Abstract
The Zika virus (ZIKV), discovered in Africa in 1947, swiftly spread across continents, causing significant concern due to its recent association with microcephaly in newborns and Guillain-Barré syndrome in adults. Despite a decrease in prevalence, the potential for a resurgence remains, necessitating urgent therapeutic interventions. Like other flaviviruses, ZIKV presents promising drug targets within its replication machinery, notably the NS3 helicase (NS3Hel) protein, which plays critical roles in viral replication. However, a lack of structural information impedes the development of specific inhibitors targeting NS3Hel. Here we applied high-throughput crystallographic fragment screening on ZIKV NS3Hel, which yielded structures that reveal 3D binding poses of 46 fragments at multiple sites of the protein, including 11 unique fragments in the RNA-cleft site. These fragment structures provide templates for direct design of hit compounds and should thus assist the development of novel direct-acting antivirals against ZIKV and related flaviviruses, thus opening a promising avenue for combating future outbreaks.
Keywords: Zika virus, helicase, NS3, fragment-screening, flavivirus
Introduction
Since its discovery in Africa in 1947, the Zika virus (ZIKV) has traversed Asia and Polynesia before making its way to Brazil between May and December of 2013 (1, 2). From there, it swiftly spread across the globe (2). Initially, the symptoms of Zika fever were deemed too benign to raise significant concern within the medical community. However, this perception quickly shifted following the revelation of the correlation between ZIKV infection and the occurrence of microcephaly in newborns, as well as Guillain-Barré syndrome in adults (1, 3). Subsequently, additional neurological issues observed in newborns, including cortical malformations, ventriculomegaly, retinal damage, and arthrogryposis, were collectively termed Zika Congenital Syndrome (ZCS) (4–7). Although ZIKV has become less prevalent in recent times, cases continue to persist, raising concerns about the potential for a new pandemic outbreak. The absence of available vaccines or therapeutic agents for ZIKV underscores the urgent medical necessity, prompting ongoing efforts to identify potential drug candidates.
Such as Dengue virus (DENV) and Yellow Fever (YFV), the ZIKV belongs to the Flaviviridae family as a flavivirus (8). Its genome comprises a positive-sense, single-stranded RNA of approximately 10 kbp. (9). This is translated into a unique polyprotein which is proteolytically cleaved to originate 10 proteins separated in two groups: three structural proteins (Capsid (C), precursor-Membrane (prM/M) and Envelope (E)) and seven non-structural proteins - NSs - (NS1, NS2A, NS2B, NS3, NS4A NS4B and NS5) (9). Structural proteins form viral particles and nonstructural proteins participate in viral replication, virion assembly and evasion of the host immune response arboviruses (9). NS3 is composed by two domains: An N-terminal serine-protease (NS3Pro) and the C-terminal NTP-dependent RNA-helicase (NS3Hel). The NS3Pro assemblies with the NS2B cofactor, forming the NS2B-NS3Pro, to cleave portions of the virion polyprotein after translation, whereas the NS3Hel domain is responsible for double-stranded RNA-unwinding prior to RNA-polymerization (10). In addition to its enzymatic activity, NS3 engages in direct interactions with the viral largest membrane protein NS4B and NS5, leading to the formation of the viral replication complex (11, 12).
Considering the pivotal roles played by both enzymatic activities and interactions within the replication machinery in the process of viral replication, NS3 stands out as an exceptionally promising target for drug discovery (13, 14). Notably, currently ongoing clinical trial investigate a compound targeting NS3-NS4B interactions as a potential treatment for DENV (15, 16) Despite its pivotal role, the lack of structural information detailing specific interactions with NS3Hel and small molecules poses a hindrance to the advancement of novel direct-acting antivirals aimed at targeting RNA-unwind activity. To address this gap, we employed high-throughput (HTP) crystallographic fragment screening to enable ZIKV NS3Hel as a drug target, providing valuable insights for the development of a first-in-class RNA-unwind flaviviruses inhibitor.
Results
Overall fragment screening data processing
The initial ZIKV NS3Hel crystals obtained were used to determine crystal structure with a good stereochemistry with 99.6 % of the residues in the most favored regions of the Ramachandran plot, after being refined to 1.9 Å with Rwork/Rfree of 0.17/0.20 (PDB 6MH3). A single molecule was observed in the asymmetric unit, consistent with previous reported structures (17–19). Upon detailed inspection of crystal packing, significant solvent channels were observed providing access to essential sites within the protein structure. Soaking of crystals 20% DMSO for 2h showed minor effect on diffraction quality of the crystals, yielding datasets with resolution between 1.8 to 2.2 Å. Using these parameters, a total of 1,567 fragments were soaked from various library collections, resulting in 1,204 usable datasets. Impressively, 75% of these datasets diffracted above 2.5 Å, with an average resolution of 2.03 Å overall. PanDDA analysis revealed 451 events in distinct subsites of the protein. Following meticulous modeling, data curation, and cross-reviewing, a total of 45 ligand-bound and 1 ground-state structures of ZIKV NS3Hel ranging from 1.36 to 2.1 Å resolution, were validated and subsequently deposited in the Protein Data Bank (PDB). PDB identifiers, names, ligand descriptors and data collection and refinement statistics are available in Supplementary Table 1.
ZIKV NS3Hel overall structure and fragments screening results
ZIKV NS3Hel showcases a prototypical flavivirus RNA helicase insert, belonging to the helicase superfamily 2 (SF2), as evidenced by sequence comparisons and conserved motifs. It falls into the DEAH/RHA subfamilies, distinguished by the signature sequence within the Walker B motif (WB) (20–22). The structure of ZIKV NS3Hel reveals a composition comprising approximately 16% β-sheets, 33% α-helices, and 51% coils, among other elements. These secondary structures delineate three distinct subdomains: Sub-domain I (DI), spanning residues 183–332; Sub-domain II (DII), encompassing residues 333–481, which exhibit characteristics akin to RecA-like domains; and Sub-domain III (DIII), spanning residues 482–617, presenting a Flavi-DEAD domain. The arrangement of these subdomains creates conserved clefts between them, referred to as RNA-cleft sites, which have been experimentally demonstrated to facilitate binding to single-strand RNAs. At the interface of DI and DII, another notably conserved feature is the presence of a cavity responsible for NTP-binding, which drives NTPase activity (10, 23).
