Abstract
Glycosaminoglycans (GAGs) on the surface of cultured cells are important in the first step of efficient respiratory syncytial virus (RSV) infection. We evaluated the importance of sulfation, the major biosynthetic modification of GAGs, using an improved recombinant green fluorescent protein-expressing RSV (rgRSV) to assay infection. Pretreatment of HEp-2 cells with 50 mM sodium chlorate, a selective inhibitor of sulfation, for 48 h prior to inoculation reduced the efficiency of rgRSV infection to 40%. Infection of a CHO mutant cell line deficient in N-sulfation was three times less efficient than infection of the parental CHO cell line, indicating that N-sulfation is important. In contrast, infection of a cell line deficient in 2-O-sulfation was as efficient as infection of the parental cell line, indicating that 2-O-sulfation is not required for RSV infection. Incubating RSV with the purified soluble heparin, the prototype GAG, before inoculation had previously been shown to neutralize its infectivity. Here we tested chemically modified heparin chains that lack their N-, C6-O-, or C2-O-sulfate groups. Only heparin chains lacking the N-sulfate group lost the ability to neutralize infection, confirming that N-sulfation, but not C6-O- or C2-O-sulfation, is important for RSV infection. Analysis of heparin fragments identified the 10-saccharide chain as the minimum size that can neutralize RSV infectivity. Taken together, these results show that, while sulfate modification is important for the ability of GAGs to mediate RSV infection, only certain sulfate groups are required. This specificity indicates that the role of cell surface GAGs in RSV infection is not based on a simple charge interaction between the virus and sulfate groups but instead involves a specific GAG structural configuration that includes N-sulfate and a minimum of 10 saccharide subunits. These elements, in addition to iduronic acid demonstrated previously (L. K. Hallak, P. L. Collins, W. Knudson, and M. E. Peeples, Virology 271:264–275, 2000), partially define cell surface molecules important for RSV infection of cultured cells.
Respiratory syncytial virus (RSV) encodes 11 proteins, 3 of which are transmembrane surface glycoproteins found in the viral envelope: the G (glycoprotein) (18, 40), F (fusion) (11), and SH (small hydrophobic) (7, 12) proteins. The G protein is a type II transmembrane protein, with an N-terminal transmembrane domain. The G protein is also found in a smaller secreted form that lacks the transmembrane region, generated by translation initiation at a second AUG in the mRNA and subsequent proteolytic trimming (33). The G protein was identified as the major viral attachment protein (27), but the isolation of an infectious RSV mutant, cp-52, lacking its G and SH genes (24), suggests that an attachment function can be provided by the sole remaining glycoprotein, the F protein. However, while the G protein appears to be dispensable for replication in vitro, the highly attenuated nature of the cp-52 mutant in vivo supports the idea that G protein is important for infection in vivo (24). The classic role of the F protein is to initiate infection by fusing the virion envelope with the target cell plasma membrane. It is also responsible for cell-to-cell fusion. The role of the SH protein is not clear. Its presence has been shown to enhance fusion induced by the F protein when expressed from plasmids (20). However, deleting the SH gene from recombinant virus did not diminish its growth in cell culture (5) and conferred only a small degree of attenuation in the respiratory tract of chimpanzees (41).
Glycosaminoglycans (GAGs) are unbranched polysaccharide chains associated with most mammalian cells. They are composed of repeating disaccharide units of hexuronic acid and hexosamine. The hexuronic acid is either d-glucuronate (GlcA) or its epimerized form, l-iduronate (IdoA), and the hexosamine is either N-acetylglucosamine (GlcNAc) or N-acetylgalactosamine (GalNAc), depending on the type of GAG. The GAG types that appear to be important for RSV infection in HEp-2 cells are heparin sulfate (HS) and chondroitin sulfate B (CS-B) (19). HS and CS-B are found on the surface of almost all animal cells as components of proteoglycans. They are covalently linked to the proteoglycan core proteins through O-glycosidic linkages to serine (44). Like most GAGs, HS and CS-B are sulfated, though the level and position of the sulfate groups vary among GAG types and even within the same GAG type in different tissues. The GAG heparin, which is found only in granulae of connective-tissue-type mast cells, has a very similar structure to that of HS but it is more heavily sulfated, especially at the N position of GlcN, and it contains more IdoA. Heparin is frequently used as a convenient analog of HS for experimental purposes (28), as in the present study.
