Abstract
Cholangiocarcinoma often remains undetected until advanced stages due to the lack of reliable diagnostic markers. Our goal was to identify a unique secretory protein for cholangiocarcinoma diagnosis and differentiation from other malignancies, benign hepatobiliary diseases, and chronic liver conditions. We conducted bulk RNA‐seq analysis to identify genes specifically upregulated in cholangiocarcinoma but not in most other cancers, benign hepatobiliary diseases, and chronic liver diseases focusing on exocrine protein‐encoding genes. Single‐cell RNA sequencing examined subcellular distribution. Immunohistochemistry and enzyme‐linked immunosorbent assays assessed tissue and serum expression. Diagnostic performance was evaluated via receiver‐operating characteristic (ROC) analysis. Inter‐alpha‐trypsin inhibitor heavy chain family member five (ITIH5), a gene encoding an extracellular protein, is notably upregulated in cholangiocarcinoma. This elevation is not observed in most other cancer types, benign hepatobiliary diseases, or chronic liver disorders. It is specifically expressed by malignant cholangiocytes. ITIH5 expression in cholangiocarcinoma tissues exceeded that in nontumorous bile duct, hepatocellular carcinoma, and nontumorous hepatic tissues. Serum ITIH5 levels were elevated in cholangiocarcinoma compared with controls (hepatocellular carcinoma, benign diseases, chronic hepatitis B, and healthy individuals). ITIH5 yielded areas under the ROC curve (AUCs) from 0.839 to 0.851 distinguishing cholangiocarcinoma from controls. Combining ITIH5 with carbohydrate antigen 19‐9 (CA19‐9) enhanced CA19‐9's diagnostic effectiveness. In conclusion, serum ITIH5 may serve as a novel noninvasive cholangiocarcinoma diagnostic marker.
Keywords: cholangiocarcinoma, diagnosis, ELISA, ITIH5, serum marker
Serum ITIH5 levels serve as a noninvasive diagnostic biomarker for cholangiocarcinoma (CCA), especially in the case of intrahepatic cholangiocarcinoma (iCCA). Combining plasma ITIH5 with serum CA19‐9 enhanced diagnostic accuracy.
Abbreviations
- AUC
the area under the receiver‐operating characteristic curve
- CA19‐9
carbohydrate antigen 19‐9
- CCA
cholangiocarcinoma
- dCCA
distal cholangiocarcinoma
- DEGs
differential expression genes
- ELISA
enzyme‐linked immunosorbent assay
- EP‐upDEGs
exocrine protein‐upregulated differentially expressed genes
- GEO
Gene Expression Omnibus
- HCC
hepatocellular carcinoma
- iCCA
intrahepatic cholangiocarcinoma
- IHC
immunohistochemistry
- ITIH5
inter‐alpha‐trypsin inhibitor heavy chain family member five
- NASH
nonalcoholic steatohepatitis
- NCCA
the total patients without CCA
- PCA
principal components analysis
- pCCA
perihilar cholangiocarcinoma
- PSC
primary sclerosing cholangitis
- ROC
receiver‐operating characteristic
- scRNA‐seq
single‐cell RNA sequencing
- TCGA
The Cancer Genome Atlas
- UMAP
Uniform Manifold Approximation and Projection
1. INTRODUCTION
Cholangiocarcinoma (CCA) ranks as the second most common primary liver cancer, following hepatocellular carcinoma (HCC), constituting around 10–15% of all liver cancer cases. 1 Diagnosing CCA early is challenging due to its anatomical location, growth patterns, “silent” clinical presentation, and the absence of clear diagnostic criteria. Because of its aggressive nature, the majority of patients with CCA are typically diagnosed in advanced stages, often missing the opportunity for optimal surgical intervention. 2 Carbohydrate antigen 19‐9 (CA19‐9) serves as the primary serum biomarker in the diagnosis of CCA. 3 Nevertheless, CA19‐9 does not demonstrate high accuracy. It is unable to differentiate CCA from pancreatic or gastric cancer, and it may also show elevated levels in nonmalignant conditions such as obstructive jaundice and severe hepatic injury. 4 Furthermore, approximately 10% of individuals are unable to produce CA19‐9 due to the absence of the Lewis antigen. 4 Hence, it is urgently necessary to identify a biomarker that is both sensitive and specific to aid in the detection of CCA.
Different types of tumors have been diagnosed with the help of secretory proteins. 5 , 6 Inter‐alpha‐trypsin inhibitor heavy chain family member five (ITIH5), a plasma protease inhibitor, is the fifth heavy‐chain member of the inter‐α‐trypsin inhibitor (ITI) family. 7 Normally, the expression level of ITIH5 is high in the placenta, while it is moderate in various organs. 7 It has been shown that ITIH5 dysfunction is associated with obesity 8 and inflammatory skin diseases, 9 suggesting it may act as a metabolic regulator. During tumor progression, ITIH5 has been demonstrated as a suppressor gene in a variety of tumors, including pancreatic cancer (PAAD), 10 gastric cancer, 11 lung cancer, 12 breast cancer, 13 melanoma, 14 bladder cancer, 15 and cervical cancer. 16 Through The Cancer Genome Atlas (TCGA) pan‐cancer analysis, we observed ITIH5 was very significantly elevated in CCA, but not in most of the other cancer types.
In the present study, with bioinformatics methods, we have presented the initial evidence that ITIH5, a gene coding for an extracellular protein, shows a significant upregulation in CCA compared with its expression in most other cancer types, benign hepatobiliary conditions, and chronic liver diseases. This notable elevation is specifically attributed to its expression in malignant cholangiocytes. Additionally, ITIH5 exhibits a significant overexpression in CCA tissues when compared with nontumorous bile duct, HCC, and nontumorous hepatic tissues. Furthermore, ITIH5 can be detected with high specificity and sensitivity in the serum of individuals with CCA when compared with HCC patients, as well as those with benign hepatobiliary diseases, chronic hepatitis B, and healthy individuals. In a receiver‐operating characteristic (ROC) analysis, ITIH5 showed substantial diagnostic potential for CCA, particularly in the case of intrahepatic cholangiocarcinoma (iCCA) patients, a crucial subtype of CCA. In addition, combining plasma ITIH5 with serum CA19‐9 enhanced the diagnostic performance. Consequently, ITIH5 has the potential to serve as a valuable diagnostic biomarker for patients with CCA.
2. MATERIALS AND METHODS
2.1. Patient samples
A total of 65 CCA and 30 HCC serum samples were collected at the Second Hospital of Hebei Medical University, taken before surgical procedures or chemoradiotherapy. The diagnoses for these patients were established through histological and clinical examinations. Furthermore, we also collected samples from 55 patients with benign hepatobiliary diseases (including 13 hepatic hemangiomas, 15 cases of bile duct stones, 15 with benign biliary tract diseases, and 12 hepatic cysts), 30 patients with chronic hepatitis B, and 32 healthy individuals. These patients' baseline characteristics are described in Table 1. In addition, we acquired tissue sections from 34 CCA, 17 nontumorous bile duct, 17 HCC, and 28 nontumorous hepatic tissues from the Second Hospital of Hebei Medical University. Ethical approval was granted by the Research Ethics Committee of the Second Hospital of Hebei Medical University.
TABLE 1.
