Abstract
The imbalance between pathogenic and beneficial species of the intestinal microbiome and metabolism in rheumatoid arthritis (RA) remains unclarified. Here, using shotgun-based metagenome sequencing for a treatment-naïve patient cohort and a “quasi-paired cohort” method, we observed a deficiency of butyrate-producing species and an overwhelming number of butyrate consumers in RA patients. These outcomes mainly occurred in patients with positive ACPA, with a mean AUC of 0.94. This panel was also validated in established RA with an AUC of 0.986 in those with joint deformity. In addition, we showed that butyrate promoted Tregs, while suppressing Tconvs and osteoclasts, due to potentiation of the reduction in HDAC expression and down-regulation of proinflammatory cytokine genes. Dietary butyrate supplementation conferred anti-inflammatory benefits in a mouse model by rebalancing TFH cells and Tregs, as well as reducing antibody production. These findings reveal the critical role of butyrate-metabolizing species and suggest the potential of butyrate-based therapies for RA patients.
Butyrate-metabolizing species influence the balance of follicular helper T cells and regulatory T cells in rheumatoid arthritis.
INTRODUCTION
Rheumatoid arthritis (RA) is an autoimmune disease characterized by joint destruction, systemic involvement, and the presence of autoantibodies, especially anti-citrullinated peptide antibodies (ACPAs). Clinically, the presence of ACPAs indicates an increased likelihood of severe impairment in cartilage and bone, as well as a worse prognosis with intense joint deformation (1). Insights into the disease pathophysiology highlight that various inflammatory pathways can lead to aberrant systemic immune responses in RA. It has been proposed that the origin of a dysregulated immune response occurs in genetically predisposed individuals with a substantial contribution from the environment. Among environmental factors, the intestinal microbiota has emerged as a possible candidate for the aberrant systemic immune responses in RA (2–4).
The gut microbiome is essential in maintaining immune homeostasis and perturbing the balance of the gut microbiota composition, which can alter local adaptive immune responses and modulate systemic inflammation. In addition, gut microbiota–derived metabolites, in particular short-chain fatty acids (SCFAs), can induce metabolic alterations in T cells by regulating their glucose metabolism. Multiple recent lines of evidence highlight the involvement of intestinal dysbiosis in the pathogenesis of RA. A pioneering study reported that increases in Prevotella abundance correlated with a reduction in Bacteroides and a loss of beneficial microbes in new-onset untreated RA (NORA) patients (2). Subsequent studies suggested that gut dysbiosis in RA patients correlates with a depletion of Gram-negative bacteria and enrichment of Gram-positive bacteria (3, 4).
While oral and fecal dysbiosis have been described in RA, research obstacles remain because of high interindividual diversity, which is influenced by a wide range of factors such as genetics, age, diet, and health conditions (5). The individual diversity is often so great and difficult to control that it even overwhelms disease-associated alterations and profoundly affects the identification of disease-associated microbiome features. Thus, the results of metagenomic studies are highly dependent on the samples collected and often include stochastic false positives or negatives (6, 7).
Genetics research has long exploited the opportunity of twin studies to identify causal genetic features (8). This concept has been extended into metagenomics research on “paired” animal studies, in which paired samples are acquired from cohoused animals of distinct phenotypes or from longitudinal sampling (6, 9). On the basis of this concept, we previously described a novel strategy for metagenomic analysis, the “quasi-paired cohort,” in which we paired samples that displayed a similar species profile but distinct phenotypes (10). Briefly, this method begins with a matrix containing species abundance inferred from sequencing-based metagenome data of samples exhibiting a binary phenotype (e.g., diseased versus healthy control). This matrix is used to populate a high-dimensional space with each detected species as a dimension. Making the assumption that two samples sharing a similar species profile yet with opposite phenotypes should theoretically reside near the boundary that delineates the two phenotype states in the Euclidean space (11), we recruited the concept of “boundary samples”—samples whose nearest neighbors are of the opposite phenotype, and paired each boundary sample with one of its nearest neighbors of the opposite side to form a “twin.” To avoid bias, we selected multiple parallel boundary samples for both sides and then compared the abundance or presence/absence of each species across twinned samples to identify candidate species for each of the two phenotypes (see Materials and Methods for details). This approach allowed us to transform the original group cohort into a paired cohort, which not only controls for individual diversity but also increases statistical power (10).
In the present study, by using the quasi-paired cohort strategy, we reported a deficiency of butyrate-producing species and enrichment of butyrate consumers in RA patients, which was associated with ACPA production and joint deformity. We further showed that dietary butyrate supplementation conferred the anti-inflammatory benefits of rebalancing follicular helper T (TFH) cells and regulatory T cells (Tregs). Our findings revealed a critical role of butyrate-metabolizing species in the pathogenesis of RA and suggested a butyrate-based therapeutic potential in RA.
RESULTS
Composition of butyrate-metabolizing species is associated with RA
In comparing the 25 treatment-naïve RA patients with the 29 age- and gender-matched healthy individuals (table S1 and Fig. 1), the two groups did not exhibit significant differences in microbial diversity (fig. S1). A univariance comparison of all bacterial species between the two groups did not identify any significantly deviating species [Mann-Whitney U test, false discovery rate (FDR) < 0.05], which was not surprising given the substantial interindividual diversity in metagenome data. Dealing with these high-dimensional datasets is of great difficulty, as the numerous cofactors are hard to control (12, 13). A common way to control diversity is to perform a meta-analysis that often includes a large number of samples.
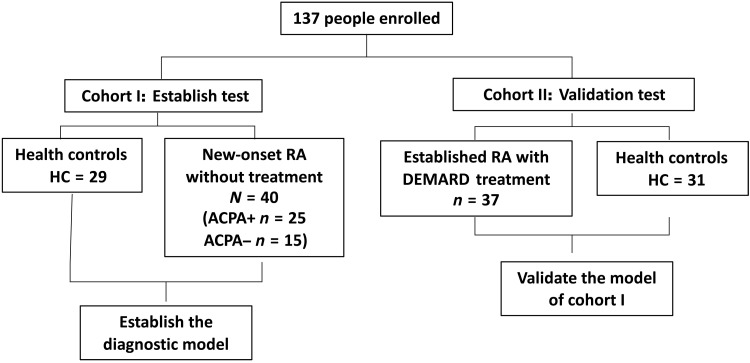
Fig. 1. Flow of participants in this study.
Diagram depicting the enrollment and analysis of patients with RA and healthy individuals. We recruited two cohorts of RA patients with or without treatment and performed shotgun sequencing of the intestinal metagenome for each subject. Cohort I was treatment-naïve new-onset RA (NORA) and healthy individuals. Cohort II was established RA patients and healthy individuals.