The biosynthesis of protein-bound GAGs such as HS and CS-B begins with the transfer of xylose from UDP-xylose to the hydroxyl group of a serine residue present in a serine-glycine motif in the core protein. This is elongated into the tetrasaccharide, βGlcA-1,3→βGal-1,3→βGal-1,4→βXyl-1-O-Ser (44). The nonreducing end of this tetrasaccharide becomes a primer for GAG chain elongation. For example, heparin and HS are synthesized as polymers of alternating GlcA and GlcNAc subunits. Following GAG synthesis, some of the N-acetyl groups are removed from the GlcNAc subunits and replaced with N-sulfate groups. The N-deacetylation and N-sulfation reactions are catalyzed by a single enzyme, N-acetyltransferase (30, 39), and the sulfation step involves the sulfate donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) (13). The N-sulfation reaction is required for the next step: epimerization of some of the GlcA to IdoA at C5. 2-O-sulfation on the newly formed IdoA and 6-O-sulfation on GlcN are common. Less common sulfation sites are at positions C3 of GlcN and C2 of GlcA. Sulfation at C3 of GlcN is required for the formation of one of the functional receptors for HSV (35). The biosynthesis of CS-B is similar, except that the hexosamine is GalNAc instead of GlcNAc and there are differences in the postsynthetic modifications of the polysaccharide chain.
Many proteins bind heparin and other GAGs, including viral proteins, enzymes, growth factors, and proteins of the extracellular matrix (34). Binding is facilitated by the negative electrostatic charges on GAG chains provided by the sulfate groups and by the flexibility of IdoA, allowing positioning of the binding site. Many viruses depend on cell surface GAG chains to efficiently initiate infection of cultured cells. The first virus shown to require cell surface HS was herpes simplex virus (HSV) (42). The group of GAG-binding viruses has now expanded to include RSV (4, 19, 26), pseudorabies virus (38), Sindbis virus (6, 25), dengue virus (8), vaccinia virus (9, 22), adeno-associated virus type 2 (AAV2) (37), and foot-and-mouth disease virus (23).
Previously we described a sensitive system to study the role of cell surface GAGs in infection using recombinant green fluorescent protein (GFP)-expressing (rgRSV). Cultured cells that were deficient in surface GAGs due to mutation or enzymatic treatment were shown to be infected with reduced efficiency. We also found that only those soluble GAGs that contained IdoA were able to block infection (19). Finally, a growth factor that specifically binds to cell surface IdoA reduced the efficiency of RSV infection. These observations indicated that efficient RSV infection requires cell surface GAGs and, furthermore, that this requirement involved GAGs that contain IdoA, specifically HS and CS-B. In the present study, we used an improved version of rgRSV to further examine the role of GAGs, determining the importance of their sulfate modifications in mediating RSV infection. We found that N-sulfation of GAGs is particularly important in rgRSV infection, that 2-O- and 6-O-sulfations are not important, and that the minimal GAG length that is effective in inhibiting rgRSV infection is 10 saccharides.
MATERIALS AND METHODS
Cell lines and media.
The human epithelial cell line, HEp-2, was maintained in OptiMEM (Life Technologies, Inc.) supplemented with 2% fetal bovine serum (FBS). CHO cell lines K1, pgsE-606 (3), and pgsF-17 (2) were provided by Jeff Esko (University of California at San Diego) and maintained in Dulbecco modified Eagle medium-F12 medium supplemented with 10% FBS. Cells were incubated at 37°C in 5% CO2.
Preparation of an improved rgRSV.
The rgRSV(125) used in our previous study (19) replicated in cell culture but produced peak titers that were approximately 10-fold lower than its parent RSV. In an attempt to enhance its replication, we modified the 3′ terminus of the full-length cDNA used to rescue rgRSV(125) so that it would contain the first 75 nucleotides (nt) of the wild-type RSV rather than only 54 nt. The additional 21 nt came from the untranslated region of the native first gene, NS1. The new rgRSV(224) replicates more efficiently, resulting in near-parental RSV titers and more rapid development of fluorescence. The rgRSV(224) also produced syncytia at a rate similar to the parental RSV and faster than rgRSV(125).