Baseline characteristics of patients in this study.
| Baseline characteristics | CCA (N = 65) | HCC (N = 30) | Benign (N = 65) | CHB (N = 30) | Healthy (N = 32) | All patients (N = 212) | p |
|---|---|---|---|---|---|---|---|
| Age (years) | |||||||
| <65 | 16 (24.6) | 20 (66.7) | 36 (65.5) | 23 (76.7) | 22 (68.8) | 117 (55.2) | <0.05 |
| ≥65 | 49 (75.4) | 10 (33.3) | 19 (34.5) | 7 (23.3) | 10 (31.3) | 95 (44.8) | |
| Gender | |||||||
| Male | 46 (70.8) | 24 (80) | 30 (36.4) | 20 (66.7) | 13 (40.6) | 123 (58) | <0.05 |
| Female | 19 (29.2) | 6 (20) | 35 (63.6) | 10 (33.3) | 19 (59.4) | 89 (42) | |
| TBil (umol/L) | 152.10 (51.77, 265.50) | 13.75 (9.91, 23.33) | 12.1 (9.10, 24.20) | 17.05 (11.80, 32.88) | 11.87 (8.85, 15.38) | 16.50 (10.42, 86.33) | <0.05 |
| AFP (ng/mL) | 3.27 (2.50, 3.98) | 23.44 (3.94,1210.00) | 2.88 (2.17, 4.09) | 2.94 (1.86, 6.37) | 3.34 (2.53, 4.40) | 3.37 (2.44, 4.93) | <0.05 |
| CEA (ng/mL) | 3.21 (1.71, 4.64) | 2.85 (1.58, 4.21) | 1.95 (1.23, 3.67) | 1.78 (0.93, 2.39) | 1.58 (0.97, 2.43) | 2.16 (1.33, 3.62) | <0.05 |
| CA‐125 (U/mL) | 15.20 (9.85, 27.45) | 14.84 (9.63, 34.15) | 13.30 (6.94, 24.50) | 33.45 (12.74, 156.75) | 9.89 (6.32, 12.05) | 13.77 (9.26, 26.68) | <0.05 |
| CA19‐9 (U/mL) | 156.00 (46.40, 562.50) | 19.60 (7.50, 57.15) | 20.30 (8.07, 90.05) | 15.95 (7.82, 28.18) | 10.37 (5.11, 16.08) | 22.73 (9.66, 128.50) | <0.05 |
| HBsAg | |||||||
| N | 62 (95.4) | 8 (26.7) | 63 (96.4) | 0 (0) | 32 (100) | 165 (74.1) | <0.05 |
| P | 3 (4.6) | 22 (73.3) | 2 (3.6) | 30 (100) | 0 (0) | 57 (26.9) | |
| TNM stage | |||||||
| I–II | 24 (36.9) | 5 (16.7) | / | / | / | / | <0.05 |
| III–IV | 41 (63.1) | 25 (83.3) | / | / | / | / | |
| Tumor size (cm) | |||||||
| ≤5 | 53 (81.6) | 11(36.7) | / | / | / | / | <0.05 |
| >5 | 12 (18.5) | 19(63.3) | / | / | / | / | |
| Location | |||||||
| iCCA | 13 (20.0) | / | / | / | / | / | |
| pCCA | 30 (46.2) | / | / | / | / | / | |
| dCCA | 22 (33.8) | / | / | / | / | / | |
| Diagnosis | |||||||
| BBTD | / | / | 11 (20.0) | / | / | / | |
| Bileduct stone | / | / | 18 (32.7) | / | / | / | |
| Hepatic cyst | / | / | 12 (21.8) | / | / | / | |
| Hepatic hemangioma | / | / | 14 (25.5) | / | / | / | |
Abbreviations: BBTD, benign biliary tract diseases; Benign, benign hepatobiliary disease; CCA, cholangiocarcinoma; CHB, chronic hepatitis B; dCCA, distal cholangiocarcinoma; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; Healthy, healthy individuals; iCCA, intrahepatic cholangiocarcinoma; N, negative; P, positive; pCCA, perihilar cholangiocarcinoma; TBil, total bilirubin.
2.2. The Cancer Genome Atlas pan‐cancer analysis
We sourced transcriptome data for the pan‐cancer study from TCGA via UCSC Xena (https://xena.ucsc.edu/). Genes responsible for encoding exocrine proteins were downloaded from the Human Protein Atlas (https://www.proteinatlas.org/) 17 (Table S1). Our initial step involved conducting a differential gene analysis for the TCGA‐CHOL (bile duct cancer) cohorts, utilizing the limma package. We set the cutoff criteria at an absolute log fold change (|LogFC|) > 1 and p‐value < 0.05. This process led us to identify 207 exocrine protein‐upregulated differentially expressed genes (EP‐upDEGs), derived by intersecting the upregulated genes with those encoding exocrine proteins. The overlap was visually represented through a Venn plot, constructed using the eulerr package.
Further, a t‐test was performed using the tinyarray package, focusing on the intersection of EP‐upDEGs with genes not upregulated in other cohorts, specifically liver hepatocellular carcinoma (LIHC), PAAD, rectum adenocarcinoma (READ), colon adenocarcinoma (COAD), stomach cancer (STAD), esophageal cancer (ESCA), lung squamous cell carcinoma (LUSC), and lung adenocarcinoma (LUAD). After an initial gene selection, further filtering was applied with criteria for differential expression set at logFC > 2 and adjusted p‐value < 0.05. The resulting genes meeting these refined criteria were compared for their expression levels across diverse cancer types in TCGA. This analysis highlighted ITIH5 as our target gene, revealing it as the most significantly upregulated gene in the study.
To deepen our understanding, we utilized TIMER2.0 (http://timer.comp‐genomics.org/timer/) 18 for an extensive exploration of ITIH5 expression across various cancer types. The expression of ITIH5 and other tumor markers in TCGA‐CHOL and TCGA‐LIHC was elucidated using the pheatmap package in R. Additionally, the diagnostic accuracy of ITIH5 at the mRNA level was evaluated employing the pROC package.
2.3. Single‐cell RNA‐seq analysis
ScRNA‐seq data (GSE138709) 19 were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds). They included five CCA and three adjacent tissues. The Seurat package (V4.3.0) 20 in R software was used to conduct the scRNA‐seq analysis. Briefly, the eight samples were converted into a Seurat object. The cells that expressed fewer than 300 genes, as well as the genes that expressed less than three cells, were removed. Additionally, we filtered the cells that expressed less than 20% mitochondrial genes, more than 3% ribosomal genes, and less than 1% hemoglobin genes. Subsequently, we performed a principal component analysis (PCA) on the filtered data after normalization. The harmony package was employed to address and correct batch effects. Following that, Uniform Manifold Approximation and Projection (UMAP) were used to reduce dimensionality. 21 Using a resolution of 0.8, we identified 23 clusters. The hub gene, ITIH5, was visualized via the ggplot2 and Seurat packages on a UMAP representation of the expression data in tumor and adjacent samples, respectively. Our analysis of cell clusters was based on the expression of known cell markers and information presented in previous articles on clustering. 19 The expression of cell markers in the cluster expressing ITIH5 has been visualized.
2.4. Gene Expression Omnibus data analysis
Data from other cohorts (GSE32225, GSE67764, GSE11819, GSE139602, GSE62029, and GSE159676) were obtained from the GEO database. Detailed sample information for the GEO datasets is presented in Table 2. Differential gene expression was calculated with the limma package in R. A cutoff criterion of |LogFC| > 1 and an adjusted p‐value of 0.05 were employed.
TABLE 2.
GEO cohorts in this study.
| GEO datasets | Samples | PubMed identifier |
|---|---|---|
| GSE32225 |
149 Intrahepatic cholangiocarcinoma 6 Control |
23295441 |
| GSE67764 | 3 Hepatic carcinoma tissues | 31385458 |
| 6 Control | ||
| GSE11819 | 4 Inflammatory hepatocellular adenomas | 19020503 |
| 4 Control | ||
| GSE62029 | 7 Liver angioma | 26459852 |
| 10 Control | ||
| GSE139602 | 5 Fibrosis (early chronic liver diseases) | 35540106 |
| 8 Compensated cirrhosis | ||
| 12 Decompensated cirrhosis | ||
| 8 Acute‐on‐chronic liver failure | ||
| 6 Control | ||
| GSE159676 | 12 Primary sclerosing cholangitis | 35278227 |
| 7 Nonalcoholic steatohepatitis | ||
| 8 Other liver diseases (i.e., primary biliary cholangitis, autoimmune hepatitis, alcoholic liver disease, and haemochromatosis) | ||
| 5 Control |
2.5. Immunohistochemistry (IHC)
According to the previously established protocol, we conducted an IHC staining against ITIH5. 22 The samples were subsequently incubated with anti‐ITIH5 (1:50, Solarbio, K111754P) and corresponding secondary antibodies (ZSGB‐BIO, PV‐9000), developed with DAB, and stained with hematoxylin. Finally, the assessment of staining outcomes was conducted in alignment with previously established methods. 23 Each field received one of four staining intensity scores: 0 indicated no staining; 1 denoted weak positivity; 2 represented moderate positivity; and 3 signified strong positivity. Additionally, the proportion of positively stained cells was categorized into five levels (percentage scores): 0–10% (0), 11–25% (1), 26–50% (2), 51–75% (3), and above 75% (4). The presence of ITIH5 was assessed through IHC scoring. This scoring involved a calculation formula: IHC score = (percentage score) multiplied by (intensity score). Consequently, the categorization of the target protein expression was divided into four distinct groups: (i) IHC score ≤ 3, indicating no staining; (ii) 3 < IHC score ≤ 6, suggesting weak staining; (iii) 6 < IHC score ≤ 9, denoting moderate staining; and (iv) IHC score > 9, reflecting strong staining. This immunostaining analysis was conducted by two independent pathologists who were not informed about the findings of the study.
