Abstract
Cells are equipped with asymmetrically localised and functionally specialised components, including cytoskeletal structures and organelles. Positioning these components to specific intracellular locations in an asymmetric manner is critical for their functionality and affects processes like immune responses, tissue maintenance, muscle functionality, and neurobiology. Here, we provide an overview of strategies to actively move, position, and anchor organelles to specific locations. By conceptualizing the cytoskeletal forces and the organelle-to-cytoskeleton connectivity, we present a framework of active positioning of both membrane-enclosed and membrane-less organelles. Using this framework, we discuss how different principles of force generation and organelle anchorage are utilised by different cells, such as mesenchymal and amoeboid cells, and how the microenvironment influences the plasticity of organelle positioning. Given that motile cells face the challenge of coordinating the positioning of their content with cellular motion, we particularly focus on principles of organelle positioning during migration. In this context, we discuss novel findings on organelle positioning by anchorage-independent mechanisms and their advantages and disadvantages in motile as well as stationary cells.
Keywords: Organelle Positioning, Cytoskeletal Forces, Motile and Branched Cells, Cytoplasmic Streaming, Organelle-Cytoskeleton Interface
Subject terms: Cell Adhesion, Polarity & Cytoskeleton; Organelles
Positioning of organelles to specific intracellular locations is critical for their functionality. This review provides an overview of strategies to actively move and anchor organelles to specific locations and how this is coordinated with cellular motion.
Introduction
Eukaryotic cells possess a set of membrane-enclosed organelles such as mitochondria, lysosomes, the Golgi, the endoplasmic reticulum, and the nucleus. Although their intracellular localisation is sometimes depicted in a simplified and random manner in textbooks and review articles, advancements in the last decade have revealed that many membrane-enclosed organelles are positioned at specific intracellular sites through active mechanisms (Keys et al, 2024; Garde et al, 2022; Zheng et al, 2022; Roman et al, 2021; Moore et al, 2021; Gundersen and Worman, 2013). Similarly, increasing evidence suggests that also organelles without an enclosing membrane, such as the centrosome (Jimenez et al, 2021) or biomolecular condensates (Liao et al, 2019; Alberti and Hyman, 2021), can be positioned in a non-random manner within cells.
This asymmetric organelle distribution is not merely a passive outcome of the cytoskeletal force distribution but can be actively employed by cells to target organelles to specific intracellular locations, enabling local functions using their compartmentalised and specialised molecular content. Examples of functional organelle positioning include diverse biological processes such as non-motile muscle cells that precisely position and space their nuclei within the muscle cell syncytium (Roman et al, 2021; Azevedo and Baylies, 2020; Roman and Gomes, 2018; Gache et al, 2017; Metzger et al, 2012), proliferating neuroepithelia and radial glia cells that move their nuclei towards an apical position for an apically-localised cell division in a process termed ‘interkinetic nuclear migration’ (Strzyz et al, 2015; Taverna and Huttner, 2010; Norden et al, 2009), and motile C.elegans anchor cells that position their mitochondria to the cell front for local ATP production during translocation through the basement membrane (Garde et al, 2022; Kelley et al, 2019). Moreover, motile cells actively position the membrane-less centrosome to utilise it as a steering organelle (Kopf et al, 2020) and to utilise it as a distribution platform for localised secretion of extracellular proteases to facilitate the opening of tight pores in the extracellular matrix (ECM) (Infante et al, 2018).
In this review, we provide a conceptual framework of how different organelles are actively positioned in diverse cell types and biological processes. In particular, we focus on our current understanding of the functions, principles, and molecular mechanisms of organelle positioning in both stationary and motile cells. We discuss the forces that act onto organelles during their movement, including forces from actin polymerisation, myosin contractility, microtubules and their motors, the cell’s cortex, as well as intracellular flows and pressure gradients. Further, we discuss the emerging concept of anchorage-independent mechanisms for organelle positioning, such as intracellular flows, which may be particularly beneficial for highly dynamic cell types such as fast-migrating cells. While our current knowledge of organelle positioning in motile cells often derives from research on the nucleus and the centrosome, we also discuss the positioning of other organelles such as mitochondria, lysosomes, and the Golgi. We particularly emphasise mechanisms in mammalian cells, including knowledge from fibroblasts, neurons, muscle cells, immune cells, and cancer cells, while also incorporating principles from C. elegans nematode worms, Xenopus amphibians, intracellular pathogens, as well as single-cell eukaryotes such as the amoebae Dictyostelium discoideum and baker’s yeast Saccharomyces cerevisiae.
Asymmetry in organelle positioning
In this section, we provide an overview of organelle positioning and emphasise its relevance for cellular and organismal functionality. We provide examples from highly ramified and extended cells like neurons, as well as highly motile cells like immune cells, to discuss the principles of active organelle positioning, as these cells constantly face the challenge of positioning their organelles either within large or highly dynamic cell bodies.
Organelle positioning
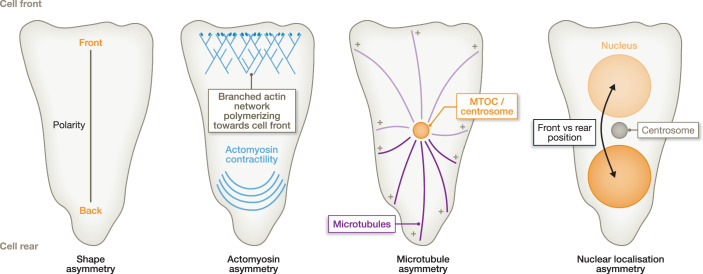
Most cells exhibit an asymmetric organisation and thus possess a polarity, such as front-back polarity in motile cells or apical-basal polarity in epithelial cells (Bornens, 2008). This cellular polarity is particularly pronounced in specialised epithelial cells that line organs, like in the intestine or the lung epithelium, where microvilli or cilia form a highly specialised apical cellular side (Rodriguez-Boulan and Macara, 2014). In these cells, the overall polarity remains highly stable and can be maintained throughout the cell’s lifetime. However, many other differentiated cells also display polarity, resulting in an asymmetric distribution of intracellular components. This asymmetry often arises from the actin and microtubule cytoskeleton, as actin and tubulin monomers themselves exhibit an asymmetric protein structure (Li and Gundersen, 2008). Upon polymerisation, these monomers form polymeric filaments with functionally distinct ends that have distinct assembly and disassembly rates. By nucleating these asymmetric filaments at specific intracellular sites, cells generate forces with specific directionality to move cargo or membranes.
Given the connectivity of many organelles to the cytoskeleton (Gurel et al, 2014), it is not surprising that the asymmetry of the actin and microtubule cytoskeleton causes an asymmetric distribution of organelles. In many cells, the membrane-less centrosome provides a reference for the relative organelle location within a cell, as it often locates at the centre of a cell due to the specific geometry of the actomyosin network (Fig. 1; see also ‘Centreing forces’) (Bornens, 2008). As the centrosome functions as a microtubule-organising centre, it orchestrates the microtubule cytoskeleton, which itself provides tracks for microtubule-dependent motor proteins that move and anchor organelle cargos like mitochondria, lysosomes, and the Golgi (see below). Consequently, minus-directed transport along microtubule tracks towards the centrosome by dynein motors causes colocalisation with the centrosome, such as observed for the membrane-surrounded Golgi (Thyberg and Moskalewski, 1999) and lysosomes (Cabukusta and Neefjes, 2018). While the nucleus is also often depicted centrally in textbooks, it is often actively moved to specific intracellular locations by anchorage-dependent and -independent mechanisms, depending on the cellular function and cell type (Gundersen and Worman, 2013). The endoplasmic reticulum (ER) is another large—or even the largest—membrane-surrounded organelle, which typically distributes throughout the entire cytoplasm with complex morphology while being directly connected to the outer nuclear membrane and the cytoskeleton (Nixon-Abell et al, 2016; Westrate et al, 2015). Yet, as we discuss below, also the ER is asymmetrically and actively positioned in different cell types.
Figure 1. Asymmetry in motile cells.
Motile cells have a distinct front-back polarity, in which the cell front (such as protrusions or lamellipodia) comprises an actin network (blue) that pushes against the cell membrane to achieve forward movement of the entire cell, and a cell rear that often contains high myosin contractility. As in non-motile cells, the centrosome/MTOC (orange) typically locates in the cell centre, providing a reference for the positioning of other organelles like the nucleus. In mesenchymal cells, the nucleus typically locates at the rear of the centrosome, whereas the nucleus typically locates at the front of the centrosome in fast amoeboid migrating cells.
Challenges to position organelles in long and branched cells
Organelle positioning to distinct subcellular locations is particularly evident but also challenging in very long or highly ramified cell types like neurons, neuroepithelia, and some immune cells. A highly extended cell type is neuroepithelial cells, which are very long and thin cells with a defined apical-basal polarity. During cell division of these cells, the nucleus moves towards the apical side, where also the centrosome is located, to undergo mitosis (Taverna and Huttner, 2010). Research on this type of movement of the nucleus, also termed interkinetic nuclear movement, provided multiple important concepts, which we will discuss below, of how cells solve the challenge of moving organelles within very long cells embedded in the tissue microenvironment.