The full-length RSV cDNA clone, MP224, used to rescue rgRSV(224) was constructed to contain GFP as its first gene essentially by inserting a BstXI fragment containing the gene start, the NS1 untranslated region, the Green Lantern Protein (Life Technologies, Inc.) gene, and the L gene end, in that order. This BstXI fragment was generated by transferring the BstXI/BamHI fragment from the minigenome C41-GFP (37a) into another GFP minigenome, MP129, to create MP166 and transferring the XhoI/NcoI fragment from MP166 into MP90, a bipartite minigenome containing the GFP gene flanked by BstXI sites and followed by the luciferase gene, to create MP169. The BstXI fragment from MP169 was moved into the full-length RSV cDNA clone, D46, to generate MP224 in two steps as described for MP125 (18), except that an AatII/XhoI fragment was used in the final step.
MP224 was rescued by transfecting it along with four plasmids expressing the N, P, L, and M2-1 support proteins into HEp-2 cells, as described previously (10). Transcription from these plasmids was driven by T7 RNA polymerase provided by the recombinant vaccinia virus MVA-T7 (43). Released infectious virus, rgRSV(224), was amplified in HEp-2 cells. For simplicity, rgRSV(224) is identified throughout this report as rgRSV.
Chemicals.
The following chemicals were purchased from the Sigma Chemical Company: dextran from Leuconostoc mesenteroides, average molecular mass of ∼10 kDa (D-9260); dextran sulfate, 5 kDa (D-7037); dextran sulfate, 10 kDa (D-6924), d-glucosamine 2-sulfate (G-7889); d-glucosamine 6-sulfate (G-8641); d-glucosamine 3,4,6-trisulfate (G-5533); d-glucosamine 3-sulfate (G-4267); d-glucosamine 2,6-disulfate (G-7514); d-glucosamine 2,3-disulfate (G-7639); d-glucosamine N2,3,6-trisulfate, and heparin disaccharide III-S (H-9392). Neoparin, Inc. (San Leandro, Calif.), was the commercial supplier for bovine intestinal heparin as well as derivatives that had been chemically modified: N-desulfated, fully N-sulfated, 6-O-desulfated, or 2-O-desulfated.
Fragments of bovine lung heparin oligosaccharides of 4-, 6-, 10-, 14-, 16-, 18- to 20-, and 22-mer lengths were prepared by limited deaminative cleavage at pH 1.5 and size fractionated as described previously (17, 36).
Neutralization assays.
Soluble molecules were diluted twofold serially in serum-free OptiMEM medium. An equal volume of rgRSV was added to each dilution, and the mixtures were incubated 45 min at 20°C to allow binding. Mixtures were then used to inoculate 24-h-old HEp-2 cell monolayers in 12-well plates, with periodic agitation. The inoculum was added at a multiplicity of infection (MOI) of 1 to 2 PFU per cell (PFU was determined on HEp-2 cells). Unbound virus was removed after 2 h, and cells were washed once with phosphate-buffered saline (PBS). Complete medium was added to cells, and they were incubated for an additional 24 h. Cells were then trypsinized by 1× Trypsin-EDTA (Gibco-BRL), fixed with 2% paraformaldehyde, and analyzed by flow cytometry to detect GFP-expressing cells as described elsewhere (19).
Infection of HEp-2 cells under conditions of sulfate depletion.
HEp-2 cells were seeded on six-well plates in OptiMEM–2% FBS and incubated overnight to form monolayers. They were then washed twice with PBS and incubated in sulfate-free medium, Joklik-modified S-MEM, (Gibco-BRL catalog no. 22300) supplemented with 10% dialyzed FBS (Gibco-BRL catalog no. 26300) in the presence or absence of 50 mM sodium chlorate (Aldrich catalog no. 403016), or magnesium sulfate as a control to replenish sulfate. After 24 h, cells were washed with PBS and inoculated with rgRSV as described above. After a 2-h adsorption period, the cells were washed with complete medium and incubated for 24 h in complete medium. Cells were then processed for flow cytometry as described above. In addition, an aliquot of each harvested monolayer was assayed for cell number.
RESULTS
Effect of dextran sulfate on rgRSV infection.
Dextran, a bacterial product, is a branched glucose polysaccharide that can be chemically sulfated to generate dextran sulfate. Up to three sulfate groups modify each glucose unit of dextran sulfate, composing 17% of its weight. Dextran sulfate has been shown to neutralize several viruses, including RSV (21).