2.6. Enzyme‐linked immunosorbent assay
Plasma ITIH5 levels were measured via a human ITIH5 ELISA kit (CSB‐EL011898HU), according to the manufacturer's directions. With a microplate reader, we measured the optical density (OD) at 450 nm for each well. Based on the standard concentrations and their corresponding OD values, a linear regression equation was estimated using the CurveExpert software. The concentrations of the samples were then calculated based on this equation.
2.7. Statistical analysis
R statistical software (version 4.2.2) was used to analyze the data. The appropriate R packages and statistical methods were described throughout the study. SPSS (version 26) was employed for statistical analysis, while GraphPad Prism (version 9.5.1) was utilized for data visualization. Data that did not follow a normal distribution were analyzed by the Kruskal–Wallis one‐way nonparametric test (k independent samples) or Mann–Whitney U‐test (two independent samples), and Bonferroni correction was applied to all multiple comparisons. The chi‐square test was used for qualitative variables. Correlation was calculated via Spearman and Pearson correlation analysis, as appropriate. DeLong's test was used to compare ROC curves. p < 0.05 was considered statistically significant, denoted as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
3. RESULTS
3.1. Identification of a potential CCA marker
The analysis flowchart is shown in Figure 1. The gene expression profiles across various cancer types in TCGA pan‐cancer data were analyzed to identify potential diagnostic markers for CCA patients. Initially, we identified 2279 upregulated differential expression genes (DEGs) in the TCGA‐CHOL dataset. Subsequently, we screened a total of 207 EP‐upDEGs by intersecting the upregulated genes with those encoding exocrine proteins (Figure 2A). In our quest to identify genes uniquely upregulated in CCA, we conducted a comparative analysis with other common solid tumors and cancer types requiring differential diagnosis from CCA: HCC, PAAD, gastric cancer, ESCA, colorectal cancer, and lung cancer. This analysis focused on identifying genes from EP‐upDEGs that were not upregulated in each of these cancers. In our study, we identified 23 genes not upregulated in STAD, ESCA, LUSC, and LUAD, while 12 such genes were found in LIHC and PAAD. Additionally, eight genes were not upregulated in READ and COAD. Overall, this led to the identification of a consolidated set of 39 non‐upregulated genes across these cancer types. The genes identified were displayed on a differential gene expression volcano plot using the TCGA‐CHOL dataset (Figure 2B). By further refining the criteria for differential gene expression to logFC > 2 and adjusted p‐value < 0.05, nine genes were retained: FDCSP, PRSS22, LIF, CXCL8, IL18, ITIH5, FOLR1, LGALS9, and KLK11. Notably, ITIH5 exhibited the most significant logFC and adjusted p‐values. Compared with normal tissues, ITIH5 expression in CCA tissues was elevated 4.15‐fold. We used TIMER 2.0 to investigate the expression of ITIH5 across various cancer types (pan‐cancer analysis; Figure 2C). ITIH5 exhibited significant upregulation in CHOL and pheochromocytoma and paraganglioma (PCPG), while in other cancer types, it either showed downregulation or no significant change. Notably, the expression level of ITIH5 in CHOL was higher than that in PCPG and had greater significance. Conversely, the remaining eight genes showed elevated expression in tumor groups across various cancer types (Figure S1), lacking cancer specificity. Consequently, ITIH5 was chosen for further analysis due to its distinctiveness. In addition, the expression of ITIH5 in CCA and HCC was verified in GSE32225 and GSE67764, respectively. In the case of CCA, the tumor group displayed significantly higher ITIH5 expression compared with the normal group (Figure 2D). However, in the case of HCC, there was no significant difference between the tumor and normal groups in terms of ITIH5 expression (Figure 2E). The findings suggested that ITIH5 exhibited a significant upregulation in CCA but did not show similar upregulation in most other cancer types.
FIGURE 1.
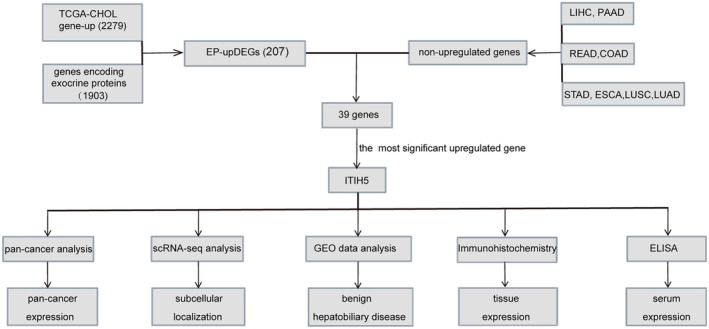
Experimental analysis flowchart. COAD, colon adenocarcinoma; CHOL, bile duct cancer; EP‐upDEGs, exocrine protein‐upregulated differentially expressed genes; ESCA, esophageal cancer; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic cancer; READ, rectum adenocarcinoma; scRNA‐seq, single‐cell RNA sequencing; STAD, stomach cancer.
FIGURE 2.
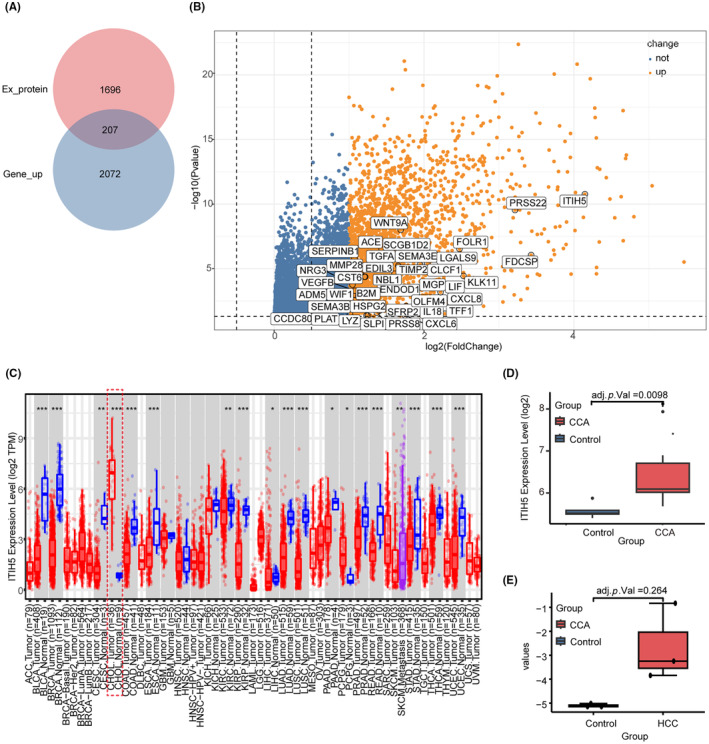
Identification of a potential cholangiocarcinoma (CCA) marker. (A) Venn diagram of intersecting genes of upregulated differential expression genes in TCGA‐CHOL and genes encoding secreted proteins. (B) The volcano plot of upregulated differentially expressed genes in TCGA‐CHOL. The genes in the white box represent 39 exocrine protein‐upregulated differentially expressed genes (EP‐upDEGs), among which ITIH5 is the most prominently elevated gene. (C) Expression of ITIH5 across various cancer types. Red dotted boxes represent ITIH5 expression in CCA. (D) The tumor group displayed significantly higher ITIH5 expression than the normal CCA group. (E) There was no significant difference in ITIH5 expression between tumor and nontumor groups in hepatocellular carcinoma (HCC). CHOL, bile duct cancer.