In addition to being very long, some cell types, like neurons, have a highly ramified cell morphology, generating additional challenges for organelle positioning. For instance, Golgi outposts in neurons can localise far distantly from the cell body, where the main Golgi ribbon is located (Valenzuela et al, 2020; Ori-McKenney et al, 2012). As the Golgi can act as a non-centrosomal microtubule-organising centre (MTOC) (Chabin-Brion et al, 2001), these Golgi outposts act as non-centrosomal MTOCs (Akhmanova and Kapitein, 2022). Thereby, they enable microtubule nucleation at cellular sites distant to the location of the centrosome, providing microtubule tracks for long-distance cargo delivery in these long and branched cells. Interestingly, the centrosome itself can also leave its central position in the cell body to move into cellular branches of microglia (Möller et al, 2022), a tissue-resident phagocytic cell type in the brain that has a highly ramified cellular morphology. Centrosome migration into one of the multiple cellular branches correlates with productive phagocytosis of apoptotic neurons, suggesting that effective movement and positioning of the centrosome can support phagocytosis in large and extended cells (Möller et al, 2022). Next to the Golgi and the centrosome, also the ER can localise to specific positions in the large cytoplasm of neurons, where it is enriched at dendritic branched points, and functionally implicated in the formation of new branches (Cui-Wang et al, 2012). These examples highlight that organelles can be repositioned to fulfil functions at specific intracellular sites, a concept that we discuss in more detail below. Importantly, intracellular organelle positioning is also influenced by the properties of the microenvironment, such as the positioning of the Golgi and the centrosome (Pouthas et al, 2008), as we will discuss in the section ‘Plasticity in organelle positioning’.
Interestingly, some motile cells like amoeboid immune cells face a similar challenge to naturally-branched cell types: they typically extend multiple simultaneous cell fronts to explore their immediate microenvironment, leading to a complex ramified cell morphology with multiple highly dynamic cell fronts (Hadjitheodorou et al, 2021; Kopf et al, 2020; Driscoll et al, 2019; Renkawitz et al, 2019; Fritz-Laylin et al, 2017; Leithner et al, 2016; Andrew and Insall, 2007). Once a migratory path has been selected along a dominant explorative cell front, large cytoplasmic parts from the other branches (‘losing’ cell fronts) have to be retracted. This necessitates substantial repositioning of large cytoplasmic parts from the retracting cell front, which may include organelles (Fig. 2D). Indeed, long-distance repositioning events of the nucleus from ‘losing’ into ‘winning’ protrusions occur during the migration of dendritic cells, T cells, and the amoebae Dictyostelium discoideum (Kroll et al, 2023). This long-distance repositioning of the nucleus is efficiently mediated by actomyosin contractility, providing migrating cells the flexibility to efficiently adjust their path and intracellular content towards an emerging guidance cue, such as when chemotactic cues overrule mechanical pore size cues (Kroll et al, 2023). While many immune cells like T cells and dendritic cells migrate with an amoeboid mode that is characterised by fast velocities, low-adhesiveness to the substrate, and dynamic cell shape changes, other immune cells like macrophages (Paterson and Lämmermann, 2021) and mast cells (Kaltenbach et al, 2023) employ a more mesenchymal migration mode that is slower and characterised by stronger adhesions to the substrate (Kameritsch and Renkawitz, 2020). Nevertheless, also macrophages are highly branched cells, thereby challenging organelle positioning, a concept that remains largely unexplored in these cells. Yet, recent findings showed that macrophages receive and phagocytose dysfunctional mitochondria during the intercellular transfer from stressed cardiomyocytes, which release mitochondria within membrane-surrounded particles (Nicolás-Ávila et al, 2020).
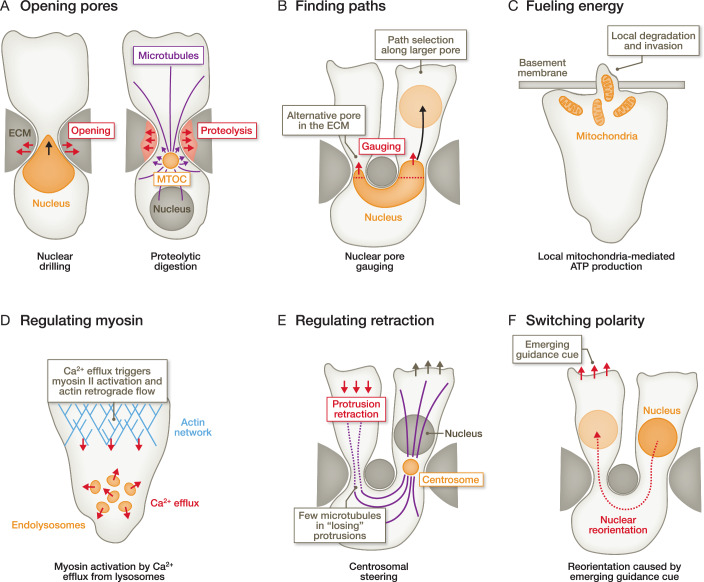
Figure 2. Functions of organelle positioning during cell migration.
(A) Motile cells can employ their nucleus (orange) as a wedge or driller to open tight spaces in their local extracellular environment (ECM) in a non-proteolytic manner (left). Alternatively, cells can use the microtubule-organising centre (MTOC; which is often the centrosome) as a platform that transports vesicles loaded with proteases via microtubules (purple) to the narrow gap in the ECM, opening it in a proteolytic manner (right). Both mechanisms lead to the opening of ECM pores and thereby facilitate cellular and nuclear translocation. (B) To avoid translocation through narrow ECM gaps, cells can use their nucleus as a mechanical gauge, probing for the sizes of close-by pores, and thereby selecting larger pores to enable their migration along the path of least resistance. (C) When cells have to translocate through extremely dense and narrow ECM-like basement membranes, they can position their mitochondria close to the invasion site as a platform for local ATP production, thereby fuelling energy into the invasion process. (D) Ca2+ efflux from endo-lysosomes through TRPML1 channels regulates the activity of myosin II and, thus actomyosin contractility in motile cells. (E) Correlative data show that protrusions can turn into ‘winning’ protrusions once the centrosome localises into the protrusion, likely by stabilising the microtubule cytoskeleton within the ‘winning’ protrusion, whereas microtubule filaments in retracting protrusions without the centrosome are disassembled. (F) Many fast-migrating cells explore their local microenvironment with different cell fronts simultaneously. Once they decide on the cell path along the ‘winning’ protrusion, the remaining cell fronts have to be retracted, and the organelles within the retracting protrusions have to be repositioned. This mechanism allows cells to readjust their path and their organelle positioning towards emerging guidance cues.
Challenges to position organelles in highly dynamic cells
Additional challenges for organelle positioning arise when cells transition between different states, such as during the transition between cell cycle or cell polarity phases. For instance, during cell budding of the yeast Saccharomyces cerevisiae, mitochondria, peroxisomes, and the ER are transported by the motor myosin V along actin cables into the bud that will develop into the new daughter cell (Knoblach and Rachubinski, 2015), enabling organelle inheritance. Furthermore, dividing mammalian cells typically acquire a round shape during mitosis (Lancaster et al, 2013), necessitating the proper distribution and positioning of organelles in the individual daughter cells (as reviewed in e.g., Carlton et al, 2020).
Independent of the cell cycle, a challenge for organelle positioning arises when cells are not stationary but motile, requiring the coordination of organelle positioning and cellular polarity along the migratory path. Directionally migrating cells establish a defined front-rear polarity, characterised by a protrusive front that generates force through actin polymerisation (Fig. 1). The protrusive front contains polymerising actin filaments, typically organised in branched actin networks nucleated by Arp2/3 (Fig. 1). Due to the high actin density in the protrusive front and the backward flow of actin towards the cell centre—which is most prominent in cells that are only low or even non-adhesive (Paluch et al, 2016; Maiuri et al, 2015; Renkawitz and Sixt, 2010; Renkawitz et al, 2009)—organelles are relatively rarely localised in the immediate protrusive cell front, especially during high-velocity movements. Yet, one can find organelles closely behind the protrusive cell front. For instance, fast-moving amoeboid cells, such as mammalian T cells, dendritic cells, and single-cell amoeba, position their nucleus directly behind the protrusive front (Renkawitz et al, 2019; Ishikawa-Ankerhold et al, 2022). This forward positioning of the nucleus is mediated by actomyosin contractility (Kroll et al, 2023) and is best described in relation to the centrosome, which typically localises to the cell centre of motile cells and behind the nucleus of fast-moving amoeboid cells (Fig. 1). In contrast, slower migrating mesenchymal cells position their nucleus behind the centrosome towards the cell rear (Gundersen and Worman, 2013; Dupin et al, 2011).
Prominent examples of rearward nuclear positioning include epithelial cells (Tsai and Meyer, 2012), astrocytes placed on micropatterns (Dupin et al, 2009), immortalised retinal pigment epithelial (RPE) cells on fibronectin-coated one-dimensional lines (Nastały et al, 2020), and motile fibroblasts moving into cell-free wounds (Luxton et al, 2010). These observations lead to the concept that more adhesive mesenchymal cells position their nucleus rearward of the centrosome, whereas low-adhesive amoeboid cells position their nucleus in front of the centrosome. It is important, however, to note that nuclear positioning may also be regulated by the microenvironment, as we will discuss below in the section ‘Plasticity in Organelle Positioning’, explaining exceptions to this rule.
Although the positioning principles of the nucleus and centrosome are increasingly well understood in various model systems, the positioning principles of other cellular organelles remain less broadly explored in motile cells. Recently, the ER in migrating fibroblasts has been shown to indirectly regulate nuclear positioning by asymmetrically decorating actin fibres when cells are moving on an adhesive surface: ventral stress fibres are decorated by the ER, preventing the coupling to the nucleus, while non-decorated dorsal actin fibres connect to the nucleus to mediate its movement (Janota et al, 2022). Notably, mitochondria are known to be moved and positioned by forces from the actin- and the microtubule cytoskeletons (López‐Doménech et al, 2018) and have recently been shown to be positioned to the protrusive cell front of C. elegans anchor cells that breach the basement membrane (Kelley et al, 2019). In contrast, mammalian T cells migrating on coated 2D substrates localise their mitochondria to the cell rear in a microtubule-dependent manner (Campello et al, 2006). Whether these differences in mitochondrial localisation are due to mechanistic differences in different cell types, or whether they are caused by different properties of the microenvironment remains to be investigated.