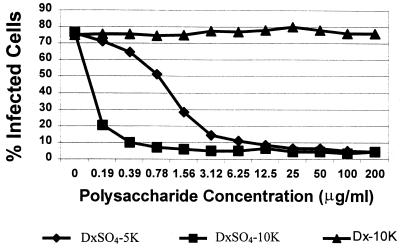
To confirm this finding and to determine the contribution of sulfate and dextran size to this neutralizing effect, we treated rgRSV with unsulfated dextran (average size, 10 kDa) and with dextran sulfate (average size, 5 or 10 kDa). Both sulfated dextrans neutralized rgRSV in a dose-dependent manner, and neutralization was more efficient with the larger polysaccharide (Fig. 1). In contrast, unsulfated dextran was completely inactive in neutralizing rgRSV. These findings confirmed that dextran sulfate can neutralize RSV infectivity very efficiently but showed that this activity was completely dependent on the presence of sulfate groups.
FIG. 1.
Effects of dextran (Dx) and dextran sulfate (DxSO4) on the efficiency of rgRSV infection of HEp-2 cells. Virus (MOI = 2) was mixed with the indicated concentrations of DxSO4-5K (average molecular mass, 5 kDa), DxSO4-10K, or Dx-10K, incubated for 45 min, and used to inoculate cells. Cells were analyzed for GFP expression at 24 h postinoculation.
Treatment with sodium chlorate reduces the susceptibility of cells to rgRSV infection.
Sodium chlorate acts as a sulfate analog, replacing the sulfate group in the sulfate donor for GAG synthesis, PAPS (14). The resulting 3′-phosphoadenosine-5′-phosphochlorate donates unstable chlorate groups to GAG chains that are spontaneously hydrolyzed (1). If GAG sulfation were critical for RSV infection, then sodium chlorate treatment of HEp-2 cells would be predicted to inhibit subsequent rgRSV infection. To test this possibility, cells were preincubated in sulfate-free medium to reduce the availability of sulfate (Fig. 2, sample 2). A second set was preincubated in sulfate-free medium containing 50 mM sodium chlorate to inhibit the incorporation of sulfate into cellular GAGs (sample 3). A third set of cells was incubated in sulfate-free medium in which the sulfate was replenished by the addition of MgSO4 (sample 1). Following a preincubation of 48 h, the cells were inoculated with rgRSV, incubated for an additional 24 h in complete medium, trypsinized, fixed, and analyzed for the expression of GFP. The efficiency of rgRSV infection in cells treated with chlorate was 40% that of control cells incubated in medium containing MgSO4 (compare sample 3 with sample 1). These results supported the idea that sulfation is important for efficient rgRSV infection.
FIG. 2.
Effects of sodium chlorate on rgRSV infection. HEp-2 cells were incubated in sulfate-free medium, without (samples 1 and 2) or with (sample 3) 50 mM sodium chlorate and with (sample 1) or without (samples 2 and 3) 0.8 mM MgSO4. After 48 h, cells were inoculated with rgRSV (MOI = 1), washed, and incubated in complete medium. At 24 h postinoculation, the medium was removed and cells were analyzed for GFP expression.
Sensitivity of sulfation-deficient CHO cell lines to rgRSV infection.
The most common positions of sulfate addition in heparin, HS, and CS-B are the N and C6 positions of GlcN and the C2 position of IdoA. To determine whether N-sulfation of GAGs is required for efficient RSV infection, we tested the sensitivity of the CHO pgsE-606 cell line to infection with rgRSV. This mutant cell line expresses normal levels of GAGs but has three- to fivefold less N-acetyltransferase activity (3). As a result, its GAGs are undersulfated on GlcN. These mutant cells and their CHO K1 parental cells were inoculated with rgRSV and incubated for 36 h at 37°C, after which infected cells were counted by flow cytometry. CHO pgsE-606 cells were threefold less sensitive to infection than the parental K1 cells (Fig. 3), indicating that N-sulfation is important for efficient infection. However, since the formation of IdoA in heparan sulfate requires the presence of N-sulfate on GlcN (Introduction), it was not clear from this result alone whether the reduction of infection in this experiment was directly due to the absence of N-sulfate groups or was a consequence of a lower content of IdoA in the cell surface GAGs. This point is addressed below.
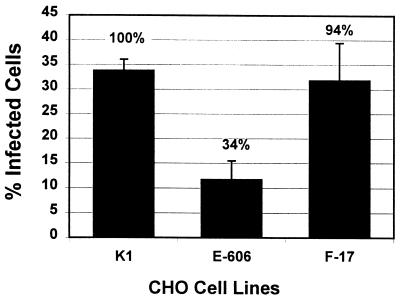
FIG. 3.
