Abstract
Background
Inflammatory bowel disease (IBD) and Parkinson’s disease (PD) are chronic disorders that have been suggested to share common pathophysiological processes. LRRK2 has been implicated as playing a role in both diseases. Exploring the genetic basis of the IBD-PD comorbidity through studying high-impact rare genetic variants can facilitate the identification of the novel shared genetic factors underlying this comorbidity.
Methods
We analyzed whole exomes from the BioMe BioBank and UK Biobank, and whole genomes from a cohort of 67 European patients diagnosed with both IBD and PD to examine the effects of LRRK2 missense variants on IBD, PD and their co-occurrence (IBD-PD). We performed optimized sequence kernel association test (SKAT-O) and network-based heterogeneity clustering (NHC) analyses using high-impact rare variants in the IBD-PD cohort to identify novel candidate genes, which we further prioritized by biological relatedness approaches. We conducted phenome-wide association studies (PheWAS) employing BioMe BioBank and UK Biobank whole exomes to estimate the genetic relevance of the 14 prioritized genes to IBD-PD.
Results
The analysis of LRRK2 missense variants revealed significant associations of the G2019S and N2081D variants with IBD-PD in addition to several other variants as potential contributors to increased or decreased IBD-PD risk. SKAT-O identified two significant genes, LRRK2 and IL10RA, and NHC identified 6 significant gene clusters that are biologically relevant to IBD-PD. We observed prominent overlaps between the enriched pathways in the known IBD, PD, and candidate IBD-PD gene sets. Additionally, we detected significantly enriched pathways unique to the IBD-PD, including MAPK signaling, LPS/IL-1 mediated inhibition of RXR function, and NAD signaling. Fourteen final candidate IBD-PD genes were prioritized by biological relatedness methods. The biological importance scores estimated by protein–protein interaction networks and pathway and ontology enrichment analyses indicated the involvement of genes related to immunity, inflammation, and autophagy in IBD-PD. Additionally, PheWAS provided support for the associations of candidate genes with IBD and PD.
Conclusions
Our study confirms and uncovers new LRRK2 associations in IBD-PD. The identification of novel inflammation and autophagy-related genes supports and expands previous findings related to IBD-PD pathogenesis, and underscores the significance of therapeutic interventions for reducing systemic inflammation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01335-2.
Keywords: Crohn’s disease, Genetic pleiotropy, Inflammatory bowel disease, Parkinson’s disease, Ulcerative colitis, Whole genome sequencing
Background
Inflammatory bowel disease (IBD) comprises a group of chronic inflammatory diseases that primarily affect the gastrointestinal tract, including Crohn’s disease (CD) and ulcerative colitis (UC). CD is distinguished by segmental transmural inflammation affecting the ileum and colon, whereas UC is usually limited to the colon, and characterized by mucosal inflammation [1]. Parkinson’s disease (PD) is one of the most common neurodegenerative disorders, presenting with bradykinesia, rigidity, resting tremor, and postural instability [2]. It is characterized by the progressive loss of dopaminergic neurons in the substantia nigra and the presence of intracellular protein aggregates known as Lewy bodies [3]. Emerging evidence suggests a link between these two apparently unrelated disorders, indicating shared risk factors and underlying pathophysiology that is consistent with the “gut-brain axis” hypothesis [4–8]. IBD-PD co-occurrence has been attributed to neurodegeneration driven by chronic intestinal inflammation and pleiotropic genetic factors [4]. A recent meta-analysis, involving 12 million patients from 9 observational studies, provided further support for previous findings, indicating that both CD and UC are associated with an increased risk of PD diagnosis [9]. This increased risk was particularly prominent in older patients (> 65 years old), irrespective of gender. Moreover, exposure to anti-inflammatory tumor necrosis factor-α inhibitors in IBD patients has been linked to a significant reduction in PD risk [6, 9]. These findings suggest the involvement of inflammation-mediated processes and/or potentially shared genetic factors underlying both IBD and PD.
The most well-established gene implicated in the IBD-PD pleiotropy is leucine-rich repeat kinase 2 (LRRK2). Polymorphisms in LRRK2 have been shown to be associated with both PD and CD, suggesting the impact of impaired autophagy in the pathogenesis of both conditions [5]. Variants that result in increased activity of LRRK2 have been shown to be associated with an elevated risk of developing both PD and CD, whilst a haplotype with a deactivating LRRK2 mutation, R1398H, has been found to be associated with protection against CD [5] and PD [10–12]. However, despite genetic pleiotropy for some of the LRRK2 variants (i.e., G2019S, N2081D, N551K, and R1398H) [5], each of these conditions is associated with specific LRRK2 variants. For example, G2019S is the major genetic risk for PD [13], whereas N2081D is considered a risk for CD [5, 14]. Moreover, other strong genetic predictors of PD, such as R1441G/C/H, Y1699C, R1628P, G2385R, and I2020T, have been shown to be associated exclusively with PD [15], whereas M2397T was not linked to PD [16]. Therefore, it is not immediately clear whether any of these or other LRRK2 variants may lead to IBD-PD comorbidity. Other than LRRK2, several other pleiotropic loci, including MAPT, HLA, MHC, ATP6V0A1, and NOD2, have been identified to be associated with PD, CD, UC, and other autoimmune disorders [4, 7, 8]. Previous studies that have examined the genetic pleiotropy between IBD and PD have primarily estimated the genetic correlation between these two conditions by means of genome-wide association study (GWAS) data from separate analyses of IBD and PD [7, 8]. However, conducting a joint analysis of individuals affected by both IBD and PD would provide important insights into the underlying mechanisms shared by these two conditions.
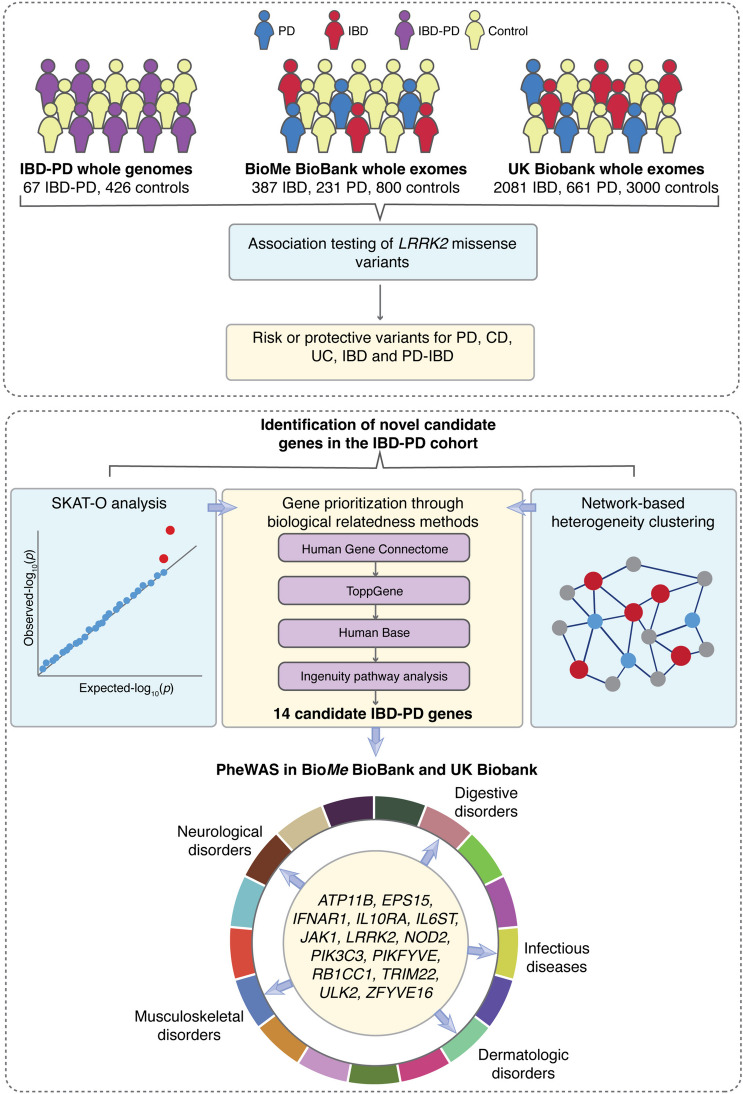
Here, we investigated the effect of LRRK2 missense variants on PD only, CD only, UC only, IBD only, and the co-occurrence of IBD and PD risk (IBD-PD) using sequencing data from the Danish Registry, the Mount Sinai BioMe BioBank, and the UK Biobank. Furthermore, we performed a series of gene-level association and network-based analyses using high-impact rare variants in the IBD-PD cohort and prioritized candidate physiologically-relevant genes associated with this comorbidity. Finally, we conducted phenome-wide association studies (PheWAS) in BioMe BioBank and UK Biobank to evaluate the pleiotropic effects of the candidate IBD-PD genes, as well as IBD-PD comorbidities (Fig. 1).
Fig. 1.
Study workflow. Parkinson’s disease (PD) and inflammatory bowel disease (IBD) cases were identified based on ICD-10 codes in the UK Biobank and BioMe BioBank. The IBD-PD cohort comprised 67 individuals diagnosed with both PD and IBD and 426 healthy controls from the 1000 Genomes Project (1KGP). Variant-level association testing was performed separately in each cohort using LRRK2 variants observed in the whole exome (UK Biobank and BioMe BioBank) and whole genome sequencing (IBD-PD cohort) data (upper panel). Discovery of novel candidate genes was performed using the sequence kernel association test (SKAT-O) and network-based heterogeneity clustering (NHC) methods; the genes were further prioritized using pathway enrichment and biological relatedness methods. Fourteen prioritized candidate genes were then investigated in the BioMe BioBank and UK Biobank using phenome-wide association analysis (PheWAS) to validate identified associations (lower panel)
Methods
Study cohorts
IBD-PD cohort
We identified 76 patients diagnosed with both IBD and PD in the Danish National Biobank resource (https://www.danishnationalbiobank.com/) using the International Classification of Diseases (ICD) codes from the Danish National Patient Registry (Additional file 1: Identification of the IBD-PD samples in the Danish National Biobank). Owing to the regulations of the Danish Data Protection Agency, we were precluded from gathering demographic and clinical data regarding patients’ specific IBD subtypes or their ages at the time of biosample collection. Whole-genome sequencing (WGS) was performed using the Illumina HiSeq X platform with 2 × 150 bp paired-end reads. Alignment of the raw reads and variant calling were performed following the best practice guidelines of the Genome Analysis Toolkit (GATK) [17] (Additional file 1: Whole genome sequencing of the IBD-PD samples).
