Abstract
The discovery of a pathogenic variant in the alpha-synuclein (SNCA) gene in the Contursi kindred in 1997 indisputably confirmed a genetic cause in a subset of Parkinson’s disease (PD) patients. Currently, pathogenic variants in one of the seven established PD genes or the strongest known risk factor gene, GBA1, are identified in ∼15% of PD patients unselected for age at onset and family history. In this Debate article, we highlight multiple avenues of research that suggest an important - and in some cases even predominant - role for genetics in PD aetiology, including familial clustering, high rates of monogenic PD in selected populations, and complete penetrance with certain forms. At first sight, the steep increase in PD prevalence exceeding that of other neurodegenerative diseases may argue against a predominant genetic etiology. Notably, the principal genetic contribution in PD is conferred by pathogenic variants in LRRK2 and GBA1 and, in both cases, characterized by an overall late age of onset and age-related penetrance. In addition, polygenic risk plays a considerable role in PD. However, it is likely that, in the majority of PD patients, a complex interplay of aging, genetic, environmental, and epigenetic factors leads to disease development.
Keywords: Parkinson’s disease, genetics, monogenic, polygenic, familial, GBA1, LRRK2, PRKN, environment, gene-environment interactions, epigenetic, progression, pandemic
INTRODUCTION
Parkinson’s disease (PD) is a clinically heterogeneous disorder [1, 2], with multiple genetic causes or contributors as well as environmental risk and protective factors identified [3, 4]. In most cases, it is likely that a complex interplay of a combination of genetic and environmental factors results in the disease and influences disease progression [5], and these factors may be different in different parts of the world [6] or exert differential effects depending on context [7, 8]. Here, we argue that the evidence supporting a genetic basis for PD is relatively more compelling than that for environmental factors.
EVIDENCE FOR AN IMPORTANT GENETIC ROLE IN PD
Many patients with PD have a positive family history
A substantial proportion of PD patients, perhaps up to ∼20%, have a positive family history [9]. Although it could be debated that this reflects not only genetic factors but also shared environmental exposure(s), some observations favor the former. Indeed, the discovery of SNCA, the first gene implicated in PD, was prompted when clinicians who were initially convinced that genetics played “no significant role in the etiology of PD” and “must be considered to be acquired” [10], encountered a very large kindred with an autosomal dominant pattern of PD transmission [11, 12]. Supportive evidence for the strong role of genetics in familial PD has been repeatedly demonstrated over the past three decades (e.g., references [13, 14], and in all regions of the world where this has been studied (http://www.mdsgene.org and [15]; Fig. 1). Recently, for example, in a study of Malaysian early-onset PD (EOPD), the rate of “solved” monogenic cases (i.e., where the cause of PD is attributed to pathogenic variant[s] in a single gene) increased from 21.7% overall to 48.5% when considering only the subgroup of EOPD patients with a positive family history [16]. Conversely, while it has been argued that the lack of a positive family history in the majority of PD patients points to environmental causation, many such individuals have, in fact, been shown to harbor a pathogenic genetic variant (e.g., 53.4% of the pathogenic variant-positive Malaysian EOPD patients had no history of PD in either the immediate or extended family [16]).
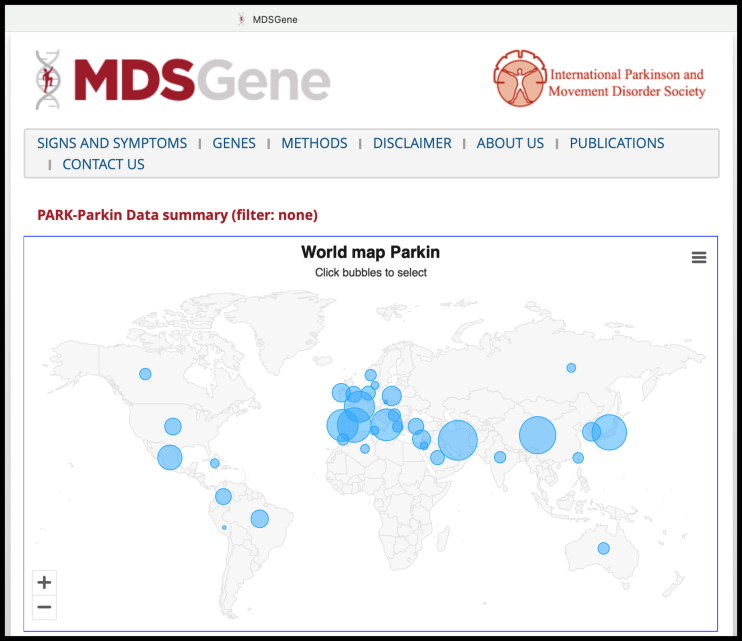
Fig. 1.
Screenshot from the MDSGene website (https://www.mdsgene.org, accessed on 5 November 2023), here depicting, as an example, the updated statistics on PARK-PRKN cases reported worldwide with individual-level data (almost 1,500 cases). The website also provides curated data regarding patients’ clinical characteristics and the pathogenicity classification of genetic variants.
A positive family history of neurodegeneration in related parkinsonian conditions such as progressive supranuclear palsy and multiple system atrophy, which have overlapping clinical features, pathology, and molecular mechanisms with PD [17–21], is reported in up to 20.4–33.0% [18, 22–24] and 40% [25] of cases, respectively, with genetic factors sometimes implicated [17, 18, 20, 26]. In contrast, environmental factors are still very poorly defined in these conditions [20, 27, 28].
Pathogenic variants in some PD genes are highly penetrant for the disease
There are several established PD genes that cause monogenic/Mendelian forms of PD: SNCA, LRRK2, VPS35, and RAB32, which cause autosomal dominant PD, and PRKN, PINK1, and PARK7/DJ-1, which cause autosomal recessive PD [13–15, 29]. In some cases, pathogenic variant(s) in a single gene (e.g., whole-gene triplication of SNCA, or homozygous or compound heterozygous pathogenic missense and/or copy number variants in PRKN) is sufficient by itself to cause PD (i.e., demonstrating full penetrance) [12–15]. Remarkably, in some populations, these monogenic forms may even account for the majority of PD patients (e.g., >50% of PD patients attending a tertiary-care neurology clinic in the Malaysian state of Sabah are EOPD, of whom >50% have homozygous or compound-heterozygous PRKN exon deletions) [30]. Very high rates (sometimes exceeding 40%) of monogenic PD, or involving the strongest known risk factor gene GBA1, are also seen in selected populations such as the North African Arab-Berbers, Ashkenazi Jews, and Spanish Basques, especially involving LRRK2 and GBA1 [31–34], which have more variable (and age-related) penetrance and overall late age of onset [15].
We acknowledge, however, that the prevalence of monogenic PD in specialty clinics may not be generalizable to the population at large [35], and further studies are needed to accurately estimate the true prevalence of monogenic PD globally. On the other hand, the frequently quoted overall rates of monogenic PD (ranging from 2–3% [13], to 5–10% [36–38] or 15% [39]) may continue to increase due to new discoveries and broadening application of genetic technologies [40, 41], with inclusion of under-represented populations [42, 43]. The latter has recently been greatly bolstered by regional [44–46] and global collaborations such as the Global Parkinson’s Genetics Program (GP2; Fig. 2) [47–50]. It is worth noting here that in China and India (by far the two most populous countries in the world, being home to ∼2.9 billion people, and also with extensive diasporas globally [16]), the vast majority of PD patients with family pedigrees compatible with autosomal dominant inheritance remain “unsolved” (e.g., 95% of the 242 probands studied by Zhao et al. [37], and 100% of 44 probands in the study by Punia et al. which tested specifically for pathogenic LRRK2 variants [51]), suggesting that additional genetic determinants of PD remain to be discovered in these large populations [52, 53].
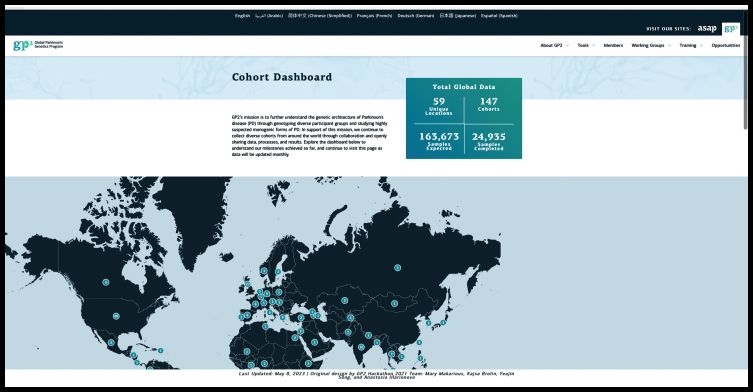
Fig. 2.
Screenshot from the Global Parkinson’s Genetics Program (GP2) website (https://gp2.org, accessed on 5 November 2023), depicting the updated statistics on an ever-growing number of contributing sites and samples.
Is there clustering of PD from environmental causes?
Conversely to the situation with genetics, to our knowledge, there have been extremely few reports of tight clustering of PD occurrence that have been convincingly demonstrated to be caused by environmental factors [54, 55]. Even for what is, to date, the most persuasive evidence for geographical linkage with PD (in Camp Lejeune, a military base in the USA), attributed to water contaminated with the solvent trichloroethylene (TCE) and other volatile organic compounds, the prevalence of PD was still “only” 0.33% (279 out of 84,824 exposed individuals; vs. 0.21% in veterans from a comparable but uncontaminated site) [28]. TCE has widespread and myriad uses, including clothes dry-cleaning [56], but to our knowledge, there have been no reports of clustering of PD among laundry or textile workers [57], other than an early study nearly 20 years ago involving three individuals working together in the office of a garment-manufacturing factory, one of whom also had a father with PD [58]. Thus, there is no environmental factor that is fully (or anywhere close to being fully) “penetrant” for PD. The highly toxic compound MPTP, which is the prototypical toxic cause for parkinsonism, was associated with seven human cases of parkinsonism [59], out of an estimated >400 (i.e., <2%) individuals thought to have been exposed to the synthetic heroin contaminant [60]. Moreover, it does not cause PD as we know it (in which disease processes take place gradually over many years, if not decades [3]), but rather results in a once-off “hit” with a severe but relatively selective nigral injury that is mostly non-progressive [61].
The study of cohabiting couples can identify environmental risk factors for disease because such couples are usually not genetically related but share residential exposures over a prolonged period. However, instances of conjugal PD appear to be very uncommon [62] and are usually considered to be coincidental [63].
Taken together, the above observations suggest that environmental factors are neither sufficient, nor necessary, to induce PD, and therefore cannot be considered a “predominant” cause for the condition.
What about sporadic (so-called “idiopathic”) PD?
In addition to pathogenic variants with high penetrance in PD genes, genomic DNA variation is now well established to contribute substantially to the risk of sporadic PD. To date, genome-wide association studies (GWASs) have identified >100 risk signals associated with PD at the population level [50, 64–67]. These common variants individually have relatively small effect sizes but in combination, can be associated with substantially elevated PD risk (e.g., with disease odds ratios [OR] of 3.4–6.1 among those with the highest decile of polygenic risk scores [PRSs] compared with the lowest-risk decile, in recent studies [64, 68, 69]). Outside the PD field, it has been suggested that PRSs for common diseases such as coronary artery disease and type 2 diabetes, utilizing much larger sample sizes and improved algorithms, can identify individuals with risk equivalent to monogenic mutations [70] (although others have argued that the true effect size is likely to be much more modest [71–74]). Recently, research on genetic resilience factors that mitigate the effects of risk/pathogenic loci and reduce the susceptibility to PD is also emerging from GWA analyses [75].