3.2. Single‐cell RNA‐seq analysis
To ascertain which cell type expresses ITIH5 in CCA, we utilized scRNA‐seq analysis. The UMAP visualization revealed 23 distinct cell clusters (Figure 3A). The distribution of ITIH5 expression in the adjacent and tumor samples is depicted in Figure 3B,C, respectively. It was mainly expressed in clusters 1, 3, 5, and 6 of tumor samples. Furthermore, the dot plot illustrated simultaneous expression of both bile duct cell markers (FXYD2, TM4SF4, ANXA4) and malignant cell markers (KRT19, KRT7, EPCAM) in these four clusters, suggesting that these clusters may originate from malignant cholangiocytes (Figure 3D). These results suggest that ITIH5 is primarily secreted by malignant cholangiocytes and is present at high expression levels in the tumor samples.
FIGURE 3.
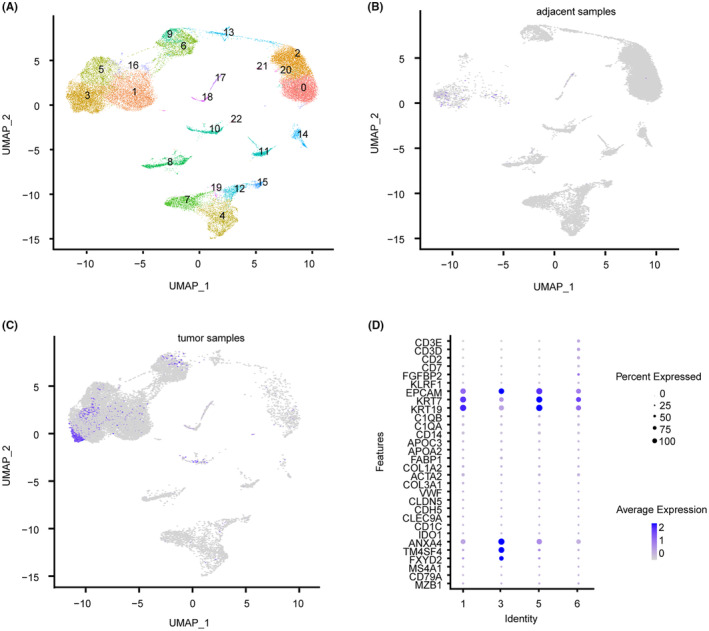
Single‐cell RNA sequencing (scRNA‐seq) analysis. (A) The Uniform Manifold Approximation and Projection (UMAP) visualization revealed 23 distinct cell clusters. (B) The distribution of ITIH5 expression in the adjacent samples. (C) The distribution of ITIH5 expression in the tumor samples. (D) The dot plot illustrated the expression of cell‐type‐specific marker genes for clusters 1, 3, 5, and 6.
3.3. The diagnostic value of ITIH5 at the mRNA level for CCA
In order to assess the specificity of ITIH5 expression levels in diverse cancers, we calculated the positive rate and fold change of ITIH5 expression in various cancer types using TCGA RNA‐seq data. The positive‐expression cases were defined as those with ITIH5 expression in the tumor samples exceeding the upper limit of normal samples. In CHOL, the positive rate of ITIH5 was 0.94%, which was the highest among the various cancer types analyzed (Figure 4A). The fold change in ITIH5 expression was 4.15 in CCA, and this value was higher than that observed in PCPG and other cancer types (Figure 4B). The high specificity of ITIH5 expression in CCA supports its potential as a valuable diagnostic marker for this condition.
FIGURE 4.
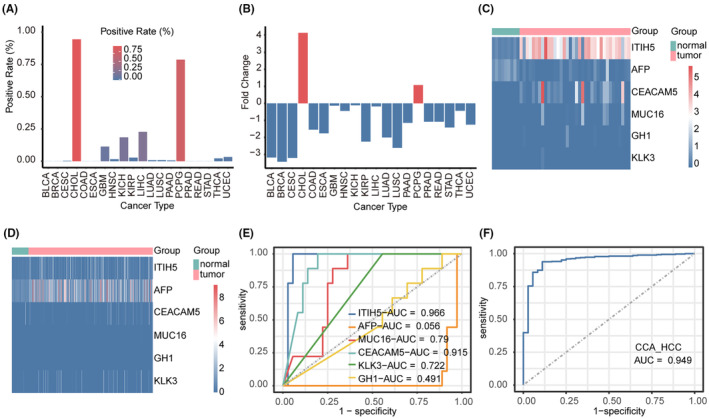
The diagnostic value of ITIH5 at the mRNA level for cholangiocarcinoma (CCA). (A) The positive rate of ITIH5 in the various cancer types. (B) Fold change in ITIH5 expression in the various cancer types. (C) Heatmap illustrating the expression level of ITIH5 and the tumor markers in TCGA‐CHOL. (D) Heatmap illustrating the expression level of ITIH5 and the tumor markers in TCGA‐LIHC. (E) ROC curve illustrating the diagnostic significance of ITIH5 and the tumor markers in CCA. (F) ROC curve illustrating the diagnostic significance of ITIH5 in distinguishing between CCA and hepatocellular carcinoma (HCC). CHOL, bile duct cancer; LIHC, liver hepatocellular carcinoma.
To further evaluate whether ITIH5 could serve as a potential diagnostic marker to identify CCA from HCC, the sensitivity and specificity of ITIH5 and other tumor markers' expression levels were analyzed in the TCGA‐CHOL and TCGA‐LIHC. The tumor markers consisted of AFP (encoded by AFP), 24 CA125 (encoded by MUC16), 25 CEA (encoded by CEACAM5), 26 PSA (encoded by KLK3) 27 and GH (encoded by GH1), 28 all of which have been commonly used in clinical settings. The heatmap illustrated ITIH5 and the tumor markers expression levels in TCGA‐CHOL (Figure 4C) and TCGA‐LIHC (Figure 4D). Our observations indicated that ITIH5 expression was elevated in the majority of CCA samples, with only a minor fraction of HCC samples exhibiting slightly increased expression. Compared with the other tumor markers, ITIH5 showed an extremely high positive rate in CCA.
Next, we conducted a ROC curve analysis with a pROC package in R to assess the diagnostic value of ITIH5 and tumor markers in CCA and HCC based on TCGA RNA‐seq data. The results revealed that ITIH5 had an AUC value of 0.966 when discriminating CCA tissues from normal tissues (Figure 4E). The value was higher than that of other tumor markers. In addition, the ROC curve analysis of ITIH5 in distinguishing between CCA and HCC yielded an AUC of 0.949 (Figure 4F). As a result, the sensitivity and specificity of ITIH5 outperformed other tumor markers at the mRNA level, suggesting that ITIH5 may be considered a novel and promising diagnostic marker for CCA.
3.4. ITIH5 expression in benign hepatobiliary diseases and chronic hepatic diseases
To determine whether ITIH5 could differentiate CCA from benign hepatobiliary diseases and chronic hepatic diseases, we conducted an analysis using the GEO datasets, which included GSE11819 (inflammatory hepatocellular tumor), GSE139602 (chronic liver diseases), GSE 62029 (liver angioma) and GSE159676 (primary sclerosing cholangitis, nonalcoholic steatohepatitis [NASH], and other liver diseases; Table 2). The expression levels of ITIH5 between the control and case groups in the aforementioned four GEO datasets are depicted in Figure 5A–F, respectively. Neither of the diseases had significant differences in ITIH5 gene expression between the control and case groups. Given the notably high expression of ITIH5 in CCA, while its expression remains unaltered in benign hepatobiliary diseases and chronic hepatic diseases, ITIH5 may serve as a distinguishing marker to separate CCA from benign hepatobiliary diseases and chronic hepatic diseases.
FIGURE 5.
