Summary
Mitochondrial function relies on the coordinated transcription of mitochondrial and nuclear genomes to assemble respiratory chain complexes. Across species, the SIN3 coregulator influences mitochondrial functions, but how its loss impacts mitochondrial homeostasis and metabolism in the context of a whole organism is unknown. Exploring this link is important because SIN3 haploinsufficiency causes intellectual disability/autism syndromes and SIN3 plays a role in tumor biology. Here we show that loss of C. elegans SIN-3 results in transcriptional deregulation of mitochondrial- and nuclear-encoded mitochondrial genes, potentially leading to mito-nuclear imbalance. Consistent with impaired mitochondrial function, sin-3 mutants show extensive mitochondrial fragmentation by transmission electron microscopy (TEM) and in vivo imaging, and altered oxygen consumption. Metabolomic analysis of sin-3 mutant animals revealed a mitochondria stress signature and deregulation of methionine flux, resulting in decreased S-adenosyl methionine (SAM) and increased polyamine levels. Our results identify SIN3 as a key regulator of mitochondrial dynamics and metabolic flux, with important implications for human pathologies.
Subject areas: Cell biology, Systems biology, Omics
Graphical abstract

Highlights
-
•
Loss of SIN-3 coregulator causes transcriptional deregulation of mitochondrial genes
-
•
C. elegans sin-3 mutants show mitochondrial fragmentation and altered respiration
-
•
Sin-3 mutants also display reduced levels of SAM and accumulation of polyamines
Cell biology; Systems biology; Omics
Introduction
Mitochondria, the main energy providers within cells, produce ATP through oxidative phosphorylation (OXPHOS) and control the levels of many metabolites essential for various cellular functions. The OXPHOS system consists of four multimeric complexes, coenzyme Q and cytochrome c that form the mitochondrial respiratory chain (I–IV) and couple redox reactions, creating an electrochemical gradient leading to the creation of ATP through a fifth complex, the F1F0 ATPase.1 Assembly of functional mitochondria relies on coordinated transcription and translation of mitochondrial and nuclear genomes,2 and both mitochondrial and nuclear DNA mutations affecting the accumulation and function of OXPHOS enzymes are the most common cause of mitochondrial diseases and are associated with neurodegeneration and aging.3,4,5 The mitochondrial genome, which is highly conserved among species, comprises 37 genes coding for two ribosomal RNAs, 22 transfer RNAs, and 13 protein subunits (12 for C. elegans) of the mitochondrial respiratory chain performing OXPHOS.6 The rest of the mitochondrial proteome, comprising over a thousand proteins, is encoded in the nucleus.7,8,9 The bidirectional regulation between mitochondria and the nucleus, referred to as mito-nuclear communication, maintains homeostasis and regulates stress responses.10 Mitochondrial damage or alterations in mitochondrial function trigger specific quality control mechanisms. Of these, the best characterized is the mitochondrial unfolded protein response (UPRmt), a cellular stress response that leads to increased transcription of mitochondrial chaperones and proteases.11 In addition to an imbalance between mitochondrial and nuclear protein quantities,12 reduced levels of TCA cycle components,13 reduction of β-oxidation and lipid biosynthesis14,15 and defective mitochondrial import,16 can all trigger UPRmt. Recent data have shown that mitochondrial stress leads to extensive chromatin reorganization,17,18 and chromatin regulatory factors contribute to mitochondrial gene expression.12 Conversely, metabolites originating from the mitochondria can initiate modifications in the nucleus.12
Depletion experiments in various models have shown that the highly conserved SIN3/HDAC coregulator plays a role in mitochondrial functions and metabolism.19 Yeast sin3 null mutants grow poorly on non-fermentable carbon sources, have lower ATP levels, and reduced respiration rates.20 In Drosophila cultured cells, reduction of SIN3 levels also resulted in altered ATP levels and deregulation of genes encoded by the mitochondrial genome.20,21 Lower levels of ATP and increased sensitivity to oxidative stress were also observed in C. elegans animals carrying the sin-3(tm1276) partial loss of function allele.22 In mice, Sin3a was detected in a transcriptional complex with the MafA pancreatic β cell-specific activator,23 and its inactivation reduced the fitness of β cells and altered glucose production in the liver.24 Significantly, in mammalian cells, SIN3 and its associated protein SUDS3 were identified in a screen for modifiers of drug-induced mitochondrial dysfunction.25 Altogether, these data from different systems support a conserved role for SIN3 in mitochondrial homeostasis and metabolism, but how SIN3 affects mitochondrial dynamics and metabolic pathways in the context of a whole organism remains largely unknown. This is particularly important given that heterozygous loss-of-function variants, as well as point mutations in SIN3 were recently identified as the underlying cause of intellectual disability (ID)/autism syndromes,26,27,28 and SIN3 levels play an important role in tumor biology.29,30,31,32
Previous studies in C. elegans have shown that knockdown of sin-3, the single SIN3 homolog in this organism, results in a decreased lifespan, altered mitochondrial membrane potential, enhanced autophagy and increased oxidative stress.22,33 How these changes affect mitochondrial morphology and function was not investigated. Here, using a sin-3 CRISPR-Cas9 knock-out allele, we show that loss of SIN-3 results in the deregulation of both mitochondria- and nuclear-encoded genes. Transmission electron microscopy (TEM) and in vivo imaging revealed extensive fragmentation of mitochondria in all tissues examined, including muscle, intestine, hypodermis, and the germline. Consistent with severe defects in mitochondrial function, both basal and maximal oxygen consumption are increased in the absence of SIN3, while spare respiratory capacity is decreased. Metabolomic analysis identified a signature of mitochondria stress and deregulation of methionine flux, resulting in reduced levels of SAM and a shift toward higher polyamine levels. Together our data identify SIN3 as an important regulator of mitochondrial dynamics in an organismal context, and reveal a SIN-3- dependent connection between expression of OXPHOS subunits, mitochondrial homeostasis, and metabolic fluxes.
Results
Loss of sin-3 results in altered expression of genes with mitochondrial functions
Previous transcriptomic analysis in young adults carrying the sin-3(tm1276) partial loss-of-function mutation revealed deregulation of the germline transcriptome, including metabolic genes.34 In order to identify high-confidence genes regulated by SIN-3, we extended our analysis to genes commonly misregulated in sin-3(tm1276) and sin-3(syb2172) young adults that carry a complete loss of function allele obtained by CRISPR-Cas9,34 generating a list of 892 genes (Table S1). Using Worm Cat,35 within this set we identified a common class of genes with functions related to mitochondria (Figure 1A; Table 1).
Figure 1.
Loss of sin-3 alters the expression of mitochondrial genes
(A) WormCat visualization of categories enriched in genes commonly misregulated in sin-3(tm1276) and sin-3(syb2172) mutants at the young adult stage34 (Table S1). The legend for bubble charts is indicated on the right, with size referring to the number of genes in each category and color referring to the p value.
(B) Respiratory complex subunits commonly misregulated in sin-3(tm1276) and sin-3(syb2172) mutants are indicated, along with their mammalian counterparts. Subunits encoded by the mitochondrial genome are shown in bold.
Table 1.
List of genes with mitochondrial function commonly misregulated in both sin-3(syb2172) and sin-3(tm1276) germlines from young adults
| WB_ID | sequence_ID | category | misregulation |
|---|---|---|---|
| WBGene00010042 | bcs-1 | Metabolism: mitochondria: chaperone | DOWN |
| WBGene00016442 | C35D10.5 | Metabolism: mitochondria: chaperone | DOWN |
| WBGene00009139 | F25H9.7 | Metabolism: mitochondria: citric acid cycle | DOWN |
| WBGene00010317 | idh-1 | Metabolism: mitochondria: citric acid cycle | DOWN |
| WBGene00016844 | sucg-1 | Metabolism: mitochondria: citric acid cycle | DOWN |
| WBGene00017759 | mrps-18B | Metabolism: mitochondria: citric acid cycle | DOWN |
| WBGene00009712 | ndub-2 | Metabolism: mitochondria: complex I | UP |
| WBGene00010958 | ndfl-4 | Metabolism: mitochondria: complex I | UP |
| WBGene00010959 | nduo-1 | Metabolism: mitochondria: complex I | UP |
| WBGene00010961 | nduo-2 | Metabolism: mitochondria: complex I | UP |
| WBGene00010967 | nduo-5 | Metabolism: mitochondria: complex I | UP |
| WBGene00021849 | ndua-8 | Metabolism: mitochondria: complex I | UP |
| WBGene00012158 | ucr-2.1 | Metabolism: mitochondria: complex III | UP |
| WBGene00017121 | cyc-2.1 | Metabolism: mitochondria: complex III | DOWN |
| WBGene00020757 | ucr-2.3 | Metabolism: mitochondria: complex III | DOWN |
| WBGene00009161 | cox-7C | Metabolism: mitochondria: complex IV | DOWN |
| WBGene00010419 | atp-1 | Metabolism: mitochondria: complex V | DOWN |
| WBGene00011273 | R53.4 | Metabolism: mitochondria: complex V | DOWN |
| WBGene00020275 | atp-4 | Metabolism: mitochondria: complex V | DOWN |
| WBGene00022336 | Y82E9BR.3 | Metabolism: mitochondria: complex V | DOWN |
| WBGene00001425 | fis-2 | Metabolism: mitochondria: morphology | UP |
| WBGene00022159 | mppa-1 | Metabolism: mitochondria: protease | UP |
| WBGene00007712 | mrpl-34 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00011759 | mrps-18B | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00011883 | mrpl-50 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00012992 | mrpl-20 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00018961 | mrps-16 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00020499 | mrps-18.C | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00020625 | mrrf-1 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00021350 | Y37E3.8 | Metabolism: mitochondria: ribosome | DOWN |
| WBGene00009305 | metl-17 | Metabolism: mitochondria: RNA methyltransferase | DOWN |
| WBGene00007686 | tomm-40 | Metabolism: mitochondria: translocase | DOWN |
| WBGene00013462 | micu-1 | Metabolism: mitochondria: transporter | DOWN |
| WBGene00000763 | coq-3 | Metabolism: mitochondria: ubiquinone | DOWN |
| WBGene00003967 | pdr-1 | Metabolism: mitochondria: unassigned | DOWN |
| WBGene00009187 | F27D4.1 | Metabolism: mitochondria: unassigned | DOWN |
| WBGene00011527 | cchl-1 | Metabolism: mitochondria: unassigned | DOWN |
| WBGene00014176 | ZK1010.2 | Metabolism: mitochondria: unassigned | DOWN |
| WBGene00020511 | immt-1 | Metabolism: mitochondria: unassigned | UP |
| WBGene00077500 | C27H6.9 | Metabolism: mitochondria: unassigned | DOWN |
The list of all commonly misregulated genes was generated by crossing the set of genes obtained for each allele with an FDR <0.05 following DESeq2 analysis (Table S1), and the list of mitochondrial genes extracted from WormCat.35 Genes encoded by the mitochondrial genome are shown in bold. See also Tables S1 and S2.