Currently, the identified risk loci explain an estimated 16–36% of the heritable risk of PD [64], and efforts are ongoing to very substantially ramp up the recruitment of patients (to reach 200,000, including patients from diverse populations) [47–50], which will further advance understanding of the genetic determinants of PD on a global scale [13–15, 65].
Environmental factors act on a background of genetic vulnerability
Even for environmental exposures with an “established” role in PD etiopathogenesis, such as pesticide exposure, caffeine intake, and smoking, their downstream effects are likely to be mediated in part or strongly influenced by genetic factors [5, 8]. We list several examples here: Intriguingly, there is a suggestion in the literature that the lack of caffeine intake, which overall is associated with a ∼2-fold increased risk of developing PD, may be especially detrimental in patients with certain genetic forms of PD, such as those harboring the common Asian LRRK2 risk variants p.G2385R or p.R1628P (present in ∼5–10% of Asian PD patients [6, 76–78]), where odds ratios for PD were 8.6 and 4.6, respectively [79]. Findings consistent with caffeine’s greater association with resistance to genetic forms of PD were also reported among an international cohort of LRRK2-PD patients, most of whom harbored the p.G2019S LRRK2 variant seen in whites and North Africans [80]. A study exploring the neuroprotective effect of tobacco (containing nicotine) found that this may be mediated in part by SIRT6, which may have a pathogenic and pro-inflammatory role in PD and whose expression is strongly influenced by several SIRT6 single nucleotide polymorphisms (SNPs) [81]. Another study of the smoking effect on PD risk found that this varied by SV2 C genotype, ranging from being highly protective to neutral to even being harmful [45, 82]. Finally, a study found that the odds for developing PD from paraquat use was 1.5 in the presence of functional GSTT1 (encoding glutathione S-transferase T1 that provides cellular protection against oxidative stress) but increased to 11.1 with homozygous deletions of the gene which were present in a substantial proportion (22%) of patients [83].
Alterations in the gut microbiome (“dysbiosis”) have consistently been reported in PD patients over the past decade [84, 85]. While the microbiome is known to be significantly influenced by environmental factors such as diet and place of living [84, 86], genetic factors likely play key, if not essential, roles in determining if and how, various aspects of the gut-brain axis (e.g., gut dysbiosis or inflammation [87]) influence disease risk. For example, apart from rotenone models [88, 89], currently the most convincing animal models of gut dysbiosis in PD involve an induced genetic aberration, causing synuclein overexpression, mitochondrial dysfunction, or LRRK2 overexpression, suggesting that an underlying genetic vulnerability is usually needed for gut dysbiosis to trigger disease expression [84, 87, 90–92].
Besides causation/development of disease, genetic factors can also have a significant influence on the disease trajectory
An increasingly highlighted example of this is the overall more rapid deterioration of motoric and cognitive-behavioral function and poorer survival in patients harboring GBA1 variants [93–95]. Some GBA1-PD patients also appear to respond less favorably to treatment with deep brain stimulation (DBS) [94, 96–98]. The converse is true for PARK-PRKN, where, on the whole, patients continue to show dopa-responsiveness even in the long term, exhibit less cognitive decline, and respond relatively favorably to DBS [96, 99, 100]. These genetic factors are increasingly taken into consideration in the clinical management of patients, e.g., in the selection or counseling of patients for DBS [101, 102], and bring the field one step closer to realizing a personalized precision medicine approach for people living with PD [1, 3, 103–105]. In sporadic PD, besides their association with disease risk, PRSs have also shown predictive value in PD phenotype or clinical outcomes [14, 106], such as age at disease onset [107]; motor progression [108]; development of dyskinesias [109], impulse control disorders [110] or cognitive decline (e.g., with one recent study reporting a hazard ratio of 4.8 for progression to PD dementia with the RIMS2 progression locus [111]); and response to medical (pharmacogenomics) [112] and surgical therapies (“surgicogenomics”) [113, 114].
In contrast, the impact of environmental factors, such as exercise [115–118] or dietary patterns (e.g., caffeine or alcohol intake [84]) on disease progression is much less well defined. That said, much remains to be understood with respect to genotype-phenotype correlations, e.g., patients with the exact same point mutation in genes linked to monogenic PD [98, 119–122] can sometimes still exhibit very different clinical courses, suggesting the presence of modifiers (genetic, epigenetic, and/or environmental) [5]. Even here, however, emerging evidence suggests a possibly more significant modifying role for genetic over environmental factors [123].
Understanding of genetic causation is paving the way for targeted therapies in PD
Genetic factors (and their related pathways) involved in the causation and progression of PD are prime targets for biomarker development and disease modification studies [14, 93, 124, 125]. For example, pathogenic variants in GBA1 or SNCA are known to be strongly linked to alpha-synuclein pathology [39], and novel biomarker approaches that enable in vivo interrogation of PD pathology (e.g., alpha-synuclein seeding assays [126–129]) are being exploited in new paradigms for biological classification/stratification of PD which will maximize the likelihood of finding successful disease-modifying therapies (e.g., targeting alpha-synuclein [130]). This has been the case in oncology, where implementation of precision medicine and a focus on genetically defined subtypes of disease have seen remarkable success in developing new and effective therapeutics [124, 131]. Most recently, a new monogenic cause, a pathogenic variant in the RAB32 gene has been found and independently confirmed to cause PD in a dominant fashion [29]. Interestingly the RAB32 protein interacts with LRRK2, and it is likely that additional genetic causes of PD will be found using new sequencing technologies. Finally, there is also a growing appreciation of a convergence of mechanisms underlying monogenic and complex forms of PD, with several of the genes discussed earlier causing monogenic PD also implicated in GWASs (e.g., SNCA and LRRK2), and likely causing relatively subtle changes in protein expression [132–137]. Thus, future genetics-targeted therapies could also potentially be deployed in (the much larger group of) sporadic PD patients.
Conversely, it is currently difficult to envision a scenario where, for example, knowledge of a patient’s remote history of neurotoxicant exposure could be directly “actionable”. Nevertheless, experimental work prompted by findings from epidemiological studies has revealed many valuable mechanistic insights (e.g., MPTP and other neurotoxicants such as paraquat or TCE causing mitochondrial dysfunction [138, 139] and more recently even pathological activation of LRRK2 kinase activity [140]). New avenues continue to be explored to understand the molecular mechanisms of PD based on epidemiological observations, for example with the study of caffeine [79, 80, 141] or nicotine [81, 142] and related metabolites.
Research on environmental and lifestyle factors is in need of technological advances and better tools
Scientific advances depend not only on new ideas and paradigm shifts but also, to a large extent, on technological advances that make these leaps possible [143]. It has to be acknowledged that a crucial reason for the preponderance of evidence linking genetic status with disease causation and progression of PD is that genetics/genomics are much more tractable/easily ascertained with currently available technologies [65, 103, 144], whereas environmental exposures are still difficult to assess and measure [140, 145] and such studies may be just “scratching the surface” [146]. Because of a lack of biological markers of exposure [140], studies of environmental factors often rely on patient retrospective self-report, which are “noisy” and prone to biases and confounding (e.g., recall bias and reverse causation), especially in a condition like PD where there is usually a long lag time spanning one or more decades between the exposure of interest and PD diagnosis [28, 147]. (Conversely, reverse causation is not an issue with genetics [which are fixed at conception], and indeed, this has been utilized for epidemiological studies using, for example, Mendelian randomization approaches [148, 149]). (That said, there is mounting evidence that somatic genetic changes, which take place in every person during development and tissue maintenance, are relevant in PD and related neurodegenerative disorders and can sometimes alter clinical presentation, for example, the age at disease onset [150–152]).
Moreover, people are often unaware of toxicants they are exposed to, as seen in the Camp Lejeune experience [28]. Epidemiological studies sometimes also lack sufficient granularity to be able to hone down to the individual level. In the Camp Lejeune study, for instance, exposure to TCE was inferred based on camp location, but the investigators could not be certain that all residents were exposed to biologically meaningful levels of contaminants [28]. Another recent study suggesting an association between air pollution and PD risk was based on participants’ residential addresses at the district level [153]. This is also exemplified in the maps presented by Professors Dorsey and Bloem (Fig. 2 in [138]), depicting geographic areas of overlap across the United States in PD incidence vis-à-vis use of/exposure to paraquat, TCE and other chlorinated solvents, and particulate matter.
Emerging studies utilizing a more comprehensive investigation of the “exposome” (i.e., the sum of all exogenous and endogenous environmental influences on the human body over the lifespan) and its impact on PD risk and progression will no doubt be an important area for future progress [140, 145, 154, 155]. These will investigate environmental exposures in a more holistic way, looking at multiple factors rather than reductionist approaches studying the effect of only a single environmental factor or class of factors [140, 154, 156]).
The elephant in the room: the “Parkinson’s pandemic”
It has been argued that the rapid increase in PD cases worldwide, since its first formal description by James Parkinson slightly over 200 years ago (“from six to six million”) [55], must be driven by environmental factors (given the relative stasis of innate biology over the same time frame), particularly industrialization and the widespread use of toxicants [55, 138]. While there is little doubt that PD is common and poses an increasingly heavy burden on healthcare systems worldwide [3, 6, 157], the argument may be more multi-faceted than it appears at first glance. One major issue relates to the increasing longevity of world populations with a concomitant increase in age-related disorders. On the surface, this phenomenon should similarly affect conditions like Alzheimer’s disease (AD) as much as it affects PD incidence and prevalence [138]. Still, there may be important differences between these age-related neurodegenerative disorders. For example, late-onset monogenic forms are well recognized in PD (e.g., LRRK2-PD), with disease penetrance being highly dependent on age [7, 158]; however, to our knowledge, late-onset monogenic AD has only very rarely been described [159, 160].
Another crucial point is the improved diagnosis of PD over time [161–164]. In the absence of reliable in vivo diagnostic biomarkers [3, 129], PD remains a clinical diagnosis, which can often be challenging [161–163, 165]. Indeed, Professors Dorsey and Bloem highlighted that approximately half of PD cases detected during an extensive multicenter epidemiological survey conducted in China in the late 1990s had been undiagnosed prior to the study [138, 166] (with rates of non-diagnosis as high as 90+% in rural communities [167]). It has been suggested that China’s rapid increase in PD cases (more than doubling in age-adjusted PD prevalence between 1990 and 2016) is due to rapid industrialization [168, 169], but it is likely that accompanying improvements in economic status and healthcare literacy, access to better healthcare (with improved diagnosis [161–164], as well as survival [164], of PD patients), and scientific interest in PD [6, 170, 171], have contributed substantially to the increased prevalence of PD. Advances in technology (e.g., multimodal biomarker testing for genetic variants, synucleinopathy and/or neurodegeneration [39, 130], and artificial intelligence/machine learning-based diagnostic algorithms [171–173]), applied widely and non-invasively, are likely to further increase the rates of diagnosis of PD, including people in prodromal or even pre-symptomatic stages of the disease [126, 129, 174].
Further research on gene-environment interactions is needed
Ultimately, gene-environment interactions are probably crucial in causing most cases of common “complex” human diseases, including PD [175], and the factors may be additive or synergistic [83]. Gene-environment interaction studies are therefore critically needed [5, 8, 140], including further research on how environmental exposures disrupt epigenetic regulators of gene expression (without changing the underlying DNA sequence) [175]. Currently, research on gene-environment interactions [5, 8, 140] and epigenetics in PD [176–178] is still in its infancy. This will be an exciting area to follow in the coming years, and developments here will be crucial to developing predictive and preventive approaches [5].
