Abstract
HMG I/Y appears to be a multifunctional protein that relies on in its ability to interact with DNA in a structure-specific manner and with DNA, binding transcriptional activators via distinct protein-protein interaction surfaces. To investigate the hypothesis that HMG I/Y may have a role in human immunodeficiency virus type 1 (HIV-1) expression, we have analyzed whether HMG I/Y interacts with the 5′ long terminal repeat and whether this interaction can modulate transcription factor binding. Using purified recombinant HMG I, we have identified several high-affinity binding sites which overlap important transcription factor binding sites. One of these HMG I binding sites coincides with an important binding site for AP-1 located downstream of the transcriptional start site, in the 5′ untranslated region at the boundary of a positioned nucleosome. HMG I binding to this composite site inhibits the binding of recombinant AP-1. Consistent with this observation, using nuclear extracts prepared from Jurkat T cells, we show that HMG I (but not HMG Y) is strongly induced upon phorbol myristate acetate stimulation and this induced HMG I appears to both selectively inhibit the binding of basal DNA-binding proteins and enhance the binding of an inducible AP-1 transcription factor to this AP-1 binding site. We also report the novel finding that a component present in this inducible AP-1 complex is ATF-3. Taken together, these results argue that HMG I may play a fundamental role in HIV-1 expression by determining the nature of transcription factor-promoter interactions.
It is firmly established that chromatin (histones plus a wealth of nonhistone proteins of largely unknown function) plays a fundamental role in regulating the transcriptional activity of a gene by establishing highly specialized structures that can either promote or inhibit transcription factor binding. How such specialized structures, including the role of many well-characterized nonhistone proteins, are established to regulate the transcription process is poorly understood.
One important function of chromatin is to repress inappropriate transcription in either a reversible or permanent manner (48). This is achieved by compacting eukaryotic DNA, in a hierarchical fashion, into inaccessible complex three-dimensional structures. For transcription to occur, histone-DNA interactions in underlying nucleosomes must be disrupted to enable the binding of transcriptional activators to important regulatory elements. This appears to be achieved in a number of different ways. Large chromatin remodeling machines exist in the nucleus of eukaryotic cells that can, in an ATP-dependent manner, facilitate transcription factor binding (35). Posttranslational modification of histones, like histone acetylation, also plays an essential role in the gene activation process (28). The existence of cofactors that can increase the affinity and stability of transcriptional activators for their DNA binding site provides an alternative strategy by which DNA-binding proteins can effectively compete with histones for naked DNA. The ability of HMG I/Y to stimulate the DNA-binding activity of a wide variety of promoter-specific activators indicates that these chromatin-associated proteins may play such a role.
HMG I and Y are isoforms which are produced by alternative splicing, whereas the other member of the family, HMG I-C, is expressed as a separate gene product (6). HMG I/Y can directly interact via protein-protein interactions, employing different interaction surfaces, with a number of different transcriptional activators, including NF-κB (42, 50, 51), ATF-2 (13), SRF (8), NF-Y (10), Oct 2A (1), Elf (24), and c-Rel (22), increasing their affinity for DNA. Typically, target genes for this HMG I/Y enhancement of factor binding are inducible and include genes for cytokines such as beta interferon (42), interleukin-2 receptor α chain (24), E-selectin (30), interleukin-2, and macrophage colony-stimulating factor (22). More recently, it was also shown that expression of the nitric oxide synthase gene may be regulated by HMG I/Y (36). Enhancement of factor binding, in some cases, also requires interaction of HMG I/Y with DNA. One explanation for this finding is that the ability of HMG I/Y to bend DNA may create a more favorable DNA conformation for factor binding (15). For the beta interferon promoter, higher-order transcription factor complex formation can be further stabilized by HMG I/Y-factor interactions (14). Paradoxically, in the case of NF-κB, the protein-interacting domain of HMG I/Y includes part of the same domain that interacts with DNA (51). The biological importance of HMG I/Y in regulating gene expression is further implied by the finding that the expression of HMG I/Y is upregulated in rapidly proliferating cells, including early embryonic cells (7) and neoplastic tissues (19, 25).
HMG I/Y is characterized by three tandemly organized basic DNA-binding modules separated by a flexible linker; each individual module is capable of interacting with the minor groove of DNA in a structurally specific manner, with the second basic repeat being responsible for high-affinity binding (4, 18, 49, 51). These consensus basic repeats adopt a defined crescent-shaped planar structure, resembling the drugs netropsin and distamycin (6). These basic DNA-binding modules are referred to as AT-hooks because in most cases, but not all, they preferentially bind to the minor groove of AT-rich sequences (6). These proteins can also bind, with high affinity, to non-B-form DNA, such as synthetic four-way junctions and supercoiled plasmid substrates (6, 21, 34). These binding characteristics support the proposal that HMG I/Y may have additional roles in other DNA-dependent processes.
All stages of the human immunodeficiency virus type 1 (HIV-1) life cycle are dependent on host-specific cellular factors. Recently, HMG I/Y was shown to be a host-specific factor required for the integration of HIV-1 preintegration complexes (16, 23). To investigate the possibility that HMG I/Y may also be involved in HIV-1 transcription, we have analyzed the interaction of HMG I/Y with the viral long terminal repeat (LTR) using purified recombinant proteins and nuclear extracts prepared from living cells. We find that HMG I/Y can function to modulate both the efficiency and selectivity of AP-1 binding to HIV-1 promoter DNA. These results suggest that HMG I/Y may play an important role at all key stages in the life cycle of HIV-1.
MATERIALS AND METHODS
Protein purification and nuclear extract preparation.
Recombinant HMG I and the DNA-binding mutant form of HMG I were purified as described (39). This mutant form has four proline-to-alanine substitutions introduced at residues 57 and 61 (located in DNA-binding domain 2) and at residues 83 and 87 (located in DNA-binding domain 3). Freeze-dried proteins were resuspended in buffer A (20 mM HEPES [pH 7.9], 100 mM NaCl, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 0.1% NP-40). His-tagged c-Fos and c-Jun (33) were purified by the method described by Thanos and Maniatis (43). To make the Fos-Jun heterodimer, equimolar quantities of c-Fos and c-Jun were codialyzed against buffer A. Typically, the final concentration of Fos-Jun was around 300 ng/μl. Individual preparations varied considerably in terms of DNA-binding activity, with usually less than 10% of the total protein being active (33). As shown in Fig. 3A, maximal binding to site AP1-3 was achieved with 300 ng of total protein. Nuclear extracts were made from Jurkat T cells according to the method described earlier (11). Half the cells were induced for 2 h, prior to harvesting, with phorbol ester 12-myristate 13-acetate (PMA). To separate HMG I from the bulk of nuclear proteins, nuclear proteins were precipitated using 60% ammonium sulfate. The supernatant and the nuclear precipitate were dialyzed against buffer D (100 mM KCl) (11). It is worth noting that among the different nuclear extract preparations used (this investigation used four different preparations), the DNA-protein complexes produced did not differ qualitatively but differences occurred quantitatively.
