Abstract
Background
Natural propolis has been used since decades owing to its broad-spectrum activities. Burn injuries are a global health problem with negative impacts on communities. Bacterial infections usually accompany burns, which demand implementation of antibiotics. Antibiotics abuse led to emergence of microbial drug resistance resulting in poor treatment outcomes. In such instances, the promising alternative would be natural antimicrobials such as propolis.
Objective
Full chemical profiling of propolis and evaluation of in vitro antibacterial, antioxidant and anti-inflammatory activities as well as in vivo burn healing properties.
Methods
Chemical profiling of propolis was performed using Liquid chromatography (UHPLC/MS-PDA and HPLC-PDA). In vitro assessment was done using Disc Diffusion susceptibility test against Staphylococcus aureus and infected burn wound mice model was used for in vivo assessment. In vitro antioxidant properties of propolis were assessed using DPPH, ABTS and FRAP techniques. The anti-inflammatory effect of propolis was assessed against lipopolysaccharide/interferon-gamma mediated inflammation.
Results
UHPLC/MS-PDA results revealed identification of 71 phytochemicals, mainly flavonoids. Upon flavonoids quantification (HPLC–PDA), Pinocembrin, chrysin and galangin recorded high content 21.58±0.84, 22.73±0.68 and 14.26±0.70 mg/g hydroalcoholic propolis extract, respectively. Propolis showed concentration dependent antibacterial activity in vitro and in vivo burn healing via wound diameter reduction and histopathological analysis without signs of skin irritation in rabbits nor sensitization in guinea pigs. Propolis showed promising antioxidant IC50 values 46.52±1.25 and 11.74±0.26 μg/mL whereas FRAP result was 445.29±29.9 μM TE/mg. Anti-inflammatory experiment results showed significant increase of Toll-like receptor 4 (TLR4), interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α) mRNA levels. Nitric oxide and iNOS were markedly increased in Griess assay and western blot respectively. However, upon testing propolis against LPS/IFN-γ-mediated inflammation, TLR4, IL-6 and TNF-α expression were downregulated at transcriptional and post-transcriptional levels.
Conclusion
Propolis proved to be a promising natural burn healing agent through its antibacterial, antioxidant and anti-inflammatory activities.
Introduction
Propolis (honeybee glue) is a natural resinous mixture of botanical balsams and resin with digestive enzymes of bees collected by honeybees Apis mellifera, from various plant sources. Propolis enriched with a myriad of natural pharmacologically active constituents such as polyphenols, terpenoids, steroids, and amino acids [1]. Propolis displays a broad spectrum of pharmacological and biological features viz. antimicrobial, anti-inflammatory, immunomodulatory, antioxidant, anticancer, antiulcer, antitumor, cardioprotective, neuroprotective and hepatoprotective actions [2]. The beneficial properties and chemical constitution of propolis differ greatly depending on the geographical origin, seasonal collection time, and botanical source [3].
Propolis has been used since ancient times in traditional medicine to keep good health, owing to its broad-spectrum activities [4]. Nowadays, it is used as a therapeutic agent to avoid different diseases and improve health [5].
Burn injuries are a major global public health problem given their high incidence and potentially devastating physical, psychosocial, and financial impacts on individuals, households, and communities [6–8]. Burns from fire, heat, and hot substances are the fourth most common type of civilian trauma worldwide, following road traffic incidents, falls, and interpersonal violence [9]. It is estimated that there are between 7 and 12 million people (up to 33,000 each day) who sustain burn injuries that require medical care, leading to prolonged absence from work or school, or result in death each year [10].
Initial burn wounds are sterile, but within a few days burns destroy the barrier between internal sterile tissue and the external environment leading to microbial penetration. Microbes can easily colonize burnt tissue as it represents a protein-rich environment favorable for microbial growth [11]. Gram positive bacteria start to colonize the burn sites from deeper structures (hair follicles and glands) leading to infection [12], if left untreated, this may be followed by a second phase, where Gram-negative bacteria such as Pseudomonas aeruginosa, Escherichia coli, Acinetobacter baumannii, and Klebsiella spp. become the predominant bacteria [13].
Healing of burn wounds is a complex, multi-step process prone to internal and external factors and implementation of antimicrobial therapy is often demanded. Antibiotics abuse has led over years to the emergence of microbial drug resistance leading to poor treatment efficiency. In such instances, the promising alternative is the use of natural antimicrobial, anti-inflammatory and antioxidant compounds, including plant extracts and those of bee origin, such as propolis [14–16].
The aim of this study is to assess the antibacterial effect and burn healing activity of hydroalcoholic extract of propolis in vitro and in vivo using infected burn-induced mouse model. In addition to studying the effect of propolis on TLR4 pathway through gene expression studies of key inflammatory intermediaries such as TLR4, TNF-α, IL-6 and iNOS. As antioxidant activity; propolis has been assesed in vitro using different techniques viz. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) and 2,2′-azinobis (3- ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) and ferric reducing antioxidant potential (FRAP) techniques.
A full chemical profiling of the total hydroalcoholic propolis extract was investigated by ultra-high performance liquid chromatography/mass spectroscopy-photodiode array detector (UHPLC/MS-PDA) in addition to standardization of the hydroalcoholic propolis extract using phenolics viz. gallic acid, quercetin, chrysin and galangin via high performance liquid chromatography–PDA (HPLC–PDA).
Materials and methods
Raw propolis botanical source was from South Asia and it was obtained as dried raw powder from Imtenan Health Co., Cairo, Egypt. The authentics used in the standardization of the hydroalcoholic extract of propolis viz. pinocembrin, chrysin and galangin have been obtained from Sigma-Aldrich (St. Louis, Mo., USA). All the solvents used for the extraction and analysis met the quality criteria and in accordance with the international standards.
LPS (Escherichia coli 0111: B4) was obtained from Sigma Chemical Co. (St. Louis, MO, USA) (Cat No. L2630). Murine interferon-γ was purchased from PeproTech (Rocky Hill, NJ, USA) (Cat No. 315–05). High-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Cat No. 41965–039) and fetal bovine serum (FBS) (Cat No. 10270–106) were obtained from Gibco. Isopropanol (HPLC grade; Cat No. BP26324), chloroform (HPLC grade; Cat No. C607SK-1), Dimethyl sulfoxide (DMSO; Cat No. 67-68-5) and ethanol (HPLC grade; Cat No. 64-17-5) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Revert Aid cDNA kit (Cat No. K1621), Maxima SYBR Green qPCR (Cat No. K0251) were all obtained from Thermo Fisher Scientific (Waltham, MA, USA) as well. The Griess assay kit (Cat No. G7921) was bought from Invitrogen (Carlsbad, CA, USA). Phosphate-buffered saline (PBS; 10X; Cat No. 17-516Q) and penicillin–streptomycin mixture (Cat No. 09-757F), and DMEM with 4.5 g/L glucose, without L-glutamine and without phenol red (Cat No. 12-917F) were obtained from Lonza Bioscience (Basel, Switzerland). MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (CT01-5 Sigma-Aldrich). The murine macrophage-like cell lines RAW 264.7 (ATCC-TIB71, Rockville, MD, USA) were obtained by the National Research Center (NRC) (Cairo, Egypt). QiAzol lysis buffer (Cat No. 79306) was purchased from Qiagen (Hilden, Germany). Staphylococcus aureus; gram positive bacteria; an isolated clinical strain known to localize burn wound infections was used in mouse model experiments.
Extraction procedure for chromatographic analysis and biological study
Dried raw propolis powder (75 g) was extracted with hydro methanol (80%). The obtained extract was further concentrated on rotary evaporator to yield 45 g and then it was kept in -20°C for further analysis.
UHPLC/MS-PDA analysis of propolis hydroalcoholic extract
The UHPLC/MS-PDA method of identification of secondary metabolites has been adopted from previous literature [17]. The negative HR-ESI and CID (collision induced dissociation) obtained from the mass spectrometer adjusted at 300 and 250°C of capillary and source heart temperatures, respectively, while the resolution of Fourier Transform Mass Spectrometry (FTMS) was 30.000 using inert N2 gas. The CID spectra (He gas as buffer) was recorded using normalized collision energy (NCE) of 35%. The separation using UHPLC was performed at 40°C, the injection volume was 2 μL and the flow rate was 150 μL/min. The gradient elution was carried out using solvent (A) water and (B) acetonitrile with 0.1% formic acid as follows: 0–1 min isocratic 5% (B), 1–11 min linear from 5 to 100% (B), 11–19 min, isocratic 100% (B) and 19–30 min, isocratic 5% (B).
The mass data were identified by XCalibur 2.2 SP1 software. Chemical constituents were identified by their UV–VIS spectra, retention times relative to external standards, mass spectra were compared to phytochemical library of natural products database mass bank (CRC press) and reference literature.
HPLC-PDA standardization of propolis hydroalcoholic extract
The major flavonoids viz. pinocembrin, chrysin and galangin have been identified and quantified in the hydroalcoholic extract of propolis using Waters 2690 Alliance High performance liquid chromatography (HPLC) system equipped with a Waters 996 photodiode array detector (PDA). A Kromasil Eternity reversed phase column (C18) was used with specifications (4.6 mm, 250 mm, 5μm particle size). Elution of major flavonoids in the propolis extract was achieved using 0.1% formic acid in water as solvent (A), and methanol 100% as solvent (B). The total run time was 65 min where gradient elution was adopted. The elution run started by 80% of solvent (A) 0–5 min, 40% (5–45 min), then 20% (45–60 min), then finally back to 80% (60–65 min). The flow rate was 1 mL/min with injection volume 10 μL. The UV detection was set at 290 nm for all flavonoids [18].
Standard calibration curves of propolis extract
The three authentic flavonoids were prepared separately viz. pinocembrin, chrysin and galangin in concentration 1 mg/mL (100% methanol). Serial dilutions (1–200 μg/mL) were then prepared for calibration curves. Quantification of the previous mentioned flavonoids in propolis extract was assembled using peak area (Y) and concentration (X) calculated as μg/mL, mean value (n = 3) ± standard deviation. The propolis extract was prepared in 1 mg/mL methanol using the following equations for the previous mentioned flavonoids: Y = 1.3184x – 1.4266 (R2 0.999), Y = 1.2417x-0.3396 (R2 0.999), Y = 0.9425x-1.7331 (R2 0.998), respectively.
In vitro antimicrobial evaluation of propolis hydroalcoholic extract
Two propolis concentrations were used in all in vitro and in vivo experiments. The two concentrations prepared contained 100% and 50% of propolis extract; where the 100% concentration was pure propolis extract with no water added and the second concentration 50% propolis was diluted with 50% water w/w.
Disc diffusion susceptibility test
To test the antibacterial activity of the 100% and 50% w/w propolis; Disc Diffusion antibiotic susceptibility test (Kirby Bauer) was performed [19] using Mueller Hinton agar (Oxoid, UK). Bacterial suspension was prepared corresponding to 0.5 McFarland standard solution, where 5–6 bacterial isolate colonies were emulsified in sterile distilled water and the turbidity was adjusted to 1.5 × 108 CFU/mL (equivalent to 0.5 McFarland standard) [20]. Following this step, a sterile cotton swab was used to inoculate the standardized bacterial suspension on the Muller Hinton agar plates evenly, then plates were left 5 min to dry. Filter paper discs (6 mm diameter—prepared according to Cheesbrough method [21] were immersed in both concentrations of propolis until saturation, then placed on the agar plates with light pressure to ensure complete contact with agar. A space of 15 mm was kept from the plates’ external margin to prevent overlapping of inhibition zones, a positive control was used (oxacillin disk 15 g). Plates were left for 15 min then incubated at 37°C for 3–5 days. After incubation, examination of plates took place and the zones of inhibition diameter were recorded.
