Abstract
Altered regulatory interactions during development likely underlie a large fraction of phenotypic diversity within and between species, yet identifying specific evolutionary changes remains challenging. Analysis of single-cell developmental transcriptomes from multiple species provides a powerful framework for unbiased identification of evolutionary changes in developmental mechanisms. Here, we leverage a “natural experiment” in developmental evolution in sea urchins, where a major life history switch recently evolved in the lineage leading to Heliocidaris erythrogramma, precipitating extensive changes in early development. Comparative analyses of scRNA-seq developmental time courses from H. erythrogramma and Lytechinus variegatus (representing the derived and ancestral states respectively) reveals numerous evolutionary changes in embryonic patterning. The earliest cell fate specification events, and the primary signaling center are co-localized in the ancestral dGRN but remarkably, in H. erythrogramma they are spatially and temporally separate. Fate specification and differentiation are delayed in most embryonic cell lineages, although in some cases, these processes are conserved or even accelerated. Comparative analysis of regulator-target gene co-expression is consistent with many specific interactions being preserved but delayed in H. erythrogramma, while some otherwise widely conserved interactions have likely been lost. Finally, specific patterning events are directly correlated with evolutionary changes in larval morphology, suggesting that they are directly tied to the life history shift. Together, these findings demonstrate that comparative scRNA-seq developmental time courses can reveal a diverse set of evolutionary changes in embryonic patterning and provide an efficient way to identify likely candidate regulatory interactions for subsequent experimental validation.
INTRODUCTION
Most metazoan life cycles contain intermediate stages that are ecologically distinct from adults. In many clades, this has resulted in the evolution of contrasting anatomical, physiological, and behavioral traits between stages in the life cycle (Garstang 1928; Thorson 1950; Strathmann 1985; Nielsen 1998; Raff and Byrne 2006; Formery and Lowe 2023). Host-specific stages of parasites, insect larvae, amphibians, and diverse marine invertebrates are often so different from adults that they are unrecognizable from the earlier stages of the same life cycle. In some clades, the evolution of these intermediate stages is remarkably labile, such that closely related species with very similar adult morphology differ profoundly earlier in the life cycle. These cases likely reflect shifts in natural selection that operate on intermediate phases of the life cycle but not on adults. Numerous adaptations related to larval dispersal, feeding, predator avoidance, and abiotic factors have been documented. Yet, it remains largely unknown how developmental mechanisms known to pattern body organization at two distinct stages of the life cycle can become decoupled to allow effective responses to changing selective regimes.
The sea urchin genus Heliocidaris provides a valuable system for studying how developmental patterning becomes decoupled across life stages due to a combination of three salient features. First, the genus contains closely related species with highly divergent life histories and a known polarity of change. Second, the selective changes responsible for the life history shift are clear. And third, developmental mechanisms responsible for patterning the ancestral life history are well defined and organized into a developmental gene regulatory network (dGRN). Taken together, these features have made Heliocidaris a productive model for understanding genomic and developmental responses to large changes in stage-specific natural selection and their impact on life history evolution (Wang et al. 2019; Davidson et al. 2022a; Davidson et al. 2022b; Devens et al 2023).
Heliocidaris illustrates how a shift in selective regimes can rapidly drive extensive changes in intermediate stages (Figure 1A). Representing the ancestral state, H. tuberculata produces small (~100 μm diameter) eggs that develop into complex larvae that feed on phytoplankton for several weeks before achieving sufficient mass to complete metamorphosis. Representing the derived state, H. erythrogramma produces much larger eggs (~430 μm diameter) in greatly reduced numbers, a classic life history trade-off (Stearns 1992). While this ~100-fold increase in maternal provisioning might seem simple, its impact on other traits has been profound. The larva of H. erythrogramma is anatomically highly divergent from H. tuberculata (Williams and Anderson 1975; Figure 1D). Unsurprisingly, it has lost the ability to feed: the gut and feeding structures are vestigial, presumably due to relaxed selection. In addition, metamorphosis occurs in just 3.5 days (Williams and Anderson 1975), a reduction of >75% in the duration of the pre-metamorphic phase of the life cycle. This enormous acceleration of early development seems unlikely to be the result of relaxed selection. Instead, the combination of high mortality in the plankton, coupled with greatly reduced fecundity due to the egg size-fecundity trade-off, likely imposes strong directional selection to reduce time to metamorphosis (Wray 2022).
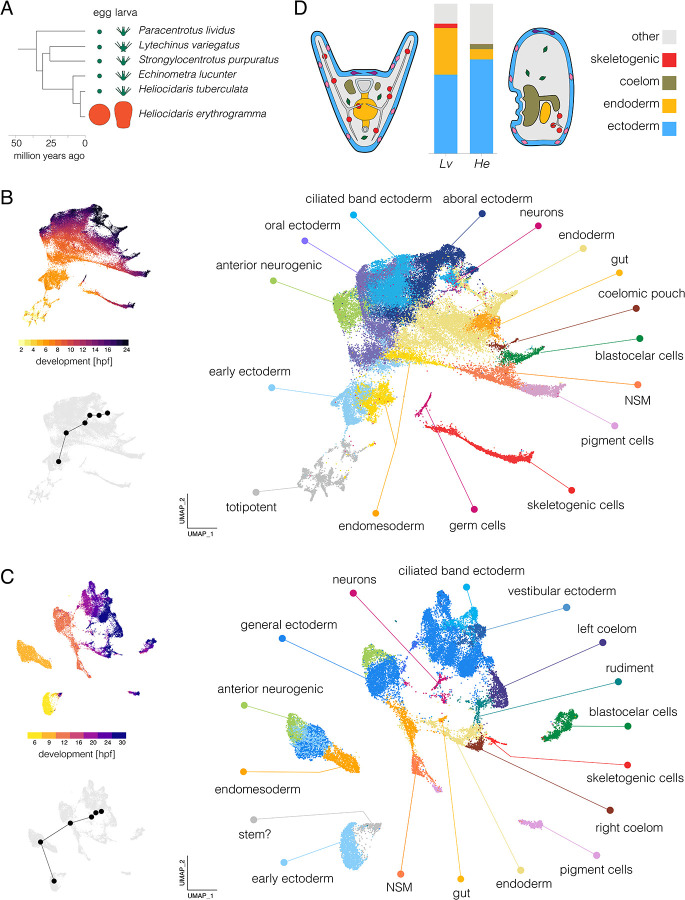
Figure 1. Single-cell developmental transcriptomes.
A. Time-tree of sea urchin species with high quality reference genomes. Egg and larva sizes approximately to scale; green indicates the ancestral life history with small eggs and feeding larvae (planktotrophy) and orange the derived life history with large eggs and non-feeding larvae (lecithotrophy). Egg and larva sizes from Mortensen 1921, Emlet et al. 1987, Williams and Anderson 1975; topology from Láruson 2016; divergence times from Ziegler et al. 2003 and Láruson 2016. B. UMAPs of scRNA-seq developmental time course for Lv. The large plot shows cells color-coded by cluster and labelled according to inferred cell types; the two smaller plots show cells color coded by time point (upper) and the centroids of the six time points common to both species (lower plot). C. UMAPS of scRNA-seq developmental time course for He. Organization parallels panel B. For individual marker gene expression, see Figure S2. Clusters in panels B and C are colored with the same encoding to facilitate comparison between species (some cell types are present only in one species or the other). D. Comparison of cell type proportions and larval morphology. Proportions of four cell types in 24 hpf larvae (see Tables S1 and S2 for cell counts at all stages). Simplified diagrams of larvae are not to scale; colors match bar plot.
In this study, we use single-cell transcriptome analysis (scRNA-seq) to investigate how extensive changes in larval anatomy and a >75% reduction in time to metamorphosis were achieved. We evaluated the presence and relative proportion of larval cell types, the timing of cellular differentiation, trajectories of transcriptional states as a proxy for cell lineages, and the co-expression of transcription factors and targets as indicators of specific regulatory interactions. Our results identify a broad delay in the divergence of transcriptional states during early development; changes in the timing, location, and order of cell fate specification and differentiation; and large shifts in the composition of cell types in the larva. In addition, some ancestral interactions within the dGRN are likely conserved in the derived life history, although most show changes in timing or location, and a few appear to have been lost entirely. Together, these analyses reveal evolutionary changes in embryonic patterning mechanisms and larval biology that were not apparent from morphological comparisons or from bulk RNA-seq analyses.
RESULTS
Transcriptional states in He accurately reflect the evolution of larval morphology
We began by constructing an atlas of early development in H. erythrogramma (hereafter, He) for comparison with our previous analysis of L. variegatus (hereafter, Lv) that spanned early cleavage through early larva (Massri et al. 2021). In order to minimize confounds when comparing between species, our approach to generating data followed the earlier study as closely as possible, including rearing embryos at the same temperature, dissociating cells using only slightly species-optimized protocols, and employing the same generation of library construction and sequencing chemistry (see Methods). We collected seven time points of He development from a single cross of outbred adults, from late cleavage (6 hours post-fertilization; hpf) through early larva (30 hpf) (see Methods). We recovered sequences from a total of 23,169 cells after filtering (average ~3,310 cells/time point). Across samples, we obtained reads from ~1000 genes/cell and ~2,000–3,000 UMIs per cell (Figure S1). The number of genes detected per cell drops across the stages sampled, likely reflecting differentiation.
We independently applied clustering and dimensional reduction to the published Lv data (Massri et al. 2021) and new He data using the same workflow. The resulting UMAPs (Figure 1B,C) are colored by cell cluster (larger plot) and developmental time (upper inset). In both cases, early stages are in the lower left (hot colors) and development proceeds up and right to later stages (dark colors); small gray UMAPs show centroids of stages common to both species. As expected, the spread of points increases during development as cells take on distinct transcriptional states. The distribution of cells is nearly continuous for Lv, while that of He is more fragmented, likely due to less dense sampling (hourly in Lv and every 3 hours in He). To identify cell clusters in He, we drew on published in situ hybridization studies and dGRN genes with conserved expression in specific cell types to annotate clusters with provisional identities (Figure S2; see Massri et al. 2021 for marker genes and supporting literature).
Several cell clusters in the He larvae (24 and 30 hpf) could be confidently assigned to a corresponding cluster in Lv (24 hpf): pigment cells, blastocoelar (immune) cells, skeletogenic cells, endoderm, coelomic pouch, ciliated band ectoderm, generalized ectoderm, anterior neurogenic ectoderm, and neurons (Figure S2). Each was previously shown to be present in early larvae of He based on morphology and marker genes (Mortensen 1921; Williams and Anderson 1975; Parks et al. 1988; Bisgrove and Raff 1989; Wilson et al. 2005a; Love et al. 2008; Koop et al. 2017).
Several differences in the UMAPs reflect the highly derived morphology of the nonfeeding He larva relative to the ancestral feeding larvae of most sea urchins, including Lv (Mortensen 1921; Williams and Anderson 1975; Wray and Raff 1989). Two clusters present in the 24 hour larva of Lv appear to be absent from He: primary germ cells and stomodeum (mouth). The absence of germ cells is consistent with the evolutionary loss in He of unequal cleavage divisions that found the primary germ cells lineage in the ancestral state (Pehrson and Cohen 1986; Oulhen et al. 2019). The lack of stomodeal cells corresponds to the absence of a larval mouth (Mortensen 1921; Williams and Anderson 1975). Conversely, some cell clusters in He larvae are not present Lv. These clusters are more challenging to identify: their apparent absence in well studied species with the ancestral life history means that there are no described marker genes. One of these clusters may represent a population of persistent pluripotent cells, based on continued proximity in UMAP space to 6 hpf cells even at 30 hpf (labeled “stem?” in Figure 1C). No corresponding cluster is evident in Lv (Figure 1B). Other clusters likely consist of cells that contribute to the adult body (vestibular ectoderm and rudiment, Figure 1C), which develops much earlier in He (Williams and Anderson 1975; Wray and Raff 1989; Koop et al. 2017). We also found that ectoderm in He does not express markers for the oral and aboral territories present ancestrally, consistent with prior studies based on in situ hybridization (Haag and Raff 1998; Love and Raff 2006; Koop et al. 2017). Instead, ectodermal gene expression is organized into a somewhat different set of clusters (Figures 1C and S3) that are not obvious 1:1 homologues of ectodermal territories in Lv based.
Substantial differences in the proportions of some cell types are also apparent. Because dissociation protocols can result in biased representation of cell types in scRNA-seq libraries, such findings need to be interpreted with caution. We therefore examined these results in light of prominent morphological differences between the larvae of two species, and highlight three differences that likely reflect true evolutionary changes in cell type proportions in early larvae (Figure 1D; Tables S1 and S2). First, endoderm makes up a much smaller fraction of cells in He than Lv (6.8 vs 31.2%), consistent with its reduced and undifferentiated endoderm (Williams and Anderson 1975; Love et al. 2008). Second, the coelomic pouches contain many more cells in He than Lv (3.4 vs 0.01%). This likely reflects the greatly accelerated development of the imaginal adult rudiment, a large fraction of which is composed of the left coelom (Williams and Anderson 1975; Wray and Raff 1989). Third, a much smaller proportion of skeletogenic cells are present in He than Lv (0.8 vs 2.9%). This is consistent with its greatly reduced larval skeleton (Emlet 1995) and antibody localization of the marker protein Msp130 (Parks et al. 1988). The differences in proportions of the last two cell types are so extreme that they are barely visible in one or the other species in Figure 1D.
