Abstract
Objectives:
Antimicrobial resistant (AMR) Campylobacter is a global health threat; however, there is limited information on genomic determinants of resistance in low- and middle-income countries. We evaluated genomic determinants of AMR using a collection of whole genome sequenced Campylobacter jejuni and C. coli isolates from Iquitos, Peru.
Methods:
Campylobacter isolates from two paediatric cohort studies enriched with isolates that demonstrated resistance to ciprofloxacin and azithromycin were sequenced and mined for AMR determinants.
Results:
The gyrA mutation leading to the Thr86Ile amino acid change was the only gyrA mutation associated with fluoroquinolone resistance identified. The A2075G mutation in 23S rRNA was present, but three other 23S rRNA mutations previously associated with macrolide resistance were not identified. A resistant-enhancing variant of the cmeABC efflux pump genotype (RE-cmeABC) was identified in 36.1% (35/97) of C. jejuni genomes and 17.9% (12/67) of C. coli genomes. Mutations identified in the CmeR-binding site, an inverted repeat sequence in the cmeABC promoter region that increases expression of the operon, were identified in 24/97 C. jejuni and 14/67 C. coli genomes. The presence of these variants, in addition to RE-cmeABC, was noted in 18 of the 24 C. jejuni and 9 of the 14 C. coli genomes.
Conclusions:
Both RE-cmeABC and mutations in the CmeR-binding site were strongly associated with the MDR phenotype in C. jejuni and C. coli. This is the first report of RE-cmeABC in Peru and suggests it is a major driver of resistance to the principal therapies used to treat human campylobacteriosis in this setting.
Keywords: Campylobacteriosis, Antibiotic resistance, Gastroenteritis, Iquitos, Whole genome sequencing
1. Introduction
Antimicrobial-resistant Gram-negative gastrointestinal pathogens are a global public health concern. The Centers for Disease Control and Prevention classified antimicrobial-resistant (AMR) Campylobacter as a pathogen of serious threat in 2019. Although the rates of azithromycin resistance have been stable for the past 10 years (2%–3% of isolates), there has been a continued rise in ciprofloxacin resistance [1], surpassing 30% in 2019 in the United States [2]. Macrolide and fluoroquinolone resistance poses a practical challenge in the clinic. Specifically, these are the two most effective oral antimicrobial treatment strategies for campylobacteriosis, the most common cause of bacterial enteritis globally [3, 4]. Although antimicrobials are not advised for treatment for uncomplicated gastroenteritis, antimicrobial therapy is advised for patients with severe symptoms, dysentery, and persistent infections and patients who are immunocompromised or pregnant. In a recent scoping review of 17 studies that described the risk of AMR Campylobacter, all studies described an elevated risk of drug resistance in individuals who reported foreign travel [5].
Sequencing clinical isolate genomes is an increasingly accessible technology, even in low- and middle-income settings (LMIC). This has the potential to guide antimicrobial therapy by characterizing the genetic variation that underlies resistance, theoretically faster than traditional culture and phenotypic resistance testing. However, Campylobacter genomes from LMICs, including Peru, are critically underrepresented in publicly available datasets, representing less than 5% of publicly available genomes (PubMLST Campylobacter jejuni/coli database; accessed 14 December 2023). As a result, monitoring of AMR determinants from Campylobacter genomes from these areas of the world is also limited. In Peru, increasing trends of ciprofloxacin and azithromycin resistance have been reported in clinical isolates [6,7]. However, there is a lack of genomic characterization of AMR determinants [8].
We examined human isolates of C. jejuni and C. coli collected from 2010 to 2022 from the Peruvian Amazon. The isolate collection included Campylobacter isolates from two paediatric cohort studies with specimen selection enriched to include strains with demonstrated phenotypic resistance to fluoroquinolones and azithromycin or clinical persistence. Through whole genome analysis, we identified genomic determinants associated with resistance to fluoroquinolones and macrolides, and other antibiotics with established CLSI breakpoints.
2. Methods
2.1. Sample selection
C. jejuni and C. coli isolates were collected as part of two paediatric cohorts in Iquitos, Peru. Stool samples in both cohorts were collected every time a child experienced diarrhoea, in addition to a routine monthly sample (even in the absence of diarrhoeal symptoms) for surveillance purposes [9]. Campylobacter was cultured using standard microbiology techniques [6,10]. The first cohort consisted of a collection of 917 Campylobacter isolates collected from children between 2009 and 2016 [11]. From this first cohort, a selection criterion was used to assure the presence of isolates resistant to both azithromycin and ciprofloxacin, two first-line antimicrobial agents [12], with 11 C. jejuni and 27 C. coli isolates selected and sequenced. We additionally selected isolates identified from persistent infections since these would be the most likely to need antibiotic treatment for campylobacteriosis in this population [13,14]. Persistent infection was defined as three consecutive infections detected by culture from monthly surveillance samples (46 C. jejuni and 26 C. coli) [15]. The second cohort is an ongoing birth cohort in which enrolment began in 2021 and had no selection, with 40 C. jejuni and 14 C. coli genomes sequenced.
2.2. Antimicrobial phenotypic profiles
Phenotypic antimicrobial susceptibility patterns were assessed using standard disk-diffusion methods [6]. Resistance to the following antibiotics was tested: ciprofloxacin (CIP), erythromycin (ERY), azithromycin (AZM), tetracycline (TET), gentamicin (GEN), amoxicillin and clavulanic acid (AMC), ampicillin (AMP), chloramphenicol (CHL) and imipenem (IMP). Zone diameter breakpoints (mm) for Campylobacter spp. from the Clinical and Laboratory Standards Institute (CLSI M45) were applied to assess CIP, ERY, AZM and TET resistance. The CLSI zone diameter breakpoints (mm) for Enterobacteriaceae were used for GEN, AMC, AMP, CHL and IMP, given that there are no established breakpoints for Campylobacter spp.
2.3. Molecular diagnostics, sequencing and bioinformatic analysis
DNA was extracted from all bacterial cultures using the Pure-Link Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA) as specified by the manufacturer’s instructions. A duplex qPCR targeting the 16S rRNA and the Campylobacter adhesion to fibronectin (cadF) genes was performed to confirm all bacterial cultures as Campylobacter spp. and C. jejuni/C. coli [16].
Libraries from Campylobacter isolates from the first paediatric cohort were prepared with the Nextera XT kit (Illumina, San Diego, CA), and batches of 24 isolates gDNA were barcoded. Genomes were sequenced using an Illumina MiSeq generating 250 bp paired-end reads. For strains from the second paediatric cohort, libraries were prepared using the Illumina DNA Prep Tagmentation kit following the manufacturer’s instructions with the following changes: decreasing the first and second volumes of the Sample Purification Beads to 40 μL and 11 μL, respectively, and a final elution in 10 μL Illumina resuspension buffer. Illumina-DNA/RNA UD Plate A, B, C and D dual index adapters were ordered from Integrated DNA Technologies (Coralville, IA) and used at 1 μM final concentration. Individual libraries were quantified using the KAPA Library Quantification Kit (Roche) in 10 μL volume reactions and 90 s annealing/extension PCR, pooled and normalized to 4 nM. Pooled libraries were requantified by ddPCR on a QX200 system (Bio-Rad, Hercules, CA), using the Illumina TruSeq ddPCR Library Quantification Kit following the manufacturer’s protocols. Libraries were sequenced using a 2 × 250 bp paired end (500-cycle, v2) reagent kit on a MiSeq instrument. Short-read data are available at NCBI SRA and are associated with BioProject PRJNA912682.
