Abstract
Background
Immunotherapy has revolutionized cancer treatment. Recent studies have suggested that the efficacy of immunotherapy can be further enhanced by the influence of gut microbiota. In this study, we aimed to investigate the impact of bacteria on the effectiveness of cancer immunotherapy by combining analysis of clinical samples with validation in animal models.
Methods
In order to characterize the diversity and composition of microbiota and its relationship with response to immune checkpoint inhibitors (ICIs), 16S ribosomal RNA (rRNA) and GC–MS sequencing was performed on 71 stool samples from patients with advanced non‐small cell lung cancer (NSCLC) prior to treatment with immune checkpoint blockade (ICB). Furthermore, fecal microbiota transplantation (FMT) was performed from different patients into mice and a subcutaneous tumor model established using the Lewis lung cancer cell line to evaluate the therapeutic effect of PD‐1 on mice with varying gut microbiota.
Results
The results demonstrated a significant association between elevated gut microbiota diversity and response to treatment with ICIs, p < 0.05. Faecalibacterium was markedly increased in the gut microbiota of responders (R), accompanied by increased short‐chain fatty acid (SCFA) levels, especially butanoic acid, acetic acid and hexanoic acid, p < 0.05. Additionally, FMT from R and nonresponders (NR) could promote an anticancer effect and reduce the expression of Ki‐67 cells in tumors in mice, p < 0.05. Moreover, R and NR FMT did not alter PD‐L1 expression in the tumor tissues of mice, p > 0.05. The diversity of gut microbiota consistently correlated with an optimistic prognosis in NSCLC patients with immunotherapy, which could be functionally mediated by SCFAs.
Conclusion
The findings of the present study indicated that the diversity of gut microbiota and SCFAs is related to the efficacy of immunotherapy. FMT can effectively delay tumor progression, and enhance the effect of immunotherapy, thus providing evidence for improving the efficacy of immunotherapy in NSCLC patients.
Keywords: biomarker, fecal microbiota transfer (FMT), gut microbiota, immunotherapy, non‐small cell lung cancer (NSCLC)
Studies have found the diversity of gut microbiota and short‐chain fatty acids (SCFAs) is related to the efficacy of immunotherapy. Fecal microbiota transplantation (FMT) can effectively delay tumor progression, and enhance the effect of immunotherapy, providing evidence for improving the efficacy of immunotherapy in non‐small cell lung cancer (NSCLC) patients.
INTRODUCTION
The development of immune checkpoint inhibitor (ICI) interaction has transformed the therapeutic landscape of patients with advanced non‐small cell lung cancer (NSCLC). 1 , 2 , 3 Immunotherapy, particularly ICIs, has shown significant efficacy and a low incidence of overall toxic and side effects, making it a promising treatment option for advanced NSCLC. 4 Although a durable benefit indicates the acquisition of long‐term immunological memory, some patients who have an initial response to ICIs and then have a relapse and develop acquired resistance have clinical and biological immunotherapy failure. 5 Primary and acquired drug resistance pose significant challenges in improving the prognosis of patients with advanced or metastatic NSCLC. 6 The tumor microenvironment (TME) plays a crucial role in the immune response to ICIs. 7 , 8 , 9 The gut microbiota has been shown to reprogram the TME, and the gut microbiome and its metabolites can influence the response to antitumor treatment. 10 Furthermore, the gut microbiota has been associated with the effects and complications of cancer therapy due to its immunoregulatory properties. 11 , 12 , 13 The microbiota can affect tissue metabolism, the differentiation and function of immune cells in the TME, and promote or inhibit cancer development. The composition and abundance of the bacterial flora at the epithelial barrier surface can simultaneously affect inflammation, immune response, and the homeostasis of epithelial and stromal cells. 14 , 15 Studies have confirmed the close relationship between gut microbiota and the response of tumor patients to immunotherapy, making it a potential indicator for predicting the efficacy of tumor immunotherapy. 16 The differentiation of T helper 17 (Th17) cells is dependent on colonization with distinct commensal bacteria, possibly through cytokines such as IL‐6, IL‐21, and IL‐23. 17 Intestinal flora can also synergize with various chemotherapy drugs to induce oxidative stress damage in tumor cells and regulate the immune response of immune cells. 18 Recent studies have shown that gut microbiota can not only directly interact with tumor cells, but also exert an indirect tumor‐suppressing or cancer‐promoting effect by producing metabolites. Metabolites such as lipopolysaccharide, bile acids, hydrogen sulfide, and short‐chain fatty acids (SCFAs) can promote or inhibit tumor progression through various molecular mechanisms. 19 SCFAs and their receptors have been found to affect various immune cells, including regulatory T cells, dendritic cells, and macrophages, thereby activating host antitumor immune responses. Probiotic supplementation has been shown to increase the abundance of butyric acid‐producing bacteria in the gut, and increased circulating butyric acid can upregulate the expression of chemokine (CC motif) ligand 20 in pulmonary endothelial cells, recruiting helper T cells 17 to the lungs and inhibiting the occurrence of lung metastasis in melanoma. 20
Fecal microbiota transplantation (FMT) has emerged as a potential strategy to improve immunotherapy for tumor patients by restoring a healthy gut microbiota, and it has become a focus of both basic and clinical research. 21 FMT has the ability to reverse immune tolerance during tumor treatment. Baruch et al. demonstrated that patients with immune‐resistant metastatic melanoma who received FMT from complete responders combined with programmed death‐1 (PD‐1) inhibitors treatment showed beneficial outcomes. 22 A complex relationship between the gut microbiota and the efficacy of tumor immunotherapy has been established, although the underlying mechanisms are not fully understood. 23 In this study, we aimed to investigate the role of fecal microbiota in immunotherapy for patients with advanced NSCLC. We examined the fecal microbiota of patients with different responses to immunotherapy and collected stool samples at different time points during the course of treatment. Additionally, metabolomic analysis was conducted to study the changes in fecal metabolites. Furthermore, we utilized animal models to study the impact of FMT on immunotherapy. Overall, this study aimed to provide insights into the role of fecal microbiota in immunotherapy for advanced NSCLC patients and explore the potential of fecal transplantation as a therapeutic strategy to enhance the efficacy of immunotherapy.
METHODS
Patient cohort and clinical characteristics
This study enrolled a total of 41 patients diagnosed with stage IIIB–IV NSCLC who received treatment with platinum‐based chemotherapy and ICIs between November 2021 and August 2023. Among them, 31 patients received first‐line treatment and 10 patients received second‐line treatment. In addition, three patients accepted PD‐1 inhibitors plus bevacizumab and chemotherapy, all of whom have harbored epidermal growth factor receptor (EGFR), and six patients had Kirsten rat sarcoma viral oncogene homolog (KRAS) gene mutations within the tumors, and the rest of patients had no genetic mutations. Based on the absolute number of white blood cells (leukocytes, lymphocytes, neutrophils, and monocytes), determined by routine blood testing, the vast majority of patients presented an adequate immune status to receive immunotherapy. Evaluation of ICI treatment efficacy was done according to Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1) by two independent radiologists at the Affiliated Hospital of Qingdao University according to the results of imaging and blood indexes of patients after receiving four cycles of immunotherapy. The patients were divided into two groups: responders (R, n = 21) and nonresponders (NR, n = 20), based on the objective response in target lesions. Responders (R) were defined as patients who experienced complete response (CR) and partial response (PR), while nonresponders (NR) were defined as patients who experienced stable disease (SD) and progressive disease (PD). 24 Stool samples were collected from 41 patients at baseline before ICIs. Additionally, stool samples were collected from 30 patients at baseline and after 3 months of immunotherapy. This study was conducted in accordance with the Declaration of Helsinki for research involving human subjects. All samples were obtained with informed consent at the Affiliated Hospital of Qingdao University, and the study was approved by the institutional review board in accordance with the Declaration of Helsinki (QYFY WZLL 27468).
