Abstract
Trichorhinophalangeal syndrome 1 (TRPS1) is a nuclear protein highly expressed in breast epithelial cells. TRPS1 immunohistochemistry (IHC) has been suggested as a breast cancer marker. To determine the diagnostic and prognostic utility of TRPS1 IHC, tissue microarrays containing 19,201 samples from 152 different tumor types and subtypes were analyzed. GATA3 IHC was performed in a previous study. TRPS1 staining was seen in 86 of 152 tumor categories with 36 containing at least one strongly positive case. TRPS1 staining predominated in various types of breast carcinomas (51%-100%), soft tissue tumors (up to 100%), salivary gland tumors (up to 46%), squamous cell carcinomas (up to 35%), and gynecological cancers (up to 40%). TRPS1 positivity occurred in 1.8% of 1083 urothelial neoplasms. In invasive breast carcinoma of no special type, low TRPS1 expression was linked to high grade (P = 0.0547), high pT (P < 0.0001), nodal metastasis (P = 0.0571), loss of estrogen receptor and progesterone receptor expression (P < 0.0001 each), and triple-negative status (P < 0.0001) but was unrelated to patient survival (P = 0.8016). In squamous cell carcinomas from 11 different sites, low TRPS1 expression was unrelated to tumor phenotype. Positivity for both TRPS1 and GATA3 occurred in 47.4% to 100% of breast cancers, up to 30% of salivary gland tumors, and 29 (0.3%) of 9835 tumors from 134 other cancer entities. TRPS1 IHC has high utility for the identification of cancers of breast (or salivary gland) origin, especially in combination with GATA3. The virtual absence of TRPS1 positivity in urothelial neoplasms is useful for the distinction of GATA3-positive urothelial carcinoma from breast cancer.
Key Words: TRPS1, tissue microarray, immunohistochemistry, diagnostic marker, breast cancer
Trichorhinophalangeal syndrome 1 (TRPS1), also termed transcriptional repressor GATA binding 1, is an atypical GATA nuclear transcription factor that mainly acts as a transcriptional repressor protein that can prevent the expression of GATA1-6-regulated genes (summarized1). For example, Runx1 and Sox9, which both are GATA3 regulated and required for cartilage formation, are suppressed by TRPS1.2,3 Multiple germline mutations of the TRPS1 gene were found to cause craniofacial and skeletal malformations.4 TRPS1 can also induce the expression of cancer-related genes, such as, for example, FOXA1, which is a negative regulator of epithelial-mesenchymal transition.5 Functional studies in cancer cells have suggested a tumor-suppressive activity of TRPS1 by preventing epithelial-mesenchymal transition,5,6 as well as a role of TRPS1 in breast cancer angiogenesis7 and in multidrug resistance of breast cancer8 and osteosarcoma.9
In normal tissues, TRPS1 is expressed in numerous cell types but expression is highest in breast epithelial cells (https://www.proteinatlas.org/ENSG00000104447-TRPS1). Because TRPS1 expression is often retained in cells that undergo malignant transformation, TRPS1 immunohistochemistry (IHC) has been suggested to represent a useful tool for the distinction of breast cancer from other cancer types in metastatic tissue.10,11 However, TRPS1 expression has recently also been described in various salivary gland tumors,10,12–14 endometrial carcinoma,15 hepatocellular carcinoma,16 colorectal cancer,17 gastric cancer,18 squamous cell carcinoma of the skin,19 synovial sarcoma,20 and osteosarcoma,21 as well as in lung22 and prostate cancer cell lines.23,24 In carcinomas of the breast,25 colon,17 and stomach,18 as well as in osteosarcoma,21 TRPS1 expression levels were associated with parameters of cancer aggressiveness. Altogether, the available data suggest a diagnostic and prognostic potential of TRPS1 IHC analysis which may be enhanced by a combination with GATA3, another commonly used breast cancer marker.10,26,27 However, many tumor entities have, so far, not been examined for TRPS1 by IHC and a systematic evaluation of different tumor entities for TRPS1 protein expression is so far lacking.
To better understand the prevalence and potential role of TRPS1 protein expression in tumors and to elucidate the potential diagnostic utility of TRPS1 IHC alone and in combination with GATA3, a survey of TRPS1 immunostaining in a broad range of tumor types is needed. In this study, we, therefore, evaluated TRPS1 expression in more than 19,000 tumor samples from 152 tumor types and subtypes for which GATA3 data were available in a tissue microarray (TMA) format.
MATERIALS AND METHODS
Tissue Microarrays
Our normal tissue TMA was composed of 8 samples from 8 different donors for each of 76 different normal tissue types (608 samples on one slide). The tumor TMAs contained a total of 19,201 primary tumors from 152 tumor types and subtypes. Detailed histopathological data on grade, pathologic tumor stage, pathologic lymph node status, and molecular data were available from 3173 tumors (1680 invasive breast carcinomas of no special type, 40 endometrioid and 369 serous ovarian carcinomas, 182 endometrioid endometrium carcinomas, and 902 squamous cell carcinomas). Clinical follow-up data were available from 877 invasive breast carcinomas of no special type with a median follow-up time of 49 months (range: 1 to 88 mo). Data on GATA3 were available for 15,964 tumors from an earlier study evaluating a large subset of our TMAs.28 The composition of both normal and tumor TMAs is described in detail in the results section. Samples were from the archives of the Institutes of Pathology, University Hospital of Hamburg, Germany, the Institute of Pathology, Clinical Center Osnabrueck, Germany, and Department of Pathology, Academic Hospital Fuerth, Germany. Tissues were fixed in 4% buffered formalin and then embedded in paraffin. TMA tissue spot diameter was 0.6 mm. The use of archived remnants of diagnostic tissues for manufacturing of TMAs and their analysis for research purposes, as well as patient data analysis, has been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics Commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Immunohistochemistry
Freshly cut TMA sections were immunostained on 1 day and in 1 experiment. Slides were deparaffinized with xylol at room temperature for 3×5 minutes, rehydrated through a graded alcohol series, and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121°C in pH 7.8, Dako Target Retrieval Solution (Agilent; #S2367). Endogenous peroxidase activity was blocked with Dako Peroxidase Blocking Solution (Agilent; #52023) for 10 minutes. A primary antibody specific for TRPS1 (rabbit recombinant monoclonal, MSVA-512R, MS Validated Antibodies; 5676-512R) was applied at 37°C for 60 minutes at a dilution of 1:20 for our normal tissue TMA and 1:300 for cancer tissues. For the purpose of validating and comparing antibodies, the normal tissue TMA and a subset of 2214 cancer tissues (666 breast cancers and 1191 non-breast cancers, including 622 non-breast adenocarcinomas) were also analyzed by the rabbit recombinant monoclonal TRPS1 antibody EPR16171 (#ab209664, Abcam) at a dilution of 1:2000 and by the rabbit polyclonal TRPS1 antibody PA5-84874 (Invitrogen/ThermoFisher) at a dilution of 1:100 with an otherwise identical protocol. Bound antibody was then visualized using the EnVision Kit (Agilent; #K5007) according to the manufacturer’s directions. The sections were counterstained with haemalaun. For tumor tissues, the percentage of positive neoplastic cells was estimated, and the staining intensity was semiquantitatively recorded (0, 1+, 2+, and 3+). For statistical analyses, the staining results were categorized into 4 groups. Tumors without any staining were considered negative. Tumors with 1+ staining intensity in ≤70% of tumor cells and 2+ intensity in ≤30% of tumor cells were considered weakly positive. Tumors with 1+ staining intensity in >70% of tumor cells, 2+ intensity in 31% to 70%, or 3+ intensity in ≤30% were considered moderately positive. Tumors with 2+ intensity in >70% or 3+ intensity in >30% of tumor cells were considered strongly positive. This scoring system has been used in numerous TMA studies and led to the identification of numerous known and novel prognostic molecular features in various tumor types.29–32
Statistics
Statistical calculations were performed with JMP 17 software (SAS Institute Inc.). Contingency tables and the χ² test were performed to search for associations between TRPS1 immunostaining and tumor phenotype. Survival curves were calculated according to Kaplan-Meier. The log-rank test was applied to detect significant differences between groups. Sensitivity and specificity for the detection of breast cancers were calculated according to the following formulas: sensitivity=number of true positives divided by the number of true positives plus number of false negatives; specificity=number of true negatives divided by the number of true negatives plus number of false positives.
RESULTS
Technical Issues
A total of 16,818 (87.6%) of 19,201 tumor samples were interpretable in our TMA analysis. Noninterpretable samples demonstrated either a lack of unequivocal tumor cells or loss of the tissue spots during technical procedures. At least 4 samples of each normal tissue type were evaluated.
TRPS1 in Normal Tissues
TRPS1 staining was always nuclear and strongest in luminal epithelial cells of breast glands. A moderate to strong nuclear staining was also observed in epithelial cells of the endometrium (stroma cells were also positive, but weaker), glial cells of the brain, sebaceous glands, and suprabasal cells of non-keratinizing squamous epithelium. A weak to moderate TRPS1 positivity was observed in various other epithelial (salivary glands, gallbladder, fallopian tube, respiratory epithelium, renal tubuli, amnion, thyroid gland, and parathyroid gland), muscular (smooth muscle and myometrium), neuronal (ganglion cells in the intestine), and germinal (spermatogonia) cell types. In other organs, TRPS1 staining of epithelial cells was only occasionally detected, such as, for example, in atrophic acinar cells of the prostate. A nuclear TRPS1 staining of variable intensity was also regularly seen in stromal cells of various tissues, especially in the case of tissue reparation (probably fibroblasts). Representative images are shown in Figure 1. A similar nuclear staining was observed in all these cell types when the anti-TRPS1 antibody EPR16171 was used (Supplemental Fig. 1, Supplemental Digital Content 1, http://links.lww.com/PAS/B785). As compared with MSVA-512R, EPR16171 resulted in additional cytoplasmic staining in basal cells of several non-keratizing squamous epithelia (Supplemental Fig. 1, Supplemental Digital Content 1, http://links.lww.com/PAS/B785), whereas PA-84874 showed significant additional cytoplasmic staining in many different normal tissues, including, for example, smooth muscle cells, kinocilia of the epididymis, parietal cells of the stomach, and lymphocytes, as well as nuclear staining in Leydig cells of the testis (Supplemental Fig. 2, Supplemental Digital Content 2, http://links.lww.com/PAS/B786). The additional stainings observed with EPR16171 and PA-84874 were considered antibody-specific cross-reactivities.
