Abstract
A systematic method was developed to assemble functional full-length genomes of large RNA and DNA viruses. Coronaviruses contain the largest single-stranded positive-polarity RNA genome in nature. The ∼30-kb genome, coupled with regions of genomic instability, has hindered the development of a full-length infectious cDNA construct. We have assembled a full-length infectious construct of transmissible gastroenteritis virus (TGEV), an important pathogen in swine. Using a novel approach, six adjoining cDNA subclones that span the entire TGEV genome were isolated. Each clone was engineered with unique flanking interconnecting junctions which determine a precise systematic assembly with only the adjacent cDNA subclones, resulting in an intact TGEV cDNA construct of ∼28.5 kb in length. Transcripts derived from the full-length TGEV construct were infectious, and progeny virions were serially passaged in permissive host cells. Viral antigen production and subgenomic mRNA synthesis were evident during infection and throughout passage. Plaque-purified virus derived from the infectious construct replicated efficiently and displayed similar plaque morphology in permissive host cells. Host range phenotypes of the molecularly cloned and wild-type viruses were similar in cells of swine and feline origin. The recombinant viruses were sequenced across the unique interconnecting junctions, conclusively demonstrating the marker mutations and restriction sites that were engineered into the component clones. Full-length infectious constructs of TGEV will permit the precise genetic modification of the coronavirus genome. The method that we have designed to generate an infectious cDNA construct of TGEV could theoretically be used to precisely reconstruct microbial or eukaryotic genomes approaching several million base pairs in length.
Molecular genetic analysis of the structure and function of RNA virus genomes has been profoundly advanced by the availability of full-length cDNA clones, the source of infectious RNA transcripts that replicate efficiently when introduced into permissive cell lines (2, 9). Recombinant DNA technology has allowed the isolation of infectious cDNA clones from a number of positive-stranded RNA viruses, including picornaviruses, caliciviruses, alphaviruses, flaviviruses, and arteriviruses, whose RNA genomes range in size from ∼7 to 15 kb in length (1, 13, 32, 34, 35, 47, 48, 54). The availability of these clones has enhanced our understanding of the molecular mechanisms of viral replication and pathogenesis and resulted in new approaches for heterologous gene expression and vaccine development.
The order Nidovirales includes mammalian positive-polarity single-stranded RNA viruses in the arterivirus and coronavirus families (10, 16). The Coronaviridae family includes the Coronavirus and Torovirus genera (10, 46). Despite significant size differences (∼13 to 32 kb), the polycistronic genome organization and regulation of gene expression from a nested set of subgenomic mRNAs are similar for all members of the order (16, 46). The family Coronaviridae contains the largest RNA viral genomes in nature (26, 44). Transmissible gastroenteritis virus (TGEV), a group I coronavirus, contains a ∼28.5-kb genomic RNA that is packaged into a helical nucleocapsid structure and surrounded by an envelope that contains three virus-specific glycoprotein spikes, including the S glycoprotein, membrane glycoprotein (M), and a small envelope glycoprotein (E) (17, 18, 33, 36). The TGEV genome is polycistronic and encodes eight large open reading frames (ORFs), which are expressed from full-length or subgenome-length mRNAs during infection (17, 42, 43). The 5′-most ∼20 kb encode the RNA replicase genes, which are encoded in two large ORFs, designated 1a and 1b, the latter of which is expressed by ribosomal frameshifting (3, 17). ORF1a encodes at least two viral proteases and several other nonstructural proteins, while ORF1b contains polymerase, helicase, and metal-binding motifs typical of an RNA polymerase (3, 17, 19). In the 3′-most ∼9 kb of the TGEV genome, each of the downstream ORFs is preceded by a highly conserved intergenic sequence element, which directs the synthesis of each of the six or seven subgenomic RNAs (11, 17, 18, 52). These subgenomic mRNAs are arranged in a nested set structure from the 3′ end of the genome, and each contains a leader RNA sequence derived from the 5′ end of the genome (26, 29, 42, 43). Subgenomic mRNAs are generated by a discontinuous transcription mechanism, the details of which are somewhat controversial (4, 40, 42, 43). In addition to the viral mRNAs, full-length and subgenome-length negative-strand RNAs are implicated in mRNA synthesis (4, 26, 40, 42, 43). Another unique feature of coronavirus replication is the high RNA recombination frequencies associated with infection (6, 25, 26).
The large size of the coronavirus genome, coupled with the inability to clone portions of the polymerase gene in microbial vectors, has hampered the ability to perform precise manipulations and reverse genetics in members of the Coronaviridae (17, 18, 26, 44). Recently these problems were overcome when a full-length cDNA of TGEV was stably cloned in bacterial artificial chromosome (BAC) vectors (3). In this report, we describe a simple and rapid approach for systematically assembling a full-length, infectious cDNA construct of TGEV using a series of smaller subclones and a novel strategy which theoretically may allow the assembly of large microbial or eukaryotic chromosomes approaching several million base pairs in length.
MATERIALS AND METHODS
Virus and cells.
The Purdue strain (ATCC VR-763) of TGEV was obtained from the American Type Culture Collection (ATCC) and passaged once in the swine testicular (ST) cell line. ST cells were obtained from the ATCC (ATCC 1746-CRL) and maintained in minimal essential medium (MEM) containing 10% fetal clone II (HyClone) and supplemented with 0.5% lactalbumin hydrolysate, 1× nonessential amino acids, 1 mM sodium pyruvate, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). Baby hamster kidney cells (BHK) were maintained in alpha-MEM containing 10% fetal calf serum supplemented with 10% tryptose phosphate broth, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). Feline kidney cells (CRFK) were maintained in Eagle's MEM with nonessential amino acids, Earle's balanced salt solution, and 10% fetal bovine serum at 37°C. Wild-type TGEV or viruses (icTGEV) derived from the full-length construct were plaque purified twice, and stocks were grown in ST cells as described (42, 43). To measure the growth rate of different viruses, cultures of ST cells (5 × 105) were infected with wild-type TGEV or various molecularly cloned isolates at a multiplicity of infection (MOI) of 5 for 1 h. The cells were washed twice with phosphate-buffered saline (PBS) to remove residual virus and incubated at 37°C in complete medium. At different times postinfection, progeny virions were harvested and assayed by plaque assay in ST cells. To study the host range phenotype, cultures of ST or CRFK cells (105) were infected with wild-type or molecularly cloned icTGEV at an MOI of 5 for 1 h and fixed at 12 h postinfection for fluorescent analysis (FA).
Mutagenesis, cloning, and sequencing of the TGEV genome.