Minimum Inhibitory concentration (MIC) and Minimum Bactericidal concentration (MBC)
Broth dilution method was used to estimate the MIC of the tested propolis, where serial dilutions of propolis stock solution (2, 1, 0.5, 0.25 and 0.125 mg/mL) were prepared in ethanol followed by adding a drop of bacterial standard inoculum prepared using Mueller Hinton Broth (DifcoTM). Then the test tubes were incubated at 37°C for 24 h followed by checking for turbidity to determine the MIC.
0.1 mL of the MIC test tubes showing no visible growth were sub-cultured on Mueller Hinton agar (DifcoTM) plates to estimate the MBC. After 24 h incubation, the plates were examined where the lowest concentration with no observable growth on subculture was regarded as the MBC.
In vivo antibacterial assessment and burn healing evaluation
Burn wound animal model was used for assessment of burn healing effect of the two propolis concentrations. 10 albino mice (weight 30–35 g), 8 weeks old, were used. Mice were separately kept in clean polyethylene cages under standard experimental conditions at 23±2°C. Mice were anaesthetized using isoflurane inhalation anesthesia device then shaved on the back using an electric clipper followed by a shaving cream. Ethanol (70% v/v) was used to disinfect the shaved area. Burn wounds were induced using a 10 mm diameter cylindrical metal rod, pre-heated over an open flame for 20 seconds then pressed for 10 seconds on the dorsal mouse skin surface under anesthesia, leading to the formation of a burn wound of diameter 10 mm. Finally, a total of 4 burn sites were generated on the dorsal skin of each mouse. All animal work was performed in agreement with the guidelines stated in the Guide for the Care and Use of Laboratory Animals and was approved by the Ethics Committee for Experimental, Clinical and Chemical Studies at Faculty of Pharmacy, The British University in Egypt (Serial number: EX-2204). At the end of in vivo burn healing evaluation experiment mice were humanely euthanized with CO2 inhalation according to institutional guidelines then dorsal skin samples were collected then subjected to histopathological analysis. After skin autopsy euthanized mice were frozen till incineration.
Bacterial infection induction
Bacterial infection of the induced burn wounds was done by inoculation of 1.5 x 107 colony forming unit (CFU) of bacteria. Using sterile cotton swab, 1 mL bacteria suspension was inoculated evenly by direct rubbing onto the fresh wound.
Treatments
Each mouse received four treatments: one treatment per burn wound position. The first burn position was treated with propolis 100%, the second position was treated with propolis 50%, the third position was treated with normal saline acting as a negative control (N), and finally the fourth position was treated with a commercial burn healing cream (silver sulfadiazine) as a positive control (P). All treatments were applied topically by rubbing on the respective mouse position once daily using a sterile cotton swab and continued daily until apparent skin healing in any of the positions. All mice were fed on commercial pellet and water ad libitum through the study.
Measurement of burn area
Burn healing evaluation was estimated by measuring the decrease in the induced original burn diameter. A digital caliper was used to measure the burn diameter (mm) daily before treatment application. Recorded measurements were saved for more analysis.
Assessment of bacterial load of the infected burns
Swabs from the surface of burn positions were collected daily before the application of treatments. Sterile cotton swabs were rubbed over the total burn area; especially where the degree of burn is maximum; for 10 seconds with slight pressure. For estimating the bacterial load in each position; serial dilutions of homogenized samples in 1 mL sterile saline were plated on sterile agar media. Plates were incubated aerobically at 37°C for 24 h. Subsequently, counting of bacterial colonies was done by visual inspection. Colony forming unit (CFU) was used to determine the total bacterial count on each plate.
Skin irritation test
Skin irritation test was done using rabbits after removal of fur on a section of the back. Propolis 100% and 50% were administered on the shaved skin then kept in contact for 4h. Skin irritation grade was scored for erythema, edema formation, eschar and corrosive effect. Observations continued for 24 h. Rabbits were housed for use in further research after a washout period.
Skin sensitization test
Skin sensitization test was done using guinea pigs after removal of fur on a section of the back. Propolis 100% and 50% were administered daily to the skin for 14 days. Two weeks after the final administration, the guinea pigs were challenged by application of propolis 100% and 50%, and the appearance of erythematous reaction was assessed. Guinea pigs were housed for use in further research after a washout period.
Histopathological analysis
Autopsy dorsal skin samples were collected from mice after observing apparent skin healing. A total of 40 skin samples were collected: four from each mouse demonstrating the locations of treatments with a total of 10 samples for each treatment. The Collected skin samples were fixed in 10% formalin solution for 24 h. Samples were washed with tap water followed by dehydration using serial dilutions of alcohol (methyl alcohol, ethyl alcohol and absolute ethyl alcohol). Skin samples were cleared in Xylene then embedded in paraffin at 56°C in hot air oven for 24 h. Paraffin bees wax tissue blocks were prepared for sectioning at 4 microns thickness by rotary LEITZ microtome. The obtained tissue sections were collected on glass slides, de-paraffinized and stained by hematoxylin & eosin stain (H&E) for observation using the light electric microscope [22]. Digital photomicrographs were taken, and the burns were assessed for the degree of acanthosis, formation and regeneration of new blood capillaries.
In vitro antioxidant activity of propolis hydroalcoholic extract
Different techniques have been assessed to evaluate the antioxidant power of the hydroalcoholic extract of propolis. The spectrophotometric techniques applied in this study adopted the principle of electron transfer as well as reduction of the colored oxidant as DPPH, ABTS and FRAP. In DPPH and ABTS assays, change in color is due to the reduction power of the antioxidant sample whereas in FRAP, the change in color is due to the change of ferric ion to ferrous by the influence of the antioxidant sample. Implementation of different antioxidant assays would be effective in confirmation of the results [23].
Propolis hydroalcoholic extract and trolox standard preparations
Propolis extract was prepared in ethanol at serial dilution (25–100 μg/mL), (5–25 μg/mL) and 0.8 mg/mL concentration. Trolox standard (20 μg/mL in methanol) was prepared in methanol in dilutions ranged from 1.25–6.25 μg/mL, 2.5–8.75 μg/mL and 25–3000 μM in the antioxidant assays DPPH, ABTS and FRAP, respectively. The procedures have been applied as previously mentioned in detail in previous literature [24]. A microplate reader FluoStar Omega has been used for data analysis and IC50 was calculated via GraphPad Prism 6 for DPPH and ABTS assays. The antioxidant activity of FRAP assay has been expressed in trolox equivalents (TE). All the results expressed as mean ± standard deviation of triplet measurements.
Cell culture for anti-inflammatory activity of propolis hydroalcoholic extract
Murine macrophage RAW 264.7 cell line was cultured at 37°C in incubator with 5% CO2. The cells were kept in high-glucose DMEM containing 1% Pen-Strep (100 units/mL penicillin and 100 μg/mL streptomycin) and 10% heat inactivated fetal bovine serum (FBS). For MTT and Griess assay, the cells were seeded at density of 2×105 in 96-well plates or at a density of 2×106 cells/well in 6-well plate for RNA extraction for 2 h. RAW 264.7 cells were stimulated by LPS (10 ng/mL) and murine interferon-γ (10 U/mL) for 24 h with or without propolis. DMSO (0.1% v/v) was used as vehicle control.
Cell viability
The colorimetric MTT assay was done to determine the nontoxic concentration of propolis as previously described [25]. RAW 264.7 macrophages were exposed for 24 h to propolis at increasing concentrations of (3.125 to 400 μM) alone or with LPS/ IFN-γ (10 ng/mL/10 U/mL). After 24 h, a new serum-free medium with 1 mg/mL MTT was added. The formed formazan crystals were dissolved with isopropanol and the OD was measured at 540 nm using Nano SPECTROstar microplate reader (BMG LABTECH, Ortenberg, Germany). Cell viability was calculated as the percentage of viable macrophages relative to the control.
Nitrite assay
The Griess method was used to determine nitrite concentration as an indicator of NO production as previously described [26]. Cells were seeded for 2 h, activated by LPS/ IFN-γ (10 ng/mL/10 U/mL), and incubated with different concentrations of propolis (3.125 to 400 μM). Supernatant medium (150 μL) was diluted with deionized water (130 μL) and Griess reagent (20 μL) using Nano SPECTROstar microplate reader, the OD was measured at 550 nm. Nitrite concentration was calculated with reference to the standard sodium nitrite curve.
Quantitative RT-PCR
RAW 264.7 macrophages were seeded at a density of 2×106 in a 6 well plate for 2 h. Then activated by 10 ng/mL LPS plus 10 U/mL IFN-γ and incubated with different concentrations of propolis (3.125 to 400 μM). Total RNA was isolated using QiAzol according to the manufacturer’s instructions. RevertAid First Strand cDNA Synthesis kit was used to synthesize cDNA from 1 μg of total RNA. Relative expressions of mRNA (TLR4, TNF-α and IL-6) were carried on ABI 7500 real-time PCR system (Applied Biosystems) with SYBR green PCR master mix. NCBI primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used for primer design and Thermo Fisher for synthesis Table 1. The relative expression of the target genes was compared to the untreated cells, normalized by GAPDH level, using the 2−ΔΔCT method [27]. The conditions used for RT-PCR mRNA expression: 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Table 1. Primers used for qPCR analyses.
| Primer sequence | Target Gene |
|---|---|
| 5’-CTTTGTCAAGCTCATTTCCTGG- 3’ | GAPDH-F |
| 5’-TCTTGCTCAGTGTCCTTGC- 3’ | GAPDH-R |
| 5’-GATGCTACCAAACTGGATATAATCAG-3’ | IL6-F |
| 5’-CTCTGAAGGACTCTGGCTTTG-3’ | IL6-R |
| 5’-TTCAGAACTTCAGTGGCTGG-3’ | TLR4-F |
| 5’-TGTTAGTCCAGAGAAACTTCCTG-3’ | TLR4-R |
| 5’-GAACTCCAGGCGGTGCCTAT-3’ | TNFa-F |
| 5’-TGAGAGGGAGGCCATTTGGG-3’ | TNFa-R |
Western blot
RAW 264.7 cells were seeded into 6-well plates (2 × 106 cells/well) for 2 h. Then activated by 10 ng/mL LPS plus 10 U/mL IFN-γ and incubated with two concentrations of propolis (50 and 100 μg/mL) for 24 h. Indomethacin (IM, 0.25 mM) was used as an anti-inflammatory positive control.
RAW 264.7 cells were washed using cold PBS and scrapped in cold lysis buffer [RIPA buffer containing protease inhibitor cocktail-100 μl/well]. Cell lysates were incubated on ice for 15 min with occasional vortex every 5 min, then centrifuged at 10000 ×g for 10 min (4°C). BCA kit (Thermofisher Scientific, USA) was used for protein quantification. Samples (20 μg total proteins) were resolved using polyacrylamide gel electrophoresis on a 10% acrylamide/bis acrylamide gel using OmniPAGE TETRAD system (Cleaver scientific, UK) at 150 Volts for 1.5 h. Resolved proteins were electro-transferred onto nitrocellulose membrane (100 Volts for 70 min). Membranes were blocked using skim milk (5% w/v in Tris buffer saline Tween 20, TBST) for 1 h at room temperature. Membranes were probed overnight (4°C) with a rabbit iNOS antibody (Biomatik, Canada) and β-actin (Thermofisher Scientific, USA) under gentle shaking. Probed membranes were washed with TBST 3 times, 5 min each with shaking. Horseradish peroxidase-conjugated secondary antibodies were added to the membranes then incubated with shaking for 1 h at room temperature. For washing TBST (3 × 5 min) was used, and the protein bands were visualized using enzyme chemiluminescence kit (Clarity, Bio-Rad, USA).
Statistical analysis
Our results are presented as mean ± SE of the performed experiments. For western blot, the intensity of each band was measured by using ImageJ software. Comparisons are made with ANOVA followed by Dunnett’s multiple comparison test; *, P < 0.05, compared with control. GraphPad Prism Software (Inc. San Diego, CA, version 5.0) was used.