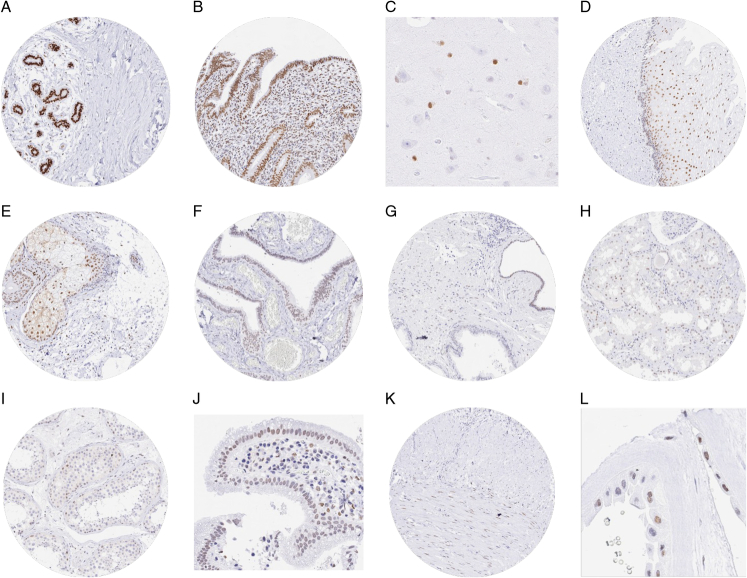
FIGURE 1.
TRPS1 immunostaining of normal tissues. The panels show a strong nuclear staining of breast epithelial cells (A) epithelial cells of the endometrium (B), and glial cells in the brain (C). Variable nuclear staining also occurred in suprabasal cells of esophageal squamous epithelium (D), sebaceous glands of the skin (E), epithelial cells of the fallopian tube (F), atrophic epithelial cells of the prostate (G), some tubular cells of the kidney (H), spermatogonia of the testis (I), epithelial cells of the gallbladder (J), smooth muscle cells of the appendix (K), and amnion cells (L).
TRPS1 in Tumor Tissues
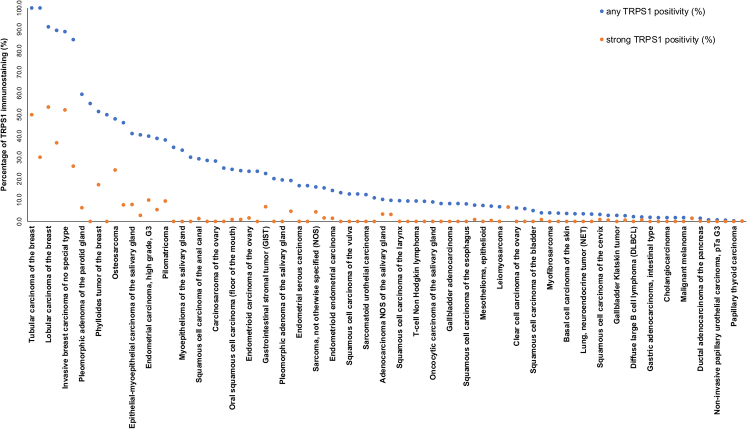
TRPS1 immunostaining was detectable in 2482 (14.8%) of the 16,818 analyzable tumors, including 614 (3.7%) with weak, 800 (4.8%) with moderate, and 1068 (6.4%) with strong immunostaining. Overall, 86 (56.6%) of 152 tumor categories showed TRPS1 positivity with 36 (23.7%) tumor categories including at least one case with strong staining (Table 1). The highest rate of TRPS1 positivity was found in various types of breast cancers (51.4% to 100%), soft tissue tumors (up to 100%), salivary gland tumors (up to 46.2%), squamous cell carcinomas of various sites of origin (up to 34.7%), and in diverse gynecological cancers (up to 40.0%). TRPS1 positivity only occurred in 1.8% of 1083 urothelial neoplasms. Representative images are shown in Figure 2. A ranking order of TRPS1-positive and strongly positive tumors is given in Figure 3. The relationship between TRPS1 expression and clinically important histopathological and molecular tumor features in breast cancer, ovarian cancer, endometrial carcinoma, and squamous cell carcinomas from different sites is shown in Table 2. In invasive breast cancer of no special type, low TRPS1 expression was linked to high grade (P = 0.0547), advanced pathologic tumor stage (P < 0.0001), nodal metastasis (P = 0.0571), loss of estrogen receptor expression (P < 0.0001), loss of progesterone receptor expression (P < 0.0001), and triple-negative status (P < 0.0001) but was unrelated to overall patient survival (P = 0.8016; Supplemental Fig 4, Supplemental Digital Content 3, http://links.lww.com/PAS/B787). A combined analysis of 677 squamous cell carcinomas from 11 different sites did not reveal associations between TRPS1 expression and tumor phenotype. TRPS1 expression was also unrelated to human papillomavirus infection status (P = 0.1159; Supplemental Table 1, Supplemental Digital Content 4, http://links.lww.com/PAS/B788). In our prostate cancers, the rate of TRPS1 positivity increased from Gleason grade 3+3 (0%) to Gleason grade 5+5 (3.8%) and recurrent adenocarcinomas under therapy (7.6%; P = 0.0164). TRPS1 immunostaining was unrelated to the histopathological tumor phenotype in ovarian and endometrial cancer.
TABLE 1.
TRPS1 Immunostaining in Human Tumors
| TRPS1 immunostaining result | |||||||
|---|---|---|---|---|---|---|---|
| Tumor entity | On TMA (n) | Analyzable (n) | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | |
| Tumors of the skin | Pilomatricoma | 35 | 21 | 61.9 | 23.8 | 4.8 | 9.5 |
| Basal cell carcinoma of the skin | 89 | 54 | 96.3 | 1.9 | 1.9 | 0.0 | |
| Benign nevus | 29 | 26 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the skin | 145 | 127 | 84.3 | 14.2 | 0.0 | 1.6 | |
| Malignant melanoma | 65 | 59 | 98.3 | 1.7 | 0.0 | 0.0 | |
| Malignant melanoma Lymph node metastasis | 86 | 72 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Merkel cell carcinoma | 48 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 93 | 90.3 | 9.7 | 0.0 | 0.0 |
| Squamous cell carcinoma of the pharynx | 60 | 49 | 65.3 | 30.6 | 4.1 | 0.0 | |
| Oral squamous cell carcinoma (floor of the mouth) | 130 | 107 | 75.7 | 22.4 | 0.9 | 0.9 | |
| Pleomorphic adenoma of the parotid gland | 50 | 47 | 40.4 | 14.9 | 38.3 | 6.4 | |
| Warthin tumor of the parotid gland | 104 | 98 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma, NOS (papillary cystadenocarcinoma) | 14 | 10 | 70.0 | 30.0 | 0.0 | 0.0 | |
| Salivary duct carcinoma | 15 | 12 | 75.0 | 16.7 | 8.3 | 0.0 | |
| Acinic cell carcinoma of the salivary gland | 181 | 146 | 97.9 | 0.7 | 0.7 | 0.7 | |
| Adenocarcinoma NOS of the salivary gland | 109 | 87 | 89.7 | 3.4 | 3.4 | 3.4 | |
| Adenoid cystic carcinoma of the salivary gland | 180 | 106 | 59.4 | 18.9 | 18.9 | 2.8 | |
| Basal cell adenocarcinoma of the salivary gland | 25 | 21 | 81.0 | 14.3 | 0.0 | 4.8 | |
| Basal cell adenoma of the salivary gland | 101 | 86 | 87.2 | 10.5 | 2.3 | 0.0 | |
| Epithelial-myoepithelial carcinoma of the salivary gland | 53 | 51 | 58.8 | 9.8 | 23.5 | 7.8 | |
| Mucoepidermoid carcinoma of the salivary gland | 343 | 295 | 92.9 | 3.7 | 3.1 | 0.3 | |
| Myoepithelial carcinoma of the salivary gland | 21 | 18 | 61.1 | 16.7 | 16.7 | 5.6 | |
| Myoepithelioma of the salivary gland | 11 | 9 | 66.7 | 0.0 | 33.3 | 0.0 | |
| Oncocytic carcinoma of the salivary gland | 12 | 11 | 90.9 | 0.0 | 9.1 | 0.0 | |
| Polymorphous adenocarcinoma, low grade, of the salivary gland | 41 | 26 | 53.8 | 19.2 | 19.2 | 7.7 | |
| Pleomorphic adenoma of the salivary gland | 53 | 36 | 80.6 | 11.1 | 8.3 | 0.0 | |
| Tumors of the lung, pleura and thymus | Adenocarcinoma of the lung | 196 | 189 | 97.4 | 2.1 | 0.0 | 0.5 |
| Squamous cell carcinoma of the lung | 80 | 74 | 90.5 | 8.1 | 1.4 | 0.0 | |
| Small cell carcinoma of the lung | 16 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mesothelioma, epithelioid | 40 | 27 | 92.6 | 7.4 | 0.0 | 0.0 | |
| Mesothelioma, biphasic | 77 | 48 | 91.7 | 4.2 | 4.2 | 0.0 | |
| Thymoma | 29 | 28 | 96.4 | 3.6 | 0.0 | 0.0 | |
| Lung (NET) | 29 | 28 | 96.4 | 0.0 | 3.6 | 0.0 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 78 | 61 | 90.2 | 3.3 | 3.3 | 3.3 |
| Squamous cell carcinoma of the vulva | 157 | 140 | 87.1 | 12.9 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the cervix | 136 | 124 | 96.8 | 1.6 | 0.8 | 0.8 | |
| Adenocarcinoma of the cervix | 23 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Endometrioid endometrial carcinoma | 338 | 269 | 85.5 | 9.7 | 3.3 | 1.5 | |
| Endometrial serous carcinoma | 86 | 60 | 83.3 | 15.0 | 1.7 | 0.0 | |
| Carcinosarcoma of the uterus | 57 | 47 | 76.6 | 17.0 | 6.4 | 0.0 | |
| Endometrial carcinoma, high grade, G3 | 13 | 10 | 60.0 | 30.0 | 0.0 | 10.0 | |
| Endometrial clear cell carcinoma | 9 | 8 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Endometrioid carcinoma of the ovary | 130 | 119 | 76.5 | 9.2 | 12.6 | 1.7 | |
| Serous carcinoma of the ovary | 580 | 537 | 76.2 | 14.5 | 8.4 | 0.9 | |
| Mucinous carcinoma of the ovary | 101 | 88 | 96.6 | 1.1 | 2.3 | 0.0 | |
| Clear cell carcinoma of the ovary | 51 | 48 | 93.8 | 2.1 | 4.2 | 0.0 | |
| Carcinosarcoma of the ovary | 47 | 46 | 71.7 | 21.7 | 6.5 | 0.0 | |
| Granulosa cell tumor of the ovary | 44 | 37 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the ovary | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the ovary | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli Leydig cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Steroid cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Brenner tumor | 41 | 38 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 1764 | 1533 | 11.1 | 7.1 | 29.5 | 52.3 |
| Lobular carcinoma of the breast | 363 | 304 | 8.9 | 6.6 | 30.9 | 53.6 | |
| Medullary carcinoma of the breast | 34 | 27 | 14.8 | 25.9 | 33.3 | 25.9 | |