Challenges to position organelles that exist several times
Cells face the additional challenge of positioning multiple numbers of the same organelle. This aspect is naturally the case for some organelles that exist in multiple numbers within a single cell, including mitochondria and lysosomes. Clustering of these multiple organelle numbers to a specific intracellular location can simply be achieved when they are transported by the same molecular machinery, such as for lysosome vesicles that are moved along microtubule tracks by dynein motors towards their minus ends at the MTOC (Matteoni and Kreis, 1987).
Conceptually more challenging is the proper distribution of organelles to different intracellular localisation in a spaced manner: for example, in the multinucleated muscle, nuclei are accurately and uniformly distributed and spaced within the syncytium but are enriched underneath the neuromuscular junction (Cadot et al, 2015; Ghasemizadeh et al, 2021; Roman et al, 2021), potentially facilitating the local protein translation of specialised mRNAs. Also, during exercise-induced muscle repair, nuclei move towards the damage site to deliver mRNA (Roman et al, 2021). Notably, muscle cells also contain large numbers of mitochondria. The total amount of the mitochondrial network appears to correlate with the ATP demand of the specific muscle type, and the mitochondrial morphology is influenced by the muscle architecture (Katti et al, 2022; Avellaneda et al, 2021; Mishra et al, 2015). Another challenge arises when organelles present in several numbers are prone to fusion. For example, membrane-less organelles, in particular biomolecular condensates, can fuse and thus may actively be spatially segregated to keep them apart, as has been recently shown for nuclear speckles and paraspeckles (Takakuwa et al, 2023).
In addition to these naturally occurring examples, some cell types, such as certain cancer cells (Jaiswal and Singh 2021) and occasionally dendritic cells (Weier et al, 2022), possess an amplified number of centrosomes. These multiple centrosomes typically cluster to enable bipolar cell division (Marthiens et al, 2012). Thus, when multiple numbers of the same organelle are present within a cell, additional mechanisms are needed to spatially arrange organelles by clustering, distribution, or segregation. In summary, organelles are frequently actively positioned, and we will next discuss their functional roles.
Functional roles of asymmetric organelle positioning
Organelles can be asymmetrically distributed, but does this merely follow the asymmetry of the cytoskeleton, or does it serve specific functions at distinct intracellular locations?
Functions of nuclear positioning in motile and non-motile cells
Substantial evidence supporting active organelle positioning for functionality derives from the discovery of specific linker proteins physically coupling organelles to the cytoskeleton, such as the LINC (linker of nucleoskeleton and cytoskeleton) complex that spans the inner and outer nuclear membrane, connecting the nucleus to the actin and microtubule cytoskeletons (Sosa et al, 2012; Starr and Han, 2002). Notably, genetic mutations in LINC complex proteins like nesprin-1, nesprin-2, and nesprin-4 have been linked to human diseases like cardiomyopathy and muscular dystrophy (Kalukula et al, 2022). Moreover, mutations in proteins responsible for moving the nucleus during interkinetic nuclear migration in the central nervous system, such as the dynein regulator Lis-1 and the microtubule-regulator doublecortin, cause lissencephaly in humans, characterised by a smooth brain surface and severe psychomotor retardation (Markus et al, 2020). These examples of nuclear mispositioning in disease underscore the functional importance of accurate nuclear positioning (Gundersen and Worman, 2013) and nuclear mechanics (Zwerger et al, 2011).
In motile cells, precise organelle positioning plays a crucial functional role in localised chemical and physical activities. When a cell migrates through a tissue microenvironment, it encounters pores in the ECM that are frequently smaller in diameter than the nucleus (Stoitzner et al, 2002; Starborg et al, 2008; Wolf et al, 2009; Weigelin et al, 2012; Kameritsch and Renkawitz, 2020). Due to the size and stiffness of the nucleus (Kalukula et al, 2022), cellular translocation through pores substantially smaller than the nucleus causes migration slowdown as the nucleus squeezes to fit through the ECM pore (Calero-Cuenca et al, 2018; McGregor et al, 2016; Thiam et al, 2016; Harada et al, 2014; Wolf et al, 2013). Thus, the nucleus acts as a bottleneck for cell migration (Wolf et al, 2013). However, the nucleus can also serve as a ‘wedge or driller’ to open ECM pores (Fig. 2A), as demonstrated in mammalian T cells translocating through an epithelial layer and Drosophila border cells migrating in the fly ovary (Penfield and Montell, 2022; Barzilai et al, 2017). Moreover, the positioning of the nucleus forward of the centrosome towards the protrusive front allows immune cells to gauge neighbouring pore sizes in the ECM (Renkawitz et al, 2019). This mechanical function of the nucleus enables immune cells to circumvent extremely narrow ECM pores (Fig. 2B), preventing the slowdown of motility (Renkawitz et al, 2019) and potentially reducing nuclear damage caused by extreme nuclear squeezing through pores (Denais et al, 2016; Raab et al, 2016). While these different functions of the nucleus during cell motility have been studied individually, they likely work in conjunction: when migrating cells encounter multiple ECM pores in front of them along their trafficking paths, the nucleus can act as a mechanical gauge for the different pores to choose the path of least resistance; but cells can also decide for narrow pores along strong chemotactic signals that can override the dismissive mechanical pore size cue (Kroll et al, 2023), which may be facilitated by the opening of the pore by nuclear wedging or proteolytic digestions.
Functions of centrosome, lysosome, and mitochondria positioning in motile cells
Different types of motile cells likely employ these physical functions of nuclear positioning to varying extents: For instance, amoeboid migrating immune cells constantly patrol the tissue without generating tissue damage along their extensive trafficking paths. This is achieved by migration mechanisms, which typically do not include proteolytic digestion of the extracellular matrix, and makes pore size gauging by the nucleus an attractive strategy (Fig. 2B), which is facilitated by frontward positioning of the nucleus (Renkawitz et al, 2019). In contrast, mesenchymal cells, which can be highly proteolytically active, open ECM pores in dense microenvironments by secreting proteases that digest ECM components (Fig. 2A), such as by actin-based invadopodia enriched with ECM degrading enzymes (Monteiro et al, 2013). Notably, at least some cancer cells store the matrix metalloproteinase MT1-MMP around their centrosome and release it upon encountering narrow pores, facilitating the rearward-positioned nucleus to move through the ECM (Infante et al, 2018). Hence, the relative positioning of the centrosome is important for its function as a local protease secretion site and for the function of the nucleus as a mechanical gauge. Notably, while the positioning of the nucleus can dynamically change within a cell (see below), the centrosome typically maintains its position at the cell centre, as shown in motile fibroblasts (Gomes et al, 2005), Dictyostelium amoebae (Ishikawa-Ankerhold et al, 2022), and immune cells (Kroll et al, 2023). Notably, also the coordination of retraction appears to be mediated by organelle positioning: once the centrosome enters the winning protrusion of migrating dendritic cells, the remaining protrusions start to retract (Fig. 2E), mediated by myosin contractility induced by the RhoA exchange factor Lfc (Kopf et al, 2020). Together, these correlative data suggest that the positioning of the centrosome is important for its function as a steering organelle during fast amoeboid cell migration.
These examples of organelle positions in different cell types highlight the direct link between the positioning of organelles and the tasks of the respective cell type. Interestingly, in another example of cell migration through tight microenvironments, C.elegans anchor cells translocate through a basement membrane, using front-localised mitochondria as a local ATP production site (Fig. 2C), likely fuelling the protrusive front with ATP for extensive F-actin formation (Kelley et al, 2019; Garde et al, 2022). Thus, the asymmetric positioning of organelles can also function as a local production site of chemicals where they are most needed. Along these lines, reports identifying asymmetric positioning of RNAs towards the protrusive front of migrating cells may indicate local protein translation at subcellular sites of high protein turnover (Moriarty et al, 2022; Dermit et al, 2020; Moissoglu et al, 2020, 2019; Mardakheh et al, 2015; Mili et al, 2008). The forces driving the locomotion of motile cells are further regulated by lysosomes. Studies in amoeboid migrating dendritic cells, T cells, and Dictyostelium amoeba identified the release of Ca2+ from lysosomes via the channel TRPML1 (transient receptor potential cation channel, mucolipin subfamily, member 1) as a regulator of actin organisation and myosin II activity, and thus migration velocity (Bretou et al, 2017; Dehio et al, 2024).
In summary, active organelle positioning is functionally critical for chemical as well as mechanical functions in both motile and non-motile cells.
Forces and anchors for organelle positioning
In this section, we will discuss the principles and molecular mechanisms of how organelles are moved to their specific intracellular sites to solve the above-mentioned challenges. In particular, we discuss the forces and anchoring strategies that move and position organelles.
Cytoskeletal tracks
Many organelles are actively positioned via directly targeted forces, such as pulling and pushing forces (Marks and Petrie, 2022). Most intuitively, cytoskeletal filaments such as microtubules and actin fibres serve as tracks for their respective motor proteins, which are themselves connected via adaptor proteins to various cargos, including organelles (Fig. 3A). For instance, kinesins and dyneins move along microtubules towards their microtubule-plus and -minus ends, respectively (Hirokawa, 1998). Thereby, these motors transport and position organelle cargos, such as dynein positioning the Golgi (Yadav and Linstedt, 2011). Hence, the organisation of the microtubule cytoskeleton serves as a platform for organelle distribution and positioning (Akhmanova and Kapitein, 2022). The detailed mechanisms and functions of motor-based transport along microtubule tracks have been reviewed elsewhere, especially in the context of organelle transport within neurons (Cason and Holzbaur, 2022). Instead, we here focus on recently emerging principles of organelle positioning independent of motor-based movement along cytoskeletal tracks.