Sensitivity of sulfate-deficient CHO cell lines to rgRSV infection. CHO pgsE-606, deficient in N-sulfation, and pgsF-17, deficient in 2-O-sulfation, were inoculated with rgRSV (MOI = 1) and analyzed for GFP expression at 36 h postinoculation. The average percent infected cells relative to the parental CHO K1 is shown above each bar.
IdoA in heparin and HS (and to a lesser extent in CS-B) is sulfated at its C2 position. To determine whether this 2-O-sulfation is important for mediating RSV infection, we measured the sensitivity of the CHO pgsF-17 cell line to rgRSV infection. This mutant cell line is deficient in 2-O-sulfotransferase (2), the enzyme required for transfer of sulfate to the 2-O position in IdoA. rgRSV was able to infect these cells nearly as efficiently as the CHO K1 parental cells (Fig. 3), indicating that 2-O-sulfation is not required for efficient RSV infection.
Effects of soluble, chemically modified heparin on rgRSV infection.
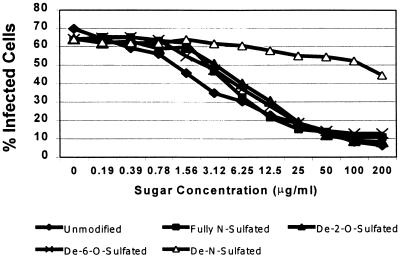
As mentioned above, preincubation of RSV with soluble heparin reduces the infectivity of the virus, presumably by saturating GAG-binding sites needed for efficient infection. To further test the importance of GAG sulfation, especially that of N-sulfation, we compared the neutralization activity of unmodified soluble heparin with that of derivatives that had been modified chemically as follows: (i) to remove N-sulfate groups, (ii) to remove the 6-O-sulfate groups, (iii) to remove the 2-O-sulfate groups, or (iv) to add N-sulfate groups to all GlcNAc subunits. These heparins were serially diluted, and each dilution was incubated with rgRSV before inoculating the HEp-2 cells. Unmodified heparin efficiently inhibited rgRSV infection (50% inhibitory concentration [IC50] of 6 μg/ml), a result consistent with results in previous studies (4, 19, 26). Heparin that had been treated to add N-sulfates to all remaining unmodified GlcN residues was not significantly different from the native form (Fig. 4), which was not surprising given the high degree of N-sulfation of native heparin. In contrast, heparin lacking N-sulfation inhibited infection very poorly (IC50 of >200 μg/ml). The neutralizing ability of heparin, therefore, appears to be highly dependent on N-sulfation. Conversely, both 6-O-desulfated and 2-O-desulfated heparin retained nearly their full inhibitory activity, indicating that sulfate at position C6 of GlcN or position C2 of GlcA or IdoA are not required for efficient binding by rgRSV.
FIG. 4.
Neutralization activity of chemically modified full-length soluble heparin chains against rgRSV. Virus (MOI = 1) was mixed with various modified heparins, as indicated, incubated for 45 min, and used to inoculate HEp-2 cells. Cells were analyzed for GFP expression at 24 h postinoculation.
Importance of heparin chain length in rgRSV neutralization.
To further define the GAG structural features that are important for binding to RSV, we examined the effect of chain length on the ability of heparin to neutralize rgRSV. This analysis first examined the free monosaccharides GlcA and GlcN. Neither monosaccharide significantly inhibited rgRSV infection (Table 1). We also tested glucosamine monosaccharides with sulfate groups at positions 2, 3, 2,3, 2,6, and N2,3,6. None of these sugars inhibited rgRSV infection even at concentrations up to 400 μg/ml (Table 1).
TABLE 1.
Inhibition efficiency of different saccharides and oligosaccharides
| Compound | IC50 (μg/ml) | Compound | IC50 (μg/ml) | |
|---|---|---|---|---|
| Heparin fragments | Modified heparin | |||
| Sulfated GlcNa | NIb | Heparin | 0.8 | |
| ΔHexA2S-GlcNS | NI | 2-O-Desulfated | 6.25 | |
| Heparin 4-mer | NI | 6-O-Desulfated | 5.0 | |
| Heparin 6-mer | NI | N-Desulfated | >200.0 | |
| Heparin 8-mer | NI | N-Sulfated | 4.6 | |
| Heparin 10-mer | >200.0 | |||
| Heparin 14-mer | 200.0 | |||
| Heparin 16-mer | 100.0 | Modified dextran | ||
| Heparin 18/20-mer | 50.0 | Dextran | NI | |
| Heparin ≥22-mer | 1.0 | Dextran sulfate (5K) | 1.2 | |
| Dextran sulfate (10K) | 0.1 |
GlcN-2-SO4, GlcN-3-SO4, GlcN-2,3-SO4, GlcN-2,6-SO4, GlcN-2,3,6-SO4, GlcA.