Stool sample collection
In this study, a total of 41 patients with NSCLC were instructed to self‐sample their stool following detailed printed instructions. Each patient was provided with stool sample containers to collect their samples. The patients were instructed to bring their collected stool samples to the laboratory within 2 h after collection. Upon arrival at the laboratory, the samples were immediately stored at −80°C until further processing.
DNA extraction and 16S rRNA gene sequencing
According to the E.Z.N.A. soil DNA kit (Omega Bio‐tek) instructions, the total DNA of the microbial community was extracted. The quality of the extracted DNA was detected by 1% agarose gel electrophoresis. DNA concentration and purity were measured by NanoDrop2000 (Thermo Fisher Scientific). To amplify the V3–V4 region of the 16S rRNA gene, the primers 338F (5′‐ACTCCTACGGGAGGCAGCAG‐3′) and 806R (5′‐GGACTACHVGGGTWTCTAAT‐3′) were used in the polymerase chain reaction (PCR). The PCR products of the same sample were compared. The formal PCR test was performed using TransGen AP221‐02, TransStart Fastpfu DNA polymerase and 20 μL reaction system: 5 × FastPfu buffer (4 μL), 2.5 mM dNTPs (2 μL), forward primer, (0.8 μL), reverse primer (0.8 μL), FastPfu polymerase (0.4 μL), BSA (0.2 μL), template DNA (10 ng) and DdH2O supplement to 20 μL. The following PCR conditions were used: initial denaturation at 95°C for 3 min, 25 cycles of denaturation (95°C for 30 s), annealing (55°C for 30 s) and extension (72°C for 30 s), and final extension at 72°C for 10 min. The PCR products were purified with AMPure XP Beads (Beckman Coulter). The purified products were recovered from a 2% agarose gel using the AxyPrep DNAGel extraction kit (Axygen Biosciences). The recovered products were detected and determined by the Quantus fluorometer (Promega) quantity. The NEXTFLEX Rapid DNA‐Seq Kit (Bioo Scientifi) was used for library construction. Using Miseq (Illumina Inc.), the PE300 (paired‐end 300 bp) platform was used for sequencing. The original sequencing data underwent quality control using the Fastp software, and the paired‐end reads were merged using the Flash software. The Uparse software was used for operational taxonomic unit (OTU) clustering of the sequences and removal of chimeras, with a 97% similarity threshold. The Silva 16S rRNA database (version 138) was used for taxonomic annotation of the OTU representative sequences, and the RDP classifier was used with a confidence threshold of 0.7 to obtain species‐level taxonomic annotation results.
Short‐chain fatty acid analysis (SCFA)
In this study, 50 mg of solid sample was accurately weighed into a 2 mL grinding tube and 500 μL methanol water solution (CH3OH: H2O, v: v = 4:1) added. The samples were further fixed using this solution. The gas chromatography‐mass spectrometry (GC–MS) analysis was performed using an Agilent 8890B gas chromatography coupled with an Agilent 5977B mass selective detector (Majorbio Bio‐Pharm Technology Co. Ltd). The instrument was equipped with an inert electron impact (EI) ionization source with an ionization voltage of 70 eV (Agilent). A DB‐5MS (40 m × 0.25 mm × 0.25 μm) capillary column was used for sample separation, with 99.999% helium as the carrier gas at a constant flow rate of 1 mL/min. The temperature program for the GC column was set to hold at 60°C for 30 s and then increase to 310°C at a rate of 8°C/min, with a total run time of 6 min. The injection volume of the samples was 1 μL, introduced in splitting mode (15:1), with an inlet temperature of 260°C. For mass spectrometry, the ion source temperature was set to 230°C, and the quadrupole temperature was set to 150°C. The scanning mode used was full scan mode, with a mass scanning range of m/z 50–500. The raw data obtained from the mass spectrometer detection of GC/MS was preprocessed using the MassHunter workstation quantitative analysis software (version 10.0.707.0). The software generated a three‐dimensional data matrix in CSV format. Metabolite identification was performed by searching databases, including public databases such as NIST (version 2017), Fiehn (version 2013), and MS‐DIAL (version 2021).
Cell culture
Lewis lung carcinoma (LLC) cells were obtained from Procell Life Science & Technology Co. Ltd. The LLC cell line was authenticated using DNA fingerprint analysis and were negative for mycoplasma contamination. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) medium (Gibco, Thermo Fisher Scientific) containing 10% (v/v) fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific) and 1% penicillin–streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Mice
Specific pathogen‐free (SPF) C57Bl/6 mice were purchased from Shandong Academy of Medical Sciences Laboratories. The mice was age and sex matched and housed in independently vented cages at the animal facility of the Affiliated Hospital of Qingdao University Laboratories. All experiments involving the mice were conducted when they were 6 weeks old. The mouse were reared in groups according to SPF conditions and provided with sterilized diet and water. The animal experiments reported in this study were approved by the Animal Care and Ethics Committee of Qingdao University (AHQU‐MAL20230331).
A total of 36 mice from different litters were equally divided into six groups: blank group (treated with phosphate buffered saline [PBS]), PD‐1 monoclonal antibody (mAb) (200 μg/mouse, clone: RMP1‐14, Bio X Cell), antibiotic (ATB) plus PD‐1 group, PD‐1 plus R group FMT, PD‐1 plus NR group FMT and PBS plus ATB. The sample size for the experiment was determined by the requirements of both metagenomic and metabolomic analyses. The resource equation method was also used to verify the sample size. According to the formula of this method, the E‐value (Evalue) is calculated as the total number of animals minus the total number of groups. In this case, the Evalue is (6 × 6)–6 = 30, which is more than 20 (the limitation of E‐value). 25 To deplete the gut microbiota, the mice were treated with a broad‐spectrum antibiotic cocktail (ABX) in their drinking water for one week. The ABX cocktail consisted of ampicillin (1 g/L), neomycin (1 g/L), and metronidazole (0.5 g/L). 26 Mice in the R and NR groups were treated with antibiotics followed by 7‐day fecal transplantation followed by LLC inoculation. The administration of bacteria continued throughout the experimental period. Donor feces were dissolved in PBS and centrifuged several times to separate the precipitate and supernatant, where the supernatant was further filtered through a 0.22 μm sterile nylon filter. The recipient mice from the FMT + PD‐1 group received supernatants by one‐to‐one gavage. Approximately 5 × 105 LLC cells were subcutaneously inoculated into the right dorsal flanks of mice. Tumor size was measured three times a week until the end point, and the tumor volume was calculated as length × width2 × 0.5. After tumor inoculation, the tumor‐bearing mice were intraperitoneally injected with anti‐PD‐1 monoclonal antibody (mAb; clone RMP1‐14, BioXCell) in PBS on specific days. The mice in the blank group and PBS plus ABT group were injected with PBS. The mice were sacrificed on day 23, and the tumors were surgically separated, weighed, and preserved for further examinations (Figure 4a). Fecal samples were collected from mice in different groups before tumor inoculation and at the time of sacrifice. The extraction of mouse stool RNA and DNA and qPCR analysis were performed for further analysis.
FIGURE 4.