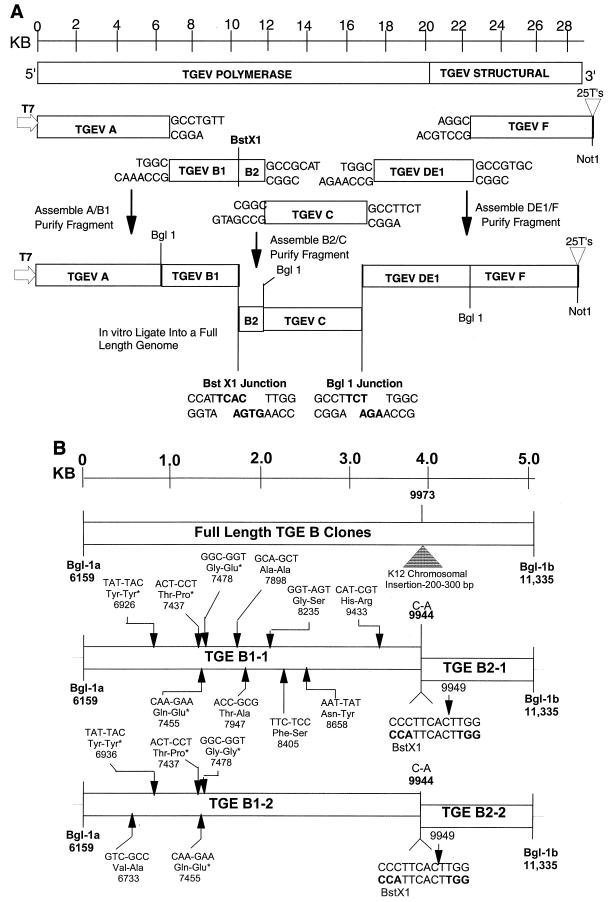
The cloning strategy for a full-length TGEV construct is illustrated in Fig. 1 and is based on the observation that the BglI restriction endonuclease cleaves at a specific sequence palindrome (GCCNNNN↓NGGC) but leaves highly variable 3-nucleotide ends that do not randomly self-assemble. Rather, these DNAs will only anneal with fragments containing the complementary 3-nucleotide overhang generated at identical BglI sites. The TGEV genome was cloned from infected ST cell intracellular RNA by reverse transcription-PCR (RT-PCR) using primer pairs directed against the Purdue strain of TGEV or a Taiwanese isolate (11, 17, 33) (Table 1). To create unique junction sites for assembly of a full-length TGEV cDNA construct, primer-mediated PCR mutagenesis was used to insert unique BglI restriction sites at the 5′ and 3′ ends of each subclone (Table 1, Fig. 1). These primer pairs do not alter the coding sequence and result in RT-PCR amplicons ranging in size from ∼5.0 to 6.9 kb in length. Total intracellular RNA was isolated from TGEV-infected cells using RNA STAT-60 reagents according to the manufacturer's directions (Tel-TEST B, Inc.). To isolate the TGEV subclones, reverse transcription was performed using Superscript II and oligodeoxynucleotide primers according to the manufacturer's recommendations (Gibco-BRL). Following cDNA synthesis at 50°C for 1 h, the cDNA was denatured for 2 min at 94°C and amplified by PCR with Expand Long Taq polymerase (Boehringer Mannheim Biochemicals) for 25 cycles at 94°C for 30 s, 58°C for 25 to 30 s, and 68°C for 1 to 7 min depending on the size of the amplicon. The PCR amplicons were isolated from agarose gels and cloned into Topo II TA (Invitrogen) or pGem-TA cloning vectors (Promega) according to the manufacturer's directions.
FIG. 1.
Strategy for directionally assembling a TGEV infectious construct. (A) The TGEV genome is a linear positive-polarity RNA of about 28,500 nucleotides. Using RT-PCR and unique oligonucleotide primer mutagenesis, five clones spanning the entire TGEV genome were isolated using standard recombinant DNA techniques. Unique BglI sites were inserted at the junctions between each clone, a unique T7 start site was inserted at the 5′ end of clone A, and a 25-nucleotide T tail and downstream NotI site were inserted at the 3′ end of clone F. The approximate location of each site is shown. (B) Cloning the TGEV B amplicon. Because of chromosomal instability in E. coli, it was noted that two B clones (TGEV B1 and B3) contained large insertions at nucleotide 9973 in the TGEV genome (17). Other TGEV B clones had deletions across these sequences. Assuming that these insertions and deletions were “detoxifying” poison TGEV sequences in E. coli, we bisected the B fragment by inserting a BstXI site at position 9949 and cloning two separate clones designated TGEV B1-1,2 and TGEV B2-1,2. Quasispecies variation in the sequence of each independent plasmid clones is shown, with conserved changes denoted with asterisks. These conserved changes differed from the published sequence reported by Eleouet et al. (17) but were identical to the sequence reported by Almazan et al. (3). To reconstruct a wild-type B1 fragment, an SfiI-PflI fragment from B1-1 was isolated and inserted into the TGEV B1-2 backbone to produce a consensus B1 clone.
TABLE 1.
Primer pairs for assembly of the TGEV infectious constructa
| Primer | Sequence | Position in genome | Orientation | Purpose |
|---|---|---|---|---|
| TGEV 5′ T7 | 5′-GTCGGCCTCTTAATACGACTCACTATAGACTTTTAAAGTAAAGTGAG-3′ | 1–19 | + | Insert T7 start TGEV A, 5′ end |
| TGEV A 3,642 | 5′-TGTTGAAGAAATCAAAGGCCTG-3′ | 3621–3642 | − | Remove T7 stop |
| TGEV A 3,621 | 5′-CAGGCCTTTGATTTCTTCAAC-3′ | 3621–3641 | + | Remove T7 stop, overlap RT-PCR |
| T | ||||
| TGEV A 6,180 | 5′-CTTGGAGATGTTGAAAATCAGC-3′ | 6180–6201 | − | TGEV A, 3′ end |
| TGEV B1 6,134 | 5′-TAAAGTCTGCAGTCTGTGGC-3′ | 6115–6134 | + | TGEV B1, 5′ end |
| TGEV B1 9,957 | 5′-TAATCCAAGTGAATGGTGTGTAC-3′ | 9935–9957 | − | TGEV B1, 3′ end, insert BstXI site |
| TGEV B2 9,939 | 5′-ACACCATTCACTTGGATTAATCC-3′ | 9939–9961 | + | TGEV B2, 5′ end, insert BstXI site |
| C | ||||
| TGEV B2 11,342 | 5′-GTAGCCGATGCGGCTGGAATG-3′ | 11342–11362 | − | TGEV B2, 3′ end |
| TGEV C 11,345 | 5′-TCCAGCCGCATCGGCTACAAG-3′ | 11345–11365 | + | TGEV C, 5′ end, insert BglI site |
| T A | ||||
| TGEV C 13,605 | 5′-GTGCAAAGAAGAAGTGTTTTAATG-3′ | 13605–13628 | − | Remove T7 stop |
| TGEV C 13,609 | 5′-AAAACACTTCTTCTTTGCACAGG-3′ | 13609–13631 | + | Remove T7 stop, overlap RT-PCR |
| T T | ||||
| TGEV C 16,580 | 5′-TGTGCCAAGAAGGCCTTGACAAC-3′ | 16580–16602 | − | TGEV C, 3′ end |
| TGEV DE 16,585 | 5′-CAAGGCCTTCTTGGCACATAATC-3′ | 16585–16607 | + | TGEV DE, 5′ end, insert BglI site |
| T A | ||||
| TGEV DE 3,133 | 5′-GTCTAGCCTGCACGGCTACTGC-3′ | 23475–23496 | − | TGEV DE, 3′ end |
| TGEV F 3,112 | 5′-GCAGTAGCCGTGCAGGCTAGAC | 23475–23496 | + | TGEV F, 5′ end, insert BglI site |
| A T | ||||
| TGEV 3′ end | 5′-NNNNNNGCGGCCGCTTTTTTTTTTTTTTTTTTTTTTTTTGGTGTATCACTATCAAAAGG-3′ | 3′ end, poly(A) tail | − | TGEV F 3′ end, Poly(A) tail, NotI |
Three to seven independent clones of each TGEV amplicon were isolated and sequenced using a panel of primers located about 0.5 kb from each other on the TGEV insert and an ABI model automated sequencer. A consensus sequence for each of the cloned fragments was determined, and when necessary (i.e., pTGEV A, pTGEV B1, pTGEV C, and pTGEV F), a consensus clone was assembled using restriction enzymes and standard recombinant DNA techniques to remove unwanted amino acid changes associated with reverse transcription or naturally occurring quasispecies variation.
Assembly of a full-length TGEV infectious construct.