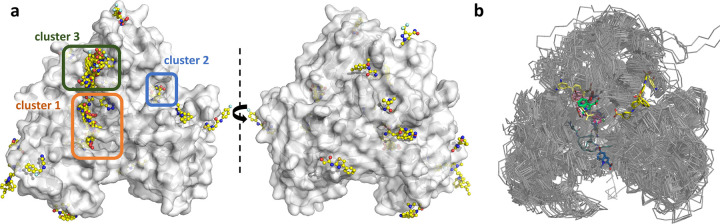
Fragments were noted to be present throughout all surfaces and cavities of ZIKV NS3Hel (Fig. 1a). Particularly noteworthy are three clusters that demonstrate promising characteristics and will be further discussed. To underscore the efficacy of HTP fragment screening, we conducted an alignment of 111 Flaviviridae PDB structures and visualized all small molecules present within them (Fig. 1b). The alignment reveals that much of the structural data available is redundant and primarily focuses on superficial ligands located at the NTP and RNA-entry sites (Fig. 1b). This comparison highlights how HTP fragment screening has successfully delved into chemical spaces that were previously unexplored for the SF2 helicase, shedding new light on the development of RNA binding competitors.
Fig. 1. Fragment screening reveal multiple binding sites.
(a) Surface view of ZIKV NS3Hel fragment screening results in two orientations. Surface is colored in grey, and fragments are depicted as sticks with yellow carbons. Selected cluster of fragments are marked with orange (1), blue (2) and green (3) boxes. (b) Overlaid structures of 111 PBDs containing superfamily 2 helicases from Flaviviridae family. Small molecules are depicted as sticks.
Fragments reveal new binding modes for the RNA-cleft site
In our HTP fragment screening initiative, we successfully elucidated the structure of ZIKV NS3Hel bound to 11 fragments in the RNA-cleft site, marked as Cluster 1: Z235341991, Z1262246195, Z44584886, Z18618496, Z2678251369, POB0128, POB0008, Z3241250482, Z56772132, Z729726784 and Z203039992 (Fig 2a). These complexes are cataloged under the PDB codes 5RHG, 7G9K, 7G9M, 7G9T, 7G9Y, 7GA1, 7GA2, 7GA3, 8UM3, 8V7R, and 8V7U, respectively. Detailed information including PDB identifiers, fragment names, ligand descriptors, and data collection/refinement statistics can be found in Supplementary Table 1.
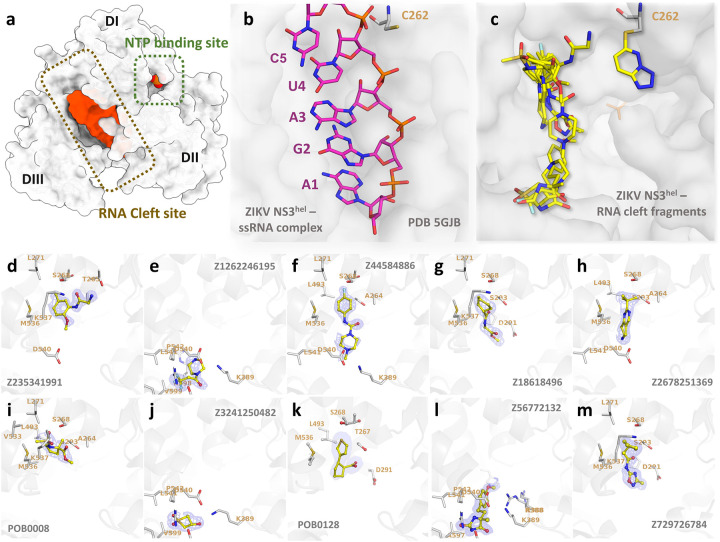
Fig. 2. Overview of RNA cleft fragments obtained.
(a) Surface view of the ZIKV NS3Hel (depicted in grey) reveals distinct features: the NTP and RNA clefts are highlighted with green and brown boxes, respectively. Additionally, the RNA and phosphate surfaces are depicted in orange to accentuate the RNA binding cleft. (b) Detailed view of ZIKV Helicase from PDB 5GJB in complex with RNA. RNA is depicted as sticks. (c) ZIKV Helicase in complex with multiple fragments obtained for the RNA-cleft binding site. Fragments are depicted as sticks. Bellow, detailed view of structures with electron densities and structural details of fragments Z235341991 (d), Z1262246195 (e), Z44584886 (f), Z18618496 (g), Z2678251369 (h), POB0008 (i), Z3241250482 (j), POB0128 (k), Z56772132 (l) and Z729726784 (m). Fragments are show as sticks with yellow carbons, while protein interacting residues are depicted as grey sticks carbon. 2Fo-Fc map is depicted as blue mesh, showed here with σ = 1.0.
The alignment of all fragments from Cluster 1 elucidates their significant overlap with the RNA-bound structure from PDB 5GJB, particularly in the region responsible for base recognition (Fig 2b and c). Z235341991 is a N-(2-methoxy-5-methylphenyl)glycinamide that forms hydrogen bound with the OG1 from the highly conserved T262 and NZ from K537 (Fig. 2d). Z1262246195 is a 3,3,3-Tris(fluoranyl)-1-Piperazin that interacts via fluorine atoms 1 with S608 and R617 (Fig. 2e). Fragment Z44584886 is a 4-Fluorophenyl-4-Methyl-Piperazine that is maintained by weak interactions with A264, S293, K389, D540 and L541 (Fig. 2f). Z18618496 is a Methyl n-(Cyclohexanecarbonyl)glycinate that is maintained by weak interactions with L271, D291, P292, S293, M536, K537 and V543 (Fig. 2g). Fragment Z2678251369 is a 2-tert-butyl-5,6,7,8-tetrahydropyrido[4,3-d]pyrimidine that weakly interact with residues A264, S268, S293, L493, M536, D540, L541 and V543 (Fig. 2h). Fragment POB0008 is a Methyl 1-(Methanesulfonyl)-2-Methyl-D-Prolinate that forms hydrogen bond with S268 and weakly interact with residues A264, S268, L271, S293, L493, V533, M536 and K53 (Fig. 2i). Fragment Z3241250482 is a 1,1-bis(oxidanylidene)thietan-3-ol that form hydrogen bounds with K389 and R617, as well as weak interactions with K389, D540, L541, P542, V599, A605, S608, F609 and R617 (Fig. 2j). Fragment POB0128 is a (1s,2r)-2-(Thiophen-3-Yl)cyclopentane-1-Carboxylic acid that form hydrogen bonds with S293 and weakly interacts A264, T267, D291, P292, S293, L493, L541 and V543 (Fig. 2k).
