Abstract
Anti‐programmed death‐ligand 1 (PD‐L1) Ab‐based therapies have demonstrated potential for treating metastatic urothelial cancer with high PD‐L1 expression. Urinary exosomes are promising biomarkers for liquid biopsy, but urine's high variability requires normalization for accurate analysis. This study proposes using the PD‐L1/Alix ratio to normalize exosomal PD‐L1 signal intensity with Alix, an internal exosomal protein less susceptible to heterogeneity concerns than surface protein markers. Extracellular vesicles were isolated using ExoDisc and characterized using various methods, including ExoView to analyze tetraspanins, PD‐L1, and Alix on individual exosomes. On‐disc ELISA was used to evaluate PD‐L1 and Alix‐normalized PD‐L1 in 15 urothelial cancer patients during the initial treatment cycle with Tecentriq. Results showed that Alix signal range was relatively uniform, whereas tetraspanin marker intensity varied for individual exosome particles. On‐disc ELISA was more reliable for detecting exosomal PD‐L1 expression than standard plate ELISA‐based measurement. Using exosomal Alix expression for normalization is a more reliable approach than conventional methods for monitoring patient status. Overall, the study provides a practical and reliable method for detecting exosomal PD‐L1 in urine samples from patients with urothelial cancer.
Keywords: exosome, normalization, PD‐L1, urine, urothelial cancer
This paper suggests using exosomal Alix as a reliable normalization method for detecting programmed death‐ligand 1 (PD‐L1) in urine. The study shows that on‐disc ELISA can accurately detect exosomal PD‐L1 without surface immobilization. This innovative approach addresses the limitations of traditional normalization methods and offers practical solutions for identifying urinary biomarkers and improving diagnostic and therapeutic approaches for urothelial cancer patients.
Abbreviations
- CCS
cell cultured supernatant
- CT
computed tomography
- EV
extracellular vesicle
- HD
healthy donor
- NTA
nanoparticle tracking analysis
- PD‐1
programmed death‐1
- PD‐L1
programmed death‐ligand 1
- PNUH
Pusan National University Hospital
- RT
room temperature
- TMB
3,3′,5,5′‐tetramethylbenzidine
- UC
urothelial cancer
1. INTRODUCTION
PD‐L1, which is an immunosuppressive transmembrane protein, is upregulated on tumor cells, enabling them to evade immune surveillance by interacting with programmed death‐1 (PD‐1) receptors on immune cells. Anti‐PD‐L1 Ab‐based therapy (MPDL3280A) inhibits the interaction of PD‐L1 with PD‐1 and B7.1 (CD80). The therapy was examined for the treatment of metastatic urothelial bladder cancer, which has high PD‐L1 expression in immunohistochemistry results, and reportedly led to better prognosis and tolerance than conventional chemotherapy. 1
Recently, EVs have emerged as a biomarker for liquid biopsies. 2 , 3 These vesicles, which range in size from 40 to 1000 nm, are enclosed by a lipid bilayer and secreted by most cell types for intercellular communications. Biogenesis of EVs is not fully understood, while the biogenetic regulation of the small size of EVs, so‐called exosomes, depends on Alix‐mediated signaling. 4 These exosomes are generated and secreted through the endosomal‐sorting complex required for transport machinery. In the sorting machinery, Alix has been known to have a role for packaging cargo to enter vesicles and trigger vesicle formation. 4 Interestingly, Alix was reported as a regulator for tumor‐mediated immunosuppression by controlling PD‐L1 presentation. 5 Compared to microvesicle PD‐L1 and free soluble PD‐L1, exosomal PD‐L1 isolated from plasma led to a better correlation with anti‐PD‐1 response and contribution as a biomarker for prediction of anti‐PD‐1. 6 , 7
As exosomes are prevalent in most body fluids, we hypothesized that the exosomes isolated from urine are a reliable source for PD‐L1 detection in UC patients. Urinary EVs have also gained interest in bladder cancer biomarker investigations due to their direct contact with bladder cancer and their potential to contain large amounts of EVs from the tumor environment. 3 , 8 , 9 Several studies have reported the presence of PD‐L1 in urine samples and its correlation with the development and prediction of UC. 10 , 11 Although promising, unlike other biofluids, urine can show considerable variability, which can lead to inconsistency during the analysis. 12 , 13 Changes in glomerular filtration rate can affect the concentration of solutes in urine, leading to variations in urine osmolarity. To normalize urinary biomarkers, creatinine or osmolarity‐based analysis has been evaluated. 14 , 15 Nevertheless, utilizing osmolarity for normalization might not be the optimal choice as it reflects the concentration of solutes in the sample rather than the biomarker of interest. 16 In the context of normalization, the essential aspect is to assess changes in the target molecule's concentration relative to the internal standard of the biomarker.
Here, we present a normalization method using exosomal Alix for the detection of PD‐L1 on exosomes isolated from the urine of UC patients. Compared to other surface protein markers, Alix, being an internal protein of the exosome, is more likely to face steric hindrance limiting the binding of labeling Abs. Therefore, we hypothesized that Alix would show a more uniform intensity on individual exosome particles than other surface protein markers, such as PD‐L1 and tetraspanins. For this, we isolated small EVs from the 0.2 μm filtered cell culture supernatant and urine. It has been observed in multiple research studies that these small EVs show PD‐L1 enrichment on their surface. 6 , 17 , 18 We suggest using Alix, an internal exosomal protein, to normalize PD‐L1 signal intensity in exosomes. This approach would enable consistent assessment of exosomal PD‐L1 expression in urine samples, despite the potential variations in sample concentration due to factors such as dehydration or lifestyle patterns.
