) and p140III (mutated in the ion-binding site), and kinase-positive mutants p140pro (mutated in both SH3-binding sites) and p136I/II (deleted in amino acids 106 to 178). pJL11 is a bacterial expression vector for the ICP10 mutant pp29la1 that consists of amino acids 1 to 283 and has threonine (Thr)-specific kinase activity (49). Constructions in which the ICP10 promoter or a promoter mutated in both AP-1 sites were inserted 5′ to the chloramphenicol acetyltransferase (CAT) gene were as described previously (86, 90).
TABLE 1.
ICP10 mutants used in binding assaysa
| Protein | ICP10 position | PK activity |
|---|---|---|
| ICP10 | Wild type | + (Ser-Thr) |
| p139TM | Δ86–105 | − |
| p140lys | Lys176 and Lys259 | − |
| p140III | Glu209 | − |
| p140pro | Pro-rich150 and 395 | + (Ser-Thr) |
| p136I/II | Δ106–178 | + (Ser-Thr) |
| pp29la1 | 1–283 | +(Thr) |
The construction and properties of the ICP10 and mutant eukaryotic expression vectors and the establishment of constitutively expressing cell lines were described earlier (49, 50, 60). p139TM is deleted in the transmembrane domain (50). p140lys is mutated in Lys176 and Lys259, p140III is mutated in Glu209, and p140pro is mutated in SH3 binding sites at positions 150 and 395 (60). p136I/II is deleted in amino acids 106 to 178 (50). pp29la1 is expressed by the bacterial vector pJL11 (49).
Cells and virus infection.
African green monkey (Vero) cells were grown in Eagle minimal essential medium (EMEM) with 10% fetal calf serum (FCS). They were infected with HSV-2 or HSV-2(R) in medium containing 1% FCS and with ICP10ΔPK in medium containing 10% FCS (73). The multiplicity of infection was 5 PFU/cell. In experiments with the MEK inhibitor PD98059 (1, 56), cells were pretreated (1 h, 37°C) with 25 or 50 μM PD98059 and infected in medium containing the same drug concentration. Cell lines that constitutively express ICP10 or its mutants were used in in vitro binding assays. They were grown in EMEM with 1% l-glutamine, 1% sodium pyruvate, 1% nonessential amino acids, and 10% FCS (50, 60).
Antibodies.
ICP10 antibody was raised in rabbits against a synthetic peptide consisting of ICP10 residues 13 to 26 (4). The following antibodies were purchased. Polyclonal antibodies to c-Fos (H-125) (Santa Cruz Biotechnology, Santa Cruz, Calif.), MAPK (recognizes MAPK1/2) (Oncogene Research Products, Cambridge, Mass.), active MAPK (recognizes the dually phosphorylated active form of MAPK1/2) (Promega Corp., Madison, Wis.), and monoclonal antibodies (MAbs) to Ras-GAP (B4F8) and Ras (F132-62 and Y13-259) (Oncogene Science, Manhasett, N.Y.).
Peptides.
ICP10 phosphopeptides pT117 (112VALGGpTSGPSA122), pT130 (125SVGTQpTSGEFL135), and pT141 (136HGNPRpTPEPQG146) were synthesized by Research Genetics, Huntsville, Ala. They are phosphorylated on Thr residues 117 (pThr117), 130 (pThr130), and 141 (pThr141), respectively. ICP10 peptides Pro1 (149AVPPPPPPPFPWGH162), WD40 (160WGHECCARRDARGG173), and 27 (CAESRRDDLESDSS) that corresponds to a sequence in the HSV-2 protein ICP27 were synthesized by the UMAB Biopolymer Facility. Phosphopeptides were dephosphorylated by treatment (2 h, 37°C) with 15 U of calf intestine alkaline phosphatase (CIAP; Boehringer-Mannheim, Indianapolis, Ind.) in a buffer supplied by the manufacturer; CIAP was removed by ultrafiltration through Centriprep-20 filters (Amicon, Beverly, Mass.). Heat-denatured CIAP (95°C, 1 h) served as a control.
Immunocomplex PK-immunoblotting assays.
Cell extracts in PK lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 100 kallikrein U/ml of aprotinin) were immunoprecipitated with ICP10 antibody and protein A-Sepharose beads. The beads were washed with wash buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.5]) and incubated (15 min, 30°C) in PK buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM MnCl2) with 10 μCi of [γ-32P]ATP (0.1 μM, 3,000 Ci/mmol) (New England Nuclear, Boston, Mass.). After an extensive washing in wash buffer, the protein-bead complexes were denatured by boiling, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7% polyacrylamide gels, and visualized by autoradiography (12, 50, 60, 69–74). For immunoblotting, proteins were transferred to nitrocellulose membranes. They were blocked for 1 h at room temperature with 5% nonfat milk in TNT buffer (Tris-buffered saline, 0.1% Tween 20 [pH 7.4]) for ICP10 and c-Fos antibodies and for 1 h at 37°C with 1% bovine serum albumin (BSA) in TNT buffer for MAPK antibodies. Immunoblotting was done at room temperature with primary antibodies (1 h for ICP10 and c-Fos antibodies; 2 h for MAPK antibodies), followed by 1 h with protein A-peroxidase (Sigma). Detection was with enhanced chemiluminescence reagents (NEN) according to the manufacturer's instructions.
Ras-GAP binding and peptide competition assays.
Ras-GAP protein interaction domains subcloned into pGEX-2T vector (8, 29) were obtained from T. Smithgall (University of Nebraska, Omaha) (see Fig. 2A). Following induction with IPTG (0.1 mM, 4 h), bacteria were resuspended in NETN buffer (20 mM Tris-HCl [pH 8.0], 0.5% NP-40, 100 mM NaCl, 1 mM EDTA), lysed by sonication, and clarified of cell debris by centrifugation at 14,000 × g for 30 min. They were immobilized on glutathione-Sepharose beads (Pharmacia, Piscataway, N.J.) by incubation for 2 h at 4°C, and the beads were extensively washed with phosphate-buffered saline (PBS) (pH 7.5) containing 1 mM PMSF and 5 mM benzamidine. Extracts of pJL11-containing bacteria induced (at 42°C) to express pp29la1 (49) were prepared in a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM EDTA, 1 mM PMSF, 5 μM pepstatin A, 10 mM benzamidine, 100 kallikrein U/ml of aprotinin, 2 mg of lysozyme per ml, and n-octyl-β-d-glucopyranoside. The extracts were then mixed with 5 M NaCl (0.2 volume), sonicated, and clarified by centrifugation (22,000 × g, 1 h). Extracts of eukaryotic cells that express ICP10 or its mutants were prepared in a lysis buffer consisting of 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 100 mM sodium fluoride, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, and 100 kallikrein U/ml of aprotinin. They were clarified of cell debris by centrifugation (20,000 × g, 30 min). For binding assays, beads coated with Ras-GAP fusion proteins (20 μg) were incubated (2 h, 4°C) with lysates of cells that express ICP10 or its mutants (500 μg), and the beads were washed three times in buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1% NP-40 and then resuspended and denatured in 100 μl of denaturing buffer at 95°C for 5 min. Proteins were resolved by SDS-PAGE on 7% polyacrylamide gels, transferred to nitrocellulose membranes, and immunoblotted with ICP10 antibody (12, 50, 60, 69–74). For peptide competition, extracts of ICP10-expressing cells (375 μg) were incubated (1 h, 4°C) with increasing concentrations of peptides prior to the addition of beads coated with Ras-GAP fusion proteins.
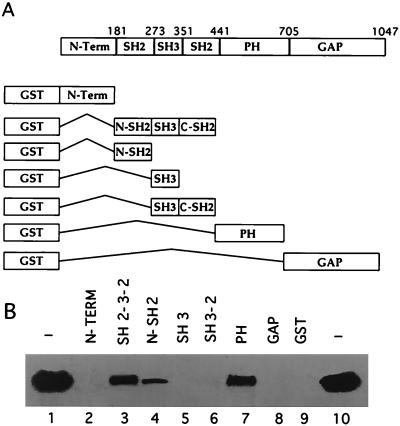
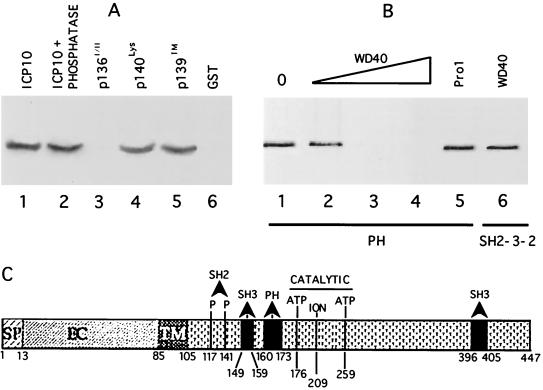
FIG. 2.
ICP10 binds Ras-GAP N-SH2 and PH fusion proteins in vitro. (A) Schematic representation of GST fusion proteins of Ras-GAP protein-protein binding domains. (B) GST beads coated with Ras-GAP fusion proteins N-term (lane 2), SH2-SH3-SH2 (lane 3), N-SH2 (lane 4), SH3 (lane 5), SH3+C-SH2 (lane 6), PH (lane 7), GAP (lane 8), or uncoated (lane 9) (20 μg) were mixed (2 h, 4°C) with extracts of cells that constitutively express the autophosphorylated ICP10 (input; lanes 1 and 10). Bound proteins were resolved by SDS-PAGE and immunoblotted with ICP10 antibody.
Kinetics of GTP hydrolysis.
GTP hydrolysis was assayed as described elsewhere (70). Extracts of cells (107) in lysis buffer (50 mM Tris-HCl [pH 7.5], 1% NP-40, 150 mM NaCl, 1 mM PMSF) were kept on ice for 15 min and clarified by centrifugation (30 min, 16,000 × g). Supernatants were incubated (1 h, 4°C) with MAb F132-62 (1 μg) that recognizes a Ras epitope distant from the Ras-GAP binding site, followed (30 min, 4°C) by protein A-Sepharose CL4B (100 μl). Beads were washed in lysis buffer and incubated (30 min, 4°C) in 100 μl of radioimmunoprecipitation assay (RIPA) buffer containing 0.1 μM [α-32P]GTP (specific activity, 3,000 Ci/mmol; NEN). Unbound nucleotide was removed by extensive washing in RIPA buffer, and the amount of bound [α-32P]GTP was determined. The samples were resuspended in 50 μl of GTPase buffer (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2) and incubated at 37°C for 0 to 60 min. At 10-min intervals, the protein A-Sepharose immunocomplex pellet was gently resuspended, and 2 μl of the reaction mixture was spotted onto a polyethyleneimmine (PEI) plate (E. Merck, Darmstadt, Germany) and chromatographed in 0.75 M KH2PO4 (pH 3.4). Plates were visualized by autoradiography, and the locations of GTP and GDP were determined by assessing the migration of 32P-labeled standards. Quantitation was done by aligning the autoradiograms with the PEI plates and scraping the GDP and GTP spots directly into scintillation vials. Results are expressed as the percent GTP hydrolysis, where percent hydrolysis = [cpmGDP/(cpmGTP + cpmGDP)] × 100.