In the structure of PDB 8V7R, two instances of Z56772132, namely 5-methyl-hydantoin, were detected, with the copy designated as chain D occupying the RNA-cleft site. This fragment engages in two hydrogen bonds: one forms between the nitrogen atom (N11) and the oxygen atom of the highly conserved residue D540, measured at 2.8 Å, while the other bond emerges between the oxygen atom (O13) and the NH2 group of R617 at 3.2 Å distance (Fig. 2i). Fragment Z729726784, a 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, demonstrates interactions with various residues including L271, D291, P292, S293, M536, K537, and V543 (Fig. 2m).
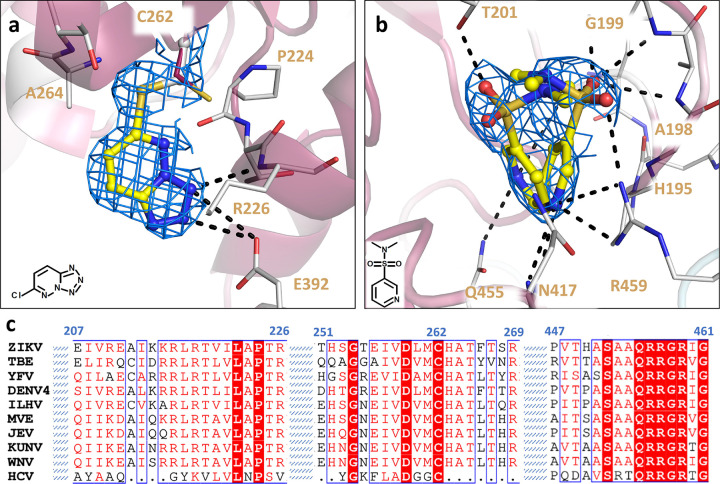
Furthermore, in the structure of ZIKV NS3Hel complexed with Z203039992 (PDB 8UM3), identified as 6-chlorotetrazolo-pyridazine, an intriguing binding pattern emerges. This fragment demonstrates an ability to target the nucleophilic thiol group of C262 by interacting with the main chain nitrogen of R226 and the OE2 atom of G392 via its N1 and N2 atoms (Fig 3a). This ligation of Z203039992 causes a 180° twist of C262 side chain conserved conformation (Fig 3a). It’s noteworthy that C262 represents a highly conserved residue across Flaviviridae family, including distant species like HCV (Fig 3c). Positioned within a sub-pocket adjacent to the NTP binding site, C262’s involvement in these interactions could be a unique opportunity for designing novel covalent allosteric or competitive inhibitors targeting Flaviviridae helicases.
Fig. 3. Details of covalent and ATP-binding fragments.
(a) Structure of ZIKV NS3Hel in complex with Z203039992. C262 covalently bound is show is depicted with gray carbon, while native apo C262 from PDB 6MH3 is showed in purple (b) Structure of ZIKV NS3Hel in complex with Z751811134 in two overlapping forms. Fragments are shown as sticks with yellow carbons, while protein interacting residues are depicted as gray sticks carbon. Cartoon is colored according to conservancy score calculated with Consurf, where purple indicated more conserved regions and white the less conserved regions. 2Fo-Fc map is depicted as blue mesh, showed here with σ = 1.0. (c) Flaviviridae sequence alignment showing selected regions of helicase that are discussed above. Sequence alignment was generated with ESPript and colored with default parameters.
Fragments for the NTP-binding site
The NTP-binding site represents a highly conserved cavity pivotal for ATP hydrolysis. In line with numerous structures in PDB, our crystal structure of Zika NS3Hel reveals the presence of a phosphate molecule intrinsically occupying the binding site. This molecule likely originates from the expression source and is presumed to compete with any incoming fragment for binding. Therefore, only fragment Z751811134, a N,N-dimethylpyridine-3-sulfonamide was observed in this site. This fragment was found in two mirrored conformations, with the sulfonamide group occupying the position of phosphate (Fig 3b). Fragment Z751811134 interacts with highly conserved residues G199, K200, P196, G197, A198, G199, K200, T201, D285, E286, A317, Q455, R459 and R462, and it’s deposited on PDB under the code 7G9Q. Detailed information including PDB identifiers, fragment names, ligand descriptors, and data collection/refinement statistics can be found in Supplementary Table 1.
Cluster 3 and surface fragments are crystal artifacts.
In Cluster 3 of ZIKV NS3Hel, we observed a promiscuous region where we were able to clearly elucidate the structure of 17 fragments, Z198194396, Z126932614, Z425387594, Z1444783243, Z235449082, Z274555794, Z291279160, Z1703168683, Z31222641, Z111782404, Z57821475, Z133716556, Z1619958679, Z1216822028, Z119989094, Z1509711879 and Z85933875, respectively deposited on PDB under the codes 5RHI, 5RHJ, 5RHL, 5RHO, 5RHQ, 5RHS, 5RHT, 5RHU, 5RHW, 7G9L, 7G9N, 7G9O, 7G9P, 7G9Z, 7GA5, 7GA6 and 7GA7. Careful analysis of Cluster 3 indicates that this region is an artifact of crystal packing, and therefore these fragments are too unreliable for seeding a drug design campaign. The same can be said for the 11 surface fragments, Z55222357, Z383202616, Z444860982, Z2048325751, Z56823075, Z1454310449, Z2856434938, Z1324080698, Z385450668, Z758198920 and Z905065822, deposited on PDB under the codes 5RHH, 5RHK, 5RHR, 5RHV, 7G9U, 7G9V, 7G9W, 7GA0, 7GA4, 5RHM, 5RHP, 5RHX, 7G9R, 7G9S and 7G9X. Despite the promising potential of surface fragments in the development of novel protein interaction blockers, our analysis reveals that these fragments are sustained not solely through interaction with the asymmetric unit but also through crystal packing constraints. Consequently, they may not be optimal candidates for design purposes. Detailed information including PDB identifiers, fragment names, ligand descriptors, and data collection/refinement statistics can be found in Supplementary Table 1.