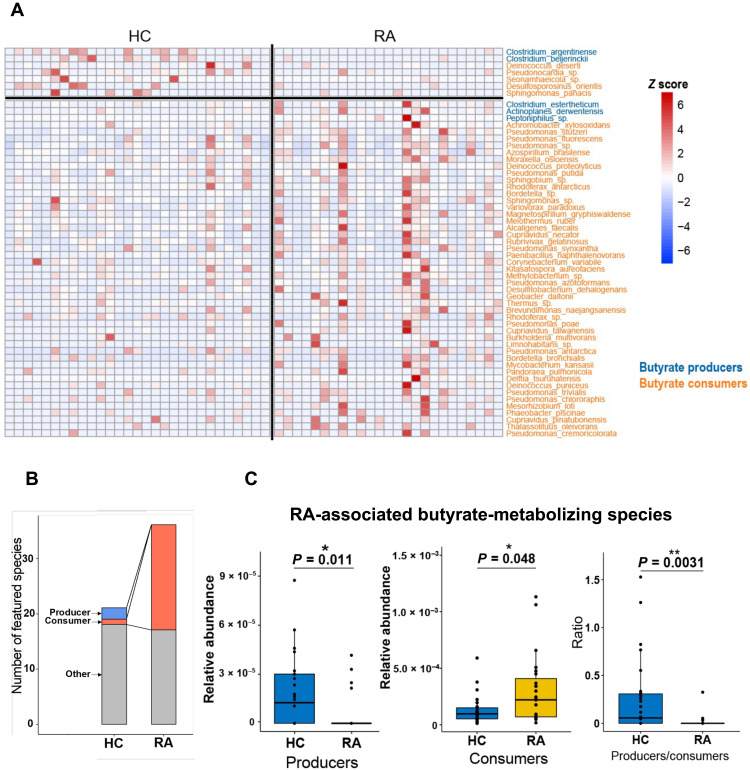
In this study, we used a novel quasi-paired cohort strategy (10) based on a small cohort with 25 treatment-naïve patients and 29 paired controls. This strategy was inspired by the concept of a paired study (6, 9). Briefly, we reconstructed a quasi-paired cohort where samples with a similar species profile, but opposite phenotypes, are twinned, and then the species with RA-associated features were identified with statistical tests from the paired samples. Hence, 186 RA-associated species were found, with 149 overrepresented and 37 deficient in RA (table S2; Wilcoxon signed-rank test for paired samples, P < 0.0001). Unexpectedly, a considerable proportion of these species, 55 of 186 (29.6%), can metabolize butyrate based on previous data (table S3) (14, 15). Notably, 51 of 55 (92.7%) of the RA-associated butyrate metabolizers were butyrate consumers, 46 of which were enriched in RA patients compared with only five consumers enriched in controls, indicating an enhanced butyrate-consuming microbiome in the gut of RA patients (Fig. 2A). A stricter test on the representative frequency (presence or absence in a sample) of each species between paired samples identified a more stringent panel of 57 RA-associated species (Kolmogorov-Smirnov test, P < 0.0001; table S4), including 38.6% (22 of 57) butyrate-metabolizing species. According to this panel, more remarkable enrichment of butyrate consumers was found in RA patients (RA, 19 species versus healthy control, 1 species) (Fig. 2B). The total abundances of butyrate producers and butyrate consumers in this RA-associated panel also significantly decreased and significantly increased, respectively, in RA patients (Mann-Whitney U test, P < 0.05; Fig. 2C), which suggests an obviously disordered butyrate metabolism in RA.
Fig. 2. Microbial butyrate metabolism in RA patients and controls of the NORA cohort.
(A) Abundance of RA-associated butyrate-metabolizing species identified by the quasi-paired cohort method (Wilcoxon signed-rank test, P < 0.0001) in each subject. The color bar indicates the Z score–normalized abundance (scaled by row); blue and orange names represent butyrate producers and butyrate consumers, respectively. (B) Number of butyrate producers and butyrate consumers in RA-associated species identified by the quasi-paired cohort (Kolmogorov-Smirnov test, P < 0.0001). (C) Gross abundance of butyrate producers, butyrate consumers, and their ratio in patients and controls. Healthy control group (HC), n = 29; rheumatoid arthritis group (RA), n = 25. Wilcoxon rank sum test, *P < 0.05 and **P < 0.01.
Core butyrate-metabolizing species associated with systemic inflammation and autoantibodies production in RA
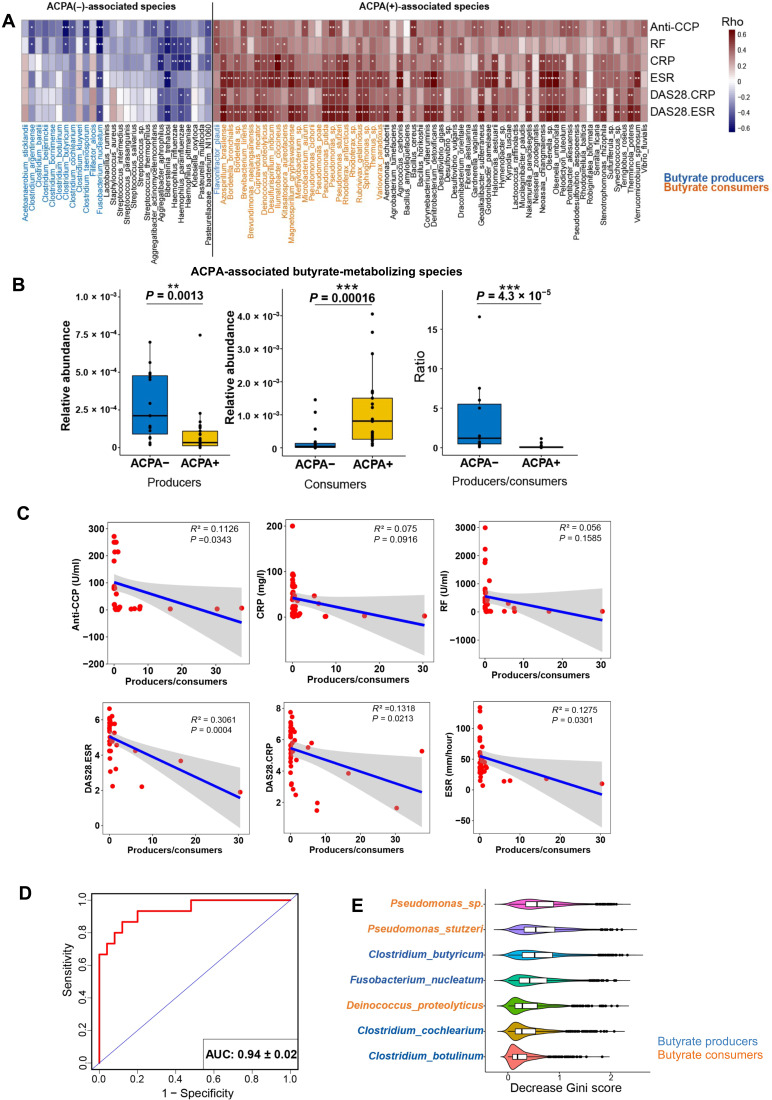
The presence of ACPA is associated with more severely deformed joints and poor prognosis (1). We thus asked whether butyrate-metabolizing species associated with autoantibody production. Using the quasi-paired cohort method to compare the 25 treatment-naïve ACPA-positive RA patients with 15 treatment-naïve ACPA-negative RA patients from the NORA cohort (table S1), we identified 92 ACPA-associated species (table S5; Wilcoxon signed-rank test for paired samples, P < 0.0001); 37 of 92 (40.2%) were butyrate metabolizers. Among them, 64.9% (24 of 37) of butyrate consumers and only 2.7% (1 of 37) of butyrate producers were enriched in ACPA(+) group, whereas 32.4% (12 of 37) of butyrate producers were enriched in ACPA(−) group with no enriched butyrate consumers (Fig. 3A and fig. S2). The proportion of butyrate-metabolizing species (37 of 92, 40%) was much higher than their usual proportion in the intestinal metagenome. Furthermore, the ACPA-associated butyrate producers and consumers in the panel showed diametrically opposite correlations to clinical features (Spearman’s correlation; Fig. 3A). The gross abundance of these butyrate producers and consumers also exhibited significant differences between ACPA(−) and ACPA(+) patients (Fig. 3B; P < 0.005). Therefore, we defined the 12 butyrate producers and 24 butyrate consumers from the ACPA-associated species panel as a core panel of RA-associated species (table S5), based on the above observations and the anti-inflammatory effect of butyrate evaluated by other studies (16–18).
Fig. 3. The intestinal butyrate metabolism correlates with ACPA status.
(A) List of ACPA-associated species and their correlated clinical markers of systemic inflammation and titers of autoantibodies. The colored bar indicates the Spearman‘s coefficients, ρ > 0, positive correlation. ρ < 0, negative correlation. *ρ > 0.4 or ρ < −0.4; **ρ > 0.5 or ρ < −0.5; ***ρ > 0.6 or ρ < −0.6. Blue, orange, and black species names represent butyrate producer, butyrate consumer, and non–butyrate-metabolizing species. ACPA, anti-citrullinated protein antibodies; anti-CCP, anti-cyclic citrullinated peptide; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; RF, rheumatoid factor; DAS28, disease activity score calculated with ESR, DA28-ESR, CRP, and DAS28-CRP. (B) Total abundances of butyrate producers and butyrate consumers in the ACPA-associated species panel and their ratio (producer/consumer) in ACPA(−) and ACPA(+) subgroups. Wilcoxon rank sum test, **P < 0.01 and ***P < 0.001. (C) Negative correlations of the producer/consumer ratio to clinical markers of systemic inflammation and titers of autoantibodies. Each dot indicates a subject. Solid blue line represents linear regression curve. (D) Performance of the random forest classifier for ACPA status based on abundance of the 36 butyrate-metabolizing species in the ACPA-associated species panel evaluated by receiver operating curve (ROC) with 1000 bootstrap replicates. AUC, area under the curve. (E) Top contributing species to the random forest model. The violins indicate the decreased Gini score in 1000 bootstrap replicates, and the middle lines in the box represent the mean of decreased Gini score. Blue and orange names represent butyrate producers and butyrate consumers, respectively.
