Abstract
Vpr, encoded by the human immunodeficiency virus type 1 genome, contains 96 amino acids and is a multifunctional protein with features which include cell cycle arrest at G2, nuclear localization, participation in transport of the preintegration complex, cation channel activity, oligomerization, and interaction with cellular proteins, in addition to its incorporation into the virus particles. Recently, structural studies based on nuclear magnetic resonance and circular dichroism spectroscopy showed that Vpr contains a helix (HI)-turn-helix (HII) core at the amino terminus and an amphipathic helix (HIII) in the middle region. Though the importance of helical domains HI and HIII has been defined with respect to Vpr functions, the role of helical domain HII is not known. To address this issue, we constructed a series of mutants in which the HII domain was altered by deletion, insertion, and/or substitution mutagenesis. To enable the detection of Vpr, the sequence corresponding to the Flag epitope (DYKDDDDK) was added, in frame, to the Vpr coding sequences. Mutants, expressed through the in vitro transcription/translation system and in cells, showed an altered migration corresponding to deletions in Vpr. Substitution mutational analysis of residues in HII showed reduced stability for VprW38S-FL, VprL42G-FL, and VprH45W-FL. An assay involving cotransfection of NLΔVpr proviral DNA and a Vpr expression plasmid was employed to analyze the virion incorporation property of Vpr. Mutant Vpr containing deletions and specific substitutions (VprW38S-FL, VprL39G-FL, VprL42G-FL, VprG43P-FL, and VprI46G-FL) exhibited a negative virion incorporation phenotype. Further, mutant Vpr-FL containing deletions also failed to associate with wild-type Vpr, indicating a possible defect in the oligomerization feature of Vpr. Subcellular localization studies indicated that mutants VprΔ35-50-H-FL, VprR36W-FL, VprL39G-FL, and VprI46G-FL exhibited both cytoplasmic and nuclear localization, unlike other mutants and control Vpr-FL. While wild-type Vpr registered cell cycle arrest at G2, mutant Vpr showed an intermediary effect with the exception of VprΔ35-50 and VprΔ35-50-H. These results suggest that residues in the HII domain are essential for Vpr functions.
Members of the lentivirus family of retroviruses have been shown to contain nonstructural proteins of viral origin in addition to the structural proteins in the virus particles, a feature noted with several DNA viruses (10, 15, 21, 27). Specifically, the virus particles produced by human immunodeficiency virus type 1 (HIV-1) have been shown to contain three nonstructural proteins, designated Vif, Vpr, and Nef (6, 10, 54). A recent study, however, has questioned the specific incorporation of Vif into the virus particles (11). The virion-associated protein Vpr has been an area of intensive research with respect to understanding Vpr's role in virus infection and a potential carrier molecule to transport peptides and proteins to the assembling and mature virus particles (10, 13, 17, 22, 27, 38, 41, 44, 47, 55, 56). In addition to its ability to incorporate into virus particles (9, 10, 20, 21, 35, 45, 50), induction of apoptosis (1, 2) and differentiation (26), cell cycle arrest at G2 stage (18, 30, 41, 43), nuclear localization (12, 13, 16, 28, 34, 37, 57, 58), transport of the preintegration complex to the nucleus (19, 37), transcriptional activation (8), cation-selective channel activity (40), and interaction with several candidate cellular proteins (4, 5, 14, 16, 18, 42, 49, 50, 52, 58) are some of the features of Vpr. With regard to the number of molecules of Vpr present in the virus particles, it was reported earlier that Vpr is present in amounts similar to that of Gag (7) or reverse transcriptase (22). Utilizing an epitope-tagging approach, our laboratory showed that Vpr is present in small amounts (14 to 18 molecules per virion) in the virus particles (48). Further, it was also shown that the extent of incorporation of Vpr into the virus particles can be influenced by the expression level of Vpr in cells (24).
Despite several studies, a correlation between the structure-function relationship of Vpr at the molecular level remains to be defined. Mutational analysis of Vpr, based on the secondary structure predicted by several algorithms, identified potential helical domains comprising residues 17 to 34 and 53 to 72 which are required for virion incorporation, nuclear localization, stability, and oligomerization (12, 31–35, 37, 57). Though the carboxyl-terminal region of Vpr did not have a predicted structure (residues 79 to 96), this region plays a crucial role in the cell cycle arrest function and also contributes to the stability of Vpr (10, 13). The predicted secondary structure of Vpr was also supported by circular dichroism spectroscopy studies of generated peptides corresponding to the helical domains (29). Studies by Roques and coworkers recently have provided information regarding the structure of Vpr utilizing nuclear magnetic resonance (NMR) (45, 53). It was shown that the Vpr molecule contains three helical domains, HI, HII, and HIII, involving residues 17 to 29, 35 to 46, and 53 to 78, respectively.
To address the role of helical domain HII, corresponding to the residues 35 to 46, on Vpr functions, a strategy involving deletion, insertion, and/or substitution mutagenesis was utilized. The data generated in this study indicate that HII is essential for the incorporation of Vpr into the virus particles. Further, Vpr harboring mutations in this domain failed to associate with wild-type Vpr, suggesting a role in the oligomerization function of Vpr.
MATERIALS AND METHODS
Cell lines.
RD, a human rhabdomyosarcoma cell line, and HeLa, a human cervix epithelioid carcinoma cell line, were obtained from the American Type Culture Collection (Manassas, Va.). Cells were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL Laboratories, Grand Island, N.Y.) supplemented with 1% l-glutamine, penicillin-streptomycin, and 10% fetal bovine serum at 37°C in 5% CO2.
Construction of recombinant plasmids containing variant Vpr.