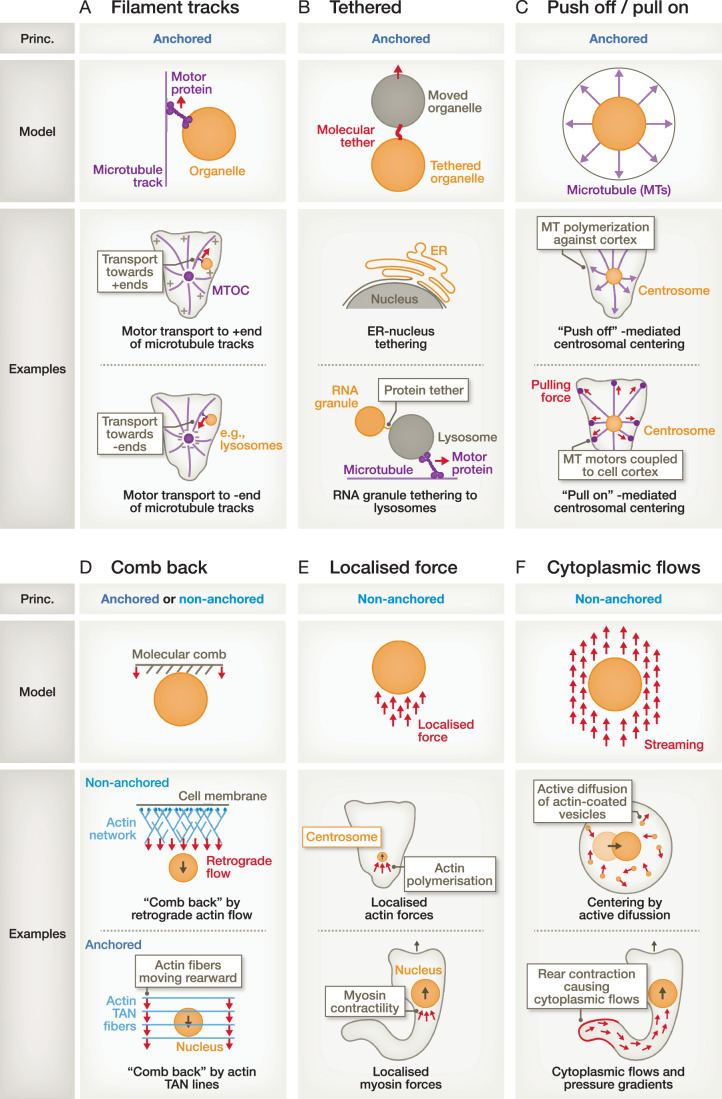
Figure 3. Forces and mechanisms to move and position organelles.
(A) Microtubule filaments provide tracks for motor-based transport of organelles towards the plus- and minus-ends of microtubules. (B) Organelles can be indirectly moved and positioned, when they are directly connected (‘tethered’) to other organelles that move, like in the case of the direct connection between the nucleus and the ER, or by a protein tether connecting RNA granules to the lysosome. (C) Radially growing microtubules (MTs) can serve as a centring structure by pushing their plus ends against the cell cortex and/or by pulling them towards the cell periphery by motors that are coupled to the cell cortex. (D) The actin cytoskeleton can serve as a ‘molecular comb’, combing back intracellular components like organelles. This mechanism is active in fibroblasts that start to migrate in cell-free areas, in which actin fibres (called TAN lines) that are attached to the nucleus via the LINC complex, comb back the nucleus towards the cell rear. Hypothetically, also the rearward-directed retrograde flow of branched actin networks could comb back cellular organelles from the cell front towards the cell body. Notably, such a mechanism would function without directly anchoring the organelles towards the flowing actin network. (E) Similarly, a localised force at the back of an organelle could push the organelle forward without the need for anchoring structures, as it may likely be the case for nuclear positioning by rearward localised myosin activity or centrosome positioning by rearward localised actin polymerisation. (F) Generally, organelles may be moved by advection, using intracellular fluid streaming in an anchorage-independent manner, which may, for example, be caused by the contraction of the cell rear.
Tethering organelles
Organelles can also be displaced by other intracellular structures. In particular, larger and stiffer organelles like the nucleus are intuitively able to physically displace other organelles, but also smaller intracellular structures like fat-filled lipid droplets can deform the nucleus (Ivanovska et al, 2023) and even push the nucleus away towards the cellular periphery (Gundersen and Worman, 2013). More actively, organelles could be indirectly moved by being connected to other, transported organelles by inter-organelle contact sites (Guo et al, 2018), such as the direct membrane connection between the ER and the nucleus (Fig. 3B) and contact sites between the ER and endosomes, lysosomes, peroxisomes, and mitochondria (Phillips and Voeltz, 2016; Raiborg et al, 2015). In epithelial sheets, cell-cell junctions provide another way to tether organelles, as revealed by the association of the ER to desmosomes, a specialised and adhesive intercellular junction structure (Bharathan et al, 2023). Notably, also the existence of contact sites between membrane-less ribonucleoprotein (RNP) granules and the membrane-surrounded ER has been recently revealed, which regulate the fission of the membrane-less granules (Lee et al, 2020). In addition to tethering mechanisms by inter-organelle contact sites, membrane-less RNA granules have recently been observed to hitchhike lysosomal transport along microtubule tracks, using the protein annexin A11 as a tether between the membrane-less RNA granule and membrane-enclosed lysosome, mediated by a lysosomal membrane binding domain and a low-complexity domain that incorporates into RNA granules (Liao et al, 2019) (Fig. 3B).
Anchoring organelles
To couple the force that has been generated by the cytoskeleton to the organelle, classical organelle movement strategies typically involve anchoring proteins that are located on the outer surface of the organelle and interact with the cytoskeleton. Anchoring is, for example, required for nuclear movement in diverse cell types. The well-studied LINC complex mediates the anchorage of the nucleus to the cytoskeleton, being composed of SUN proteins in the inner nuclear membrane and KASH/nesprin proteins in the outer nuclear membrane that interact together and thereby bridge the perinuclear space (Janota et al, 2017; Ungricht and Kutay, 2017; Sosa et al, 2012). To couple to the cytoskeleton, KASH domain proteins interact with microtubules, actin filaments, and intermediate filaments, often mediated by adaptors, such as the microtubule motor proteins dynein and kinesin. Yet, the nucleus can also couple to the microtubule cytoskeleton in a LINC-independent manner via nuclear pore complexes (Janota et al, 2017). A prominent example of nuclear movement by anchorage to the cytoskeleton occurs in fibroblasts migrating towards the cell-free space of a wound scratch assay: the nucleus is moved towards the cell rear by actin fibres called TAN (transmembrane actin-associated nuclear) lines that move rearward and transmit the force via the LINC complex directly to the nucleus (Luxton et al, 2010). Thus, the cytoskeleton is here not employed as a railway track for motors but as a ‘comb’, combing back the nucleus towards the rear (Fig. 3D). As the rearward-moving TAN lines are anchored to the nucleus and not to the centrosome, this mechanism ensures the rearward movement of the nucleus while the centrosome maintains its relative position. Whether this TAN line-based mechanism also exists in 3D tissue microenvironments remains to be elucidated.
Fibroblasts migrating in highly confining and crosslinked ECM have been shown to employ a lobopodial migration mode (Petrie and Yamada, 2015; Petrie et al, 2014), in which fibroblasts adhere to the ECM and generate actomyosin-based pulling forces onto the nucleus, as demonstrated by the observation of a nuclear rearward drift upon local inhibition of actomyosin contractility at the cellular front (Petrie et al, 2014). Nuclear anchorage in lobopodial fibroblasts is achieved by nesprin-3 and vimentin, leading to a piston-like movement of the nucleus through the cytoplasm, pressurising the front and thereby generating bleb-like protrusions. Furthermore, it has been shown that the LINC complex protein nesprin-2 accumulates in a polarised manner at the tip of the nucleus when fibroblasts translocate through narrow pores, indicating forces from the front pulling the nucleus through narrow microenvironmental gaps (Davidson et al, 2020). Additionally, other adaptor proteins have been described, such as amphiphysin 2 (BIN1), which acts as a linker between the cytoskeleton and the nucleus via nesprins (Falcone et al, 2014; D’Alessandro et al, 2015).
While these examples highlight anchorage strategies of membrane-surrounded organelles, also organelles without a surrounding membrane can be anchored. The best example is the centrosome, as positioning organelles by microtubules (Schmidt and Stehbens, 2024) or their motor proteins require that microtubules are anchored to the centrosome. Thereby, the generated force does not lead to the displacement of the microtubule itself, but of the cargo. At the centrosome, microtubule minus-ends are anchored by anchoring proteins like ninein and γ-TuRC that provide linkage to the stable centriolar pair at the core of the centrosome (Akhmanova and Kapitein, 2022; Delgehyr et al, 2005; Bornens, 2002; Piel et al, 2000). Thus, anchorage does not occur at the outer surface but at the inner core of the organelle.
Centring forces
The membrane-less centrosome often localises approximately to the cell centre. But how does it find the centre? One mechanism of centring employs the activity of the centrosome as a microtubule-organising centre (MTOC): by nucleating microtubules from the centrosome, microtubules can grow until they hit a resistance, such as the cell’s cortex or plasma membrane. Thereby, these pushing forces can centre the centrosome (Fig. 3C), provided the microtubules are sufficiently stiff to tolerate the necessary pushing force before they buckle (Wühr et al, 2009), as suggested, for example, in fission yeast (Tran et al, 2001). Alternatively, motors within the cell’s cortex could pull on microtubules, when they are attached to limited sites in the cell’s cortex, thereby centring the centrosome by pulling forces (Fig. 3C) (Grill and Hyman, 2005), a mechanism demonstrated to be active in C. elegans (Grill et al, 2001) and S. cerevisiae (Adames and Cooper, 2000). Yet, in very large cells like in Xenopus eggs, these mechanisms are unlikely to be functional as microtubules are too short to reach the cellular periphery at all cell sides (Wühr et al, 2009). However, the centrosome still finds the cell centre by mechanisms involving the actin cytoskeleton as a boundary (as we will discuss in the next paragraph) and anchorage-independent mechanisms based on intracellular dynamics and diffusion (as we will discuss in the section ‘anchorage-independent organelle positioning’). This caused substantial discussions about the mechanisms of centrosome centring, probably with different mechanisms functioning in different model systems, as reviewed previously (Deshpande and Telley, 2021).