Each of the plasmids was grown to high concentration, isolated, and digested or double-digested with BglI, BstXI, or NotI according to the manufacturer's direction (NEB) (Fig. 1A). The TGEV A clone was digested with ApaI, treated with calf intestine alkaline phosphatase, and subsequently digested with BglI, resulting in a ∼6.3-kb fragment. The TGEV F clone was NotI digested, treated with calf alkaline phosphatase, and then BglI digested. All other vectors were digested with BglI or BstXI. The appropriately sized cDNA inserts were isolated from 0.8 to 1.2% agarose gels in TAE buffer (Tris, acetate EDTA) containing 5 mM cytidine (Fluka) and extracted using Qiaex II gel extraction kits according to the manufacturer's directions (Qiagen Inc., Valencia, Calif.). Cytidine was incorporated to reduce DNA damage associated with cumulative UV exposure during visualization in agarose gels (21). Appropriate cDNA subsets (A+B1, B2+C, and DE-1+F) were pooled into 100- to 300-μl aliquots, and equivalent amounts of each DNA were ligated with T4 DNA ligase (15 U/100 μl) at 16°C overnight in 30 mM Tris-HCl (pH 7.8)–10 mM MgCl2–10 mM dithiothreitol–1 mM ATP. Appropriately sized products (A/B1, B2/C, and DE-1/F) were separated in 0.7% agarose gels containing 5 mM cytidine as described, isolated, and religated as described above. The final products were purified by phenol-chloroform-isoamyl alcohol (1:1:24) and chloroform extraction and precipitated under ethanol prior to in vitro transcription reactions. The full-length TGEV construct is designated TGEV 1000.
The nucleocapsid protein may function as part of the transcriptional complex (7, 15, 26). To provide N protein in trans, the TGEV N gene was amplified from the TGEV F clone using primer pairs flanking the N gene ORF. The upstream primer contained an SP6 site (5′-TCGGCCTCGATTTAGGTGACACTATAGATGGCCAACCAGGGACAACG-3′), while the downstream primer introduced a 14-nucleotide oligo(T) stretch, providing a poly(A) tail following in vitro transcription (5′-TTTTTTTTTTTTTTAGTTCGTTACCTCGTCAATC-3′). The TGEV leader RNA sequence, 3′-most ORF, and noncoding sequences were not present in this construct. The PCR product was purified from gels and used directly for in vitro transcription.
RNA transfection.
Full-length transcripts of the TGEV cDNA, TGEV 1000, were generated in vitro as described by the manufacturer (mMessage mMachine; Ambion, Austin, Tex.) with certain modifications. For 2 h at 37°C, several 30-μl reactions were performed that were supplemented with 4.5 μl of a 30 mM GTP stock, resulting in a 1:1 ratio of GTP to cap analog. Similar reactions were performed using 1 μg of PCR amplicons encoding the TGEV N gene sequence or Sindbis virus noncytopathic replicons encoding green fluorescent protein (pSin-GFP; kindly provided by Charlie Rice, Washington University) and a 2:1 ratio of cap analog to GTP (1). The transcripts were treated with DNase I, denatured, and separated in 0.5% agarose gels in TAE buffer containing 0.1% sodium dodecyl sulfate. Alternatively, the transcripts were either treated with 50 ng of RNase A for 15 min at room temperature, DNase I treated, or directly electroporated into BHK cells.
BHK or ST cells were grown to subconfluence, trypsinized, washed twice with PBS, and resuspended in PBS at a concentration of 107 cells/ml. RNA transcripts were added to 800 μl of the cell suspension in an electroporation cuvette, and three electrical pulses of 850 V at 25 μF were given with a Bio-Rad Gene Pulser II electroporator. The BHK cells were seeded with 106 uninfected ST cells in a 75-cm2 flask and incubated at 37°C for 3 to 4 days. Virus progeny were then passaged in ST cells in 75-cm2 flasks at 2-day intervals and purified twice by plaque assay.
Immunofluorescence assays.
Cells were grown on LabTek chamber slides (four or eight wells) and infected with wild-type TGEV or molecularly cloned viruses (icTGEV-1, icTGEV-2, and icTGEV-3) generated from the infectious construct. At 12 h postinfection, cells were fixed in acetone-methanol (1:1) and stored at 4°C. Fixed cells were rehydrated in PBS (pH 7.2) and incubated with a 1:100 dilution of mouse anti-TGEV polyclonal antiserum for 30 min at room temperature. After three washes in PBS, the cells were incubated with a 1:100 dilution of goat anti-mouse immunoglobulin G-fluorescein isothiocyanate conjugate (Sigma) for 30 min at room temperature. After three additional washes with PBS, the cells were visualized and photographed under a Zeiss LSM110 confocal fluorescence microscope. Images were digitized and assembled in Photoshop 5.5 (Adobe Systems Inc.).
RT-PCR to detect marker mutations and sequence analysis.
Cultures of ST cells were infected for 1 h at room temperature with wild-type TGEV or plaque-purified icTGEV-1 and icTGEV-3 viruses that were derived from the infectious construct. Intracellular RNA was isolated at 12 h postinfection and used as the template for RT-PCRs using four different primer pair sets that asymmetrically flank each of the interconnecting BglI or BstXI junctions that were used in the assembly of TGEV 1000. RT reactions were performed using Superscript II reverse transcriptase for 1 h at 50°C as described by the manufacturer (Gibco-BRL) prior to PCR amplification with the reverse primer that flanked a particular interconnecting junction. To amplify across the B1-B2 junction, forward (5′-GCATCGTAAGACTCAACAAGG-3′) and reverse (5′-GTCACAGCAAGTGAGAACCATG-3′) primers were located at nucleotides 9738 to 9759 and 10248 to 10270, respectively, and resulted in a 532-bp amplicon (17). In virus derived from the infectious construct, BstXI digestion should result in 321- and 211-bp fragments. To amplify across the B2-C junction, forward (5′-TTGAGCGCGAAGCATCAGTGC-3′) and reverse (5′-TTCCACTGCCGAAAGCTTCACC-3′) primers were located at nucleotides 11231 to 11151 and 11634 to 11655, respectively, and resulted in an amplicon of 424 bp (17). In molecularly cloned virus, BglI digestion should result in products of 300 and 124 bp. To amplify across the C–DE-1 junction, forward (GAATGTGCACACTAGGACCTG) and reverse (AGCAGGTGGTATGTATTGTTCG) primers were located at nucleotides 16380 to 16400 and 16936 to 16957, respectively (17). If a BglI site is present in this 577-bp amplicon, digestion should result in products of 370 and 207 bp. To amplify across the DE-1–F junction, forward (CGTTGTACAGGTGGTTATGAC) and reverse (CTCCGCTTGTCTGGTTAGAGTC) primers were located at nucleotides 23304 to 23324 and 23852 to 23873 in the S gene, respectively (3, 33). Following BglI digestion of this 569-bp amplicon, 386- and 183-bp fragments should be visualized in icTGEV virus derived from the infectious construct. Following 28 cycles of amplification with Taq polymerase (Expand Long Kit; Roche Biochemical), the PCR products were separated and isolated from agarose gels. PCR amplicons were either subcloned directly into pGemT cloning vectors for sequencing or digested with BglI or BstXI restriction endonuclease according to the manufacturer's directions (NEB). The digested DNAs were then separated in 1.5% agarose gels in TAE buffer and visualized under UV light. All sequence comparisons were performed using the Vector Suite II (Informax Inc.) Align X program.
RESULTS
Theoretical framework.
Conventional restriction enzymes, such as PstI and EcoRI, leave sticky ends that assemble with similarly cut DNA fragments in the presence of DNA ligase (39) (Table 2). Assuming a random sequence, the rare cutters (NotI, etc.) recognize an 8-nucleotide palindrome sequence and cleave DNA on average every 65,000 bp (39). This class of restriction enzymes leave compatible ends that randomly concatamerize or reassemble with other DNA molecules having a similar compatible end. In contrast, a subclass of restriction enzymes (BglI, BstXI, and SfiI) also recognize palindrome sequences but leave random sticky ends of 1 to 4 nucleotides in length that are not complementary to most other sticky ends generated with the same enzyme at other sites in the DNA. The BglI restriction endonuclease recognizes the palindrome sequence GCCNNNN↓NGGC and is predicted to cleave the DNA every ∼4,096 bp in a random DNA sequence (39). Because a 3-nucleotide variable overhang is generated following cleavage, 64 (43) different variable ends can be generated, which efficiently assemble only with the appropriate 3-nucleotide complementary overhang generated at an identical BglI site (Table 2). Consequently, identical BglI sites are repeated every ∼262,144 bp in a random sequence of DNA.