| Tubular carcinoma of the breast | 29 | 18 | 0.0 | 5.6 | 44.4 | 50.0 | |
| Mucinous carcinoma of the breast | 65 | 38 | 10.5 | 13.2 | 39.5 | 36.8 | |
| Phyllodes tumor of the breast | 50 | 35 | 48.6 | 8.6 | 25.7 | 17.1 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 |
| Adenomatous polyp, high-grade dysplasia | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma of the colon | 2483 | 2247 | 99.9 | 0.1 | 0.0 | 0.0 | |
| Gastric adenocarcinoma, diffuse type | 215 | 198 | 99.5 | 0.0 | 0.5 | 0.0 | |
| Gastric adenocarcinoma, intestinal type | 215 | 202 | 98.0 | 1.5 | 0.5 | 0.0 | |
| Gastric adenocarcinoma, mixed type | 62 | 62 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma of the esophagus | 83 | 70 | 98.6 | 0.0 | 0.0 | 1.4 | |
| Squamous cell carcinoma of the esophagus | 76 | 61 | 91.8 | 8.2 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the anal canal | 91 | 75 | 70.7 | 26.7 | 1.3 | 1.3 | |
| Cholangiocarcinoma | 58 | 57 | 98.2 | 1.8 | 0.0 | 0.0 | |
| Gallbladder adenocarcinoma | 51 | 48 | 91.7 | 2.1 | 6.3 | 0.0 | |
| Gallbladder Klatskin tumor | 42 | 36 | 97.2 | 2.8 | 0.0 | 0.0 | |
| Hepatocellular carcinoma | 312 | 274 | 99.3 | 0.7 | 0.0 | 0.0 | |
| Ductal adenocarcinoma of the pancreas | 659 | 638 | 98.6 | 1.1 | 0.2 | 0.2 | |
| Pancreatic/ampullary adenocarcinoma | 98 | 97 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Acinar cell carcinoma of the pancreas | 18 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| GIST | 62 | 58 | 77.6 | 13.8 | 1.7 | 6.9 | |
| Appendix (NET) | 25 | 20 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Colorectal (NET) | 12 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ileum (NET) | 53 | 52 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas (NET) | 101 | 97 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Colorectal (NEC) | 14 | 12 | 91.7 | 8.3 | 0.0 | 0.0 | |
| Ileum (NEC) | 8 | 8 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Gallbladder (NEC) | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas (NEC) | 14 | 14 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the urinary system | Noninvasive papillary urothelial carcinoma, pTa G2 low grade | 177 | 174 | 100.0 | 0.0 | 0.0 | 0.0 |
| Noninvasive papillary urothelial carcinoma, pTa G2 high grade | 141 | 140 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Noninvasive papillary urothelial carcinoma, pTa G3 | 219 | 138 | 99.3 | 0.7 | 0.0 | 0.0 | |
| Urothelial carcinoma, pT2-4 G3 | 735 | 631 | 97.1 | 1.7 | 0.5 | 0.6 | |
| Squamous cell carcinoma of the bladder | 22 | 20 | 95.0 | 5.0 | 0.0 | 0.0 | |
| Small cell NEC of the bladder | 23 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcomatoid urothelial carcinoma | 25 | 16 | 87.5 | 12.5 | 0.0 | 0.0 | |
| Urothelial carcinoma of the kidney pelvis | 62 | 52 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Clear cell renal cell carcinoma | 1286 | 1131 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Papillary renal cell carcinoma | 368 | 316 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Clear cell (tubulo) papillary renal cell carcinoma | 26 | 23 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chromophobe renal cell carcinoma | 170 | 151 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Oncocytoma of the kidney | 257 | 222 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3+3 | 83 | 78 | 100.0 | 0.0 | 0.0 | 0.0 |
| Adenocarcinoma of the prostate, Gleason 4+4 | 80 | 69 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma of the prostate, Gleason 5+5 | 85 | 79 | 96.2 | 3.8 | 0.0 | 0.0 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 237 | 92.4 | 5.1 | 1.7 | 0.8 | |
| Small cell NEC of the prostate | 19 | 7 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Seminoma | 682 | 665 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Embryonal carcinoma of the testis | 54 | 48 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the testis | 31 | 23 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the testis | 2 | 2 | 50.0 | 0.0 | 50.0 | 0.0 | |
| Sex cord-stromal tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Spermatocytic tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Yolk sac tumor | 53 | 46 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Teratoma | 53 | 49 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the penis | 92 | 67 | 86.6 | 10.4 | 3.0 | 0.0 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 113 | 110 | 100.0 | 0.0 | 0.0 | 0.0 |
| Papillary thyroid carcinoma | 391 | 353 | 99.7 | 0.3 | 0.0 | 0.0 | |
| Follicular thyroid carcinoma | 154 | 145 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Medullary thyroid carcinoma | 111 | 105 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Parathyroid gland adenoma | 43 | 27 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Anaplastic thyroid carcinoma | 45 | 42 | 90.5 | 7.1 | 2.4 | 0.0 | |
| Adrenal cortical adenoma | 50 | 46 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical carcinoma | 28 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pheochromocytoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of hematopoietic and lymphoid tissues | Hodgkin’s lymphoma | 103 | 93 | 100.0 | 0.0 | 0.0 | 0.0 |
| B-SLL/B-CLL | 50 | 39 | 100.0 | 0.0 | 0.0 | 0.0 | |
| DLBCL | 113 | 93 | 97.8 | 2.2 | 0.0 | 0.0 | |
| Follicular lymphoma | 88 | 71 | 100.0 | 0.0 | 0.0 | 0.0 | |
| T-cell non-Hodgkin’s lymphoma | 25 | 21 | 90.5 | 4.8 | 4.8 | 0.0 | |
| Mantle cell lymphoma | 18 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Marginal zone lymphoma | 16 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| DLBCL in the testis | 16 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Burkitt lymphoma | 5 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of soft tissue and bone | Tendosynovial giant cell tumor | 45 | 29 | 44.8 | 37.9 | 17.2 | 0.0 |
| Granular cell tumor | 53 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyosarcoma | 94 | 88 | 93.2 | 5.7 | 1.1 | 0.0 | |
| Liposarcoma | 145 | 123 | 95.9 | 2.4 | 0.8 | 0.8 | |
| MPNST | 15 | 14 | 71.4 | 7.1 | 21.4 | 0.0 | |
| Myofibrosarcoma | 26 | 25 | 96.0 | 4.0 | 0.0 | 0.0 | |
| Angiosarcoma | 74 | 55 | 98.2 | 0.0 | 1.8 | 0.0 | |
| Angiomyolipoma | 91 | 88 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 17 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ganglioneuroma | 14 | 14 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Kaposi sarcoma | 8 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 117 | 98.3 | 1.7 | 0.0 | 0.0 | |
| Sarcoma (NOS) | 74 | 68 | 83.8 | 11.8 | 0.0 | 4.4 | |
| Paraganglioma | 41 | 41 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ewing sarcoma | 23 | 15 | 93.3 | 0.0 | 0.0 | 6.7 | |
| Rhabdomyosarcoma | 7 | 6 | 83.3 | 16.7 | 0.0 | 0.0 | |
| Schwannoma | 122 | 121 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Synovial sarcoma | 12 | 10 | 0.0 | 20.0 | 50.0 | 30.0 | |
| Osteosarcoma | 44 | 25 | 52.0 | 16.0 | 8.0 | 24.0 | |
| Chondrosarcoma | 40 | 18 | 88.9 | 11.1 | 0.0 | 0.0 | |
| Rhabdoid tumor | 5 | 5 | 80.0 | 20.0 | 0.0 | 0.0 | |
| Solitary fibrous tumor | 17 | 17 | 94.1 | 0.0 | 5.9 | 0.0 | |
B-CLL indicates chronic lymphocytic leukemia, B-cell type; B-SLL, small lymphocytic lymphoma, B-cell type; DLBCL, diffuse large B-cell lymphoma; GIST, gastrointestinal stromal tumor; MPNST, malignant peripheral nerve sheath tumor; NEC, neuroendocrine carcinoma; NET, neuroendocrine tumor; NOS, not otherwise specified.