All genomes were assembled using the Spades assembler plugin for Geneious Prime v. 2022.2.2 (https://www.geneious.com). Genome quality metrics included <300 contigs and N50 >10 000 bp (Supplementary Table S1). Genomes were assessed using CheckM v. 1.1.3 software to determine completeness and/or contamination. CheckM parameters for genome quality control included completeness of more than 98.9% and contamination of less than 4.0%. Genomes with contamination greater than 4% by CheckM were analysed using the Kraken Taxonomic Sequence Classification System which used Kraken2 v. 2.1.2 software with the standard database (created on 18 August 22) to remove any sample in which bacterial genomes other than Campylobacter spp. or more than one Campylobacter species were detected. All genomes were assessed for heterogeneity through CheckM. Additionally, samples identified as heterogeneous (mixed strains, between 9% and 70%) by CheckM were validated as mixed using Geneious Prime v. 2022.2.2 to identify single nucleotide polymorphisms (SNPs) that occur in >10% of trimmed reads mapping to a nucleotide of the de novo assembly. Samples were validated as heterogeneous when more than five different SNPs from reads that mapped to the de novo assembly were identified. All assembled genomes are accessible in the public Campylobacter PubMLST database.
Sequence types (ST) and associated clonal complexes (CC) were determined through the PubMLST allelic database. Genomes were mined for antimicrobial resistance chromosomal point mutations and antibiotic resistance genes using the Comprehensive Antibiotic Resistance Database (CARD), ResFinder and PointFinder, and NCBI AMRFinder databases. A positive match was determined when a gene had more than 80% nucleotide identity and more than 60% coverage. Additionally, mutations in the gyrA gene [17–19], the cmeABC efflux system operon [20–22], 23S rRNA gene [23], L4 and L22 ribosomal protein encoding genes [24] and blaOXA [25] gene and promoter region were determined using BLASTN plugin in Geneious Prime v.2022.2.2.
A core genome was generated for the C. jejuni (n = 97) and C. coli (n = 67) genomes in this study using Roary v.3.12.0 with a 90% identity cutoff and creating a multiFASTA alignment of core genes using PRANK and a fast core gene alignment using MAFFT v.7.475 (concatenated alignment found in Supplementary Data File S1). The best-fit model of evolution was determined using ModelTest-NG v.0.1.7 software, and then the maximum likelihood phylogenies were constructed using RAxML v.8.2.12 using the General Time Reversible (GTR) with gamma distributed rates and invariable sites (GTRGAMMAIX model) and bootstrapped 1000 times. Visualization was done in iTOL v.6.6. The presence and absence of resistance genes, point mutations, lipooligosaccharide class, sequence type and clonal complex were added as metadata to the Newick file and visualized using Microreact. References for all bioinformatic tools are available in Supplementary Data File S2).
2.4. Statistical analysis
Frequency of detection of point mutations and genes conferring antimicrobial resistance were tabulated for C. jejuni and C. coli separately. Phenotypic resistance profiles are taken as the benchmark. Through comparison to this, the positive predictive value (PPV), negative predictive value (NPV), and accuracy (Accuracy= [True Positive + True Negative]/[True Positive + True Negative + False Positive + False Negative]) of genomic determinants was calculated for C. jejuni and C. coli separately. The total number of genes and point mutations were calculated for each genome. The number of antibiotic classes with one or more antibiotic resistance genes or point mutation per genome were also calculated for each genome. Data management, statistical analysis and graphical representation were done in Stata 17 (Stata Corp 2021, College Station, TX) and R v.4.2.1.
3. Results
C. jejuni and C. coli strains were isolated from paediatric individuals who were part of two cohort studies (details shown in Table 1). Of the 97 C. jejuni strains, 28 (28.9%) were CIP and AZM susceptible, 56 (57.8%) were CIP resistant and AZM susceptible, 12 (12.3%) were both CIP and AZM resistant, and 1 (1.0%) was CIP susceptible and AZM resistant. Of the 67 C. coli strains, 15 (22.4%) were CIP and AZM susceptible, 24 (35.8%) were CIP resistant and AZM susceptible, and 28 (41.8%) were both CIP and AZM resistant. CIP resistance was prevalent in both C. jejuni and C. coli strains at 70.1% (68/97) and 77.6% (52/67), respectively. AZM resistance was found in 12.4% (12/97) C. jejuni and 41.8% (28/67) in C. coli. Including ERY resistance increased macrolide resistance to 16.5% (16/97) in C. jejuni and 44.8% (30/67) in C. coli (see Supplementary Table S2A and Supplementary Table S2B).
Table 1.
Campylobacter genomes selected for analysis of antimicrobial resistance genes and point mutations
| Characteristic | C. jejuni (N = 97)a | C. coli (N = 67) | ||||
|---|---|---|---|---|---|---|
| CIP resistant AZM resistant (n = 12) | CIP resistant AZM susceptible (n = 56) | CIP susceptible AZM susceptible (n = 28) | CIP resistant AZM resistant (n = 28) | CIP resistant AZM susceptible (n = 24) | CIP susceptible AZM susceptible (n = 15) | |
| Sex (n) | ||||||
| Female | 6 | 32 | 13 | 15 | 19 | 7 |
| Male | 6 | 24 | 15 | 13 | 5 | 8 |
| Type of Sample (n) Diarrhoea | 3 | 25 | 15 | 5 | 6 | 2 |
| Asymptomatic | 9 | 31 | 13 | 23 | 18 | 13 |
| Age (months) | ||||||
| Median | 11.1 | 12.8 | 8.3 | 16.7 | 16.5 | 9.0 |
| IQR | 8.0–15.7 | 7.6–16.7 | 6.3–15.8 | 14.0–20.0 | 10.5–20.1 | 7.9–17.9 |
| Min | 3.1 | 1.0 | 2.1 | 0.4 | 4.8 | 2.6 |
| Max | 35.4 | 34.0 | 32.0 | 24.0 | 38.9 | 23.0 |
Selection for inclusion enriched to include isolates with phenotypic resistance upon antimicrobial resistance testing to the two first-line antimicrobials ciprofloxacin and azithromycin.
One isolate (n = 1) is classified as CIP susceptible and AZM resistant. It is from an asymptomatic faecal sample from a female participant.
These C. jejuni and C. coli strains were sequenced, and genome sizes ranged between 1.59 Mb and 1.89 Mb, with an average size of 1.73 Mb. Genome assemblies resulted in 17–295 contigs, with a median of 82 contigs per genome assembly. Supplementary Table S1 shows quality metrics for each individual genome. CheckM identified 113 samples with 0% heterogeneity. For 51 genomes, heterogeneity was reported between 9% and 70%. These 51 assemblies were manually examined using reference assembly within Geneious Prime v.2022.2.2, and the genomes were determined to represent single Campylobacter isolates.
3.1. Population structure
Multilocus sequence typing (MLST) assigned sequence types (ST) for 96/97 C. jejuni strains from 13 known different clonal complexes (CC) and 49 STs. One C. jejuni genome was not assigned a ST due to incomplete aspA allele. C. coli genomes were assigned to two different clonal complexes (CC828 and CC1150) and a total of 29 different STs. Overall, phylogenetic analysis of the strains used in this study were diverse, and AMR phenotypes or genotypes did not cluster within a specific ST or CC. A complete list of allele numbers, STs and CC for each isolate is presented in Supplementary Table S1.
3.2. Antimicrobial resistance genotypes
Antimicrobial resistance genes and point mutations detected in C. jejuni and C. coli isolate genomes are shown in Table 2A and Table 2B, respectively, alongside the associated antibiotic susceptibility profiles. C. coli genomes harboured a median number of four (interquartile range [IQR]: 2–5) antimicrobial genes and point mutations, whereas C. jejuni harboured a median of one (IQR: 0–4) (Fig. 1C). Forty-nine percent (44/97) of C. jejuni genomes and 67.1% (45/67) of C. coli genomes contained antimicrobial resistance genes and point mutations that conferred resistance to three or more antibiotic classes. C. coli isolates had a significantly higher number of AMR genes compared to C. jejuni (Mann-Whitney U test, P < 0.0001). Similarly, C. coli isolates had a significantly higher number of AMR-related SNPs compared to C. jejuni (Mann-Whitney U test, P < 0.0001) (Fig. 1C). Generally, C. coli isolates exhibited increased phenotypic resistance compared to C. jejuni isolates for all major antibiotic classes tested (Fig. 1D). The presence of resistance genes and point mutations as well as the phenotypic profile of each sample is presented in Supplementary Table 2A and 2B. Internal validity metrics comparing specific antibiotic phenotypes and antibiotic resistance genes or point mutations and associated contingency tables are shown in Supplementary Table 3A and Supplementary Table 3B.