TABLE 2.
Restriction enzymes that cleave at specific sites and leave variable sticky ends
| Restriction enzyme | Restriction sites | No. of sticky ends | Cutting frequencya | Theoretical end redundanceb | Actual no. of restriction sitesc (% with nonunique ends)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | VZV | EBV | FP | MG | HH6 | CJ | TGEV | |||||
| BglI | GCCNNNN↓NGGC | 3 | 4,096 (8,640 ± 23,266) | 262,144 | 1 (0) | 32 (38) | 198 (ND) | 3 (0) | 8 (0) | 19 (26) | 57 (79) | 1 (0) |
| CGGN↑NNNNCCG | ||||||||||||
| BstXI | CCANNNNN↓NTGG | 4 | 4,096 | 1,048,576 | 3 (0) | 31 (6) | 80 (41) | 16 (13) | 103 (ND) | 33 (12) | 135 (ND) | 3 (0) |
| GGTN↑NNNNNACC | ||||||||||||
| SfiI | GGCCNNNN↓NGGCC | 3 | 65,536 | 4,194,304 | 0 | 2 (0) | 68 (62) | 0 | 0 | 2 (0) | 0 | 0 |
| CCGGN↑NNNNCCGG | ||||||||||||
| SapI | GCTCTTCN↓NNN | 3 | 16,384 | 1,048,576 | 2 (0) | 11 (18) | 34 (35) | 10 (60) | 27 (30) | 14 (29) | 220 (ND) | 4 (0) |
| CGAGAAGNNNN↑ | ||||||||||||
| EcoRI | G↓AATTC | 0 | 4,096 | 4,096 | 3 | 15 | 15 | 71 | 74 | 52 | 283 | 7 |
| CTTAA↑G | ||||||||||||
Mean distance between BglI sites (in base pairs) in the genomes of these organisms (fragment sizes ranged from 9 to 191,414 bp).
Frequency of end compatibility with a random sequence.
MV, Marburg virus (19,104-bp genome; NC001608); VZV, varicella-zoster virus (124,884-bp genome; X04370); EBV, Epstein-Barr virus (172,281-bp genome; V01555); FP, fowl pox virus (288,539-bp genome; AF198100); MG, M. genitalium (580,074-bp genome; NC000908); HH6, human herpesvirus 6B strain Z29 (162,114-bp genome; AF157706); CJ, C. jejuni (1,641,481-bp genome; NC002163); TGEV, 28,586-bp genome (3). ND, not done.
As DNA and RNA sequences are not random, the actual distribution of these restriction sites will vary considerably and be heavily influenced by the sequence of the genome, percent base pair composition, and the presence of duplications, inversions, and repetitive sequences. To address these questions, we determined the frequency of BglI, SapI, BstXI, SfiI, and EcoRI sites in the genome of a variety of microbial and viral pathogens, including Marburg virus, TGEV, various herpesviruses, fowlpox virus, and Campylobacter jejuni and Mycoplasma genitalium (Table 2). These data clearly demonstrate that the expected repeat distance of identical BglI sites in a given genome may be far less than or greater than once every 262,144 bp (Table 2). For example, the large genomes of C. jejuni and M. genitalium are devoid of SfiI sites, yet the genome of Epstein-Barr virus contains 68 SfiI sites because of its high GC content and the presence of duplications in the sequence. Potential problems of identical-end duplicity, however, can be circumvented if the DNA pieces are cleverly sorted using recursive techniques, allowing the assembly of approximately 264 fragments of various sizes that contain different BglI ends. Importantly, these data suggest that the genomes of many microbial organisms can be engineered and then assembled by in vitro ligation from a series of smaller subclones. As the TGEV genome contains a single BglI site (Table 2), we hypothesized that a sequential series of smaller DNA subclones, each flanked by unique BglI junctions, could be systematically and precisely assembled into an intact full-length TGEV cDNA construct from which in vitro transcription will result in an infectious RNA (Fig. 1). To test this hypothesis, we assembled a full-length infectious construct of a coronavirus, thereby demonstrating the method's potential application for assembling other large genomes or chromosomes in vitro.
Assembly of a full-length TGEV construct.
Initially, we isolated five cDNA subclones spanning the entire TGEV genome (designated TGEV A, B, C, DE, and F). Each cDNA subclone was flanked by unique BglI sites and will only anneal with the appropriate adjacent subclone, resulting in a full-length TGEV cDNA construct (Fig. 1A). To RT-PCR clone the 6.2-kb TGEV A fragment located at the 5′ end of the TGEV genome, the forward primer included a T7 start site and the 5′-most TGEV leader RNA sequences, while the reverse primer was located at nucleotide 6180, just downstream from a naturally occurring BglI site (GCCTGTT↓TGGC) in the TGEV genome (3, 17) (Table 1). The 5.2-kb B fragment was amplified using a forward primer upstream of the BglI site at position 6159 and a reverse primer which introduced a unique BglI site (GCCGCAT↓CGGC) at position 11355 (Fig. 1). The 5.2-kb C fragment was amplified using a forward primer which introduced the same BglI site at nucleotide 11355 and a reverse primer which introduced another unique BglI site (GCCTTCT↓TGGC) at position 16595. While our original cloning strategy called for separate D and E fragments, it became evident that a single 6.9-kb fragment was stable in microbial vectors. Therefore, a single DE-1 fragment was amplified using a forward primer that introduced the same BglI site at position 16595 and a reverse primer which introduced a new BglI site (GCCGTGC↓AGGC) in the S glycoprotein gene at nucleotide 23487. The F fragment was cloned with a forward primer that introduced the same BglI site at position 23487 and a reverse primer that contained the 3′-most nucleotides of the TGEV genome, including an additional 25 T's prior to terminating at a NotI site. A list of the primers used to mutagenize the TGEV genome and to isolate each of the TGEV subclones is shown in Table 1. These primer pairs did not alter the amino acid coding sequence of the virus. The sequence of each unique interconnecting junction is shown in Fig. 1.
The pTGEV A, C, DE, and F clones were stable in plasmid DNAs in Escherichia coli. The B fragment, however, was unstable, and only a few slow-growing isolates were obtained, all of which contained deletions or insertions in the wild-type sequence. During two different cloning attempts, a 200- to 300-nucleotide fragment from the E. coli chromosome was inserted at position 9973, which is in a region of instability in the TGEV genome noted by other investigators (Fig. 1B) (3, 17). In addition, some clones contained a ∼500-bp deletion across this region. We reasoned that breaks in the TGEV B sequence at or around nucleotide 9973 might ablate fragment instability and allow the cloning of these sequences into E. coli. Since the TGEV B fragment does not contain a BstXI site, we used primer-mediated mutagenesis (C-A change at position 9944) to bisect the B fragment into TGEV B1 and TGEV B2 amplicons with an adjoining BstXI (CCATTCAC↓TTGG) site located at position 9949 in the TGEV genome (Fig. 1, Table 1). The 4-nucleotide overhang generated by BstXI would also provide additional specificity and sensitivity in systematically assembling the TGEV subclones. After these modifications, pTGEV B1 and B2 plasmid subclones which were stable and grew efficiently in E. coli were rapidly identified. The location of each of the subclones used in the assembly of the TGEV full-length construct is shown in relationship to important motifs or cis-acting sequences in the viral genome (Fig. 2). Data from our lab and others suggest that sequences in and around the TGEV poliovirus 3C-like protease (3-Clpro) motif are either bactericidal or unstable in microbial vectors (3, 17).