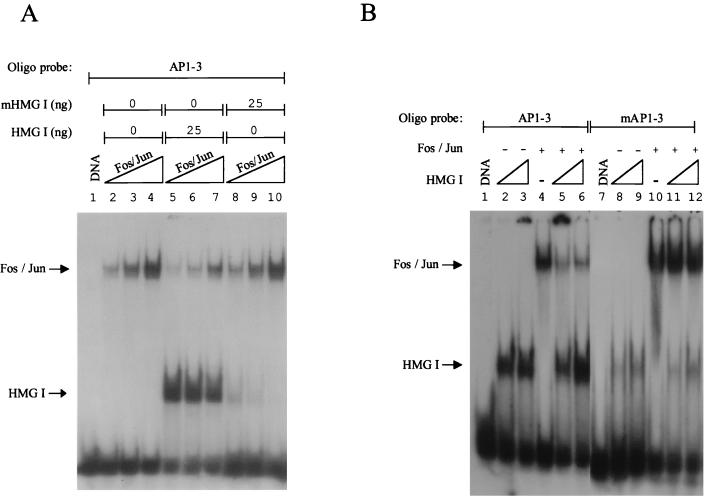
FIG. 3.
HMG I inhibits the binding of Fos-Jun to site AP1-3. (A) To examine the effect of HMG I on the binding of Fos-Jun to site AP1-3, a Fos-Jun titration (see Materials and Methods) was carried out in which either no protein, 25 ng of HMG I, or 25 ng of a mutant DNA-binding form of HMG I (mHMG I) was added to binding reactions containing a 25-bp probe. (B) The experiment in panel A was repeated except that the HMG I/Y binding site was mutated. Lanes 2, 5, 8, and 11 received 12 ng of HMG I, while lanes 3, 6, 9, and 12 received 25 ng of HMG I.
Gel mobility shift assays.
The mobility shift assays were carried out as described previously (33). The oligonucleotide used in gel mobility shift assays was 5′-CCCTTTTAGTCAGTGTGGAAAATCT-3′. The final buffer (buffer R) contained 6 mM HEPES (pH 7.9), 10 mM Tris (pH 8.0), 1 mM MgCl2, 1 mM EDTA (pH 8.0), 10 mM dithiothreitol, 5% glycerol, 1% sucrose, 0.1% NP-40, and 40 mM NaCl in a final volume of 20 μl. In the case of DNA-binding reactions using nuclear extracts, KCl replaced NaCl. Poly(dG-dC) and/or poly(dI-dC) (Amersham-Mannheim) were also included in reactions using both recombinant proteins (100 ng) and nuclear extracts (2 μg). Reactions were run on preelectrophoresed 4.5% nondenaturing polyacrylamide gels (0.5× Tris-borate-EDTA [TBE]) at 15 V/cm (4°C). Gels were dried, exposed to X-ray film, and quantitated by phosphoimaging.
DNase I footprinting assays.
DNase I footprinting analysis was carried out as described previously (33). DNA-binding reactions were carried out as for gel shift assays. Two PCR primers, upstream HIV-LTR (−187 to +15) and downstream HIV-LTR (−5 to +230), were synthesized with restriction sites at both ends to allow footprinting on both strands. Typically, 0.05 U of DNase I (Boehringer-Mannheim) was used in reactions involving purified recombinant proteins. Purified digestion products were run on a 7 M urea–8% polyacrylamide gel (1× TBE) at 40 V/cm, transferred to DEAE paper (Whatman), dried, and exposed to X-ray film or to a phosphoimage screen.
Western blotting and immunoprecipitation.
For Western blot analysis of HMG I/Y in nuclear extracts, affinity-purified rabbit polyclonal antibodies were used following the method described by Reeves and Nissen (39). Immunoprecipitation of HMG I/Y in nuclear extracts was carried out using the method described before (12). Typically, 8.5 μg of antibody was added to 100 μl of nuclear extract (7.5 mg/ml). Immunodepletion of HMG I/Y was confirmed by Western blot and gel shift analyses. AP-1/cyclic AMP response element binding protein (CREB) antibodies were purchased from Santa Cruz Biotechnology and used according to their instructions.
Immobilized-template assays.
Dynabeads (M280 streptavidin; Dynal) (200 μg) were washed twice in buffer T (10 mM Tris [pH 7.5], 1 mM EDTA, 1 M NaCl). Beads were then resuspended in 20 μl of buffer T containing 10 pmol of biotinylated oligonucleotide probe and agitated gently at room temperature for 30 min. After washing several times in buffer T to remove any unbound probe, beads were equilibrated in buffer R for 20 min. The magnetic beads were concentrated in a magnetic particle concentrator (Dynal) before being resuspended in buffer R containing 250 μg of nuclear protein and 40 ng of poly(dG-dC) per μl in a final volume of 120 μl and agitated gently for 20 min at room temperature. Binding reactions were then washed three times in buffer R containing 10 ng of poly(dG-dC) per μl before the beads were concentrated and resuspended in sodium dodecyl sulfate (SDS) loading dye, loaded on SDS–12% polyacrylamide gel electrophoresis (PAGE) gels, and electrophoresed at 15 V/cm for 1.5 h.
RESULTS
Characterization of HMG I/Y binding sites within the 5′-UTR of the HIV-1 promoter.
To begin to address the question of whether HMG I/Y may have a functional role in HIV-1 expression, we first examined whether these chromosomal proteins can interact with the LTR by determining the location of potential HMG I/Y binding sites using the DNase I footprinting assay. The region of the LTR that we focused on centred around the transcriptional initiation site (−187 to +230). This is a particularly interesting region of the promoter, which includes part of the 5′-UTR, because it contains a number of important regulatory elements that bind both inducible and constitutive transcription factors essential for efficient viral transcription and replication (2, 47). In addition, the 5′-UTR interacts with a specifically positioned nucleosome that is displaced or disrupted upon transcriptional activation (46). It is believed that the binding of AP-1 to the 5′-UTR may be involved in this disruption process (33, 47).
Representative footprints are shown in Fig. 1A to C, and a summary of the position of both high- and low-affinity HMG I/Y binding sites is shown in Fig. 2. In these experiments, highly purified recombinant HMG I was used (Fig. 1D); identical results were obtained with recombinant HMG Y (data not shown). Figure 1A displays high-affinity (footprint region 3) and low-affinity (footprint region 2) binding sites. The entire HMG I/Y footprinted region can extend over relatively large DNA distances (over 30 bp) because they are a composite of smaller individual footprints which reflect the binding of individual AT-hooks to DNA. The binding of at least two AT-hook peptide motifs is required for high-affinity binding (see Introduction). Figures 1B and C also displays high-affinity binding sites (footprint regions 5 and 3, respectively); only 5 ng of protein (21 nM) is required to produce a clear protected region.
FIG. 1.
5′-UTR of the HIV-1 promoter contains multiple high-affinity HMG I binding sites. To determine the location of potential HMG I binding sites, DNase I footprinting reactions were carried out as described in the text. Analysis of three segments within the 5′-UTR is shown. These include regions −105 to +1 (A), +56 to +128 (B), and +112 to +194 (C). The numbering is relative to the start site of transcription at +1. Solid boxes represent high-affinity binding sites, whereas shaded boxes illustrate the position of low-affinity HMG I binding sites. Since HMG I contains three distinct individual DNA-binding domains, each capable of producing a small footprint, the combination of these individual footprints has been described as a footprint region. (D) SDS–15% polyacrylamide gel showing 2 μg of recombinant HMG I.