Returning to the earlier discussion on the Parkinson’s pandemic, it is instructive to learn from other non-communicable diseases. In the case of obesity, for example, an even larger pandemic has been documented [179], with the global prevalence tripling over the past four decades, now affecting >600 million individuals [180]. There is no doubt that changes in dietary patterns, such as the wide availability of calorie-dense processed food and drinks and physical inactivity, have contributed to this phenomenon. However, for what is considered by many to be a prototypical “lifestyle”-related condition, the heritability for obesity is estimated to be ∼40–70% [180–182], with the brain (which controls hunger and systemic energy metabolism) harboring most of the gene products and pathways that have been linked to obesity in hundreds of genetics studies [182]. Analyses of genome-wide gene-by-environment interactions over the past decade have revealed important insights and informed understanding of disease pathophysiology, although they remain challenging and require sample sizes in the hundreds of thousands or more [180]. As a side note, revolutionary anti-obesity drugs have very recently come to market [182]; these had their beginnings in research on genes related to the glucagon pathway [183].
CONCLUSION
Although we have collated and presented here what we believe to be robust evidence, accumulated over the past 30 years, in support of a strong genetic basis for PD, at the end of the day, we also salute, unreservedly endorse, and stand in solidarity with the advocacy efforts of our esteemed colleagues Professors Dorsey and Bloem, and others [55, 138, 140], to curtail the widespread use of toxicants, to help curb the global burden of PD, and also for a healthier future overall [184, 185]. We whole-heartedly concur with a quote they recently highlighted from Jerry Ensminger, one of the personnel who served at Camp Lejeune who developed PD in his retirement (and whose young daughter died of leukemia while he was serving there), that: “The benefit of the doubt should go to the people, not the chemical” [56].
ACKNOWLEDGMENTS
We would like to thank Andrew Singleton and Huw Morris for insightful discussions on the role of genetics in the “Parkinson’s disease pandemic”.
FUNDING
SYL has received research grant funding from the Ministry of Higher Education Malaysia Fundamental Research Grant Scheme (FRGS/1/2020/SKK0/UM/01/2); University of Malaya Parkinson’s Disease and Movement Disorders Research Program (PV035-2017); and the Global Parkinson’s Genetics Program (GP2).
CK has received research grant funding from the Global Parkinson’s Genetics Program (GP2). The GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation (MJFF) for Parkinson’s Research (https://gp2.org). CK is also supported by the DFG and MJFF (outside GP2).
CONFLICT OF INTEREST
SYL has received consultancy or lecturing honoraria from the GP2 (the GP2 is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org)); Lundbeck International Neuroscience Foundation (Neurotorium) Editorial Board; International Parkinson and Movement Disorder Society (MDS); Eisai; Lundbeck; and Medtronic.
CK is a medical advisor to Centogene and Retromer Therapeutics and received Speakers’ honoraria from Bial and Desitin.
REFERENCES
- [1]. Lim SY, Tan AH, Fox SH, Evans AH, Low SC (2017) Integrating patient concerns into Parkinson’s disease management. Curr Neurol Neurosci Rep 17, 3. [DOI] [PubMed] [Google Scholar]
- [2]. Greenland JC, Williams-Gray CH, Barker RA (2019) The clinical heterogeneity of Parkinson’s disease and its therapeutic implications. Eur J Neurosci 49, 328–338. [DOI] [PubMed] [Google Scholar]
- [3]. Bloem BR, Okun MS, Klein C (2021) Parkinson’s disease. Lancet 397, 2284–2303. [DOI] [PubMed] [Google Scholar]
- [4]. Wüllner U, Borghammer P, Choe CU, Csoti I, Falkenburger B, Gasser T, Lingor P, Riederer P (2023) The heterogeneity of Parkinson’s disease. J Neural Transm (Vienna) 130, 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Periñán MT, Brolin K, Bandres-Ciga S, Blauwendraat C, Klein C, Gan-Or Z, Singleton A, Gomez-Garre P, Swanberg M, Mir P, Noyce A (2022) Effect modification between genes and environment and Parkinson’s disease risk. Ann Neurol 92, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Lim SY, Tan AH, Ahmad-Annuar A, Klein C, Tan LCS, Rosales RL, Bhidayasiri R, Wu YR, Shang HF, Evans AH, Pal PK, Hattori N, Tan CT, Jeon B, Tan EK, Lang AE (2019) Parkinson’s disease in the Western Pacific Region. Lancet Neurol 18, 865–879. [DOI] [PubMed] [Google Scholar]
- [7]. Hentati F, Trinh J, Thompson C, Nosova E, Farrer MJ, Aasly JO (2014) LRRK2 parkinsonism in Tunisia and Norway: A comparative analysis of disease penetrance. Neurology 83, 568–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Reynoso A, Torricelli R, Jacobs BM, Shi J, Aslibekyan S, Norcliffe-Kaufmann L, Noyce AJ, Heilbron K (2023) Gene-environment interactions for Parkinson’s disease. Ann Neurol, doi: 10.1002/ana.26852. [DOI] [PubMed] [Google Scholar]
- [9]. Mehanna R, Smilowska K, Fleisher J, Post B, Hatano T, Pimentel Piemonte ME, Kumar KR, McConvey V, Zhang B, Tan EK, Savica R; International Parkinson and Movement Disorder Society Task Force on Early Onset Parkinson’s Disease (2022) Age cutoff for early-onset Parkinson’s disease: Recommendations from the International Parkinson and Movement Disorder Society Task Force on Early Onset Parkinson’s Disease. Mov Disord Clin Pract 9, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Duvoisin RC (1984) Is Parkinson’s disease acquired or inherited? Can J Neurol Sci 11, 151–155. [DOI] [PubMed] [Google Scholar]
- [11]. Golbe LI, Di Iorio G, Bonavita V, Miller DC, Duvoisin RC (1990) A large kindred with autosomal dominant Parkinson’s disease. Ann Neurol 27, 276–282. [DOI] [PubMed] [Google Scholar]
- [12]. Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- [13]. Domingo A, Klein C (2018) Genetics of Parkinson disease. Handb Clin Neurol 147, 211–227. [DOI] [PubMed] [Google Scholar]
- [14]. Day JO, Mullin S (2021) The genetics of Parkinson’s disease and implications for clinical practice. Genes (Basel) 12, 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Vollstedt EJ, Schaake S, Lohmann K, Padmanabhan S, Brice A, Lesage S, Tesson C, Vidailhet M, Wurster I, Hentati F, Mirelman A, Giladi N, Marder K, Waters C, Fahn S, Kasten M, Brüggemann N, Borsche M, Foroud T, Tolosa E, Garrido A, Annesi G, Gagliardi M, Bozi M, Stefanis L, Ferreira JJ, Correia Guedes L, Avenali M, Petrucci S, Clark L, Fedotova EY, Abramycheva NY, Alvarez V, Menéndez-González M, Jesús Maestre S, Gómez-Garre P, Mir P, Belin AC, Ran C, Lin CH, Kuo MC, Crosiers D, Wszolek ZK, Ross OA, Jankovic J, Nishioka K, Funayama M, Clarimon J, Williams-Gray CH, Camacho M, Cornejo-Olivas M, Torres-Ramirez L, Wu YR, Lee-Chen GJ, Morgadinho A, Pulkes T, Termsarasab P, Berg D, Kuhlenbäumer G, Kühn AA, Borngräber F, de Michele G, De Rosa A, Zimprich A, Puschmann A, Mellick GD, Dorszewska J, Carr J, Ferese R, Gambardella S, Chase B, Markopoulou K, Satake W, Toda T, Rossi M, Merello M, Lynch T, Olszewska DA, Lim SY, Ahmad-Annuar A, Tan AH, Al-Mubarak B, Hanagasi H, Koziorowski D, Ertan S, Genç G, de Carvalho Aguiar P, Barkhuizen M, Pimentel MMG, Saunders-Pullman R, van de Warrenburg B, Bressman S, Toft M, Appel-Cresswell S, Lang AE, Skorvanek M, Boon AJW, Krüger R, Sammler EM, Tumas V, Zhang BR, Garraux G, Chung SJ, Kim YJ, Winkelmann J, Sue CM, Tan EK, Damásio J, Klivényi P, Kostic VS, Arkadir D, Martikainen M, Borges V, Hertz JM, Brighina L, Spitz M, Suchowersky O, Riess O, Das P, Mollenhauer B, Gatto EM, Petersen MS, Hattori N, Wu RM, Illarioshkin SN, Valente EM, Aasly JO, Aasly A, Alcalay RN, Thaler A, Farrer MJ, Brockmann K, Corvol JC, Klein C; MJFF Global Genetic Parkinson’s Disease Study Group (2023) Embracing monogenic Parkinson’s disease: The MJFF Global Genetic PD Cohort. Mov Disord 38, 286–303. [DOI] [PubMed] [Google Scholar]
- [16]. Tay YW, Tan AH, Lim JL, Lohmann K, Ibrahim KA, Abdul Aziz Z, Chin YT, Mawardi AS, Lim TT, Looi I, Chia YK, Ooi JCE, Cheah WK, Dy Closas AMF, Lit LC, Hor JW, Toh TS, Muthusamy KA, Bauer P, Skrahin V, Rolfs A, Klein C, Ahmad-Annuar A, Lim SY (2023) Genetic study of early-onset Parkinson’s disease in the Malaysian population. Parkinsonism Relat Disord 111, 105399. [DOI] [PubMed] [Google Scholar]
- [17]. Jabbari E, Koga S, Valentino RR, Reynolds RH, Ferrari R, Tan MMX, Rowe JB, Dalgard CL, Scholz SW, Dickson DW, Warner TT, Revesz T, Höglinger GU, Ross OA, Ryten M, Hardy J, Shoai M, Morris HR; PSP Genetics Group (2021) Genetic determinants of survival in progressive supranuclear palsy: A genome-wide association study. Lancet Neurol 20, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Lim SY, Dy Closas AMF, Tan AH, Lim JL, Tan YJ, Vijayanathan Y, Tay YW, Abdul Khalid RB, Ng WK, Kanesalingam R, Martinez-Martin P, Ahmad Annuar A, Lit LC, Foo JN, Lim WK, Ng ASL, Tan EK (2023) New insights from a multi-ethnic Asian progressive supranuclear palsy cohort. Parkinsonism Relat Disord 108, 105296. [DOI] [PubMed] [Google Scholar]
- [19]. Lim SY, Lim JL, Ahmad-Annuar A, Lohmann K, Tan AH, Lim KB, Tay YW, Shing YL, Muthusamy KA, Bauer P, Rolfs A, Klein C (2020) Clinical phenotype ofR1441C in 2 Chinese sisters. Neurodegener Dis 20, 39–45. [DOI] [PubMed] [Google Scholar]
- [20]. Fanciulli A, Wenning GK (2015) Multiple-system atrophy. N Engl J Med 372, 249–263. [DOI] [PubMed] [Google Scholar]
- [21]. Robinson JL, Lee EB, Xie SX, Rennert L, Suh E, Bredenberg C, Caswell C, Van Deerlin VM, Yan N, Yousef A, Hurtig HI, Siderowf A, Grossman M, McMillan CT, Miller B, Duda JE, Irwin DJ, Wolk D, Elman L, McCluskey L, Chen-Plotkin A, Weintraub D, Arnold SE, Brettschneider J, Lee VM, Trojanowski JQ (2018) Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 141, 2181–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Donker Kaat L, Boon AJ, Azmani A, Kamphorst W, Breteler MM, Anar B, Heutink P, van Swieten JC (2009) Familial aggregation of parkinsonism in progressive supranuclear palsy. Neurology 73, 98–105. [DOI] [PubMed] [Google Scholar]