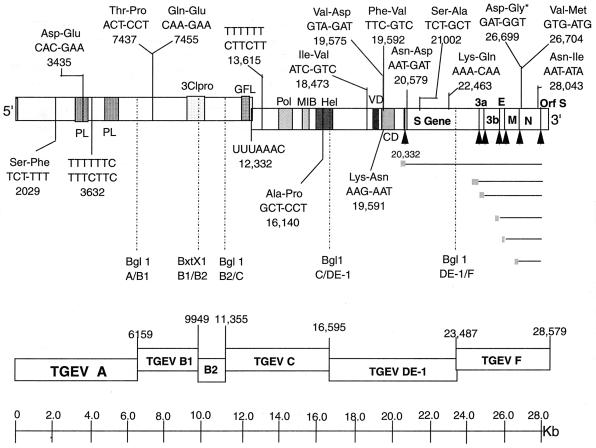
FIG. 2.
Sequence and chromosomal location of the TGEV subclones. The consensus amino acid changes that differ from the published sequence are shown in each of the final clones used to assemble a full-length TGEV construct (17). Each of these changes in the TGEV sequence has also been noted by Almazan et al. (3) at all indicated positions except those denoted by an asterisk. The relative locations of the different TGEV motifs were taken from Eleouet et al. (17). Abbreviations: PL, papain-like protease; GFL, growth factor-like domain; Pol, polymerase motif; MIB, metal-binding motif; Hel, helicase motif; VD, variable domain; CD, conserved domain; ↑, intergenic starts.
Inserts from three to six independent subclones from each fragment were sequenced, and a consensus TGEV subclone was assembled using standard recombinant DNA techniques. The consensus sequence of our Perdue TGEV full-length construct contained 15 amino acid changes and numerous silent changes compared with the published sequence (Fig. 2) (17). These changes were also noted by Almazan et al. (3). T7 termination sites might also prevent efficient in vitro transcription of infectious full-length TGEV transcripts from the construct. Two types of sites are known to cause pausing and/or termination by bacteriophage T7 RNA polymerase (28, 37). A type I termination site consists of a stable stem-loop structure that terminates transcription in adjacent stretches of T residues, while a type II pause site consists of a specific 7-bp sequence (ATCTGTT) (28, 37). Transcription termination will occur when a stretch of T's is located 6 to 8 nucleotides downstream of this sequence (28, 37). Type I stops had prevented transcription of vesicular stomatitis virus full-length negative-strand RNAs in vitro with T7 polymerase (58). To preempt potential problems in the generation of full-length TGEV transcripts, putative type I T7 RNA polymerase termination sites (long runs of six T's) were identified in the TGEV consensus sequence that starts at nucleotide 3632 in the pTGEV A subclone and 13615 in the pTGEV C subclone (3, 17). Mutations were introduced by primer-mediated overlapping PCR mutagenesis without altering the coding sequence. Putative type II pause sites were also identified at nucleotides 17551, 19957, and 23103 in the TGEV genome (3), but did not contain the prerequisite downstream T-rich stretch necessary for efficient T7 termination.
To assemble a full-length cDNA construct of TGEV, plasmids were digested with BglI, BstXI, or NotI, and the appropriate sized inserts were isolated from agarose gels (Fig. 3A). The TGEV A and B1, B2, and C, and DE-1 and F fragments were ligated overnight in the presence of T7 DNA ligase. Systematically assembled products were isolated from agarose gels (Fig. 3B to D), and the TGEV A-B1, B2-C3, and DE-1–F fragments were religated overnight. The final products were purified by phenol-chloroform-isoamyl alcohol and chloroform extraction, precipitated under ethanol, and then separated in agarose gels (Fig. 4A and B). Clearly, an appropriately sized full-length TGEV cDNA of about 29 kb in length (TGEV 1000) was generated as well as some assembly intermediates. Capped T7 transcripts were synthesized, treated with DNase, and analyzed in 0.5% agarose gels in parallel with the TGEV 1000 assembled product. These data demonstrate that low levels of high-molecular-weight transcripts were evident following T7 transcription in vitro (Fig. 4C). Using transcripts driven from various pSin replicons (encoding GFP or T7 polymerase) as a control, we predict that these TGEV transcripts were likely full length (data not shown). DNase treatment removed the TGEV full-length cDNA as well as the incomplete assembly intermediates (Fig. 4C).
FIG. 3.
Assembly of the TGEV full-length construct. (A) The various TGEV plasmid DNAs were digested with BglI, BstXI, or NotI, and the appropriate-sized products were isolated from agarose gels as described in the text. The TGEV A and B1 fragments, TGEV B2 and C3 fragments, or TGEV DE-1 and F fragments were ligated at 16°C overnight in separate reactions. Appropriate-sized products were isolated from agarose gels. (B) A+B1. (C) B2+C. (D) DE-1+F. Following purification from agarose gels, the purified products are shown in panel A as well.
FIG. 4.
In vitro transcription from full-length TGEV constructs. The A-B1, B2-C, and DE-1–F contigs were ligated in vitro as described in the text. (A and B) DNA positions after 8 and 30 h of electrophoresis, respectively. Lane 1, purified A-B1 product; lane 2, purified B2-C product; lane 3, purified DE-1–F product; lane 4, 1-kb ladder; lane 5, in vitro-ligated products; lane 6, high-molecular-weight markers. (C) The in vitro-ligated products were transcribed in vitro, and the products were digested with DNase for 15 min at room temperature and separated in agarose gels. Lane 1, high-molecular-weight DNA markers; lane 2, 1-kb DNA ladder; lane 3, in vitro-ligated TGEV products; lane 4, DNase-treated in vitro transcripts. Arrow indicates high-molecular-weight transcripts.
Transfection and recovery of infectious virus.
Synthesis of full-length TGEV transcripts was difficult but resulted in high-molecular-weight RNA product (Fig. 4C). To enhance transfection efficiencies, we tested several different strategies to maximize infectivity of the putative full-length transcripts in vitro. Under identical conditions of treatment, about 10 to 20% of ST cells are efficiently transfected with Sindbis replicons encoding GFP (1), compared with about 60 to 80% of the BHK cells (data not shown). As coronavirus host range specificity occurs primarily at entry and the genomic RNA is infectious in a variety of permissive and nonpermissive cells (5, 14, 41, 44, 51), we reasoned that BHK cells might be more sensitive primary hosts because of the intrinsically higher transfection efficiency. In addition, several reports have suggested that the coronavirus N nucleocapsid protein may function as part of the transcription complex and may influence the translation efficiency of viral mRNAs (7, 15, 49). Because these data suggested that N might enhance the infectivity of full-length transcripts, four different transfection strategies were tested in BHK cells. We transfected BHK cells with TGEV transcripts alone, TGEV plus TGEV N gene transcripts, or just TGEV N transcripts. In a parallel experiment, TGEV and TGEV N gene transcripts were treated with RNase A prior to transfection. Following electroporation, the BHK cells were seeded with 106 ST cells to serve as appropriate permissive hosts for progeny virus amplification.
Three days posttransfection, supernatants were passaged into ST cells, and cytopathic effect was observed within 36 h postinfection only with supernatants derived from the TGEV-N gene-transfected cultures. Supernatants were harvested at 48 h postinfection and passaged twice more at 48-h intervals in fresh ST cell cultures. Cytopathic effect typical of TGEV infection was evident at each passage (data not shown). Fluorescent antibody staining with mouse anti-TGEV serum demonstrated that viral antigen was clearly present in each passage (data not shown). Using RT-PCR with primer pairs located within the leader RNA sequence and at the 3′ end of the TGEV genome, leader-containing subgenomic mRNA transcripts (mRNAs 6 and 7) encoding the N and hydrophobic membrane proteins were also evident in passage 1 ST cells (data not shown). Furthermore, virus derived from the TGEV 1000 transcripts was diluted and inoculated into ST cells, where plaques developed after 48 h (Fig. 5). No significant differences in plaque morphology were noted between the molecularly cloned recombinant viruses and wild type, suggesting that the cDNA construct did not contain debilitating mutations. Transcripts of TGEV plus N gene treated with RNase A prior to electroporation did not result in the production of infectious virus.