Table 2A.
Distribution of antimicrobial resistance genes and point mutations identified in Campylobacter jejuni genomes according to their phenotypes as determined by disk diffusion testing
| Campylobacter jejuni (N = 97) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistance genes and point mutations | Quinolonesa (N = 97) | Macrolidesb (N = 97) | Tetracyclinesc (N = 97) | Aminoglycosidesd (N = 97) | Amoxicillin-clavulanic acid (N = 91) | Ampicillin (N = 57) | ||||||
| Resistant (N = 68) | Susceptible (N = 29) | Resistant (N = 16) | Susceptible (N = 81) | Resistant (N = 50) | Susceptible (N = 47) | Resistant (N = 10) | Susceptible (N = 87) | Resistant (N = 3) | Susceptible (N = 88) | Resistant (N = 27) | Susceptible (N = 30) | |
| MDR | ||||||||||||
| RE-cmeABC | 44.0% (33/68) | 6.9% (2/29) | 62.5% (10/16) | 30.9% (25/81) | 64.0% (32/50) | 6.4% (3/47) | 70.0% (7/10) | 32.0% (28/87) | 66.7% (2/3) | 35.2% (31/88) | 51.9% (14/27) | 0% (0/30) |
| Quinolones | ||||||||||||
| gyrA Thr86Ile | 75.0% (51/68) | 13.8% (4/29) | 68.8% (11/16) | 54.3% (44/81) | 82.0% (41/50) | 29.8% (14/47) | 80.0% (8/10) | 54.0% (47/87) | 66.7% (2/3) | 62.5 (51/88) | 55.6% (15/27) | 33.3% (10/30) |
| Macrolides | ||||||||||||
| 23S rRNA A2075G | 7.4% (5/68) | 3.5% (1/29) | 31.3% (5/16) | 1.2% (1/81) | 12.0% (6/50) | 0% (0/47) | 0% (0/10) | 11.5% (10/87) | 0% (0/3) | 6.8% (6/88) | 18.5% (5/27) | 0% (0/30) |
| Tetracyclines | ||||||||||||
| tet(O) | 57.4% (39/68) | 17.2% (5/29) | 68.8% (11/16) | 40.7% (33/81) | 76.0% (38/50) | 12.8% (6/47) | 90.0% (9/10) | 40.2% (35/87) | 66.7% (2/3) | 45.5% (40/88) | 55.6% (15/27) | 6.7% (2/30) |
| Aminoglycosides | ||||||||||||
| Any gene | 32.4% (22/68) | 6.9% (2/29) | 12.5% (2/16) | 27.2% (22/81) | 42.0% (21/50) | 6.4% (3/47) | 70.0% (7/10) | 19.5% (17/87) | 33.3% (1/3) | 23.9% (21/88) | 3.7% (1/27) | 0% (0/30) |
| aph(2′)-If | 13.2% (9/68) | 0% (0/29) | 6.3% (1/16) | 9.9% (8/81) | 18.0% (9/50) | 0% (0/47) | 60.0% (6/10) | 3.5% (3/87) | 0% (0/3) | 8.0% (7/88) | 0% (0/27) | 0% (0/30) |
| aph(3′)-IIIa | 33.3% (20/68) | 6.9% (2/29) | 6.3% (1/16) | 25.9% (21/81) | 38.0% (19/50) | 6.4% (3/47) | 60.0% (6/10) | 18.4% (16/87) | 33.3% (1/3) | 21.6% (19/88) | 0% (0/27) | 0% (0/30) |
| aad6 | 1.5% (1/68) | 0% (0/29) | 0% (0/16) | 1.2% (1/81) | 2.0% (1/50) | 0% (0/47) | 0% (0/10) | 1.1% (1/87) | 0% (0/3) | 1.1% (1/88) | 0% (0/27) | 0% (0/30) |
| aad9 | 30.9% (21/68) | 6.9% (2/29) | 6.3% (1/16) | 27.2% (22/81) | 40.0% (20/50) | 6.4% (3/47) | 70.0% (7/10) | 18.4% (16/87) | 33.3% (1/3) | 22.7% (20/88) | 0% (0/27) | 0% (0/30) |
| aadE | 19.1% (13/68) | 6.9% (2/29) | 0% (0/16) | 18.5% (15/81) | 24.0% (12/50) | 6.4% (3/47) | 10% (1/10) | 16.1% (14/87) | 33.3% (1/3) | 14.8% (13/88) | 0% (0/27) | 0% (0/30) |
| ant6-Ia | 20.6% (14/69) | 6.9% (2/29) | 0% (0/16) | 19.8% (16/81) | 26.0% (13/50) | 6.4% (3/47) | 10% (1/10) | 17.2% (6/87) | 33.3% (1/3) | 7.8% (14/88) | 0% (0/27) | 0% (0/30) |
| SAT4 | 1.5% (1/68) | 0% (0/29) | 0% (0/16) | 1.2% (1/81) | 2.0% (1/50) | 0% (0/47) | 10% (1/10) | 1.1% (1/87) | 0% (0/3) | 1.1% (1/88) | 0% (0/27) | 0% (0/30) |
| rpsL | 1.5% (1/68) | 0% (0/29) | 6.3% (1/16) | 0% (0/81) | 2.0% (1/50) | 0% (0/47) | 0% (0/10) | 1.1% (1/87) | 0% (0/3) | 1.1% (1/88) | 3.7% (1/27) | 0% (0/30) |
| Beta-lactams | ||||||||||||
| blaOXA genee | 73.5% (50/68) | 37.9% (11/29) | 75.0% (12/16) | 60.5% (49/81) | 86.0% (43/50) | 38.3% (18/47) | 80.0% (8/10) | 60.9% (53/87) | 66.7% (2/3) | 62.5% (55/88) | 74.1% (20/27) | 30.0% (9/30) |
| Active TATA boxf | 55.9% (38/68) | 17.2% (5/29) | 50.0% (8/16) | 43.2% (35/81) | 74.0% (37/50) | 12.8% (6/47) | 80.0% (8/10) | 40.2% (35/87) | 66.7% (2/3) | 42.0% (37/88) | 48.1% (13/27) | 3.3% (1/30) |
NOTE: Other mutations in the gyrA gene (Asp90Asn, Thr86Lys, Thr86Val, Thr86Ala and Asp90Tyr substitutions, and double mutations Thr86Ile-Pro104Ser and Thr86Ile-Asp90Asn), 23S rRNA genes (A2074G, A2075C), mutations in L4 and L22 ribosomal proteins, and ermB resistance gene were not found. bolded = resistance genes associated with phenotypic profile MDR, multidrug-resistant.
Includes susceptibility to CIP.
Includes susceptibility to AZM and ERY.
Includes susceptibility to TET.
Includes susceptibility to GEN.
All blaOXA genes belong to the blaOXA61 and blaOXA184 family.
G to T transversion represents an activation of the TATA box.
Table 2B.