Analysis of GDP and GTP bound to Ras.
Analysis of GDP and GTP bound to Ras was done as described elsewhere (68, 70). Cells (5 × 106) were labeled with [32P]orthophosphate (500 μCi/ml; NEN) in phosphate-free EMEM with 0.1 mM nonessential amino acids–1 mM sodium pyruvate–dialyzed FCS for 18 h at 37°C and infected with HSV-2, ICP10ΔPK, or HSV-2(R). At 0, 2, 4, 8, and 12 h postinfection (p.i.), cells were lysed in a buffer consisting of 50 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF, and 100 kallikrein U/ml of aprotinin and then incubated on ice for 15 min; extracts were clarified by centrifugation at 16,000 × g for 30 min. To reduce free nucleotides, 0.1 ml of a 10% charcoal solution (previously treated with 10 mg of BSA per ml) was added to the supernatant, mixed vigorously, and clarified by centrifugation at 8,000 × g for 10 min. Supernatants (3 × 107 to 1 × 108 cpm) were immunoprecipitated (1 h, 4°C) with 1 μg (10 μl) of MAb Y13-259, followed by treatment with 100 μl of protein A-Sepharose CL4B beads (50% [vol/vol], 30 min, 4°C) which had been previously coated with rabbit anti-rat immunoglobulin G. Beads were washed three times with RIPA buffer and twice with PBS (pH 7.1) containing 20 mM MgCl2 and then resuspended in 20 μl of 20 mM Tris-HCl (pH 7.5), 20 mM EDTA, 2% SDS, 0.5 mM GTP, and 0.5 mM GDP. They were heated (5 min, 65°C) and centrifuged (5 min, 8,000 × g). The supernatants were spotted onto PEI plates and chromatographed in 0.75 M KH2PO4 (pH 3.4). The radioactivity was quantitated by densitometric scanning using a Bio-Rad GS700 densitometer. Results are expressed as the percent bound GTP = [GTP/(GTP + GDP)] × 100.
DNA transfection and CAT assays.
Cells (105/well) were plated 24 h prior to transfection into six-well cluster dishes (6 by 35 mm; Costar, Corning, N.Y.). DNA transfection was done by the calcium phosphate precipitation method and employed a glycerol boost. Transfection mixtures contained 1 μg of target plasmid DNA. Infection with HSV-2, ICP10ΔPK, or HSV-2(R) (5 PFU/cell) was done 24 h after transfection, and CAT assays were performed 40 to 48 h posttransfection. Routine assay conditions employed [14C]chloramphenicol (0.2 μCi) substrate and a 60-min incubation (22, 86, 87, 90). For quantitative estimates of CAT activity, the appropriate sections were cut from thin-layer chromatography plates and placed in toluene 2,5-diphenyloxazole-12,4-bis(5-phenyloxazole) benzene scintillation fluid and the radioactivity was counted in a Beckman LS6800 liquid scintillation counter. Quantitative comparisons were made by measuring the amounts of chloramphenicol-acetate product with enzyme levels on the linear part of the curve for product formation versus the extract concentration and time. pSV2CAT was used as a control for transfection efficiency, and the parent pCATB′ was used as a negative control (22, 86, 87, 90).
Gel retardation assay.
The 61-bp DNA fragments that contain the ICP10 promoter (AP-wt) or its mutant in both AP-1 cis response elements (AP-mu) and nuclear extracts of Vero cells mock infected (with PBS) or infected with HSV-2 or ICP10ΔPK were used as previously described (22, 86, 87, 90). DNA fragments were end labeled with [α-32P]dCTP and separated by 8% PAGE. Double-stranded oligonucleotides AP (5′-GATCCAAGCTATGACTCATCCGGTCTAGAA-3′) and ΔAP (5′-GATCCAAGCTATGAACCATCCGGTCTAGAA-3′) were used in competition assays. Retardation mixtures were incubated with 2 μg of nuclear protein and 4 fmol of radiolabeled DNA fragment for 40 min on ice. Electrophoresis was done at 10 V/cm and at 4°C through a 4% native polyacrylamide gel (29:1 bisacrylamide) in 0.25× TBE that had been prerun for 2 h. For supershift assays, c-Fos antibody (Oncogene Science, 1:25 dilution) was included in the incubation mixture.
Single-step growth assays.
Vero cells were pretreated (1 h, 37°C) with 50 μM MEK inhibitor PD98059 (Calbiochem, LaJolla, Calif.) or were mock treated with reconstitution solution free of PD98059 and then infected with HSV-2 in the presence or absence of PD98059. Adsorption was done for 1 h (0 h in the growth curve). Cells and supernatants were harvested 2 to 48 h after adsorption and then frozen, thawed, and assayed for the virus titers by plaque assay (73).
RESULTS
Ras-GAP coprecipitates with ICP10 PK from HSV-2-infected cells.
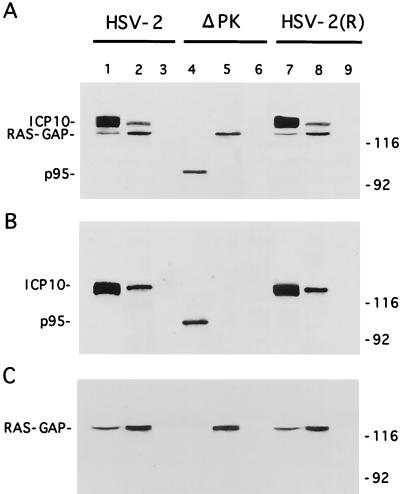
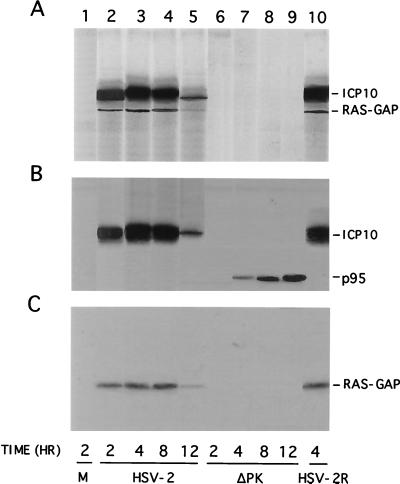
We have previously shown that Ras-GAP coprecipitates with ICP10 from constitutively expressing cells (70). To determine whether this also happens during virus infection, Vero cells were infected with HSV-2, ICP10ΔPK, or HSV-2(R) labeled with [35S]methionine at 0 to 8 h p.i., and the extracts were used in immunoprecipitation-immunoblotting with antibodies to ICP10 or Ras-GAP. These time points were chosen because we had previously shown that ICP10 is optimally expressed at 4 to 8 h p.i. (12) and a relatively long labeling interval is required for the detection of the methionine-poor Ras-GAP (data not shown). Two proteins (140 and 120 kDa, respectively) were coprecipitated from HSV-2-infected cells by the ICP10 (Fig. 1A, lane 1) and Ras-GAP (Fig. 1A, lane 2) antibodies. They were also coprecipitated from HSV-2(R)-infected cells (Fig. 1A, lanes 7 and 8). Coprecipitation was not seen in ICP10ΔPK-infected cells. Here, p95 was precipitated by the ICP10 antibody (Fig. 1A, lane 4) and the 120-kDa protein by the Ras-GAP antibody (Fig. 1A, lane 5). The 140-kDa protein in precipitates from HSV-2 (Fig. 1B, lanes 1 and 2)- and HSV-2(R) (Fig. 1B, lanes 7 and 8)-infected cells and the p95 protein in precipitates from ICP10ΔPK-infected cells (Fig. 1B, lane 4) were recognized by the ICP10 antibody in immunoblotting. Ras-GAP antibody recognized the 120-kDa protein in the ICP10 immunocomplexes (Fig. 1C, lanes 1, 2, 5, 7, and 8). Preimmune serum was negative (Fig. 1A, B, and C, lanes 3, 6, and 9). The data indicate that Ras-GAP interacts with ICP10 in HSV-2-infected cells and that the interaction occurs at sequences within the ICP10 PK domain.
FIG. 1.
Ras-GAP coprecipitates with ICP10 but not p95 from virus-infected cells. (A) Vero cells infected with HSV-2 (lanes 1 to 3), ICP10ΔPK (lanes 4 to 6), or HSV-2(R) (lanes 7 to 9) were labeled with [35S]methionine at 0 to 8 h p.i. Cell extracts obtained at this time were precipitated with antibody to ICP10 (lanes 1, 4, and 7) or RAS-GAP (lanes 2, 5, and 8) or with preimmune serum (lanes 3, 6, and 9). (B) Immunoblotting of precipitates shown in panel A with antibody to ICP10 (recognizes ICP10 and p95). (C) Immunoblotting of precipitates shown in panel A with antibody to Ras-GAP.
ICP10 binds to the Ras-GAP N-terminal SH2 (N-SH2) and PH domains.
To confirm that Ras-GAP forms a complex with ICP10 and to identify the domains involved in complexation, we used in vitro binding assays with the panel of GST fusion proteins shown in Fig. 2A. Beads coated with the Ras-GAP fusion proteins (or GST control) were mixed with extracts of cells that constitutively express ICP10, and the proteins in the complexes were resolved by SDS-PAGE and immunoblotted with ICP10 antibody. ICP10 was seen in complexes obtained with Ras-GAP fusion proteins SH2-SH3-SH2 (Fig. 2B, lane 3) N-SH2 (Fig. 2B, lane 4), and PH (Fig. 2B, lane 7). It was not seen in complexes obtained with the Ras-GAP fusion proteins N-term (Fig. 2B, lane 2), SH3 (Fig. 2B, lane 5), SH3+C-SH2 (Fig. 2B, lane 6), or GAP (Fig. 2B, lane 8), nor with GST alone (Fig. 2B, lane 9). ICP10 did not bind to beads coated with extracts of uninduced bacteria (data not shown). Densitometric scanning analysis indicated that 34 and 33% of the input ICP10, respectively, bound the PH and SH2-SH3-SH2 domains (densitometric units = 2,110 and 2,007 for PH and SH2-SH3-SH2, respectively). Consistent with previous reports (13), the levels of ICP10 bound by the N-SH2 fusion protein were lower (14% of input) (densitometric units = 850 and 6,071 for N-SH2 and input, respectively), presumably reflecting a lower binding affinity and/or stability due to the suboptimal conformation of the fusion protein. However, because ICP10 did not bind fusion proteins SH3 and SH3+C-SH2, we conclude that binding involves the N-SH2 module.
ICP10 residues pThr117 and pThr141 bind the Ras-GAP N-SH2 domain.