The interaction of microtubules with other organelles, such as the nucleus, adds complexity to interpreting centrosome centreing mechanisms. Recent findings using enucleated cytoplasts attached to defined micropatterned shapes showed that the centrosome typically localises to the cell centre when the cell has a round shape. However, its position can be off-centred when the cell has an asymmetric shape and asymmetric distribution of actin, where the actin network appears to act as a boundary for microtubule growth (Jimenez et al, 2021). In vitro reconstruction with purified actin and microtubules support this mechanism, showing that an asymmetrical actin cortex leads to an asymmetrical MTOC position (Yamamoto et al, 2022). Together, these data suggest that the architecture of the actin cytoskeleton itself contributes to centrosome centreing. It will be interesting to explore such an actin-based mechanism in different cell types with different architectures and properties of the actin network, such as in lowly or non-adhesive cells like immune cells that typically do not possess thick and stable actin fibres but dynamic actin retrograde flows.
Anchorage-independent organelle positioning
As described above, many models of organelle positioning are based on movement via anchoring linkers to the cytoskeleton. These anchoring mechanisms are also employed to immobilise the organelle to counteract diffusion. However, substantial evidence suggests that cells may employ mechanisms for organelle positioning that are independent of anchorage. In Drosophila nurse cells, actin cables polymerise from the plasma membrane towards the nucleus (Huelsmann et al, 2013), generating a pushing force that does not necessarily require anchorage to the moved object. Conceptually, the to-be-moved organelle can also be itself not anchored but carry an anchored nucleator for actin polymerisation. This principle of an anchored nucleator is employed by the intracellular pathogen Listeria (Lambrechts et al, 2008), in which the Listeria surface-anchored protein ActA directly activates the Arp2/3 complex of the host cell, generating a forward-pushing actin comet behind the pathogen (Welch et al, 1998), and the intracellular bacterium Rickettsia (Haglund et al, 2010), where formin- and Arp2/3-based nucleation generates the comet-like movement. Notably, this principle has recently been shown to also be employed by organelles, as mitochondria use twin comet tails, induced by an unknown nucleator, to drive their movement in a comet-like manner (Moore et al, 2021).
In addition to these mechanisms, increasing evidence suggests that organelles are also moved by advection—which is the transport of material by fluid flow (Illukkumbura et al, 2020)—using intracellular fluid streaming as an anchorage-independent movement strategy. Indeed, experiments in the 1970s showed that plasma membrane receptors accumulate at the cellular rear of migrating lymphocytes when externally crosslinked by antibodies (Taylor et al, 1971; Bray and White, 1988). This process, known as ‘antigen capping’ or ‘receptor capping’, drags crosslinked transmembrane receptors by the retrograde actin flow towards the cellular back. But do intracellular flows and diffusion also impact the positioning of organelles? Interestingly, in mouse oocytes which lack centrosomes, the positioning of the nucleus is driven by the diffusion of actin-coated vesicles that generate a propulsive force to move the nucleus to the cell centre in an anchorage-independent manner (Almonacid et al, 2015). Further, there is increasing evidence that a localised contractile activity of myosin may lead to the movement of organelles without requiring the anchorage of the organelle. In muscle cells, individual nuclei in the syncytium can be moved by actomyosin activity even when the LINC complex is not functional (Roman et al, 2017), while the accurate spacing between the individual nuclei requires the LINC complex (Roman et al, 2017). Furthermore, myosin II enriches behind the nucleus of migrating dendritic cells when the nucleus has to be repositioned from a ‘losing’ towards a ‘winning’ protrusion (Kroll et al, 2023), suggesting a mechanism in which a rearward localised contractility pushes the nucleus forward (Fig. 3E). Horizontal cells in the zebrafish retina move their nucleus with rearward localised actin activity and frequent nuclear shape changes (Amini et al, 2022). Similarly, nuclear movement in interneurons is accompanied by actin localised behind the nucleus and depends on myosin activity (Silva et al, 2018). Moreover, recent data showed that some cells, like HT1080 fibrosarcoma cells, generate pushing forces from the cell rear by contracting the cell’s rear cortex, leading to high cytosolic pressures in the back to push the nucleus through narrow gaps (Keys et al, 2024). Generally, a rearward localised contractility mechanism likely causes streaming within the cytoplasm, which cells can employ to move their organelles by advection (Fig. 3F). This mechanism will likely function particularly well on large organelle cargos within cells that are confined by the ECM, providing a large organelle surface being exposed to the streaming force and preventing the streaming force from bypassing the organelle on its sides, causing an intracellular pressure difference in the cytoplasm before and behind the organelle. This model is supported by findings in mouse oocytes, in which the diffusion of actin-coated vesicles leads to centring movement of the large nucleus and large inert oil droplets but not of smaller fluorescent particles (Colin et al, 2020).
Interestingly, the mechanisms of anchorage-independent organelle movement show similarities with the mechanisms driving the movement of amoeboid cells. Hallmarks of amoeboid cells are rearward localised myosin activity, their constantly changing cell shape, and their movement without tightly anchoring to the extracellular microenvironment (Yamada and Sixt, 2019; Paluch et al, 2016). In analogy and intracellularly, the nucleus can be moved by a rearward localised contraction, presumably without directly anchoring the nucleus to the cytoskeleton (Kroll et al, 2023; Amini et al, 2022; Keys et al, 2024; Roman et al, 2017). Such an anchorage-independent mechanism has the advantage of functioning in a fast and flexible manner, rapidly pushing the nucleus forward by rearward contractility, without the need to constantly dismantle and reassemble anchorage structures between the nucleus and the cytoskeleton. Thereby, this principle may be particularly well-suited for highly dynamic cells such as immune cells, amoeba, and some cancer cell types, where the cell shape constantly changes, and organelles like the nucleus have to be constantly moved to new intracellular positions. For instance, immune cells like dendritic cells, T cells, and the single-cell amoeba Dictyostelium frequently adapt their nuclear position between different alternative cell fronts when migrating in 3D microenvironments (Kroll et al, 2023). As the nucleus has to travel for long intracellular distances from a ‘losing’ towards a ‘winning’ protrusion (Fig. 2F), which is associated with two rapid switches in the nucleus-centrosome polarity (Kroll et al, 2023), the anchorage-independent nuclear movement would provide the necessary dynamics and velocity for this nuclear behaviour. Thus, it might not be surprising that neutrophil granulocytes, a white blood cell type of the innate immune system that moves in an amoeboid manner with high velocities (Nourshargh et al, 2010), have been described to be LINC-less (Olins et al, 2009), possibly suggesting a missing tight linkage between the nucleus and the cytoskeleton. Notably, anchorage-independent mechanisms for intracellular organelle positioning would likely also alter the degree of mechanical stress that is exerted onto the nucleus. Considering that gene expression is regulated by the mechanical properties of the microenvironment via the linkage of cytoskeleton forces to the nucleus (Kechagia et al, 2019), an anchorage-independent mechanism of nuclear movement may have important implications for gene expression as it may relieve mechanical stress from the nucleus and thereby reduce events of mechanically triggered nuclear envelope rupture. Nevertheless, whether there is indeed no anchorage or whether there are alternative anchorage mechanisms in fast-migrating cells has to be experimentally addressed. In particular, the proximity between the centrosome and the nucleus in amoeboid migrating cells suggests a direct linkage between these organelles, presumably via the microtubule cytoskeleton. Yet, even if the centrosome is tethered to the nucleus, the nucleus could still be moved by the above proposed anchorage-independent mechanism, and thereby would rather drag along the centrosome with its movement. Such a strategy may allow coupling between the nucleus and the centrosome to ensure that both organelles move relatively simultaneously into the cellular branch along the ‘winning’ cellular path while allowing flexible movement of the organelles without tight anchoring structures. (Lämmermann and Sixt, 2009; Moreau et al, 2020; Kameritsch and Renkawitz, 2020).
Current knowledge of anchorage-independent organelle positioning mechanisms derives from studies on nuclear positioning. Thus, investigations on this mechanism for the positioning of other organelles, including organelles of different sizes and interactions with the cytoskeleton, will characterise the utilisation of anchorage-independent mechanisms to position organelles. Generally, it is important to note that the different types of forces and anchorage strategies described in this section may also oppose each other, such as myosin forces opposing microtubule forces (Lin et al, 2016; Kapitein et al, 2013; McIntosh et al, 2015), until the final organelle localisation is established by a force equilibrium. Similarly, forces can act in parallel to each other in a complementary manner, such as for the establishment and maintenance of the large and dynamic ER network, which is moved by motors along the microtubule network, moved by coupling to growing microtubule tips, and moved by its contact to other organelles such as lysosomes (Lu et al, 2020; Zajac et al, 2013; Friedman et al, 2010; Waterman-Storer et al, 1995; Lee and Chen, 1988). Considering that anchorage-dependent mechanisms likely achieve a more precise intracellular position of an organelle than a more diffuse anchorage-independent pushing mechanism towards the cell front, it will be interesting to further observe how these two principles may act in a complementary manner to achieve organelle positioning in a fast and flexible but also robust manner.
Plasticity in organelle positioning
Importantly, the specific positions of organelles described earlier are not necessarily fixed. They can change according to the cellular activity and the properties of the extracellular microenvironment. An example of organelle repositioning due to changes in cellular activity derives from T cells: T cells typically migrate with a nucleus-forward and centrosome-rearward configuration, but this configuration reverts when T cells engage in an immune synapse with an antigen-presenting cell or target cells, causing the centrosome to relocate towards the immune synapse (Pineau et al, 2023; Krummel and Macara, 2006). This reorientation depends on dynein, CDC42, and formins (Stowers et al, 1995; Combs et al, 2006; Gomez et al, 2007), and is required for T-cell responses and target cell killing (Kuhn and Poenie, 2002; Stinchcombe et al, 2006). Also, dendritic cells reorganise their actomyosin architecture and organelle positioning when they mature upon encounter with danger signals such as bacterial cell wall components like lipopolysaccharides (LPS). Upon maturation, the actin cytoskeleton reorganises from a preferential frontward to a rearward localisation (Vargas et al, 2016), which is regulated by the release of calcium from rearward-located lysosomes through Trpml1 channels (Bretou et al, 2017).