We further investigated the correlations of core panel and clinical markers of RA. The ratio of butyrate producers to butyrate consumers in the core species showed significantly negative correlations with various clinical manifestations and titers of autoantibodies in RA (Fig. 3C). Then, we constructed a random forest classifier with the abundance of the 36 species in the panel as input, which displayed excellent performance in distinguishing ACPA(−) patients from ACPA(+) patients with a mean area under the curve (AUC) of 0.94 in the receiver operating characteristic (ROC) curve evaluation (Fig. 3D). When ranked with the decreased Gini score, which indicates the contribution of each species to the model, the top species included both butyrate producers and butyrate consumers (Fig. 3E), suggesting that they were both implicated in determining ACPA status.
Core butyrate-metabolizing species indicate bone erosion and joint deformity in RA
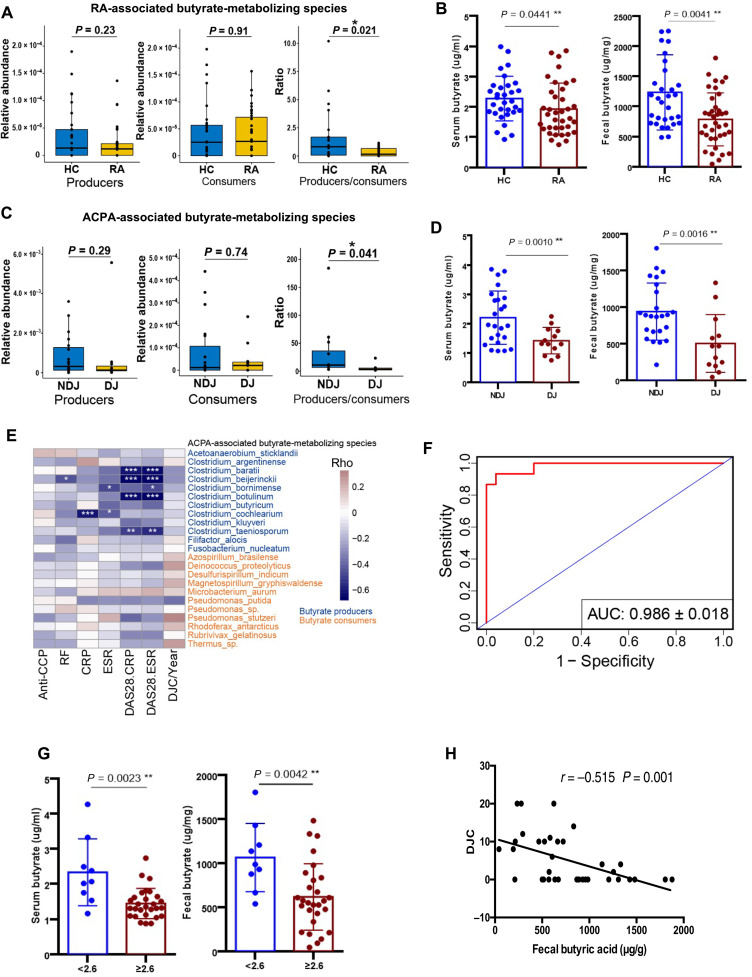
To validate the effect of the core panel of butyrate-metabolizing species in RA, we enrolled another cohort of 37 established RA patients and well-matched healthy individuals (n = 31; table S6 and Fig. 1), especially those with joint erosion. As shown in Fig. 4A, the ratio of the butyrate producer to consumer significantly decreased in established RA patients compared to controls, which was confirmed by significant reductions in both serum and fecal butyrate levels in RA patients when compared to controls in the targeted metabolic profiling of SCFAs in all subjects (Mann-Whitney U test, P < 0.05; Fig. 4B and fig. S3). Next, we subgrouped the established patients into DJ (deformed joints; n = 13) and NDJ (no deformed joint; 2 years after diagnosis, n = 20) groups and compared their gross abundances of ACPA-associated butyrate producer or consumer species. The producer/consumer ratio was significantly decreased in DJ patients (Fig. 4C) and consistent with significantly elevated concentrations of serum and fecal butyrate in the targeted measurement (Mann-Whitney U test, P < 0.05; Fig. 4D).
Fig. 4. Gut butyrate metabolism in the validation cohort of established RA patients.
(A) Gross abundance of RA-associated butyrate producer, butyrate consumer, and the producer/consumer ratio in subjects of both groups. RA, established RA patients; HC, healthy controls. (B) Serum and fecal butyrate concentrations in RA and HC subjects. (C) Gross abundance of butyrate producer and butyrate consumer in the ACPA-associated panel as well as the producer/consumer ratio in NDJ and DJ patients. DJ, patients with deformed joints; NDJ, patients with no deformed joints. (D) Serum and fecal butyrate concentrations in NDJ and DJ patients. (E) Correlations of the abundance of butyrate-metabolizing species in the ACPA-associated panel to clinical markers of systemic inflammation, titers of autoantibodies, and the count of deformed joints. The colored bar indicates the Spearman’s coefficients, ρ > 0, positive correlation. ρ < 0, negative correlation. *ρ > 0.4 or ρ < −0.4; **ρ > 0.5 or ρ < −0.5; ***ρ > 0.6 or ρ < −0.6. Blue and orange species names represent butyrate producer and butyrate consumer. DJC/year, the count of deformed joints standardized by disease duration. (F) Performance of the random forest model trained by ACPA status (refer to Fig. 2D) in predicting the development of joint deformity in established patients evaluated by ROC with 1000 bootstrap replicates. (G) Comparison of serum and fecal butyrate concentration between patients with high (DAS28-ESR ≥ 2.6) and low disease activity (DAS28-ESR < 2.6). (H) Correlation between the DJC and the level of fecal butyrate. Solid line indicates the curve of the linear regression.
Butyrate production influenced the inflammatory status in RA
In the validation cohort, butyrate producers and butyrate consumers in the ACPA-associated panel also exhibited a reverse correlation to inflammation markers and the number of deformed joints standardized by disease duration (Spearman’s correlation; Fig. 4E). The random forest model, which was trained to distinguish ACPA status in the NORA cohort with the abundance of the 36 butyrate-metabolizing species in the ACPA-associated panel, was tested by the validation cohort for predicting the outcome of established RA patients, i.e., whether or not they developed deformed joints. The model discriminated patients in the DJ group from NDJ groups with an accuracy of 0.986 when evaluated with ROC (Fig. 4F), indicating that both ACPA generation and joint erosion were associated with intestinal butyrate metabolism. Consistently, measurements of fecal and serum butyrate concentrations revealed significantly higher levels of butyrate in RA patients with low disease activity (assessed by disease activity score, DAS28-ESR < 2.6) than those with high disease activity (DAS28-ESR ≥ 2.6) (Mann-Whitney U test, P < 0.005; Fig. 4G), and fecal butyrate levels were negatively correlated with the number of deformed joints in RA patients (P < 0.001; Fig. 4H).
Butyrate treatment suppressed the osteoclast differentiation in RA patients
As the butyrate producer/consumer ratio was shown to decrease significantly in DJ patients, we then examined whether butyrate affected bone destruction in RA. In this study, human data indicated that levels of fecal and serum butyrate were lower in patients with RA with increased levels of Rankl, an essential component of osteoclastic signaling (Mann-Whitney U test, P < 0.05; fig. S4, A and B). In vitro, osteoclast differentiation assays showed that butyrate suppressed the differentiation of osteoclasts (fig. S4, C and D), which is consistent with a recent study (19), showing the potential implications of gut microbial metabolite supplementation for bone restoration. Mechanically, butyrate administration led to changes of the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways in osteoclast differentiation (fig. S5) and histone deacetylase (HDAC) reduction (fig. S4, E and F). Together, these results demonstrated that butyrate changes the differentiation of osteoclasts in RA and prevents bone destruction.