Moreover, the measurement of exosomal PD‐L1 through standard plate ELISA could vary depending on the choice of exosome capture marker selected to detect PD‐L1. 17 To address the challenges, researchers have reported the development of an anti‐PDL1 immuno‐biochip utilizing a label‐free detection method such as surface plasmon resonance (SPR) and total internal reflection fluorescence (TIRF). 19 , 20 , 21 However, this approach remains challenged by the limited surface area of the plate and unavailability of a method for measuring the expression level of PD‐L1 on each vesicle. In contrast, on‐disc ELISA allows for measuring exosomal PD‐L1 expression without a limited binding surface area. This approach has three distinctive advantages:
Unrestricted by surface limitations. Instead of capturing EVs using anti‐PD‐L1 or tetraspanin markers, small EVs are captured in the chamber, and the intensity of exosomal PD‐L1 is measured. Increasing the target number is crucial because it has been reported that a significantly smaller proportion of abundant proteins is detected at the single EV level, highlighting the scarcity of tumor‐defining biomarkers and inequality between the total number of particles and number of target molecules. 22
Quantifying total PD‐L1 intensity. This allows for the measurement of both the overall PD‐L1 intensity in terms of the input sample volume and the normalization of these measurements to determine PD‐L1 expression per vesicle. Assessing the level of PD‐L1 expression is crucial, as changes in exosomal PD‐L1 expression can potentially diagnose cancer and predict treatment response. 6 , 23 , 24
Label‐free capture. This approach eliminates the need for selecting an Ab pair to capture and detect exosomes. As the detection domain of Abs and their correlated therapies are varied, 25 , 26 the different combinations of PD‐L1 capture and detection will lead to different results. 27 This study also examined clinical samples by analyzing urinary exosomes from UC patients and showed that exosomal PD‐L1 could be a potentially useful biomarker for UC diagnostics and treatment monitoring.
2. MATERIALS AND METHODS
2.1. Cell lines and culture for EV production
The cell lines used in this study (UMUC3, T24, RWPE, and A549) were obtained from ATCC. To produce EVs from the cell culture, 2 × 106 cells were seeded in 100 mm cell culture dishes and incubated for 2 days in Advanced RPMI (Thermo Fisher Scientific) supplemented with glutamine, 0.1% exo‐free FBS (Thermo Fisher Scientific) for UMUC3, T24, and A549, or keratinocyte serum‐free medium (K‐SFM) for RWPE and 1% antibiotics/antimycotics under 5% CO2 at 37°C. Cell cultured supernatant solutions were collected and centrifuged at 300 g for 10 min, and supernatants at 2000 g for 10 min. The clear supernatants were stored at −80°C until analysis.
2.2. Clinical samples
Urine samples were collected from UC patients at PNUH (Table S1). The study protocol was reviewed and approved by the Institutional Review Board of PNUH (IRB 1702‐008‐051). To check the background level of PD‐L1, urine samples from healthy volunteers were included. Written informed consent was obtained from all subjects. Ten milliliters of the collected urine samples was centrifuged at 500 g for 10 min to remove cellular debris and stored at −80°C until analysis. To isolate exosomes, urine samples were then thawed and centrifuged at 2500 g for 15 min, and the clear supernatants were used immediately for the EV isolation step.
In this study, we analyzed a total of 38 urine samples, which included 26 samples from cancer patients and 12 from healthy donors. Among 26 patient samples, 15 had both pretreatment and T1 samples available, and these 15 samples were specifically used for the treatment monitoring analysis. Due to limited sample volumes, our single exosome analysis focused exclusively on urine samples obtained from four HD and 14 UC patients, utilizing ExoView, as illustrated in Figure S1e,f. The pearson's r correlation between PD‐L1 and exosomal markers (CD9, CD63, CD81, and Alix) were analyzed from five cancer patients and five healthy donors.
2.3. Exosome isolation
To analyze the expression level of PD‐L1, exosomes were isolated using ExoDisc containing a filter with pore size of 20 nm. In brief, after the prefiltration step using a filter with a pore size of 0.22 μm (Acrodisc 25 mm syringe filter, product number 4612; Pall Corporation), 0.5–4 mL of clear supernatant of CCS or urine was injected onto the disc. The concentrated EVs were washed with 0.1 μm prefiltered PBS and used for subsequent analysis.
2.4. Nanoparticle tracking analysis
For quantitative measurement of EV amount, isolated EVs were diluted in 0.1 μm prefiltered PBS to obtain the recommended concentration (25–100 particles/frame) for NTA measurement (Nanosight NS500; Malvern Instruments). Experiment videos were analyzed using the NTA 3.1 software (Malvern). All NTA measurements were carried out under identical conditions for consistent results.
