Abstract
Autism spectrum disorders (ASDs) are characterized by social, communication, and behavioral challenges. UBE3A is one of the most common ASD genes. ASDs display a remarkable sex difference with a 4:1 male to female prevalence ratio; however, the underlying mechanism remains largely unknown. Using the UBE3A-overexpressing mouse model for ASD, we studied sex differences at behavioral, genetic, and molecular levels. We found that male mice with extra copies of Ube3A exhibited greater impairments in social interaction, repetitive self-grooming behavior, memory, and pain sensitivity, whereas female mice with UBE3A overexpression displayed greater olfactory defects. Social communication was impaired in both sexes, with males making more calls and females preferring complex syllables. At the molecular level, androgen receptor (AR) levels were reduced in both sexes due to enhanced degradation mediated by UBE3A. However, AR reduction significantly dysregulated AR target genes only in male, not female, UBE3A-overexpressing mice. Importantly, restoring AR levels in the brain effectively normalized the expression of AR target genes, and rescued the deficits in social preference, grooming behavior, and memory in male UBE3A-overexpressing mice, without affecting females. These findings suggest that AR and its signaling cascade play an essential role in mediating the sexually dimorphic changes in UBE3A-dependent ASD.
Introduction
Autism Spectrum Disorder (ASD) is a neurodevelopmental disorder characterized by impairments in sociability and communication, as well as the presence of restricted and repetitive behaviors 1. According to reports from the Centers for Disease Control and Prevention (CDC) of the USA, the prevalence of ASD has been increasing over the past decades. The global prevalence of the disease is estimated at 1%, and approximately 1 in 36 children have been diagnosed with ASD in the United States 2. However, the cause of ASD remains less clear despite the identification of various risk factors. Environmental risk factors include parental age, maternal pregnancy, and pregnancy complications 3. Population screening studies have shown that genetics play a significant role in the development of the disease, supported by a heritability estimated at 90% in ASD 4. Recent research has discovered hundreds of genetic variations, both common and rare, that contribute to the development of ASD 5.
Remarkably, ASD has a four-fold higher occurrence in males than in females 6. To this day, the exact mechanisms and genetic determinants underlying the sex-bias in autism are still largely unknown. One possibility is related to male-biased risk factors, such as the influence of sex steroid hormone signaling and prenatal programming on brain development, while another possibility pertains to a female-biased protective mechanism that results in a higher mutation “burden” for ASD risk in females. However, the etiology of these possibilities is not well-understood 7–9. In addition, different phenotypic presentations of ASD in males and females have been reported in human patients. Males with ASD tend to exhibit more externalizing behaviors such as aggression, hyperactivity, reduced social behavior, and increased restricted and repetitive behaviors 7,9–12, whereas females with ASD show greater internalizing symptoms such as anxiety, depression, and other emotional problems 7,12,13. Interestingly, sex chromosomes, sex hormone signaling pathways, and sexually dimorphic factors such as differential gene expression in neurons and glial cells, have all been proposed as potential causes of these sex differences 7,9. Despite these possibilities, however, the exact mechanisms underlying sexually dimorphic expression of autistic phenotypes remain unclear.
UBE3A is a significant ASD risk gene located on chromosome 15q11–13. The protein product of UBE3A functions as a HECT family E3 ligase regulating protein ubiquitination and degradation, as well as a transcription coactivator regulating gene expression 14. The UBE3A gene is imprinted in the brain, with only the maternal copy of the gene being expressed in neurons, whereas the paternal copy is silenced 14. The UBE3A gene plays essential roles in brain development, and it is well established that deletions of the UBE3A gene result in Angelman syndrome 15. In contrast, excessive UBE3A expression and activity resulting from duplication or triplication of maternal chromosome 15q11–13 14,16, or de novo hyperactivating missense mutations of UBE3A 17–19, are implicated in the development of ASD. The 15q11.2 microduplication encompassing only the UBE3A gene has been reported to cause learning disability and autistic features in human patients, highlighting the significance of the UBE3A gene in ASD development 20. ASD diagnoses due to genetic alterations in UBE3A account for 1–2% of all autism cases, making it one of the strongest genetic risk factors for autism 21,22.
Ube3A 2x transgenic (Tg) mice carry a tripled Ube3A gene dosage that mimics the disease condition, and exhibit typical autistic-like behaviors such as impaired social behavior and communication, as well as increased repetitive behavior 23,24. Using the Ube3A 2xTg ASD mice, we examined whether sexually dimorphic phenotypes manifest in Ube3A-dependent ASD and investigated the potential mechanisms that underlie the observed sex differences in this disorder. To summarize, we observed male-biased behavioral abnormalities in social interaction, repetitive self-grooming behavior, memory function, and pain sensitivity, which were associated with sexually dimorphic changes in the androgen receptor (AR) signaling pathway. We found that AR was decreased in 2xTg male mice due to enhanced degradation, and more importantly, restoration of AR expression in the brain was able to rescue a large portion of the behavioral deficits in male 2xTg mice.
Results
Sexually dimorphic behavioral impairments in Ube3A 2xTg ASD mice
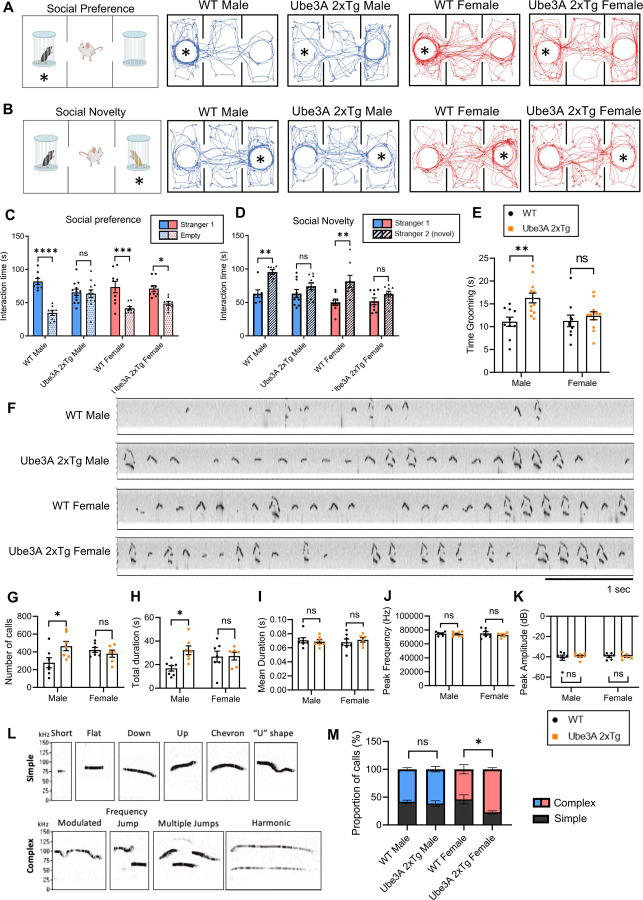
To investigate whether sexually dimorphic changes are present in UBE3A-dependent ASD, we examined the three core autism-relevant behaviors, including impaired social interaction, abnormal social communication, and the presence of restricted and repetitive behaviors in male and female Ube3A 2xTg mice. To examine their social behaviors, we used the three-chamber social test, in which a stranger mouse was placed into a cage at either side chamber and a test mouse of the same sex was allowed to move freely in the apparatus to examine its social preference (Fig. 1A). The wild type (WT) male and female mice displayed normal preference to interact with the stranger mice, whereas the 2xTg male mice lost this preference and spent a similar amount of time interacting with the two cages (Fig. 1A, 1C). Surprisingly, female Tg mice preserved the preference to the stranger mouse (Fig. 1A, 1C), indicating social disruptions in Tg male, but not Tg female, mice. We then introduced a second mouse of the same sex (novel mouse) into a cage on the remaining side chamber, and the test mouse was allowed to interact with both mice (Fig. 1B). Quantification of the interaction time in WT animals showed a normal preference to the novel mouse as expected, but both male and female Tg mice spent a similar amount of time interacting with the two conspecifics, indicating that social novelty was impaired in Tg animals regardless of sex (Fig. 1B, 1D). Furthermore, to assess repetitive self-grooming behavior, each test mouse was placed individually into a regular housing cage, and the amount of time spent grooming was quantified. Elevated grooming time was observed in male but not female Ube3A 2xTg mice, suggesting that male Tg mice are preferentially affected in their repetitive grooming behaviors (Fig. 1E).
Fig. 1: Sexually dimorphic deficits in social interaction, repetitive self-grooming behaviors, and ultrasonic vocalizations in male and female Ube3A 2xTg mice.
(A,B) The paradigm for the three-chamber social test and traces of track paths. For social preference (A), an unfamiliar mouse was placed into either of the side chambers and the test mouse was allowed to move freely in the apparatus. For social novelty (B), a second mouse (novel mouse) was placed into the remaining empty chamber, and the test mouse was allowed to interact with both mice.
(C,D) Quantification of the interaction time showed a decrease in preference for the stranger mouse (C) and novel mouse (D) in male Ube3A 2xTg mice, while female Ube3A 2xTg mice showed mild change in social preference (C) but clear impairment in social novelty (D).
(E) Male but not female Ube3A 2xTg mice showed increased repetitive self-grooming behavior.
(F) Representative vocalization recordings from P5 WT and Ube3A 2xTg mice.
(G-K) Quantification of the number of calls (G), total call duration (H), mean call syllable duration (I), peak frequency (J), and peak amplitude (K) for P5 WT and Ube3A 2xTg mice.
(L) Representative calls of each type used in syllable characterization.
(M) Ube3A 2xTg female animals showed an increase in Complex type calls and a decrease in Simple type calls at P5, while Ube3A 2xTg males are similar to WT males.
In (C), n=8 WT male; n=12 Tg male; n=9 WT female; n=9 Tg female. In (D), n=7 WT male; n=10 Tg male; n=8 WT female; n=9 Tg female. In (E), n=10 WT male; n=12 Tg male; n=10 WT female; n=12 Tg female. In (G-M) n=7 WT male; n=7 Tg male; n=7 WT female; n=6 Tg female. Mean ± SEM. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant. In (C), (D), (M) Three-way ANOVA with Bonferroni’s multiple comparisons test; In (E), (G-K) Two-way ANOVA with Bonferroni’s multiple comparisons test.
Impairments in social communication have been established as one of the three main ASD hallmarks. To examine the presence of communication deficits in male and female Ube3A 2xTg animals, we recorded and analyzed their ultrasonic vocalizations (USVs). Individual pups isolated from the mother and littermates at P5, P7 and P9 were placed in a recording chamber for 5 min in which a microphone was present to record vocalization signals (Fig. 1F). At P5, but not at P7 and P9, we observed an increase in the number of calls and the total call duration in Ube3A 2xTg males compared to WT males (Fig. 1G, 1H, S4A, S4B), a trend also noted in other ASD models 25–29. Conversely, no change was observed in the number of calls and total call duration in Ube3A 2xTg females at P5, P7 and P9 (Fig. 1G, 1H, S4A, S4B). Furthermore, both male and female Tg animals displayed a normal mean call duration, peak frequency of calls, and peak amplitude of calls at P5, P7 and P9 (Fig. 1I-1K, S4C-E). Further analysis of the call syllable types (Fig. 1L) revealed that at P5, female Ube3A 2xTg animals made more complex USV calls than WT females (Fig. 1M), characterized by a significant increase in harmonic calls and a decrease in chevron calls (Fig. S1B). In contrast, male Tg mice used call syllable types similar to WT males at P5 (Fig. 1M, S1A). At P7 and P9, both male and female Tg mice exhibited call syllable types comparable to those of their WT counterparts (Fig. S4F, S4G). These findings suggest that UBE3A over-dosage results in altered communication in both male and female animals at P5, but in different manners, with male Tg mice increasing their number of calls and total call duration, while female Tg mice using more complex syllables.