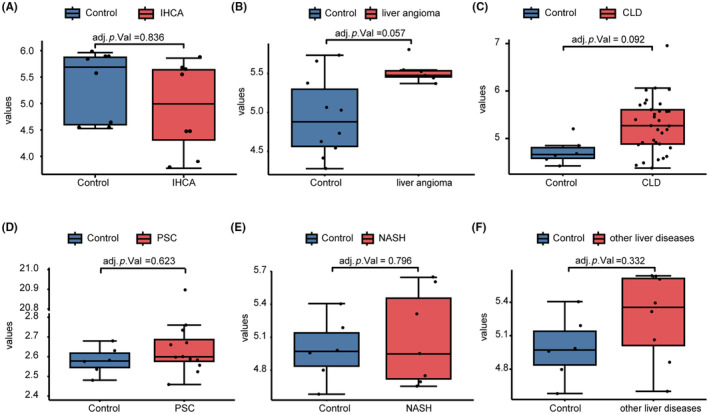
The expression of ITIH5 in benign hepatobiliary diseases and chronic hepatic diseases. (A–F) The expression levels of ITIH5 between the control and case groups in IHCA (GSE11819), liver angioma (GSE62029), PSC (GSE159676), NASH (GSE159676), and other liver diseases (GSE159676), respectively. CLD, chronic liver diseases; IHCA, inflammatory hepatocellular tumor; NASH, nonalcoholic steatohepatitis; PSC, primary sclerosing cholangitis.
3.5. ITIH5 immunohistochemistry analysis
In our study, we assessed ITIH5 expression levels in CCA tissues compared with control groups, which included nontumorous bile duct tissues, HCC, and nontumorous hepatic tissues, through immunohistochemistry analysis. The findings revealed that in CCA tissues, there was a predominant occurrence of strong staining in 25 out of 34 cases (73.5%), moderate staining in 8 of 34 cases (23.5%), and weak staining in 1 of 17 cases (5.9%). Notably, ITIH5 was primarily expressed on the cell membranes of CCA epithelial cells, followed by cytoplasmic expression (see Figure 6A). In contrast, nontumorous bile duct tissues exhibited moderate staining in 7 out of 17 cases (41.2%), weak staining in 5 of 17 cases (29.4%), and no staining in 5 of 17 cases (29.4%; Figure 6B). In the HCC samples, moderate staining was observed in 3 out of 17 cases (17.6%), while a majority, 11 out of 17 cases (64.7%), showed no staining (as depicted in Figure 6C). Among nontumorous hepatic tissues, moderate staining was present in 8 of 28 cases (28.6%), weak staining in 14 of 28 cases (50%), and no staining in 6 of 28 cases (21.4%) (illustrated in Figure 6D). Significantly, the IHC scores for ITIH5 were higher in CCA tissues compared with nontumorous bile duct tissues, HCC, and nontumorous hepatic tissues, indicating a statistically significant difference (Kruskal–Wallis one‐way nonparametric test, Figure 6E, p < 0.001). Collectively, these findings provide strong evidence that ITIH5 expression levels were markedly elevated in CCA tissues.
FIGURE 6.
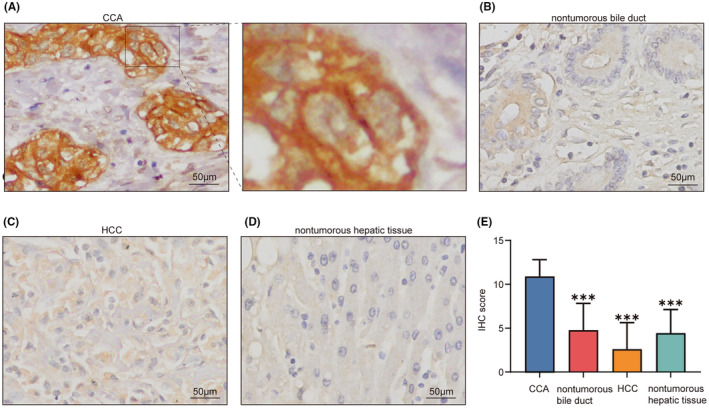
ITIH5 immunohistochemistry analysis. (A) Cholangiocarcinoma (CCA) tissues exhibited positive cytoplasm and membrane staining for ITIH5. The right panel displays an enlarged view of the area highlighted by the black box in the left panel. (B–D) ITIH5 expression in nontumorous bile duct, hepatocellular carcinoma (HCC), and nontumorous hepatic tissues, respectively. (E) The immunohistochemistry (IHC) score of ITIH5 expression levels in CCA, nontumorous bile duct, HCC, and nontumorous hepatic tissues. ***p < 0.001. Scale, 50 μm.
3.6. Serum ITIH5 expression levels may serve as a potential diagnostic marker for CCA
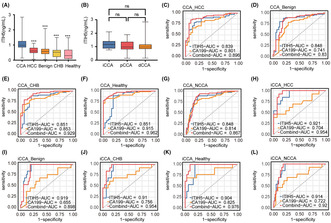
Serum ITIH5 expression levels were assessed using the ELISA method in a cohort of 65 patients with CCA, 30 patients with HCC, 55 patients with benign hepatobiliary diseases, 30 individuals with chronic hepatitis B, and 32 healthy individuals. The plasma ITIH5 concentration was 0.979 (0.854, 1.249) ng/mL, 0.607 (0.484, 0.803) ng/mL, 0.576 (0.420, 0.732) ng/mL, 0.522 (0.226, 0.657) ng/mL, 0.301 (0.097, 0.672) ng/mL in CCA, HCC, benign hepatobiliary diseases, chronic hepatitis B, and healthy individuals, respectively (Figure 7A). The results demonstrated a significant elevation of ITIH5 levels in the serum of the CCA group compared with each of the other group. (Kruskal–Wallis one‐way nonparametric test, p < 0.001, Figure 7A). In addition, we conducted an analysis of the correlation between serum ITIH5 levels and clinicopathological characteristics in CCA (Table 3). The results indicated that ITIH5 showed no significant correlation with patient gender, age, TBil, AFP, CEA, CA125, CA19‐9, tumor location, TNM stage, or tumor size. Notably, a significant positive correlation was observed with IHC scores (Pearson correlation, R = 0.620, p < 0.001). The correlation with IHC scores suggests that the elevated ITIH5 levels in the serum can be attributed to CCA tissues.
FIGURE 7.
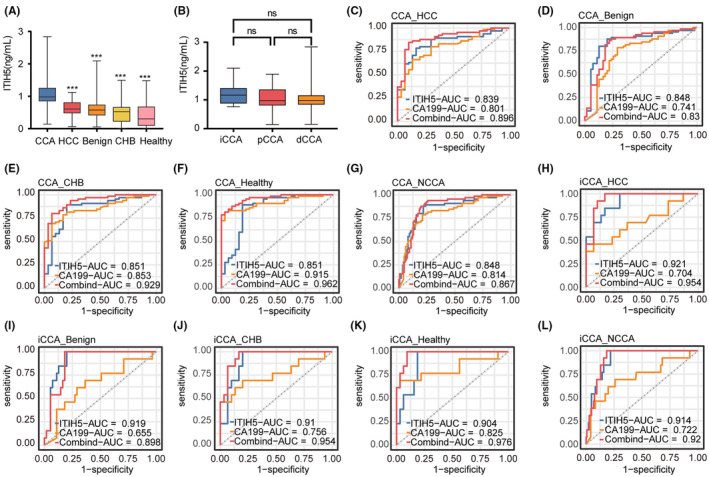
Serum ITIH5 levels and diagnostic accuracy assessment. (A) Serum ITIH5 levels in CCA, HCC, Benign, CHB, and Healthy. (B) Serum ITIH5 levels in iCCA, pCCA, and dCCA. (C–G) The ROC analysis results for ITIH5, CA19‐9, and the ITIH5/CA19‐9 panel in discriminating CCA patients from those with HCC, Benign, CHB, Healthy and NCCA, respectively. (H–L) The ROC analysis results for ITIH5, CA19‐9, and the ITIH5/CA19‐9 panel in discriminating iCCA patients from those with HCC, Benign, CHB, Healthy, and NCCA, respectively. ***p < 0.001. ns, no statistical differences. Benign, benign hepatobiliary diseases; CHB, chronic hepatitis B; CCA, cholangiocarcinoma; dCCA, distal cholangiocarcinoma; Healthy, healthy individuals; HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; pCCA, perihilar cholangiocarcinoma; NCCA, the total patients without CCA.