Cell fate specification is broadly delayed in He
Examination of the UMAPs at earlier stages of development reveals additional differences (Figure 1B,C). We first identified cell clusters corresponding to two functionally significant territories: the anterior neurogenic domain and the primary signaling center. In the ancestral state, the anterior neurogenic domain is located at the animal pole and develops into the primary sensory organ of the larva (Angerer et al. 2011). The anterior neurogenic domain is clear in He, with overlapping expression of dGRN regulators six3, foxQ2, nkx3.2, zic1, and acsc (Figure S3). The primary signaling center is located at the vegetal pole, and produces ligands that initiate a cascade of signaling events that pattern the animal-vegetal axis (Davidson et al. 1998; McClay 2011). In the ancestral state, the primary signaling center is established ~3 hpf in the precursors of the skeletogenic cells; they expresses genes encoding ligands, including wnt8, wnt1, and delta (Sherwood and McClay 1999; Sweet et al. 2002; Wikramanayake et al. 2004; Wei et al. 2012). In He, these genes are expressed together within a single cluster (Figure 1C, S2), but beginning much later (6–9 hpf). These cells also express foxA and other markers of endoderm (Figure S2). These results suggest that the primary signaling center has become physically separated from specification of skeletogenic cells, a surprising reorganization of pivotal early patterning events in the embryo.
Some other clusters in the He embryo could not be confidently assigned to corresponding clusters in Lv. The earliest time point sampled (6 hpf) consists of a single cluster lacking any distinctive transcriptional signature (provisionally labeled “early ectoderm” in Figure 1C; the adjacent cluster labeled “stem?” consists of cells from later stages). Remarkably, there is no indication of an early population of either skeletogenic mesenchyme or germ cells in He, although these are the first two cell types specified in the ancestral state. Not until 16 hpf in He is a population of skeletogenic cells evident, a remarkable delay relative to the ancestral state. While a distinct germ cell cluster is clear in Lv (Figure 1B), at no time is a distinct group of cells expressing germ cell markers evident in He. nanos2 and vasa, two early regulators of germ cells species with feeding larvae (Juliano et al. 2010; Oulhen et al. 2019), are co-expressed at 9 and 12 hpf in He in the presumptive endoderm (Figure S2), but expression disappears at later stages. These observations suggest that some early fate specification events are delayed in He relative to Lv.
To better understand these differences quantitatively, we systematically analyzed the timing of transcriptional states in the two species. We combined reads without integration prior to dimensional reduction (Figure 2A), which allows us to visualize the relative distribution of cells from each species separately within the same manifold. As expected, reads separated entirely by species on the first dimension (UMAP1), while the second dimension (UMAP2) corresponds to developmental trajectories that run in parallel from bottom to top in both species. These trajectories differ in two informative ways. First, the Lv trajectory is initially narrow and spreads into a branch-like structure across UMAP1 as transcriptional states progressively diverge during cellular differentiation (Massri et al. 2021), while the He trajectory is also initially narrow but expands only modestly across UMAP1 over time. Given that the transcriptomes of the two species are projected from the same manifold, this contrast suggests overall less divergence in transcriptional states at later stages in He. Second, the centroids of embryonic time points in He correspond most closely to those of earlier time points in Lv along UMAP2 (Figure 2B,C; e.g., 6 hpf in He is similar to 2–4 hpf in Lv), while later stages occupy similar positions. This suggests that transcriptional states in He are initially delayed relative to those in Lv but eventually “catch up” during gastrulation. This result is consistent with earlier studies that suggested cell fate specification might be delayed in He (Wray and Raff 1989; Wray and Raff 1990; Wang et al. 2020; Davidson et al. 2022a).
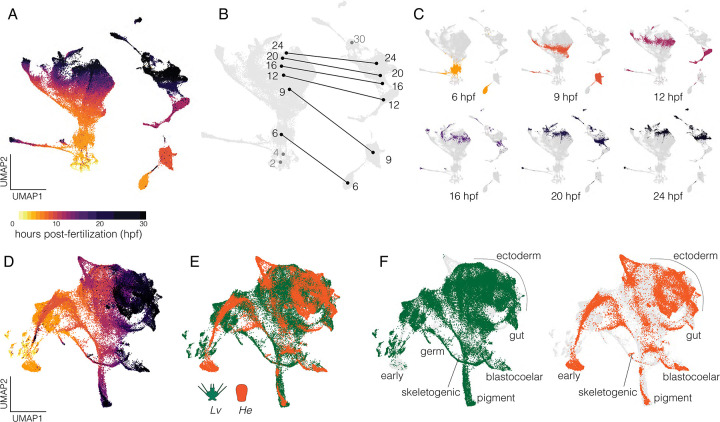
Figure 2. Combined single-cell transcriptomes.
A. UMAP of combined data from all gene models from both species without integration and cells colored according to developmental stage. The mass on the left is composed exclusively of cells from Lv while the more fragmented clumps on the right are composed exclusively of cells from He. B. Same plot, showing centroids of cells across developmental stages with lines connecting the same time points between species. Numbers correspond to hours post-fertilization (hpf); grey numbers refer to stages sampled in one species only. C. Individual UMAPs of shared time points with cells colored according to stage (same encoding as panel A). D. UMAP of integrated data from both species incorporating gene models of 1:1 orthologues only. Cells colored by stage (same encoding as panel A). E. Same plot with cells colored by species. F. Same plot as panel E with each species shown separately and cell identities labeled; light gray cells are from the complementary species.
Next, we integrated reads from 1:1 orthologues of Lv and He using canonical correlation analysis (Butler et al. 2018) prior to dimensional reduction. The resulting UMAP is presented colored by time (Figure 2D) and by species (Figure 2E). Note that cells from the same time point in the two species generally do not overlap early in development (e.g., “early” in Figure 2F), but that differentiated cells later in development generally do overlap (e.g., “blastocoelar” and “pigment” in Figure 2F). These observations reinforce the inference that diversification of transcriptional states during development take place on different schedules in the two species, with He generally lagging at early time points, but later aligning as cells undergo differentiation.
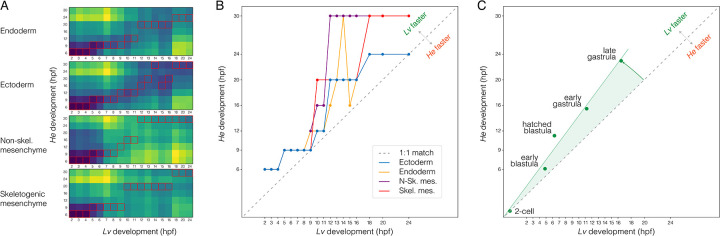
To investigate these evolutionary shifts in timing quantitatively, we used Waddington OT (Schiebinger et al. 2019) to compute transcriptional trajectories for four cell types based on an optimal transport algorithm. We then measured the overall distance between transcriptomes in the two species within each cell lineage (Figure 3A). The most similar time points are indicated by red boxes and plotted in 1:1 aspect ratio in Figure 3B. Points lie predominantly above a line of slope = 1 in Figure 3B in all four cell lineages, indicating that progression through transcriptional states is broadly delayed throughout embryonic development in He. We also used a meta-clustering method implemented in CIDER (Hu et al. 2021) to measure overall, rather than lineage-specific, similarity of transcriptomes (Figure S4). Again, results indicate that transcriptional states in He lag behind those in Lv. This broad transcriptional delay is consistent with a comparison of developmental stages based on morphology, which also shows a delay in He (Figure 3C). Because the two species were reared at the same temperature, these rate differences are likely genetically based.
Figure 3. Temporal shifts in transcriptomes.
A. Heatmaps showing degree of similarity among scRNA-seq transcriptomes (1:1 orthologues only) for four different embryonic cell lineages. Assignment of cells to lineages is based on optimal transport (see Methods). Red boxes indicate the most similar time point of He for each time point of Lv. B. Line plots showing developmental time of best matches among transcriptomes in panel A. Note the overall delay in He, with most points above the line defined by a slope of 1. C. Line plot showing developmental time of morphogenetic events. Again, there is an overall delay in He.
Differentiation is broadly delayed in He
To better understand the timing of expression changes during differentiation, we plotted transcriptional trajectories towards defined differentiated cell states (Figures 4 and S5) following Schiebinger et al. 2019. In Figure 4, each dot corresponds to a cell, with purple and red representing 70% probability of differentiating into a blastocoelar or skeletogenic cell fate, respectively; grey represents transcriptomes predictive of other cell fates and blue indicates uncommitted cells. The triangle is a flattened projection of a high dimensional space and the location of each cell indicates the degree of similarity between its transcriptome and that of two specific differentiated states (top and right apexes) and all other differentiated states (bottom apex). The transcription profile that defines each apex is based on gene expression at 24 hours, when many cell types in Lv are approaching a fully differentiated state (Massri et al. 2021). Immediately below each triangle plot is a UMAP showing the location of the same cells.
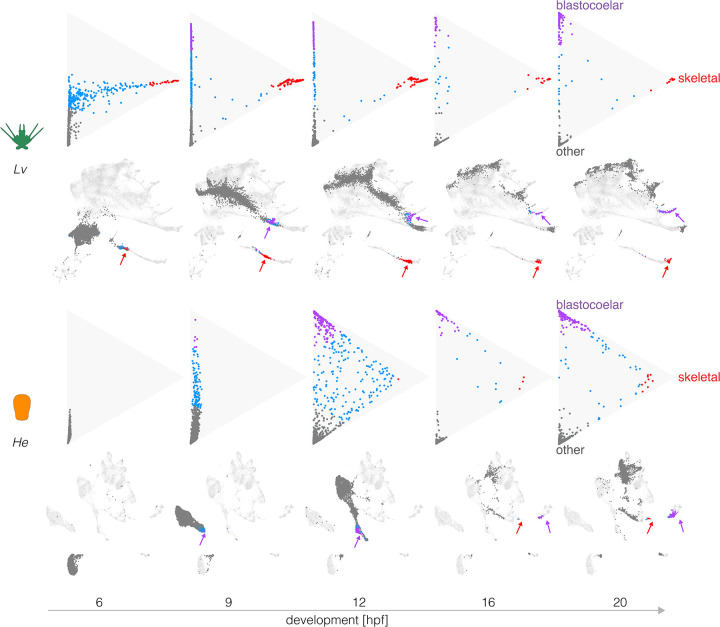
Figure 4. Evolutionary changes in timing of differentiation.
Optimal transport was used to predict the likely fate for each cell at five stages, based on transcriptomes at 24 hpf (see Methods). Triangle plots show transcriptomes predictive of blastocoelar cell (green), skeletogetogenic cell (red), or any other cell fate (dark gray); cells with undifferentiated transcriptomes are shown in blue. Corresponding UMAPs are shown below. Note the much earlier differentiation of skeletogenic cells in Lv and the slightly earlier differentiation of blastocoelar cells in He. See text for additional interpretation.
Figure 4 shows that in Lv, many cells take on a transcriptional state predictive of differentiating into a skeletogenic cell as early as 6 hpf (top left triangle plot, red dots). It is not until 9 hpf that a subset of cells are predicted to differentiate into blastocoelar cells (purple dots), consistent with the order of differentiation of these cells in Lv (McClay 2011).
Several informative differences are evident in He. First, dots are much more tightly bunched at 9 hpf, indicating less divergence in transcriptional states among cells than is the case for Lv at the same time and consistent with the delay in diversification of transcriptional states noted earlier (Figures 2 and 3). Second, the order of differentiation is reversed. Blastocoelar cell transcriptomes appear well before those of skeletogenic cells in He, whereas skeletogenic cells begin differentiating long before blastoceolar cells in Lv. This appears to be due primarily to a shift in skeletogenic cell differentiation, since blastocoelar cells are evident at 9 hpf in both species, while skeletogenic cells appear ~6 hours later in He than Lv. Third, at 20 hpf many dots remain far from any apex in He, but most dots are at or near an apex in Lv. This indicates that more cells remain uncommitted to any specific cell fate in the early larva of He than that of Lv. Finally, the degree of skeletogenic cell differentiation differs between species. None of the skeletogenic cell transcriptomes reaches the apex in He, while many do so in Lv, and they begin to arrive much earlier in development (9 hpf). A rather different pattern is seen with blastocoelar cells, where many reach the apex in both species, and this begins earlier in He (12 hpf) than Lv (20 hpf).
