Abstract
Glioblastoma (GBM) is the most common malignant diffuse glioma of the brain. Although immunotherapy with immune checkpoint inhibitors (ICIs), such as programmed cell death protein (PD)‐1/PD ligand‐1 inhibitors, has revolutionized the treatment of several cancers, the clinical benefit in GBM patients has been limited. Lymphocyte‐activation gene 3 (LAG‐3) binding to human leukocyte antigen‐II (HLA‐II) plays an essential role in triggering CD4+ T cell exhaustion and could interfere with the efficiency of anti‐PD‐1 treatment; however, the value of LAG‐3–HLA‐II interactions in ICI immunotherapy for GBM patients has not yet been analyzed. Therefore, we aimed to investigate the expression and regulation of HLA‐II in human GBM samples and the correlation with LAG‐3+CD4+ T cell infiltration. Human leukocyte antigen‐II was highly expressed in GBM and correlated with increased LAG‐3+CD4+ T cell infiltration in the stroma. Additionally, HLA‐IIHighLAG‐3High was associated with worse patient survival. Increased interleukin‐10 (IL‐10) expression was observed in GBM, which was correlated with high levels of HLA‐II and LAG‐3+ T cell infiltration in stroma. HLA‐IIHighIL‐10High GBM associated with LAG‐3+ T cells infiltration synergistically showed shorter overall survival in patients. Combined anti‐LAG‐3 and anti‐IL‐10 treatment inhibited tumor growth in a mouse brain GL261 tumor model. In vitro, CD68+ macrophages upregulated HLA‐II expression in GBM cells through tumor necrosis factor‐α (TNF‐α). Blocking TNF‐α‐dependent inflammation inhibited tumor growth in a mouse GBM model. In summary, T cell–tumor cell interactions, such as LAG‐3–HLA‐II, could confer an immunosuppressive environment in human GBM, leading to poor prognosis in patients. Therefore, targeting the LAG‐3–HLA‐II interaction could be beneficial in ICI immunotherapy to improve the clinical outcome of GBM patients.
Keywords: glioblastoma, HLA‐II, IL‐10, immune checkpoint inhibitor, LAG‐3
T cell–tumor cell interactions, such as lymphocyte‐activation gene 3 (LAG‐3)– human leukocyte antigen‐II (HLA‐II), could confer an immunosuppressive environment in human glioblastoma (GBM), leading to poor prognosis in patients. Therefore, targeting the LAG‐3–HLA‐II interaction could be beneficial in immune checkpoint inhibitor immunotherapy to improve the clinical outcome of GBM patients.
Abbreviations
- APC
antigen‐presenting cell
- Arg‐1
Arginase‐1
- CGGA
Chinese Glioma Genome Atlas
- FCM
flow cytometry
- GBM
glioblastoma
- GEPIA
Gene Expression Profiling Interaction Analysis
- GFAP
glial fibrillary acidic protein
- HLA
human leukocyte antigen
- ICI
immune checkpoint inhibitor
- IHC
immunohistochemistry
- IL‐10
interleukin‐10
- LAG‐3
lymphocyte‐activation gene 3
- OS
overall survival
- PD‐1
programmed cell death protein 1
- PD‐L1
programmed cell death ligand 1
- PGE2
prostaglandin E2
- PMA
phorbol 12‐myristate 13‐acetate
- ssGSEA
single‐sample gene set enrichment analysis
- sTNFR:Fc
soluble TNF receptor:Fc fusion protein
- TCGA
The Cancer Genome Atlas
- TIL
tumor‐infiltrating lymphocyte
- TIM‐3
T cell immunoglobulin domain and mucin domain‐3
- TNF‐α
tumor necrosis factor‐α
1. INTRODUCTION
Glioblastoma is the most common malignant diffuse glioma and is a highly aggressive brain tumor. With standard treatments, such as surgery, temozolomide chemotherapy, radiotherapy, and corticosteroid therapy, the median survival rates for GBM patients remain low. 1 Although immunotherapy utilizing ICIs, such as PD‐1/PD‐L1 inhibitors, has revolutionized the treatment of several cancers, its clinical benefit in GBM patients is limited. 2 In GBM, the lack of clinically meaningful responses to ICI has been attributed to the specialized immune environment of the brain, which includes mechanical obstruction at the blood–brain barrier, T cell exhaustion, and a generally immunosuppressive environment. 3 A comprehensive understanding of ICI regulation within the brain tumor could reveal potential methods for improving the efficiency of immunotherapeutic interventions. Therefore, preventing cancer immunosuppression by targeting ICI could provide a suitable strategy for immunotherapy in GBM patients. 1 , 4 , 5
Lymphocyte‐activation gene 3 is a well‐known immune checkpoint, and many studies have concluded that LAG‐3, PD‐1, or TIM‐3 are markers of T cell exhaustion in glioma and are associated with poor prognosis. 4 , 6 , 7 Lymphocyte‐activation gene 3 in activated T cells primarily binds to MHC‐II molecules in APCs, reducing T cell proliferation and increasing T cell exhaustion. 8 , 9 , 10 , 11 The benefits of anti‐LAG‐3 therapy in different malignancies have been reported. 12 , 13 Upregulation of LAG‐3 limits the antitumor activity of PD‐1/PD‐L1 inhibitors by attenuating T cell activation and preventing tumor cell destruction. 14 , 15 A preclinical study using a mouse model for glioma showed that treatment with anti‐LAG‐3 Abs, either alone or in combination with a PD‐1 inhibitor, resulted in prolonged survival. 16 The prognostic value of LAG‐3 expression in solid tumors has been monitored as an important immune checkpoint 17 ; however, the role of LAG‐3 ICI therapy in GBM remains to be elucidated.
The MHC, encoding the major histocompatibility antigens of animals, is also known as the HLA in humans. Human leukocyte antigen‐II is encoded by three different loci: HLA‐DR, HLA‐DQ, and HLA‐DP. 18 , 19 Human leukocyte antigen‐II expression in tumor cells is a predictive biomarker of the preferred anti‐PD‐1 therapy in melanoma, Hodgkin's lymphoma, and lung adenocarcinoma. 20 , 21 , 22 Lymphocyte‐activation gene 3 can directly and specifically interact with HLA‐II heterodimers, providing new insights into the mechanism by which LAG‐3 initiates T cell inhibition. 23 Human leukocyte antigen‐II has been shown to inhibit T cell antitumor activity in cutaneous melanoma cells through their LAG‐3 receptor. 24 Lymphocyte‐activation gene 3 and its ligand HLA‐II are highly expressed in high‐risk uveal melanoma and could be a target for adjuvant immunotherapy. 25 Lymphocyte‐activation gene 3 binding to HLA‐II plays an essential role in triggering CD4+ T cell exhaustion and might interfere with the efficiency of anti‐PD‐1 treatment in classical Hodgkin's lymphoma. 26 However, the expression of HLA‐II in human GBM requires elucidation, and the value of LAG‐3–MHC‐II interactions in ICI immunotherapy for patients with GBM has not been analyzed.
We, therefore, aimed to explore the expression of HLA‐II and its correlation with LAG‐3+CD4+ T cell infiltration in human GBM samples. Moreover, we investigated the mechanism involved in the regulation of HLA‐II in GBM cells in vitro. This study provides novel insights into the theoretical basis for the development of LAG‐3–MHC‐II immune‐related intervention targets for GBM.
2. MATERIALS AND METHODS
2.1. Materials
Information regarding materials is provided in Appendix S1.
2.2. Human case selection
Fifty‐one cases of paraffin‐embedded GBM specimens and 27 cases of paraffin‐embedded normal brain specimens were archived in the Department of Pathology, Second Hospital of Hebei Medical University (collected between January 2011 and August 2016) with a median follow‐up period of 18 months (range, 8–27 months). Clinical histopathological data of patients and information on OS of patients were collected until death or the end of the investigation. Twenty‐one cases of fresh GBM and paired paraneoplastic brain tissues for protein and RNA extraction, and 30 cases of frozen GBM specimens and 6 cases of frozen control brain specimens for immunofluorescence staining were provided by the Department of Neurosurgery, Second Hospital of Hebei Medical University. All the cases above were diagnosed by at least two board‐certified neuropathologists according to the 2021 WHO guidelines. The expression of IDH1‐R132H protein was determined by IHC. The normal brain tissues used as negative controls were obtained from paraneoplastic brain tissues who underwent glioma surgery or nontumor patients who underwent traumatic brain injury decompression surgery. Patients who agreed to have their samples used for scientific research provided written informed consent. The study was approved by the Ethics Committee of Hebei Medical University, and the treatment of the collected data and specimens was in accordance with ethical and legal standards.
The Cancer Genome Atlas of glioma and survival data was downloaded from TCGA: Oncomine database (https://www.oncomine.org) and GEPIA (http://gepia.cancer‐pku.cn/) online platforms. Furthermore, 301 glioma samples were downloaded from the CGGA (http://cgga.org.cn/).
2.3. Immunofluorescence staining
The tumor tissues from the GBM patients were immersed in 4% formalin, embedded in an optimal cutting temperature compound, and then subjected to immunofluorescence staining per the manufacturer's protocol. 27
2.4. Immunohistochemical score in human GBM
The IHC staining was processed as previously described. 27 The degree of immunostaining was scored independently by stained tumor cells and staining intensity according to a previous study. 27 The details of score are provided in Appendix S1.