Vpr coding sequences, amplified through PCR using proviral NL4-3 DNA as a template, were cloned into the expression vector pCDNA3 (Invitrogen, Carlsbad, Calif.). The deletion, insertion, and substitution of amino acid residues in the HII domain of Vpr were carried out by using PCR methodologies (31, 47). The details of the primers used for the generation of deletion and substitution mutants are available upon request. Sequences corresponding to the Flag epitope (DYKDDDDK) were added to the 3′ end of the Vpr coding sequence to enable the detection of Vpr (46). Chimeric enhanced green fluorescent protein (EGFP)-Vpr expression plasmids were generated by fusing EGFP coding sequences at the 5′ end of the Vpr coding sequence. The integrity of plasmid DNAs was tested by application of a restriction enzyme followed by DNA sequence analysis.
In vitro transcription/translation and RIPA of Vpr.
The coupled T7 transcription/translation system (Promega, Madison, Wis.) was used for assessing the expression of the protein directed by the Vpr clones. Incubation conditions were monitored according to the manufacturer's instructions. Radioimmunoprecipitation analysis (RIPA) of in vitro-translated proteins was carried out using polyclonal antiserum to the Flag epitope (Santa Cruz Biotechnology, Santa Cruz, Calif.) as described previously (47).
Expression of Vpr in cells.
HeLa cells (106) in 35-mm-diameter petri dishes were infected with recombinant vaccinia virus vTF7-3 expressing T7 RNA polymerase at a multiplicity of infection of 10 for 1 h. At the end of incubation, the virus inoculum was removed and the cells were washed with phosphate-buffered saline (PBS). Vpr expression plasmids were transfected into cells using FuGENE 6 transfection reagent (Roche, Indianapolis, Ind.). Forty-eight hours after transfection the cells were washed with PBS and lysed in RIPA buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride). Cell lysate was centrifuged to remove the cell debris. Estimation of the protein content of the cell lysate was carried out using Bradford reagent (Bio-Rad, Richmond, Calif.), and 150 μg equivalent of cellular proteins was subjected to immunoblot analysis.
Immunofluorescence assay.
HeLa cells seeded onto poly-l-lysine-coated coverslips in a 35-mm-diameter petri dish were infected with vaccinia virus vTF7-3 and were transfected with Vpr expression plasmid DNA as described above. Twenty-four hours after transfection, the cells were washed with PBS and fixed in 4% paraformaldehyde at room temperature for 30 min. After being washed three times with PBS, the cells were incubated with anti-Flag M2 monoclonal antibody-fluorescein isothiocyanate (FITC) conjugate (Sigma, St. Louis, Mo.) at 37°C in a humidified incubator for 90 min. Following several washes with PBS, the cells were incubated with 4,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml) to counterstain the nuclei, washed three times with PBS, and mounted on glass slides using a Slow Fade antifade reagent (Molecular Probes, Eugene, Oreg.). Immunofluorescence was detected using a Zeiss Axiovert 100 inverted fluorescence microscope with an attached Bio-Rad MRC 600 laser scanning confocal imaging system. To produce a merged image, each fluorochrome was recorded and the superimposed images were generated with Image-Pro software (Media Cybernetics, Silver Spring, Md.).
Cell cycle studies.
To assess the effect of mutant Vpr on the cell cycle, we utilized a chimeric protein approach in which EGFP was fused to the amino terminus of Vpr. HeLa cells were transfected with EGFP-Vpr-encoding plasmids by the calcium phosphate precipitation method (48). At 48 h posttransfection, the cells were washed with PBS, trypsinized, diluted with PBS, and pelleted. The cells were resuspended in PBS and were gated on the fluorescence-activated cell sorter (FACScan; Coulter Apex Elite, Hialeah, Fla.) for both the EGFP-positive and -negative populations. The EGFP-positive and -negative cells were pelleted and resuspended in 80% ice-cold ethanol for 30 min. Following an additional wash with PBS, the cells were incubated in PBS containing RNase A (50 μg/ml) and propidium iodide (40 μg/ml) for 60 min at 4°C. The cellular DNA content was analyzed with a FACScan apparatus. The DNA profile was analyzed by the Multicycle AV program (Phoenix Flow System, San Diego, Calif.).
Transfection and generation of virus particles.
HIV-1 proviral DNA (pNL4-3) was modified to disrupt the expression of Vpr by an insertion (AATT) between residues 63 and 64 within the Vpr coding region (NLΔVpr). To generate virus particles containing wild-type or mutant Vpr, NLΔVpr proviral DNA was cotransfected with the respective Vpr expression plasmid by calcium phosphate coprecipitation method into RD cells (47). Similarly, cotransfection of NL4-3, containing an intact open reading frame for Vpr, with Vpr expression plasmids was carried out to generate virus particles for assessing the association of mutant Vpr with wild-type Vpr. The virus particles released into the culture supernatant were collected 120 h after transfection. The culture supernatants were precleared for 10 min at 10,000 rpm and subsequently spun at 40,000 rpm for 3 h using sucrose density gradient centrifugation. Virus pellets were lysed in lysis buffer (62.5 mM Tris-HCl [pH 6.8], 0.2% sodium dodecyl sulfate, 1% β-mercaptoethanol, 10% glycerol), and a p24 antigen assay was used to quantitate the amount of protein present in the virus particles.
Immunoblot analysis.