2.5. Enzyme‐linked immunosorbent assay
For standard curves to calculate the arbitrary amount, recombinant human CD9 (cat#11029‐H08H; Sino Biological), CD63 (cat#11271‐H08H; Sino Biological), CD81 (cat#14244‐H07H; Sino Biological), Alix (cat#TP303735; Origene), and PD‐L1 (cat#156‐B7; R&D Systems) were loaded on 96‐well plates. After washing with 200 μL of 0.1% BSA in PBS, plates were blocked with 200 μL/well of 1% BSA for 2 h at RT. After washing with 200 μL of 0.1% BSA in PBS, 50 μL/well of detection Abs (anti‐CD9, HI9A, biotin‐conjugated, cat#312112, BioLegend; anti‐CD63, H5C6, biotin‐conjugated, cat#353017, BioLegend; anti‐CD81, JS‐81, cat555675, BD Biosciences; anti‐PD‐L1, MIH1, biotin‐conjugated, cat#13‐5983‐82, Invitrogen; and anti‐Alix, cat#ab76608, Abcam) in PBS buffer (500 ng/mL) was loaded and incubated at RT for 1 h. After washing twice with 200 μL of 0.1% BSA in PBS, the plate was incubated with 50 μL/well of HRP‐conjugated streptavidin (cat#DY998; R&D Systems) in PBS (1:500) or anti‐rabbit‐HRP in PBS (1:10,000) at RT for 30 min. Plates were washed thoroughly three times with 200 μL of 0.1% BSA in PBS, then 50 μL/well of TMB solution was added and incubated at RT for 30 min. Reactions were stopped by adding 50 μL/well of stop solution, and the absorbance of each well was measured at 450 nm/570 nm on a plate reader (Tecan Group).
2.6. On‐disc ELISA
To analyze the total expression of CD9, CD63, CD81, Alix, and PD‐L1 in a designated volume of sample, on‐disc ELISA was conducted using ExoDisc. The procedures for on‐disc ELISA were further optimized in our previous study. 28 In brief, the sample chamber and filter chamber were introduced by adding 1 mL of 1% BSA and spinning at 3000 rpm for 30 s. Then, all surfaces containing 1% BSA were blocked in an incubated shaker (100 rpm) (Lab Companion; JeioTech) at 37°C for 1 h, followed by washing with 1 mL PBS by spinning the disc at 3000 rpm for 5 min. The clear supernatant of CCS and urine was injected into the disc, and concentrated EVs were washed with 1 mL PBS. To label the internal proteins of EVs, EVs were fixed and permeabilized under 4% of paraformaldehyde at RT for 20 min and 0.2% Triton X at RT for 10 min. The concentrated EVs were incubated on a disc with a solution of detection Abs (anti‐CD9, HI9A, biotin‐conjugated, cat#312112, BioLegend; anti‐CD63, H5C6, biotin‐conjugated, cat#353017, BioLegend; anti‐CD81, JS‐81, cat555675, BD Biosciences; anti‐PD‐L1, MIH1, biotin‐conjugated, cat#13‐5983‐82, Invitrogen; and anti‐Alix, cat#ab76608, Abcam) in PBS (500 ng/mL) at 37°C for 10 min in an incubated shaker (100 rpm). After washing twice with 1 mL PBS, the EVs were incubated with secondary Abs (HRP‐conjugated streptavidin [1:500], or anti‐rabbit‐HRP [1:10,000]) in 0.1% BSA in PBS at 37°C for 10 min in an incubated shaker (100 rpm). After washing twice with 1 mL PBS, the concentrated and labeled EVs were eluted in 100 μL PBS. Fifty microliters of samples was loaded in 96‐well plates and incubated with 100 μL TMB at 37°C for 15 min in an incubated shaker (100 rpm). Reactions were stopped by adding 50 μL/well of stop solution, and the absorbance of each well was measured at 450 nm/570 nm on a plate reader (Tecan Group).
2.7. ExoView
The experimental details are given in Appendix S1.
3. RESULTS
3.1. Normalization using exosomal Alix expression
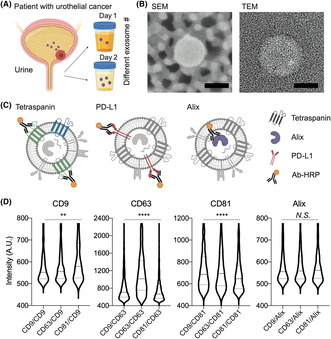
Precise monitoring of urinary exosomal PD‐L1 relies on proper normalization of PD‐L1 expression, as the number of exosomes present in a urine sample can fluctuate on a daily basis, as depicted in Figure 1A. With the properly normalized PD‐L1 expressions, it enables an evaluation of the effectiveness of treatments and tracks disease progression.
FIGURE 1.
Normalization method using exosomal Alix expression. (A) Exosomes (20–200 nm) are secreted from urothelial cancer (UC) and exist in urine in different numbers depending on the condition of the patient. (B) Exosomes were isolated and detected in scanning electron microscope (SEM) and transmission electron microscope (TEM) images. Scale bar, 50 nm. (C) Internal markers such as Alix may have less space for Ab binding compared to surface markers. (D) Intensity analysis of single exosomes for their surface tetraspanin markers (e.g. CD9, CD63, and CD81) and Alix. The intensity of tetraspanin markers (CD9, CD63, and CD81) of particles, isolated from T24 CCS using ExoDisc, attached on the surface coated with CD9, CD63, and CD81 showed varying intensity ranges, whereas Alix showed a uniform intensity range. One‐way AVOVA test was used for statistical analysis. *p < 0.05; **p < 0.005; ****p < 0.0001; A.U., arbitrary unit; N.S., not significant; PD‐L1, programmed death‐ligand 1.
Here, we isolated exosomes smaller than 200 nm from the urine of UC patients to limit the size range of exosomes. The scanning electron microscope and transmission electron microscope images represent the exosomes isolated from the ExoDisc (Figure 1B).