Analysis of other cell types reveals additional evolutionary changes in the timing and degree of differentiation, as well as an example of conservation in timing (Figure S5). Two cell types are noteworthy because changes in their rate of differentiation may be related to the life history shift. Endoderm shows a particularly large delay in the onset of differentiation in He: endodermal cells are evident at 9 hpf in Lv, but even at 20 hpf none are present in He (Figure S5A,B). Coelom shows a less dramatic delay in initial differentiation in He, but the number of coelomic cells in He overtakes those in Lv (Figure S5A). In contrast, blastocoelar and pigment cells show similar overall trajectories in the two species (Figure S5C), indicating that the pace of differentiation within some cell lineages remains relatively unchanged.
Together, these analyses reveal a complex mosaic of evolutionary changes in the timing, order, and degree of differentiation among the two species. While the onset of differentiation is generally delayed in He, individual cell types have evolved in distinct ways: blastocoelar cells differentiate earlier in He relative to Lv, some others are delayed to different degrees (coelom less than skeletogenic cells and gut), and some differentiate at about the same time (pigment cells).
The order of cell fate specification is altered in He
The evolutionary differences in the timing of differentiation noted above are consistent with cell lineage tracing studies in He (Wray and Raff 1989; Wray and Raff 1990). However, those studies also suggested that the order of cell fate specification decisions might differ. We therefore reconstructed transcriptional trajectories during development (Chen et al. 2018; Kester and van Oudenaarden 2018; Forrow and Schiebinger 2021) using optimal transport (Schiebinger et al. 2019; Forrow and Schiebinger 2021).
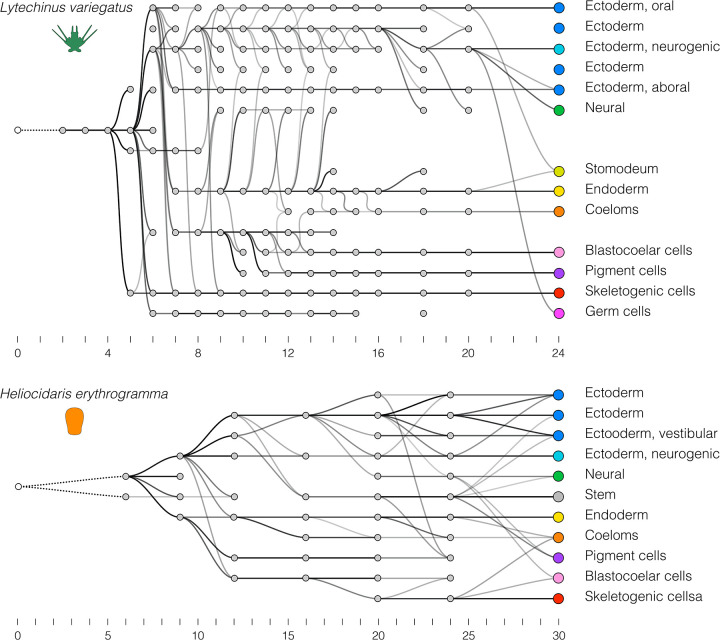
As a positive control, we first evaluated how well transcriptional trajectories based on scRNA-seq data recapitulate actual cell lineages using the published Lv time course (Massri et al. 2021), where the cell lineage is well defined by independent methods (McClay 2011). In the resulting directed graph (Figure 5A), nodes correspond to cell clusters and edges connect nodes to their inferred “ancestor” (darker edges indicate higher confidence). This graph contains several features consistent with published analyses of embryonic cell lineages in Lv and other sea urchins with feeding larvae (Hörstadius 1973; Pehrson and Cohen 1986; Cameron et al. 1987; Cameron et al. 1990; Ruffins and Ettensohn 1996; Martik and McClay 2017). In particular: skeletogenic and primary germ cells diverge very early; pigment and coelomic cells share a common source population that is distinct from other endomesodermal cells; coeloms, stomodaeum, and gut share a common origin; and neurons derive from both gut and from the anterior neurogenic domain.
Figure 5. Evolutionary changes in overall transcriptional trajectories.
Optimal transport was used to identify “ancestors” for each cell cluster, starting with the final time point (unlike Figure 4, where transcriptomes of individual cells are measured against those of differentiated cells). These trajectories reflect the progressive divergence of transcriptomes among cells during development, and thus are an indirect reflection of cell lineages. A. Transcriptional trajectory of Lv. B. Transcriptional trajectory of He.
Some inconsistencies are present, however. Most notably, there is no cluster that corresponds to the four micromeres, the direct ancestors of the skeletogenic and primary germ cell clonal founders. This is likely because cell lineage-specific zygotic transcription is extremely limited at the time the micromeres are present (Ernst et al. 1980) and thus overwhelmed by uniformly distributed maternal transcripts. In addition, the germ cell lineage is discontinuous and shows a late contribution from ectoderm; these are likely artifacts due to their tiny number (8 cells) in proportion to the rest of the embryo at later stages (>1000 cells at 24 hpf). Finally, it should be noted that the ancestor-descendant linkages are in general rather noisy, with several spurious connections. Despite these inconsistencies, the overall topology of the graph resembles the cell lineage as defined by more direct forms of evidence (see citations above).
We then applied the same approach to the He scRNA-seq time course (Figure 5B). This graph shares some similarities with that of Lv: gut and coeloms are closely related, as are ectodermal territories including the anterior neurogenic domain. Several other features are notably different, however. In He, skeletogenic cells are among the last cell clusters to become transcriptionally distinct and are most closely related to blastocoelar cells, while in Lv they are one of the first to become transcriptionally distinct and are not related to blastocoelar cells. In addition, pigment cells in He are not closely related to blastocoelar cells and become transcriptionally distinct well before they do, while in Lv pigment cells and blastocoelar cells derive from a unique common precursor population and simultaneously diverge transcriptionally (Massri et al. 2021). These differences are consistent with the triangle plots (Figures 4 and S5.5). They also imply evolutionary changes in the temporal order and spatial location of fate specification among mesodermal cell lineages (see Discussion).
Other differences in the two graphs are associated with structures or cell types that are present in the larva of one species but not the other. This is apparent in the ectoderm, which is organized anatomically and transcriptionally into somewhat different territories in He relative to the ancestral state (Figure S3), with the hugely accelerated appearance of a distinct vestibular ectoderm territory being the most prominent difference (Haag and Raff 1998; Love and Raff 2006; Koop et al. 2017) (Figure 1C). Other notable differences include the apparent absence of endodermally-derived neurons and primary germ cells in He.
scRNA-seq data accurately reflects known regulatory interactions in Lv
Results presented above indicate that cells in the embryo of He traverse rather different transcriptional trajectories (Figure 4) relative to Lv, and that some differentiating cells emerge from distinct precursor populations in the two species (Figure 5). These observations hint at evolutionary changes in underlying regulatory interactions. Two previous studies used scRNA-seq results to infer that specific regulatory interactions present in the sea urchin S. purpuratus are absent in the seastar Patiria miniata (Foster et al. 2022; Spurrell et al. 2023). We built on this approach, defining criteria for inferring four different evolutionary scenarios: conserved interaction, conserved interaction but with a timing or spatial shift, novel interaction, and loss of interaction (Figure S6).
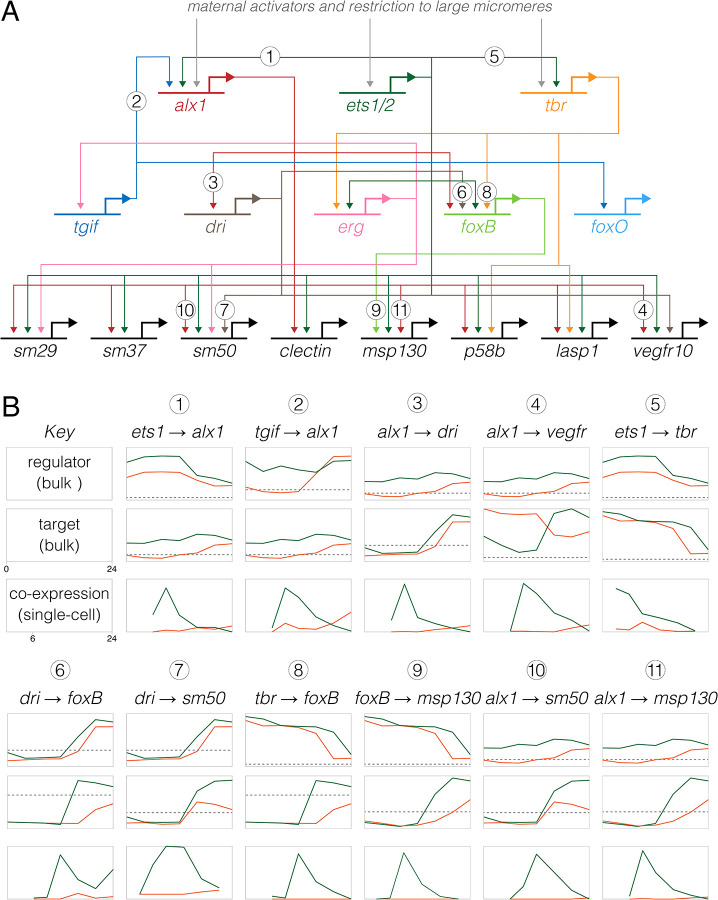
As positive controls, we first examined experimentally validated regulatory interactions in Lv, focusing on the well-studied skeletogenic cell lineage. Figure 6A shows a simplified version of the skeletogenic cell portion of the ancestral dGRN (Kurokawa et al. 1999; Ettensohn et al. 2003; Oliveri et al. 2008; Wahl et al. 2009; Yamazaki et al. 2009; Sharma and Ettensohn 2010; Rafiq et al. 2012; Rafiq et al. 2014). Across the top are the three primary activators of skeletogenic cell-specific transcription, and across the bottom a few of the many effector genes of differentiated skeletogenic cells; between them lie some of the transcription factors that reinforce the differentiated state. We initially focused on alx1, which encodes the master regulator of skeletogenic cell specification (Ettensohn et al. 2003; Sharma and Ettensohn 2010), examining interactions involving the two known activators of its transcription (ets1 and tgif) and some of its many known targets (e.g., dri, vegfr, sm50, and msp130). We analyzed co-expression of regulators and targets in two ways: as the proportion of cells with co-expression over developmental time (Figure 6B) and by the location of cells with co-expression within the first two dimensions of UMAP space (Figures 7 and S7).
Figure 6. Inference of evolutionary changes in regulatory interactions based on proportion of co-expressing cells.
A. Simplified version of the skeletogenic portion of the ancestral dGRN present in camarodont sea urchins with feeding larvae (based on Kurokawa et al. 1999; Oliveri et al. 2002, Ettensohn et al. 2003; Oliveri et al. 2008; Rafiq 2012; Rafiq 2014). The three primary activators of skeletogenic-specific transcription (top) feed directly or indirectly into a large set of effector genes, some of which are illustrated (bottom). B. Co-expression analysis of 11 experimentally validated regulatory interactions, where Lv = green lines and He = orange lines. Numbers correspond to interactions in panel A) The top two plots for each interaction show expression of regulator and target based on bulk RNAseq (Israel et al., 2016), with a log2 y-axis and the dashed line indicating very low expression (an average of 5 counts per million reads across time points). The plot directly below shows the proportion of cells that co-express both genes based on scRNA-seq, with a linear y-axis; these time-points begin a 6 hpf, the first time point common to both data sets. Note that y-axes are not equivalently scaled because genes have a wide range of expression and co-expression levels. Most gene pairs show a strong peak of co-expression at 9 hpf in Lv, which then drops as skeletogenic cells stop dividing while other cell lineages continue to proliferate. In contrast, this peak is notably absent in He; instead, co-expression is initially zero or very low at 9 hpf and rises modestly 16–24 hpf.
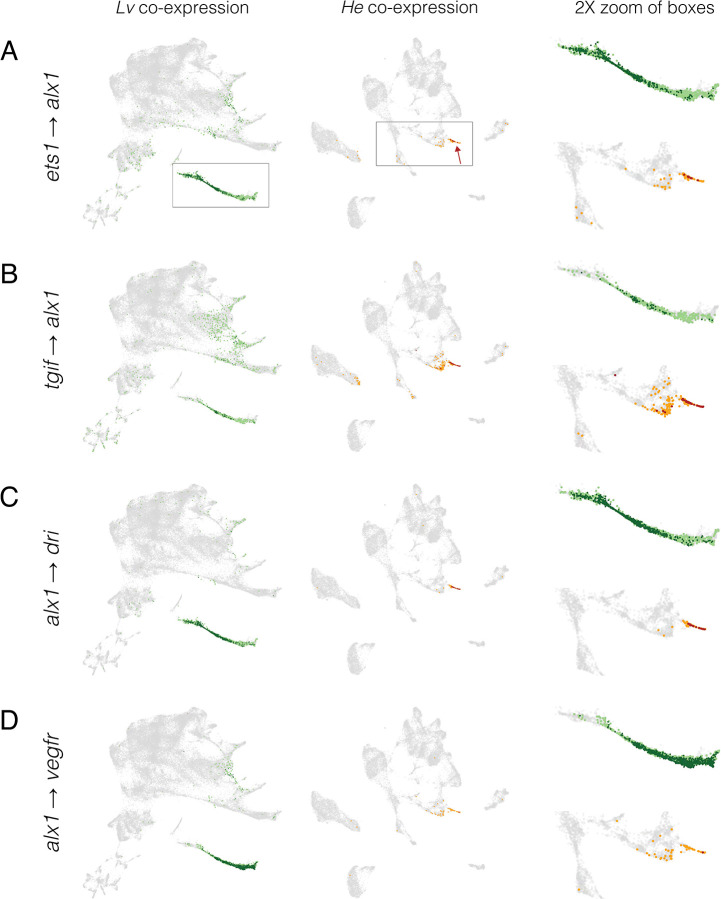
Figure 7. Inference of evolution changes in regulatory interactions based on distribution of co-expressing cells.