For the control group, high-coverage WGS data of 1000 Genomes Project Phase 3 (1KGP) individuals were downloaded on March 20, 2023, from the following URL: http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20190425_NYGC_GATK/raw_calls_updated/ [18]. This dataset comprised 427 samples from non-Finnish European populations. Joint genotyping and quality control (QC) steps were performed on this combined case–control dataset (Additional file 1: Genotyping and quality control of the IBD-PD cohort and Additional file 2: Table S1). After QC filtration, which led to the exclusion of 9 IBD-PD cases and 1 control, the final dataset comprised a total of 20,158,023 variants across 67 IBD-PD cases and 426 controls. Of these, 40 (59.7%) IBD-PD cases and 214 (50.2%) controls were males. Genetic ancestries of the IBD-PD cases were determined using principal component analysis (PCA) and ADMIXTURE [19] by utilizing 1KGP populations as a reference, which confirmed their European ancestry (Additional file 1: Determining the genetic ancestries of the IBD-PD cases, Fig. S1 and S2).
BioMe BioBank
The Mount Sinai BioMe BioBank comprises whole exome sequencing (WES) data from 31,250 participants who were recruited from Mount Sinai primary care clinics (https://icahn.mssm.edu/research/ipm/programs/biome-biobank). No specific criteria were used during the recruitment; therefore, the BioMe participants should be a representative sample of New York City and its neighborhood. The genetic data of the participants are linked to their electronic health records (EHR), which we used to select IBD, CD, UC, and PD cases and controls. WES was performed by Regeneron (NY, USA) using the IDT xGen capture kit on an Illumina v4 HiSeq 2500 platform (a total of 8,761,478 sites across 31,250 samples). A QC check was performed to remove contaminated, low coverage, gender discordant, genotype-exome discordant, and duplicate samples. Variant sites with a missingness rate > 0.02, or allelic balance < 0.30 or > 0.80 were excluded from the dataset. The final dataset included 30,813 samples and 8,890,425 variants.
UK Biobank
The UK Biobank dataset is a comprehensive collection of health-related information gathered from over 500,000 participants across the United Kingdom [20]. WES data of 200,643 UK Biobank participants were used in the current study. A QC check was performed to select samples and variants of high quality, using the following steps using PLINK v.1.9 [21]. Samples with a missingness rate exceeding 5%, displaying gender discrepancies and duplicated samples were excluded. Second-degree or closer relatives were determined using KING and excluded to retain unrelated samples in the dataset [22]. Variants were removed if their missingness rate exceeded 20% or if they demonstrated a significant deviation from Hardy–Weinberg equilibrium (HWE) with a P < 1 × 10−6. Consequently, the final dataset comprised 189,448 samples 17,402,345 variants.
In both BioMe BioBank and UK Biobank, individuals of European descent were genetically determined using HapMap3 populations [23] as the reference through an analysis with fastSTRUCTURE [24]. CD, UC, IBD, and PD cases and controls were identified based on ICD-9 and ICD-10 diagnoses of BioMe BioBank and UK Biobank participants (Additional file 1: Identification of IBD and PD samples with European descent from BioMe and UK Biobank). The final counts of PD, IBD, CD, and UC cases, as well as controls, are detailed in Table 1.
Table 1.
Cohorts used in the variant-level analysis of LRRK2
| Cohort | Phenotype | Case# | Control# | Sex, cases (F/M) | Sex, controls (F/M) |
|---|---|---|---|---|---|
| IBD-PD | IBD-PD | 67 | 426 | 27/40 | 214/212 |
| BioMe BioBank | PD | 231 | 800 | 93/138 | 441/359 |
| IBD | 387 | 800 | 205/182 | ||
| CD | 209 | 800 | 116/93 | ||
| UC | 141 | 800 | 67/74 | ||
| UK Biobank | PD | 661 | 3000 | 270/391 | 1734/1266 |
| IBD | 2081 | 3000 | 1069/1012 | ||
| CD | 603 | 3000 | 350/253 | ||
| UC | 1282 | 3000 | 613/669 |
Variant annotation and filtration
To assess predicted impacts on gene products and population frequencies of the variants identified in the IBD-PD cohort, as well as in BioMe Biobank and UK Biobank, variants were annotated using Variant Effect Predictor (VEP) v.106, CADD scores (v.1.6), and allele frequencies in gnomAD v2 and 1KGP populations [18, 25–27]. To control the false-negative rate of predicted deleterious mutations by CADD, a Mutation Significance Cutoff (MSC, https://lab.rockefeller.edu/casanova/MSC) was applied for each gene [28]. Additionally, genes were annotated with the Gene Damage Index (GDI, https://lab.rockefeller.edu/casanova/GDI), which serves as an indicator of genes that exhibit high polymorphism in the general healthy population and hence are less likely to be disease-associated [29].
For the identification of novel candidate genes associated with IBD-PD, variants were filtered based on their consequences as obtained from VEP annotation. High-impact variants (“start lost,” “stop lost,” “stop gained,” “splice_acceptor_variant,” “splice_donor_variant,” “protein_altering_variant,” “start_retained_variant,” “stop_retained_variant,” and ‘frameshift_variant”) and moderate-impact variants (“missense variants,” “inframe_insertion,” “inframe_deletion”) were extracted. Moderate-impact variants were further filtered if their CADD scores were greater than the lower boundary of the 95% confidence interval of the corresponding gene’s MSC. Further, variants in genes with a GDI value of less than 13.84 (the cutoff proposed for human diseases under the generalized model) were included [29]. Finally, variants with a maximum minor allele frequency (MAF) below 1% in gnomAD and 1KGP subpopulations were retained to identify the final set of presumably deleterious variants for further analyses. The same filtering steps according to MAF and GDI were applied to obtain synonymous variants for the neutral model, which estimates the degree of inflation due to population substructure or possible technical artifacts.
Analysis of LRRK2 variants
To conduct an in-depth characterization of the role of LRRK2 missense variants in IBD-PD comorbidity, as well as to assess their effect on CD, UC, IBD, and PD, we extracted all LRRK2 missense variants from the IBD-PD, BioMe BioBank, and UK Biobank cohorts. PCA was performed using PLINK v1.9 with linkage disequilibrium (LD)-pruned variants (r2 = 0.2) with a MAF greater than 5% and not exceeding HWE with a P < 1 × 10−6 [21]. The selection of the number of PCs to be included as covariates in the analyses was based on the scree test [30] (Additional file 1: Fig. S3). The first two genetic principal components (PCs), sex and age, were used as covariates in the BioMe BioBank analyses while PC1, sex, and age were employed as covariates in the UK Biobank analyses. For the IBD-PD cohort, PC1 and sex were used as covariates. For association testing of LRRK2 missense variants with a minor allele count (MAC) ≥ 3 and a case MAC ≥ 1, we used Firth’s logistic regression method implemented in PLINK v.2.0 [21, 31]. Pairwise LDs of LRRK2 missense variants were calculated using PLINK v.1.9.
Sequence Kernel Association Test analysis and network-based heterogeneity clustering of the IBD-PD cohort
To identify novel genes associated with the IBD-PD comorbidity, we used the optimized method of the Sequence Kernel Association Test (SKAT-O) to accommodate rare variants with potentially different directions of effect [32]. SKAT-O is an optimal combination of the burden test and SKAT, aiming to enhance statistical power. The burden test combines minor alleles at variants in a region, assuming they have the same direction of effect, and compares the difference in allele frequencies between cases and controls. On the other hand, SKAT employs a regression framework and a variance-component test, allowing it to account for variants with different directions of effect. SKAT-O dynamically operates as the burden test when it surpasses SKAT in power, and functions as SKAT when SKAT exhibits greater power than the burden test [32]. Initially, we employed SKAT-O using a set of synonymous variants of 72 IBD-PD cases and 426 controls (Fig. 1), incorporating PC1 and sex as covariates [33]. The analysis revealed genes with significantly inflated P values (Additional file 1: Fig. S4A). Similarly, SKAT-O using presumably deleterious variants also displayed genes with inflated P values (Additional file 1: Fig. S4B). Further investigation into the cause of this inflation resulted in the removal of 5 IBD-PD cases that exhibited an excess number of heterozygous variants in these genes. Subsequent analysis with 67 IBD-PD cases and 426 controls resolved the inflation issue (Additional file 1: Fig. S4C and Fig. S4D). Among the analyzed genes, only EPHA4 exhibited a significant burden of synonymous variants (P = 3.26 × 10−6). Subsequently, SKAT-O was performed using the set of presumably deleterious variants in the 67 IBD-PD cases and 426 controls, while employing the same covariates. The Bonferroni correction method was used to adjust P values so as to allow for multiple testing.
Additionally, we used the network-based heterogeneity clustering (NHC) algorithm to cluster cases and genes that harbor presumably deleterious variants by performing a gene cluster-based burden test. NHC was specifically tailored for small-sized cohorts to increase statistical power by utilizing a background protein–protein interaction (PPI) network to identify gene clusters significantly enriched in cases compared to controls [34]. In addition, NHC performs several pathway and ontology enrichment analyses for the identified gene clusters. The same set of presumably deleterious variants and covariates was used in this analysis to identify and prioritize candidate gene clusters that displayed significant enrichment in IBD-PD cases compared to controls.
Gene prioritization via pathway and gene enrichment analysis
To capture additional potential gene candidates associated with IBD-PD, we performed pathway and gene enrichment analyses, relaxing the P value threshold to < 0.01 based on the SKAT-O results, resulting in 84 genes (Fig. 1). Additionally, we selected genes in significant clusters identified by NHC (cluster P < 0.05), except for those enriched for previously-identified control-embedded pathways, leaving us with 38 genes [34]. The resulting gene set comprised 120 unique genes, which were considered potential candidates associated with IBD-PD. To examine the genes that have a close biological relationship to the known IBD and PD genes, we generated sets of known genes. The set of 157 known IBD genes was generated based on studies of IBD, CD, and UC [35, 36] following the methodology described in our previous study [37]. We included 142 genes from the merged signals of the genome-wide significant loci (P < 5 × 10−8) from Jostins et al. [36], as well as 15 genes from the 95% credible set of the fine-mapping results by Huang et al. [35]. For the set of known PD genes, we extracted genes associated with PD from the Human Gene Mutation Database (HGMD) Professional v. 2022.4 [38], which is the largest database currently available of high-quality and manually curated pathogenic variants. We specifically used the HGMD disease-causing mutations with the highest confidence only based on experimental evidence (DM), resulting in the identification of 103 known PD genes. We employed Ingenuity Pathway Analysis (IPA, QIAGEN Inc., version 01–21-03 https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) to compare significantly enriched pathways in the three gene sets.
Then, by utilizing the human gene connectome (HGC, https://lab.rockefeller.edu/casanova/HGC) [39], we calculated the average biological distance of 120 IBD-PD-associated genes (Dcandidate) to 157 known IBD genes and 103 known PD genes. We compared Dcandidate with Drandom, which was derived from randomly sampled gene sets of equivalent size (n = 120) to calculate empirical P values using 10,000 resampling iterations (Additional file 1: Methods used in biological relatedness, pathway, and gene enrichment analysis).