Discussion
After the onset of the COVID-19 pandemic, it became increasingly evident that there is a critical need to prioritize the development of broad-spectrum direct-acting antiviral capable of effectively combating viruses with pandemic potential (24, 25). By applying HTP fragment-screening combined machine learning methods, our prelude group was able to develop a novel oral non-peptidomimetic inhibitor of SARS-CoV-2 Main protease in the open-science format (25–27). Our derived group named AI-driven Structure-enabled Antiviral Platform (ASAP), backed by the NIH Antiviral Drug Discovery (AViDD) program, is committed to employing a similar strategy and ethos to accelerate the development of broad-spectrum, clinic-ready antiviral agents in anticipation of future pandemics, including those caused by flaviviruses.
Despite its promise for therapeutic design, targeting flaviviruses NS3Hel poses significant challenges, particularly due to the scarcity of structural data available for conserved sites. Only a handful of NS3Hel inhibitors, such as benzimidazoles, benzothiazole, pyrrolone, ivermectin, ST-610, and suramin, have been documented in the literature (28–30). However, none of these have been substantiated with comprehensive structural data to support design of new candidates. In the case of HCV, another member of the Flaviviridae family, a few structures of NS3Hel in complex with inhibitors have been reported. These include a triphenylmethane series of NTP-site competitors (31) and an indole series that competes with the +4 site in the RNA exit tunnel (32). However, it’s worth noting that ATP competitors can potentially lead to significant toxicological effects, while the pocket accommodating the indole derivatives is wide and groove-like, with inherent flexibility of the protein in this region. Therefore, it’s evident that acquiring new structural data is imperative to facilitate drug design targeting NS3Hel.
The ZIKV NS3Hel is segmented into three distinct sub-domains, namely DI, DII, and DIII. This domain exhibits distinctive characteristics with two binding sites: one that selectively engages with single-strand RNA, and the other that binds to a nucleoside triphosphate (NTP) (18). These binding sites are situated within conserved clefts formed between the three NS3Hel subdomains (17, 23, 33). Consequently, these clefts emerge as intriguing areas for exploration in drug design and development (33).
In a prior fragment screening endeavor employing x-ray crystallography on ZIKV NS3Hel, a total of 154 fragments were tested, resulting in the detection of only four hits. Notably, all four hits were located at the surface, distantly from both the NTP and the RNA-cleft binding sites or any other conserved region (34). In a comparable study using a thermal-shift assay, researchers screened 500 fragments against DENV helicase but found no hits (35). By applying HTP crystallographic fragment screening, we successfully revealed the structures of 45 fragments at multiple sites for ZIKV NS3Hel. Notably, this encompassed the identification of 11 distinct fragments specifically binding to the RNA-cleft site, including a unique covalent hit of the conserved C262 (Fig. 2). A recent publication showed an allosteric inhibitor capable of binding to C441 and C444 on the SARS-CoV-2 helicase (36). The proximity of Z203039992 to both the RNA cleft and NTP site, as well as the other fragments discovered in Cluster 1, suggests its potential in serving as a structural template for the design of novel orthostatic or allosteric inhibitors targeting flaviviruses. These findings pave the way for the rapid development of unique inhibitors, specifically targeting the RNA-unwinding activity of flavivirus helicases. Such advancements hold promise for the creation of a new class of direct-acting antivirals.
MATERIALS AND METHODS
Cloning, Expression and Purification
The coding region of ZIKV NS3Hel from the MR766 strain (GB: AY632535) was codon optimized for E. coli expression and synthesized by Genscript. Region encoding primers CAGGGCGCCATGAGTGGCGCCCTCT-3’ and 5’-GACCCGACGCGGTTACTTTTTCCAGCGG-3’ was amplified and cloned into pETTrx-1a/LIC by Ligation independent cloning method (LIC) (37).
Transformed Rosetta (DE3) E. coli (Novagen) cells were grown in ZYM 5052 autoinduction medium (38)supplemented with 50 μg.ml-1 kanamycin and 34 μg.ml-1 chloramphenicol at 37 °C and 200 r.p.m. until OD600 of 1.0, followed by 36h shaking at 18 °C. Cells were harvested by centrifugation (4,000 g for 40 min, at 4 °C) and cell pellet was resuspended in 20 mM Bis-Tris pH 7.0, 500 mM NaCl, 40 mM Imidazole, 10% glycerol supplemented with 4.0 mM dithiothreitol, 2.0 mM phenylmethyl sulfonyl fluoride, 200 μg.L−1 lysozyme, 10 U.ml−1 benzonase and incubated for 30 min in ice bath. After sonication, cell debris was separated by centrifugation (20,000 g, 20 min, 4°C) and clarified material was loaded into HisTrap HP 5.0 mLpre-equilibrated with 20 mM Bis-Tris pH 7.0, 500 mM NaCl, 40 mM Imidazole, 10% glycerol. After washing, protein was eluted with a gradient of the same buffer containing 500 mM imidazole. Protein 6xHis-Thioredoxin tag was cleaved with TEV protease during O.N. dialysis in 20 mM Bis-Tris pH 7.0, 500 mM NaCl, 40 mM Imidazole, 10% glycerol supplemented with 4.0 mM dithiothreitol. A second passage through HisTrap HP 5.0 mL was used to remove cleaved tag and TEV. Protein was concentrated and inject into size-exclusion chromatography on a Superdex 16/60 75 column (GE Healthcare) pre-equilibrated in buffer 20 mM Bis-Tris pH 7.0, 500 mM NaCl, 10% glycerol for polishing. For crystallization, samples where concentrated to 5.0 mg.ml−1 based on theoretical extinction coefficient of 64,775 M−1.cm−1 at 280 nm as reference (39).