NI, no inhibition at concentrations of up to 200 μg/ml (heparin 4-mer, 6-mer, or 8-mer and dextran) or 400 μg/ml (sulfated GlcN and ΔHexA2S-GlcNS).
We then tested the RSV-neutralizing activity of the heparin disaccharide ΔHexA2S1-4GlcNS (III-S), resulting from the digestion of heparin or HS with heparinases I and II. Like the monosaccharides, it was unable to inhibit rgRSV infection at the tested concentrations. These results suggested that the ability to bind to and neutralize RSV depended on a larger GAG size and/or additional structural components found on heparin chains.
We then tested bovine lung heparin fragments of increasing length, namely, 4-, 6-, 8-, 10-, 14-, 16-, 18- to 20-, and 22-mer saccharide subunits, for the ability to neutralize rgRSV infection (Fig. 5). Even at the highest concentration tested (200 μg/ml), the 4-, 6-, and 8-mer fragments did not significantly neutralize rgRSV infectivity. The minimum chain length that provided detectable neutralization of rgRSV was a 10-mer, although inhibition was relatively weak (IC50 of >200 μg/ml). Inhibition of infection increased with increasing heparin chain length, with a mixture of 22-mer and longer chains neutralizing rgRSV as efficiently as native heparin.
FIG. 5.
Effects of heparin chain length on its ability to neutralize rgRSV. Virus (MOI = 1) was mixed with heparin chains of increasing length, as indicated, incubated for 45 min, and used to inoculate HEp-2 cells. Cells were analyzed for GFP expression at 24 h postinoculation.
DISCUSSION
We previously showed that HS and CS-B, two GAGs that are found on cell surface proteoglycans, are important for efficient RSV infection (19). HS, CS-B, and heparin, the latter being a structural analog of HS, were each able to neutralize rgRSV infectivity when preincubated with the virus. All three of these GAGs contain IdoA, whereas three other GAGs that do not contain IdoA did not neutralize rgRSV. In the present study, the presence of sulfate groups was found to be important for polysaccharide binding to RSV and for RSV infection of cultured cells. Furthermore, we examined the importance of sulfation at specific GAG positions by using mutant cell lines that are defective in specific sulfotransferases and by using soluble heparin and modified heparins.
Dextran sulfate strongly inhibited rgRSV infection, as is evident from the IC50 of 0.1 μg/ml for 10K dextran sulfate, compared to that of heparin (1.0 μg/ml), HS (12.5 μg/ml), and CS-B (200 μg/ml) (19). The very strong binding of dextran sulfate indicated by these data is dependent on its heavy sulfation, since desulfated dextran sulfate was completely inactive in neutralizing rgRSV. Although the very high sulfate content and branched nature of dextran sulfate makes it an inexact model for cell surface GAGs such as HS and CS-B, the strong inhibition observed shows that the binding of RSV to sulfated polysaccharides can be very strong indeed and that sulfation is important.
Chlorate treatment of cells reduces the overall level of GAG sulfation and has been shown to reduce infection by vaccinia virus (9) and human immunodeficiency virus type 1 (31), but not by AAV2 (32), despite the fact that HS is thought to act as a receptor for AAV2 (37). In our experiments, sodium chlorate pretreatment of cells reduced the efficiency of rgRSV infection to 40% that of untreated cells, suggesting a role for sulfation.
Most of the GlcN residues in heparin and approximately half of the GlcN residues in HS (29) are modified by substitution of their N-acetyl groups with N-sulfate, generating N-sulfated GlcN. There are other sulfation sites in heparin and HS, most notably the C6 position of GlcN and the C2 position of IdoA. Infrequent sulfation also occurs at the C3 position of GlcN and the C2 position of GlcA (28). Here we provided evidence that N-sulfation, but not C6 or C2 sulfation, is required for efficient RSV infection. This conclusion is supported by two observations. (i) The pgsE-606 CHO-cell line, deficient in N-sulfation but expressing normal levels of cell surface HS and CS, was only 34% as permissive to rgRSV infection as the parent cell line. This reduction in virus infectivity is similar in magnitude to the decrease (three- to fivefold) in N-sulfation enzymatic activity (3). On the other hand, CHO pgsF-17, a cell line deficient in C2-O-sulfation, was fully permissive for rgRSV infection. (ii) Chemically modified, full-length N-desulfated heparin lost its neutralizing activity. These results indicate that the N-sulfation is important in rgRSV binding. Both 6-O-desulfated and 2-O-desulfated heparin chains retained their neutralizing activity, indicating that these sulfate groups are not important for binding to rgRSV.