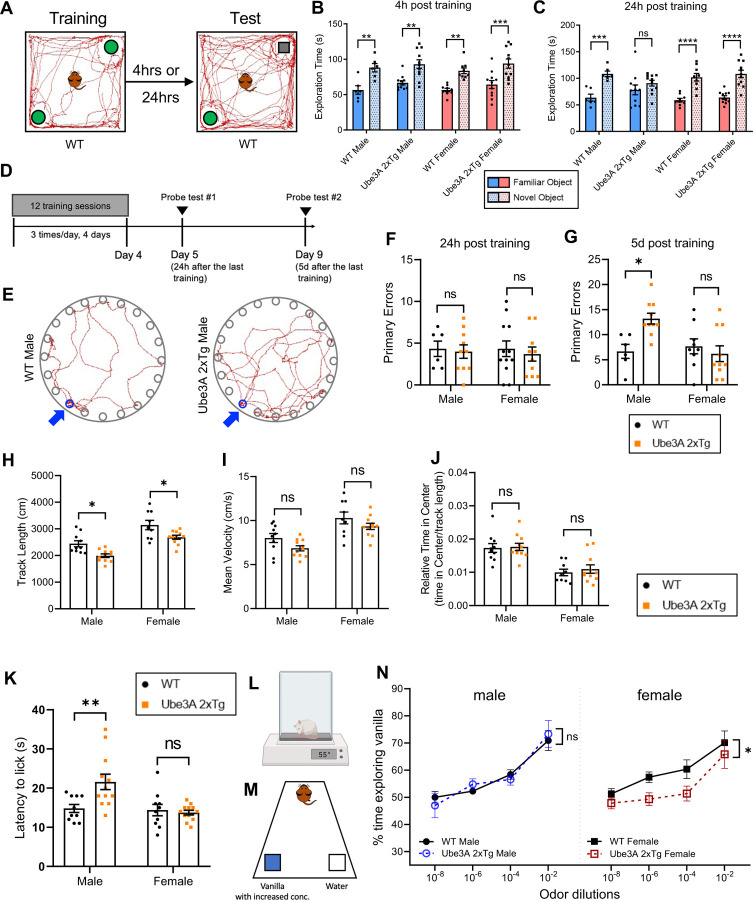
Intellectual and cognitive abnormalities are one of the most common comorbidities seen in human ASD 30,31. We therefore sought to examine whether memory deficits are differentially affected in male and female Ube3A 2xTg animals using the Novel Object Recognition (NOR) test and Barnes maze test. For the NOR test, two identical objects were introduced to the test mouse, and 4 hours or 24 hours later, one of the familiarized objects was replaced with a novel object (Fig. 2A). The Tg animals showed normal short-term memory 4 h after training, regardless of sex (Fig. 2B). However, while Tg females displayed intact recognition capabilities after 24 h, Tg males failed to recognize the familiar object, indicating that long-term memory was impaired specifically in Ube3A 2xTg males (Fig. 2C). In the Barnes maze test for spatial memory, mice were trained over a period of 4 days to learn the location of a target escape hole on a circular board containing 20 holes along its circumference, with spatial cues placed around the testing room to help animals navigate and locate the target hole (Fig. 2D, 2E). 24 hours and 5 days after training sessions, the test mouse was returned to the board with the target hole covered, and the number of errors made before reaching the escape hole was quantified (Fig. 2D, 2E). In the 24 h probe test, neither male nor female Tg animals showed significant differences in the number of errors as compared to WT mice (Fig. 2F). However, probing long-term memory retention 5 d after training showed that male, but not female, Tg mice made significantly more primary errors than their WT counterparts (Fig. 2G), Together with the results from the NOR test, these findings indicate that Ube3A 2xTg males are preferentially affected in long-term memory in contrast to Ube3A 2xTg females.
Fig. 2: Sexually dimorphic deficits in memory, sensory functions, but not locomotion in male and female Ube3A 2xTg mice.
(A) The paradigm for novel object recognition test.
(B,C) Exploration time of familiar object vs. novel object 4 h (B) and 24 h (C) post training. Ube3A 2xTg animals displayed intact short time memory (B), and only Tg males showed impaired long-term memory (C).
(D) The timeline of Barnes spatial memory maze.
(E) Traces of track paths during the 5 d memory probe for WT and Ube3A 2xTg males. Blue arrows point to the escape hole.
(F,G) Primary errors made to find the escape hole 24 h (F) and 5 d (G) after the last training. Ube3A 2xTg animals displayed intact short time memory (F), and only Tg males showed impaired long-term memory (G). (H-J) Both male and female Ube3A 2xTg mice showed decreased track lengths (H), normal mean velocities (I) and no change in relative time in center (J) in the open-field test.
(K,L) Test mice were put on 55℃ hot plate and their latency to lick a hindpaw was recorded. Male Ube3A 2xTg mice showed reduced pain sensitivity compared to WT males, while Tg females were intact.
(M) The paradigm for the Olfactory Detection Threshold Test. Test mice were exposed to increased concentration of vanilla. Time spent exploring vanilla vs. water was recorded.
(N) Male Ube3A 2xTg mice showed intact olfactory sensitivity compared to WT males, whereas Tg females showed reduced olfactory sensitivity compared to WT females.
In (B-C), n=6 WT male; n=11 Tg male; n=9 WT female; n=11 Tg female. In (F), n=6 WT male; n=10 Tg male; n=12 WT female; n=10 Tg female. In (G), n=6 WT male; n=10 Tg male; n=9 WT female; n=10 Tg female. In (H-J), n=10 WT male; n=11 Tg male; n=9 WT female; n=11 Tg female. In (K), n=10 WT male; n=12 Tg male; n=10 WT female; n=12 Tg female. In (N), n=10 WT male; n=10 Tg male; n=9 WT female; n=10 Tg female. Mean ± SEM. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant. In (B), (C), (N) Three-way ANOVA with Bonferroni’s multiple comparisons test; In (F-K) Two-way ANOVA with Bonferroni’s multiple comparisons test.
Next, in the Open-field test, although the 2xTg animals displayed normal locomotion, both sexes showed a significant decrease in their total track length (Fig. 2H). Quantification of the average velocity and the time spent in the center of the open field normalized to the total distance traveled showed no difference between WT and Tg mice within each sex (Fig. 2I, 2J). However, in both genotypes, female mice generally exhibited a longer track length, higher mean velocity, and spent less relative time in the center compared to male mice (Fig. 2H-2J). Together, these results indicate that Ube3A 2xTg animals are less active in general, without signs of enhanced anxiety.
In addition, atypical sensory functioning has often been found in ASD patients and animal models, such as impaired peripheral somatosensory functions 32–34 and odor perception 35,36. We first performed the hot plate assay to evaluate the pain sensitivity of Ube3A 2xTg mice by measuring the latency to lick their hind paw after being placed on a surface heated to 55 °C (Fig. 2L). Male Tg mice displayed reduced sensitivity to the hot temperature, whereas female Tg had intact pain sensitivity (Fig. 2K). We then examined their olfactory sensitivity using the olfactory detection threshold test, in which the mice were exposed to increased concentrations of vanilla, and the time spent exploring the vanilla odor versus the water control was recorded (Fig. 2M). We observed normal detection of the vanilla odor in 2xTg male mice, but the female Tg animals were unable to perceive the odor until the highest concentration was applied (Fig. 2N), indicating a reduced olfactory sensitivity specific to female, but not male, 2xTg animals.
The androgen receptor signaling pathway is dysregulated mainly in male Ube3A 2xTg ASD mice
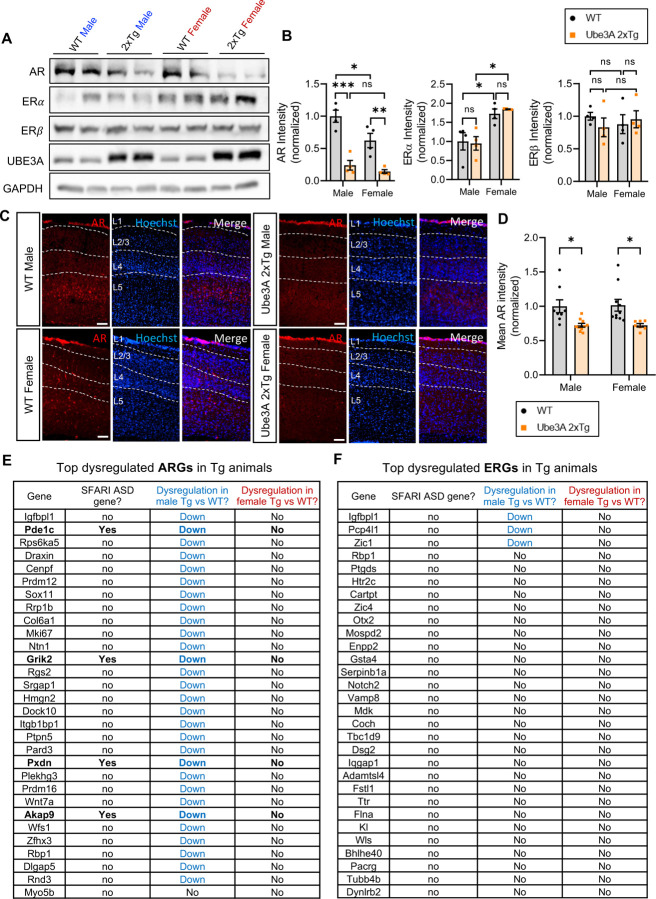
Sex hormones act throughout the entire brain of both males and females, regulating many neural and behavioral functions 37–40. Sex hormones bind to and activate sex hormone steroid receptors, either androgen receptors (AR) or estrogen receptors (ER), and the complexes will then translocate into the nucleus to regulate gene expression 37,41. Importantly, the sex differences observed across several brain functions are mediated by sex hormone signaling pathways 37. Both AR and ER are known to be regulated by E3 ubiquitin ligases, which target them for ubiquitination and degradation 42–45. Given that UBE3A functions as an E3 ligase, it is intriguing to consider whether UBE3A regulates the turnover of AR and ER, which may contribute to the sexually dimorphic changes in Ube3A 2xTg animals. To this end, we examined the expression of AR, and ER alpha (ERα) and beta (ERβ) in prefrontal cortical lysates of 2xTg brains by western blotting. Surprisingly, quantification analysis revealed decreased expression of AR in both male and female Ube3A 2xTg animals, whereas ERα and ERβ expression were largely intact compared to their WT counterparts, regardless of sex (Fig. 3A, 3B). Consistently, by immunostaining of AR in cortical brain slices, we observed a reduction of AR immunointensity in both male and female 2xTg brains (Fig. 3C, 3D). Given that at basal conditions, AR activity is significantly lower in female brains due to low androgen hormone levels compared to males, we predicted that AR reduction should affect downstream gene expression mainly in males 46,47. To test the idea, we performed RNAseq assays using 2xTg mouse brain tissues and analyzed the androgen-regulated genes (ARGs) and estrogen-regulated genes (ERGs) in comparison with their WT counterparts. As expected, ARGs were found to be markedly downregulated in male Ube3A 2xTg brains, but not in female 2xTg brains (Fig. 3E), indicating a minimal downstream disturbance caused by AR reduction in female 2xTg brains. On the other hand, little to no ERGs were dysregulated in Tg animals, regardless of sex (Fig. 3F). These findings suggest that the AR signaling pathway is disrupted in Ube3A 2xTg animals, which leads to strong genetic disturbances specifically in males.
Fig. 3: Androgen receptor expression is reduced in Ube3A 2xTg animals, leading to dysregulation of androgen responsive genes specifically in males.
(A,B) Androgen receptors (AR) were reduced in both Tg male and Tg female prefrontal cortical lysates at P7. Estrogen receptors alpha (ERα) and beta (ERβ) didn’t change in Ube3A 2xTg animals compared to WT counterparts. n=4 animals/group.
(C,D) Immunohistochemistry of WT and Tg cortical slices at P60 showed a marked decrease in AR immunointensity in Tg animals. n = 8 WT male; n = 10 Tg male; n = 10 WT female; n = 7 Tg female. Scale bars, 100 μm.
(E,F) Top dysregulated androgen responsive genes (ARGs, E) and estrogen responsive genes (ERGs, F) in Ube3A 2xTg male and female brains, compared to their WT counterparts (n=3 animals/group). Blue fonts indicate dysregulation in male Ube3A 2xTg brains compared to WT males. Red fonts indicate dysregulation in female Ube3A 2xTg brains compared to WT females. Bold fonts indicate known SFARI ASD genes. ARGs were strongly dysregulated in Tg males but not Tg females. Little to no ERGs were dysregulated in Tg animals. Mean ± SEM. *p<0.05; **p<0.01; ***p<0.001. ns, not significant. Two-way ANOVA with Bonferroni’s multiple comparisons test.