Crystal growing and optimization for screening
Initial crystals were grow in a crystallization solution derived from Morpheus HT Crystallization Screen (Molecular Dimensions), comprising 0.12 M NPS Mix (consisting of 0.3 M Sodium phosphate dibasic dihydrate, 0.3 M Ammonium sulphate, and 0.3 M Sodium nitrate from Molecular Dimensions), along with 0.1 M MES/Imidazole at pH 6.5 (Molecular Dimensions), and 33% Precipitant Mix 4 (11% MPD, 11% PEG 1,000, and 11% PEG 3,350 from Sigma Aldrich), maintained at 20°C. An initial model was obtained at a resolution of 1.9 Å using a Rigaku Micromax-007 HF X-ray diffractometer, processed within the P21 space group using XDS and Aimless (40, 41), and subsequently solved through molecular replacement employing the PDB entry 5GJB as a reference model with Phaser (42) and refined with phenix.refine (43). The resulting structure was deposited under the code 6MH3 (Supplementary Table 1).
For fragment screening, crystals were treated by aspirating and vortexed them with 100 μL of the crystallization solution along with glass beads. Subsequently, 0.05 μL of seed stock was introduced into each drop, maintaining a 1:1 ratio of protein to crystallization solution. This process led to the acquisition of viable crystals in approximately 60–70% of the drops within 48 hours of the plate setup. To determine solvent tolerability, crystals were incubated with dimethyl sulfoxide (DMSO) in concentrations of 10–30% and incubated for periods of 1–3h at room temperature.
Fragment screening and data collection and processing and analysis
The fragment screening was performed using the XCHEM infrastructure available at Diamond Light Source. The soaking was done by transferring fragments from DSi-Poised Library (44), Minifrags (45), Fraglites (46), Peplites (47), York 3D (48), SpotXplorer (49), and Essential Fragment Library (Enamine) to the crystal drop using ECHO Liquid Handler at proportion of 20%. Crystals were incubated for 2h at room temperature and harvested using Crystal Shifter (Oxford Lab Technologies), mounted on loops and flash frozen in liquid nitrogen.
All data was collected at I04–1 beamline at Diamond Light Source at 100 K and processed with automated pipelines using combined softwarès, with resolution cutoff defined algorithmically (40, 50–52). All further analysis was performed using XChemExplorer (53). Initial density maps were generated using DIMPLE (52) and PDB 6MH3 as template model. Ligand restrains were generated with ACEDRG and GRADE (54, 55). A ground state model was constructed utilizing PanDDA by amalgamating 100 apo datasets (56). This model, deposited under the code 5RHY (Supplementary Table 1), served as the template for molecular replacement for the full analysis. Event maps were calculated with PanDDA (56), and ligands were modeled using Coot (57) Models were refined with Refmac (58), Buster (54) and phenix.refine (43), and models and quality annotations cross-reviewed. Coordinates, structure factors and PanDDA event maps for all data sets are deposited in the Protein Data Bank (PDB). Data collection and refinement statistics, PBD codes and ligand identifier are available in Supplementary Table 1. Figures were made using PyMOL (Schrödinger), ChimeraX (59), Consurf (60) and E/ENDScript (61). Protein alignment in Fig. 1b used PDBs 5JPS, 5Y4Z, 6ADW, 6ADX, 6ADY, 6MH3, 5VI7, 5GJB, 5GJC, 5JMT, 5JRZ, 5JWH, 5K8I, 5K8L, 5K8T, 5K8U, 5TXG, 5Y6M, 5Y6N, 7V2Z, 8CXG, 8CXH, 8CXI, 5MFX, 6RWZ, 6S0J, 7WD4, 2BHR, 2BMF, 8GZQ, 8GZR, 2JLS, 2VBC, 2WHX, 2WZQ, 2JLR, 2JLU, 2JLV, 2JLW, 2JLX, 2JLZ, 5XC6, 5YVJ, 5YVU, 5YVV, 5YVW, 5YVY, 5YW1, 2JLQ, 2JLY, 5XC7, 7XT0, 2V8O, 2WV9, 2QEQ, 2Z83, 8JIX, 2V6I, 2V6J, 1YKS, 5FFM, 7BLV, 7BM0, 7NXU, 7OJ4, 7JNO, 7V4Q, 7V4R, 6M40, 1HEI, 4OJQ, 4OK3, 4OK5, 4OK6, 4OKS, 4WXR, 1A1V, 5GVU, 5WSO, 3KQH, 3KQK, 3KQL, 3KQN, 3KQU, 4CBL, 5E4F, 2ZJO, 4CBG, 4CBH, 4CBI, 5MZ4, 5WX1, 1CU1, 2F55, 3O8B, 3O8C, 3O8D, 3O8R, 3RVB, 4A92, 4B6E, 4B6F, 4B71, 4B73, 4B74, 4B75, 4B76, 4CBM, 4WXP, 5FPS, 5FPT, 5FPY, 8OHM and 5WDX.
Supplementary Material
Acknowledgements
Research reported in this publication was supported in part by NIAID of the National Institute of Health under award number U19AI171399. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. GO and ASG acknowledge Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP project 2013/07600-3). Authors acknowledge Diamond Light Source access to I04-1 beamline and XCHEM facilities through proposals lb20289 and lb32627.
Data availability
Any other data can be supplied upon reasonable request. Crystallographic coordinates and structure factors for all structures have been deposited in the PDB with the following accession codes 6MH3, 5RHG, 7G9K, 7G9M, 7G9T, 7G9Y, 7GA1, 7GA2, 7GA3, 8UM3, 8V7R, 8V7U, 7G9Q, 5RHI, 5RHJ, 5RHL, 5RHO, 5RHQ, 5RHS, 5RHT, 5RHU, 5RHW, 7G9L, 7G9N, 7G9O, 7G9P, 7G9Z, 7GA5, 7GA6, 7GA7, 5RHH, 5RHK, 5RHR, 5RHV, 7G9U, 7G9V, 7G9W, 7GA0, 7GA4, 5RHY, 5RHM, 5RHP, 5RHX, 7G9R, 7G9S and 7G9X.