Co-expression analysis of experimentally validated regulatory interactions. UMAPs show the location of cells with co-expression of indicated regulator and target. Lv = green dots and He = orange dots; dark colors indicate cells with >2 UMIs for both regulator and target gene; pale colors indicate low co-expressing cells, where one or both genes have 1 or 2 UMIs. Boxes indicate areas shown at 2X in the right-hand column and arrows indicate skeletogenic cells.
Two general points stand out from the Lv data (green lines and dots in Figures 6B, 7, S7). First, co-expression of regulator and target occurs at the expected developmental stages. For the six alx1 interactions shown in Figure 6B, the proportion of cells expressing both regulator and target rises rapidly between 6 and 9 hpf, then declines over time as the skeletogenic cell precursors stop dividing while most other cell lineages continue to proliferate (Martik and McClay 2017). Note that the peaks of co-expressing cells are not evident in the bulk expression of the respective genes (plots immediately above). Many targets of alx1 show delayed co-expression, with some not yet co-expressed at 6 hpf (e.g., alx1 → foxB) or peaking after 9 hpf (e.g., alx1 → sm50) (Figures 6B, 7, S7). The delay in onset of structural gene expression is consistent with the gap of many hours between skeletogenic cell fate specification and differentiation (Rafiq et al. 2012; Rafiq et al. 2014).
Second, most co-expressing cells are restricted to the skeletogenic cell lineage and the majority of cells in the skeletogenic lineage express both genes (Figures 7, S7). This indicates that co-expression is readily detected despite the sparseness of scRNA-seq data. Both restriction to skeletogenic cells and presence in the vast majority of skeletogenic cells are consistent across many activator → target gene pairs involving alx1 (Figure 6B, 7, S7) as well as interactions involving other transcription factors within skeletogenic cells (Figure S8). When a transcriptional activator is broadly expressed, co-expression typically involves a specific subset of the cells within its expression domain. For instance, ets1 and tbr are expressed in the endomesoderm as well as in the skeletogenic cell lineage, but co-expression of ets1-sm32 and tbr-foxB are limited to skeletogenic cells (Figure S9).
Overall, these results are consistent with the developmental times and restriction to the skeletogenic cell lineage for these particular regulatory interactions in Lv. Figure S10 shows a sampling of co-expression related to regulatory interactions in other embryonic territories. These are also largely consistent with the expected times and locations of experimentally validated regulatory interactions within the ancestral life history. For example, expression of otx is quite broad in the embryo, but shows distinct patterns of co-expression with two different targets: in the endoderm, non-skeletogenic mesenchyme, and blastocoelar cells for gataE, but just in the endoderm for endo16 (Figure S10).
A subset of regulatory interactions may be altered in He
Next, we examined the same regulatory interactions in an evolutionary context (Figures 6, 7, 8, S7, S8, and S10; Lv = green and He = orange). In most cases, co-expression occurs in the same cell lineage in both species. For instance, the ancestral interactions involving alx1 are reflected as co-expression primarily within skeletogenic cells in He. Similarly, gataE and pks1 are co-expressed primarily in the pigment cells (Figure S10). These results are consistent with conservation of ancestral regulatory interactions in He. Several differences between species, however, point to evolutionary changes in specific regulatory interactions, including: changes in timing (earlier or later), location (extent or cell lineage), and presence/absence (Figure S6).
Figure 8. Inference of evolutionary loss of a regulatory interaction.
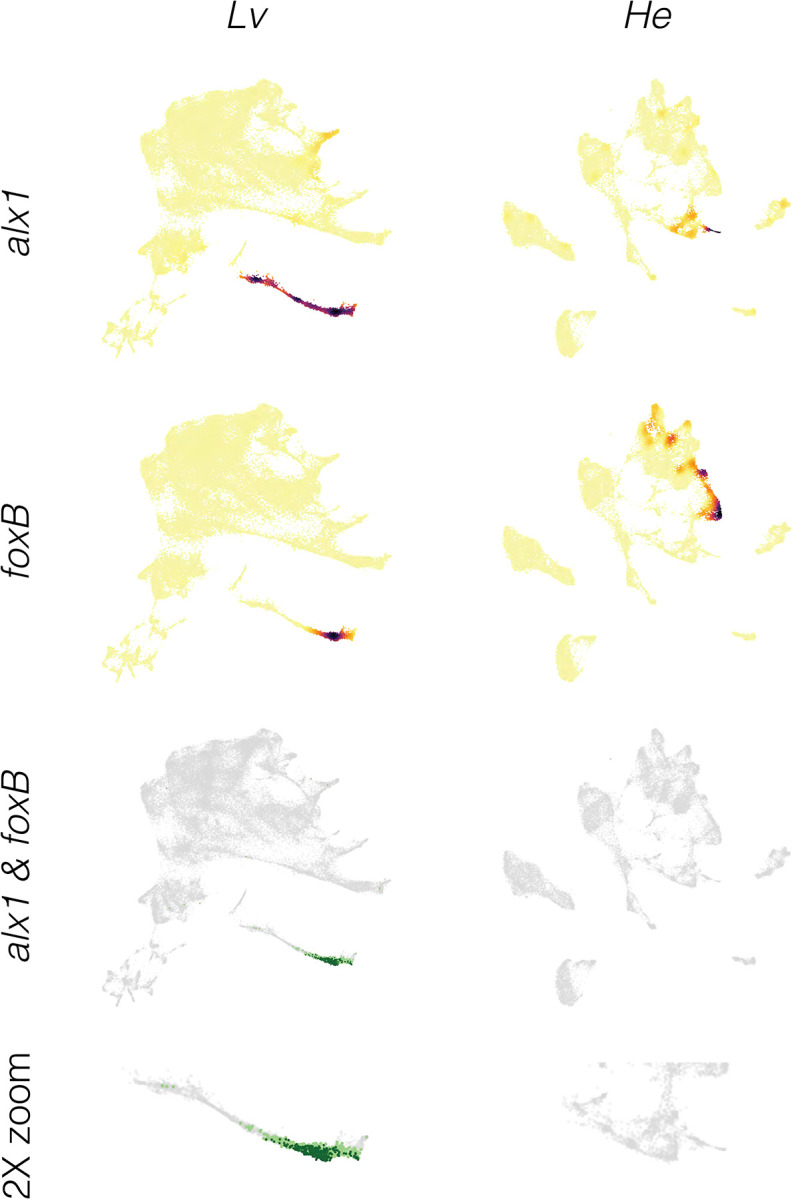
A. Density plots showing expression of regulator (alx1) and target (foxB) genes in both species. Note that both alx1 and foxB transcripts are readily detected in both species. B. Co-expression plots. The complete absence of co-expression in He suggests that the ancestral alx1 → foxB regulatory interaction has been lost in this species.
The most common differences are in timing of co-expression. The prominent early peak of alx1 interactions in Lv is reduced or entirely absent in He (Figure 6B); instead, the proportion of co-expressing cells rises later in He, reflecting much later differentiation. This is largely due to a late rise in alx1 expression (Figure 6B; dashed line indicates < 5 transcripts per million across 3 replicates). Among targets of alx1 expression, dri shows a similar expression profile among species, while vegfr shows highly divergent expression; nonetheless, the co-expression time courses and UMAPs are very similar for both interactions. The simplest explanation for these results is that some regulatory interactions take place in He but that they are considerably delayed relative to Lv. Earlier observations indicating a delay in both specification and differentiation of skeletogenic cells in He (Figures 1, 3, 4, 5) are consistent with this interpretation.
Less commonly, an interaction appears to be absent in He. For the ancestral interaction alx1 → foxB, no cells at any time contain reads from both genes in He (Figure 8). Since both genes are robustly expressed at other times and locations in the He embryo, the complete absence of co-expression is not simply a technical issue with detection. Another example is the ancestral interaction alx1 → foxO, where in He only one cell across all time points expresses both genes and it contains low UMI counts from each gene (indicated by light orange) (Figure S7). Given that co-expression often occurs in a small number of scattered cells outside the region where a specific interaction is thought to occur (Figures 7, S7, S8, S10), this low level of co-expression of alx1 and foxO in He is likely not functionally significant. Other examples involves tbr, which is not expressed in skeletogenic cells in He, despite being expressed elsewhere in the embryo (Figure S8). The simplest interpretation is that these regulatory interactions do not take place in He.
Although the focus here has been on skeletogenic cells, the same general findings are evident in other territories (Figure S10). Again, most co-expression of regulator and target in Lv corresponds to expected times and locations. When comparing species, most co-expression occurs in the same embryonic territory or cell lineage, but with notable exceptions that suggest specific kinds of evolutionary change (Figure S6). Among these, differences in timing or location are the most common. For instance, co-expression of otx and gataE occurs throughout the endomesoderm of both species, but is largely confined to 9–12 hpf in He while present from 6 to 24 hpf in Lv. Similarly, co-expression of bra and foxA is largely endodermal in both species, but in addition is more extensive in the ectoderm of He than in Lv. A few additional likely losses of regulatory interactions are also evident. These include bra → apobec within the endoderm, which may be absent in He (Figure S10). Due to the large number of documented interactions within the ancestral dGRN, a comprehensive co-expression analysis throughout the dGRN is beyond the scope of the present study.
DISCUSSION
Comparisons of single-cell transcriptomes between species have been used to document the presence or absence of cell types (e.g., Levy et al. 2021; Tarashansky et al. 2021; Wang et al. 2021; Alvarez-Campos et al. 2023; Mah and Dunn 2023), but less commonly to understand how developmental mechanisms evolve and contribute to organismal traits. This study used scRNA-seq to examine the evolution of cell fate specification and differentiation in Heliocidaris erythrogramma (He), a sea urchin with a recently modified life history (Figure 1A) (Raff 1992; Ziegler et al. 2003; Wray 2022). The goal was to gain insights into the developmental basis for massively modified larval morphology and hugely abbreviated pre-metamorphic development. We generated a developmental time course of scRNA-seq data from He and carried out comparative analyses with our published data for Lytechinus variegatus (Lv) (Massri et al. 2021), representing the ancestral life history in sea urchins (McEdward and Miner 2001; Raff and Byrne 2006). This discussion is organized around three broad themes revealed by comparative analyses of the scRNA-seq time courses.
Evolution of embryonic patterning
The earliest indication that embryonic patterning might be modified in He came from observations of cleavage divisions, which differ from the stereotypical pattern in the ancestral life history. In sea urchins with feeding larvae, unequal vegetal cleavage divisions establish the clonal founders of two distinct cell lineages: the germ line (Pehrson and Cohen 1986; Oulhen et al. 2019) and the skeletogenic cells, which also become the primary signaling center of the embryo (Hörstadius 1973; Sherwood and McClay 1999; Sweet et al. 2002; Wikramanayake et al. 2004; Wei et al. 2012). These processes appear to be conserved in H. tuberculata, a close relative of He (Figure 1A) (Wray and Raff 1988; Love et al. 2007; Morris et al. 2019). Over the next few hours, a series of inductive interactions initiated by the primary signaling center specify other embryonic cell lineages (reviewed in McClay 2011). These critical early patterning events are broadly among sea urchin species with the ancestral life history (McClay 2011; Thompson et al. 2015; Minokawa et al. 2017; Yamazaki et al. 2021) (Figure 1A). In contrast, He lacks any early unequal cleavage divisions (Williams and Anderson 1975). Dye-tracer studies reveal a general delay in specification and, specifically, that no early blastomeres are clonal founders of either the germ line or skeletogenic cells (Wray and Raff 1989; Wray and Raff 1990).
The scRNA-seq results reported here confirm this delay and add new information. At 6 hpf, the embryo of Lv contains four transcriptionally distinct populations of cells: germ cell precursors, skeletogenic cell precursors, early ectoderm, and endomesoderm (Massri et al. 2021) (Figure 1B). In contrast, the 6 hpf embryo of He contains a single population of cells producing transcripts characteristic of undifferentiated epithelium (Figures 1C and S2).