The conclusion most consistent with these data is that particular sulfate groups on GAGs, rather than the total amount of sulfate, are important for promoting rgRSV infection. Besides the involvement of N-sulfate groups, polyvalence appears to be important. Neither highly sulfated mono- or disaccharides nor heparin fragments shorter than 10-mer were effective in neutralizing rgRSV. This indicates that there is a need for longer ligands, either due to specific features within the binding site or a requirement of polyvalent interaction with virion surface components.
The finding that efficient binding of RSV to GAGs requires IdoA, N-sulfation, and a minimum saccharide chain length of 10 indicates that the binding is not simply a function of charge but instead has greater specificity. In the case of dengue virus, a 10-saccharide heparin fragment was required to inhibit virus binding to host cells (8), whereas HSV was inhibited by a 12-saccharide fragment (17). This 12-mer must contain at least one 2-O- and one 6-O-sulfate group, and yet no N-sulfate is necessary. This difference between the HSV specificity and the RSV requirement for only N-sulfation demonstrated in the present here clearly indicates that different viruses have different GAG-binding specificities. These viruses have subtle but distinct differences in the specificities of their interaction with cell surface GAGs.
While it is clear that cell surface GAGs are required for efficient rgRSV infection of cultured cells, it is not clear whether this binding represents attachment in toto, as appears to be the case for AAV2 (37), or whether the interaction between RSV and GAGs represents a low-affinity first step in attachment followed by a second step that remains to be identified. This latter case would resemble the situation with HSV, where binding to GAGs is an initial step that enhances the ability of the virus to find its specific receptor (42). It is also unclear which of the three RSV glycoproteins is responsible for binding to GAGs to initiate infection. Synthetic peptides representing the consensus regions in the G protein of subgroups A and B of RSV were found to bind to Vero cells and inhibit RSV infection. Heparin prevented this peptide from binding to Vero cells (16), suggesting that this segment of the G protein binds to cell surface GAGs. But the ability of the RSV mutant cp-52 to infect cultured cells, even though it lacks the genes for both the G and SH proteins (24), would suggest that the F protein can also function as an attachment protein.
Recently, Feldman et al. (15) reported that the F protein released from RSV-infected cells by detergent lysis also binds to heparin, that cp-52 is neutralized by heparin, and that pretreatment of cells with heparinase inhibits cp-52 infection. We have made similar observations with rgRSV lacking both the SH and G genes (S. Techaarpornkul, N. Barretto, P. L. Collins, and M. E. Peeples, manuscript in preparation). These results suggest that the F protein also binds to GAGs on the cell surface to initiate infection in the absence of the G protein. It is not yet clear whether the GAG-binding specificities of the F and G proteins are identical; what the relative contributions of the F, G, and SH proteins are to initiating infection; or whether these proteins act cooperatively.
ACKNOWLEDGMENTS
We thank Greg Spear and Alan Landay for use of the flow cytometer, Jeff Esko and Patricia Spear for providing the CHO cell lines, Ada Cole for use of the inverted fluorescence microscope, and Barbara Newton for technical support.
This work was supported by a grant from the Rush University Committee on Research.