Apart from the dynamic repositioning of organelles for functional adaptation, properties of the microenvironment can impact the spatial positioning of organelles. This influence of the microenvironment has been demonstrated for the positioning of the centrosome, using adhesive two-dimensional (2D) patterns of different shapes (Théry et al, 2006; Hale et al, 2011). Further, investigations of Golgi positioning using adhesive one-dimensional (1D) lines, where cells typically behave similarly to 3D microenvironments but differently than on 2D surfaces, showed a preferential localisation of the Golgi behind the nucleus on 1D lines but in front of the nucleus on 2D surfaces (Pouthas et al, 2008). Moreover, imaging of mitochondria during the migration of breast cancer cells through tunnels within collagen matrices showed a more preferential localisation of mitochondria towards the cell front in more confining tunnels (Mosier et al, 2023).
Considering that microenvironmental adhesiveness and the degree of cellular confinement directly regulate the degree of myosin-mediated contractility (Lomakin et al, 2020; Liu et al, 2015; Lämmermann and Sixt, 2009), it makes intuitive sense that organelle distribution is influenced by the microenvironment. This plasticity is likely particularly important for highly motile cells that encounter various microenvironments on their trafficking routes. It is attractive to speculate that organelle movement may be largely driven by anchorage-independent mechanisms when fast-migrating cells move in loose environments. However, when they encounter denser environments or tight obstacles, they may switch to an anchorage-dependent pulling mechanism, or even use pushing and pulling by anchorage-dependent and -independent mechanisms simultaneously to generate sufficient forces to overcome tight barriers. Thus, the above-described forces are non-mutually exclusive but likely support each other to adapt to the physiological situation. Given that centrosome positioning also changes with the contractility of the cell type (Jimenez et al, 2021), the findings that the nucleus senses the degree of cellular confinement, leading to increased cellular contractility in highly confining environments (Lomakin et al, 2020; Venturini et al, 2020), also directly suggests that organelle positioning depends on the degree of cellular confinement. Together, these findings suggest that plasticity in organelle positioning caused by the microenvironment is mostly dictated by the degree of (i) organelle anchorage strength, (ii) cellular contractility, (iii) and microenvironmental confinement.
Conclusions
We are only at the beginning of understanding the mechanisms and functions of active organelle positioning and how they are influenced by the cell type, cell state, and the tissue microenvironment (see Box 1). While the positioning of the centrosome and the nucleus are increasingly well explored, it is tempting to speculate that many more organelles, including membrane-less organelles and biomolecular condensates, are actively positioned to either provide metabolites and cellular building blocks to the subcellular localisation where they are needed, and to counteract cytoplasmic mixing during dynamic morphological changes, or to minimise their exposure to mechanical stresses. Regarding the latter, it will be important to discover how organelles shield themselves to maintain their integrity while they are exposed to the forces that move and anchor them to specific cellular sites. Moreover, studying the positioning of membrane-less organelles may uncover novel principles for movement and anchorage, given the lack of possibility to anchor membrane-less organelles to the cytoskeleton via membrane-spanning proteins. Considering that an increasing number of reports identify the importance of proper spatial organelle arrangement for cellular functions, future research may also uncover additional roles of organelle positioning in human diseases.
Next to these conceptual gaps of knowledge, research on the underexplored topic of organelle positioning may well lead to surprises in the employed molecular mechanisms, such as findings showing that different cytoskeletal protein isoforms may have different roles in organelle positioning (Roman et al, 2017) and recent findings suggesting that the LINC complex protein ANC-1 in C- elegans is not only required for nuclear positioning but also for positioning of the ER, mitochondria, and lipid droplets (Hao et al, 2021; Fischer et al, 2022). Additionally, we are only at the beginning of understanding how asymmetric patterns of posttranslational modifications of cytoskeleton components like microtubules (Lavrsen et al, 2023; Janke and Magiera, 2020) influence organelle localisation, such as the recently identified role of polyglutamylated microtubules in ER and lysosome positioning (Zheng et al, 2022). From a conceptual perspective, current research on organelle positioning often focuses on how the cytoskeleton moves and anchors organelles. Yet, how the localisation of organelles to specific sites itself impacts the organisation of the cytoskeleton is less well explored and may uncover important reciprocal relationships between cytoskeletal architecture and organelle positioning. Along these lines, research visualising multiple organelles simultaneously will unravel reciprocal relationships between their localisation.
Recent methodological advances like the optogenetic control of organelle transport (Bergeijk et al, 2015, 2016), the precise control of microenvironmental properties (Théry et al, 2006; Lautenschläger and Piel, 2013; Garcia-Arcos et al, 2019; Kroll et al, 2022), the usage of extremely long and ramified cells like neurons (Tas et al, 2017) or spatiotemporally extremely dynamic cells like immune cells (Moreau et al, 2020; Clausen et al, 2022) as cellular models, the subcellular control of cytoskeletal activity (Meiring et al, 2022; Wittmann et al, 2020; Borowiak et al, 2015; Ballister et al, 2015), and the advances in computational analysis (Schmied et al, 2023; Berg et al, 2019) have the potential to uncover fundamental principles and their underlying mechanisms in organelle positioning. Studying organelle positioning in a quantitative high throughput manner across different cell types, organisms, stimuli, microenvironmental contexts, and diseases, could also reveal quantitatively whether some organelles maintain a rather fixed position that is conserved throughout different organisms, cell types, and contexts, or may more dynamically adapt to the cellular and physiological requirement. These approaches may also lead to the characterisation of whether organelle positioning evolved with cell motility strategies and the complexity of multicellular organisms (Brunet and Booth, 2023; Fritz-Laylin, 2020) and whether cancer cells or intracellular parasites, such as the obligate intracellular parasite Toxoplasma gondii, hijack mechanisms of precise, efficient, and robust organelle positioning.
Box 1 In need of answers.
-
(i)
How does the microenvironment regulate organelle positioning?
-
(ii)
Do membrane-less organelles utilise specific mechanistic principles for their active movement, positioning, and anchorage?
-
(iii)
Do organelles shield themselves to maintain their integrity while being exposed to intracellular positioning and anchoring forces?
-
(iv)
Do cancer cells or intracellular parasites hijack the principles of active organelle positioning to their benefit?
Acknowledgements
We thank all members of the laboratory of J.R. for the discussions and acknowledge the usage of ‘Grammarly’ for a final grammar check. This study was supported by the Peter Hans Hofschneider Professorship of the foundation ‘Stiftung Experimentelle Biomedizin’ (to J.R.), the LMU Institutional Strategy LMU-Excellent within the framework of the German Excellence Initiative (to J.R.), and the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation; CRC914 project A12, to J.R.).