We then employed four complementary methods of pathway and biological relatedness approaches: ToppGene [40], IPA, HumanBase [41], and the Human Gene Connectome (HGC) [39] to obtain the final 14 IBD-PD associated candidate genes that were selected by all four methods (Additional file 1: Methods used in biological relatedness, pathway, and gene enrichment analysis). Additionally, DM variants in the final 14 IBD-PD-associated genes were explored in the HGMD.
The biological importance scores
We evaluated the biological importance of candidate genes by counting the number of IBD and PD genes in significantly enriched pathways, PPI networks, and ontologies with a P value < 0.01. We utilized several tools and databases to calculate the scores, including IPA core pathway analysis, InnateDB pathway analysis, InnateDB Gene Ontology (GO) analysis, and STRING interactome from NetworkAnalyst [42, 43]. We calculated the IBD-specific, PD-specific, and combined IBD-PD scores by summing the scaled scores from each data source.
PheWAS
Gene-level PheWAS was conducted in both the BioMe BioBank and UK Biobank to validate the phenotypes associated with the 14 candidate IBD-PD genes, as well as to evaluate their pleiotropic effects (Fig. 1). The BioMe BioBank includes samples from diverse ancestral origins (Additional file 2: Table S2), whereas the UK Biobank is primarily composed of individuals of European ancestry. ICD-9 (n = 1056 and 3344 in BioMe Biobank and UK Biobank, respectively) and ICD-10 diagnoses (n = 19,085 and 11,727 in BioMe Biobank and UK Biobank, respectively) of Biobank participants were converted into 1856 clinically relevant phenotypes (phecodes) using Phecode Map v1.2 [44]. 1376 and 1415 phecodes with at least 20 cases were used in the BioMe and UK Biobank analyses, respectively. We used a combination of PheWAS and SKAT-O binary robust methods, employing a case–control ratio not exceeding 1:99 for each disease and trait [45, 46]. Only variants identified within the IBD-PD cohort were included. PCA was conducted using the same parameters as those used in the IBD-PD cohort. Age, biological sex, and PC1-10 were used as covariates in the analysis. The Bonferroni-adjusted phenome-wide P value was determined as 3.63 × 10−5 (0.05/1376) and 3.53 × 10−5 (0.05/1415) for BioMe and UK Biobank analyses, respectively.
Results
Analysis of LRRK2 variants in the IBD-PD, BioMe, and UK Biobank cohorts
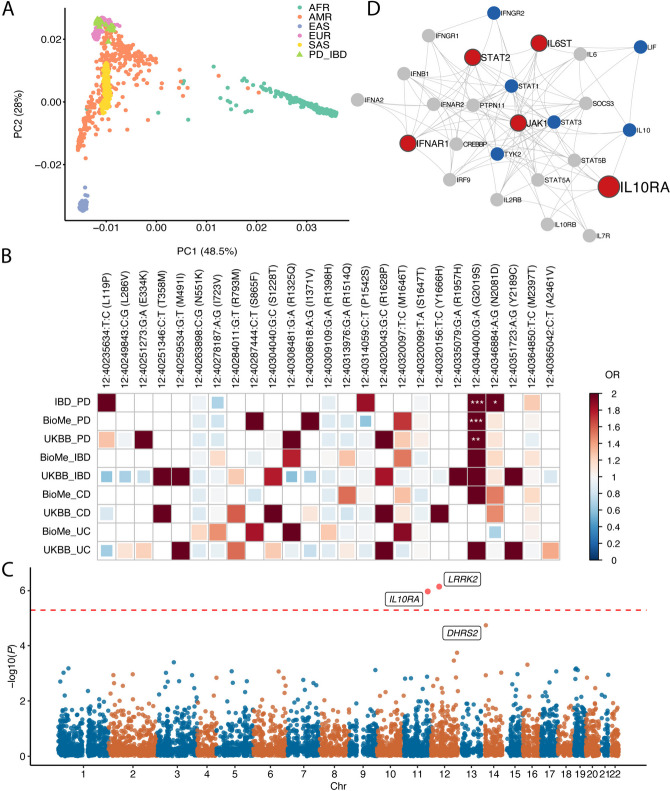
The IBD-PD cohort comprised 67 individuals of Northwestern and Southern European ancestry diagnosed with both IBD and PD, along with 426 non-Finnish Europeans from the 1KGP serving as controls (Fig. 2A and Additional file 1: Fig. S1 and Fig. S2). We performed variant-level association tests to assess the impact of 9 LRRK2 missense variants identified in the IBD-PD dataset on the IBD-PD phenotype. Additionally, we extracted a total of 14 LRRK2 missense variants from the BioMe cohort and 28 missense variants from the UK Biobank cohort and examined their associations with PD, IBD, CD, and UC (Fig. 2B, Additional file 2: Table S3 and Table S4). Our analysis identified the well-known PD and CD-associated variant, G2019S, as being significantly associated with the IBD-PD comorbidity (P = 1.59 × 10−4, Bonferroni-corrected P = 5.56 × 10−3) [5, 14, 47]. Moreover, G2019S showed an increased risk for PD in both the BioMe and UK Biobank cohorts (P = 6.56 × 10−5 and 7.66 × 10−3, respectively). Additionally, the N2081D variant was associated with the IBD-PD comorbidity at a nominal significance level (P = 0.027).
Fig. 2.
LRRK2-centered and genome-wide analysis of the IBD-PD comorbidity. A PCA plot demonstrating the distribution of IBD-PD cases among 1000 Genomes Project (1KGP) superpopulations. B Correlogram displaying the associations of LRRK2 missense variants with Parkinson’s disease (PD), inflammatory bowel disease (IBD), Crohn’s disease (CD), and ulcerative colitis (UC) in the Danish Registry, BioMe BioBank, and UK Biobank (UKBB) cohorts. The colors indicate odds ratios (ORs), whereas asterisks denote the significance level of the P values (* P < 0.05, ** P < 0.01, ***P < 0.001). Empty cells indicate that the variant had a minor allele count (MAC) < 3 in the respective cohort or MAC < 1 in cases, and hence the association could not be assessed. C Manhattan plot displaying the results of the SKAT-O analysis. The two genes that passed the level of genome-wide significance (LRRK2 and 1L10RA) are highlighted in red. The dashed line denotes the genome-wide significance level. D STRING protein–protein interaction (PPI) network generated using genes identified in the most significant cluster (cluster 7) in the NHC analysis (in red). The size of the nodes is proportional to the number of variant carriers. Known IBD-associated genes in the extended PPI network are highlighted in blue
We further investigated the odds ratios (ORs) derived from Firth’s regression for all 25 variants identified in the IBD-PD, BioMe, and UK Biobank cohorts. The G2019S variant demonstrated an increase for PD and both IBD subtypes, whereas the N2081D and L119P variants were associated with an elevated risk for increased risk for PD and CD. In addition to G2019S, N2081D, and L119P, the P1542S and M2397T variants exhibited an increased risk for the combined IBD-PD phenotype. Interestingly, the P1542S variant was exclusively associated with an increased risk of IBD-PD comorbidity, whereas it exhibited a decreased risk of PD in the BioMe BioBank analysis, despite displaying odds ratios close to 1 in the remaining analyses. Additionally, E334K, S865F, I1371V, R1325Q, R1628P, and M1646T variants displayed an increased risk for both PD and one or both subtype(s) of IBD based on the BioMe and UK Biobank analyses. S1647T, previously considered as a neutral variant [48], displayed ORs close to 1 for both PD and IBD subtypes, as expected.
Afterwards, we searched these 25 LRRK2 variants in HGMD and found that 16 variants had been reported to be associated with PD at varying confidence levels (Additional file 2: Table S5) [38]. Among those, 6 variants (E334K, R793M, S1228T, R1325Q, G2019S, and Y2189C) were associated with PD and classified under the DM category. Interestingly, the R793M, S1228T, and Y2189C variants demonstrated increased ORs for IBD but not for PD in the UK Biobank cohort, although the association of R793M and Y2189C with PD could not be assessed due to low MAC. The R1398H, N551K variants (the only variant pair in strong LD, r2 = 0.93) and I723V exhibited low ORs for the IBD-PD comorbidity. However, they displayed a positive relationship with UC and IBD risk in the BioMe BioBank dataset. Overall, these results provide further evidence in support of the role of LRRK2 variants in the pathogenesis of IBD-PD comorbidity. We note that while the reported OR results implicate trends of risk or protection, the majority of the P values in the aforementioned results (Fig. 2B and Additional file 2: Table S4) did not reach study-wide statistical significance, most likely due to the limited sample size of the study cohorts.
Gene-level association testing using the IBD-PD cohort
To identify novel genes associated with the IBD-PD comorbidity, SKAT-O analyses were performed using 9561 genes containing 34,640 presumably deleterious rare variants (Fig. 2C, Additional file 2: Table S6). As a result, LRRK2 (P = 7.41 × 10−7) and IL10RA (P = 1.11 × 10−6) attained the genome-wide significance level (Bonferroni-corrected P = 5.23 × 10−6). Consistent with its well-established association with both PD and IBD, LRRK2 was identified as the most significant gene in the analyses [5]. Following LRRK2 and IL10RA, the third most significant gene according to the SKAT-O results was DHRS2 (P = 1.85 × 10−5). These findings confirmed that LRRK2 is an IBD-PD-associated gene and revealed IL10RA (and potentially DHRS2) as candidate genes.
Network-based heterogeneity clustering in the IBD-PD cohort
Genetic heterogeneity is a common feature in many human diseases, including IBD and PD [49, 50]. To overcome the challenges caused by genetic heterogeneity in identifying disease-associated genes through conventional frequency-based case–control studies that assume genetic homogeneity, we employed NHC to identify candidate gene clusters associated with IBD-PD [34]. NHC is optimized for case–control analyses of small cohorts by performing pathway-level aggregations of genes carrying candidate variants, employing a clustering approach of gene–gene biological relatedness metric. NHC revealed 12 gene clusters with a cluster P < 0.05 (Additional file 2: Table S7). Four significant gene clusters (clusters 68, 70, 71, and 76) were deprioritized because the significantly enriched pathways in these gene clusters had been previously found to be highly enriched in healthy European controls [34]. Two significant gene clusters (clusters 7 and 73) included genes previously associated with Parkinsonism (FIG4 and VAC14) [51, 52] and IBD (IFNAR1, IL6ST, ATG16L1, and NOD2) [36] (Fig. 2D and Additional file 1: Fig. S5).