Results and discussion
Identification of secondary metabolites using UHPLC/MS-PDA
In a previous study by Arafa, et. al. 2018, UPLC-PDA-HRMS identifcation of the major flavonoids in the same propolis sample was depiceted. Only the major flavonoids viz. pinobanksin, chrysin, pinocembrin and galangin have been identified through their mass fragmentation pattern [28]. Hereby in this study, a full chemical profiling of the whole propolis sample was performed using UHPLC/MS-PDA to understand the influence of all phytochemicals on the antibacterial, antioxidant and anti-inflammatory activities of propolis.
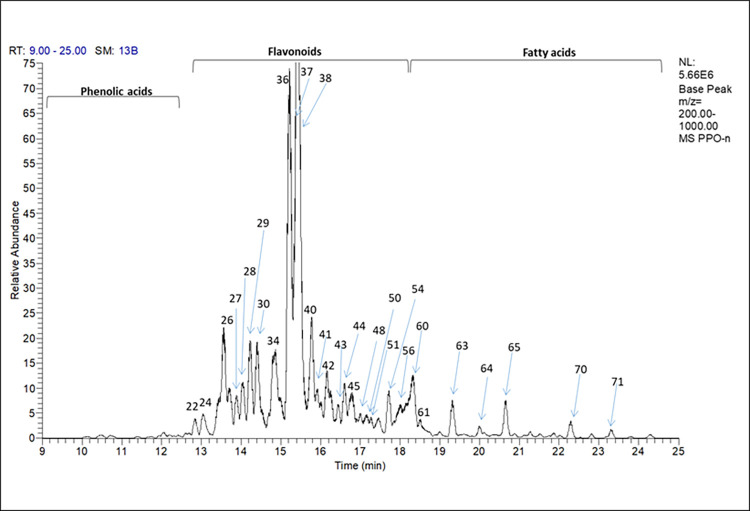
The interpretation of the peaks was carried out on the negative mode since it is highly sensitive for the identification of the phenolic compounds [29]. Table 2 summarizes the compounds identified in the propolis sample. The identification depends on the detection of the [M-H]- and further analysis of the MS2 fragments. The identified compounds were compared to the MS/MS fragmentation of literature data. A total of 71 compounds were identified including sugars, phenolic acids and their derivatives, flavonoids “free, methylated and ester forms” and fatty acids. The total ion chromatogram is illustrated in (Fig 1).
Table 2. Identified propolis hydroalcoholic extract secondary metabolites by UHPLC/MS-PDA in the negative ionization mode.
| Peak | Rt. (min) | UV | (M-H) | Formula-H | MS-MS | Error | ID | Class |
|---|---|---|---|---|---|---|---|---|
| P-1 | 1.09 | 254, 366 | 179.0558 | C6H11O6 | 131, 161 | -1.906 | Hexose | Sugar |
| P-2 | 1.35 | 363, 370 | 221.0661 | C8H13O7 | 161 | -2.515 | Di-O-methyl-hexuronic acid | Sugar acid |
| P-3 | 1.51 | 274 | 169.0141 | C7H5O5 | 125 | -1.045 | Gallic acid | Phenolic acid |
| P-4 | 2.10 | 279 | 131.0348 | C5H7O4 | 87 | -1.084 | Deoxy-arabinolactone | Sugar |
| P-5 | 5.12 | 372 | 181.0489 | C9H9O4 | 137 | -0.167 | Unknown acid | Acid |
| P-6 | 9.06 | 323 | 200.0091 | C11H4O4 | 110 | -11.934 | unknown | _ |
| P-7 | 10.24 | 319 | 193.0503 | C10H9O4 | 149 | -1.772 | Ferulic acid | Phenolic acid |
| P-8 | 10.34 | 319 | 287.0537 | C15H11O6 | 177 | -8.47 | unknown | _ |
| P-9 | 10.45 | 319 | 315.0863 | C17H15O6 | 161, 179, 271 | -1.853 | Caffeic acid derivative | Phenolic acid |
| P-10 | 10.87 | 298, 323 | 609.1472 | C27H29O16 | 301, 464 | -4.035 | Rutin | Flavonol glycoside |
| P-11 | 10.90 | 298, 323 | 267.0849 | C13H15O6 | 193 | -9.403 | 1-O-Feruloyl-glycerol | Phenolic acid |
| P-12 | 10.91 | 244 | 165.0538 | C9H9O3 | 121, 150 | -11.86 | unknown acid | Acid |
| P-13 | 11.28 | 280 | 445.1486 | C23H25O9 | 255, 265, 341, 385 | -4.146 | Pinocembrin derivative | Flavanone |
| P-14 | 11.39 | 270, 319 | 247.0969 | C14H15O4 | 147, 187, 203 | -2.802 | Dihydroxy-prenylcinnamic acid | Prenylated Phenolic acid |
| P-15 | 11.67 | 279, 319 | 457.1711 (503.18) | C21H29O11 | 293, 457 | -1.061 | unknown | _ |
| P-16 | 11.90 | 279, 364 | 283.0613 | C16H11O5 | 240, 268 | 0.294 | Galangin-5-methyl ether | Flavonol |
| P-17 | 11.75 | 279 | 405.1181 (451.12) | C20H21O9 | 341 | -2.408 | Trihydroxy-oxododecanoic acid. | Fatty acid |
| P-18 | 12.14 | 279, 364 | 313.0716 | C17H13O6 | _ | -0.643 | 3′,7-Dihydroxy-5′,6-dimethoxyisoflavone | Isoflavone |
| P-19 | 12.24 | 297, 322 | 301.0706 | C16H13O6 | 165 | -3.791 | 3’, 4’,6-Trihydroxy-7-methoxyflavanone | Flavanone |
| P-20 | 12.74 | 279, 364 | 301.0347 | C15H9O7 | 151, 179 | -2.245 | Quercetin | Flavonol |
| P-21 | 12.77 | 323 | 463.1018 | C25 H19 O9 | 309, 353 | -3.682 | Phenolic acid derivative | Phenolic acid |
| P-22 | 12.85 | 296 | 285.076 | C16H13O5 | 139, 241, 253, 267 | -3.216 | Methyl Pinobanksin | Dihydroflavonol |
| P-23 | 13.00 | 273 | 229.0863 | C14H13O3 | _ | -3.176 | 3-(2,3-Dihydro-2-isopropenyl-5-benzofuranyl)-2-propenoic acid | Benzofuran |
| P-24 | 13.04 | 277, 363 | 315.0502 | C16H11O7 | 300 | -2.526 | Quercetin methyl ether | Flavonol |
| P-25 | 13.31 | 279, 323 | 427.1387 | C23H23O8 | 135, 179, 247, 255, 273, 341 | -2.741 | Caffeic acid isoprenyl ester derivative | Phenolic acid |
| P-26 | 13.56 | 292 | 271.0606 | C15H11O5 | 253 | -2.386 | Pinobanksin | Dihydroflavonol |
| P-27 | 13.88 | 269, 362 | 299.0555 | C16H11O6 | 284 | -2.011 | Kampferol methyl ether | Flavonol |
| P-28 | 14.05 | 269, 362 | 329.066 | C17H13O7 | 314 | -2.115 | Quercetin di methyl ether | Flavonol |
| P-29 | 14.22 | 290 | 435.1072 | C24H19O8 | 135, 179, 255, 281 | -2.99 | Caffeoyl pinocembrin | Flavanone-Phenolic acid |
| P-30 | 14.44 | 266, 362 | 283.0606 | C16H11O5 | 239, 268 | -2.179 | Galangin -3-methyl ether | Flavonol |
| P-31 | 14.65 | 274 | 447.1074 | C25H19O8 | 337, 403 | -2.641 | Flavonoid derivative | Flavonoid |
| P-32 | 14.65 | 274 | 407.1124 | C23H19O7 | 297 | -3.086 | 5-hydroxy, 3,7, dimethoxy flavone derivative | Flavone |
| P-33 | 14.73 | 273 | 419.1129 | C24H19O7 | 255, 375 | -2.782 | p-coumaroyl trihydroxyflavone | Flavone-phenolic acid |
| P-34 | 14.83 | 269, 362 | 329.0658 | C17 H13O7 | 299, 314 | -2.662 | Quercetin di methyl ether isomer | Flavonol |
| P-35 | 14.95 | 279, 289 | 449.1231 | C25H21O8 | 339, 417, 434 | -2.473 | unknown | _ |
| P-36 | 15.22 | 269, 312 | 253.0502 | C15H9O4 | 209 | -1.628 | Chrysin | Flavone |
| P-37 | 15.32 | 290 | 255.0658 | C15H11O4 | 151, 213 | -1.773 | Pinocembrin | Flavanone |
| P-38 | 15.42 | 267, 360 | 269.0452 | C15H9O5 | 241, 227 | -1.289 | Galangin | Flavonol |
| P-39 | 15.68 | 271, 368 | 431.0761 (477.1179) | C24H15O8 | 162, 269, 367 | -2.553 | Galangin derivative | Flavonol |
| P-40 |
15.78 | 267, 290, 341 | 417.0973 (463.1393) | C24H17O7 | 162, 255, 353 | -1.573 | Pinocembrin derivative | Flavanone |
|
P-41 |
15.88 |
272, 368 |
415.0818 (461.1231) |
C24H15O7 |
269, 373, 387 |
-1.219 |
Galangin derivative |
Flavonol |
| P-42 | 16.16 | 279, 319 | 389.1027 | C23H17O6 | 295, 374 | -1.032 | unknown | _ |
| P-43 | 16.38 | 277, 291 | 327.0866 | C18H15O6 | _ | -2.45 | 5,7-Dihydroxy-3-propanoyloxyflavanone. (3-O-Propanoylpinobanksin) | Flavanone |
| P-44 | 16.59 | 276, 319 | 389.1026 | C23H17O6 | 295, 374 | -1.032 | unknown | _ |
| P-45 | 16.79 | 287 | 405.1335 | C24H21O6 | 255, 281, 295, 390 | -2.176 | Pinocembrin derivative | Flavanone |
| P-46 | 16.94 | 277 | 403.1179 | C24H19O6 | 255, 267, 388 | -2.112 | 7-Hydroxy-5-methoxyflavone derivative | Flavone |
| P-47 | 17.04 | 287 | 433.1282 | C25H21O7 | 403, 418 | -2.554 | 8-Cinnamoyl-3,3′,5′,7-tetrahydroxy-5-methoxyflavan. | Flavan |
| P-48 | 17.06 | 289 | 341.1021 | C19H17O6 | 253 | -2.702 | Pinobanksin butyrate [(3-O-(2-Methylpropanoyl) pinobanksin)] | Dihydroflavonol |
| P-49 | 17.16 | 288 | 451.1399 | C25H23O8 | _ | 0.042 | Propolisbenzofuran A | Benzofuran |
| P-50 | 17.33 | 270, 296sh | 387.0865 | C23H15O6 | 255, 343, 369 | -2.458 | Pinocembrin derivative | Flavone |
| P-51 | 17.57 | 279 | 455.2429 | C27H35O6 | 393, 411, 437 | -2.245 | Unknown |
_ |
| P-52 | 17.60 | 277, 360 | 399.0868 (445.1281) | C24H15O6 | 269 | -1.557 | Galangin derivative | Flavonol |
| P-53 | 17.69 | 291 | 355.1179 | C20H19O6 | 271, 253 | -2.314 | Pinobanksin valerate [(3-O-(2-methylbutanoyl) pinobanksin)] | Dihydroflavonol |
| P-54 | 17.88 | 289 | 385.1074 (403.0811) | C24H17O5 | 255 | -2.043 | Pinocembrin derivative | Flavanone |
| P-55 | 17.96 | 286 | 323.1284 | C20H19O4 | 164, 219, 268, 281 | -1.431 | unknown | _ |
| P-56 | 18.01 | _ | 471.3465 | C30H47O4 | _ | -3.083 | 3,7-Dihydroxycycloart-24-en-28-oic acid | Cycloartan- terpenoids |
| P-57 | 18.06 | 270 | 337.1075 | C20H17O5 | 253, 282 | -1.889 | Chrysin derivative | Flavone |
| P-58 | 18.11 | 269 | 369.1123 | C24H17O4 | _ | -2.417 | 6-Cinnamyl chrysin | Flavone-Phenolic acid |
| P-59 | 18.32 | 272, 297sh | 385.107 | C24H17O5 | _ | -3.082 | unknown | _ |
| P-60 | 18.50 | _ | 297.2427 | C18H33O3 | 171 | -2.618 | Hydroxy linoleic acid | Fatty acid |
| P-61 | 18.82 | _ | 325.183 | C21H25O3 | 325 | 6.249 | Unknown sterol | sterol |
| P-62 | 19.32 | _ | 343.2843 | C20H39O4 | 283, 325 | -3.067 | Dihydroxy eicosenoic acid | Fatty acid |
| P-63 | 20.01 | _ | 357.3001 | C21H41O4 | 297, 339 | -2.499 | unknown | _ |
| P-64 | 20.66 | _ | 371.3155 | C22H43O4 | 311, 353 | -3.132 | Dihydroxy docosahexenoic acid | Fatty acid |
| P-65 | 21.68 | _ | 353.3049 | C22H41O3 | 225 | -3.534 | Hydroxy-docosenoic acid | Fatty acid |
| P-66 | 21.71 | _ | 313.2735 | C19H37O3 | 142, 180 | -4.304 | Hydroxynonadecanoic acid | Fatty acid |
| P-67 | 21.80 | _ | 399.3474 | C24H47O4 | 339, 381 | -1.36 | Dihydroxytetracosanoic acid | Fatty acid |
| P-68 | 22.09 | _ | 413.3243 | C24H45O5 | 353 | -7.035 | Dimethoxy hydroxy-docosenoic acid | Fatty acid |
| P-69 | 22.28 | _ | 327.2896 | C20H39O3 | 267, 281 | -2.806 | Hydroxyeicosanoic acid | Fatty acid |
| P-70 | 22.77 | _ | 341.3049 | C21H41O3 | 253, 264, 326 | -3.57 | Methoxyeicosanoic acid. | Fatty acid |
| P-71 | 23.18 | _ | 355.3209 | C22H43O3 | 198, 232, 287 | -2.388 | Hydroxydocosanoic acid | Fatty acid |
Fig 1. UHPLC/MS-PDA total ion chromatogram of propolis hydroalcoholic extract.