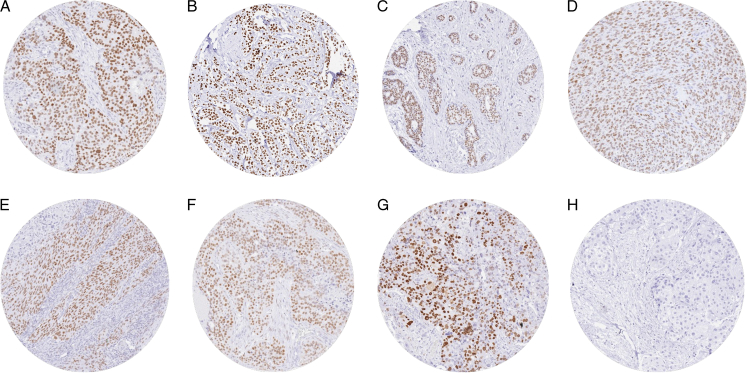
FIGURE 2.
TRPS1 immunostaining in cancer. The panels show a nuclear TRPS1 positivity of variable intensity in an invasive breast carcinoma of no special type (A) a lobular breast carcinoma (B), an adenoid cystic carcinoma of the parotid gland (C), a synovial sarcoma (D), a squamous cell carcinoma of oral cavity (E), a high-grade serous carcinoma of the ovary (F), and a recurrent adenocarcinoma of the prostate (G). TRPS1 staining is lacking in urothelial carcinoma of the urinary bladder (H).
FIGURE 3.
Ranking order of TRPS1 immunostaining in tumors. Both the percentage of positive cases (blue dots) and the percentage of strongly positive cases (orange dots) are shown.
TABLE 2.
TRPS1 and Tumor Phenotype
| TRPS1 immunostaining result | ||||||
|---|---|---|---|---|---|---|
| n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | P | |
| Invasive breast carcinoma of no special type | ||||||
| pT1 | 697 | 6.5 | 5.3 | 30.0 | 58.2 | <0.0001 |
| pT2 | 600 | 13.2 | 8.7 | 29.0 | 49.2 | — |
| pT3-4 | 120 | 18.3 | 6.7 | 31.7 | 43.3 | — |
| G1 | 166 | 4.8 | 6.0 | 36.1 | 53.0 | 0.0547 |
| G2 | 758 | 10.3 | 7.0 | 27.8 | 54.9 | — |
| G3 | 533 | 12.0 | 7.1 | 30.8 | 50.1 | — |
| pN0 | 635 | 9.0 | 7.1 | 30.7 | 53.2 | 0.0571 |
| pN1 | 316 | 11.4 | 6.3 | 31.6 | 50.6 | — |
| pN2 | 112 | 14.3 | 11.6 | 30.4 | 43.8 | — |
| pN3 | 68 | 20.6 | 11.8 | 22.1 | 45.6 | — |
| pM0 | 177 | 5.1 | 6.2 | 37.3 | 51.4 | 0.0072 |
| pM1 | 111 | 15.3 | 9.9 | 37.8 | 36.9 | — |
| HER2 negative | 832 | 7.8 | 6.7 | 29.1 | 56.4 | 0.5888 |
| HER2-positive | 114 | 7.9 | 7.0 | 35.1 | 50.0 | — |
| ER-negative | 197 | 18.3 | 8.6 | 39.1 | 34.0 | <0.0001 |
| ER-positive | 706 | 4.7 | 6.2 | 27.2 | 61.9 | — |
| PR-negative | 374 | 14.2 | 9.1 | 34.5 | 42.2 | <0.0001 |
| PR-positive | 564 | 3.4 | 5.0 | 26.8 | 64.9 | — |
| Non-triple negative | 742 | 5.0 | 6.5 | 28.8 | 59.7 | <0.0001 |
| Triple negative | 133 | 22.6 | 9.0 | 33.1 | 35.3 | — |
| Endometrioid endometrial carcinoma | ||||||
| pT1 | 79 | 83.5 | 11.4 | 2.5 | 2.5 | 0.9265 |
| pT2 | 22 | 81.8 | 9.1 | 4.5 | 4.5 | — |
| pT3-pT4 | 23 | 82.6 | 13.0 | 4.3 | 0.0 | — |
| pN0 | 37 | 75.7 | 16.2 | 5.4 | 2.7 | 0.1875 |
| pN+ | 21 | 85.7 | 4.8 | 0.0 | 9.5 | — |
| Endometrioid carcinoma of the ovary | ||||||
| pT1 | 25 | 68.0 | 12.0 | 20.0 | 0.0 | 0.2860 |
| pT2 | 5 | 80.0 | 0.0 | 20.0 | 0.0 | — |
| pT3 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | — |
| pN0 | 22 | 68.2 | 13.6 | 18.2 | 0.0 | 0.0609 |
| pN1 | 9 | 100.0 | 0.0 | 0.0 | 0.0 | — |
| Serous carcinoma of the ovary | ||||||
| pT1 | 33 | 81.8 | 6.1 | 9.1 | 3.0 | 0.0650 |
| pT2 | 45 | 71.1 | 8.9 | 17.8 | 2.2 | — |
| pT3 | 266 | 80.8 | 13.5 | 5.3 | 0.4 | — |
| pN0 | 84 | 77.4 | 10.7 | 10.7 | 1.2 | 0.8677 |
| pN1 | 169 | 78.7 | 12.4 | 7.7 | 1.2 | — |
| Squamous cell carcinomas of different sites* | ||||||
| pT1 | 225 | 87.1 | 10.7 | 1.8 | 0.4 | 0.5931 |
| pT2 | 226 | 85.8 | 12.8 | 0.9 | 0.4 | — |
| pT3 | 117 | 82.9 | 16.2 | 0.9 | 0.0 | — |
| pT4 | 109 | 89.0 | 10.1 | 0.0 | 0.9 | — |
| pN0 | 266 | 86.8 | 11.7 | 0.8 | 0.8 | 0.2684 |
| pN+ | 260 | 84.2 | 14.6 | 1.2 | 0.0 | — |
| G1 | 26 | 84.6 | 11.5 | 3.8 | 0.0 | 0.2907 |
| G2 | 370 | 87.8 | 11.1 | 0.3 | 0.8 | — |
| G3 | 234 | 83.3 | 14.5 | 1.7 | 0.4 | — |
Oral, pharynx, larynx, esophagues, cervix, vagina, vulva, penis, anal, skin, and lung.
ER indicates estrogen receptor; G, grade; pM, pathologic status of distant metastasis; pN, pathologic lymph node status; PR, progesterone receptor; pT, pathologic tumor stage.
TRPS1 Versus GATA3 Expression
A comparative description of TRPS1 versus GATA3 expression is given in Figure 4 and in Supplemental Table 2 (Supplemental Digital Content 5, http://links.lww.com/PAS/B789) for 11,891 tumors with data on both markers. The data show that positivity for both markers is frequent (47.4% to 100%) in breast cancer, and also occurs in a fraction of salivary gland tumors (up to 30.0%) while it is exceedingly rare in all other cancer entities. Of 1159 tumors positive for TRPS1 and GATA3, 97.5% were derived from either the breast or salivary glands. Considering that TRPS1 should stain positive in breast and salivary gland tumors while GATA3 should label breast, urothelial, and salivary gland tumors the numbers of positive cases outside of these tumor categories were assessed. TRPS1 positivity occurred in 4.7% of 9835 tumors from 55 of 114 “additional categories.” GATA3 staining was seen in 2.3% of 8980 tumors from 50 of 109 “additional categories.” Positivity for both TRPS1 and GATA3 occurred in 47.4% to 100% of breast cancers, and up to 30% of salivary gland tumors, but in only 29 (0.3%) of 9835 tumors from 134 other cancer entities.
FIGURE 4.