Closer examination of each of the three early patterning events reveals striking changes in early patterning. (1) Skeletogenic cells. Both species express alx1, which encodes the master regulator of skeletogenic cell fate (Ettensohn et al. 2003), but with a large delay from ~3 hpf in Lv to later than 12 hpf in He (Figures 1C, S2 and S7). A distinct skeletogenic transcriptional state is apparent by ~6 hpf in Lv but not until ~16 hpf in He. Even at 30 hpf, skeletogenic cells of He are not as differentiated as they are at 24 hpf in Lv (Figure 4). (2) Germ cells. At no point up to 30 hpf in He is there a distinct cell population expressing germ cell markers. Species with the ancestral life history express nanos2 broadly, but transcripts and protein accumulate exclusively within the small micromeres (Oulhen et al. 2019). Expression of nanos2 also occurs in He, but does not become localized, remaining widespread in endomesoderm up to 30 hpf (Figure S2). The same is true of vasa, another germ line marker (Figure S2). (3) Primary signaling center. In Lv, the skeletogenic precursors between 3 and 6 hpf express genes that encode signaling ligands, including wnt1, wnt8, and delta (Massri et al. 2021). In He, however, these genes are co-expressed with markers of endomesoderm (foxA, ism, blimp1; Figure S2) and never with skeletogenic markers, suggesting that the primary signaling center is spatially separated from skeletogenic cell fate specification. In addition, the timing of expression differs in He: only wnt8 is expressed at 6 hpf, while all three transcripts show peak expression at 9 hpf (Figure S2). These observations indicate that the three earliest embryonic patterning events in the ancestral state are all delayed in He, and that they have become spatially and temporally separated from each other. This delay is reflected more broadly in the embryo, with transcriptional states in multiple territories diverging later on average (Figures 3, 4, and 5).
The simplest model to explain these observations is that embryonic patterning mechanisms are conserved in He but activated later in development. Three lines of evidence suggest that the situation may actually be more complicated. First, the three earliest patterning events are nearly simultaneous in the ancestral state but occur at widely separated times in He: the primary signaling center is established prior to 9 hpf, skeletogenic cell fate specification takes place between 12 and 16 hpf, and germ cell fate specification occurs some time after 30 hpf. Second, in a previous study, we showed through perturbation experiments that the earliest regulatory interactions responsible for skeletogenic cell fate specification have been lost in He (Davidson et al. 2022a). Germ cell fate specification has not been experimentally investigated in He, but foxY, which encodes a key regulator of nanos2 transcription in the ancestral life history (Oulhen et al. 2019), is not tightly co-expressed with it. Third, some populations of larval cell types in He derive from different founder cells than in the ancestral condition (Figure 5). In particular, pigment cells and blastocoelar (immune) cells derive from a uniquely shared population of non-skeletogenic mesenchyme cells in Lv (Figure 1B) and other species with the ancestral life history (McClay 2011); in contrast in He these two cell types derive from spatially and temporally distinct source populations, and instead it is blastocoelar and skeletogenic cells that share a common origin (Figures 1C and 5). Thus, embryonic patterning and cell fate specification appear to be rearranged in a manner inconsistent with a simple conservation-with-delay model.
Importantly, not all embryonic patterning events are delayed in He. A striking counter-example is the breaking of left-right symmetry, which occurs before first cleavage (Henry and Raff 1990; Henry et al. 1991). In contrast, the first indication of left-right asymmetry in the ancestral developmental mode occurs in the late gastrula (Duboc et al. 2005; Bessodes et al. 2012). Another accelerated patterning event in He involves the early establishment of the imaginal adult rudiment, which begins at about 30 hpf in He (Williams and Anderson 1975; Wray and Raff 1989; Koop et al. 2017) but not until several days post-fertilization in the ancestral condition (Lowe et al. 2002; Formery et al. 2022).
In sum, patterning mechanisms in the early embryo of He appear to represent a complex mosaic of changes. Three critical early patterning events that are tightly associated with a set of unequal cleavages in the very early embryo of the ancestral state are delayed in He, and in addition are dissociated from each other in time and location. In contrast, some other pivotal patterning events are accelerated in He. Furthermore, the origins of some larval cell types have been rearranged, likely reflecting changes in embryonic cell lineages. Together, these changes suggest that several modifications have evolved in interactions within the dGRN, as discussed next.
Evolution of regulatory interactions during development
The ability to assay transcription from single cells provides exciting opportunities to investigate the evolution of transcriptional regulation. In particular, the interaction between a transcriptional activator and a regulatory target should be reflected by co-expression within the same cell. It is important to emphasize that co-expression does not by itself constitute direct evidence: it can reveal a pattern consistent with a regulatory interaction, but experimental evidence is needed to confirm. For this reason, we restrict attention here to gene pairs representing experimentally documented regulatory interactions in the ancestral state, rather than attempting to identify previously unknown interactions. Our current understanding of ancestral dGRN interactions in sea urchins comes primarily from three species: Strongylocentrotus purpuratus, Lytechinus variegatus, and Paracentrotus lividus, all of which have the ancestral life history and diverged ~35–50 million years ago (Figure 1A). Most regulatory interactions that have been experimentally tested in multiple species appear well conserved, as are expression domains of most of genes (McClay, 2011; Gildor and Ben-Tabou De-leon 2015; Israel et al. 2016; Massri et al. 2023).
We first assessed how well scRNA-seq is able to capture previously documented regulatory interactions in Lv by analyzing the distribution of regulator and target gene co-expression during development (Figures 7, S7, S8, S10; green dots and lines). In each case examined, co-expression corresponds to known developmental times and locations of specific regulatory interactions. For instance, ets1 and tbr are expressed throughout the endomesoderm, but ets1-sm32 and tbr-foxB are co-expressed exclusively within the skeletogenic cell lineage and only beginning at ~12 hpf (Figure S9). Co-expression is readily detected for all gene pairs, despite the sparseness of scRNA-seq data. For most interactions, the majority of cells in the expected territory express multiple transcripts: dark green dots in the UMAPs indicate individual cells containing at least 2 unique molecular indices (UMIs) from each gene, while light green indicates just 1 UMI for one or both genes. Overall, co-expression plots are consistent with results from prior experimental studies and are sufficiently sensitive that absence of co-expression is biologically meaningful.
Based on this information, it is possible to make inferences about evolutionary conservation and change by examining co-expression of gene pairs among species (Figure S6). Comparisons of co-expression are shown in Figures 6, 7, S7, S8, S9, and S10 (Lv = green, He = orange). (1) Conservation of an interaction. Most gene pairs show co-expression in the same embryonic territories or cell types in both species. Co-expression in a completely distinct location from Lv was not observed in He for any of the gene pairs examined. The most straightforward interpretation of this pattern is that the ancestral regulatory interaction occurs during development in He. (2) Temporal and/or spatial shift in a conserved interaction. Although the location of co-expression was largely conserved in He, its timing and extent typically was not. Most shifts in timing involved a delay in the appearance of co-expression in He relative to Lv. Among many examples are alx1-dri (Figure S7) and ets1-delta (Figure S8). These cases are consistent with the general delay in specification and differentiation in He shown in Figures 3 and 4. While the timing of developmental gene expression can differ among sea urchin species with the ancestral life history, those shifts are typically smaller in magnitude and not biased in direction (Gildor and Ben-Tabou deLeon 2015; Israel et al. 2016; Massri et al. 2023). Clear examples of evolutionary differences in the extent of co-expression include all interactions specific to the gut and skeletogenic cells, both of which involve proportionally far fewer cells in He (Figures 7, S7, S8, S9, and S10). (3) Loss of an interaction. A minority of gene pairs that are co-expressed in the expected location in Lv show no or barely detectable co-expression in He. Examples include alx1-foxB (Figure S7) and tbr-lasp1 (Figure S8). In these and other cases, lack of co-expression is not due to a technical issue with detection, as transcripts from both genes are detected elsewhere in the embryo (Figure S2). The most straightforward interpretation is that the specific regulatory interaction likely does not occur in He.
Inferred evolutionary changes in regulatory interactions in He are not randomly distributed across the developmental gene regulatory network, but instead concentrated around particular developmental processes. As discussed earlier, a very early patterning event in the ancestral dGRN is the establishment of cells that are both the founders of the skeletogenic cell lineage and the primary signaling center. In Lv, genes encoding the key transcriptional activators of the skeletogenic cell lineage, alx1 and ets1, are first expressed at about the same time as genes encoding signaling ligands (wnt1, wnt8, and delta) (Massri et al. 2021). In contrast, expression of these genes occurs in two distinct phases and locations in He: an earlier phase in the archenteron involving genes that encode ligands (peaking at 9 hpf and greatly reduced by 12 hpf), and a later phase in the mesenchyme involving genes specific to skeletogenic cells (begins ~16 hpf) (Figure 6B and S2). These results suggest that two key patterning events that are co-localized in the ancestral state have become independently regulated during the origin of the derived life history. This is remarkable, given the prior conservation of the ancestral state for over 230 million years (Thompson et al. 2015; Erkenbrack et al. 2018; Yamazaki et al. 2021).
The most obvious way a regulatory interaction could be lost during evolution is if the regulator is simply not expressed in the appropriate cell lineage or territory within the embryo. This is the case for two transcription factors, tbr and foxB. Both are expressed within the skeletogenic cell lineage of Lv (Saunders and McClay 2014), but the scRNA-seq data from He do not reveal any expression within these cells despite clear expression elsewhere in the embryo (Figure S2). Indeed, of the eleven genes known to encode transcription factors that activate expression within the skeletogenic cell lineage of species with the ancestral life history (Oliveri et al. 2008; Saunders and McClay 2014; Rafiq et al. 2012), tbr and foxB are only two that are not expressed in these cells in He. The absence of tbr expression may have limited impact on the expression of effector genes in skeletogenic cells, as Tbr appears to have far fewer targets than Alx1 and Ets1, the two primary activators of skeletogenic-specific transcription (Rafiq et al. 2012). The four known effector gene targets of Tbr are all expressed in skeletogenic cells of He, likely because they also receive input from other transcriptional activators, including Alx1 and Ets1 (Rafiq et al. 2012). tbr was previously proposed to be a more recent evolutionary addition to the skeletogenic cell GRN due to having fewer regulatory targets than Alx1 and Ets1 (Rafiq et al. 2012). The evolutionary loss of tbr expression within skeletogenic cells may have been possible for the same reason, coupled with the fact that nine other genes encoding transcription factors with roles in activating effector gene expression are also expressed within skeletogenic cells, thus providing some degree of regulatory redundancy.
Evolution of morphology and life history
The evolution of massive maternal provisioning in He also precipitated changes in larval morphology and life history traits (Raff and Byrne 2006; Wray 2022). The most obvious are loss of feeding structures and a functional digestive tract (Williams and Anderson 1975), which are no longer needed with a richly provisioned egg (Hoegh-Guldberg and Emlet 1997; Byrne et al. 1999; Davidson et al. 2019). Another set of changes was likely driven by selection to reduce larval mortality by shortening pre-metamorphic development, including earlier left-right symmetry breaking, differentiation of coeloms, and formation of the adult imaginal rudiment (Williams and Anderson 1975; Wray and Raff 1989; Henry et al. 1991; Koop et al. 2017).
The scRNA-seq data reflect both sets of changes in the proportions of cell types in the early larva (Figure 1D). In He, far fewer cells are allocated to endoderm, which is non-functional until after metamorphosis, and to skeletogenic cells, which produce a vestigial larval skeleton (Williams and Anderson 1975; Emlet 1995). Conversely, more cells are allocated to coeloms and ectoderm, both of which contribute substantially to accelerated development of the post-metamorphic juvenile (Williams and Anderson 1975; Wray and Raff 1989; Koop et al. 2017).
In sea urchins with the ancestral life history involving feeding larvae, four territories of ectodermal cells are evident from anatomy and gene expression: a ciliated band used for feeding and locomotion, an anterior neurogenic domain, and generalized ectoderm with distinct oral and aboral domains. These territories are recovered as separate clusters with scRNA-seq in Lv (Figures 1B and S3). Previous studies examining ectodermal gene expression in He found no evidence of conserved oral and aboral territories, and suggested instead that the ancestral ectodermal domains are reorganized (Haag and Raff 1998; Love and Raff 2006; Koop et al. 2017). The scRNA-seq data reveal a well-defined anterior neurogenic domain in He (Figures S2 and S3). However, the other ancestral ectodermal domains are more difficult to recognize in the He larva. Markers of oral and aboral ectoderm the ancestral state are not consistently co-localized in He (Figure S3). The ciliated band, which is used for feeding, has been lost in He (Williams and Anderson, 1975). The only regions of dense cilia in the He larva likely correspond instead to the epaulettes of late larvae in the ancestral state, which are used exclusively for swimming (Emlet 1995). The other derived trait in He that likely contributes to changes in expression of regulatory genes within the ectoderm is the greatly accelerated development of the imaginal adult rudiment (Williams and Anderson 1975; Wray and Raff 1989; Emlet 1995; Koop et al. 2017). Vestibular ectoderm is a distinct gene expression territory within the ectoderm of He by 24 hpf (Koop et al. 2017). Comparative analysis of gene expression in the epaulettes and vestibule will require extending the Lv scRNA-seq time course to late larval stages, as these structures have not yet developed in the early larva.