- [23]. Borroni B, Goldwurm S, Cerini C, Cosseddu M, Meucci N, Mariani C, Pezzoli G, Padovani A (2011) Familial aggregation in progressive supranuclear palsy and corticobasal syndrome. Eur J Neurol 18, 195–197. [DOI] [PubMed] [Google Scholar]
- [24]. Geut H, Hepp DH, Foncke E, Berendse HW, Rozemuller JM, Huitinga I, van de Berg WDJ (2020) Neuropathological correlates of parkinsonian disorders in a large Dutch autopsy series. Acta Neuropathol Commun 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Leys F, Eschlböck S, Campese N, Mahlknecht P, Peball M, Goebel G, Sidoroff V, Granata R, Bonifati V, Zschocke J, Kiechl S, Poewe W, Seppi K, Wenning GK, Fanciulli A (2022) Family history for neurodegeneration in multiple system atrophy: Does it indicate susceptibility? Mov Disord 37, 2310–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Lim SY, Tan AH, Foo JN, Tan YJ, Chew EG, Annuar AA, Closas AMD, Pajo A, Lim JL, Tay YW, Nadhirah A, Hor JW, Toh TS, Lit LC, Zulkefli J, Ngim SJ, Lim WK, Morris HR, Tan EK, Ng AS (2024) Loss-of-function SMPD1 gene variant in Progressive Supranuclear Palsy-Richardson Syndrome patients of Chinese ancestry. J Mov Disord, doi: 10.14802/jmd.24009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Park HK, Ilango SD, Litvan I (2021) Environmental risk factors for progressive supranuclear palsy. J Mov Disord 14, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Goldman SM, Weaver FM, Stroupe KT, Cao L, Gonzalez B, Colletta K, Brown EG, Tanner CM (2023) Risk of Parkinson disease among service members at Marine Corps Base Camp Lejeune. JAMA Neurol 80, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Gustavsson EK, Follett J, Trinh J, Barodia SK, Real R, Liu Z, Grant-Peters M, Fox JD, Appel-Cresswell S, Stoessl AJ, Rajput A, Rajput AH, Auer R, Tilney R, Sturm M, Haack TB, Lesage S, Tesson C, Brice A, Vilariño-Güell C, Ryten M, Goldberg MS, West AB, Hu MT, Morris HR, Sharma M, Gan-Or Z, Samanci B, Lis P, Tocino T, Amouri R, Ben Sassi S, Hentati F, Global Parkinson’s Genetics Program (GP2), Tonelli F, Alessi DR, Farrer MJ (2024) A pathogenic variant in RAB32 causes autosomal dominant Parkinson’s disease and activates LRRK2 kinase. medRxiv, doi: 10.1101/2024.01.17.24300927 [Preprint]. Posted January 18, 2024. [DOI] [Google Scholar]
- [30]. Tay YW, Ooi JCE, Lim SY, Chia YK, Sukri A, Toh TS, Lim JL, Tan AH, Ahmad-Annuar A (2023) Very high frequency of early-onset Parkinson’s disease with homozygous PRKN exon 3 deletion amongst indigenous populations in Sabah, Malaysia [abstract]. Mov Disord 38(Suppl 1), 1115.37475614 [Google Scholar]
- [31]. Lesage S, Houot M, Mangone G, Tesson C, Bertrand H, Forlani S, Anheim M, Brefel-Courbon C, Broussolle E, Thobois S, Damier P, Durif F, Roze E, Tison F, Grabli D, Ory-Magne F, Degos B, Viallet F, Cormier-Dequaire F, Ouvrard-Hernandez AM, Vidailhet M, Lohmann E, Singleton A, Corvol JC, Brice A; French Parkinson disease Genetics Study Group (PDG) (2020) Genetic and phenotypic basis of autosomal dominant Parkinson’s disease in a large multi-center cohort. Front Neurol 11, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Tolosa E, Vila M, Klein C, Rascol O (2020) LRRK2 in Parkinson disease: Challenges of clinical trials. Nat Rev Neurol 16, 97–107. [DOI] [PubMed] [Google Scholar]
- [33]. Vinagre-Aragón A, Campo-Caballero D, Mondragón-Rezola E, Pardina-Vilella L, Hernandez Eguiazu H, Gorostidi A, Croitoru I, Bergareche A, Ruiz-Martinez J (2021) A more homogeneous phenotype in Parkinson’s disease related to R1441G mutation in the LRRK2 gene. Front Neurol 12, 635396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen CM, Clark LN, Condroyer C, De Marco EV, Dürr A, Eblan MJ, Fahn S, Farrer MJ, Fung HC, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen GJ, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan EK, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu YR, Zabetian CP, Zhao Y, Ziegler SG (2009) Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med 361, 1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Tan MMX, Malek N, Lawton MA, Hubbard L, Pittman AM, Joseph T, Hehir J, Swallow DMA, Grosset KA, Marrinan SL, Bajaj N, Barker RA, Burn DJ, Bresner C, Foltynie T, Hardy J, Wood N, Ben-Shlomo Y, Grosset DG, Williams NM, Morris HR (2019) Genetic analysis of Mendelian mutations in a large UK population-based Parkinson’s disease study. Brain 142, 2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Bandres-Ciga S, Diez-Fairen M, Kim JJ, Singleton AB (2020) Genetics of Parkinson’s disease: An introspection of its journey towards precision medicine. Neurobiol Dis 137, 104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Zhao Y, Qin L, Pan H, Liu Z, Jiang L, He Y, Zeng Q, Zhou X, Zhou X, Zhou Y, Fang Z, Wang Z, Xiang Y, Yang H, Wang Y, Zhang K, Zhang R, He R, Zhou X, Zhou Z, Yang N, Liang D, Chen J, Zhang X, Zhou Y, Liu H, Deng P, Xu K, Xu K, Zhou C, Zhong J, Xu Q, Sun Q, Li B, Zhao G, Wang T, Chen L, Shang H, Liu W, Chan P, Xue Z, Wang Q, Guo L, Wang X, Xu C, Zhang Z, Chen T, Lei L, Zhang H, Wang C, Tan J, Yan X, Shen L, Jiang H, Zhang Z, Hu Z, Xia K, Yue Z, Li J, Guo J, Tang B (2020) The role of genetics in Parkinson’s disease: A large cohort study in Chinese mainland population. Brain 143, 2220–2234. [DOI] [PubMed] [Google Scholar]
- [38]. Jia F, Fellner A, Kumar KR (2022) Monogenic Parkinson’s disease: Genotype, phenotype, pathophysiology, and genetic testing. Genes (Basel) 13, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Höglinger GU, Adler CH, Berg D, Klein C, Outeiro TF, Poewe W, Postuma R, Stoessl AJ, Lang AE (2024) A biological classification of Parkinson’s disease: The SynNeurGe research diagnostic criteria. Lancet Neurol 23, 191–204. [DOI] [PubMed] [Google Scholar]
- [40]. Cook L, Schulze J, Kopil C, Hastings T, Naito A, Wojcieszek J, Payne K, Alcalay RN, Klein C, Saunders-Pullman R, Simuni T, Foroud T (2021) Genetic testing for Parkinson disease: Are we ready? Neurol Clin Pract 11, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Tan AH, Cornejo-Olivas M, Okubadejo N, Pal PK, Saranza G, Saffie-Awad P, Ahmad-Annuar A, Schumacher-Schuh AF, Okeng’o K, Mata IF, Gatto EM, SY Lim (2024) Genetic testing for Parkinson’s disease and movement disorders in less privileged areas: Barriers and opportunities. Mov Disord Clin Pract 11, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Schumacher-Schuh AF, Bieger A, Okunoye O, Mok KY, Lim SY, Bardien S, Ahmad-Annuar A, Santos-Lobato BL, Strelow MZ, Salama M, Rao SC, Zewde YZ, Dindayal S, Azar J, Prashanth LK, Rajan R, Noyce AJ, Okubadejo N, Rizig M, Lesage S, Mata IF; Global Parkinson’s Genetics Program (GP2) (2022) Underrepresented populations in Parkinson’s genetics research: Current landscape and future directions. Mov Disord 37, 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]. Appelbaum PS, Burke W, Parens E, Zeevi DA, Arbour L, Garrison NA, Bonham VL, Chung WK (2022) Is there a way to reduce the inequity in variant interpretation on the basis of ancestry? Am J Hum Genet 109, 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Foo JN, Tan LC, Liany H, Koh TH, Irwan ID, Ng YY, Ahmad-Annuar A, Au WL, Aung T, Chan AY, Chong SA, Chung SJ, Jung Y, Khor CC, Kim J, Lee J, Lim SY, Mok V, Prakash KM, Song K, Tai ES, Vithana EN, Wong TY, Tan EK, Liu J (2014) Analysis of non-synonymous-coding variants of Parkinson’s disease-related pathogenic and susceptibility genes in East Asian populations. Hum Mol Genet 23, 3891–3897. [DOI] [PubMed] [Google Scholar]
- [45]. Foo JN, Chew EGY, Chung SJ, Peng R, Blauwendraat C, Nalls MA, Mok KY, Satake W, Toda T, Chao Y, Tan LCS, Tandiono M, Lian MM, Ng EY, Prakash KM, Au WL, Meah WY, Mok SQ, Annuar AA, Chan AYY, Chen L, Chen Y, Jeon BS, Jiang L, Lim JL, Lin JJ, Liu C, Mao C, Mok V, Pei Z, Shang HF, Shi CH, Song K, Tan AH, Wu YR, Xu YM, Xu R, Yan Y, Yang J, Zhang B, Koh WP, Lim SY, Khor CC, Liu J, Tan EK (2020) Identification of risk loci for Parkinson disease in Asians and comparison of risk between Asians and Europeans: A genome-wide association study. JAMA Neurol 77, 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Loesch DP, Horimoto ARVR, Heilbron K, Sarihan EI, Inca-Martinez M, Mason E, Cornejo-Olivas M, Torres L, Mazzetti P, Cosentino C, Sarapura-Castro E, Rivera-Valdivia A, Medina AC, Dieguez E, Raggio V, Lescano A, Tumas V, Borges V, Ferraz HB, Rieder CR, Schumacher-Schuh A, Santos-Lobato BL, Velez-Pardo C, Jimenez-Del-Rio M, Lopera F, Moreno S, Chana-Cuevas P, Fernandez W, Arboleda G, Arboleda H, Arboleda-Bustos CE, Yearout D, Zabetian CP; 23andMe Research Team; Cannon P, Thornton TA, O’Connor TD, Mata IF; Latin American Research Consortium on the Genetics of Parkinson’s Disease (LARGE-PD) (2021) Characterizing the genetic architecture of Parkinson’s disease in Latinos. Ann Neurol 90, 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Lange LM, Avenali M, Ellis M, Illarionova A, Keller Sarmiento IJ, Tan AH, Madoev H, Galandra C, Junker J, Roopnarain K, Solle J, Wegel C, Fang ZH, Heutink P, Kumar KR, Lim SY, Valente EM, Nalls M, Blauwendraat C, Singleton A, Mencacci N, Lohmann K, Klein C; Global Parkinson’s Genetic Program (GP2) (2023) Elucidating causative gene variants in hereditary Parkinson’s disease in the Global Parkinson’s Genetics Program (GP2). NPJ Parkinsons Dis 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Towns C, Richer M, Jasaityte S, Stafford EJ, Joubert J, Antar T, Martinez-Carrasco A, Makarious MB, Casey B, Vitale D, Levine K, Leonard H, Pantazis CB, Screven LA, Hernandez DG, Wegel CE, Solle J, Nalls MA, Blauwendraat C, Singleton AB, Tan MMX, Iwaki H, Morris HR; Global Parkinson’s Genetics Program (GP2) (2023) Defining the causes of sporadic Parkinson’s disease in the global Parkinson’s genetics program (GP2). NPJ Parkinsons Dis 9, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Rizig M, Bandres-Ciga S, Makarious MB, Ojo OO, Crea PW, Abiodun OV, Levine KS, Abubakar SA, Achoru CO, Vitale D, Adeniji OA, Agabi OP, Koretsky MJ, Agulanna U, Hall DA, Akinyemi RO, Xie T, Ali MW, Shamim EA, Ani-Osheku I, Padmanaban M, Arigbodi OM, Standaert DG, Bello AH, Dean MN, Erameh CO, Elsayed I, Farombi TH, Okunoye O, Fawale MB, Billingsley KJ, Imarhiagbe FA, Jerez PA, Iwuozo EU, Baker B, Komolafe MA, Malik L, Nwani PO, Daida K, Nwazor EO, Miano-Burkhardt A, Nyandaiti YW, Fang ZH, Obiabo YO, Kluss JH, Odeniyi OA, Hernandez DG, Odiase FE, Tayebi N, Ojini FI, Sidranksy E, Onwuegbuzie GA, D’Souza AM, Osaigbovo GO, Berhe B, Osemwegie N, Reed X, Oshinaike OO, Leonard HL, Otubogun FM, Alvarado CX, Oyakhire SI, Ozomma SI, Samuel SC, Taiwo FT, Wahab KW, Zubair YA, Iwaki H, Kim JJ, Morris HR, Hardy J, Nalls MA, Heilbron K, Norcliffe-Kaufmann L; Nigeria Parkinson Disease Research Network; International Parkinson’s Disease Genomics Consortium Africa; Black and African American Connections to Parkinson’s Disease Study Group; 23andMe Research Team; Blauwendraat C, Houlden H, Singleton A, Okubadejo NU; Global Parkinson’s Genetics Program (2023) Identification of genetic risk loci and causal insights associated with Parkinson’s disease in African and African admixed populations: A genome-wide association study. Lancet Neurol 22, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Kim JJ, Vitale D, Otani DV, Lian MM, Heilbron K; 23andMe Research Team; Iwaki H, Lake J, Solsberg CW, Leonard H, Makarious MB, Tan EK, Singleton AB, Bandres-Ciga S, Noyce AJ; Global Parkinson’s Genetics Program (GP2); Blauwendraat C, Nalls MA, Foo JN, Mata I (2024) Multi-ancestry genome-wide association meta-analysis of Parkinson’s disease. Nat Genet 56, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Punia S, Behari M, Govindappa ST, Swaminath PV, Jayaram S, Goyal V, Muthane UB, Juyal RC, Thelma BK (2006) Absence/rarity of commonly reported LRRK2 mutations in Indian Parkinson’s disease patients. Neurosci Lett 409, 83–88. [DOI] [PubMed] [Google Scholar]
- [52]. Skipper L, Shen H, Chua E, Bonnard C, Kolatkar P, Tan LC, Jamora RD, Puvan K, Puong KY, Zhao Y, Pavanni R, Wong MC, Yuen Y, Farrer M, Liu JJ, Tan EK (2005) Analysis of LRRK2 functional domains in nondominant Parkinson disease. Neurology 65, 1319–1321. [DOI] [PubMed] [Google Scholar]
- [53]. Foo JN, Liu J, Tan EK (2015) CHCHD2 and Parkinson’s disease. Lancet Neurol 14, 681–682. [DOI] [PubMed] [Google Scholar]
- [54]. Gash DM, Rutland K, Hudson NL, Sullivan PG, Bing G, Cass WA, Pandya JD, Liu M, Choi DY, Hunter RL, Gerhardt GA, Smith CD, Slevin JT, Prince TS (2008) Trichloroethylene: Parkinsonism and complex 1 mitochondrial neurotoxicity. Ann Neurol 63, 184–192. [DOI] [PubMed] [Google Scholar]
- [55]. Dorsey R, Sherer T, Okun MS, Bloem BR (2020) Ending Parkinson’s disease: A prescription for action. PublicAffairs [Google Scholar]
- [56]. Dorsey ER, Greenamyre JT, Willis AW (2023) The water, the air, the Marines-Camp Lejeune, trichloroethylene, and Parkinson disease. JAMA Neurol 80, 663–665. [DOI] [PubMed] [Google Scholar]
- [57]. Checkoway H, Ilango S, Li W, Ray RM, Tanner CM, Hu SC, Wang X, Nielsen S, Gao DL, Thomas DB (2018) Occupational exposures and parkinsonism among Shanghai women textile workers. Am J Ind Med 61, 886–892. [DOI] [PubMed] [Google Scholar]
- [58]. Kumar A, Calne SM, Schulzer M, Mak E, Wszolek Z, Van Netten C, Tsui JK, Stoessl AJ, Calne DB (2004) Clustering of Parkinson disease: Shared cause or coincidence? Arch Neurol 61, 1057–1060. [DOI] [PubMed] [Google Scholar]
- [59]. Ballard PA, Tetrud JW, Langston JW (1985) Permanent human parkinsonism due to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP): Seven cases. Neurology 35, 949–956. [DOI] [PubMed] [Google Scholar]
- [60]. Pankratz N, Foroud T (2007) Genetics of Parkinson disease. Genet Med 9, 801–811. [DOI] [PubMed] [Google Scholar]
- [61]. Langston JW (2017) The MPTP story. J Parkinsons Dis 7, S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Willis AW, Sterling C, Racette BA (2010) Conjugal Parkinsonism and Parkinson disease: A case series with environmental risk factor analysis. Parkinsonism Relat Disord 16, 163–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Rajput AH, Ferguson LW, Robinson CA, Guella I, Farrer MJ, Rajput A (2016) Conjugal parkinsonism is coincidental. Parkinsonism Relat Disord 33, 149–150. [DOI] [PubMed] [Google Scholar]
- [64]. Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrøm L, Siitonen A, Iwaki H, Leonard H, Faghri F, Gibbs JR, Hernandez DG, Scholz SW, Botia JA, Martinez M, Corvol JC, Lesage S, Jankovic J, Shulman LM, Sutherland M, Tienari P, Majamaa K, Toft M, Andreassen OA, Bangale T, Brice A, Yang J, Gan-Or Z, Gasser T, Heutink P, Shulman JM, Wood NW, Hinds DA, Hardy JA, Morris HR, Gratten J, Visscher PM, Graham RR, Singleton AB; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium (2019) Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol 18, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Blauwendraat C, Nalls MA, Singleton AB (2020) The genetic architecture of Parkinson’s disease. Lancet Neurol 19, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Bustos BI, Billingsley K, Blauwendraat C, Gibbs JR, Gan-Or Z, Krainc D, Singleton AB, Lubbe SJ; International Parkinson’s Disease Genomics Consortium (IPDGC) (2023) Genome-wide contribution of common short-tandem repeats to Parkinson’s disease genetic risk. Brain 146, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67]. Billingsley KJ, Ding J, Jerez PA, Illarionova A, Levine K, Grenn FP, Makarious MB, Moore A, Vitale D, Reed X, Hernandez D, Torkamani A, Ryten M, Hardy J; UK Brain Expression Consortium (UKBEC); Chia R, Scholz SW, Traynor BJ, Dalgard CL, Ehrlich DJ, Tanaka T, Ferrucci L, Beach TG, Serrano GE, Quinn JP, Bubb VJ, Collins RL, Zhao X, Walker M, Pierce-Hoffman E, Brand H, Talkowski ME, Casey B, Cookson MR, Markham A, Nalls MA, Mahmoud M, Sedlazeck FJ, Blauwendraat C, Gibbs JR, Singleton AB (2023) Genome-wide analysis of structural variants in Parkinson disease. Ann Neurol 93, 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Jacobs BM, Belete D, Bestwick J, Blauwendraat C, Bandres-Ciga S, Heilbron K, Dobson R, Nalls MA, Singleton A, Hardy J, Giovannoni G, Lees AJ, Schrag AE, Noyce AJ (2020) Parkinson’s disease determinants, prediction and gene-environment interactions in the UK Biobank. J Neurol Neurosurg Psychiatry 91, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Koch S, Laabs BH, Kasten M, Vollstedt EJ, Becktepe J, Brüggemann N, Franke A, Krämer UM, Kuhlenbäumer G, Lieb W, Mollenhauer B, Neis M, Trenkwalder C, Schäffer E, Usnich T, Wittig M, Klein C, König IR, Lohmann K, Krawczak M, Caliebe A (2021) Validity and prognostic value of a polygenic risk score for Parkinson’s disease. Genes (Basel) 12, 1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70]. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S (2018) Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Curtis D (2019) Clinical relevance of genome-wide polygenic score may be less than claimed. Ann Hum Genet 83, 274–277. [DOI] [PubMed] [Google Scholar]
- [72]. Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P, Tzoulaki I (2020) Predictive accuracy of a polygenic risk score-enhanced prediction model vs. a clinical risk score for coronary artery disease. JAMA 323, 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Wald NJ, Old R (2019) The illusion of polygenic disease risk prediction. Genet Med 21, 1705–1707. [DOI] [PubMed] [Google Scholar]
- [74]. Hingorani AD, Gratton J, Finan C, Schmidt AF, Patel R, Sofat R, Kuan V, Langenberg C, Hemingway H, Morris JK, Wald NJ (2023) Performance of polygenic risk scores in screening, prediction, and risk stratification: Secondary analysis of data in the Polygenic Score Catalog. BMJ Med 2, e000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75]. Liu H, Dehestani M, Blauwendraat C, Makarious MB, Leonard H, Kim JJ, Schulte C, Noyce A, Jacobs BM, Foote I, Sharma M; International Parkinson’s Disease Genomics Consortium; Comprehensive Unbiased Risk Factor Assessment for Genetics and Environment in Parkinson’s Disease Consortium; Nalls M, Singleton A, Gasser T, Bandres-Ciga S (2022) Polygenic resilience modulates the penetrance of Parkinson disease genetic risk factors. Ann Neurol 92, 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Xie CL, Pan JL, Wang WW, Zhang Y, Zhang SF, Gan J, Liu ZG (2014) The association between the LRRK2 G2385R variant and the risk of Parkinson’s disease: A meta-analysis based on 23 case-control studies. Neurol Sci 35, 1495–1504. [DOI] [PubMed] [Google Scholar]