There were four additional potentially relevant significant gene clusters identified by NHC. Cluster 24 contained ATP6V0A1, ATP6V0A2, ATP6V0D2, ATP6V1B1, and ATP6V1H, genes encoding components of the vacuolar-ATPase complex, which plays a role in phagosome acidification. The vacuolar-ATPase complex has been implicated in the pathophysiology of several neurodegenerative diseases, including PD [53]. Additionally, previous GWAS have identified ATP6V0A1 as a candidate for PD [54]. Cluster 20 consisted of genes involved in GMP biosynthesis, GMPS, IMPDH1, and IMPDH2. IMPDH1 has been suggested to play a role in PD pathogenesis through protein misfolding and accumulation [55]. Furthermore, mutations in IMPDH2 have been associated with a dominant-type juvenile-onset dystonia-tremor disorder [56]. Cluster 57 encompassed EPS15, FCHO1, FCHO2, and HGS, genes related to endocytosis. Deficiency of FCHO1 has been shown to be associated with both IBD and Guillain Barré syndrome [57]. Additionally, EPS15 encodes an endocytic accessory protein that interacts with parkin and might contribute to the pathogenesis of PD [58]. Lastly, cluster 40 contained LIN9, MYBL2, RBL1, and TFDP1. TFDP1 has been found to be associated with IBD in a previous GWAS [59]. These findings highlight the potential relevance of these gene clusters to the pathogenesis of IBD and PD.
Comparison of IBD-PD-associated genes with established IBD and PD genes
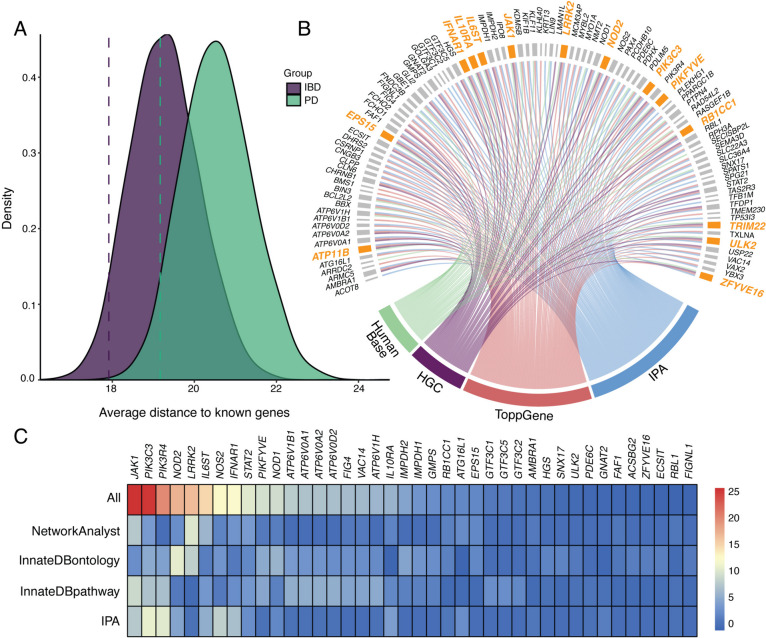
To compare the genes identified in the IBD-PD cohort with genes known to be associated with IBD and PD, we combined genes with nominal significance (P < 0.01) from SKAT-O results and genes from significant NHC gene clusters into a single gene set (n = 120). We then compared it to the sets of known IBD (n = 157) and PD (n = 103) genes (Additional file 2: Table S8). Only LRRK2 overlapped in known IBD and known PD gene sets. When comparing these gene sets with the IBD-PD-associated gene set, only 7 (5.83%) genes overlapped with known IBD genes and 4 (3.33%) genes overlapped with known PD genes (Additional file 2: Table S8). To evaluate the functional relevance of the 120 IBD-PD-associated genes to IBD and PD, we calculated their average biological distance (Dcandidate) to 157 known IBD genes and 103 known PD genes and found that Dcandidate was 17.9 to known IBD genes whereas it was 19.2 to known PD genes (Fig. 3A). The average distances within the known IBD (DIBD) and PD (DPD) genes were 13.9 and 16.8, respectively. Then, we compared Dcandidate to randomly sampled gene sets of equivalent size (n = 120) in 10,000 resampling iterations and obtained P values of 0.044 for IBD and 0.045 for PD, indicating that the biological relatedness of the candidate genes to IBD and PD was not random (Fig. 3A).
Fig. 3.
Candidate gene prioritization and validation using a phenotype-wide association study (PheWAS). A Density plots displaying the distribution of the average distances of 120 random genes (Drandom) to known inflammatory bowel disease (IBD, in purple) and known Parkinson’s disease (PD, in green) genes obtained from 10,000 resampling iterations. Dashed lines represent the average distance of the candidate gene set (Dcandidate, n = 120) to known IBD and PD genes. B Circos plot showing the candidate genes identified by four complementary pathway and biological relatedness approaches. Final candidate genes that were identified by all four methods are highlighted in orange. C Heatmap of the combined biological importance scores of the IBD-PD candidate genes. A higher score indicates a higher number of shared pathways, ontologies, or modules with known IBD and PD genes. The colors represent the magnitude of the scaled scores calculated by each method. The highest-scaled scores are depicted in red, whereas the lowest scores are shown in blue. The top 40 genes are shown in the plot
Pathway enrichment analysis with IPA showed that among the 104 significantly enriched pathways in the IBD-PD gene set, 6 pathways, “Iron homeostasis signaling pathway,” “Th1 Pathway,” “Autophagy,” “Th1 and Th2 Activation Pathway,” “Docosahexaenoic Acid (DHA) Signaling,” and “Th2 Pathway,” were significantly enriched in all three gene sets (Additional file 2: Table S9). A total of 75 significantly enriched pathways were shared between the IBD-PD and known IBD gene sets, primarily related to the inflammatory response and infections. By contrast, only 4 pathways were significantly enriched in both the IBD-PD and known PD gene sets, including those related to autophagy, endocytosis, and lysosomal biogenesis. Interestingly, 19 pathways were unique to the IBD-PD gene set including “Purine Nucleotides De Novo Biosynthesis II”, “Assembly of RNA Polymerase III Complex” and “Role of MAPK Signaling in Promoting the Pathogenesis of Influenza”. Additionally, 18 pathways were enriched in both known IBD and PD gene sets but not in the IBD-PD gene set, including “ERBB4 Signaling”, “Immunogenic Cell Death Signaling Pathway” and “Neuroinflammation Signaling Pathway”. These results suggested the potential role of inflammation and autophagy in the combined IBD-PD phenotype.
Gene prioritization through biological relatedness, pathway, and gene enrichment analyses
To further identify and prioritize candidate genes associated with IBD-PD from the set of SKAT-O and NHC significant genes, we employed biological relatedness and pathway enrichment. As a result, 14 genes (ATP11B, EPS15, IFNAR1, IL10RA, IL6ST, JAK1, LRRK2, NOD2, PIK3C3, PIKFYVE, RB1CC1, TRIM22, ULK2, and ZFYVE16) were prioritized based on the overlapping genes from HumanBase, HGC, ToppGene and IPA results (Fig. 3B and Additional file 2: Table S10). Among 14 candidate genes, four (IFNAR1, IL6ST, LRRK2, and NOD2) were known IBD-associated genes [36], whereas LRRK2 is also a well-known PD gene. We then explored HGMD for pathogenic mutations within candidate genes and found that 11 genes harbor “DM” (disease-causing) variants in various diseases (EPS15 in congenital heart disease, IFNAR1 and JAK1 in immunodeficiency, IL10RA and TRIM22 in IBD, IL6ST in hyper-IgE syndrome, LRRK2 in PD, NOD2 in CD, PIKFYVE in corneal dystrophy, ULK2 in neurodevelopmental disorder and ZFYVE16 in brain arteriovenous malformation).
In addition, we assessed the biological significance of the SKAT-O and NHC significant genes by calculating scores for shared interactions, pathways, and terms with known IBD and PD genes using PPI networks, pathway, and gene ontology enrichment analyses (Additional file 2: Tables S9, S11 and S12). The results revealed that genes involved in JAK-STAT and interferon alpha signaling, as well as autophagy-related genes, were particularly noteworthy as candidate genes for IBD-PD (Fig. 3C and Additional file 2: Table S13). Specifically, immunity-related genes, such as JAK1, IL6ST, STAT2, and IFNAR1, were more prominent in the IBD-specific analysis, whereas LRRK2 and other phagosome-related genes including PIK3C3, PIKFYVE, PIK3R4, and VAC14 were more significant in the PD-specific analysis (Additional file 1: Fig. S6, Additional file 2: Tables S14 and S15). Then, we investigated the tissue RNA expression patterns of the 14 prioritized genes using the consensus RNA tissue expression dataset from the Human Protein Atlas (https://www.proteinatlas.org/) [60]. We observed that 11 out of the 14 genes were ubiquitously expressed, whereas 3 exhibited tissue enrichment or enhancement in addition to being expressed in almost all tissues including the gastrointestinal tract, brain and bone marrow and lymphoid tissues: LRRK2 showed tissue enrichment in the lung, IL10RA displayed tissue enhancement in the bone marrow and lymphoid tissue, and NOD2 in bone marrow, esophagus, skin, and vagina. These findings further supported the potential role of these genes in the pathogenesis of IBD-PD comorbidity.