FIG. 5.
Plaque morphology of icTGEV viruses. Cultures of ST cells were infected with wild-type (WT) TGEV, icTGEV-1, and icTGEV-3. Cells were stained with neutral red at 48 h postinfection, and images were digitized and prepared using PhotoShop 5.5.
Plaque-purified stocks prepared from passages 1 through 3 of icTGEV (icTGEV-1, icTGEV-2, and icTGEV-3, respectively) were used in growth curves and compared to the parental TGEV strain in ST cells. Cultures were infected with virus at an MOI of 5 for 1 h, and samples were harvested at selected times over the next 44 h. No significant differences in the replication of wild-type TGEV- or TGEV 1000-derived viruses icTGEV-1, icTGEV-2, and icTGEV-3 were noted in ST cells, and all viruses replicated to titers that approached 108 PFU/ml within 28 h (Fig. 6). These data indicate that viruses derived from the infectious cDNA construct had phenotypes indistinguishable from those of wild-type TGEV in swine cells. TGEV efficiently utilizes the feline and porcine aminopeptidase N receptors for docking and entry and can replicate efficiently in feline CRFK cells (14, 51) (data not shown). To determine the host range phenotype of these viruses, cultures of ST and CRFK cells were infected with the molecularly cloned icTGEV-1 or icTGEV-3 virus at an MOI of 5 and fixed at 12 h postinfection. Viral antigen expression was measured by FA (Fig. 7). Efficient virus docking and entry were evidenced by significant levels of antigen expression in swine and feline cells infected with the molecularly cloned viruses. These data demonstrate that the molecularly cloned viruses had a host range phenotype similar to that of the wild type.
FIG. 6.
Growth curves of plaque-purified molecularly cloned viruses. Plaque-purified wild-type TGEV and recombinant TGEV viruses (icTGEV-1, icTGEV-2, and icTGEV-3) derived from the infectious construct were inoculated into ST cells at an MOI of 5 for 1 h at room temperature. The virus was removed, and the cultures were incubated in complete medium at 37°C. Samples were harvested at the indicated times and assayed by plaque assay in ST cells.
FIG. 7.
Host range phenotype of molecularly cloned icTGEV. Cultures of ST or CRFK cells (105) were inoculated with molecularly cloned viruses icTGEV-1 and icTGEV-3 at an MOI of 5 for 1 h at room temperature. The inocula were removed, and the cells were incubated in complete medium at 37°C for 12 h. Medium was removed, and the cells were fixed in a 50% methanol–acetone mix, washed, and stained by FA as described in the text. (A) Mock-infected ST. (B) icTGEV-1-infected ST cells. (C) icTGEV-3-infected ST cells. (D) Mock-infected CRFK cells. (E) icTGEV-1-infected CRFK cells. (F) icTGEV-3-infected CRFK cells.
Identification of marker mutations.
Infectious virus derived from transfected cultures should contain the four unique interconnecting junction sequences used in the construction of the infectious TGEV 1000 construct (Fig. 1 and 2). If these noncoding mutations produce a neutral phenotype on virus replication, they should also be stable during passage. Consequently, wild-type TGEV, icTGEV-1, and icTGEV-3 were inoculated into ST cells, and intracellular RNA was isolated at 12 h postinfection. Using RT-PCR and primer pairs that asymmetrically flank each of the B1-B2, B2-C, C–DE-1, and DE-1–F junctions, we amplified products of ∼400 to 600 bp (Fig. 8). Results using restriction fragment length polymorphism analysis demonstrated that none of the marker mutations were present in the wild-type TGEV genome (Fig. 8A). However, the icTGEV-1 and icTGEV-3 viruses both contained the unique marker mutation profiles used to create the unique BglI and BstXI restriction sites within the TGEV 1000 construct (Fig. 8B and C). The PCR products were subcloned, and the reverse complement of the sequence is shown, demonstrating that the appropriate mutations were present in the viruses isolated from the infectious construct (Fig. 9). Clearly, TGEV 1000 transcripts were infectious and produced virus which contained the appropriate marker mutations. These data illustrate that infectious constructs of coronaviruses can be systematically and precisely assembled from a series of smaller subclones in vitro.
FIG. 8.
Marker mutations are present in virus derived from the infectious construct. Cultures of ST cells were infected with wild-type (WT) TGEV or plaque-purified icTGEV isolates derived from the infectious construct. Intracellular RNA was isolated, and RT-PCR was performed using primer pairs that asymmetrically flank each of the unique BglI-BstXI junctions inserted into the infectious construct. (A) Wild-type TGEV. (B) icTGEV-1 (passage 1). (C) icTGEV-3 (passage 3). In panel C, a larger ∼1.6-kb wild-type TGEV amplicon spanning the B1-B2 junction was also treated with BstXI as a control (the amplicon was derived from primer pairs located between nucleotides 9730 and 9750 and 11342 and 11362 in the TGEV sequence [3]). Arrows indicate cleaved DNA intermediates.
FIG. 9.
Sequence analysis of icTGEV-3. The uncut RT-PCR amplicons shown in Fig. 8 were isolated from gels and subcloned into Topo II TA cloning vectors. Inserts were sequenced using universal primers and an automated sequencer. (A) icTGEV-3 B2-C junction. (B) icTGEV-3 C-DE junction. (C) icTGEV-3 DE-F junction. Note: sequences are the reverse complement of the genomic TGEV sequence. The wild-type virus sequence is also noted in each panel.
DISCUSSION
The complete ∼30-kb nucleotide sequence for a number of coronaviruses has been available for about 10 years (8, 17, 22, 27), yet until recently, a full-length infectious clone has not been assembled because of size constraints and regions of coronavirus genomic instability in bacterial vectors, the requirement for a vector system which allows simple reverse genetic applications, and the inability to synthesize full-length transcripts in vitro. Each of these inherent restrictions must be circumvented to assemble infectious coronavirus constructs and at the same time allow easy reverse genetic applications. In a landmark achievement, a full-length TGEV infectious clone was recently engineered into BAC vectors using standard DNA techniques (3). Following DNA transfection into ST cells, full-length transcripts were initially transcribed from a cytomegalovirus (CMV) promoter and then amplified by virus replication in the cytoplasm of the cell. In this paper, we describe a rapid approach to systematically assembling a full-length infectious TGEV cDNA from a panel of six smaller subclones using in vitro ligation. These methods will provide a powerful complementary approach to systematically assemble new large cDNAs from a variety of microbial pathogens into BAC or other vectors that stably maintain large DNA inserts (30). Importantly, RNA or DNA genomes which are too large, circular, or unstable in these cloning vectors can still be assembled using this in vitro ligation technique. As coronaviruses contain the largest RNA genome, these approaches should permit reverse genetic studies for all RNA viruses.
Evidence from several experiments demonstrated that transcripts of the TGEV 1000 genomic construct were infectious. Transcripts treated with RNase were not infectious, indicating that infection was likely initiated from the RNA transcripts synthesized in vitro. Medium from transfected cultures could be used to propagate infection, with corresponding cytopathology and viral antigen expression in fresh cultures of cells. Progeny virions formed plaques in monolayers of permissive cells, and plaque-purified molecularly cloned virus grew efficiently to levels equivalent to those of wild-type virus in permissive host cells. The host range phenotypes of molecularly cloned viruses and wild-type virus were similar in vitro, although additional experiments are needed to determine if these viruses utilize the feline aminopeptidase receptor for docking and entry into feline cells (51). Most importantly, plaque-purified virus contained the expected BglI and BstXI marker mutations, providing definitive evidence that transcripts driven from the TGEV 1000 construct were infectious in vitro. The presence of these neutral mutations did not restrict the ability of icTGEV to replicate efficiently in ST cells.