FIG. 2.
Summary of the positions of HMG I binding sites within the 5′-LTR. Shown is the sequence of the 5′-LTR from −187 to +230. Previously identified transcription factor binding sites are also shown (2, 44). Solid and hatched rectangles below the sequence represent high- and low-affinity binding sites for HMG I, respectively. The shaded region (+10 to +155) indicates the approximate location of the positioned nucleosome identified by in vivo footprinting (43). The three AP-1 binding sites within this nucleosome are labeled AP1-1, AP1-2, and AP1-3. The two T residues that were mutated to G residues in footprint region 5 are highlighted with stars. Bracketed is the DNA probe used for gel shift analysis. R, start site of transcription.
The positions of these and other characterized footprints are located at potentially significant regions (Fig. 2). In other reported examples, typically HMG I/Y binds to sites that lie adjacent to or are part of a transcription factor binding site (see Introduction). Footprint region 5 overlaps an AP-1 and an NF-AT site, while footprint regions 1, 2, and 3 overlap binding sites for USF, NF-κB, and TATA-binding protein (TBP), respectively. Interestingly, footprint region 4 is located at or near the dyad of the positioned nucleosome, whereas footprint regions 3 and 5 are located at the boundaries. To begin an investigation of whether these HMG binding sites may have a biological function, in this study we have focused on whether the binding of HMG I to footprint region 5 can regulate the binding of AP-1.
HMG I can inhibit the binding of Fos-Jun.
The 5′-UTR of the HIV-1 promoter contains three AP-1 binding sites (AP1-1, AP1-2, and AP1-3) (Fig. 2). The first two sites are located within the positioned nucleosome, whereas the third site is located at the boundary. Given that the binding site for HMG I and this third AP-1 binding site overlap (footprint region 5), the effect of HMG I binding on the binding of Fos-Jun to this site was analyzed by carrying out a Fos-Jun titration in both the presence and absence of HMG I. Figure 3A clearly shows that the binding of HMG I to the labeled probe inhibits the binding of Fos-Jun to site AP1-3 (compare lanes 5 to 7 with lanes 2 to 4). We estimate that a 5- to 10-fold molar excess of HMG I is required to inhibit Fos-Jun binding by more than 80%. Furthermore, this inhibition of Fos-Jun binding is dependent on the ability of HMG I to bind to DNA, because when we used a DNA-binding mutant, this inhibition of binding was no longer observed (compare lanes 8 to 10 with lanes 5 to 7).
To confirm that this observed inhibition of Fos-Jun binding was due to specific HMG I binding, the binding site of HMG I was mutated, and this probe was used in DNA-binding assays. The mutation involved changing two A residues that lie immediately outside the core AP-1 binding site to C residues (TTTTAGTCAG to CCTTAGTCAG), (Fig. 2). Figure 3B shows that this mutation markedly inhibits HMG I binding to the labeled probe (compare lanes 8 and 9 with 2 and 3), and inhibition of Fos-Jun binding is no longer observed (compare lanes 11 and 12 with 5 and 6). Importantly, this mutation does not alter the binding of Fos-Jun (compare lane 10 with 4). These results have also been verified by using DNase I footprinting assays (data not shown). We therefore conclude that the binding of HMG I and Fos-Jun to this composite site is mutually exclusive. It is also worth noting that changing these two T residues to G residues alters the mobility of the free probe, implying that these T residues may be involved in DNA bending.
Interaction between endogenous HMG I and the 5′-UTR.
Next, we investigated whether the binding by HMG I to the composite site AP1-3 (footprint region 5) could be reproduced in a complex nuclear protein extract prepared from living cells. Given that phorbol esters can dramatically induce HIV-1 transcription (46), we addressed the question of whether HMG I/Y may be involved in this induction process by examining whether HMG I/Y present in PMA-induced Jurkat nuclear extracts can interact with site AP1-3. To study this possibility, gel mobility shift assays were performed using the same labeled probe as used in Fig. 3A.
Figure 4A shows that, because HMG I/Y is a minor groove DNA-binding protein, an interaction between HMG I/Y and DNA in nuclear extracts is observed but only when poly(dG-dC) and not poly(dI-dC) is used as competitor DNA in binding reactions. Most significantly, the specific association of HMG I/Y with site AP1-3 when poly(dG-dC) instead of poly(dI-dC) is used is correlated with the generation of a completely new protein-DNA binding profile (compare lanes 8 to 10 with lanes 3 to 5 in panel A of Fig. 4).
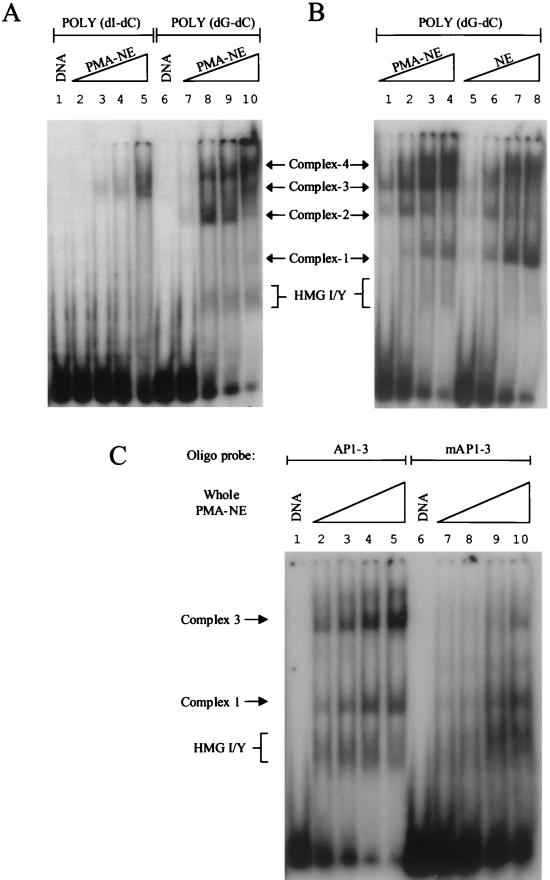
FIG. 4.
Endogenous HMG I binds to site AP1-3. (A) DNA-binding reactions were carried out using a labeled probe containing site AP1-3 and increasing amounts of a nuclear extract (NE) prepared from PMA-induced Jurkat cells. As indicated, reactions received either poly(dI-dC) or poly(dG-dC) (2 μg). Lanes 2 and 7, 3 and 8, 4 and 9, and 5 and 10 received 1, 5, 10, and 20 μg of nuclear extract, respectively. A mobility gel shift assay was performed to identify assembled nucleoprotein complexes. (B) DNA-binding reactions, as indicated, were carried out using nuclear extracts prepared from uninduced and PMA-induced Jurkat cells. The nucleoprotein complexes generated are shown as complexes 1 to 4. The formation of HMG I-DNA complexes is also highlighted. Lanes 1 and 5, 2 and 6, 3 and 7, and 4 and 8 received 2, 5, 10, and 15 μg of nuclear extract, respectively. (C) Mobility gel shift assay was carried out using labeled DNA probes that contained either a mutated or an unmodified HMG I/Y binding site. Lanes 2 to 5 and lanes 7 to 10 received 15, 20, 25, and 30 μg of nuclear extract, respectively.