- [77]. Zhang Y, Sun Q, Yi M, Zhou X, Guo J, Xu Q, Tang B, Yan X (2017) Genetic analysis of LRRK2 R1628P in Parkinson’s disease in Asian populations. Parkinsons Dis 2017, 8093124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78]. Gopalai AA, Lim SY, Chua JY, Tey S, Lim TT, Mohamed Ibrahim N, Tan AH, Eow GB, Abdul Aziz Z, Puvanarajah SD, Viswanathan S, Looi I, Lim SK, Tan LP, Chong YB, Tan CT, Zhao Y, Tan EK, Ahmad-Annuar A (2014) LRRK2 G2385R and R1628P mutations are associated with an increased risk of Parkinson’s disease in the Malaysian population. Biomed Res Int 2014, 867321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Ong YL, Deng X, Li HH, Narasimhalu K, Chan LL, Prakash KM, Au WL, Ratnagopal P, Tan LCS, Tan EK (2023) Caffeine intake interacts with Asian gene variants in Parkinson’s disease: A study in 4488 subjects. Lancet Reg Health West Pac 40, 100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Crotty GF, Maciuca R, Macklin EA, Wang J, Montalban M, Davis SS, Alkabsh JI, Bakshi R, Chen X, Ascherio A, Astarita G, Huntwork-Rodriguez S, Schwarzschild MA (2020) Association of caffeine and related analytes with resistance to Parkinson disease among LRRK2 mutation carriers: A metabolomic study. Neurology 95, e3428–e3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81]. Nicholatos JW, Francisco AB, Bender CA, Yeh T, Lugay FJ, Salazar JE, Glorioso C, Libert S (2018) Nicotine promotes neuron survival and partially protects from Parkinson’s disease by suppressing SIRT6. Acta Neuropathol Commun 6, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Hill-Burns EM, Singh N, Ganguly P, Hamza TH, Montimurro J, Kay DM, Yearout D, Sheehan P, Frodey K, McLear JA, Feany MB, Hanes SD, Wolfgang WJ, Zabetian CP, Factor SA, Payami H (2013) A genetic basis for the variable effect of smoking/nicotine on Parkinson’s disease. Pharmacogenomics J 13, 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Goldman SM, Kamel F, Ross GW, Bhudhikanok GS, Hoppin JA, Korell M, Marras C, Meng C, Umbach DM, Kasten M, Chade AR, Comyns K, Richards MB, Sandler DP, Blair A, Langston JW, Tanner CM (2012) Genetic modification of the association of paraquat and Parkinson’s disease. Mov Disord 27, 1652–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Tan AH, Lim SY, Lang AE (2022) The microbiome-gut-brain axis in Parkinson disease – from basic research to the clinic. Nat Rev Neurol 18, 476–495. [DOI] [PubMed] [Google Scholar]
- [85]. Müller-Nedebock AC, Dekker MCJ, Farrer MJ, Hattori N, Lim SY, Mellick GD, Rektorová I, Salama M, Schuh AFS, Stoessl AJ, Sue CM, Tan AH, Vidal RL, Klein C, Bardien S (2023) Different pieces of the same puzzle: A multifaceted perspective on the complex biological basis of Parkinson’s disease. NPJ Parkinsons Dis 9, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Toh TS, Chong CW, Lim SY, Bowman J, Cirstea M, Lin CH, Chen CC, Appel-Cresswell S, Finlay BB, Tan AH (2022) Gut microbiome in Parkinson’s disease: New insights from meta-analysis. Parkinsonism Relat Disord 94, 1–9. [DOI] [PubMed] [Google Scholar]
- [87]. Herrick MK, Tansey MG (2021) Is LRRK2 the missing link between inflammatory bowel disease and Parkinson’s disease? NPJ Parkinsons Dis 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Klingelhoefer L, Reichmann H (2015) Pathogenesis of Parkinson disease–the gut-brain axis and environmental factors. Nat Rev Neurol 11, 625–636. [DOI] [PubMed] [Google Scholar]
- [89]. Perez-Pardo P, Dodiya HB, Engen PA, Forsyth CB, Huschens AM, Shaikh M, Voigt RM, Naqib A, Green SJ, Kordower JH, Shannon KM, Garssen J, Kraneveld AD, Keshavarzian A (2019) Role of TLR4 in the gut-brain axis in Parkinson’s disease: A translational study from men to mice. Gut 68, 829–843. [DOI] [PubMed] [Google Scholar]
- [90]. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK (2016) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Matheoud D, Cannon T, Voisin A, Penttinen AM, Ramet L, Fahmy AM, Ducrot C, Laplante A, Bourque MJ, Zhu L, Cayrol R, Le Campion A, McBride HM, Gruenheid S, Trudeau LE, Desjardins M (2019) Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1-/- mice. Nature 571, 565–569. [DOI] [PubMed] [Google Scholar]
- [92]. Lin CH, Lin HY, Ho EP, Ke YC, Cheng MF, Shiue CY, Wu CH, Liao PH, Hsu AY, Chu LA, Liu YD, Lin YH, Tai YC, Shun CT, Chiu HM, Wu MS (2022) Mild chronic colitis triggers parkinsonism in LRRK2 mutant mice through activating TNF-α pathway. Mov Disord 37, 745–757. [DOI] [PubMed] [Google Scholar]
- [93]. Senkevich K, Rudakou U, Gan-Or Z (2022) New therapeutic approaches to Parkinson’s disease targeting GBA, LRRK2 and Parkin. Neuropharmacology 202, 108822. [DOI] [PubMed] [Google Scholar]
- [94]. Lim JL, Lohmann K, Tan AH, Tay YW, Ibrahim KA, Abdul Aziz Z, Mawardi AS, Puvanarajah SD, Lim TT, Looi I, Ooi JCE, Chia YK, Muthusamy KA, Bauer P, Rolfs A, Klein C, Ahmad-Annuar A, Lim SY (2022) Glucocerebrosidase (GBA) gene variants in a multi-ethnic Asian cohort with Parkinson’s disease: Mutational spectrum and clinical features. J Neural Transm (Vienna) 129, 37–48. [DOI] [PubMed] [Google Scholar]
- [95]. Lanore A, Casse F, Tesson C, Courtin T, Menon PJ, Sambin S, Mangone G, Mariani LL, Lesage S, Brice A, Elbaz A, Corvol JC; French Clinicians Network for Parkinson’s Disease Genetics (the PDG Group) (2023) Differences in survival across monogenic forms of Parkinson’s disease. Ann Neurol 94, 123–132. [DOI] [PubMed] [Google Scholar]
- [96]. Artusi CA, Dwivedi AK, Romagnolo A, Pal G, Kauffman M, Mata I, Patel D, Vizcarra JA, Duker A, Marsili L, Cheeran B, Woo D, Contarino MF, Verhagen L, Lopiano L, Espay AJ, Fasano A, Merola A (2019) Association of subthalamic deep brain stimulation with motor, functional, and pharmacologic outcomes in patients with monogenic Parkinson disease: A systematic review and meta-analysise. JAMA Netw Open 2, 187800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Pal G, Mangone G, Hill EJ, Ouyang B, Liu Y, Lythe V, Ehrlich D, Saunders-Pullman R, Shanker V, Bressman S, Alcalay RN, Garcia P, Marder KS, Aasly J, Mouradian MM, Link S, Rosenbaum M, Anderson S, Bernard B, Wilson R, Stebbins G, Nichols WC, Welter ML, Sani S, Afshari M, Verhagen L, de Bie RMA, Foltynie T, Hall D, Corvol JC, Goetz CG (2022) Parkinson disease and subthalamic nucleus deep brain stimulation: Cognitive effects in GBA mutation carriers. Ann Neurol 91, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98]. Dy Closas AMF, Tan AH, Tay YW, Hor JW, Toh TS, Lim JL, Lew CY, Cham CY, Yim CCW, Chee KY, Ng CG, Lit LC, Khairul Anuar AN, Lange LM, Fang ZH, Bandres Ciga S, Lohmann K, Klein C, Ahmad-Annuar A, Muthusamy KA, Lim SY (2023) Real-world experience of deep brain stimulation surgery in a developing Southeast Asian country. medRxiv, doi: 10.1101/2023.08.23.23294286 [Preprint]. Posted August 25, 2023. [DOI] [Google Scholar]
- [99]. Kasten M, Hartmann C, Hampf J, Schaake S, Westenberger A, Vollstedt EJ, Balck A, Domingo A, Vulinovic F, Dulovic M, Zorn I, Madoev H, Zehnle H, Lembeck CM, Schawe L, Reginold J, Huang J, König IR, Bertram L, Marras C, Lohmann K, Lill CM, Klein C (2018) Genotype-phenotype relations for the Parkinson’s disease genes Parkin, PINK1, DJ1: MDSGene systematic review. Mov Disord 33, 730–741. [DOI] [PubMed] [Google Scholar]
- [100]. Lesage S, Lunati A, Houot M, Romdhan SB, Clot F, Tesson C, Mangone G, Toullec BL, Courtin T, Larcher K, Benmahdjoub M, Arezki M, Bouhouche A, Anheim M, Roze E, Viallet F, Tison F, Broussolle E, Emre M, Hanagasi H, Bilgic B, Tazir M, Djebara MB, Gouider R, Tranchant C, Vidailhet M, Le Guern E, Corti O, Mhiri C, Lohmann E, Singleton A, Corvol JC, Brice A; French Parkinson Disease Genetics Study Group (2020) Characterization of recessive Parkinson disease in a large multicenter study. Ann Neurol 88, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101]. Weill C, Gallant A, Lintsky E, Dienstag A, Israel Z, Arkadir D (2022) Cognitive effects of deep brain stimulation in GBA-related Parkinson disease. Ann Neurol 92, 344–345. [DOI] [PubMed] [Google Scholar]
- [102]. Chan GH (2022) The role of genetic data in selecting device-aided therapies in patients with advanced Parkinson’s disease: A mini-review. Front Aging Neurosci 14, 895430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Klein C, Schlossmacher MG (2007) Parkinson disease, 10 years after its genetic revolution: Multiple clues to a complex disorder. Neurology 69, 2093–2104. [DOI] [PubMed] [Google Scholar]
- [104]. Titova N, Chaudhuri KR (2017) Personalized medicine in Parkinson’s disease: Time to be precise. Mov Disord 32, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105]. Tosin MH, Goetz CG, Stebbins GT (2024) Patient with Parkinson disease and care partner perceptions of key domains affecting health-related quality of Life. Neurology 102, e208028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106]. Dehestani M, Liu H, Gasser T (2021) Polygenic risk scores contribute to personalized medicine of Parkinson’s disease. J Pers Med 11, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107]. Escott-Price V; International Parkinson’s Disease Genomics Consortium; Nalls MA, Morris HR, Lubbe S, Brice A, Gasser T, Heutink P, Wood NW, Hardy J, Singleton AB, Williams NM; IPDGC consortium members (2015) Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann Neurol 77, 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. Paul KC, Schulz J, Bronstein JM, Lill CM, Ritz BR (2018) Association of polygenic risk score with cognitive decline and motor progression in Parkinson disease. JAMA Neurol 75, 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Martinez-Carrasco A, Real R, Lawton M, Iwaki H, Tan MMX, Wu L, Williams NM, Carroll C, Hu MTM, Grosset DG, Hardy J, Ryten M, Foltynie T, Ben-Shlomo Y, Shoai M, Morris HR (2023) Genetic meta-analysis of levodopa induced dyskinesia in Parkinson’s disease. NPJ Parkinsons Dis 9, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Weintraub D, Posavi M, Fontanillas P, Tropea TF, Mamikonyan E, Suh E, Trojanowski JQ, Cannon P, Van Deerlin VM; 23andMe Research Team; Chen-Plotkin AS (2022) Genetic prediction of impulse control disorders in Parkinson’s disease. Ann Clin Transl Neurol 9, 936–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111]. Liu G, Peng J, Liao Z, Locascio JJ, Corvol JC, Zhu F, Dong X, Maple-Grødem J, Campbell MC, Elbaz A, Lesage S, Brice A, Mangone G, Growdon JH, Hung AY, Schwarzschild MA, Hayes MT, Wills AM, Herrington TM, Ravina B, Shoulson I, Taba P, Kõks S, Beach TG, Cormier-Dequaire F, Alves G, Tysnes OB, Perlmutter JS, Heutink P, Amr SS, van Hilten JJ, Kasten M, Mollenhauer B, Trenkwalder C, Klein C, Barker RA, Williams-Gray CH, Marinus J; International Genetics of Parkinson Disease Progression (IGPP) Consortium; Scherzer CR (2021) ) Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson’s disease. Nat Genet 53, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112]. Cha PC, Satake W, Ando-Kanagawa Y, Yamamoto K, Murata M, Toda T (2020) Genome-wide association study identifies zonisamide responsive gene in Parkinson’s disease patients. J Hum Genet 65, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113]. Visanji NP, Ghani M, Yu E, Kakhki EG, Sato C, Moreno D, Naranian T, Poon YY, Abdollahi M, Naghibzadeh M, Rajalingam R, Lozano AM, Kalia SK, Hodaie M, Cohn M, Statucka M, Boutet A, Elias GJB, Germann J, Munhoz R, Lang AE, Gan-Or Z, Rogaeva E, Fasano A (2022) Axial impairment following deep brain stimulation in Parkinson’s disease: A surgicogenomic approach. J Parkinsons Dis 12, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Yoon E, Ahmed S, Li R, Bandres-Ciga S, Blauwendraat C, Dustin I, Scholz S, Hallett M, Ehrlich D (2023) Association of polygenic risk score with response to deep brain stimulation in Parkinson’s disease. BMC Neurol 23, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115]. Bloem BR, de Vries NM, Ebersbach G (2015) Nonpharmacological treatments for patients with Parkinson’s disease. Mov Disord 30, 1504–1520. [DOI] [PubMed] [Google Scholar]