PheWAS
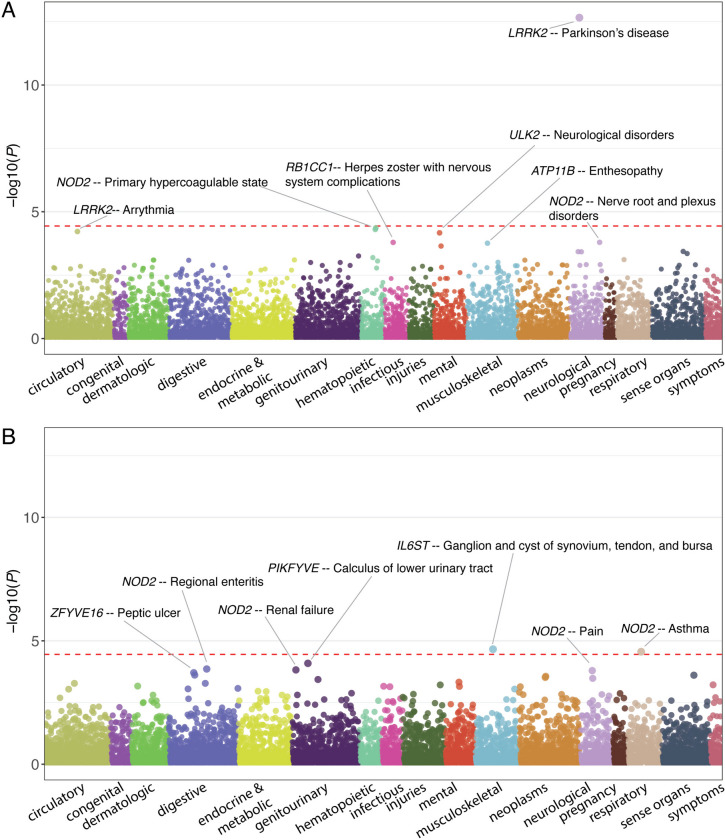
Lastly, we conducted a gene-level PheWAS in BioMe BioBank and UK Biobank using presumably deleterious variants located in the 14 candidate genes that were identified in the IBD-PD cohort (Table S16) to validate the associations of candidate genes and explore their pleiotropic effects. The analysis in BioMe BioBank revealed that the association between LRRK2 and PD (phecode, 332) was the most significant result (P = 2.19 × 10−13), surpassing the phenome-wide significance level (Fig. 4A and Additional file 2: Table S17). Moreover, LRRK2 was associated with torsion dystonia (333.4, P = 3.79 × 10−4) and “Extrapyramidal disease and abnormal movement disorders” (333, P = 1.29 × 10−3) in BioMe BioBank. We further observed supporting results for the prioritized genes associated with IBD and PD in BioMe with a nominal significance level. Specifically, NOD2 was associated with CD (Regional enteritis, 555.1, P = 1.38 × 10−4 in UK Biobank and P = 2.56 × 10−2 in BioMe BioBank) and IBD (P = 4.49 × 10−2 in UK Biobank and P = 2.29 × 10−2 in BioMe BioBank) (Fig. 4B and Additional file 2: Table S18). TRIM22 was associated with UC-related phecodes (556, P = 3.36 × 10−3, 578, P = 1.14 × 10−2, 556.1, P = 1.63 × 10−2 and 555.21, P = 2.89 × 10−2) and PD (332, P = 2.15 × 10−2) and “Myoclonus” (333.2, P = 2.51 × 10−2) in UK Biobank. It was also associated with “Extrapyramidal disease and abnormal movement disorders” and essential tremor in BioMe BioBank (333, P = 1.79 × 10−2 and 333.1 2.87 × 10−2, respectively). IL10RA was found to be associated with UC (555.21 P = 2.09 × 10−3 in BioMe and 3.69 × 10−2 in UK Biobank) and gastrointestinal hemorrhage (P = 1.67 × 10−2 in BioMe and 1.67 × 10−2 in UK Biobank). Interestingly, IL10RA was also found to be associated with “Abnormality of gait” (350.2, P = 2.79 × 10−2) and “Abnormal movement” (350, P = 3.87 × 10−2) in BioMe BioBank. EPS15 exhibited associations with essential tremor (333.1, P = 1.03 × 10−2) in UK Biobank and “Abnormal movement” (350, P = 9.95 × 10−3) and “Abnormality of gait” (350.2, P = 3.99 × 10−2) in BioMe BioBank. Furthermore, PIKFYVE showed an association with “Abnormal involuntary movements” in UK Biobank (350.1, P = 2.26 × 10−2), whereas it was associated with “Ulceration of intestine” (556.1, P = 2.91 × 10−2) and “Ulceration of the lower GI tract” (556, P = 4.67 × 10−2) in BioMe. ULK2 was associated with PD in UK Biobank (332, P = 3.5 × 10−2) and noninfectious gastroenteritis and “Hemorrhage of gastrointestinal tract” in BioMe (P = 1.38 × 10−2 and 3.12 × 10−2). Additionally, JAK1 was associated with essential tremor (333.1, P = 1.03 × 10−2) and RB1CC1 displayed an association with cholangitis (575.1, P = 1.02 × 10−2) and IL6ST with noninfectious gastroenteritis in UK Biobank (558, P = 3.14 × 10−2). Lastly, we observed associations between IFNAR1 and noninfectious gastroenteritis (558, P = 3.96 × 10−3), ZFYVE16 and UC (555.21 P = 1.96 × 10−2), ATP11B and “Abnormality of gait” (350.2, P = 3.27 × 10−2), PIK3C3 and PD and “Abnormal involuntary movements” (332, P = 3.49 × 10−2 and 350.1, P = 3.69 × 10−2) in BioMe BioBank.
Fig. 4.
Gene-level PheWAS of 14 candidate genes in A BioMe BioBank and B UK Biobank. The red dashed lines denote the study-wide significance levels
PheWAS results also revealed interesting pleiotropic effects of these genes, which have been shown to exert relevant functions in the identified phenotypes, such as the associations of NOD2 and hypercoagulability (286.81 and 286.8) [61] and asthma (495) [62], IL6ST and joint disease (727.4) [63] and systemic and cutaneous lupus erythematosus (695.4, 695.42, and 695.41) [64], and IFNAR1 and viral hepatitis [65]. Moreover, candidate genes displayed associations with various other autoimmune and allergic conditions, including ankylosing spondylitis, enthesopathy, rheumatoid arthritis, erythematous skin lesions, eosinophilia, and polyarteritis nodosa. These findings provide additional evidence for the functional significance of these genes in the development and manifestation of diverse phenotypes beyond the primary conditions of interest.
Discussion
In the present study, we successfully replicated the well-established associations of the LRRK2 G2019S and N2081D variants with IBD-PD. Both variants are located in the kinase domain of LRRK2 and have been linked to increased kinase activity [5, 48]. Furthermore, we identified 11 additional candidate LRRK2 variants that may contribute to IBD-PD comorbidity. Previous studies have already reported L119P, S1228T, R1628P, M1646T, and Y2189C as PD risk variants [66–69], while I1371V has been shown to cause increased phosphorylation and aggregation of α-synuclein in neurons [70]. Additionally, P1542S is a common variant (gnomAD MAF = 0.03) and has been listed as a CD-associated polymorphism in HGMD [38]. E334K, R1325Q, and R1628P have been previously shown to increase the LRRK2- dependent Rab10 Thr73 phosphorylation, which is used as an indicator of increased LRRK2 kinase activity conferring higher risk of PD [48]. This observation indicates that the carriers of these variants potentially benefit from LRRK2 kinase inhibitor therapy [48]. However, it is important to note that the remaining candidate LRRK2 variants identified in the current study have been previously shown to have no impact on LRRK2-dependent rab10 Thr73 phosphorylation, autophosphorylation of LRRK2Ser1292 or autophosphorylation of LRRK2Ser935. This suggests the possibility of the involvement of alternative mechanisms in the pathogenesis of IBD-PD comorbidity for these specific variants and further investigations are warranted to elucidate their precise effect.
On the other hand, we observed that the R1398H, N551K, and I723V variants were protective against IBD-PD but showed a trend to increase risk in the UC-specific and combined IBD analyses in BioMe BioBank. The protective effect of the R1398H variant on PD and CD has been previously documented, which was linked to GTPase activation leading to a reduction in LRRK2 kinase activity. Also, N551K is in strong LD with R1398H [5, 38]. Our results may indicate no protective effect of these variants on UC, which probably contributed to the lack of association also observed in the combined IBD (CD + UC) analyses.
In the second part of the study, by examining the IBD-PD cohort using different computational approaches, we discovered both previously known and novel genes associated with PD and IBD. Although the sample size was relatively small, LRRK2 and IL10RA attained genome-wide statistical significance in SKAT-O analysis and the P value of DHRS2 was just below the Bonferroni-corrected significance threshold. IL10 plays a major role in anti-inflammatory processes, and variants in the genes encoding IL10 and IL10 receptors have been shown to be associated with very early onset IBD [71–74]. Impairments in IL10 production and signaling have also been implicated in neurodegenerative diseases [75] including PD [76–78]. DHRS2 encodes a short-chain dehydrogenase/reductase that is involved in lipid metabolism and redox homeostasis. Under ischemic conditions, DHRS2 exerts cytoprotective effects by reducing the accumulation of dicarbonyl compounds and reactive oxygen species (ROS) and implicated in the PD pathogenesis [79, 80].
As a complementary approach to SKAT-O, we utilized NHC, which is particularly advantageous for prioritizing candidate gene clusters in small cohorts for diseases with high genetic heterogeneity, like PD and IBD [49, 50], as conventional frequency-based case–control studies assume genetic homogeneity [34]. Through NHC analysis, we identified 6 biologically-relevant gene clusters, two of which contained previously known IBD- and/or PD-associated genes. We then examined significant genes obtained from SKAT-O analysis (P < 0.01) and genes within significant gene clusters from the NHC analyses. The results indicated that the average distance of these 120 significant genes to known IBD genes (Dcandidate = 17.9) was slightly shorter than that to known PD genes (Dcandidate = 19.2). However, when considering the shorter average biological distance within known IBD genes (DIBD = 13.9) compared to that within known PD genes (DPD = 16.8), these findings suggest similar contributions from IBD and PD to the IBD-PD-associated genes. Despite a minor overlap between IBD-PD-associated genes with known IBD and PD gene sets, the pathway enrichment results revealed a notable overlap in enriched pathways, especially between IBD and IBD-PD gene sets. In addition to 6 pathways related to autophagy and immune response enriched in all three gene sets, there were 10 pathways that were unique to IBD-PD-associated genes. Among these pathways were those involving three of the final 14 candidate genes, namely PIK3C3, PIKFYVE, and JAK1, along with several other genes. Of especial note, the genes involved in purine nucleotide biosynthesis, MAPK signaling, ephrin A signaling, LPS/IL-1 mediated inhibition of RXR function, and the NAD signaling pathway [81] could also be of particular interest for future studies, given their distinctive signatures for IBD-PD comorbidity, as well as their established or suggested roles in intestinal inflammation and neuronal metabolism [82–89]. By applying four distinct pathway enrichment and biological relatedness methods using both known IBD and known PD gene sets to reduce the likelihood of IBD-only or PD-only associations contributing to the observed comorbidity, we further identified and prioritized candidate genes associated with IBD-PD. This approach led us to identify 14 candidate genes, all of which have been reported to display functions that are relevant to IBD and PD pathogenesis (Additional file 1: Supplementary information regarding 14 prioritized genes). By calculating the biological importance scores using PPI networks and pathway and ontology enrichment, we showed that inflammation and autophagy-related genes likely play a significant role in IBD-PD pathogenesis.