Virus samples, normalized on the basis of p24 antigen values obtained using an enzyme-linked immunosorbent assay (Organon Teknika, Durham, N.C.) were immunoprecipitated with polyclonal antiserum to Flag epitope (Santa Cruz Biotechnology) and protein A-Sepharose CL-4B (Amersham Pharmacia Biotech, Piscataway, N.J.) at 4°C overnight. The Sepharose beads were then washed and boiled in sample buffer for 5 min, and immunoprecipitated proteins were separated on NuPAGE 10% N,N-methylenebisacrylamide–Tris gel followed by transfer onto a nitrocellulose membrane. Membranes were blocked with 5% nonfat dry milk and incubated with rabbit polyclonal antiserum to Flag epitope for 2 h. The membranes were washed three times for 10 min each with TBST (20 mM Tris [pH 7.5], 500 mM NaCl, 0.05% Tween-20) and then probed with secondary antibody (anti-rabbit immunoglobulin AP conjugate; Promega), washed again with TBST, and developed with CDP-Star as the chemiluminescent substrate (Promega).
RESULTS
Structural features and generation of mutant Vpr.
The predicted secondary structure as indicated by several algorithms combined with site-specific mutagenesis studies showed that Vpr contains helical domains with a basic amino acid enriched C terminus (12, 31–35, 37, 57). Recently, Wecker and Roques (53) reported the structure of Vpr utilizing NMR spectroscopy (Fig. 1). The amino-terminal segment of Vpr comprising amino acids 1 to 51 has been shown to have three turns around the first three proline residues P5, P10, and P14. This is followed by a long helix-turn-helix motif encompassing residues 17 to 46 with another turn extending from residues 47 to 49. The helix-turn-helix motif corresponds to residues 17 to 29 (helical domain; HI), 30 to 34 (β-turn type IV), and 35 to 46 (helical domain; HII). HII is less amphipathic than HI. The studies involving the C-terminal fragment of Vpr corresponding to residues 52 to 96 showed a long amphipathic helix (residues 53 to 78; HIII) followed by a less-defined domain extending from residues 79 to 96.
FIG. 1.
Schematic representation of wild-type and mutant Vpr. The sequences corresponding to the Flag epitope were added to the 3′ end of the Vpr coding sequence. (A) The residues deleted from the HII domain and the adjoining region and the designations of mutants are indicated. (B) Substitutional mutational analysis of residues in the HII domain.
With respect to the structure-function relationship of Vpr, molecular analyses involving site-specific mutagenesis have provided useful information (12, 31–35, 57). However, there is no information available regarding the role of residues present in the HII domain of Vpr. To evaluate the role of the residues in this domain, we have considered an approach involving a combination of deletion and site-specific mutagenesis. PCR-based methods were used to generate Vpr mutants lacking 1, 5, 9, and 14 residues in the HII domain and the adjoining region (Fig. 1A). In addition, a variant containing a hinge region (GGSSG) in place of the deleted residues in the HII domain was also generated. Further, to enable the detection of Vpr, sequences corresponding to the Flag epitope were fused in frame to the 3′ end of the Vpr coding sequence. Substitution Vpr mutants (Fig. 1B) were also generated utilizing similar methods.
Effect of mutations in helical domain II on Vpr expression.
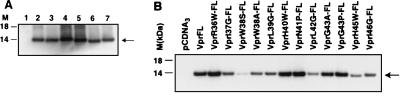
As the Vpr expression plasmid contains the T7 promoter upstream of the coding sequences, the protein directed by each plasmid was tested using an in vitro transcription-coupled translation system (TNT; Promega). In vitro-translated proteins were immunoprecipitated with polyclonal Flag antiserum. The mutant Vpr protein was detected at the same level as the wild-type Vpr-FL protein (data not shown). As expected, the deletion of various numbers of amino acid residues (1 to 14) resulted in mutant proteins with mobilities different from that of the wild-type Vpr-FL. We also utilized recombinant vaccinia virus vTF7-3 expressing T7 polymerase to study the effect of mutations in the HII domain on the expression of Vpr in cells. vTF7-3-infected HeLa cells were transfected with wild-type or mutant Vpr expression plasmids by FuGENE 6 transfection reagent. Cell lysates, prepared 48 h after transfection, were subjected to immunoblot analysis using Flag antibodies. Transfection with each of the deletion mutants resulted in detectable levels of Vpr-FL in cell lysate (Fig. 2A). The electrophoretic mobilities of the mutant Vpr proteins were similar to those in the data for the corresponding proteins translated in vitro. Analysis of substitution mutants indicated that the protein directed by VprW38S-FL was highly unstable (Fig. 2B). In addition, mutants VprW38A-FL, VprL39G-FL, VprL42G-FL, and VprI46G-FL showed an altered stability in comparison to Vpr-FL.
FIG. 2.
Expression of wild-type and mutant Vpr. (A) Immunoblot analysis of Vpr in cells. HeLa cells were infected with vaccinia virus vTF7-3 and transfected with wild-type and mutant Vpr expression plasmids. Cell lysates were processed for immunoblot analysis as described in Materials and Methods. M, molecular mass markers (in kilodaltons). Lanes: 1, pCDNA3; 2, Vpr-FL; 3, VprΔ44-FL; 4, VprΔ42-46-FL; 5, VprΔ40-48-FL; 6, VprΔ37-50-FL; 7, VprΔ37-50-H-FL. Arrow, position of proteins. (B) Expression of Vpr harboring substitutions in HII domain in HeLa cells.
Incorporation of mutant Vpr into virus particles.