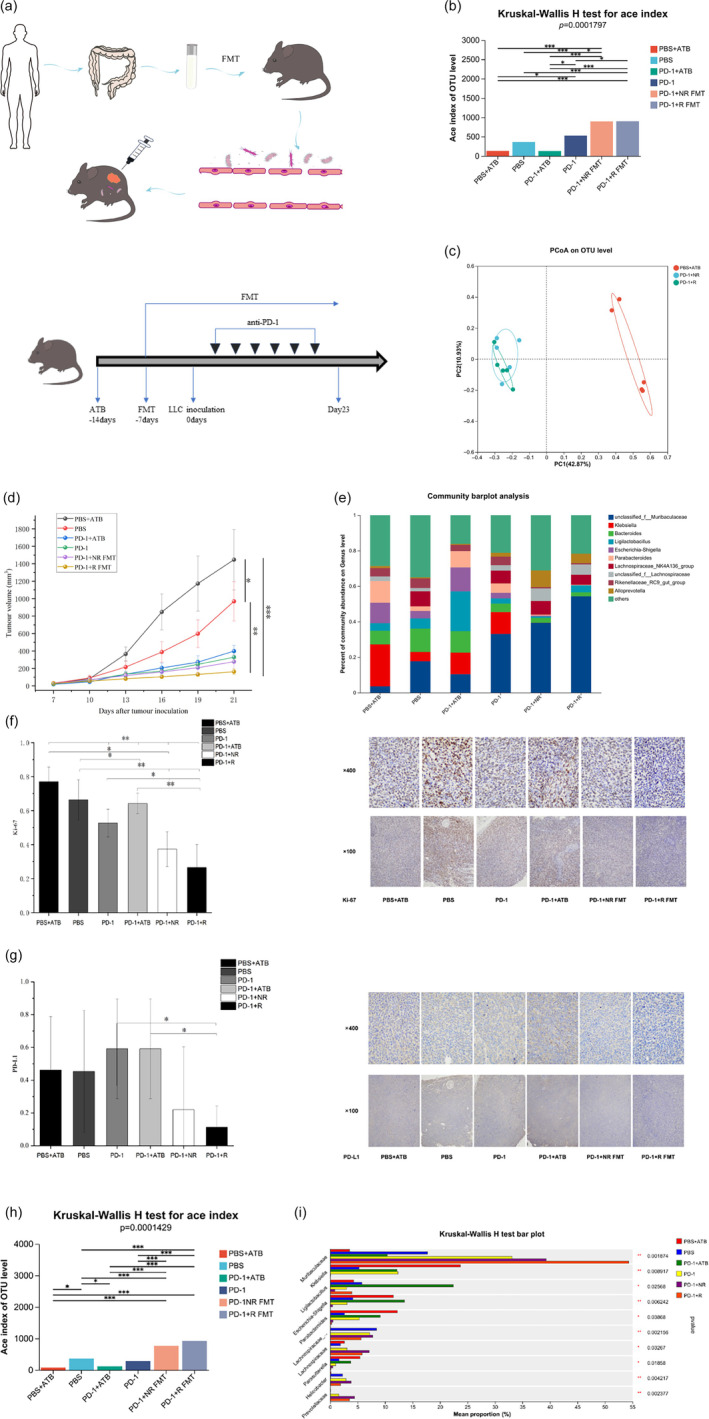
Transcriptomic and metabolomic analysis of syngeneic tumor models treated with responders (R) and nonresponders (NR). (a) Experimental protocol. (b, c) Characteristics of intestinal flora of the phosphate buffered saline (PBS) + antibiotic (ATB), PBS, PD‐1, PD‐1 + ATB, PD‐1 + NR FMT, and PD‐1 + responders fecal microbiota transplantation (R‐FMT) groups, there was a significant difference between the groups in the level of diversity ace index, p < 0.05. (d) Growth curves of subcutaneous tumors in C57BL mice. (e) Characteristics of intestinal flora at the level of alpha diversity ace index of the PBS + ATB, PBS, PD‐1, PD‐1 + ATB, PD‐1 + NR, and PD‐1 + R groups at the baseline of treatment. (f, g) Representative immunohistochemical (IHC) analysis. Compared with six groups, the R‐FMT group comprised significantly decreased numbers of Ki‐67 positive cells, p < 0.05. Compared with the PD‐1 + ATB group and PD‐1 group, the R‐FMT group comprised partly decreased numbers of PD‐L1 positive cells, p < 0.05. (h) The bar plot visually shows the changes of different species in six groups. Characteristics of intestinal flora at the level of alpha diversity ace index of the PBS + ATB, PBS, PD‐1, PD‐1 + ATB, PD‐1 + NR FMT, and PD‐1 + R FMT groups after different treatments. (i) Analysis of species differences between the six groups at the genus level. Significant differences were evaluated using the Kruskal‐Wallis rank sum test (b, h, i), ordinary one‐way analysis of variance (ANOVA) (d, f, g) test. Data are expressed as mean ± SEM. *p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001. PD‐1, programmed death‐1.
Statistical analysis
Statistical analyses were performed using SPSS 24.0 software (IBM Corp.), Microsoft Office Excel 2007 (Microsoft Inc.) or R software version 4.0.3 (R Foundation for Statistical Computing). Chi‐squared test and nonparametric tests (Mann–Whitney U or Kruskal–Wallis) were applied to evaluate associations between patient clinicopathological characteristics and microbiota composition. Permutational multivariate analysis of variance (PERMANOVA) was used to compare compositional differences between groups. Linear discriminant analysis (LDA) effect size (LEfSe) was performed to study the differential abundance analysis of gut microbiota composition. Hazard ratios and 95% confidence intervals were calculated using the Cox regression model. All results tested were two‐tailed and were considered statistically significant if the p‐value was less than 0.05.
RESULTS
Baseline gut microbiota diversity is associated with favorable responses to immunotherapy
In order to investigate the role of the gut microbiota as a biomarker for immunotherapy, we conducted a study involving 41 advanced NSCLC patients starting treatment with ICIs (Figure 1a). The detailed clinical information for patients is provided in Table 1. The median age in this cohort was 65. According to response, the patients with CR and PR were classified as the responder group (R), accounting for 21 patients (51.2%), and the patients with SD and PD were classified as the nonresponder group (NR), accounting for 20 patients (48.8%). A total of 31 patients received first‐line immunotherapy and 10 patients received second‐line immunotherapy. Correlation analysis revealed that age, gender, smoking status, body mass index, stage, histology, bone metastasis, PD‐L1 expression and treatment lines were clinical factors not associated with response to immune checkpoint blockade (ICB) in this cohort. We performed 16S rRNA gene sequencing on stool samples from these patients and obtained a total of 4 098 536 sequence reads and 1 694 221 698 base reads. The differences in alpha diversity indexes between the R group and the NR group were explored through intergroup difference tests shown in Figure 1b–e, such as Ace, Chao, Coverage and Sobs diversity. Among them, the levels of Ace, Chao and Sobs indices represent the microbial diversity, while Coverage index represents the authenticity of microbial samples. We found that the alpha diversity and richness of intestinal microbes were higher in the R group compared to the NR group (p < 0.05). We also compared the diversity of gut microbiomes between the two groups and a significant difference in β‐diversity was observed (p < 0.05). Taxonomic analysis revealed that the richness of Bacteroidetes and Actinobacteriota was higher in the R group than that in the NR group (Figure 1g).
FIGURE 1.