It is remarkable that two entirely different approaches can be exploited to engineer infectious constructs of large RNA and DNA viruses. Our assembly strategy for coronavirus infectious constructs is simple and straightforward and does not depend on the availability of an existing viral defective interfering cDNA clone as a foundation for building the infectious construct (3). In contrast to infectious clones of other positive-strand RNA viruses (1, 2, 3, 13, 32, 35, 54), the TGEV 1000 construct must be assembled de novo and does not exist intact in bacterial vectors, circumventing problems in sequence instability. This did not restrict its applicability for reverse genetic applications, but rather allowed genetic manipulation of independent subclones, which will minimize the introduction of spurious mutations elsewhere in the genome during recombinant DNA manipulation. Another advantage of our approach is that different combinations of restriction sites can be used that generate highly variable 5′ or 3′ overhangs of 1 to 4 nucleotides in length, further increasing the specificity and sensitivity of the assembly cascade (Table 2). Because of insert toxicity in E. coli, infectious clones of yellow fever virus and Japanese encephalitis virus were assembled by in vitro ligation from two subclones but used conventional restriction enzymes like BamHI, ApaI, and AatI (34, 48). Our strategy, however, prevents spurious self-assembly of subclones and will provide a strong complementary approach to engineering large RNA or DNA genomes into BAC vectors or other vectors that stably maintain large DNA inserts (3).
It is interesting that in both TGEV infectious constructs assembled to date, sequences in or around the TGEV 3-Clpro motif were unstable in E. coli. Our studies, coupled with the findings by Almazan et al. (3), suggest that the unstable sequences can be disabled by bisecting the sequence between nucleotides 9758 and 9949 in the TGEV genome. This information may permit the isolation of larger TGEV A-B1, B2-C, and DE-F subclones and allow the assembly of infectious cDNAs following a single DNA isolation-ligation step. It is not clear whether similar unstable sequences are located at this position in other group 1 and group 2 coronaviruses.
Synthesizing ∼29-kb transcripts in vitro is problematic and the greatest impediment to generating infectious RNA from the assembled TGEV 1000 construct. Using a DNA launch platform and transcription of TGEV RNAs from a CMV promoter, transfection resulted in ∼36 infectious units/10 μg of DNA (3). Using an RNA launch platform, similar results were obtained in our laboratory. Compared with Sindbis virus replicons encoding GFP, we synthesized ∼100-fold less full-length TGEV transcripts in vitro, probably due to the extreme size of the viral genome (data not shown). Using transcripts driven from the ∼28.5-kb TGEV full-length construct alone, viral structural gene expression was not noted in 105 cells. In BHK cultures cotransfected with TGEV and N gene transcripts, ∼100 to 500 cells per 105 cells expressed viral structural proteins under identical conditions (data not shown). At 16 h posttransfection, little if any structural protein expression was noted in BHK cells electroporated with N gene transcripts alone or transcripts treated with RNase A. This compares with transfection efficiencies of greater than 60% using the 11- to 12-kb Sindbis virus noncytopathic replicons encoding GFP. Although less dramatic, similar problems were reported with the ∼13-kb infectious arterivirus cDNA clone (54). These problems may be circumvented somewhat by constructing BHK cell lines that simultaneously express the swine aminopeptidase N receptor and T7 RNA polymerase, allowing DNA transfection and transcription in vivo, and direct selection of progeny virus amplification in susceptible BHK cell lines (1, 14, 51). Alternatively, CMV promoters can be inserted at the 5′ end of the TGEV A clone, allowing DNA launch of infectious RNA (3).
In our studies, we could not generate infectious full-length transcripts until the putative T7 polymerase stop signals were removed from the TGEV genome, cytidine was included in agarose gels to reduce UV damage to DNA fragments, and BHK cells were used as recipient hosts (21). At this time, we have no direct evidence that the T stretches in the TGEV A and C fragments might act as T7 termination sites, as the RNA structure in these regions has not been characterized biochemically. Inclusion of capped N gene transcripts during the transfection process also enhanced the infectivity of the TGEV full-length construct in three separate trials. It is not completely clear whether these results were simply serendipitous or whether N transcripts were simply protecting the full-length transcripts from degradation by competitively interfering with RNase activity in cells or culture medium. The N protein may also protect the genome-length RNA in a ribonucleoprotein structure in the cell, enhance infectivity directly by stabilizing or functioning as part of an intact replication complex (7, 15, 26), or enhance the expression of viral mRNAs (49). Interestingly, TGEV engineered into BAC vectors did not require the presence of nucleocapsid protein to enhance transcript infectivity, suggesting an ancillary role for N transcripts in our system (3).
Prior to these and earlier studies (3), targeted RNA recombination using defective interfering donor RNAs was the best method for introducing precise alterations into the structural genes of the group II coronavirus mouse hepatitis virus, but this approach has been essentially limited to the 3′-most 9 kb of the mouse hepatitis virus genome (24, 25, 53). The availability of TGEV infectious constructs will obviously benefit studies of all aspects of TGEV biology and pathogenesis, including analysis of the coronavirus replicase and the somewhat controversial transcription processes which govern expression of the subgenome-length mRNAs (17, 40, 42, 43). The future development of TGEV vaccines and expression vectors is a particularly intriguing application, as the polycistronic genome organization and synthesis of subgenome-length mRNAs may allow the simultaneous expression of multiple foreign genes (18). It will also be relatively easy to target TGEV to other species by simple replacements of the S glycoprotein gene (14, 25, 51). In contrast to arterivirus expression vectors, the coronavirus intergenic sequences rarely overlap upstream ORFs, simplifying the design and expression of foreign genes from downstream intergenic promoters (11, 17, 52). Several TGEV downstream ORFs also appear to encode luxury functions that can be deleted from the viral genome without affecting infectivity in vitro (18, 29, 56, 57). Finally, the helical TGEV nucleocapsid structure may minimize packaging constraints and allow the expression of multiple large genes from a single construct (18, 26, 36).
The theoretical limits of our technique may approach several million base pairs of DNA and provide a rapid approach for inserting large cDNAs into BAC vectors (20, 30, 45). The systematic assembly method should be appropriate for constructing full-length infectious constructs of other large RNA viruses, including coronaviruses (27 to 32 kb), toroviruses (24 to 27 kb), and filoviruses like the Ebola and Marburg viruses (19 kb) (10, 26, 31). Viral genomes which are unstable in prokaryotic vectors might also be successfully cloned using these methods (9, 34, 48). Moreover, full-length infectious double-stranded DNA genomes of adenoviruses and herpesviruses promise to be a powerful tool in vaccination, gene transfer, and gene therapy (30, 45, 50, 55). Historically, full-length infectious constructs of these DNA viruses have been generated by ligation of DNA fragments, by homologous recombination (the more widely used method), or as full-length clones in BAC vectors (30, 38, 45, 50, 55). Direct ligation of DNA fragments has been restricted by the low efficiency of large-fragment ligations and the scarcity of unique restriction sites that make the approach technically challenging. Systematic and precise assembly using rare cutters (SfiI and SapI) that leave variable ends and can be purposely engineered into a sequence should simplify assembly of large double-stranded DNA viruses (Table 2). This will alleviate the difficulties associated with typical restriction enzymes or recombination approaches, which often result in second-site alterations (38, 45, 50, 55). This method may also circumvent other restrictions inherent in recombination-based methods which are limited to specific regions in the viral genome and which often result in recombinant viruses which are not wild type while allowing the introduction or removal of only a few genes in the virus vectors.