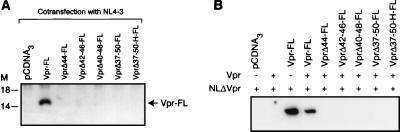
To address the role of the HII domain in the virion incorporation property of Vpr, we employed an assay system involving the cotransfection of HIV-1 proviral DNA containing a frameshift mutation in Vpr coding sequences (NLΔVpr) and the Vpr expression plasmid into cells to generate virus particles. The rationale for the assay is that Vpr, expressed in trans, will be incorporated into the virus particles directed by HIV-1 proviral DNA. The virus particles released into the culture medium were centrifuged and quantitated by a p24 antigen assay. The virus particles were normalized on the basis of p24 antigen values and subjected to immunoblot analysis to monitor the extent of incorporation of mutant Vpr into virus particles. The results showed that virion incorporation of Vpr deletion mutants is completely abolished (Fig. 3A). On the other hand, VprR36W-FL, VprI37G-FL, VprW38A-FL, VprH40W-FL, VprG43A-FL, and VprH45W-FL showed a positive virion incorporation phenotype. Interestingly, VprW38S-FL, VprL39G-FL, VprL42G-FL, VprG43P-FL, and VprI46G-FL exhibited a reduction in virion incorporation in comparison to Vpr-FL (Fig. 3B).
FIG. 3.
Immunoblot analysis of virus particles using antibodies against Flag epitope. Viral lysates normalized on the basis of p24 antigen values were subjected to analysis following separation on Nu-PAGE 10% N,N-methylenebisacrylamide-Tris gel and transfer to nitrocellulose membrane. (A) Virus particles generated through cotransfection of NLΔVpr and Vpr-FL and mutant Vpr-FL expression plasmids. M, molecular mass markers (in kilodaltons). Lanes: 1, NLΔVpr; 2, NLΔVpr + Vpr-FL; 3, NLΔVpr + VprΔ44-FL; 4, NLΔVpr + VprΔ42-46-FL; 5, NLΔVpr + VprΔ40-48-FL; 6, NLΔVpr + VprΔ37-50-FL; 7, NLΔVpr + VprΔ37-50-H-FL. (B) Virion incorporation phenotypes of Vpr substitution mutants.
It was earlier reported (59) that the oligomerization property of Vpr may involve HI and downstream residues. This implies that residues in the HII domain may contribute to the dimerization/oligomerization feature of Vpr. To address this, we utilized an assay system in which the association of Vpr-FL mutants with wild-type Vpr was measured. This is an indirect assay for monitoring the oligomerization capabilities of Vpr in cells. Vpr mutants that lack or exhibit a low level of virion incorporation are ideal candidates for this assay. For this purpose, HIV-1 proviral DNA NL4-3 was cotransfected with either a Vpr-FL- or Vpr-FL-encoding mutant plasmid. Since the Flag epitope is present only in Vpr directed by the expression plasmid and is absent in the Vpr directed by the proviral DNA, the detection of Flag epitope-containing Vpr in the virus particles would indicate that Vpr-FL or a Vpr-FL mutant is incorporated into the virus particles by itself and/or in association with wild-type Vpr. The immunoblot analysis of virus particles generated by cotransfection of NL4-3 and Vpr-FL showed that a protein with a molecular mass of 14 kDa was detectable with Flag antibodies. On the other hand, a Vpr-FL mutant was not detectable in the virus particles, indicating that Vpr mutants failed to associate with wild-type Vpr (Fig. 4A). Since the number of molecules of Vpr present in the virus particles is low (14 to 18 molecules per virion), it is likely that overexpression of mutant Vpr through a heterologous promoter may mask the wild-type Vpr incorporation expressed through HIV-1 proviral DNA. This may result in the absence of mutant Vpr-FL in the virus particles. Considering this, we also utilized a cotransfection approach in which HIV-1 proviral DNA lacking Vpr expression (NLΔVpr) and wild-type and mutant Vpr expression plasmids were transfected into cells. This was based on our earlier work showing that the expression of Vpr in trans leads to efficient incorporation into virus particles (392 to 550 Vpr molecules per virion). Hence, the expression of wild-type Vpr and mutant Vpr-FL in trans may provide an opportunity to detect mutant Vpr-FL in the virus particles through its association with wild-type Vpr. The immunoblot analysis of virus particles derived from cells cotransfected with Vpr-FL detected a band reactive to Flag antisera. However, virus particles derived from cotransfection of NLΔVpr, wild-type Vpr, and mutant Vpr-FL containing deletions did not show a band (Fig. 4B). These results suggest that mutant Vpr-FL molecules containing deletions are defective for oligomerization of Vpr. The possibility that a transdominant effect by mutant Vpr-FL on wild-type Vpr could also result in the failure to detect mutant Vpr-FL in the virus particles existed. To investigate this, virus particles derived from cotransfection were analyzed using antibodies against Vpr. Such an analysis showed similar levels of Vpr except where NLΔVpr was cotransfected with the pCDNA3 vector control (data not shown), ruling out an effect on virion incorporation of wild-type Vpr.
FIG. 4.
Dimerization or oligomerization of Vpr in cells. (A) Extent of incorporation of mutant Vpr-FL and wild-type Vpr-FL into the virus particles directed by NL4-3 proviral DNA. Vpr was expressed in the context of HIV-1 proviral DNA. (B) Incorporation of mutant Vpr-FL in association with wild-type Vpr. Vpr was expressed through a heterologous promoter.
Subcellular localization of Vpr.
It is likely that the lack of incorporation of Vpr mutant into the virus particles may result from altered subcellular localization of the mutant protein in cells. In order to verify this, we used Vpr constructs containing the Flag epitope. Transfected HeLa cells were incubated with anti-Flag M2 monoclonal antibody-FITC conjugate followed by incubation with DAPI to stain the nucleus. As a control, we used cells transfected with the backbone pCDNA3 plasmid. As noted earlier, Vpr expressed in cells was localized to the nuclear region (Fig. 5). The observed patterns include an intense signal at the rim of the nucleus and diffuse and focal staining in the nucleus. The specificity was demonstrated by the absence of staining in the cells transfected with pCDNA3 and mock-transfected cells (data not shown). All Vpr mutants, except VprΔ37-50-H-FL, showed a localization pattern similar to that of Vpr-FL. Vpr directed by the mutant VprΔ37-50-H-FL showed an intense signal at the rim of the nucleus, and a considerable amount of protein was also present in the cytoplasm (Fig. 5). The substitution mutants designated VprR36W-FL, VprL39G-FL, and VprI46G-FL showed both nuclear and cytoplasmic localization, unlike the other substitution mutants, which localized in the nucleus (Table 1).