2.5. Mouse s.c. tumor model
Female C57BL/6 mice (6–8 weeks; 18–20 g) were maintained in specific pathogen‐free conditions at animal facilities. Experimental protocols were approved by the Institutional Animal Care and Use Committee of Hebei Medical University. All animals were managed and treated according to the guidelines of Hebei Medical University. Briefly, C57BL/6 mice were inoculated with 2 × 106 GL261 tumor cells s.c. into the flank region. When visible tumors appeared (day 12), those mice were divided into four groups, and injected with anti‐LAG‐3 or anti‐IL‐10 or combined anti‐LAG‐3 and anti‐IL‐10 (10 μg/kg i.p., given four times). In some experiments, the mice were injected with sTNFR:Fc every 2 days (100 mg/kg body weight i.p., given four times). Another 10 days later, the animals were killed, and the tumor weight derived from each group was measured at day 22. The tumor volumes were measured every 2 days from day 12 to 22 using the following formula:
2.6. Cell culture and treatment
Human U87, U251, and THP‐1 cells were purchased from Peking Union Cell Resource Center. U87 and U251 were cultured in DMEM containing 10% FBS and penicillin/streptomycin at 37°C and 5% CO2 humidified atmosphere. All experiments were carried out with mycoplasma‐free cells. U87 and U251 cells were treated with different doses of TNF‐α, and the expression of HLA‐II was measured by FCM and western blot analysis. THP‐1 cells were cultured in RPMI‐1640 supplemented with antibiotics and 10% FCS and matured by PMA (Sigma). To differentiate into M0 macrophages, THP‐1 cells (3.5 × 105 cells/well) were induced by 50 ng/mL PMA for 48 h as previously described. The obtained M0 cells were treated with lipopolysaccharide by 100 ng/mL to induce their differentiation into M1 macrophages. The conditioned medium from M1 cells was collected, with or without blocking by 15 μg/mL anti‐TNF‐α, and then used to stimulate U87 or U251 cells for 24 h. The HLA‐DP or HLA‐DQ expression in U87 or U251 cells was quantified through western blot assays. The experiments were undertaken at least three times.
2.7. Western blot analyses
Tissues or cells were harvested and homogenized in 50 μL lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X‐100, cocktail) as previously described. 27 The experiments were processed according to a previous study. 27
2.8. Quantitative real‐time PCR
The experiments were processed according to a previous study. 27 The details of scores are provided in Appendix S1. The primer sequences used for PCR amplification are shown in Table 1.
TABLE 1.
Primer sets used for quantitative real‐time PCR.
| Human | Forward primer | Reverse primer |
|---|---|---|
| HLA‐DP | AGGCAGCATTCAAGTCCGAT | GGGGGTCATTTCCAGCATCA |
| HLA‐DQ | ATGTCCAGTAACACAGAAGCCA | AAGGGCAGACGGTATCCATTA |
| IL‐10 | GCTCTTGCAAAACCAAACCACA | TCTCGAAGCATGTTAGGCAG |
| TGF‐β | CGACTCGCCAGAGTGGTTAT | CGGTAGTGAACCCGTTGATGT |
| PGE2 (PTGS2) | CTGCGCCTTTTCAAGGATGG | CCCCACAGCAAACCGTAGAT |
| TNF‐α | TGTCCCTTTCACTCACTGGC | CATCTTTTGGGGGAGTGCCT |
| IL‐1β | TCCCCAGCCCTTTTGTTGAG | GGAGCGAATGACAGAGGGTT |
| IL‐6 | TGCAATAACCACCCCTGACC | GTGCCCATGCTACATTTGCC |
Abbreviations: HLA, human leukocyte antigen; IL, interleukin; PGE2, prostaglandin E2; TGF‐β, transforming growth factor‐β; TNF‐α, tumor necrosis factor‐α.
2.9. Statistical analysis
The results were analyzed using SPSS 24.0 (IBM Corp.). Data from clinical samples were analyzed using the Mann–Whitney U‐test and Spearman's correlation. Overall survival was measured from the date of operation to the date of death, or the last clinical follow‐up time before September 2022, according to the Kaplan–Meier method. The log‐rank test was used to compare the survival distribution. All in vitro data were tested using one‐way ANOVA. Data are expressed as mean ± SD obtained from at least three independent experiments. Data were analyzed using the nonparametric Mann–Whitney U‐test (significance level p < 0.05).
3. RESULTS
3.1. Expression of HLA‐II in human GBM
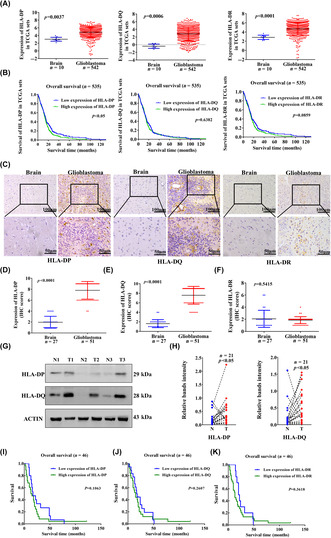
Among the 552 specimens from TCGA (542 cases of GBM and 10 cases of normal brain tissues), the mRNA expression levels of HLA‐DP, HLA‐DQ, and HLA‐DR were higher in GBM than in normal brain (Figure 1A). The CGGA datasets showed the mRNA expression levels of HLA‐DP, HLA‐DQ, and HLA‐DR in GBM WHO IV were higher than those in WHO II (Figure S1A). The HLA‐DQ and HLA‐DR expression was not associated with shorter OS, whereas HLA‐DP was associated with shorter OS in patients with GBM (Figure 1B).
FIGURE 1.
Expression of human leukocyte antigen‐II (HLA‐II) molecules in human glioblastoma (GBM). (A) Evaluation of HLA‐II expression from The Cancer Genome Atlas (TCGA). (B) Association between HLA‐DP, HLA‐DQ, and HLA‐DR expression and survival follow‐up time in TCGA datasets. (C) Immunohistochemical (IHC) images indicate the expression of HLA‐II in paraffin GBM specimens. (D–F) Comparison of HLA‐II molecules expression in control and GBM samples was analyzed (51 cases of GBM and 27 control tissues). (G, H) Twenty‐one pairs of fresh tumor tissues and peritumoral brain tissues were collected, and the expression of HLA‐II was measured by western blot. The comparison of HLA‐II between GBM (T) and control (N) tissues is shown (n = 21). (I–K) Overall survival was measured according to the expression of HLA‐II in paraffin‐embedded GBM (n = 46).
To further validate the findings from TCGA and CGGA, we explored HLA‐II expression in 51 cases of paraffin‐embedded GBM specimens and 27 control specimens. Immunohistochemical staining showed positive expression of HLA‐DP, HLA‐DQ, and HLA‐DR in the GBM (Figure 1C). The expression of HLA‐DP and HLA‐DQ was significantly higher in human GBM than in the control (Figure 1D,E). There was no difference in HLA‐DR expression between GBM and control (Figure 1F). In addition, a comparison of 21 pairs of fresh GBM and adjacent control samples revealed that HLA‐DP and HLA‐DQ were upregulated in GBM (Figures 1G,H and S1C). We also detected HLA‐DP and HLA‐DQ expression in GFAP+ GBM cells using immunofluorescence staining (Figure S1D,F). The numbers of HLA‐DP+GFAP+ and HLA‐DQ+GFAP+ cells were significantly higher in human GBM than in the control (Figure S1E,G). Using seven pairs of fresh GBM and adjacent control samples, we also detected HLA‐II expression in human GFAP+ GBM cells by FCM staining. Flow cytometry analysis indicated the percentage of HLA‐II+GFAP+ cells in human GBM was higher than in control brain tissues (Figure S2A,B). However, high levels of HLA‐II were not associated with OS according to HLA‐II expression in paraffin‐embedded GBM specimens (Figure 1I–K), suggesting that HLA‐II in tumor cells might not be an independent factor in the prognosis of GBM patients.
3.2. Expression of HLA‐II is related to infiltration of LAG‐3 + CD4 + T cells in GBM
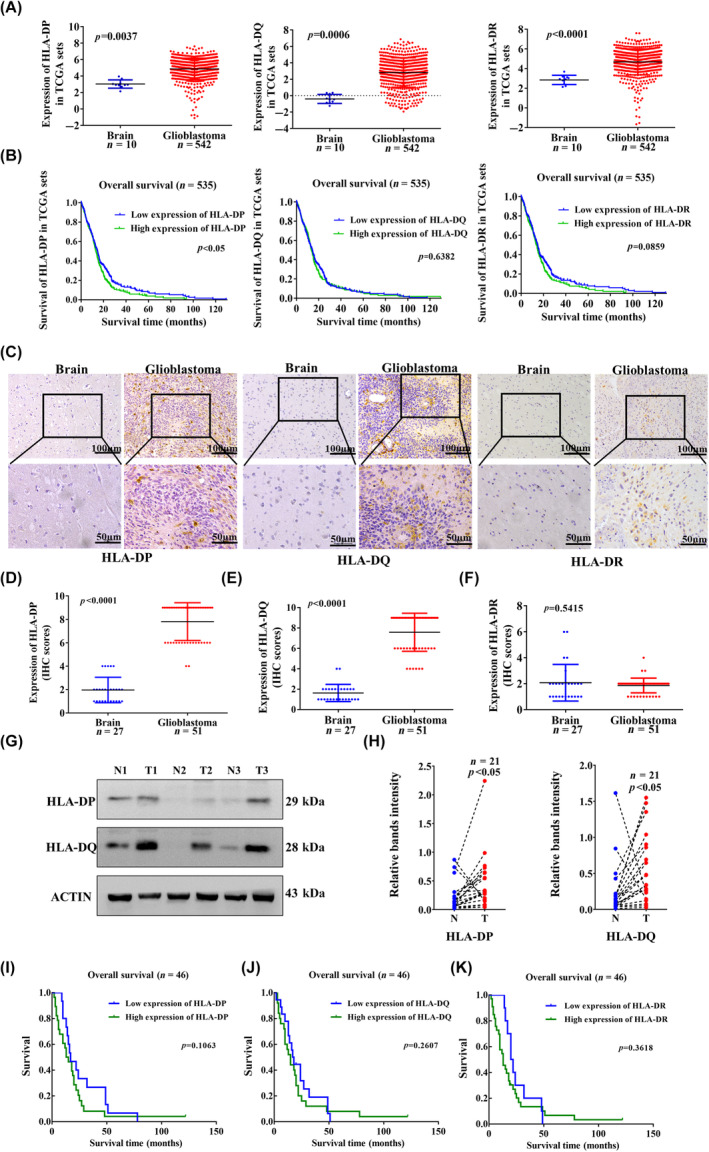
Based on TCGA, the mRNA expression of LAG‐3, TIM‐3, and PD‐1 was significantly higher in GBM than in controls (Figure 2A). The CGGA datasets showed the mRNA expression levels of LAG‐3, TIM‐3, and PD‐1 in GBM WHO IV were higher than those in WHO II (Figure S3A). The TCGA from the Oncomine and GEPIA databases as well as CGGA databases showed that high mRNA expression of LAG‐3, TIM‐3, and PD‐1 was not associated with OS in GBM patients (Figure S3B–D). In seven pairs of fresh GBM and adjacent control samples, FCM staining showed the percentage of LAG‐3+CD4+ T cells in human GBM was higher than that in control brain tissues (Figure S2C,D). Based on IHC staining in 51 cases of paraffin‐embedded GBM specimens, we found LAG‐3, TIM‐3, and PD‐1 were mainly expressed in tumor stroma (Figure S4A–C). The infiltration of LAG‐3+, TIM‐3+, and PD‐1+ cells in GBM stroma was greater than that in controls (Figure S4D–F). The expression of LAG‐3, TIM‐3, and PD‐1 was upregulated in infiltrating lymphocytes. To analyze patient survival, we used the median expression levels of LAG‐3+, TIM‐3+, and PD‐1+ cells as cut‐off points to divide the GBM into high‐ and low‐expression groups. High expression of LAG‐3, TIM‐3, and PD‐1 was not associated with shorter OS of patients with GBM (Figure S4G–I).