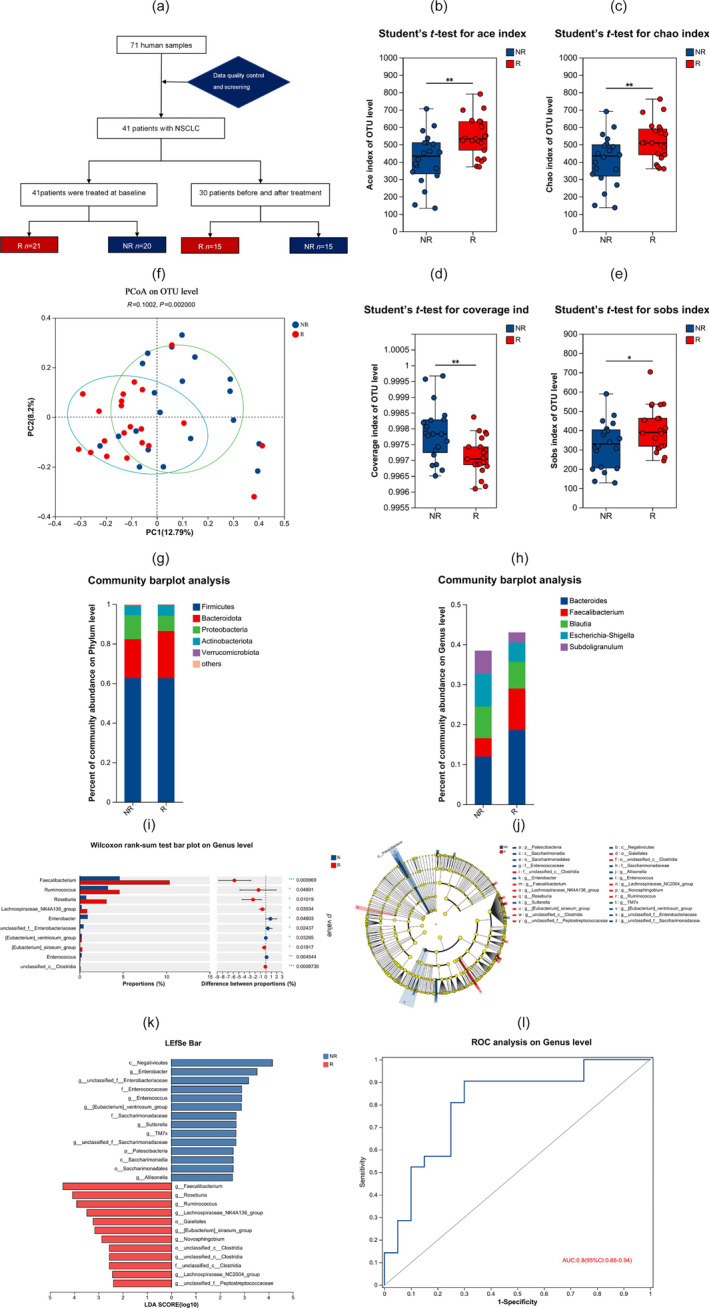
Marker gene sequence 16S rRNA profiling of the gut microbiome in patients with non‐small cell lung cancer (NSCLC). (a) Patient enrolment, treatment and response details. A total of 71 human samples (41 patients with NSCLC) were enrolled. The 41 patients with NSCLC (stage IIIB–IV) were divided into responders (R) (CR + PR, n = 21) and nonresponders (NR) (SD + PD, n = 20). Among 30 patients, samples were collected simultaneously at baseline and after treatment. (b–e) Box‐and‐whisker plot illustrating the α‐diversity. The figures show the significant differences between the two selected groups of samples, with the group name on the abscissa and the index range for each group on the ordinate. The differences in Ace, Chao, Coverage, and Sobs diversity indexes between the R group and the NR groups are shown. Ace, Chao and Sobs estimate microbial diversity in the sample. Coverage reflects the real situatuin of microorganisms in the sample. (f) Principal coordinates analysis (PCoA). The β‐diversity of gut microbiomes in NR (n = 20) and R (n = 21) patients with NSCLC. (g, h) Trends of microbes and their relative abundance in different groups. Bar plots show the microbial communities of all samples. (g) At phylum, (h) At genus. (i, j, k) Cladogram generated from linear discriminant analysis effect size (LEfSe) and the linear discriminant analysis (LDA) score. A plot of LDA scores from the LEfSe method illustrating the differential abundance of the indicated taxa in the gut microbiomes of NR and R. The results of linear discriminant analysis effect size (LEfSe) further confirmed that both Faecalibacterium (LDA = 4.466), Roseburia (LDA = 4.068) and Ruminococcus (LDA = 3.899) were higher in the feces of R patients compared to NR patients. (l) Based on the analysis of Faecalibacterium with significant differences between the R and NR groups, the OUT data set of Faecalibacterium was established to find the microbial biomarker of the R and NR groups, after which we constructed the receiver operating characteristic (ROC) curve. Operational taxonomic units (OTU) is a method used in microbial ecology to group sequences from a sample into taxonomic units based on their similarity. The area under the curve (AUC) was 0.8 (95% CI: 0.66–0.94). *p < 0.05; **p < 0.01; ***p < 0.001.
TABLE 1.
Baseline clinicopathological characteristics of patients and their association with ICB treatment response.
| Patient characteristics | Total (n = 41) | R | NR | p‐value |
|---|---|---|---|---|
| Age | ||||
| ≤65 | 21 (51.2%) | 13 (61.9%) | 12 (38.1%) | 0.395 |
| >65 | 20 (48.8%) | 18 (40%) | 12 (60%) | |
| Gender | ||||
| Male | 33 (80.5%) | 15 (71.4%) | 18 (90.0%) | 0.238 |
| Female | 8 (19.5%) | 6 (28.6%) | 2 (10.0%) | |
| Smoking status | ||||
| Smoker | 16 (42.9%) | 9 (38.1%) | 7 (35.0%) | 0.606 |
| Nonsmoker | 25 (57.1%) | 12 (61.9%) | 13 (65.0%) | |
| Body mass index | ||||
| High (≥25) | 21 (34.1%) | 10 (47.6%) | 11 (55.0%) | 0.636 |
| Low (<25) | 20 (65.9%) | 11 (52.4%) | 9 (45.0%) | |
| Stage | ||||
| III | 14 (34.1%) | 7 (33.3%) | 7 (35.0%) | 0.910 |
| IV | 27 (65.9%) | 14 (66.7%) | 13 (65.0%) | |
| Histology | ||||
| Adenocarcinoma | 22 (53.7%) | 10 (47.6%) | 12 (60.0%) | |
| Squamous carcinoma | 18 (43.9%) | 10 (47.6%) | 8 (40.0%) | 0.510 |
| Sarcomatoid carcinoma | 1 (2.4%) | 1 (4.8%) | 0 (0) | |
| PD‐L1 | ||||
| Nonspecified | 2 (4.9%) | 2 (9.5%) | 0 (0) | 0.288 |
| Negative (TPS < 1%) | 17 (41.5%) | 7 (33.3%) | 10 (50.0%) | |
| Positive (1% < PD‐L1 < 49%) | 15 (36.6%) | 7 (33.3%) | 8 (40.0%) | |
| Positive (PD‐L1 ≥ 50%) | 7 (17.1%) | 5 (23.8%) | 2 (10.0%) | |
| Bone metastasis | ||||
| Yes | 6 (14.6%) | 2 (9.5%) | 4 (20.0%) | 0.410 |
| No | 35 (85.4%) | 19 (90.5%) | 16 (80.0%) | |
| Treatment lines | ||||
| 1 | 31 (75.6%) | 14 (66.7%) | 17 (85.0%) | 0.177 |
| 2 | 10 (24.4%) | 7 (33.3%) | 3 (15.0%) | |
Abbreviations: ICB, immune checkpoint blockade; NR, nonresponders; PD‐1, programmed death‐1; R, responders.
At the genus level, we also observed differences in species composition between the two groups (Figure 1h). Five bacterial genera including Faecalibacterium (p = 0.000969), Ruminococcus (p = 0.04891), Ruminococcus (p = 0.01019), Lachnospiraceae_NK4A136_group (p = 0.03034), [Eubacterium]_siraeum_group (p = 0.04603) and unclassified_c__Clostridia (p = 0.0008736) were significantly enriched in R group patients compared to the NR group (Figure 1i). Using linear discriminant analysis of effect size (LEfSe), we also observed that Faecalibacterium was enriched in the R group (Figure 1j). The results of LEfSe further demonstrated that Faecalibacterium (LDA = 4.466) were higher in the feces of R group compared to the NR group (Figure 1k). We also analyzed the correlation between the abundances of Faecalibacterium and the clinical characteristics of NSCLC patients. Based on the analysis of Faecalibacterium with significant differences between the R and NR groups, we developed a prognostic prediction model for NSCLC based on gut microbiota profile. The analysis of Faecalibacterium showed an area under the curve (AUC) of 0.8 (95% CI: 0.66–0.94) in the receiver operating characteristic (ROC) curve (Figure 1l). These findings suggested that the high diversity of gut microbiota and abundance of Faecalibacterium may play an important role in the efficacy of immunotherapy in patients with advanced NSCLC.