TABLE 3.
The potential association between clinicopathology characteristics and serum ITIH5 levels in cholangiocarcinoma patients.
| Characteristics | R | p |
|---|---|---|
| Gender | −0.202 | 0.107 |
| Age | 0.177 | 0.158 |
| TBil | −0.068 | 0.593 |
| AFP | 0.011 | 0.929 |
| CEA | −0.069 | 0.586 |
| CA125 | 0.009 | 0.943 |
| CA199 | 0.019 | 0.879 |
| Location | −0.128 | 0.309 |
| TNM stage | 0.060 | 0.633 |
| Tumor size | −0.036 | 0.776 |
| IHC score | 0.620 | <0.001*** |
Note: Data were analyzed using Spearman and Pearson correlation analysis, as appropriate. R represents the correlation coefficient.
Abbreviation: IHC, immunohistochemistry.
p < 0.001.
Furthermore, we also compared the expression levels of ITIH5 in different anatomical locations of CCA. The concentration of ITIH5 in serum was 1.164 (0.888, 1.397) ng/mL, 0.971 (0.817, 1.351) ng/mL, and 0.979 (0.851, 1.145) ng/mL in iCCA, perihilar cholangiocarcinoma (pCCA), and distal cholangiocarcinoma (dCCA), respectively (Figure 7B). There were no significant differences in ITIH5 expression among the three groups.
To assess the diagnostic utility of ITIH5 for CCA, we conducted a ROC curve analysis. In this analysis, CCA patients were designated as true positive samples, while the combined control groups served as true negative samples. The AUC values were 0.839, 0.848, 0.851, 0.851, and 0.848 for discriminating CCA patients from those with HCC, benign hepatobiliary diseases, chronic hepatitis B, healthy individuals, and the total patients without CCA (NCCA), respectively (Figure 7C–G). To distinguish between CCA patients and NCCA, we established an optimal cutoff value of 0.753 ng/mL using the Youden index method. Serum ITIH5 levels demonstrated a diagnostic sensitivity and specificity of 87.7% and 78.2%, respectively. Therefore, serum ITIH5 levels may serve as a diagnostic biomarker for CCA.
Additionally, previous studies have established a strong association between CA19‐9 and CCA. 29 To assess whether the combined utilization of ITIH5 and CA19‐9 could enhance the diagnostic potential for CCA compared with CA19‐9 alone, logistic regression analysis was conducted. The ITIH5/CA19‐9 panel for distinguishing CCA from HCC, benign hepatobiliary diseases, chronic hepatitis B, healthy individuals, and NCCA yielded AUC values of 0.896, 0.831, 0.929, 0.962, and 0.867, respectively, all of which surpassed the AUC of CA19‐9 alone (Figure 7C–G, Table 4). Notably, the AUC values of the panel in differentiating CCA from HCC, benign hepatobiliary diseases, chronic hepatitis B, and healthy individuals surpassed those of CA19‐9 alone (Table 5, DeLong test). Specifically, in the discrimination of benign hepatobiliary diseases, ITIH5 exhibited superior diagnostic performance compared with CA19‐9. However, the combined use of ITIH5 did not improve diagnostic performance in distinguishing NCCA, suggesting the potential need for an expanded sample size. According to the results, combining ITIH5 may enhance the diagnostic efficacy of CA19‐9 in diagnosing CCA.
TABLE 4.
AUC calculations of ROC analysis for patients with CCA and iCCA versus HCC, benign hepatobiliary diseases, chronic hepatitis B, and healthy individuals.
| n | ITIH5 | CA19‐9 | ITIH5 + CA19‐9 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 95%CI | AUC | 95%CI | AUC | 95%CI | |||||
| CCA versus controls | ||||||||||
| CCA‐HCC | 65/30 | 0.839 | 0.756 | 0.923 | 0.801 | 0.710 | 0.892 | 0.896 | 0.827 | 0.965 |
| CCA‐Benign | 65/55 | 0.848 | 0.771 | 0.925 | 0.741 | 0.647 | 0.835 | 0.831 | 0.752 | 0.909 |
| CCA‐CHB | 65/30 | 0.851 | 0.762 | 0.940 | 0.853 | 0.778 | 0.928 | 0.929 | 0.876 | 0.982 |
| CCA‐Healthy | 65/32 | 0.851 | 0.758 | 0.945 | 0.915 | 0.860 | 0.969 | 0.962 | 0.931 | 0.994 |
| CCA‐NCCA | 65/147 | 0.848 | 0.789 | 0.907 | 0.814 | 0.709 | 0.879 | 0.867 | 0.817 | 0.918 |
| iCCA versus controls | ||||||||||
| iCCA‐HCC | 13/30 | 0.921 | 0.841 | 1.000 | 0.704 | 0.513 | 0.895 | 0.954 | 0.897 | 1.000 |
| iCCA‐Benign | 13/55 | 0.919 | 0.854 | 0.984 | 0.655 | 0.480 | 0.829 | 0.898 | 0.825 | 0.971 |
| iCCA‐CHB | 13/30 | 0.910 | 0.825 | 0.996 | 0.756 | 0.571 | 0.942 | 0.954 | 0.898 | 1.000 |
| iCCA‐Healthy | 13/32 | 0.904 | 0.818 | 0.990 | 0.825 | 0.655 | 0.994 | 0.976 | 0.941 | 1.000 |
| iCCA‐NCCA | 13/147 | 0.914 | 0.862 | 0.966 | 0.722 | 0552 | 0.891 | 0.920 | 0.874 | 0.966 |
Abbreviations: AUC, area under the receiver‐operating characteristic curve; Benign, benign hepatobiliary diseases; CCA, cholangiocarcinoma; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma; Healthy, healthy individuals; iCCA, intrahepatic cholangiocarcinoma; NCCA, the total patients without CCA.
TABLE 5.
Comparison of the individual and combined AUCs for ITIH5 and CA199 in patients with CCA and iCCA versus HCC, benign hepatobiliary diseases, chronic hepatitis B, and healthy individuals.
| ITIH5 versus CA19‐9 | ITIH5 versus ITIH5 + CA19‐9 | CA19‐9 versus ITIH5 + CA19‐9 | |
|---|---|---|---|
| CCA versus controls | |||
| CCA‐HCC | 0.437 | 0.023* | 0.007** |
| CCA‐Benign | 0.039* | 0.212 | 0.041* |
| CCA‐CHB | 0.958 | 0.007** | 0.008** |
| CCA‐Healthy | 0.247 | 0.006** | 0.034* |
| CCA‐NCCA | 0.396 | 0.284 | 0.073 |
| iCCA versus controls | |||
| iCCA‐HCC | 0.057 | 0.314 | 0.009** |
| iCCA‐Benign | 0.007** | 0.283 | 0.012* |
| iCCA‐CHB | 0.158 | 0.102 | 0.039* |
| iCCA‐Healthy | 0.467 | 0.077 | 0.073 |
| iCCA‐NCCA | 0.053 | 0.781 | 0.029* |
Abbreviations: AUC, area under the receiver‐operating characteristic curve; Benign, benign hepatobiliary diseases; CCA, cholangiocarcinoma; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma; Healthy, healthy individuals; iCCA, intrahepatic cholangiocarcinoma; NCCA, the total patients without CCA.
p < 0.05.
p < 0.01.
Distinguishing iCCA from HCC can be challenging in clinical practice. To evaluate the diagnostic utility of the ITIH5 and ITIH5/CA19‐9 panels for iCCA patients, we conducted ROC analysis and logistic regression. We found an AUC of 0.921 for discriminating between iCCA and HCC (Figure 7H, Table 4). Furthermore, we observed an AUC of 0.954 for the ITIH5/CA19‐9 panel, which outperformed ITIH5 and CA19‐9 when used individually (Figure 7H, Tables 4 and 5). Moreover, the ITIH5/CA19‐9 panel demonstrated AUC values of 0.898, 0.954, 0.976, and 0.920 for distinguishing iCCA from benign hepatobiliary diseases, chronic hepatitis B, and NCCA, respectively. In all cases, these AUC values were superior to those obtained with CA19‐9 alone (Figure 7I,J,L, Tables 4 and 5). However, the combined use of ITIH5 did not improve the diagnostic performance of CA19‐9 in distinguishing iCCA from healthy individuals (Figure 7K, Table 5). A large‐sample study may be necessary to assess its diagnostic efficacy more comprehensively. In conclusion, the ITIH5/CA19‐9 panel may have the potential to enhance the diagnostic performance of iCCA.