Reduction of the androgen receptor in Tg mice is caused by UBE3A-mediated ubiquitination and degradation
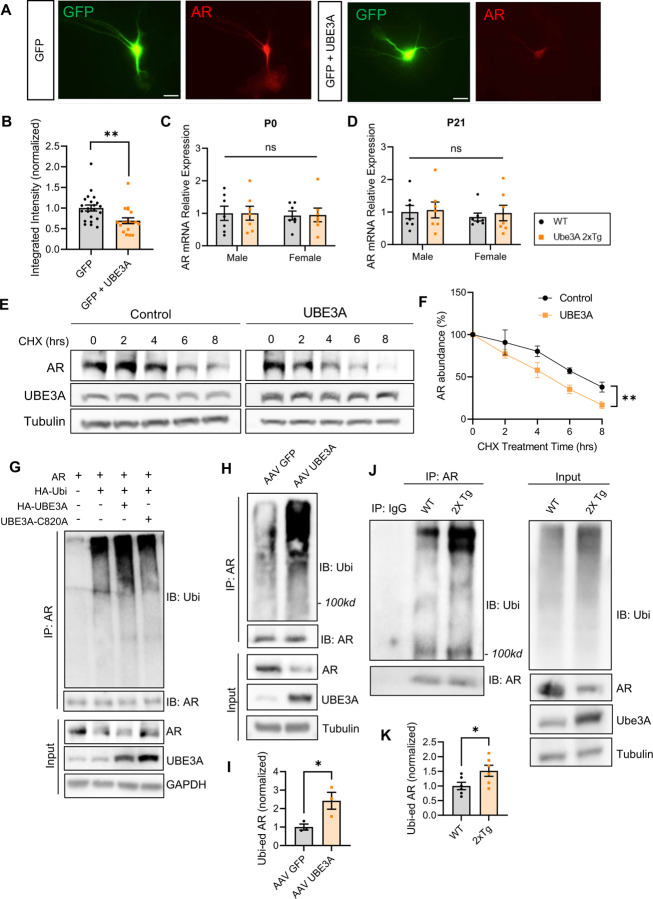
To verify whether AR reduction is dependent on an elevated dosage of UBE3A, we transfected primary cortical neurons with UBE3A and analyzed AR expression via immunocytochemistry (ICC) at DIV 8 (Fig. 4A). We found that the AR level decreased significantly in neurons overexpressing UBE3A as compared to the control (Fig. 4B), indicating that UBE3A overexpression causes AR reduction.
Fig. 4: UBE3A overexpression can independently induce AR reduction by increasing ubiquitination and degradation of AR.
(A) Representative images of immunocytochemistry for AR (red) at DIV 8 in rat cortical neuron cultures after transfected with GFP alone or together with UBE3A at DIV4. Scale bars, 25 μm.
(B) UBE3A overexpression reduced AR intensity in primary culture neurons. GFP: n=22, GFP+UBE3A: n=19. (C,D) AR mRNA expression detected by qPCR was not altered in Tg males and Tg females at postnatal day 0 (C) and postnatal day 21 (D). P0: n=7 animals/group; P21: n=7 animals/group.
(E) Degradation assay of AR with or without UBE3A. Transfected HEK cells were treated with cycloheximide (CHX) for various time points and cell lysates were collected to examine AR levels by Western blot.
(F) Quantification of the degradation rate of AR over time; n=3 independent experiments.
(G) AR ubiquitination assay. HEK293T cells were transfected with AR, HA-ubiquitin (Ubi), and either a vector control, UBE3A, or the E3 ligase dead mutant UBE3A C820A for 2 d. AR was immunoprecipitated and probed for ubiquitin. Cell lysates (input) were also probed to detect total protein levels.
(H,I) AR ubiquitination assays using lysates of neurons infected with AAV2 GFP or AAV2 UBE3A virus for 10 d (H). Increased intensity of ubiquitination signals on AR was detected (I). n=3 independent experiments. (J,K) AR was immunoprecipitated from brain lysates and probed for ubiquitin signals. An elevated level in AR ubiquitination was detected in Ube3A 2xTg mice compared to WT. n=6 animals/group.
Mean ± SEM. *p<0.05; **p<0.01. ns, not significant. In (C), (D), (F) Two-way ANOVA with Bonferroni’s multiple comparisons test; In (B), (I), (K) Unpaired two-tailed t test.
The UBE3A-induced reduction in AR could result from facilitated degradation or altered transcription. To clarify this, we performed real-time PCR (qPCR) on WT and Tg brain lysates. At either postnatal day 0 or postnatal day 21, no significant changes in AR mRNA level were observed in Tg animals, regardless of sex (Fig. 4C, 4D).
To determine whether the degradation of AR is altered by UBE3A, we transfected HEK cells with AR alone or together with UBE3A for 48 h. Cells were then incubated with the protein translation inhibitor cycloheximide for 0, 2, 4, 6, 8 h before being harvested for western blot analysis. We found that while the AR levels decreased over time due to degradation, the rate of decay became faster in cells co-transfected with UBE3A (Fig. 4E, 4F), indicating that UBE3A expression enhanced the degradation of AR.
Given that UBE3A is a ubiquitin protein ligase, the change in AR turnover by UBE3A could result from changes in its ubiquitination status. To test this possibility, we transfected HEK cells with AR, HA-Ubi, together with either HA-UBE3A, or the ligase dead mutant UBE3A C820A, for 2 d. AR was immunoprecipitated and probed for ubiquitin. Indeed, the ubiquitin signals of AR were significantly increased in cells co-transfected with UBE3A as compared to the control (Fig. 4G). In contrast, no changes in AR ubiquitination were observed in cells co-transfected with the ligase dead mutant UBE3A C820A (Fig. 4G). Moreover, a significant decrease in AR was detected in UBE3A-overexpressing cells (Fig. 4G). To further confirm the role of UBE3A on AR ubiquitination, we performed a ubiquitination assay in neurons infected with an AAV2 virus of either GFP or UBE3A for 10 d. Indeed, compared with GFP, the UBE3A virus caused a significant increase in AR ubiquitination (Fig. 4H, 4I). In addition, ubiquitination assays using lysates from mouse brain tissues also showed increased AR ubiquitination in UBE3A over-dosage brains as compared to WT brains (Fig. 4J, 4K). These results suggest that AR is targeted by UBE3A for ubiquitination and degradation.
AR restoration in Ube3A 2xTg brains normalizes the expression of ASD-associated ARGs
Following binding with androgens, the activated ARs will translocate into the nucleus to regulate gene expression. In addition to mediating sex hormone signaling and participating in important brain functions 37,39,40,46,48, AR and many AR-regulated genes are known to be associated with ASD 5,49–61. Indeed, AR itself and at least seven ARGs are listed in the SFARI ASD gene database 5. Additionally, studies have shown that dysregulation of AR is associated with the development of several autistic features, including altered social behaviors 51,62, impaired spatial learning and memory 63,64, and hyperactivity symptoms such as attention deficit hyperactivity disorder (ADHD) 52,65. Therefore, the AR system may play an important role in mediating the sexually dimorphic alterations in Ube3A 2xTg ASD mice.
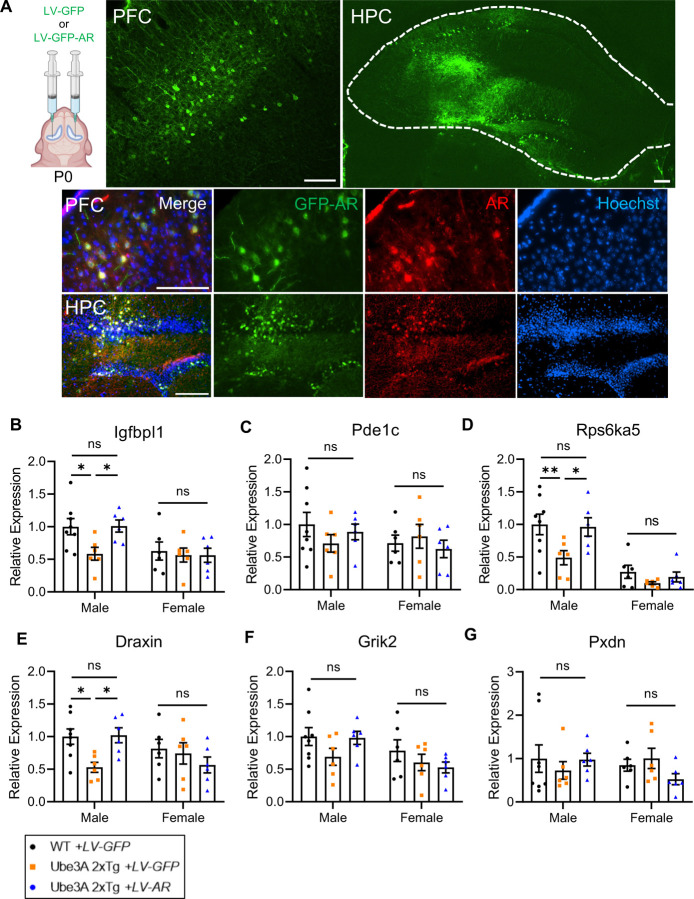
To test this possibility, we performed bilateral intracerebroventricular (ICV) injections of a GFP-AR virus (LV-GFP-AR) at postnatal day 0 to replenish AR in Ube3A 2xTg brains, or a GFP virus (LV-GFP) as a control (2 μl per side) in WT and Tg pups (Fig. 5A). Animals were later subjected to different behavioral tests at postnatal day 30 to 55. The infected animal brains were processed for biochemical analysis at postnatal day 30 to 60. Imaging of brain slices confirmed the expression of GFP-AR and GFP control virus in the prefrontal cortex (PFC) and hippocampus (HPC). Immunohistochemistry (IHC) of AR also showed co-localization of elevated AR immunosignals with GFP-AR virus-infected green cells in the PFC and HPC (Fig. 5A).
Fig. 5: AR restoration in Ube3A 2xTg mouse brains rescues expression of androgen responsive genes in males, without affecting females.
(A) Intracerebroventricular injection of GFP (LV-GFP) or GFP-fused AR (LV-GFP-AR) lentivirus at postnatal day 0. The mice were perfused at postnatal day 30 to 60 for cryostat to validify viral expression. LV-GFP-AR infection significantly increased AR expression in PFC and HPC. PFC, prefrontal cortex. HPC, hippocampus. Scale bars, 100 μm.
(B-G) Expressions of six ASD-associated androgen responsive genes were rescued in Ube3A 2xTg males by ICV injection of LV-GFP-AR at postnatal day 0, detected by qPCR in postnatal day 40 prefrontal cortical lysates. WT male+LV-GFP: n=8, Tg male+LV-GFP: n=6, Tg male+LV-GFP-AR: n=6, WT female+LV-GFP: n=6, Tg female+LV-GFP: n=6, Tg female+LV-GFP-AR: n=6. Mean ± SEM. *p<0.05; **p<0.01; ns, not significant. Two-way ANOVA with Bonferroni’s multiple comparisons test.
We next wanted to determine whether AR restoration could rescue the dysregulation of ARGs in Ube3A 2xTg males. To test this idea, we selected 6 candidates out of the top dysregulated ARGs in male Ube3A 2xTg brains (Fig. 3E) which are known to be associated with ASD 5,53–61, and examined whether their expression was rescued by LV-GFP-AR.
Of the 6 ARG candidates, dysregulated candidate lgfbpl1 encodes insulin-like growth factor-binding protein like 1, which acts as a PI3K-AKT pathway inhibitor 66. Upregulation of this pathway is involved in the pathogenesis of autism, and has become a common symptom in human patients as well as a focal point in autism studies 53,54,67. Inherited missense variants and intronic single nucleotide polymorphisms (SNP) within the second candidate gene, Pde1c, have been associated with ASD 55–57. The Rps6ka5 gene is located on 14q32.11, and its microdeletion has been found to cause developmental and language delay in human patients58. Draxin is involved in the regulation of axon extension, and several human neurological disorders including intellectual disability and ASD are known to be associated with aberrant axon development 59. The last two dysregulated candidate ARGs are Grik2, whose mutations have also been associated with ASD 60,68,69, and Pxdn, duplication of which has been observed in human autism patients 61.