In summary, the scRNA-seq data presented here reveal numerous features of development in He that are likely conserved and others that are likely modified since its divergence from other sea urchins that share the ancestral life history. While scRNA-seq data alone do not provide direct evidence about molecular mechanisms, they can produce detailed information about specific developmental processes that are not evident from bulk RNA-seq and would otherwise require gene-by-gene expression analyses. Here we report a close correlation between evolutionary changes in the timing and location of regulatory genes and evolutionary changes in larval morphology and life history. Many specific regulatory interactions that are widely conserved among sea urchins with the ancestral life history appear to be conserved but delayed in He, while a small number may have been lost entirely. These results provide specific predictions that can be tested efficiently using perturbation experiments, greatly facilitating the daunting challenge of understanding which connections within developmental gene regulatory networks are conserved, altered, or lost during the course of evolution.
Methods
Spawning and embryo culture
Adult H. erythrogramma were collected under permit near Sydney, Australia during October and November. Crosses were initially established for the purpose of optimizing dissociation protocols; subsequently, a single cross was used to source samples for this study. Adults were spawned by injecting 0.5 ml 0.5 M KCl intracoelomically. Unfertilized eggs were allowed to float and washed 3X in filtered natural sea water (FNSW). Eggs were fertilized with sperm in FNSW containing 0.02 g PABA / 100 ml. Zygotes were washed an additional 3X in FNSW to remove residual sperm and PABA and embryos cultured at 23 °C in FNSW. At each time point, embryos were visually verified to be morphologically similar prior to dissociation. Throughout, methods closely matched our previous scRNA-seq analysis of L. variegatus (Massri et al. 2021), including only slightly species-optimized dissociation protocols, same rearing temperature, timematched samples, and same versions of 10X library kits and Illumina sequencing chemistry.
Time points sampled
Embryos / larvae were sampled at seven time points: 6, 9, 12, 16, 20, 24, and 30 hpf (late cleavage through early larva). Time points were chosen to align with Massri et al. 2021, with two additional considerations. First, due to the large egg size of He (~430 μm diameter), blastomeres exceed the diameter of the microfluidics on the 10X platform until the 512-cell stage (6 hpf), which became our first time point. Second, prior studies suggested that activation of the zygotic genome in He is somewhat delayed relative to Lv (Wang et al. 2020; Davidson et al. 2022); thus, we collected one additional time point (30 hpf) beyond the last sampled in Lv (24 hpf). Comparative analyses, drew on published data from Massri et al. 2021 for Lv and from the present study for He.
Cell dissociation and fixation
At each time point, the culture was sub-sampled and embryos washed two times in CalciumFree Artificial SeaWater (CFASW). ~3ml embryos in CFASW were added to 7ml of dissociation buffer (1.0M glycine and 0.25 mM EDTA, pH 8.0 with HCl) at 4 °C, and then placed on a rocker for 4 minutes. Following incubation, samples were triturated 10–15X, then 10ml of ice-cold methanol was added, and incubated for 4 additional minutes on a rocker. Following incubation, samples were triturated 10–15X additionally, and visually inspected under a microscope for a homogenous single cell suspension. To fix cells, 40ml of ice-cold methanol was added to a final concentration of 80%. Samples were then placed on a rocker for one hour prior to storage at −20 °C.
Library preparation and sequencing
Fixed cells were washed once in methanol, then rehydrated by washing in 3X sodium citrate buffer. Cell concentrations were determined using a hemacytometer. Seven libraries were prepared using the 10X Genomics 3 ‘v3 gene expression kit and the 10X Chromium platform to encapsulate single cells within droplets. Library quality was verified using an Agilent 2100 Bioanalyzer. Libraries were titered and pooled at Duke University’s Sequencing and Genomic Technologies Core Facility, then sequenced in one S1 flow cell on an Illumina NovaSeq 6000 with 28 × 8 × 91 bp.
Computational Analysis
Initial processing and production of raw csv count files
Following sequencing, Cellranger 3.1.0 was used to convert Illumina-generated BCL files to fastq files using the Cellranger “mkfastq” command. The “mkref” command was then applied to index the H. erythrogramma 1.0 Genome (Davidson et al. 2022a). The “count” command was used to demultiplex and quantify reads mapping to the reference He genome. The “mat2csv” command was used to generate CSV RNA count matrix files for each time point for downstream analysis.
Data filtering and normalization
All 19 Lv and 7 He CSV RNA count matrix files were uploaded to R, and a merged Seurat object (Hao et al. 2022) was generated for each species. The Lv Seurat object was filtered to remove lower quality cells with nFeature_RNA > 200, nFeature_RNA < 7000, and nCount_RNA < 10000. In total, 50,638 Lv cells remained. The He Seurat object was filtered with nFeature_RNA > 200, nFeature_RNA < 4000, and nCount_RNA < 10000. In total, 23,156 He cells remained. SCTransform was then independently applied to the Lv merged filtered object and the He merged filtered object to perform normalization, and regression of ribosomal and cell cycle related genes using the command: vars.to.regress = c(“percent.Rb”, “cell.cycle”). These metacolumns were added with the following commands: PercentageFeatureSet(merged, pattern = “\\b\\w*Rp[sl]\\w*\\b”, col.name = “percent.Rb”) and PercentageFeatureSet(merged, pattern = “\\b\\w*C[d|y]c\\w*\\b”, col.name = “cell.cycle”).
Dimensionality reduction, visualization, and clustering
We next independently performed Principal Component Analysis on the SCTransformed Lv Seurat object and He Seurat object, and found the nearest neighbors and clusters (Hao et al. 2022). UMAP was then applied to each species to visualize the multidimensional scRNA-seq in a two-dimensional space. Each species clusters were annotated using co-expression of dGRN genes, and published in situ hybridization patterns as markers. Echinobase (Arshinoff et al. 2021) was used to identify gene function. See Massri et al. 2021 for a list of marker genes and supporting literature.
Multi-species integrated analysis
Orthologroups were identified using OrthoFinder v 2.5.4 (Ems and Kelly 2019) and used to generate a list of 1:1 orthologues in Lv and He. In total, 7,349 genes were identified as 1:1 orthologues and expressed in the combined data set of 72,445 cells. The standard Seurat/SCTransform pipeline was performed, and then integrated by species using the Canonical Correlation Analysis (CCA) workflow (Butler et al. 2018) using Lv as the reference.
Waddington-OT developmental trajectories
To infer developmental trajectories in He, we used Waddington-OT (Schiebinger et. 2019). To execute we used the SCTransform normalized expression matrix obtained after running Seurat, a table of cell barcodes with cell-type annotations, and a growth rates table that was estimated from expected changes in lineage proportions over time using the model implemented in Waddington-OT. To estimate cell division rates, we used the best estimate of the expected number of cells at key developmental time points. We assumed that cell divisions were uniform between estimates of expected cell numbers. Next we recalculated transport maps using the modeled cell division rates, optimization parameters ε=0.05, λ1=1, and λ2=50, and 20 iterations of growth rate learning. We used the transport map model throughout our analysis that included triangle plots and lineage trees.
Waddington-OT time alignment
To estimate timing differences between the Lv and He datasets, we used optimal transport combined with the gene orthology tables. First, we used the previously calculated transport maps for both datasets to obtain fate probabilities for the cells at each time point. Fate probabilities were computed relative to cell types found in the last time point of their respective dataset. Next, we restricted the normalized counts for Lv and He to known gene orthologues using the previously generated gene orthologue table. Then, for each cell type, a time point by time point matrix of earth mover distances between the two datasets was computed. In the calculations for each cell type and pair of times, cells were weighted by their fate probabilities to the cell type in question. Finally, for each Lv time point, the He time point corresponding to the minimum earth mover distance to it was found. These pairs were found for each cell type. We then take these pairs of time points to be the optimal developmental time alignments for the cell type.
Waddington-OT triangle plots
To construct triangle plots, we used transport maps calculated before to compute fate probabilities with respect to the last common time point in our dataset (24 hpf) and visualized them by computing the barycentric coordinates of cell fates between two different cell types and at a threshold of 0.7.
Developmental lineage trees
To infer cell lineage trees, we used our modeled transport maps to find connections between cell clusters by calculating the fraction of descendants that end up in cluster/cell-type j at time ti+1 from cluster/cell-type i. The minimum number of cells for a cluster to be represented set to 10 and the minimal edge weight cutoff was set to 0.15. Once the unwanted edges were removed, the data was written in a format that is usable by d3.js.
Co-expression analyses
The number of cells expressing any two specified genes was tallied from the count tables and normalized by the total number of cells for each sample using a custom Python script, and used to generate time courses for both species. To visualize co-expression in UMAP space, a custom R script was used.
Supplementary Material
Figure S1. Metrics of scRNA-seq transcriptomes over development in He. Violin plots showing the frequency distribution of four informative metrics. A. Distinct genes/cell from which transcripts were detected (nFeature_RNA in Seurat). The modest decline over development likely reflects the transition of a more restricted transcriptome during differentiation. B. Distinct UMIs recovered per cell (nCount_RNA in Seurat). C. Percentage of transcripts mapping to genes encoding ribosomal proteins. The increase over development reflects the maternal-to-zygotic transition in protein expression. D. Percentage of transcripts mapping to genes encoding cell cycle control proteins. The slight decrease in the last two stages reflects the slowing of cell division as differentiation commences in the early larva.
Figure S2. Expression of marker genes in He. Density plots showing the distribution in UMAP space of cells expressing marker genes for specific embryonic territories and larval cell types. See Massri et al. 2021 for supporting literature.
Figure S3. Expression of marker genes for ectodermal territories in Lv and He. Density plots showing the distribution in UMAP space of cells expressing marker genes for specific embryonic territories and larval cell types. See Massri et al. 2021 for supporting literature.
Figure S4. Overall similarity of transcriptomes in Lv and He over development. Heatmap showing overall similarity of expression of 1:1 orthologues among all developmental stages examined. Similarity measured using CIDER (Hu et al. 2021).
Figure S5. Evolutionary changes in timing of differentiation, additional examples. Optimal transport was used to predict the likely fate for each cell at five stages, based on transcriptomes at 24 hpf (see Methods). The organization of this figure parallels that of main text Figure 4. Triangle plots show transcriptomes predictive of two specific cell types. A. Coelom (red) and endoderm (yellow). B. Blastocoelar cells (purple) and endoderm (yellow). C. Blastocoelar cells (purple) and pigment cells (pink). Any other cell fate is indicated in dark gray; cells with undifferentiated transcriptomes are shown in blue. See text for interpretation.
Figure S6. Logic underlying inference of distinct types of evolution change in regulatory interactions. On the extreme left, hypothetical data from Lv illustrating an experimentally validated regulatory interaction: transcriptional activator, gene A (blue) and its target, gene B (purple), are co-expressed in the same cells. Four possible scenarios in He for the same pair of genes are shown to the right, with interpretations regarding the regulatory interaction shown below. Scenario 1 is consistent with conservation. Scenario 2 is consistent with conservation, but with a delay in the timing of the interaction. Scenario 3 implies that the interaction no longer occurs and that some other transcription factor must activate expression of gene B. Scenario 4 is consistent with conservation, but additionally implies that some other transcription factor must operate earlier to initiate expression of gene B.
Figure S7. Co-expression of alx1 and several of its interactors within the skeletogenic dGRN. See caption to Figure 7.
Figure S8. Co-expression of additional regulators and targets within the skeletogenic dGRN. See caption to Figure 7.
Figure S9. Comparison of regulator expression and co-expression with a specific target gene. Five validated transcription factor → target gene interactions are illustrated. Left panels show density plots of expression for the regulator while right panels show cells expressing both the regulator and its target. Note that co-expression domains are often a subset of the cells expressing the regulator.
Figure S10. Co-expression of several regulators and targets in non-skeletogenic territories of the dGRN. See caption to Figure 7.
Table S1. Cell counts over development in Lv.
Table S2. Cell counts over development in He.
ACKNOWLEDGEMENTS
We thank the staff of the Sydney Institute for Marine Sciences for skilled support and the staff of Duke’s Sequencing and Genomic Technologies core facility for expert advice and assistance. Echinobase was critical to the success of this project. Matthew Clements, Phillip Davidson, Hannah Devens, Allison Edgar, Brennan McDonald, Carl Manner, Esther Miranda, Zach Pracher, Paulina Selvakumaraswamy, Jane Swart, and Emma Wallace provided invaluable advice. This research was financially supported by NIH Training Grant HD040372 and NSF Graduate Research Fellowship DGE-1644868 to AJM; NIH grant HD014483 to DRM; and NSF grant IOS 1929934 to GAW.