Butyrate directly promoted Treg polarization and suppressed Tconvs and proinflammatory cytokines
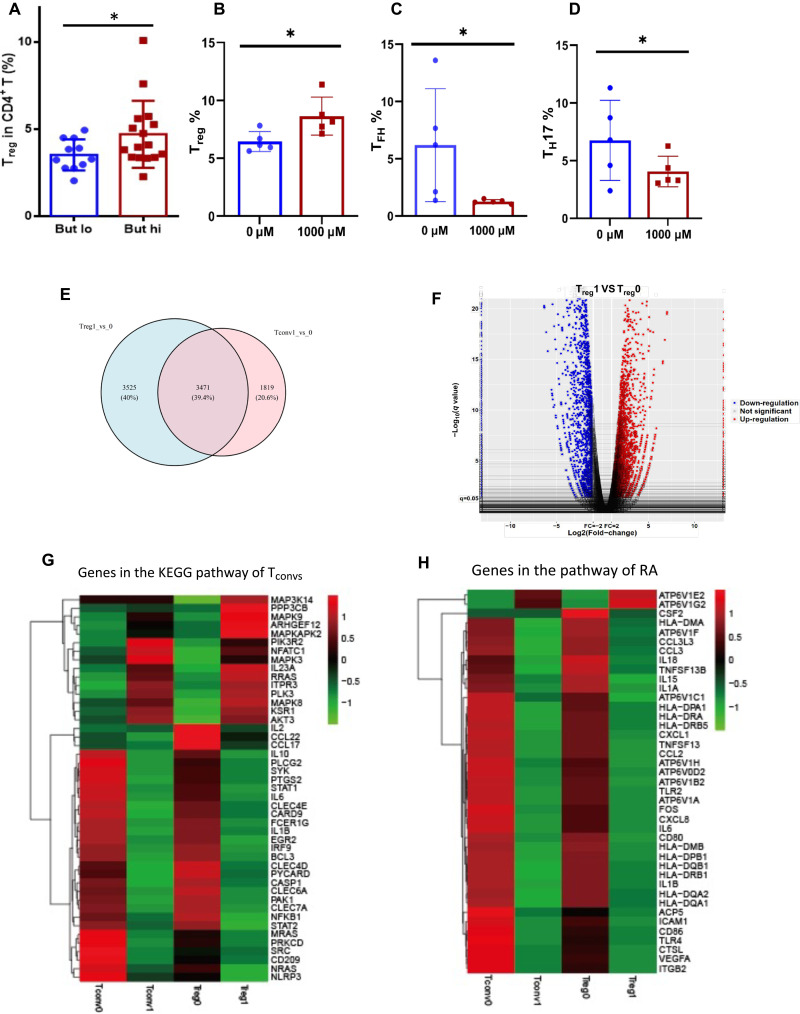
To further explore the mechanisms through which intestinal butyrate metabolism imposes on autoantibody generation, we evaluated the subtypes of circulating CD4+ T cells [including Tregs, TFH, T helper 17 (TH17), TH1, and TH2] in patients with RA. We observed significantly higher proportions of circulating Tregs in RA patients associated with a high levels of stool butyrate (Mann-Whitney U test, P = 0.0437; Fig. 5A). We also cocultured butyrate with peripheral blood mononuclear cells (PBMCs) from RA patients and found that the percentage of Tregs was increased (Fig. 5B) and TFH and TH17 cells decreased significantly (Fig. 5, C and D), but not TH1 and TH2 cells.
Fig. 5. Butyrate up-regulates Tregs and down-regulates TH17 and TFH cells.
(A) Circulating Tregs (%) in patients with high or low fecal butyrate concentration [the cutoff value was 1825 μg/mg, according to the 95% confidence interval of the fecal butyrate levels (1429 to 2244)]. (B to D) Change of the percentage of Tregs (B), TFH cells (C), and TH17 (D) after coculture of butyrate with PBMCs from RA patients. (E) Venn diagram of known differentially expressed genes in freshly isolated Tregs and conventional TH cells (Tconvs). (F) Volcano plot depicting transcript fold change before and after butyrate stimulated PBMCs (n = 3) versus P value. (G) Heat map of Tconvs differentiation-related genes in Tregs and Tconvs before (0) and after (1) butyrate administrations. (H) Heat map of RA development–related genes in Tregs and Tconvs before and after butyrate administrations. 0, before butyrate administration; 1, after butyrate administration.
Moreover, to understand how butyrate supports Treg differentiation, CD4+ T cells were isolated from PBMCs of RA patients and treated with butyrate in vitro for 4 days, and the difference in regulation between Tregs and effector Tconvs (conventional TH cells) was evaluated with RNA sequencing (RNA-seq). Altered genes between Tregs and Tconvs were unexpectedly different (Fig. 5E), and only 39.4% of the genes were shared. Differential expression is shown in a volcano plot (Fig. 5F). As expected, Tregs after butyrate administration have a unique gene expression profile compared to Tregs after butyrate administration, with down-regulation of linked immune factors in Tconv differentiation (Fig. 5G) and RA development (Fig. 5H). These genes are known TFH, TH17, TH1, TH2, and associated autoimmune genes (figs. S6 and S7), suggesting that butyrate could not only promote Tregs but also suppress Tconvs and inflammatory cytokines, probably helping lead to the hemostasis of the immune system. Specifically, certain HDAC subtypes were reduced in Tregs due to administration of butyrate (fig. S8). Together, these data suggest that butyrate selectively promoted Tregs and inhibited Tconvs, suggesting a substantial impact on immune balance in RA.
Butyrate suppresses arthritis in CIA mice by modulating cellular and humoral immune responses
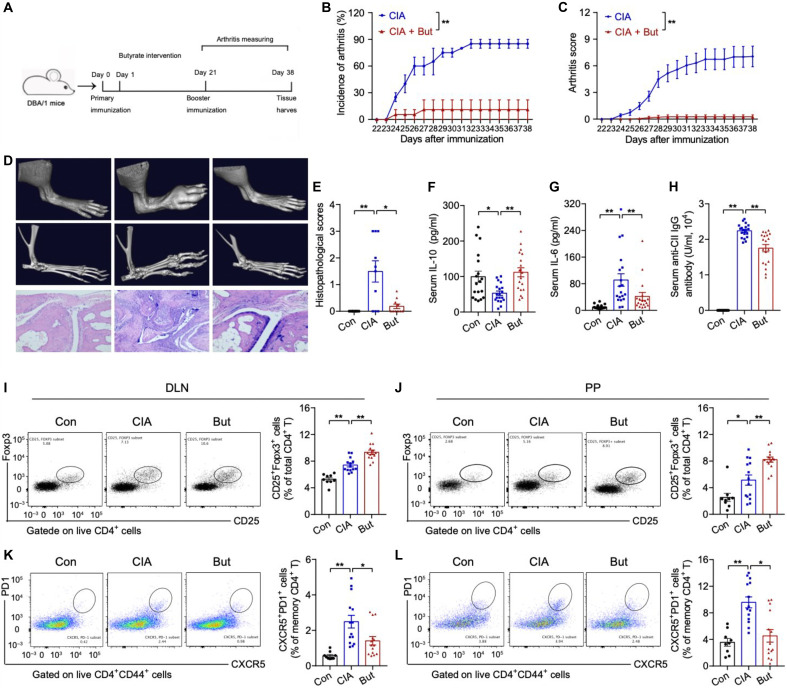
Next, we used the collagen-induced arthritis (CIA) model to investigate whether butyrate in vivo can confer protective effects against RA. A butyrate-rich diet was started on the first day of collagen immunization (Fig. 6A). Compared to control CIA mice fed on normal chow, butyrate supplementation significantly increased butyrate levels in stool and blood (P < 0.05; fig. S10), accompanied by the significantly lower overall incidence of arthritis (11% versus 85%, P < 0.01), reduced severity of joint inflammation (P < 0.05), and milder arthritis characterized by a lack of bone destruction (Fig. 6, B to E).
Fig. 6. Butyrate suppresses immune responses and reduces joint inflammation and bone destruction in mice.