4. DISCUSSION
Cholangiocarcinoma is a rare malignant tumor originating from the epithelial cells within the biliary tree. 30 Radical surgical resection is a viable treatment approach. However, due to its concealed anatomical location and the lack of early, noticeable symptoms in the early stages of CCA, patients often receive a diagnosis when the disease has already advanced or metastasized, missing the window for surgical intervention, resulting in lower survival rates and a poor prognosis. 31 Currently, the incidence and mortality rates of CCA are on the rise. Despite significant advancements in the fields of radiology and pathology, early diagnosis remains a challenging endeavor. It is widely recognized that early cancer detection can significantly improve survival rates, especially for certain types of cancer. 32 To enhance treatment outcomes for CCA patients, there is a pressing need for more effective early diagnostic and screening tests. Unfortunately, as of now, there are no specific serum tumor markers that can selectively identify CCA. It is believed that secreted proteins play a crucial role in cell‐to‐cell signaling, communication, and growth. Furthermore, they have the potential to reflect different stages of pathology, making them a promising source for biomarker discovery and novel findings. 33 Therefore, we aim to discover an exocrine protein that exhibits specific overexpression in CCA, capable of distinguishing CCA from other malignancies as well as benign hepatic and biliary diseases, for noninvasive screening of CCA.
Extensive research has illuminated the pivotal role of ITIH5, a secreted protein, in exerting a potent inhibitory effect on tumor growth and metastasis across a spectrum of cancer types. 10 , 14 , 34 In our study, a meticulous examination of TCGA data through a pan‐cancer analysis unveiled a compelling narrative: ITIH5 exhibited a conspicuous upregulation in CCA tumor tissues, boasting a remarkable positive rate of up to 94%. Notably, the mRNA expression levels of ITIH5 within the confines of CCA tumor tissues soared to a remarkable 4.15 times higher than their nontumor counterparts, an observation that set CCA apart from a majority of other malignancies. Leveraging single‐cell data, we further discerned that ITIH5 was discretely secreted by the malignant epithelial cells that originate from the bile duct, underlining its tissue specificity. The diagnostic potential of ITIH5 in CCA was underscored by its serum expression levels, which exhibited robust diagnostic performance with an impressive AUC of 0.848. Moreover, when combined with the clinically employed tumor marker CA19‐9, ITIH5 augmented diagnostic efficiency, further strengthening its candidacy as a promising diagnostic marker for CCA.
According to anatomical location, CCA can be subdivided into iCCA, pCCA, and dCCA. iCCA, situated proximal to the secondary bile ducts within the hepatic parenchyma, ranks as the second most common primary liver cancer following HCC. 35 In its early stages, iCCA typically presents with no symptoms, often being serendipitously detected through imaging studies during health checkups or other medical examinations. Characteristically, iCCA manifests as an intraparenchymal liver mass, requiring differentiation from HCC and benign hepatic tumors. Notably, the therapeutic strategies for iCCA and HCC diverge. For instance, liver transplantation is currently considered a treatment option for HCC but remains controversial for iCCA. 36 More than that, recent research has shed light on the presence of specific mutations, such as IDH1/2 or FGFR2 fusions in small‐duct iCCA, paving the way for targeted therapies. 37 , 38 Furthermore, the prognosis for iCCA is notably worse than that of HCC. Therefore, the accurate diagnosis of the primary liver cancer type is of paramount importance, as it significantly impacts treatment decisions and prognostic assessments. In this study, through bluk RNA‐seq data analysis, we observed that ITIH5 did not exhibit elevated expression in HCC, hepatic hemangiomas, or inflammatory hepatocellular adenomas. IHC further substantiated that ITIH5 is expressed at higher levels in CCA than in HCC. Importantly, the serum levels of ITIH5 can be effectively employed to distinguish between iCCA, HCC, and benign hepatic diseases, showcasing a high diagnostic efficiency.
Perihilar CCA and distal CCA predominantly present as obstructive conditions, often necessitating clinical differentiation from conditions caused by inflammatory narrowing of the bile duct or obstructive diseases resulting from bile duct stones. Furthermore, nonalcoholic fatty liver disease, 39 HBV infection, 40 and PSC 41 stand as risk factors influencing the incidence of cholangiocarcinoma. In this study, our transcriptomic analysis revealed that ITIH5 exhibited no differential expression in NASH, chronic liver diseases, or PSC. Notably, the serum levels of ITIH5 in CCA surpassed those in chronic hepatitis B, benign biliary strictures, and bile duct stones, with statistically significant distinctions. When distinguishing CCA from chronic hepatitis B, ITIH5 displayed an impressive AUC of 0.851, and in conjunction with CA19‐9, the AUC soared to 0.929. Additionally, we confirmed the utility of ITIH5 in discriminating between CCA and benign biliary tract disorders.
Current research or development is focused on identifying possible CCA diagnosis markers, including spermatogenesis‐associated protein 20 (SSP411), 30 mucin 1 (KL‐6), 31 S100 calcium‐binding protein A6 (S100A6), 32 matrix metalloproteinase (MMP)‐7, 33 and so on. As many of these require the examination of CCA tissues, their use as a screening tool before treatment is less appropriate. In contrast, ITIH5, as a serum‐based test, offers a simple, transferrable, and minimally invasive procedure that is well suited for regulatory approval.
In conclusion, this study has demonstrated that serum ITIH5 levels serve as a noninvasive diagnostic biomarker for CCA, especially in the case of iCCA. It was also found that combining plasma ITIH5 with serum CA19‐9 enhanced diagnostic accuracy. While the sensitivity of 87.7% and specificity of 78.2% may be insufficient for screening the low incidence of bile duct cancer, the indicator can be employed for the initial screening of suspicious populations. In the presence of elevated level of the indicator, a combination with other laboratory parameters, imaging studies, cholangioscopy, and pathology can enhance the precision of diagnosis. To provide more comprehensive diagnostic accuracy information, further studies with a larger patient population are warranted.
AUTHOR CONTRIBUTIONS
Meiru Chen: Conceptualization; data curation; funding acquisition; resources; software; writing – original draft. Jinghan Ma: Data curation; methodology; writing – review and editing. Xiaoli Xie: Methodology; software. Miao Su: Supervision. Dongqiang Zhao: Resources; supervision; writing – review and editing.
FUNDING INFORMATION
This work was supported by the Natural Science Foundation of Hebei Province (grant number H2022206539).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Review Board: The study was censored and approved by the Ethics Committee of the Second Hospital of Hebei Medical University (approval no. 2023‐R485), and all the participants signed an informed consent form. All procedures performed in this study involving human participants were guided by the Helsinki Declaration.
Informed Consent: N/A.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: N/A.
Supporting information
Table S1. The genes responsible for encoding exocrine proteins.
Figure S1. (A–H) Fold changes of excluded genes in the various cancer types, respectively.
ACKNOWLEDGMENTS
The authors would like to thank the TCGA and GEO databases for the availability of the data.