FIGURE 2.
Human leukocyte antigen‐II (HLA‐II) expression is related to the infiltration of lymphocyte‐activation gene 3 (LAG‐3)+CD4+ T cells in glioblastoma (GBM). (A) Evaluation of LAG‐3, T cell immunoglobulin domain and mucin domain‐3 (TIM‐3), and programmed cell death protein 1 (PD‐1) expression from the TCGA. (B) Thirty cases of frozen GBM samples and six cases of control brain samples were collected, and immunofluorescence staining showed that LAG‐3 was colocalized with CD4, and TIM‐3 and PD‐1 were colocalized with CD3 in the tumor stroma. White arrows indicate colocalized cells. (C) The number of LAG‐3+CD4+, TIM‐3+CD3+, and PD‐1+CD3+ cells in GBM and control samples is shown (p < 0.05). (D) Thirty cases of frozen GBM and six control samples were collected, and immunofluorescence staining indicated that LAG‐3+CD4+ cells in the tumor stroma, which is associated with HLA‐DP and HLA‐DQ expression. White arrows indicate colocalized cells. (E) The correlation between number of CD4+LAG‐3+ T cell infiltration and HLA‐DP is shown. (F) The correlation between HLA‐DP expression and the number of CD4+LAG‐3+ T cells is shown. (G, H) Based on both HLA‐II and LAG‐3 expression in paraffin‐embedded GBM specimens, overall survival was analyzed according to HLA‐IIHighLAG‐3High and HLA‐IILowLAG‐3Low (n = 46). (I) Kaplan–Meier curves for survival outcomes of patients are shown according to low expression of HLA‐DP and LAG‐3 (n = 46).
We collected 30 frozen GBM specimens and six frozen control specimens for immunofluorescence staining, and detected CD4+LAG‐3+, CD3+TIM‐3+, and CD3+PD‐1+ T cell infiltration in the tumor stroma (Figure 2B). The infiltration of CD4+LAG‐3+, CD3+TIM‐3+, and CD3+PD‐1+ T cells was higher in GBM specimens than those in the controls (Figure 2C). Immunofluorescence images indicated the infiltration of LAG‐3+CD4+ cells in tumor stroma, which was associated with HLA‐DP and HLA‐DQ expression in the same field (Figures 2D and S5). Glioblastoma patients with high HLA‐DP expression showed increased CD4+LAG‐3+ T cell infiltration (Figure 2E). The number of CD4+LAG‐3+ T cells closely correlated with HLA‐DP expression based on immunofluorescence staining (Figure 2F). In seven pairs of fresh GBM and adjacent control samples, FCM staining showed the percentage of LAG‐3+CD8+ T cells in human GBM was higher than that in control brain tissues (Figure S2C,E). Immunofluorescence staining indicated that the infiltration of CD8+LAG‐3+ T cells was higher in GBM specimens than in controls (Figure S6A,B). However, the number of CD8+LAG‐3+ T cells was not correlated with HLA‐DP or HLA‐DQ expression based on immunofluorescence staining (Figure S6C,D). Thus, the results clearly indicate that increased HLA‐II expression is related to the infiltration of LAG‐3+CD4+ T cells in GBM.
In addition, data from TCGA (Figure S7A) and CGGA (Figure S7B) showed that HLA‐DP/HLA‐DQ upregulation was positively correlated with LAG‐3 expression in GBM. Immunohistochemical staining showed that positive expression of HLA‐DP, HLA‐DQ, CD4, and LAG‐3 was observed in the same field of paraffin‐embedded GBM samples (Figure S7C). Based on both HLA‐II and LAG‐3 expression in paraffin‐embedded GBM specimens, the case numbers of HLA‐IIHighLAG‐3High or HLA‐IILowLAG‐3Low is shown in Table 2. Spearman's correlation analysis showed that HLA‐DP and HLA‐DQ expression was positively correlated with LAG‐3+ lymphocyte infiltration in 51 cases of GBM samples (Figure S7D,E). Glioblastoma patients with high HLA‐DP expression showed increased LAG‐3+ T cell infiltration (Figure S7F,G). We analyzed the OS of patients according to HLA‐DPHighLAG‐3High versus HLA‐DPLowLAG‐3Low. Patients with HLA‐DPLowLAG‐3Low had a longer OS than those with HLA‐DPHighLAG‐3High (Figure 2G). Patients with HLA‐DQHighLAG‐3High were not associated with poor survival (Figure 2H). Interestingly, patients with HLA‐DPLowLAG‐3Low had a better prognosis (Figure 2I). Therefore, HLA‐DPHigh and LAG‐3High are independent factors, which could confer an immunosuppressive environment in GBM.
TABLE 2.
Correlations among human leukocyte antigen (HLA)‐DP, HLA‐DQ, lymphocyte‐activation gene 3 (LAG‐3), and interleukin‐10 (IL‐10) levels in glioblastoma.
| HLA‐DP | r | p value | HLA‐DQ | r | p value | LAG‐3 | r | p value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | High | Low | |||||||
| LAG‐3 | 0.3507 | <0.05* | 0.3107 | <0.05* | – | – | ||||||
| High | 19 | 7 | – | – | 18 | 8 | – | – | – | – | – | – |
| Low | 13 | 12 | – | – | 13 | 12 | – | – | – | – | – | – |
| IL‐10 | 0.299 | <0.05* | 0.313 | <0.05* | 0.3935 | <0.05* | ||||||
| High | 16 | 5 | – | – | 14 | 7 | – | – | 16 | 5 | – | – |
| Low | 16 | 14 | – | – | 17 | 13 | – | – | 10 | 20 | – | – |
p < 0.05 indicates statistically significant difference.
3.3. Human leukocyte antigen‐II is related to IL‐10 expression in GBM
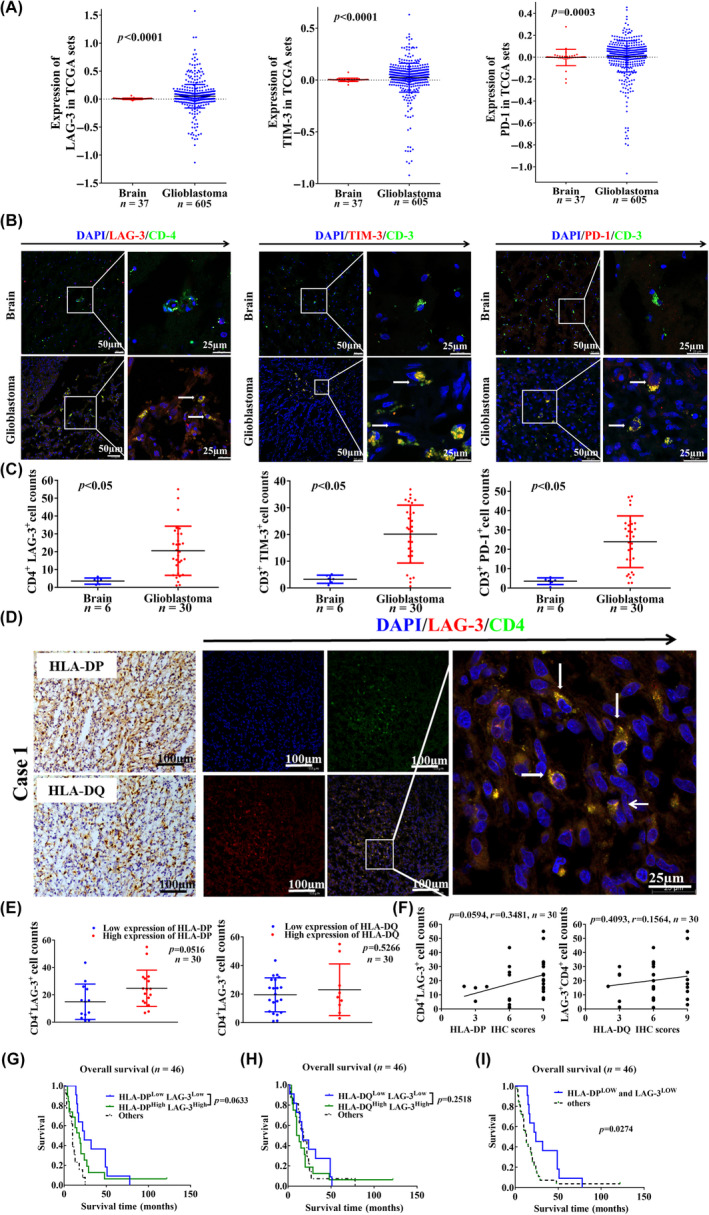
Based on TCGA, the mRNA expression of anti‐inflammatory cytokines IL‐10, TGF‐β, and PGE2 and pro‐inflammatory cytokines IL‐1β and IL‐6 was upregulated in GBM compared to that in normal brain tissues (Figure S8A). We isolated 21 fresh GBM samples, and measured HLA‐II and cytokine expression. The expression of HLA‐DP was upregulated in 13 cases of GBM samples, which showed upregulation of IL‐10, TNF‐α, and PGE2 (Figures 3A,B and S8B). Upregulation of HLA‐DQ was observed in 14 cases of GBM samples, which showed high expression of IL‐10 and IL‐6 (Figures 3A,C and S8B).