(A) Scheme of the experimental design for a butyrate-based preventive regimen using the classic CIA mouse model of RA. (B and C) Arthritis incidence and scores in CIA mice with or without butyrate (But) supplementation. CIA, n = 19; But, n = 19. (D) Arthritis and bone erosion of the paws by images of micro-CT (top and middle) and hematoxylin and eosin (H&E)–stained (bottom) analysis. (E) Histopathological scoring of the paws. Con, control. Scale bars, 200 μm. n = 10 per group. (F to H) Concentrations of serum IL-10 (F), IL-6 (G), and anti–collagen type II (CII) antibody (H). n = 19 per group. (I to L) Representative flow cytometry plots with graphs showing the frequency of CD4+CD44+CD25+Foxp3+ Tregs (I and J) and CD4+CD44+CXCR5+PD1+Bcl6+ TFH cells (K and L). Con, n = 9; CIA, n = 8 to 14; But, n = 8 to 14. Data were pooled from two independent experiments and are here expressed as means ± SEM. Significance was determined using Kaplan-Meier analysis with log-rank test (B), two-way ANOVA followed by Tukey’s multiple comparisons test (C), or Mann-Whitney U test (E-L). *P < 0.05 and **P < 0.01.
Compared with control CIA mice, the butyrate supplemented mice had significantly increased serum interleukin-10 (IL-10) levels (111.9 ± 6.099 pg/ml versus 53.1 ± 12.71 pg/ml, P < 0.01) as well as significantly decreased levels of serum IL-6 (41.88 ± 11.74 pg/ml versus 91.14 ± 18.75 pg/ml, P < 0.01) and autoantibodies (1.752 ± 0.1009 × 104 U/ml versus 2.239 ± 0.04373 × 104 U/ml, P < 0.01) (Fig. 6, F to H). Flow cytometry analysis of T cells revealed that butyrate supplementation significantly increased the proportion of CD4+CD25+Foxp3+ Tregs in both joint draining lymph nodes (DLNs) (P < 0.01; Fig. 6I) and Peyer’s patches (P < 0.01; Fig. 6J) and reduced the proportion of CD4+CD44+CXCR5+PD-1+Bcl6+ TFH cells in both joint draining lymph nodes (P < 0.01; Fig. 6K) and Peyer’s patches (P < 0.01; Fig. 6L), consistent with previous reports of butyrate effects on T cell subset modulation (18, 20, 21). Follicular regulatory T (TFR) cells are known to suppress TFH cell differentiation and germinal center formation (22, 23). The butyrate supplementation in CIA mice also significantly increased the number of CD4+CD44+CXCR5+PD-1+Foxp3+ TFR cells, especially in the DLN (fig. S10, A and B). Further, we also found a significant decrease in the proportion of B220+CD4−CD95+GL-7+ germinal center B (GCB) cells (P < 0.005; fig. S10, C and D). These results confirmed the protective effects of intestinal butyrate in the development of RA.
DISCUSSION
Our study provided evidence that the adjusted gut microbiome with increased butyrate-producing species and reduced butyrate consumers could increase butyrate yield and is associated with mild inflammatory condition and good prognosis.
Here, by applying a hypersensitive analytical method—quasi-paired cohort, we confirmed the essential role of butyrate-metabolizing species in a clinical RA cohort and uncovered the contribution of gut butyrate consumers to the final butyrate concentration, which highlighted the effects of both sides of butyrate metabolism—producer and consumer—on the deregulation (or dysregulation) of Tregs and effector Tconvs. We also established a panel of species that specifically distinguished patients that were ACPA positive and had joint deformation. Although numerous studies have identified molecular mechanisms of how butyrate affects intestinal epithelia and immune system that may be beneficial in RA or other autoimmune diseases (2–4, 10, 24–26), there remains a compelling need for evidence of its protective role in clinical research and the delineation of the process.
Previous studies reported that fecal butyrate in RA patients decreased significantly. The interaction between butyrate producers and butyrate consumers in a person’s gut maintains a dynamic equilibrium. Butyrate consumer species, such as Pseudomonas spp., are often robust and able to survive in harsh environments even without carbohydrate supply. These species can live on various carbon sources, such as butyrate, lactate, acetate, and so on. Butyrate is one of the best nutrients for higher calories, but it is not necessary for their survival. Intestinal butyrate is primarily supplied by butyrogenic species, but if it is not sufficient, butyrate consumers can use other nutrients to survive.
Our results point to obviously increased abundance of bacteria that produce butyrate in RA patients with lower incidence of DJC and ACPA(+), suggesting potential roles of butyrate in alleviating inflammation. We found that net butyrate yield, deduced from the ratio between producer and consumer and supported by metabolite measurements, exhibits negative correlations to disease activity, antibody production, and joint deformity, which further consolidates the protective effects of butyrate in RA.
Butyrate is essential to inhibit CIA, likely via the Treg/IL-10/TH17 axis (27) or through B cell regulation (28). Our study showed higher butyrate associated with increased Tregs in RA patients. Another aspect of our finding is that a butyrate diet alone can protect CIA mice against arthritis, accompanied with the promotion of Tregs and suppression of TFH cells in both Peyer’s patches and joint draining lymph nodes. In that regard, we and others have reported that the presence of circulating TFH cells in the blood suggests autoimmune conditions and disease activation, including RA and systemic lupus erythematosus (SLE) (23). We would like to emphasize here that, in addition to the fact that butyrate has strong promoting effects on Tregs, the impact on effector Tconvs, such as TFH cells, is rather specific. The primary function of TFH cells is to activate GCB cells to differentiate into plasma and memory B cells, leading to autoantibody production in autoimmune disease, such as ACPA in RA patients. Thus, we ultimately determined that gut butyrate exposure might restore the immune balance and dampen autoimmunity locally, such as the intestinal tract or the distant tissue target like arthritis, by influencing T cell differentiation to hemostasis. All these facts demonstrated that butyrate can extinguish the inflammation in RA patients and imply future therapeutic applications. Meanwhile, butyrate restrains osteoclast differentiation, which aggravates the joint deformity of RA. Thus, our data demonstrated the multiple efficiency of butyrate in protecting from joint deformity, consistent with the results of a previous study (28).
However, the relative abundances of ACPA-associated butyrate-metabolizing species are very low. The abundance of butyrate producers gradually decreases along the colon from their major habitat in the ileocecum. As we used stool samples in this study, the butyrate producers we detected were only a portion of their abundant populations in the gut. However, the differences in fecal butyrate producers between groups might indicate differences in the upper colon where these species are a considerable proportion of the microbial flora and contribute to a shift in serum and fecal butyrate in a physiologically relevant way. Unfortunately, the current metagenome study was not able to determine the absolute number of butyrate-metabolizing bacteria in each individual as internal references were not added in routine metagenomic research, and it is not convenient to obtain microbiome samples of the ileocecum.
Together, metagenomic and metabolic analyses of RA patients identified a common butyrate-metabolizing species panel mechanistically connected to both ACPA status and outcomes of joint deformity, which supports a protective role of microbial-derived intestinal butyrate in systemic immune modulation in RA. These findings also provide a strong rationale for future clinical investigations on the manipulation of intestinal butyrate metabolism to improve the treatment of RA patients through dietary or microbiome-based therapy.
MATERIALS AND METHODS
Study design and sample collection
The study recruited male and female patients aged 18 to 70 years with RA, diagnosed according to the 2010 American College of Rheumatology and European League Against Rheumatism (EULAR) classification criteria. Individuals were excluded if they were complicated by any other acute or chronic disease. Cohort I (NORA) included 40 treatment-naïve RA patients (25 ACPA-positive RA patients and 15 ACPA-negative RA patients) with the 29 age- and gender-matched healthy individuals (table S1). Cohort II (established RA) included 37 established RA patients undergoing standard treatment and 31 age- and gender-matched healthy controls (table S6 and Fig. 1). The study was performed in accordance with the provisions of the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice guidelines and was approved by the Peking University People’s Hospital Ethics Committee. The protocol and all relevant study forms for the trial were approved by the Peking University People’s Hospital institutional review board (approval no. 2016PHB200). All patients provided written informed consent.