Figure 4 also clearly shows a broad band that runs in the same position as an HMG I/Y-DNA complex in the induced extract but not in the uninduced extract (compare lanes 8 to 10 in panel A and lanes 2 to 4 in panel B with lanes 6 to 8 in panel B). Confirmation that this protein-DNA complex indeed contains HMG I/Y is shown in Fig. 4C. With the mutated HMG binding site used as a labeled probe at relatively high concentrations of nuclear extract, HMG I/Y binding is no longer observed (compare lanes 7 and 8 with lanes 2 and 3). It is worth noting that at the high nuclear extract concentrations used in this experiment, complex 2 is not seen (compare lane 4 with lane 2 in Fig. 4B). At even higher concentrations of nuclear extract, in the absence of HMG I/Y binding, nonspecific DNA-binding proteins like histones associate with the DNA probe (data not shown and lanes 9 and 10 in panel C). The conclusion that endogenous HMG I/Y binds to site AP1-3 is also supported by immunoprecipitation experiments (see below). At this stage we do not know why HMG I/Y protein-DNA complexes migrate as a diffuse band, but one possibility is that HMG I and HMG Y may be heavily modified in these extracts (see Discussion).
In addition to HMG I-DNA complexes, three additional protein-DNA complexes are observed when induced nuclear extracts are used. Complexes 1, 2, and 4 are also observed when uninduced extracts are employed, implying that these DNA-binding proteins are basal factors. On the other hand, AP-1 complex 3 is specifically induced upon PMA induction (compare lanes 1 to 4 with lanes 5 to 8 in Fig. 4B). We conclude that previous studies that have used poly(dI-dC) in their binding reactions have potentially missed important factor-DNA interactions at the 5′-UTR of the HIV-1 promoter (45).
Mutation of the HMG binding site not only inhibits HMG I/Y binding, but also abolishes the formation of the inducible AP-1 complex 3 and to a lesser extent complex 1 (compare lanes 7 and 8 with lanes 2 and 3 in Fig. 4C). Therefore, it is possible that, in contrast to the results in Fig. 3B, this mutation not only inhibits HMG I/Y binding but may also inhibit the formation of these nucleoprotein complexes, perhaps by altering the DNA-bending properties of the DNA fragment. Since the formation of inducible complex 3 correlates with the binding of HMG I/Y, the following experiments were designed to examine the interplay between HMG I/Y binding and complex 3 formation and whether the ability of HMG I to inhibit factor binding (Fig. 3A) has a role in this process.
HMG I is induced upon PMA stimulation.
The results in Fig. 4 indicate that HMG I/Y associates with site AP1-3 upon PMA induction. To investigate this further, a Western blot analysis was carried out examining the abundance of HMG I and HMG Y in uninduced and induced Jurkat extracts (Fig. 5A) (the SDS protein gel above the Western blot demonstrates that appropriate induced and uninduced lanes received an equivalent amount of protein; see the figure legend). Most interestingly, HMG I is markedly induced (about fivefold), whereas the induction of HMG Y was modest (compare lane 2 with lane 3 and lane 6 with lane 7). In an attempt to separate HMG I/Y from the bulk of nuclear protein, (NH4)2SO4 was added to the nuclear extract (final concentration, 60%). Most unexpectedly, HMG I fractionated to the supernatant, whereas HMG Y was precipitated with the bulk of nuclear protein (compare lanes 5 and 6 with lanes 3 and 4).
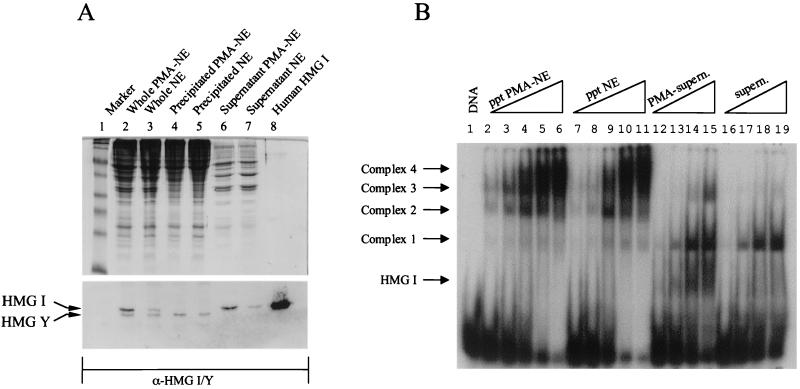
FIG. 5.
PMA induction of Jurkat T cells induces HMG I. (A) Nuclear extract (NE) proteins prepared from uninduced and PMA-induced Jurkat T cells were precipitated using 60% (NH4)2SO4 (see Materials and Methods). Western blot analysis of precipitated and supernatant fractions was carried out using affinity-purified polyclonal antibodies raised against HMG I/Y. Lanes 2 to 5 and 6 and 7 received 40 and 10 μg of total protein, respectively. (B) Mobility gel shift assay carried out using site AP1-3 and (NH4)2SO4-precipitated (ppt) and supernatant (supern.) fractions derived from uninduced and induced nuclear extracts. Lanes 2 to 6 and 7 to 11, received 1, 2, 5, 10, and 15 μg of total protein, respectively. Lanes 12 to 15 and 16 to 19 received 2, 5, 10, and 15 μg of total protein, respectively.
Having separated HMG I from HMG Y, it was of interest to investigate whether the HMG binding observed in unfractionated nuclear extracts (Fig. 4A) was due to the interaction of HMG I or HMG Y, or both, to site AP1-3. DNA-binding assays were carried out using induced and uninduced nuclear pellet and supernatant fractions, and the protein-DNA complexes formed were analyzed by mobility gel shift assays. Figure 5B clearly shows that HMG I present in the induced supernatant fraction can bind strongly to the labeled probe. Very little binding is observed in the uninduced supernatant fraction (compare lanes 12 to 15 with lanes 16 to 19). Interestingly, HMG Y present in the induced nuclear pellet fraction does bind to the labeled DNA fragment (lanes 2 to 6). We therefore conclude that HMG I is responsible for the binding observed in unfractionated nuclear extracts. Figure 5B also shows that basal complex 1 and a new complex (which migrates faster than complex 3) are also present in the supernatant fractions.
HMG I can selectively determine which nuclear DNA-binding protein binds to the 5′-UTR.
The above results have clearly demonstrated that using recombinant proteins, HMG I can inhibit AP-1 binding to site AP1-3 on the 5′-UTR (Fig. 3). On the other hand, using induced Jurkat nuclear extracts, an inducible AP-1 factor can bind to DNA in the presence of HMG I (Fig. 4). An attractive hypothesis that can reconcile these observations is that the induced HMG I can both selectively inhibit the binding of basal DNA-binding proteins (which are present in both uninduced and induced nuclear extracts) and promote the binding of the inducible AP-1 transcription factor. To test this hypothesis, we took advantage of our ability to separate HMG I from the inducible AP-1 complex (Fig. 5A). A titration was performed in which HMG I, present in the (NH4)2SO4 supernatant fraction, was added to a constant amount of precipitated nuclear protein which contains the PMA-inducible AP-1 factor (complex 3) (Fig. 6B). The supernatant fraction was immunoprecipitated with either HMG I antibodies or control antibodies against histone H2A.