FIG. 5.
Subcellular localization of wild-type and mutant Vpr. HeLa cells 24 h after transfection were fixed and stained with anti-Flag M2 monoclonal antibody-FITC conjugate followed by DAPI. Cells were analyzed using a confocal microscope at ×60 magnification. To produce a merged image, each fluorochrome was recorded and the superimposed images were generated with Image-Pro software. (A) Anti-Flag M2-FITC conjugate; (B) DAPI; (C) superimposed images.
TABLE 1.
Effect of mutations on Vpr functionse
| Designation of Vpr | Virion incorporationa | Stabilityb | Subcellular localizationc | Oligomerizationd |
|---|---|---|---|---|
| Vpr-FL | + | + | N | + |
| VprΔ44-FL | − | + | N | − |
| VprΔ42-46-FL | − | + | N | − |
| VprΔ40-48-FL | − | + | N | − |
| VprΔ37-50-FL | − | + | N | − |
| VprΔ37-50-H-FL | − | + | N + C | − |
| VprR36W-FL | + | + | N + C | ND |
| VprI37G-FL | + | + | N | ND |
| VprW38S-FL | − | Reduced | N (diffuse) | ND |
| VprW38A-FL | + | + | N (diffuse) | ND |
| VprL39G-FL | − | + | N + C | − |
| VprH40W-FL | + | + | N | ND |
| VprN41P-FL | +/− | + | ND | ND |
| VprL42G-FL | − | Reduced | N | − |
| VprG43A-FL | + | + | N | ND |
| VprG43P-FL | − | + | ND | − |
| VprH45W-FL | + | Reduced | N (diffuse) | ND |
| VprI46G-FL | − | + | N + C | − |
Virion incorporation was carried out using cotransfection of NLΔVpr and Vpr expression plasmids.
Stability of the protein was determined using recombinant vaccinia virus expressing T7 polymerase.
FITC-conjugated Flag antibodies were used for subcellular localization studies.
Incorporation of mutant Vpr-FL in association with wild-type Vpr was assessed in the virus particles.
+, positive; −, negative; +/−, intermediate; N, nuclear; C, cytoplasmic; ND, not done.
Effect of mutations in helical domain II on cell cycle functions of Vpr.
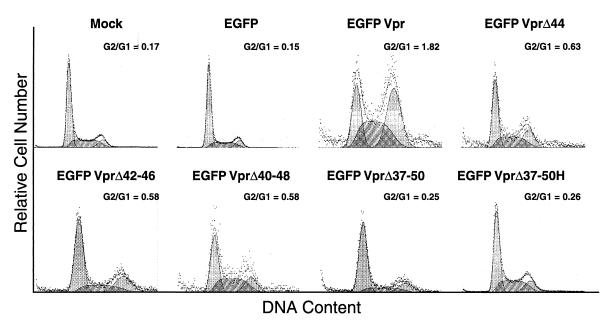
It was reported earlier that Vpr induces an arrest of cells at G2 phase of the cell cycle (18, 30, 41, 43). Though the C terminus of Vpr containing basic amino acids has been implicated in the cell cycle arrest function (12), mutations in the amino terminus have also been shown to have an effect in this regard. This has prompted us to evaluate the effect of mutation in the HII domain on cell cycle arrest. As the addition of residues at the C terminus of Vpr may result in loss of the cell cycle arrest function (12), we have utilized a chimeric Vpr without Flag epitope at the C terminus. To visualize the cells for expression and cell cycle arrest, the EGFP coding region was fused to the 5′ end of the Vpr coding region. Cells transfected with EGFP- and EGFP-Vpr-encoding plasmids were sorted initially based on EGFP expression. The EGFP-positive and -negative cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. Mock- and EGFP-transfected cells showed G2/G1 ratios of 0.17 and 0.15, respectively (Fig. 6). Cells expressing EGFP-Vpr registered cell cycle arrest with a G2/G1 ratio of 1.82. EGFP-VprΔ44, EGFP-VprΔ42-46, and EGFP-VprΔ40-48 mutants showed an intermediary level of cell cycle arrest with G2/G1 ratios of 0.63, 0.58, and 0.58, respectively. On the other hand, EGFP-VprΔ37-50 and EGFP-VprΔ37-50-H mutants exhibited loss of cell cycle arrest function as the G2/G1 ratios are close to the values obtained with the negative control. All the EGFP-negative cells showed nearly identical DNA profiles, similar to that of the control (data not shown).
FIG. 6.
Effect of wild-type and mutant Vpr on the cell cycle. HeLa cells were transfected with EGFP and EGFP-Vpr expression plasmids. At 48 h posttransfection, cells were sorted for EGFP-positive and -negative populations. The cells were stained for DNA content with propidium iodide and analyzed by flow cytometry. The G2/G1 ratio, as determined by the Multicycle AV program, of each cell population is indicated.
DISCUSSION
Vpr is a protein expressed late in HIV-1-infected cells (10, 27). Since its identification, there has been an enormous interest in understanding the functions of this protein. The demonstration that a related protein, Vpx, is present in HIV-2 and simian immunodeficiency virus virions in amounts equal to that of Gag (20) has suggested that the virion-associated proteins may have a role in the events related to virus infection analogous to the nonstructural proteins present in the virus particles directed by DNA viruses (15). The observations in support of such a role for Vpr include a positive effect on infection of macrophages by HIV-1, its role in the transport of the viral preintegration complex to the nucleus, and an effect at the level of transcription (10, 27). Furthermore, it has been shown that Vpr induces cell cycle arrest at the G2 phase depending on the cell type (13) and is cytotoxic to cells through the induction of apoptosis (1, 2). With regard to the amount of Vpr, recent studies from our laboratory showed that HIV-1 Vpr is present in small amounts (14 to 18 molecules per virion) in the virus particles (48), contrary to the data reported earlier (7, 22).