Exosomes isolated using ExoDisc were characterized for their tetraspanin content by examining the positive particles for CD9, CD63, and CD81 using ExoView. The bright field reflection images for the attached particles on the coated surfaces are shown in Figure S1A. The size of isolated exosomes was measured using NTA (Figure S1B) and ExoView (Figure S1C), with major size ranges varying between 100–200 nm and 50–100 nm, respectively. The tetraspanin‐positive exosomes were counted, and CD9+ exosomes were found to be the most prevalent, while CD81+ exosomes were the least prevalent in the exosomes isolated from urothelial cell lines such as RWPE, UMUC3, and T24. Conversely, CD81+ exosomes were the most prevalent in A549, a lung cancer cell line, while CD9+ exosomes were the least prevalent (Figure S1D). It is important to note that the tetraspanin expression type can vary depending on the exosome's origin, making it difficult to select them as a normalizing marker. Furthermore, we isolated exosomes from urine (n = 18) using ExoDisc. It showed the highest population of CD9+ exosomes and the lowest population of CD81+ exosomes (Figure S1E). We further compared PD‐L1 colocalization from the exosomes isolated from urine samples of healthy donors (n = 4) and UC patients (n = 14) (Figure S1F). Interestingly, EVs showing dual positivity, CD9+ and PD‐L1+, could distinguish between UC patients and healthy donors. However, the effectiveness of discrimination was not as notable when EVs with PD‐L1 colocalized with CD63 or CD81 were used (Figure S1F). Although promising, we approximated that the signal intensity of surface tetraspanins, such as CD9, would differ on each exosome.
Here, we hypothesized that the Alix would have a more uniform expression level than other surface protein markers due to their limited space for Ab attachment inside of the small size of exosomes (<200 nm). The proposed hypothesis suggests that Alix could be a dependable normalization factor for exosome measurements. This is crucial in accurately evaluating patients' status, particularly when exosome quantities might fluctuate due to conditions such as urine (Figure 1C). To test this hypothesis, we measured the intensity of individual particles for CD9, CD63, CD81, PD‐L1, and Alix using ExoView (Figures 1D and S2). It should be noted that fixation and permeabilization procedures are carried out before measuring Alix. Using a tetraspanin marker (CD9, CD63, or CD81) coated plate, the intensity of single exosomes was measured for CD9, CD63, CD81, PD‐L1, and Alix. While the surface marker intensity, including CD9, CD63, CD81, and PD‐L1, showed a broad range of intensities for individual particles, Alix, an internal protein marker, displayed a relatively uniform intensity. This finding suggests that Alix is a more stable marker for exosome normalization compared to other surface markers.
3.2. Characterization of exosomal PD‐L1, Alix, and tetraspanin expression
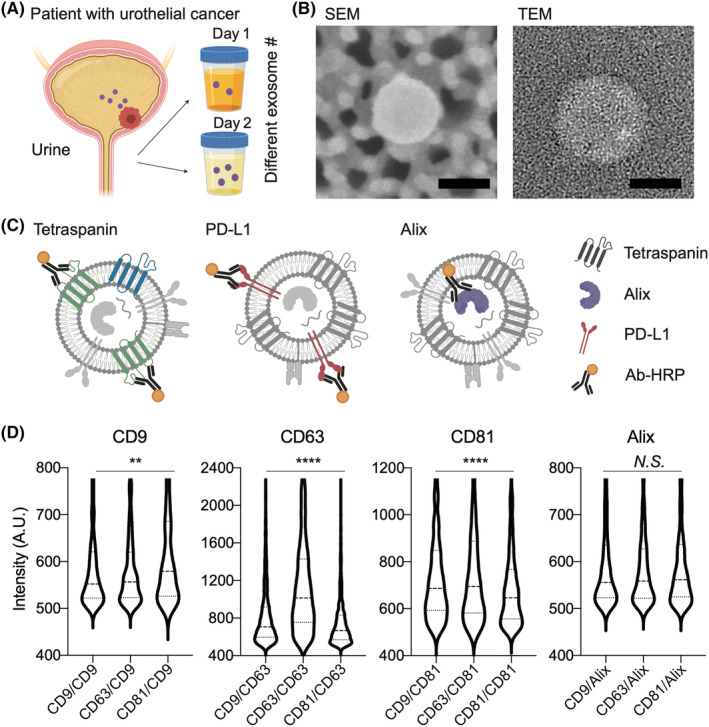
Exosomes were isolated from urine samples of UC patients and analyzed for the expression of PD‐L1, Alix, or tetraspanins using ExoDisc. The isolated exosomes are labeled in each chamber with one of the Abs for tetraspanins (e.g. CD9, CD63, or CD81), PD‐L1, or Alix, and an excess of Abs was washed on the ExoDisc with the protocols we previously reported (Figure 2A). 28 , 29 Alix labeling required fixation and permeabilization procedures after isolation due to its internal localization in exosomes. The quantification of exosomal PD‐L1 was verified using different input volumes of CCS from two cell types: UMUC3, which has high PD‐L1 expression, and A549, which has low PD‐L1 expression (Figure 2B). To compare the expression levels of exosomal markers and PD‐L1, the expression of tetraspanin, PD‐L1, and Alix was measured individually in 2 mL CCS for UMUC3 and A549 (Figure 2C). Despite the high expression of tetraspanin markers, the expressions of PD‐L1 and Alix were relatively low. Furthermore, the expressions of these markers were evaluated in exosomes isolated from 250 μL urine samples from HD and UC patients, and the results showed a higher expression level of PD‐L1 in UC patients compared to HD (Figure 2D).
FIGURE 2.