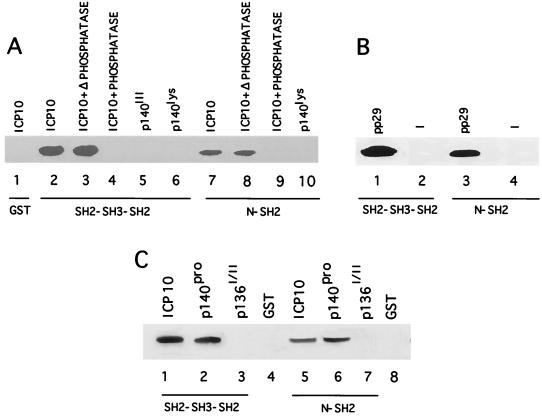
ICP10 binding of the Ras-GAP N-SH2 domain is consistent with recent findings which extend the range of potential SH2-protein interactions to include pSer and/or pThr residues (13, 51, 52, 59, 64). To examine whether the ICP10/N-SH2 interaction is phosphorylation dependent, binding assays were done with extracts of cells that constitutively express the autophosphorylated ICP10 untreated or treated (1 h, 37°C) with CIAP or heat-denatured CIAP and with extracts of cells that constitutively express PK negative ICP10 mutants p140lys and p140III (Table 1). The SH2-SH3-SH2 (Fig. 3A, lane 2) and N-SH2 (Fig. 3A, lane 7) fusion proteins bound the autophosphorylated ICP10 in untreated cells and in cells treated with the heat-denatured phosphatase (Fig. 3A, lanes 3 and 8). They did not bind the dephosphorylated ICP10 in extracts of phosphatase-treated cells (Fig. 3A, lanes 4 and 9) nor the kinase-negative mutants (Fig. 3A, lanes 5, 6, and 10), suggesting that binding is phosphorylation dependent. Binding occurs at pThr residues, as evidenced by the observation that SH2-SH3-SH2 (Fig. 3B, lane 1) and N-SH2 (Fig. 3B, lane 3) bound the ICP10 mutant pp29la1, the PK activity of which is Thr specific (49). Because the fusion proteins did not bind the ICP10 mutant p136I/II (Fig. 3C, lanes 3 and 7), which is deleted in amino acids 106 to 178 but retains PK activity (Table 1), the data suggest that binding involves ICP10 pThr residues at positions 106 to 178. Binding to Ras-GAP fusion proteins was also seen with p140pro, which is mutated in SH3-binding sites (60) (Fig. 3C, lanes 2 and 6). Binding was not seen with uninduced bacteria (Fig. 3B, lanes 2 and 4) or GST alone (Fig. 3A, lane 1, and C, lanes 4 and 8).
FIG. 3.
Ras-GAP N-SH2 binds ICP10 pThr residues at positions 106 to 178. (A) In vitro binding assays with GST beads (lane 1) or beads coated with Ras-GAP fusion proteins SH2-SH3-SH2 (lanes 2 to 6) or N-SH2 (lanes 7 to 10) and untreated extracts of cells that express the autophosphorylated ICP10 (lanes 1, 2, and 7) or extracts of the same cells treated for 2 h at 37°C with 15 U of CIAP (lanes 4 and 9) or 15 U of CIAP that had been denatured by heating (1 h, 95°C) (lanes 3 and 8). Extracts of cells that express the PK negative ICP10 mutants p140III (lane 5) or p140lys (lanes 6 and 10) were studied in parallel. (B) In vitro binding assays with Ras-GAP fusion proteins SH2-SH3-SH2 (lanes 1 and 2) or N-SH2 (lanes 3 and 4) and extracts of bacteria induced to express the ICP10 mutant pp29la1 which has pThr-specific PK activity (lanes 1 and 3) or uninfected bacteria (lanes 2 and 4). (C) In vitro binding assays with Ras-GAP fusion proteins SH2-SH3-SH2 (lanes 1 to 3) or N-SH2 (lanes 5 to 7) or uncoated beads (lanes 4 and 8) and extracts of cells that constitutively express autophosphorylated ICP10 (lanes 1 and 5) or the PK positive ICP10 mutants p140pro (lanes 2 and 6) or p136I/II (lanes 3 and 7).
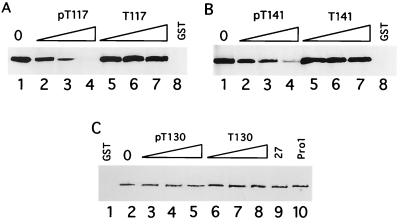
Computer-assisted analysis identified two motifs at ICP10 positions 106 to 178 which are similar to the site in the PDGFβ receptor (770YMAP773) that binds the Ras-GAP N-SH2 domain (20, 40). As with the platelet-derived growth factor β receptor, both ICP10 motifs contain a Thr residue (Thr117 and Thr141) that is followed at the +3 position by Pro. A third Thr residue (Thr130) is followed at the +3 position by Glu. To examine whether these sequences are involved in ICP10 binding of the Ras-GAP N-SH2 domain, we used competition assays as described in Materials and Methods. Dose-dependent inhibition of ICP10 binding was seen with phosphopeptide pT117. Densitometric scanning analyses indicated that the levels of ICP10 were 85, 33, and 1% of the input (Fig. 4A, lane 1) at 10, 50, and 100 μM, respectively (Fig. 4A, lanes 2 to 4) (densitometric units = 1,990, 1,695, 657, and 20 for input, 10, 50, and 100 μM, respectively). ICP10 binding was also competed for by phosphopeptide pT141 to a largely similar extent. In densitometric analyses, the levels of ICP10 were 87, 67, and 8% of the input (Fig. 4B, lane 1) at 10, 50, and 100 μM, respectively (Fig. 4B, lanes 2 to 4) (densitometric units = 2,075, 1,805, 1,390, and 181 for input, 10, 50, and 100 μM, respectively). ICP10 binding was not significantly reduced by phosphopeptide pT130, as evidenced by the finding of 90 to 99% of the ICP10 input (Fig. 4C, lane 2) at 10, 50, and 100 μM (Fig. 4C, lanes 3 to 5). Binding was also not reduced by dephosphorylated peptides T117 (Fig. 4A, lanes 5 to 7) and T141 (Fig. 4B, lanes 5 to 7) nor by peptides T130 (Fig. 4C, lanes 6 to 8), Pro1 (Fig. 4C, lane 10), and 27 (Fig. 4C, lane 9). We interpret the data to indicate that the Ras-GAP N-SH2 domain binds ICP10 at residues pThr117 and pThr141 singly or in tandem.
FIG. 4.
SH2 binds ICP10 pThr117 and pThr141. (A) Extracts of cells that constitutively express the autophosphorylated ICP10 (lane 1) were incubated (1 h, 37°C) with peptide pT117 (lanes 2 to 4 h) or T117 (lanes 5 to 7) at 10 μM (lanes 2 and 5), 50 μM (lanes 3 and 6), or 100 μM (lanes 4 and 7) and exposed (2 h, 4°C) to GST beads coated with Ras-GAP fusion protein SH2-SH3-SH2 (lanes 2 to 7) or left uncoated (lane 8). Bound proteins were resolved by SDS-PAGE and immunoblotted with ICP10 antibody. (B) Competition experiments were done as in panel A but with peptides pT141 (lanes 2 to 4) or T141 (lanes 5 to 7). Uncoated beads are shown in lane 8. (C) Competition experiments were done as in panel A but with peptides pT130 (lanes 3 to 5), T130 (lanes 6 to 8), 27 (100 μM; lane 9), or Pro1 (100 μM; lane 10). Uncoated beads are shown in lane 1, and input ICP10 is shown in lane 2.
The Ras-GAP PH domain binds a WD40-like motif in ICP10 PK.
In vitro binding and peptide competition assays were also used to examine the ICP10 interaction with the Ras-GAP PH domain. The PH fusion protein bound the autophosphorylated (Fig. 5A, lane 1) and dephosphorylated (Fig. 5A, lane 2) ICP10 and the kinase negative mutants p140lys (Fig. 5A, lane 4) and p139TM (Fig. 5A, lane 5). It did not bind p136I/II that is deleted at positions 106 to 178 (Fig. 5A, lane 3) and GST was negative (Fig. 5A, lane 6). The data suggest that binding is phosphorylation independent and involves ICP10 residues at positions 106 to 178. Computer-assisted analysis identified an ICP10 sequence at positions 160 to 173 that is consistent with the core of WD40 motifs known to bind PH domains (28, 63). A peptide (WD40) that represents this sequence caused dose-dependent inhibition of ICP10 binding to the PH fusion protein (Fig. 5B, lanes 2 to 4). Densitometric scanning analyses indicated that the levels of bound ICP10 relative to input (Fig. 5B, lane 1) were 65, <1, and <1% for 10, 50, and 100 μM concentrations of the peptide, respectively (densitometric units = 720, 470, 7, and 5 for input, 10, 50, and 100 μM, respectively). Inhibition was specific. It was not seen with peptide Pro1 (Fig. 5B, lane 5), and WD40 peptide did not inhibit ICP10 binding to SH2-SH3-SH2 (Fig. 5B, lane 6). Based on these and previous (60) findings, we conclude that the N-SH2, PH, and SH3 binding sites in ICP10 PK are located in close proximity to the core catalytic domain, which includes an ion-binding site and two ATP-binding sites (Fig. 5C).
FIG. 5.
Ras-GAP PH binds a WD-40 like site in ICP10 PK. (A) In vitro binding assays with Ras-GAP fusion protein PH (lanes 1 to 5) or uncoated beads (lane 6) and extracts of cells that constitutively express the autophosphorylated ICP10 untreated (lanes 1 and 6) or treated (2 h, 37°C) with 15 U of CIAP (lane 2) or extracts of cells that constitutively express ICP10 mutants p136I/II (lane 3), p140lys (lane 4), or p139TM (lane 5). Uncoated beads are shown in lane 6. (B) Peptide competition assays using beads coated with Ras-GAP fusion protein PH (lanes 1 to 5) or SH2-SH3-SH2 (lane 6), extracts of cells that constitutively express ICP10 (lane 1), and peptide WD40 at 10 μM (lane 2), 50 μM (lane 3), or 100 μM (lanes 4 and 6) or peptide Pro1 (100 μM; lane 5). (C) Schematic model of known sites in ICP10 PK includes signal peptide (SP; residues 1 to 13), extracellular domain (EC), transmembrane domain (TM; residues 85 to 105), binding sites for RAS-GAP N-SH2 (residues pThr117 and pThr141), and PH (residues 160 to 173) domains, Grb2 SH3 binding site (residues 149 to 159 and 396 to 405), and catalytic core (ATP-binding Lys176 and Lys259 and ion-binding Glu209).
Ras-GAP complexed to ICP10 PK is phosphorylated.