- [116]. Schootemeijer S, Darweesh SKL, de Vries NM (2022) Clinical trial highlights - aerobic exercise for Parkinson’s disease. J Parkinsons Dis 12, 2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Tsukita K, Sakamaki-Tsukita H, Takahashi R (2022) Long-term effect of regular physical activity and exercise habits in patients with early Parkinson disease. Neurology 98, e859–e871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Lewis A, Galetta S (2022) Editors’ note: Long-term effect of regular physical activity and exercise habits in patients with early Parkinson disease. Neurology 99, 131. [DOI] [PubMed] [Google Scholar]
- [119]. Tan AH, Lohmann K, Tay YW, Lim JL, Ahmad-Annuar A, Ramli N, Chin YT, Mawardi AS, Azmi K, Aziz ZA, Puvanarajah SD, Bauer P, Klein C, Rolfs A, Lim SY (2020) PINK1 p.Leu347Pro mutations in Malays: Prevalence and illustrative cases. Parkinsonism Relat Disord 79, 34–39. [DOI] [PubMed] [Google Scholar]
- [120]. Jagota P, Lim SY, Pal PK, Lee JY, Kukkle PL, Fujioka S, Shang H, Phokaewvarangkul O, Bhidayasiri R, Mohamed Ibrahim N, Ugawa Y, Aldaajani Z, Jeon B, Diesta C, Shambetova C, Lin CH (2023) Genetic movement disorders commonly seen in Asians. Mov Disord Clin Pract 10, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121]. Morales-Briceno H, Ong TL, Duma SR, Murray N, Pepper EM, Ha A, Tchan MC, Fung VSC (2022) Recurrent biallelic p.L347P PINK1 variant in Polynesians with parkinsonism and isolated dopa-responsive dystonia. Mov Disord Clin Pract 9, 696–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122]. Mencacci NE, Isaias IU, Reich MM, Ganos C, Plagnol V, Polke JM, Bras J, Hersheson J, Stamelou M, Pittman AM, Noyce AJ, Mok KY, Opladen T, Kunstmann E, Hodecker S, Münchau A, Volkmann J, Samnick S, Sidle K, Nanji T, Sweeney MG, Houlden H, Batla A, Zecchinelli AL, Pezzoli G, Marotta G, Lees A, Alegria P, Krack P, Cormier-Dequaire F, Lesage S, Brice A, Heutink P, Gasser T, Lubbe SJ, Morris HR, Taba P, Koks S, Majounie E, Raphael Gibbs J, Singleton A, Hardy J, Klebe S, Bhatia KP, Wood NW; International Parkinson’s Disease Genomics Consortium and UCL-exomes consortium (2014) Parkinson’s disease in GTP cyclohydrolase 1 mutation carriers. Brain 137, 2480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123]. Cukier HN, Kim H, Griswold AJ, Codreanu SG, Prince LM, Sherrod SD, McLean JA, Dykxhoorn DM, Ess KC, Hedera P, Bowman AB, Neely MD (2022) Genomic, transcriptomic, and metabolomic profiles of hiPSC-derived dopamine neurons from clinically discordant brothers with identical PRKN deletions. NPJ Parkinsons Dis 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124]. Sjögren M, Huttunen HJ, Svenningsson P, Widner H (2021) Genetically targeted clinical trials in Parkinson’s disease: Learning from the successes made in oncology. Genes (Basel) 12, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125]. Prasuhn J, Brüggemann N (2021) Genotype-driven therapeutic developments in Parkinson’s disease. Mol Med 27, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126]. Siderowf A, Concha-Marambio L, Lafontant DE, Farris CM, Ma Y, Urenia PA, Nguyen H, Alcalay RN, Chahine LM, Foroud T, Galasko D, Kieburtz K, Merchant K, Mollenhauer B, Poston KL, Seibyl J, Simuni T, Tanner CM, Weintraub D, Videnovic A, Choi SH, Kurth R, Caspell-Garcia C, Coffey CS, Frasier M, Oliveira LMA, Hutten SJ, Sherer T, Marek K, Soto C; Parkinson’s Progression Markers Initiative (2023) Assessment of heterogeneity among participants in the Parkinson’s Progression Markers Initiative cohort using α-synuclein seed amplification: A cross-sectional study. Lancet Neurol 22, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127]. Kluge A, Bunk J, Schaeffer E, Drobny A, Xiang W, Knacke H, Bub S, Lückstädt W, Arnold P, Lucius R, Berg D, Zunke F (2022) Detection of neuron-derived pathological α-synuclein in blood. Brain 145, 3058–3071. [DOI] [PubMed] [Google Scholar]
- [128]. Okuzumi A, Hatano T, Matsumoto G, Nojiri S, Ueno SI, Imamichi-Tatano Y, Kimura H, Kakuta S, Kondo A, Fukuhara T, Li Y, Funayama M, Saiki S, Taniguchi D, Tsunemi T, McIntyre D, Gérardy JJ, Mittelbronn M, Kruger R, Uchiyama Y, Nukina N, Hattori N (2023) Propagative α-synuclein seeds as serum biomarkers for synucleinopathies. Nat Med 29, 1448–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Cardoso F, Goetz CG, Mestre TA, Sampaio C, Adler CH, Berg D, Bloem BR, Burn DJ, Fitts MS, Gasser T, Klein C, de Tijssen MAJ, Lang AE, Lim SY, Litvan I, Meissner WG, Mollenhauer B, Okubadejo N, Okun MS, Postuma RB, Svenningsson P, Tan LCS, Tsunemi T, Wahlstrom-Helgren S, Gershanik OS, Fung VSC, Trenkwalder C (2024) A statement of the MDS on biological definition, staging and classification of Parkinson’s disease. Mov Disord 39, 259–266. [DOI] [PubMed] [Google Scholar]
- [130]. Simuni T, Chahine LM, Poston K, Brumm M, Buracchio T, Campbell M, Chowdhury S, Coffey C, Concha-Marambio L, Dam T, DiBiaso P, Foroud T, Frasier M, Gochanour C, Jennings D, Kieburtz K, Kopil CM, Merchant K, Mollenhauer B, Montine T, Nudelman K, Pagano G, Seibyl J, Sherer T, Singleton A, Stephenson D, Stern M, Soto C, Tanner CM, Tolosa E, Weintraub D, Xiao Y, Siderowf A, Dunn B, Marek K (2024) A biological definition of neuronal α-synuclein disease: Towards an integrated staging system for research. Lancet Neurol 23, 178–190. [DOI] [PubMed] [Google Scholar]
- [131]. Tsuboi M, Herbst RS, John T, Kato T, Majem M, Grohé C, Wang J, Goldman JW, Lu S, Su WC, de Marinis F, Shepherd FA, Lee KH, Le NT, Dechaphunkul A, Kowalski D, Poole L, Bolanos A, Rukazenkov Y, Wu YL; ADAURA Investigators (2023) Overall survival with Osimertinib in resected EGFR-mutated NSCLC. N Engl J Med 389, 137–147. [DOI] [PubMed] [Google Scholar]
- [132]. Singleton A, Hardy J (2011) A generalizable hypothesis for the genetic architecture of disease: Pleomorphic risk loci. Hum Mol Genet 20, R158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133]. Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R (2016) Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134]. Reed X, Bandrés-Ciga S, Blauwendraat C, Cookson MR (2019) The role of monogenic genes in idiopathic Parkinson’s disease. Neurobiol Dis 124, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135]. Pihlstrøm L, Blauwendraat C, Cappelletti C, Berge-Seidl V, Langmyhr M, Henriksen SP, van de Berg WDJ, Gibbs JR, Cookson MR; International Parkinson Disease Genomics Consortium; North American Brain Expression Consortium; Singleton AB, Nalls MA, Toft M (2018) A comprehensive analysis of SNCA-related genetic risk in sporadic Parkinson disease. Ann Neurol 84, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136]. Grenn FP, Kim JJ, Makarious MB, Iwaki H, Illarionova A, Brolin K, Kluss JH, Schumacher-Schuh AF, Leonard H, Faghri F, Billingsley K, Krohn L, Hall A, Diez-Fairen M, Periñán MT, Foo JN, Sandor C, Webber C, Fiske BK, Gibbs JR, Nalls MA, Singleton AB, Bandres-Ciga S, Reed X, Blauwendraat C; International Parkinson’s Disease Genomics Consortium (IPDGC) (2020) The Parkinson’s disease genome-wide association study locus browser. Mov Disord 35, 2056–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137]. Kalogeropulou AF, Purlyte E, Tonelli F, Lange SM, Wightman M, Prescott AR, Padmanabhan S, Sammler E, Alessi DR (2022) Impact of 100 LRRK2 variants linked to Parkinson’s disease on kinase activity and microtubule binding. Biochem J 479, 1759–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138]. Dorsey ER, Bloem BR (2024) Parkinson’s is predominantly an environmental disease. J Parkinsons Dis, doi: 10.3233/JPD-230357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139]. Tanner CM, Kamel F, Ross GW, Hoppin JA, Goldman SM, Korell M, Marras C, Bhudhikanok GS, Kasten M, Chade AR, Comyns K, Richards MB, Meng C, Priestley B, Fernandez HH, Cambi F, Umbach DM, Blair A, Sandler DP, Langston JW (2011) Rotenone, paraquat, and Parkinson’s disease. Environ Health Perspect 119, 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140]. De Miranda BR, Goldman SM, Miller GW, Greenamyre JT, Dorsey ER (2022) Preventing Parkinson’s disease: An environmental agenda. J Parkinsons Dis 12, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141]. Fujimaki M, Saiki S, Li Y, Kaga N, Taka H, Hatano T, Ishikawa KI, Oji Y, Mori A, Okuzumi A, Koinuma T, Ueno SI, Imamichi Y, Ueno T, Miura Y, Funayama M, Hattori N (2018) Serum caffeine and metabolites are reliable biomarkers of early Parkinson disease. Neurology 90, e404–e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142]. Quik M, O’Leary K, Tanner CM (2008) Nicotine and Parkinson’s disease: Implications for therapy. Mov Disord 23, 1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143]. Editor (2000) The importance of technological advances. Nat Cell Biol 2, E37. [DOI] [PubMed] [Google Scholar]