Both nuclear- and mitochondrial-encoded genes contribute to the assembly of mitochondrial respiratory chain complexes (MRC) I-V.1 Five of the 7 MRC complex I subunits encoded by the mitochondrial genome were strongly upregulated in both sin-3 mutants: ndub-2, ndfl-4, nduo-1, nduo-2, and nduo-5 (Figure 1B). By contrast, nuclear-encoded MRC subunits identified in our dataset were mostly downregulated (Figure 1B), as were the majority of additional nuclear-encoded genes with mitochondria-related functions (31 out of 37, Table 1). These include mitochondrial ribosomal proteins, components of the citric acid cycle, the tomm-40 translocase and the coq-3 coenzyme Q3. Comparison of our list of misregulated genes associated with mitochondrial functions to a list of SIN3 targets on chromatin36 revealed that 23 of these 37 genes have SIN-3 binding at their promoter region, and the majority (18/23) are downregulated in sin-3 mutants, including ribosomal protein genes, the tomm-40 mitochondrial translocase, the ATP synthases atp-1 and atp-4, and the cytochrome c oxidase cox-7C (Table S2). SIN-3 may therefore directly promote expression of these genes. Together, our analyses suggest that loss of sin-3 perturbs the coordinated expression of mitochondrial genes encoded by nuclear and mitochondrial genomes, possibly resulting in mito-nuclear imbalance and affecting mitochondrial homeostasis. Consistent with mitochondrial dysfunction, sin-3 mutants show reduced fertility, increased sensitivity to oxidative stress22,33,34,36 and altered lifespan22,33 (Figure S1), as observed in other mitochondrial mutants.37
Transmission electron microscopy reveals extensive mitochondrial fragmentation in sin-3 mutant animals in all tissues examined
Mitochondrial morphology is tightly linked to mitochondrial function, and its steady state is determined by the balance between fission and fusion events that may be disrupted under conditions of mitochondrial stress.38 To test whether mitochondrial morphology is affected in sin-3 mutant animals we performed electron microscopy analyses on wild type and mutant young adults. Mitochondria in the body wall muscles of wild type animals vary in size reflecting dynamic morphological changes, and tend to be cylindrical- or ovular-shaped with regular outer membranes and dense cristae throughout each organelle (Figure 2 panel a, white arrows). In animals carrying the loss of function allele sin-3(tm1276), or the sin-3(syb2172) null allele, mitochondria were instead highly fragmented and appeared more numerous, with a stronger effect in syb2172 animals (Figure 2 panels b and c, white arrows). For both mutants, highly fragmented mitochondria were visible in all of the tissues examined, including intestine, pharynx, hypodermis, and germline (Figures 2A and S2A). We also observed the presence of enlarged or “giant” mitochondria, most prominent in muscle and intestinal cells, and often containing electron-dense material (Figure 2A panels b, c, and e; Figure 2B panels a-d, black arrows) that may indicate iron deposits39,40,41 or protein aggregates.42,43,44,45 Mitochondria morphology is most easily studied in muscle cells on the outer body surface: these are relatively large and flat, making it possible to easily evaluate mitochondria size, shape, and distribution.46 Our analysis revealed that for both mutants, the perimeter and surface area of individual mitochondria in these cells decreased, and the effect was greater for the sin-3(syb2172) allele (Figures 2C and S2B). We also observed an increase in the average circularity index, although this was only statistically significant for the sin-3(syb2172) null allele (Figure 2C), and an increase in the number of individual mitochondria (Figure S2C). These findings suggest that loss of SIN-3 decreases mitochondrial fusion and increases fission events.
Figure 2.
TEM microscopy of mitochondria in sin-3 mutant animals
(A) Transverse sections captured with electron microscopy of mitochondria from the body wall muscles, intestine, pharynx and germline of young adult wild type, sin-3(tm1276) and sin-3(syb2172) mutant animals. Images are representative of a total of 6 animals imaged for wild type, 4 for sin-3(syb2172) and 5 for sin-3(syb2172). Further examples for each genotype are displayed in Figure S1. Single mitochondria are indicated by arrows.
(B) Detailed view of enlarged mitochondria with aggregates. White arrows in A (panels a–j) indicate examples of individual mitochondria, black arrows in A (panels b, c, and e) and B indicate mitochondria with densely stained aggregates. Scale bars are indicated for each image.
(C) Quantification of perimeter, circularity and area of individual mitochondria from TEM images. For each worm, muscle mitochondria were analyzed on several TEM micrographs (at least 2–3 mitochondria on 25 to 60 images per condition), allowing the analysis of 72 mitochondria for wild type N2 (n = 4), 63 for sin-3(tm1276) (n = 4) and 180 for sin-3(syb2172) (n = 4). See also Figure S2.
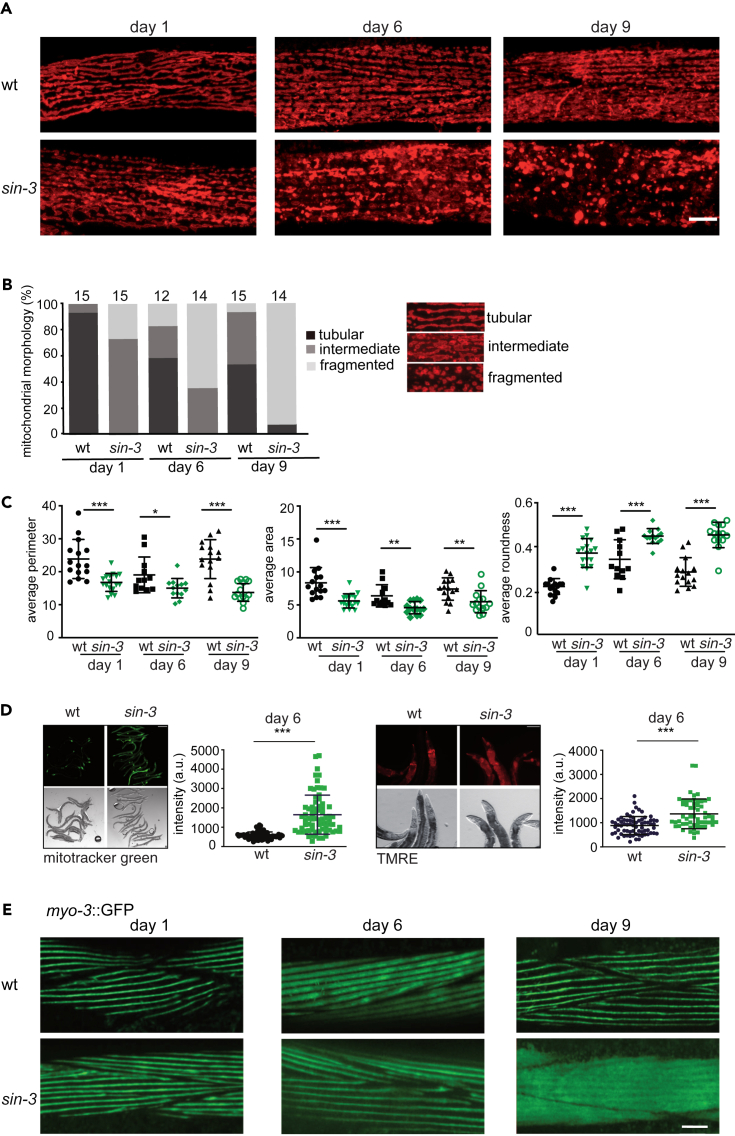
Time-dependent increase in mitochondrial fragmentation in sin-3 muscle cells
To further examine mitochondria dynamics in live animals, we used a transgenic strain expressing a red fluorescent protein (RFP) fused at the N terminus to the TOMM-20 translocase of the outer mitochondrial membrane and expressed under the control of a promoter in muscle cells (myo-3p::20Nter::wrmScarlet)47, where mitochondria are abundant and easily visible. We used tm1276 mutant animals because although less fertile than wild type, they can be maintained as homozygotes, while syb2172 animals are fully sterile.34 In wild type, the majority of body-wall muscle cells have longitudinally arrayed tubular mitochondria, while a smaller percentage show either elongated mitochondria in an interconnected mesh-like network or fragmented mitochondria (Figure 3A). We arbitrarily classified mitochondrial morphology into three classes-tubular, intermediate, or fragmented,48,49 and scored animals falling into each of these at days 1, 6, and 9 of adulthood. In wild-type day 1 adults, the large majority of mitochondria showed a tubular morphology (Figure 3B). Mitochondria of sin-3(tm1276) mutants of the same age were smaller and rounder compared to wild type, consistent with increased fragmentation and TEM results. At day 6, intermediate and fragmented mitochondria also appeared in wild type, when the majority of mitochondria in sin-3 mutants were highly fragmented. At day 9, mitochondrial fragmentation further increased in sin-3(tm1276) mutants, but remained more or less constant in wild type (Figure 3B). As an independent measure of mitochondrial morphology we used ImageJ software analysis (Figure 3C). In agreement with the TEM results, we observed a decrease in the average perimeter and surface area of individual mitochondria in sin-3 mutants, and an increase in roundness (Figure 3C).50,51 Using a mex-5p::tomm-20:mKate2 reporter expressed in the germline, we also observed changes in mitochondrial morphology in this tissue (Figure S2D). Excessive mitochondrial fragmentation results from an imbalance between fusion and fission that are mediated primarily by two classes of GTPases structurally related to dynamins: mitofusins (MFNs)/Fzo1/FZO-1 and Drp1/DRP-1, respectively.38,46,52 Expression of neither gene was altered in sin-3 mutants34 (Table S1). Because of the absence of C. elegans specific antibodies, we were unable to look at protein levels of mitofusin or DRP-1 proteins.
Figure 3.
Altered mitochondria morphology and microfilament structure in sin-3(tm1276) muscle cells
(A) Representative confocal images of mitochondria morphology in body wall muscles expressing myo-3p::tomm-20Nter::wrmScarlet in wild type and sin-3(tm1276) mutant animals. Images were taken at day 1, 6 or 9 of adulthood. Scale bar: 10μm.
(B) Quantification of animals with the indicated muscle mitochondria phenotypes at day 1, 6, or 9 of adulthood. Total number of worms scored for each condition is indicated above bars.
(C) Quantification of morphological parameters of segmented mitochondria from wild type and sin-3(tm1276) at day 1, 6, or 9 of adulthood. Measured parameters are average perimeter, average area and average roundness (see STAR Methods for specifications). Between 12 and 15 animals were analyzed per condition. Data represents mean ± SD. Statistical differences were calculated using unpaired t test or Mann Whitney test (∗p < 0.05, ∗∗p < 0.007, ∗∗∗p < 0.0001).
(D) Quantification of Mitotracker Green and TMRE staining of wild type and sin-3 (tm1276) mutants. Representative images of Mitotracker Green (left panel) and TMRE staining (right panel) are shown accompanied by the corresponding bright field images. Scale bar: 400 μm for Mitotracker Green and 100 μm for TMRE. Data represent the mean ± SD of 2–3 independent replicates, where each data point represents one worm. Statistical differences were calculated using Mann Whitney test (∗∗∗p < 0.0001).
(E) Representative confocal images of myofilament structure in body wall muscles expressing myo-3::GFP in wild type or sin-3(tm1276) mutant animals. Images were taken at day 1, 6, or 9 of adulthood. Scale bar: 10μm.
To measure whether mitochondrial mass increases in sin-3(tm1276) mutants we used MitoTracker Green (MTG), a widely used fluorescence dye that accumulates in mitochondria independent of mitochondrial membrane potential.44 MTG staining was significantly increased in day 6 mutant animals (Figure 3D, left panel). Increased staining in mutant animals was also observed using the membrane potential sensitive dye tetramethylrhodamine, ethyl ester (TMRE) that accumulates in active mitochondria44 (Figure 3D, right panel). Together, these results suggest that loss of SIN-3 leads to an increase in total mitochondrial mass, as suggested by TEM results in young adults.