REFERENCES
- Alvarez-Campos P., Garcia-Castro H., Emili E., Perez-Posada A., Salamanca-Diaz D.A., Mason V., Metzger B., Bely A.E., Kenny N., Ozpolat B.D., Solana J., 2023. Annelid adult cell type diversity and their pluripotent cellular origins. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer L.M., Yaguchi S., Angerer R.C., Burke R.D., 2011. The evolution of nervous system patterning: insights from sea urchin development. Development 138: 3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshinoff B.I., Cary G.A., Karimi K., Foley S., Agalakov S., Delgado F., Lotay V.S., Ku C.J., Pells T.J., Beatman T.R., Kim E., Cameron R.A., Vize P.D., Telmer C.A., Croce J.C., Ettensohn C.A., Hinman V.F., 2021. Echinobase: leveraging an extant model organism database to build a knowledgebase supporting research on the genomics and biology of echinoderms. Nucleic Acids Res 50(D1): D970–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessodes N., Haillot E., Duboc V., Rottinger E., Lahaye F., Lepage T., 2012. Reciprocal signaling between the ectoderm and a mesendodermal left-right organizer directs left-right determination in the sea urchin embryo. PLoS Genet 8: e1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove B.W., Raff R.A., 1989. Evolutionary conservation of the larval serotonergic nervous system in a direct developing sea urchin. Dev Growth Differ 31: 363–370. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M., Villinski J.T., Cisternas P., Siegel R.K., Popodi E., Raff R.A., 1999. Maternal factors and the evolution of developmental mode: evolution of oogenesis in Heliocidaris erythrogramma. Dev Genes Evol 209: 275–283. [DOI] [PubMed] [Google Scholar]
- Cameron R.A., Fraser S.E., Britten R.J., Davidson E.H., 1990. Segregation of oral from aboral ectoderm precursors is completed at fifth cleavage in the embryogenesis of Strongylocentrotus purpuratus. Dev Biol 137: 77–85. [DOI] [PubMed] [Google Scholar]
- Cameron R.A., Hough-Evans B.R., Britten R.J., Davidson E.H., 1987. Lineage and fate of each blastomere of the eight-cell sea urchin embryo. Genes Dev 1: 75–85. [DOI] [PubMed] [Google Scholar]
- Cary G.A., Hinman V.F., 2017. Echinoderm development and evolution in the post-genomic era. Dev Biol 427: 203–211. [DOI] [PubMed] [Google Scholar]
- Cary G.A., McCauley B.S., Zueva O., Pattinato J., Longabaugh W., Hinman V.F., 2020. Systematic comparison of sea urchin and sea star developmental gene regulatory networks explains how novelty is incorporated in early development. Nat Commun 11: 6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Renia L., Ginhoux F., 2018. Constructing cell lineages from single-cell transcriptomes. Mol Aspects Med 59: 95–113. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., 2010. Emerging properties of animal gene regulatory networks. Nature 468: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H., Cameron R.A., Ransick A., 1998. Specification of cell fate in the sea urchin embryo: summary and some proposed mechanisms. Development 125: 3269–3290. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Otim O., Brown C.T., Livi C.B., Lee P.Y., Revilla R., Rust A.G., Pan Z., Schilstra M.J., Clarke P.J., Arnone M.I., Rowen L., Cameron R.A., McClay D.R., Hood L., Bolouri H., 2002a. A genomic regulatory network for development. Science 295: 1669–1678. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., Rast J.P., Oliveri P., Ransick A., Calestani C., Yuh C.H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Otim O., Brown C.T., Livi C.B., Lee P.Y., Revilla R., Schilstra M.J., Clarke P.J., Rust A.G., Pan Z., Arnone M.I., Rowen L., Cameron R.A., McClay D.R., Hood L., Bolouri H., 2002b. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol 246: 162190. [DOI] [PubMed] [Google Scholar]
- Davidson P.L., Byrne M., Wray G.A., 2022a. Evolutionary changes in the chromatin landscape contribute to reorganization of a developmental gene regulatory network during rapid life history evolution in sea urchins. Mol Biol Evol 39: msac172. doi: 10.1093/molbev/msac172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.L., Guo H., Swart J.S., Massri A.J., Edgar A., Wang L., Berrio A., Devens H.R., Koop D., Cisternas P., Zhang H., Zhang Y., Byrne M., Fan G., Wray G.A., 2022b. Recent reconfiguration of an ancient developmental gene regulatory network in Heliocidaris sea urchins. Nat Ecol Evol 6: 1907–1920. [DOI] [PubMed] [Google Scholar]
- Davidson P.L., Guo H., Wang L., Berrio A., Zhang H., Chang Y., Soborowski A.L., McClay D.R., Fan G., Wray G.A., 2020. Chromosomal-Level genome assembly of the sea urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biol Evol 12: 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driesch H., 1891. Entwicklungsmechanische Studien. I. Der Werth der beiden ersten Furchungszellen der Echinodermenementwicklung. Experementelle Erzeugung von Thielund Doppelbildungen. II. Über die Beziehungen des Lichtes zur ersten Etappe der thierischen Formbildung. Z. Wiss. Zool. 53: 160. [Google Scholar]
- Duboc V., Rottinger E., Lapraz F., Besnardeau L., Lepage T., 2005. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell 9: 147158. [DOI] [PubMed] [Google Scholar]
- Emlet R.B., 1995. Larval spicules, cilia, and symmetry as remnants of indirect development in the direct developing sea urchin Heliocidaris erythrogramma. Dev Biol 167: 405–415. [DOI] [PubMed] [Google Scholar]
- Emlet R.B., Hoegh-Guldberg O., 1997. Effects of Egg Size on Postlarval performance: Experimental evidence from a sea urchin. Evolution 51: 141–152. [DOI] [PubMed] [Google Scholar]
- Emlet R.B., McEdward L.R., Strathmann R.A., 1987. Echinoderm larval ecology viewed from the egg, in: Jangoux M., Lawrence J.M. (Eds.), Echinoderm Studies. Balkema, Rotterdam, pp. 55–136. [Google Scholar]
- Emms D.M., Kelly S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkenbrack E.M., Davidson E.H., Peter I.S., 2018. Conserved regulatory state expression controlled by divergent developmental gene regulatory networks in echinoids. Development 145: dev167288. doi: 10.1242/dev.167288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S.G., Hough-Evans B.R., Britten R.J., Davidson E.H., 1980. Limited complexity of the RNA in micromeres of sixteen-cell sea urchin embryos. Dev Biol 79: 119–127. [DOI] [PubMed] [Google Scholar]
- Erwin D.H., Davidson E.H., 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet 10: 141–148. [DOI] [PubMed] [Google Scholar]
- Ettensohn C.A., Illies M.R., Oliveri P., De Jong D.L., 2003. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130: 2917–2928. [DOI] [PubMed] [Google Scholar]
- Ettensohn C.A., McClay D.R., 1988. Cell lineage conversion in the sea urchin embryo. Dev Biol 125: 396–409. [DOI] [PubMed] [Google Scholar]
- Ettensohn C.A., Ruffins S.W., 1993. Mesodermal cell interactions in the sea urchin embryo: properties of skeletogenic secondary mesenchyme cells. Development 117: 1275–1285. [DOI] [PubMed] [Google Scholar]
- Formery L., Wakefield A., Gesson M., Toisoul L., Lhomond G., Gilletta L., Lasbleiz R., Schubert M., Croce J.C., 2022. Developmental atlas of the indirect-developing sea urchin Paracentrotus lividus: From fertilization to juvenile stages. Front Cell Dev Biol 10: 966408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formery L. and Lowe C. J.. 2023. Integrating complex life cycles in comparative developmental biology. Ann Rev Genetics 57: 321–339. [DOI] [PubMed] [Google Scholar]
- Forrow A., Schiebinger G., 2021. LineageOT is a unified framework for lineage tracing and trajectory inference. Nat Commun 12: 4940. doi: 10.1038/s41467-021-25133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S., Oulhen N., Fresques T., Zaki H., Wessel G., 2022. Single-cell RNA-sequencing analysis of early sea star development. Development 149: dev200982. doi: 10.1242/dev.200982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S., Oulhen N., Wessel G., 2020. A single cell RNA sequencing resource for early sea urchin development. Development 147: dev191528. doi: 10.1242/dev.191528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garstang W. J. 1928. The origin and evolution of larval forms. British Assoc Adv Sci 1929: 77–98. [Google Scholar]
- Gildor T., Ben-Tabou de-Leon S., 2015. Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLoS Genet 11: e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K.L., Ruvinsky I., 2012. Tempo and mode in evolution of transcriptional regulation. PLoS Genet 8, e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J.A., Scialdone A., Marioni J.C., 2018. Using single-cell genomics to understand developmental processes and cell fate decisions. Mol Syst Biol 14: e8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E.S., Raff R.A., 1998. Isolation and characterization of three mRNAs enriched in embryos of the direct-developing sea urchin Heliocidaris erythrogramma: evolution of larval ectoderm. Dev Genes Evol 208: 188–204. [DOI] [PubMed] [Google Scholar]
- Hao Y, Stuart T., Kowalski M., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C., and Satija R. 2022. Dictionary learning for integrative, multimodal, and scalable single-cell analysis. bioRxiv doi: 10.1101/2022.02.24.481684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J.J., Raff R.A., 1990. Evolutionary change in the process of dorsoventral axis determination in the direct developing sea urchin, Heliocidaris erythrogramma. Dev Biol 141: 55–69. [DOI] [PubMed] [Google Scholar]
- Henry J.J., Wray G.A., Raff R.A., 1990. The dorsoventral axis is specified prior to first cleavage in the direct developing sea urchin Heliocidaris erythrogramma. Development 110: 875–884. [DOI] [PubMed] [Google Scholar]
- Hinman V.F., Nguyen A., Davidson E.H., 2007. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol 312: 584–595. [DOI] [PubMed] [Google Scholar]
- Hinman V.F., Nguyen A.T., Cameron R.A., Davidson E.H., 2003. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA 100: 13356–13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O., Emlet R.B., 1997. Energy use during the development of a lecithotrophic and a planktotrophic echinoid. Biol Bull 192: 27–40. [DOI] [PubMed] [Google Scholar]
- Hörstadius S., 1973. Experimental Embryology of Echinoderms. Clarendon, Oxford. [Google Scholar]
- Hu Z., Ahmed A.A., Yau C., 2021. CIDER: an interpretable meta-clustering framework for single-cell RNA-seq data integration and evaluation. Genome Biol 22: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel J.W., Martik M.L., Byrne M., Raff E.C., Raff R.A., McClay D.R., Wray G.A., 2016. Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus Heliocidaris. PLoS Biol 14: e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C.E., Yajima M., Wessel G.M., 2010. Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev Biol 337, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester L., van Oudenaarden A., 2018. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23: 166–179. [DOI] [PubMed] [Google Scholar]
- Koga H., Matsubara M., Fujitani H., Miyamoto N., Komatsu M., Kiyomoto M., Akasaka K., Wada H., 2010. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Dev Genes Evol 220: 107–115. [DOI] [PubMed] [Google Scholar]
- Koop D., Cisternas P., Morris V.B., Strbenac D., Yang J.Y., Wray G.A., Byrne M., 2017. Nodal and BMP expression during the transition to pentamery in the sea urchin Heliocidaris erythrogramma: insights into patterning the enigmatic echinoderm body plan. BMC Dev Biol 17: 4. doi: 10.1186/s12861-017-0145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D., Kitajima T., Mitsunaga-Nakatsubo K., Amemiya S., Shimada H., Akasaka K., 1999. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech Dev 80: 41–52. [DOI] [PubMed] [Google Scholar]
- Láruson Á.J., 2016. Rates and relations of mitochondrial genome evolution across the Echinoidea, with a special focus on the superfamily Odontophora. Ecology and Evolution 7: 4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Elek A., Grau-Bove X., Menendez-Bravo S., Iglesias M., Tanay A., Mass T., Sebe-Pedros A., 2021. A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 184: 2973–2987 e2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A.C., Lee A.E., Andrews M.E., Raff R.A., 2008. Co-option and dissociation in larval origins and evolution: the sea urchin larval gut. Evol Dev 10: 74–88. [DOI] [PubMed] [Google Scholar]
- Love A.C., Raff R.A., 2006. Larval ectoderm, organizational homology, and the origins of evolutionary novelty. J Exp Zool B Mol Dev Evol 306: 18–34. [DOI] [PubMed] [Google Scholar]
- Lowe C.J., Issel-Tarver L., Wray G.A., 2002. Gene expression and larval evolution: changing roles of distal-less and orthodenticle in echinoderm larvae. Evol Dev 4: 111–123. [DOI] [PubMed] [Google Scholar]
- Mah J.L., Dunn C.W., 2023. Reconstructing cell type evolution across species through cell phylogenies of single-cell RNA seq data. bioRxiv. [Google Scholar]
- Martelaz F., Couloux A., Poulain J., Labadie K., Da Silva C., et al. 2023. Analysis of the P. lividus sea urchin genome highlights contrasting trends of genomic and regulatory evolution in deuterostomes. Cell Genom 3:100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik M.L., McClay D.R., 2017. New insights from a high-resolution look at gastrulation in the sea urchin, Lytechinus variegatus. Mech Dev 148: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massri A.J., Greenstreet L., Afanassiev A., Berrio A., Wray G.A., Schiebinger G., McClay D.R., 2021. Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo. Development 148: dev198614. doi: 10.1242/dev.198614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massri A.J., McDonald B., Wray G.A., McClay D.R., 2023. Feedback circuits are numerous in embryonic gene regulatory networks and offer a stabilizing influence on evolution of those networks. Evodevo 14: 10. doi: 10.1186/s13227-023-00214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna S.C., Davidson E.H., 2012. A comprehensive analysis of Delta signaling in pregastrular sea urchin embryos. Dev Biol 364: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley B.S., Weideman E.P., Hinman V.F., 2010. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol 340: 200–208. [DOI] [PubMed] [Google Scholar]