References
- 1.Barreto ML, Barral-Netto M, Stabeli R, Almeida-Filho N, Vasconcelos PFC, Teixeira M, Buss P, Gadelha PE. 2016. Zika virus and microcephaly in Brazil: a scientific agenda. The Lancet 387:919–921. [DOI] [PubMed] [Google Scholar]
- 2.Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. 2017. An update on Zika virus infection. The Lancet 390:2099–2109. [DOI] [PubMed] [Google Scholar]
- 3.de Araújo TVB, Ximenes RA de A, Miranda-Filho D de B, Souza WV, Montarroyos UR, de Melo APL, Valongueiro S, de Albuquerque M de FPM, Braga C, Filho SPB, Cordeiro MT, Vazquez E, Cruz D di CS, Henriques CMP, Bezerra LCA, Castanha PM da S, Dhalia R, Marques-Júnior ETA, Martelli CMT, Rodrigues LC, Dhalia C, Santos M, Cortes F, Kleber de Oliveira W, Evelim Coelho G, Cortez-Escalante JJ, Campelo de Albuquerque de Melo CF, Ramon-Pardo P, Aldighieri S, Mendez-Rico J, Espinal M, Torres L, Nassri Hazin A, Van der Linden A, Coentro M, Santiago Dimech G, Siqueira de Assuncao R, Ismael de Carvalho P, Felix Oliveira V. 2018. Association between microcephaly, Zika virus infection, and other risk factors in Brazil: Final report of a case-control study. Lancet Infect Dis 18:328–336. [DOI] [PubMed] [Google Scholar]
- 4.Barreto ML, Barral-Netto M, Stabeli R, Almeida-Filho N, Vasconcelos PF, Teixeira M, Buss P, Gadelha PE. 2016. Zika virus and microcephaly in Brazil: a scientific agenda. Lancet 387:919–921. [DOI] [PubMed] [Google Scholar]
- 5.Ventura C V, Maia M, Bravo-Filho V, Gois AL, Belfort R. Jr. 2016. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387:228. [DOI] [PubMed] [Google Scholar]
- 6.de Paula Freitas B, de Oliveira Dias JR, Prazeres J, Sacramento GA, Ko AI, Maia M, Belfort R. Jr. 2016. Ocular Findings in Infants With Microcephaly Associated With Presumed Zika Virus Congenital Infection in Salvador, Brazil. JAMA Ophthalmol 10.1001/jamaophthalmol.2016.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.França GVA, Schuler-Faccini L, Oliveira WK, Henriques CMP, Carmo EH, Pedi VD, Nunes ML, Castro MC, Serruya S, Silveira MF, Barros FC, Victora CG. 2016. Congenital Zika virus syndrome in Brazil: a case series of the first 1501 livebirths with complete investigation. Lancet 388:891–897. [DOI] [PubMed] [Google Scholar]
- 8.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. 1998. Phylogeny of the genus Flavivirus. J Virol 72:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus Genome Organization, Expression, and Replication. Annu Rev Microbiol 44:649–688. [DOI] [PubMed] [Google Scholar]
- 10.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J 27:3209–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osawa T, Aoki M, Ehara H, Sekine S ichi. 2023. Structures of dengue virus RNA replicase complexes. Mol Cell 83:2781–2791.e4. [DOI] [PubMed] [Google Scholar]
- 12.Klema VJ, Padmanabhan R, Choi KH. 2015. Flaviviral Replication Complex: Coordination between RNA Synthesis and 51-RNA Capping. Viruses 2015/August/20. 7:4640–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo D, Vasudevan SG, Lescar J. 2015. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res 2015/April/07. 118:148–158. [DOI] [PubMed] [Google Scholar]
- 14.de Godoy AS, Sachetto Fernandes R, Campos Aguiar AC, Vieira Bueno R, de Moraes Roso Mesquita NC, Carvalho Guido RV, Oliva G. 2019. Structural and mechanistic insight from antiviral and antiparasitic enzyme drug targets for tropical infectious diseases. Curr Opin Struct Biol 59. [DOI] [PubMed] [Google Scholar]
- 15.Kaptein SJF, Goethals O, Kiemel D, Marchand A, Kesteleyn B, Bonfanti JF, Bardiot D, Stoops B, Jonckers THM, Dallmeier K, Geluykens P, Thys K, Crabbe M, Chatel-Chaix L, Münster M, Querat G, Touret F, de Lamballerie X, Raboisson P, Simmen K, Chaltin P, Bartenschlager R, Van Loock M, Neyts J. 2021. A pan-serotype dengue virus inhibitor targeting the NS3–NS4B interaction. Nature 2021 598:7881 598:504–509. [DOI] [PubMed] [Google Scholar]
- 16.Goethals O, Kaptein SJF, Kesteleyn B, Bonfanti J-F, Van Wesenbeeck L, Bardiot D, Verschoor EJ, Verstrepen BE, Fagrouch Z, Putnak JR, Kiemel D, Ackaert O, Straetemans R, Lachau-Durand S, Geluykens P, Crabbe M, Thys K, Stoops B, Lenz O, Tambuyzer L, De Meyer S, Dallmeier K, McCracken MK, Gromowski GD, Rutvisuttinunt W, Jarman RG, Karasavvas N, Touret F, Querat G, de Lamballerie X, Chatel-Chaix L, Milligan GN, Beasley DWC, Bourne N, Barrett ADT, Marchand A, Jonckers THM, Raboisson P, Simmen K, Chaltin P, Bartenschlager R, Bogers WM, Neyts J, Van Loock M. 2023. Blocking NS3–NS4B interaction inhibits dengue virus in non-human primates. Nature 615:678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian H, Ji X, Yang X, Xie W, Yang K, Chen C, Wu C, Chi H, Mu Z, Wang Z, Yang H. 2016. The crystal structure of Zika virus helicase: basis for antiviral drug design. Protein Cell 7:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian H, Ji X, Yang X, Zhang Z, Lu Z, Yang K, Chen C, Zhao Q, Chi H, Mu Z, Xie W, Wang Z, Lou H, Yang H, Rao Z. 2016. Structural basis of Zika virus helicase in recognizing its substrates. Protein Cell 7:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R, Coloma J, García-Sastre A, Aggarwal AK. 2016. Structure of the NS3 helicase from Zika virus. Nat Struct Mol Biol 23:752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowsky E, Fairman ME. 2007. RNA helicases - one fold for many functions. Curr Opin Struct Biol 17:316–324. [DOI] [PubMed] [Google Scholar]