In an effort to gain information about the structure-function relationship of Vpr, we have utilized the structural data that were recently reported for Vpr by Roques and coworkers (45, 53). NMR studies of the synthetic N- and C-terminal peptides comprising residues 1 to 51 and 52 to 96 of Vpr revealed a structure with three helical domains. Residues 17 to 29, 35 to 46, and 53 to 78 correspond to HI, HII, and HIII domains, respectively. Both HI and HIII domains have also been predicted by several algorithms, and mutational analyses have been carried out to determine the role of these domains in Vpr functions (32, 56, 57). In comparison to HI and HIII, the HII domain was unknown till the structural data came about, and hence there is no information available regarding the role of the HII domain in Vpr functions. The HII domain consists of 12 amino acids. The residues with hydrophilic properties, R36, N41, and Q44, are located on one side of the helix. The hydrophobic amino acids W38, L39, L42, H45, and I46 are located on the other side of the helix (53). The location of residues I37 and H40 on the former side hinders the formation of a perfect amphipathic helix (53). We have constructed several Vpr mutants involving deletion, insertion, and substitution mutagenesis approaches to understand the contribution of the HII domain to Vpr functions. The rationale for this approach is that the HII domain may be critical for maintaining the biological properties of Vpr. On the basis of this assumption, it is hypothesized that a Vpr mutant with an altered HII may not behave like a wild-type Vpr. It is likely that HII may be involved in stabilizing interactions between the HI and HIII domains or contribute to binding to Gag for its incorporation into the virus particles. The parameters that were used for assessing the effect of the mutations in the HII domain of Vpr include protein expression, stability, virion incorporation, subcellular localization, and cell cycle arrest. The results presented here indicate that the HII domain is critical for the Vpr functions. While the expression and stability of wild-type Vpr and mutants except VprW38S-FL, VprL42G-FL, and VprH45W-FL remain the same in transfected cells, the alterations in the HII domain exerted a drastic effect on the incorporation of Vpr into the virus particles. Vpr harboring mutations in the HII domain failed to get incorporated into the virus particles. A deletion of even one residue at the C terminus of the HII domain (Q44) abolished virion incorporation, similar to what was found for other deletion mutants. The deletion in VprΔ42-46-FL is confined to the HII domain comprising four residues. On the other hand, mutants VprΔ40-48-FL, VprΔ37-50-FL, and VprΔ37-50-H-FL involved deletion of residues in HII and also adjoining residues 47 to 50, which have been shown to form a Υ turn. The substitution mutational analysis provided evidence supporting a crucial role for the hydrophobic residues in the virion incorporation function. Specifically, substitutions targeting W38, L39, L42, and I46 resulted in a drastic reduction in the virion incorporation of Vpr with the exception of H45. Similar studies involving the residues located on the side of the helix opposite to the hydrophobic residues (R36, I37, H40, and N41) showed that substitution did not abrogate the virion incorporation function.
In addition to lack of virion incorporation, mutant Vpr also failed to associate with wild-type Vpr. This was arrived at by using an indirect assay in which virus particles were generated through cotransfection of either HIV-1 proviral DNA NL4-3 or NLΔVpr and Vpr expression plasmids. The presence of a Flag epitope in the mutant Vpr and its absence from Vpr encoded by NL4-3 provide the premise for analyzing mutant Vpr in the virus particles using Flag antibodies. As the mutant Vpr-FL exhibits a negative virion incorporation phenotype, the association of mutant Vpr-FL with Vpr is the only mechanism by which mutant Vpr-FL will be incorporated into the virus particles. Since wild-type Vpr was shown to be present in the virus particles derived from cotransfection by using antibodies against Vpr, it is reasonable to suggest that the absence of mutant Vpr-FL may be due to a defect at the level of dimerization or oligomerization. Recently, Schuler et al. (45) reported that a synthetic peptide corresponding to the C terminus of Vpr (residues 52 to 96) exhibited the dimerization property. However, Vpr mutants used in this study which have mutations only in HII with an intact C terminus failed to associate with wild-type Vpr. This indicates that the observation noted with the synthetic peptide (residues 52 to 96) is not applicable to full-length Vpr. The lack of incorporation of the Vpr HII domain mutant into the virus particles could result from the lack of the protein available in a sufficient amount. However, this does not seem to be the case, as equal amounts of wild-type and mutant Vpr are shown to be present in cells. Alternatively, the residues present in the HII domain may play a crucial role in terms of facilitating the interactions between Vpr and Gag (25, 44). Such a view is indeed supported by our studies, as the substitution of hydrophobic residues W38, L39, L42, and I46 resulted in a drastic reduction in the incorporation of Vpr into the virus particles. The failure to incorporate into virus particles could be due to a result of the altered subcellular localization of mutant Vpr proteins. This possibility was tested by analyzing the cellular localization of mutant Vpr in comparison to wild-type Vpr. The studies in this regard showed that most of the mutant Vprs and wild-type Vpr-FL exhibited perinuclear and diffuse staining of the nucleus. On the other hand, cells transfected with VprΔ37-50-H-FL, VprR36W-FL, VprL39G-FL, and VprI46G-FL indicated a staining pattern involving both the rim of the nucleus and cytoplasm. As cell cycle arrest is a characteristic feature of Vpr in HIV-1-infected and Vpr expression plasmid-transfected cells, Vpr mutants harboring deletions were evaluated for this function. Though the cell cycle arrest property has been attributed to the C terminus of Vpr, mutation at the amino terminus of Vpr has an effect on the cell cycle (12). While wild-type Vpr exhibited a typical cell cycle arrest at G2, several mutants showed an intermediary level of cell cycle arrest. VprΔ37-50 and VprΔ37-50-H did not have any effect on the cell cycle. Overall, the experimental data described in this study point out that the hydrophobic residues of the HII domain are essential for Vpr functions.