Effect of immunotherapy on gut microbiota composition
To further investigate the changes in the gut microbiota after immunotherapy, we analyzed the sequencing data of fecal samples. We sequenced the 16S rRNA gene in fecal samples obtained from 30 patients with advanced NSCLC before and after. ICI treatment to observe the effect of immunotherapy on the gut microbiota. Samples prior to immunotherapy were defined as "B". Samples following immunotherapy were defined as "A". We analyzed the changes in the level of the microbiota diversity index after immunotherapy. The Ace, Chao and Sobs indexes were used to assess the diversity of gut microbes. We observed an increase in the diversity of gut microbes after immunotherapy, as indicated by these diversity indexes (p < 0.05) (Figure 2a–d). At the phylum level, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia were found to be the major bacterial communities in the fecal samples (Figure 2e). However, at the phylum level, there was no significant difference in species composition before and after immunotherapy. After that, we performed group differences analysis at the genus level. Faecalibacterium (p = 0.02813) showed significant differences at the genus level (Figure 2f,g). Faecalibacterium increased after immunotherapy (p < 0.05).
FIGURE 2.
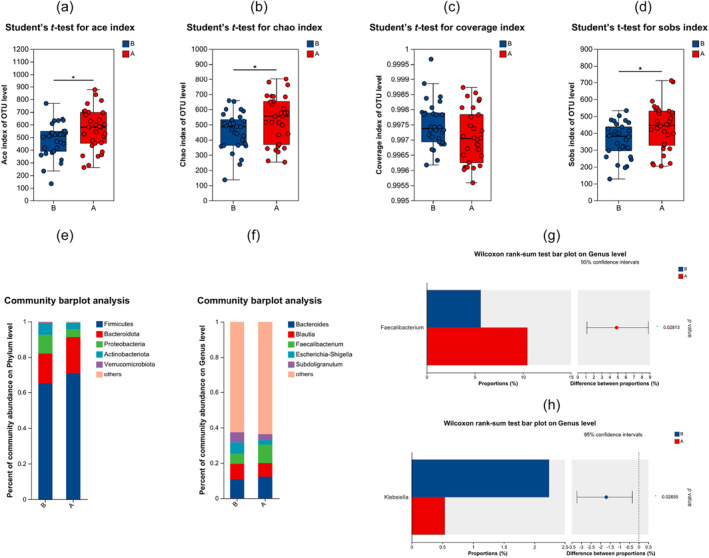
Differences in the composition of the gut microbiome before and after immunotherapy. Samples prior to immunotherapy were defined as “B.” Samples following immunotherapy were defined as “A.” (a–d) Ace, Chao, coverage and Sobs index score of the gut microbiota in patients stratified according to time point of treatment, by the Mann–Whitney U rank sum test. (e, f) At the phylum and genus level, the species composition of the gut microbiota differed between the 30 patients before and after treatment. (g, h) There were significant differences in the genus level of Faecalibacterium (p = 0.02813) and Klebsiella (p = 0.02655) before and after treatment. Faecalibacterium increased after immunotherapy, while Klebsiella decreased after treatment.*p < 0.05.
Intestinal flora plays an antitumor role through SCFA
Based on the above studies, we found that Faecalibacterium were the dominant genus for immunotherapy. Faecalibacterium is one of the main bacterial groups that produce butyric acid. 27 Next, we further explored the influence by which metabolites of gut microbiota contribute to immunotherapy. We targeted SCFAs in the feces of 30 patients after immunotherapy using GC–MS. The metabolite butyric acid in the gut of the R group was significantly higher than that of the NR group, p < 0.05 (Figure 3a). We also analyzed the differences of other SCFAs between the two groups. The contents of acetic acid, butanoic acid, isovaleric acid and isobutyric acid were significantly higher in the R group compared to the NR group (Figure 3b, p < 0.05). We further analyzed the correlation between different SCFAs. Positive correlations between propanoic acid and acetic acid, butanoic acid and acetic acid, and isovaleric acid and butanoic acid, were found (Figure 3c, p < 0.05). When comparing the two groups, the content of SCFA was generally lower in the NR group, especially in butanoic, acetic and hexanoic acid (Figure 3d, p < 0.05). We calculated the AUC values for each SCFA and constructed ROC curves to assess their predictive value for the prognosis of patients with advanced NSCLC. The results showed that except for isohexanoic acid, the other seven SCFAs had good predictive value for the prognosis of patients with advanced NSCLC (Figure 3e). In the correlation analysis between gut microbiota and SCFAs, Faecalibacterium was positively correlated with butyrate (Figure 3f).
FIGURE 3.
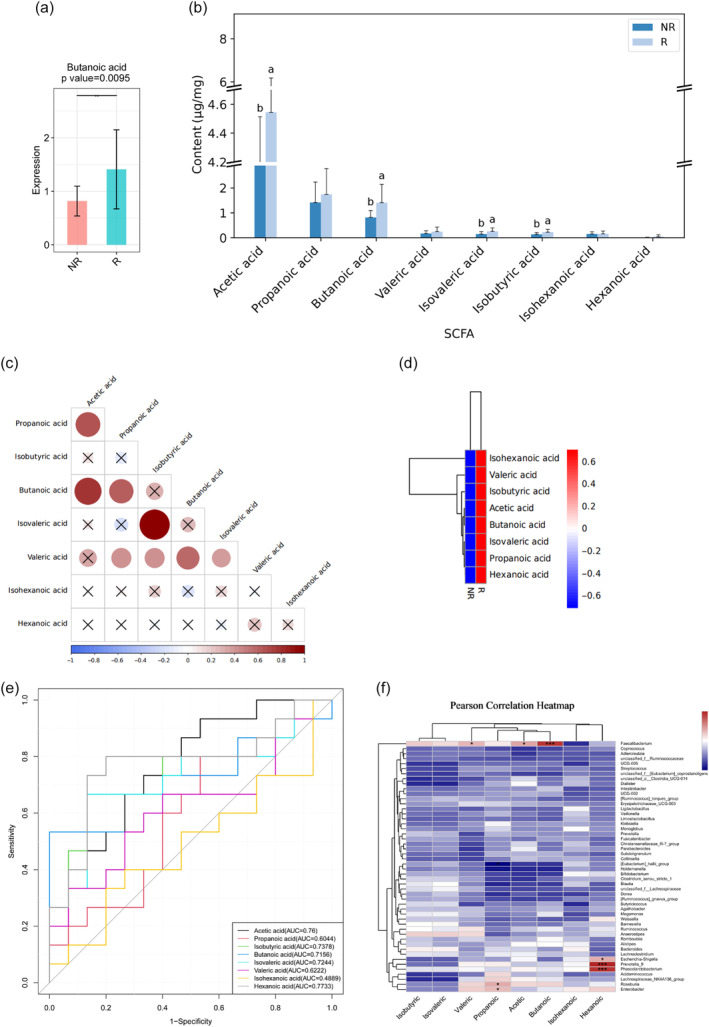
Targeted short‐chain fatty acids (SCFAs) in the feces of 30 patients after immunotherapy were detected by gas chromatography‐mass spectrometry. (a) The metabolite butyric acid in the gut of responders (R) was significantly higher than that of nonresponders (NR). (b) The contents of acetic acid, butanoic acid, isovaleric acid and isobutyric acid in the R group were significantly higher than those in the NR group. According to the analysis of variance, the difference of SCFAS in different groups was calculated, and the post hoc multiple comparisons were made. For the same index, if the letters of the two groups were the same, it meant that there was no significant difference in the content of the index in the two groups. If the letters of the two groups were different, it meant that there was a significant difference in the content of the index in the two groups. (c) The correlation between SCFAs. Circles represent the correlation between two substances, larger circles indicate higher correlation, blue represents negative correlation, red represents positive correlation, and × represents no significance at the test level. (d) Compared with group R, the content of SCFAs in group NR was generally lower. (e) The results showed that except for the receiver operating characteristic (ROC) of isohexanoic acid, the ROC models of the other seven SCFAs had good predictive value for the prognosis of advanced NSCLC. The x‐axis is 1‐specificity, and the coordinate axis is 0–1. The y‐axis is sensitivity, and the coordinate axis is 0–1. The point marked on the curve is the best critical value (the value in parentheses is the specificity and sensitivity corresponding to the point). (f) In the correlation analysis between gut microbiota and SCFAs, Faecalibacterium was positively correlated with butyrate, acetic acid and valeric acid. Escherichia‐Shigella, Prevotella_9, and Phascolarctobacterium was positively correlated with hexanoic acid. Roseburia and Enterobacter was positively correlated with propanoic acid. Statistical analysis was performed by one‐way analysis of variance (ANOVA) (a), Mann‐Whitney (b, d) or Spearman's correlation (c, f) tests. Data are expressed as mean ± SEM. *0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001.