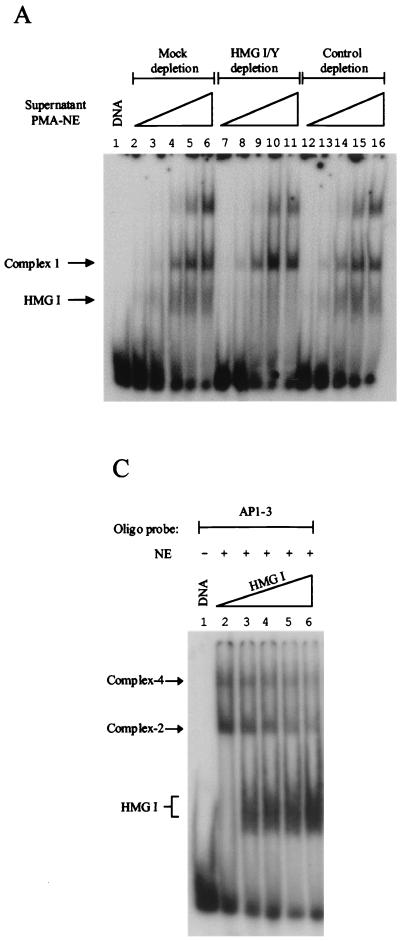
FIG. 6.
HMG I can both selectively inhibit and promote endogenous factor binding to site AP1-3. (A) The 60% (NH4)2SO4 supernatant fraction, derived from PMA-induced nuclear extracts (NE), was mock immunodepleted (lanes 2 to 6) or immunodepleted with affinity-purified antibodies against HMG I/Y (lanes 7 to 11) or H2A (control, lanes 12 to 16). A mobility gel shift assay, using site AP1-3, was carried out using these immunodepleted supernatant fractions. Lanes 2 to 6, 7 to 11, and 12 to 16 received 2, 5, 10, 15, and 20 μg of the supernatant fraction, respectively. (B) PMA-induced supernatant fractions that were immunodepleted with either control (H2A) or HMG I affinity-purified antibodies were titrated back to a PMA-induced nuclear precipitated fraction. Lanes 2 to 12 received 7 μg of nuclear (NH4)2SO4-precipitated proteins. Lanes 3 to 7 and lanes 8 to 12 received 5, 10, 20, 30, and 40 μg of H2A- or HMG I/Y-depleted supernatant fractions, respectively. (C) Increasing amounts of recombinant HMG I were added to DNA-binding reactions that received 10 μg of uninduced nuclear precipitated extract and a labeled oligonucleotide that contained site AP1-3. Lanes 2 to 6 received 0, 25, 50, 100, and 200 ng of HMG I, respectively.
Figure 6A shows that HMG I/Y immunodepletion, and not mock or immunodepletion of H2A, of the induced supernatant fraction removes only HMG I and not the other DNA-binding proteins present in this fraction, including basal factor 1. Lane 2 of Fig. 6B shows the formation of basal complex 2 and the inducible AP-1 complex 3 in the induced nuclear precipitated fraction. When control immunodepleted supernatant (containing HMG I) is titrated back to the (NH4)2SO4-precipitated fraction, at the lowest concentration of supernatant, formation of basal complex 2 is inhibited (compare lane 3 with lane 2). As the concentration of HMG I is increased in the binding reaction, the formation of inducible AP-1 complex 3 is enhanced. Concurrently, the production of basal complex 1 is also observed (this is because this basal factor is present in the supernatant fraction). This binding profile mimics the situation when the unfractionated nuclear extract is used in binding reactions (compare lane 7 of Fig. 6B with lane 4 of Fig. 4B).
Strikingly, when HMG I is immunodepleted, the production of the inducible AP-1 complex 3 is markedly reduced, while the formation of basal complex 1 is enhanced (Fig. 6B, compare lanes 9 to 12 with lanes 4 to 7). This result clearly shows that the inducible AP-1 factor is in competition with the basal DNA-binding protein for the same binding site and that HMG I selectively facilitates the formation of the inducible AP-1–DNA complex. This result can also be mimicked when the uninduced supernatant fraction is added back (data not shown). In addition, at the lowest concentration of HMG I-depleted supernatant added, the formation of basal complex 2 is no longer inhibited (compare lane 8 with lane 3). However, as the concentration of the HMG I-immunodepleted supernatant increases, basal factor 1 outcompetes basal factor 2 for DNA binding. Clearly, there is a complex interplay between the inducible AP-1 factor, the different basal factors, and HMG I for DNA binding, but this result clearly shows that in this complex mixture of proteins, HMG I selectively enhances the binding of the inducible AP-1 factor to site AP1-3.
To confirm that HMG I can indeed inhibit the DNA-binding activity of basal factor 2, an HMG I titration was carried out in which recombinant HMG I was added to uninduced nuclear precipitated extracts. Lane 2 of Fig. 6C shows the formation of DNA-protein complexes 2 and 4. As increasing amounts of HMG I are added to the binding reactions, formation of both of these complexes is inhibited, which coincides with the binding of HMG I to the labeled probe. This result clearly shows that HMG I can inhibit the binding not only of recombinant AP-1, but also of basal DNA-binding proteins present in uninduced nuclear extracts. Taken together, these results support the above hypothesis by showing that HMG I can both inhibit and facilitate factor binding in a selective manner and provide a potential mechanism for how this nonhistone chromosomal protein may play an important role in HIV-1 expression by determining which transcription factor associates with important DNA regulatory elements.
PMA-inducible AP-1 complex contains ATF-3.
Having determined that HMG I plays an important role in facilitating the binding of inducible AP-1 complex 3 to DNA, the next important question to be addressed was to identify the key AP-1 components present in this inducible complex. Previously, it was reported that by employing supershift assays using antibodies raised against different AP-1 members, c-Fos, Jun D, CREB, ATF-1, and ATF-2 interacted with site AP1-3 (37, 38). However, employing this assay, we were unable to reproduce these results. Furthermore, close examination of these reported studies revealed that these transcription factors are only minor components of the inducible complex. We therefore adopted an alternative approach to determine the major components of this inducible complex. Biotinylated site AP1-3 was incubated with PMA-induced and uninduced nuclear extracts under the same conditions as used for gel shift experiments (Fig. 4). The DNA-bound transcription factors were then purified from the bulk of nuclear proteins by using magnetic streptavidin beads (Fig. 7B). After several washes with binding buffer, the isolated DNA-bound proteins were eluted from the DNA and analyzed by Western blotting using a battery of different ATF/CREB family members (Fig. 7A). In crude nuclear extracts, all of the tested ATF/CREB family members were present. As expected, treatment of Jurkat T cells with PMA, under the conditions used here, induced the synthesis of c-Fos, CREB-1, c-Jun, and ATF-3. Most interestingly, neither CREB-1, c-Jun, nor c-Fos was purified by site AP1-3 attached to magnetic streptavidin beads. On the other hand, we find that ATF-3 is the major component purified from the crude nuclear extract. A minor component that binds to site AP1-3 is ATF-2. Importantly, as a control, these transcription factors were not detected when magnetic beads alone were incubated with nuclear extracts (data not shown). The specificity of this assay is also highlighted by the observation that despite a strong antibody-Jun D reaction in crude nuclear extracts, Jun D does not bind to the purified DNA probe. In addition to ATF-3, Fig. 7B shows that Fra-1 is a factor present in uninduced nuclear extracts which binds strongly to site AP1-3. Under the induction conditions used here, the level of DNA binding does not change between the induced and uninduced states.