The gene-level PheWAS in two different Biobanks further supported the PD and/or IBD associations of the candidate genes. Since we aimed to investigate the potential pathogenicity of the rare variants identified in the IBD-PD cohort, we exclusively used these variants to examine the associations of the candidate genes in the BioMe BioBank and UK Biobank, which might have resulted in higher P values than anticipated when replicating some previously known associations. As expected, the most significant association was observed between LRRK2 and PD. TRIM22, IL10RA, PIKYVE, and ULK2 were found to be associated with both IBD- and PD-related phecodes, whereas NOD2, RB1CC1, IL6ST, IFNAR1, and ZFYVE16 were associated specifically with IBD-related phecodes. Additionally, LRRK2, JAK1, ATP11B, and PIK3C3 were associated with PD-related phecodes. Although not all candidate genes showed significant associations directly with IBD and PD diagnoses, we observed other relevant phenotypes that can be related to these diseases. For example, one of the earliest and most common motor changes observed in PD is the gait disturbance, which is caused by the impaired basal ganglia function [90]. Since IL10RA, EPS15, and ATP11B were associated with “Abnormality of gait,” they might be associated with PD. Further, RB1CC1 was found to be associated with cholangitis, a disease closely associated with IBD [91]. These results support the potential connection between immune dysregulation, gut inflammation, and motor symptoms observed in PD and are in line with previous findings regarding the reduced incidence of PD in IBD patients receiving anti-inflammatory therapy [6, 9].
Previous studies investigating shared genetic factors between IBD and PD have mainly focused on common variants, leading to the identification of several loci associated with both diseases. Recently, a report examining the genetic correlation between IBD subtypes and PD using summary statistics of previous GWAS identified 23 novel loci in addition to the 9 loci reported previously [8]. Interestingly, ATP6V0A1, a gene that was identified in our NHC analysis, was among the novel pleiotropic loci shared between PD and CD and between PD and UC in that study. However, two recent Mendelian randomization studies, which also used summary statistics from large-scale GWAS of PD and IBD, failed to find a causal relationship between the two conditions [92, 93]. These conflicting findings might be attributable to the possibility that common genetic variants contributing to the IBD-PD comorbidity account for only a small fraction of the overall cases [50, 93]. Instead, the risks are driven by rarer variants. Other factors, such as chronic low-grade inflammation and environmental factors might cumulatively contribute to the development of PD in some IBD cases [6, 92].
However, in contrast to these previous studies that investigated the shared genetic factors between PD and IBD using separate cohorts with only one of these diseases phenotyped, our approach involved the analysis of WGS data from patients diagnosed with both IBD and PD. We used high-impact rare variants to identify novel genetic defects with larger effect sizes, which were not properly captured by GWAS studies due to their design. Applying our approach, we identified novel candidate genes implicated in inflammation and autophagy, which may play a role in the pathogenesis of IBD-PD comorbidity. However, it is important to consider the potential impact of incomplete penetrance, a common feature of many genetic diseases, including PD [94], on the manifestation of the disease in variant carriers. A thorough evaluation of the penetrance of our detected putative disease-causing variants can be more effectively performed in large case–control studies [95]. It will be crucial to continue investigating the genetic determinants of IBD-PD comorbidity to gain a comprehensive understanding of the underlying pathogenesis and develop effective risk stratification and personalized treatment strategies.
We believe that our study’s major strength is that, despite the limited sizes of the study cohorts to accommodate stringent variant-level analyses especially when considering rare variants, we applied a framework of analyses that were specifically designed to handle small sample sizes, thereby enabling novel gene discoveries for unique clinical phenotypes. It is nevertheless important to acknowledge certain limitations of our study. First, detailed clinical data were missing for the IBD-PD cohort owing to the strict regulation of the Danish Data Protection Agency for linking patient-level data with biospecimens, thereby preventing us from assessing the age of onset of these conditions or IBD subtypes (CD versus UC). Moreover, due to the lack of access to genetic data for IBD-only or PD-only patients, and healthy controls in the Danish Registry, we utilized existing datasets with available clinical information and performed extensive QC checks and genetic ancestry analyses to reduce the potential ascertainment bias. In addition, the disease cohorts were generated based on ICD codes, which may inherently lack granularity for certain disease subtypes. Also, we could not rule out genetic contributions from other immune-mediated diseases. Extensive evidence points towards a link of both IBD and PD with several organ-specific and multi-organ autoimmune disorders [96–101], making it challenging to exclude IBD and PD cases with concomitant immune-mediated diseases from the BioMe BioBank and UK Biobank cohorts. Although adjusting association tests for polygenic risk scores for autoimmune diseases could reduce the potential confounding effect, there is a lack of a consensus on representative conditions that should be adjusted for. Moreover, such an approach might bias the results towards the null, especially those related to IBD, which itself is an immune-mediated condition. Nevertheless, an in-depth investigation of the potential confounding by autoimmune diseases is still warranted. An important aspect to consider when interpreting the results is that all IBD-PD cases included in this study share a common European ancestry and the results, therefore, may not be generalizable to more ethnically and racially diverse populations. Despite leveraging the ethnically-diverse BioMe BioBank to validate our findings within the IBD-PD cohort, it is still crucial to validate these findings across diverse populations.
Furthermore, the relatively small cohorts used in the LRRK2 coding sequence analysis limited our ability to observe and analyze very rare functional variants, as large sample sizes are pivotal for effectively detecting disease-associated rare variants and facilitating their replication across diverse cohorts [32]. Moreover, most of the P values in the variant-level analysis did not attain genome-wide statistical significance, which prevented us from drawing definitive conclusions. However, despite this limitation, we were able to replicate the well-known associations of the LRRK2 G2019S and N2081D variants with both IBD and PD and identify potential novel causal variants. Furthermore, even though analyses to detect novel candidate genes associated with IBD-PD comorbidity were based on sequencing data from 67 IBD-PD cases, we used the NHC algorithm to address the low statistical power resulting from the small sample size and genetic heterogeneity, and identified six functionally-relevant gene clusters. In addition, by employing several pathway enrichment and biological relatedness approaches, we were able to prioritize genes from both SKAT-O and NHC results with high confidence. Lastly, the focus on high-impact rare variants during the discovery of novel candidate genes may have limited the detection of potential associations with common variants or variants with smaller effect sizes. Future studies incorporating rare and common variants, larger sample sizes in addition to analyses specific to the subtypes of IBD could provide a more comprehensive understanding of the genetic architecture underlying the IBD-PD comorbidity.
Conclusions
In conclusion, our study highlights the significance of shared genetic factors in the IBD-PD overlap by both supporting previous findings and introducing novel candidate genes and variants. Future investigation of the interplay between inflammation and autophagy could in principle provide a better understanding of the shared etiology of IBD and PD and potential therapeutic targets for drug development and repurposing.
Supplementary Information
Additional file 1. Supplementary Information. Supplementary Methods. Supplementary information regarding 14 prioritized genes. Supplementary references. Fig. S1. Cross-validation errors of the ADMIXTURE analysis of IBD-PD cases and the 1KGP populations. Fig. S2. ADMIXTURE analysis of IBD-PD cases and the 1KGP populations. Fig. S3. Scree plot of principal components. Fig. S4. QQ plots of the SKAT-O results. Fig. S5. STRING PPI network of the Cluster73 from NHC analysis. Fig. S6. Heatmap of the combined biological importance scores.
Additional file 2. Table S1. Sample-level QC metrics of the IBD-PD cohort. Table S2. BioMe participants by ancestry. Table S3. Minor allele frequencies of the LRRK2 variants. Table S4. Variant-level association tests of LRRK2 variants. Table S5. HGMD classification of the LRRK2 missense variants. Table S6. SKAT-O results. Table S7. NHC results. Table S8. Known IBD and PD genes and 120 significant genes from SKAT-O and NHC analysis of IBD-PD cohort. Table S9. IPA canonical pathway enrichment analysis of 120 SKAT-O and NHC significant IBD-PD genes, known IBD genes and known PD genes. Table S10. Gene prioritization according to 4 biological relatedness and pathway approaches. Table S11. InnateDB pathway enrichment analysis. Table S12. InnateDB GO enrichment analysis. Table S13. Biological importance scores for IBD-PD. Table S14. Biological importance scores for PD. Table S15. Biological importance scores for IBD. Table S16. Presumably deleterious rare variants located in the 14 candidate genes identified in the IBD-PD cohort. Table S17. PheWAS of 14 candidate genes in BioMe BioBank. Table S18. PheWAS of 14 candidate genes in UK Biobank.
Acknowledgements
Not applicable.
Abbreviations
- 1KGP
1000 Genomes Project Phase 3
- CD
Crohn’s disease
- DM
Disease-causing mutation
- EHR
Electronic health records
- GATK
Genome Analysis Toolkit
- GDI
Gene Damage Index
- GO
Gene Ontology
- HGC
Human Gene Connectome
- HGMD
Human Gene Mutation Database
- HWE
Hardy-Weinberg equilibrium
- IBD
Inflammatory bowel disease
- IPA
Ingenuity Pathway Analysis
- LD
Linkage disequilibrium
- MAC
Minor allele count
- MAF
Minor allele frequency
- MSC
Mutation Significance Cutoff
- NHC
Network-based heterogeneity clustering
- OR
Odds ratio
- PC
Principal component
- PCA
Principal component analysis
- PD
Parkinson’s disease
- PheWAS
Phenome-wide association studies
- QC
Quality control
- SKAT-O
Optimized sequence kernel association test
- UC
Ulcerative colitis
- VEP
Variant effect predictor
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Authors’ contributions
YI and IP conceived and supervised the study. MEK and YW performed the analyses. JB, PDS, and DNC contributed to the data utilized. MEK and YI wrote the manuscript with input from IP, DNC, and JB. All authors read and approved the final manuscript.
Funding
YI, MEK, and YW were funded by the Charles Bronfman Institute for Personalized Medicine. This work was funded by the Michael J Fox Foundation and National Institute of Health’s National Institute of Neurological Disorders and Stroke grant P20NS123220 (to IP). This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences.
Availability of data and materials
The WGS data of the IBD-PD cohort and the WES data of the BioMe BioBank cohorts used and/or analyzed in this study are available from the corresponding authors upon reasonable request within one week. This research was conducted using the UK Biobank resource under application number 53074. UK Biobank WES datasets are accessible upon approval of application. The 1KGP WGS data used in the current study is available at http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20190425_NYGC_GATK/raw_calls_updated/ [18]. The gene- and variant-level association statistics generated in this study are provided in the supplementary data. The Mutation Significance Cutoff and the Gene Damage Index databases for selecting high-impact variants and genes are available at https://lab.rockefeller.edu/casanova/MSC [28] and https://lab.rockefeller.edu/casanova/GDI [29]. The Human Gene Connectome is available for download at https://lab.rockefeller.edu/casanova/HGC [39]. Other tools and methods used in the analysis are described in the Methods section with references.
Declarations
Ethics approval and consent to participate
This study was approved by the Danish National Center for Ethics (reference number 1606029). Approval for working with data from the Copenhagen Hospital Biobank was granted by the Danish Data Protection Agency (reference number 2007–58-0015). Informed consent was obtained from all study participants. This research conformed to the principles of the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Inga Peter and Yuval Itan share equal contribution.