Distribution of antimicrobial resistance genes and point mutation in Campylobacter coli genomes according to their phenotypes as determined by disk diffusion testing
| Campylobacter coli (N = 67) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resistance genes and point mutations | Quinolonesa (N = 67) | Macrolidesb (N = 67) | Tetracyclinesc (N = 67) | Aminoglycosidesd (N = 67) | Amoxicillin-clavulanic acid (N = 64) | Ampicillin (N = 53) | ||||||
| Resistant (N = 52) | Susceptible (N = 15) | Resistant (N = 30) | Susceptible (N = 37) | Resistant (N = 44) | Susceptible (N = 23) | Resistant (N = 25) | Susceptible (N = 42) | Resistant (N = 9) | Susceptible (N = 55) | Resistant (N = 36) | Susceptible (N = 17) | |
| MDR | ||||||||||||
| RE-cmeABC | 21.1% (11/52) | 6.7% (1/15) | 26.7% (8/30) | 10.8% (4/37) | 25.0% (11/44) | 4.3% (1/23) | 24.0% (6/25) | 14.3% (6/42) | 0% (0/9) | 21.8% (12/55) | 22.2% (8/36) | 0% (0/17) |
| Quinolones | ||||||||||||
| gyrA Thr86Ile | 96.2% (50/52) | 40.0% (6/15) | 100% (30/30) | 70.3% (26/37) | 95.5% (42/44) | 60.9% (14/23) | 100% (25/25) | 73.8% (31/42) | 100% (9/9) | 80.0% (44/55) | 94.4% (34/36) | 58.8% (10/17) |
| Macrolides | ||||||||||||
| 23S rRNA A2075G | 55.8% (29/52) | 26.7% (4/15) | 90.0% (27/30) | 16.2% (6/37) | 70.5% (31/44) | 8.7% (2/23) | 96% (24/25) | 21.4% (9/42) | 66.7% (6/9) | 43.6% (24/55) | 69.4% (25/36) | 0% (0/17) |
| Tetracyclines | ||||||||||||
| tet(O) | 75.0% (39/52) | 33.3% (5/15) | 100% (30/30) | 37.8% (14/37) | 93.2% (41/44) | 13.0% (3/23) | 100% (25/25) | 45.2% (19/42) | 88.9% (8/9) | 60.0% (33/55) | 86.1% (31/36) | 11.8% (2/17) |
| Aminoglycosides | ||||||||||||
| Any gene | 55.8% (29/52) | 26.7% (4/15) | 86.7% (26/30) | 18.9% (1/33) | 70.5% (31/44) | 8.7% (2/23) | 100% (25/25) | 19.0% (8/42) | 66.7% (6/9) | 43.6% (33/55) | 63.9% (23/36) | 0% (0/17) |
| aph(2′)-If | 5.8% (3/52) | 13.3% (2/15) | 3.3% (1/30) | 10.8% (4/37) | 6.8% (3/44) | 8.7% (2/23) | 8.0% (2/25) | 16.7% (7/42) | 0% (0/9) | 3.6% (2/55) | 0% (0/36) | 0% (0/17) |
| aph(3′)-IIIa | 53.8% (28/52) | 26.7% (4/15) | 83.3% (25/30) | 18.9% (7/37) | 68.2% (30/44) | 8.7% (2/23) | 100% (25/25) | 16.7% (7/42) | 66.7% (6/9) | 41.8% (23/55) | 61.1% (22/36) | 0% (0/17) |
| aad6 | 9.6% (5/52) | 26.7% (4/15) | 6.7% (2/30) | 18.9% (7/37) | 15.9% (7/44) | 8.7% (2/23) | 8.0% (2/25) | 16.7% (7/42) | 22.2% (2/9) | 7.3% (4/55) | 0% (0/36) | 0% (0/17) |
| aad9 | 7.7% (4/52) | 13.3% (2/15) | 6.7% (2/30) | 10.8% (4/37) | 9.1% (4/44) | 8.7% (2/23) | 12.0% (3/25) | 7.1% (3/42) | 11.1% (1/9) | 3.6% (2/55) | 0% (0/36) | 0% (0/17) |
| aadE | - | - | - | - | - | - | - | - | - | - | - | - |
| ant6-Ia | 9.6% (5/52) | 26.7% (4/15) | 6.7% (2/30) | 18.9% (7/37) | 15.9% (7/44) | 8.7% (2/23) | 8.0% (2/25) | 16.7% (7/42) | 22.2% (2/9) | 7.3% (4/55) | 0% (0/36) | 0% (0/17) |
| SAT4 | 7.7% (4/52) | 26.7% (4/15) | 3.3% (1/30) | 18.9% (7/37) | 13.6% (6/44) | 8.7% (2/23) | 4.00% (1/25) | 16.7% (7/42) | 11.1% (1/9) | 7.3% (4/55) | 0% (0/36) | 0% (0/17) |
| rpsL | 40.4% (21/52) | 0% (0/15) | 70.0% (21/30) | 0% (0/30) | 47.7% (21/44) | 0% (0/23) | 80.0% (20/25) | 2.4% (1/42) | 44.4% (4/9) | 30.9% (17/57) | 55.6% (20/36) | 0% (0/17) |
| Beta-lactams | ||||||||||||
| blaOXA genee | 96.2% (50/52) | 60.0% 9/15) | 100% (30/30) | 78.4% (29/37) | 97.7% (43/44) | 69.6% (16/23) | 100.0% (25/25) | 81.0% (34/42) | 100% (9/9) | 85.5% (47/55) | 97.2% (35/36) | 58.5% (10/17) |
| Active TATA boxf | 61.5% (32/52) | 33.3% (5/15) | 80.0% (24/30) | 35.1% (13/37) | 83.8% (31/37) | 26.1% (6/23) | 80.0% (20/25) | 40.5% (17/42) | 88.9% (8/9) | 47.3% (26/55) | 72.2% (26/36) | 0% (0/17) |
NOTE: Other mutations in the gyrA gene (Asp90Asn, Thr86Lys, Thr86Val, Thr86Ala and Asp90Tyr substitutions, and double mutations Thr86Ile-Pro104Ser and Thr86Ile-Asp90Asn), 23S rRNA genes (A2074G, A2075C), mutations in L4 and L22 ribosomal proteins, and ermB resistance gene were not found. bolded = resistance genes associated with phenotypic profile MDR, multidrug-resistant.
Includes susceptibility to CIP and NAL.
Includes susceptibility to AZM and ERY.
Includes susceptibility to TET.
Includes susceptibility to GEN.
All blaOXA genes belong to the blaOXA61 and blaOXA184 family.
G to T transversion represents an activation of the TATA box.
Fig. 1.
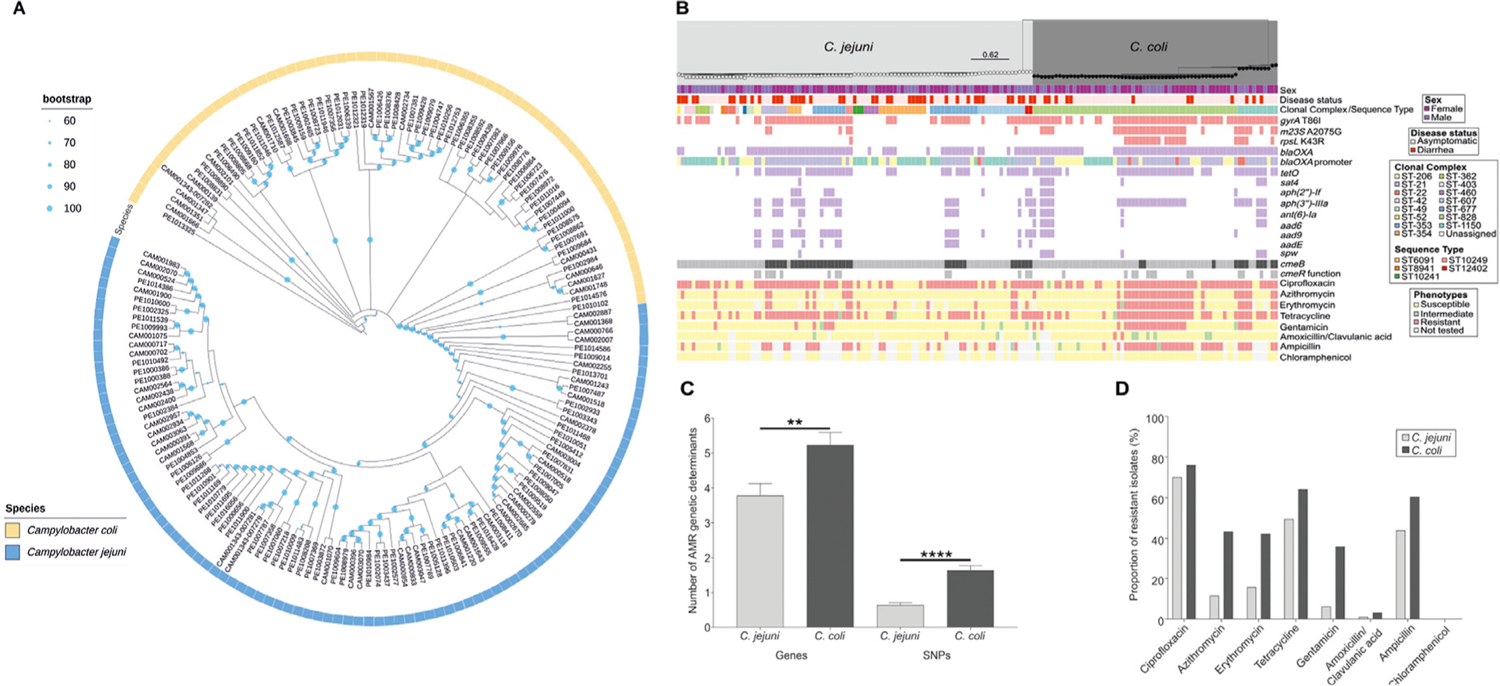
Population structure and antimicrobial resistance profiles of 97 C. jejuni and 67 C. coli isolates used in our study. (A) C. jejuni isolates (yellow) and C. coli isolates (blue) are shown on a circular phylogenetic tree reconstructed using an approximation of the maximum-likelihood algorithm implemented in RAxML, with the isolate names indicated next to the associated tip of the tree. The scale represents the number of substitutions per site. (B) The same phylogenetic tree is shown in rectangular form with epidemiologic, clinical and microbiologic data reported. The sex (male: purple, female: pink) associated with each isolate is shown in the top row, followed by disease status (symptomatic: dark red, asymptomatic: light red) in the second row of the data. The third rows show the clonal complexes and sequence types. The following three rows indicate the genotypic AMR-associated SNPs (red), and the next 13 rows indicate the presence/absence (purple) or allelic variation at known resistance gene loci (efflux pumps: black/grey). The remaining eight rows illustrate the phenotypic resistance (susceptible: yellow, intermediate: green, resistant: red, not tested: light grey) for the eight major antibiotic classes used in our study. N1 = New Clonal Complex 1, N2 = New Clonal Complex 2. (C) Number of AMR genetic determinants (genes and SNPs) identified in silico for C. jejuni (light grey) and C. coli (black) isolates, respectively. The error bar represents the standard error of the mean (SEM). Significance was tested using Mann–Whitney U test: *P < 0.05, **P < 0.01 and ****P < 0.001. (D) The proportion (%) of phenotypically resistant C. jejuni (light grey) and C. coli (black) isolates, for seven major antibiotic classes used in our study.