Real-time PCR quantification of mRNA from prefrontal cortical tissue of virus-infected animals revealed that the expression of all candidate genes trended towards reduction in male, but not female, Ube3A 2xTg brains (Fig. 5B-5G). Among these genes, expression of lgfbpl1, Rps6ka5 and Draxin was significantly decreased in Tg males (Fig. 5B, 5D, 5E). Pde1c, Grik2 and Pxdn exhibited a trend of reduced expression, although the changes were not statistically significant (Fig. 5F, 5G). More importantly, ICV injection of LV-GFP-AR in Tg males effectively rescued the expression of these candidate genes to a level comparable to that of WT males (Fig. 5B-5G).
Together, these results showed that AR restoration rescued the expression of ASD-associated ARGs, suggesting that the altered transcriptional activities of AR in the brain may be the key molecular event leading to the male-biased behavioral deficits.
AR reduction is responsible for male-biased impairments in social preference, repetitive self-grooming and memory
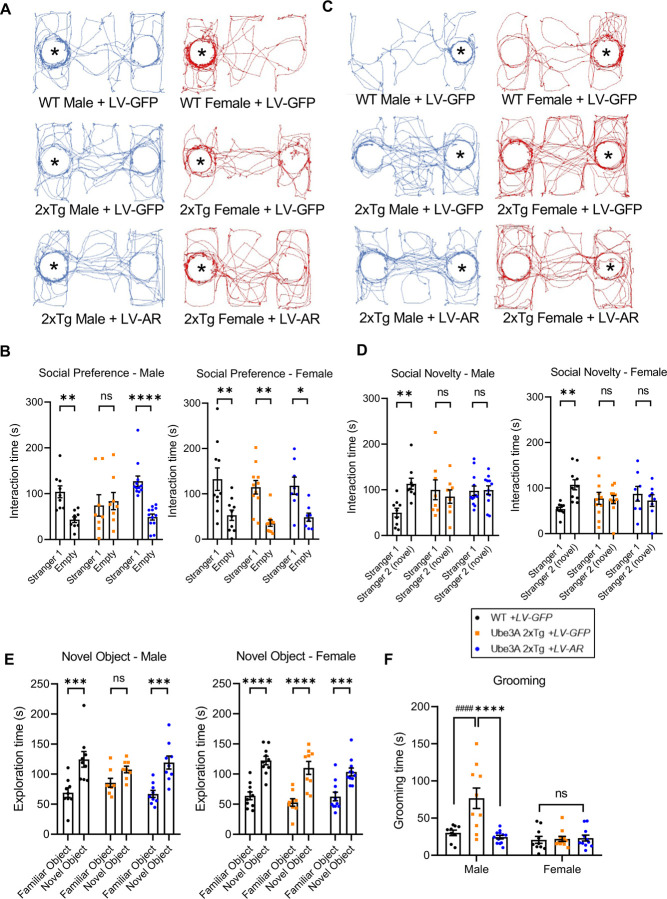
Previous research has established a link between dysregulation of AR and changes in multiple autistic features, such as social behaviors 51,62, learning and memory 63,64, and hyperactivity symptoms 52,65. Therefore, it is intriguing to hypothesize that AR plays a crucial role in mediating the sexually dimorphic autism-relevant behaviors observed in Ube3A 2xTg mice. We first sought to determine whether AR restoration by ICV injection of LV-GFP-AR at postnatal day 0 can rescue the impaired social behaviors in Ube3A 2xTg mice. In the three-chamber sociability test, when testing social preference by examining interaction time with a stranger mouse versus an empty cage, impaired preference was found only in Tg males not Tg females (Fig. 1C). Similarly, Tg males with LV-GFP injection lost the preference to the stranger mouse compared to WT males with LV-GFP, whereas Tg females with LV-GFP showed normal social preference as compared to WT females with LV-GFP (Fig. 6A, 6B). Surprisingly, ICV injection of LV-GFP-AR rescued social preference in male Ube3A 2xTg mice, but had minimal effect on the social preference of female Ube3A 2xTg animals (Fig. 6A, 6B). For social novelty, where the test mouse was examined on its preference between the novel and the familiar mouse, LV-GFP-AR failed to rescue the abnormal preference observed in male and female Tg animals (Fig. 6C, 6D). These results indicate that AR reduction plays an essential role in the male-biased deficit in social preference, but does not contribute to deficits in social novelty, which is similarly affected in both male and female Tg animals.
Fig. 6: AR restoration in Ube3A 2xTg mouse brains rescues social preference, memory, and repetitive self-grooming behaviors in males, without affecting females.
(A-D) Social behaviors in mice following ICV injection of LV-GFP-AR at postnatal day 0. Three chamber tests were performed at postnatal day 30 to 50. (A) Traces of track paths for social preference test. (B) LV-GFP-AR infection in Tg male rescued its preference for stranger mouse. Tg female showed normal social preference which was not affected by LV-GFP-AR infection. (C) Traces of track paths for social novelty test. (D) LV-GFP-AR infection failed to rescue impaired social novelty in Tg male and Tg female. WT male+LV-GFP: n=9, Tg male+LV-GFP: n=8, Tg male+LV-GFP-AR: n=12, WT female+LV-GFP: n=10, Tg female+LV-GFP: n=11, Tg female+LV-GFP-AR: n=8.
(E) Long term memory in mice following ICV injection of LV-GFP-AR at postnatal day 0, detected by novel object recognition test at postnatal day 30 to 50. Tg males with LV-GFP-AR displayed better discrimination between the familiar vs. novel object compared to the LV-GFP infected Tg males, indicating successful rescue of memory. Tg female showed normal memory which was not affected by LV-GFP-AR. WT male+LV-GFP: n=9, Tg male+LV-GFP: n=8, Tg male+LV-GFP-AR: n=9, WT female+LV-GFP: n=10, Tg female+LV-GFP: n=9, Tg female+LV-GFP-AR: n=11.
(F) Repetitive self-grooming behavior was impaired only in Tg males but not females. ICV injection of LV-GFP-AR successfully rescued grooming behaviors in Ube3A 2xTg males with no significant effect on Ube3A 2xTg females. WT male+LV-GFP: n=9, Tg male+LV-GFP: n=10, Tg male+LV-GFP-AR: n=12, WT female+LV-GFP: n=10, Tg female+LV-GFP: n=11, Tg female+LV-GFP-AR: n=11.
Mean ± SEM. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant. Two-way ANOVA with Bonferroni’s multiple comparisons test.
We then examined whether AR restoration by LV-GFP-AR could rescue long-term memory by assessing novel object recognition, which was impaired mainly in male Ube3A 2xTg mice (Fig. 2C). Quantification of exploration time displayed successful rescue of memory by LV-GFP-AR in Ube3A 2xTg males, indicated by a significant increase in the time spent exploring the novel object (Fig. 6E). Tg female mice showed normal memory and were not affected by LV-GFP-AR injection (Fig. 6E). Further, repetitive self-grooming behavior was also differentially affected in Ube3A 2xTg animals, with only male Tg mice showing increased grooming (Fig. 1E). Similarly, LV-GFP-AR successfully reversed the grooming behavior back to WT levels, and did not alter the grooming time in Tg females (Fig. 6F).
Together, these results demonstrate that AR restoration in the brain at P0 was able to rescue several male-biased behavioral deficits in Tg animals, without affecting normal behaviors in female Tg mice, indicating that reduced AR signaling in the brain contributes to the expression of autism-relevant behaviors in male mice.
Discussion
As one of the most common genetic factors in ASD etiology, UBE3A overdosage or hyperactivity correlates with ASD in humans and autistic-like behaviors in mouse models 14,16,17,19,23,24. By studying the differential effects of UBE3A overexpression in male and female mice, we found that male-predominant behavioral impairments, including deficits in social preference, memory and repetitive grooming behaviors, are associated with the downregulation of androgen receptors. Our findings demonstrate the existence of sexually dimorphic behavioral abnormalities and provide new insight into the molecular underpinnings of pathogenesis in UBE3A-dependent ASD.
Behaviorally, male Ube3A 2xTg mice showed deficits in both social preference and social novelty, and were more severely affected in their repetitive self-grooming behaviors, long-term memory, and pain sensitivity as compared to Tg females. Conversely, female Ube3A 2xTg mice displayed impairments in social novelty but not social preference, and were more severely impacted in their olfactory sensitivity. Interestingly, both male and female Tg mouse pups showed alterations in communication as indicated by their mother-seeking USVs induced by maternal separation. While the male Tg mice tend to call more, female Tg mice tend to use more complex syllables. These behaviors are consistent with the syndromes observed in patients with excessive UBE3A activity, including typical autism-relevant behaviors such as social and communication deficits, intellectual disability, as well as sensory disorders 16.
Recent studies have proposed diverse mechanisms underlying the sexually dimorphic expression of ASD. Sex-differential factors such as sex chromosomes and sex hormone signaling may function as sex-specific risk factors or protective factors 7,70. For instance, fetal testosterone during development has been proposed as one potential factor that may contribute to the male-biased features, like impaired social behaviors and restricted interests 71,72. Recent research has also identified non-sex-differential factors, such as transcriptomic alterations. The downregulation of neuronal genes and the upregulation of glial genes in males with autism may contribute to the sex imbalance in ASD 73. Some studies suggest that females require a higher mutation load to exhibit the diagnostic level of autism-relevant phenotypes, leading to the development of the “female protective model” 8,74,75. Furthermore, sexually dimorphic mechanisms related to ASD may involve not only transcriptomic changes, but also other mechanisms, such as synaptic transmission and neuronal activity 76–78. Despite these findings, understanding of the biological mechanisms remains limited.
By compiling case reports, we find that approximately 70% of dup 15q11–13 ASD cases are males 79–85. The male-predominant behavioral deficits in Ube3A 2xTg mice are consistent with this higher incidence of ASDs reported in males with the 15q11–13 duplication than in females. Previous studies on Ube3A 2xTg mice often limit their data analysis to comparing the differences between WT and Tg, but not male vs. female animals 23,24. Interestingly, upon combination of the male and female data with a near 1:1 ratio, our behavioral data are in line with the findings from previous research 23,24. Specifically, Ube3A 2xTg animals exhibit notable alterations in social preference, social novelty, vocalization, and repetitive self-grooming behaviors (Fig. S2). Of note, memory functions and pain sensitivity appear to be largely intact when the data from both sexes are combined (Fig. S3D, S3F-S3I), indicating that deficits in memory and pain sensitivity observed in male Tg mice (Fig. 2C, 2G, 2K) can become indiscernible when that data is diluted by the normality in female Tg mice (Fig. 2C, 2G, 2K). Similarly, when male and female results were combined, olfactory sensitivity, which was mainly impaired in Tg females (Fig. 2N), did not show a significant difference between WT and Ube3A 2xTg mice (Fig. S3E). Obviously, sex-orientated data analysis is important to reveal subtle phenotypes hidden in a general population of both sexes.
The crucial role of AR in brain development and function has been well documented. AR is strongly expressed in the brain, and in fact, male and female mouse brains have comparable levels of ARs at most time points during development, except postnatal day 7 86. However, the ligands of ARs, androgens, differ significantly between male and female brains 87. Both testosterone and 5𝛼-dihydrotestosterone are markedly more abundant in male brains than in female brains, resulting in much higher activities of AR signaling in male brains 87. In line with this, we find that a similar reduction of AR caused more severe disturbances in male compared to female mice in gene regulation and behaviors.
The androgen system regulates a range of behaviors, including male sexual behaviors, social behaviors, as well as learning and memory 51,62,64,88,89. In addition, ARs have been shown to increase the neural progenitor cell pool in the developing cortex, further indicating its vital role in neurodevelopment 48. Therefore, dysfunction of ARs is expected to interfere with brain development. In fact, several studies have linked AR dysregulation to ASD. Specifically, AR itself and at least seven AR target genes have been identified as ASD genes 5. Altered AR activity has also been implicated in the hyperactivity symptoms of ASD, such as ADHD 52,65, which is a common comorbidity found in 30% of autism patients 52. Our findings further confirmed the crucial role of AR in regulating autism-relevant behaviors, including social preference, repetitive self-grooming and memory. Continued investigation into the role of ARs in neurodevelopment and neurological diseases may lead to the development of novel therapeutics, such as the potential of using AR agonists as a treatment strategy for UBE3A-dependent ASD.