Metagenomics sequencing and annotation
All fecal samples were collected at Peking University People’s Hospital, transported frozen to the Beijing Institute of Genomics, Chinese Academy of Sciences, and stored at −80°C for subsequent metagenome sequencing. We extracted DNA from fecal samples using the E.Z.N.A. Soil DNA Kit (D5625-02, OMEGA) and performed paired-end metagenomic sequencing on an Illumina Hiseq2000/2500 platform [100 base pairs (bp)/150 bp × 2; insert size, 350 bp]. Raw reads were next applied to quality control where ambiguous sequences and adapters were filtered by FastQC (version 0.11.5) and low-quality bases and reads by Trimmomatic (29) (version 0.33, option: SLIDINGWINDOW:4:20 MINLEN:40) or FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html, version 0.0.13, option: -q 20 -p 80). Then, reads of human origin that mapped to Hs37 human genomes were removed by BWA (mem module with default parameters) (http://bio-bwa.sourceforge.net/), and polymerase chain reaction (PCR) duplicates were removed by PRINSEQ (http://prinseq.sourceforge.net/) with the options -verbose -derep 1 -derep_min 2. The final clean reads were applied to taxonomy annotation. Microbial classification was performed with Kraken (http://ccb.jhu.edu/software/kraken/, version 1.0, module Kraken-translate, option --mpa-format) for the clean data of all sequenced samples, whereby total relative abundance of all species in a sample was normalized to 1. Then, species of extremely low abundance (relative abundance < 10−5) that were present in very few samples (<10% total samples) were removed to avoid stochastic associations. Functional profiling of every sample was performed with HUMAnN2 (http://huttenhower.sph.harvard.edu/humann2), which maps reads to a default database of functionally annotated pangenomes (ChocoPhlAn database) and identifies the gene family (UniRef database) and potential metabolic pathway (MetaCyc database collection, https://biocyc.org/) of a microbial community.
Construction of the quasi-paired cohort
The quasi-paired cohort was constructed as previously described (10), excepting a few modifications. Briefly, a high-dimensional space was first constructed where the abundance of each species represented a dimension, and all subjects were positioned in the space according to their species profiles. Thus, the samples formed a Euclidean distance–based similarity network. The next step was to find boundary samples, which are critical in defining the boundaries between phenotype groups. Boundary samples are those exhibiting a higher species profile similarity to samples of an opposite group than to samples in the same group, i.e., longer distance to neighbors than members of an opposite group in the high-dimensional space. To find boundary samples, the similarity of each sample to its nearest k neighbors was first calculated as KNN (the average Euclidean distance to k nearest neighbors), where k was the square root of the sample size. Then, outliers that were too far (KNN > mean KNN + SD) and redundancies that were too near (KNN < mean KNN-SD) to their neighbors were removed to avoid stochastic impacts. Last, samples of intragroup KNN > intergroup KNN were selected as boundary samples for both groups. Each boundary sample was used to construct k sample pairs with its k nearest neighbors of the opposite side, and these pairs were used to constitute a cohort of paired samples where samples of each pair bear a similar species profile (nearest neighbors in the high-dimensional space) but opposite phenotypes. After removing redundant pairs, the quasi-paired cohort was finally reconstructed.
Identification of butyrate-metabolizing species
For butyrate producers, Vital et al. (14) searched a total of 3184 bacterial genomes and identified 225 bacteria with the potential to produce butyrate according to the presence of all the essential genes in the butyrogenic pathways and the synteny of these genes. These 225 bacteria belong to 131 species and show apparent consistency in butyrogenic potentials in strains of the same species. Thus, we classified the annotated species in our samples as butyrate producers if they were within the species list of the 131 butyrogenic species provided by this previous publication.
For butyrate consumers, there is no such report that provides a clear list of microbial species of butyrate consumption. Ziels et al. (15) identified the essential genes of microbial butyrate degradation that catalyze the reactions of butyrate➔crotonyl–coenzyme A (CoA)➔3-hydroxybutyryl-CoA. The corresponding genes are acyl-CoA dehydrogenase or butyryl-CoA dehydrogenase for the first step and enoyl-CoA hydratase or crotonyl-CoA hydratase for the second step. Thus, we screened publicly deposited genomes of all species annotated in our samples for the presence of the four genes according to the KEGG database, and identified those genomes carrying enzymes of both steps as potential butyrate consumers, which belong to 185 species. We noticed that many butyrate producers also contain butyrate-consuming genes, suggesting their abilities of using the butyrate they synthesize; these species were still classified as producers. All the butyrate-producing and butyrate-consuming species identified in samples of this study are listed in table S3.
Random forest model in evaluating diagnostic power
Random forest model with the abundance of the 12 butyrate-producing species and 24 butyrate-consuming species as input was constructed with the caret package (https://cran.r-project.org/web/packages/caret/) and the randomForest R package (https://cran.r-project.org/web/packages/randomForest/index.html). Then, the diagnostic capacity of the butyrate species in the panel was evaluated with AUC and accuracy through the R packages pROC (https://cran.r-project.org/web/packages/pROC/index.html) and ROCR (https://cran.r-project.org/web/packages/ROCR/index.html) in discriminating the ACPA status and DJ from NDJ patients. The model was trained by 40% of samples through twofold cross-validation, and all samples were used as a testing dataset to assess the diagnostic capability. The procedure was repeated for 1000 bootstrap replicates and species, and decreased Gini score of each replicate was recorded.
CIA induction and butyrate intervention
Male DBA/1 mice (6 to 8 weeks old) were purchased from Huafukang Co. Ltd. (Beijing, China) and fed under specific pathogen–free conditions. All experiments were carried out in accordance with guidelines prescribed by the Animal Care and Use Committee of Peking University People’s Hospital. CIA induction was performed according to a previously published protocol (30). Briefly, DBA/1 mice were immunized intradermally at the base of the tail with 150 μg of bovine type II collagen (CII; Chondrex, Redmond, WA, USA) emulsified in complete Freund’s adjuvant (Sigma-Aldrich, St. Louis, MO, USA) in equal volumes. Twenty-one days later, a booster immunization was performed using 75 μg of CII emulsified in Freund’s incomplete adjuvant (Sigma-Aldrich). Clinical score was assessed after the booster immunization using the following system detailed previously (30): 0, normal; 1, erythema and swelling of one or several digits; 2, erythema and moderate swelling extending from the ankle to the mid-foot (tarsals); 3, erythema and severe swelling extending from the ankle to the metatarsal joints; 4, complete erythema and swelling encompassing the ankle, foot, and digits, resulting in deformity and/or ankyloses. The scores of all four limbs were summed, yielding total scores of 0 to 16 per mouse.
We randomized mice into two groups, butyrate diet and control. All mice were fed with conventional fodder containing 20% (w/w) butyrate [20% (w/w), butyrate uniformly mixed in fodder, HFK Bioscience Co. Ltd.] or conventional fodder (HFK Bioscience Co. Ltd) after the initial immunization (day 1) and throughout the experiment. At the study endpoints, mice were euthanized and serum samples were collected for cytokine and autoantibody detection. The spleens, joint draining lymph nodes (popliteal and axillary lymph nodes, DLN), Peyer’s patches, and mesenteric lymph nodes were obtained from mice, sieved through a 70-μm cell strainer (Corning) in RPMI 1640 medium with 10% fetal bovine serum (FBS), and single-cell suspensions (106 cells/100 μl) were prepared for flow cytometry.
Radiography evaluations and histological analyses
The paws from each mouse were collected and fixed in 4% paraformaldehyde for 48 hours and then scanned using a Micro-CT scanner (Quantum FX, Caliper, USA). Then, the paws were decalcified in 5% EDTA, paraffin-embedded, sectioned, and stained with hematoxylin and eosin. A microscopic assessment of sagittal sections was performed, and histopathological changes were scored on the basis of the following previously reported parameters (31): 0, normal synovium; 1, synovial membrane hypertrophy and cell infiltrates; 2, pannus and cartilage erosion; 3, major erosion of cartilage and subchondral bone; 4, loss of joint integrity and ankylosis. The scores of all four limbs were summed and divided by four, yielding average scores of 0 to 4 per mouse.
Flow cytometric analyses
Cell surface markers were first stained, and the cells were then fixed, permeabilized with an intracellular staining buffer set (Thermo Fisher Scientific) following the manufacturer’s protocol, and stained with intracellular or intranuclear markers. As shown in table S7, antibodies were purchased from BD Biosciences (San Jose, CA, USA), eBioscience (Thermo Fisher Scientific), or BioLegend (San Jose, CA, USA). Tregs were defined as CD4+CD25+FOXP3+, TFH cells as CD4+CD44+CXCR5+PD-1+Bcl6+, TFR cells as CD4+CD44+CXCR5+PD-1+Foxp3+, and GCB cells as B220+CD4-CD95+GL-7+. See fig. S11 for the detailed gating strategies. Flow cytometry was performed using FACSAria II (BD Biosciences), and the data were analyzed using FlowJo v10.0.7 software (Tree Star Inc., Ashland, OR, USA).