Having shown that Ras-GAP binds to ICP10 PK, we wanted to know whether it is phosphorylated. Vero cells were infected with HSV-2, ICP10ΔPK or HSV-2(R), and extracts obtained at 2, 4, 8, and 12 h p.i. were used in immunocomplex PK assays with ICP10 antibody. The phosphorylated proteins in the immunoprecipitates were electrophoretically separated by SDS-PAGE and immunoblotted with ICP10 and Ras-GAP antibodies. Phosphorylated proteins were not seen in mock-infected cells (Fig. 6A, lane 1). In contrast, two phosphorylated proteins consistent with ICP10 and Ras-GAP (140 and 120 kDa, respectively) were seen in immunocomplex PK assays of extracts from HSV-2-infected cells at 2 h (Fig. 6A, lane 2), 4 h (Fig. 6A, lane 3), 8 h (Fig. 6A, lane 4), and 12 h (Fig. 6A, lane 5) p.i. Densitometric scanning indicated that the levels of both phosphoproteins increased between 2 and 8 h p.i. and decreased thereafter (densitometric units = 1,875, 5,610, 4,766, and 682 for ICP10 and 301, 942, 815, and 110 for Ras-GAP at 2, 4, 8, and 12 h p.i., respectively). Similar results were obtained for HSV-2(R)-infected cells as shown for 4-h-infected cells in Fig. 6A, lane 10. By contrast, phosphorylated proteins were not seen in immunocomplex PK assays of extracts from ICP10ΔPK-infected cells (Fig. 6A, lanes 6 to 9). Immunoblotting confirmed the presence of ICP10 (Fig. 6B, lanes 2 to 5 and 10) and Ras-GAP (Fig. 6C, lanes 2 to 5 and 10) in the precipitates from HSV-2- and HSV-2(R)-infected cells. It also identified the nonphosphorylated p95 in precipitates from cells infected with ICP10ΔPK for 4 h (Fig. 6B, lane 7), 8 h (Fig. 6B, lane 8), and 12 h (Fig. 6B, lane 9), but the precipitates did not contain Ras-GAP (Fig. 6C, lanes 7 to 9). Consistent with previous findings for p95 expression (73), neither p95 nor Ras-GAP were precipitated from cells infected with ICP10ΔPK for 2 h (Fig. 6B, and C, lanes 6). The data indicate that Ras-GAP in the ICP10 complex is phosphorylated. Phosphoamino acid analysis, done as previously described (12, 71), indicated that phosphorylation is on Ser-Thr residues (data not shown).
FIG. 6.
Ras-GAP complexed to ICP10 in HSV-2-infected cells is phosphorylated. (A) Immunocomplex PK assays of extracts from cells mock infected (lane 1) or infected with HSV-2 at 2 h (lane 2), 4 h (lane 3), 8 h (lane 4), or 12 h (lane 5) p.i.; ICP10ΔPK at 2 h (lane 6), 4 h (lane 7), 8 h (lane 8), or 12 h (lane 9) p.i.; or HSV-2(R) at 4 h (lane 10) p.i. (B) Immunoblotting of gel in panel A with antibody to ICP10. (C) Immunoblotting of gel in panel A with antibody to Ras-GAP.
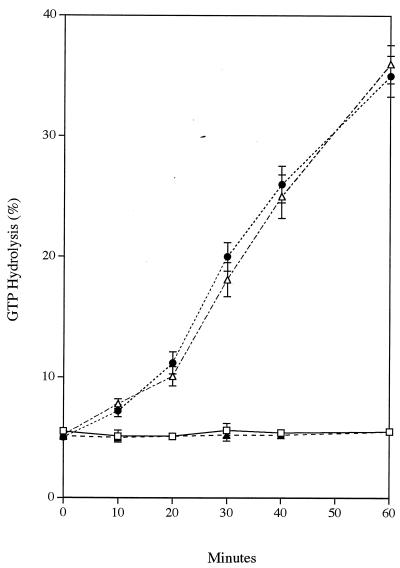
GTPase activity is lower in HSV-2- than in ICP10ΔPK-infected cells.
Because the GTPase activity of Ras-GAP phosphorylated on Ser-Thr residues is reduced (11, 24), we next asked whether GTPase activity is lower in cells infected with HSV-2 than in cells infected with ICP10ΔPK. Extracts from cells infected with HSV-2, ICP10ΔPK, or HSV-2(R) or that were mock infected (with PBS) in medium containing 1 or 10% FCS for 4 h were immunoprecipitated with MAb F132-62, and the immunoprecipitates were assayed for GTP hydrolysis as described in Materials and Methods. This time point was chosen because Ras-GAP binding and phosphorylation is maximal at this time (Fig. 6). As shown in Fig. 7 for three independent experiments, the proportion of RAS.GDP in ICP10ΔPK- and mock-infected Vero cells increased as a function of time, reaching maximal levels (36 to 39%) at 60 min. On the other hand, the rates of GTP hydrolysis (which reflects GTPase activity) were significantly lower (4 to 6%) in HSV-2- and HSV-2(R)-infected cells throughout the incubation period.
FIG. 7.
GTPase activity is lower in HSV-2-infected cells than in ICP10ΔPK-infected cells. Extracts of cells infected with HSV-2 (□), HSV-2(R) (▴), or ICP10ΔPK (●) or mock infected with PBS in 1% FCS (▵) for 4 h were immunoprecipitated with anti RAS antibody (F123-62). GTPase reactions were performed on the immunoprecipitates with [α-32P]GTP and chromatographed on PEI plates. Data represent the percent GTP hydrolyzed at each time point ± the standard error of the mean. Similar results were obtained in ICP10ΔPK- and mock-infected cells in 10% FCS.
Ras is activated in HSV-2-infected cells but not ICP10ΔPK-infected cells.
Having shown that GTPase activity is lower in HSV-2-infected cells than in ICP10ΔPK-infected cells, the question arose as to whether this translates into Ras activation. Vero cells were labeled with [32P]orthophosphate (18 h, 37°C) and either infected with HSV-2, ICP10ΔPK, or HSV-2(R) or mock infected with PBS (in medium containing 1 or 10% FCS). The cells were analyzed for Ras activation as described in Materials and Methods. The results of three independent experiments are summarized in Table 2. Low levels of Ras.GTP were seen in mock-infected cells (at both serum concentrations) and in HSV-2-, HSV-2(R)-, or ICP10ΔPK-infected cells at 0 h p.i. These levels increased significantly at 2 to 8 h p.i. with HSV-2 or HSV-2(R) and decreased thereafter, reaching background levels at 12 h p.i. Ras.GTP levels did not increase in cells infected with ICP10ΔPK. The kinetics of Ras activation mimic those seen for Ras-GAP binding and phosphorylation (Fig. 6) and are consistent with the increased levels of GTPase activity in HSV-2- and HSV-2(R)-infected cells but not in ICP10ΔPK-infected cells at this time.
TABLE 2.
Ras-GTP levels in virus infected cellsa
| Infection | Mean Ras-GTP level ± SEM at time (h) p.i.:
|
||||
|---|---|---|---|---|---|
| 0 | 2 | 4 | 8 | 12 | |
| HSV-2 | 3.0 ± 0.1 | 12.1 ± 0.2 | 19.1 ± 0.4 | 15.0 ± 0.6 | 3.3 ± 0.5 |
| ICP10ΔPK | 4.1 ± 0.5 | 3.5 ± 0.4 | 3.5 ± 0.3 | 3.1 ± 0.5 | 3.9 ± 0.2 |
| HSV-2(R) | 2.8 ± 0.2 | ND | 18.4 ± 0.6 | 13.2 ± 0.4 | 3.0 ± 0.1 |
| Mock (10% FCS) | 3.9 ± 0.7 | 3.4 ± 0.4 | 3.9 ± 0.3 | 3.5 ± 0.6 | 3.2 ± 0.4 |
| Mock (1% FCS) | 3.6 ± 0.6 | 3.1 ± 0.3 | 4.0 ± 0.1 | 3.1 ± 0.2 | 3.5 ± 0.3 |
Vero cells were labeled with [32P]orthophosphate for 18 h in medium containing 1 or 10% FCS and infected with HSV-2 or HSV-2(R) (in 1% FCS), or ICP10ΔPK (in 10% FCS) (73) or mock infected with PBS (in 1 or 10% FCS). Extracts obtained at various times p.i. were precipitated with Ras antibody and the levels of Ras.GTP and Ras.GDP in the precipitates were determined after separation on PEI cellulose thin-layer plates.
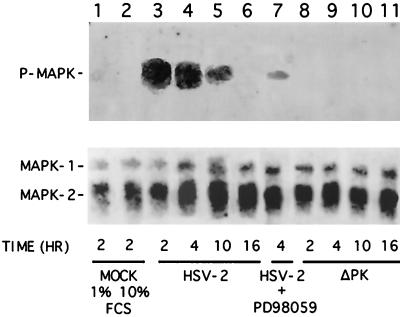
MAPK is activated in HSV-2-infected cells but not in ICP10ΔPK-infected cells.
Having shown that Ras is activated in cells infected with HSV-2 but not cells infected with ICP10ΔPK, we wanted to determine whether this leads to activation of MEK and MAPK. Cells were infected with HSV-2, ICP10ΔPK, or HSV-2(R), and extracts obtained at 2, 4, 10, and 16 h p.i. were analyzed for MAPK activation by immunoblotting with phosphorylation state-specific antibody. Mock-infected cells (in 1 or 10% FCS) and cells infected with HSV-2 for 4 h in the presence of a 50 μM concentration of the MEK-specific inhibitor PD98059 (1, 56) were studied in parallel. Phosphorylated MAPK (P-MAPK) was not seen in mock-infected cells (Fig. 8, lanes 1 and 2). It was also not seen in cells infected with ICP10ΔPK (Fig. 8, lanes 8 to 11), but it was seen in cells infected with HSV-2 at 2 h (Fig. 8, lane 3), 4 h (Fig. 8, lane 4), and 10 h (Fig. 8, lane 5). It was no longer seen at 16 h p.i. (Fig. 8, lane 6). PD98059 caused a significant (90%) decrease in P-MAPK levels in HSV-2-infected cells (Fig. 8, lane 7) (densitometric units = 1,750 and 201 for untreated and treated cells, respectively). Similar results were obtained with HSV-2(R) (data not shown). The levels of P-MAPK reflect different activation states, because similar quantities of protein were seen with a MAPK antibody (Fig. 8, bottom bands). The data indicate that MEK and MAPK are activated in HSV-2-infected cells but not in ICP10ΔPK-infected cells at the same time as Ras-GAP phosphorylation and Ras activation.
FIG. 8.
MAPK is activated in HSV-2-infected cells but not ICP10ΔPK-infected cells. Vero cells were mock infected with PBS in 1% (lane 1) or 10% (lane 2) FCS or were infected with HSV-2 for 2 h (lane 3), 4 h (lane 4), 10 h (lane 5), or 16 h (lane 6); HSV-2 in the presence of 50 μM PD98059 for 4 h (lane 7); or ICP10ΔPK for 2 h (lane 8), 4 h (lane 9), 10 h (lane 10), or 16 h (lane 11). Extracts obtained at these times were immunoblotted with antibody to the phosphorylated MAPK (P-MAPK; top panel), stripped, and reblotted with antibody to MAPK1/2 (bottom panel).
Ras/MEK/MAPK activation in HSV-2-infected cells but not in ICP10ΔPK-infected cells causes c-Fos induction and stabilization.