FIG. 7.
Inducible AP-1 complex 3 contains ATF-3. Biotinylated site AP1-3 was incubated with induced or uninduced nuclear extract (NE) (250 μg of total protein), and the DNA-bound AP-1 factors were isolated as described in Materials and Methods. The isolated proteins were run on an SDS–12% polyacrylamide gel and either probed with commercial antibodies raised against different AP-1/CREB family members (A) or stained with silver (B). To determine which AP-1/CREB family members are present initially in uninduced and induced nuclear extracts, 15 μg of unpurified nuclear extract was also probed with the different antibodies (A) and stained with silver (B). NE, uninduced Jurkat T cells; PMA-NE, Jurkat T cells stimulated with PMA.
DISCUSSION
The focus of this study was to examine whether HMG I can interact with an important regulatory region in the HIV-1 promoter, the 5′-UTR, and whether this interaction could play a role in modulating the binding of AP-1 to key binding sites in this region. The approach adopted was to define HMG I interactions, in combination with AP-1, with the 5′-UTR using recombinant proteins. Then we tested the significance of these findings by employing nuclear extracts containing endogenous HMG I/Y prepared from PMA-induced Jurkat extracts, since phorbol esters are strong inducers of HIV-1 expression.
In the 5′-UTR, we have identified several high-affinity sites for HMG I/Y, and most of these sites overlap important transcription factor binding sites. Interestingly, similar uncharacterized footprints can be seen in a genomic footprint of integrated HIV-1 (see Fig. 2 in reference 5). A number of previous studies have shown that such an overlap can have a critical role in regulating gene expression (22, 24, 30, 36). We have focused on one of these overlapping binding sites, site AP1-3. The location of this AP-1 binding site, at the boundary of a positioned nucleosome, suggests that it may play an important role in modulating chromatin architecture. We have observed enhanced HMG I binding to site AP1-3 using PMA-induced nuclear extracts but only under in vitro conditions that allow HMG I binding, i.e., when poly(dG-dC) rather than poly(dI-dC) is used as competitor DNA. Most importantly, PMA also induces the synthesis of a new AP-1 complex, and strong binding of this transcription factor to site AP1-3 is dependent upon HMG I. Because of this new finding, previous in vitro binding studies should be reevaluated. Concerning the role of HMG I/Y, we cannot rule out the possibility that at least a subset of these identified binding sites may have a role in the integration process.
A major unanswered question concerning the function of these chromosomal proteins is whether HMG I and Y play the same role or have different roles. Interestingly, we observed that HMG I and not HMG Y is strongly induced upon PMA stimulation of Jurkat cells. This suggests that these proteins may indeed have different functions. Given that these two proteins originate from alternative splicing of the same RNA transcript, this differential regulation must occur via a posttranscriptional mechanism. Such differential control could, for example, operate at the level of the splicing event itself, result from differences in the stability or translational efficiency of the alternatively spliced transcripts, or, perhaps, result from intrinsic differences in the stability of the HMG I and HMG Y proteins (although both proteins appear to be extremely stable in living cells) (6). A similar selective enhancement of HMG I protein by tetradecanoyl phorbol acetate was observed in transformation-resistant JB6 murine cell lines (9). Most interestingly, this same study raised the possibility that HMG Y may have a role in the transformation process, since they observed that HMG Y is induced by tumor promoters only in transformation-sensitive cells. Potentially, the conserved 11-amino-acid segment absent in HMG Y might alter the quality or the specificity of protein-protein interactions with target proteins. Concerning their DNA-binding activity in Jurkat nuclear extracts, we also report here another difference between HMG I and HMG Y. HMG Y is present in both uninduced and induced nuclear extracts and, at least as shown by mobility gel shift assays, does not bind to DNA when both of these nuclear extracts are used. Although this may be due to low abundance, another possible explanation for this is that HMG Y is postranslationally modified in vivo in a differential manner from HMG I (Banks and Reeves, unpublished data), and it is this constellation of secondary biochemical modifications (including phosphorylation and others) that inhibits the binding of the HMG Y isoform protein to DNA (32). Clearly, this hypothesis will need to be tested.
Many transcription factor families, like AP-1, comprise different individual members which are able to dimerize with each other to form a diverse range of transcription factor complexes, each capable of recognizing the same or similar DNA-binding sequences. The mechanisms that operate in the nucleus to determine which factor actually binds to a regulatory site in a promoter are poorly understood. The results presented here suggest that HMG I may play an important role in this selection process. We found that in a complex nuclear extract consisting of a wealth of DNA-binding proteins, a situation analogous to the environment within the nucleus, HMG I can selectively promote the binding of an inducible AP-1 factor, ATF-3, to the AP1-3 site (complex 3), relative to the binding of competing basal DNA-binding proteins. In such a competition mechanism, the ability of HMG I to increase the affinity or stability of ATF-3 for site AP1-3 would enable this factor to compete more effectively with basal factors. The binding of this inducible AP-1 factor would be further enhanced if HMG I could selectively and directly inhibit the binding of basal factors. Indeed, such an inhibition of binding was observed when HMG I was added to uninduced extracts. Therefore, HMG I can either selectively interfere with or enhance protein-DNA binding, and which of these events occurs appears to be partially dependent on the nature of the DNA-binding protein involved (see below). Precedents exist for such a mode of differential regulation by the HMG I protein. Previously, using two different recombinant isoforms of ATF-2, ATF-2195 and ATF-2192, it was shown that while HMG I could stimulate ATF-2195 DNA binding, it actually inhibited the binding of ATF-2192 to the beta interferon promoter. ATF-2192 lacks important amino acid residues involved in HMG I–ATF-2 interactions (13). Such a striking differential effect was also observed when the interaction of Oct-1 and Oct-2 with the octamer sequence was compared (1). HMG I selectively enhanced Oct-2 binding while at the same time inhibiting Oct-1–DNA interactions.
There are at least two, nonmutually exclusive, mechanisms by which HMG I could enhance ATF-3 binding to site AP1-3 in a PMA-induced nuclear extract. These HMG proteins have been described as being architectural transcription factors because they can bend DNA, thereby establishing a particular DNA conformation that may be more favorable for transcription factor binding (6). The binding of several HMG molecules along a stretch of DNA may even create a DNA scaffold that can facilitate the formation of large nucleoprotein complexes via cooperative protein-protein interactions. Such a mechanism operates in the induction of human beta interferon gene expression (44). Individually, the binding of two HMG I molecules to positive regulatory domains IV and II enhances ATF-2/c-Jun and NF-κB binding, respectively, by reversing intrinsic DNA bends at these factor-binding sites. In combination, these two HMG proteins facilitate the formation of a highly stable and sterospecific transcription factor complex. The selectivity observed with regard to the enhanced binding of PMA-inducible ATF-3 to site AP1-3 could, in part, be explained by the generation of a highly specific DNA structure by HMG I. Analogous to the human beta interferon gene, it will also be interesting to examine whether the location of multiple HMG I binding sites in the 5′-UTR contributes to the formation of a large stable nucleoprotein complex.