Chen M, Ma J, Xie X, Su M, Zhao D. Serum ITIH5 as a novel diagnostic biomarker in cholangiocarcinoma. Cancer Sci. 2024;115:1665‐1679. doi: 10.1111/cas.16143
DATA AVAILABILITY STATEMENT
The transcriptome data for TCGA pan‐cancer analysis was available in UCSC Xena (https://xena.ucsc.edu/). The gene list responsible for encoding exocrine proteins was available in the Human Protein Atlas (https://www.proteinatlas.org/). The datasets (GSE32225, GSE67764, GSE11819, GSE139602, GSE62029, GSE159676, and GSE138709) analyzed during the current study are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The data and R script that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745‐761. [DOI] [PubMed] [Google Scholar]
- 2. Jarnagin WR, Fong Y, DeMatteo RP, et al. Staging, Resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg. 2001;234:507‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Charatcharoenwitthaya P, Enders FB, Halling KC, Lindor KD. Utility of serum tumor markers, imaging, and biliary cytology for detecting cholangiocarcinoma in primary sclerosing cholangitis. Hepatology. 2008;48:1106‐1117. [DOI] [PubMed] [Google Scholar]
- 4. Khan SA, Davidson BR, Goldin RD, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012;61:1657‐1669. [DOI] [PubMed] [Google Scholar]
- 5. Lawrenson K, Grun B, Lee N, et al. NPPB is a novel candidate biomarker expressed by cancer‐associated fibroblasts in epithelial ovarian cancer: tumor stroma biomarkers for ovarian cancer. Int J Cancer. 2015;136:1390‐1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonney GK, Craven RA, Prasad R, Melcher AF, Selby PJ, Banks RE. Circulating markers of biliary malignancy: opportunities in proteomics? Lancet Oncol. 2008;9:149‐158. [DOI] [PubMed] [Google Scholar]
- 7. Himmelfarb M, Klopocki E, Grube S, et al. ITIH5, a novel member of the inter‐alpha‐trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004;204:69‐77. [DOI] [PubMed] [Google Scholar]
- 8. Anveden Å, Sjöholm K, Jacobson P, et al. ITIH‐5 expression in human adipose tissue is increased in obesity. Obesity (Silver Spring). 2012;20:708‐714. [DOI] [PubMed] [Google Scholar]
- 9. Huth S, Heise R, Vetter‐Kauczok CS, et al. Inter‐α‐trypsin inhibitor heavy chain 5 (ITIH5) is overexpressed in inflammatory skin diseases and affects epidermal morphology in constitutive knockout mice and murine 3D skin models. Exp Dermatol. 2015;24:663‐668. [DOI] [PubMed] [Google Scholar]
- 10. Young ED, Manley SJ, Beadnell TC, et al. Suppression of pancreatic cancer liver metastasis by secretion‐deficient ITIH5. Br J Cancer. 2021;124:166‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mai C, Zhao J, Tang X, et al. Decreased ITIH5 expression is associated with poor prognosis in primary gastric cancer. Med Oncol. 2014;31:53. [DOI] [PubMed] [Google Scholar]
- 12. Dötsch MM, Kloten V, Schlensog M, et al. Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients' outcome. Epigenetics. 2015;10:903‐912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Veeck J, Chorovicer M, Naami A, et al. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node‐negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene. 2008;27:865‐876. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Cao F, Li X, et al. ITIH5, a p53‐responsive gene, inhibits the growth and metastasis of melanoma cells by downregulating the transcriptional activity of KLF4. Cell Death Dis. 2021;12:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose M, Gaisa NT, Antony P, et al. Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high‐grade urothelial tumours. Carcinogenesis. 2014;35:727‐736. [DOI] [PubMed] [Google Scholar]
- 16. Dittmann J, Ziegfeld A, Jansen L, et al. Gene expression analysis combined with functional genomics approach identifies ITIH5 as tumor suppressor gene in cervical carcinogenesis. Mol Carcinog. 2017;56:1578‐1589. [DOI] [PubMed] [Google Scholar]
- 17. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 18. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor‐infiltrating immune cells. Nucleic Acids Res. 2020;48:W509‐W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang M, Yang H, Wan L, et al. Single‐cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020;73:1118‐1130. [DOI] [PubMed] [Google Scholar]
- 20. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 2020.
- 22. Gao B, Lin J, Jiang Z, et al. Upregulation of chemokine CXCL10 enhances chronic pulmonary inflammation in tree shrew collagen‐induced arthritis. Sci Rep. 2018;8:9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang Z, Zhou C, Cheng L, et al. Inhibiting YAP expression suppresses pancreatic cancer progression by disrupting tumor‐stromal interactions. J Exp Clin Cancer Res. 2018;37:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El‐Serag HB, Kanwal F, Davila JA, Kramer J, Richardson P. A new laboratory‐based algorithm to predict development of hepatocellular carcinoma in patients with hepatitis C and cirrhosis. Gastroenterology. 2014;146:1249‐1255.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183‐203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu S, Zaidi S, Hassan MI, et al. Mutated CEACAMs disrupt transforming growth factor Beta signaling and Alter the intestinal microbiome to promote colorectal carcinogenesis. Gastroenterology. 2020;158:238‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lilja H, Ulmert D, Vickers AJ. Prostate‐specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268‐278. [DOI] [PubMed] [Google Scholar]
- 28. Fahlbusch R, Keller BV, Ganslandt O, Kreutzer J, Nimsky C. Transsphenoidal surgery in acromegaly investigated by intraoperative high‐field magnetic resonance imaging. Eur J Endocrinol. 2005;153:239‐248. [DOI] [PubMed] [Google Scholar]
- 29. Hatzaras I, Schmidt C, Muscarella P, Melvin WS, Ellison EC, Bloomston M. Elevated CA 19‐9 portends poor prognosis in patients undergoing resection of biliary malignancies. HPB (Oxford). 2010;12:134‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lumachi F, Lo Re G, Tozzoli R, et al. Measurement of serum carcinoembryonic antigen, carbohydrate antigen 19‐9, cytokeratin‐19 fragment and matrix metalloproteinase‐7 for detecting cholangiocarcinoma: a preliminary case‐control study. Anticancer Res. 2014;34:6663‐6667. [PubMed] [Google Scholar]
- 31. Alvaro D. The challenge of cholangiocarcinoma diagnosis: the turning point is in extracellular vesicles? Hepatology. 2017;66:1029‐1031. [DOI] [PubMed] [Google Scholar]
- 32. Early detection: a long road ahead. Nat Rev Cancer. 2018;18:401. [DOI] [PubMed] [Google Scholar]
- 33. Stastna M, Van Eyk JE. Secreted proteins as a fundamental source for biomarker discovery. Proteomics. 2012;12:722‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rose M, Meurer SK, Kloten V, et al. ITIH5 induces a shift in TGF‐β superfamily signaling involving endoglin and reduces risk for breast cancer metastasis and tumor death. Mol Carcinog. 2018;57:167‐181. [DOI] [PubMed] [Google Scholar]
- 35. Brindley PJ, Bachini M, Ilyas SI, et al. Cholangiocarcinoma. Nat Rev Dis Primers. 2021;7:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee DD, Croome KP, Musto KR, et al. Liver transplantation for intrahepatic cholangiocarcinoma. Liver Transpl. 2018;24:634‐644. [DOI] [PubMed] [Google Scholar]
- 37. Abou‐Alfa GK, Macarulla T, Javle MM, et al. Ivosidenib in IDH1‐mutant, chemotherapy‐refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double‐blind, placebo‐controlled, phase 3 study. Lancet Oncol. 2020;21:796‐807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mazzaferro V, El‐Rayes BF, Droz dit Busset M, et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion‐positive intrahepatic cholangiocarcinoma. Br J Cancer. 2019;120:165‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khan SA, Tavolari S, Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019;39:19‐31. [DOI] [PubMed] [Google Scholar]
- 40. Li M, Li J, Li P, et al. Hepatitis B virus infection increases the risk of cholangiocarcinoma: a meta‐analysis and systematic review. J Gastroenterol Hepatol. 2012;27:1561‐1568. [DOI] [PubMed] [Google Scholar]
- 41. Boonstra K, Weersma RK, Van Erpecum KJ, et al. Population‐based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology. 2013;58:2045‐2055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The genes responsible for encoding exocrine proteins.
Figure S1. (A–H) Fold changes of excluded genes in the various cancer types, respectively.
Data Availability Statement
The transcriptome data for TCGA pan‐cancer analysis was available in UCSC Xena (https://xena.ucsc.edu/). The gene list responsible for encoding exocrine proteins was available in the Human Protein Atlas (https://www.proteinatlas.org/). The datasets (GSE32225, GSE67764, GSE11819, GSE139602, GSE62029, GSE159676, and GSE138709) analyzed during the current study are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The data and R script that support the findings of this study are available from the corresponding author upon reasonable request.