3.3. Fluoroquinolone resistance
A chromosomal point mutation of the gyrA gene that causes a threonine to isoleucine change at amino acid 86 (Thr86Ile) in the gyrase protein was the only point mutation identified in gyrA that would confer quinolone resistance in either C. jejuni or C. coli. Its presence supported resistance to ciprofloxacin in 75.0% (51/68) of CIP-resistant C. jejuni isolates, whereas 13.8% (4/29) of CIP-susceptible C. jejuni isolates also harboured the mutation. Of the 97 C. jejuni isolates with phenotypic and genomic data, the state of codon 86 correctly identified 51 resistant isolates and 25 susceptible isolates. For C. coli, 50 of the 52 resistant isolates and 9 of the 15 susceptible isolates were correctly classified. Additionally, amino acid substitutions Asp90Asn, Thr86Lys, Thr86Val, Thr86Ala and Asp90Tyr as well as double amino acid substitutions Thr86Ile-Pro104Ser and Thr86Ile-Asp90Asn in the GyrA protein previously identified to confer quinolone resistance were not found in this set of genomes [17–19,23,26,27].
3.4. Macrolide resistance
The A2075G point mutation in the 23S rRNA gene was present in 31.3% (5/16) of C. jejuni isolates and in 90.0% (27/30) of C. coli isolates that were macrolide resistant. This mutation was identified in three macrolide-susceptible C. jejuni isolate and six macrolide-susceptible C. coli isolates. Of significance, the emerging ermB gene conferring macrolide resistance was not identified in this set of genomes, nor were mutations in the genes encoding L4 and L22 ribosomal proteins previously associated with macrolide resistance.
3.5. Resistance to other antibiotics
Although less important in the clinical LMIC setting, we identified the genotypic resistant determinants for aminopenicillins, aminoglycosides and tetracyclines. The presence of resistance genes associated with an aminopenicillin was confined to blaOXA being present in 62.9% (61/97) of C. jejuni and of 88.1% (59/67) of C. coli samples. The genotypic databases divided the blaOXA into families including blaOXA-61 and blaOXA-184; however, SNP-based analysis revealed few differences (Supplementary Fig. S1), so only blaOXA is reported. Genotypic databases did not report the presence of a functional TATA box in the promoter of blaOXA gene that is linked to high-level beta-lactam resistance [28]. The only tetracycline resistance determinant identified was the tet(O) gene. Genotypic databases identified the tet(O) gene in 76.0% (38/50) tetracycline-resistant C. jejuni isolates and 93.2% (41/44) tetracycline-resistant C. coli. Six tetracycline-susceptible C. jejuni and two susceptible C. coli harboured tet(O). Finally, the aminoglycoside-resistant genes identified were aph(2’)-If, aph(3’)-IIIa, aad6, aad9, aadE, ant6-Ia, SAT4 and rpsL. The presence of any aminoglycoside resistance gene explained resistance in 100% (25/25) of C. coli isolates and 70% (7/10) of C. jejuni isolates, with aph(3’)-IIIa being the main resistance gene identified in both species.
3.6. Multidrug efflux pump
The universally present and constitutively expressed Campylobacter multidrug efflux pump (CmeABC) is a tripartite energy-dependent transmembrane pump belonging to the resistance nodulation division (RND) that plays a role in resistance to bile salts as well as multiple antimicrobials, including macrolides and quinolines [20,29,30]. The cmeABC operon is regulated by repressor proteins CmeR and CosR [31]. Mutations in two distinct genomic regions related to the operon and its regulatory elements result in clearly defined changes in the MICs of target antibiotics: major mutations within the cmeB gene and mutations in the CmeR-binding site (an inverted repeat (IR) region) [21,32].
Previously described atypical cmeB genes result in an altered CmeABC efflux pump (RE-cmeABC) that increases the MIC to ciprofloxacin by nine-fold and to erythromycin by four-fold [21, 33]. Among cmeABC, the cmeB gene from C. jejuni was most often identified as absent by the CARD database. However, screening for the cmeB gene in most of these samples identified atypical cmeB variants that should result in production of RE-cmeABC [21]. The RE-cmeABC was identified in 36.1% (35/97) C. jejuni genomes and 17.9% (12/67) C. coli genomes (Table 2A and Table 2B). RE-cmeABC was identified in 66.7% (10/15) of C. jejuni isolates resistant to both macrolides and quinolones, and in 27.6% (8/29) of C. coli isolates resistant to both macrolides and quinolones. That said, there is a significant relationship between the presence of RE-cmeABC and C. jejuni isolates that have a multidrug-resistant phenotype (X2 (1,N = 97) = 17.23, P < 0.001) and genotype (X2 (1,N = 97) = 59.2, P < 0.001). Specifically, 75.0% (18/24) of C. jejuni isolates with a MDR genotype contained RE-cmeABC, and 62.5% (15/24) of C. jejuni isolates with a MDR phenotype contained RE-cmeABC. Of the 45 C. coli isolates with an MDR genotype, 11 had RE-cmeABC (X2 (1, N = 67) = 3.98, P < 0.046), and of the 35 C. coli isolates with an MDR phenotype, eight had RE-cmeABC (X2 (1, N = 67) = 1.21, P < 0.269). Table 3 shows the presence of the RE-cmeABC according to the number of antibiotics to which the Campylobacter isolates are phenotypically resistant or contain a genomic determinant of resistance.
Table 3.