FIGURE 3.

Human leukocyte antigen‐II (HLA‐II) molecules are related to the expression of interleukin‐10 (IL‐10) in glioblastoma (GBM). (A) Based on the RT‐PCR results of 21 pairs of fresh tumor tissues, the expression of HLA‐DP was upregulated in 13 cases of GBM, and HLA‐DQ was upregulated in 14 cases of GBM. (B, C) GBM cases were divided into HLA‐II upregulation and HLA‐II downregulation groups. The expression of IL‐10, transforming growth factor‐β (TGF‐β), and prostaglandin E2 (PGE2) in fresh GBM samples was compared between those two groups (p < 0.05). (D) Representative immunohistochemical (IHC) images show the expression of IL‐10 in paraffin‐embedded GBM specimens. (E) Expression of IL‐10 was compared between GBM and control specimens (p < 0.001). (F) Survival outcomes of patients were shown according to the expression of IL‐10 in paraffin GBM specimens. (G) Representative IHC images indicate the expression of HLA‐DP, HLA‐DQ, and IL‐10 in paraffin GBM specimens. (H, I) Spearman's correlation analysis indicates the relationship between the expression of HLA‐II and IL‐10 in paraffin GBM specimens (n = 51). (J, K) Survival outcomes of patients were shown according to both HLA‐II and IL‐10 expression in paraffin GBM specimens, HLA‐IILowIL‐10Low versus HLA‐IIHighIL‐10High (n = 46). N, Normal control brain; T, Tumor.
Data from TCGA and CGGA showed HLA‐DP/DQ expression was also correlated with IL‐10 expression in GBM (Figure S8C,D). Immunohistochemical staining showed positive expression of IL‐10 in 51 cases of paraffin‐embedded GBM specimens, and IL‐10 expression was increased in GBM compared to that in control specimens (Figure 3D,E). A trend toward worse OS was indicated in patients with IL‐10 upregulation (Figure 3F). In paraffin‐embedded GBM specimens, IHC images showed positive staining of HLA‐DP, HLA‐DQ, and IL‐10 in the same field (Figure 3G). The case numbers of HLA‐IIhighIL‐10high or HLA‐IIlowIL‐10low are shown in Table 2. Both HLA‐DP and HLA‐DQ expression was positively correlated with IL‐10 upregulation (Figure 3H,I). Based on both HLA‐II and IL‐10 expression in paraffin‐embedded GBM specimens, multivariate analysis indicated the patients with HLA‐IILowIL‐10Low had a longer OS than those with HLA‐IIHighIL‐10High (Figure 3J,K).
3.4. Blocking of LAG‐3 and IL‐10 inhibits GBM progression
We also analyzed the relationship between IL‐10 expression and LAG‐3+ cell infiltration in 51 paraffin‐embedded GBM specimens. An increased number of LAG‐3+ cells was positively correlated with IL‐10 upregulation (Figure 4A). Furthermore, we divided the paraffin‐embedded GBM samples into four groups based on the combined expression of HLA‐II and IL‐10: HLA‐IIHighIL‐10High, HLA‐IIHighIL‐10Low, HLA‐IILowIL‐10Low, and HLA‐IILowIL‐10High to analyze LAG‐3+ cell infiltration. The number of LAG‐3+ cells in HLA‐DP/DQHighIL‐10High GBM stroma was significantly higher than that in HLA‐DP/DQLowIL‐10Low GBM stroma (Figure 4B,C). The case numbers of LAG‐3highIL‐10high or LAG‐3lowIL‐10low are shown in Table 2. Patients with LAG‐3HighIL‐10High had shorter OS than those with LAG‐3LowIL‐10Low (Figure 4D). According to combined expression of HLA‐II, LAG‐3, and IL‐10 in paraffin‐embedded GBM specimens, multivariate analysis indicated that patients with HLA‐IIHighLAG‐3HighIL‐10High had a worse OS (Figure 4E,F). Patients with HLA‐II+IL‐10+ associated with increased LAG‐3+ T cell infiltration synergistically leads to poor prognosis. We established an in vivo subcutaneous GBM tumor model using brain GL261 tumor cells, and treated the tumor‐bearing mice with anti‐LAG‐3 and IL‐10 (Figure 4G). We found anti‐LAG‐3 and IL‐10 combined treatment significantly inhibited tumor growth compared to anti‐LAG‐3 or anti‐IL‐10 alone (Figure 4H,I). Combined anti‐LAG‐3 and anti‐IL‐10 inhibition significantly decreased the tumor weight compared to anti‐LAG‐3 or anti‐IL‐10 alone (Figure 4J). Furthermore, immunofluorescence staining analysis showed that combined anti‐LAG‐3 and anti‐IL‐10 also reduced CD4+LAG‐3+ (Figure 4K,L) and CD8+LAG‐3+ cell infiltration in tumor tissues (Figure 4M,N). Therefore, blocking of LAG‐3 and IL‐10 might be beneficial for improving the clinical outcome of GBM patients.
FIGURE 4.
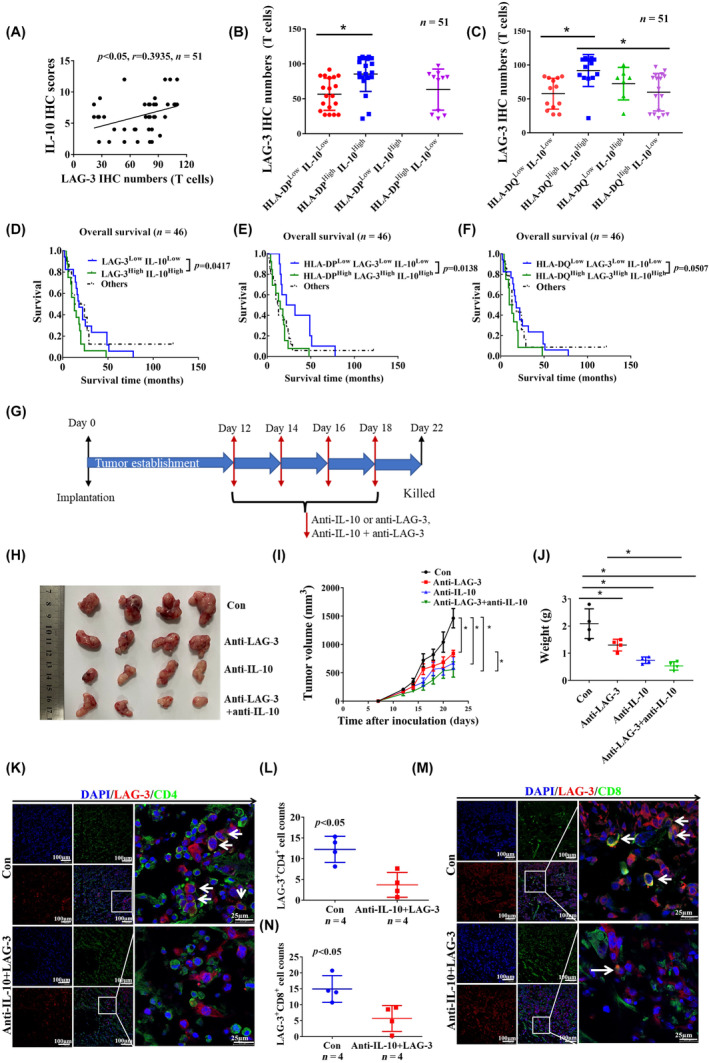
Targeting of lymphocyte‐activation gene 3 (LAG‐3) and interleukin‐10 (IL‐10) inhibits glioblastoma (GBM) progression. (A) Based on the immunohistochemical (IHC) staining in paraffin GBM specimens, the correlation between LAG‐3+ cell number and IL‐10 expression is shown (n = 51). (B, C) The number of LAG‐3+ cells in patients with human leukocyte antigen‐II (HLA‐II)HighIL‐10High was compared to that in patients with HLA‐IILowIL‐10Low (n = 51). (D) Survival outcomes of patients are shown according to both LAG‐3 and IL‐10 expression in paraffin‐embedded GBM specimens, LAG‐3LowIL‐10Low versus LAG‐3HighIL‐10High (n = 46). (E, F) Survival outcomes of patients are shown according to combined expression of HLA‐II, LAG‐3, and IL‐10 together in paraffin‐embedded GBM specimens, HLA‐IILowLAG‐3LowIL‐10Low versus HLA‐IIHighLAG‐3HighIL‐10High (n = 46). (G) Schematic of experimental design. (H) Images of tumors from control, anti‐IL‐10, anti‐LAG‐3, and anti‐IL‐10 + LAG‐3 groups are shown at day 22 after injection. (I) Tumor volumes were measured every 2 days from day 12 to 22. Data are shown as mean ± SD (n = 4). (J) Tumor weights were measured at day 22. Data are shown as mean ± SD (n = 4). (K, M) Immunofluorescence staining indicated that CD4+LAG‐3+ and CD8+LAG‐3+ cells were infiltrated in the tumor tissues. White arrows show colocalized cells. (L, N) Data are shown as mean ± SD (n = 4). *p < 0.05. Con, control; TNF‐α, tumor necrosis factor‐α.