FMT delays the progression of LLC mice
To investigate the influence of gut microbiota on the tumor growth, we established a subcutaneous tumor model of C57BL mice and PD‐1 was administered via intraperitoneal injection. We then administered antibiotic (ATB) compounds to deplete gut microbiota for one week. Stools from NR and R group patients were orally gavaged into LLC recipient mice. A total of 36 mice were divided into six groups with six mice in each group. The mice in each group were gavaged with the fecal supernatant of six NSCLC patients to perform FMT (Figure 4a).
After antibiotic treatment, the diversity indicators and OTUs of gut microbiota were significantly decreased. Fecal transplantation restored the diversity of the gut microbiota caused by antibiotics (Figure 4b,c). The diversity of gut microbiota in mice treated with FMT combined with PD‐1 monoclonal antibody was significantly increased, p < 0.05. Then, we evaluated the effect of FMT on the tumor burden in LLC mice. The result showed that, to a certain extent, the gut microbiota from LLC mice, especially in the R FMT group, delayed the tumor progression (Figure 4d). The study findings showed that the lower the intestinal diversity of mice, the faster the tumor progression. Immunotherapy can delay the tumor progression, and FMT further enhances the effect of immunotherapy. The number of Ki‐67 tumor cells were reduced in the NR FMT and R FMT groups, p < 0.05 (Figure 4e). FMT also led to a reduction in PD‐L1 expression compared with blank (PBS) group, although the difference was not significant, p > 0.05 (Figure 4f).
We sequenced the 16S rRNA of the colon contents and found that the composition of the gut microbiota differed among the six groups of mice. The functional prediction of the gut microbiota in mice showed similar results to those observed in human samples (Figure 4g). Moreover, the results demonstrated that the diversity of the gut microbiota was significantly reduced in mice that received antibiotics, which disrupted the gut microbiota and accelerated tumor development. Antibiotics also attenuated the therapeutic effect of PD‐1 inhibitors. After analyzing the mice following changes in the gut microbiota, we found that the diversity of the gut microbiota in mice receiving FMT from R patients was improved compared to baseline. However, the results were not significantly different when compared with the FMT in the NR group, p > 0.05 (Figure 4h). At the genus level, we observed differences in species composition between each group, although not in the same way as observed in clinical samples (Figure 4i). Muribaculaceae and Ligilactobacillus were significantly enriched in the R FMT group compared to the NR FMT group. Escherichia‐Shigella, Parabacteroides, Lachnospiraceae_NK4A136_group, Helicobacter and Blautia were significantly enriched in the NR FMT group. Overall, our findings suggest that diversity of gut microbiota can influence the response to PD‐1 inhibitor treatment. FMT from patients can improve the diversity of the gut microbiota and delay tumor progression in animal models.
DISCUSSION
NSCLC is a type of lung cancer that is characterized by its aggressive nature and high mortality rate. 28 , 29 The occurrence and progression of NSCLC are often associated with immune evasion, which is related to immune suppression. Although immunotherapy has achieved promising results in the treatment of patients with advanced NSCLC, it is only effective in a subset of tumor types, and the incidence of immune‐related complications is also high due to individual variations. 30 , 31 The challenges of immunotherapy for NSCLC are numerous and complex. One of the primary challenges is the heterogeneity of NSCLC, which means that different patients may respond differently to immunotherapy. This heterogeneity can be attributed to various factors such as tumor genotype, immune status, and tumor microenvironment. Another challenge is the development of resistance to immunotherapy. While immunotherapy has shown promising results in some NSCLC patients, many patients eventually develop resistance to the treatment, which can lead to tumor recurrence. Understanding the mechanisms of resistance and finding ways to overcome it are crucial for improving the long‐term outcomes of immunotherapy. 32 Advancements in genomics and metabolomics technology have shed light on the role of gut microbiota and their metabolites in tumor occurrence, development, and treatment. 33 It is now increasingly recognized that the microbiota can influence tumor immunotherapy by modulating the host immune system and TME. 34 , 35 Microbiota‐derived metabolites can directly or indirectly regulate host immunity and metabolism, and play a crucial role in tumor initiation and progression. 36
Sivan et al. demonstrated that bifidobacterium species can inhibit tumor growth by activating dendritic cells (DCs), which in turn enhances effector function of tumor‐specific CD8 + T cells. However, the specific mechanism by which bifidobacterium activates DCs to improve the antitumor effect of CD8 + cells require further investigation. 21 In a study by Routy et al., which included data from patients with advanced NSCLC, renal and urothelial cancer, it was found that patients who received antibiotics before or immediately after immunotherapy had shorter progression‐free and overall survival with PD‐1 inhibitor immunotherapy. Shotgun sequencing of the gut microbiota revealed that responders to PD‐1 inhibitor immunotherapy were enriched in Akkermansia, suggesting that Akkermansia may enhance the response to PD‐1 inhibitor immunotherapy. However, the underlying mechanism by which Akkermansia enhances PD‐1 inhibitor immunotherapy remains unknown. 37 In addition to the composition of gut microbiota, microbes' metabolites may also play a key role. 38 SCFAs are major metabolites produced by intestinal microbial fermentation of dietary fiber. SCFAs can directly activate G protein coupled receptor (GPCR) and inhibit histone deacetylases (HDACs). 39 , 40 SCFAs can also regulate tumor immunity by modulating the differentiation and function of various immune cells, increasing the sensitivity of tumor cells to apoptosis, reducing hypoxia‐inducible factors (HIFs), and enhancing the immunogenicity of tumor cells to improve the efficacy of chemotherapy and immunotherapy. 27 , 41
In this study, we conducted a prospective investigation of the gut microbiota profiles in 41 patients with advanced NSCLC starting ICI therapy using 16S rRNA sequencing. Our findings indicate that patients who exhibited better clinical responses had higher diversity and a distinct gut microbiome composition compared to NR. The alpha diversity index, represented by Ace, Chao, Coverage, and Sobs indexes, was significantly higher in the R group, indicating a greater number of species in the gut microbiota. Ace, Chao and Sobs estimate microbial diversity in the sample. Coverage reflects the real situatuin of microorganisms in the sample. This observation was consistent with the differential composition of the gut microbiome between R and NR patients. Beta diversity analysis through PCoA also revealed distinct clusters based on clinical responses to ICB treatment. These results suggest that regardless of genetic background and geographic location, the diversity of the gut microbiota may be a common factor associated with a favorable response to ICB treatment. In addition to diversity, the composition of the gut microbiota was also related to favorable responses to ICI treatment. Bacteroides was the most frequent genus observed in our cohort of patients with advanced NSCLC, which is consistent with previous studies demonstrating an abundance of Bacteroides in the human gut microbiota. 42 Further analysis revealed differences in the gut microbiota composition between R and NR. Faecalibacterium, Ruminococcus, Lachnospiraceae_NK4A136_group, [Eubacterium]_siraeum_group, and unclassified_c__Clostridia were enriched in the R group. LEfSe further confirmed a higher abundance of Faecalibacterium in the feces of the R group compared to the NR group. To further investigate the changes in the gut microbiota after immunotherapy, we analyzed the sequencing data of fecal samples. A higher alpha diversity index represented by Ace, Chao and Sobs indexes after immunotherapy was found. Interestingly, we found that Blautia and E. Shigella decreased after immunotherapy, but the difference was not significant. Studies have shown that Blautia levels are significantly higher in patients with lung cancer relative to healthy people. The results showed that Blautia was the prominent gene level biomarkers for the early‐stage NSCLC group. 43 Studies have found harmful effects of cancer cachexia on patients with advanced NSCLC treated with ICIs. The intestinal microbiota profile shows that E. Shigella is highly associated with cachexia in patients at the genus level. 44 This also suggests that E. Shigella may be associated with a poor prognosis of immunotherapy in patients with NSCLC. 45 However, more research data may be needed to confirm the specific mechanism of action of the unique gut microbiota in lung cancer immunotherapy. Further analysis revealed differences in gut microbiota composition between the prior to immunotherapy (B) and the following immunotherapy (A). Our results show that Faecalibacterium increased after immunotherapy. A previous clinical investigation found that Faecalibacterium were positively associated with the efficacy of PD‐1 and CTLA‐4 blockade. 46 This is also consistent with the report that patients are more sensitive to ICI treatment.