Our systematic assembly approach is not limited to manipulating the chromosomes of large RNA and DNA viruses. Over the past decade, the genome sequence of a large number of prokaryotic and eukaryotic chromosomes has provided significant insight into gene organization, structure, and function and likely identified the minimal set of genes required for prokaryotic life (12, 23; TIGR home page http://www.tigr.org). Reconstruction of a minimal genome from the bottom up is technically challenging and requires systematically assembling large DNA fragments and then inserting the reconstructed genome into an environment that allows metabolic activity and replication (12). Using a recursive approach, the systematic assembly of large chromosomes or minichromosomes from the bottom up is theoretically feasible (Table 2). Technical challenges will likely include the isolation of large DNA fragments and accompanying assembly intermediates from gels and the introduction of large DNA genomes into environments that permit replication. Our approach, however, may provide a means to address the function of large blocks of DNA, like pathogenesis islands, or to directly engineer chromosomes that contain large gene cassettes of interest (12). Additional studies will be needed to test the application of these methods in other viral, prokaryotic, and eukaryotic genomes.
ACKNOWLEDGMENTS
We thank Robert E. Johnston, Nancy Davis, Patrick Harrington, Mary Schaad, Mark Denison, and Lawrence Park for helpful discussion and encouragement during the course of these studies.
This work was supported by a research grant from the National Institutes of Health (AI 23946).
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlquist P, French R, Janda M, Loesch-Fries L S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almazan F, Gonzalez J M, Penzes Z, Izeta A, Calvo E, Plana-Duran J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric R S, Yount B. Subgenomic negative-strand function during mouse hepatitis virus infection. J Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baric R S, Sullivan E, Hensley L, Yount B, Chen W. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J Virol. 1999;71:638–649. doi: 10.1128/jvi.73.1.638-649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baric R S, Schaad M C, Stohlman S. Establishing a genetic recombination map for the murine coronavirus strain A59 complementation groups. Virology. 1990;177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baric R S, Nelson G W, Fleming J O, Lai M M C, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boursnell M E, Brown T D, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 9.Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh D, Horzinek M C. Genus Torovirus assigned to the Coronaviridae. Arch Virol. 1993;128:395–396. doi: 10.1007/BF01309450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C M, Cavanagh D, Britton P. Cloning and sequencing of a 8.4 kb region from the 3′ end of a Taiwanese virulent isolate of the coronavirus transmissible gastroenteritis virus. Virus Res. 1997;38:83–89. doi: 10.1016/0168-1702(95)00046-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M K, Magnus D, Caplan A L, McGee the Ethics of Genomics Group. Ethical considerations in synthesizing a minimal genome. Science. 1999;286:2087–2090. doi: 10.1126/science.286.5447.2087. [DOI] [PubMed] [Google Scholar]
- 13.Davis N L, Willis L V, Smith J F, Johnston R E. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology. 1989;171:189–204. doi: 10.1016/0042-6822(89)90526-6. [DOI] [PubMed] [Google Scholar]
- 14.Delmas B, Gelfi J, L'Haridon R, Vogel L K, Sjostrom H, Noren O, Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison M R, Spaan w J, van der Meer Y, Gibson C A, Sims A C, Prentice B, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vries A A F, Horzinek M C, Rottier P J M, de Groot R I. The genome organization of the Nidovirales: similarities and differences between arteriviruses, toroviruses and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. The complete sequence (20kb) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enjuanes L, Van der Zeijst B A M. Molecular basis of transmissible gastroenteritis coronavirus (TGEV) epidemiology. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 337–376. [Google Scholar]
- 19.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the nonstructural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes B, Cooke H. Engineering mammalian chromosomes. Hum Mol Genet. 1998;7:1635–1640. doi: 10.1093/hmg/7.10.1635. [DOI] [PubMed] [Google Scholar]
- 21.Grundemann D, Schomig E. Protection of DNA during preparative agarose gel electrophoresis against damage induced by ultraviolet light. Biotechniques. 1996;21:898–903. doi: 10.2144/96215rr02. [DOI] [PubMed] [Google Scholar]
- 22.Herold J, Raabe T, Schelle-Prinz B, Siddell S G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchison C A, Peterson S N, Gill S R, Cline R T, White O, Fraser C M, Smith H O, Venter J C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 24.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo L, Godeke G-J, Raamsman J B M, Masters P S, Rottier P J M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H J, Shieh C K, Gorbalenya A E, et al. The complete nucleotide sequence of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyakhov D L, He B, Zhang X, Studier F W, Dunn J J, McAllister W T. Pausing and termination by bacteriophage T7. J Mol Biol. 1998;280:201–213. doi: 10.1006/jmbi.1998.1854. [DOI] [PubMed] [Google Scholar]
- 29.McGoldrick A, Lowing J P, Paton D J. Characterization of a recent virulent TGEV from Britain with a deleted ORF3a. Arch Virol. 1999;4:763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters C J, Sanchez A, Rollin P E, Ksiazek T G, Murphy F A. Filoviridae: Marburg and Ebola viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott Williams and Wilkens; 1996. pp. 1161–1176. [Google Scholar]
- 32.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 33.Rasschaert D, Laude H. The predicted primary structure of the peplomer protein E2 of the porcine coronavirus transmissible gastroenteritis virus. J Gen Virol. 1987;68:1883–1890. doi: 10.1099/0022-1317-68-7-1883. [DOI] [PubMed] [Google Scholar]
- 34.Rice C M, Grakovig A, Galler R, Chambers T J. Transcription of yellow fever RNA from full length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- 35.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risco C, Anton I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg A H, Lade B N, Chui D-s, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld M A, Siegfried W, Yoshimura K, Yoneyama K, Fukayama M, Stier L F, Paako P K, Gilardi P, Stratford-Perricaudet L D, Perricaudet M, Jallat S, Pavirani A, Lecocq J P, Crystal R G. Adenovirus-mediated transfer of a recombinant α1 antitrypsin gene to the lung epithelium in vitro. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 5.1–5.31. [Google Scholar]
- 40.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schochetman G, Stevens R H, Simpson R W. Presence of infectious polyadenylated RNA in the coronavirus avian infectious bronchitis virus. Virology. 1977;77:772–782. doi: 10.1016/0042-6822(77)90498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethna P B, Hung S L, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 45.Smith G A, Enquist L W. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97:4873–4979. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snijder E J, Horzinek M C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J Gen Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- 47.Sosnovtsev S, Green K Y. RNA transcripts derived from a cloned full length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 48.Sumyoshi H, Hoeke C M, Trent D W. Infectious Japanese encephalitis virus RNA can be synthesized from in vitro-ligated cDNA templates. J Virol. 1992;66:5425–5431. doi: 10.1128/jvi.66.9.5425-5431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tahara S M, Dietlin T A, Bergmann C C, Nelson G W, Kyuwa S, Anthony R P, Stohlman S A. Coronavirus translational regulation: leader effects mRNA efficiency. Virology. 1994;202:621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong-Chuan H, Zhou S, Da Costa L T, yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tresnan D B, Levis R, Holmes K V. Feline aminopeptidase N serves as a receptor for feline, porcine, and human coronaviruses in serogroup 1. J Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tung F Y T, Abraham S, Sethna M, Hung S-L, Sethna P B, Hogue B G, Brian D A. The 9.1 kilodalton hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Most R G, Heijnen L, Spaan W J, de Groot R J. Homologous recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992;20:3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious retrovirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Zijl M, Quint W, Briaire J, de Rover T, Gielkens A, Berns A. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J Virol. 1988;62:2191–2195. doi: 10.1128/jvi.62.6.2191-2195.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaughn E M, Halbur P G, Paul P S. Sequence comparisons of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J Virol. 1995;69:3176–3184. doi: 10.1128/jvi.69.5.3176-3184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wesley R D, Woods R D, Cheung A K. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J Virol. 1991;65:3369–3373. doi: 10.1128/jvi.65.6.3369-3373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whelan S, Ball A, Barr J, Wertz G. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]