Culture of osteoclast
Monocytes were isolated from blood obtained from normal, healthy donors and patients with RA by Ficoll separation. Collected cells were resuspended in α–minimum essential medium (Thermo Fisher Scientific) containing 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml) and plated in 24-well flat-bottom plates for 7 to 9 days. To determine the effect of butyrate on osteoclast development, monocytes were cultivated with 1000 μM butyrate or without it. Monocytes were differentiated and matured in osteoclasts with macrophage colony-stimulating factor (M-CSF) (30 ng/ml) (PeproTech, Suzhou, China) and receptor activator of nuclear factor-kappa B ligand (RANKL) (50 ng/ml; R&D Systems, Shanghai, China). The media and cytokines were fully removed and replaced every 3 days. By the end of the culture period, the cells were collected and transferred to TRIzol in preparation for PCR quantitation.
In vitro cell culture and butyrate stimulation
PBMCs were isolated using density gradient centrifugation performed in 24-well plates. Subsequently, cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin with 5% CO2 at 37°C. Butyrate in 1000 μg/ml was added for cell stimulation. Three days later, cells were washed in phosphate-buffered saline (PBS) with 2% FBS and stained with conjugated antibodies as follows: anti-CD3–peridinin chlorophyll protein (PerCP), anti-CD4–fluorescein isothiocyanate (FITC), anti-CD25–phycoerythrin (PE), and anti-CD127–Brilliant Violet 605 (BV605). After incubating for 30 min in the dark, Tregs (CD25+CD127−) and Tconvs (CD4+CD25−) were sorted and purified using an Astrios EQ flow cytometer (Beckman Coulter, USA).
RNA-seq analyses
RNA-seq was performed on isolated RNA from fresh Tregs (CD25+CD127−) and Tconvs (CD4+CD25−) from the same donor generated before and after butyrate coculture. Briefly, RNA-seq libraries were generated using Illumina TruSeq Stranded Total RNA Kits (Illumina, San Diego, CA, USA). Raw sequencing reads were preprocessed by filtering ribosomal RNA (rRNA) reads, sequencing adapters, short fragment reads, and other low-quality reads. We used Hisat2 (version: 2.0.4) to map the clean reads to the human hg38 reference genome with two mismatches. After genomic mapping, StringTie (version 1.3.0) was run with a reference annotation to produce FPKM (reads per kilobase of exon model per million mapped reads) values for known gene models. Differentially expressed genes were identified using edge R. The significance threshold for the P value in multiple tests was set by the FDR.
Real-time quantitative PCR
A total of 1 × 105 PBMC-derived Tregs or isolated osteoclast precursors were sorted, and RNA was harvested using the manufacturer’s instructions (Tiangen). Total RNA (0.5 μg) was used to generate first-strand complementary DNA (cDNA) as the initial step of a two-step reverse transcription PCR protocol with the RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific). For real-time PCR, SYBR Green Master Mix was used with the ViiA 7 System (Applied Biosystems). The following primers were used: HDAC1, ATCCGCATGACTCATAATTTGC (sense) and GGATGGAGCGCAAGAATTTAAT (antisense); HDAC2, AAAGTCTGCTACTACTACGACG (sense) and TTATGGGTCATGCGGATTCTAT (antisense); HDAC3, TTCAATATCCCTCTACTCGTGC (sense) and AGGTTTTCAAAGATTGTCTGGC (antisense); HDAC4, GCCAAAGATGACTTCCCTCTTA (sense) and TTTCGGCCACTTTCTGCTTTAG (antisense); HDAC5, CTACGACACGTTCATGCTAAAG (sense) and CACTGTCTGGATCTCATCTAGC (antisense); HDAC6, GTGTCACTTCGAAGCGAAATAT (sense) and CCACGATTAGGTCTTCTTCCAT (antisense); HDAC7, CTCAAACTGGACAACGGGAAG (sense) and AATGAAGCTCATTCCAGATGGT (antisense); HDAC8, TGCTGTCCTGGGAATATTACGATTGC (sense) and AACTGAATGCGTCTTCTACACCATCTC (antisense); HDAC9, ACCTCTCCTTCTTTGCCCAACATTAC (sense) and GGAACACCTTGCCTAAGCGTCTG (antisense); HDAC10, GCTTGTGTACCATGAGGACAT (sense) and CAGTGAAAGGTACTCTGTGCA (antisense); glyceraldehyde-3-phosphate dehydrogenase (GADPH), TGCACCACCAACTGCTTAGC (sense) and GGCATGGACTGTGGTCATGAG (antisense).
Metabolomics analysis
Feces and blood were processed for SCFA analysis using gas chromatography. The analysis of SCFA included the following fatty acids: acetic acid (C 2:0), propionic acid (C 3:0), isobutyric acid (C 4:0 i), butyric acid (C 4:0 n), isovaleric acid (C 5:0 i) valeric acid (C 5:0 n), isocaproic acid (C 6:0 i), caproic acid (C 6:0 n), and heptanoic acid (C 7:0). Chromatographic analysis was carried out using an Agilent Technologies 1260 A GC system with a flame ionization detector. A fused-silica capillary column with a free fatty acid phase (30 m × 0.53 mm × 0.5 μm) was used. Hydrogen was supplied as the carrier gas at a flow rate of 14.4 ml/min. The initial temperature was 100°C, which was maintained for 0.5 min, then raised to 180°C at 8°C/min, and held for 1 min. Next, the temperature was increased to 200°C at 20°C/min and finally held at 200°C for 5 min. The injection volume was 1 μl, and the run time for each analysis was 17.5 min.
Integrated peak areas corresponding to metabolite concentrations were further analyzed using MetaboAnalyst (www.metaboanalyst.ca). Metabolite abundance was expressed relative to the internal standard.
Cytokine and autoantibody detection
The concentrations of cytokines (IL-6 and IL-10) were measured using enzyme-linked immunosorbent assay (ELISA) kits (Multisciences), and the titer of collagen-specific antibody was analyzed using a mouse anti-bovine type II collagen IgG antibody assay kit (Chondrex). All measurements followed kit protocols as described in the manufacturer’s instructions.
Statistical analysis
The R package Vegan (https://cran.r-project.org/web/packages/vegan/index.html) was used to analyze ɑ diversity and β diversity of the microbiome. The Simpson index, Shannon index, and evenness (Pielou index) were calculated to indicate ɑ diversity. Principal coordinate analysis (PcoA) with Bray-Curtis dissimilarity distance was analyzed using ADONIS [permutational multivariate analysis of variance (PERMANOVA)] (https://cran.r-project.org/web/packages/PERMANOVA/index.html) to indicate β diversity. The significance of differences in abundance between groups was tested with Mann-Whitney U test for each species or pathways with P values adjusted by Benjamini and Hochberg’s approach. For paired samples in the quasi-paired cohort, statistical significance in species abundance was tested with a Wilcoxon signed-rank test, and a one-sided Kolmogorov-Smirnov test was used to test the difference in species’ presence frequency. Correlations between species abundance and clinical markers were evaluated with a Spearman rank correlation test or linear regression.
In the experiment, differences in arthritis scores and incidence between groups were determined by two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test and Kaplan-Meier analysis with log-rank test, respectively. The difference of cytokines, antibodies, or lymphocytes between two groups was analyzed by Mann-Whitney U nonparametric test. Statistical analyses were performed in GraphPad Prism v7.00 software (GraphPad, San Diego, CA, USA). A two-sided P value of <0.05 was considered statistically significant.
Acknowledgments
We thank Y. Zhou (Department of Medicine, University of Connecticut Health Center, USA) and H. Qi (Department of Basic Biomedical Sciences, School of Medicine, Laboratory of Dynamic Immunobiology, Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing, China) for manuscript revisions.