Exosomal marker correlation with programmed death‐ligand 1 (PD‐L1) to identify a normalizing marker for PD‐L1. (A) Exosomes were isolated from cell culture supernatant (CCS) or urine by using ExoDisc. Following isolation, exosomes were labeled with individual Abs of tetraspanins (CD9, CD63, or CD81), PD‐L1, or Alix on the filter chamber of the disc, and their expression levels were quantified. (B) Exosomes isolated from different volumes of A549‐ and UMUC3‐CCS solutions were quantified for PD‐L1 expression using on‐disc ELISA. (C, D) On‐disc ELISA results show CD9, CD63, CD81, Alix, and PD‐L1 expression levels in exosomes isolated from (C) 2 mL of A549‐ and UMUC3‐CCS solution, as well as (D) 250 μL of urine from healthy donors (HD) and patients with urothelial cancer (UC). Error is denoted as SD. (E) PD‐L1 correlation tests for the clinical samples (HD, n = 5; UC, n = 5). There was a weak positive correlation between PD‐L1 and CD9, with Pearson's r value of 0.13. However, no significant correlation was observed between PD‐L1 and CD63 (Pearson's r = 0.014) or CD81 (Pearson's r = −0.17). In contrast, Alix showed a strong positive correlation with PD‐L1, with a Pearson's r value of 0.95. Pearson's r value for the correlation analysis were provided by Prism software. A.A., arbitrary amount; A.U., arbitrary unit; conc., concentration; OD, optical density.
In addition, we investigated the correlation between PD‐L1 and exosomal markers (CD9, CD63, CD81, and Alix) in 10 urine samples (Figure 2E). Pearson's correlation coefficient analysis revealed a small positive correlation between CD9 and PD‐L1 with a value of r = 0.13, while CD63 and CD81 showed no correlation with PD‐L1, with r values of 0.014 and −0.17, respectively. However, Alix showed a significant positive correlation with PD‐L1 with a Pearson's r value of 0.95, indicating that Alix can be used for normalizing PD‐L1 expressions.
3.3. Urinary exosome analysis reveals higher concentration of PD‐L1 and Alix particles in UC patients compared to HD
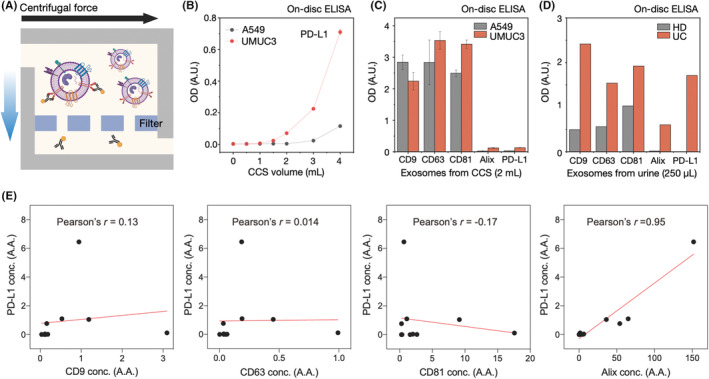
Exosomes were isolated from the urine of HD (n = 12) and UC patients (n = 26) using ExoDisc. On‐disc ELISA was carried out to measure the expression of PD‐L1 and Alix proteins, and NTA was used to measure the number of exosomes. The results showed a higher concentration of PD‐L1 and Alix positive particles in exosomes isolated from UC patients than those isolated from HD (Figure 3A).
FIGURE 3.
Exosomal programmed death‐ligand 1 (PD‐L1) and Alix expression were quantified from extracellular vesicles isolated from clinical samples (healthy donors [HD], n = 12; urothelial cancer patients [UC], n = 26). (A) Exosomes in 1 mL urine were isolated using ExoDisc, and their exosomal PD‐L1 and Alix expression levels were analyzed with the corresponding particle number measured by nanoparticle tracking analysis (NTA). Relative quantification was calculated by standard curves and marked as an arbitrary amount (A.A.). (B) Correlation analysis between PD‐L1 and Alix expression shows a strong positive correlation with a Pearson's correlation coefficient of 0.96. (C) Pearson's correlation coefficient of 0.28 indicates a weak positive correlation between PD‐L1 and the number of particles measured by NTA. conc., concentration.
However, no significant difference was observed in the number of particles measured by NTA (Figure S3). The relative quantification was calculated using standard curves (Figure S4) and presented as an arbitrary amount (A.A.). Like Figure 2E, a positive correlation between PD‐L1 expression and Alix positive particles was observed with Pearson's r = 0.96 (Figure 3B). Additionally, we examined the correlation between PD‐L1 expression and the number of particles and found a weak correlation with Pearson's r = 0.28 (Figure 3C).
We further examined the correlation between exosomal PD‐L1 concentration and immune cell score of PD‐L1 (Figure S5). Exosomal PD‐L1 was quantified from EVs isolated from clinical samples (HD, n = 12; UC, n = 19, excluding tissue data from unavailable patients [n = 7] from total patients [n = 26]). Exosomal PD‐L1 was detected in all cohort patients, while only 47% of tumor tissues were positive for PD‐L1.
3.4. Exosomal PD‐L1/Alix ratio as a potential biomarker for monitoring Tecentriq treatment in UC
During the initial stage of treatment with Tecentriq, the urine of UC patients (n = 15) was collected, and their exosomal PD‐L1, Alix, and PD‐L1/Alix ratio levels were quantified. The patients were classified into responders and nonresponders based on their disease stability, 30 and their exosomal PD‐L1/Alix ratio levels were compared. Patients were categorized as responders if they showed stable disease, indicating that there was a change of less than 20% in the sum of diameters of tumor lesions on follow‐up CT imaging carried out 3 months after the initiation of Tecentriq treatment. Patients were classified as nonresponders if there was an increase of at least 20% in the sum of diameters of tumor lesions or if they had died as a result of cancer.
Among the 15 patients, six showed stable disease, while nine showed a recurrence, progressive disease, or death caused by cancer (Figures 4A,B, S6, and S7). An increasing PD‐L1/Alix ratio rate was found in the responder group (p‐value = 0.0312), while a single PD‐L1 intensity pattern had no correlation (p = 0.4219). Similarly, a decreasing PD‐L1/Alix ratio rate was found in the nonresponder group (p = 0.0371), while a single PD‐L1 intensity pattern had no correlation (p = 0.3262). With the small patient cohort due to the limited number of patients under Tecentriq for UC treatment, we still found the ratio of PD‐L1/Alix ratio is a more suitable biomarker than PD‐L1 or Alix expression alone for monitoring the response of Tecentriq treatment in UC. Note that the patient marked as NR1 in Figure 4B includes CT images for pretreatment, posttreatment, and progression of UC of the urinary bladder (Figure S7). This patient initially showed no interval change of tumor size after T1 but the progression of disease later (we put this patient in the nonresponder group), while the other patients in the responder/nonresponder groups had no changes. Representative CT images of a responder and nonresponder are included in Figure 4C,D, and the CT images of the other patients were shown in Figures S6 and S7. The responder showed a slightly reduced tumor size after 3 months (Figure 4C), while the nonresponder showed an increased tumor size after 1 month (Figure 4D).
FIGURE 4.
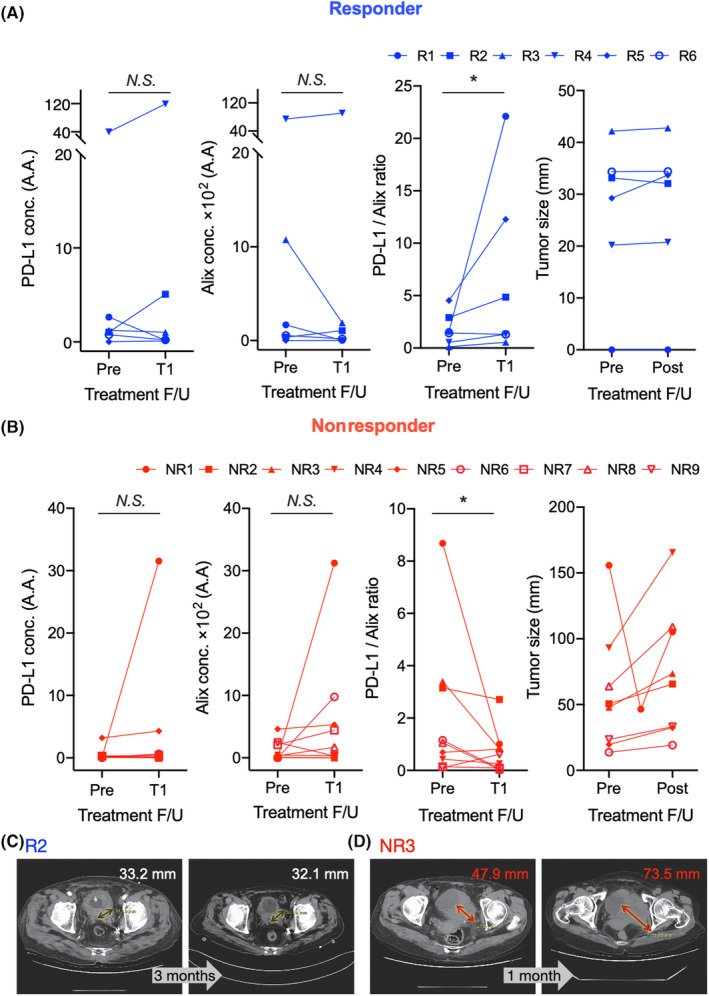
During the initial treatment cycle of Tecentriq in 15 patients with urothelial cancer, programmed death‐ligand 1 (PD‐L1), Alix, and the PD‐L1/Alix ratio of exosomes were quantified. Tumor size changes were observed for all patients. (A) Of the 15 patients, six showed stable disease from December 17, 2019 to the present (responder [R]1), from July 22, 2020 to the present (R2), from August 19, 2020 to the present (R3), from November 9, 2020 to the present (R4), from August 14, 2020 to the present (R5), and from July 16, 2020 to the present (R6). (B) Of the 15 patients, nine experienced recurrence (nonresponder [NR]1), progressive disease (NR4, NR5, NR6, NR8, and NR9), or cancer‐related death (NR2, NR3, and NR7 (computed tomography [CT] image not available due to cancer‐related death before obtaining follow‐up CT). During the first cycle of Tecentriq treatment, responders showed nonsignificant (N.S.) changes in the concentrations of PD‐L1 and Alix, while the PD‐L1/Alix ratio increased (*). Nonresponders showed an opposite trend of PD‐L1/Alix ratio (*) during the same period. (C) Representative CT image of responders (R2) showing the slightly reduced tumor size from 33.2 mm to 32.1 mm after 3 months. (D) Representative CT image of nonresponders (NR3) showing the increased tumor size from 47.9 to 73.5 mm after 1 month. Wilcoxon test was used for statistical analysis. *p < 0.05; A.A., arbitrary amount; N.S., not significant; conc., concentration; F/U, follow‐up.
We further undertook a detailed analysis of urine samples collected during the long‐term follow‐up of a patient (R4) undergoing PD‐L1 Ab therapy (Figure S8). At the third treatment cycle (after 9 weeks), the PD‐L1/Alix ratio reached its peak. However, the PD‐L1 concentration showed fluctuating patterns, alternating between decreases and increases, that could potentially be attributed to fluctuations in exosome levels. Similar observations have been reported in other studies, 6 , 17 showing a large increase in exosomal PD‐L1 levels between 3 and 6 weeks after the initial treatment, suggesting a successful elicitation of antitumor immunity, typically observed in responders to the treatment. Conversely, nonresponders tended to experience notable tumor progression approximately 4.5 months posttreatment, leading to an increase in the secretion levels of exosomal PD‐L1. 17 , 31
4. DISCUSSION
Measuring biomarkers in urine can be challenging due to difficulties in normalization. Conventional normalization methods (e.g., creatinine amount measurement, collection time, urine specific gravity) 16 rely on the urine osmolality parameter rather than the internal biomarker standards. Here, we suggest an Alix‐based exosome normalization method to quantify the changes of exosomal PD‐L1 in urine. Combining the on‐disc ELISA method, this approach quantifies the expression of PD‐L1 in a total injected volume and the ratio of PD‐L1 expression to Alix‐positive exosomes. We further found that only Alix has uniform intensity on each exosome particle while other tetraspanins were not, indicating that other surface markers are difficult to apply as normalization markers. We then exploited different cell lines to examine their exosomal tetraspanin profile, resulting in their different profiles depending on the original cell types. Both varied intensity profiles of tetraspanins and varied populations depending on the cell type (e.g., high CD9 population in bladder cell lines [RWPE, UMUC3, T24] but high CD81 population in lung cancer cell line A549) made an extra challenge in their normalization to select tetraspanin as normalization marker.
Here, we found that exosome from urine of UC patients is a reliable source for PD‐L1 detection. Furthermore, the increasing rate of PD‐L1/Alix was found in responders (n = 6), while the decreasing rate of PD‐L1/Alix was found in nonresponders (n = 9) during the initial stage of the Tecentriq treatment. It is worthwhile to highlight that the PD‐L1/Alix ratio resulted in significant differences, while a single measurement of PD‐L1 was not. This is the first report to use Alix, an internal marker in exosomes, as a normalizing parameter for exosomal PD‐L1 expression, enabling the precise monitoring of exosomal PD‐L1 in the urine of UC patients. The result is also consistent with the report from Chen et al., 6 as they mentioned a higher increment of plasma exosomal PD‐L1 during the initial stage of anti‐PDL1 Ab treatment. While our findings are promising, we must acknowledge that our investigation only involved 15 UC patients due to the limited insurance coverage for Tecentriq treatment. Therefore, further investigation with a larger number of patients is necessary to determine the clinical role of urinary exosomal PD‐L1 in monitoring treatment response.
AUTHOR CONTRIBUTIONS
Hyun‐Kyung Woo: Conceptualization; data curation; formal analysis; investigation; methodology; software; validation; visualization; writing – original draft. Juhee Park: Data curation; formal analysis; investigation; validation. Kyung Hwan Kim: Data curation; investigation; resources; writing – original draft. Ja Yoon Ku: Data curation; investigation; resources. Hong Koo Ha: Investigation; methodology; project administration; resources; supervision; writing – review and editing. Yoon‐Kyoung Cho: Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; visualization; writing – review and editing.
FUNDING INFORMATION
This work was mainly supported by a grant from the Institute for Basic Science of Korea (grant no. IBS‐R020‐D1).
CONFLICT OF INTEREST STATEMENT
UNIST has filed patents on ExoDisc, and Yoon‐Kyoung Cho is named as an inventor, which are licensed to LabSpinner, Inc. Yoon‐Kyoung Cho is a stockholder of LabSpinner, Inc. No potential conflicts of interest were disclosed by the other authors.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Review Board: The biospecimens and data used for this study were provided by the Biobank of Pusan National University Hospital, a member of the Korea Biobank Network, which is supported by the Ministry of Health and Welfare (PNUH IRB 1702‐008‐051).
Informed Consent: Written informed consent has been obtained from all subjects.
Registry and the Registration No. of the study/trial: N/A.
Animal Studies: N/A.
Supporting information
Appendix S1
Figures S1–S8
Table S1
ACKNOWLEDGMENTS
We express our gratitude to all patients and healthy volunteers who provided urine samples for this study.
Woo H‐K, Park J, Kim KH, Ku JY, Ha HK, Cho Y‐K. Alix‐normalized exosomal programmed death‐ligand 1 analysis in urine enables precision monitoring of urothelial cancer. Cancer Sci. 2024;115:1602‐1610. doi: 10.1111/cas.16106
Hyun‐Kyung Woo, Juhee Park, and Kyung Hwan Kim contributed equally to this work.
Contributor Information
Hong Koo Ha, Email: hongkooha@pusan.ac.kr, Email: hongkooha@naver.com.
Yoon‐Kyoung Cho, Email: ykcho@unist.ac.kr.
REFERENCES
- 1. Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti‐PD‐L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558‐562. [DOI] [PubMed] [Google Scholar]
- 2. Zhou B, Xu K, Zheng X, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Urabe F, Kimura T, Ito K, et al. Urinary extracellular vesicles: a rising star in bladder cancer management. Transl Androl Urol. 2021;10(4):1878‐1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baietti MF, Zhang Z, Mortier E, et al. Syndecan‐syntenin‐ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677‐685. [DOI] [PubMed] [Google Scholar]
- 5. Monypenny J, Milewicz H, Flores‐Borja F, et al. ALIX regulates tumor‐mediated immunosuppression by controlling EGFR activity and PD‐L1 presentation. Cell Rep. 2018;24(3):630‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen G, Huang AC, Zhang W, et al. Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature. 2018;560(7718):382‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li C, Zhi C, Liang W, et al. Clinical significance of PD‐L1 expression in serum‐derived exosomes in NSCLC patients. J Transl Med. 2019;17(1):355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu YR, Ortiz‐Bonilla CJ, Lee YF. Extracellular vesicles in bladder cancer: biomarkers and beyond. Int J Mol Sci. 2018;19(9):2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomiyama E, Matsuzaki K, Fujita K, et al. Proteomic analysis of urinary and tissue‐exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. 2021;112(5):2033‐2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tosev G, Wahafu W, Reimold P, et al. Detection of PD‐L1 in the urine of patients with urothelial carcinoma of the bladder. Sci Rep. 2021;11(1):14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vikerfors A, Davidsson S, Frey J, Jerlström T, Carlsson J. Soluble PD‐L1 in serum and urine in urinary bladder cancer patients. Cancers (Basel). 2021;13(22):5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blijdorp CJ, Tutakhel OAZ, Hartjes TA, et al. Comparing approaches to normalize, quantify, and characterize urinary extracellular vesicles. J Am Soc Nephrol. 2021;32(5):1210‐1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erdbrügger U, Blijdorp CJ, Bijnsdorp IV, et al. Urinary extracellular vesicles: a position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J Extracell Vesicles. 2021;10(7):e12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waikar SS, Sabbisetti VS, Bonventre JV. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010;78(5):486‐494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mervant L, Tremblay‐Franco M, Jamin EL, et al. Osmolality‐based normalization enhances statistical discrimination of untargeted metabolomic urine analysis: results from a comparative study. Metabolomics. 2021;17(1):2. [DOI] [PubMed] [Google Scholar]
- 16. Gunasekaran PM, Luther JM, Byrd JB. For what factors should we normalize urinary extracellular mRNA biomarkers? Biomol Detect Quantif. 2019;17:100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu ZL, Liu JY, Chen G. Small extracellular vesicle PD‐L1 in cancer: the knowns and unknowns. NPJ Precis Oncol. 2022;6(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Li J, Zhang R, et al. Suppression of PD‐L1 release from small extracellular vesicles promotes systemic anti‐tumor immunity by targeting ORAI1 calcium channels. J Extracell Vesicles. 2022;11(12):e12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Kannisto E, Yu G, Reid ME, Patnaik SK, Wu Y. An immuno‐biochip selectively captures tumor‐derived exosomes and detects exosomal RNAs for cancer diagnosis. ACS Appl Mater Interfaces. 2018;10(50):43375‐43386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu C, Zeng X, An Z, et al. Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens. 2018;3(8):1471‐1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J, Zhu Y, Guan M, et al. Isolation of circulating exosomes and identification of exosomal PD‐L1 for predicting immunotherapy response. Nanoscale. 2022;14(25):8995‐9003. [DOI] [PubMed] [Google Scholar]
- 22. Ferguson S, Yang KS, Weissleder R. Single extracellular vesicle analysis for early cancer detection. Trends Mol Med. 2022;28(8):681‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang M, Yang J, Wang T, et al. Homogeneous, low‐volume, efficient, and sensitive quantitation of circulating exosomal PD‐L1 for cancer diagnosis and immunotherapy response prediction. Angew Chem Int Ed Engl. 2020;59(12):4800‐4805. [DOI] [PubMed] [Google Scholar]
- 24. de Miguel‐Perez D, Russo A, Arrieta O, et al. Extracellular vesicle PD‐L1 dynamics predict durable response to immune‐checkpoint inhibitors and survival in patients with non‐small cell lung cancer. J Exp Clin Cancer Res. 2022;41(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahoney KM, Sun H, Liao X, et al. PD‐L1 antibodies to its cytoplasmic domain most clearly delineate cell membranes in immunohistochemical staining of tumor cells. Cancer Immunol Res. 2015;3(12):1308‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Powles T, Smith K, Stenzl A, Bedke J. Immune checkpoint inhibition in metastatic urothelial cancer. Eur Urol. 2017;72(4):477‐481. [DOI] [PubMed] [Google Scholar]
- 27. Cha YJ, Kim D, Bae SJ, et al. PD‐L1 expression evaluated by 22C3 antibody is a better prognostic marker than SP142/SP263 antibodies in breast cancer patients after resection. Sci Rep. 2021;11(1):19555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Woo HK, Sunkara V, Park J, et al. ExoDisc for rapid, size‐selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017;11(2):1360‐1370. [DOI] [PubMed] [Google Scholar]
- 29. Woo HK, Park J, Ku JY, et al. Urine‐based liquid biopsy: non‐invasive and sensitive AR‐V7 detection in urinary EVs from patients with prostate cancer. Lab Chip. 2018;19(1):87‐97. [DOI] [PubMed] [Google Scholar]
- 30. Hodi FS, Ballinger M, Lyons B, et al. Immune‐modified response evaluation criteria in solid tumors (imRECIST): refining guidelines to assess the clinical benefit of cancer immunotherapy. J Clin Oncol. 2018;36(9):850‐858. [DOI] [PubMed] [Google Scholar]
- 31. Cordonnier M, Nardin C, Chanteloup G, et al. Tracking the evolution of circulating exosomal‐PD‐L1 to monitor melanoma patients. J Extracell Vesicles. 2020;9(1):1710899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Figures S1–S8
Table S1