The identities, retention time values (Rt), UV and mass data are listed in Table 2. Numbers represent peak number of each metabolite.
Phenolic acids
Few phenolic acids have been detected as gallic acid P-3 [m/z 169.0147 (C7H5O5)-] S1 Fig; ferulic acid P-7 [m/z 193.0503 (C10H9O4)-] with their UVmax at 274 and 319, respectively and common fragmentation of COO- moiety loss (-44 Da) resulting in peaks (125 and 149 Da, respectively) S2 Fig. A Phenolic glycerol viz. 1-O-feruloyl glycerol P-11 m/z 267.0849 (C13H15O6)-], UVmax at 298, 323 with major MS2 peak of ferulic acid (m/z 193) and a glycerol moiety of m/z 74 S5 Fig. In previous literatures, isolation and identification of different phenolic glycerides have been observed in poplar type propolis [30] as well as Chinese one [31]. A dihydroxy-prenyl cinnamic acid P-14 [m/z 247.0969 (C14H15O4)-], UVmax 270, 319 with MS2 peaks of m/z 187 owing to the loss of carboxylic and hydroxy groups (-60 Da), m/z 203 due to loss of carboxylic group (COO-, 44 Da) and 147 m/z of cinnamic acid S8 Fig. Presence of derivatives of phenolic acids as caffeic acid isoprenyl ester derivative P-25 [m/z 427.1387 (C23H23O8)-], UVmax 279, 323 whereas appearance of fragments as caffeic acid isoprenyl ester [m/z 247 (C14H15O4)-] and caffeic acid [m/z 179 (C9H7O4)-] S16 Fig. Other peaks as P-9 and P-21, their MS2 fragmentation revealed the loss of (-44 Da, COO- group) which represents loss of carboxylic group as shown in Table 2. P-9, a caffeic acid derivative [m/z 315.0863 (C17H15O6)-] S3 Fig showed a fragment at m/z 179.03 which stands for molecular formula (C9H7O4)- of caffeic acid and fragment [m/z 271.10 (C16H15O4)- which represents loss of COO- group (-44 Da)]. P-21, is a phenolic acid derivative [m/z 463.1018 (C25H19O9)-] S13 Fig where a fragment at m/z 309.08 due to loss of COO- moiety from m/z 353.07 (C19H13O7)-. The identified phenolic acids are commonly present in propolis obtained from different origins [32,33] except for gallic acid where this is the first report of its identification in such sample. A peak P-12 [m/z 165 (C9H9O3)] was assigned as unknown acid due to loss of COO- group (-44 Da) S6 Fig. Another peak P-51 [m/z 455.2429 (C27H35O6)] was assigned to have an acid moiety due to the loss of (-44 Da, COO- group) and m/z 60 which represents loss of both carboxylic and hydroxyl groups S34 Fig.
Flavonoids
Flavonoids are considered a major class of this propolis sample [34]. Different common classes have been previously identified as flavones of UVmax 270 viz, chrysin [28]; flavonols of UVmax 360 viz, galangin [28] and quercetin; flavanones and dihydroflavonols (UVmax 292) as pinocembrin and pinobanksin [28] besides flavans UVmax 287 as in Table 2. Flavonoids represented in this propolis sample are aglycones, esters, methylated flavonoids and glycoside derivatives. Peaks of flavonoid aglycones were identified as 3’, 7 dihydroxy-5’, 6’ di methoxy isoflavone P-18 [m/z 313.0716 (C17H13O6)-] and 3’, 4’, 6 trihydroxy-7-methoxy flavanone P-19 [m/z 301.0706 (C16H13O6)-] which has been previously isolated from Nepalese propolis [35] S10 and S11 Figs respectively. Peak P-47 [m/z 433.1282 (C25H21O7)-] S31 Fig was identified as 8-Cinnamoyl-3,3′,5′,7-tetrahydroxy-5-methyoxyflavan whereas it has been isolated previously from Chinese propolis [36]. Common flavonoid aglycones in propolis as quercetin P-20 [m/z 301.0347 (C15H9O7)-] S12 Fig [33]. Pinobanksin P-26 [271.0606 (C15H11O5)-], chrysin P-36 [m/z 253.0502 (C15H9O4)-] pinocembrin P-37 [m/z 255.0658 (C15H11O4)-] and galangin P-38 [m/z 269.0452 (C15H9O5)-] in which those four aglycones were identified and reported for the same propolis sample in previous literature [28] as well as in Jordanian propolis [37]. Flavonoid esters as 5, 7-dihydroxy-3-propanoyloxyflavanone commonly named 3-O-propanoyl pinobanksin P-43 [m/z 327.0866 (C18H15O6)-] and [(3-O-(2-Methylpropanoyl) pinobanksin)] known as pinobanksin butyrate P-48 [m/z 341.1021 (C19H17O6)-]. S28 and S32 Figs, respectively, were also detected and previously identified in Chilean propolis [38]; (3-O-(2-Methylbutanoyl) pinobanksin) known as Pinobanksin valerate P-53 [m/z 355.1179 (C20H19O6)-] S36 Fig [33]. Methylated flavonoids as methyl pinobanksin P-22 [m/z 285.0759 (C16H13O5)-] S14 Fig, quercetin methyl ether P-24 [m/z 315.0502 (C16H11O7)-] S15 Fig, Kaempferol methyl ether P-27 [m/z 299.0555 (C16H11O6)-] S17 Fig, quercetin di-methyl ether P-28 S18 Fig and P-34 [m/z 329.0660 (C17H13O7)-], galangin-5-methyl ether [m/z 283.0606 (C16H11O5)-] P-16 S9 Fig and galangin-3-methyl ether P-30 [m/z 283.0606 (C16H11O5)-] S20 Fig, where the identification and fragmentation of these methylated flavonoids Table 2 agreed with previous literature [33].
Identification of hydrophilic flavonoid glycosides is considered unique in such sample due to the lipophilic nature of propolis, still previous identification of glycosides of quercetin and kaempferol derivatives was reported previously in [39] which have been classified in such work as uncommon flavonoid glycosides in Portuguese propolis. In this propolis sample, identification of rutin P-10 [m/z 609.1472 (C27H29O16)-] as detection of peak at m/z 301.03 which represents quercetin aglycone and loss of (-308 Da) which represents the sugar moiety, another peak at m/z 464.43 which stands for loss of deoxy-sugar rhamnose (-145 Da) S4 Fig [37,40].
Derivatives of both galangin and pinocembrin flavonoids have been predicted in peaks P-39 [m/z 431.0761 (C24H15O8)-], P-41 [m/z 415.0818 (C24H15O7)-] and P-52 [m/z 399.0868 (C24H15O6)-]. Galangin aglycon peak at m/z 269.04 (C15H9O5)- was identified where loss of (-162 Da) in peak P-39 S24 Fig and (-146 Da) in P-41 S26 Fig, as well as loss of (-130 Da) in peak P-52 S35 Fig, respectively, have been observed. Pinocembrin flavonoid derivatives identified in peaks P-40 [m/z 417.0973 (C24H17O7)-] S25 Fig, P-50 [m/z 387.0865 (C23H15O6)-] S33 Fig and P-54 [m/z 403.0811 (C24H17O5)-] S37 Fig with pinocembrin aglycone [m/z 255.07 (C15H11O4)] with loss of (-162 Da), (-132 Da) and (-130 Da) for P-40, P-50 and P-54, respectively. Another pinocembrin derivatives could be detected for P-13 [m/z 445.1486 (C23H25O9)-] S7 Fig and P-45 [m/z 405.1335 (C24H21O6)-] where a peak at m/z 255.06 (C15H11O4)- of pinocembrin was observed S29 Fig. A peak P-46 [m/z 403.1179 (C24H19O6)- could be a hydroxy-methoxy flavone derivative where a peak of pinocembrin m/z 255.07 (C15H11O4)- was predicted in addition to another peak at m/z 388.10 (C23H16O6)- due to loss of methyl group (-15 Da) S30 Fig. Flavonoid derivative P-31 [m/z 447.1074 (C25H19O8)-] S21 Fig could be structurally related to P-32 [m/z 407.1124 (C23H19O7)-] S22 Fig which could be a hydroxy dimethoxyflavone derivative as both showed the loss of C6H6O2 (-110 Da). It is worth mentioning that the isolation of hydroxy methoxy flavones have been carried out in a previous study from Argentinean propolis [41]. P-57 [m/z 337.1075 (C20H17O5)-] S39 Fig could be chrysin derivative as a significant peak appeared at m/z 253.05 (C15H9O4)- which stands for chrysin.
Another class of flavonoids with phenolic acids within their structures was identified in this propolis sample as caffeoyl pinocembrin P-29 [m/z 435.1072 (C24H19O8)-] S19 Fig as detection of peaks at m/z 255.07 (C15H11O4)- and 179.04 (C9H7O4)- which present pinocembrin and caffeic acid moieties, respectively. p-coumaroyl trihydroxy-flavone P-33 [m/z 419.1129 (C24H19O7)-] S23 Fig as a peak at m/z 255.07 (C15H11O4)- was identified as trihydroxy-flavone [M-163]- where loss of (-163 Da) occurred which presents p-coumaroyl moiety and m/z 375.12 which presents [M-44]- due to loss of (-44 Da) of COO- moiety of p-coumaroyl part. A 6-cinnamyl chrysin P-58 [m/z 369.1123 (C24H17O4)-] S40 Fig may be predicted where it has been isolated before from a Chinese propolis sample [42]. A propolis Benzofuran compound P-49 [m/z 451.1399 (C25H23O8)-] could be identified as it has been previously isolated from Brazilian propolis [43].
Fatty acids
Fatty acid class was identified at the end of the run. Identified fatty acids were P-60 [m/z 297.2427 (C18H33O3)-], hydroxy linoleic acid; P-62 [m/z 343.2843 (C20H39O4)-], dihydroxy eicosenoic acid; P-64 [m/z 371.3155 (C22H43O4)-]; dihydroxy docosahexenoic acid; P-65 [m/z 353.3049 (C22H41O3)-], hydroxy-docosenoic acid; P-66 [m/z 313.2735 (C19H37O3)-], hydroxynonadecanoic acid; P-67 [m/z 399.3474 (C24H47O4)-], dihydroxytetracosanoic acid; P-68 [m/z 413.3243 (C24H45O5)-], dimethoxy hydroxy-docosenoic acid; P-69 [m/z 327.2896 (C20H39O3)-], hydroxyeicosanoic acid; P-70 [m/z 341.3049 (C21H41O3)-], Methoxyeicosanoic acid; P-71 [m/z 355.3209 (C22H43O3)-], hydroxydocosanoic acid and an unknown sterol P-61 [m/z 325.183 (C21H25O3)-]. Another peak could be detected P-56 [m/z 471.3465 (C30H47O4)-] S38 Fig as 3,7-Dihydroxycycloart-24-en-28-oic acid where it has been isolated previously from Cretan propolis [44] also a group of cycloartane terpenes have been previously isolated from propolis obtained from Myanmar [45].
From the analysis of the propolis sample using UHPLC/MS-PDA, it could be predicted that it is a poplar type of propolis due to the presence of major specific flavonoids viz. chrysin, galangin, pinocembrin and pinobanksin with their derivatives [46,47]. The antibacterial activity of propolis could be related mainly to the enrichment of the sample with flavonoid class Table 2 and (Fig 1), in addition to the phenolic acids mainly caffeic acid and its derivatives [48]. In the next section, standardization of the hydroalcoholic extract of propolis would be crucial for quantification of its phenolic ingredients using HPLC-PDA.
Standardization of phenolics in propolis extract using HPLC-PDA
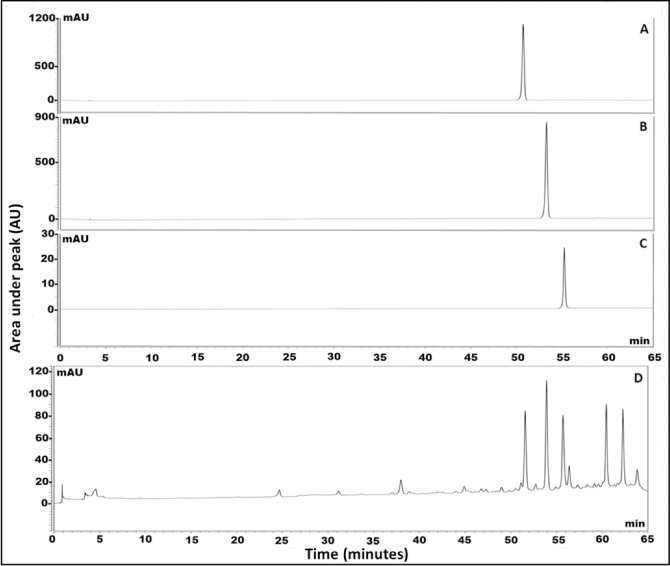
Standardization of the hydroalcoholic extract of propolis versus major flavonoids identified in the UHPLC/MS-PDA chromatographic run Table 2 have been assessed using HPLC-PDA. Identification of the major flavonoids at retention time (Rt.) viz. pinocembrin, chrysin and galangin were 51.00, 53.30, and 55.00 min, respectively (Fig 2). The results showed the enrichment of propolis sample by chrysin, followed by pinocembrin then galangin recording 22.73±0.68, 21.58±0.84 and 14.26±0.70 mg/g propolis extract, respectively. It is worth to mention the enrichment of propolis alcoholic extract of this current study with high content of flavonoids when compared to other propolis of different geographical sources [49]. In the previous work by Bozkuş et al. 2021, the content of both chrysin and galangin were recorded 641.33 and 534.11 μg/25 mg of Turkish propolis extract, respectively that would enhance its biological activities [49].
Fig 2. HPLC chromatogram of authentics.
(A) pinocembrin, (B) chrysin, (C) galangin and (D) total propolis hydroalcoholic extract.
In vitro antimicrobial evaluation of hydroalcoholic extract of propolis
Minimum Inhibitory concentration (MIC) and Minimum Bactericidal concentration (MBC)
The MIC of propolis could not be identified due to its dark color and turbidity where visualization would have been inaccurate in the prepared test tubes. This problem was resolved by going forward to identification of MBC through plating all dilutions on Mueller hinton agar to identify the bactericidal concentration of propolis. Plate number (5) showed that the MBC of propolis was 0.2 mg/mL.
Disc diffusion susceptibility test
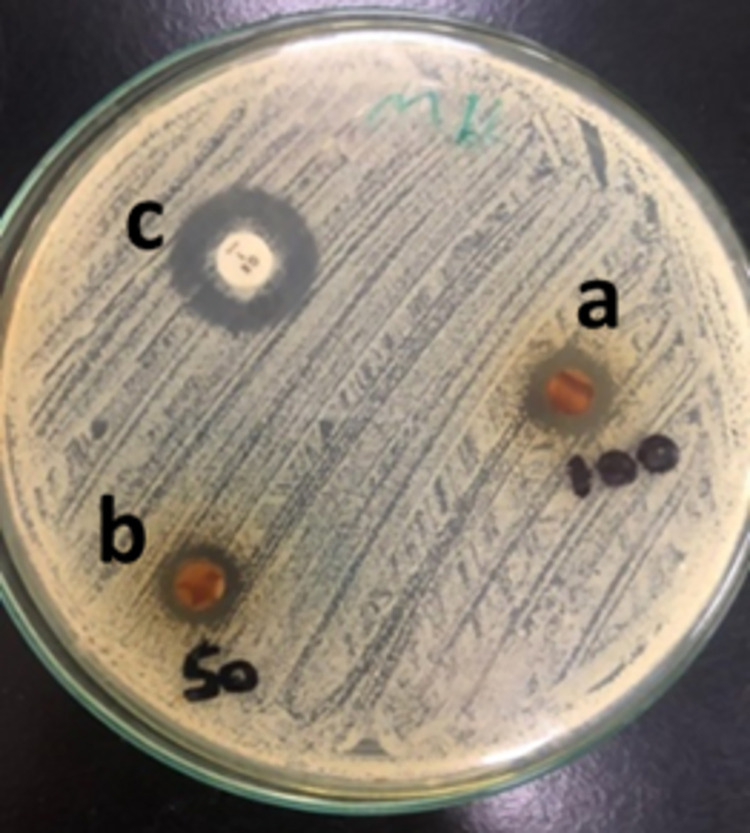
This test was to assess the antibacterial effect of the two propolis concentrations. Propolis 100%, 50% and oxacillin showed diameters of zones of inhibition measuring 14, 11 and 18 mm respectively (Fig 3) The test showed that Staphylococcus aureus was sensitive to 100% propolis and showed intermediate susceptibility to 50% propolis compared to the standard antibiotic. This shows the concentration dependent activity of propolis that is due to the higher concentration of constituents in the 100% concentration.
Fig 3. Disc Diffusion susceptibility test of the propolis against Staphylococcus aureus.
a: 100% propolis; b: 50% propolis c: Control; Oxacillin antibiotic disc.
Generally, it appears that the antibacterial activity of propolis is greater against Gram-positive than Gram-negative bacteria. This could be attributed to the species-specific structure of the outer membrane of the Gram-negative bacteria and the production of hydrolytic enzymes which break down the active ingredients of propolis [50,51]. Other studies demonstrated that the broad spectrum biological properties of propolis, including antimicrobial activities, anti-inflammatory, immunomodulatory, antioxidant and radical scavenging actions are mostly due to phenolic compounds, terpenes, caffeic, ferulic and coumaric acids, esters, and flavonoids [52–54].
In vivo antibacterial evaluation and burn healing assessment
Burn wound mouse model was used to assess the antibacterial and burn healing properties of the tested two propolis concentrations. Both concentrations were applied until complete healing was observed.
Burn healing assessment
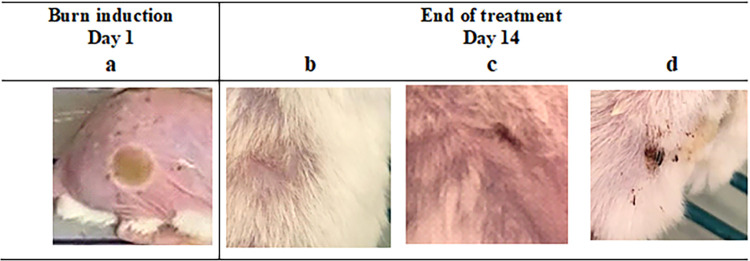
The main criterion for indication of ongoing healing was measurement of burn wound diameter, where contraction of burn border expressed reduction of the original diameter of burn. The readings were documented daily through the duration of treatment. The positions treated on all mice with both concentrations of propolis; 100% and 50%, demonstrated on-going burn border contraction along the duration of treatment. The end of the treatment period was decided by visual observation of complete skin healing which was reached by 100% propolis by day 14 (Fig 4) and Table 3. It was found that the antibacterial activity of propolis was due to bacterial cell membrane damage and cell lysis [55]. Furthermore, it has the ability to inhibit cell division, protein synthesis and bacterial motility by affecting RNA-polymerase [56].
Fig 4. Burn wound on mouse dorsal skin before and after treatments application.
a: Mouse burn (diameter 10 mm) before treatment application at day 1. b, c, d: Mouse burn after end of treatment at day 14, of 100% propolis, 50% propolis and silver sulfadiazine respectively.
Table 3. Burn wound diameter percent reduction of the applied treatments at day 14.
|
Mean percent reduction ± SD |
Treatment | |||
| Propolis 100% | Propolis 50% | Cream (positive control) | Saline (negative control) | |
| 98.9 ± 1 | 89 ± 3.1 | 80.5 ± 4.8 | 65 ± 4.2 | |
Assessment of bacterial load of the infected burns
Swabs were collected daily from the burn sites to assess the antibacterial activity of the tested propolis compared to the commercial product; silver sulfadiazine cream; as a positive control. All swabs from all positions showed reduction in bacterial load with time. By day 7 no bacteria were found in burn sites treated with both propolis concentrations. While burn sites treated with positive control continued to show bacterial growth till day 9, finally superficial bacterial growth was detected in the saline treated burn sites till the end of the treatment period (day 14) Table 4.
Table 4. Bacterial load percent reduction of the applied treatments at day 7.
|
Mean percent reduction ± SD |
Treatment | |||
| Propolis 100% | Propolis 50% | Cream (positive control) | Saline (negative control) | |
| 99 ± 1.1 | 99 ± 1.1 | 85 ± 3.7 | 45 ± 4 | |
Percent reduction =
Where:
A: Bacterial count before treatment
B: Bacterial count after treatment
Skin irritation and sensitization tests
The possible effect of propolis to produce skin irritation and sensitization was tested and it was found that it did not yield any undesirable side effects when applied on the skin either once (irritation in rabbits) or repeatedly (sensitization in guinea pigs) for both concentrations.