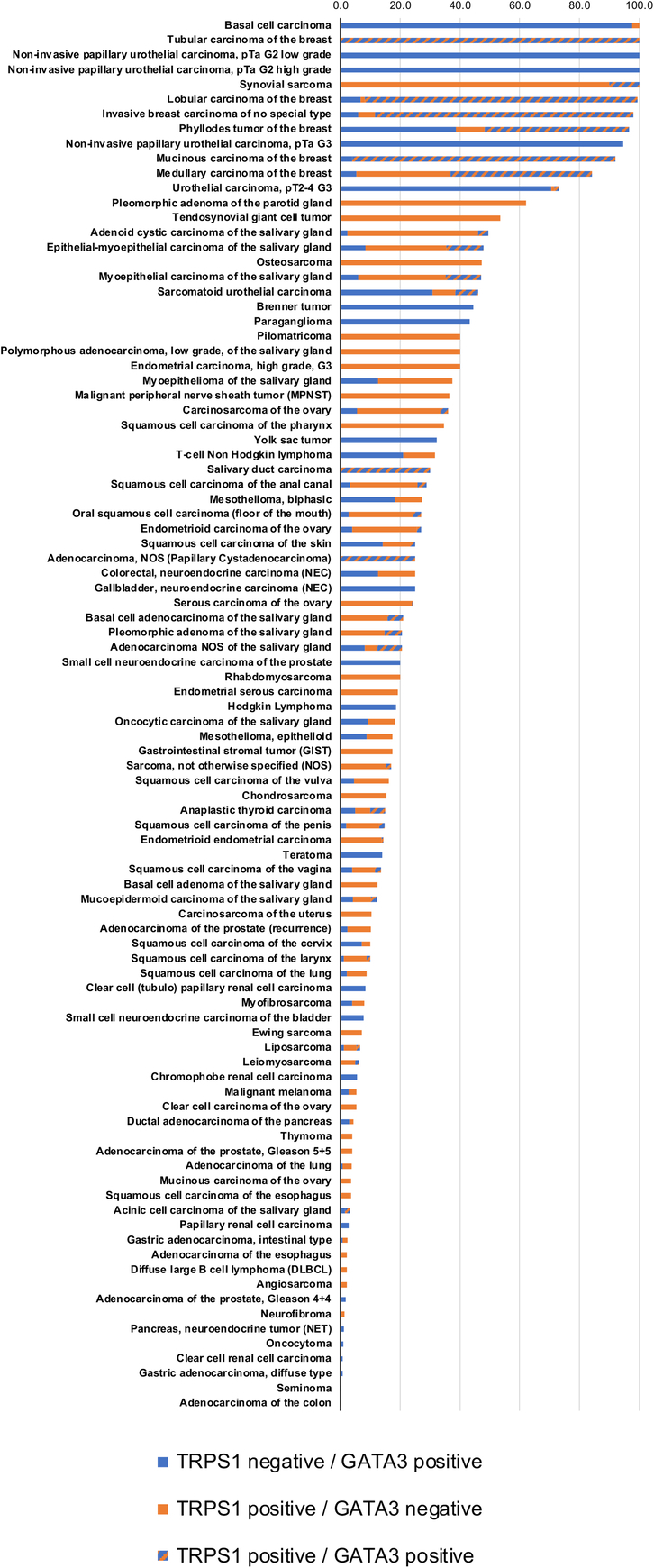
TRPS1 and GATA3 immunostaining in tumors.
Comparison of Sensitivity and Specificity by Using Different TRPS1 Assays
Staining results for the antibodies MSVA-512R, EPR16171, and PA-84874 on 2214 cancers and the respective sensitivities and specificities for the detection of breast cancers and triple-negative breast cancers are given in Table 3. Examples of tumors stained with the 3 antibodies are given in Supplemental Figure 3 (Supplemental Digital Content 6, http://links.lww.com/PAS/B790). At the selected conditions, sensitivity for the detection of breast cancer/triple-negative breast cancer was higher for EPR16171 (85%/86%) and PA5-84874 (85%/86%) as for MSVA-512R (83%/76%). Specificity for the distinction between breast cancers and non-breast adenocarcinomas/non-breast cancers was highest for MSVA-512R (92%/89%), followed by PA5-84874 (67%/47%) and EPR16171 (58%/44%).
TABLE 3.
Sensitivity and Specificity of Anti-TRPS1 Antibodies
| TRPS1 MSVA-512R | TRPS1 EPR16171 | TRPS1 PA5-84874 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor entity | Category for sensitivity/specificity calculation | n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | n | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | |
| Tumors of the skin | Basal cell carcinoma | Non-breast cancer | 14 | 100.0 | 0.0 | 0.0 | 0.0 | 26 | 38.5 | 57.7 | 3.8 | 0.0 | 25 | 52.0 | 36.0 | 12.0 | 0.0 |
| Squamous cell carcinoma of the skin | Non-breast cancer | 30 | 83.3 | 16.7 | 0.0 | 0.0 | 37 | 24.3 | 45.9 | 27.0 | 2.7 | 36 | 25.0 | 38.9 | 33.3 | 2.8 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | Non-breast cancer | 50 | 86.0 | 14.0 | 0.0 | 0.0 | 53 | 22.6 | 34.0 | 34.0 | 9.4 | 55 | 16.4 | 47.3 | 34.5 | 1.8 |
| Squamous cell carcinoma of the pharynx | Non-breast cancer | 49 | 65.3 | 30.6 | 4.1 | 0.0 | 55 | 25.5 | 32.7 | 32.7 | 9.1 | 57 | 24.6 | 33.3 | 35.1 | 7.0 | |
| Oral squamous cell carcinoma (floor of the mouth) | Non-breast cancer | 60 | 65.0 | 33.3 | 0.0 | 1.7 | 70 | 14.3 | 31.4 | 47.1 | 7.1 | 73 | 12.3 | 35.6 | 49.3 | 2.7 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | Non-breast cancer | 28 | 89.3 | 7.1 | 3.6 | 0.0 | 28 | 46.4 | 35.7 | 7.1 | 10.7 | 28 | 53.6 | 32.1 | 3.6 | 10.7 |
| Squamous cell carcinoma of the vulva | Non-breast cancer | 74 | 89.2 | 10.8 | 0.0 | 0.0 | 74 | 20.3 | 37.8 | 31.1 | 10.8 | 75 | 16.0 | 44.0 | 25.3 | 14.7 | |
| Squamous cell carcinoma of the cervix | Non-breast cancer | 77 | 98.7 | 0.0 | 0.0 | 1.3 | 78 | 66.7 | 16.7 | 14.1 | 2.6 | 76 | 56.6 | 27.6 | 13.2 | 2.6 | |
| Carcinosarcoma of the uterus | Non-breast adenocarcinoma | 19 | 68.4 | 26.3 | 5.3 | 0.0 | 15 | 33.3 | 26.7 | 40.0 | 0.0 | 14 | 28.6 | 28.6 | 14.3 | 28.6 | |
| Endometrioid carcinoma of the ovary | Non-breast adenocarcinoma | 32 | 65.6 | 12.5 | 18.8 | 3.1 | 21 | 28.6 | 28.6 | 19.0 | 23.8 | 21 | 23.8 | 23.8 | 23.8 | 28.6 | |
| Serous carcinoma of the ovary | Non-breast adenocarcinoma | 79 | 58.2 | 25.3 | 13.9 | 2.5 | 71 | 25.4 | 26.8 | 21.1 | 26.8 | 70 | 18.6 | 30.0 | 15.7 | 35.7 | |
| Mucinous carcinoma of the ovary | Non-breast adenocarcinoma | 24 | 95.8 | 4.2 | 0.0 | 0.0 | 16 | 75.0 | 0.0 | 18.8 | 6.3 | 15 | 80.0 | 0.0 | 20.0 | 0.0 | |
| Clear cell carcinoma of the ovary | Non-breast adenocarcinoma | 21 | 85.7 | 4.8 | 9.5 | 0.0 | 16 | 25.0 | 31.3 | 31.3 | 12.5 | 16 | 43.8 | 25.0 | 18.8 | 12.5 | |
| Carcinosarcoma of the ovary | Non-breast adenocarcinoma | 17 | 76.5 | 17.6 | 5.9 | 0.0 | 12 | 41.7 | 25.0 | 16.7 | 16.7 | 10 | 30.0 | 40.0 | 10.0 | 20.0 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | Breast cancer | 462 | 19.7 | 7.8 | 26.6 | 45.9 | 422 | 5.5 | 5.5 | 9.0 | 80.1 | 430 | 6.0 | 8.4 | 5.3 | 80.2 |
| Lobular carcinoma of the breast | Breast cancer | 142 | 10.6 | 6.3 | 31.0 | 52.1 | 118 | 2.5 | 4.2 | 9.3 | 83.9 | 119 | 3.4 | 4.2 | 4.2 | 88.2 | |
| Medullary carcinoma of the breast | Breast cancer | 8 | 0.0 | 25.0 | 25.0 | 50.0 | 7 | 0.0 | 0.0 | 14.3 | 85.7 | 7 | 0.0 | 0.0 | 14.3 | 85.7 | |
| Tubular carcinoma of the breast | Breast cancer | 2 | 0.0 | 0.0 | 50.0 | 50.0 | 2 | 0.0 | 0.0 | 0.0 | 100.0 | 2 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Mucinous carcinoma of the breast | Breast cancer | 6 | 0.0 | 0.0 | 33.3 | 66.7 | 6 | 0.0 | 0.0 | 0.0 | 100.0 | 7 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Hormone receptor-positive breast cancer * | — | 425 | 16.9 | 7.1 | 28.0 | 48.0 | 402 | 3.0 | 3.5 | 8.2 | 85.3 | 410 | 3.7 | 5.9 | 4.9 | 85.6 | |
| Triple-negative breast cancer * | Triple-negative breast cancer | 58 | 32.8 | 17.2 | 22.4 | 27.6 | 58 | 13.8 | 15.5 | 10.3 | 60.3 | 59 | 13.6 | 20.3 | 5.1 | 61.0 | |
| Tumors of the digestive system | Adenocarcinoma of the colon | Non-breast adenocarcinoma | 79 | 100.0 | 0.0 | 0.0 | 0.0 | 69 | 94.2 | 5.8 | 0.0 | 0.0 | 70 | 94.3 | 5.7 | 0.0 | 0.0 |
| Gastric adenocarcinoma, diffuse type | Non-breast adenocarcinoma | 67 | 100.0 | 0.0 | 0.0 | 0.0 | 40 | 75.0 | 15.0 | 10.0 | 0.0 | 43 | 86.0 | 4.7 | 2.3 | 7.0 | |
| Gastric adenocarcinoma, intestinal type | Non-breast adenocarcinoma | 69 | 95.7 | 2.9 | 1.4 | 0.0 | 42 | 54.8 | 33.3 | 2.4 | 9.5 | 42 | 52.4 | 35.7 | 4.8 | 7.1 | |
| Adenocarcinoma of the esophagus | Non-breast adenocarcinoma | 70 | 98.6 | 0.0 | 0.0 | 1.4 | 43 | 58.1 | 23.3 | 16.3 | 2.3 | 45 | 57.8 | 24.4 | 8.9 | 8.9 | |
| Squamous cell carcinoma of the esophagus | Non-breast cancer | 61 | 91.8 | 8.2 | 0.0 | 0.0 | 39 | 15.4 | 46.2 | 23.1 | 15.4 | 35 | 25.7 | 40.0 | 22.9 | 11.4 | |
| Squamous cell carcinoma of the anal canal | Non-breast cancer | 30 | 80.0 | 20.0 | 0.0 | 0.0 | 37 | 5.4 | 54.1 | 35.1 | 5.4 | 38 | 5.3 | 34.2 | 44.7 | 15.8 | |
| Ductal adenocarcinoma of the pancreas | Non-breast adenocarcinoma | 80 | 98.8 | 1.3 | 0.0 | 0.0 | 64 | 79.7 | 14.1 | 6.3 | 0.0 | 68 | 82.4 | 14.7 | 2.9 | 0.0 | |
| Pancreatic/ampullary adenocarcinoma | Non-breast adenocarcinoma | 28 | 100.0 | 0.0 | 0.0 | 0.0 | 23 | 78.3 | 17.4 | 4.3 | 0.0 | 23 | 87.0 | 13.0 | 0.0 | 0.0 | |
| Acinar cell carcinoma of the pancreas | Non-breast adenocarcinoma | 5 | 100.0 | 0.0 | 0.0 | 0.0 | 4 | 75.0 | 25.0 | 0.0 | 0.0 | 4 | 75.0 | 0.0 | 25.0 | 0.0 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3+3 | Non-breast adenocarcinoma | 78 | 100.0 | 0.0 | 0.0 | 0.0 | 64 | 84.4 | 10.9 | 4.7 | 0.0 | 65 | 96.9 | 3.1 | 0.0 | 0.0 |
| Adenocarcinoma of the prostate, Gleason 4+4 | Non-breast adenocarcinoma | 69 | 100.0 | 0.0 | 0.0 | 0.0 | 55 | 45.5 | 27.3 | 27.3 | 0.0 | 57 | 71.9 | 19.3 | 8.8 | 0.0 | |