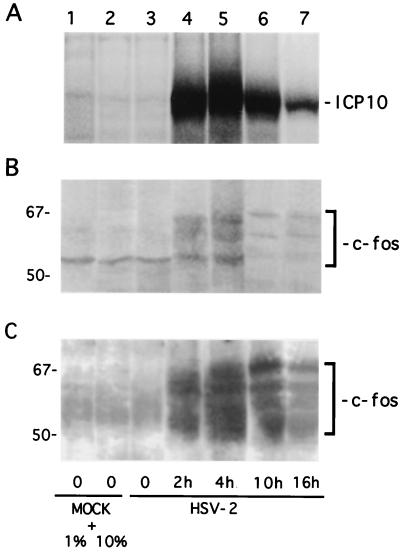
Having shown that the Ras/MEK/MAPK pathway is activated in HSV-2-infected cells, we wanted to know whether this is related to c-Fos induction (26). In a first series of experiments, we examined the temporal relationship between expression of c-Fos and ICP10. Vero cells were infected with HSV-2 or were mock infected with PBS (in 1 or 10% FCS). They were pulse-labeled with 150 μCi of [35S]methionine per ml for 30 min at 0, 2, 4, 10, and 16 h p.i., and cell extracts obtained at these times were immunoprecipitated with ICP10 antibody or immunoprecipitated and/or immunoblotted with c-Fos antibody. Consistent with previous findings (12), ICP10 was first seen at 2 h p.i. (Fig. 9A, lane 4). Its levels increased somewhat at 4 h p.i. (Fig. 9A, lane 5) and decreased thereafter, but synthesis still occurred at 10 h (Fig. 9A, lane 6) and 16 h (Fig. 9A, lane 7) p.i. (densitometric units = 9,160, 10,460, 6,250, and 1,970 for 2, 4, 10, and 16 h, respectively). Mock-infected cells were negative (Fig. 9A, lanes 1 and 2).
FIG. 9.
c-Fos is induced and stabilized in HSV-2-infected cells. (A) Vero cells were mock infected with PBS in 1% FCS (lane 1) or 10% FCS (lane 2) or were infected with HSV-2 for 30 min (lane 3), 2 h (lane 4), 4 h (lane 5), 10 h (lane 6), or 16 h (lane 7). At these times they were pulse-labeled with [35S]methionine for 30 min, and the extracts were immunoprecipitated with ICP10 antibody. (B) Duplicate samples of the cell extracts in panel A were immunoprecipitated with c-Fos antibody. (C) Immunoprecipitates in panel B were immunoblotted with c-Fos antibody.
The intrinsically unstable c-Fos protein was undetectable in mock-infected cells, as determined by immunoprecipitation of [35S]methionine-labeled cells which identified multiple (56 to 72 kDa) species (45) (Fig. 9B, lanes 1 and 2). Their identity was confirmed by immunoblotting of the precipitates with c-Fos antibody (Fig. 9C, lanes 1 and 2). c-Fos levels were still low at 30 min p.i. (Fig. 9B and C, lane 3), but expression was significantly increased at 2 h (Fig. 9B, and C, lane 4) and at 4 h (Fig. 9B and C, lane 5) p.i. The levels of c-Fos precipitated from [35S]methionine-labeled cells decreased at 10 h (Fig. 9B, lane 6) and at 16 h (Fig. 9B, lane 7) p.i., suggesting that c-Fos synthesis is maximal at 2 to 10 h p.i. (densitometric units = 201, 640, 910, 162, and 131 for 0, 2, 4, 10, and 16 h p.i., respectively). In contrast, the levels of c-Fos were not significantly decreased in immunoblots of the precipitates from cells infected for 10 h (Fig. 9C, lane 6) or for 16 h (Fig. 9C, lane 7) (densitometric units = 2,085, 3,498, 4,500, 2,751, and 2,406 for 30 min and 2, 4, 10, and 16 h, respectively). We interpret the data to indicate that c-Fos expression, as well as its metabolic stability, is increased in HSV-2-infected cells, a finding consistent with previous reports that c-Fos is metabolically stabilized by phosphorylation on C-terminal Ser-Thr residues (34, 62).
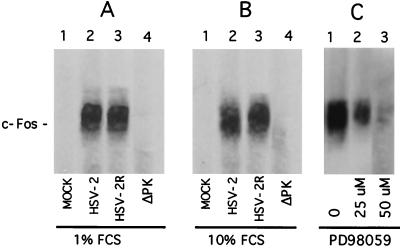
In a second series of experiments, we asked whether c-Fos induction and stabilization is related to Ras/MEK/MAPK activation. Vero cells were infected with HSV-2 (with or without PD98059), ICP10ΔPK, or HSV-2(R). Cells mock infected with PBS (in 1 or 10% FCS) served as a control. Extracts obtained at 18 h p.i. were immunoprecipitated or immunoblotted with the c-Fos antibody. c-Fos was seen in cells infected with HSV-2 (Fig. 10A and B, lanes 2) or HSV-2(R) (Fig. 10A and B, lanes 3) but not in cells infected with ICP10ΔPK (Fig. 10A and B, lanes 4) or in mock-infected cells (Fig. 10A and B, lanes 1). The levels of c-Fos were significantly reduced by a 25 μM concentration of PD98059 (Fig. 10C, lane 2), and c-Fos was barely detectable in cells treated with 50 μM PD98059 (Fig. 10C, lane 3) (densitometric units = 6,000, 1,900, and 350 for untreated cells and 25 and 50 μM treated cells, respectively). We interpret the data to indicate that c-Fos induction and stabilization likely result from Ras/MEK/MAPK activation by ICP10 PK.
FIG. 10.
ICP10 PK and activation of the Ras/MEK/MAPK pathway are required for c-Fos induction and stabilization. (A) Immunoprecipitation-immunoblotting assay with c-Fos antibody of extracts from cells mock infected (lane 1) or infected (18 h) with HSV-2 (lane 2), HSV-2(R) (lane 3), or ICP10ΔPK (lane 4) in medium containing 1% serum. (B) Immunoprecipitation-immunoblotting assay was done as in panel A but with extracts from cells infected in medium containing 10% serum. (C) Immunoprecipitation-immunoblotting assay was done as in panel A but with extracts from cells infected with HSV-2 (18 h) in medium containing 1% FCS without (lane 1) or with 25 μM (lane 2) or 50 μM (lane 3) PD98059. Blots shown represent long (15-min) exposure times.
ICP10 expression is regulated by a positive feedback loop that involves c-Fos.
Having shown that c-Fos is induced and/or stabilized in HSV-2-infected cells, we wanted to know whether this affects ICP10 expression. The question arises because basal expression from the ICP10 promoter is AP-1 dependent (90). In a first series of experiments, Vero cells were transfected with chimeric constructions consisting of the ICP10 or the double AP-1 mutant promoters driving the CAT gene (86, 87, 90). They were infected with HSV-2, ICP10ΔPK, or HSV-2(R) or were mock infected (with PBS in 1% FCS) 24 h later and then assayed for CAT activity after 40 to 48 h. Basal expression from the ICP10 promoter was low in mock-infected cells (3.3% conversion). It was significantly increased by infection with HSV-2 (72% conversion) or HSV-2(R) (65% conversion), but not by ICP10ΔPK infection (3.9% conversion) or HSV-2 infection in the presence of 50 μM PD98059 (3.5% conversion). Conversion in mock-infected cells and in cells infected with ICP10ΔPK or in the presence of PD98059 presumably reflects the low levels of c-Fos in these cells (Fig. 9), since the AP-1 mutant promoter was negative (0.4 to 1% conversion). The data suggest that ICP10 expression is increased by c-Fos induced and/or stabilized as a result of Ras/MEK/MAPK activation.
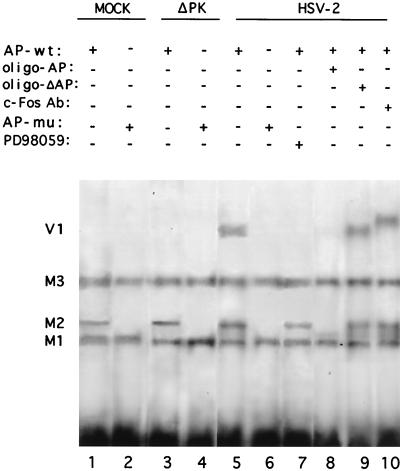
To test the validity of this interpretation, we used electrophoretic mobility shift assays with wild-type (AP-wt) and AP-1 mutant (AP-mu) DNA fragments and nuclear extracts from infected cells. Three complexes designated M1 through M3 in order of increasing electrophoretic mobility were formed by AP-wt DNA with all of the nuclear extracts (Fig. 11, lanes 1, 3, 5, and 7). The M2 complex was not formed by AP-mu DNA (Fig. 11, lanes 2, 4, and 6), and it was competed for by the AP (Fig. 11, lane 8) but not by the ΔAP (Fig. 11, lane 9) oligonucleotide, suggesting that it is AP-1 specific. However, c-Fos is not involved in its formation, since M2 was not supershifted by c-Fos antibody (Fig. 11, lane 10). A lower mobility complex (V1) was formed by AP-wt DNA (Fig. 11, lane 5) but not by AP-mu DNA (Fig. 11, lane 6) with extracts from HSV-2-infected cells. V1 was not formed with extracts of ICP10ΔPK-infected cells (Fig. 11, lane 3) or when MEK/MAPK activation was inhibited by PD98059 (Fig. 11, lane 7). Its formation was competed for by the AP (Fig. 11, lane 8) but not by the ΔAP (Fig. 11, lane 9) oligonucleotide, and its migration was retarded by c-Fos antibody (Fig. 11, lane 10), indicating that c-Fos is involved in its formation. Taken in toto, the data suggest that ICP10 expression in HSV-2-infected cells is regulated by a positive feedback loop which involves c-Fos induction or stabilization.
FIG. 11.
c-Fos in HSV-2-infected cells but not in ICP10ΔPK-infected cells complexes with the ICP10 promoter. The 61-bp AflIII/PvuI DNA fragments from the wild-type ICP10 promoter (AP-wt, lanes 1, 3, 5, and 7 to 10) or the double AP-1 mutant promoter (AP-mu, lanes 2, 4 and 6) were incubated with nuclear extracts from Vero cells mock infected (lanes 1 and 2) or infected for 20 h with ICP10ΔPK (lanes 3 and 4), HSV-2 (lanes 5, 6, and 8 to 10), or HSV-2 with 50 μM PD98059 (lane 7). In some experiments, 50-fold excess cold synthetic oligonucleotide competitors AP-1 (oligo-AP, lane 8) or mutant AP-1 (oligo-ΔAP, lane 9) or c-Fos antibody (lane 10) were added.
Ras/MEK/MAPK activation is required for timely onset of HSV-2 growth.