- 21.Fairman-Williams ME, Guenther U-P, Jankowsky E. 2010. SF1 and SF2 : family matters 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tani H. 2017. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase / nucleoside triphosphatase at a … Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase / nucleoside triphosphatase at a resolution of 10.1016/j.virol.2007.12.018. [DOI] [PubMed]
- 23.Cao X, Li Y, Jin X, Guo F, Jin T. 2016. Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design. Nucleic Acids Res 44:10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Delft A, Hall MD, Kwong AD, Purcell LA, Saikatendu KS, Schmitz U, Tallarico JA, Lee AA. 2023. Accelerating antiviral drug discovery: lessons from COVID-19. Nature Reviews Drug Discovery 2023. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Delft F, Calmiano M, Chodera J, Griffen E, Lee A, London N, Matviuk T, Perry B, Robinson M, von Delft A. 2021. A white-knuckle ride of open COVID drug discovery. Nature 2021 594:7863 594:330–332. [DOI] [PubMed] [Google Scholar]
- 26.C J, L AA, L N, von D F. 2020. Crowdsourcing drug discovery for pandemics. Nat Chem 12:581. [DOI] [PubMed] [Google Scholar]
- 27.Boby ML, Fearon D, Ferla M, Filep M, Koekemoer L, Robinson MC, Consortium‡ TCM, Chodera JD, Lee AA, London N, von Delft A, von Delft F, Achdout H, Aimon A, Alonzi DS, Arbon R, Aschenbrenner JC, Balcomb BH, Bar-David E, Barr H, Ben-Shmuel A, Bennett J, Bilenko VA, Borden B, Boulet P, Bowman GR, Brewitz L, Brun J, BVNBS S, Calmiano M, Carbery A, Carney DW, Cattermole E, Chang E, Chernyshenko E, Clyde A, Coffland JE, Cohen G, Cole JC, Contini A, Cox L, Croll TI, Cvitkovic M, De Jonghe S, Dias A, Donckers K, Dotson DL, Douangamath A, Duberstein S, Dudgeon T, Dunnett LE, Eastman P, Erez N, Eyermann CJ, Fairhead M, Fate G, Fedorov O, Fernandes RS, Ferrins L, Foster R, Foster H, Fraisse L, Gabizon R, García-Sastre A, Gawriljuk VO, Gehrtz P, Gileadi C, Giroud C, Glass WG, Glen RC, Glinert I, Godoy AS, Gorichko M, Gorrie-Stone T, Griffen EJ, Haneef A, Hassell Hart S, Heer J, Henry M, Hill M, Horrell S, Huang QYJ, Huliak VD, Hurley MFD, Israely T, Jajack A, Jansen J, Jnoff E, Jochmans D, John T, Kaminow B, Kang L, Kantsadi AL, Kenny PW, Kiappes JL, Kinakh SO, Kovar B, Krojer T, La VNT, Laghnimi-Hahn S, Lefker BA, Levy H, Lithgo RM, Logvinenko IG, Lukacik P, Macdonald HB, MacLean EM, Makower LL, Malla TR, Marples PG, Matviiuk T, McCorkindale W, McGovern BL, Melamed S, Melnykov KP, Michurin O, Miesen P, Mikolajek H, Milne BF, Minh D, Morris A, Morris GM, Morwitzer MJ, Moustakas D, Mowbray CE, Nakamura AM, Neto JB, Neyts J, Nguyen L, Noske GD, Oleinikovas V, Oliva G, Overheul GJ, Owen CD, Pai R, Pan J, Paran N, Payne AM, Perry B, Pingle M, Pinjari J, Politi B, Powell A, Pšenák V, Pulido I, Puni R, Rangel VL, Reddi RN, Rees P, Reid SP, Reid L, Resnick E, Ripka EG, Robinson RP, Rodriguez-Guerra J, Rosales R, Rufa DA, Saar K, Saikatendu KS, Salah E, Schaller D, Scheen J, Schiffer CA, Schofield CJ, Shafeev M, Shaikh A, Shaqra AM, Shi J, Shurrush K, Singh S, Sittner A, Sjö P, Skyner R, Smalley A, Smeets B, Smilova MD, Solmesky LJ, Spencer J, Strain-Damerell C, Swamy V, Tamir H, Taylor JC, Tennant RE, Thompson W, Thompson A, Tomásio S, Tomlinson CWE, Tsurupa IS, Tumber A, Vakonakis I, van Rij RP, Vangeel L, Varghese FS, Vaschetto M, Vitner EB, Voelz V, Volkamer A, Walsh MA, Ward W, Weatherall C, Weiss S, White KM, Wild CF, Witt KD, Wittmann M, Wright N, YahalomRonen Y, Yilmaz NK, Zaidmann D, Zhang I, Zidane H, Zitzmann N, Zvornicanin SN. 2024. Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors. Science (1979) 382:eabo7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Res MC. 2016. Benzothiazole and Pyrrolone Flavivirus Inhibitors Targeting the Viral Helicase 13:1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borowski P, Deinert J, Schalinski S, Bretner M, Ginalski K, Kulikowski T, Shugar D. 2003. Halogenated benzimidazoles and benzotriazoles as inhibitors of the NTPase/helicase activities of hepatitis C and related viruses. Eur J Biochem 270:1645–1653. [DOI] [PubMed] [Google Scholar]
- 30.García LL, Padilla L, Castaño JC. 2017. Inhibitors compounds of the flavivirus replication process. Virol J 14:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CS, Chiou CT, Chen GS, Chen SC, Hu CY, Chi WK, Chu YD, Hwang LH, Chen PJ, Chen DS, Liaw SH, Chern JW. 2009. Structure-based discovery of triphenylmethane derivatives as inhibitors of hepatitis C virus helicase. J Med Chem 52:2716–2723. [DOI] [PubMed] [Google Scholar]
- 32.LaPlante SR, Padyana AK, Abeywardane A, Bonneau P, Cartier M, Coulombe R, Jakalian A, Wildeson-Jones J, Li X, Liang S, McKercher G, White P, Zhang Q, Taylor SJ. 2014. Integrated Strategies for Identifying Leads That Target the NS3 Helicase of the Hepatitis C Virus. J Med Chem 57:2074–2090. [DOI] [PubMed] [Google Scholar]