Histopathological analysis
Skin autopsy samples treated with 100% propolis showed few inflammatory cells infiltration and oedema between the hair follicles at the dermis underneath the epidermal layer (Fig 5A). The tissue between the deep dermis and musculature showed few focal inflammatory cells infiltration. The skin positions treated with 50% propolis showed focal thickening acanthosis in the epidermis associated with fibrosis and inflammatory cells infiltration in the underlying dermis (Fig 5B). The deep layer of the dermis showed oedema with dilatation in the blood vessels, while the deep layer of the dermis and musculature had inflammatory cells infiltration. Perivascular inflammatory cells infiltration was detected surrounding the dilated blood vessels between the dermis and musculature. On the other hand, the positions treated with silver sulfadiazine showed focal invagination of the epidermis into the dermis, associated with fibrosis with inflammatory cells infiltration in the dermis underneath the epidermis. Oedema infiltrated with inflammatory cells was observed in between the dermis and musculature (Fig 5C). The tissue between the dermis and the adjacent musculature also showed oedema, congested blood vessels and inflammatory cells infiltration. Sever suppuration in the subcutaneous tissue was found in the group treated with saline, while this was not seen with other groups. The severity of histopathological alteration in skin autopsy of different experimental groups has been summarized in Table 5.
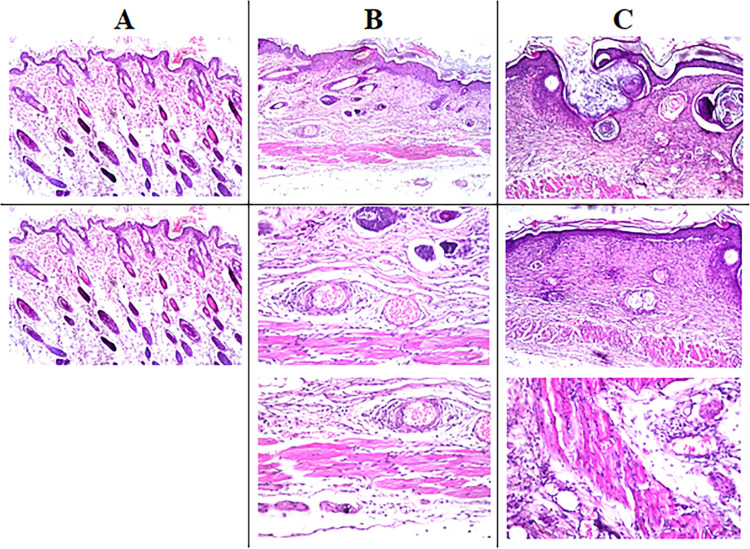
Fig 5. Representative photomicrographs of tissue sections collected at day 14 after burn stained with H&E.
A: Skin of mice treated with 100% propolis showing few inflammatory cells and oedema in between the hair follicles at the dermal layer underneath the epidermis and between the dermis and muscular layer. (16x mag.) B: Skin of mice treated with 50% propolis showing acanthosis in the epidermis with underlying fibrosis and inflammatory cells in the dermis and musculature accompanied by oedema with dilation of blood vessels (16x mag.; 40x mag.; 40x mag.). C: Skin of mice treated with positive control showing invagination of the epidermal layer in the dermis with fibrosis and inflammatory cells infiltration accompanied by oedema and congested blood vessels. (16x mag.; 16x mag.; 40x mag.).
Table 5. The severity of histopathological alteration in skin autopsy of different experimental groups.
| Histopathological alterations | Groups*/Score | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Acanthosis in the epidermis | - | ++ | ++ | - | |
| Inflammatory reaction in subcutaneous tissue | + | ++ | ++ | +++ | |
| Suppuration in subcutaneous tissue | - | - | - | +++ | |
| Fibrosis and edema | - | ++ | +++ | +++ | |
*Group 1: Treated with 100% propolis, Group 2: Treated with 50% propolis, Group3: Treated with silver sulfadiazine, Group 4: Treated with saline. Score: +++ Sever, ++ Moderate, + Mild,—Nil.
Gregory et al., 2002 compared healing activity of Brazilian propolis cream with silver sulfadiazine on patients who suffer minor second-degree burns, where propolis showed to be more effective than silver sulfadiazine in terms of inflammation reduction [57].
In another study, it was shown that 5% propolis ointment administered twice daily reduced the wound size of diabetic foot ulcer during a four-week long observation healing process [58]. In another randomized trial, foot ulcer areas in diabetic patients were also significantly reduced with the use of aqueous propolis in liquid form. This reduction, accompanied with improved healing rate, was detected regardless of whether the patient administered an antibiotic or not. Furthermore, wound fluids in the group treated with propolis showed decreased bacterial count [59].
Antioxidant activity in vitro using different techniques (DPPH, ABTS and FRAP)
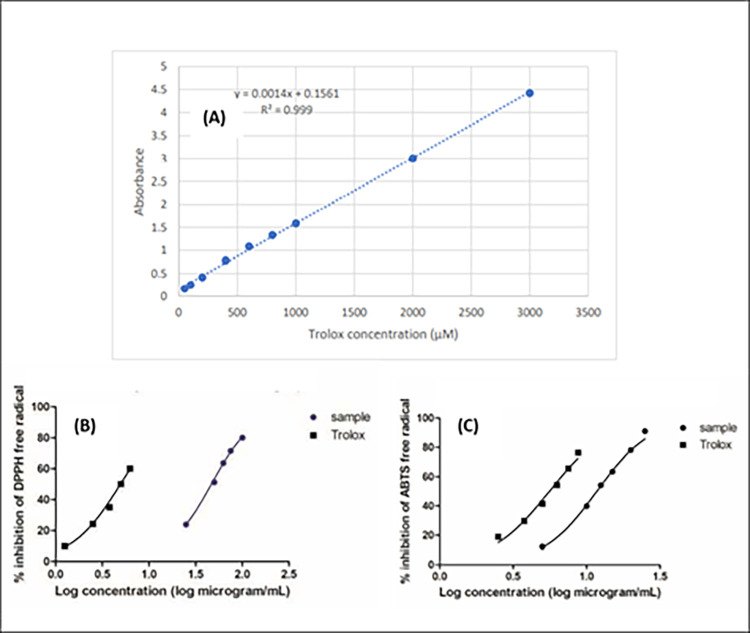
The antioxidant activity of the hydroalcoholic extract of propolis has been assessed via three different assays viz. DPPH, ABTS and FRAP. All the antioxidant assays results revealed the promising antioxidant effect of the propolis sample used in this study. The DPPH IC50 of propolis extract was 46.52±1.25 μg/mL (standard trolox 4.89±0.26 μg/mL) where the results matched those previously published in [60] of Taiwanese propolis from different districts. The ABTS assay revealed IC50 result 11.74±0.26 μg/mL (standard trolox 5.58±0.10 μg/mL) whereas the FARP assay recorded 445.29±29.9 μM TE/mg of propolis extract. The results are summarized in (Fig 6).
Fig 6. Antioxidant activity of propolis extract using different assays.
(A): Trolox calibration curve used in FRAP assay (B): DPPH assay, (C): ABTS assay.
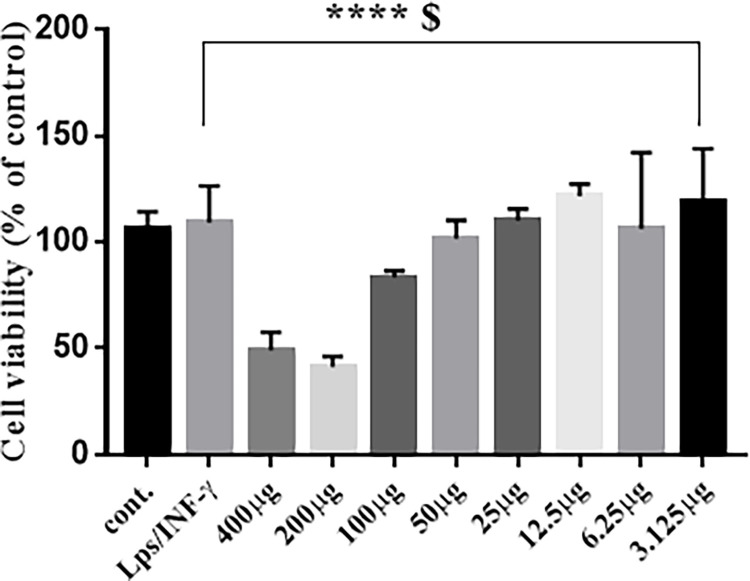
Concentration‑dependent effect of propolis on the viability of RAW 264.7 macrophages
MTT assay was used to determine the nontoxic concentration of propolis, RAW 264.7 cells were treated for 24 h with increasing concentrations of propolis (3.125 to 400 μg) alone or with LPS (10 ng/mL) plus IFN-γ (10 U/mL). (Fig 7) shows that exposure to propolis inhibited cell viability in a concentration dependent manner. When cells were stimulated with LPS (10 ng/mL) plus IFN-γ (10 U/mL) alone, no change in cell viability was observed. The 50 μg concentration did not significantly affect cell viability compared to LPS/ IFN-γ control and accordingly was used in all downstream experiments.
Fig 7. Effects of propolis on cell viability.
RAW 264.7 macrophages were exposed for 24 h to propolis at increasing concentrations of (3.125 to 400 μM) alone or with LPS (10 ng/mL) plus IFN-γ (10 U/mL). Cell cytotoxicity was determined using MTT assay. Data are expressed as a percentage of untreated control (which is set at 100%) ± S.E. (n = 4).
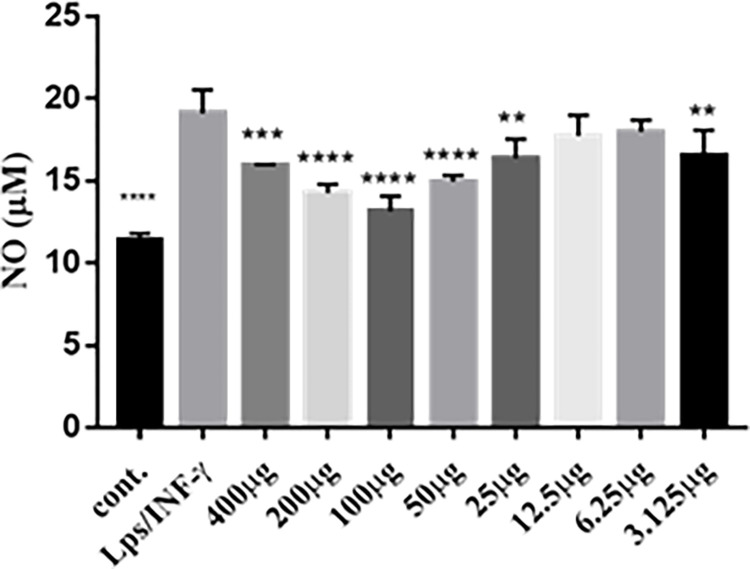
Concentration‑dependent effects of propolis on nitrite formation in RAW 264.7 macrophages
Nitrite concentrations were measured using the Griess assay. RAW 264.7 cells were subjected for 24 h to propolis (3.125 to 400 μg) alone or with LPS /IFN-γ (10 ng/mL/10 U/mL). (Fig 8) shows that LPS/IFN-γ have increased nitrite level by 160% compared to control. Cells exposed to propolis (concentration 100–6.25 μg) and LPS/IFN-γ, did not affect cell viability and also inhibited nitrite production in concentration dependent manner.
Fig 8. Concentration-dependent effects of LPS and IFN-γ on nitrite production in RAW 264.7 macrophages.
RAW 264.7 macrophages were exposed to LPS/IFN-γ (10 ng/mL/10 U/mL) and different propolis concentrations for 24 h. Data are expressed as mean ± S.E. (n = 8).
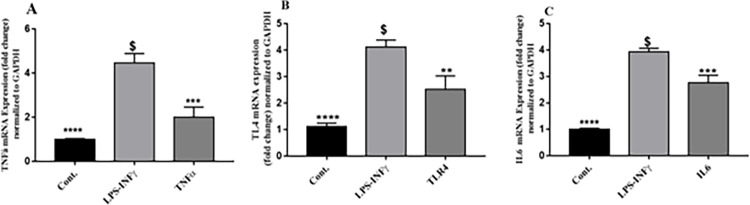
Effect of propolis on TNF-α, TLR4 and IL-6 mRNA expression levels in LPS/IFN‑γ‑stimulated RAW 264.7 macrophages
TNF-α, TLR4 and IL-6 mRNA expression levels were estimated by real-time PCR. In (Fig 9), cells treated with LPS/IFN-γ (10 ng/mL-10 U/mL) for 6 h significantly induced TNF-α, TLR4 and IL-6 expression by approximately 400, 450 and 400%, respectively, compared to untreated control. RAW cells activated with LPS/IFN-γ and co-treated with 50 μg propolis for 24 h showed reduction in mRNA expression of TNF-α, TLR4 and IL-6 by 55, 40, 39% respectively.