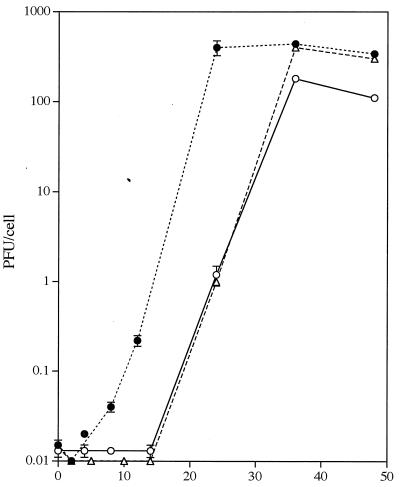
Having documented that ICP10 PK is involved in the activation of the RAS/MEK/MAPK mitogenic pathway, we wanted to know whether this activation is important for virus replication. To address this question, single-step growth curves were done in cells infected with HSV-2 in the absence or presence of PD98059 (50 μM) and compared to those of ICP10ΔPK. HSV-2 replication in untreated cells began at 2 h p.i. Maximal titers (400 PFU/cell) were reached at 24 h p.i. and remained relatively stable until 48 h p.i. In contrast, in the presence of PD98059, HSV-2 growth began at 14 h p.i., and titers did not reach levels similar to those in untreated cells until 36 h p.i. The ICP10ΔPK growth pattern was similar to that of HSV-2 in the presence of PD98059 (Fig. 12). We interpret the data to indicate that the Ras/MEK/MAPK mitogenic pathway must be activated for timely onset of HSV-2 growth and that activation is mediated by ICP10 PK.
FIG. 12.
Onset of HSV-2 growth is delayed by PD98059. Vero cells were pretreated (1 h, 37°C) with 50 μM PD98059 (○) or PD98059 reconstitution medium (●) and infected with HSV-2. They were reincubated in medium containing 1% FCS with or without PD98059, respectively. Adsorption was for 1 h at 37°C (0 h of growth curve). Virus titers were determined at 2 to 48 h after adsorption, and the results are expressed as the burst size (PFU/cell ± the standard error of the mean). Cells infected with ICP10ΔPK in medium containing 10% FCS (▵) were studied in parallel.
DISCUSSION
The salient feature of the data presented here is the finding that the timely onset of HSV-2 replication depends on the activation of the Ras/MEK/MAPK mitogenic pathway by a strategy which involves Ras-GAP inactivation by ICP10 PK. The following comments seem pertinent with respect to these findings.
Starting with the observation that in cells labeled with [35S]methionine at 0 to 8 h p.i., Ras-GAP coprecipitates with ICP10 but not is PK-deleted mutant (p95), we used in vitro binding and peptide competition assays to demonstrate that ICP10 binds the Ras-GAP N-SH2 and PH modules. SH2 modules bind phosphotyrosine (pTyr) residues contained within a specific sequence context (43, 53, 75, 76), the most relevant determinant of which is the +3 residue relative to pTyr (19, 82, 83). Their interaction was recently extended to include pSer and pThr (13, 51, 52, 59, 64), and this is consistent with our findings for ICP10 PK. Thus, the Ras-GAP N-SH2 fusion protein bound the autophosphorylated ICP10 in constitutively expressing cells and in cells treated with heat-inactivated phosphatase, but binding was not seen for extracts of phosphatase-treated cells or cells that express the PK negative mutants p140III or p140lys. We conclude that binding was specific because (i) cell lines that express ICP10 or its PK negative mutants were established in the same 293 cells (50, 60), (ii) p140III and p140lys have different site mutations (60) and are therefore unlikely to have lost a specific binding site for N-SH2, (iii) the antibody is specific for ICP10 (4, 12, 33, 50, 60, 69–73), and (iv) both dephosphorylated ICP10 and the PK negative mutants bound the Ras-GAP PH fusion protein. ICP10 binding of the N-SH2 fusion protein was competed by phosphopeptides pT117 and pT141, but not by the dephosphorylated peptides T117 and T141 or unrelated peptides, suggesting that binding occurs at these sites. Competition was not due to the generation of nonspecific localized interactive clusters by pThr residues (13, 51, 52, 64) since it was not seen with phosphopeptide pT130 that lacks the +3 Pro residue (relative to pThr) which imparts binding specificity (20, 40, 82, 83).
Consistent with previous reports about protein interactions involving PH domains (63, 81), ICP10 binding to the PH fusion protein was phosphorylation independent (seen with dephosphorylated ICP10 and the PK negative mutants) and it was competed for by a peptide that represents a WD40-like sequence at ICP10 positions 106 to 173. Densitometric analyses indicated that the same proportions (33 and 34%) of the input ICP10 is bound by the Ras-GAP PH and SH2-SH3-SH2 fusion proteins. However, it is still unclear whether either or both Ras-GAP domains are bound in HSV-2-infected cells and by the same or different ICP10 PK molecules. We conclude that the PH domain is necessary and sufficient for binding, because Ras-GAP binds the PK negative mutant p139TM in constitutively expressing cells (70). However, binding of both Ras-GAP sites may stabilize the interaction and/or improve Ras-GAP presentation to the adjacent ICP10 PK catalytic core (Fig. 5C). Indeed, immunocomplex PK-immunoblotting assays with Ras-GAP and ICP10 antibodies indicated that Ras-GAP is complexed to ICP10 as early as 2 h p.i. and that it is phosphorylated. Maximal levels of phosphorylated Ras-GAP were seen at the time of maximal ICP10 synthesis (4 and 8 h p.i.). Ras-GAP was neither bound nor phosphorylated by p95 expressed in ICP10ΔPK-infected cells.
Viruses use various strategies to activate the mitogenic Ras/MEK/MAPK pathway. Vaccinia virus encodes a protein that mimics epidermal growth factor (EGF) in terms of its ability to stimulate cognate receptors (42). MAPK is incorporated into HIV virions and is required for the translocation of the reverse transcriptase complex to the nucleus (35). SV40 small T antigen binds protein phosphatase 2A, thereby preventing it from dephosphorylating MEK and MAPK2 and prolonging their activated state (77). A herpesvirus saimiri protein (STP-C488) associates with Ras and activates it (38) and a coxsackievirus protein (Sam68) binds Ras-GAP and inactivates it, thereby activating Ras (32). Our findings suggest that HSV-2 uses a strategy similar to that of coxsackievirus in order to activate Ras/MEK/MAPK. Thus, it is generally believed that Ras-GAP is a negative regulator of Ras (27, 31, 48, 57) and that its inactivation, for example by phosphorylation on Ser-Thr residues (11, 24, 66, 89), promotes Ras activation. Consistent with this interpretation, we found significantly reduced GTPase activity in HSV-2-infected cells, in which Ras-GAP was bound and phosphorylated by ICP10 PK, compared to ICP10ΔPK-infected cells in which Ras-GAP was not similarly altered. Conversely, the levels of activated Ras were significantly increased in HSV-2-infected cells at the time of maximal (2 to 8 h p.i.) ICP10 synthesis and Ras-GAP phosphorylation. Although serum stimulates the Ras/MEK/MAPK pathway in previously starved cells, the HSV-2-induced stimulation was unrelated to serum concentration, since cells were grown in 10% FCS and infected in medium containing 1 or 10% FCS [HSV-2/HSV-2(R) and ICP10ΔPK, respectively]. The levels of activated Ras were equally low in cells mock infected (with PBS) in the presence of 1 or 10% FCS. They were not increased in cells infected with ICP10ΔPK, although its growth is optimal in 10% FCS (73). HSV-2(R) behaved like HSV-2, indicating that ICP10 PK is required for Ras activation.
Immunoblotting with antibody specific to the phosphorylated state indicated that MAPK is also activated in cells infected with HSV-2 ([and HSV-2(R)] but no in ICP10ΔPK-infected cells. The kinetics of MAPK activation were similar to those of Ras-GAP phosphorylation and Ras activation, with maximal levels of P-MAPK seen at 2 to 10 h p.i. We noticed that only one band was resolved with this antibody, although it recognizes P-MAPK1/2 and both species were seen with antibody to MAPK1/2. This may reflect poor resolution or the activation of only one MAPK species, the identity of which is still unclear. However, P-MAPK was virtually undetectable in cells infected with HSV-2 in the presence of the specific MEK inhibitor PD98059 (1, 56), indicating that activation is related to Ras/MEK. We conclude that Ras-GAP binding and/or phosphorylation by ICP10 PK is involved, since MAPK was not activated in cells infected with ICP10ΔPK. In this context it is important to point out that growth factor receptors activated by ligand binding, recruit a complex consisting of a nucleotide releasing factor and an adaptor protein (viz. Sos1-Grb2) in order to activate Ras (9, 10, 47). ICP10 has a functional transmembrane domain, and it is located on the cell surface (5, 12, 33, 50, 70). In constitutively expressing cells, ICP10 PK recruits Sos1-Grb2 (binds SH3 modules in Grb2) and phosphorylates Ras-GAP (5, 33, 60, 70). EGF uses a similar strategy to activate the Ras/MEK/MAPK pathway in cells transfected with a chimera consisting of the EGF ligand-binding domain and ICP10 PK (71). Because both Ras-GAP and the Sos1-Grb2 complex bind the ICP10 PK domain which is deleted in ICP10ΔPK, their respective contribution toward Ras activation in HSV-2-infected cells remains unclear. Studies of a HSV-2 mutant that expresses p136I/II which does not bind Ras-GAP but retains the ability to recruit Sos1 are needed in order to address this question.
We considered the possibility that activation of the Ras/MEK/MAPK pathway is a nonspecific response to the mechanics of infection or due to growth factors or cytokines present in the inoculum. This is particularly relevant because an HSV receptor is a member of the tumor necrosis factor receptor family which, upon ligand binding, can generate a signal that regulates NF-κB and AP-1 activation (54). However, we conclude that this is unlikely, because the pathway was not activated by ICP10ΔPK, the adsorption and/or penetration properties of which are identical to those of the wild-type virus (73). The following observations indicate that HSV-2 must enter the cell and express ICP10 PK in order to activate the Ras/MEK/MAPK pathway: (i) activation was not seen at 30 min after infection; (ii) activation was concomitant with de novo ICP10 synthesis; and (iii) Ras was not activated in cells treated with antibody neutralized HSV-2, which can bind but does not penetrate the cells (data not shown). It is unclear whether the ICP10 protein in the virion tegument (72) contributes to pathway activation.
Activation of the Ras/MEK/MAPK pathway is involved in increased transcription of the c-fos gene (14, 39) and metabolic stabilization of the c-Fos protein by phosphorylation on C-terminal Ser-Thr residues (34, 62). Immunoprecipitation studies using cells pulse-labeled with [35S]methionine indicated that c-Fos expression is increased at 2 to 10 h p.i. with HSV-2 [or HSV-2(R)] and is maximal at 4 h p.i., concomitant with Ras-GAP phosphorylation and Ras/MAPK activation. However, immunoblotting experiments indicated that the levels of c-Fos are still elevated at 16 to 18 h p.i., suggesting that protein stability was also increased. Increased expression and stability is due to Ras/MEK/MAPK activation by ICP10 PK, because c-Fos levels were not increased in cells infected with ICP10ΔPK or in cells infected with HSV-2 in the presence of PD98059. Significantly, our data suggest that c-Fos functions in a positive feedback loop to regulate ICP10 expression. Thus, expression from the ICP10 promoter (but not its AP-1 mutant) was increased by infection with HSV-2 [or HSV-2(R)] but not ICP10ΔPK. The increase was ablated by PD98059. In gel retardation assays ICP10, but not AP-1 mutated DNA formed a complex (V1) with nuclear extracts of HSV-2-infected cells that was supershifted by c-Fos antibody and was not formed with extracts from cells infected in the presence of PD98059 or with ICP10ΔPK. These findings may explain the biphasic kinetics of ICP10 expression (18, 30, 84, 86, 87).