Resistance to multiple antimicrobial classes is strongly associated with the presence of the RE-cmeABC in Campylobacter jejuni and Campylobacter coli genomes
| Campylobacter jejuni | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number antimicrobial-resistant classes | Genotype | Phenotype | ||||||
| Total number of isolates | RE-cmeABC (% (n/N)) | Overexpressed cmeABC (% (n/N))a | RE-cmeABC and overexpressed cmeABC (% (n/N))a | Total number of isolates | RE-cmeABC (% (n/N)) | Overexpressed cmeABC (% (n/N))a | RE-cmeABC and overexpressed cmeABC (% (n/N))a | |
| 0 | 25 | 0% (0/25) | 0% (0/23) | 0% (0/23) | 20 | 0% (0/20) | 5.0% (1/20) | 0% (0/20) |
| 1 | 25 | 0% (0/25) | 4.2% (1/24) | 0% (0/24) | 24 | 12.5% (3/24) | 13.0% (3/23) | 8.7% (2/23) |
| 2 | 3 | 33.3% (1/3) | 33.3% (1/3) | 33.3% (1/3) | 25 | 52.0% (13/25) | 33.3% (8/24) | 25.0% (6/24) |
| 3 | 19 | 63.2% (12/19) | 38.9% (7/18) | 33.3% (6/18) | 14 | 64.3% (9/14) | 25.0% (3/12) | 25.0% (3/12) |
| 4 | 23 | 87.0% (20/23) | 56.5% (13/23) | 47.8% (11/23) | 13 | 69.2% (9/13) | 61.5% (8/13) | 61.5% (8/13) |
| 5 | 2 | 100% (2/2) | 100% (2/2) | 100% (2/2) | 1 | 100% (1/1) | 100% (1/1) | 100% (1/1) |
| Total | 97 | 97 | 93 | 93 | 97 | 97 | 93 | 93 |
| Campylobacter coli | ||||||||
| Number antimicrobial resistant classes | Genotype | Phenotype | ||||||
| Total number of isolates | RE-cmeABC (% (n/N)) | Overexpressed cmeABC (% (n/N))b | RE-cmeABC and overexpressed cmeABC (% (n/N))b | Total number of isolates | RE-cmeABC (% (n/N)) | Overexpressed cmeABC (% (n/N))b | RE-cmeABC and overexpressed cmeABC (% (n/N))b | |
| 0 | 6 | 0% (0/6) | 0% (0/6) | 0% (0/6) | 11 | 0% (0/11) | 9.1% (1/11) | 0% (0/11) |
| 1 | 7 | 0% (0/7) | 14.3% (1/7) | 0% (0/7) | 9 | 11.1% (1/9) | 0% (0/9) | 0% (0/9) |
| 2 | 9 | 11.1% (1/9) | 0% (0/9) | 0% (0/9) | 12 | 25.0% (3/12) | 8.3% (1/12) | 8.3% (1/12) |
| 3 | 10 | 0% (0/10) | 20/0% (2/10) | 0% (0/10) | 6 | 0% (0/6) | 0% (0/6) | 0% (0/6) |
| 4 | 5 | 60.0% (3/5) | 20.0% (1/5) | 20.0% (1/5) | 6 | 50.0% (3/6) | 66.7% (4/6) | 33.3% (2/6) |
| 5 | 30 | 26.7% (8/30) | 35.7% (10/28) | 14.3% (4/28) | 23 | 21.7% (5/23) | 38.1% (8/21) | 18.2% (2/21) |
| Total | 67 | 67 | 65 | 65 | 67 | 67 | 64 | 64 |
NOTE: n = number of isolates with the genomic determinant; N = total number of isolates for which the genomic determinant could be evaluated.
N differs from total number of isolates given that for four samples the status of the cmeR binding site could not be determined.
N differs from total number of isolates given that for two samples the status of the cmeR binding site could not be determined.
The second cmeABC group of genomic variants includes mutations identified in the CmeR-binding site, an inverted repeat sequence in the cmeABC promoter region which increases expression of the operon and, in turn, leads to higher levels of resistance in C. jejuni isolates [32,34]. Twenty-four C. jejuni genomes had a disrupted IR which could lead to overexpression of the CmeABC pump. Nineteen C. jejuni genomes had both the RE-cmeABC and functional mutation in the IR region. Of these, 18 had an MDR genotypes. Among C. coli genomes, 14 had a functional mutation in the IR region, and five had both the RE-cmeABC form of the pump and functional IR mutation, all of which corresponded to MDR genotypes (Table 3). The presence of IR variants that could lead to an overexpression of the cmeABC pump had a significant relationship with the occurrence of an MDR genotype (X2 (1,N = 97) = 26.9, P < 0.001) and MDR phenotype (X2 (1,N = 97) = 7.8, P = 0.005) among C. jejuni isolates. This association was also identified among C. coli isolates (MDR genotype (X2 (1,N = 67) = 5.68, P = 0.017); MDR phenotype (X2 (1,N = 67) = 8.72, P = 0.003). Table 4 presents mutations and deletions in the specified IR sequence, including what the potential functional meaning of these could be and its association with a MDR genotype and phenotype in both C. jejuni and C. coli. We identified 10 variations in the IR region in C. jejuni, seven of which would hypothetically lead to the overexpression of the CmeABC pump. Among C. coli, 14 variations were identified, with only one predicted to cause overexpression, and 10 C. coli genomes were missing the IR region, which should lead to overexpression of the cmeABC. Although the presence of the genes encoding the CmeABC efflux pump were identified by the CARD database, none of the databases reported the presence or absence of cmeABC genotypes associated with increased multidrug resistance [32, 34].
Table 4.
Characterization of the Cme repressor (cmeR) binding site in Campylobacter jejuni and Campylobacter coli demonstrates a strong association between both genetic mechanisms predicting overexpression of the efflux pump cmeABC with a multidrug-resistant genotype and phenotype
| N | MDR genotype | MDR phenotype | |
|---|---|---|---|
| Campylobacter jejuni | 97 | 45.4% (44/97) | 28.9% (28/97) |
| Not repressed | 24 | 91.7% (22/24) a | 50.0% (12/24) b |
| TGTAATAA-TATTACA | 2 | 100% (2/2) | 0% (0/2) |
| TGTAATAAA-ATTACA | 4 | 100% (4/4) | 100% (4/4) |
| TGT | 1 | 100% (1/1) | 0% (0/1) |
| TGTAATAAATATCACA | 1 | 0% (0/1) | 0% (0/1) |
| TGTAATAAATATGACA | 1 | 100% (1/1) | 100% (1/1) |
| TGTAATAAATATTGCA | 8 | 87.5% (7/8) | 62.5% (5/8) |
| TGTCA | 7 | 100% (7/7) | 28.6% (2/7) |
| Repressed | 69 | 30.4% (21/69) a | 20.3% (14/69) b |
| TGCCA | 2 | 100% (2/2) | 100% (2/2) |
| TGTCA | 11 | 0% (0/11) | 18.2% (2/11) |
| TGTCA (c) | 45 | 20.0% (9/45) | 17.8% (8/45) |
| TGT | 11 | 90.9% (10/11) | 18.2% (2/11) |
| Undetermined | 4 | 25.0% (1/4) | 50.0% (2/4) |
| Campylobacter coli | 67 | 67.2% (45/67) | 52.2% (35/67) |
| Not repressed | 14 | 92.9% (13/14) d | 85.7% (12/14) e |
| No CmeR binding site | 10 | 100% (10/10) | 90.0% (9/10) |
| TGTAATAAATATTGCA | 4 | 75.0% (3/4) | 75.0% (3/4) |
| Repressed | 51 | 58.8% (30/51) d | 41.2% (21/51) e |
| TGCCA | 14 | 57.1% (8/14) | 50.0% (7/14) |
| TGCCA | 10 | 90.0% (9/10) | 90.0% (9/10) |
| TGTCA | 2 | 0% (0/2) | 0% (0/2) |
| TGTCA (c) | 25 | 52.0% (13/25) | 20.0% (5/25) |
| Undetermined | 2 | 50.0% (1/2) | 50.0% (1/2) |
Relationship between the cmeR function and a MDR genotype among Campylobacter jejuni isolates (X2 (1, N = 97) = 26.9, P < 0.001).
Relationship between the cmeR function and a MDR phenotype among Campylobacter jejuni isolates (X2 (1, N = 97) = 7.8, P = 0.005).
Wild type, conserved inverted repeat region.
Relationship between the cmeR function and a MDR genotype among Campylobacter coli isolates (X2 (1, N = 67) = 5.68, P = 0.017).