- 33.Jain R, Coloma J, García-Sastre A, Aggarwal AK. 2016. Structure of the NS3 helicase from Zika virus. Nat Struct Mol Biol 23:752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munawar A, Beelen S, Munawar A, Lescrinier E, Strelkov S V. 2018. Discovery of Novel Druggable Sites on Zika Virus NS3 Helicase Using X-ray Crystallography-Based Fragment Screening. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coutard B, Decroly E, Li C, Sharff A, Lescar J, Bricogne G, Barral K. 2014. Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res 106:61–70. [DOI] [PubMed] [Google Scholar]
- 36.Ramsey JR, Shelton PMM, Heiss TK, Olinares PDB, Vostal LE, Soileau H, Grasso M, Casebeer SW, Adaniya S, Miller M, Sun S, Huggins DJ, Myers RW, Chait BT, Vinogradova E V, Kapoor TM. 2024. Using a Function-First “Scout Fragment”-Based Approach to Develop Allosteric Covalent Inhibitors of Conformationally Dynamic Helicase Mechanoenzymes. J Am Chem Soc 146:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18:6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. [DOI] [PubMed] [Google Scholar]
- 39.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook 571–607. [Google Scholar]
- 40.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 2010/February/04. 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine P V, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox OB, Krojer T, Collins P, Monteiro O, Talon R, Bradley A, Fedorov O, Amin J, Marsden BD, Spencer J, Von Delft F, Brennan PE. 2016. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem Sci 7:2322–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Reilly M, Cleasby A, Davies TG, Hall RJ, Ludlow RF, Murray CW, Tisi D, Jhoti H. 2019. Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design. Drug Discov Today 24:1081–1086. [DOI] [PubMed] [Google Scholar]
- 46.Wood DJ, Lopez-Fernandez JD, Knight LE, Al-Khawaldeh I, Gai C, Lin S, Martin MP, Miller DC, Cano C, Endicott JA, Hardcastle IR, Noble MEM, Waring MJ. 2019. FragLites—Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation. J Med Chem 62:3741–3752. [DOI] [PubMed] [Google Scholar]
- 47.Douangamath A, Fearon D, Gehrtz P, Krojer T, Lukacik P, Owen CD, Resnick E, Strain-Damerell C, Aimon A, Ábrányi-Balogh P, Brandão-Neto J, Carbery A, Davison G, Dias A, Downes TD, Dunnett L, Fairhead M, Firth JD, Jones SP, Keeley A, Keserü GM, Klein HF, Martin MP, Noble MEM, O’Brien P, Powell A, Reddi RN, Skyner R, Snee M, Waring MJ, Wild C, London N, von Delft F, Walsh MA. 2020. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downes TD, Jones SP, Klein HF, Wheldon MC, Atobe M, Bond PS, Firth JD, Chan NS, Waddelove L, Hubbard RE, Blakemore DC, De Fusco C, Roughley SD, Vidler LR, Whatton MA, Woolford AJ-A, Wrigley GL, O’Brien P. 2020. Design and Synthesis of 56 Shape-Diverse 3D Fragments. Chemistry – A European Journal 26:8969–8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bajusz D, Wade WS, Satała G, Bojarski AJ, Ilaš J, Ebner J, Grebien F, Papp H, Jakab F, Douangamath A, Fearon D, von Delft F, Schuller M, Ahel I, Wakefield A, Vajda S, Gerencsér J, Pallai P, Keserű GM. 2021. Exploring protein hotspots by optimized fragment pharmacophores. Nature Communications 2021. 12:1 12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G. 2018. DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr D Struct Biol 74:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. 2011. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 67:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krojer T, Talon R, Pearce N, Collins P, Douangamath A, Brandao-Neto J, Dias A, Marsden B, Von Delft F. 2017. The XChemExplorer graphical workflow tool for routine or large-scale protein-ligand structure determination. Acta Crystallogr D Struct Biol 73:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. 2012. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 68:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long F, Nicholls RA, Emsley P, Gražulis S, Merkys A, Vaitkus A, Murshudov GN. 2017. AceDRG: a stereochemical description generator for ligands. Acta Crystallographica Section D 73:112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pearce NM, Krojer T, Bradley AR, Collins P, Nowak RP, Talon R, Marsden BD, Kelm S, Shi J, Deane CM, Von Delft F. 2017. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat Commun 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.P EF, G TD, H CC, M EC, C GS, C TI, M JH, F TE. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any other data can be supplied upon reasonable request. Crystallographic coordinates and structure factors for all structures have been deposited in the PDB with the following accession codes 6MH3, 5RHG, 7G9K, 7G9M, 7G9T, 7G9Y, 7GA1, 7GA2, 7GA3, 8UM3, 8V7R, 8V7U, 7G9Q, 5RHI, 5RHJ, 5RHL, 5RHO, 5RHQ, 5RHS, 5RHT, 5RHU, 5RHW, 7G9L, 7G9N, 7G9O, 7G9P, 7G9Z, 7GA5, 7GA6, 7GA7, 5RHH, 5RHK, 5RHR, 5RHV, 7G9U, 7G9V, 7G9W, 7GA0, 7GA4, 5RHY, 5RHM, 5RHP, 5RHX, 7G9R, 7G9S and 7G9X.