Fig 9. Effect of Total propolis extract on TNF-α, TLR4 and IL-6 mRNA in LPS/IFN-γ-mediated RAW 264.7 macrophages.
RAW 264.7 cells were treated for 24 h with propolis (50 μg) in the presence of LPS (10 ng/mL) plus IFN-γ (10 U/mL). TNF-α (A), TLR4 (B) and IL-6 (C) mRNA levels were quantified using qRT-PCR and were normalized to GAPDH. Data are expressed as mean ± S.E. (n = 3).
In the current study, the effect of propolis on TLR4 signaling pathway was explored through gene expression studies of key inflammatory intermediaries as TLR4, TNF-α, IL-6 and iNOS. Upon activation of RAW 264.7 macrophages with LPS/IFN-γ, a significant increase in these mediators was detected. Propolis effectively attenuated LPS/IFN-γ induction of RAW 264.7 macrophage through TLR4 pathway by downregulating the expression of TLR4, TNF-α, IL-6 and iNOS at the transcriptional level and post-transcriptional level for iNOS. Furthermore, upon LPS/IFN-γ activation of RAW 264.7 macrophage, a significant increase in iNOS protein level and nitrite was detected as previously proved by Fresta et al. [61]. The significant overexpression of TLR4, IL-6 and TNF-α mRNA in LPS/IFN-γ activated RAW 264.7 macrophage (Fig 9), confirmed the relation between LPS/TLR4 signal transduction and proinflammatory cytokine stimulation from macrophages [62]. These results are in agreement with other studies which show that propolis suppresses prostaglandin and leukotriene generation by inhibiting the expression and activities of cyclooxygenases (COX-1 and COX-2) and lipoxygenases (LOX), retarding the gene expression of inducible nitric oxide synthase (iNOS), blocking tumor necrosis factor-α (TNF-α)-mediated NF-қB activation and reducing immune response in T cells. Chrysin, pinocembrin and galangin counted as selective inhibitors of NF-қB activation as previously mentioned in previous literatures, which may provide the molecular basis for their anti-inflammatory activity [63–65].
Effects of propolis on iNOS protein levels inLPS/IFN‑γ‑stimulated RAW 264.7 macrophages
iNOS protein levels in LPS/IFN‑γ‑stimulated RAW 264.7 macrophages were evaluated by western blot. As illustrated in (Figs 10 and S41), RAW 264.7 macrophages treated with LPS/IFN-γ (10 ng/mL–10 U/mL) shows an increase in the expression level of iNOS protein approximately by 1500% compared to the untreated cells. Upon treatment with propolis 50 and 100 μg, significant decrease in iNOS protein expression by 80 and 87%, respectively, compared to LPS/IFN-γ treated control.
Fig 10. Concentration-dependent effect of propolis on iNOS protein expression on LPS and IFN-γ induced RAW 264.7 macrophages.
RAW 264.7 macrophages were exposed to LPS (10 ng/mL) in the presence of IFN-γ (10 U/mL) for 24 h. RAW264.7 cells pretreated with 50 and 100 μg propolis. Anti-iNOS monoclonal antibodies were used for iNOS protein detection and actin was used as a loading control.
Conclusion
Burns are a global public health problem, accounting for an estimated 180 000 deaths annually. Despite a variety of burn treatment products there is still no consensus regarding better performance. Propolis; a natural product; enriched with a myriad of natural phytochemicals through which possess therapeutic potential in the treatment of infected burns including inflammatory ones.
In this current study, propolis proved to be a potent natural antibacterial agent that aid in burn healing. The antibacterial action against Staphylococcus aureus was tested in vitro; using Disc Diffusion susceptibility test and in vivo; using burn-induced infected mouse model. Propolis showed in vitro concentration dependent antibacterial activity and in vivo highest burn healing rate compared to a commercial product as shown by burn diameter reduction and histopathological analysis with no signs of skin irritation in rabbits, nor sensitization in guinea pigs.
In addition to the antibacterial and burn healing properties of propolis, this study unveiled its antioxidant and anti-inflammatory actions in vitro. Our results showed that propolis successfully modulates TLR4 signaling activation in RAW 264.7 macrophages by inhibiting LPS/IFN-γ-induced NO, iNOS, TNF-α, and IL-6 gene expression. Our data proves the anti-inflammatory potential of propolis at the transcriptional and posttranscriptional levels, and thus imply that propolis has a potential immunomodulatory agent against LPS/IFN-γ-mediated inflammation. The chemical profiling of propolis was conducted through full mapping of the phytochemical constituents using UHPLC/MS-PDA as well as standardization of the major flavonoids identified in this propolis sample using HPLC-PDA to relate the propolis biological activity to its phytochemical constituents through full identification as well as quantification of the major flavonoids.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Acknowledgments
Dina M. El-Kersh and Rania F. Abou El-Ezz are grateful to Prof. Mohamed A. Farag, Pharmacognosy Department, College of Pharmacy, Cairo University, Kasr El Aini St., 11562, Cairo, Egypt for his generous assistance in analysis of propolis sample on UHPLC/MS-PDA.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Anjum SI, Ullah A, Khan KA, Attaullah M, Khan H, Ali H, et al. Composition and functional properties of propolis (bee glue): A review. Saudi Journal of Biological Sciences. 2019;26(7):1695–703. doi: 10.1016/j.sjbs.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooqui AA, Farooqui T. Beneficial effects of propolis on human health and neurological diseases. Frontiers in Bioscience-Elite. 2012;4(2):779–93. doi: 10.2741/e418 [DOI] [PubMed] [Google Scholar]
- 3.Hossain R, Quispe C, Khan RA, Saikat ASM, Ray P, Ongalbek D, et al. Propolis: An update on its chemistry and pharmacological applications. Chinese medicine. 2022;17(1):1–60. doi: 10.1186/s13020-022-00651-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daugsch A, Moraes CS, Fort P, Park YK. Brazilian red propolis—chemical composition and botanical origin. Evidence-based complementary and alternative medicine. 2008;5(4):435–41. doi: 10.1093/ecam/nem057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salatino A. Perspectives for uses of propolis in therapy against infectious diseases. Molecules. 2022;27(14):4594. doi: 10.3390/molecules27144594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forjuoh SN. Burns in low-and middle-income countries: a review of available literature on descriptive epidemiology, risk factors, treatment, and prevention. Burns. 2006;32(5):529–37. doi: 10.1016/j.burns.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Peck MD, Kruger GE, Van Der Merwe AE, Godakumbura W, Ahuja RB. Burns and fires from non-electric domestic appliances in low and middle income countries: Part I. The scope of the problem. Burns. 2008;34(3):303–11. doi: 10.1016/j.burns.2007.08.014 [DOI] [PubMed] [Google Scholar]
- 8.Organization WH. World health statistics 2008: World Health Organization; 2008. [Google Scholar]
- 9.Odondi RN, Shitsinzi R, Emarah A. Clinical patterns and early outcomes of burn injuries in patients admitted at the Moi Teaching and Referral Hospital in Eldoret, Western Kenya. Heliyon. 2020;6(3):e03629. doi: 10.1016/j.heliyon.2020.e03629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James SL, Lucchesi LR, Bisignano C, Castle CD, Dingels ZV, Fox JT, et al. Epidemiology of injuries from fire, heat and hot substances: global, regional and national morbidity and mortality estimates from the Global Burden of Disease 2017 study. Injury prevention. 2020;26(Suppl 2):i36–i45. doi: 10.1136/injuryprev-2019-043299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladhani HA, Yowler CJ, Claridge JA. Burn wound colonization, infection, and sepsis. Surgical infections. 2021;22(1):44–8. doi: 10.1089/sur.2020.346 [DOI] [PubMed] [Google Scholar]
- 12.Cambiaso-Daniel J, Gallagher JJ, Norbury WB, Finnerty CC, Herndon DN, Culnan DM. Treatment of infection in burn patients. Total burn care: Elsevier; 2018. p. 93–113. e4. doi: [DOI] [Google Scholar]
- 13.Trupkovic T, Gille J, Fischer H, Kleinschmidt S. Antimicrobial treatment in burn injury patients. Der Anaesthesist. 2012;61:249–58. doi: 10.1007/s00101-012-1994-4 [DOI] [PubMed] [Google Scholar]
- 14.El-Kased RF. Natural antibacterial remedy for respiratory tract infections. Asian Pacific Journal of Tropical Biomedicine. 2016;6(3):270–4. doi: 10.1016/j.apjtb.2015.12.002 [DOI] [Google Scholar]
- 15.El-Kased RF, Amer RI, Attia D, Elmazar MM. Honey-based hydrogel: In vitro and comparative In vivo evaluation for burn wound healing. Scientific reports. 2017;7(1):9692. doi: 10.1038/s41598-017-08771-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medellin-Luna MF, Castaneda-Delgado JE, Martínez-Balderas VY, Cervantes-Villagrana AR. Medicinal plant extracts and their use as wound closure inducing agents. Journal of Medicinal Food. 2019;22(5):435–43. doi: 10.1089/jmf.2018.0145 [DOI] [PubMed] [Google Scholar]
- 17.Farag MA, Ali SE, Hodaya RH, El-Seedi HR, Sultani HN, Laub A, et al. Phytochemical profiles and antimicrobial activities of Allium cepa red cv. and A. sativum subjected to different drying methods: a comparative MS-based metabolomics. Molecules. 2017;22(5):761. doi: 10.3390/molecules22050761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sha N, Huang H-L, Zhang J-Q, Chen G-T, Tao S-J, Yang M, et al. Simultaneous quantification of eight major bioactive phenolic compounds in Chinese propolis by high-performance liquid chromatography. Natural Product Communications. 2009;4(6):1934578X0900400615. doi: 10.1177/1934578X0900400615 [DOI] [PubMed] [Google Scholar]
- 19.Bauer A, Kirby W, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966;45(4_ts):493–6. doi: 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- 20.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. Journal of pharmaceutical analysis. 2016;6(2):71–9. doi: 10.1016/j.jpha.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheesbrough M. District laboratory practice in tropical countries, part 2: Cambridge university press; 2005. [Google Scholar]
- 22.Suvarna KS, Layton C, Bancroft JD. Bancroft’s theory and practice of histological techniques E-Book: Elsevier health sciences; 2018. [Google Scholar]
- 23.Floegel A, Kim D-O, Chung S-J, Koo SI, Chun OK. Comparison of ABTS/DPPH assays to measure antioxidant capacity in popular antioxidant-rich US foods. Journal of food composition and analysis. 2011;24(7):1043–8. doi: 10.1016/j.jfca.2011.01.008 [DOI] [Google Scholar]
- 24.Emad AM, Rasheed DM, El-Kased RF, El-Kersh DM. Antioxidant, Antimicrobial Activities and Characterization of Polyphenol-Enriched Extract of Egyptian Celery (Apium graveolens L., Apiaceae) Aerial Parts via UPLC/ESI/TOF-MS. Molecules. 2022;27(3):698. doi: 10.3390/molecules27030698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahuguna A, Khan I, Bajpai VK, Kang SC. MTT assay to evaluate the cytotoxic potential of a drug. ||| Bangladesh Journal of Pharmacology. 2017;12(2):115–8. doi: 10.3329/bjp.v12i2.30892 [DOI] [Google Scholar]