Contributor Information
Inga Peter, Email: inga.peter@mssm.edu.
Yuval Itan, Email: yuval.itan@mssm.edu.
References
- 1.Nikolaus S, Schreiber S. Diagnostics of inflammatory bowel disease. Gastroenterology. 2007;133(5):1670–1689. doi: 10.1053/j.gastro.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Tolosa E, Garrido A, Scholz SW, Poewe W. Challenges in the diagnosis of Parkinson's disease. Lancet Neurol. 2021;20(5):385–397. doi: 10.1016/S1474-4422(21)00030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Lee HS, Lobbestael E, Vermeire S, Sabino J, Cleynen I. Inflammatory bowel disease and Parkinson's disease: common pathophysiological links. Gut. 2021;70(2):408–417. doi: 10.1136/gutjnl-2020-322429. [DOI] [PubMed] [Google Scholar]
- 5.Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, Hsu NY, et al. Functional variants in the LRRK2 gene confer shared effects on risk for Crohn's disease and Parkinson's disease. Sci Transl Med. 2018;10(423):eaai7795. doi: 10.1126/scitranslmed.aai7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter I, Dubinsky M, Bressman S, Park A, Lu C, Chen N, et al. Anti-tumor necrosis factor therapy and incidence of Parkinson disease among patients with inflammatory bowel disease. JAMA Neurol. 2018;75(8):939–946. doi: 10.1001/jamaneurol.2018.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witoelar A, Jansen IE, Wang Y, Desikan RS, Gibbs JR, Blauwendraat C, et al. Genome-wide pleiotropy between Parkinson disease and autoimmune diseases. JAMA Neurol. 2017;74(7):780–792. doi: 10.1001/jamaneurol.2017.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang X, Ploner A, Wang Y, Ludvigsson JF, Williams DM, Pedersen NL, et al. Genetic overlap between Parkinson's disease and inflammatory bowel disease. Brain Commun. 2023;5(1):fcad002. doi: 10.1093/braincomms/fcad002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Yuan M, Liu Y, Yang F, Chen WZ, Xu ZZ, et al. Association between inflammatory bowel diseases and Parkinson's disease: systematic review and meta-analysis. Neural Regen Res. 2022;17(2):344–353. doi: 10.4103/1673-5374.317981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heckman MG, Elbaz A, Soto-Ortolaza AI, Serie DJ, Aasly JO, Annesi G, et al. Protective effect of LRRK2 p.R1398H on risk of Parkinson's disease is independent of MAPT and SNCA variants. Neurobiol Aging. 2014;35(1):266e5–14. doi: 10.1016/j.neurobiolaging.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heckman MG, Soto-Ortolaza AI, Aasly JO, Abahuni N, Annesi G, Bacon JA, et al. Population-specific frequencies for LRRK2 susceptibility variants in the Genetic Epidemiology of Parkinson's Disease (GEO-PD) Consortium. Mov Disord. 2013;28(12):1740–1744. doi: 10.1002/mds.25600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, et al. Association of LRRK2 exonic variants with susceptibility to Parkinson's disease: a case-control study. Lancet Neurol. 2011;10(10):898–908. doi: 10.1016/S1474-4422(11)70175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billingsley KJ, Bandres-Ciga S, Saez-Atienzar S, Singleton AB. Genetic risk factors in Parkinson's disease. Cell Tissue Res. 2018;373(1):9–20. doi: 10.1007/s00441-018-2817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivas MA, Avila BE, Koskela J, Huang H, Stevens C, Pirinen M, et al. Insights into the genetic epidemiology of Crohn's and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 2018;14(5):e1007329. doi: 10.1371/journal.pgen.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rui Q, Ni H, Li D, Gao R, Chen G. The role of LRRK2 in neurodegeneration of Parkinson disease. Curr Neuropharmacol. 2018;16(9):1348–1357. doi: 10.2174/1570159X16666180222165418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake J, Reed X, Langston RG, Nalls MA, Gan-Or Z, Cookson MR, et al. Coding and noncoding variation in LRRK2 and Parkinson's disease risk. Mov Disord. 2022;37(1):95–105. doi: 10.1002/mds.28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.0.1–.0.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szustakowski JD, Balasubramanian S, Kvikstad E, Khalid S, Bronson PG, Sasson A, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet. 2021;53(7):942–948. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International HapMap Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197(2):573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13(2):109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Vélez M, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112(44):13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattell RB. The scree test for the number of factors. Multivariate Behav Res. 1966;1(2):245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 31.Ma C, Blackwell T, Boehnke M, Scott LJ, GoT2D investigators Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37(6):539–50. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Cobat A, Lee YS, Wu Y, Bayrak CS, Boccon-Gibod C, et al. A computational approach for detecting physiological homogeneity in the midst of genetic heterogeneity. Am J Hum Genet. 2021;108(6):1012–1025. doi: 10.1016/j.ajhg.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Fang M, Jostins L, Umicevic Mirkov M, Boucher G, Anderson CA, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547(7662):173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Gettler K, Kars ME, Giri M, Li D, Bayrak CS, et al. Identifying high-impact variants and genes in exomes of Ashkenazi Jewish inflammatory bowel disease patients. Nat Commun. 2023;14(1):2256. doi: 10.1038/s41467-023-37849-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, et al. The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet. 2020;139(10):1197–1207. doi: 10.1007/s00439-020-02199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itan Y, Zhang SY, Vogt G, Abhyankar A, Herman M, Nitschke P, et al. The human gene connectome as a map of short cuts for morbid allele discovery. Proc Natl Acad Sci U S A. 2013;110(14):5558–5563. doi: 10.1073/pnas.1218167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–33. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10(6):823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 44.Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019;7(4):e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Z, Bi W, Zhou W, VandeHaar P, Fritsche LG, Lee S. UK Biobank whole-exome sequence binary phenome analysis with robust region-based rare-variant test. Am J Hum Genet. 2020;106(1):3–12. doi: 10.1016/j.ajhg.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76(4):672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalogeropulou AF, Purlyte E, Tonelli F, Lange SM, Wightman M, Prescott AR, et al. Impact of 100 LRRK2 variants linked to Parkinson's disease on kinase activity and microtubule binding. Biochem J. 2022;479(17):1759–1783. doi: 10.1042/BCJ20220161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye H, Robak LA, Yu M, Cykowski M, Shulman JM. Genetics and pathogenesis of Parkinson's syndrome. Annu Rev Pathol. 2023;18:95–121. doi: 10.1146/annurev-pathmechdis-031521-034145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loddo I, Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol. 2015;6:551. doi: 10.3389/fimmu.2015.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmermann M, Schuster S, Boesch S, Korenke GC, Mohr J, Reichbauer J, et al. FIG4 mutations leading to parkinsonism and a phenotypical continuum between CMT4J and Yunis Varon syndrome. Parkinsonism Relat Disord. 2020;74:6–11. doi: 10.1016/j.parkreldis.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 52.de Gusmao CM, Stone S, Waugh JL, Yang E, Lenk GM, Rodan LH. VAC14 gene-related parkinsonism-dystonia with response to deep brain stimulation. Mov Disord Clin Pract. 2019;6(6):494–497. doi: 10.1002/mdc3.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boland B, Yu WH, Corti O, Mollereau B, Henriques A, Bezard E, et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat Rev Drug Discov. 2018;17(9):660–688. doi: 10.1038/nrd.2018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet. 2017;49(10):1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aherne A, Kennan A, Kenna PF, McNally N, Lloyd DG, Alberts IL, et al. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum Mol Genet. 2004;13(6):641–650. doi: 10.1093/hmg/ddh061. [DOI] [PubMed] [Google Scholar]
- 56.Kuukasjarvi A, Landoni JC, Kaukonen J, Juhakoski M, Auranen M, Torkkeli T, et al. IMPDH2: a new gene associated with dominant juvenile-onset dystonia-tremor disorder. Eur J Hum Genet. 2021;29(12):1833–1837. doi: 10.1038/s41431-021-00939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aydemir S, Islek A, Nepesov S, Yaman Y, Baysoy G, Beser OF, et al. Inflammatory bowel disease and Guillain Barre syndrome in FCHO1 deficiency. J Clin Immunol. 2021;41(6):1406–1410. doi: 10.1007/s10875-021-01042-2. [DOI] [PubMed] [Google Scholar]
- 58.Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8(8):834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 59.Ostrowski J, Paziewska A, Lazowska I, Ambrozkiewicz F, Goryca K, Kulecka M, et al. Genetic architecture differences between pediatric and adult-onset inflammatory bowel diseases in the Polish population. Sci Rep. 2016;6:39831. doi: 10.1038/srep39831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Zhang S, Hu L, Zhai L, Xue R, Ye J, et al. Nucleotide-binding oligomerization domain 2 receptor is expressed in platelets and enhances platelet activation and thrombosis. Circulation. 2015;131(13):1160–1170. doi: 10.1161/CIRCULATIONAHA.114.013743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boute M, Ait Yahia S, Fan Y, Alvarez-Simon D, Vorng H, Balsamelli J, et al. NOD2 signaling circuitry during allergen sensitization does not worsen experimental neutrophilic asthma but promotes a Th2/Th17 profile in asthma patients but not healthy subjects. Int J Mol Sci. 2022;23(19):11894. [DOI] [PMC free article] [PubMed]
- 63.Srirangan S, Choy EH. The role of interleukin 6 in the pathophysiology of rheumatoid arthritis. Ther Adv Musculoskelet Dis. 2010;2(5):247–256. doi: 10.1177/1759720X10378372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ripley BJ, Goncalves B, Isenberg DA, Latchman DS, Rahman A. Raised levels of interleukin 6 in systemic lupus erythematosus correlate with anaemia. Ann Rheum Dis. 2005;64(6):849–853. doi: 10.1136/ard.2004.022681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duncan CJA, Randall RE, Hambleton S. Genetic lesions of type I interferon signalling in human antiviral immunity. Trends Genet. 2021;37(1):46–58. doi: 10.1016/j.tig.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bright JM, Carlisle HJ, Toda AMA, Murphy M, Molitor TP, Wren P, et al. Differential inhibition of LRRK2 in Parkinson's disease patient blood by a G2019S selective LRRK2 inhibitor. Mov Disord. 2021;36(6):1362–1371. doi: 10.1002/mds.28490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Covy JP, Yuan W, Waxman EA, Hurtig HI, Van Deerlin VM, Giasson BI. Clinical and pathological characteristics of patients with leucine-rich repeat kinase-2 mutations. Mov Disord. 2009;24(1):32–39. doi: 10.1002/mds.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berg D, Schweitzer KJ, Leitner P, Zimprich A, Lichtner P, Belcredi P, et al. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease*. Brain. 2005;128(Pt 12):3000–3011. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 69.Bryant N, Malpeli N, Ziaee J, Blauwendraat C, Liu Z, Consortium AP, et al. Identification of LRRK2 missense variants in the accelerating medicines partnership Parkinson’s disease cohor. Hum Mol Genet. 2021;30(6):454–66. doi: 10.1093/hmg/ddab058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jagtap S, Potdar C, Yadav R, Pal PK, Datta I. Dopaminergic neurons differentiated from LRRK2 I1371V-induced pluripotent stem cells display a lower yield, alpha-synuclein pathology, and functional impairment. ACS Chem Neurosci. 2022;13(17):2632–2645. doi: 10.1021/acschemneuro.2c00297. [DOI] [PubMed] [Google Scholar]