Relationship between the cmeR function an MDR phenotype among Campylobacter coli isolates (X2 (1, N = 67) = 8.72, P = 0.003).
4. Discussion
Current trends in rising resistance to ciprofloxacin and azithromycin in clinical isolates of Campylobacter compromise treatment using first-line orally administered therapies. In this study, we analysed the genomic determinants of C. jejuni and C. coli clinical isolates derived from children under two years of age in Iquitos, Peru. Reports on genomic determinants of Campylobacter spp. in Latin America are generally concentrated on strains of animal origin, with few reports from human clinical isolates in Brazil [35–37], Peru [8] and Chile [38].
Increasing trends in quinolone resistance have previously been detected in Campylobacter [6,39,40]. Quinolone resistance is predominantly associated with mutations in gyrA [17–19,23,26,27]. In this study, as well as in others from Latin America, the C to T transitional mutation at position 257 of gyrA (Thr86Ile) was the only substitution identified in gyrA of the Campylobacter isolates. Other mutations that have been previously reported to confer quinolone resistance were not identified. With regard to macrolide resistance, the A2075G mutation in 23S rRNA was the only mutation identified that would exclusively confer resistance to these antibiotics.
The mutations affecting the structure (RE-cmeABC) or expression (IR polymorphisms) of the CmeABC efflux pump have been demonstrated to affect resistance to both quinolones, macrolides and other classes of antibiotics in C. jejuni [21,24,32,41]. Both genotypic changes were informative in relation to quinolone and macrolide resistance phenotypes. Moreover, it is possible that an altered CmeABC is associated with phenotypic resistance to other antibiotics that were shown to be genotypically susceptible. RE-cmeABC was significantly linked with multidrug resistance among this collection of C. jejuni and C. coli and helped explain macrolide resistance among C. jejuni isolates. Although this resistant form of the pump has been identified globally in C. jejuni since at least 2014, this study is one of the first reports of the RE-cmeABC form of the pump in C. coli within the Latin American region [32].
Both permeases and efflux pumps are relatively poorly reflected in CARD, ResFinder and PointFinder, which focus mostly on point mutations in enzyme-modifying antibiotics and/or mutations causing conformational changes at known antibiotic binding sites. None of the databases currently include RE-cmeABC or functional mutations in the IR, hindering the performance of these tools. Despite the global importance of this enteric pathogen, there is a relative paucity of studies exploring highly resistant Campylobacter isolates among large-scale population studies with epidemiologically linked specimens. More studies of this nature are needed to contribute to these AMR mining tools to improve their performance.
Phenotype to genotype association was imperfect for both C. coli and C. jejuni for almost all antimicrobials. This finding is atypical compared to other studies [42–45] and could be a result of heterogenous culturable Campylobacter populations, even though quality control metrics for genomes included in the analysis were considered optimal. Phenotypic resistance data obtained from a mixed culture of Campylobacter could be associated with discrepant phenotype to genotype correlations. That said, there is a need to integrate additional information in the most commonly used genomic databases prior for the direct use for clinical decision making or policy based on from genomic testing alone for Campylobacter.
Historically, most therapy directed at Campylobacter would also be adequate for shigellosis, the other major bacterial aetiology of severe inflammatory enteritis [46]. Emerging resistance will challenge our ability to treat both pathogens concurrently with an oral antimicrobial that is currently licenced and widely available. Rapid turnaround of point-of-care testing for resistance determinants is a high-value target for the appropriate regional surveillance and the management of individual patients [47]. This would be especially attractive in LMIC settings, where heterogeneous protocols in clinical microbiology predominate across healthcare institutions. To achieve this, a more complete understanding of genomic determinants of resistance in human-derived Campylobacter from low-resource settings is required.
Supplementary Material
Acknowledgements:
We thank Keith Arora Williams for his support during early investigations on the genomic determinants of observed antimicrobial resistance and Drs. Paul Auwaerter and Shmuel Shoham for discussions on clinical management.
Funding:
Funding for this study was provided by the Bill and Melinda Gates Foundation (OPP1066146 and OPP1152146 to MNK) and the National Institutes of Health of the United States (R01AI158576 and R21AI163801 to MNK and CP; D43TW010913 to MNK; K43TW012298 to FS). This research was also supported in part by USDA-ARS CRIS project 2030-42000-055-00D (to CP).
Footnotes
Competing interests: None to declare.
Ethical approval: Human fecal samples used in this study are part of two studies approved by the Institutional Review Board of the Johns Hopkins School of Public Health, the Ethics Committee of Asociacion Benefica PRISMA, the Regional Health Department of Loreto, and the University of Virginia (Charlottesville, VA, United States). Written consent to participate in the study was obtained from the parents or legal guardians of children. Participants of both studies consented for further use of biological specimens.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jgar.2024.01.009.
References
- [1].Sproston EL, Wimalarathna HML, Sheppard SK. Trends in fluoroquinolone resistance in Campylobacter. Microb Genom 2018;4. doi: 10.1099/mgen.0.000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].CDC’s Antibiotic Resistance Threats in the United States; 2019. https://www.cdc.gov/drugresistance/biggest-threats.html.
- 3.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, et al. 2017 Infectious Diseases Society of America Clinical Practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017;65:1963–73. doi: 10.1093/cid/cix959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor DN, Hamer DH, Shlim DR. Medications for the prevention and treatment of travellers’ diarrhea. J Travel Med 2017;24(Suppl 1):S17–22. doi: 10.1093/jtm/taw097. [DOI] [PubMed] [Google Scholar]
- 5.Neustaedter CM, Robertson K, Tschritter D, Reid-Smith RJ, MacKinnon MC, Murphy CP, et al. A scoping review of factors associated with antimicrobial-resistant Campylobacter species infections in humans. Epidemiol Infect 2023;151:e100. doi: 10.1017/S0950268823000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiaffino F, Colston JM, Paredes-Olortegui M, Francois R, Pisanic N, Burga R, et al. Antibiotic resistance of Campylobacter species in a pediatric cohort study. Antimicrob Agents Chemother 2019;63. doi: 10.1128/AAC.01911-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollett S, Rocha C, Zerpa R, Patino L, Valencia A, Camina M, et al. Campylobacter antimicrobial resistance in Peru: a ten-year observational study. BMC Infect Dis 2012;12. doi: 10.1186/1471-2334-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quino W, Caro-Castro J, Hurtado V, Flores-Leon D, Gonzalez-Escalona N, Gavilan RG. Genomic analysis and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli in Peru. Front Microbiol 2021;12:802404. doi: 10.3389/fmicb.2021.802404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard SA, Barrett LJ, Guerrant RL, Checkley W, Miller MA, Investigators MEN. Disease surveillance methods used in the 8-site MAL-ED cohort study. Clin Infect Dis 2014;59(Suppl 4):S220–4. doi: 10.1093/cid/ciu435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson G, Aitchison LB. The use of a combined enrichment-filtration technique for the isolation of Campylobacter spp. from clinical samples. Clin Microbiol Infect 2007;13:643–4. doi: 10.1111/j.1469-0691.2007.01712.x. [DOI] [PubMed] [Google Scholar]
- 11.Yori PP, Lee G, Olortegui MP, Chavez CB, Flores JT, Vasquez AO, et al. Santa Clara de Nanay: the MAL-ED cohort in Peru. Clin Infect Dis 2014;59(Suppl 4):S310–16. doi: 10.1093/cid/ciu460. [DOI] [PubMed] [Google Scholar]
- 12.Riddle MS, DuPont HL, Connor BA. ACG Clinical Guideline: diagnosis, treatment, and prevention of acute diarrheal infections in adults. Am J Gastroenterol 2016;111:602–22. doi: 10.1038/ajg.2016.126. [DOI] [PubMed] [Google Scholar]
- 13.Auwaerter P. Campylobacter and Helicobacter Species. Johns Hopkins ABX Guide, The Johns Hopkins University, 2020 Johns Hopkins Guides; 2020. www.hopkinsguides.com/hopkins/view/Johns_Hopkins_ABX_Guide/540073/all/Campylobacter_and_helicobacter_species. [Google Scholar]
- 14.World Health Organization. Campylobacter. https://www.who.int/news-room/fact-sheets/detail/campylobacter [accessed 24 August 2021].