Author contributions
Jörg Renkawitz: Conceptualisation; Writing—original draft; and Writing—review and editing. Janina Kroll: Writing—original draft and Writing—review and editing.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Disclosure and competing interests statement
The authors declare no competing interests.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Kapitein LC. Mechanisms of microtubule organization in differentiated animal cells. Nat Rev Mol Cell Bio. 2022;23:541–558. doi: 10.1038/s41580-022-00473-y. [DOI] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Bio. 2021;22:196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- Almonacid M, Ahmed WW, Bussonnier M, Mailly P, Betz T, Voituriez R, Gov NS, Verlhac M-H. Active diffusion positions the nucleus in mouse oocytes. Nature cell biology. 2015;17:470–479. doi: 10.1038/ncb3131. [DOI] [PubMed] [Google Scholar]
- Amini R, Bhatnagar A, Schlüßler R, Möllmert S, Guck J, Norden C. Amoeboid-like migration ensures correct horizontal cell layer formation in the developing vertebrate retina. Elife. 2022;11:e76408. doi: 10.7554/eLife.76408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- Avellaneda J, Rodier C, Daian F, Brouilly N, Rival T, Luis NM, Schnorrer F. Myofibril and mitochondria morphogenesis are coordinated by a mechanical feedback mechanism in muscle. Nat Commun. 2021;12:2091. doi: 10.1038/s41467-021-22058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo M, Baylies MK. Getting into position: nuclear movement in muscle cells. Trends Cell Biol. 2020;30:303–316. doi: 10.1016/j.tcb.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Ayloo S, Chenoweth DM, Lampson MA, Holzbaur ELF. Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Curr Biol. 2015;25:R407–R408. doi: 10.1016/j.cub.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai S, Yadav SK, Morrell S, Roncato F, Klein E, Stoler-Barak L, Golani O, Feigelson SW, Zemel A, Nourshargh S, et al. Leukocytes breach endothelial barriers by insertion of nuclear lobes and disassembly of endothelial actin filaments. Cell Rep. 2017;18:685–699. doi: 10.1016/j.celrep.2016.12.076. [DOI] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. ilastik: interactive machine learning for (bio)image analysis. Nat Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- Bergeijk P, van, Adrian M, Hoogenraad CC, Kapitein LC. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeijk P, van, Hoogenraad CC, Kapitein LC. Right time, right place: probing the functions of organelle positioning. Trends Cell Biol. 2016;26:121–134. doi: 10.1016/j.tcb.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Bharathan NK, Giang W, Hoffman CL, Aaron JS, Khuon S, Chew T-L, Preibisch S, Trautman ET, Heinrich L, Bogovic J, et al. Architecture and dynamics of a desmosome–endoplasmic reticulum complex. Nat Cell Biol. 2023;25:823–835. doi: 10.1038/s41556-023-01154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- Borowiak M, Nahaboo W, Reynders M, Nekolla K, Jalinot P, Hasserodt J, Rehberg M, Delattre M, Zahler S, Vollmar A, et al. Photoswitchable inhibitors of microtubule dynamics optically control mitosis and cell death. Cell. 2015;162:403–411. doi: 10.1016/j.cell.2015.06.049. [DOI] [PubMed] [Google Scholar]
- Bray D, White JG. Cortical flow in animal cells. Science. 1988;239:883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- Bretou M, Sáez PJ, Sanséau D, Maurin M, Lankar D, Chabaud M, Spampanato C, Malbec O, Barbier L, Muallem S, et al. Lysosome signaling controls the migration of dendritic cells. Sci Immunol. 2017;2:eaak9573. doi: 10.1126/sciimmunol.aak9573. [DOI] [PubMed] [Google Scholar]
- Brunet T, Booth DS. Cell polarity in the protist-to-animal transition. Curr Top Dev Biol. 2023;154:1–36. doi: 10.1016/bs.ctdb.2023.03.001. [DOI] [PubMed] [Google Scholar]
- Cabukusta B, Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic. 2018;19:761–769. doi: 10.1111/tra.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B, Gache V, Gomes ER. Moving and positioning the nucleus in skeletal muscle – one step at a time. Nucleus. 2015;6:373–381. doi: 10.1080/19491034.2015.1090073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Cuenca FJ, Janota CS, Gomes ER. Dealing with the nucleus during cell migration. Curr Opin Cell Biol. 2018;50:35–41. doi: 10.1016/j.ceb.2018.01.014. [DOI] [PubMed] [Google Scholar]
- Campello S, Lacalle RA, Bettella M, Mañes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Jones H, Eggert US. Membrane and organelle dynamics during cell division. Nat Rev Mol Cell Biol. 2020;21:151–166. doi: 10.1038/s41580-019-0208-1. [DOI] [PubMed] [Google Scholar]
- Cason SE, Holzbaur ELF. Selective motor activation in organelle transport along axons. Nat Rev Mol Cell Biol. 2022;23:699–714. doi: 10.1038/s41580-022-00491-w. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Poüs C. The golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Amon L, Backer RA, Berod L, Bopp T, Brand A, Burgdorf S, Chen L, Da M, Distler U, et al (2022) Guidelines for mouse and human DC functional assays. Eur J Immunol 53:e2249925 [DOI] [PubMed]
- Colin A, Letort G, Razin N, Almonacid M, Ahmed W, Betz T, Terret M-E, Gov NS, Voituriez R, Gueroui Z, et al. Active diffusion in oocytes nonspecifically centers large objects during prophase I and meiosis I. J Cell Biol. 2020;219:1667. doi: 10.1083/jcb.201908195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs J, Kim SJ, Tan S, Ligon LA, Holzbaur ELF, Kuhn J, Poenie M. Recruitment of dynein to the Jurkat immunological synapse. Proc Natl Acad Sci USA. 2006;103:14883–14888. doi: 10.1073/pnas.0600914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro M, Hnia K, Gache V, Koch C, Gavriilidis C, Rodriguez D, Nicot A-S, Romero NB, Schwab Y, Gomes E, et al. Amphiphysin 2 orchestrates nucleus positioning and shape by linking the nuclear envelope to the actin and microtubule cytoskeleton. Dev Cell. 2015;35:186–198. doi: 10.1016/j.devcel.2015.09.018. [DOI] [PubMed] [Google Scholar]
- Davidson PM, Battistella A, Déjardin T, Betz T, Plastino J, Borghi N, Cadot B, Sykes C. Nesprin‐2 accumulates at the front of the nucleus during confined cell migration. EMBO Rep. 2020;123:740. doi: 10.15252/embr.201949910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio P, Michard C, Yam-Puc JC, Líndez A-AMI, Fabre L, Schaefer T, Wymann MP, Okkenhaug K, Soldati T, Mehling M, et al (2024) An evolutionary-conserved VPS34-PIKfyve-TRPML1-Myosin II axis regulates the speed of amoeboid cell migration. Preprint at bioRxiv: 2024.01.22.575998
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, Lindert M, te, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermit M, Dodel M, Lee FCY, Azman MS, Schwenzer H, Jones JL, Blagden SP, Ule J, Mardakheh FK. Subcellular mRNA localization regulates ribosome biogenesis in migrating cells. Dev Cell. 2020;55:298–313.e10. doi: 10.1016/j.devcel.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande O, Telley IA (2021) Nuclear positioning during development: pushing, pulling and flowing. Semin Cell Dev Biol 120:10–21 [DOI] [PubMed]
- Driscoll MK, Welf ES, Jamieson AR, Dean KM, Isogai T, Fiolka R, Danuser G. Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat Methods. 2019;141:1–8. doi: 10.1038/s41592-019-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I, Dupin I, Etienne-Manneville S, Etienne-Manneville S. Nuclear positioning: mechanisms and functions. Int J Biochem Cell Biol. 2011;43:1698–1707. doi: 10.1016/j.biocel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Falcone S, Roman W, Hnia K, Gache V, Didier N, Lainé J, Auradé F, Marty I, Nishino I, Charlet‐Berguerand N, et al. N‐WASP is required for Amphiphysin‐2/BIN1‐dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol Med. 2014;6:1455–1475. doi: 10.15252/emmm.201404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer NC, Friedman V, Martinez-Reyes MA, Hao H, Chowdhury TA, Starr DA, Quinn CC. The ANC-1 (Nesprin-1/2) organelle-anchoring protein functions through mitochondria to polarize axon growth in response to SLT-1. PLoS Genet. 2022;18:e1010521. doi: 10.1371/journal.pgen.1010521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER–mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK. The evolution of animal cell motility. Curr Biol. 2020;30:R477–R482. doi: 10.1016/j.cub.2020.03.026. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Riel-Mehan M, Chen B-C, Lord SJ, Goddard TD, Ferrin TE, Nicholson-Dykstra SM, Higgs H, Johnson GT, Betzig E, et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. Elife. 2017;6:e26990. doi: 10.7554/eLife.26990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gache V, Gomes ER, Cadot B. Microtubule motors involved in nuclear movement during skeletal muscle differentiation. Mol Biol Cell. 2017;28:865–874. doi: 10.1091/mbc.e16-06-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arcos JM, Chabrier R, Deygas M, Nader G, Barbier L, Sáez PJ, Mathur A, Vargas P, Piel M. Reconstitution of cell migration at a glance. J Cell Sci. 2019;132:jcs225565. doi: 10.1242/jcs.225565. [DOI] [PubMed] [Google Scholar]
- Garde A, Kenny IW, Kelley LC, Chi Q, Mutlu AS, Wang MC, Sherwood DR. Localized glucose import, glycolytic processing, and mitochondria generate a focused ATP burst to power basement-membrane invasion. Dev Cell. 2022;57:732–749.e7. doi: 10.1016/j.devcel.2022.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemizadeh A, Christin E, Guiraud A, Couturier N, Abitbol M, Risson V, Girard E, Jagla C, Soler C, Laddada L, et al. MACF1 controls skeletal muscle function through the microtubule-dependent localization of extra-synaptic myonuclei and mitochondria biogenesis. eLife. 2021;10:e70490. doi: 10.7554/eLife.70490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Gomez TS, Kumar K, Medeiros RB, Shimizu Y, Leibson PJ, Billadeau DD. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Gönczy P, Stelzer EHK, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li D, Zhang S, Yang Y, Liu J-J, Wang X, Liu C, Milkie DE, Moore RP, Tulu US, et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell. 2018;175:1430–1442.e17. doi: 10.1016/j.cell.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Gurel PS, Hatch AL, Higgs HN. Connecting the cytoskeleton to the endoplasmic reticulum and golgi. Curr Biol. 2014;24:R660–R672. doi: 10.1016/j.cub.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjitheodorou A, Bell GRR, Ellett F, Shastry S, Irimia D, Collins SR, Theriot JA. Directional reorientation of migrating neutrophils is limited by suppression of receptor input signaling at the cell rear through myosin II activity. Nat Commun. 2021;12:6619. doi: 10.1038/s41467-021-26622-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CM, Chen W-C, Khatau SB, Daniels BR, Lee JSH, Wirtz D. SMRT analysis of MTOC and nuclear positioning reveals the role of EB1 and LIC1 in single-cell polarization. J Cell Sci. 2011;124:4267–4285. doi: 10.1242/jcs.091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Kalra S, Jameson LE, Guerrero LA, Cain NE, Bolivar J, Starr DA. The Nesprin-1/-2 ortholog ANC-1 regulates organelle positioning in C. elegans independently from its KASH or actin-binding domains. Elife. 