| Adenocarcinoma of the prostate, Gleason 5+5 | Non-breast adenocarcinoma | 79 | 96.2 | 3.8 | 0.0 | 0.0 | 67 | 20.9 | 52.2 | 17.9 | 9.0 | 62 | 62.9 | 22.6 | 8.1 | 6.5 | |
| Squamous cell carcinoma of the penis | Non-breast cancer | 55 | 87.3 | 9.1 | 3.6 | 0.0 | 72 | 30.6 | 23.6 | 30.6 | 15.3 | 71 | 15.5 | 39.4 | 36.6 | 8.5 | |
| Breast cancers vs non-breast adenocarcinomas | Sensitivity | 0.83 | Sensitivity | 0.95 | Sensitivity | 0.95 | |||||||||||
| Specificity | 0.92 | Specificity | 0.58 | Specificity | 0.67 | ||||||||||||
| Breast cancers vs non-breast adenocarcinomas/non-breast cancers | Sensitivity | 0.83 | Sensitivity | 0.95 | Sensitivity | 0.95 | |||||||||||
| Specificity | 0.89 | Specificity | 0.44 | Specificity | 0.47 | ||||||||||||
| Triple-negative breast cancers vs non-breast adenocarcinomas | Sensitivity | 0.76 | Sensitivity | 0.86 | Sensitivity | 0.86 | |||||||||||
| Specificity | 0.92 | Specificity | 0.58 | Specificity | 0.67 | ||||||||||||
| Triple-negative breast cancers vs non-breast adenocarcinomas/non-breast cancers | Sensitivity | 0.76 | Sensitivity | 0.86 | Sensitivity | 0.86 | |||||||||||
| Specificity | 0.89 | Specificity | 0.44 | Specificity | 0.47 | ||||||||||||
*Included in the other subtypes of breast cancers.
DISCUSSION
Considering conflicting previous results on TRPS1 IHC in cancer and the large scale of our study, emphasis was placed on a thorough validation of our assay. The International Working Group for Antibody Validation has proposed that an acceptable antibody validation for IHC on formalin-fixed tissues must include either a comparison of the findings obtained by 2 different independent antibodies or a comparison with expression data obtained by another independent method.33 Both methods were applied in this project. The rather ubiquitous expression of TRPS1 RNA in normal tissues which was suggested by compiled RNA data (https://www.proteinatlas.org) from 3 public databases34–36 could largely be confirmed if the MSVA-512R antibody was highly concentrated (1:20). Given the expression in virtually all organs and cell type specificity of TRPS1 expression, a comparison with a method based on disaggregated tissue is suboptimal for TRPS1 antibody validation, however. Pivotal evidence for the validity of our assay thus comes from a confirmation of all TRPS1-positive cell types seen by MSVA-512R by the independent antibodies EPR16171 and PA5-84874. The additional cytoplasmic staining of basal cells in squamous epithelia that were only seen by EPR16171 and the additional cytoplasmic staining of several cell types by PA5-84874 were considered antibody-specific cross-reactivities. It is of note that the use of a very broad selection of normal tissue categories (n = 76) for antibody validation maximizes the probability of detecting cross-reactivities because virtually all proteins occurring in normal human adult cells are included in the validation process.
Our initial tumor screening revealed that TRPS1 expression levels were highest in breast cancer but other tumors were also TRPS1-positive. For this study, our staining protocol was thus adjusted to the highest possible dilution that still resulted in a strong TRPS1 staining of a large fraction of breast cancers but as low as possible staining of other tumors and normal tissues. Under these conditions, our analysis of 19,201 tumors from 152 different tumor categories demonstrated a strong predominance of TRPS1 staining in breast cancers. This was expected based on RNA expression data from different tumor types which are described in The Cancer Genome Atlas database (https://www.cancer.gov/tcga) and data from previous IHC studies.10,15 Of tumors, 94% with strong and 74% with moderate TRPS1 positivity were breast neoplasms in this study. Our TRPS1 positivity rate of 75% across all breast cancer subtypes is in the middle range of the 41% to 92% TRPS1 positivity rates that have previously been described in studies employing IHC on cohorts of 50 to 1061 breast cancers.7,26,37 The comparison of our data with two different TRPS1 assays suggests that a higher TRPS1 positivity rate in breast cancer goes along with a marked decrease in specificity for the detection of both breast cancer and triple-negative breast cancer.
Several studies have suggested that TRPS1 IHC could complement GATA3 in its role as a breast cancer marker in case of tumor masses of uncertain derivation.10,11,26,27 The significantly higher positivity rate for TRPS1 (77%) than for GATA3 (55%) in our triple-negative breast cancers is consistent with data from several earlier reports and underscores the diagnostic utility of TRPS1 in these tumors. Other studies have reported 79% to 91% TRPS1 positivity in triple-negative breast cancer.10,26,38,39 These figures are again higher than the 43% to 56% positivity rates described for GATA3 in these tumors.10,26,27
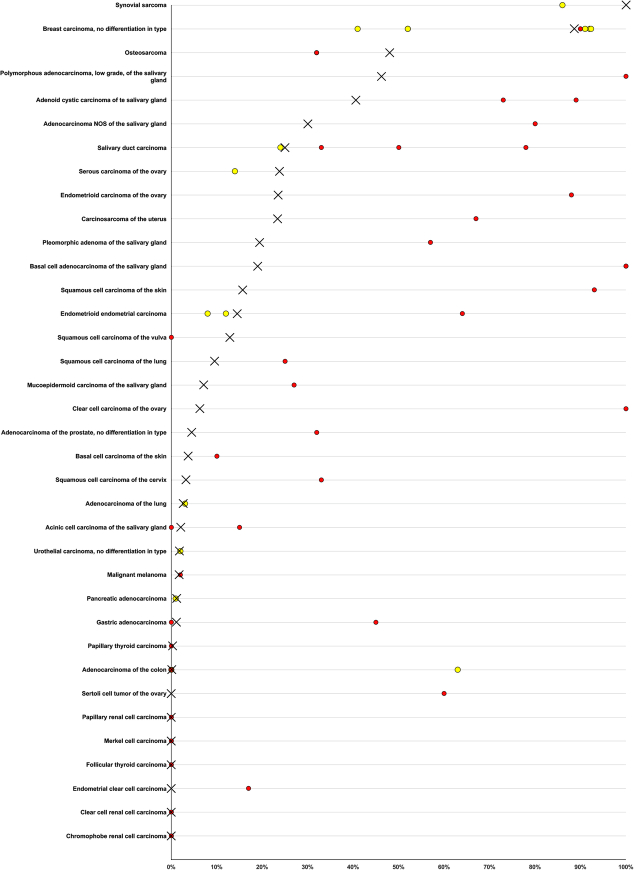
A comprehensive overview of TRPS1 expression across human tumors represents the key result of our study. TRPS1 immunostaining in various further tumor entities, often at a lower level than in breast cancer, parallels the expression of TRPS1 in a broad range of different normal cell types. Proteins expressed in normal cell types are mostly retained after malignant transformation.40 The highest rates of TRPS1 positivity among non-breast tumors were seen in subtypes of soft tissue tumors, salivary gland tumors, squamous cell carcinomas, and gynecological cancers. This is in line with individual reports describing TRPS1 positivity in these entities (Fig. 5). Several authors recently described frequent TRPS1 positivity in salivary gland tumors. In line with our data, these authors described the highest TRPS1 positivity rates in tumors with ductal differentiation, such as pleomorphic adenomas, adenoid cystic carcinomas, or epithelial-myoepithelial carcinomas while TRPS1 expression was low in acinus cell carcinomas or mucoepidermoid carcinomas.41,42 Based on a TRPS1 positivity in 13 of 14 squamous cell carcinomas but in only 1 of 10 basal cell carcinomas of the skin, Liu et al19 suggested that the distinction of these tumors may represent another application for TRPS1 IHC. Although our assay was designed to have low sensitivity in non-breast cancers, the 16% positivity in our squamous cell carcinomas as compared with 4% in basal cell carcinomas of the skin is consistent with this notion.