A large number of studies have shown that most SCFAs in the human body, including butyrate, are derived from the metabolism of intestinal bacteria. Faecalibacterium is known to have anti‐inflammatory properties, improve metabolic disorders, and is a major producer of butyrate in the gut. 47 In this study, the results showed that R patients had a significantly higher fecal butyric acid content than NR patients. In comparing the two groups, the content of SCFAs was generally lower in the NR group. It has previously been reported that NSCLC R patients exhibited higher levels of serum acetic acid, propionic acid, and butyric acid than NR patients. 48 The correlation between gut microbiota and SCFAs was analyzed, and it was found that faecalibacterium was positively correlated with butyrate. In recent studies, Clostridium butyricum MIYAIRI has been reported to have an optimistic association with ICI efficacy in NSCLC. 49 Although the results showed a positive association between butyric acid and immunotherapy response, a significant difference in Clostridium butyricum prevalence between responders and nonresponders was not found. One possibility lies in the number limitation in this study. Overall, this study highlights the association between gut microbiota composition, diversity, and the response to ICI therapy in patients with advanced NSCLC. The enrichment of specific bacterial taxa and SCFAs, particularly Faecalibacterium and butyrate, in responders suggests their potential role in enhancing the efficacy of ICB therapy. It is not out of expectation that systemic antitumor immune responses are diverse in patients with different gut microbiome composition.
Given these findings, we investigated whether combining ICIs with FMT could reverse the nonresponse state. Further research is needed to explore the therapeutic potential of combining ICB with FMT to modulate the gut microbiota and improve treatment outcomes in nonresponsive patients. In this study, we performed FMT by transferring stools from three R patients and three NR patients to C57BL mice with LLC tumors. We were excited to find that the mice gavaged with fecal supernatant from NSCLC patients showed restoration of gut microbiota diversity after antibiotic treatment. However, diversity was also not different in mice with FMT from the R and NR groups. We considered that only a portion of the patient‐derived gut microbiota colonized the gut of mice. We observed that the mice treated with antibiotics had a poor response to PD‐1 treatment, and FMT from the effective group enhanced the antitumor effect of PD‐1. Additionally, the diversity of the gut microbiota was significantly reduced in mice that received antibiotics, which disrupted the gut microbiota and accelerated tumor development. Antibiotics also attenuated the therapeutic effect of PD‐1 inhibitors. This finding indicated that the disordered microbiota affected the progression of the disease. Analysis of tumor tissues showed that FMT effectively reduced Ki‐67 expression, but had no significant effect on PD‐L1 expression, which may contribute to the ability of FMT to slow tumor growth. However, in this study, after administration of FMT, LLC mice showed no significant difference in progression of the disease between R FMT and NR FMT, which indicated that the antitumor effect of R FMT was limited. We sequenced the 16S rRNA of the colon contents and found differences in the composition of the gut microbiota among the six groups of mice, with similar functional predictions as observed in human samples. The results of this study indicated that FMT could effectively enrich the diversity of gut microbiota in mice. However, there was no statistically significant difference in antitumor effect among R and NR FMT. This finding suggests that while FMT could indeed modulate the gut microbiota, its antitumor effect may not be solely determined by the diversity of gut microbiota. Other factors, such as the specific composition of the microbiota, the interaction between microbiota and the immune system, and the genetic background of the host may also play important roles in determining the antitumor effect of FMT. Nevertheless, our findings do not rule out the potential of FMT in immunotherapy. Future studies are needed to further explore the mechanisms underlying the anti‐tumor effect of FMT and identify the optimal conditions for its application in immunotherapy.
However, there were several limitations to our study. First, we used a mouse model of LLC with the technical limitations of direct cellular implantation, which may not perfectly replicate the anatomy of lung cancer. Nonetheless, we believe that our study convincingly demonstrates the influence of the gut microbiota on lung cancer and immune checkpoint blockade. Second, tumor growth is influenced by various complex factors, and immunity is just one aspect. The individual differences in the composition of the gut microbiota between animal models and humans should be considered, and the microbial spectrum of the gut microbiota in different individuals explored further, which is crucial for developing personalized strategies for optimizing the gut microbiota in precision medicine. Lastly, a small sample size may not be representative of the broader population, limiting the generalizability of the study findings. The research is currently limited to horizontal clinical studies, and more clinical samples are needed for clinical studies. It is believed that there are many undiscovered potential connections between the complex gut microbiota and the immune system waiting to be explored.
In conclusion, research has demonstrated that the gut microbiota and its metabolites play a role in the development of various malignancies and can also impact the clinical outcomes of tumor immunotherapy. Gut microbiota diversity and SCFAs are significantly related to the response to NSCLC immunotherapy. The higher the gut diversity and SCFAs the better the effect of immunotherapy. Faecalibacterium shows a correlation with immunotherapy efficacy. Further exploration of the relationship between gut microbiota metabolites and the immune system could provide new avenues for the treatment of advanced NSCLC patients. Additionally, investigating whether interventions targeting the gut microbiota, such as antibiotics, probiotics, and fecal microbiota transplantation (FMT), can influence the cancer‐promoting or cancer‐suppressing effects of metabolites is an important area for future research.
AUTHOR CONTRIBUTIONS
Shengnan Ren performed the data analysis, wrote the manuscript and performed the formal analysis. Lingxin Feng performed the data analysis. Haoran Liu collected clinical samples. Yuke Mao performed the validation. Zhuang Yu performed the writing‐review and supervision.
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
The present study was supported by Shandong Provincial Natural Science Foundation (ZR202102240880), Qingdao Natural Science Foundation(23‐2‐1‐189‐zyyd‐jch), Chinese Society of Clinical Oncology Research foundation (Y‐QL202101‐0258 and Y‐pierrefabre202101‐0074), Wu Jieping Medical Foundation (320.6750.2021‐02‐92), ShanDong Provincial Medical Association (YXH2022ZX02020), and National Natural Science Foundation of China (82373170).
Ren S, Feng L, Liu H, Mao Y, Yu Z. Gut microbiome affects the response to immunotherapy in non‐small cell lung cancer. Thorac Cancer. 2024;15(14):1149–1163. 10.1111/1759-7714.15303
DATA AVAILABILITY STATEMENT
Data are available on request to the authors.