Limitations of the study
Intracerebroventricular injection of LV-GFP-AR virus was conducted in P0 mice, which limits the ability to assess the effects of AR replenishment on prenatal brain development and its potential influence on sex differences and autistic behaviors. Future studies involving prenatal replenishment of AR expression will help determine the roles AR reduction may play during embryonic stages.
Materials and Methods
Antibodies
The following antibodies were used in the indicated dilution or amount: Mouse anti-AR (WB: 1:500; IF: 1:200; IP: 1.5 µg; Santa Cruz Biotechnology; Cat# sc-7305). Mouse anti-ERα (WB: 1:500; Invitrogen; Cat# MA1-80216). Rabbit anti-ERβ (WB: 1:500; Invitrogen; Cat# PA1-311). Mouse anti-UBE3A (E6AP; WB: 1:1000; IF: 1:200; Sigma-Aldrich; Cat# E8655). Mouse anti-Ubiquitin (WB: 1:500; Invitrogen; Cat# 14-6078-82). Mouse anti-GAPDH (WB: 1:5000; EMD Millipore; Cat# MAB374). Mouse anti-α tubulin (WB: 1:5000; Sigma-Aldrich; Cat# T9026).
Animal care and use
Ube3A conventional BAC overexpressing mouse models on the FVB background (Ube3A-Tg 2x, strain #019730) 23,24 and FVB/NJ WT mice (strain #001800) were used in this study. All animal procedures were conducted in compliance with the policies of the Institutional Animal Care and Use Committee (IACUC) at Boston University. Ube3A 2x transgenic (2xTg) animals were obtained by mating heterozygous males and heterozygous females, expected to produce a 1:2:1 (WT: 1xTg: 2xTg) offspring ratio. WT and homozygous Ube3A 2xTg mice from the litters were used for the experiments, whereas the 1xTg mice were kept for further breeding. Therefore, the number of independent litters for the WT group and that for the 2xTg group used in the experiments is similar. Both male and female mice were used for behavioral tests and tissue collection. Most mice underwent multiple behavioral tests, with individual tests being separated by at least three days to prevent interference. Animals naïve to behavioral tests were used for the majority of the brain work, except those used for confirmation of virus expression and the rescue effect on ARGs.
Behavioral tests
For all behavioral tests, male and female mice were trained and tested in parallel. During all behavioral tests, except the Barnes maze spatial memory test, the testing room was dimly lit by a small desk lamp in the corner to enable the experimenter to maneuver. For the Barnes maze spatial memory test, bright ceiling lamps were used as an aversive stimulus.
Three-chamber social test
The three-chamber social test was performed as described in our previous study 90. A three-chambered box was made from plastic board that is 0.75 inches thick and measures 65 × 28 × 28 cm. The walls to the center chamber had cut-out doors that are 10 × 10 cm in size allowing movement between chambers. The two side chambers contained small wire cages to house the social stranger and/or novel mice. Before the test, mice were habituated to the apparatus with empty cages in each side chamber for 3 days, with each session lasting 5 min, allowing them to move freely within the apparatus. Social Preference Test: mice were singly placed into the middle chamber, with the doors blocked by plastic boards and a stranger mouse was placed into one of the side chambers under the wire cage. The doors were unblocked, and the test mouse was allowed to move freely within the apparatus for 5 min. Social Novelty Test: the test mouse was then returned to the middle chamber, the doors were blocked again. A second mouse (novel mouse) was introduced into the other side chamber. The center doors were unblocked and the test mouse was allowed to move freely within the apparatus for another 5 min. The entire apparatus was wiped with 70% ethanol between mice to eliminate odor cues between animals. Video recordings were captured with a Logitech c920 webcam during the test and the time spent interacting with each mouse or empty cage (nose ≤ 2 cm) and locomotion tracks were scored using TrackMo, an open-source animal tracking toolbox available at https://github.com/zudi-lin/tracking_toolbox.git, with the analyzer blinded to the genotype of the animals.
Ultrasonic vocalization (USV) recording
Ultrasonic vocalization recording of pup isolation calls was performed as described in our previous work 90. On the test day, pups were separated from the parents and littermates, and isolated in a sound-proof cylindrical plastic container. Mouse vocalizations were recorded on P5, P7 and P9, in a random order for each litter, using a CM16/CMPA microphone (Avisoft Bioacoustics) positioned 15 cm above the pups. The recording chamber was cleaned with 70% ethanol and dried between each pup. USV signals were recorded for 5 min at a sampling rate of 300 kHz in 16-bit format. The microphone was connected to a preamplifier UltraSoundGate 116Hb (Avisoft Bioacoustics) and digitized sonograms were stored in a computer. Recordings were analyzed using SASLab Pro 5.2.12 (Avisoft Bioacoustics). Spectrograms were generated using fast Fourier transform (256 FFT length, 100% frame, FlatTop window, and 50% window overlap) and a high-pass filter was applied to eliminate background noise <25 kHz. USVs were detected automatically (threshold: −47 ± 10 dB, hold time: 7 ms) followed by manual inspection to ensure accuracy. Several acoustic parameters of calls were measured including number of calls, mean call duration, total time spent calling, peak frequency, and peak amplitude. Call syllables were classified based on their acoustic features: simple (short, flat, upward, downward, chevron, U shape) and complex (modulated, frequency jump, multiple jumps, harmonic).
Self-grooming behavior test
The self-grooming behavior test was performed as described in our previous work 91. Each test mouse was placed individually into a regular housing cage with a fresh thin layer (∼0.5 cm) of wood chip beddings and was allowed to acclimate for 20 mins. The mice were then video recorded for 10 mins from the side view of the cage by a camera. The total time spent on grooming was measured.
Barnes maze spatial memory test
The Barnes maze spatial memory test was performed as described in our previous work 90. The Barnes maze is a circular maze constructed from a plastic board with a diameter of 48 inches and a thickness of 0.75 inches. Twenty holes with a diameter of 2 inches were evenly spaced around the perimeter of the maze, each with a distance of 1 inch to the edge. The maze was mounted on a 30-inch-high pedestal and could rotate at its center. The escape cage was a black plastic box with a ramp connected beneath the escape hole for easy access. During testing, four bright ceiling lamps and an alarm were used as aversive stimuli. To prevent olfactory cues, the maze and escape cage were cleaned with 70% ethanol between each testing session, and the maze was randomly rotated after each mouse. During the test, the mice were habituated to the maze on day 1. For each session, one mouse was placed in the center of the maze, covered with an opaque cardboard chamber for 15 s, and then slowly guided to the escape hole with a glass beaker. Each mouse was given 3 min to enter the escape hole on its own, but if they failed to do so, they were nudged gently into the hole. Afterward, the mouse was allowed to stay in the escape hole for 2 min. The ceiling lights and white noise remained on while the mouse was exploring the maze and turned off immediately after they entered the escape hole. This procedure was repeated for all mice. On days 2–5, the mice were trained to ensure a strong memory for the escape hole (4 times/mouse/day for 4 days, 16 times in total) and to learn to enter the escape hole by themselves. During each training trial, the mice were allowed to stay in the escape cage for 1 min. Memory of the escape hole location was probed 24 hours and 5 days later. During the memory retention tests, the escape hole was sealed and spatial memory for the subject mice was tested. Video recordings were captured for 4min. The number of errors committed prior to locating the target hole (primary errors) and traces of locomotion were scored by TrackMo.
Novel object recognition (NOR) test
The novel object recognition test was performed as described in our previous work 92. The NOR protocol comprises three sessions: habituation, training, and test sessions. During habituation, each mouse exposed to an open-field apparatus twice a day for 3 consecutive days, with each session lasting 10 min. On day 4, two identical objects (falcon tissue culture flask filled with beddings) were placed in two corners of the arena and mice were allowed to explore freely for 10 min. 4 hours or 24 hours later, during the test sessions, one object was replaced by a novel object (tower of Lego bricks). Video recordings were captured for 5 min, and exploration time for each object and traces of locomotion were scored by TrackMo.
Hot plate test
To assess the pain sensitivity of mice, the hot plate test was conducted following the protocol detailed in our previous study 92. Prior to the test, mice were habituated to the non-heated hot plate surface and a covered 5L glass beaker for 2 days (15 min/day). During day 3, the hot plate temperature was set to 55°C. Mice were placed on the hot plate and covered with a 5L glass beaker. The latency to response was recorded when the hind paw lick occurred.
Olfactory detection threshold test
The olfactory detection threshold test was performed as described in previous protocols with minor modifications 93. On day 1, two filter papers were placed at two corners of a regular housing cage, one with a vanilla solution at the lowest concentration and the other with a filter paper soaked in water (vehicle). The test mouse was allowed to freely explore for 3 min, and the time spent on each filter paper was recorded. During day 2–4, the session was repeated once a day, but using vanilla solution with a 100 times higher concentration each day. If the mouse cannot detect the odorant, it was expected to spend an equal amount of time sniffing the vanilla paper and the vehicle. The percentage of odorant sniffing was calculated using the following equation: the percentage of odorant sniffing = time spent on odorant / (time spent on odorant + time spent on vehicle) X 100.
Open-field test
The open-field test was performed as described in our previous work 90. The open-field test was conducted in a chamber that measured 28 × 28 × 28 cm and was made of 0.75-inch-thick plastic board. The chamber was placed on a large black plastic board, and the lights in the testing room were turned off except for a small desk lamp in the corner allowing the experimenter to see. Mice were habituated to the testing room for 5 min/day for 3 days. On the test day, each mouse was placed individually in the center of the box and allowed to freely explore the chamber. Video recordings were captured during the test and the time spent in the center, velocity and track lengths were quantified blinded to the genotype of the animals.
Primary neuron culture
Primary cortical neurons were prepared from embryonic day 18 rat fetuses as described previously 90. Timed pregnant Sprague–Dawley rats were purchased from Charles River Laboratories Inc. Cortical brain tissues were dissected and digested with papain (0.5 mg/mL in Hanks balanced salt solution, Sigma-Aldrich; cat. #4762) for 20 min at 37°C, then gently triturated in a trituration buffer containing 0.1% DNase (cat. #PA5–22017 RRID: AB_11153259), 1% ovomucoid (Sigma-Aldrich; cat. #T2011), 1% bovine serum albumin (Sigma-Aldrich; cat. #05470) in DMEM to fully dissociate neurons. Dissociated neurons were counted and plated onto 18mm circular coverslips (Carolina Biology Supply, cat. #633013). Coverslips had been coated in poly-l-lysine (Sigma-Aldrich; cat. #P2636; 100 μg/ml in borate buffer) overnight at 37°C, then washed three times with sterile deionized water and left in plating medium [DMEM supplemented with 10% fetal bovine serum (Atlanta Biologicals; cat. #S11550), 5% horse serum (Atlanta Biologicals; cat. #S12150), 31 mg of l-cysteine, 1% penicillin/streptomycin (Corning; cat. #30–002-Cl), and l-glutamine (Corning; cat. #25–005-Cl)] before cell plating. The day after plating, plating medium was replaced by feeding medium [neurobasal medium (Gibco; cat. #21103049) supplemented with 2% Neurocult SM1 Neuronal Supplement (StemCell Technologies; cat. #05711), 1% horse serum (Atlanta Biologicals; cat. #S12150), 1% penicillin/streptomycin (Corning; cat. #30–002-CI), and l-glutamine (Corning; cat. # 25–005-CI)]. One week after plating, 5-fluorodeoxyuridine (10μM; Sigma-Aldrich; cat. #F0503) was added to the medium to inhibit glial growth. All cells were maintained in a humidified incubator containing 5% CO2 at 37°C.