ACKNOWLEDGMENTS
We express our thanks to Sashi Reddy for his help in the preparation of the manuscript. Kimmel Cancer Center Confocal Microscopy, Flow Cytometry, and Nucleic Acid core facilities provided services for the studies.
This work was supported by funds from the National Institutes of Health (AI29306) and a grant from the Commonwealth of Pennsylvania to the Biotechnology Foundation, Inc.
REFERENCES
- 1.Arunagiri C, Macreadie I, Hewish D, Azad A. A C-terminal domain of HIV-1 accessory protein Vpr is involved in penetration, mitochondrial dysfunction and apoptosis of human CD4+ lymphocytes. Apoptosis. 1997;2:69–76. doi: 10.1023/a:1026487609215. [DOI] [PubMed] [Google Scholar]
- 2.Ayyavoo V, Mahboubi A, Mahalingham S, Ramalingam R, Kudchodkar S, Williams W V, Green D R, Weiner D B. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor κB. Nat Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 3.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of human immunodeficiency virus type 1 accessory genes vpr, vpu and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 4.BouHamdan M, Benichou S, Rey F, Navarro J M, Agstini I, Spore B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.BouHamdan M, Xue Y, Baudat Y, Hu B, Sire J, Pomerantz R J, Duan L X. Diversity of HIV-1 Vpr interactions involves usage of the WXXF motif of host cell proteins. J Biol Chem. 1998;273:8009–8016. doi: 10.1074/jbc.273.14.8009. [DOI] [PubMed] [Google Scholar]
- 6.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E A, Terwilliger E F, Jalinoos Y, Proulx J, Sodroski J G, Haseltine W A. Identification of HIV-1 Vpr product and function. J Acquir Immune Defic Syndr. 1990;1:11–18. [PubMed] [Google Scholar]
- 9.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R. HIV-1 auxillary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 11.Dettenhofer M, Yu X-F. Highly purified human immunodeficiency virus type 1 reveals virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerman M. HIV-1 Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 14.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, Vpr, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. 1 and 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 16.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Albright A V, Gonzalesz-Scarano F, Malim M M. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle—a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 18.Gragerov A, Kino T, IIyina-Gragerova G, Chrousos G P, Pavlakis G N. HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology. 1998;245:323–330. doi: 10.1006/viro.1998.9138. [DOI] [PubMed] [Google Scholar]
- 19.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson L E, Sowder T D, Copeland R E, Beneveniste R E, Oroszlan S. Isolation and characterization of a novel protein (X-ORF product) from SIV and HIV-2. Science. 1988;241:199–201. doi: 10.1126/science.3388031. [DOI] [PubMed] [Google Scholar]
- 21.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 22.Kobinger G P, Borsetti A, Nie Z, Mercier J, Daniel N, Gottlinger H G, Cohen A. Virion-targeted viral inactivation of human immunodeficiency virus type 1 by using Vpr fusion proteins. J Virol. 1998;72:5441–5448. doi: 10.1128/jvi.72.7.5441-5448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo E, Mammano F, Cohen E A, Gottlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous virus particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai D, Singh S P, Cartas M, Murali R, Kalyanaraman V S, Srinivasan A. Extent of incorporation of HIV-1 Vpr into the virus particles is flexible and can be modulated by expression level in cells. FEBS Lett. 2000;469:191–195. doi: 10.1016/s0014-5793(00)01264-3. [DOI] [PubMed] [Google Scholar]
- 25.Lavallee C, Yao X J, Ladha A, Gottlinger H, Haseltine W, Cohen E. Requirement of the Pr55gag precursor for incorporation of the Vpr product into human immunodeficiency virus type 1 viral particles. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy D N, Fernandes L S, Williams W V, Weiner D B. Induction of cell differentiation by human immunodeficiency virus 1 vpr. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-y. [DOI] [PubMed] [Google Scholar]
- 27.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Z, Butcher D J, Murali R, Srinivasan A, Huang Z. Structural studies of synthetic peptide fragments derived from the HIV-1 Vpr protein. Biochem Biophys Res Commun. 1998;244:732–736. doi: 10.1006/bbrc.1998.8330. [DOI] [PubMed] [Google Scholar]
- 30.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azard A A. A domain of human immunodeficiency virus type-1 Vpr containing repeated H(S/F)RIG amino acid causes cell growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahalingam S, Khan S A, Jabbar M A, Monken C E, Collman R, Srinivasan A. Identification of residues in the N-terminal acidic domain of HIV-1 Vpr essential for virion incorporation. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 32.Mahalingam S, Khan S A, Murali R, Jabbar M A, Monken C E, Collman R, Srinivasan A. Mutagenesis of the putative a-helical domain of the Vpr protein of human immunodeficiency virus type 1: effect on stability and virion incorporation. Proc Natl Acad Sci USA. 1995;92:3794–3798. doi: 10.1073/pnas.92.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahalingam S, Collman R G, Monken C E, Patel M, Srinivasan A. Role of the conserved dipeptide Gly75 and Cys76 on HIV-1 Vpr function. Virology. 1995;210:495–500. doi: 10.1006/viro.1995.1368. [DOI] [PubMed] [Google Scholar]