FIGURE 5.
Comparison with previous TRPS1 literature. An “X” indicates the fraction of TRPS1-positive tumors in the present study, and dots indicate the reported frequencies from the literature for comparison; red dots mark studies with <100 analyzed tumors and yellow dots mark studies with ≥100 analyzed tumors.
TRPS1 expression in non-breast gynecological tumors—as seen in our study—has also been described by other authors.10,15 Skvarca et al39,43 found TRPS1 staining in 8% of 184 endometrioid G3 tumors of the endometrium and identified a significant association between high TRPS1 expression and unfavorable disease outcome. Rammal et al39,43 reported TRPS1 positivity from 71% of 69 endometrial endometrioid adenocarcinomas, 1 out of 5 endometrioid carcinomas, and 17% of 250 ovarian tumors. Chen and colleagues have shown TRPS1 staining in 20% of 152 ovarian and 12% of 152 endometrial carcinomas, and Ai and colleagues found 8% to 14% TRPS1 staining in 251 ovarian carcinomas of different subtypes. Our TRPS1 positivity in 100% of our synovial sarcomas is consistent with a recent report by Cloutier et al20 showing 86% TRPS1-positive cases among 165 synovial sarcomas. Based on published RNA expression data and their ChIP-seq (chromatin immunoprecipitation DNA sequencing) findings these authors suggested that TRPS1 overexpression may be mediated by the enrichment of several transcriptionally activating histone modifications in these tumors. TRPS1 positivity in 29% of our 14 malignant peripheral nerve sheath tumors while there was a complete lack of TRPS1 staining in 121 Schwannomas could potentially indicate a role of TRPS1 in the malignant transformation of nerve sheath tumors and suggest that detectable TRPS1 staining could represent a feature of malignancy in these tumors. Earlier studies also reported a high sensitivity and specificity of TRPS1 for primary extramammary Paget disease,44 and differential staining of TRPS1 in various types of malignant and benign cutaneous sweat glands.45
Several studies have proposed oncogenic46,47 or tumor suppressive5,6 roles for TRPS1. The availability of clinic-pathologic data for several tumor cohorts enabled us to interrogate the potential clinical significance of aberrant TRPS1 expression in vivo. That a reduced TRPS1 expression could be observed in invasive breast carcinomas with no special type with unfavorable histopathological tumor features is in line with the more aggressive behavior of TRPS1-negative cancers. Other studies evaluating TRPS1 in 152 and 341 breast carcinomas had earlier reported a significant link between low TRPS1 expression and poor prognosis15,25 or unfavorable tumor features.25 Two further studies on 152 and 180 cancers have failed to find evidence for a clinical role of TRPS1 expression in breast cancer, however.15,48 The link between low rates of TRPS1 expression and unfavorable tumor features in cancers derived from TRPS1 expressing normal tissues is consistent with either a tumor suppressive role of TRPS1 or a progressive loss of TRPS1 expression during cellular dedifferentiation that is unrelated to cell functions required for tumor progression. It is well known that neoplastic cells continuously lose the expression of nonessential proteins during cancer progression.49,50 In line with a possible oncogenic role of TRPS1, a relationship between high TRPS1 expression and unfavorable histologic tumor features was recorded for prostate cancer in this study. This is consistent with recent data by Bachert et al12 describing high TRPS1 expression in metastatic prostate cancers.
Our data confirm the recently proposed utility of the combination TRPS1 and GATA3 IHC for the assessment of tumor masses of unknown origin10,26,27 and for the distinction of breast cancer from other specific tumor entities.51–53 A major advantage of this combination lies in the virtual absence of TRPS1 positivity in urothelial carcinomas, the tumor entity with the second highest GATA3 positivity rate after breast cancer. In our analysis, a combined GATA3 and TRPS1 positivity was almost exclusively seen in neoplasms of the breast and the salivary glands. Only 2.5% of 1159 GATA3/TRPS1 dual-positive tumors were of non-breast and non-salivary gland origin in this study. That only 1.4% of 842 urothelial neoplasms were TRPS1-positive underscores the utility of TRPS1 for the distinction of breast cancer from urothelial carcinoma. That “TRPS1 alone” positivity was more common than “GATA3 alone” positivity again reflects the common low-level expression of TRPS1 in many different normal cell types.
CONCLUSION
The results of our study demonstrate that TRPS1 is expressed in a broad range of normal cell types with the highest expression levels in normal breast epithelial cells. Because the average TRPS1 expression in cancers is highest in breast neoplasms, immunostaining protocols can be defined that result in high sensitivity and considerable specificity of TRPS1 staining for breast (or salivary gland) cancer although various other tumor entities can show—a usually less intense—TRPS1 positivity.
Supplementary Material
Footnotes
M.L., S.S., N.L., R.S., M.K., and G.S.: conception, design, data collection, data analysis, and manuscript writing. M.L., D.H., S.D.R., Cv.B., S.K., V.R., F.V., F.L., V.B., C.F., N.G., S.W., N.C.B., A.M., R.U., T.K., A.H., E.B., A.H.M., P.L., D.D., S.M., F.J., T.S.C., and C.B.: participated in pathology data analysis, data interpretation, and collection of samples. R.S., M.K., and C.H.M.: data analysis. S.S., R.S., and G.S.: study supervision.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website www.ajsp.com.
Conflicts of Interest and Source of Funding: The rabbit recombinant TRPS1 antibody, clone MSVA-512R was provided from MS Validated Antibodies GmbH (owned by a family member of G.S.). The company MS Validated Antibodies is owned by a relative of G.S. For the remaining authors none were declared.
Contributor Information
Maximilian Lennartz, Email: m.lennartz@uke.de.
Neele Löhr, Email: neeleloehr@gmail.com.
Doris Höflmayer, Email: d.hoeflmayer@uke.de.
Sebastian Dwertmann Rico, Email: s.dwertmann-rico@uke.de.
Clara von Bargen, Email: c.von-bargen@uke.de.
Simon Kind, Email: s.kind@uke.de.
Viktor Reiswich, Email: v.reiswich@uke.de.
Florian Viehweger, Email: f.viehweger@uke.de.
Florian Lutz, Email: f.lutz@uke.de.
Veit Bertram, Email: v.bertram@uke.de.
Christoph Fraune, Email: c.fraune@uke.de.
Natalia Gorbokon, Email: n.gorbokon@uke.de.
Sören Weidemann, Email: s.weidemann@uke.de.
Niclas C. Blessin, Email: n.blessin@uke.de.
Claudia Hube-Magg, Email: c.hube@uke.de.
Anne Menz, Email: a.menz@uke.de.
Ria Schlichter, Email: r.uhlig@uke.de.
Till Krech, Email: t.krech@uke.de.
Andrea Hinsch, Email: a.hinsch@uke.de.
Eike Burandt, Email: e.burandt@uke.de.
Guido Sauter, Email: g.sauter@uke.de.
Ronald Simon, Email: r.simon@uke.de.
Martina Kluth, Email: m.kluth@uke.de.
Andreas H. Marx, Email: Andreas.Marx@klinikum-fuerth.de.
Patrick Lebok, Email: p.lebok@uke.de.
David Dum, Email: d.dum@uke.de.
Sarah Minner, Email: s.minner@uke.de.
Frank Jacobsen, Email: f.jacobsen@uke.de.
Till S. Clauditz, Email: t.clauditz@uke.de.
Christian Bernreuther, Email: c.bernreuther@uke.de.
Stefan Steurer, Email: s.steurer@uke.de.