- McClay D.R., 2011. Evolutionary crossroads in developmental biology: sea urchins. Development 138: 2639–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEdward L.R., Miner B.G., 2001. Larval and life-cycle patterns in echinoderms. Canadian Journal of Zoology 79: 1125–1170. [Google Scholar]
- Minokawa T., 2017. Comparative studies on the skeletogenic mesenchyme of echinoids. Dev Biol 427: 212–218. [DOI] [PubMed] [Google Scholar]
- Morris V.B., Kable E., Koop D., Cisternas P., Byrne M., 2019. Early development of the feeding larva of the sea urchin Heliocidaris tuberculata: role of the small micromeres. Dev Genes Evol 229: 1–12. [DOI] [PubMed] [Google Scholar]
- Mortensen T., 1921. Studies of the development and larval forms of echinoderms. G.E.C. Gad, Copenhagen. [Google Scholar]
- Nielsen C. 2009. How did indirect development with planktotrophic larvae evolve? Biol Bull 216: 203–215. [DOI] [PubMed] [Google Scholar]
- Nielsen M.G., Wilson K.A., Raff E.C., Raff R.A., 2000. Novel gene expression patterns in hybrid embryos between species with different modes of development. Evol Dev 2: 133–144. [DOI] [PubMed] [Google Scholar]
- Oliveri P., Carrick D.M., Davidson E.H., 2002. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol 246: 209–228. [DOI] [PubMed] [Google Scholar]
- Oliveri P., Davidson E.H., 2004. Gene regulatory network analysis in sea urchin embryos. Methods Cell Biol 74: 775–794. [DOI] [PubMed] [Google Scholar]
- Oliveri P., Tu Q., Davidson E.H., 2008. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA 105: 5955–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N., Swartz S.Z., Wang L., Wikramanayake A., Wessel G.M., 2019. Distinct transcriptional regulation of Nanos2 in the germ line and soma by the Wnt and delta/notch pathways. Dev Biol 452: 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Parr B.A., Chin J.-E., Leaf D.S., Raff R.A., 1988. Molecular analysis of heterochronic changes in the evolution of direct developing sea urchins. J Evol Biol 1: 27–44. [Google Scholar]
- Pehrson J.R., Cohen L.H., 1986. The fate of the small micromeres in sea urchin development. Dev Biol 113: 522–526. [DOI] [PubMed] [Google Scholar]
- Peter I.S., Davdison E.H. 2011. A gene regulatory network controlling the embryonic specification of endoderm. Nature 474:635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff R.A., Byrne M., 2006. The active evolutionary lives of echinoderm larvae. Heredity 97: 244–252. [DOI] [PubMed] [Google Scholar]
- Rafiq K., Cheers M.S., Ettensohn C.A., 2012. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development 139: 579–590. [DOI] [PubMed] [Google Scholar]
- Rafiq K., Shashikant T., McManus C.J., Ettensohn C.A., 2014. Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins. Development 141: 950–961. [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Patel N.H., Hinman V.F., 2015. Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development. Ann Rev Genomics Hum Genet 16: 103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-i-Domingo R., Oliveri P., Davidson E.H., 2007. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA 104: 12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F., Fernandez-Serra M., Squarzoni P., Archimandritis A., Arnone M.I., 2006. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev Biol 300: 35–48. [DOI] [PubMed] [Google Scholar]
- Ruffins S.W., Ettensohn C.A., 1996. A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus) mesenchyme blastula. Development 122: 253–263. [DOI] [PubMed] [Google Scholar]
- Saudemont A., Haillot E., Mekpoh F., Bessodes N., Quirin M., Lapraz F., Duboc V., Rottinger E., Range R., Oisel A., Besnardeau L., Wincker P., Lepage T., 2010. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet 6: e1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders L.R., McClay D.R., 2014. Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development 141: 1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebinger G., Shu J., Tabaka M., Cleary B., Subramanian V., Solomon A., Gould J., Liu S., Lin S., Berube P., Lee L., Chen J., Brumbaugh J., Rigollet P., Hochedlinger K., Jaenisch R., Regev A., Lander E.S., 2019. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176: 928–943 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A.J., Angerer R.C., Angerer L.M. 2009. Gene regulatory network interactions in sea urchin endomesoderm induction. PLoS Biol 7: e1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T., Ettensohn C.A., 2010. Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development 137: 1149–1157. [DOI] [PubMed] [Google Scholar]
- Sherwood D.R., McClay D.R., 1999. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 126: 1703–1713. [DOI] [PubMed] [Google Scholar]
- Strathmann R. R. 1985. Feeding and nonfeeding larval development and life-history evolution in marine invertebrates. Ann Rev Ecol Syst 16: 339–361. [Google Scholar]
- Sweet H.C., Gehring M., Ettensohn C.A., 2002. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development 129: 1945–1955. [DOI] [PubMed] [Google Scholar]
- Tarashansky A.J., Musser J.M., Khariton M., Li P., Arendt D., Quake S.R., Wang B., 2021. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Petsios E., Davidson E.H., Erkenbrack E.M., Gao F., Bottjer D.J., 2015. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci Rep 5: 15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorson G. 1950. Reproductive and larval ecology of marine bottom invertebrates. Biol Rev 25: 1–45. [DOI] [PubMed] [Google Scholar]
- Wahl M.E., Hahn J., Gora K., Davidson E.H., Oliveri P., 2009. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev Biol 335: 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun H., Jiang M., Li J., Zhang P., Chen H., Mei Y., Fei L., Lai S., Han X., Song X., Xu S., Chen M., Ouyang H., Zhang D., Yuan G.C., Guo G., 2021. Tracing cell-type evolution by cross-species comparison of cell atlases. Cell Rep 34: 108803. [DOI] [PubMed] [Google Scholar]
- Wang L., Israel J.W., Edgar A., Raff R.A., Raff E.C., Byrne M., Wray G.A., 2020. Genetic basis for divergence in developmental gene expression in two closely related sea urchins. Nat Ecol Evol 4, 831–840. [DOI] [PubMed] [Google Scholar]
- Wikramanayake A.H., Peterson R., Chen J., Huang L., Bince J.M., McClay D.R., Klein W.H., 2004. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis 39: 194–205. [DOI] [PubMed] [Google Scholar]
- Williams D.H.C., Anderson D.T., 1975. The reproductive system, embryonic development, larval development and metamorphosis of the sea urchin Heliocidaris erythrogramma (Val.) (Echinoidea : Echinometridae). Austr J Zoology 23: 371–403 [Google Scholar]
- Wilson K.A., Andrews M.E., Raff R.A., 2005. Dissociation of expression patterns of homeodomain transcription factors in the evolution of developmental mode in the sea urchins Heliocidaris tuberculata and H. erythrogramma. Evol Dev 7: 401–415. [DOI] [PubMed] [Google Scholar]
- Wilson K.A., Andrews M.E., Rudolf Turner F., Raff R.A., 2005. Major regulatory factors in the evolution of development: the roles of goosecoid and Msx in the evolution of the direct-developing sea urchin Heliocidaris erythrogramma. Evol Dev 7: 416–428. [DOI] [PubMed] [Google Scholar]
- Woych J., Ortega Gurrola A., Deryckere A., Jaeger E.C.B., Gumit E., Merello G., Gu J., Joven Araus A., Leigh N.D., Yun M., Simon A., Tosches M.A., 2022. Cell-type profiling in salamanders identifies innovations in vertebrate forebrain evolution. Science 377: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G., 1996. Parallel evolution of nonfeeding larvae in echinoids. Syst Biol 45: 15. [Google Scholar]
- Wray G.A., 2022. Extreme phenotypic divergence and the evolution of development. Curr Top Dev Biol 146: 79–112. [DOI] [PubMed] [Google Scholar]
- Wray G.A., Raff R.A., 1989. Evolutionary modification of cell lineage in the direct-developing sea urchin Heliocidaris erythrogramma. Dev Biol 132: 458–470. [DOI] [PubMed] [Google Scholar]
- Wray G.A., Raff R.A., 1990. Novel origins of lineage founder cells in the direct-developing sea urchin Heliocidaris erythrogramma. Dev Biol 141: 41–54. [DOI] [PubMed] [Google Scholar]
- Yamazaki A., Minokawa T., 2015. Expession patterns of mesenchyme specification genes in two distantly related echinoids, Glyptocidaris crenularis and Echinocardium cordatum. Gene Expr Patterns 17: 87–97. [DOI] [PubMed] [Google Scholar]
- Yamazaki A., Yamakawa S., Morino Y., Sasakura Y., Wada H., 2021. Gene regulation of adult skeletogenesis in starfish and modifications during gene network co-option. Sci Rep 11: 20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh C.H., Bolouri H., Davidson E.H., 1998. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279: 1896–1902. [DOI] [PubMed] [Google Scholar]
- Yuh C.H., Dorman E.R., Davidson E.H., 2005. Brn1/2/4, the predicted midgut regulator of the endo16 gene of the sea urchin embryo. Dev Biol 281: 286–298. [DOI] [PubMed] [Google Scholar]
- Zigler K.S., Raff E.C., Popodi E., Raff R.A., Lessios H.A., 2003. Adaptive evolution of binding in the genus Heliocidaris is correlated with the shift to direct development. Evolution 57: 2293–2302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Metrics of scRNA-seq transcriptomes over development in He. Violin plots showing the frequency distribution of four informative metrics. A. Distinct genes/cell from which transcripts were detected (nFeature_RNA in Seurat). The modest decline over development likely reflects the transition of a more restricted transcriptome during differentiation. B. Distinct UMIs recovered per cell (nCount_RNA in Seurat). C. Percentage of transcripts mapping to genes encoding ribosomal proteins. The increase over development reflects the maternal-to-zygotic transition in protein expression. D. Percentage of transcripts mapping to genes encoding cell cycle control proteins. The slight decrease in the last two stages reflects the slowing of cell division as differentiation commences in the early larva.
Figure S2. Expression of marker genes in He. Density plots showing the distribution in UMAP space of cells expressing marker genes for specific embryonic territories and larval cell types. See Massri et al. 2021 for supporting literature.
Figure S3. Expression of marker genes for ectodermal territories in Lv and He. Density plots showing the distribution in UMAP space of cells expressing marker genes for specific embryonic territories and larval cell types. See Massri et al. 2021 for supporting literature.
Figure S4. Overall similarity of transcriptomes in Lv and He over development. Heatmap showing overall similarity of expression of 1:1 orthologues among all developmental stages examined. Similarity measured using CIDER (Hu et al. 2021).
Figure S5. Evolutionary changes in timing of differentiation, additional examples. Optimal transport was used to predict the likely fate for each cell at five stages, based on transcriptomes at 24 hpf (see Methods). The organization of this figure parallels that of main text Figure 4. Triangle plots show transcriptomes predictive of two specific cell types. A. Coelom (red) and endoderm (yellow). B. Blastocoelar cells (purple) and endoderm (yellow). C. Blastocoelar cells (purple) and pigment cells (pink). Any other cell fate is indicated in dark gray; cells with undifferentiated transcriptomes are shown in blue. See text for interpretation.
Figure S6. Logic underlying inference of distinct types of evolution change in regulatory interactions. On the extreme left, hypothetical data from Lv illustrating an experimentally validated regulatory interaction: transcriptional activator, gene A (blue) and its target, gene B (purple), are co-expressed in the same cells. Four possible scenarios in He for the same pair of genes are shown to the right, with interpretations regarding the regulatory interaction shown below. Scenario 1 is consistent with conservation. Scenario 2 is consistent with conservation, but with a delay in the timing of the interaction. Scenario 3 implies that the interaction no longer occurs and that some other transcription factor must activate expression of gene B. Scenario 4 is consistent with conservation, but additionally implies that some other transcription factor must operate earlier to initiate expression of gene B.
Figure S7. Co-expression of alx1 and several of its interactors within the skeletogenic dGRN. See caption to Figure 7.
Figure S8. Co-expression of additional regulators and targets within the skeletogenic dGRN. See caption to Figure 7.
Figure S9. Comparison of regulator expression and co-expression with a specific target gene. Five validated transcription factor → target gene interactions are illustrated. Left panels show density plots of expression for the regulator while right panels show cells expressing both the regulator and its target. Note that co-expression domains are often a subset of the cells expressing the regulator.
Figure S10. Co-expression of several regulators and targets in non-skeletogenic territories of the dGRN. See caption to Figure 7.
Table S1. Cell counts over development in Lv.
Table S2. Cell counts over development in He.