- 26.Xie C, Li X, Zhu J, Wu J, Geng S, Zhong C. Magnesium isoglycyrrhizinate suppresses LPS-induced inflammation and oxidative stress through inhibiting NF-κB and MAPK pathways in RAW264. 7 cells. Bioorganic & medicinal chemistry. 2019;27(3):516–24. doi: 10.1016/j.bmc.2018.12.033 [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 28.Arafa MG, Ghalwash D, El-Kersh DM, Elmazar MM. Propolis-based niosomes as oromuco-adhesive films: A randomized clinical trial of a therapeutic drug delivery platform for the treatment of oral recurrent aphthous ulcers. Scientific reports. 2018;8(1):1–14. doi: 10.1038/s41598-018-37157-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Fernández O, Domínguez R, Pateiro M, Munekata PE, Rocchetti G, Lorenzo JM. Determination of polyphenols using liquid chromatography–tandem mass spectrometry technique (LC–MS/MS): A review. Antioxidants. 2020;9(6):479. doi: 10.3390/antiox9060479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankova V, Popova M, Bogdanov S, Sabatini A-G. Chemical composition of European propolis: expected and unexpected results. Zeitschrift für Naturforschung C. 2002;57(5–6):530–3. doi: 10.1515/znc-2002-5-622 [DOI] [PubMed] [Google Scholar]
- 31.Shi H, Yang H, Zhang X, Sheng Y, Huang H, Yu L. Isolation and characterization of five glycerol esters from Wuhan propolis and their potential anti-inflammatory properties. Journal of agricultural and food chemistry. 2012;60(40):10041–7. doi: 10.1021/jf302601m [DOI] [PubMed] [Google Scholar]
- 32.Almutairi S, Edrada-Ebel RA, Fearnley J, Igoli JO, Alotaibi W, Clements CJ, et al. Isolation of diterpenes and flavonoids from a new type of propolis from Saudi Arabia. Phytochemistry letters. 2014;10:160–3. doi: 10.1016/j.phytol.2014.08.022 [DOI] [Google Scholar]
- 33.Saleh K, Zhang T, Fearnley J, Watson DG. A Comparison of the Constituents of Propolis from Different Regions of the United Kingdom by Liquid Chromatograpghy-high Resolution Mass Spectrometry Using a Metabolomics Approach Current Metabolomics. 2015;3:42–53. doi: 10.2174/2213235X03666150328000505 [DOI] [Google Scholar]
- 34.Xu W, Lu H, Yuan Y, Deng Z, Zheng L, Li H. The Antioxidant and Anti-Inflammatory Effects of Flavonoids from Propolis via Nrf2 and NF-κB Pathways. Foods. 2022;11(16):2439. doi: 10.3390/foods11162439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrestha SP, Narukawa Y, Takeda T. Chemical constituents of Nepalese propolis (II). Chemical and pharmaceutical bulletin. 2007;55(6):926–9. doi: 10.1248/cpb.55.926 [DOI] [PubMed] [Google Scholar]
- 36.Sha N, Guan S-H, Lu Z-Q, Chen G-T, Huang H-L, Xie F-B, et al. Cytotoxic constituents of Chinese propolis. Journal of Natural products. 2009;72(4):799–801. doi: 10.1021/np900118z [DOI] [PubMed] [Google Scholar]
- 37.Naik RR, Shakya AK, Oriquat GA, Katekhaye S, Paradkar A, Fearnley H, et al. Fatty acid analysis, chemical constituents, biological activity and pesticide residues screening in Jordanian propolis. Molecules. 2021;26(16):5076. doi: 10.3390/molecules26165076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castro C, Mura F, Valenzuela G, Figueroa C, Salinas R, Zuñiga MC, et al. Identification of phenolic compounds by HPLC-ESI-MS/MS and antioxidant activity from Chilean propolis. Food Research International. 2014;64:873–9. doi: 10.1016/j.foodres.2014.08.050 [DOI] [PubMed] [Google Scholar]
- 39.Falco SI, Vale L, Gomes P, Domingues MRM, Freire C, Cardoso SM, et al. Phenolic Profiling of Portuguese Propolis by LC-MS Spectrometry: Uncommon Propolis Rich in Flavonoid Glycosides. Phytochem Anal. 2013;24:309–18. doi: 10.1002/pca.2412 [DOI] [PubMed] [Google Scholar]
- 40.Cao Y, Wang Y, Yuan Q. Analysis of flavonoids and phenolic acid in propolis by capillary electrophoresis. Chromatographia. 2004;59(1):135–40. doi: 10.1365/s10337-003-0138-z [DOI] [Google Scholar]
- 41.Vera N, Solorzano E, Ordoñez R, Maldonado L, Bedascarrasbure E, Isla MI. Chemical composition of Argentinean propolis collected in extreme regions and its relation with antimicrobial and antioxidant activities. Natural Product Communications 2011;6(6):1934578X1100600618. doi: 10.1177/1934578X1100600618 [DOI] [PubMed] [Google Scholar]
- 42.Usia T, Banskota AH, Tezuka Y, Midorikawa K, Matsushige K, Kadota S. Constituents of Chinese propolis and their antiproliferative activities. Journal of natural products. 2002;65(5):673–6. doi: 10.1021/np010486c [DOI] [PubMed] [Google Scholar]
- 43.Salatino A, Teixeira ÉW, Negri G. Origin and chemical variation of Brazilian propolis. Evidence-Based Complementary and Alternative Medicine. 2005;2(1):33–8. doi: 10.1093/ecam/neh060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popova MP, Chinou IB, Marekov IN, Bankova VS. Terpenes with antimicrobial activity from Cretan propolis. Phytochemistry. 2009;70(10):1262–71. doi: 10.1016/j.phytochem.2009.07.025 [DOI] [PubMed] [Google Scholar]
- 45.Li F, Awale S, Zhang H, Tezuka Y, Esumi H, Kadota S. Chemical constituents of propolis from Myanmar and their preferential cytotoxicity against a human pancreatic cancer cell line. Journal of Natural Products. 2009;72(7):1283–7. doi: 10.1021/np9002433 [DOI] [PubMed] [Google Scholar]
- 46.Šuran J, Cepanec I, Mašek T, Radić B, Radić S, Tlak Gajger I, et al. Propolis extract and its bioactive compounds—From traditional to modern extraction technologies. Molecules. 2021;26(10):2930. doi: 10.3390/molecules26102930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falcão SI, Vilas-Boas M, Estevinho LM, Barros C, Domingues MRM, Cardoso SM. Phenolic characterization of Northeast Portuguese propolis: usual and unusual compounds. Analytical Bioanalytical Chemistry. 2010;396:887–97. doi: 10.1007/s00216-009-3232-8 [DOI] [PubMed] [Google Scholar]
- 48.Przybyłek I, Karpiński TM. Antibacterial properties of propolis. Molecules. 2019;24(11):2047. doi: 10.3390/molecules24112047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bozkuş TN, Değer O, Yaşar A. Chemical characterization of water and ethanolic extracts of Turkish propolis by HPLC-DAD and GC-MS. Journal of Liquid Chromatography & Related Technologies. 2021;44(1–2):77–86. doi: 10.1080/10826076.2021.1883648 [DOI] [Google Scholar]
- 50.Sforcin JM. Biological properties and therapeutic applications of propolis. Phytotherapy research. 2016;30(6):894–905. doi: 10.1002/ptr.5605 [DOI] [PubMed] [Google Scholar]
- 51.Drago L, Mombelli B, Vecchi ED, Tocalli MFL, Gismondo M. In vitro antimicrobial activity of propolis dry extract. Journal of chemotherapy. 2000;12(5):390–5. doi: 10.1179/joc.2000.12.5.390 [DOI] [PubMed] [Google Scholar]
- 52.Inui S, Hatano A, Yoshino M, Hosoya T, Shimamura Y, Masuda S, et al. Identification of the phenolic compounds contributing to antibacterial activity in ethanol extracts of Brazilian red propolis. Natural product research. 2014;28(16):1293–6. doi: 10.1080/14786419.2014.898146 [DOI] [PubMed] [Google Scholar]
- 53.Veiga R, De Mendonça S, Mendes P, Paulino N, Mimica M, Lagareiro Netto A, et al. Artepillin C and phenolic compounds responsible for antimicrobial and antioxidant activity of green propolis and Baccharis dracunculifolia DC. Journal of Applied Microbiology. 2017;122(4):911–20. doi: 10.1111/jam.13400 [DOI] [PubMed] [Google Scholar]
- 54.Farooqui T, A Farooqui A. Molecular mechanism underlying the therapeutic activities of propolis: a critical review. Current nutrition & food science. 2010;6(3):186–99. doi: 10.2174/157340110792389136 [DOI] [Google Scholar]
- 55.Bryan J, Redden P, Traba C. The mechanism of action of Russian propolis ethanol extracts against two antibiotic‐resistant biofilm‐forming bacteria. Letters in applied microbiology. 2016;62(2):192–8. doi: 10.1111/lam.12532 [DOI] [PubMed] [Google Scholar]
- 56.Mirzoeva O, Grishanin R, Calder P. Antimicrobial action of propolis and some of its components: the effects on growth, membrane potential and motility of bacteria. Microbiological research. 1997;152(3):239–46. doi: 10.1016/S0944-5013(97)80034-1 [DOI] [PubMed] [Google Scholar]
- 57.Gregory SR, Piccolo N, Piccolo MT, Piccolo MS, Heggers JP. Comparison of propolis skin cream to silver sulfadiazine: a naturopathic alternative to antibiotics in treatment of minor burns. The Journal of Alternative & Complementary Medicine. 2002;8(1):77–83. doi: 10.1089/107555302753507203 [DOI] [PubMed] [Google Scholar]
- 58.Afkhamizadeh M, Aboutorabi R, Ravari H, Fathi Najafi M, Ataei Azimi S, Javadian Langaroodi A, et al. Topical propolis improves wound healing in patients with diabetic foot ulcer: a randomized controlled trial. Natural product research. 2018;32(17):2096–9. doi: 10.1080/14786419.2017.1363755 [DOI] [PubMed] [Google Scholar]
- 59.Henshaw FR, Bolton T, Nube V, Hood A, Veldhoen D, Pfrunder L, et al. Topical application of the bee hive protectant propolis is well tolerated and improves human diabetic foot ulcer healing in a prospective feasibility study. Journal of Diabetes and its Complications. 2014;28(6):850–7. doi: 10.1016/j.jdiacomp.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 60.Lu L-C, Chen Y-W, Chou C-C. Antibacterial and DPPH free radical-scavenging activities of the ethanol extract of propolis collected in Taiwan. Journal of Food and Drug Analysis. 2003;11(4):277–82. doi: 10.38212/2224-6614.2676 [DOI] [Google Scholar]
- 61.Fresta CG, Fidilio A, Lazzarino G, Musso N, Grasso M, Merlo S, et al. Modulation of pro-oxidant and pro-inflammatory activities of m1 macrophages by the natural dipeptide carnosine. International journal of molecular sciences. 2020;21(3):776. doi: 10.3390/ijms21030776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu Ain, Batool M, Choi S. TLR4-targeting therapeutics: structural basis and computer-aided drug discovery approaches. Molecules. 2020;25(3):627. doi: 10.3390/molecules25030627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeo H, Lee YH, Koh D, Lim Y, Shin SY. Chrysin inhibits NF-κB-dependent CCL5 transcription by targeting IκB kinase in the atopic dermatitis-like inflammatory microenvironment. International Journal of Molecular Sciences. 2020;21(19):7348. doi: 10.3390/ijms21197348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borriello M, Iannuzzi C, Sirangelo I. Pinocembrin protects from AGE-induced cytotoxicity and inhibits non-enzymatic glycation in human insulin. Cells. 2019;8(5):385. doi: 10.3390/cells8050385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu H, Yao H, Zou R, Chen X, Xu H. Galangin suppresses renal inflammation via the inhibition of NF-κB, PI3K/AKT and NLRP3 in uric acid treated NRK-52E tubular epithelial cells. BioMed Research International. 2019;2019. doi: 10.1155/2019/3018357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.