REFERENCES
- 1.Yang L, Gong X, Wang J, et al. Functional mechanisms of TRPS1 in disease progression and its potential role in personalized medicine. Pathol Res Pract. 2022;237:154022. [DOI] [PubMed] [Google Scholar]
- 2.Kanno S, Gui T, Itoh S, et al. Aberrant expression of the P2 promoter-specific transcript Runx1 in epiphyseal cartilage of Trps1-null mice. Exp Mol Pathol. 2011;90:143–148. [DOI] [PubMed] [Google Scholar]
- 3.Fantauzzo KA, Kurban M, Levy B, et al. Trps1 and its target gene Sox9 regulate epithelial proliferation in the developing hair follicle and are associated with hypertrichosis. PLoS Genet. 2012;8:e1003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ergoren MC, Akcan N, Manara E, et al. Characterization of a novel frameshift mutation within the TRPS1 gene causing trichorhinophalangeal syndrome type 1 in a kindred cypriot family. Appl Immunohistochem Mol Morphol. 2022;30:635–639. [DOI] [PubMed] [Google Scholar]
- 5.Huang JZ, Chen M, Zeng M, et al. Down-regulation of TRPS1 stimulates epithelial-mesenchymal transition and metastasis through repression of FOXA1. J Pathol. 2016;239:186–196. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Su P, Jiao M, et al. TRPS1 suppresses breast cancer epithelial-mesenchymal transition program as a negative regulator of SUZ12. Transl Oncol. 2018;11:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Su P, Jia M, et al. TRPS1 expression promotes angiogenesis and affects VEGFA expression in breast cancer. Exp Biol Med (Maywood). 2014;239:423–429. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Zhang H, Liu L, et al. TRPS1 confers multidrug resistance of breast cancer cells by regulating BCRP expression. Front Oncol. 2020;10:934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia M, Hu J, Li W, et al. Trps1 is associated with the multidrug resistance of osteosarcoma by regulating MDR1 gene expression. FEBS Lett. 2014;588:801–810. [DOI] [PubMed] [Google Scholar]
- 10.Ai D, Yao J, Yang F, et al. TRPS1: a highly sensitive and specific marker for breast carcinoma, especially for triple-negative breast cancer. Mod Pathol. 2021;34:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelwahed M, Yurtsever N, Savant D, et al. Utility of TRPS-1 immunohistochemistry in diagnosis of metastatic breast carcinoma in cytology specimens. J Am Soc Cytopathol. 2022;11:345–351. [DOI] [PubMed] [Google Scholar]
- 12.Bachert SE, Di J, Zhang S, et al. TRPS1 expression in primary and metastatic prostatic adenocarcinoma, muscle invasive bladder urothelial carcinoma, and breast carcinoma: Is TRPS1 truly specific and sensitive for a breast primary? Hum Pathol. 2023;143:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakkar A Sriram S Mahendru R, et al. Transcriptional Repressor GATA Binding 1 (TRPS1) Immunoexpression in Salivary Gland Neoplasms. United States and Canadian Academy of Pathology (USCAP); 2023 March 11-16 Laboratory Investigation, USCAP 112th Annual Meeting Abstracts, Volume 103, Issue 3, Supplement, 2023.
- 14.Tjendra Y, Kerr DA, Gomez-Fernandez C, et al. TRPS1 immunohistochemical expression in salivary gland tumors: a pilot study. Am J Clin Pathol. 2023;160:633–639. [DOI] [PubMed] [Google Scholar]
- 15.Chen JQ, Litton J, Xiao L, et al. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer. 2010;1:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu SP, Yan RH, Gao J, et al. Significance of TRPS1 in the development and clinicopathologic of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2020;24:9325–9332. [DOI] [PubMed] [Google Scholar]
- 17.Hong J, Sun J, Huang T. Increased expression of TRPS1 affects tumor progression and correlates with patients’ prognosis of colon cancer. Biomed Res Int. 2013;2013:454085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Huang Z, Huang M, et al. Clinical significance and biological function of transcriptional repressor GATA binding 1 in gastric cancer: a study based on data mining, RT-qPCR, immunochemistry, and vitro experiment. Cell Cycle. 2020;19:2866–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YA Aung PP Ding Q, et al. TRPS1 may be an effective discriminatory marker for basal cell carcinoma and squamous cell carcinoma: an immunohistochemical analysis of TRPS1 in non-melanocytic cutaneous neoplasms. United States and Canadian Academy of Pathology (USCAP); Laboratory Investigation, USCAP 112th Annual Meeting Abstracts, Volume 103, Issue 3, Supplement, 2023.
- 20.Cloutier JM, Ingram DR, Wani K, et al. Frequent TRPS1 expression in synovial sarcoma is associated with SS18-SSX fusion oncoprotein activity. Hum Pathol. 2022;130:88–94. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Jia M, Wu X, et al. Overexpression of Trps1 contributes to tumor angiogenesis and poor prognosis of human osteosarcoma. Diagn Pathol. 2015;10:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Liao Y, Tang M, et al. Trps1 is associated with the multidrug resistance of lung cancer cell by regulating MGMT gene expression. Cancer Med. 2018;7:1921–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang GT, Jhamai M, van Weerden WM, et al. The TRPS1 transcription factor: androgenic regulation in prostate cancer and high expression in breast cancer. Endocr Relat Cancer. 2004;11:815–822. [DOI] [PubMed] [Google Scholar]
- 24.Chang GT, Gamble SC, Jhamai M, et al. Proteomic analysis of proteins regulated by TRPS1 transcription factor in DU145 prostate cancer cells. Biochim Biophys Acta. 2007;1774:575–582. [DOI] [PubMed] [Google Scholar]
- 25.Chen JQ, Bao Y, Lee J, et al. Prognostic value of the trichorhinophalangeal syndrome-1 (TRPS-1), a GATA family transcription factor, in early-stage breast cancer. Ann Oncol. 2013;24:2534–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du T, Pan L, Zheng C, et al. Matrix Gla protein (MGP), GATA3, and TRPS1: a novel diagnostic panel to determine breast origin. Breast Cancer Res. 2022;24:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon EC, Wang G, Parkinson B, et al. TRPS1, GATA3, and SOX10 expression in triple-negative breast carcinoma. Hum Pathol. 2022;125:97–107. [DOI] [PubMed] [Google Scholar]
- 28.Reiswich V, Schmidt CE, Lennartz M, et al. GATA3 expression in human tumors: a tissue microarray study on 16,557 tumors. Pathobiology. 2023:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoflmayer D, Steinhoff A, Hube-Magg C, et al. Expression of CCCTC-binding factor (CTCF) is linked to poor prognosis in prostate cancer. Mol Oncol. 2020;14:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buscheck F, Sulimankhil M, Melling N, et al. Loss of cytoplasmic survivin expression is an independent predictor of poor prognosis in radically operated prostate cancer patients. Cancer Med. 2020;9:1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraune C, Yehorov S, Luebke AM, et al. Upregulation of PTTG1 is associated with poor prognosis in prostate cancer. Pathol Int. 2020;70:441–451. [DOI] [PubMed] [Google Scholar]
- 32.Bonk S, Kluth M, Jansen K, et al. Reduced KLK2 expression is a strong and independent predictor of poor prognosis in ERG-negative prostate cancer. Prostate. 2020;80:1097–1107. [DOI] [PubMed] [Google Scholar]
- 33.Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium GT. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lizio M, Abugessaisa I, Noguchi S, et al. Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47(D1):D752–D758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 37.Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almasi S, Kuthi L, Sejben A, et al. TRPS1 expression in cytokeratin 5 expressing triple-negative breast cancers, its value as a marker of breast origin. Virchows Arch. 2023;482:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rammal R, Goel K, Elishaev E, et al. Utility of TRPS1 immunohistochemistry in confirming breast carcinoma: Emphasis on staining in triple-negative breast cancers and gynecologic tumors. Am J Clin Pathol. 2023;160:425–434. [DOI] [PubMed] [Google Scholar]
- 40.Danielsson F, Skogs M, Huss M, et al. Majority of differentially expressed genes are down-regulated during malignant transformation in a four-stage model. Proc Natl Acad Sci USA. 2013;110:6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachert SE Di J McDonald R, et al. Intermediate and high expression of TRPS1 is frequently observed in many primary salivary gland neoplasms. United States and Canadian Academy of Pathology (USCAP); 2023 March 11-16; New Orleans Convention Center and Hilton New Orleans Riverside: USCAP; Abstract 105; 2023.
- 42.Tjendra Y Gomez-Fernandez C Kryvenko ON, et al. TRPS1 immunohistochemical expression in salivary gland tumors: a pilot study. United States and Canadian Academy of Pathology (USCAP); 2023 March 11-16; Laboratory Investigation, USCAP 112th Annual Meeting Abstracts, Volume 103, Issue 3, Supplement, 2023.
- 43.Skvarca L Kousar A Rammal R, et al. High TRPS1 expression predicts distant recurrence of early-stage endometrial endometroid adenocarcinoma (EMACA). United States and Canadian Academy of Pathology (USCAP); 2023 March 11-16; Laboratory Investigation, USCAP 112th Annual Meeting Abstracts, Volume 103, Issue 3, Supplement, 2023.
- 44.Cook EE, Harrison BT, Hirsch MS. TRPS1 expression is sensitive and specific for primary extramammary Paget disease. Histopathology. 2023;83:104–108. [DOI] [PubMed] [Google Scholar]
- 45.Zengin HB, Bui CM, Rybski K, et al. TRPS1 is differentially expressed in a variety of malignant and benign cutaneous sweat gland neoplasms. Dermatopathology (Basel). 2023;10:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serandour AA, Mohammed H, Miremadi A, et al. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene. 2018;37:5281–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Liu X, Huang Y, et al. TRPS1 drives heterochromatic origin refiring and cancer genome evolution. Cell Rep. 2021;34:108814. [DOI] [PubMed] [Google Scholar]
- 48.Su P, Hu J, Zhang H, et al. Association of TRPS1 gene with different EMT markers in ERalpha-positive and ERalpha-negative breast cancer. Diagn Pathol. 2014;9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. 2021;12:636568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 51.Baban F, Koepplin JW, Ahmad M, et al. TRPS1 outperforms GATA3 in pleural effusions with metastatic breast carcinoma versus mesothelioma. Diagn Cytopathol. 2023;51:488–492. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Peng Y, Sun H, et al. TRPS1 and GATA3 expression in invasive breast carcinoma with apocrine differentiation. Arch Pathol Lab Med. 2023;148:200–205. [DOI] [PubMed] [Google Scholar]
- 53.Parkinson B, Chen W, Shen T, et al. TRPS1 expression in breast carcinomas: focusing on metaplastic breast carcinomas. Am J Surg Pathol. 2022;46:415–423. [DOI] [PubMed] [Google Scholar]