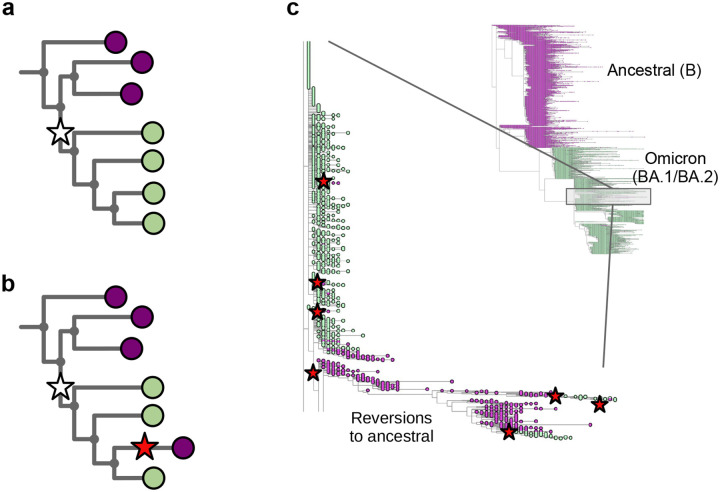
Figure 1: Assemblers which wrongly default to the reference base in the absence of data cause reversions in the phylogeny.
a) Cartoon phylogeny built from perfect genomes, with leaves coloured by genotype at a specific position X (purple - ancestral base, green - derived base). Just one mutation at this site, shown as a white star, is needed to explain the data. b) Cartoon showing the effect of assembly software assuming that a genome is identical to the reference genome when there is no data - here the amplicon containing position X is dropped in the lowest-but-one genome on the tree, creating one lone purple leaf. The tool which infers the phylogeny looks for a parsimonious explanation for this colour distribution, and concludes it was caused by a mutation (white star) followed by a “reversion” back to the ancestral base (red star). Errors in assembly caused by reference-bias tend to create enrichments of reversions. c) Part of the current UShER SARS-CoV-2 phylogeny, coloured by genotype at genome position 22813 (spike codon 417). Blow-up shows multiple reversions back to the ancestral purple. A non-exhaustive set of artefactual mutations (reversions, unreversions, re-reversions etc) are shown with red stars, where there is a flip back and forth from green to/from purple.