Potentially, HMG I could also selectively enhance the binding of one transcription factor over another by its ability to interact directly with transcription factors themselves via specific protein-protein interaction surfaces. Indeed, stabilization of the assembled nucleoprotein complex on the human beta interferon gene enhancer appears to require specific protein-protein interactions between HMG I and transcriptional activators (50). Such protein-protein interactions can also enhance factor binding in a mechanism that is independent of HMG I's interacting with DNA. For example, protein-protein interactions between HMG I and NF-Y (10) and SRF (8) are sufficient for enhancement of their DNA-binding activity, presumably by inducing an active conformation. It has also been shown that HMG I can, in part, stimulate the binding of ATF-2/c-Jun to positive regulatory domain IV by promoting the dimerization reaction. Interestingly, we have observed that HMG I can enhance Fos-Jun binding to site AP1-1 in nuclear extracts even though HMG I does not bind to this AP-1 site (unpublished data).
Our results, using recombinant and nuclear proteins, also suggest that the second part of the mechanism that promotes specific binding of the inducible AP-1 complex to the composite site AP1-3 is the selective inhibition of binding of basal DNA-binding proteins by HMG I. Since the binding sites for HMG I and AP-1 overlap, this competitive inhibition could be achieved either by direct steric hindrance or by HMG I altering the conformation of the DNA to a structure that is not compatible for basal factor binding. Interestingly, HMG I does not inhibit the binding of recombinant NF-AT to an adjacent binding site (Fig. 2 and data not shown). As discussed above, HMG I can selectively inhibit the DNA-binding activity of ATF-2192 and Oct-1. It has also been reported that HMG I can inhibit the binding of NF-AT factors to the interleukin-4 promoter (26). This inhibition of binding, which is reversed by phosphorylation of HMG I, is believed to be important for development of Th2 cells. Similarly, HMG I (and HMG I-C) can interfere with the binding of homeodomain proteins to target sequences (these sequences contain TAAT as a core motif) (3). This inhibition appears to be due to HMG I-induced conformational changes in the DNA. Therefore, it appears that HMG I can modulate gene expression by functioning either as an activator or as a repressor. Moreover, we show here that HMG I can enhance the binding of one DNA-binding protein and inhibit the binding of another factor even on the same transcription factor-binding site. We also conclude that the final outcome of whether HMG I selectively inhibits or stimulates transcription factor binding is dependent not only on the nature of the transcriptional activator, but also on a complex interplay between relative DNA affinities and protein concentrations, the location of the HMG binding site with respect to the factor-binding site, and presumably, the biochemical modification status of HMG I.
Transfection-cocultivation experiments with wild-type and mutant HIV-1 proviral DNAs have show that the three AP-1 sites encompassing the downstream-positioned nucleosome play a fundamental role in the life cycle of the virus. Sites AP1-1 and AP1-2 (Fig. 2) play a critical role on HIV-1 replication, while site AP1-3 appears to be important for transcriptional activation in response to a broad range of external stimuli under different physiological conditions (37, 38, 41, 45). This site is highly conserved among HIV-1 isolates. Studies employing supershift analysis have reported that AP-1 complexes that interact with this site contain c-Fos and Jun D as well as CREB, ATF-1, and ATF-2 (38, 41). In contrast to these published results, using a more stringent assay, we have found that ATF-3 is a major component that interacts with site AP1-3. One possible reason for this discrepancy is that under our DNA-binding conditions, HMG I is able to interact with the DNA, thereby influencing the composition of bound transcription factors.
The association of ATF-3 with site AP1-3 is consistent with the observation that this site responds to a broad range of extracellular stimuli. ATF-3 mRNA is induced in cultured cells within 2 h by many treatments like different growth-stimulating factors, PMA, and cytokines as well as physiological stresses, including tissue damage (reviewed in reference 20). Consistent with these observations, analysis of the 5′-flanking region of the ATF-3 gene revealed inducible AP-1, ATF/CRE, and NF-κB binding sites. Interestingly, E2F and Myc/Max binding sites were also identified, raising the possibility that ATF-3 may be regulated in a cell cycle-dependent manner (31). ATF-3 functions as an activator when it heterodimerizes with Jun family members (20). The results of this study show that neither c-Jun nor Jun D partners ATF-3 to assemble PMA-inducible complex 3. To date, no physiologically important target genes mediating the activation of signaling pathways involving ATF-3 have been identified. Interestingly, we also have identified the Fos-related antigen Fra-1 as a factor that binds to site AP1-3 in uninduced nuclear extracts. Consistent with this observation, it has been reported that, in contrast to c-Fos, the fra-1 gene is expressed at high levels in proliferating cells (27).
The results of this study raise the possibility that HMG I may play an important role in HIV-1 expression. Preliminary experiments have shown that antisense HMG I RNA can reduce the expression of HIV reporter constructs (data not shown). Other viruses might employ this strategy to ensure their propagation in host cells, especially if critical transcription factors have a low affinity for important viral promoter elements. HMG I has been shown to stimulate Tst-1/Oct-6 binding to an important regulatory element that mediates the activation of human papovavirus JC virus gene expression (29). The latency-active promoter 2 in herpes simplex virus type 1 (which becomes a nucleosomal episome) contains a stretch of 23 thymidine residues critical for promoter activity, which interacts with HMG I/Y. In vitro, the binding of HMG I/Y to this promoter element enhances the binding of SP-1 to neighboring binding sites (17). With regard to chromatin disruption and the activation of HIV-1 transcription, based on the location of site AP1-3 at the boundary of the positioned nucleosome, we postulate that HMG I may play a fundamental role in the chromatin remodeling process. The association of HMG I with the nucleosomal dyad may also contribute to this disruption process (40). We are currently testing this hypothesis.
ACKNOWLEDGMENTS
The first two authors contributed equally to the manuscript.
We thank Mark Nissen for providing recombinant HMG proteins, Adele Holloway for assistance in setting up the immobilized-template assay, and Frances Shannon for many helpful discussions.