- 34.Mahalingam S, Collman R G, Patel M, Monken C E, Srinivasan A. Functional analysis of HIV-1 Vpr: identification of determinants essential for subcellular localization. Virology. 1995;212:331–339. doi: 10.1006/viro.1995.1490. [DOI] [PubMed] [Google Scholar]
- 35.Mahalingam S, Patel M, Collman R G, Srinivasan A. The carboxy terminal domain is essential for stability and not for virion incorporation of HIV-1 Vpr into virus particles. Virology. 1995;214:647–652. doi: 10.1006/viro.1995.0079. [DOI] [PubMed] [Google Scholar]
- 36.Myers G, Korber B, Wain-Hobson S, Smith R F. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1993. [Google Scholar]
- 37.Nie Z, Bergeron D, Subbramanian R A, Yao X-J, Checroune F, Rougeau N, Cohen E A. The putative alpha helix 2 of human immunodeficiency virus type 1 Vpr contains a determinant which is responsible for the nuclear translocation of proviral DNA in growth-arrested cells. J Virol. 1998;72:4104–4115. doi: 10.1128/jvi.72.5.4104-4115.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okui N, Kobayashi N, Kitamura Y. Production of uninfectious human immunodeficiency virus type 1 containing viral protein R fused to a single-chain antibody against viral integrase. J Virol. 1998;72:6960–6964. doi: 10.1128/jvi.72.8.6960-6964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piller S C, Ewart G D, Premkumar A, Cox G B, Gage P W. Vpr protein of human immunodeficiency virus type 1 forms cation-selective channels in planar lipid bilayers. Proc Natl Acad Sci USA. 1996;93:5281–5286. doi: 10.1073/pnas.93.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon B, Grovit-Feerbas K, Stewart S A, Chen I S Y. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science. 1998;281:266–269. doi: 10.1126/science.281.5374.266. [DOI] [PubMed] [Google Scholar]
- 42.Refaeli Y, Levy D N, Weiner D B. The glucocorticoid receptor type II complex is a target of the HIV-1 Vpr gene product. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato A, Yoshimoto J, Isaka Y, Miki S, Suyama A, Hayami M, Fujiwara T, Yoshie O. Evidence for direct association of Vpr and Matrix protein p17 within the HIV-1 virion. Virology. 1996;220:208–212. doi: 10.1006/viro.1996.0302. [DOI] [PubMed] [Google Scholar]
- 45.Schuler W, Wecker K, de Rocquigny H, Baudat Y, Sire J, Roques B P. NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions. J Mol Biol. 1999;285:2105–2117. doi: 10.1006/jmbi.1998.2381. [DOI] [PubMed] [Google Scholar]
- 46.Serio D, Weber I T, Harrison R W, Louis J M, Srinivasan A. Epitope-based assay to determine the efficiency of cleavage by HIV-1 protease. BioTechniques. 1999;26:242–246. doi: 10.2144/99262bm13. [DOI] [PubMed] [Google Scholar]
- 47.Serio D, Rizvi T A, Cartas M, Kalyanaraman V S, Weber I T, Koprowski H, Srinivasan A. Development of a novel anti-HIV-1 agent from within: effect of chimeric Vpr containing protease cleavage site residues on virus replication. Proc Natl Acad Sci USA. 1997;94:3346–3351. doi: 10.1073/pnas.94.7.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh S P, Lai D, Cartas M, Serio D, Murali R, Kalyanaraman V S, Srinivasan A. Epitope-tagging approach to determine the stoichiometry of the structural and nonstructural proteins in the virus particles: amount of Vpr in relation to Gag in HIV-1. Virology. 2000;268:364–371. doi: 10.1006/viro.2000.0191. [DOI] [PubMed] [Google Scholar]
- 49.Stark L A, Hay R T. Human immunodeficiency virus type 1 (HIV-1) viral protein R (Vpr) interacts with Lys-tRNA synthetase: implications for priming of HIV-1 reverse transcription. J Virol. 1998;72:3037–3044. doi: 10.1128/jvi.72.4.3037-3044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tung H Y L, De Rocquigny H, Zhao L J, Cayla X, Roques B P, Ozon R. Direct activation of protein phosphatase-2A0 by HIV-1 encoded protein complex NCp7:Vpr. FEBS Lett. 1997;401:197–201. doi: 10.1016/s0014-5793(96)01470-6. [DOI] [PubMed] [Google Scholar]
- 51.Wang J J, Lu Y L, Ratner L. Particle assembly and Vpr expression in human immunodeficiency virus type 1 infected cells demonstrated by immunoelectron microscopy. J Gen Virol. 1994;75:2607–2614. doi: 10.1099/0022-1317-75-10-2607. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 53.Wecker K, Roques B P. NMR structure of the (1-51) N-terminal domain of the HIV-1 regulatory protein Vpr. Eur J Biochem. 1999;266:359–369. doi: 10.1046/j.1432-1327.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- 54.Welker R, Kotler H, Kalbitzer H R, Krausslich H G. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 55.Wu X, Liu H, Xiao H, Kim J, Seshaiah S, Natsoulis G, Boeke J D, Hahn B H, Kappes J C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao X, Mouland A J, Subbramanian R A, Forget J, Rougeau N, Bergeron D, Cohen E A. Vpr stimulates viral expression and induces cell killing in human immunodeficiency virus type 1-infected dividing Jurkat T cells. J Virol. 1998;72:4686–4693. doi: 10.1128/jvi.72.6.4686-4693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao X, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao L-J, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function: specific interaction with a cellular protein. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 59.Zhao L-J, Wang L, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Oligomerization mediated by the N-terminal domain. J Biol Chem. 1994;269:32131–32137. [PubMed] [Google Scholar]