- 15.Rouhani S, Griffin NW, Yori PP, Olortegui MP, Salas MS, Trigoso DR, et al. Gut microbiota features associated with Campylobacter burden and postnatal linear growth deficits in a Peruvian birth cohort. Clin Infect Dis 2020;71:1000–7. doi: 10.1093/cid/ciz906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francois R, Yori PP, Rouhani S, Salas MS, Olortegui MP, Trigoso DR, et al. The other Campylobacters: not innocent bystanders in endemic diarrhea and dysentery in children in low-income settings. PLoS Negl Trop Dis 2018;12:e0006200. doi: 10.1371/journal.pntd.0006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIver C, Hogan T, White P, Tapsall J. Patterns of quinolone susceptibility in Campylobacter jejuni associated with different gyrA mutations. Pathology 2004;36:166–9. doi: 10.1080/00313020410001672019. [DOI] [PubMed] [Google Scholar]
- 18.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist 2001;7:257–61. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- 19.Piddock LJ, Ricci V, Pumbwe L, Everett MJ, Griggs DJ. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J Antimicrob Chemother 2003;51:19–26. doi: 10.1093/jac/dkg033. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 2002;46:2124–31. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, et al. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. MBio 2016;7. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cagliero C, Cloix L, Cloeckaert A, Payot S. High genetic variation in the multidrug transporter cmeB gene in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 2006;58:168–72. doi: 10.1093/jac/dkl212. [DOI] [PubMed] [Google Scholar]
- 23.Payot S, Bolla JM, Corcoran D, Fanning S, Megraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect 2006;8:1967–71. doi: 10.1016/j.micinf.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Cagliero C, Mouline C, Cloeckaert A, Payot S. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 2006;50:3893–6. doi: 10.1128/AAC.00616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casagrande Proietti P, Guelfi G, Bellucci S, De Luca S, Di Gregoria S, Pieramati C, et al. Beta-lactam resistance in Campylobacter coli and Campylobacter jejuni chicken isolates and the association between blaOXA-61 gene expression and the action of beta-lactamase inhibitors. Vet Microbiol 2020;241:108553. doi: 10.1016/j.vetmic.2019.108553. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Yuanshu Z, Yuhan Z, Yaojie Yingxia L. Mutant prevention concentrations of fluoroquinolones against Campylobacter jejuni isolated from chicken. Vet Microbiol 2010;144:409–14. doi: 10.1016/j.vetmic.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz J, Moreno A, Jimenez de Anta MT, Vila J. A double mutation in the gyrA gene is necessary to produce high levels of resistance to moxifloxacin in Campylobacter spp. clinical isolates. Int J Antimicrob Agents 2005;25:542–5. doi: 10.1016/j.ijantimicag.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Zeng X, Brown S, Gillespie B, Lin J. A single nucleotide in the promoter region modulates the expression of the beta-lactamase OXA-61 in Campylobacter jejuni. J Antimicrob Chemother 2014;69:1215–23. doi: 10.1093/jac/dkt515. [DOI] [PubMed] [Google Scholar]
- 29.Corcoran D, Quinn T, Cotter L, O’Halloran F, Fanning S. Characterization of a cmeABC operon in a quinolone-resistant Campylobacter coli isolate of Irish origin. Microb Drug Resist 2005;11:303–8. doi: 10.1089/mdr.2005.11.303. [DOI] [PubMed] [Google Scholar]
- 30.Payot S, Cloeckaert A, Chaslus-Dancla E. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb Drug Resist 2002;8:335–43. doi: 10.1089/10766290260469606. [DOI] [PubMed] [Google Scholar]
- 31.Grinnage-Pulley T, Mu Y, Dai L, Zhang Q. Dual repression of the multidrug efflux pump CmeABC by CosR and CmeR in Campylobacter jejuni. Front Microbiol 2016;7:1097. doi: 10.3389/fmicb.2016.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao H, Zhao W, Jiao D, Schwarz S, Zhang R, Li XS, et al. Global distribution, dissemination and overexpression of potent multidrug efflux pump RE-CmeABC in Campylobacter jejuni. J Antimicrob Chemother 2021;76:596–600. doi: 10.1093/jac/dkaa483. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Lizer N, Wu Z, Morgan CE, Yan Y, Zhang Q, et al. Cryo-electron microscopy structures of a Campylobacter multidrug efflux pump reveal a novel mechanism of drug recognition and resistance. Microbiol Spectr 2023:e0119723. doi: 10.1128/spectrum.01197-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J, Akiba M, Sahin O, Zhang Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 2005;49:1067–75. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes CN, Campioni F, Barker DOR, Che EV, da Silva Duque S, et al. Antimicrobial resistance genotypes and phenotypes of Campylobacter coli isolated from different sources over a 16-year period in Brazil. J Glob Antimicrob Resist 2023;33:109–13. doi: 10.1016/j.jgar.2023.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Sierra-Arguello YM, Perdoncini G, Morgan RB, Salle CT, Moraes HL, Gomes MJ, et al. Fluoroquinolone and macrolide resistance in Campylobacter jejuni isolated from broiler slaughterhouses in southern Brazil. Avian Pathol 2016;45:66–72. doi: 10.1080/03079457.2015.1120272. [DOI] [PubMed] [Google Scholar]
- 37.Frazao MR, Cao G, Medeiros MIC, Duque SDS, Allard MW, Falcao JP. Antimicrobial resistance profiles and phylogenetic analysis of Campylobacter jejuni strains isolated in Brazil by whole genome sequencing. Microb Drug Resist 2021;27:660–9. doi: 10.1089/mdr.2020.0184. [DOI] [PubMed] [Google Scholar]
- 38.Bravo V, Katz A, Porte L, Weitzel T, Varela C, et al. Genomic analysis of the diversity, antimicrobial resistance and virulence potential of clinical Campylobacter jejuni and Campylobacter coli strains from Chile. PLoS Negl Trop Dis 2021;15:e0009207. doi: 10.1371/journal.pntd.0009207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zachariah OH, Lizzy MA, Rose K, Angela MM. Multiple drug resistance of Campylobacter jejuni and Shigella isolated from diarrhoeic children at Kapsabet County referral hospital, Kenya. BMC Infect Dis 2021;21:109. doi: 10.1186/s12879-021-05788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouftah SF, Pascoe B, Calland JK, Mourkas E, Tonkin N, Lefevre C, et al. Local accessory gene sharing among Egyptian Campylobacter potentially promotes the spread of antimicrobial resistance. Microb Genom 2022;8. doi: 10.1099/mgen.0.000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan M, Sahin O, Lin J, Zhang Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother 2006;58:1154–9. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 42.Rokney A, Valinsky L, Vranckx K, Feldman N, Agmon V, Moran-Gilad J, et al. WGS-based prediction and analysis of antimicrobial resistance in Campylobacter jejuni isolates from Israel. Front Cell Infect Microbiol 2020;10:365. doi: 10.3389/fcimb.2020.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahl LG, Joensen KG, Osterlund MT, Kiil K, Nielsen EM. Prediction of antimicrobial resistance in clinical Campylobacter jejuni isolates from whole-genome sequencing data. Eur J Clin Microbiol Infect Dis 2021;40:673–82. doi: 10.1007/s10096-020-04043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 2019;63. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Painset A, Day M, Doumith M, Rigby J, Jenkins C, Grant K, et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Campylobacter jejuni and Campylobacter coli isolated from cases of diarrhoeal disease in England and Wales, 2015–16. J Antimicrob Chemother 2020;75:883–9. doi: 10.1093/jac/dkz539. [DOI] [PubMed] [Google Scholar]
- 46.Williams PCM, Berkley JA. Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence. Paediatr Int Child Health 2018;38(Suppl):S50–65. doi: 10.1080/20469047.2017.1409454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tribble DR. Antibiotic therapy for acute watery diarrhea and dysentery. Mil Med 2017;182(S2):17–25. doi: 10.7205/MILMED-D-17-00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