REFERENCES
- 1.Baeuerle P A, Huttner W B. Chlorate: a potent inhibitor of protein sulfation in intact cells. Biochem Biophys Res Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 2.Bai X, Esko J D. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 3.Bame K J, Esko J D. Undersulfated heparan sulfate in a Chinese hamster ovary cell mutant defective in heparan sulfate N-sulfotransferase. J Biol Chem. 1989;264:8059–8065. [PubMed] [Google Scholar]
- 4.Bourgeois C, Bour J B, Lidholt K, Gauthray C, Pothier P. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol. 1998;72:7221–7227. doi: 10.1128/jvi.72.9.7221-7227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cane P A, Pringle C R. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene) J Gen Virol. 1991;72:349–357. doi: 10.1099/0022-1317-72-2-349. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 9.Chung C S, Hsiao J C, Chang Y S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins P L, Huang Y T, Wertz G W. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1984;81:7683–7687. doi: 10.1073/pnas.81.24.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins P L, Olmsted R A, Johnson P R. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J Gen Virol. 1990;71:1571–1576. doi: 10.1099/0022-1317-71-7-1571. [DOI] [PubMed] [Google Scholar]
- 13.Conard H E. Heparin binding proteins. San Diego, Calif: Academic Press; 1998. Structures of heparinoids; pp. 7–60. [Google Scholar]
- 14.Conard H E. Heparin binding proteins. San Diego, Calif: Academic Press; 1998. The cellular metabolism of heparan sulfate; pp. 137–182. [Google Scholar]
- 15.Feldman S A, Audet S, Beeler J A. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman S A, Hendry R M, Beeler J A. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999;73:6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feyzi E, Trybala E, Bergstrom T, Lindahl U, Spillmann D. Structural requirement of heparan sulfate for interaction with herpes simplex virus type 1 virions and isolated glycoprotein C. J Biol Chem. 1997;272:24850–24857. doi: 10.1074/jbc.272.40.24850. [DOI] [PubMed] [Google Scholar]
- 18.Gruber C, Levine S. Respiratory syncytial virus polypeptides. III. The envelope-associated proteins. J Gen Virol. 1983;64:825–832. doi: 10.1099/0022-1317-64-4-825. [DOI] [PubMed] [Google Scholar]
- 19.Hallak L K, Collins P L, Knudson W, Peeples M E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271:264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- 20.Heminway B R, Yu Y, Tanaka Y, Perrine K G, Gustafson E, Bernstein J M, Galinski M S. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology. 1994;200:801–805. doi: 10.1006/viro.1994.1245. [DOI] [PubMed] [Google Scholar]
- 21.Hosoya M, Balzarini J, Shigeta S, De Clercq E. Differential inhibitory effects of sulfated polysaccharides and polymers on the replication of various myxoviruses and retroviruses, depending on the composition of the target amino acid sequences of the viral envelope glycoproteins. Antimicrob Agents Chemother. 1991;35:2515–2520. doi: 10.1128/aac.35.12.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiao J C, Chung C S, Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J Virol. 1998;72:8374–8379. doi: 10.1128/jvi.72.10.8374-8379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson T, Ellard F M, Ghazaleh R A, Brookes S M, Blakemore W E, Corteyn A H, Stuart D I, Newman J W, King A M. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krusat T, Streckert H J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 27.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl U, Lidholt K, Spillmann D, Kjellen L. More to “heparin” than anticoagulation. Thromb Res. 1994;75:1–32. doi: 10.1016/0049-3848(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 29.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. Domain structure of heparan sulfates from bovine organs. J Biol Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 30.Mandon E, Kempner E S, Ishihara M, Hirschberg C B. A monomeric protein in the Golgi membrane catalyzes both N-deacetylation and N-sulfation of heparan sulfate. J Biol Chem. 1994;269:11729–11733. [PubMed] [Google Scholar]
- 31.Ohshiro Y, Murakami T, Matsuda K, Nishioka K, Yoshida K, Yamamoto N. Role of cell surface glycosaminoglycans of human T cells in human immunodeficiency virus type-1 (HIV-1) infection. Microbiol Immunol. 1996;40:827–835. doi: 10.1111/j.1348-0421.1996.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 32.Qiu J, Handa A, Kirby M, Brown K E. The interaction of heparin sulfate and adeno-associated virus 2. Virology. 2000;269:137–147. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- 33.Roberts S R, Lichtenstein D, Ball L A, Wertz G W. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J Virol. 1994;68:4538–4546. doi: 10.1128/jvi.68.7.4538-4546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmivirta M, Lidholt K, Lindahl U. Heparan sulfate: a piece of information. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 35.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 36.Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. J Biol Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 37.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voigt A, Sawitzky D, Zeichhardt H, Habermehl K O. Cellular receptor structures for pseudorabies virus are blocked by antithrombin III. Med Microbiol Immunol. 1995;184:97–103. doi: 10.1007/BF00221393. [DOI] [PubMed] [Google Scholar]
- 39.Wei Z, Swiedler S J, Ishihara M, Orellana A, Hirschberg C B. A single protein catalyzes both N-deacetylation and N-sulfation during the biosynthesis of heparan sulfate. Proc Natl Acad Sci USA. 1993;90:3885–3888. doi: 10.1073/pnas.90.9.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitehead S S, Bukreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Esko J D. Amino acid determinants that drive heparan sulfate assembly in a proteoglycan. J Biol Chem. 1994;269:19295–19299. [PubMed] [Google Scholar]