3.5. TNF‐α+ CD68 + macrophage infiltration associated with HLA‐II upregulation in GBM
Based on TCGA, we analyzed the immune infiltration patterns according to the HLA‐II scores in GBM using ssGSEA. Heat map analysis revealed that the expression of HLA‐II was positively correlated with the infiltration of macrophages and neutrophils, and immune checkpoint molecules (Figure S9).
To explore whether infiltration of immune cells contributes to HLA‐II upregulation in GBM, we measured the relationship between HLA‐II expression and macrophage infiltration in 51 paraffin‐embedded GBM specimens. Positive expression of CD68 and TNF‐α associated with HLA‐DP, HLA‐DQ, and HLA‐DR was observed in the same field of GBM (Figure 5A). Tumor necrosis factor‐α expression and CD68+ macrophage infiltration were higher in GBM specimens than in control specimens (Figure 5B,C). Increased CD68+ macrophage infiltration positively correlated with TNF‐α upregulation in GBM samples (Figure 5D). Increased TNF‐α expression was also positively correlated with CD68+ macrophage infiltration and HLA‐II upregulation (Figure 5E–G). CD68+ macrophage infiltration was positively correlated with HLA‐II upregulation (Figure 5H,I). These results suggest that CD68+ macrophages expressing TNF‐α could be related to HLA‐II upregulation in GBM.
FIGURE 5.
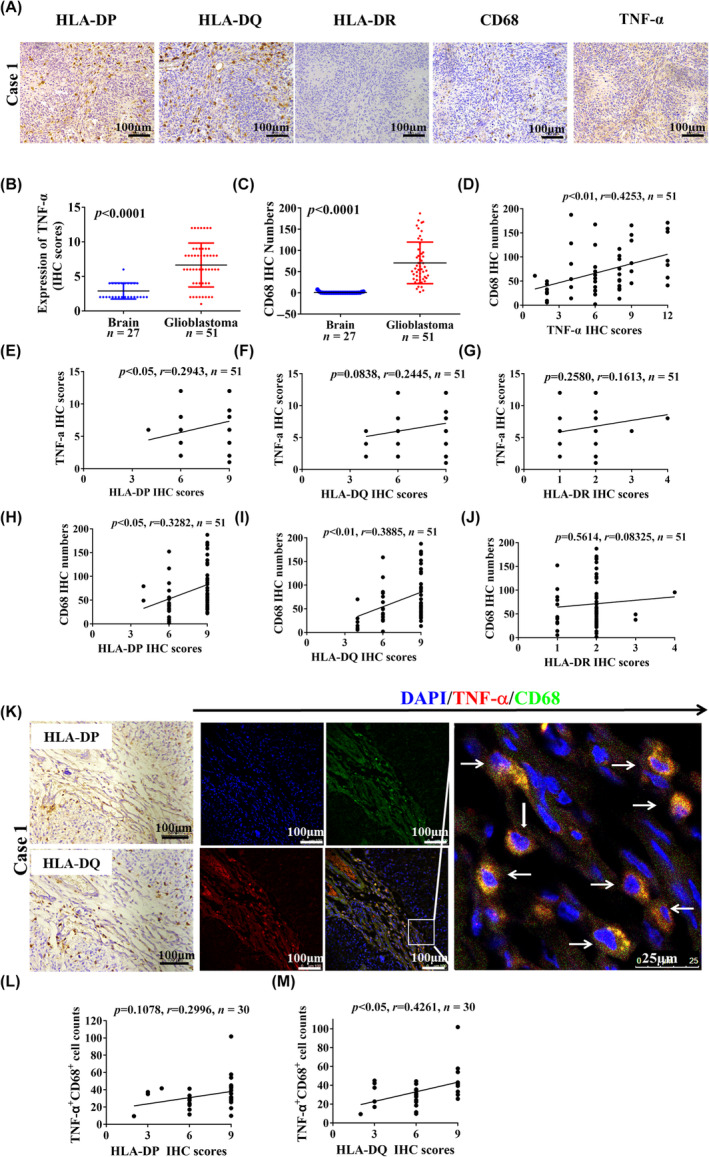
Tumor necrosis factor‐α (TNF‐α)+CD68+ macrophage infiltration is associated with human leukocyte antigen‐II (HLA‐II) upregulation in glioblastoma (GBM). (A) Representative immunohistochemical (IHC) images indicate the expression of HLA‐ II, TNF‐α, and CD68 in the same field in paraffin‐embedded GBM samples. (B, C) Expression of TNF‐α and the CD68+ macrophage infiltration were compared between GBM and control specimens (p < 0.05). (D) Based on the IHC staining in paraffin‐embedded GBM samples, the correlation between CD68+ macrophage infiltration and TNF‐α expression is shown (n = 51). (E–G) Correlation between TNF‐α expression and HLA‐II expression (n = 51). (H–J) Correlation between CD68+ macrophage infiltration and HLA‐II expression (n = 51). (K) Immunofluorescence staining shows that TNF‐α and CD68 is colocalized in the tumor stroma, which is associated with HLA‐DP and HLA‐DQ expression in the same field. White arrows indicate colocalized cells. (L, M) Based on immunofluorescence staining of TNF‐α and CD68, the correlation between the number of infiltrated TNF‐α+CD68+ cells and HLA‐II expression is shown (n = 30).
Based on immunofluorescence staining in 30 frozen GBM specimens, the relationship between TNF‐α+CD68+ macrophage infiltration and HLA‐II expression was further investigated. Tumor necrosis factor‐α and CD68 colocalization was observed in the GBM stroma, which was associated with HLA‐DP and HLA‐DQ expression in the same field (Figures 5K and S10A). The increased number of TNF‐α+CD68+ macrophages was correlated with HLA‐II upregulation in GBM specimens (Figure 5L,M). These results indicate that CD68+ macrophages producing TNF‐α could contribute to HLA‐II upregulation in GBM. We also detected IL‐10 expression in Arg‐1+ M2 macrophages in frozen GBM stroma. Immunofluorescence staining showed the infiltration of Arg‐1+ M2 (Figure S10B,C) and the number of IL‐10+Arg‐1+ macrophages (Figure S10D,E) was increased in GBM stroma. Thus, M2 macrophages infiltrated the GBM stroma, and might also contribute to IL‐10 upregulation.
3.6. Tumor necrosis factor‐α upregulating HLA‐II expression contributes to GBM progression
To further confirm that macrophages secrete TNF‐α to upregulate the expression of HLA‐II in GBM cells, human GBM U87 and U251 cell lines were used as in vitro models. After stimulation with TNF‐α for 24 h, we detected HLA‐II expression in human GBM cells by FCM staining and western blot analysis. Flow cytometry analysis indicated TNF‐α increased HLA‐II expression in U87 and U251 cells (Figure 6A,B). Western blot results showed the expression of HLA‐DP and HLA‐DQ was significantly upregulated in GBM cells treated with TNF‐α (Figure 6C,D). Human monocyte THP‐1 cells were cultured and differentiated into M1‐type macrophages, the supernatant was then collected to stimulate the U87 and U251 cells. Supernatants from M1‐type THP‐1 macrophages upregulated the expression of HLA‐DP and HLA‐DQ in tumor cells (Figure 6E,F). Inhibiting TNF‐α in the medium from M1‐type THP‐1 cells prevented the upregulation of HLA‐DP and HLA‐DQ in tumor cells (Figure 6G,H). These results indicate that macrophages secreting TNF‐α upregulate HLA‐II expression in GBM cells.
FIGURE 6.
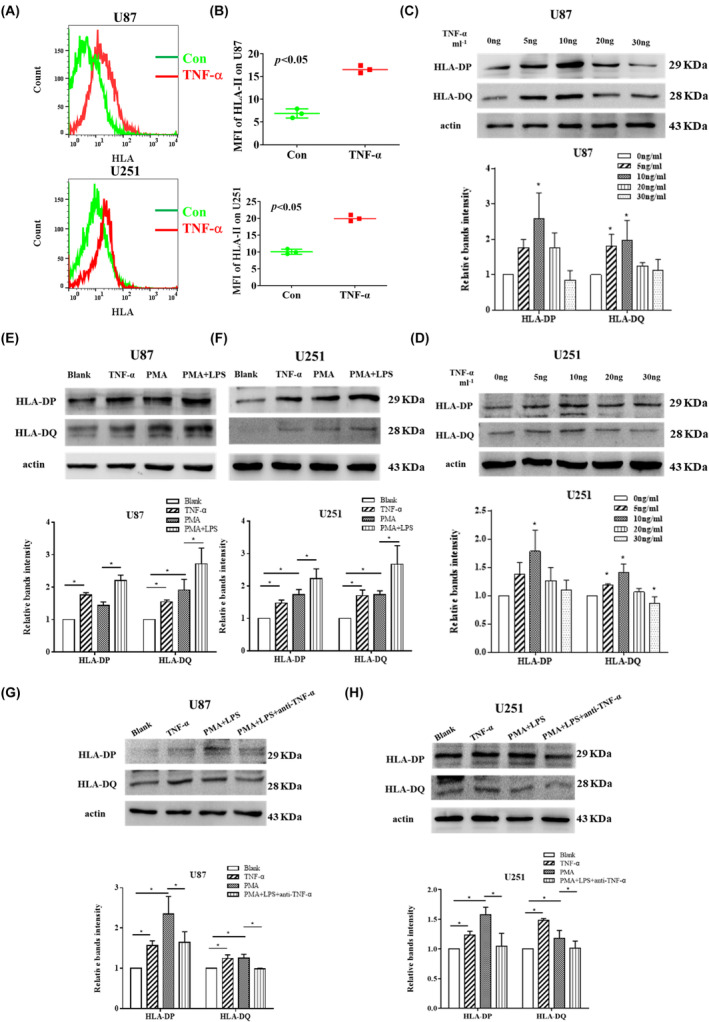
THP‐1 macrophages secreting tumor necrosis factor‐α (TNF‐α) upregulate the expression of human leukocyte antigen (HLA)‐DP and HLA‐DQ in U87 and U251 cells. (A, B) U87 and U251 cells were treated with 10 ng/mL TNF‐α, and the expression of HLA‐DP and HLA‐DQ was measured by flow cytometry. Data were shown as mean ± SD (n = 3, *p < 0.05 vs. Control [Con]). (C, D) U87 and U251 cells were treated with different doses of TNF‐α, and the expression of HLA‐DP and HLA‐DQ was measured by western blot analysis. Data are shown as mean ± SD (n = 3, *p < 0.05 vs. Con). (E, F) The supernatant from phorbol 12‐myristate 13‐acetate (PMA)‐induced THP‐1 (M0) and lipopolysaccharide (LPS)‐activated PMA‐induced THP‐1 (M1) was collected to stimulate U87 and U251 cells, and the expression of HLA‐DP and HLA‐DQ were measured by western blot analysis. Data were shown as mean ± SD (n = 3, *p < 0.05). (G, H) Anti‐TNF‐α neutralizing Ab was used to neutralize the TNF‐α in the supernatant, and the expression of HLA‐DP and HLA‐DQ were measured by western blot. Data are shown as mean ± SD (n = 3, *p < 0.05). The experiments were repeated three times. MFI, mean fluorescence intensity.
We established AN in vivo subcutaneous GBM tumor model using brain GL261 tumor cells, and treated the tumor‐bearing mice with sTNFR:Fc fusion protein to block TNF‐α (Figure S11A). We found sTNFR:Fc treatment significantly inhibited tumor growth (Figure S11B–D). Immunofluorescence staining analysis showed sTNFR:Fc decreased mouse MHC‐II (IAIE) expression in tumor tissues (Figure S11E,F). In addition, blocking TNF‐α also reduced CD4+LAG‐3+ cell infiltration in tumor tissues (Figure S11G,H). Therefore, the results support that TNF‐α‐dependent inflammation contributes to GBM progression by upregulating HLA‐II.
4. DISCUSSION
Given the extensive heterogeneity of glioma cells, it has proven difficult to develop small‐molecule inhibitors for GBM because blocking a single oncogenic signaling pathway might not be sufficient to induce tumor apoptosis. 28 The current understanding of the immune microenvironment of gliomas is gradually improving and the central nervous system immune system is no longer considered an “immune‐privileged” site. 29 , 30 Blockade of PD‐1 has shown significant efficacy in improving the survival of patients with melanoma brain metastases 31 ; however, ICI immunotherapy options for GBM require continued development, despite the failure of PD‐1 blockade in patients. 32 In terms of the diversity of ICI systems in GBM, several receptor–ligand systems, such as PD‐1–PD‐L1, TIM‐3–galectin‐9, and LAG‐3–MHC‐II, could act in combination to inhibit T cell‐mediated immunity. 28 Therefore, this study mainly focused on T cell–tumor cell interactions, LAG‐3–MHC‐II, in human GBM, which could reveal additional inhibitory strategies for immunotherapy.
The MHC‐II of APCs, such as macrophages, dendritic cells, or B cells, present antigenic peptides to CD4+ T cells, which are central to adaptive immune responses. 33 However, tumor cells with MHC‐II expression and lower levels of Th1 cytokines showed resistance to anti‐PD‐1. 21 This study supports that expression of MHC‐II in cancer cells, nonspecific APCs, are related to T cell exhaustion in the tumor environment. 34 Abnormal expression of HLA‐DR promoted the migration and invasion of melanoma cells 27 and caused tumor escape by recruiting inflammatory tumor antigen‐specific CD4+ T cells. 35 In this study, we analyzed the expression of HLA‐II based on TCGA and clinical specimens. Using immunofluorescence staining and FCM analysis, we clearly observed that HLA‐II expression was increased in GFAP+ GBM cells. Human leukocyte antigen II was highly expressed in human GBM and correlated with CD4+ T cell infiltration in the GBM microenvironment. High levels of HLA‐II in tumor cells have been associated with increased intratumoral CD4+ T cell density, whereas tumor MHC‐II‐stimulated CD4+ T cell subsets show dysfunctions in immunity. 36 , 37 Therefore, HLA‐II+ GBM might be related to CD4+ T cell exhaustion.
Lymphocyte‐activation gene 3, an early marker of T cell exhaustion, is expressed in TILs and tumor‐associated perivascular lymphocytes in human GBM. 16 The combination of anti‐LAG‐3 with PD‐1 inhibitor achieved better treatment outcomes in a mouse glioma model. 8 The inhibition of CD4+ T cell activation through LAG‐3 is dependent on stable pMHC‐II recognition. 8 Major histocompatibility complex‐II interacts with LAG‐3 through residues on the membrane‐distal region, the top face of the LAG‐3 D1 domain. 38 Mair et al. reported LAG‐3 was rarely expressed on TILs in human GBM by scoring cell density, 39 while Harris‐Bookman et al. observed higher expression of LAG‐3 in GBM, in which 66% of GBM samples showed variable LAG‐3 expression on perivascular lymphocytes. 16 In our study, LAG‐3, TIM‐3, and PD‐1 quantification was calculated by counting the number of positive lymphocytes per high‐powered field of view. Here, the infiltration of LAG‐3+, TIM‐3+, and PD‐1+ cells in GBM stroma was more than those in the controls. Lymphocyte‐activation gene 3 was mostly expressed in the GBM stroma, not in tumor cells or macrophages. In addition, increased CD4+LAG‐3+ T cell infiltration in GBM stroma was confirmed by LAG‐3 and CD4 colocalization in the tumor stroma by immunofluorescence staining, which are consistent with Harris‐Bookman et al.'s findings. Although LAG‐3 or MHC‐II expression was not associated with OS, the combination of HLA‐DQHigh and LAG‐3High was associated with poor OS in GBM patients. A previous study showed that MHC‐II+ tumors with increased LAG‐3 expression suppress MHC‐II‐mediated antigen presentation, which preferentially results in acquired resistance to PD‐1 therapy. 14 Therefore, interactions between LAG‐3 and MHC‐II might confer an immunosuppressive environment in GBM patients. Interference of the LAG‐3–MHC‐II interaction could be a potential therapeutic target for GBM.
T cell activation requires antigen recognition by MHC molecules binding to T cell receptors (signal 1), costimulatory signaling (signal 2), and cytokine priming (signal 3). 40 , 41 Cytokines have received more attention as important predictive biomarkers for prognosis and adverse reactions to ICI therapy. Immunosuppressive cytokines, such as IL‐10, PEG2, and TGF‐β, are produced by immune cells and tumor cells, impairing the efficiency of ICI therapy. 42 , 43 , 44 , 45 In this study, the upregulation of IL‐10, PEG2, and TGF‐β was observed in GBM. Interleukin‐10+Arg‐1+ macrophages were increased in GBM stroma compared to that in the controls, suggesting M2 macrophages were also an important producer of IL‐10 in GBM. Interleukin‐10 plays crucial immunosuppressive roles, promoting tumor progression and immune evasion in GBM. 44 Interleukin‐10 has broad immunosuppressive functions through the downstream STAT3 pathway and inhibits APC‐mediated T cell activation, directly causing CD4+ T cell anergy. 46 An IL‐10 inhibitor was shown to generate a T cell‐mediated carcinoma cell death, which was reversed by MHC‐II blockade, confirming the essential role of MHC‐II in IL‐10‐induced CD4+ T cell dysfunction in the tumor environment. 47 In this study, HLA‐II upregulation and increased number of LAG‐3+ cells in the GBM stroma were also positively correlated with IL‐10 upregulation in GBM, and the combination of IL‐10High and HLA‐IIHigh was associated with poor OS in patients. A combination of IL‐10 and IL‐35 has been shown to induce LAG‐3 expression in memory T cells, dampening the immune response. 48 Our study showed that an increased number of LAG‐3+ cells in the GBM stroma was also positively correlated with IL‐10 upregulation. Glioblastoma with HLA‐IIHighIL‐10High showed increased LAG‐3+ T cell infiltration, which synergistically led to poor prognosis in patients. Therefore, IL‐10 might be a strong factor in promoting MHC‐II–LAG‐3‐induced CD4+ T cell exhaustion. To show direct evidence that targeting of the LAG‐3–HLA‐II axis and IL‐10 could be beneficial in immunotherapy in GBM patients, we established an in vivo therapeutic model using anti‐LAG‐3 and anti‐IL‐10 Ab. We found combined anti‐LAG‐3 and IL‐10 treatment significantly inhibited tumor growth in tumor‐bearing mice. Furthermore, combined anti‐LAG‐3 and anti‐IL‐10 also reduced CD4+LAG‐3+ and CD8+LAG‐3+ cell infiltration in tumor tissues. Human leukocyte antigen‐II comprises multiple MHC‐II isotypes in humans, which are critical for antigen presentation to CD4+ T lymphocytes, not for CD8+ T cells. 40 , 41 Except for CD4+LAG‐3+ cells, we found CD8+LAG‐3+ cell infiltration was increased in human GBM tissues. However, the number of CD8+LAG‐3+ cells was not correlated with HLA‐II expression. Interleukin‐10, an important immunosuppressive cytokine, plays a critical role in CD8+ T cell exhaustion by promoting inhibitory receptor expression (PD‐1, TIM‐3, and LAG‐3). 49 Blocking LAG‐3–IL‐10 could inhibit CD4+ and CD8+ T cell exhaustion, which contributes to antitumor efficacy. Human leukocyte antigen‐II on the surface of tumor‐associated IL‐10 might be sufficient to induce CD4+ T cell suppression through LAG‐3 in GBM. Thus, targeting these pathways could be beneficial for improving the clinical outcome of GBM patients.
The immune infiltration patterns associated with HLA‐II expression in GBM were analyzed using ssGSEA. The expression of HLA‐II is positively correlated with macrophages and neutrophils. In GBM, the most abundant stromal cell type is macrophages, which can comprise more than 30% of infiltrating cells 50 , 51 ; the majority (80%) of GBM‐associated macrophages differentiates from recruited bone marrow macrophages. Macrophages adjust their phenotype and function according to the local microenvironment and participate in tumor development. 52 , 53 Increased macrophage recruitment to the tumor area induces epithelial cell activation by secreting TNF‐α, which contributes to tumor progression. 50 Serum levels of TNF‐α are significantly enhanced in patients with GBM. 54 Macrophages and microglia in GBM are also major producers of TNF‐α. 55 In the current study, we observed that TNF‐α is not only expressed in GBM tumor cells but also in CD68+ macrophages in the tumor stroma. We recently discovered that TNF‐α‐dependent lung inflammation increases MHC‐II expression in lung adenocarcinoma cells of AT‐II cellular origin, contributing to regulatory T cell expansion. 34 Tumor necrosis factor‐α can mediate aberrant expression of MHC‐II‐like molecules in melanoma cells, thereby limiting the tumor‐suppressive activity of CD4+ T cells. 35 In this study, HLA‐II expression in GBM was correlated with TNF‐α+CD68+ macrophage infiltration in the stroma. Furthermore, mononuclear THP‐1 cells secreted TNF‐α, which could increase HLA‐II expression in U87 and U251 cells in vitro. CD68+ macrophages upregulate HLA‐II expression in GBM cells through TNF‐α. In order to address the contribution of TNF‐α in GBM progression, we established an in vivo therapeutic model using sTNFR:Fc to neutralize TNF‐α. 34 We found sTNFR:Fc treatment significantly inhibited tumor growth. In addition, blocking TNF‐α decreased MHC‐II expression in mice GBM tumor cells, and reduced CD4+LAG‐3+ cell infiltration. These results indicate that TNF‐α upregulating HLA‐II expression contributes to GBM progression.
In conclusion, we found that HLA‐II and IL‐10 were highly expressed in GBM and correlated with increased CD4+LAG‐3+ cell infiltration in the stroma. Patients with HLA‐II+IL‐10+ associated with increased LAG‐3+ T cell infiltration synergistically have poor prognosis. In addition, combined anti‐LAG‐3 and anti‐IL‐10 treatment inhibited tumor growth in a mouse brain GL261 tumor model. CD68+ macrophages in the tumor microenvironment upregulated HLA‐II expression in GBM cells through TNF‐α. Blocking TNF‐α‐dependent inflammation inhibited tumor growth in a mouse GBM model. T cell–tumor cell interactions, LAG‐3–HLA‐II, could confer an immunosuppressive environment in GBM patients. Interleukin‐10 could be a strong factor that promotes MHC‐II–LAG‐3‐induced CD4+ T cell exhaustion in GBM, leading to poor prognosis (Figure 7). This study provides new insights into targeting the LAG‐3–MHC‐II interaction that could be beneficial for improving the clinical outcomes of GBM patients.
FIGURE 7.
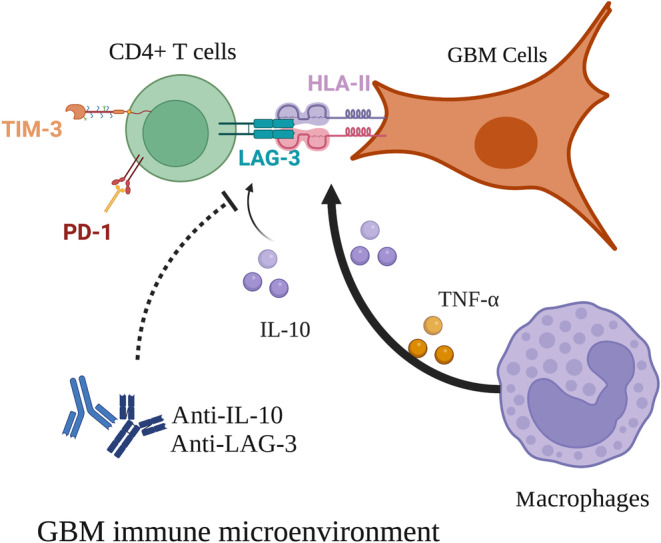
Macrophages in the tumor microenvironment upregulated human leukocyte antigen‐II (HLA‐II) expression in glioblastoma (GBM) cells through tumor necrosis factor‐α (TNF‐α). HLA‐II interacted with lymphocyte‐activation gene 3 (LAG‐3) confers an immunosuppressive environment in GBM. Interleukin‐10 (IL‐10) could be a strong factor that promotes HLA‐II–LAG‐3‐induced CD4+ T cell exhaustion in GBM, leading to poor prognosis. PD‐1, programmed cell death protein 1; TIM‐3, T cell immunoglobulin domain and mucin domain‐3.
AUTHOR CONTRIBUTIONS
Wenli Guo: Data curation; investigation; methodology; writing – original draft. Daijun Peng: Conceptualization; data curation; investigation; methodology; writing – original draft. Yuee Liao: Formal analysis; methodology. Lei Lou: Formal analysis; methodology. Moran Guo: Formal analysis; methodology. Chen Li: Formal analysis; resources. Wangyang Yu: Formal analysis; software. Xiaoxi Tian: Formal analysis; methodology. Guohui Wang: Formal analysis; software. Ping Lv: Conceptualization; investigation. Jing Zuo: Conceptualization; investigation. Haitao Shen: Conceptualization; data curation; funding acquisition; investigation; project administration; supervision; writing – original draft; writing – review and editing. Yuehong Li: Conceptualization; funding acquisition; investigation; project administration; supervision; writing – original draft; writing – review and editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
Approval of the research protocol by an institutional review board: The study of human sample was approved by the Ethics Committee of Hebei Medical University, the Second Hospital, Hebei, China.
Informed consent: Informed consent was obtained from all patients in accordance with a protocol approved by the ethics review board of Hebei Medical University (approval no. 2019‐R260). Written informed consent was obtained from each subject.
Registry and the registration no. of the study/trial: N/A.
Animal studies: Experimental protocols were approved by the Institutional Animal Care and Use Committee of Hebei Medical University. All animals were managed and treated according to the guidelines of Hebei Medical University.
Supporting information
Figure S1.
Appendix S1.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31570894; 81670939; 81672706), Natural Science Foundation of Hebei Province, China (H2019206709; H2021206351; H2022206551), and Science and Technology Program of Hebei Province, China (216Z7707G; 216Z7701G).
Guo W, Peng D, Liao Y, et al. Upregulation of HLA‐II related to LAG‐3+CD4+ T cell infiltration is associated with patient outcome in human glioblastoma. Cancer Sci. 2024;115:1388‐1404. doi: 10.1111/cas.16128
Wenli Guo and Daijun Peng contributed equally to this work.
Contributor Information
Haitao Shen, Email: haitaoshen78@hotmail.com.
Yuehong Li, Email: liyuehong1993@126.com.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article and its additional files. The analyzed datasets and used materials in the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: state of the art and future directions[J]. CA Cancer J Clin. 2020;70(4):299‐312. [DOI] [PubMed] [Google Scholar]
- 2. Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti‐PD‐1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma[J]. Nat Med. 2019;25(3):477‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors[J]. Cancer Cell. 2017;31(3):326‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu W, Wang W, Li H, et al. Single‐cell atlas reveals complexity of the immunosuppressive microenvironment of initial and recurrent glioblastoma[J]. Front Immunol. 2020;11:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma[J]. Nat Rev Neurol. 2018;14(8):482‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion[J]. Nat Rev Immunol. 2015;15(8):486‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurachi M. CD8(+) T cell exhaustion[J]. Semin Immunopathol. 2019;41(3):327‐337. [DOI] [PubMed] [Google Scholar]
- 8. Maruhashi T, Okazaki IM, Sugiura D, et al. LAG‐3 inhibits the activation of CD4(+) T cells that recognize stable pMHCII through its conformation‐dependent recognition of pMHCII[J]. Nat Immunol. 2018;19(12):1415‐1426. [DOI] [PubMed] [Google Scholar]
- 9. Maruhashi T, Sugiura D, Okazaki IM, Okazaki T. LAG‐3: from molecular functions to clinical applications[J]. J Immunother Cancer. 2020;8(2):e001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huard B, Prigent P, Tournier M, Bruniquel D, Triebel F. CD4/major histocompatibility complex class II interaction analyzed with CD4‐ and lymphocyte activation gene‐3 (LAG‐3)‐Ig fusion proteins[J]. Eur J Immunol. 1995;25(9):2718‐2721. [DOI] [PubMed] [Google Scholar]
- 11. Wang J, Sanmamed MF, Datar I, et al. Fibrinogen‐like protein 1 is a major immune inhibitory ligand of LAG‐3[J]. Cell. 2019;176:334‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ascierto PA, Melero I, Bhatia S, et al. Initial efficacy of anti‐lymphocyte activation gene‐3 (anti–LAG‐3; BMS‐986016) in combination with nivolumab (nivo) in pts with melanoma (MEL) previously treated with anti–PD‐1/PD‐L1 therapy [M]. Am Soc Clin Oncol. 2017;35:9520. [Google Scholar]
- 13. Duhoux FP, Jager A, Dirix LY, et al. Combination of paclitaxel and a LAG‐3 fusion protein (eftilagimod alpha), as a first‐line chemoimmunotherapy in patients with metastatic breast carcinoma (MBC): final results from the run‐in phase of a placebo‐controlled randomized phase II [M]. Am Soc Clin Oncol. 2018;36:1050. [Google Scholar]
- 14. Johnson DB, Nixon MJ, Wang Y, et al. Tumor‐specific MHC‐II expression drives a unique pattern of resistance to immunotherapy via LAG‐3/FCRL6 engagement[J]. JCI Insight. 2018;3(24):e120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews LP, Somasundaram A, Moskovitz JM, et al. Resistance to PD1 blockade in the absence of metalloprotease‐mediated LAG3 shedding[J]. Sci Immunol. 2020;5(49):eabc2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harris‐Bookman S, Mathios D, Martin AM, et al. Expression of LAG‐3 and efficacy of combination treatment with anti‐LAG‐3 and anti‐PD‐1 monoclonal antibodies in glioblastoma[J]. Int J Cancer. 2018;143(12):3201‐3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lecocq Q, Keyaerts M, Devoogdt N, Breckpot K. The next‐generation immune checkpoint LAG‐3 and its therapeutic potential in oncology: third Time's a charm[J]. Int J Mol Sci. 2020;22(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson J, Waller MJ, Fail SC, et al. The IMGT/HLA database[J]. Nucleic Acids Res. 2009;37:D1013‐D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Lith M, Mcewen‐Smith RM, Benham AM. HLA‐DP, HLA‐DQ, and HLA‐DR have different requirements for invariant chain and HLA‐DM[J]. J Biol Chem. 2010;285(52):40800‐40808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma[J]. J Clin Oncol. 2018;36(10):942‐950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson AM, Bullock BL, Neuwelt AJ, et al. Cancer cell‐intrinsic expression of MHC class II regulates the immune microenvironment and response to anti‐PD‐1 therapy in lung adenocarcinoma[J]. J Immunol. 2020;204(8):2295‐2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson DB, Estrada MV, Salgado R, et al. Melanoma‐specific MHC‐II expression represents a tumour‐autonomous phenotype and predicts response to anti‐PD‐1/PD‐L1 therapy[J]. Nat Commun. 2016;7:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maclachlan BJ, Mason GH, Greenshields‐Watson A, et al. Molecular characterization of HLA class II binding to the LAG‐3 T cell co‐inhibitory receptor[J]. Eur J Immunol. 2021;51(2):331‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hemon P, Jean‐Louis F, Ramgolam K, et al. MHC class II engagement by its ligand LAG‐3 (CD223) contributes to melanoma resistance to apoptosis[J]. J Immunol. 2011;186(9):5173‐5183. [DOI] [PubMed] [Google Scholar]
- 25. Souri Z, Wierenga APA, Kroes WGM, et al. LAG3 and its ligands show increased expression in high‐risk uveal melanoma[J]. Cancers (Basel). 2021;13(17):4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagasaki J, Togashi Y, Sugawara T, et al. The critical role of CD4+ T cells in PD‐1 blockade against MHC‐II‐expressing tumors such as classic Hodgkin lymphoma[J]. Blood Adv. 2020;4(17):4069‐4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Costantini F, Barbieri G. The HLA‐DR mediated signalling increases the migration and invasion of melanoma cells, the expression and lipid raft recruitment of adhesion receptors, PD‐L1 and signal transduction proteins[J]. Cell Signal. 2017;36:189‐203. [DOI] [PubMed] [Google Scholar]
- 28. Marx S, Godicelj A, Wucherpfennig KW. A conceptual framework for inducing T cell‐mediated immunity against glioblastoma[J]. Semin Immunopathol. 2022;44(5):697‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang K, Wu Z, Zhang H, et al. Glioma targeted therapy: insight into future of molecular approaches[J]. Mol Cancer. 2022;21(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application[J]. Cancer Lett. 2020;476:1‐12. [DOI] [PubMed] [Google Scholar]
- 31. Bander ED, Yuan M, Carnevale JA, et al. Melanoma brain metastasis presentation, treatment, and outcomes in the age of targeted and immunotherapies[J]. Cancer. 2021;127(12):2062‐2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reardon DA, Brandes AA, Omuro A, et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the CheckMate 143 phase 3 randomized clinical trial[J]. JAMA Oncol. 2020;6(7):1003‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Unanue ER, Turk V, Neefjes J. Variations in MHC class II antigen processing and presentation in health and disease[J]. Annu Rev Immunol. 2016;34:265‐297. [DOI] [PubMed] [Google Scholar]
- 34. Guo N, Wen Y, Wang C, et al. Lung adenocarcinoma‐related TNF‐α‐dependent inflammation upregulates MHC‐II on alveolar type II cells through CXCR‐2 to contribute to Treg expansion[J]. FASEB J. 2020;34(9):12197‐12213. [DOI] [PubMed] [Google Scholar]
- 35. Donia M, Andersen R, Kjeldsen JW, et al. Aberrant expression of MHC class II in melanoma attracts inflammatory tumor‐specific CD4+ T‐ cells, which dampen CD8+ T‐cell antitumor reactivity[J]. Cancer Res. 2015;75(18):3747‐3759. [DOI] [PubMed] [Google Scholar]
- 36. Senosain MF, Zou Y, Novitskaya T, et al. HLA‐DR cancer cells expression correlates with T cell infiltration and is enriched in lung adenocarcinoma with indolent behavior[J]. Sci Rep. 2021;11(1):14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer S, Handke D, Mueller A, et al. Distinct molecular mechanisms of altered HLA class II expression in malignant melanoma[J]. Cancers (Basel). 2021;13(15):3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baixeras E, Huard B, Miossec C, et al. Characterization of the lymphocyte activation gene 3‐encoded protein. A new ligand for human leukocyte antigen class II antigens[J]. J Exp Med. 1992;176(2):327‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mair MJ, Kiesel B, Feldmann K, et al. LAG‐3 expression in the inflammatory microenvironment of glioma[J]. J Neuro‐Oncol. 2021;152(3):533‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han Y, Xie W, Song D‐G, Powell DJ Jr. Control of triple‐negative breast cancer using ex vivo self‐enriched, costimulated NKG2D CAR T cells[J]. J Hematol Oncol. 2018;11(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qiao Y, Qiu Y, Ding J, et al. Cancer immune therapy with PD‐1‐dependent CD137 co‐stimulation provides localized tumour killing without systemic toxicity[J]. Nat Commun. 2021;12(1):6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang M, Zhai X, Li J, et al. The role of cytokines in predicting the response and adverse events related to immune checkpoint inhibitors[J]. Front Immunol. 2021;12:670391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bertrand F, Montfort A, Marcheteau E, et al. TNFalpha blockade overcomes resistance to anti‐PD‐1 in experimental melanoma[J]. Nat Commun. 2017;8(1):2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Widodo SS, Dinevska M, Furst LM, Stylli SS, Mantamadiotis T. IL‐10 in glioma[J]. Br J Cancer. 2021;125:1466‐1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee JM, Tsuboi M, Kim ES, Mok TSK, Garrido P. Overcoming immunosuppression and pro‐tumor inflammation in lung cancer with combined IL‐1beta and PD‐1 inhibition[J]. Future Oncol. 2022;18(27):3085‐3100. [DOI] [PubMed] [Google Scholar]
- 46. Saraiva M, Vieira P, O'Garra A. Biology and therapeutic potential of interleukin‐10[J]. J Exp Med. 2020;217(1):e20190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sullivan KM, Jiang X, Guha P, et al. Blockade of interleukin 10 potentiates antitumour immune function in human colorectal cancer liver metastases[J]. Gut. 2022;72:325‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koga MM, Engel A, Pigni M, et al. IL10‐ and IL35‐secreting MutuDC lines act in cooperation to inhibit memory T cell activation through LAG‐3 expression[J]. Front Immunol. 2021;12:607315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sawant DV, Yano H, Chikina M, et al. Adaptive plasticity of IL‐10+ and IL‐35+ Treg cells cooperatively promotes tumor T cell exhaustion[J]. Nat Immunol. 2019;20(6):724‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei Q, Singh O, Ekinci C, et al. TNFalpha secreted by glioma associated macrophages promotes endothelial activation and resistance against anti‐angiogenic therapy[J]. Acta Neuropathol Commun. 2021;9(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gabrusiewicz K, Rodriguez B, Wei J, et al. Glioblastoma‐infiltrated innate immune cells resemble M0 macrophage phenotype[J]. JCI Insight. 2016;1(2):e85841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS‐) vs. alternatively activated macrophages[J]. Front Immunol. 2019;10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dan H, Liu S, Liu J, et al. RACK1 promotes cancer progression by increasing the M2/M1 macrophage ratio via the NF‐κB pathway in oral squamous cell carcinoma[J]. Mol Oncol. 2020;14(4):795‐807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Albulescu R, Codrici E, Popescu ID, et al. Cytokine patterns in brain tumour progression[J]. Mediat Inflamm. 2013;2013:979748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meisen WH, Wohleb ES, Jaime‐Ramirez AC, et al. The impact of macrophage‐ and microglia‐secreted TNFalpha on oncolytic HSV‐1 therapy in the glioblastoma tumor microenvironment[J]. Clin Cancer Res. 2015;21(14):3274‐3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Appendix S1.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files. The analyzed datasets and used materials in the current study are available from the corresponding author on reasonable request.