In the presence of PD98059, HSV-2 growth began at 14 h p.i. compared to 2 h p.i. in untreated cells, a delayed onset similar to that seen for ICP10ΔPK. These data suggest that Ras/MEK/MAPK activation is required for the timely onset of virus growth. Ongoing studies with activated MEK (MEKE) and dominant-negative Ras mutants are designed to further examine the role of Ras/MEK/MAPK pathway in HSV-2 growth. However, unlike ICP10ΔPK, which is growth impaired in 1% FCS (73), HSV-2 grew well under these conditions once replication was initiated, indicating that pathway activation is unrelated to the failure of ICP10ΔPK to grow in 1% FCS. Because the onset of IE protein synthesis, most notably ICP4 and ICP27, is also delayed in ICP10ΔPK-infected cells (73), activation of Ras/MEK/MAPK may also be required for the timely expression of HSV-2 IE genes. However, a function which is not inhibited by PD98059 (MEK independent) is ultimately induced, and it provides the cellular environment conducive to virus growth. The identity of this compensatory function and the mechanism responsible for its induction are presently unknown. Potential candidates are cellular genes functionally similar to ICP10 PK (74) and other Ras effector pathways (41, 46, 67). In this context it may be important to point out that HSV-1 does not induce c-Fos (26, 58), but it activates JNK by a Ras-independent pathway (58, 88). The role of Ras activation and the HSV-1 protein(s) responsible for pathway activation are unknown. However, the HSV-1 RR1 PK is only 38% identical to ICP10 PK (61), it is not located on the cell surface (16), and it may lack trans-phosphorylating activity (15).
What is the relationship of Ras/MEK/MAPK activation to disease pathogenesis? Because AP-1 transcription factors are induced by stimuli which cause reactivation of latent virus (21, 36, 37, 80) and ICP10 is the only known viral gene that responds to AP-1 (86, 87, 90), the expression of ICP10 is likely to be induced by reactivation-inducing stimuli. The resulting activation of the Ras/MEK/MAPK mitogenic pathway indicated by our findings provides a positive feedback amplification loop for ICP10 expression. This results in the timely expression of viral IE genes (73) and RR activity which is required for DNA synthesis in nonreplicating (neuronal) cells (25). The outcome is initiation of the lytic cascade and the production of infectious virus. Because activated Ras has antiapoptotic activity in neurons (56), its activation by ICP10 PK may also be required for latency establishment. Consistent with these interpretations, LAT is an inefficient and weak determinant of HSV-2 reactivation (85), RR1 is detected before IE transcripts during reactivation (79), and ICP10ΔPK is impaired in latency reactivation and/or establishment (6). Ongoing studies are designed to examine the validity of these interpretations.
REFERENCES
- 1.Alessi D R, Cuenda A, Cohen O, Dudley D T, Saltiel A R. PD98059 is a specific inhibitor of the activation of mitogen-activated protein kinases kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 2.Ali M A, Prakash S S, Jariwalla R J. Localization of the antigenic sites and intrinsic protein kinase domain within a 300 amino acid segment of the ribonucleotide reductase large subunit from herpes simplex virus type 2. Virology. 1992;187:360–367. doi: 10.1016/0042-6822(92)90328-m. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K P, Frink R J, Devi G B, Gaylord B H, Wagner E K. Detailed characterization of the mRNA mapping in the HindIII fragment K region of the herpes simplex virus type 1 genome. J Virol. 1981;37:1011–1027. doi: 10.1128/jvi.37.3.1011-1027.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurelian L, Terzano P, Smith C C, Chung T D, Shamsuddin A, Costa S, Orlandi C. Amino-terminal epitope of herpes simplex virus type 2 ICP10 protein as a molecular diagnostic marker for cervical intraepithelial neoplasia. Cancer Cells. 1989;7:187–191. [Google Scholar]
- 5.Aurelian L. Herpes simplex virus type 2: unique biological properties include neoplastic potential mediated by the PK domain of the large subunit of ribonucleotide reductase. Front Biosci. 1998;3:d237–d249. doi: 10.2741/a238. [DOI] [PubMed] [Google Scholar]
- 6.Aurelian L, Kokuba H, Smith C C. Vaccine potential of a herpes simplex virus type 2 mutant deleted in the PK domain of the large subunit of ribonucleotide reductase (ICP10) Vaccine. 1999;17:1951–1963. doi: 10.1016/s0264-410x(98)00470-8. [DOI] [PubMed] [Google Scholar]
- 7.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 8.Bryant S S, Briggs S, Smithgall T E, Martin G A, McCormick F, Chang J H, Parsons S J, Jove R. Two SH2 domains of p120 RasGTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J Biol Chem. 1995;270:17947–17952. doi: 10.1074/jbc.270.30.17947. [DOI] [PubMed] [Google Scholar]
- 9.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 10.Chardin P, Carnonis J H, Gale N W, VanAelst L, Schlesinger J, Willer M H, Bar-Sagi D. Human Sos1—a guanine nucleotide exchange factor for ras that binds to Grb2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 11.Choudhury S, Krishna M, Bhattacharya R K. Activation of the ras oncogenes during hepatocarcinogenesis induced by N-nitrosodiethylamine: possible involvement of PKC and GAP. Cancer Lett. 1996;109:149–154. doi: 10.1016/s0304-3835(96)04439-4. [DOI] [PubMed] [Google Scholar]
- 12.Chung T D, Wymer J P, Smith C C, Kulka M, Aurelian L. Protein kinase activity associated with the large subunit of the herpes simplex virus type ribonucleotide reductase (ICP10) J Virol. 1989;63:3389–3398. doi: 10.1128/jvi.63.8.3389-3398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleghon V, Morrison D K. Raf-1 interacts with Fyn and Src in a non-phosphotyrosine-dependent manner. J Biol Chem. 1994;269:17749–17755. [PubMed] [Google Scholar]
- 14.Cook S J, Aziz N, McMahon M. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol Cell Biol. 1999;19:330–341. doi: 10.1128/mcb.19.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conner J, Cooper J, Furlong J, Clements J B. An autophosphorylating but not transphosphorylating activity is associated with the unique N terminus of the herpes simplex virus type 1 ribonucleotide reductase large subunit. J Virol. 1992;66:7511–7516. doi: 10.1128/jvi.66.12.7511-7516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conner J, Murray J, Cross A, Clements J B, Marsden H S. Intracellular localisation of herpes simplex virus type 1 ribonucleotide reductase subunits during infection of cultured cells. Virology. 1995;213:615–623. doi: 10.1006/viro.1995.0033. [DOI] [PubMed] [Google Scholar]
- 17.Cooper J, Conner J, Clements J B. Characterization of the novel protein kinase activity present in the R1 subunit of herpes simplex virus ribonucleotide reductase. J Virol. 1995;69:4979–4985. doi: 10.1128/jvi.69.8.4979-4985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai P, Ramakrishnan R, Lin Z W, Osak B, Glorioso J C, Levine M. The RR1 gene of herpes simplex virus type 1 is uniquely trans-activated by ICP0 during infection. J Virol. 1993;67:6125–6135. doi: 10.1128/jvi.67.10.6125-6135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eck M J, Shoelson S E, Harrison S C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p561ck. Nature. 1993;362:87–91. doi: 10.1038/362087a0. [DOI] [PubMed] [Google Scholar]
- 20.Fantl W J, Escobedo J A, Martin G A, Turck C W, Rosario M, McCormick F, Williams L T. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 21.Fawl R L, Roizman B. Induction of reactivation of herpes simplex virus in murine sensory ganglia in vivo by cadmium. J Virol. 1993;67:7025–7031. doi: 10.1128/jvi.67.12.7025-7031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng C-P, Kulka M, Aurelian L. NF-κB-binding proteins induced by HSV-1 infection of U937 cells are not involved in activation of human immunodeficiency virus. Virology. 1993;192:491–500. doi: 10.1006/viro.1993.1065. [DOI] [PubMed] [Google Scholar]
- 23.Gimble J M, Duh E, Ostrove J M, Gendelman H E, Max E E, Rabson A B. Activation of the human immunodeficiency virus long terminal repeat by herpes simplex virus type 1 is associated with induction of a nuclear factor that binds to the NF-kappaB/core enhancer sequence. J Virol. 1988;62:4104–4112. doi: 10.1128/jvi.62.11.4104-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gschwendt M, Kittstein W, Harks F. Protein kinase C forms a complex with and phosphorylates the GTPase-activating protein GAP. Biochem Biophys Res Commun. 1983;194:571–576. doi: 10.1006/bbrc.1993.1858. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensible for virus growth and DNA synthesis: isolation and characterization of an ICP6lacZ insertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goswami B B. Transcriptional induction of proto-oncogene FOS by HSV-2. Biochem Biophys Res Commun. 1987;143:1055–1062. doi: 10.1016/0006-291x(87)90359-7. [DOI] [PubMed] [Google Scholar]
- 27.Hall A. ras and GAP—who's controlling whom? Cell. 1990;61:921–923. doi: 10.1016/0092-8674(90)90054-i. [DOI] [PubMed] [Google Scholar]
- 28.Harlan J E, Yoon H S, Hajduk P J, Fesik S W. Structural characterization of the interaction between a pleckstrin homology domain and phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1995;34:9859–9864. doi: 10.1021/bi00031a006. [DOI] [PubMed] [Google Scholar]
- 29.Hjermstad S J, Briggs S D, Smithgall T E. Phosphorylation of the ras GTPase-activating protein (GAP) by the p93c-fes protein-tyrosine kinase in vitro and formation of GAP-fes complexes via an SH2 domain-dependent mechanism. Biochemistry. 1993;32:10519–10525. doi: 10.1021/bi00090a031. [DOI] [PubMed] [Google Scholar]
- 30.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D C, Marshall C J, Hancock J F. Plasma membrane targeted Ras GTPase activating proteins is a potent suppressor of p21ras function. Mol Cell Biol. 1993;4:2420–2431. doi: 10.1128/mcb.13.4.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M, Watson K A, Selinka H C, Carthy C M, Klingel K, McManus B M, Kandolf R. Cleavage of RasGAP and phosphorylation of mitogen-activated protein kinase in the course of coxsackievirus B3 replication. J Virol. 1999;73:3587–3594. doi: 10.1128/jvi.73.5.3587-3594.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter J C R, Smith C C, Bose D, Kulka M, Broderick R, Aurelian L. Intracellular internalization and signaling pathways triggered by the large subunit of HSV-2 ribonucleotide reductase (ICP10) Virology. 1995;210:345–360. doi: 10.1006/viro.1995.1351. [DOI] [PubMed] [Google Scholar]