Immunocytochemistry of cultured neurons
Culture neurons were washed twice in ice-cold PBS and fixed for 10 min in a 4% paraformaldehyde solution at room temperature. Cell membranes were then permeabilized for 8 min in 0.3% Triton X-100 (Fisher Biotec; cat. #BP151–100) in PBS. Cells were rinsed three times in PBS, and blocked in 10% goat serum for 1 h. Primary antibodies (in 5% goat serum PBS) were added, and the cells were incubated overnight at 4℃, washed twice with PBS and incubated with appropriate Alexa Fluor-conjugated fluorescent secondary antibodies (1:400, Life Technologies) for an additional hour. Cells were then rinsed (with Hoechst 1:2000 in the first rinse) and mounted to microscopy glass slides with Prolong Gold anti-fade mounting reagent (Invitrogen; cat. #P36930) and stored at 4℃ in dark for subsequent imaging.
Neuronal and HEK293T cell transfection
Neurons were transfected with Lipofectamine 2000 (Invitrogen, cat. #11668019) and the target plasmids on DIV4 according to the manufacturer’s instruction. Each coverslip was transfected with 1 μg of either GFP or HA-UBE3A plasmid (Addgene, cat. #8648), and 1 μL Lipofectamine 2000. The plasmid and Lipofectamine 2000 were diluted separately in 50 μL DMEM in 1.5 mL Eppendorf tubes and incubated for 5 min. Then solutions in two tubes were mixed together and incubated at room temperature for another 20 min to form the transfection complex. Thereafter, the transfection complex was gently added into the 12-well plate with 1 coverslip/well in a drop-wise manner. The neurons were incubated with the transfection complex at 37 °C for 3 h before the medium was removed and replaced with feeding medium. On DIV8 (4 d following transfections), neurons were fixed for immunocytochemistry or lysed for biochemical analysis. HEK cell transfections were performed using the polyethylenimine (PEI) transfection reagent (Polysciences; cat. #23966) with a 3:1 PEI-to-DNA ratio when the cells reached 70% confluency. HEK cells were lysed and collected 48 h post-transfection.
Immunoprecipitation
For ubiquitination immunoprecipitation assays, cultured cortical neurons, HEK cells, or brain tissues were rinsed with cold PBS and lysed on ice in 200 μl of modified radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) supplemented with cOmplete protease inhibitor (Roche; cat. #11873580001). Cell or brain lysates were further solubilized by sonication and 1 h rotation at 4℃. After rotation, lysates were centrifuged at 12000g, 20 min, 4℃ to collect supernatant. A small portion of each sample was saved as a total cell lysate while the reminder was adjusted to 500μl with more RIPA buffer for immunoprecipitation. Protein A-Agarose beads (Santa Cruz Biotechnology; cat. #sc-2001) were added to the lysates along with antibodies against AR (Santa Cruz Biotechnology; cat. # sc-7305) and samples were incubated overnight for 12–16 h on rotation at 4°C. Agarose beads were washed three times with cold RIPA buffer, resuspended in 2X Laemmli sample buffer, and denatured at 95°C for 10 min before being subjected to Western blotting.
Western blot
Cultured neurons were lysed in 2X Laemmli sample buffer (4% SDS, 10% 2- mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 0.125 M Tris HCl) and boiled for 10 min at 95°C before being stored at −20°C for later use. Mouse brain tissues were dissected on ice immediately after sacrificing animals, and then lysed on ice using RIPA lysis buffer. Samples were further solubilized by sonication and 1 h rotation at 4°C. Lysates were then centrifuged at 12000g, 20 min, 4°C to collect supernatants, which were then subjected to a BCA assay according to the manufacturer’s protocol (Pierce, cat. #23225) to determine protein concentrations. Protein levels were normalized using RIPA lysis buffer. Samples were then mixed with 2X Laemmli sample buffer and boiled for 10 min at 95°C to prepare for SDS-PAGE.
Standard SDS-PAGE procedures were used to separate proteins of interest in the cell or brain lysates. Proteins were transferred to PVDF membranes (Bio-Rad, cat. #1620177) and blocked for 1 h in 5% milk in PBS. Membranes were then probed for different targets with the appropriate primary antibodies overnight at 4°C in Tris buffered saline supplemented with 0.05% Tween (TBST). Membranes were washed 3 times in TBST, followed by incubation with the appropriate secondary antibody for 1 h. After a further three washes with TBST, immunoblots were visualized using a chemiluminescence detection system (Sapphire Biomolecular Imager, Azure biosystems, CA, USA) and analyzed using ImageJ.
Immunohistochemistry of brain slices
As described in our previous work 90, before removing the brain from the skull, mice were subjected to transcardial perfusion with ice-cold PBS. Brains were fixed in 4% paraformaldehyde for 4–6 h and then cryoprotected in 30% sucrose/PBS until sinking to the bottom. To prepare for sectioning, the dehydrated brains were frozen in optimal cutting temperature-embedded medium (Tissue-Tek, cat. #25608–930) and sliced into 20 μm sections using a CM1850 cyrostat (LEICA Biosystems) at −20°C. Sections were immediately mounted onto SuperFrost microscope slides (Fisher Scientific, cat.# 12–550-15) and stored at −80°C for future use. Prior to immunostaining, sections were rinsed with PBS for at least 2 h. Sections were then blocked in 5% goat serum, 0.3% Triton X-100/PBS for 1 h at room temperature, followed by incubation with properly diluted primary antibodies overnight at 4°C. The following day, sections were washed three times with PBS and incubated with the appropriate Alexa Fluor-conjugated secondary antibodies (1:250, Life Technologies). Brain slices were then washed three times with PBS, with the first wash containing Hoescht (1:2000, Thermo Fisher Scientific, cat. # 62249). Sections were then mounted with ProLong-Gold mounting medium (Invitrogen, cat. #P36930), left to dry at room temperature overnight, and stored at −20°C in dark before imaging.
Viral constructs preparation and virus infection
GFP-fused AR was cut from pEGFP-C1-AR plasmid (Addgene, cat. #28235) and inserted into the pFUW lentiviral (LV) vector, to generate pFUW-GFP-AR construct. To package LV particles, HEK293T cells at 70% confluency in 15-cm culture plates were transfected with pFUW-GFP-AR, along with viral packaging and envelope proteins (pRSV/REV, pMDLg/RRE, and pVSV-G) using polyethylenimine transfection reagent (Polysciences; cat. #23966). 48 h post-transfection, cell medium was filtered through a 0.45 μm filter, and PEG-it virus precipitation solution (1:5, System Biosciences; cat. # LV810A-1) was added and kept at 4°C overnight. On the next day, the solution was centrifuged (1500 rpm, 30 min, 4°C), and the pellet was resuspended with 100 μl cold PBS. Aliquots of the virus were stored at −80°C until use. For in vitro assays, neurons were infected with lentivirus at DIV6, and the medium was replaced with feeding medium 2 d later.
Intracerebroventricular brain injection of virus in neonatal mice
Bilateral intracerebroventricular (ICV) injections were performed in mouse pups on postnatal day 0. Newborn mice were cryoanesthetized and placed on a cold metal plate. A 30-gauge needle (Hamilton, cat. # 7803–04) and 10 μL Hamilton syringe (Hamilton, cat. # 7653–01) were used for injection. To pierce the skull, a separate needle was employed at a site that is roughly two-fifths of the distance between the lamda and the eye. Each cerebral ventricle was slowly injected with 2 μL of either pFUW-GFP or pFUW-GFP-AR virus mixed with 1% Fast Green Dye. Following injection, neonatal mice were kept with their parents until they were weaned. On postnatal day 30, all mice were subject to behavioral assessments before being euthanized for biochemical and histological analysis.
Real-time PCR
Total RNA from prefrontal cortical area was extracted using the RNeasy® Mini Kit (Qiagen, cat. # 74104) following the manufacturer’s instructions. cDNA was reverse transcribed from 500 ng of total RNA for each sample using HiFiScript cDNA Synthesis Kit (CoWin Biosciences, cat. # CW2569M) according to the manufacturer’s protocol. cDNA samples were then used as a template for further mRNA level analysis.
For qPCR, 1 μL of the reverse transcribed cDNA was used as a template and mixed with primer pairs:
AR (5′- GATGGTATTTGCCATGGGTTG-3′, 5′- GGCTGTACATCCGAGACTTGTG-3′);
lgfbpl1 ((5′-CTGACCATGAGACCACATCCTG-3′, 5′-ACTGAGCCTCTCCAATGGCGTT-3′);
Pde1c (5′-CAGTCATCCTGCGAAAGCATGG-3′, 5′-CCACTTGTGACTGAGCAACCATG-3′);
Rps6ka5 (5′-TGGTCCATAGCACCTCTCAGCT-3′, 5′-CTCTCCGCCATTCAGAAGTTCC-3′);
Draxin (5′-GTGGCAGAGAACACAAGAGACG-3′, 5′-GGTCTTCAGAGGGTTCCACCTT-3′);
Grik2 (5′-CTGACTCAGGTTTGCTGGATGG-3′, 5′- CAAGGAGCTGACTGTCATCTGG-3′);
Pxdn (5′-GTTCAGCATGGCTTGATGGTGG-3′, 5′-AGCCTGACAGGTTGGCGATGAG-3′);
10 μL reactions were prepared with UltraSYBR Mixture kit (CoWin Biosciences, cat. #CW2602F) according to the manufacturer’s protocol and placed in Applied Biosystems 7900HT Fast Real-Time PCR system for real-time monitoring.
RNA Sequencing
Brain tissues from three animals in each group (WT male, WT female, Tg male, Tg female) were dissected and preserved in RNAlater™ Stabilization Solution (Invitrogen, cat. #AM7020) before being sent to commercial RNA-sequencing services provided by BGI genomics. RNA-seq datasets were generated and analyzed by BGI genomics to identify differentially expressed genes and gene ontology enrichment. Datasets from Ube3A 2xTg males were compared with those from WT males, and datasets from Ube3A 2xTg females were juxtaposed with those from WT females.
Statistics and reproducibility
Graphics and statistical analysis were performed using GraphPad Prism 8.0 (GraphPad Software). Data of genotypes (for combined results of both males and females, or in vitro assays) were analyzed using an unpaired two-tailed Student’s t-test, with genotype as a factor. Comparisons involving two independent variables, such as sexually dimorphic changes between WT and Ube3A 2xTg animals, were analyzed by two-way analysis of variance (ANOVA). Three-way ANOVAs were used for comparisons involving three independent variables, each with at least two levels: interaction items (stranger mouse vs. empty cage, stranger mouse vs. novel mouse, or familiar object vs. novel object), sex (male vs. female), and genotype (WT vs. Tg). Post hoc multiple comparisons were corrected using Bonferroni’s method. Animals that did not exhibit normal behavior or health at the onset of the experiment, or showed signs of injury and sickness during experiments, were excluded. Following statistical analysis, individual data points with a value larger or smaller than 2 standard deviations were considered outliers and removed from the data pool. All results are presented as mean ± s.e.m. (standard error of mean), and *p < 0.05 was considered statistically significant. All statistics and sample sizes are reported in Supplementary Table 1.
Supplementary Material
Acknowledgements
We would like to thank the Man Lab members for helpful discussion. We thank KathrynAnn Odamah and Dr. Mike Baum for their comments on the manuscript. This work was supported by NIH grants R01 MH079407, R01 MH130600, R21 MH133014 and the Harvard/MIT Joint Research Grants Program in Basic Neuroscience (HYM).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Data availability
Data used in the analysis of this study are available from the corresponding author upon reasonable request.