2021;10:e61069. doi: 10.7554/eLife.61069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin J-W, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Huelsmann S, Huelsmann S, Ylänne J, Ylänne J, Brown NH, Brown NH. Filopodia-like actin cables position nuclei in association with perinuclear actin in Drosophila nurse cells. Dev Cell. 2013;26:604–615. doi: 10.1016/j.devcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illukkumbura R, Bland T, Goehring NW. Patterning and polarization of cells by intracellular flows. Curr Opin Cell Biol. 2020;62:123–134. doi: 10.1016/j.ceb.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante E, Castagnino A, Ferrari R, Monteiro P, Agüera-González S, Paul-Gilloteaux P, Domingues MJ, Maiuri P, Raab M, Shanahan CM, et al. LINC complex-Lis1 interplay controls MT1-MMP matrix digest-on-demand response for confined tumor cell migration. Nat Commun. 2018;9:2443. doi: 10.1038/s41467-018-04865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa-Ankerhold H, Kroll J, Heuvel D, van den, Renkawitz J, Müller-Taubenberger A. Centrosome positioning in migrating dictyostelium cells. Cells. 2022;11:1776. doi: 10.3390/cells11111776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska IL, Tobin MP, Bai T, Dooling LJ, Discher DE. Small lipid droplets are rigid enough to indent a nucleus, dilute the lamina, and cause rupture. J Cell Biol. 2023;222:e202208123. doi: 10.1083/jcb.202208123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Singh P. Centrosome dysfunction in human diseases. Semin Cell Dev Biol. 2021;110:113–122. doi: 10.1016/j.semcdb.2020.04.019. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM. The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Bio. 2020;21:307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- Janota CS, Calero-Cuenca FJ, Costa J, Gomes ER. SnapShot: nucleo-cytoskeletal interactions. Cell. 2017;169:970–970.e1. doi: 10.1016/j.cell.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Janota CS, Pinto A, Pezzarossa A, Machado P, Costa J, Campinho P, Franco CA, Gomes ER. Shielding of actin by the endoplasmic reticulum impacts nuclear positioning. Nat Commun. 2022;13:2763. doi: 10.1038/s41467-022-30388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AJ, Schaeffer A, Pascalis CD, Letort G, Vianay B, Bornens M, Piel M, Blanchoin L, Théry M. Acto-myosin network geometry defines centrosome position. Curr Biol. 2021;31:1206–1220.e5. doi: 10.1016/j.cub.2021.01.002. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L, Martzloff P, Bambach SK, Aizarani N, Mihlan M, Gavrilov A, Glaser KM, Stecher M, Thünauer R, Thiriot A, et al. Slow integrin-dependent migration organizes networks of tissue-resident mast cells. Nat Immunol. 2023;24:915–924. doi: 10.1038/s41590-023-01493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalukula Y, Stephens AD, Lammerding J, Gabriele S (2022) Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol 23:583–602 [DOI] [PMC free article] [PubMed]
- Kameritsch P, Renkawitz J. Principles of leukocyte migration strategies. Trends Cell Biol. 2020;30:818–832. doi: 10.1016/j.tcb.2020.06.007. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, van Bergeijk P, Lipka J, Keijzer N, Wulf PS, Katrukha EA, Akhmanova A, Hoogenraad CC. Myosin-V opposes microtubule-based cargo transport and drives directional motility on cortical actin. Curr Biol. 2013;23:828–834. doi: 10.1016/j.cub.2013.03.068. [DOI] [PubMed] [Google Scholar]
- Katti P, Ajayi PT, Aponte A, Bleck CKE, Glancy B. Identification of evolutionarily conserved regulators of muscle mitochondrial network organization. Nat Commun. 2022;13:6622. doi: 10.1038/s41467-022-34445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagia JZ, Ivaska J, Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol. 2019;20:457–473. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- Kelley LC, Chi Q, Cáceres R, Hastie E, Schindler AJ, Jiang Y, Matus DQ, Plastino J, Sherwood DR. Adaptive F-actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs. Dev Cell. 2019;48:313–328.e8. doi: 10.1016/j.devcel.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys J, Cheung BCH, Elpers MA, Wu M, Lammerding J (2024) Rear cortex contraction aids in nuclear transit during confined migration by increasing pressure in the cell posterior. Preprint at bioRxiv: 2022.09.10.507419
- Knoblach B, Rachubinski RA. Sharing the cell’s bounty – organelle inheritance in yeast. J Cell Sci. 2015;128:621–630. doi: 10.1242/jcs.151423. [DOI] [PubMed] [Google Scholar]
- Kopf A, Renkawitz J, Hauschild R, Girkontaite I, Tedford K, Merrin J, Thorn-Seshold O, Trauner D, Häcker H, Fischer K-D, et al. Microtubules control cellular shape and coherence in amoeboid migrating cells. J Cell Biol. 2020;219:e201907154. doi: 10.1083/jcb.201907154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J, Hauschild R, Kuznetcov A, Stefanowski K, Hermann MD, Merrin J, Shafeek L, Müller‐Taubenberger A, Renkawitz J. Adaptive pathfinding by nucleokinesis during amoeboid migration. EMBO J. 2023;42:e114557. doi: 10.15252/embj.2023114557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J, Ruiz‐Fernandez MJA, Braun MB, Merrin J, Renkawitz J. Quantifying the probing and selection of microenvironmental pores by motile immune cells. Curr Protoc. 2022;2:e407. doi: 10.1002/cpz1.407. [DOI] [PubMed] [Google Scholar]
- Krummel MF, Macara I. Maintenance and modulation of T cell polarity. Nat Immunol. 2006;7:1143–1149. doi: 10.1038/ni1404. [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Poenie M. Dynamic polarization of the microtubule cytoskeleton during CTL-mediated killing. Immunity. 2002;16:111–121. doi: 10.1016/S1074-7613(02)00262-5. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Troys MV. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008;18:220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Lämmermann T, Sixt M. Mechanical modes of “amoeboid” cell migration. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Berre ML, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell. 2013;25:270–283. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Lautenschläger F, Piel M. Microfabricated devices for cell biology: all for one and one for all. Curr Opin Cell Biol. 2013;25:116–124. doi: 10.1016/j.ceb.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Lavrsen K, Rajendraprasad G, Leda M, Eibes S, Vitiello E, Katopodis V, Goryachev AB, Barisic M. Microtubule detyrosination drives symmetry breaking to polarize cells for directed cell migration. Proc Natl Acad Sci USA. 2023;120:e2300322120. doi: 10.1073/pnas.2300322120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cathey PI, Wu H, Parker R, Voeltz GK. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science. 2020;367:eaay7108. doi: 10.1126/science.aay7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithner A, Eichner A, Müller J, Reversat A, Brown M, Schwarz J, Merrin J, Gorter DJJ, de, Schur F, Bayerl J, et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat Cell Biol. 2016;18:1253–1259. doi: 10.1038/ncb3426. [DOI] [PubMed] [Google Scholar]
- Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- Liao Y-C, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, et al. RNA granules hitchhike on lysosomes for long-distance transport, using Annexin A11 as a molecular tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Schuster M, Guimaraes SC, Ashwin P, Schrader M, Metz J, Hacker C, Gurr SJ, Steinberg G. Active diffusion and microtubule-based transport oppose myosin forces to position organelles in cells. Nat Commun. 2016;7:11814. doi: 10.1038/ncomms11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-J, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Lomakin AJ, Cattin CJ, Cuvelier D, Alraies Z, Molina M, Nader GPF, Srivastava N, Sáez PJ, Garcia-Arcos JM, Zhitnyak IY, et al. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science. 2020;370:eaba2894. doi: 10.1126/science.aba2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Doménech G, Covill‐Cooke C, Ivankovic D, Halff EF, Sheehan DF, Norkett R, Birsa N, Kittler JT. Miro proteins coordinate microtubule‐ and actin‐dependent mitochondrial transport and distribution. EMBO J. 2018;37:321–336. doi: 10.15252/embj.201696380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Tartwijk FW, van, Lin JQ, Nijenhuis W, Parutto P, Fantham M, Christensen CN, Avezov E, Holt CE, Tunnacliffe A, et al. The structure and global distribution of the endoplasmic reticulum network are actively regulated by lysosomes. Sci Adv. 2020;6:eabc7209. doi: 10.1126/sciadv.abc7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Rupprecht J-F, Wieser S, Ruprecht V, Bénichou O, Carpi N, Coppey M, Beco SD, Gov N, Heisenberg C-P, et al. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- Mardakheh FK, Paul A, Kümper S, Sadok A, Paterson H, Mccarthy A, Yuan Y, Marshall CJ. Global analysis of mRNA, translation, and protein localization: local translation is a key regulator of cell protrusions. Dev Cell. 2015;35:344–357. doi: 10.1016/j.devcel.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PC, Petrie RJ. Push or pull: how cytoskeletal crosstalk facilitates nuclear movement through 3D environments. Phys Biol. 2022;19:021003. doi: 10.1088/1478-3975/ac45e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Marzo MG, McKenney RJ. New insights into the mechanism of dynein motor regulation by lissencephaly-1. Elife. 2020;9:e59737. doi: 10.7554/eLife.59737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Piel M, Basto R. Never tear us apart – the importance of centrosome clustering. J Cell Sci. 2012;125:3281–3292. doi: 10.1242/jcs.094797. [DOI] [PubMed] [Google Scholar]
- Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol. 1987;105:1253–1265. doi: 10.1083/jcb.105.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AL, Hsia C-R, Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr Opin Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BB, Holzbaur ELF, Ostap EM. Control of the initiation and termination of kinesin-1-driven transport by myosin-Ic and nonmuscle tropomyosin. Curr Biol. 2015;25:523–529. doi: 10.1016/j.cub.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring JCM, Grigoriev I, Nijenhuis W, Kapitein LC, Akhmanova A. Opto-katanin, an optogenetic tool for localized, microtubule disassembly. Curr Biol. 2022;32:4660–4674.e6. doi: 10.1016/j.cub.2022.09.010. [DOI] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Varuzhanyan G, Pham AH, Chan DC. Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell Metab. 2015;22:1033–1044. doi: 10.1016/j.cmet.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissoglu K, Stueland M, Gasparski AN, Wang T, Jenkins LM, Hastings ML, Mili S. RNA localization and co‐translational interactions control RAB13 GTPase function and cell migration. EMBO J. 2020;39:e104958. doi: 10.15252/embj.2020104958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissoglu K, Yasuda K, Wang T, Chrisafis G, Mili S. Translational regulation of protrusion-localized RNAs involves silencing and clustering after transport. eLife. 2019;8:e44752. doi: 10.7554/eLife.44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller K, Brambach M, Villani A, Gallo E, Gilmour D, Peri F. A role for the centrosome in regulating the rate of neuronal efferocytosis by microglia in vivo. eLife. 2022;11:e82094. doi: 10.7554/eLife.82094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P, Rossé C, Castro-Castro A, Irondelle M, Lagoutte E, Paul-Gilloteaux P, Desnos C, Formstecher E, Darchen F, Perrais D, et al. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J Cell Biol. 2013;203:1063–1079. doi: 10.1083/jcb.201306162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Coscia SM, Simpson CL, Ortega FE, Wait EC, