Abstract
Mesenchymal stem cell (MSC) differentiation can be manipulated by nanotopographic interface providing unique strategy to engineering stem cell therapy and circumvent complex cellular reprogramming. However, our understanding of the nanotopographic-mechanosensitive properties of MSCs and the underlying biophysical linkage of the nanotopography-engineered stem cell to directed commitment, remains elusive. Here, we show that osteogenic differentiation of human MSCs (hMSCs) can be largely promoted using our nanoengineered topographic glass substrates in the absence of dexamethasone, a key exogenous factor for osteogenesis induction. We demonstrate that hMSCs sense and respond to surface nanotopography, through modulation of adhesion, cytoskeleton tension and nuclear activation of TAZ (transcriptional coactivator with PDZ-binding motif), a transcriptional modulator of hMSCs. Our findings demonstrate the potential of nanotopographic surfaces as non-invasive tools to advance cell-based therapies for bone engineering and highlight the origin of biophysical response of hMSC to nanotopography.
Keywords: nanotopography, human mesenchymal stem cell, mechanosensitive, osteogenesis, TAZ
Human mesenchymal stem cells (MSCs) in bone marrow present significant plasticity by giving rise to a variety of stromal lineages, including osteoblastic (bone) lineage, and have been targeted to advance cell-therapies for bone engineering.1 Success of bone engineering related cell therapies using MSCs rely on an efficient induction of osteogenesis. However, failure of implants happens due to the high plasticity of MSCs which results in the production of soft tissues instead of bone.2,3 Therefore, precise control and high-efficient promotion of MSC differentiation towards osteogenic lineage in the stem cell niche is a key factor for the success of bone engineering. Recent studies for stem cell engineering have investigated the effect of nanotopographic cue on stem cell proliferation and differentiation.1,2,4–16 For instance, disordered nanopit arrays2 made of polycarbonates and titanium oxide nanotube4 have been observed to promote early osteogenesis of hMSCs without any addition of osteogenic inducing factors. In vivo, stem cells interact with, and interrogate their surroundings at the nanometer length scale. At the cell-extracellular matrix (ECM) interface, both in vivo ECM and cell membrane are enriched with adhesive molecules and ligands with spatial organizations and characteristic dimensions ranging from a few nanometers, to hundreds of nanometers.17 Therefore, novel biomaterials that can encode specific nanoscale biological cues offer unique opportunities for engineering hMSC functions.
Furthermore, understanding the mechanisms that control the hMSC osteogenesis is essential to develop new bone engineering related cell therapies. While researchers are still striving to identify the appropriate soluble chemical environment for hMSC culture and differentiation, it is clear that physical microenvironment in stem cell niche also influences those processes. However, the current research interests have largely focused on ECM rigidity and force related stem cell mechanobiology. For example, stiff substrate and shear stress were investigated to promote MSC osteogenesis and chondrogenesis through activating the Hippo/YAP pathway,18,19 respectively. Soft substrates have been shown to lead to a substantial increase in the active form of β1 integrin and a reduction in the cell surface distribution of β1 integrin in MSCs by enhancing the detachment of integrin-ECM protein complexes, thus induces neurogenic fate of MSC.20 The mechanism studies exploring the influence of the topography-relayed mechanosensing and transduction remains suboptimal.
Previous studies using the combination of osteogenic media and nanostructured culture substrates to accelerate osteogenesis and bone formation of MSCs in vitro, obscure the roles of the nanostructured surface topography by itself during the differentiation process.6 And those attempts of stimulating MSC osteogenesis in the absence of any osteogenic supplements were either not fully satisfied in tissue engineering or failed to determine the topography-relayed mechanosensitive mechanisms that mimic the well-documented functions of biochemical factors4. In the current osteogenic differentiation protocol, a cocktail of three key morphogens, dexamethasone (DEX), ascorbic acid (AS), and β-glycerophosphate (β-GP), is the standard treatment for the osteogenic induction of MSCs. The regulatory function of each chemical factor in this osteogenic cocktail has been broadly explored.21 It is, therefore, possible that the topography-relayed mechanosensing can replace or work synergistically with these morphogens to promote hMSCs toward osteogenesis. We assume that nanotopography-relayed stem cell signaling could share a similar molecular machinery with the osteogenic morphogens. Indeed, the recent studies on mechanisms of the osteogenic cocktail suggested regulatory effects on ECM adhesive proteins22–24 and hMSC transcriptional activation25,26 by DEX, AS and β-GP in the cocktail which might also be activated by nanotopography signaling.
Here, using a recently developed, large-scale nanofabrication technique based on reactive-ion etching (RIE),12,13 we generated random nanoscale structures (quantified as nanoroughness) on glass surfaces with high precision and reproducibility to regulate the stem cell behaviors. We demonstrate that nanoscale surface roughness can replace the osteogenic inducing factor DEX and work synergistically with the other two soluble morphogen components to promote hMSCs toward osteoblast linage. Studies conducted on the nanotopography-relayed mechanism revealed a mechanotransductive process involving cell adhesion, cytoskeleton tension, and TAZ (transcriptional coactivator with PDZ-binding motif) activation. These results suggest that nanotopography is an important biophysical cue in stimulating hMSC osteogenesis and nanotopographic substrate can thus serve as a promising osteoinductive material in the development of “cell-instructive” scaffolds for bone tissue engineering.
RESULTS & DISCUSSION
Nanotopography enhances hMSC osteogenesis
Nanotopographic surfaces and scaffolds have been developed for in vitro stem cell research.27–30 However, previous patterned nanoscale structures may not fully recapitulate the intrinsic random features of nanotopography in the in vivo cell microenvironment. In addition, some of the nanoengineering tools and fabrication methods are either complex or costly, such as electron beam and nanoimprint lithography.1,2,27,28,31,32 Here, we utilized a recently developed, large-scale nanofabrication technique based on RIE to generate random nanoscale structures on glass surfaces with high precision and reproducibility (Figure 1a & Figure S1).33 The nanoroughness was quantitatively characterized using atomic force microscope (AFM) as the root mean square (RMS) roughness Rq. The influence of nanotopographic cue on hMSC behaviors was assessed by using fibronectin-coated glass surfaces with a broad range of nanoscale roughness (Rq = 1 – 200 nm). AFM surface characterization further confirmed that the nanoroughness Rq of unprocessed smooth (with Rq = 1 nm) and nanorough glass surfaces did not significantly change (± 3 nm) before and after fibronectin coating.33 Detailed surface characterization also confirmed that the density of fibronectin absorbed on glass surfaces was independent of nanoroughness Rq.33
Figure 1. Nanotopographic substrate promotes hMSC early osteogenesis.
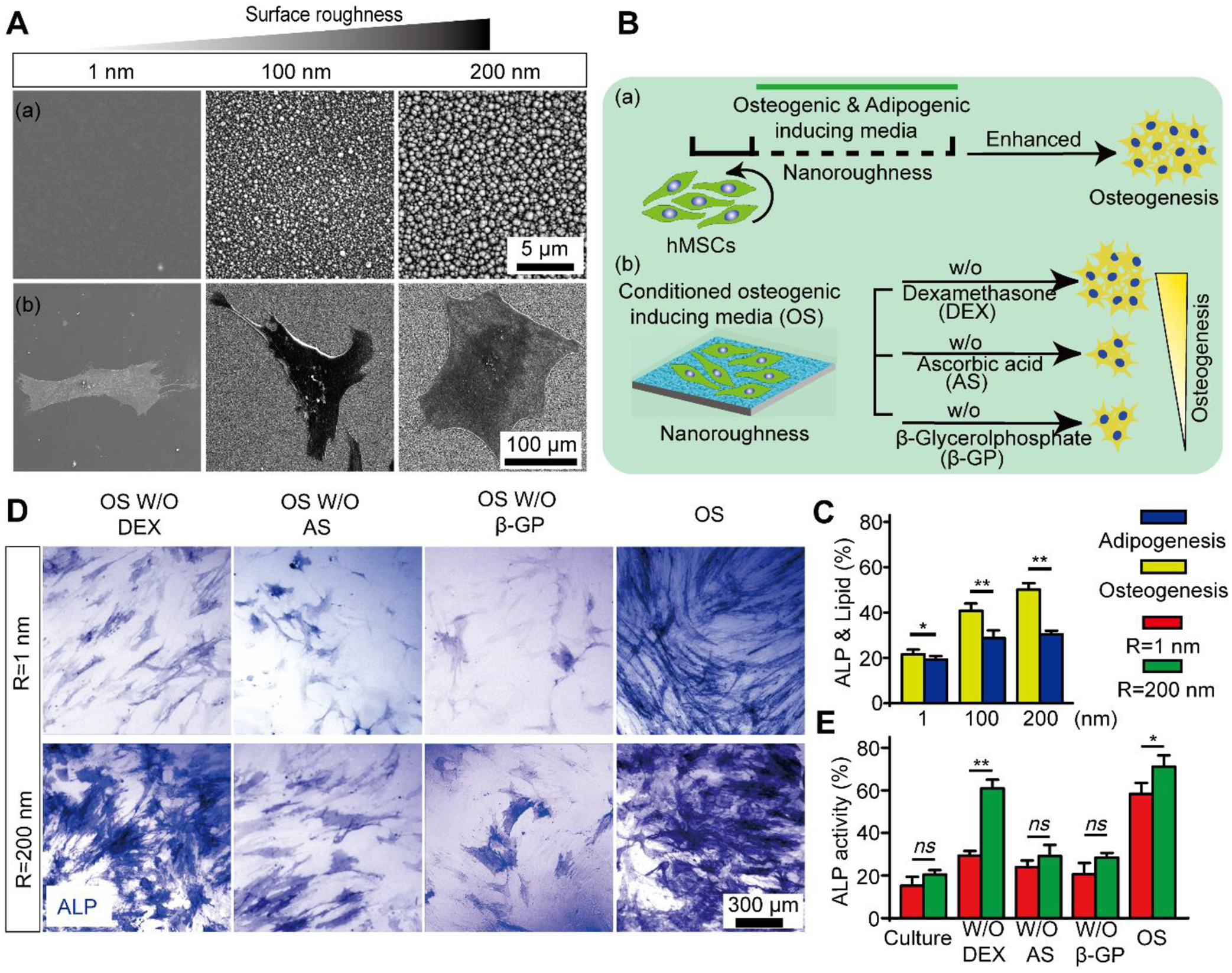
(A) Representative SEM images (a) showing nanotopography of different roughness (Rq = 1 nm, 100 nm, and 200 nm) and (b) hMSC cultured on smooth (Rq = 1 nm) and nanorough (Rq = 100 nm and 200 nm) glass surfaces as indicated. (B) Schematic diagram showing experimental design of hMSC lineage induction. (a) hMSCs were cultured for 7 days in osteogenic and adipogenic induction media on smooth (Rq = 1 nm) and nanorough (Rq = 100 nm and 200 nm) glass surfaces, respectively; (b) hMSCs were cultured in conditioned osteogenic induction media without inductive factors DEX, AS, or β-GP, respectively. (C) Quantifications of osteogenesis and adipogenesis based on ALP and oil-red lipid staining for hMSCs cultured in osteogenic or adipogenic differentiation media for 7 days, respectively. (D) Representative ALP staining and (E) quantifications of osteogenesis for hMSC differentiation in conditioned osteogenic media on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces for 14 days as indicated. Data represent the mean ± standard error of the mean (s.e.m.) with n ≥ 3. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
We firstly confirmed the biocompability of our nanorough glass substrates for hMSCs (Figure S2). Cell proliferation assays using both cell growth and osteogenic induction media indicate that while cells proliferated on both smooth (Rq = 1 nm) and nanorough (Rq = 100, 200 nm) surfaces, the cell proliferation rate (indicated by the percentage of Ki67+ cells34 and number increase in the DAPI-stained nucleus) is higher on the smooth substrates than the nanorough substrates, suggesting that the proliferation rate of hMSCs was compromised due to culture on nanorough substrates.
The high-plasticity hMSCs can give rise to a variety cell lineages, including osteoblastic, adipogenic, myoblastic, chondrogenic, and fibroblastic lineages.35,36 Directing hMSCs toward osteoblasts instead of connective tissue cell types is essential for the success of bone tissue engineering. To investigate the regulatory effect of the nanotopographic cue on the hMSC fate decision, we applied either osteogenic or adipogenic differentiation medium to hMSCs on glass surfaces with different nanoroughness (Fig. 1B). Specifically, hMSCs were firstly seeded on fibronectin-coated glass surfaces with different roughness and cultured in osteogenic or adipogenic differentiation medium for one week. Alkaline phosphatase (ALP) activity and formation of lipid droplets (Lip) were used to characterize osteogenic and adipogenc differentiation degree of hMSCs, respectively. Results show that after a 7-day differentiation, the ALP activity in hMSCs increased with an increase in substrate roughness, with ~21.6%, ~40.8%, and ~50.1% ALP positive cells on substrates with Rq = 1 nm, 100 nm, and 200 nm, respectively (Fig. 1C& Figure S3). However, there is no significant enhancement in Lip formation of hMSCs on different substrates (Rq = 1 nm: ~19.2%, Rq = 100 nm: ~28.5%, and Rq = 200 nm: 30.4%) (Fig. 1D & Figure S3). The significant difference between osteogenic and adipogenic differentiation on nanorough glass indicates that nanorough glass surface can promote osteogenic differentiation over adipogenic linage of hMSCs.
We further explored the regulatory effect of nanoroughness on topographical induction of hMSC osteogenesis to determine which molecular signal was triggered in nanoroughness-promoted osteogenesis. First, we investigated which morphogen-relayed signaling was activated by the nanoscale topographic cue to promote hMSC osteogenic linage. In doing so, we induced hMSC osteogenic differentiations on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) substrates for 14 days under different conditioned osteogenic media in absence of each key morphogens, DEX, AS, or β-GP, respectively (Fig. 1D & E). Osteogenic-inductive medium including DEX, AS, or β-GP was found to regulate ECM adhesion proteins22–24 and activate several transcriptions25,26 in MSCs, that further induce MSC osteogenesis. Surprisingly, our experiment demonstrated that nanorough substrate (Rq = 200 nm) significantly promoted ALP expression (~60%) compared to the cells on smooth substrates (Rq = 1 nm; ~29.3%) after 14 day’s differentiation in absence of DEX supplement. Such osteogenic enhancement is comparable to the differentiation supplied with full osteogenic (OS) medium (~71.1%). However, hMSCs showed insufficient ALP expression in induction with osteogenic medium without β-GP (Rq = 1 nm: ~20.6%; Rq = 200 nm: ~28.4%) or AS (Rq = 1 nm: ~24.0%; Rq = 200 nm: ~29.1%) on both smooth and nanorough substrates. Hence, it’s confirm that nanorough glass surface can replace DEX-relayed signaling during hMSC osteogenic induction.
We further confirmed hMSC late osteogenesis by using immunofluorescence staining to characterize the expression of typical bone-specific ECM proteins osteopontin (OPN) and osteocalcin (OCN) markers37 in hMSCs cultured on different substrates and conditioned osteogenic medium for 21 days (Fig. 2A & Figure S4). Our results showed that the OPN expression in hMSCs cultured on nanorough substrate (Rq = 200 nm) was significantly increased compared to cells on smooth substrate (Rq = 1 nm) under both conditioned osteogenic media without DEX (4.4-fold increase) and full osteogenic media (1.9-fold increase). While the OCN expression showed similar enhancement pattern as that of OPN for conditioned osteogenic media without DEX (5.4-fold increase), we didn’t see a difference in OCN expression for cells cultured under full osteogenic media on smooth and nanorough surface, which may occur due to the biochemical effect of DEX. The formation of calcium deposition of hMSCs is an important indicator of functional osteogenesis that is commonly used to assess the osteogenic differentiation of hMSCs. Here, Alizarin Red S (ARS) was used to assess the late osteogenesis (21 days) of hMSCs on smooth substrate (Rq = 1 nm) and nanorough substrate (Rq = 200 nm).As shown in Fig. 2B&C, there were significantly more mineralized calcium on the nanorough substrate (Rq = 200 nm) than that on the smooth substrate (Rq = 1 nm) with the conditioned osteogenic media without DEX, while there were no differences for substrates supplied with cell growth media and full osteogenic media. The results further confirm that hMSCs on the nanorough substrate (Rq = 200 nm) have an enhanced osteogenic differentiation in the absence of DEX. Together, the enhancements in the OPN an OCN expressions and calcium deposition after differentiation in absence of DEX suggest that nanorough surface can trigger and enhance the osteogenic differentiation of hMSCs by replacing DEX-relayed signaling.
Figure 2. Nanotopographic substrate promotes hMSC late osteogenesis.
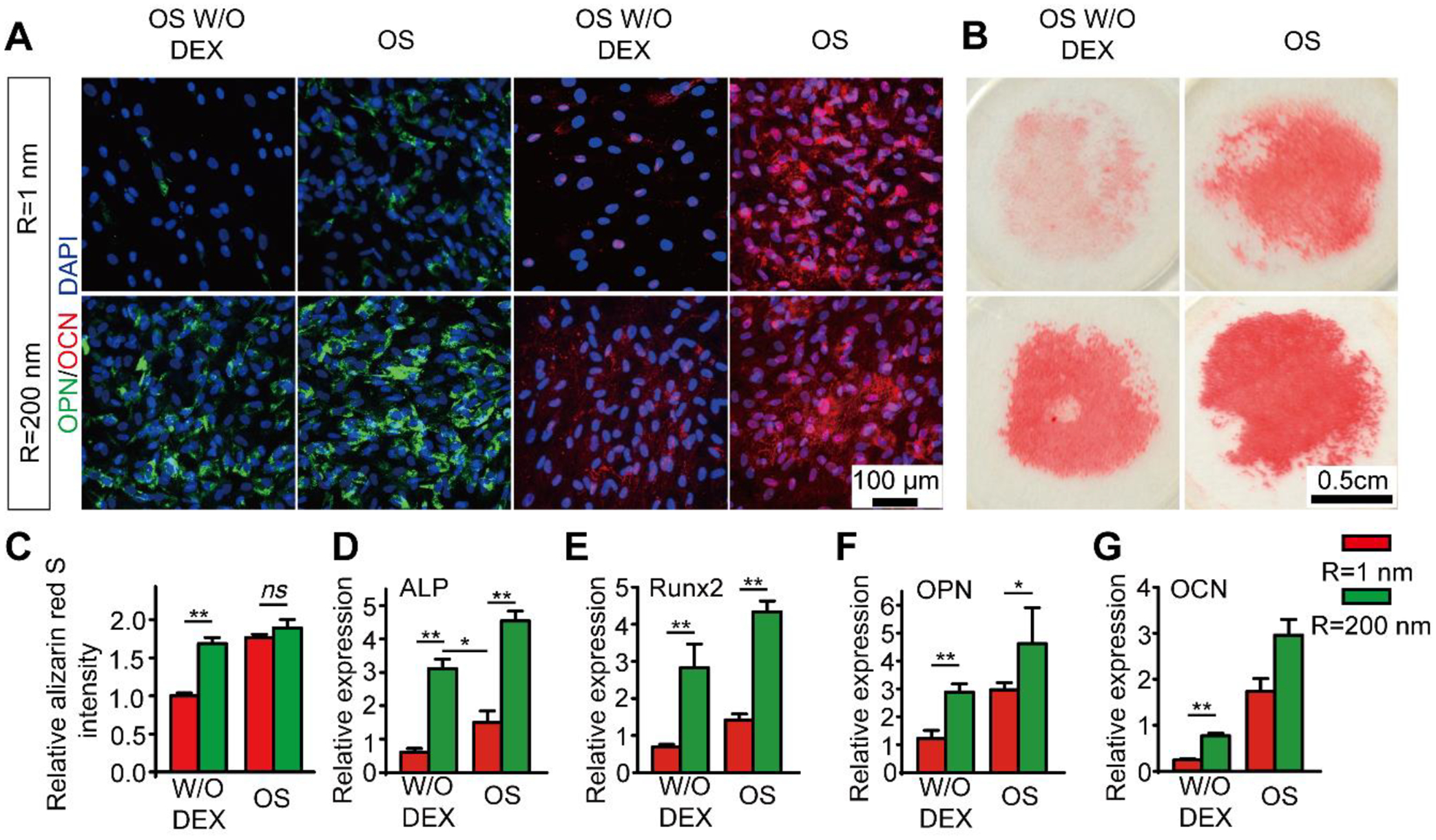
(A) Immunofluorescence images showing hMSC OPN and OCN expression after 21 days of differentiation in conditioned osteogenic differentiation media on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (B) Representative images showing Alizarin Red S (ARS) staining of hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces supplies conditioned osteogenic differentiation media after 21 d culture. (C) Bar plot showing the quantification of relative deposition of calcium of hMSCs in ARS staining assay on cultured on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces supplied with conditioned osteogenic differentiation media. (D-G) Q-PCR analysis of relative expression of osteoblast-specific genes (D) ALP, (E) Runx2, (F) OPN and (G) OCN for hMSCs after 14 days of differentiation in conditioned osteogenic differentiation media on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces. Data represent the mean ± standard error of the mean (s.e.m.) with n ≥ 3. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
Further, osteogenic differentiation markers and transcription factors at the RNA level were analyzed using quantitative polymerase chain reaction (q-PCR). Specifically, ALP, Runx2, OPN and OCN (Fig. 2D–G) were quantified as they are the key markers and transcription factors that regulate osteogenic differentiation and bone development and are commonly used as markers for osteogenesis.38 These four osteogenic marker genes were observed for the hMSCs cultured on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) substrates for 14 days in osteogenic differentiation medium without DEX and full osteogenic differentiation medium. We found that the all the four osteogenic genes, ALP (Fig. 2D; ~3.0 fold for OS media; ~5.2-fold for OS media without DEX), Runx2 (Fig. 2E; ~3.0 fold for OS media; ~4.1-fold for OS media without DEX), OPN (Fig. 2F; ~1.6 fold for OS media; ~2.4-foldfor OS media without DEX) and OCN (Fig. 2G; ~1.6 fold for OS media; ~3.2-foldfor OS media without DEX) were significantly up-regulated in hMSCs cultured on nanorough substrate compared to those on the smooth substrates both in full osteogenic inducing media and conditioned media without supplementary DEX, respectively. Among these four genes, increases in ALP, Runx2, and OPN expressions in hMSCs cultured on nanorough substrate without DEX was even greater than that of hMSCs cultured in full osteogenic inducing factors on smooth substrates (Fig. 2D–G), indicating that the nanorough surface not only activates the necessary DEX-relayed pathways in hMSC osteogenesis but further triggers ‘nanotopography-relayed’ machinery to provide an enhanced osteogenic differentiation.
Nanotopography-dependent cell adhesion, morphology, and actomyosin cytoskeleton remodeling predict hMSC osteogenic fate
Abundant evidence supports the notion that nanotopographic cues at cell-ECM interfaces control stem cell function1. However, the means through which nanotopographic signals in stem cell niche are transduced through cell-ECM interactions into biochemical and cellular functional responses (i.e., mechanotransduction) remain unclear. The bidirectional regulation of integrin-ligand binding is involving in the early stage of adherent cells interacting with surrounding ECM.39–41 During the “inside-out” and “outside-in” interaction between cells and ECM, many proteins including vinculin and tyrosine kinases such as focal adhesion kinase (FAK) play critical role in the formation of cell-ECM adhesion (also known as “focal adhesion”, or FA).42,43 Manipulating the conformation of integrin by engineering substrate topography in the scale of nanometers can further control the dynamic organization of adaptor and signaling proteins in FA.44 There is substantial evidence that suggests the possible involvement of several principal mechanotransductive pathways,45 including integrin-mediated adhesion signaling42 and actin cytoskeleton (CSK) integrity46,47 in mechanotransductive sensing and transduction of extracellular nanotopographic signals.48
To investigate the nanotopography-dependent early cell adhesion adaption, we investigated the effect of nanotopography on hMSC integrin-mediated FA formation (Figure 3A–E & Figure S5). On smooth glass surfaces with Rq = 1 nm, vinculin-containing mature FAs exhibited aligned, distributed and mature FAs throughout the cell area. However, FAs of hMSCs on nanorough glass surfaces, where Rq = 200 nm, localized primarily on cell periphery. In contrast to the mature FAs formed on smooth glass surfaces, hMSCs on the nanorough surface, exhibited greater density but smaller-size, punctate FAs (Fig. 3A–E). An important signaling axis downstream of integrin-mediated FA is the FAK-Src pathway.42,49,50 Western blot result shows that nanotopographic glass surfaces enhanced FAK phosphorylation (pFAK; Fig. 3F), supporting the assertion that FAK activation in response to nanotopographic sensing is required for the subsequent osteogenesis of hMSCs.51
Figure 3. Surface nanotopography regulates integrin-mediated cell adhesion in hMSCs.
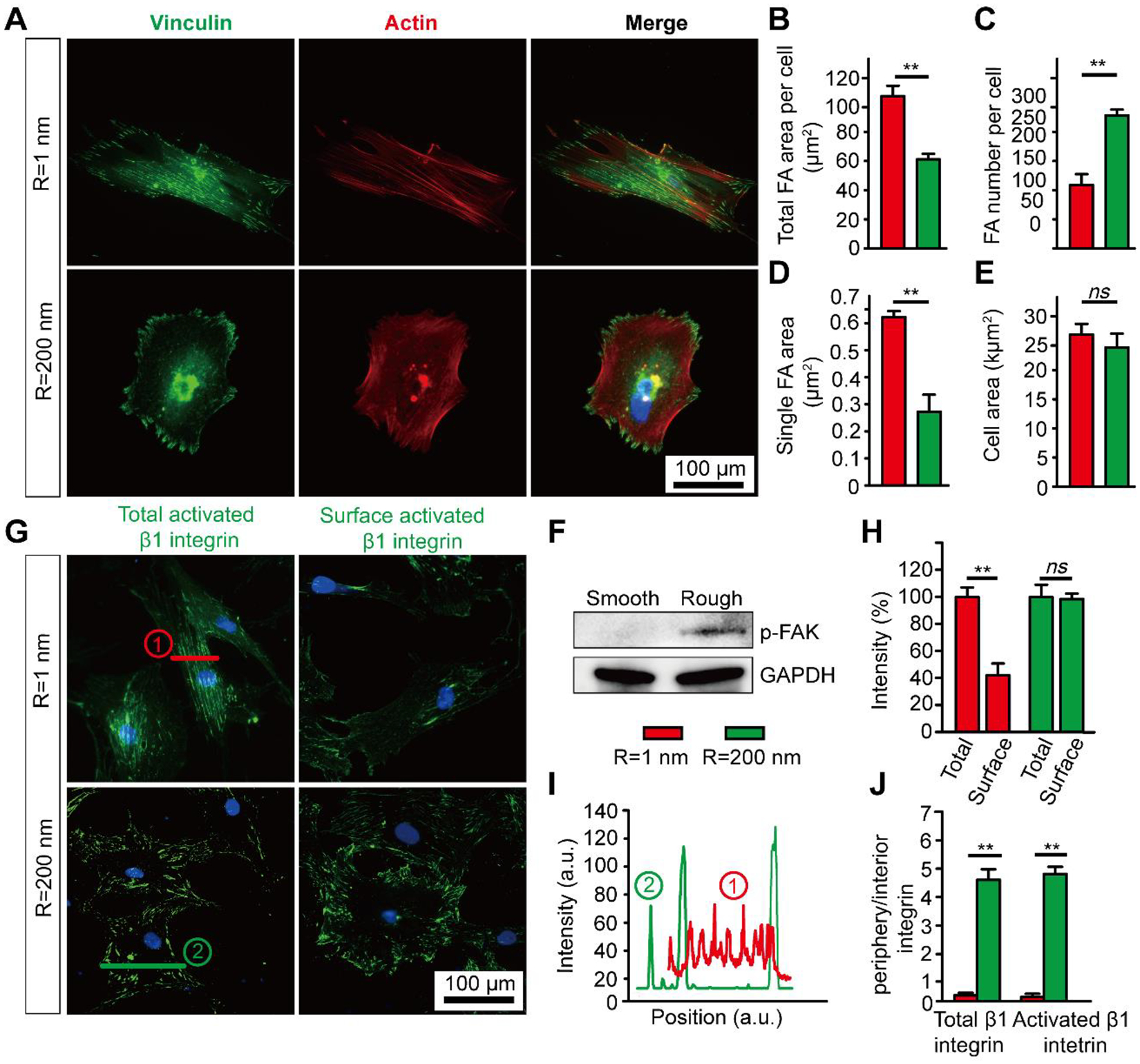
(A) Immunofluorescence images showing vinculin and F-actin in undifferentiated hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates after 24 hr of culture. (B-E) Bar graphs showing quantitative results of total FA area per cell (B), number of FAs per cell (C), average single FA area (D), cell spread area (E), and (F) Western blot analysis of pFAK for hMSCs cultured on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates after 24 hr of culture. (G) Immunofluorescence images showing total and surface activated β1 integrin in undifferentiated hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates after 24 hr of culture. (H-J) Quantification of total activated β1 integrin and surface activated β1 integrin (H), distribution of activated β1 integrin (I & J) for hMSCs cultured on substrates with different nanoroughness as indicated. Error bars represent ± s.e.m. with n > 10. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
Following the observed significant adaptions of hMSC FA to nanotopography, we examined the integrin activities, which is a major adhesion receptor for mechanosensing52 that mediates FA assembly and CSK organization. Integrin activities including activation, internalization and clustering are known to mediate MSC behavior through a cascade of downstream signal pathways53–55 and commit distinct lineages.20,44 We specifically investigated integrin activation and internalization in hMSCs by immunostaining for total β1 integrin and its active conformation (Fig. 3G–J & Figure S6). Although we did not observe an obvious difference in the level of total and activated β1 integrin on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces, we found that both total and activated β1 integrin showed significant less surface integrin and enhanced integrin internalization on smooth (Rq = 1 nm) glass surface (Fig. 3G&H & Figure S6A&B). This is consistent with the FA results. Integrin co-localized with vinculin and both the total and active β1 integrin were distributed primarily on cell periphery in case of nanorough (Rq = 200 nm) surface and located in the interior area of the entire cell on smooth surface (Fig. 3I&J & Figure S6C). The peripheral distribution of β1 integrin confirmed our observations from hMSC FAs on nanoroughness. Further, it is known that surface conservation of β1 integrin may lead to an activation of BMP/Smad signaling through co-internalization of integrin and BMP type-I receptor (i.e., BMPRIA) and promote phosphorylation of Smad 1/5/8, thus further promote hMSC differentiation.20
Cell morphology is evolving at different stages of hMSC osteogenic differentiation, with elongated and aligned morphology before differentiation and polygonal morphology after differentiation.56–58 Although cell shape evolving accompany with hMSC differentiation, it is recognized that modulating cell shape can in turn regulate the fate of stem cells as well59,60. In our study, we observed a clear morphological difference in hMSCs cultured on smooth (Rq =1 nm) and nanorough substrates (Rq = 200 nm) (Fig. 1A & Fig. 3A). hMSCs exhibited fibroblastic appearance on smooth surface while osteoblastic morphology on nanorough surface. Quantitative analysis of cellular aspect ratio suggested that the fibroblast-esque cells on smooth substrates were highly elongated (polarized morphology) whilst osteoblast-esque cells on nanorough substrates were more spread-out (polygonal morphology) (Figure S7). The morphological difference indicates a possible change in cell actomyosin CSK. Actin CSK and its integrity can integrate multiple extracellular biophysical cues including nanotopography in mechanotransduction.46 Such observed morphological and cytoskeletal change during early stages, on our nanorough surface, may lead to different differentiation behaviors and predict the osteogenic fate of hMSCs.60–62
Indeed, the actin distribution showed similar trend to cell morphology and FAs on smooth and nanorough glass surface, with elongated and aligned actin fibers across the length of the entire cell on smooth surface (Rq = 1 nm) and intense and chaotic actin fibers localized on the cell periphery on nanorough surface (Rq = 200 nm). A previous study recognized that thick actin filament bundles located at the cell periphery is a characteristic for osteogenic differentiated MSCs.62 This indicates that our nanorough surface can generate ‘osteoblast-esque’ morphology and actin structures before and at early stage of differentiation. We further characterized spatial distribution of cytoskeletal filaments in single hMSCs on surfaces of different nanoroughness. Actomyosin CSK in hMSCs supports cell structure and integrates inward the topographic cues from cell’s immediate microenvironment. Distinct cellular mechanical properties arise from different actomyosin CSK structures and could be indicative measures of cellular response to the immediate microenvironment.63,64 Specifically, we quantified fractal dimensions (Df) describing complex patterns of cytoskeletal filaments in hMSCs (see MATERIALS & METHODS for details). As shown, there is a distinct distribution (Df) of actin fibers in hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) surface (Figure 4 A&B). By looking into subcellular distribution of actin fibers, we found larger Df value in the perinuclear and cytosolic area of cells on nanorough (Rq = 200 nm) surface. Intensity of actin fiber follows the same trend as Df distribution and we found a larger grey value of actin in hMSCs on nanorough (Rq = 200 nm) surface (Fig. 4C), indicating a stronger actomyosin structure.
Figure 4. Surface nanotopography regulates hMSC actomyosin CSK and biomechanical properties.
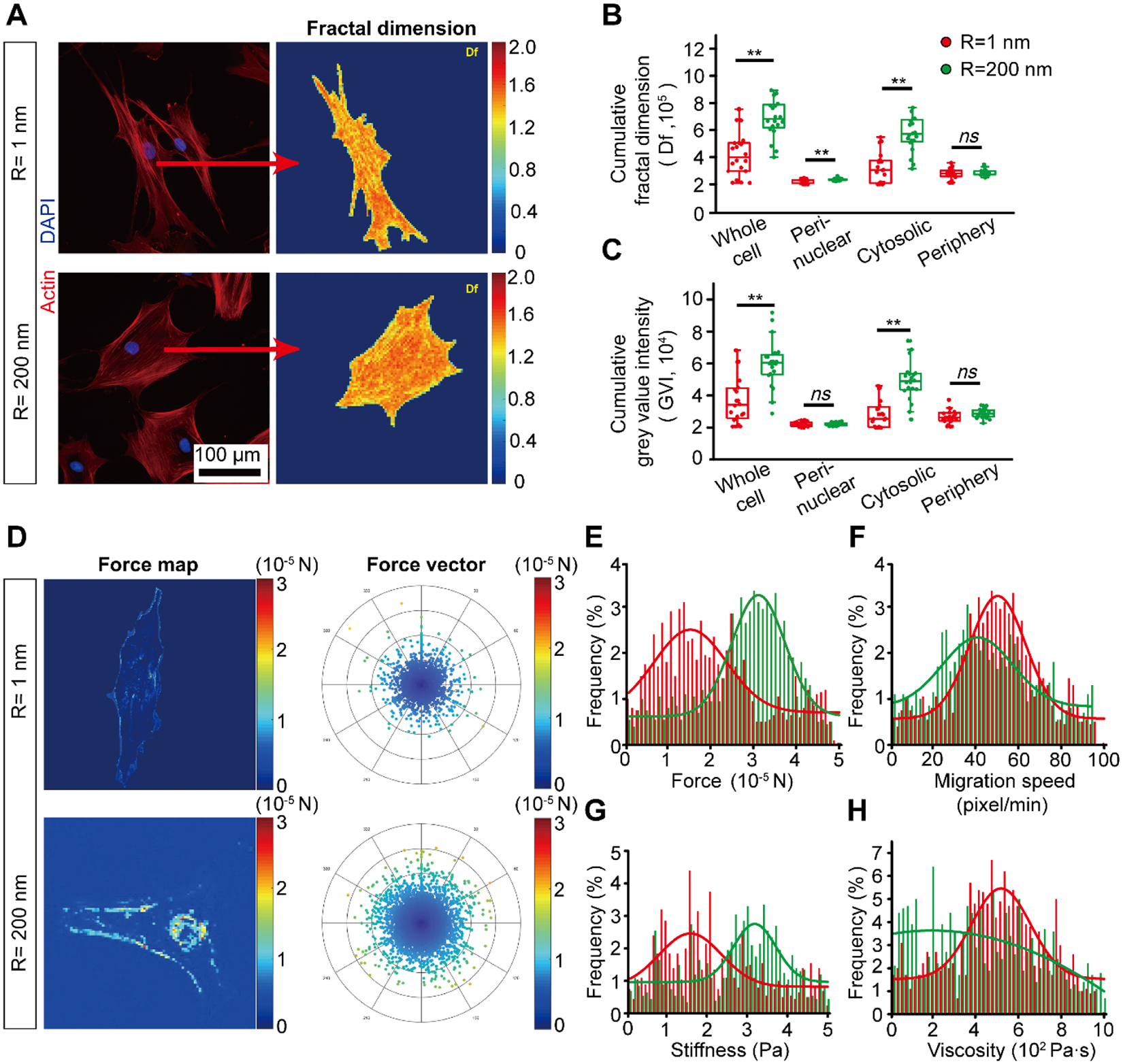
(A) Representative immunofluorescence images (left panel) of the hMSC actin CSK on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates and the results of corresponding 2D fractal dimension (Df) array (right panel). (B-C) Quantification of actin CSK arrangement (Df) (B) and grey value intensity (C) in each subcellular region of interests. (D) Representative intracellular force map (left panel) and force vector histogram (right panel) for hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates as indicated. (E-H) The distribution and probability of force (E), migration speed (F), stiffness (G) and viscosity (F) of hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates. Error bars represent ± s.e.m. with n > 10. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
In addition to investigating the CSK organization in single cells, we further examined whether CSK showed different evolving tracks from a collective view for a colony of hMSCs under osteogenic induction. Our results showed that hMSCs in colony on smooth and nanorough glass surfaces demonstrate distinct collective CSK organization during the early days of osteogenic differentiation (Figure S8). We observed that nanorough surfaces induced spontaneous collective CSK remodeling in hMSC colony during early differentiation stage. Chaotic pattern of CSK was consistent for hMSCs on nanorough glass surfaces during early days of differentiation which eventually gave way to an aggregated pattern. On smooth surfaces, CSK showed high degree of alignment that gradually subsided into a more chaotic arrangement. However, the temporal change towards chaos resulted in an arrangement that was still more ordered when compared to the CSK arrangement on nanorough surfaces. Indeed, previous characterization of CSK evolving during osteogenic differentiation recognized that intense and chaotic CSK organization, cell aggregation is essential for the formation of bone nodule2,62. The CSK adaption to nanotopography provides early clues that drive hMSC lineage commitment.
Nanotopography induces distinct hMSC biomechanical properties
The distinct nanotopography-induced cell adhesion and actomyosin CSK properties drive us to further explore the possible changes in cell mechanical properties that may serve as novel and early phenotypic stem cell markers to predict specific lineages.56,65 Previous studies demonstrated that stem cell lineages are associated with cell’s mechanical properties65. Teo et al. found that there is a decrease in cellular stiffness and increase of cellular viscosity during the process osteogenic differentiation for hMSCs.64 Different lineages of MSCs also showed diverse mechanical properties. For instance, research found that osteoblasts are stiffer than adipocytes65 and neurons showed even lower stiffness than that of adipocytes.66 Cellular traction force has been associated with cellular mechanical properties like stiffness67–69 and it plays an integral role in cell sensing ECM and trigger “outside-in” signaling.70,71 Thus cellular traction force further impacts cell functions via mechanotransduction including differentiation.71
Understanding how hMSCs sense nanotopographic cues and adjusts cytoskeletal tension to befit the microenvironment has important implications for engineering ECM properties for bone engineering. However, no such study has been done to evaluate the nanotopography-mediated cellular mechanical properties such as cell stiffness and force. Various techniques have been developed to probe cell mechanical properties reflecting cellular mechanotransduction. For example, micropost array detector (mPAD) is used to directly measure cellular traction force.72,73 Micropipette aspiration,74 atomic force microscopy (AFM),75 and optical tweezers76 have been used to examine cellular stiffness and cytoskeletal tension. However, previous methods for cellular mechanical properties investigations are invasive and thus hinder their applications in investigating cellular properties without disturbing the biomimetic microenvironments. It is also technically challenging to integrate our nanotopographic cues with these techniques. Here, integrating live cell imaging with an in house MATLAB processing method, we are able to non-invasively decipher intracellular properties for cellular mechanotransduction on our nanotopographic surface (see Methods for details).63,64
With this method, we examined the cellular biomechanics of hMSCs on surfaces of different nanoroughness (see MATERIALS & METHODS for details).64 Time-lapse images of live hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) were acquired for 12 h under standard cell culture environment for the subsequent single cell biomechanical properties analysis. Although our method is evaluating relative biomechanical properties change, the differences we found between cells on smooth (Rq = 1 nm) and nanorough (Rq = 1 nm) can reflect an intrinsic discrepancy of cell functions. On average, the intracellular forces were measured larger when hMSCs were on nanorough (Rq = 200 nm) surface compared to that on smooth (Rq = 1 nm) surface (Fig. 4D). We also found a distinct distribution of intracellular forces in a subcellular level as showed in Fig. 4E. Compared to hMSCs on smooth (Rq = 1 nm) surface, which shows little force across the whole cell area, there is a large portion of force on the periphery of hMSCs on nanorough (Rq = 200 nm) surface. Cells sense their surrounding substrates and respond by regulating their cell shape and internal CSK through mechanotransduction, which may result in distinct biomechanical properties such as stiffness.61,77 In addition, hMSCs behave as viscoelastic solid and their viscoelasticity is dependent on actin CSK strucute.78 Although we did not observe a clear difference in cell migration speed, we found an increase in cellular stiffness and a decrease in cellular viscosity when hMSCs were seeded on nanorough (Rq = 200 nm) surface (Fig. 4F–H). Our results conform to previous founding that strong CSK integrity in osteoblast-esque cells accompany with high cellular stiffness and low cellular viscosity.61,77,78 Together, our single-cell mechanical analysis showed distinct cellular biomechanical properties and increase in cellular contractility, which may likely be the result of enhancement in cytoskeletal actin integrity and strength measured above (Fig. 4 A–C). This confirms that hMSCs respond to nanotopography via a mechanosensitive adhesion-CSK pathway and results in enhanced osteogenic differentiation on nanorough surface.
Nanotopography regulates hMSC osteogenesis through TAZ activation
Our results indicate that nanotopographic cue can be effective to replace DEX and work synergistically with other introduction morphogens in the ontogenetic differentiation process. DEX is known as an key osteogenesis induction factor promoting hMSC osteogenesis via up-regulating the expression of transcriptional coactivator with TAZ in hMSCs.21,26 TAZ modulates MSC fate by activating osteogenic differentiation and inhibiting adipocyte differentiation through stimulating Runx2-mediated gene transcription.79 Nuclear accumulation of TAZ directs bias for hMSC osteogenesis.80,81 The above discussed “mechanophenotypic” adaption of hMSC in response to the nanotopographic cue indicates a mechanotransductive process that may involve activation of several transcription of target genes including TAZ. Recently, TAZ and YAP (Yes associated protein) have been characterized as effector nuclear transducers in the Hippo signaling pathway, which plays an important role in mechanotransduction82 as well as stem cell linage decisions.79,82,83 We thus seek to prove the hypothesis that the Hippo/TAZ signaling is a potential mechanosensitive pathway regulated by the nanotopographic signals from the local stem cell niche for nanotopography-induced hMSC fate decision.
We first examined whether nanoroughness regulates TAZ phosphorylation and nuclear accumulation in undifferentiated hMSCs (Figure 5A–B & Figure S9) to control early osteogenic induction of hMSCs. We measured and defined the nuclear/cytoplasm ratio of TAZ to quantify the level of nuclear accumulation of TAZ. We found that the nanorough substrates (Rq = 200 nm) significantly promoted the nuclear accumulation of TAZ (nuclear/cytoplasm ratio of TAZ = 1.86) as compared to smooth (Rq = 1 nm, 1.35) substrates (Figure S10). The nucleocytoplasmic localization of TAZ was also observed after 3-day osteogenic induction, with TAZ nuclear/cytoplasm ratio of 1.63 for smooth surface and 2.18 for nanorough surface (Fig. 5A–B), strongly suggesting nucleocytoplasmic translocation of TAZ as a critical component involved in nanotopography-dependent osteogenic induction of hMSCs. Furthermore, to confirm nuclear accumulation of TAZ directly promote the downstream transcription factor Runx2, which promote hMSC osteogenesis, we showed similar nuclear accumulation of Runx2 on nanorough substrates (Rq = 200 nm) after 3 days of osteogenic induction without DEX (Fig. 5A–B Supplementary Figure 10). Immunoblots further demonstrated that nanorough substrate decrease TAZ phosphorylation on serine 89 (ser89), a key target of Lats1/2 kinase downstream of the Hippo pathway (Fig. 5C & Figure S11).84,85 It is recognized that TAZ bind phosphorylated Smads (phosphoSmads) and control their nucleocytoplasmic shuttling in stem cells.86 Indeed, western blot results in Fig. 5C confirmed the promotion of nanotopography on phosphorylation of Smad1/5/8.
Figure 5. Nanotopography regulates hMSC osteogenesis through mechanosensitive TAZ activation.
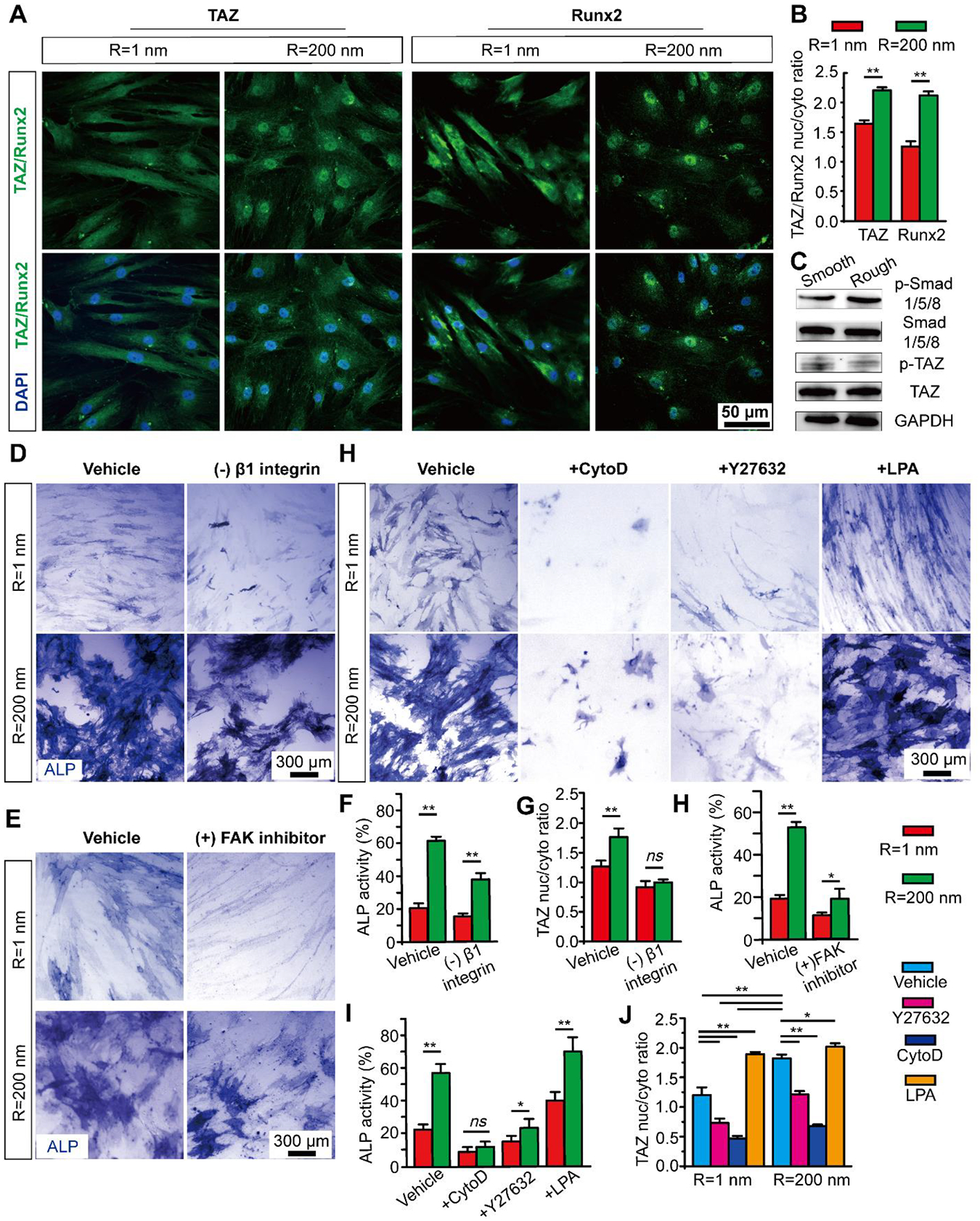
(A) Representative immunofluorescence images, and (B) Bar plot showing nanoroughness-dependent subcellular localization of TAZ and Runx2 at day 3 on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (C) Western blotting showing total and phosphorylated Smad 1/5/8 (p-Smad 1/5/8), phosphorylated TAZ on serine 89 (p-TAZ S89) and TAZ in hMSCs differentiated for 3 days on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces. (D-E) Representative ALP staining images and (F-H) bar plots showing percentages of ALP activity (F&H) and nanoroughness-dependent subcellular localization of TAZ (G) in hMSCs after 7 d osteogenic differentiation in conditioned osteogenic media without DEX supplement and under treatment of β1 integrin and FAK inhibitor on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (H) Representative ALP staining images and (I-J) bar plots showing percentages of ALP activity (H) and nanoroughness-dependent subcellular localization of TAZ (I) in hMSCs after 7 d osteogenic differentiation in conditioned osteogenic media without DEX on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces under different CSK drug treatment as indicated. Data represent the mean ± s.e.m. with n = 3. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
With knowledge of the functional involvement of Hippo/TAZ signaling in nanotopographic signaling, we may further explore how nanotopographic signals at the cell-ECM interface are transmitted to the nucleus. Integrins transmit signals via intracellular signaling proteins including integrin-linked kinase (ILK) and FAK, which have recently been shown to suppress the Hippo pathway and nuclear accumulation of YAP/TAZ.87 Recent mechanotransduction studies also reveal a critical link between YAP/TAZ activity and CSK tension and integrity.88–90 Extracellular cellular mechanical signals can be integrated by contractile force from contractile actin stress fibers and actin CSK integrity.8,71 Further, FA formation and actin CSK structure and tension are largely depend on the RhoA/ROCK signaling pathway.91 Our nanorough surface demonstrated a regulatory effect on hMSC adhesion, CSK organization and tension.
We therefore investigated the effect of nanotopography on integrin-mediated adhesion, RhoA/ROCK activity and CSK tension in regulating Hippo signaling and thus hMSC osteogenic induction. First, we inhibited β1 integrin with anti-β1 integrin antibody and observed decreased β1 integrin expression on both smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) surface, and the before observed peripheral distribution of β1 integrin on nanorough surface diminished (Figure S12A–C). We then quantified the resultant osteogenic differentiation and nuclear TAZ of hMSCs treated with β1 integrin inhibitor. Inhibiting β1 integrin reduced both ALP activity and nuclear TAZ for hMSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) surface (Fig. 5D & F–G & Figure S12D). The inhibition of β1 integrin may modulate FAs and reduced cell contractility,92,93 thus further decrease nuclear TAZ and hMSC osteogenesis. In addition, we found that inhibiting FAK largely abate the promotion of hMSCs on nanorough surface (Fig. 5E & H) and further confirmed the importance of FAK activation for hMSCs osteogenesis. We then treated hMSCs independently with Y27632 (inhibitor of Rho-associated kinase, or ROCK, a downstream effector of RhoA), which decreases actomyosin contractility while maintaining intact CSK structure; cytochalasin D (CytoD), an inhibitor of actin polymerization; or lysophosphatidic acid (LPA), which stimulates RhoA and facilitates formation of actin microfilaments (Figure S13 & S14). Compared with controls, Y27632 and CytoD promoted cytoplasmic localization of TAZ and significantly decreased the ALP expression of hMSCs both on smooth and nanorough substrates (Fig. 5H–J & Figure S15). In contrast, LPA facilitated nuclear localization of TAZ and promoted ALP expression on both smooth and nanorough substrates (Fig. 5H–J & Figure S15). These data are consistent with findings that ROCK inhibition inhibits osteogenic differentiation of MSCs.94 Together, our studies suggest that nanotopography induces an integrin-mediated adhesion-CSK-TAZ signaling. The integrin-mediated adhesion signaling acts as a mechanical signal sensor and transmits these signals through mechano-sensitive and mechano-transductive pathways such as Smad, RhoA/ROCK and Hippo/TAZ pathways, that further mediate osteogenic gene expression.
CONCLUSION
The microenvironment where hMSCs reside is a niche filled with nanostructures that are normally of the order of hundred nanometers.95,96 Investigating the regulation of stem cell behaviors from nanotopographic cues has been highlighted over the past years. It has recently become evident that stem cell fate can be regulated by topographies in stem cell microenvironment, besides various biochemical factors. However, the molecular mechanism underlying the regulation of stem cell behavior by ECM topography is yet to be established. From such studies, a better understanding of the effect of the cellular microenvironment on hMSCs behavior can be attained which will thus aid in developing better tissue engineering platforms for cell therapy. Therefore, elucidation of the effect of nanotopographies on hMSCs behaviors and exploration of the mechanism for the regulation of hMSC is needed to optimize protocol of MSC-based cell therapies.
In this study, we introduced nanoengineered glass substrates with different surface roughness and demonstrated a promotion of osteogenic differentiation of hMSCs on nanorough glass substrates in the presence of DEX, an important biochemical factor for osteogenesis induction. As we investigate how hMSC sense exogenous nanotopography and the pathways that transduce these nanotopographic cues into intracellular biochemical signals, we demonstrated that our nanotopographic substrates can replace the function of DEX morphogen in promoting hMSCs osteogenic differentiation. We proposed that our nanoroughness can activate TAZ and further mediate osteogenic gene expression akin to the function performed by DEX during hMSC osteogenesis. The mechanism for our nanotopographic induction of osteogenesis involves integrin internalization inhibition, actin CSK enhancement and TAZ activation by nanoroughness. Specifically, we found that the internalization of integrin was significantly inhibited by nanorough substrate in comparison with a smooth substrate. Being an important membrane receptor for ECM proteins, integrin directly affects FA assembly, cytoskeletal organization. Hence, we also examined cell adhesion, actomyosin CSK, and Hippo signaling pathway. We confirmed the enhanced Smad phosphorylation and increase of nuclear localization of TAZ on nanorough substrate. Taken together, it is likely that nanotopographic signals are transmitted from the cell-ECM interface toward the nucleus through a regulatory pathway involving integrin internalization, BMP/Smad signaling, and finally the Hippo pathway to control hMSC fate decision.
In conclusion, we successfully demonstrated the regulation of hMSC behavior on nanotopographic glass substrates with different surface roughness, including cell adhesion, morphology, proliferation, and differentiation. Nanoroughness promoted the osteogenic differentiation of hMSCs and demonstrate a route using the difference of surface topography to control differentiation behaviors of hMSCs. By establishing a controllable, reproducible, and scalable fully-defined synthetic culture platform employing essential nanotopographic cue at cell-ECM interface, it can potentially enable efficient production of osteoblasts from hMSCs and thus contribute significantly to future cell-based therapies for osseous defects and anomalies. The nanoroughness on biocompatible glass substrates can be easily constructed on the surface of implants, making it a versatile surface nanostructure to control the osteogenic differentiation of hMSCs. Our study further revealed nanotopography-sensitive cellular machineries including integrin internalization, actin CSK enhancement, BMP/Smad and the Hippo signaling function synergistically to control the osteogenesis of hMSCs. This may advance the understanding of how nanotopography-mediated mechanobiology is involved in regulation of hMSC behaviors and fate. Our study will contribute significantly to the understanding of how hMSCs interact with local microenvironment to develop multicellular communication systems during bone healing.
MATERIALS & METHODS
Fabrication and Surface Characterization of Nanorough Glass Samples
Glass wafers (Borofloat 33; Plan Optik) were processed with RIE (LAM 9400, Lam Research) for different periods of time to generate nanoscale surface roughness (ranging from 1 nm to 200 nm). The corresponding RIE process condition was selected as: SF6 (8 sccm), C4F8 (50 sccm), He (50 sccm), Ar (50 sccm), chamber pressure (1.33 Pa), bias voltage (100 V), and radio frequency power (500 W). All the processed glass wafers were cut into small pieces (1 cm × 1 cm or 1.5 cm × 1.5 cm) using the ADT7100 dicing saw (Advanced Dicing Technologies) before placed into standard 24-well or 12-well tissue culture plates. To promote cell attachment, glass substrates were functionalized with human fibronectin (Sigma) by immersing the substrates in a fibronectin solution (20 μg mL−1) in distilled water overnight. Glass substrates were rinsed twice with PBS before they were used for cell seeding.
Nanoroughness of the glass surfaces was measured at room temperature with the Veeco NanoMan Atomic Force Microscope (AFM, Digital Instruments) using a non-contact, tapping mode and standard Si tapping mode AFP tips. The AFM scan image size was 10 μm × 10 μm with a scan rate of 1 Hz. The resulting map of local surface height was represented using the AFM topographs. The nanoroughness of each glass sample was characterized by the root mean square (RMS) roughness Rq of the local surface height over the scanned areas collected using the AFM topographs. Unprocessed bare glass wafers had an intrinsic surface roughness Rq of 1 nm.
Cell culture and reagents
hMSCs (Lonza) were maintained in Lonza formulated growth medium. Early passages of hMSCs were used in experiments (passages 3–6). For hMSC differentiation assays, the complete osteogenic induction media consists of growth media and osteogenic supplements with 10 nM dexamethasone, 10 mM glycerophosphate and 0.2 mM ascorbic acid. Conditioned osteogenic medium was prepared without dexamethasone, glycerophosphate, and ascorbic acid, respectively. Adipogenic induction medium was Lonza formulated. Culture media were replaced every 3 d in all experiments. All the media were pre-equilibrated at 37°C in 5% CO2 humidified atmosphere before use.
For hMSC differentiation under CSK drug treatment, differentiation media were prepared with CyoD (100 nM), Y27632 (2 μM) and LPA (20 μM) respectively and cultured for 7 days. Control group was supplied with DMSO. When investigate nuclear TAZ under drug treatment, culture media were prepared with CyoD (2 μM), Y27632 (10 μM) and LPA (50 μM) respectively and cultured for 2 hours before being fixing. Control group was supplied with DMSO.
For hMSC differentiation investigation under FAK inhibition, FAK inhibitor (5 μM, Santa Cruz Biotechnology) was added to the conditioned osteogenic medium (OS without DEX) and cultured for 7 days. For hMSC differentiation investigation under β1 integrin inhibition, differentiation media were prepared with IgG1 isotype control from murine myeloma (10 μg/ml) and β1 integrin antibody (10 μg/ml) respectively and cultured for 7 days. For immunostaining of β1 integrin and TAZ under β1 integrin inhibition, hMSCs were cultured in culture media supplied with IgG1 isotype control (25 μg/ml) and β1 integrin antibody (25 μg/ml) respectively for 2 hours before being fixing.
Immunocytochemistry
hMSCs were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min and then permeabilized with 0.1% Triton X-100 (Roche Applied Science) for 20 min at room temperature. Fixed cells were then incubated with 3% bovine serum (Sigma) for 1 hr and then primary antibodies (Table S1) for 1 hr. Alexa Fluor 488 and 555 conjugated goat anti-mouse (or anti-rabbit) IgG secondary antibodies (Invitrogen) were used as secondary antibodies. Alexa Fluor 555 conjugated phalloidin (Invitrogen) and 4’,6-diamidino-2-phenylindole (DAPI; Invitrogen) were used for visualization of actin microfilaments and nucleus, respectively.
For ALP and Oil red staining, hMSCs were firstly fixed with 4% paraformaldehyde in PBS and stained for alkaline phosphatase using Fast Blue RR salt/naphthol (Sigma-Aldrich) per manufacturer instructions. To stain lipid fat droplets, cells were fixed in 4% paraformaldehyde, rinsed in PBS and 60% isopropanol, stained with 3 mg ml−1 Oil Red O (Sigma-Aldrich) in 60% isopropanol and rinsed in PBS. For total cell counts, cell nuclei were stained with DAPI. Percentage of marker-positive cells was quantified as the ratio of ALP-positive cell number and the total cell number. For Alizarin Red S staining, hMSCs were firstly fixed with 4% paraformaldehyde in PBS for 15 mins and then incubated in 40 mM ARS (EMD Millipore) at room temperature for 20 – 30 min with gentle shaking.
Western blotting
Whole cell lysates were prepared from hMSCs by using Halt Protease and Phosphatase Inhibitor Cocktail in RIPA cell lysis buffer (1:100, Thermo Scientific), separated on SDS-polyacrylamide gel (BIO-RAD) and transferred to PVDF membranes (BIO-RAD). The membranes were incubated with 5% milk in PBS for 1 hr and then incubated with primary antibodies (Table S1) overnight at 4°C. Blots were incubated with goat anti-rabbit (GAR)-HRP conjugate secondary antibodies (BIO-RAD) for 1 hr, and protein expression was detected with ChemiDoc™ Touch Imaging System (BIO-RAD).
RNA isolation and Real-time qPCR analysis
Total RNA was isolated from hMSCs grown on glass substrates using RNeasy kit (Qiagen) and reverse transcribed to cDNA with Omniscript Reverse Transcription Kit (Qiagen). Real-time PCR (RT-PCR) was performed and monitored using an Applied Biosystems StepOnePlus™ Real-Time PCR system. RT-qPCR was performed with Taqman-probes PCR mastermix. Human 18S primers (Hs03003631_g1) were used as an endogenous control for relative quantifications. Premiers used in this work include: ALP (Hs01029144_m1), Runx2 (Hs01047973_m1), OPN (Hs01587814_g1), and OCN (Hs01587814_g1). All primers are from Applied Biosystems (Life Technologies). All analyses were performed with three replicates. Relative expression levels were determined by calculating 2−ΔΔCt with corresponding s.e.m.
SEM specimen preparation
Cell samples were washed three times with 50 mM Na-cacodylate buffer (pH 7.3; Sigma-Aldrich), fixed for 1 hr with 2% glutaraldehyde (Electron Microscopy Sciences) in 50 mM Na-cacodylate buffer, and dehydrated in a graded series (30%, 50%, 70%, 80%, 90%, and 100%) of ethanol concentrations over a period of 1.5 hr. Dehydration in 100% ethanol was performed three times. Afterwards, dehydrated substrates were dried with liquid CO2 using a super critical point dryer (Samdri®-PVT-3D, Tousimis). Samples were mounted on stubs, sputtered with gold palladium for 15 s, observed and photographed under a Hitachi S-3400N Ultra-High Resolution SEM machine (Hitachi High Technologies America).
Actin CSK quantification
Automated image processing was used to perform fast and standardized quantification of the spatial distribution of the cell actin CSK.63 From the raw images, the program identifies two sets of coordinate points that defines perimeters of the entire cell and the nucleus. The located boundaries are then redefined through an offset by a predetermined distance of 5 μm. Binary masks are then generated and eventually used to extract subcellular regions of interest, namely, the perinuclear, cytosolic, and peripheral regions. Another binary image with a mix of white fragments, lines of varying lengths, bifurcations and loops, which collectively represents the actin CSK, is generated by an edge detection function performed on the raw image of the actin. The image is subsequently divided into interrogation windows using squares of user defined pixel lengths. Within each window the CSK arrangement is assigned a fractal dimension (Df) through box-counting method and the mean of the original gray value. For each pair of nuclei and CSK images, the final outputs are two dimensional (2D) arrays of Df values and corresponding mean gray value intensities (GVI), giving temporal information regarding the pattern and the amount of actin. Collective behavior of actin fiber anisotropy during osteogenic differentiation was analyzed using ImageJ (NIH).
Non-invasive single cell biomechanical analysis
An established image-morphing technique was used for non-invasive measurement of biomechanical properties of cells.64 Briefly, image morphing minimizes the differences between consecutive images of a time-lapse dataset. These differences are presented as changes in cell shape and greyscale information within the cell boundaries. Ultimately, a displacement field that describes this minimization is found. This displacement is not rigid but modeled as a viscous fluid because cell shape changes are governed by cytoskeletal changes (a polymer within fluid has viscoelastic properties), additionally the use of the model can accommodate large deformations as seen in cellular movements.
The force field f is the link between image dataset and the viscous fluid flow model. For each pair of consecutive images (S and R), f is derived from minimization of image similarity measures through
| (1) |
here x is the position of a pixel (for an image x = [xx, xy]) and u (x, t) is the displacement of particles as they move through x, according to the Eulerian framework. f is the derivative of the sum of squared difference between images and provides the driving force to the interactively solved Navier-Stokes-Duhem partial differential equation (PDE). We then derive intracellular biomechanical properties (stiffness and viscosity) by fitting the Kelvin–Voigt viscoelastic mechanical model76,97 to the strain information obtained from the interactive process.
Statistics
All experiments were conducted in n ≥ 3 biological replicates and repeated in n ≥ 3 independent experiments. For all comparisons, P < 0.05 was considered statistically significant. P-value was calculated using the student t-test function in Excel (Microsoft). All data presented in the manuscript represents the mean ± standard error of the mean (s.e.m.) with n ≥ 3.
Supplementary Material
Acknowledgements
We acknowledge financial support from the Department of Mechanical and Aerospace Engineering at New York University and the American Heart Association Scientist Development Grant (16SDG31020038).
REFERENCE
- (1).Chen W, Shao Y, Li X, Zhao G & Fu J Nanotopographical Surfaces for Stem Cell Fate Control: Engineering Mechanobiology from the Bottom. Nano Today, 2014, 9, 759–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD & Oreffo RO The Control of Human Mesenchymal Cell Differentiation Using Nanoscale Symmetry and Disorder. Nat. Mater, 2007, 6, 997–1003. [DOI] [PubMed] [Google Scholar]
- (3).Ratner BD & Bryant SJ Biomaterials: Where We Have Been and Where We Are Going. Annu. Rev. Biomed. Eng, 2004, 6, 41–75. [DOI] [PubMed] [Google Scholar]
- (4).Oh S, Brammer KS, Li YJ, Teng D, Engler AJ, Chien S & Jin S Stem Cell Fate Dictated Solely by Altered Nanotube Dimension. Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 2130–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Datta N, Pham QP, Sharma U, Sikavitsas VI, Jansen JA & Mikos AG In Vitro Generated Extracellular Matrix and Fluid Shear Stress Synergistically Enhance 3d Osteoblastic Differentiation. Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Qiu J, Li J, Wang S, Ma B, Zhang S, Guo W, Zhang X, Tang W, Sang Y & Liu H Tio2 Nanorod Array Constructed Nanotopography for Regulation of Mesenchymal Stem Cells Fate and the Realization of Location-Committed Stem Cell Differentiation. Small, 2016. [DOI] [PubMed] [Google Scholar]
- (7).Sonam S, Sathe SR, Yim EK, Sheetz MP & Lim CT Cell Contractility Arising from Topography and Shear Flow Determines Human Mesenchymal Stem Cell Fate. Sci. Rep, 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yim EK, Darling EM, Kulangara K, Guilak F & Leong KW Nanotopography-Induced Changes in Focal Adhesions, Cytoskeletal Organization, and Mechanical Properties of Human Mesenchymal Stem Cells. Biomaterials, 2010, 31, 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Faia-Torres AB, Guimond-Lischer S, Rottmar M, Charnley M, Goren T, Maniura-Weber K, Spencer ND, Reis RL, Textor M & Neves NM Differential Regulation of Osteogenic Differentiation of Stem Cells on Surface Roughness Gradients. Biomaterials, 2014, 35, 9023–9032. [DOI] [PubMed] [Google Scholar]
- (10).Abagnale G, Steger M, Nguyen VH, Hersch N, Sechi A, Joussen S, Denecke B, Merkel R, Hoffmann B & Dreser A Surface Topography Enhances Differentiation of Mesenchymal Stem Cells Towards Osteogenic and Adipogenic Lineages. Biomaterials, 2015, 61, 316–326. [DOI] [PubMed] [Google Scholar]
- (11).Mazón P, García-Bernal D, Meseguer-Olmo L, Cragnolini F & Piedad N Human Mesenchymal Stem Cell Viability, Proliferation and Differentiation Potential in Response to Ceramic Chemistry and Surface Roughness. Ceram. Int, 2015, 41, 6631–6644. [Google Scholar]
- (12).Chen W, Villa-Diaz LG, Sun Y, Weng S, Kim JK, Lam RH, Han L, Fan R, Krebsbach PH & Fu J Nanotopography Influences Adhesion, Spreading, and Self-Renewal of Human Embryonic Stem Cells. ACS Nano, 2012, 6, 4094–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen W, Sun Y & Fu J Microfabricated Nanotopological Surfaces for Study of Adhesion-Dependent Cell Mechanosensitivity. Small, 2013, 9, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kato RB, Roy B, De Oliveira FS, Ferraz EP, De Oliveira PT, Kemper AG, Hassan MQ, Rosa AL & Beloti MM Nanotopography Directs Mesenchymal Stem Cells to Osteoblast Lineage through Regulation of Microrna-Smad-Bmp-2 Circuit. J. Cell. Physiol, 2014, 229, 1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo RO & Dalby MJ Nanoscale Surfaces for the Long-Term Maintenance of Mesenchymal Stem Cell Phenotype and Multipotency. Nat. Mater, 2011, 10, 637–644. [DOI] [PubMed] [Google Scholar]
- (16).Dalby MJ, Gadegaard N & Oreffo RO Harnessing Nanotopography and Integrin-Matrix Interactions to Influence Stem Cell Fate. Nat. Mater, 2014, 13, 558–569. [DOI] [PubMed] [Google Scholar]
- (17).Kim HN, Jiao A, Hwang NS, Kim MS, Kim D-H & Suh K-Y Nanotopography-Guided Tissue Engineering and Regenerative Medicine. Adv. Drug Delivery Rev, 2013, 65, 536–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yang C, Tibbitt MW, Basta L & Anseth KS Mechanical Memory and Dosing Influence Stem Cell Fate. Nat. Mater, 2014, 13, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhong W, Tian K, Zheng X, Li L, Zhang W, Wang S & Qin J Mesenchymal Stem Cell and Chondrocyte Fates in a Multishear Microdevice Are Regulated by Yes-Associated Protein. Stem Cells Dev, 2013, 22, 2083–2093. [DOI] [PubMed] [Google Scholar]
- (20).Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng X-Q, Chien S & Yang C Integrin Activation and Internalization on Soft Ecm as a Mechanism of Induction of Stem Cell Differentiation by Ecm Elasticity. Proc. Natl. Acad. Sci. U. S. A, 2011, 108, 9466–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Langenbach F & Handschel J Effects of Dexamethasone, Ascorbic Acid and B-Glycerophosphate on the Osteogenic Differentiation of Stem Cells in Vitro. Stem Cell Res. Ther, 2013, 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jaiswal N, Haynesworth SE, Caplan AI & Bruder SP Osteogenic Differentiation of Purified, Culture-Expanded Human Mesenchymal Stem Cells in Vitro. J. Cell. Biochem, 1997, 64, 295–312. [PubMed] [Google Scholar]
- (23).Franceschi RT, Iyer BS & Cui Y Effects of Ascorbic Acid on Collagen Matrix Formation and Osteoblast Differentiation in Murine Mc3t3-E1 Cells. J. Bone Miner. Res, 1994, 9, 843–854. [DOI] [PubMed] [Google Scholar]
- (24).Kundu AK, Khatiwala CB & Putnam AJ Extracellular Matrix Remodeling, Integrin Expression, and Downstream Signaling Pathways Influence the Osteogenic Differentiation of Mesenchymal Stem Cells on Poly (Lactide-Co-Glycolide) Substrates. Tissue Eng., Part A, 2008, 15, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hamidouche Z, Haÿ E, Vaudin P, Charbord P, Schüle R, Marie PJ & Fromigué O Fhl2 Mediates Dexamethasone-Induced Mesenchymal Cell Differentiation into Osteoblasts by Activating Wnt/B-Catenin Signaling-Dependent Runx2 Expression. FASEB J, 2008, 22, 3813–3822. [DOI] [PubMed] [Google Scholar]
- (26).Hong D, Chen H-X, Xue Y, Li D-M, Wan X-C, Ge R & Li J-C Osteoblastogenic Effects of Dexamethasone through Upregulation of Taz Expression in Rat Mesenchymal Stem Cells. J. Steroid Biochem. Mol. Biol, 2009, 116, 86–92. [DOI] [PubMed] [Google Scholar]
- (27).Chen W & Ahmed H Fabrication of 5–7 Nm Wide Etched Lines in Silicon Using 100 Kev Electron-Beam Lithography and Polymethylmethacrylate Resist. Appl. Phys. Lett, 1993, 62, 1499–1501. [Google Scholar]
- (28).Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ & Nealey PF Epithelial Contact Guidance on Well-Defined Micro-and Nanostructured Substrates. J. Cell. Sci, 2003, 116, 1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gallagher J, McGhee K, Wilkinson C & Riehle M Interaction of Animal Cells with Ordered Nanotopography. IEEE Trans. Nanobiosci, 2002, 1, 24–28. [DOI] [PubMed] [Google Scholar]
- (30).Yim EK, Pang SW & Leong KW Synthetic Nanostructures Inducing Differentiation of Human Mesenchymal Stem Cells into Neuronal Lineage. Exp. Cell Res, 2007, 313, 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chou SY, Krauss PR & Renstrom PJ Imprint Lithography with 25-Nanometer Resolution. Science, 1996, 272, 85. [Google Scholar]
- (32).Schift H Nanoimprint Lithography: An Old Story in Modern Times? A Review. J. Vac. Sci. Technol., B: Microelectron. Nanometer. Struct. Process. Meas. Phenom, 2008, 26, 458–480. [Google Scholar]
- (33).Chen W, Villa-Diaz L, Sun Y, Weng S, Kim J, Lam R, Han L, Fan R, Krebsbach P & Fu J Nanotopography Influences Adhesion, Spreading, and Self-Renewal of Human Embryonic Stem Cells. ACS Nano, 2012, 6, 4094–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Krinner A, Zscharnack M, Bader A, Drasdo D & Galle J Impact of Oxygen Environment on Mesenchymal Stem Cell Expansion and Chondrogenic Differentiation. Cell Prolif, 2009, 42, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Oreffo RO, Cooper C, Mason C & Clements M Mesenchymal Stem Cells. Stem Cell Rev, 2005, 1, 169–178. [DOI] [PubMed] [Google Scholar]
- (36).Bianco P & Robey PG Stem Cells in Tissue Engineering. Nature, 2001, 414, 118–121. [DOI] [PubMed] [Google Scholar]
- (37).Li J, Mou X, Qiu J, Wang S, Wang D, Sun D, Guo W, Li D, Kumar A & Yang X Surface Charge Regulation of Osteogenic Differentiation of Mesenchymal Stem Cell on Polarized Ferroelectric Crystal Substrate. Adv. Healthc. Mater, 2015, 4, 998–1003. [DOI] [PubMed] [Google Scholar]
- (38).Wang G, Zheng L, Zhao H, Miao J, Sun C, Ren N, Wang J, Liu H & Tao X In Vitro Assessment of the Differentiation Potential of Bone Marrow-Derived Mesenchymal Stem Cells on Genipin-Chitosan Conjugation Scaffold with Surface Hydroxyapatite Nanostructure for Bone Tissue Engineering. Tissue Eng., Part A, 2011, 17, 1341–1349. [DOI] [PubMed] [Google Scholar]
- (39).Hynes R Integrins: Bidirectional, Allosteric Signaling Machines. Cell, 2002. [DOI] [PubMed] [Google Scholar]
- (40).Qin J, Vinogradova O & Plow E Integrin Bidirectional Signaling: A Molecular View. PLoS. Biol, 2004, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Askari JA, Buckley PA, Mould AP & Humphries MJ Linking Integrin Conformation to Function. J. Cell Sci, 2009, 122, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Geiger B, Spatz J & Bershadsky A Environmental Sensing through Focal Adhesions. Nat. Rev. Mol. Cell Biol, 2009, 10, 21–33. [DOI] [PubMed] [Google Scholar]
- (43).Zaidel-Bar R & Geiger B The Switchable Integrin Adhesome. J. Cell. Sci, 2010, 123, 1385–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA & Griffith LG Co-Regulation of Cell Adhesion by Nanoscale Rgd Organization and Mechanical Stimulus. J. Cell. Sci, 2002, 115, 1423–1433. [DOI] [PubMed] [Google Scholar]
- (45).Zhang Y, Gordon A, Qian W & Chen W Engineering Nanoscale Stem Cell Niche: Direct Stem Cell Behavior at Cell-Matrix Interface. Adv. Healthc. Mater, 2015, 4, 1900–1914. [DOI] [PubMed] [Google Scholar]
- (46).DuFort C, Paszek M & Weaver V Balancing Forces: Architectural Control of Mechanotransduction. Nat. Rev. Mol. Cell Biol, 2011, 12, 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang N, Tytell JD & Ingber DE Mechanotransduction at a Distance: Mechanically Coupling the Extracellular Matrix with the Nucleus. Nat. Rev. Mol. Cell Biol, 2009, 10, 75–82. [DOI] [PubMed] [Google Scholar]
- (48).Dalby M, Gadegaard N & Oreffo R Harnessing Nanotopography and Integrin-Matrix Interactions to Influence Stem Cell Fate. Nat. Mater, 2014, 13, 558–569. [DOI] [PubMed] [Google Scholar]
- (49).Mitra S, Hanson D & Schlaepfer D Focal Adhesion Kinase: In Command and Control of Cell Motility. Nat. Rev. Mol. Cell Biol, 2005, 6, 56–68. [DOI] [PubMed] [Google Scholar]
- (50).Seong J, Tajik A, Sun J, Guan JL, Humphries MJ, Craig SE, Shekaran A, Garcia AJ, Lu SY, Lin MZ, Wang N & Wang YX Distinct Biophysical Mechanisms of Focal Adhesion Kinase Mechanoactivation by Different Extracellular Matrix Proteins. Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 19372–19377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Teo BKK, Wong ST, Lim CK, Kung TY, Yap CH, Ramagopal Y, Romer LH & Yim EK Nanotopography Modulates Mechanotransduction of Stem Cells and Induces Differentiation through Focal Adhesion Kinase. ACS Nano, 2013, 7, 4785–4798. [DOI] [PubMed] [Google Scholar]
- (52).Hynes RO Integrins: Versatility, Modulation, and Signaling in Cell Adhesion. Cell, 1992, 69, 11–25. [DOI] [PubMed] [Google Scholar]
- (53).Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M & Schwartz MA Integrins in Mechanotransduction. Curr. Opin. Cell Biol, 2013, 25, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Riggs KA, Hasan N, Humphrey D, Raleigh C, Nevitt C, Corbin D & Hu C Regulation of Integrin Endocytic Recycling and Chemotactic Cell Migration by Syntaxin 6 and Vamp3 Interaction. J. Cell Sci, 2012, 125, 3827–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Bridgewater RE, Norman JC & Caswell PT Integrin Trafficking at a Glance. J. Cell. Sci, 2012, 125, 3695–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Engler AJ, Sen S, Sweeney HL & Discher DE Matrix Elasticity Directs Stem Cell Lineage Specification. Cell, 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- (57).Pockwinse SM, Stein JL, Lian JB & Stein GS Developmental Stage-Specific Cellular Responses to Vitamin D and Glucocorticoids During Differentiation of the Osteoblast Phenotype: Interrelationship of Morphology and Gene Expression by in Situ Hybridization. Exp. Cell. Res, 1995, 216, 244–260. [DOI] [PubMed] [Google Scholar]
- (58).Sikavitsas VI, Temenoff JS & Mikos AG Biomaterials and Bone Mechanotransduction. Biomaterials, 2001, 22, 2581–2593. [DOI] [PubMed] [Google Scholar]
- (59).Thomas CH, Collier JH, Sfeir CS & Healy KE Engineering Gene Expression and Protein Synthesis by Modulation of Nuclear Shape. Proc. Natl. Acad. Sci. U. S. A, 2002, 99, 1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, Chen CS & Moghe PV Cytoskeleton-Based Forecasting of Stem Cell Lineage Fates. Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).McBeath R, Pirone DM, Nelson CM, Bhadriraju K & Chen CS Cell Shape, Cytoskeletal Tension, and Rhoa Regulate Stem Cell Lineage Commitment. Dev. Cell, 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- (62).Pablo Rodríguez J, González M, Ríos S & Cambiazo V Cytoskeletal Organization of Human Mesenchymal Stem Cells (Msc) Changes During Their Osteogenic Differentiation. J. Cell. Biochem, 2004, 93, 721–731. [DOI] [PubMed] [Google Scholar]
- (63).Alhussein G, Shanti A, Farhat IA, Timraz SB, Alwahab NS, Pearson YE, Martin MN, Christoforou N & Teo J A Spatiotemporal Characterization Method for the Dynamic Cytoskeleton. Cytoskeleton, 2016, 73, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Pearson YE, Lund AW, Lin AW, Ng CP, Alsuwaidi A, Azzeh S, Gater DL & Teo JC Non-Invasive Single-Cell Biomechanical Analysis Using Live-Imaging Datasets. J. Cell Sci, 2016, 129, 3351–3364. [DOI] [PubMed] [Google Scholar]
- (65).Darling EM, Zauscher S, Block JA & Guilak F A Thin-Layer Model for Viscoelastic, Stress-Relaxation Testing of Cells Using Atomic Force Microscopy: Do Cell Properties Reflect Metastatic Potential? Biophys. J, 2007, 92, 1784–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Elkin BS, Azeloglu EU, Costa KD & Morrison Iii B Mechanical Heterogeneity of the Rat Hippocampus Measured by Atomic Force Microscope Indentation. J. Neurotrauma, 2007, 24, 812–822. [DOI] [PubMed] [Google Scholar]
- (67).Discher DE, Janmey P & Wang Y -l. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science, 2005, 310, 1139–1143. [DOI] [PubMed] [Google Scholar]
- (68).Han SJ, Bielawski KS, Ting LH, Rodriguez ML & Sniadecki NJ Decoupling Substrate Stiffness, Spread Area, and Micropost Density: A Close Spatial Relationship between Traction Forces and Focal Adhesions. Biophys. J, 2012, 103, 640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kraning-Rush CM, Califano JP & Reinhart-King CA Cellular Traction Stresses Increase with Increasing Metastatic Potential. PLoS ONE, 2012, 7, e32572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Chrzanowska-Wodnicka M & Burridge K Rho-Stimulated Contractility Drives the Formation of Stress Fibers and Focal Adhesions. J. Cell Biol, 1996, 133, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K & Chen CS Cells Lying on a Bed of Microneedles: An Approach to Isolate Mechanical Force. Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z & Chen CS Mechanical Regulation of Cell Function with Geometrically Modulated Elastomeric Substrates. Nat. Meth, 2010, 7, 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Chen CS, Tan J & Tien J Mechanotransduction at Cell-Matrix and Cell-Cell Contacts. Annu. Rev. Biomed. Eng, 2004, 6, 275–302. [DOI] [PubMed] [Google Scholar]
- (74).Kee Y-S & Robinson DN Micropipette Aspiration for Studying Cellular Mechanosensory Responses and Mechanics. Dictyostelium discoideum Protocols, 2013, 367–382. [DOI] [PubMed] [Google Scholar]
- (75).Haase K & Pelling AE Investigating Cell Mechanics with Atomic Force Microscopy. J. Royal Soc. Interface, 2015, 12, 20140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ekpenyong AE, Whyte G, Chalut K, Pagliara S, Lautenschläger F, Fiddler C, Paschke S, Keyser UF, Chilvers ER & Guck J Viscoelastic Properties of Differentiating Blood Cells Are Fate-and Function-Dependent. PLoS ONE, 2012, 7, e45237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Tee S-Y, Fu J, Chen CS & Janmey PA Cell Shape and Substrate Rigidity Both Regulate Cell Stiffness. Biophys. J, 2011, 100, L25–L27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tan SC, Pan WX, Ma G, Cai N, Leong KW & Liao K Viscoelastic Behaviour of Human Mesenchymal Stem Cells. BMC Cell Biol, 2008, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Hong J-H, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM & Sharp PA Taz, a Transcriptional Modulator of Mesenchymal Stem Cell Differentiation. Science, 2005, 309, 1074–1078. [DOI] [PubMed] [Google Scholar]
- (80).MacQueen L, Sun Y & Simmons CA Mesenchymal Stem Cell Mechanobiology and Emerging Experimental Platforms. J. Royal Soc. Interface, 2013, 10, 20130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Talele NP, Fradette J, Davies JE, Kapus A & Hinz B Expression of A-Smooth Muscle Actin Determines the Fate of Mesenchymal Stromal Cells. Stem Cell Rep, 2015, 4, 1016–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M & Bicciato S Role of Yap/Taz in Mechanotransduction. Nature, 2011, 474, 179–183. [DOI] [PubMed] [Google Scholar]
- (83).Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH & Fu J Hippo/Yap-Mediated Rigidity-Dependent Motor Neuron Differentiation of Human Pluripotent Stem Cells. Nat. Mater, 2014, 13, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Pan D The Hippo Signaling Pathway in Development and Cancer. Dev. Cell, 2010, 19, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zhao B, Tumaneng K & Guan K-L The Hippo Pathway in Organ Size Control, Tissue Regeneration and Stem Cell Self-Renewal. Nat. Cell Biol, 2011, 13, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW & Wrana JL Taz Controls Smad Nucleocytoplasmic Shuttling and Regulates Human Embryonic Stem-Cell Self-Renewal. Nat. Cell Biol, 2008, 10, 837–848. [DOI] [PubMed] [Google Scholar]
- (87).Serrano I, McDonald PC, Lock F, Muller WJ & Dedhar S Inactivation of the Hippo Tumour Suppressor Pathway by Integrin-Linked Kinase. Nat Commun, 2013, 4, 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Wada K, Itoga K, Okano T, Yonemura S & Sasaki H Hippo Pathway Regulation by Cell Morphology and Stress Fibers. Development, 2011, 138, 3907–3914. [DOI] [PubMed] [Google Scholar]
- (89).Zhao B, Li L, Wang L, Wang CY, Yu J & Guan KL Cell Detachment Activates the Hippo Pathway Via Cytoskeleton Reorganization to Induce Anoikis. Genes Dev, 2012, 26, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G & Sahai E Mechanotransduction and Yap-Dependent Matrix Remodelling Is Required for the Generation and Maintenance of Cancer-Associated Fibroblasts. Nat. Cell Biol, 2013, 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Sun Y, Chen CS & Fu J Forcing Stem Cells to Behave: A Biophysical Perspective of the Cellular Microenvironment. Annu. Rev. Biophys, 2012, 41, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Lin GL, Cohen DM, Desai RA, Breckenridge MT, Gao L, Humphries MJ & Chen CS Activation of Beta 1 but Not Beta 3 Integrin Increases Cell Traction Forces. FEBS Lett, 2013, 587, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Gershlak JR & Black LD Beta 1 Integrin Binding Plays a Role in the Constant Traction Force Generation in Response to Varying Stiffness for Cells Grown on Mature Cardiac Extracellular Matrix. Exp. Cell Res, 2015, 330, 311–324. [DOI] [PubMed] [Google Scholar]
- (94).Shih YRV, Tseng KF, Lai HY, Lin CH & Lee OK Matrix Stiffness Regulation of Integrin-Mediated Mechanotransduction During Osteogenic Differentiation of Human Mesenchymal Stem Cells. J. Bone Miner. Res, 2011, 26, 730–738. [DOI] [PubMed] [Google Scholar]
- (95).Caplan AI Mesenchymal Stem Cells. J. Orthop. Res, 1991, 9, 641–650. [DOI] [PubMed] [Google Scholar]
- (96).Crisan M, Yap S, Casteilla L, Chen C-W, Corselli M, Park TS, Andriolo G, Sun B, Zheng B & Zhang L A Perivascular Origin for Mesenchymal Stem Cells in Multiple Human Organs. Cell. Stem. Cell, 2008, 3, 301–313. [DOI] [PubMed] [Google Scholar]
- (97).Lim C, Zhou E & Quek S Mechanical Models for Living Cells—a Review. J. Biomech, 2006, 39, 195–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