References
- 1.Hodges H., Fealko C., and Soares N. (2020). Autism spectrum disorder: definition, epidemiology, causes, and clinical evaluation. Transl Pediatr 9, S55. 10.21037/TP.2019.09.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maenner M.J., Shaw K.A., Bakian A. v., Bilder D.A., Durkin M.S., Esler A., Furnier S.M., Hallas L., Hall-Lande J., Hudson A., et al. (2021). Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2018. MMWR. Surveillance Summaries 70, 1–16. 10.15585/MMWR.SS7011A1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karimi P., Kamali E., Mousavi S.M., and Karahmadi M. (2017). Environmental factors influencing the risk of autism. J Res Med Sci 22. 10.4103/1735-1995.200272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratajczak H. V. (2011). Theoretical aspects of autism: Causes—A review. 10.3109/1547691X.2010.545086 8, 68–79. 10.3109/1547691X.2010.545086. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., and Packer A. (2013). SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4, 1–3. 10.1186/2040-2392-4-36/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsabbagh M., Divan G., Koh Y.J., Kim Y.S., Kauchali S., Marcín C., Montiel-Nava C., Patel V., Paula C.S., Wang C., et al. (2012). Global Prevalence of Autism and Other Pervasive Developmental Disorders. Autism Research 5, 160–179. 10.1002/AUR.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werling D.M., and Geschwind D.H. (2013). Sex differences in autism spectrum disorders. Curr Opin Neurol 26, 146. 10.1097/WCO.0B013E32835EE548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Li N., Li C., Zhang Z., Teng H., Wang Y., Zhao T., Shi L., Zhang K., Xia K., et al. (2020). Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Translational Psychiatry 2020 10:1 10, 1–10. 10.1038/s41398-020-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferri S.L., Abel T., and Brodkin E.S. (2018). Sex Differences in Autism Spectrum Disorder: A Review. Curr Psychiatry Rep 20, 9. 10.1007/S11920-018-0874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Giambattista C., Ventura P., Trerotoli P., Margari F., and Margari L. (2021). Sex Differences in Autism Spectrum Disorder: Focus on High Functioning Children and Adolescents. Front Psychiatry 12, 1063. 10.3389/FPSYT.2021.539835/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giarelli E., Wiggins L.D., Rice C.E., Levy S.E., Kirby R.S., Pinto-Martin J., and Mandell D. (2010). Sex differences in the evaluation and diagnosis of autism spectrum disorders among children. Disabil Health J 3, 107–116. 10.1016/J.DHJO.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandy W., Chilvers R., Chowdhury U., Salter G., Seigal A., and Skuse D. (2012). Sex differences in autism spectrum disorder: Evidence from a large sample of children and adolescents. J Autism Dev Disord 42, 1304–1313. 10.1007/S10803-011-1356-0. [DOI] [PubMed] [Google Scholar]
- 13.Solomon M., Miller M., Taylor S.L., Hinshaw S.P., and Carter C.S. (2012). Autism symptoms and internalizing psychopathology in girls and boys with autism spectrum disorders. J Autism Dev Disord 42, 48–59. 10.1007/S10803-011-1215-Z/TABLES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khatri N., and Man H.Y. (2019). The autism and Angelman syndrome protein Ube3A/E6AP: The gene, E3 ligase ubiquitination targets and neurobiological functions. Front Mol Neurosci 12, 109. 10.3389/FNMOL.2019.00109/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer P.L., Hanayama R., Bloodgood B.L., Mardinly A.R., Lipton D.M., Flavell S.W., Kim T.K., Griffith E.C., Waldon Z., Maehr R., et al. (2010). The Angelman Syndrome-associated ubiquitin ligase Ube3A regulates synapse development by ubiquitinating Arc. Cell 140, 704. 10.1016/J.CELL.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finucane B.M., Lusk L., Arkilo D., Chamberlain S., Devinsky O., Dindot S., Jeste S.S., LaSalle J.M., Reiter L.T., Schanen N.C., et al. (2021). Maternal 15q Duplication Syndrome. GeneReviews®. [Google Scholar]
- 17.Yi J.J., Berrios J., Newbern J.M., Snider W.D., Philpot B.D., Hahn K.M., and Zylka M.J. (2015). An Autism-Linked Mutation Disables Phosphorylation Control of UBE3A. Cell 162, 795–807. 10.1016/J.CELL.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014 515:7526 515, 216–221. 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weston K.P., Gao X., Zhao J., Kim K.S., Maloney S.E., Gotoff J., Parikh S., Leu Y.C., Wu K.P., Shinawi M., et al. (2021). Identification of disease-linked hyperactivating mutations in UBE3A through large-scale functional variant analysis. Nature Communications 2021 12:1 12, 1–15. 10.1038/s41467-021-27156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noor A., Dupuis L., Mittal K., Lionel A.C., Marshall C.R., Scherer S.W., Stockley T., Vincent J.B., Mendoza-Londono R., and Stavropoulos D.J. (2015). 15q11.2 Duplication Encompassing Only the UBE3A Gene Is Associated with Developmental Delay and Neuropsychiatric Phenotypes. Hum Mutat 36, 689–693. 10.1002/HUMU.22800. [DOI] [PubMed] [Google Scholar]
- 21.Abrahams B.S., and Geschwind D.H. (2008). Advances in autism genetics: on the threshold of a new neurobiology. Nature Reviews Genetics 2008 9:5 9, 341–355. 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogart A., Wu D., LaSalle J.M., and Schanen N.C. (2010). The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis 38, 181–191. 10.1016/J.NBD.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S.E.P., Zhou Y.D., Zhang G., Jin Z., Stoppel D.C., and Anderson M.P. (2011). Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med 3. 10.1126/SCITRANSLMED.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan V., Stoppel D.C., Nong Y., Johnson M.A., Nadler M.J.S., Ozkaynak E., Teng B.L., Nagakura I., Mohammad F., Silva M.A., et al. (2017). Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 2017 543:7646 543, 507–512. 10.1038/nature21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Premoli M., Memo M., and Bonini S. (2021). Ultrasonic vocalizations in mice: relevance for ethologic and neurodevelopmental disorders studies. Neural Regen Res 16, 1158. 10.4103/1673-5374.300340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Premoli M., Bonini S.A., Mastinu A., Maccarinelli G., Aria F., Paiardi G., and Memo M. (2019). Specific profile of ultrasonic communication in a mouse model of neurodevelopmental disorders. Scientific Reports 2019 9:1 9, 1–12. 10.1038/s41598-019-52378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmeisser M.J., Ey E., Wegener S., Bockmann J., Stempel A.V., Kuebler A., Janssen A.L., Udvardi P.T., Shiban E., Spilker C., et al. (2012). Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 2012 486:7402 486, 256–260. 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 28.Lai J.K.Y., Sobala-Drozdowski M., Zhou L., Doering L.C., Faure P.A., and Foster J.A. (2014). Temporal and spectral differences in the ultrasonic vocalizations of fragile X knock out mice during postnatal development. Behavioural Brain Research 259, 119–130. 10.1016/J.BBR.2013.10.049. [DOI] [PubMed] [Google Scholar]
- 29.Scattoni M.L., Ricceri L., and Crawley J.N. (2011). Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav 10, 44–56. 10.1111/J.1601-183X.2010.00623.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava A.K., and Schwartz C.E. (2014). Intellectual disability and autism spectrum disorders: Causal genes and molecular mechanisms. Neurosci Biobehav Rev 46, 161–174. 10.1016/J.NEUBIOREV.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matson J.L., and Shoemaker M. (2009). Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil 30, 1107–1114. 10.1016/J.RIDD.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Orefice L.L. (2020). Peripheral Somatosensory Neuron Dysfunction: Emerging Roles in Autism Spectrum Disorders. Neuroscience 445, 120. 10.1016/J.NEUROSCIENCE.2020.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa S., Nomura K., Miyaho K., Sanada K., Iwamoto C., Naraoka M., Yoneda S., Tomizawa Y., Sawae Y., Iwanaga R., et al. (2021). Gastrointestinal symptoms and sensory abnormalities associated with behavioral problems in children with neurodevelopmental disorders. Autism Research 14, 1996–2001. 10.1002/AUR.2549. [DOI] [PubMed] [Google Scholar]
- 34.Sukoff Rizzo S.J., and Crawley J.N. (2017). Behavioral Phenotyping Assays for Genetic Mouse Models of Neurodevelopmental, Neurodegenerative, and Psychiatric Disorders. 10.1146/annurev-animal-022516-022754 5, 371–389. . [DOI] [PubMed] [Google Scholar]
- 35.Tonacci A., Billeci L., Tartarisco G., Ruta L., Muratori F., Pioggia G., and Gangemi S. (2015). Olfaction in autism spectrum disorders: A systematic review. Child Neuropsychology 23, 1–25. 10.1080/09297049.2015.1081678. [DOI] [PubMed] [Google Scholar]
- 36.Koehler L., Fournel A., Albertowski K., Roessner V., Gerber J., Hummel C., Hummel T., and Bensafi M. (2018). Impaired Odor Perception in Autism Spectrum Disorder Is Associated with Decreased Activity in Olfactory Cortex. Chem Senses 43, 627–634. 10.1093/CHEMSE/BJY051. [DOI] [PubMed] [Google Scholar]
- 37.Marrocco J., and McEwen B.S. (2016). Sex in the brain: hormones and sex differences. Dialogues Clin Neurosci 18, 373. 10.31887/DCNS.2016.18.4/JMARROCCO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mcewen B.S., ALVES Harold S.E., and Milliken M. (1999). Estrogen Actions in the Central Nervous System. Endocr Rev 20, 279–307. 10.1210/EDRV.20.3.0365. [DOI] [PubMed] [Google Scholar]
- 39.McEwen B.S., and Milner T.A. (2007). Hippocampal formation: Shedding light on the influence of sex and stress on the brain. Brain Res Rev 55, 343–355. 10.1016/J.BRAINRESREV.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinow D.R., and Schmidt P.J. (2006). Androgens, brain, and behavior. 10.1176/ajp.153.8.974 153, 974–984. . [DOI] [PubMed] [Google Scholar]
- 41.Gegenhuber B., Wu M. v., Bronstein R., and Tollkuhn J. (2022). Gene regulation by gonadal hormone receptors underlies brain sex differences. Nature 2022 606:7912 606, 153–159. 10.1038/s41586-022-04686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez M., Picard N., Sauvé K., and Tremblay A. (2012). Coordinate regulation of estrogen receptor β degradation by Mdm2 and CREB-binding protein in response to growth signals. Oncogene 2012 32:1 32, 117–126. 10.1038/onc.2012.19. [DOI] [PubMed] [Google Scholar]
- 43.Duong V., Boulle N., Daujat S., Chauvet J., Bonnet S., Neel H., and Cavaillès V. (2007). Differential Regulation of Estrogen Receptor α Turnover and Transactivation by Mdm2 and Stress-Inducing Agents. Cancer Res 67, 5513–5521. 10.1158/0008-5472.CAN-07-0967. [DOI] [PubMed] [Google Scholar]
- 44.Fan M., Park A., and Nephew K.P. (2005). CHIP (Carboxyl Terminus of Hsc70-Interacting Protein) Promotes Basal and Geldanamycin-Induced Degradation of Estrogen Receptor-α. Molecular Endocrinology 19, 2901–2914. 10.1210/ME.2005-0111. [DOI] [PubMed] [Google Scholar]
- 45.Li B., Lu W., and Chen Z. (2014). Regulation of Androgen Receptor by E3 Ubiquitin Ligases: for More or Less. Receptors Clin Investig 1. 10.14800/RCI.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T., Matsumoto T., Kawano H., Watanabe T., Uematsu Y., Sekine K., Fukuda T., Aihara K.I., Krust A., Yamada T., et al. (2004). Brain masculinization requires androgen receptor function. Proc Natl Acad Sci U S A 101, 1673–1678. 10.1073/PNAS.0305303101/SUPPL_FILE/5303FIG7.JPG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dart D.A., Waxman J., Aboagye E.O., and Bevan C.L. (2013). Visualising Androgen Receptor Activity in Male and Female Mice. PLoS One 8, e71694. 10.1371/JOURNAL.PONE.0071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Rosa P., Bartoli G., Farioli Vecchioli S., Cesari E., Pagliarini V., Sette C., Gemelli A., Correspondence Claudio Sette I., La Sapienza R., and Stefano Farioli Vecchioli I. (2021). Androgen Receptor signaling promotes the neural progenitor cell pool in the developing cortex. J Neurochem 157, 1153–1166. 10.1111/JNC.15192. [DOI] [PubMed] [Google Scholar]