- 34.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 35.Jacque J M, Mann A, Enslen H, Sharuva N, Brichacek B, Davis R J, Stevenson M. Modulation of HIV-1 infectivity by MAPK, a virion associated kinase. EMBO J. 1998;17:2607–2618. doi: 10.1093/emboj/17.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang K L, Pulverer B, Woodyeth J R, Latchman D S. Activation of the cellular transcription factor AP-1 in herpes simplex virus infected cells is dependent on the viral immediate-early protein ICP0. Nucleic Acid Res. 1991;19:4879–4883. doi: 10.1093/nar/19.18.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin P, Ringertz N R. Cadmium induces transcription of proto-oncogenes c-jun and c-myc in rat L6 myoblasts. J Biol Chem. 1990;265:14061–14064. [PubMed] [Google Scholar]
- 38.Jung J U, Desrosiers R C. Association of viral oncoprotein, STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan D R, Morrison D K, Wong G, McCormick F, Williams L T. PDGF β-receptor stimulates tyrosine phosphorylation of GAP and association of GAP with a signaling complex. Cell. 1990;61:125–133. doi: 10.1016/0092-8674(90)90220-9. [DOI] [PubMed] [Google Scholar]
- 41.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 42.King C S, Cooper J A, Moss B, Twardzik D R. Vaccinia virus growth factor stimulates tyrosine protein kinase activity of A431 cell epidermal growth factor receptors. Mol Cell Biol. 1986;6:332–336. doi: 10.1128/mcb.6.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch C A, Anderson D, Moran M F, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 44.Kovary K, Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruijer W, Cooper A J, Hunter T, Verma I M. Platelet-derived growth factor induces rapid but transient expression of c-fos gene and protein. Nature. 1984;312:711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- 46.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 47.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 48.Lowy D, Willumsen B. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 49.Luo J H, Smith C C, Kulka M, Aurelian L. A truncated protein kinase domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) expressed in Escherichia coli. J Biol Chem. 1991;266:20976–20983. [PubMed] [Google Scholar]
- 50.Luo J, Aurelian L. The transmembrane helical segment but not the invariant lysine is required for the kinase activity of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) J Biol Chem. 1992;267:9645–9653. [PubMed] [Google Scholar]
- 51.Malek S N, Desiderio S. A cyclin-dependent kinase homologue, p130PITSLRE is a phosphotyrosine-independent SH2 ligand. J Biol Chem. 1995;269:33009–33020. [PubMed] [Google Scholar]
- 52.Malek S N, Desiderio S. p150TSP, a conserved nuclear phosphoprotein that contains multiple tetratricopeptide repeats and binds specifically to SH2 domains. J Biol Chem. 1996;271:6952–6962. doi: 10.1074/jbc.271.12.6952. [DOI] [PubMed] [Google Scholar]
- 53.Margolis B, Li N, Koch A, Mohammadi M, Hurwitz D R, Zilberstein A, Ullrich A, Pawson T, Schlessinger J. The tyrosine phosphorylated carboxyterminus of the EGF receptor is a binding site for GAP and PLC-gamma. EMBO J. 1990;9:4375–4380. doi: 10.1002/j.1460-2075.1990.tb07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marsters S A, Ayers T M, Skubatch M, Gray C L, Rothe M, Ashkenazi A. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 55.Mayer B J, Baltimore D. Signaling through SH2 and SH3 domains. Trends Cell Biol. 1993;3:8–13. doi: 10.1016/0962-8924(93)90194-6. [DOI] [PubMed] [Google Scholar]
- 56.Mazzoni I E, Said F A, Aloyz R, Miller F D, Kaplan D. Ras regulates sympathetic neuron survival by suppressing the p53-mediated cell death pathway. J Neurosci. 1999;19:9716–9727. doi: 10.1523/JNEUROSCI.19-22-09716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 58.McLean T I, Bachenheimer S L. Activation of cJUN N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J Virol. 1999;73:8415–8426. doi: 10.1128/jvi.73.10.8415-8426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller A J, Pendergast A, Havlik M H, Puil L, Pawson T, Witte O N. A limited set of SH2 domains binds BCR through a high-affinity phosphotyrosine-independent interaction. Mol Cell Biol. 1992;12:5087–5093. doi: 10.1128/mcb.12.11.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson J W, Zhu J, Smith C C, Kulka M, Aurelian L. ATP and SH3 binding sites in the protein kinase of the large subunit of herpes simplex virus type 2 of ribonucleotide reductase (ICP10) J Biol Chem. 1996;271:17021–17027. doi: 10.1074/jbc.271.29.17021. [DOI] [PubMed] [Google Scholar]
- 61.Nikas I, McLauchlan J, Davison A J, Taylor W R, Clements J B. Structural features of ribonucleotide reductase. Proteins Struct Funct Genet. 1986;1:376–384. doi: 10.1002/prot.340010411. [DOI] [PubMed] [Google Scholar]
- 62.Okazaki K, Sagata N. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH3T3 cells. EMBO J. 1995;14:5048–5059. doi: 10.1002/j.1460-2075.1995.tb00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pawson T, Scott J D. Signaling through scaffold, anchoring and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 64.Pendergast A M, Muller A J, Havlik M H, Maru Y, Witte O N. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991;66:161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- 65.Peng T, Hunter J R C, Nelson J W. The novel protein kinase of the RR1 subunit of herpes simplex virus has autophosphorylation and transphosphorylation activity that differs in its ATP requirements for HSV-1 and HSV-2. Virology. 1996;216:184–196. doi: 10.1006/viro.1996.0045. [DOI] [PubMed] [Google Scholar]
- 66.Quilliam L A, Castro A F, Rogers-Graham K S, Martin C B, Der C J, Bi C. M-Ras/R-Ras3, a transforming Ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J Biol Chem. 1999;274:23850–23857. doi: 10.1074/jbc.274.34.23850. [DOI] [PubMed] [Google Scholar]
- 67.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways, Curr. Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 68.Satoh T, Endo M, Nakafuku M, Nakamura S, Kaziro Y. Platelet-derived growth factor stimulates formation of active p21ras GTP complex in Swiss mouse 3T3 cells. Proc Natl Acad Sci USA. 1990;87:5993–5997. doi: 10.1073/pnas.87.15.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith C C, Kulka M, Wymer J P, Chung T D, Aurelian L. Expression of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is required for virus growth and neoplastic transformation. J Gen Virol. 1992;73:1417–1428. doi: 10.1099/0022-1317-73-6-1417. [DOI] [PubMed] [Google Scholar]
- 70.Smith C C, Luo J H, Hunter J C R, Ordonez J V, Aurelian L. The transmembrane domain of the large subunit of HSV-2 ribonucleotide reductase (ICP10) is required for the transformation-related signaling pathways that involve ras activation. Virology. 1994;200:598–612. doi: 10.1006/viro.1994.1223. [DOI] [PubMed] [Google Scholar]
- 71.Smith C C, Luo J H, Aurelian L. The protein kinase activity of the large subunit of herpes simplex virus type 2 (ICP10) fused to the extracellular domain of the epidermal growth factor receptor is ligand inducible. Virology. 1996;217:425–434. doi: 10.1006/viro.1996.0136. [DOI] [PubMed] [Google Scholar]
- 72.Smith C C, Aurelian L. The large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is associated with the virion tegument and has PK activity. Virology. 1997;234:235–242. doi: 10.1006/viro.1997.8645. [DOI] [PubMed] [Google Scholar]
- 73.Smith C C, Peng T T, Kulka M, Aurelian L. The PK domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is involved in IE gene transcription and virus growth. J Virol. 1998;72:9131–9141. doi: 10.1128/jvi.72.11.9131-9141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith C C, Yu Y X, Kulka M, Aurelian L. A novel human gene similar to the PK coding domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) codes for a serine-threonine PK and is expressed in melanoma cells. J Biol Chem. 2000;275:25690–25699. doi: 10.1074/jbc.M002140200. [DOI] [PubMed] [Google Scholar]
- 75.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 76.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates MAP kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 78.Sze P, Herman R C. The herpes simplex virus type 1 ICP6 gene is regulated by a “leaky” early promoter. Virus Res. 1992;2:141–152. doi: 10.1016/0168-1702(92)90153-z. [DOI] [PubMed] [Google Scholar]
- 79.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Laeary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valyi-Nagy T, Deshmane S, Dillner A, Fraser N W. Induction of cellular transcription factors in trigeminal ganglia of mice by corneal scarification, herpes simplex virus type 1 infection, and explanation of trigeminal ganglia. J Virol. 1991;65:4142–4152. doi: 10.1128/jvi.65.8.4142-4152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang D S, Shaw R, Hattori M, Arai H, Inoue K, Shaw G. Binding of pleckstrin homology domains to WD40/beta-transducin repeat containing segments of the protein product of the Lis-1 gene. Biochem Biophys Res Commun. 1995;209:622–629. doi: 10.1006/bbrc.1995.1545. [DOI] [PubMed] [Google Scholar]
- 82.Waksman G, Kominos D, Robertson S C, Pant N, Baltimore D, Birge R B, Cowburn D, Hanafusa H, Mayer B J, Overduin M, et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 83.Waksman G, Shoelson S E, Pant N, Cowburn D, Kurian J. Binding of high-affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 84.Wagner E. Transcription patterns in HSV infections. In: Klein G, editor. Advances in viral oncology. New York, N.Y: Raven Press; 1983. pp. 239–276. [Google Scholar]
- 85.Wang K, Pesnicak L, Straus S E. Mutations in the 5′ end of the herpes simplex virus type 2 latency-associated transcript (LAT) promoter affect LAT expression in vivo but not the rate of spontaneous reactivation of genital herpes. J Virol. 1997;71:7903–7910. doi: 10.1128/jvi.71.10.7903-7910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wymer J P, Chung T D, Chang Y N, Hayward G S, Aurelian L. Identification of immediate-early type cis-response elements in the promoter for the ribonucleotide reductase large subunit from herpes simplex virus type 2. J Virol. 1989;63:2773–2784. doi: 10.1128/jvi.63.6.2773-2784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wymer J P, Aprhys C M J, Chung T D, Feng T C-P, Kulka M, Aurelian L. Immediate-early and functional AP-1 cis-response elements are involved in the transcriptional regulation of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) Virus Res. 1992;23:253–270. doi: 10.1016/0168-1702(92)90112-m. [DOI] [PubMed] [Google Scholar]
- 88.Zachos G, Clements B, Conner J. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J Biol Chem. 1999;274:5097–5103. doi: 10.1074/jbc.274.8.5097. [DOI] [PubMed] [Google Scholar]
- 89.Zheng L, Eckerdal J, Dimitrijevic I, Andersson T. Chemotactic peptide-induced activation of Ras in human neutrophils is associated with inhibition of p120-GAP activity. J Biol Chem. 1997;272:23448–23454. doi: 10.1074/jbc.272.37.23448. [DOI] [PubMed] [Google Scholar]
- 90.Zhu J, Aurelian L. AP-1 cis-response elements are involved in basal expression and Vmw110 transactivation of the large subunit of herpes implex virus type 2 ribonucleotide reductase (ICP0) Virology. 1997;231:301–312. doi: 10.1006/viro.1997.8522. [DOI] [PubMed] [Google Scholar]