REFERENCES
- 1.Abdulkadir S A, Krishna S, Thanos D, Maniatis T, Strominger J L, Ono S J. Functional roles of the transcription factor Oct-2A and the high mobility group protein I/Y in HLA-DRA gene expression. J Exp Med. 1995;182:487–500. doi: 10.1084/jem.182.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Harthi L, Roebuck K A. Human immunodeficiency virus type-1 transcription: role of the 5′-untranslated leader region. Int J Mol Med. 1998;1:875–881. doi: 10.3892/ijmm.1.5.875. [DOI] [PubMed] [Google Scholar]
- 3.Arlotta P, Rustighi A, Mantovani F, Manfioletti G, Giancotti V, Tell G, Damante G. High mobility group I proteins interfere with the homeodomains binding to DNA. J Biol Chem. 1997;272:29904–29910. doi: 10.1074/jbc.272.47.29904. [DOI] [PubMed] [Google Scholar]
- 4.Banks G C, Mohr B, Reeves R. The HMG-I(Y) A.T-hook peptide motif confers DNA-binding specificity to a structured chimeric protein. J Biol Chem. 1999;274:16536–16544. doi: 10.1074/jbc.274.23.16536. [DOI] [PubMed] [Google Scholar]
- 5.Brown D A, Xu X, Nerenberg M. Genomic footprinting of HTLV type I and HIV type 1 in human T cell lines. AIDS Res Hum Retroviruses. 1996;12:829–832. doi: 10.1089/aid.1996.12.829. [DOI] [PubMed] [Google Scholar]
- 6.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 7.Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai B M, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. [PubMed] [Google Scholar]
- 8.Chin M T, Pellacani A, Wang H, Lin S S, Jain M K, Perrella M A, Lee M E. Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y) J Biol Chem. 1998;273:9755–9760. doi: 10.1074/jbc.273.16.9755. [DOI] [PubMed] [Google Scholar]
- 9.Cmarik J L, Li Y, Ogram S A, Min H, Reeves R, Colburn N H. Tumor promoter induces high mobility group HMG-Y protein expression in transformation-sensitive but not -resistant cells. Oncogene. 1998;16:3387–3396. doi: 10.1038/sj.onc.1201888. [DOI] [PubMed] [Google Scholar]
- 10.Currie R A. Functional interaction between the DNA binding subunit trimerization domain of NF-Y and the high mobility group protein HMG-I(Y) J Biol Chem. 1997;272:30880–30888. doi: 10.1074/jbc.272.49.30880. [DOI] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrov S, Dasso M C, Wolffe A P. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126:591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du W, Maniatis T. The high mobility group protein HMG I(Y) can stimulate or inhibit DNA binding of distinct transcription factor ATF-2 isoforms. Proc Natl Acad Sci USA. 1994;91:11318–11322. doi: 10.1073/pnas.91.24.11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 15.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 16.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 17.French S W, Schmidt M C, Glorioso J C. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol Cell Biol. 1996;16:5393–5399. doi: 10.1128/mcb.16.10.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geierstanger B H, Volkman B F, Kremer W, Wemmer D E. Short peptide fragments derived from HMG-I/Y proteins bind specifically to the minor groove of DNA. Biochemistry. 1994;33:5347–5355. doi: 10.1021/bi00183a043. [DOI] [PubMed] [Google Scholar]
- 19.Giancotti V, Pani B, D'Andrea P, Berlingieri M T, Di Fiore P P, Fusco A, Vecchio G, Philp R, Crane-Robinson C, Nicolas R H, et al. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by cotransfection with c-mycand polyoma middle T genes. EMBO J. 1987;6:1981–1987. doi: 10.1002/j.1460-2075.1987.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hai T, Wolfgang C D, Marsee D K, Allen A E, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 21.Hill D A, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himes S R, Coles L S, Reeves R, Shannon M F. High mobility group protein I(Y) is required for function and for c-Rel binding to CD28 response elements within the GM-CSF and IL-2 promoters. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]
- 23.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K R, Lehn D A, Elton T S, Barr P J, Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y) J Biol Chem. 1988;263:18338–18342. [PubMed] [Google Scholar]
- 26.Klein-Hessling S, Schneider G, Heinfling A, Chuvpilo S, Serfling E. HMG I(Y) interferes with the DNA binding of NF-AT factors and the induction of the interleukin 4 promoter in T cells. Proc Natl Acad Sci USA. 1996;93:15311–15316. doi: 10.1073/pnas.93.26.15311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovary K, Bravo R. Existence of different Fos/Jun complexes during the G0-to-G1transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol Cell Biol. 1992;12:15–23. doi: 10.1128/mcb.12.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:15–26. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 29.Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis H, Kaszubska W, DeLamarter J F, Whelan J. Cooperativity between two NF-kappa B complexes, mediated by high-mobility-group protein I(Y), is essential for cytokine-induced expression of the E-selectin promoter. Mol Cell Biol. 1994;14:5701–5709. doi: 10.1128/mcb.14.9.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang G, Wolfgang C D, Chen B P, Chen T H, Hai T. ATF3 gene: genomic organization, promoter, and regulation. J Biol Chem. 1996;271:1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 32.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 33.Ng K W, Ridgway P, Cohen D R, Tremethick D J. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 1997;16:2072–2085. doi: 10.1093/emboj/16.8.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissen M S, Reeves R. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J Biol Chem. 1995;270:4355–4360. doi: 10.1074/jbc.270.9.4355. [DOI] [PubMed] [Google Scholar]
- 35.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 36.Perrella M A, Pellacani A, Wiesel P, Chin M T, Foster L C, Ibanez M, Hsieh C M, Reeves R, Yet S F, Lee M E. High mobility group-I(Y) protein facilitates nuclear factor-kappaB binding and transactivation of the inducible nitric-oxide synthase promoter/enhancer. J Biol Chem. 1999;274:9045–9052. doi: 10.1074/jbc.274.13.9045. [DOI] [PubMed] [Google Scholar]
- 37.Rabbi M F, Al-Harthi L, Roebuck K A. TNFalpha cooperates with the protein kinase A pathway to synergistically increase HIV-1 LTR transcription via downstream TRE-like cAMP response elements. Virology. 1997;237:422–429. doi: 10.1006/viro.1997.8798. [DOI] [PubMed] [Google Scholar]
- 38.Rabbi M F, Saifuddin M, Gu D S, Kagnoff M F, Roebuck K A. U5 region of the human immunodeficiency virus type 1 long terminal repeat contains TRE-like cAMP-responsive elements that bind both AP-1 and CREB/ATF proteins. Virology. 1997;233:235–245. doi: 10.1006/viro.1997.8602. [DOI] [PubMed] [Google Scholar]
- 39.Reeves R, Nissen M S. Purification and assays for high mobility group HMG-I(Y) protein function. Methods Enzymol. 1999;304:155–188. doi: 10.1016/s0076-6879(99)04011-2. [DOI] [PubMed] [Google Scholar]
- 40.Reeves R, Wolffe A P. Substrate structure influences binding of the non-histone protein HMG-I(Y) to free nucleosomal DNA. Biochemistry. 1996;35:5063–5074. doi: 10.1021/bi952424p. [DOI] [PubMed] [Google Scholar]
- 41.Roebuck K A, Gu D S, Kagnoff M F. Activating protein-1 cooperates with phorbol ester activation signals to increase HIV-1 expression. AIDS. 1996;10:819–826. doi: 10.1097/00002030-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 42.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 43.Thanos D, Maniatis T. In vitro assembly of enhancer complexes. Methods Enzymol. 1996;274:162–173. doi: 10.1016/s0076-6879(96)74015-6. [DOI] [PubMed] [Google Scholar]
- 44.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 45.Van Lint C, Amella C A, Emiliani S, John M, Jie T, Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol. 1997;71:6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. . (Erratum, 12:4900.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widlak P, Garrard W T. Nucleosomes and regulation of gene expression: structure of the HIV-1 5′LTR. Acta Biochim Pol. 1998;45:209–219. [PubMed] [Google Scholar]
- 48.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol Cell Biol. 1997;17:3649–3662. doi: 10.1128/mcb.17.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yie J, Merika M, Munshi N, Chen G, Thanos D. The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J. 1999;18:3074–3089. doi: 10.1093/emboj/18.11.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X M, Verdine G L. A small region in HMG I(Y) is critical for cooperation with NF-kappaB on DNA. J Biol Chem. 1999;274:20235–20243. doi: 10.1074/jbc.274.29.20235. [DOI] [PubMed] [Google Scholar]