- 71.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361(21):2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143(2):347–355. doi: 10.1053/j.gastro.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 73.Uhlig HH, Muise AM. Clinical genomics in inflammatory bowel disease. Trends Genet. 2017;33(9):629–641. doi: 10.1016/j.tig.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 74.Yanagi T, Mizuochi T, Takaki Y, Eda K, Mitsuyama K, Ishimura M, et al. Novel exonic mutation inducing aberrant splicing in the IL10RA gene and resulting in infantile-onset inflammatory bowel disease: a case report. BMC Gastroenterol. 2016;16:10. doi: 10.1186/s12876-016-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lobo-Silva D, Carriche GM, Castro AG, Roque S, Saraiva M. Balancing the immune response in the brain: IL-10 and its regulation. J Neuroinflammation. 2016;13(1):297. doi: 10.1186/s12974-016-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porro C, Cianciulli A, Panaro MA. The regulatory role of IL-10 in neurodegenerative diseases. Biomolecules. 2020;10(7):1017. doi: 10.3390/biom10071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bialecka M, Klodowska-Duda G, Kurzawski M, Slawek J, Gorzkowska A, Opala G, et al. Interleukin-10 (IL10) and tumor necrosis factor alpha (TNF) gene polymorphisms in Parkinson's disease patients. Parkinsonism Relat Disord. 2008;14(8):636–640. doi: 10.1016/j.parkreldis.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Arimoto T, Choi DY, Lu X, Liu M, Nguyen XV, Zheng N, et al. Interleukin-10 protects against inflammation-mediated degeneration of dopaminergic neurons in substantia nigra. Neurobiol Aging. 2007;28(6):894–906. doi: 10.1016/j.neurobiolaging.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 79.Iannielli A, Bido S, Folladori L, Segnali A, Cancellieri C, Maresca A, et al. Pharmacological inhibition of necroptosis protects from dopaminergic neuronal cell death in Parkinson's disease models. Cell Rep. 2018;22(8):2066–2079. doi: 10.1016/j.celrep.2018.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Z, Liu H, Bode A, Luo X. Emerging roles of dehydrogenase/reductase member 2 (DHRS2) in the pathology of disease. Eur J Pharmacol. 2021;898:173972. doi: 10.1016/j.ejphar.2021.173972. [DOI] [PubMed] [Google Scholar]
- 81.Gerner RR, Klepsch V, Macheiner S, Arnhard K, Adolph TE, Grander C, et al. NAD metabolism fuels human and mouse intestinal inflammation. Gut. 2018;67(10):1813–1823. doi: 10.1136/gutjnl-2017-314241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crittenden S, Cheyne A, Adams A, Forster T, Robb CT, Felton J, et al. Purine metabolism controls innate lymphoid cell function and protects against intestinal injury. Immunol Cell Biol. 2018;96(10):1049–1059. doi: 10.1111/imcb.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scholefield M, Church SJ, Taylor G, Knight D, Unwin RD, Cooper GJS. Multi-regional alterations in glucose and purine metabolic pathways in the Parkinson's disease dementia brain. NPJ Parkinsons Dis. 2023;9(1):66. doi: 10.1038/s41531-023-00488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Broom OJ, Widjaya B, Troelsen J, Olsen J, Nielsen OH. Mitogen activated protein kinases: a role in inflammatory bowel disease? Clin Exp Immunol. 2009;158(3):272–280. doi: 10.1111/j.1365-2249.2009.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bohush A, Niewiadomska G, Filipek A. Role of mitogen activated protein kinase signaling in Parkinson's disease. Int J Mol Sci. 2018;19(10):2973. [DOI] [PMC free article] [PubMed]
- 86.Coulthard MG, Morgan M, Woodruff TM, Arumugam TV, Taylor SM, Carpenter TC, et al. Eph/Ephrin signaling in injury and inflammation. Am J Pathol. 2012;181(5):1493–1503. doi: 10.1016/j.ajpath.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 87.Barquilla A, Pasquale EB. Eph receptors and ephrins: therapeutic opportunities. Annu Rev Pharmacol Toxicol. 2015;55:465–487. doi: 10.1146/annurev-pharmtox-011112-140226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pajares M, A IR, Manda G, Bosca L, Cuadrado A. Inflammation in Parkinson's disease: mechanisms and therapeutic implications. Cells. 2020;9(7):1687. [DOI] [PMC free article] [PubMed]
- 89.Brakedal B, Dolle C, Riemer F, Ma Y, Nido GS, Skeie GO, et al. The NADPARK study: A randomized phase I trial of nicotinamide riboside supplementation in Parkinson's disease. Cell Metab. 2022;34(3):396–407e6. doi: 10.1016/j.cmet.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Hausdorff JM. Gait dynamics in Parkinson's disease: common and distinct behavior among stride length, gait variability, and fractal-like scaling. Chaos. 2009;19(2):026113. doi: 10.1063/1.3147408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loftus EV, Jr, Harewood GC, Loftus CG, Tremaine WJ, Harmsen WS, Zinsmeister AR, et al. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54(1):91–96. doi: 10.1136/gut.2004.046615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeng R, Wang J, Zheng C, Jiang R, Tong S, Wu H, et al. Lack of causal associations of inflammatory bowel disease with Parkinson's disease and other neurodegenerative disorders. Mov Disord. 2023;38(6):1082–1088. doi: 10.1002/mds.29386. [DOI] [PubMed] [Google Scholar]
- 93.Freuer D, Meisinger C. Association between inflammatory bowel disease and Parkinson's disease: a Mendelian randomization study. NPJ Parkinsons Dis. 2022;8(1):55. doi: 10.1038/s41531-022-00318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taymans JM, Fell M, Greenamyre T, Hirst WD, Mamais A, Padmanabhan S, et al. Perspective on the current state of the LRRK2 field. NPJ Parkinsons Dis. 2023;9(1):104. doi: 10.1038/s41531-023-00544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wright CF, West B, Tuke M, Jones SE, Patel K, Laver TW, et al. Assessing the pathogenicity, penetrance, and expressivity of putative disease-causing variants in a population setting. Am J Hum Genet. 2019;104(2):275–286. doi: 10.1016/j.ajhg.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzalez-Latapi P, Marras C. Epidemiological evidence for an immune component of Parkinson's disease. J Parkinsons Dis. 2022;12(s1):S29–S43. doi: 10.3233/JPD-223180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tansey MG, Wallings RL, Houser MC, Herrick MK, Keating CE, Joers V. Inflammation and immune dysfunction in Parkinson disease. Nat Rev Immunol. 2022;22(11):657–673. doi: 10.1038/s41577-022-00684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li M, Wan J, Xu Z, Tang B. The association between Parkinson's disease and autoimmune diseases: A systematic review and meta-analysis. Front Immunol. 2023;14:1103053. doi: 10.3389/fimmu.2023.1103053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson JC, Furlano RI, Jick SS, Meier CR. Inflammatory bowel disease and the risk of autoimmune diseases. J Crohns Colitis. 2016;10(2):186–193. doi: 10.1093/ecco-jcc/jjv193. [DOI] [PubMed] [Google Scholar]
- 100.van Sommeren S, Janse M, Karjalainen J, Fehrmann R, Franke L, Fu J, et al. Extraintestinal manifestations and complications in inflammatory bowel disease: from shared genetics to shared biological pathways. Inflamm Bowel Dis. 2014;20(6):987–994. doi: 10.1097/MIB.0000000000000032. [DOI] [PubMed] [Google Scholar]
- 101.Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4(1):6. doi: 10.1186/gm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Information. Supplementary Methods. Supplementary information regarding 14 prioritized genes. Supplementary references. Fig. S1. Cross-validation errors of the ADMIXTURE analysis of IBD-PD cases and the 1KGP populations. Fig. S2. ADMIXTURE analysis of IBD-PD cases and the 1KGP populations. Fig. S3. Scree plot of principal components. Fig. S4. QQ plots of the SKAT-O results. Fig. S5. STRING PPI network of the Cluster73 from NHC analysis. Fig. S6. Heatmap of the combined biological importance scores.
Additional file 2. Table S1. Sample-level QC metrics of the IBD-PD cohort. Table S2. BioMe participants by ancestry. Table S3. Minor allele frequencies of the LRRK2 variants. Table S4. Variant-level association tests of LRRK2 variants. Table S5. HGMD classification of the LRRK2 missense variants. Table S6. SKAT-O results. Table S7. NHC results. Table S8. Known IBD and PD genes and 120 significant genes from SKAT-O and NHC analysis of IBD-PD cohort. Table S9. IPA canonical pathway enrichment analysis of 120 SKAT-O and NHC significant IBD-PD genes, known IBD genes and known PD genes. Table S10. Gene prioritization according to 4 biological relatedness and pathway approaches. Table S11. InnateDB pathway enrichment analysis. Table S12. InnateDB GO enrichment analysis. Table S13. Biological importance scores for IBD-PD. Table S14. Biological importance scores for PD. Table S15. Biological importance scores for IBD. Table S16. Presumably deleterious rare variants located in the 14 candidate genes identified in the IBD-PD cohort. Table S17. PheWAS of 14 candidate genes in BioMe BioBank. Table S18. PheWAS of 14 candidate genes in UK Biobank.
Data Availability Statement
The WGS data of the IBD-PD cohort and the WES data of the BioMe BioBank cohorts used and/or analyzed in this study are available from the corresponding authors upon reasonable request within one week. This research was conducted using the UK Biobank resource under application number 53074. UK Biobank WES datasets are accessible upon approval of application. The 1KGP WGS data used in the current study is available at http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20190425_NYGC_GATK/raw_calls_updated/ [18]. The gene- and variant-level association statistics generated in this study are provided in the supplementary data. The Mutation Significance Cutoff and the Gene Damage Index databases for selecting high-impact variants and genes are available at https://lab.rockefeller.edu/casanova/MSC [28] and https://lab.rockefeller.edu/casanova/GDI [29]. The Human Gene Connectome is available for download at https://lab.rockefeller.edu/casanova/HGC [39]. Other tools and methods used in the analysis are described in the Methods section with references.