REFERENCES
- 1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553(7689):446–454. [DOI] [PubMed] [Google Scholar]
- 2. Uprety D, Mandrekar SJ, Wigle D, Roden AC, Adjei AA. Neoadjuvant immunotherapy for NSCLC: current concepts and future approaches. J Thorac Oncol. 2020;15(8):1281–1297. [DOI] [PubMed] [Google Scholar]
- 3. Proto C, Ferrara R, Signorelli D, Lo Russo G, Galli G, Imbimbo M, et al. Choosing wisely first line immunotherapy in non‐small cell lung cancer (NSCLC): what to add and what to leave out. Cancer Treat Rev. 2019;75:39–51. [DOI] [PubMed] [Google Scholar]
- 4. Suresh K, Naidoo J, Lin CT, Danoff S. Immune checkpoint immunotherapy for non‐small cell lung cancer: benefits and pulmonary toxicities. Chest. 2018;154(6):1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Błach J, Wojas‐Krawczyk K, Nicoś M, Krawczyk P. Failure of immunotherapy‐the molecular and immunological origin of immunotherapy resistance in lung cancer. Int J Mol Sci. 2021;22(16):9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aldea M, Andre F, Marabelle A, Dogan S, Barlesi F, Soria JC. Overcoming resistance to tumor‐targeted and immune‐targeted therapies. Cancer Discov. 2021;11(4):874–899. [DOI] [PubMed] [Google Scholar]
- 7. Horvath L, Thienpont B, Zhao L, Wolf D, Pircher A. Overcoming immunotherapy resistance in non‐small cell lung cancer (NSCLC) ‐ novel approaches and future outlook. Mol Cancer. 2020;19(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giannone G, Ghisoni E, Genta S, Scotto G, Tuninetti V, Turinetto M, et al. Immuno‐metabolism and microenvironment in cancer: key players for immunotherapy. Int J Mol Sci. 2020;21(12):4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Genova C, Dellepiane C, Carrega P, Sommariva S, Ferlazzo G, Pronzato P, et al. Therapeutic implications of tumor microenvironment in lung cancer: focus on immune checkpoint blockade. Front Immunol. 2022;12:799455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai J, Sun L, Gonzalez FJ. Gut microbiota‐derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. 2022;30(3):289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong Q, Chen ES, Zhao C, Jin C. Host‐microbiome interaction in lung cancer. Front Immunol. 2021;24(12):679829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bishehsari F, Voigt RM, Keshavarzian A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat Rev Endocrinol. 2020;16(12):731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simpson RC, Shanahan ER, Batten M, Reijers ILM, Read M, Silva IP, et al. Diet‐driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat Med. 2022;28(11):2344–2352. [DOI] [PubMed] [Google Scholar]
- 14. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amoroso C, Perillo F, Strati F, Fantini MC, Caprioli F, Facciotti F. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells. 2020;9(5):1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yi M, Jiao D, Qin S, Chu Q, Li A, Wu K. Manipulating gut microbiota composition to enhance the therapeutic effect of cancer immunotherapy. Integr Cancer Ther. 2019. Jan‐Dec;18:1534735419876351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sano T, Kageyama T, Fang V, Kedmi R, Martinez CS, Talbot J, et al. Redundant cytokine requirement for intestinal microbiota‐induced Th17 cell differentiation in draining lymph nodes. Cell Rep. 2021;36(8):109608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L, Zhou X, Wang Y, Wang D, Ke Y, Zeng X. Propionate and butyrate produced by gut microbiota after probiotic supplementation attenuate lung metastasis of melanoma cells in mice. Mol Nutr Food Res. 2021;65(15):e2100096. [DOI] [PubMed] [Google Scholar]
- 21. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350(6264):1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baruch EN, Youngster I, Ben‐Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy‐refractory melanoma patients. Science. 2021;371(6529):602–609. [DOI] [PubMed] [Google Scholar]
- 23. Huang J, Liu D, Wang Y, Liu L, Li J, Yuan J, et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti‐PD‐1/PD‐L1) immunotherapy. Gut. 2022;71(4):734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 25. Charan J, Kantharia ND. How to calculate sample size in animal studies? J Pharmacol Pharmacother. 2013;4(4):303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao L, Huang Y, Lu L, Yang W, Huang T, Lin Z, et al. Saturated long‐chain fatty acid‐producing bacteria contribute to enhanced colonic motility in rats. Microbiome. 2018;6(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Silva YP, Bernardi A, Frozza RL. The role of short‐chain fatty acids from gut microbiota in gut‐brain communication. Front Endocrinol (Lausanne). 2020. Jan;31(11):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. [DOI] [PubMed] [Google Scholar]
- 29. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):463–473. [DOI] [PubMed] [Google Scholar]
- 30. Zhou F, Qiao M, Zhou C. The cutting‐edge progress of immune‐checkpoint blockade in lung cancer. Cell Mol Immunol. 2021;18(2):279–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen R, Manochakian R, James L, Azzouqa AG, Shi H, Zhang Y, et al. Emerging therapeutic agents for advanced non‐small cell lung cancer. J Hematol Oncol. 2020;13(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reck M, Remon J, Hellmann MD. First‐line immunotherapy for non‐small‐cell lung cancer. J Clin Oncol. 2022;40(6):586–597. [DOI] [PubMed] [Google Scholar]
- 33. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage‐specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. [DOI] [PubMed] [Google Scholar]
- 34. Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome‐influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology. 2021;160(2):600–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. 2022;15(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou CB, Zhou YL, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. 2021;7(7):647–660. [DOI] [PubMed] [Google Scholar]
- 37. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. [DOI] [PubMed] [Google Scholar]
- 38. Zhang S, Wang R, Li D, Zhao L, Zhu L. Role of gut microbiota in functional constipation. Gastroenterol Rep (Oxf). 2021;9(5):392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barki N, Bolognini D, Börjesson U, Jenkins L, Riddell J, Hughes DI, et al. Chemogenetics defines a short‐chain fatty acid receptor gut‐brain axis. Elife. 2022;1(11):e73777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Traisaeng S, Herr DR, Kao HJ, Chuang TH, Huang CM. A derivative of butyric acid, the fermentation metabolite of Staphylococcus epidermidis, inhibits the growth of a Staphylococcus aureus strain isolated from atopic dermatitis patients. Toxins (Basel). 2019;11(6):311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Albillos A, de Gottardi A, Rescigno M. The gut‐liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–577. [DOI] [PubMed] [Google Scholar]
- 42. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ni B, Kong X, Yan Y, Fu B, Zhou F, Xu S. Combined analysis of gut microbiome and serum metabolomics reveals novel biomarkers in patients with early‐stage non‐small cell lung cancer. Front Cell Infect Microbiol. 2023;20(13):1091825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hakozaki T, Nolin‐Lapalme A, Kogawa M, Okuma Y, Nakamura S, Moreau‐Amaru D, et al. Cancer cachexia among patients with advanced non‐small‐cell lung cancer on immunotherapy: An observational study with exploratory gut microbiota analysis. Cancers (Basel). 2022;14(21):5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He D, Li X, An R, Wang L, Wang Y, Zheng S, et al. Response to PD‐1‐based immunotherapy for non‐small cell lung cancer altered by gut microbiota. Oncol Ther. 2021;9(2):647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martín R, Rios‐Covian D, Huillet E, Auger S, Khazaal S, Bermúdez‐Humarán LG, et al. Faecalibacterium: a bacterial genus with promising human health applications. FEMS Microbiol Rev. 2023;47(4):fuad039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu X, Li K, Liu G, Wu R, Zhang Y, Wang S, et al. Microbial metabolite butyrate promotes anti‐PD‐1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microbes. 2023;15(2):2249143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paz Del Socorro T, Oka K, Boulard O, Takahashi M, Poulin LF, Hayashi A, et al. The biotherapeutic clostridium butyricum MIYAIRI 588 strain potentiates enterotropism of Rorγt+Treg and PD‐1 blockade efficacy. Gut Microbes. 2024;16(1):2315631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on request to the authors.