- [144]. Claussnitzer M, Cho JH, Collins R, Cox NJ, Dermitzakis ET, Hurles ME, Kathiresan S, Kenny EE, Lindgren CM, MacArthur DG, North KN, Plon SE, Rehm HL, Risch N, Rotimi CN, Shendure J, Soranzo N, McCarthy MI (2020) A brief history of human disease genetics. Nature 577, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145]. Chen H, Ritz B (2018) The search for environmental causes of Parkinson’s disease: Moving forward. J Parkinsons Dis 8, S9–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146]. Wadman M (2023) Solvent exposure strongly linked to Parkinson’s. Science 380, 683. [DOI] [PubMed] [Google Scholar]
- [147]. Pang SY, Ho PW, Liu HF, Leung CT, Li L, Chang EES, Ramsden DB, Ho SL (2019) The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson’s disease. Transl Neurodegener 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148]. Walker VM, Davey Smith G, Davies NM, Martin RM (2017) Mendelian randomization: A novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol 46, 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149]. Noyce AJ, Bandres-Ciga S, Kim J, Heilbron K, Kia D, Hemani G, Xue A, Lawlor DA, Smith GD, Duran R, Gan-Or Z, Blauwendraat C, Gibbs JR; 23andMe Research Team5, International Parkinson’s Disease Genomics Consortium (IPDGC); Hinds DA, Yang J, Visscher P, Cuzick J, Morris H, Hardy J, Wood NW, Nalls MA, Singleton AB (2019) The Parkinson’s Disease Mendelian Randomization Research Portal. Mov Disord 34, 1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150]. Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, Sherman MA, Vitzthum CM, Luquette LJ, Yandava CN, Yang P, Chittenden TW, Hatem NE, Ryu SC, Woodworth MB, Park PJ, Walsh CA (2018) Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151]. Lobon I, Solís-Moruno M, Juan D, Muhaisen A, Abascal F, Esteller-Cucala P, García-Pérez R, Martí MJ, Tolosa E, Ávila J, Rahbari R, Marques-Bonet T, Casals F, Soriano E (2022) Somatic mutations detected in Parkinson disease could affect genes with a role in synaptic and neuronal processes. Front Aging 3, 851039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152]. Trinh J, Lüth T, Schaake S, Laabs BH, Schlüter K, Laβ J, Pozojevic J, Tse R, König I, Jamora RD, Rosales RL, Brüggemann N, Saranza G, Diesta CCE, Kaiser FJ, Depienne C, Pearson CE, Westenberger A, Klein C (2023) Mosaic divergent repeat interruptions in XDP influence repeat stability and disease onset. Brain 146, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153]. Jo S, Kim YJ, Park KW, Hwang YS, Lee SH, Kim BJ, Chung SJ (2021) Association of NO2 and other air pollution exposures with the risk of Parkinson disease. JAMA Neurol 78, 800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154]. Ding E, Wang Y, Liu J, Tang S, Shi X (2022) A review on the application of the exposome paradigm to unveil the environmental determinants of age-related diseases. Hum Genomics 16, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155]. Talavera Andújar B, Aurich D, Aho VTE, Singh RR, Cheng T, Zaslavsky L, Bolton EE, Mollenhauer B, Wilmes P, Schymanski EL (2022) Studying the Parkinson’s disease metabolome and exposome in biological samples through different analytical and cheminformatics approaches: A pilot study. Anal Bioanal Chem 414, 7399–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156]. Sauerbeck A, Hunter R, Bing G, Sullivan PG (2012) Traumatic brain injury and trichloroethylene exposure interact and produce functional, histological, and mitochondrial deficits. Exp Neurol 234, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157]. Schiess N, Cataldi R, Okun MS, Fothergill-Misbah N, Dorsey ER, Bloem BR, Barretto M, Bhidayasiri R, Brown R, Chishimba L, Chowdhary N, Coslov M, Cubo E, Di Rocco A, Dolhun R, Dowrick C, Fung VSC, Gershanik OS, Gifford L, Gordon J, Khalil H, Kühn AA, Lew S, Lim SY, Marano MM, Micallef J, Mokaya J, Moukheiber E, Nwabuobi L, Okubadejo N, Pal PK, Shah H, Shalash A, Sherer T, Siddiqui B, Thompson T, Ullrich A, Walker R, Dua T (2022) Six action steps to address global disparities in Parkinson disease: A World Health Organization priority. JAMA Neurol 79, 929–936. [DOI] [PubMed] [Google Scholar]
- [158]. Lee AJ, Wang Y, Alcalay RN, Mejia-Santana H, Saunders-Pullman R, Bressman S, Corvol JC, Brice A, Lesage S, Mangone G, Tolosa E, Pont-Sunyer C, Vilas D, Schüle B, Kausar F, Foroud T, Berg D, Brockmann K, Goldwurm S, Siri C, Asselta R, Ruiz-Martinez J, Mondragón E, Marras C, Ghate T, Giladi N, Mirelman A, Marder K; Michael J. Fox LRRK2 Cohort Consortium (2017) Penetrance estimate of LRRK2 p.G2019S mutation in individuals of non-Ashkenazi Jewish ancestry. Mov Disord 32, 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159]. Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Foroud TM, Boeve BF, Graff-Radford NR, St Jean P, Lawson M, Ehm MG, Mayeux R, Goate AM; NIA-LOAD/NCRAD Family Study Consortium (2012) Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PloS One 7, e31039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160]. Reitz C, Pericak-Vance MA, Foroud T, Mayeux R (2023) A global view of the genetic basis of Alzheimer disease. Nat Rev Neurol 19, 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161]. Hughes AJ, Daniel SE, Ben-Shlomo Y, Lees AJ (2002) The accuracy of diagnosis of parkinsonian syndromes in a specialist movement disorder service. Brain 125, 861–870. [DOI] [PubMed] [Google Scholar]
- [162]. Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, Litvan I, Lang AE, Halliday G, Goetz CG, Gasser T, Dubois B, Chan P, Bloem BR, Adler CH, Deuschl G (2015) MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord 30, 1591–1601. [DOI] [PubMed] [Google Scholar]
- [163]. Postuma RB, Lang AE (2023) The clinical diagnosis of Parkinson’s disease-we are getting better. Mov Disord 38, 515–517. [DOI] [PubMed] [Google Scholar]
- [164]. Zheng Z, Zhu Z, Zhou C, Cao L, Zhao G (2023) Burden of Parkinson disease in China, 1990–2019: Findings from the 2019 Global Burden of Disease Study. Neuroepidemiology 57, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165]. Lim SY, Tan AH, Tay YW (2023) Deep brain stimulation in dopa-responsive parkinsonism - look out for red flags: Expert commentary. Parkinsonism Relat Disord 110, 105276. [DOI] [PubMed] [Google Scholar]
- [166]. Zhang ZX, Roman GC, Hong Z, Wu CB, Qu QM, Huang JB, Zhou B, Geng ZP, Wu JX, Wen HB, Zhao H, Zahner GE (2005) Parkinson’s disease in China: Prevalence in Beijing, Xian, and Shanghai. Lancet 365, 595–597. [DOI] [PubMed] [Google Scholar]
- [167]. Zhang ZX, Anderson DW, Huang JB, Li H, Hong X, Wei J, Yang EL, Maraganore DM (2003) Prevalence of Parkinson’s disease and related disorders in the elderly population of greater Beijing, China. Mov Disord 18, 764–772. [DOI] [PubMed] [Google Scholar]
- [168]. Dorsey ER, Sherer T, Okun MS, Bloem BR (2018) The emerging evidence of the Parkinson pandemic. J Parkinsons Dis 8, S3–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169]. GBD 2016 Parkinson’s Disease Collaborators (2018) Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 17, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170]. Li G, Ma J, Cui S, He Y, Xiao Q, Liu J, Chen S (2019) Parkinson’s disease in China: A forty-year growing track of bedside work. Transl Neurodegener 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171]. Lai H, Li XY, Xu F, Zhu J, Li X, Song Y, Wang X, Wang Z, Wang C (2023) Applications of machine learning to diagnosis of Parkinson’s disease. Brain Sci 13, 1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172]. Topol EJ (2023) As artificial intelligence goes multimodal, medical applications multiply. Science 381, adk6139. [DOI] [PubMed] [Google Scholar]
- [173]. Yang Y, Yuan Y, Zhang G, Wang H, Chen YC, Liu Y, Tarolli CG, Crepeau D, Bukartyk J, Junna MR, Videnovic A, Ellis TD, Lipford MC, Dorsey R, Katabi D (2022) Artificial intelligence-enabled detection and assessment of Parkinson’s disease using nocturnal breathing signals. Nat Med 28, 2207–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174]. Buchman AS, Shulman JM, Nag S, Leurgans SE, Arnold SE, Morris MC, Schneider JA, Bennett DA (2012) Nigral pathology and parkinsonian signs in elders without Parkinson disease. Ann Neurol 71, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175]. Wu H, Eckhardt CM, Baccarelli AA (2023) Molecular mechanisms of environmental exposures and human disease. Nat Rev Genet 24, 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176]. Labbé C, Lorenzo-Betancor O, Ross OA (2016) Epigenetic regulation in Parkinson’s disease. Acta Neuropathol 132, 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177]. Pavlou MAS, Outeiro TF (2017) Epigenetics in Parkinson’s disease. Adv Exp Med Biol 978, 363–390. [DOI] [PubMed] [Google Scholar]
- [178]. Tang X, Gonzalez-Latapi P, Marras C, Visanji NP, Yang W, Sato C, Lang AE, Rogaeva E, Zhang M (2022) Epigenetic clock acceleration is linked to age at onset of Parkinson’s disease. Mov Disord 37, 1831–1840. [DOI] [PubMed] [Google Scholar]
- [179]. The Lancet Gastroenterology Hepatology (2021) Obesity: Another ongoing pandemic. Lancet Gastroenterol Hepatol 6, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180]. Loos RJF, Yeo GSH (2022) The genetics of obesity: From discovery to biology. Nat Rev Genet 23, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181]. McPherson R (2007) Genetic contributors to obesity. Can J Cardiol 23, 23A-27A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182]. Müller TD, Blüher M, Tschöp MH, DiMarchi RD (2022) Anti-obesity drug discovery: Advances and challenges. Nat Rev Drug Discov 21, 201–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183]. Drucker DJ, Habener JF, Holst JJ (2017) Discovery, characterization, and clinical development of the glucagon-like peptides. J Clin Invest 127, 4217–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184]. Barker RA (2020) Parkinson’s disease as a preventable pandemic. Lancet Neurol 19, 813. [Google Scholar]
- [185]. Münzel T, Sørensen M, Hahad O, Nieuwenhuijsen M, Daiber A (2023) The contribution of the exposome to the burden of cardiovascular disease. Nat Rev Cardiol 20, 651–669. [DOI] [PubMed] [Google Scholar]