Muscle fibers degenerate prematurely in sin-3 mutants
Mitochondrial dysfunction impairs muscle health and causes subsequent muscle wasting, commonly referred to as sarcopenia.53 Using a muscle myosin reporter myo-3::GFP54 in wild type young adults sarcomeres appear as straight lines of GFP, and no obvious change was observed up to day 9 (Figure 3E). By contrast, in sin-3(tm 1276) mutants at day 9 we observed diffuse GFP fluorescence, suggesting that muscle integrity is affected in these mutants (Figure 3E). Degradation of muscle proteins may also occur prematurely in these animals.55 Because defects in muscle fibers are observed only in older sin-3 mutant adults, while mitochondrial defects already appear at day 1, these results are consistent with a decline in mitochondrial network structure preceding muscle decline, as previously reported.56,57
Mitochondrial UPR is dampened in the absence of SIN-3
In response to mitochondrial stress, cells can trigger the mitochondrial unfolded protein response (UPRmt) that transmits mitochondrial stress signals to the nucleus to regulate mitochondrial chaperone genes and other factors necessary for the recovery of damaged mitochondria.58 Increased mitochondrial fragmentation in sin-3 mutants suggests that SIN-3 may be required for mitochondrial homeostasis. We therefore asked whether UPRmt is properly activated in sin-3 mutant animals, using the hsp-6 chaperone as a reporter (Haeussler and Conradt 2022). sin-3(tm1276) mutants carrying hsp-6::GFP were sterile, most likely due genetic interaction with a background mutation in the hsp-6::GFP bearing strain (Caehnorhabditis Genetics Center, GCG), so we used RNAi to deplete sin-3. sin-3(RNAi) alone had no reproducible effect on hsp-6::GFP expression (Figures 4A and 4B). Induction of mitochondrial stress by RNAi knockdown of the NADH ubiquinone oxidoreductase component nuo-4, or the mitochondrial ribosomal protein mrps-5, resulted in strong hsp-6::GFP expression in the intestine, as expected.59 Simultaneous depletion of both sin-3 and nuo-4, or sin-3 and mrps-5, significantly decreased GFP expression in older animals at day 6 (Figure 4A), while no significant difference was observed at day 3 (Figure S3). RT-qPCR analysis confirmed knockdown of the respective genes in single and double RNAi experiments (Figure S3B). These results suggest that SIN-3 is required for a full response to mitochondrial stress depending of the animal’s age. Interestingly, components of the NuRD chromatin remodeling complex have been shown to relocate to the nucleus upon mitochondrial stress18 to influence the UPRmt. Using a SIN-3:RFP translational fusion, we observed no obvious change in SIN-3 localization following mitochondrial stress (Figure S4).
Figure 4.
Loss of sin-3 dampens the response to mitochondrial stress
(A) Images of wild type or sin-3(RNAi) animals expressing hsp-6p::GFP reporter under basal conditions (no stress), or following mitochondrial stress induced by RNAi knock-down of mitochondrial genes mrps-5 and nuo-4. Parent worms were grown on HT115 control or sin-3 RNAi from hatch until the adult stage. F1 offspring were then grown from hatch on control, nuo-4 or mrps-5 single RNAi, or sin-3, sin-3 + nuo-4, and sin-3 + mrps-5 double RNAi. GFP expression was measured 5 days after eggs were laid. Scale bar: 300μm.
(B) Quantification of hsp-6p::GFP expression. Data are representative of one of two independent experiments with 30 animals per condition each. Boxplots with median corresponding to the middle of the box, first and third quartiles to the edges of the box, and lines extreme values. Significance was calculated using Mann-Whitney test, p value ∗ <0.05, ∗∗∗∗ <0.0001. See also Figure S3.
Increased oxygen consumption in aged sin-3 mutant animals
To understand how altered mitochondrial structure following loss of SIN-3 relates to mitochondrial function, we measured oxygen consumption in sin-3(tm1276) mutants. In C. elegans, oxygen consumption rates (OCRs) as a measure of ETC function can be accurately assessed using a Seahorse Analyzer with intact animals.37,60 Wild type animals maintained a constant basal OCR from the young adult stage to day 6 of adulthood, although a tendency to reduce basal respiration during aging was apparent, as previously reported (Figure 5A, compare YA to D6).61,62 By contrast, as sin-3 mutants aged, basal OCR dramatically increased, so that at day six of adulthood it was more than 2-fold higher in sin-3 mutants compared to wild type (Figure 5A, D6).
Figure 5.
Oxygen consumption rate (OCR) and abundance of mitochondrial respiratory chain subunits in sin-3(tm1276) mutants
(A) Basal OCR of wild type and sin-3 mutants at the young adult (YA) stage and at day 6 of adulthood (D6).
(B) Maximal OCR of wild type and sin-3 mutants at the YA stage and at D6 of adulthood.
(C) Spare respiratory capacity of wild type and sin-3 mutants at the YA stage and at D6 of adulthood. Significance was calculated using unpaired t-test or, when appropriate, Mann-Whitney test, p value ∗ <0.05, ∗∗∗∗ <0.0001.
(D) Western blot showing SDHB-1/SDHB (complex II), CTC-1/UQCRC2 (complex IV), and ATP-2/ATP5A (complex V) protein levels in total protein extracts from wild type and sin-3(tm1276) L4-young adult worms detected by fluorescence.
(E) Levels of each protein were quantified with ImageJ and normalized to tubulin levels. Bar plot represents the mean of two to three independent biological replicates; error bars correspond to SD.
We next measured maximal respiratory capacity following mitochondrial uncoupling by the addition of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP). In sin-3 young adults, maximal OCR was similar to wild type, but we again observed a significant increase in aged sin-3 mutants compared to age-matched wild type (Figure 5B). Moreover, in old sin-3 mutants the observed increase in basal respiration was proportionally larger than the increase in maximal respiratory capacity (Figures 5A and 5B). Consequently mitochondrial spare capacity, which characterizes the ability of mitochondria to meet extra energy requirements beyond the basal level,63 was reduced compared to old wild type worms (Figure 5C). Increased OCR could be a side effect of partial mitochondrial uncoupling.64 We note, however, that ucp-4, the single mitochondrial uncoupling protein in C. elegans, was not present in the list of sin-3 misregulated genes, while expression of the ant-1.1 ADP/ATP translocator whose activity can also result in mitochondrial uncoupling was down (Table S1).65,66 Consistent with depletion of SIN-3 affecting mitochondrial integrity, in extracts from sin-3(tm1276) animals we observed a trend toward decreased levels of conserved electron transport proteins, detected using antibodies against the highly conserved mammalian complex III subunit UQCRC2 and complex II subunit SDHB1. Protein abundance of the ATP synthase ATP5 was less affected (Figures 5D and 5E).
sin-3 inactivation does not alter levels of TCA cycle metabolites, but results in a signature of mitochondrial stress
The previous results clearly establish that sin-3 knockdown or deletion leads to defective mitochondrial function. Because mitochondria are a hub for biosynthetic processes, we used metabolomic analysis to probe for SIN-3-dependent metabolic changes in animals carrying the sin-3(tm1276) loss of function allele. We used young adult animals, the same stage used for transcriptomics analysis and TEM observations. We chose this allele because the sterility of sin-3(syb2172) null mutants precluded growing the large number of animals required for this type of analysis. We observed no significant change in the abundance of TCA cycle intermediates detected, including succinyl-CoA, alpha-keto-glutaric acid, succinic acid, fumaric acid, and malic acid (Table S4; Figures S5A and S5B). Although two peaks putatively annotated as glycolytic intermediates glucose-6-P and glucose-1-P showed a significant reduction in sin-3 mutants, the downstream metabolite fructose-1,6-bis-P showed no difference (Table S4; Figure S5B). Glucose-1-P is a metabolite of gluconeogenesis and starch and sucrose metabolism. C. elegans can store excess energy in the form of trehalose and glycogen.67 However, we detected no significant change in UDP-glucose (an intermediary for glycogen and trehalose synthesis), trehalose-6-P, or trehalose in sin-3 mutant animals (Table S4; Figure S5B). Together, these results suggest that in young animals the absence of SIN-3 has little or no influence on energy metabolism. This was further corroborated by only minor changes in lipid metabolism intermediates including short-chain acyl-CoA and short-chain acyl-carnitine species (Table S5; Figure S5A). Nonetheless, it is important to note that metabolite levels are only indicative of changes in metabolic flux, defined as the turnover rate of metabolites through enzymatically controlled pathways.68 Increased mitochondrial fragmentation may lead to changes or redirection of metabolic fluxes without altering metabolite levels.
In contrast to the previous results, significant changes in amino acid metabolism were detected: aspartic acid levels significantly decreased in sin-3 mutants, while proline, threonine, lysine, serine, and citrulline increased (Figures 6A and S5C; Table S5). Interestingly, decreased aspartic acid and increased serine have been shown to be a signature of mitochondrial stress.69 No changes in glutathione (GSH) and glutathione disulfide (GSSG), both associated with oxidative stress, were detected (Table S5; Figure S5C). An increase in sarcosine and dimethylarginine was also detected (Table S5; Figure S5C).
Figure 6.
Metabolic alterations in sin-3(tm1276) mutant young adult animals and their related pathways
Changes in amino acid metabolites (A) and polyamine-related metabolites (B) in sin-3(tm1276) mutants at the young adult stage. Welch test, p-value ∗ <0.05, ∗∗ <0.01, ∗∗∗ <0.005.
(C) Schematic representation of biosynthetic pathway for arginine-derived polyamines.
(D) Biosynthetic pathway of cadaverine and lysine degradation. Arrows and colors in (C) and (D) indicate significant changes for metabolites and metabolic genes (blue = down, red = up). Areas shaded in gray indicate specific pathways, or reactions that take place in the mitochondria. See also Figure S5 and Tables S1, S2, S3, S4, and S5.
SIN-3 is required to maintain polyamine homeostasis
Both serine and sarcosine are connected to one-carbon metabolism, which encompasses both the folate and methionine cycles to generate one-carbon units (methyl groups) that are used for the biosynthesis of important precursors and for methylation reactions.70 While no changes in folate cycle intermediates folate, 5-methyl tetrahydrofolic (mTHF) or dihydrofolic (DHF) acid were found (Table S5; Figure S5D), a significant decrease in the methionine cycle metabolite S-adenosylmethionine (SAM) was observed (Figures 6B and S5D; Table S5).
SAM is the major methyl donor in the cell and is critical for the synthesis of phosphatidylcholine, an important component of cellular membranes,71 as well as for the methylation of DNA, RNA, and histones through transmethylation.70 SAM is also connected to polyamine synthesis through the activity of S-adenosylmethionine decarboxylase (adoMETDC/smd-1) that results in the production of decarboxylated S-adenosylmethioninamine (dc-SAM) (Figure 6C). Spermidine synthase (SPDS/spds-1), a key enzyme in the pathway, then catalyzes the transfer of an aminopropyl moiety of dcSAM to putrescine, resulting in the formation of spermidine and 5′-methylthioadenosine (MTA). Putrescine is provided by another key enzyme, ornithine decarboxylase (ODC/odc-1) (Figure 6C). Interestingly while the abundance of downstream metabolites of the transmethylation pathway, S-adenosyl homocysteine (SAH) and cystathionine, was not altered in sin-3 mutants (Figures 6B and S5E; Table S5), both dcSAM and MTA were strongly reduced (Table S5; Figure S5E). Levels of neither arginine nor ornithine, two precursors of putrescine, nor putrescine itself was altered in sin-3 mutants (Figures 6B and S5C; Table S5), while we detected a large increase in both spermidine and N-Acetyl-spermine, the degradation products of spermine and spermidine, respectively (Figures 6C and S5E; Table S5). Spermine could not be detected in our analysis. Together, these results suggest that in the absence of SIN-3, the metabolic flux is shifted from transmethylation to polyamine biosynthesis. Consistent with a shift in metabolic flux toward the synthesis of polyamines, the abundance of N-acetylputrescine, a degradation product of putrescine, was unaltered in our analyses (Table S5; Figure S5D). The abundance of another polyamine, cadaverine, was also increased in sin-3 mutants (Figures 6D and S5E; Table S5). Cadaverine is produced by the decarboxylation of lysine, and is also more abundant in sin-3 mutants (Figures 6D and S5C; Table S5). No dedicated lysine decarboxylase has been annotated in C. elegans to date, but odc-1 could potentially fulfill this function.72
Another lysine catabolite whose abundance increased in sin-3 mutants is saccharopine, while aminoadipic acid (L-2-Aminoadipate), the metabolite further downstream, showed a slight but not significant increase (Figures 6D and S5C; Table S5). Because lysine degradation via the formation of saccharopine is confined to the mitochondria,73 a potential blockage in its degradation due to defective mitochondria could increase its abundance (Figure 6D).
Transcriptomics analysis reveals deregulation of metabolic pathways
The previous metabolic analysis was conducted on sin-3(tm1276) mutant animals at the young adult stage. Close inspection of the previously published list of genes misregulated in these mutants animals at the same stage36 identified candidates whose misexpression correlates with the observed metabolic changes (Figure 6C). The ornithine decarboxylase odc-1, which converts ornithine to putrescine,74 was strongly upregulated in sin-3(tm1276) mutants (log2FC 1.15, p = 4.09e-06), as was the spermidine synthase spds-1 (log2FC 2.29, p = 2.88e-19). Regulation of polyamine synthesis is mainly achieved by controlling the activity of ornithine decarboxylase through its inhibitor antizyme, the binding of which disrupts ODC enzymatic activity and targets it for ubiquitin-independent degradation.75 Significantly, the C. elegans homologue of antizyme, oaz-1, was significantly downregulated (log2FC −0.86, p = 4.29e-32), potentially leading to increased ODC-1 activity and spermidine biosynthesis in these mutants. Because odc-1 may also decarboxylate lysine to produce cadaverin,72 its increased activity could contribute to the observed increase in cadaverin levels. The polyamine oxidase (PAO) hpo-15 is instead upregulated in sin-3(tm1276) mutants (log2FC 1.74, p = 6.56e-08). N-acetylspermine and N-acetylspermidine, metabolites of spermine and spermidine, respectively, are used as substrates by PAO to produce H2O2 and either spermidine or putrescine, depending on the starting substrate. An increased hpo-15 expression may therefore also contribute to both the increase in spermidine levels and increased sensitivity to oxidative stress in sin-3 mutant animals, as observed in mammalian cells.76 Interestingly, spermidine serves as the sole biosynthetic precursor for hypusination, a post-translational modification that is an integral component of eukaryotic translation initiation factor 5A (eIF5A).77 Genes encoding the two enzymes required for hypusination, the deoxyhypusine synthase dhps-1 and the deoxyhypusine hydroxylase dohh-1, were both upregulated in sin-3(tm1276) mutants (log2FC 0.7390, p = 7.89e-27 and 0.44, p = 2.60e-18, respectively). Of these genes only oaz-1 and hpo-15 are included in the list of genes commonly misregulated in the two sin-3 alleles (Table 1), most likely reflecting experimental differences between the two transcriptomics analyses.34 Altogether our data are consistent with deregulation of specific metabolic genes in sin-3(tm1276) mutants contributing to the observed changes in polyamine flux. Whether these changes are a cause or consequence of the observed changes in mitochondrial dynamics and respiration remains to be established.
Discussion
The SIN3 corepressor has been linked to mitochondrial function in several models, but how its loss alters mitochondrial dynamics and metabolism in the context of an entire organism has not been explored. Here, using two different sin-3 mutant alleles, we show that in C. elegans adult animals partial or complete loss of sin-3 function results in increased expression of genes encoded by the mitochondrial genome and an overall decrease in the expression of nuclear-encoded mitochondrial genes, potentially leading to nuclear-mitochondrial imbalance. We show that the altered expression of mitochondrial genes is associated with increased mitochondrial fragmentation in all tissues examined, increased oxygen consumption, and metabolic changes, including a pronounced shift in metabolic flux from methionine to polyamine biosynthesis. Together our data identify SIN-3 as a key regulator of mitochondrial dynamics and polyamine homeostasis.
Knockdown of SIN3 in different systems results in strikingly similar phenotypes. Lower ATP levels were observed following reduction of SIN3 activity in yeast in all media conditions, in Drosophila cultured cells in nutrient-depleted conditions, and in C. elegans adults.20,33 Both Drosophila and C. elegans adult animals show an increase in oxidative stress.22,78 Deregulation of mitochondrial genes was reported following either knockdown or overexpression of SIN3 in cultured Drosophila cells,20,21 or conditional mSin3A knock-out in primary cell culture.79 Our transcriptomics analysis on sin-3(tm1276) and sin-3(syb2178) mutant animals revealed that expression of respiratory complex I subunits encoded by the mitochondrial genome is strongly increased in both mutants, while expression of nuclear-encoded subunits of additional complexes is downregulated. The expression of additional genes with important roles in mitochondrial function, including mitochondrial ribosomal proteins and the tomm-1 translocase is also significantly decreased in sin-3 mutants. Importantly, SIN-3 binding on a number of downregulated genes suggests that SIN-3 may positively regulate their expression, as observed for SIN3 in other contexts.34,80,81,82
The nucleus and mitochondria constantly communicate to adjust their activities in order to ensure cellular homeostasis,83 and reducing the expression of a single major ribosomal nuclear protein (MRP) is sufficient to induce a stoichiometric imbalance between nuclear and mitochondrial-DNA encoded ETC subunits59 and activate the UPRmt.84 Interestingly, the C. elegans histone deacetylase hda-1, encoding a conserved component of SIN3 complexes,34,36 was shown to be required for this stress response.85 We observed that expression of the UPRmt reporter hsp-6 was dampened following induction of mitochondrial stress in sin-3 knockdown animals, but only in old adults. Therefore, it is likely that HDA-1 acts in a context other than SIN-3 in the UPRmt, most likely the NuRD histone deacetylase complex.18
One of the most striking phenotypes we observed is that either reduced SIN-3 function or its complete absence results in a dramatic increase in mitochondrial fragmentation, observed by TEM in all tissues examined, including muscle, intestine, pharynx, hypodermis, and germ cells, and through live imaging in muscle cells. The ability to transition between fission/fusion states is essential for mitochondrial function in cellular bioenergetics, regulation of intracellular Ca2+, and cellular stress responses.86 Fusion allows damaged mitochondria to mitigate stress by mixing contents, while fission contributes to quality control of damaged mitochondria by budding off deteriorating components for targeted breakdown via autophagy or mitophagy. The increased fragmentation we observe is consistent with a defect in the maintenance of mitochondrial homeostasis. We also observed examples of enlarged mitochondria, another hallmark of mitochondrial damage,87 and the accumulation of electron-dense material in mitochondria that may represent iron deposits or misfolded proteins. Similar aggregates were observed by TEM in C. elegans following acute heat stress,42 in sperm-derived mitochondria before their elimination by mitophagy,43,44 and in the mitochondria of worms containing mutations in eat-3/OPA1 and fzo-1/mfn1 fusion genes,88 supporting a link between their formation and mitochondrial morphological defects. Alternatively, these aggregates may represent iron deposits such as those observed in the mitochondria of a mouse model of Friedrich ataxia39,89 and congenital sideroblastic anemia.90 Interestingly mitochondrial iron overload promotes ROS production, as observed in sin-3 mutants (Pandey et al., 2018), and is associated with mitochondrial dysfunction and oxidative damage.91 Regardless of the underlying cause, disruption of mitochondria homeostasis, as observed in sin-3 mutant animals, is likely to have major consequences for mitochondrial health and function.
The rate of oxygen consumption is another measure of mitochondrial activity, and we observed significant differences in respiration in sin-3(tm1276) mutants. While neither basal nor maximal OCRs were affected in mutant young adults, both were significantly elevated in older mutants compared to wild type. Conversely, mitochondrial spare respiratory capacity was reduced in these mutants. Spare respiratory capacity can be viewed as a determinant of mitochondrial fitness and correlates with level of mitochondrial plasticity, allowing bioenergetic adaptability in response to pathophysiological stress conditions.63 Its decrease in older sin-3(tm1276) mutants is consistent with mitochondrial dysfunction in these animals.
Increased basal and maximal oxygen consumption in old sin-3(tm1276) animals contrasts with wild type, in which oxygen consumption has a tendency to decrease with age.60,62 Interestingly, a similar increase during aging was observed in worms lacking the mitochondrial prohibitin (PHB) complex subunits phb-1 and phb-2.92 Like sin-3 mutants, prohibiting mutants are also short-lived and show severely fragmented mitochondria.93,94 PHB plays a prominent role in the response to mitochondrial stress, quality control, biogenesis, and degradation,95 and its expression is induced by metabolic stress resulting from mito-nuclear imbalance, but not other cellular stresses.96 It was suggested that PHB may act as chaperone in respiratory complex assembly,97 with its loss affecting complex integrity. Likewise, imbalance between nuclear- and mitochondrial-encoded subunits in sin-3 mutants could result in defects in respiratory complex assembly. In this context, increased oxygen consumption may reflect compensatory activity from properly assembled supercomplexes, or mitochondrial adaptation resulting in an increase in mitochondrial mass,98 as we observe in sin-3 mutants.
Consistent with mitochondrial fragmentation being generally associated with metabolic dysfunction and disease,86 metabolomic analysis revealed dramatic changes in the levels of several metabolites in sin-3(tm1276) mutants. One of the most striking differences we observed is an increase in the polyamines spermine and cadaverine. Polyamine biosynthesis is tightly controlled, and changes in polyamine levels can have dramatic consequences on physiology, with established links between polyamine metabolism and human diseases including cancer and diabetes.99 Spermine is a potent free radical scavenger and an important antioxidant,100,101 and in C. elegans, spermidine has been shown to inhibit neurodegeneration and have pro-longevity effects.102 However, spermidine or spermine in excess can also have deleterious consequences on animal physiology,101,103,104,105 and SIN-3 inactivation or knock-down in C. elegans is associated with sterility, decreased longevity, and increased oxidative stress (Sharma et al., 2018). Our expression profiling suggests that decreased expression of the oaz-1 antizyme, a critical regulator of polyamine biosynthesis,75 may be a major contributor to the effect of SIN3 knockdown on polyamine homeostasis.
Among a broad range of functions, spermidine serves as the sole biosynthetic precursor for hypusination, a post-translational modification that is an integral component of eukaryotic translation initiation factor 5A (eIF5A).106 Polyamine levels are elevated in most cancers, and hypusinated eIF5A is a critical regulator of cell growth.104 More recently, hypusinated eIF5A was shown to promote expression of mitochondrial proteins in macrophages,107 suggesting that it may also alter mitochondrial activity in both normal and cancer cells. This raises the intriguing possibility that increased spermidine biosynthesis and eIF5A hypusylation in sin-3 mutants may be part of a mechanism to increase the expression of mitochondrial proteins in response to mitochondrial damage.
Further evidence for a shift in metabolic flux toward polyamine biosynthesis comes from the analysis of downstream metabolites in the transmethylation pathway. Conversion of SAM to dcSAM provides necessary amino-propyl groups to sustain polyamine synthesis.104 The methionine salvage pathway (MSP) recycles one carbon unit lost during polyamine synthesis back to the methionine cycle for SAM replenishment. In addition to showing a strong decrease in dcSAM, and a smaller but significant decrease in SAM, sin-3 mutants also show reduced activity of the salvage pathway, as illustrated by a decrease in levels of the MSP intermediate methyladenosine (Figure 6C). Interestingly, in Prostate cancer (PCa) cells, which have an intrinsically high polyamine metabolic flux and therefore rely heavily on the methionine salvage pathway, MSP inhibition while maintaining high polyamine flux was shown to be an effective cancer therapy by blocking the ability of the cell to mitigate this stress, leading to cell death.108 In other tumor cell types, by contrast, defects in the MSP were found to increase polyamine levels, highlighting how the stress that is generated by metabolic perturbations is often context-dependent.104 Because increasing evidence points to a causal link between mitochondrial dysfunction and pediatric neurodevelopmental disorders, our study may also suggest new lines of investigation into the interconnected links between SIN3, mitochondrial homeostasis, and metabolism in the context of SIN3-related disorders.
Limitations of the study
While changes in respiration were only observed in older sin-3 mutant animal, the transcriptomics and metabolomic studies reported here were carried out on young adults, so that relevant changes that may occur later in life could have been missed. We also do not know whether the metabolic changes reported for sin-3 mutants are a cause or consequence of mitochondrial defects. Metabolomic analysis of older animals, as well as genetic analysis, will help clarify these outstanding questions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IRDye 680RD Goat anti-Mouse IgG | LI-COR Biosciences | Cat# 926-68070, RRID:AB_10956588 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, DyLight™ 800 | Thermo Fisher Scientific | Cat# SA5-10036, RRID:AB_2556616 |
| Anti-alpha Tubulin antibody - Microtubule Marker | Abcam | Cat# ab18251, RRID:AB_2210057 |
| Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat# ab110413, RRID:AB_2629281 |
| Bacterial and virus strains | ||
| sin-3 (RNAi) | Kamath, R.S. et al.109 | https://www.dnaform.jp/en/products/clone/c_elegans_rnai_library/I-4K03 |
| mrps-5 (RNAi) | Kamath, R.S. et al.109 | https://www.dnaform.jp/en/products/clone/c_elegans_rnai_library/V-8K16 |
| nuo-4 (RNAi) | Kamath, R.S. et al.109 | https://www.dnaform.jp/en/products/clone/c_elegans_rnai_library/III-3F02 |
| E. coli OP50 | CGC | WB Cat# WBStrain00041969 |
| E. coli RNAi feeding strain L4440 HT115(DE3) | source bioscience | WB Cat# WBStrain0004107 |
| Chemicals, peptides, and recombinant proteins | ||
| Tetramethylrhodamine, ethyl ester, perchlorate (TMRE) | Invitrogen™ | Cat# T669 |
| MitoTracker™ Dyes for Mitochondria Labeling | Invitrogen™ | Cat# M7512 |
| formvar coated grid, stabilized with evaporated carbon film | Electron Microscopy Science | Cat# FCF2010-Cu |
| Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) | Sigma-Aldrich | Cat#C2920 CAS : 370-86-5 |
| Sodium azide | Sigma-Aldrich | Cat#13412 CAS: 26628-22-8 |
| Deposited data | ||
| Raw transcriptomics data | Caron et al.34 | GEO : GSE227499 |
| Raw metabolomics files | This paper |
https://www.ebi.ac.uk/metabolights/index: MTBLS8439 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain N2 Bristol | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00000001 |
| C. elegans: Strain SJ4100 zcIs13[hsp-6p::GFP+lin-15(+)] | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00034068 |
|
C. elegans: Strain PFR590 sin-3(tm1276)I |
Beurton et al.36 | N/A |
|
C. elegans: Strain PHX2172 sin-3(syb2172)/hT2[bli-4(e937)let-?(q782)qIs48]I |
Caron et al.34 | N/A |
| C. elegans: Strain EN7714 krSi134[myo-3p::tomm-20Nter::wrmScarlet] | Roy et al.,47 | N/A |
| C. elegans: Strain EGD629 eagxSi155[mex-5p::tomm-20::mKate2::pie-13’UTR+unc-119(+)]II | Fan X, et al.110 | N/A |
| C. elegans: Strain KAG420 kagIs4[gfp::myo-3]V | Mergoud Dit Lamarche et al.54 | N/A |
|
C. elegans: Strain PFR750 sin-3(tm1276)I;kagIs4[gfp::myo-3]V |
This study | N/A |
|
C. elegans: Strain PFR754 sin-3(tm1276)I;krSi134[myo-3p::tomm-20Nter::wrmScarlet] |
This study | N/A |
|
C. elegans: Strain PFR758 sin-3(tm1276);egxSi155[mex-5p::tomm-20::mKate2::pie-13’UTR+unc-119(+)]II |
This study | N/A |
|
C. elegans: Strain PFR669 sin-3::mCherry(syb521);oxIs279[pie-1p::GFP::H2B+unc-119(+)]II |
This study | N/A |
| Oligonucleotides | ||
| sin-3_for:AGGGCCAACAGTCAACAACA | This study | N/A |
| sin-3_rev:GACGATGATGGGCCAGGATT | This study | N/A |
| mrps-5_for:CAACTGGCCGAACGAAAAGG | This study | N/A |
| mrps-5_rev:CTCGGTCTTGAGTTCAGTGGA | This study | N/A |
| nuo-4_for:GGACACCATCTTCCAGAACCA | This study | N/A |
| nuo-4_rev:TCCGAATCCAAATCCAGCGA | This study | N/A |
| act-1_for:gctggacgtgatcttactgattacc | Hoogewijs D et al.111 | N/A |
| act-1_rev:gtagcagagcttctccttgatgtc | Hoogewijs D et al.111 | N/A |
| cdc-42_for:ctgctggacaggaagattacg | Hoogewijs D et al.111 | N/A |
| cdc-42_rev:ctcggacattctcgaatgaag | Hoogewijs D et al.111 | N/A |
| Software and algorithms | ||
| ImageJ | https://imagej.net/ | ImageJ (RRID:SCR_003070) |
| GraphPad Prism | http://www.graphpad.com/ | GraphPad Prism (RRID:SCR_002798) |
| viridis | https://cran.r-project.org/web/packages/viridis/vignettes/intro-to-viridis.html | viridis (RRID:SCR_016696) |
| ggsignif | https://cran.r-project.org/package=ggsignif | ggsignif (RRID:SCR_023047) |
| readxl | https://cran.r-project.org/web/packages/readxl/index.html | readxl (RRID:SCR_018083) |
| scales | https://CRAN.R-project.org/package=scales | scales (RRID:SCR_019295) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Francesca Palladino (francesca.palladino@ens-lyon.fr).
Materials availability
C. elegans strains generated in this study are available upon request.
Data and code availability
-
•
Metabolomics data have been deposited at MetaboLights with the study ID MTBLS8439 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw transcriptomics data were deposited in GEO with accession number GSE227499 (Caron et al.34). Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
C. elegans strains were cultured on Nematode Growth Medium (NGM) agarose plates with Escherichia coli OP50 and incubated at 20°C. The following strains were used: wild type N2 Bristol, PFR590 sin-3(tm1276)I, PHX2172 sin-3(syb2172)/hT2[bli-4(e937)let-?(q782)qIs48]I, KAG420 kagIs4[gfp::myo-3]V, PFR750 sin-3(tm1276)I;kagIs4[gfp::myo-3]V, EN7714 krSi134[myo-3p::tomm-20Nter::wrmScarlet], PFR754 sin-3(tm1276)I;krSi134, EGD629 egxSi155[mex-5p::tomm-20::mKate2::pie-1 3’UTR + unc-119(+)]II, PFR758 sin-3(tm1276); egxSi155, PFR669 sin-3::mCherry(syb521); oxIs279[pie-1p::GFP::H2B + unc-119(+)]II; SJ4100 zcIs13[hsp-6p::GFP+lin-15(+)]. PFR750 and PFR754 were obtained by crossing sin-3(tm1276) hermaphrodites with GFP::myo-3 or myo-3p::tomm-20Nter::wrmScarlet males, respectively.
Method details
Gene enrichment analysis
Enrichment analysis was carried out using WormCat (http://wormcat.com/), a nematode-specific gene ontology enrichment analysis and visualisation tool allowing for analysis of specific pathways. For DEGs functional analysis, significantly up- and downregulated genes were input to WormCat35 using default settings. Significance scores are reported as Fisher’s exact test P-values.
Transmission electron microscopy
Worms (young adults) were fixed by HPF with EMPACT-2 (Leica Microsystems) and then freeze substituted in anhydrous acetone containing 1% OsO4, 0.5% glutaraldehyde and 0.25% uranyl acetate for 60 h in a FS system (AFS-2, Leica Microsystems). Larvae were embedded in Epon-Araldite mix (EMS hard formula). To gain better anteroposterior orientation and sectioning, adhesive frames were used (11560294 GENE-FRAME 65 μl, Thermo Fisher Scientific) for flat-embedding, as previously described.112 Ultrathin sections were cut on an ultramicrotome (UC7; Leica Microsystems) and collected on formvar-coated slot grids (FCF2010-CU, EMS). Each larva was sectioned in five different places with ≥10 μm between each grid to ensure that different cells were observed. Each grid contained at least 5-10 consecutive sections of 70 nm. TEM grids were observed using a JEM-1400 transmission electron microscope (JEOL) operated at 120 kV, equipped with a Gatan Orius SC200 camera (Gatan) and piloted by the Digital Micrograph program. To quantify mitochondria, we used Image J. The clearly discernible outlines of the mitochondria were traced with the magic wand tool and the number of mitochondria was counted; the surface area (size of the mitochondria) is expressed in square micrometers; the perimeter in micrometers. Circularity [4π·(area/perimeter 2)] is an index with values of 1 indicating perfect spheroids.
Quantification of muscle mitochondria and imaging of myofilament structure
For mitochondria and myofilament observation in muscle cells, staged worms at the young adult stage were mounted on agarose pads in M9 solution containing 10% sodium azide. Muscles in the posterior part of worms were imaged. Z-stack images were acquired using a Zeiss LSM980 inverted confocal microscope with 63× oil immersion objective. Z-stacks were acquired every 0.27μm to image mitochondria and every 0.23μm to image myofilaments. Images shown are projections using max intensity method in Fiji. Manual quantification of mitochondrial morphology in body wall muscle cells was performed in double blind experiments according to the following criteria: cells containing long interconnected mitochondrial networks were classified as tubular, cells containing a combination of interconnected mitochondrial networks along with smaller fragmented mitochondria were classified as intermediate, and cells with sparse small round mitochondria were classified as fragmented.48 Automated quantification was based on Boch and Calvo (2019), following conversion of raw images to binary images using ImageJ. Briefly, maximum projection was applied to z-stacks. To correct for heterogeneous background, a median filter (kernel radius=20) was subtracted. To remove salt and pepper noise after subtraction, a median filter (kernel radius=1) was applied to the resulting image. Finally, images were binarised using the auto threshold Li's method.113 The "analyze particles filter" (size=1-Infinity; exclude on edges) was used prior quantification.
TMRE and Mitotracker staining
Mitochondrial membrane potential was measured using the tetramethylrhodamine ethyl ester dye (TMRE, Invitrogen) and mitochondrial content was measured using Mitotracker Green FM (MTG, Invitrogen). Synchronized animals were grown at 20°C until the appropriate stage (day 5 adulthood) and then transferred to NGM plates in the presence of the dye (2,5 μM TMRE or 100 nm MTG) for 17 h. Next, for intestinal clearing and prior to imaging, worms were transferred 1h to NGM plates without dye. Animals were then mounted on 2% agarose pads with 10 mM Levamisole and imaged using an ORCA-Flash4.0 LT Hamamatsu digital camera on a Leica M205 Stereoscope equipped with a Plan Apo 5.0x/0.50 LWD (TMRE) or Plan Apo 1x (MTG) objective. Segmentation of the head (TMRE) or the whole worm (MTG) was done with the free hand tool from ImageJ software. Emission intensity was measured on greyscale images with a pixel depth of 16 bits. Aged worms with internal organ extrusion through the vulva were censored. At least two independent assays (11-31 worms each) were performed and the combined data was analyzed using theGraphPad Prism software.
Activation of UPRmt by RNAi treatment
RNAi bacteria were induced by directly adding 1 mM IPTG directly onto the bacteria lawn 2 hours before the transfer of worms 112. Transgenic animals expressing hsp-6::GFP were synchronized by bleaching, and the L1 stage animals obtained transferred to empty vector HT115 or sin-3(RNAi) 35mm plates, and allowed to develop to egg-laying adults (P0). 4-6 P0 adults from each RNAi treatment were then individually transferred onto: 1) empty vector control; 2) single RNAi (sin-3, nuo-4, mrps-5) or double RNAi plates (sin-3/nuo-4, sin-3/mrps-5) to induce mitochondrial stress in F1 progeny. sin-3 RNAi treatment was started in P0 animals to ensure depletion of maternal SIN-3 protein in scored animals. GFP expression was assessed in F1 progeny 5 days after egg lay. For imaging, worms were immobilized using 2.5mM Levamisole in M9 buffer (22mM KH2PO4, 42mM Na2HPO4, 86mM NaCl, 1mM MgSO4). Images were acquired with a Nikon AZ100M microscope.
Oxygen consumption measurements
Worm oxygen consumption was measured using the Agilent Seahorse XFp Analyzer. Animals were synchronized by allowing gravid mothers to lay eggs for 2-3 hours before removing them from plate. Worms (20-30 per well) at the young adult stage or at day 6 of adulthood were transferred into M9-filled Seahorse plates. Working solutions were diluted in M9 at the following final concentrations: Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (Sigma-Aldrich) 250 μM, NaN3 (Sigma-Aldrich) 400 mM. Oxygen Consumption Rate (OCR) measurements were repeated 8 times in basal conditions, 10 times after FCPP injection (for maximal respiration), and 4 times after NaN3 injection (no mitochondrial respiration). Each cycle consisted of 3 min mix, 30 second wait, and 3 min measure. Values were normalized per worm. Three independent assays were carried out and the combined data was analyzed by Unpaired t-test, or, when appropriate, Mann-Whitney test, using GraphPad Prism software.
Western blot analysis
Young adult worms were collected in M9 buffer, washed 3 times, pelleted and frozen in dry ice. Pellets were resuspended in TNET buffer (50 mM Tris·HCl (pH 8), 300 mM NaCl, 1 mM EDTA, 0,5% Triton X-100 and cOmplete™ Protease Inhibitor Cocktail [Merck # 11697498001]) and lysed with zirconium beads [Lysing Matrix Y, MP Biomedicals #116960050] using a Precellys24 homogenizer [Ozyme] with the following parameters: 6000 rpm 2x20sec. Homogenates were centrifuged and supernatants aliquoted and frozen at −80°C. Total protein amount was quantified by the Bradford assay [Bio-Rad]. 10,20,30,40,50 and 80 ug of protein extracts were loaded either on 12% NuPage Novex or 12 % SDS PAGE gels for western blot analysis. After transfer, membranes were incubated overnight with total OXPHOS Rodent WB Antibody Cocktail [Abcam #ab110413] diluted at 1:500 or anti-alpha-tubulin [Abcam #ab18251] diluted at 1:5,000. The next day, membranes were incubated for 1 hour with goat anti-rabbit DyLight™ 800 [Invitrogen # SA5-10036] and IRDye® 680RD goat anti-mouse [LiCOR #926-68070] diluted at 1:10,000. Aquisition was performed on a ChemiDoc MP apparatus [Bio-Rad]. Quantification was carried out using Image J, and each protein signal was normalized to the level of tubulin. Two to three independent biological replicates and three to four technical replicates were used for quantification.
Metabolite extraction and profiling
Embryos derived from bleaching were transferred onto 90 mm plates seeded with 0P50 bacteria and left to develop to the young adult stage. Animals were collected in M9 medium, washed 3 times in 10 ml M9, and pellets quick frozen in liquid nitrogen114 prior to processing. 8 replicas, each containing approximately 2000 worms, were processed per genotype. All chemicals and solvents were of LC-MS or analytical grade. C. elegans samples were thawed on ice and 1 ml of ice-cold H2O/MeOH/CHCl3 (1/3/1, v/v/v). After suspension in solvent, samples were transferred to 2 ml MN Bead Tubes Type A (Macherey-Nagel, Düren, Germany) and lysed using a Precellys Bead Beating system with an additional Cryolys cooling module (Bertin Instruments, Montigny-le-Bretonneux, France). After lysis, samples were incubated for 10 minutes in an ice-cold ultrasonic bath, followed by centrifugation at 4°C and 13,000 rpm for 15 minutes. The supernatant was transferred to a fresh reaction tube and evaporated to dryness using a centrifugal evaporator. Proteins were extracted from residue debris pellets and quantified using a BCA kit (Sigma-Aldrich, Taufkirchen, Germany). Metabolite profiling was performed using a Sciex ExionLC AD coupled to a Sciex ZenoTOF 7600 under the control of Sciex OS 3.0 (Sciex, Darmstadt, Germany). Separation was achieved on an Agilent InfinityLab Poroshell 120 HILIC-Z column (2.1 mm x 150 mm, 2.7 μm particle size, PEEK-lined) (Agilent Technologies, Waldbronn, Germany). Different eluents and gradients were applied for positive and negative ionization modes. In positive ionization mode, eluent A consisted of 100% H2O + 10 mM ammonium formate / 0.1% formic acid, and eluent B consisted of 10% H2O / 90% ACN + 10 mM ammonium formate / 0.1% formic acid. In negative ionization mode, eluent A consisted of 100% H2O + 10 mM ammonium acetate / 2.5 μM medronic acid, pH = 9, and eluent A consisted of 15% H2O / 85% ACN + 10 mm ammonium acetate / 2.5 μM medronic acid, pH = 9. The column temperature was set to 25°C and 50°C for positive and negative ionization modes, respectively, and the flow rate was 0.25 mL/min in both cases. Gradients for both ionization modes are summarized in Table S3. Dried samples were re-dissolved in 50 μL H2O/ACN (1/3, v/v), and 40 μL were transferred to an autosampler vial and 10 μL to a pooled QC sample. Autosampler temperature was set to 5°C and 5 μl were injected for analysis. In MS1, ions in the m/z range 70 to 1500 were accumulated for 0.1s, and information-dependent acquisition (IDA) of MS2 was used with a maximum number of 6 candidate ions and a collision energy of 35 eV with a spread of 15 eV. Accumulation time for MS2 was set to 0.025 seconds, yielding a total cycle time of 0.299 seconds. ZenoTrapping was enabled with a value of 80000. QC samples were used for conditioning the column and were injected every ten samples. Automatic calibration of the mass spectrometer in MS1 and MS2 mode was performed every five injections using the ESI positive Calibration Solution for the Sciex X500 system or the ESI negative Calibration Solution for the Sciex X500 system (Sciex, Darmstadt, Germany).
Metabolite data analysis
Data analysis was performed in a targeted fashion for identified metabolites (see Tables S4 and S5). Metabolites were identified by comparison to in-house reference standards, publicly available reference spectra, and manual interpretation of fragmentation spectra. Data analysis was performed in Sciex OS 3.0.0.3339 (Sciex, Darmstadt, Germany). Peaks for all metabolites indicated in Tables S4 and S5 were integrated with an XIC width of 0.02 Da and a Gaussian smooth width of 3 points using the MQ4 peak-picking algorithm. Peak areas were exported to a .txt file and normalized according to the protein content of the respective sample. All further processing was performed in R 4.2.1 within RStudio using the following packages: tidyverse (1.3.2), readxl (1.4.1), ggsignif (0.6.4), gghalves (0.1.4), scales (1.2.1) and viridis (0.6.2). Data were plotted using ggplot with gghalves using half box- and half dot-plots. Significance was tested using a Welch test within ggsignif.
Quantification and statistical analysis
Details of all quantification methods and statistical analyses used in this paper are reported in the relevant STAR Methods sections and figure legends, and summarized hereafter. Figure 1: Significance scores are reported as Fisher’s exact test p-value; Figure 2: at least 2-3 mitochondria on 25 to 60 images per condition scored; Figure 3: total number of worms scored for each condition is indicated above bars, measured parameters are average perimeter, average area and average roundness (as specified in material and methods, 12-15 animals per condition), data represents mean ± SD calculated using unpaired t test or Mann Whitney test. Quantification of Mitotracker Green and TMRE staining represent the mean ± SD of 2-3 independent replicates, where each data point represents one worm and Statistical differences were calculated using Mann Whitney test; Figure 4: data is representative of one of two independent experiments with 30 animals per condition each, repeated 8 times in basal conditions, 10 times after FCPP injection, and 4 times after NaN3 injection, with three independent assays carried out and the combined data analyzed by Unpaired t-test, or, when appropriate, Mann-Whitney test, using GraphPad Prism software. For western blot levels of each protein were quantified with Image J. Bar plot represents the mean of two to three independent biological replicates; error bars correspond to SD; Figure 6: p-value calculated using a Welch test. All softwares used for quantification and statistical analysis are reported in the Method details and in the key resources table.
Acknowledgments
F.P. was supported by ANR (Agence Nationale de la Recherche) grant N° 19-CE12-0025-01 and the Centre National de la Recherche Scientifique. M.A.S. was funded through grants from the Ministerio de Ciencia, Innovación y Universidades, the Agencia Estatal de Investigación (AEI) PID 2019-104145GB-I00. G.M. laboratory receives institutional funding from the Centre National de la Recherche Scientifique and the Université de Rennes. TEM imaging was performed at the Microscopy Rennes imaging Center (Biosit, Rennes, France), a member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04). Thanks to Emiliano Ricci for OXPHOS antibodies, Florence Solari for strains. Special thanks to Liesbeth de Jong for the graphical abstract.
Author contributions
P.F., F.P., and M.A.S. conceived and designed experiments, P.F., M.G., M.J.R.P., M.A.S., and C.B. performed experiments and analyzed the data, G.M. conceived and supervised E.M. experiments and quantification with O.N. and M.W. conceived, performed, and analyzed metabolomic studies, F.P. wrote the paper, M.A.S., M.W., and G.M. contributed to specific sections, and all authors contributed to figures. F.P. and M.A.S. share responsibility for all aspects of this work.
Declaration of interests
The authors declare no competing interests.
Published: April 22, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109789.
Contributor Information
Marta Artal-Sanz, Email: martsan@upo.es.
Francesca Palladino, Email: francesca.palladino@ens-lyon.fr.
Supplemental information
References
- 1.Signes A., Fernandez-Vizarra E. Assembly of mammalian oxidative phosphorylation complexes I–V and supercomplexes. Essays Biochem. 2018;62:255–270. doi: 10.1042/EBC20170098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter-Dennerlein R., Oeljeklaus S., Lorenzi I., Ronsör C., Bareth B., Schendzielorz A.B., Wang C., Warscheid B., Rehling P., Dennerlein S. Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein. Cell. 2016;167:471–483.e10. doi: 10.1016/j.cell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMauro S., Schon E.A. Mitochondrial Respiratory-Chain Diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Vizarra E., Zeviani M. Mitochondrial disorders of the OXPHOS system. FEBS Lett. 2021;595:1062–1106. doi: 10.1002/1873-3468.13995. [DOI] [PubMed] [Google Scholar]
- 5.Reeve A.K., Krishnan K.J., Turnbull D. Mitochondrial DNA Mutations in Disease, Aging, and Neurodegeneration. Ann. N. Y. Acad. Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- 6.Taanman J.-W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999;1410:103–123. doi: 10.1016/S0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 7.Ali A.T., Boehme L., Carbajosa G., Seitan V.C., Small K.S., Hodgkinson A. Nuclear genetic regulation of the human mitochondrial transcriptome. Elife. 2019;8 doi: 10.7554/eLife.41927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barshad G., Blumberg A., Cohen T., Mishmar D. Human primitive brain displays negative mitochondrial-nuclear expression correlation of respiratory genes. Genome Res. 2018;28:952–967. doi: 10.1101/gr.226324.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.-M.J., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A., et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quirós P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 11.Anderson N.S., Haynes C.M. Folding the Mitochondrial UPR into the Integrated Stress Response. Trends Cell Biol. 2020;30:428–439. doi: 10.1016/j.tcb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matilainen O., Quirós P.M., Auwerx J. Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends Cell Biol. 2017;27:453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Yang R., Li Y., Wang Y., Zhang J., Fan Q., Tan J., Li W., Zou X., Liang B. NHR-80 senses the mitochondrial UPR to rewire citrate metabolism for lipid accumulation in Caenorhabditis elegans. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110206. [DOI] [PubMed] [Google Scholar]
- 14.Rolland S., Conradt B. Genetic screen identifies non-mitochondrial proteins involved in the maintenance of mitochondrial homeostasis. MicroPubl. Biol. 2022;2022 doi: 10.17912/micropub.biology.000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolland S.G., Schneid S., Schwarz M., Rackles E., Fischer C., Haeussler S., Regmi S.G., Yeroslaviz A., Habermann B., Mokranjac D., et al. Compromised Mitochondrial Protein Import Acts as a Signal for UPRmt. Cell Rep. 2019;28:1659–1669.e5. doi: 10.1016/j.celrep.2019.07.049. [DOI] [PubMed] [Google Scholar]
- 16.Lionaki E., Gkikas I., Daskalaki I., Ioannidi M.-K., Klapa M.I., Tavernarakis N. Mitochondrial protein import determines lifespan through metabolic reprogramming and de novo serine biosynthesis. Nat. Commun. 2022;13:651. doi: 10.1038/s41467-022-28272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., Meyer B.J., Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR mt. Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu D., Wu X., Zhou J., Li X., Huang X., Li J., Wu J., Bian Q., Wang Y., Tian Y. NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaubal A., Pile L.A. Same agent, different messages: insight into transcriptional regulation by SIN3 isoforms. Epigenet. Chromatin. 2018;11:17. doi: 10.1186/s13072-018-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes V.L., Strunk B.S., Lee I., Hüttemann M., Pile L.A. Loss of the SIN3 transcriptional corepressor results in aberrant mitochondrial function. BMC Biochem. 2010;11:26. doi: 10.1186/1471-2091-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pile L.A., Spellman P.T., Katzenberger R.J., Wassarman D.A. The SIN3 Deacetylase Complex Represses Genes Encoding Mitochondrial Proteins. J. Biol. Chem. 2003;278:37840–37848. doi: 10.1074/jbc.M305996200. [DOI] [PubMed] [Google Scholar]
- 22.Sharma M., Pandey R., Saluja D. ROS is the major player in regulating altered autophagy and lifespan in sin-3 mutants of C. elegans. Autophagy. 2018;14:1239–1255. doi: 10.1080/15548627.2018.1474312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scoville D.W., Cyphert H.A., Liao L., Xu J., Reynolds A., Guo S., Stein R. MLL3 and MLL4 Methyltransferases Bind to the MAFA and MAFB Transcription Factors to Regulate Islet β-Cell Function. Diabetes. 2015;64:3772–3783. doi: 10.2337/db15-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Graff S.M., Heiser C.N., Ho K.-H., Chen B., Simmons A.J., Southard-Smith A.N., David G., Jacobson D.A., Kaverina I., et al. Coregulator Sin3a Promotes Postnatal Murine β-Cell Fitness by Regulating Genes in Ca2+ Homeostasis, Cell Survival, Vesicle Biosynthesis, Glucose Metabolism, and Stress Response. Diabetes. 2020;69:1219–1231. doi: 10.2337/db19-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To T.-L., Cuadros A.M., Shah H., Hung W.H.W., Li Y., Kim S.H., Rubin D.H.F., Boe R.H., Rath S., Eaton J.K., et al. A Compendium of Genetic Modifiers of Mitochondrial Dysfunction Reveals Intra-organelle Buffering. Cell. 2019;179:1222–1238.e17. doi: 10.1016/j.cell.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramanian M., Dingemans A.J.M., Albaba S., Richardson R., Yates T.M., Cox H., Douzgou S., Armstrong R., Sansbury F.H., Burke K.B., et al. Comprehensive study of 28 individuals with SIN3A-related disorder underscoring the associated mild cognitive and distinctive facial phenotype. Eur. J. Hum. Genet. 2021;29:625–636. doi: 10.1038/s41431-020-00769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latypova X., Vincent M., Mollé A., Adebambo O.A., Fourgeux C., Khan T.N., Caro A., Rosello M., Orellana C., Niyazov D., et al. Haploinsufficiency of the Sin3/HDAC corepressor complex member SIN3B causes a syndromic intellectual disability/autism spectrum disorder. Am. J. Hum. Genet. 2021;108:929–941. doi: 10.1016/j.ajhg.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witteveen J.S., Willemsen M.H., Dombroski T.C.D., van Bakel N.H.M., Nillesen W.M., van Hulten J.A., Jansen E.J.R., Verkaik D., Veenstra-Knol H.E., van Ravenswaaij-Arts C.M.A., et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 2016;48:877–887. doi: 10.1038/ng.3619. [DOI] [PubMed] [Google Scholar]
- 29.Bansal N., Petrie K., Christova R., Chung C.-Y., Leibovitch B.A., Howell L., Gil V., Sbirkov Y., Lee E., Wexler J., et al. Targeting the SIN3A-PF1 interaction inhibits epithelial to mesenchymal transition and maintenance of a stem cell phenotype in triple negative breast cancer. Oncotarget. 2015;6:34087–34105. doi: 10.18632/oncotarget.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal N., David G., Farias E., Waxman S. Emerging Roles of Epigenetic Regulator Sin3 in Cancer. Adv. Cancer Res. 2016;130:113–135. doi: 10.1016/bs.acr.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Farias E.F., Petrie K., Leibovitch B., Murtagh J., Chornet M.B., Schenk T., Zelent A., Waxman S. Interference with Sin3 function induces epigenetic reprogramming and differentiation in breast cancer cells. Proc. Natl. Acad. Sci. USA. 2010;107:11811–11816. doi: 10.1073/pnas.1006737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis M.J., Liu J., Libby E.F., Lee M., Crawford N.P.S., Hurst D.R. SIN3A and SIN3B differentially regulate breast cancer metastasis. Oncotarget. 2016;7:78713–78725. doi: 10.18632/oncotarget.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey R., Sharma M., Saluja D. SIN-3 as a key determinant of lifespan and its sex dependent differential role on healthspan in Caenorhabditis elegans. Aging. 2018;10:3910–3937. doi: 10.18632/aging.101682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert V.J., Caron M., Gely L., Adrait A., Pakulska V., Couté Y., Chevalier M., Riedel C.G., Bedet C., Palladino F. SIN3 acts in distinct complexes to regulate the germline transcriptional program in C. elegans. Development. 2023;150 doi: 10.1242/dev.201755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holdorf A.D., Higgins D.P., Hart A.C., Boag P.R., Pazour G.J., Walhout A.J.M., Walker A.K. WormCat: An Online Tool for Annotation and Visualization of Caenorhabditis elegans Genome-Scale Data. Genetics. 2020;214:279–294. doi: 10.1534/genetics.119.302919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beurton F., Stempor P., Caron M., Appert A., Dong Y., Chen R.A.-J., Cluet D., Couté Y., Herbette M., Huang N., et al. Physical and functional interaction between SET1/COMPASS complex component CFP-1 and a Sin3S HDAC complex in C. elegans. Nucleic Acids Res. 2019;47:11164–11180. doi: 10.1093/nar/gkz880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kropp P.A., Wu J., Reidy M., Shrestha S., Rhodehouse K., Rogers P., Sack M.N., Golden A. Allele-specific mitochondrial stress induced by Multiple Mitochondrial Dysfunctions Syndrome 1 pathogenic mutations modeled in Caenorhabditis elegans. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 39.Puccio H., Simon D., Cossée M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 40.Schober F.A., Moore D., Atanassov I., Moedas M.F., Clemente P., Végvári Á., Fissi N.E., Filograna R., Bucher A.-L., Hinze Y., et al. The one-carbon pool controls mitochondrial energy metabolism via complex I and iron-sulfur clusters. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiavi A., Maglioni S., Palikaras K., Shaik A., Strappazzon F., Brinkmann V., Torgovnick A., Castelein N., De Henau S., Braeckman B.P., et al. Iron-Starvation-Induced Mitophagy Mediates Lifespan Extension upon Mitochondrial Stress in C. elegans. Curr. Biol. 2015;25:1810–1822. doi: 10.1016/j.cub.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y., Leboutet R., Largeau C., Zentout S., Lefebvre C., Delahodde A., Culetto E., Legouis R. Autophagy facilitates mitochondrial rebuilding after acute heat stress via a DRP-1–dependent process. J. Cell Biol. 2021;220 doi: 10.1083/jcb.201909139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al Rawi S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., Hajjar C., Boyd L., Legouis R., Galy V. Postfertilization Autophagy of Sperm Organelles Prevents Paternal Mitochondrial DNA Transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Zhang Y., Chen L., Liang Q., Yin X.-M., Miao L., Kang B.-H., Xue D. Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat. Commun. 2016;7 doi: 10.1038/ncomms12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q., Li H., Li H., Nakagawa A., Lin J.L.J., Lee E.-S., Harry B.L., Skeen-Gaar R.R., Suehiro Y., William D., et al. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 2016;353:394–399. doi: 10.1126/science.aaf4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. C. elegans Dynamin-Related Protein DRP-1 Controls Severing of the Mitochondrial Outer Membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/S1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 47.Roy C., Molin L., Alcolei A., Solyga M., Bonneau B., Vachon C., Bessereau J.L., Solari F. DAF-2/insulin IGF-1 receptor regulates motility during aging by integrating opposite signaling from muscle and neuronal tissues. Aging Cell. 2022;21 doi: 10.1111/acel.13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regmi S.G., Rolland S.G., Conradt B. Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging. 2014;6:118–130. doi: 10.18632/aging.100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei W., Ruvkun G. Lysosomal activity regulates Caenorhabditis elegans mitochondrial dynamics through vitamin B12 metabolism. Proc. Natl. Acad. Sci. USA. 2020;117:19970–19981. doi: 10.1073/pnas.2008021117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholtes C., Bellemin S., Martin E., Carre-Pierrat M., Mollereau B., Gieseler K., Walter L. DRP-1-mediated apoptosis induces muscle degeneration in dystrophin mutants. Sci. Rep. 2018;8:7354. doi: 10.1038/s41598-018-25727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosch A., Calvo M. In: Computer Optimized Microscopy Methods in Molecular Biology. Rebollo E., Bosch M., editors. Springer; 2019. Automated Quantitative Analysis of Mitochondrial Morphology; pp. 99–115. [DOI] [PubMed] [Google Scholar]
- 52.Chen H., Chan D.C. Mitochondrial dynamics-fusion, fission, movement, and mitophagy-in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christian C.J., Benian G.M. Animal models of sarcopenia. Aging Cell. 2020;19 doi: 10.1111/acel.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mergoud Dit Lamarche A., Molin L., Pierson L., Mariol M.-C., Bessereau J.-L., Gieseler K., Solari F. UNC-120/SRF independently controls muscle aging and lifespan in Caenorhabditis elegans. Aging Cell. 2018;17 doi: 10.1111/acel.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang V., Ullrich M., Lam H., Chew Y.L., Banister S., Song X., Zaw T., Kassiou M., Götz J., Nicholas H.R. Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell. Mol. Life Sci. 2014;71:3339–3361. doi: 10.1007/s00018-014-1558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Etheridge T., Rahman M., Gaffney C.J., Shaw D., Shephard F., Magudia J., Solomon D.E., Milne T., Blawzdziewicz J., Constantin-Teodosiu D., et al. The integrin-adhesome is required to maintain muscle structure, mitochondrial ATP production, and movement forces in Caenorhabditis elegans. Faseb. J. 2015;29:1235–1246. doi: 10.1096/fj.14-259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaffney C.J., Pollard A., Barratt T.F., Constantin-Teodosiu D., Greenhaff P.L., Szewczyk N.J. Greater loss of mitochondrial function with ageing is associated with earlier onset of sarcopenia in C. elegans. Aging. 2018;10:3382–3396. doi: 10.18632/aging.101654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y.-F., Haynes C.M. Metabolism and the UPR mt. Mol. Cell. 2016;61:677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koopman M., Michels H., Dancy B.M., Kamble R., Mouchiroud L., Auwerx J., Nollen E.A.A., Houtkooper R.H. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat. Protoc. 2016;11:1798–1816. doi: 10.1038/nprot.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higashitani A., Teranishi M., Nakagawa Y., Itoh Y., Sudevan S., Szewczyk N.J., Kubota Y., Abe T., Kobayashi T. Increased mitochondrial Ca 2+ contributes to health decline with age and Duchene muscular dystrophy in C. elegans. FASEB J. 2023;37 doi: 10.1096/fj.202201489RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang S.-H., Lin Y.-W. Bioenergetic Health Assessment of a Single Caenorhabditis elegans from Postembryonic Development to Aging Stages via Monitoring Changes in the Oxygen Consumption Rate within a Microfluidic Device. Sensors. 2018;18:2453. doi: 10.3390/s18082453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchetti P., Fovez Q., Germain N., Khamari R., Kluza J. Mitochondrial spare respiratory capacity: Mechanisms, regulation, and significance in non-transformed and cancer cells. Faseb. J. 2020;34:13106–13124. doi: 10.1096/fj.202000767R. [DOI] [PubMed] [Google Scholar]
- 64.Demine S., Renard P., Arnould T. Mitochondrial Uncoupling: A Key Controller of Biological Processes in Physiology and Diseases. Cells. 2019;8:795. doi: 10.3390/cells8080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berry B.J., Mjelde E., Carreno F., Gilham K., Hanson E.J., Na E., Kaeberlein M. Preservation of Mitochondrial Membrane Potential is Necessary for Lifespan Extension from Dietary Restriction. GeroScience. 2023;45:1573–1581. doi: 10.1007/S11357-023-00766-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertholet A.M., Chouchani E.T., Kazak L., Angelin A., Fedorenko A., Long J.Z., Vidoni S., Garrity R., Cho J., Terada N., et al. H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature. 2019;571:515–520. doi: 10.1038/s41586-019-1400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanover J.A., Forsythe M.E., Hennessey P.T., Brodigan T.M., Love D.C., Ashwell G., Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc. Natl. Acad. Sci. USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nielsen J. It Is All about MetabolicFluxes. J. Bacteriol. 2003;185:7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quirós P.M., Prado M.A., Zamboni N., D’Amico D., Williams R.W., Finley D., Gygi S.P., Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye C., Sutter B.M., Wang Y., Kuang Z., Tu B.P. A Metabolic Function for Phospholipid and Histone Methylation. Mol. Cell. 2017;66:180–193.e8. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaeffer J.M., Donatelli M.R. Characterization of a high-affinity membrane-associated ornithine decarboxylase from the free-living nematode Caenorhabditis elegans. Biochem. J. 1990;270:599–604. doi: 10.1042/bj2700599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leandro J., Houten S.M. The lysine degradation pathway: Subcellular compartmentalization and enzyme deficiencies. Mol. Genet. Metabol. 2020;131:14–22. doi: 10.1016/j.ymgme.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 74.Macrae M., Plasterk R.H., Coffino P. The ornithine decarboxylase gene of Caenorhabditis elegans: cloning, mapping and mutagenesis. Genetics. 1995;140:517–525. doi: 10.1093/genetics/140.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coffino P. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- 76.Jeong H.D., Kim J.H., Kwon G.E., Lee S.-T. Expression of Polyamine Oxidase in Fibroblasts Induces MMP-1 and Decreases the Integrity of Extracellular Matrix. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms231810487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park M.H., Nishimura K., Zanelli C.F., Valentini S.R. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barnes V.L., Bhat A., Unnikrishnan A., Heydari A.R., Arking R., Pile L.A. SIN3 is critical for stress resistance and modulates adult lifespan. Aging. 2014;6:645–660. doi: 10.18632/aging.100684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dannenberg J.-H., David G., Zhong S., van der Torre J., Wong W.H., Depinho R.A. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baymaz H.I., Karemaker I.D., Vermeulen M. Perspective on unraveling the versatility of ‘co-repressor’ complexes. Biochim. Biophys. Acta. 2015;1849:1051–1056. doi: 10.1016/j.bbagrm.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 81.van Oevelen C., Bowman C., Pellegrino J., Asp P., Cheng J., Parisi F., Micsinai M., Kluger Y., Chu A., Blais A., et al. The Mammalian Sin3 Proteins Are Required for Muscle Development and Sarcomere Specification. Mol. Cell Biol. 2010;30:5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu F., Zhu Q., Ye D., Zhang Q., Yang Y., Guo X., Liu Z., Jiapaer Z., Wan X., Wang G., et al. Sin3a–Tet1 interaction activates gene transcription and is required for embryonic stem cell pluripotency. Nucleic Acids Res. 2018;46:6026–6040. doi: 10.1093/nar/gky347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryan M.T., Hoogenraad N.J. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 84.Melber A., Haynes C.M. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shao L.-W., Peng Q., Dong M., Gao K., Li Y., Li Y., Li C.-Y., Liu Y. Histone deacetylase HDA-1 modulates mitochondrial stress response and longevity. Nat. Commun. 2020;11:4639. doi: 10.1038/s41467-020-18501-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wai T., Langer T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metabol. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 87.Vincent A.E., Ng Y.S., White K., Davey T., Mannella C., Falkous G., Feeney C., Schaefer A.M., McFarland R., Gorman G.S., et al. The Spectrum of Mitochondrial Ultrastructural Defects in Mitochondrial Myopathy. Sci. Rep. 2016;6 doi: 10.1038/srep30610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Byrne J.J., Soh M.S., Chandhok G., Vijayaraghavan T., Teoh J.-S., Crawford S., Cobham A.E., Yapa N.M.B., Mirth C.K., Neumann B. Disruption of mitochondrial dynamics affects behaviour and lifespan in Caenorhabditis elegans. Cell. Mol. Life Sci. 2019;76:1967–1985. doi: 10.1007/s00018-019-03024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whitnall M., Suryo Rahmanto Y., Huang M.L.-H., Saletta F., Lok H.C., Gutiérrez L., Lázaro F.J., Fleming A.J., St Pierre T.G., Mikhael M.R., et al. Identification of nonferritin mitochondrial iron deposits in a mouse model of Friedreich ataxia. Proc. Natl. Acad. Sci. USA. 2012;109:20590–20595. doi: 10.1073/pnas.1215349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ducamp S., Fleming M.D. The molecular genetics of sideroblastic anemia. Blood. 2019;133:59–69. doi: 10.1182/blood-2018-08-815951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ward D.M., Cloonan S.M. Mitochondrial Iron in Human Health and Disease. Annu. Rev. Physiol. 2019;81:453–482. doi: 10.1146/annurev-physiol-020518-114742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de la Cruz-Ruiz P., Hernando-Rodríguez B., Pérez-Jiménez M.M., Rodríguez-Palero M.J., Martínez-Bueno M.D., Pla A., Gatsi R., Artal-Sanz M. Prohibitin depletion extends lifespan of a TORC2/SGK-1 mutant through autophagy and the mitochondrial UPR. Aging Cell. 2021;20 doi: 10.1111/acel.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Artal-Sanz M., Tsang W.Y., Willems E.M., Grivell L.A., Lemire B.D., van der Spek H., Nijtmans L.G.J. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 2003;278:32091–32099. doi: 10.1074/jbc.M304877200. [DOI] [PubMed] [Google Scholar]
- 94.Artal-Sanz M., Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature. 2009;461:793–797. doi: 10.1038/nature08466. [DOI] [PubMed] [Google Scholar]
- 95.Hernando-Rodríguez B., Artal-Sanz M. Mitochondrial Quality Control Mechanisms and the PHB (Prohibitin) Complex. Cells. 2018;7:238. doi: 10.3390/cells7120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoneda T., Benedetti C., Urano F., Clark S.G., Harding H.P., Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 97.Nijtmans L.G., de Jong L., Artal Sanz M., Coates P.J., Berden J.A., Back J.W., Muijsers A.O., van der Spek H., Grivell L.A. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bennett C.F., Latorre-Muro P., Puigserver P. Mechanisms of mitochondrial respiratory adaptation. Nat. Rev. Mol. Cell Biol. 2022;23:817–835. doi: 10.1038/s41580-022-00506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madeo F., Eisenberg T., Pietrocola F., Kroemer G. Spermidine in health and disease. Science. 2018;359 doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 100.Murray Stewart T., Dunston T.T., Woster P.M., Casero R.A. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018;293:18736–18745. doi: 10.1074/jbc.TM118.003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pegg A.E. Toxicity of Polyamines and Their Metabolic Products. Chem. Res. Toxicol. 2013;26:1782–1800. doi: 10.1021/tx400316s. [DOI] [PubMed] [Google Scholar]
- 102.Yang X., Zhang M., Dai Y., Sun Y., Aman Y., Xu Y., Yu P., Zheng Y., Yang J., Zhu X. Spermidine inhibits neurodegeneration and delays aging via the PINK1-PDR1-dependent mitophagy pathway in C. elegans. Aging. 2020;12:16852–16866. doi: 10.18632/aging.103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar V., Mishra R.K., Ghose D., Kalita A., Dhiman P., Prakash A., Thakur N., Mitra G., Chaudhari V.D., Arora A., Dutta D. Free spermidine evokes superoxide radicals that manifest toxicity. Elife. 2022;11 doi: 10.7554/eLife.77704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakanishi S., Cleveland J.L. Polyamine Homeostasis in Development and Disease. Med. Sci. 2021;9:28. doi: 10.3390/medsci9020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Til H.P., Falke H.E., Prinsen M.K., Willems M.I. Acute and subacute toxicity of tyramine, spermidine, spermine, putrescine and cadaverine in rats. Food Chem. Toxicol. 1997;35:337–348. doi: 10.1016/S0278-6915(97)00121-X. [DOI] [PubMed] [Google Scholar]
- 106.Barba-Aliaga M., Alepuz P. Role of eIF5A in Mitochondrial Function. Int. J. Mol. Sci. 2022;23:1284. doi: 10.3390/ijms23031284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puleston D.J., Buck M.D., Klein Geltink R.I., Kyle R.L., Caputa G., O’Sullivan D., Cameron A.M., Castoldi A., Musa Y., Kabat A.M., et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metabol. 2019;30:352–363.e8. doi: 10.1016/j.cmet.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Affronti H.C., Rowsam A.M., Pellerite A.J., Rosario S.R., Long M.D., Jacobi J.J., Bianchi-Smiraglia A., Boerlin C.S., Gillard B.M., Karasik E., et al. Pharmacological polyamine catabolism upregulation with methionine salvage pathway inhibition as an effective prostate cancer therapy. Nat. Commun. 2020;11:52. doi: 10.1038/s41467-019-13950-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kamath R.S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 110.Fan X., De Henau S., Feinstein J., Miller S.I., Han B., Frøkjær-Jensen C., Griffin E.E. SapTrap Assembly of Caenorhabditis elegans MosSCI Transgene Vectors. G3 (Bethesda) 2020;10:635–644. doi: 10.1534/g3.119.400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kolotuev I., Bumbarger D.J., Labouesse M., Schwab Y. Methods in Cell Biology. Elsevier; 2012. Targeted Ultramicrotomy; pp. 203–222. [DOI] [PubMed] [Google Scholar]
- 113.Li C.H., Tam P.K.S. An iterative algorithm for minimum cross entropy thresholding. Pattern Recogn. Lett. 1998;19:771–776. doi: 10.1016/S0167-8655(98)00057-9. [DOI] [Google Scholar]
- 114.Spanier B., Laurençon A., Weiser A., Pujol N., Omi S., Barsch A., Korf A., Meyer S.W., Ewbank J.J., Palladino F., et al. Correction to: Comparison of lipidome profiles of Caenorhabditis elegans—results from an inter-laboratory ring trial. Metabolomics. 2021;17:33. doi: 10.1007/s11306-021-01784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Metabolomics data have been deposited at MetaboLights with the study ID MTBLS8439 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw transcriptomics data were deposited in GEO with accession number GSE227499 (Caron et al.34). Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.