Funding: This study was supported by the National Natural Science Foundation of China (U1903210, 31530020 to Z.L.; 31970568 to Y.K.; 82071813 to J.H.; 81901648 to J.L.; 31671350 to J.Y.), Beijing Science and Technology Program (Z191100006619114 to J.H.; PKU2020LCXQ018 to J.H.), National Science and Technology Major Project (2018ZX10712001-018-002 to Y.K.), the National Key Research and Development Program of China (2016YFC0903800 to Y.K.), Programs of the Chinese Academy of Sciences (QYZDY-SSW-SMC017 to J.Y.), and Macao Science and Technology Fund (0094/2018/A3 to Z.L.
Author contributions: Z.L., J.H., and Y.K. conceived and designed the project. J.H., J.L., Y.C., Q.M., L.S., Y.G., and Y.S. were in charge of participant enrollment and sample collection. Y.C., J.J., J.T., Y.L., J.W. Y.Z., B.H., and C.S. carried out DNA extraction, PCR, and microbiota sequencing. F.H., W.X., Y.C., Q.M., X.S., B.H., R.F., and X.Sh. completed all the bioinformatic and statistical analyses of the microbiota. J.L. Y.W., R.F., and J.T. performed all the in vitro and in vivo experiments. J.H., J.L., Y.K., and Z.L. conducted data acquiring and processing and prepared figures. J.H., J.L., Y.C., Q.M., Y.K., Z.L., and J.Y. wrote and edited the manuscript. Y.L. and J.G. revised the manuscript. All the authors have revised and approved the manuscript submission.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw metagenome sequencing data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number PRJCA000337 at http://bigd.big.ac.cn/gsa.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 to S7
Other Supplementary Material for this manuscript includes the following:
Excel file S1 to S7
REFERENCES AND NOTES
- 1.Catrina A. I., Svensson C. I., Malmstrom V., Schett G., Klareskog L., Mechanisms leading from systemic autoimmunity to joint-specific disease in rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 79–86 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Scher J. U., Sczesnak A., Longman R. S., Segata N., Ubeda C., Bielski C., Rostron T., Cerundolo V., Pamer E. G., Abramson S. B., Huttenhower C., Littman D. R., Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D., Wu X., Li J., Tang L., Li Y., Lan Z., Chen B., Li Y., Zhong H., Xie H., Jie Z., Chen W., Tang S., Xu X., Wang X., Cai X., Liu S., Xia Y., Li J., Qiao X., Al-Aama J. Y., Chen H., Wang L., Wu Q. J., Zhang F., Zheng W., Li Y., Zhang M., Luo G., Xue W., Xiao L., Li J., Chen W., Xu X., Yin Y., Yang H., Wang J., Kristiansen K., Liu L., Li T., Huang Q., Li Y., Wang J., The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Wright K., Davis J. M., Jeraldo P., Marietta E. V., Murray J., Nelson H., Matteson E. L., Taneja V., An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Price J., Abu-Ali G., Huttenhower C., The healthy human microbiome. Genome Med. 8, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surana N. K., Kasper D. L., Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayfach S., Pollard K. S., Toward accurate and quantitative comparative metagenomics. Cell 166, 1103–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hrubec Z., Robinette C. D., The study of human twins in medical research. N. Engl. J. Med. 310, 435–441 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Plantinga A. M., Chen J., Jenq R. R., Wu M. C., pldist: Ecological dissimilarities for paired and longitudinal microbiome association analysis. Bioinformatics 35, 3567–3575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Chu Y., Meng Q., Ding R., Shi X., Wang Z., He Y., Zhang J., Liu J., Zhang J., Yu J., Kang Y., Wang J., A quasi-paired cohort strategy reveals the impaired detoxifying function of microbes in the gut of autistic children. Sci. Adv. 6, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B., Chen Y. W., Chen Y. Q., The nearest neighbor algorithm of local probability centers. IEEE Trans. Syst. Man Cybern. B Cybern. 38, 141–154 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Fischbach M. A., Microbiome: Focus on causation and mechanism. Cell 174, 785–790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawinkel S., Mattiello F., Bijnens L., Thas O., A broken promise: Microbiome differential abundance methods do not control the false discovery rate. Brief. Bioinform. 20, 210–221 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Vital M., Howe A. C., Tiedje J. M., Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5, e00889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziels R. M., Nobu M. K., Sousa D. Z., Elucidating syntrophic butyrate-degrading populations in anaerobic digesters using stable-isotope-informed genome-resolved metagenomics. mSystems 4, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luu M., Visekruna A., Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 49, 842–848 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Macia L., Tan J., Vieira A. T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., Binge L., Thorburn A. N., Chevalier N., Ang C., Marino E., Robert R., Offermanns S., Teixeira M. M., Moore R. J., Flavell R. A., Fagarasan S., Mackay C. R., Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6, 6734 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Marino E., Richards J. L., McLeod K. H., Stanley D., Yap Y. A., Knight J., McKenzie C., Kranich J., Oliveira A. C., Rossello F. J., Krishnamurthy B., Nefzger C. M., Macia L., Thorburn A., Baxter A. G., Morahan G., Wong L. H., Polo J. M., Moore R. J., Lockett T. J., Clarke J. M., Topping D. L., Harrison L. C., Mackay C. R., Erratum: Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 18, 1271 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Lucas S., Omata Y., Hofmann J., Bottcher M., Iljazovic A., Sarter K., Albrecht O., Schulz O., Krishnacoumar B., Kronke G., Herrmann M., Mougiakakos D., Strowig T., Schett G., Zaiss M. M., Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun. 9, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N. N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J. M., Topping D. L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H., Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Takahashi D., Hoshina N., Kabumoto Y., Maeda Y., Suzuki A., Tanabe H., Isobe J., Yamada T., Muroi K., Yanagisawa Y., Nakamura A., Fujimura Y., Saeki A., Ueda M., Matsumoto R., Asaoka H., Clarke J. M., Harada Y., Umemoto E., Komatsu N., Okada T., Takayanagi H., Takeda K., Tomura M., Hase K., Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine 58, 102913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wing J. B., Ise W., Kurosaki T., Sakaguchi S., Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41, 1013–1025 (2014). [DOI] [PubMed] [Google Scholar]
- 23.He J., Tsai L. M., Leong Y. A., Hu X., Ma C. S., Chevalier N., Sun X., Vandenberg K., Rockman S., Ding Y., Zhu L., Wei W., Wang C., Karnowski A., Belz G. T., Ghali J. R., Cook M. C., Riminton D. S., Veillette A., Schwartzberg P. L., Mackay F., Brink R., Tangye S. G., Vinuesa C. G., Mackay C. R., Li Z., Yu D., Circulating precursor CCR7(lo)PD-1(hi) CXCR5+ CD4+ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity 39, 770–781 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kishikawa T., Maeda Y., Nii T., Motooka D., Matsumoto Y., Matsushita M., Matsuoka H., Yoshimura M., Kawada S., Teshigawara S., Oguro E., Okita Y., Kawamoto K., Higa S., Hirano T., Narazaki M., Ogata A., Saeki Y., Nakamura S., Inohara H., Kumanogoh A., Takeda K., Okada Y., Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 79, 103–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInnes I. B., Schett G., The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 365, 2205–2219 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Smolen J. S., Aletaha D., Grisar J. C., Stamm T. A., Sharp J. T., Estimation of a numerical value for joint damage-related physical disability in rheumatoid arthritis clinical trials. Ann. Rheum. Dis. 69, 1058–1064 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Hui W., Yu D., Cao Z., Zhao X., Butyrate inhibit collagen-induced arthritis via Treg/IL-10/Th17 axis. Int. Immunopharmacol. 68, 226–233 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Rosser E. C., Piper C. J. M., Matei D. E., Blair P. A., Rendeiro A. F., Orford M., Alber D. G., Krausgruber T., Catalan D., Klein N., Manson J. J., Drozdov I., Bock C., Wedderburn L. R., Eaton S., Mauri C., Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. 31, 837–851.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand D. D., Latham K. A., Rosloniec E. F., Collagen-induced arthritis. Nat. Protoc. 2, 1269–1275 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa M., Myoui A., Tomita T., Takahi K., Nampei A., Yoshikawa H., Prevention of the onset and progression of collagen-induced arthritis in rats by the potent p38 mitogen-activated protein kinase inhibitor FR167653. Arthritis Rheum. 48, 2670–2681 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S7
Excel file S1 to S7