- 49.Henningsson S., Jonsson L., Ljunggren E., Westberg L., Gillberg C., Råstam M., Anckarsäter H., Nygren G., Landén M., Thuresson K., et al. (2009). Possible association between the androgen receptor gene and autism spectrum disorder. Psychoneuroendocrinology 34, 752–761. 10.1016/J.PSYNEUEN.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Baron-Cohen S., Knickmeyer R.C., and Belmonte M.K. (2005). Sex differences in the brain: implications for explaining autism. Science 310, 819–823. 10.1126/SCIENCE.1115455. [DOI] [PubMed] [Google Scholar]
- 51.Bodo C., and Rissman E.F. (2008). The androgen receptor is selectively involved in organization of sexually dimorphic social behaviors in mice. Endocrinology 149, 4142–4150. 10.1210/EN.2008-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pivovarciova A., Durdiakova J., Babinska K., Kubranska A., Vokalova L., Minarik G., Celec P., Murin M., and Ostatnikova D. (2016). Testosterone and Androgen Receptor Sensitivity in Relation to Hyperactivity Symptoms in Boys with Autism Spectrum Disorders. PLoS One 11, e0149657. 10.1371/JOURNAL.PONE.0149657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma A., and Mehan S. (2021). Targeting PI3K-AKT/mTOR signaling in the prevention of autism. Neurochem Int 147, 105067. 10.1016/J.NEUINT.2021.105067. [DOI] [PubMed] [Google Scholar]
- 54.Chen J., Alberts I., and Li X. (2014). Dysregulation of the IGF-I/PI3K/AKT/mTOR signaling pathway in autism spectrum disorders. Int J Dev Neurosci 35, 35–41. 10.1016/J.IJDEVNEU.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 55.Krupp D.R., Barnard R.A., Duffourd Y., Evans S.A., Mulqueen R.M., Bernier R., Rivière J.B., Fombonne E., and O’Roak B.J. (2017). Exonic Mosaic Mutations Contribute Risk for Autism Spectrum Disorder. Am J Hum Genet 101, 369–390. 10.1016/J.AJHG.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaags A.K., Lionel A.C., Sato D., Goodenberger M., Stein Q.P., Curran S., Ogilvie C., Ahn J.W., Drmic I., Senman L., et al. (2012). Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet 90, 133–141. 10.1016/J.AJHG.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia (2017). Mol Autism 8, 21. 10.1186/S13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eno C.C., Graakjaer J., Svaneby D., Nizon M., Kianmahd J., Signer R., Martinez-Agosto J.A., and Quintero-Rivera F. (2021). 14q32.11 microdeletion including CALM1, TTC7B, PSMC1, and RPS6KA5: A new potential cause of developmental and language delay in three unrelated patients. Am J Med Genet A 185, 1519–1524. 10.1002/AJMG.A.62117. [DOI] [PubMed] [Google Scholar]
- 59.Nugent A.A., Kolpak A.L., and Engle E.C. (2012). Human disorders of axon guidance. Curr Opin Neurobiol 22, 837–843. 10.1016/J.CONB.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jamain S., Betancur C., Quach H., Philippe A., Fellous M., Giros B., Gillberg C., Leboyer M., Bourgeron T., Gillberg C., et al. (2002). Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry 7, 302–310. 10.1038/SJ.MP.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer K.J., Axelsen M.S., Sheffield V.C., Patil S.R., and Wassink T.H. (2012). Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatr Genet 22, 137–140. 10.1097/YPG.0B013E32834DC3F5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karlsson S.A., Studer E., Kettunen P., and Westberg L. (2016). Neural androgen receptors modulate gene expression and social recognition but not social investigation. Front Behav Neurosci 10, 41. 10.3389/FNBEH.2016.00041/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koss W.A., and Frick K.M. (2019). Activation of androgen receptors protects intact male mice from memory impairments caused by aromatase inhibition. Horm Behav 111, 96–104. 10.1016/J.YHBEH.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rizk A., Robertson J., and Raber J. (2005). Behavioral performance of tfm mice supports the beneficial role of androgen receptors in spatial learning and memory. Brain Res 1034, 132–138. 10.1016/J.BRAINRES.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 65.Comings D.E., Chen C., Wu S., and Muhleman D. (1999). Association of the androgen receptor gene (AR) with ADHD and conduct disorder. Neuroreport 10, 1589–1592. 10.1097/00001756-199905140-00036. [DOI] [PubMed] [Google Scholar]
- 66.Liu Y., Zhang M., He T., Yang W., Wang L., Zhang L., and Guo M. (2020). Epigenetic silencing of IGFBPL1 promotes esophageal cancer growth by activating PI3K-AKT signaling. Clin Epigenetics 12, 1–12. 10.1186/S13148-020-0815-X/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onore C., Yang H., van de Water J., and Ashwood P. (2017). Dynamic Akt/mTOR signaling in children with autism spectrum disorder. Front Pediatr 5, 43. 10.3389/FPED.2017.00043/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holt R., Barnby G., Maestrini E., Bacchelli E., Brocklebank D., Sousa I., Mulder E.J., Kantojärvi K., Järvelä I., Klauck S.M., et al. (2010). Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet 18, 1013–1020. 10.1038/EJHG.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shuang M., Liu J., Jia M.X., Yang J.Z., Wu S.P., Gong X.H., Ling Y.S., Ruan Y., Yang X.L., and Zhang D. (2004). Family-based association study between autism and glutamate receptor 6 gene in Chinese Han trios. Am J Med Genet B Neuropsychiatr Genet 131B, 48–50. 10.1002/AJMG.B.30025. [DOI] [PubMed] [Google Scholar]
- 70.Jeon S.J., Gonzales E.L., Mabunga D.F.N., Valencia S.T., Kim D.G., Kim Y., Adil K.J.L., Shin D., Park D., and Shin C.Y. (2018). Sex-specific Behavioral Features of Rodent Models of Autism Spectrum Disorder. Exp Neurobiol 27, 321–343. 10.5607/EN.2018.27.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knickmeyer R., Baron-Cohen S., Raggatt P., and Taylor K. (2005). Foetal testosterone, social relationships, and restricted interests in children. J Child Psychol Psychiatry 46, 198–210. 10.1111/J.1469-7610.2004.00349.X. [DOI] [PubMed] [Google Scholar]
- 72.Auyeung B., Baron-Cohen S., Ashwin E., Knickmeyer R., Taylor K., and Hackett G. (2009). Fetal testosterone and autistic traits. British Journal of Psychology 100, 1–22. 10.1348/000712608X311731. [DOI] [PubMed] [Google Scholar]
- 73.Werling D.M., Parikshak N.N., and Geschwind D.H. (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nature Communications 2016 7:1 7, 1–11. 10.1038/ncomms10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson E.B., Lichtenstein P., Anckarsäter H., Happé F., and Ronald A. (2013). Examining and interpreting the female protective effect against autistic behavior. Proc Natl Acad Sci U S A 110, 5258–5262. 10.1073/PNAS.1211070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacquemont S., Coe B.P., Hersch M., Duyzend M.H., Krumm N., Bergmann S., Beckmann J.S., Rosenfeld J.A., and Eichler E.E. (2014). A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet 94, 415–425. 10.1016/J.AJHG.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jung H., Park H., Choi Y., Kang H., Lee E., Kweon H., Roh J.D., Ellegood J., Choi W., Kang J., et al. (2018). Sexually dimorphic behavior, neuronal activity, and gene expression in Chd8-mutant mice. Nature Neuroscience 2018 21:9 21, 1218–1228. 10.1038/s41593-018-0208-z. [DOI] [PubMed] [Google Scholar]
- 77.Mottron L., Duret P., Mueller S., Moore R.D., Forgeot D’Arc B., Jacquemont S., and Xiong L. (2015). Sex differences in brain plasticity: a new hypothesis for sex ratio bias in autism. Mol Autism 6. 10.1186/S13229-015-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mercer A.A., Palarz K.J., Tabatadze N., Woolley C.S., and Raman I.M. (2016). Sex differences in cerebellar synaptic transmission and sex-specific responses to autism-linked Gabrb3 mutations in mice. Elife 5. 10.7554/ELIFE.07596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al Ageeli E., Drunat S., Delanoë C., Perrin L., Baumann C., Capri Y., Fabre-Teste J., Aboura A., Dupont C., Auvin S., et al. (2014). Duplication of the 15q11-q13 region: clinical and genetic study of 30 new cases. Eur J Med Genet 57, 5–14. 10.1016/J.EJMG.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 80.Urraca N., Cleary J., Brewer V., Pivnick E.K., Mcvicar K., Thibert R.L., Schanen N.C., Esmer C., Lamport D., and Reiter L.T. (2013). The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Res 6, 268–279. 10.1002/AUR.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen Y., Dies K.A., Holm I.A., Bridgemohan C., Sobeih M.M., Caronna E.B., Miller K.J., Frazier J.A., Silverstein I., Picker J., et al. (2010). Clinical Genetic Testing for Patients With Autism Spectrum Disorders. Pediatrics 125, e727. 10.1542/PEDS.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai G., Edelmann L., Goldsmith J.E., Cohen N., Nakamine A., Reichert J.G., Hoffman E.J., Zurawiecki D.M., Silverman J.M., Hollander E., et al. (2008). Multiplex ligation-dependent probe amplification for genetic screening in autism spectrum disorders: efficient identification of known microduplications and identification of a novel microduplication in ASMT. BMC Med Genomics 1. 10.1186/1755-8794-1-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wintle R.F., Lionel A.C., Hu P., Ginsberg S.D., Pinto D., Thiruvahindrapduram B., Wei J., Marshall C.R., Pickett J., Cook E.H., et al. (2011). A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism Research 4, 89–97. 10.1002/AUR.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Urraca N., Potter B., Hundley R., Pivnick E.K., McVicar K., Thibert R.L., Ledbetter C., Chamberlain R., Miravalle L., Sirois C.L., et al. (2016). A rare inherited 15q 11.2-q13.1 interstitial duplication with maternal somatic mosaicism renal carcinoma, and autism. Front Genet 7, 205. 10.3389/FGENE.2016.00205/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas J.A., Johnson J., Peterson Kraai T.L., Wilson R., Tartaglia N., LeRoux J., Beischel L., McGavran L., and Hagerman R.J. (2003). Genetic and clinical characterization of patients with an interstitial duplication 15q11-q13, emphasizing behavioral phenotype and response to treatment. Am J Med Genet A 119A, 111–120. 10.1002/AJMG.A.10176. [DOI] [PubMed] [Google Scholar]
- 86.Tsai H.-W., Taniguchi S., Samoza J., and Ridder A. (2015). Age- and Sex-Dependent Changes in Androgen Receptor Expression in the Developing Mouse Cortex and Hippocampus. Neurosci J 2015, 1–11. 10.1155/2015/525369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abi Ghanem C., Degerny C., Hussain R., Liere P., Pianos A., Tourpin S., Habert R., Macklin W.B., Schumacher M., and Ghoumari A.M. (2017). Long-lasting masculinizing effects of postnatal androgens on myelin governed by the brain androgen receptor. PLoS Genet 13, e1007049. 10.1371/JOURNAL.PGEN.1007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cunningham R.L., Lumia A.R., and McGinnis M.Y. (2012). Androgen Receptors, Sex Behaviour, and Aggression. Neuroendocrinology 96, 131. 10.1159/000337663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mhaouty-Kodja S. (2018). Role of the androgen receptor in the central nervous system. Mol Cell Endocrinol 465, 103–112. 10.1016/J.MCE.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 90.Gilbert J., O’Connor M., Templet S., Moghaddam M., Di Via Ioschpe A., Sinclair A., Zhu L.Q., Xu W., and Man H.Y. (2020). NEXMIF/KIDLIA Knock-out Mouse Demonstrates Autism-Like Behaviors, Memory Deficits, and Impairments in Synapse Formation and Function. J Neurosci 40, 237–254. 10.1523/JNEUROSCI.0222-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huo Y., Lu W., Tian Y., Hou Q., and Man H.-Y. (2022). Prkn knockout mice show autistic-like behaviors and aberrant synapse formation. iScience, 104573. 10.1016/j.isci.2022.104573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qiao H., Tian Y., Huo Y., and Man H.-Y. (2022). Role of the DUB enzyme USP7 in dendritic arborization, neuronal migration and autistic-like behaviors in mice. iScience 0. 10.1016/j.isci.2022.104595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zou J., Wang W., Pan Y.W., Lu S., and Xia Z. (2015). Methods to measure olfactory behavior in mice. Current protocols in toxicology / editorial board, Mahin D. (editor-in-chief) … [et al. ] 63, 11.18.1. 10.1002/0471140856.TX1118S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in the analysis of this study are available from the corresponding author upon reasonable request.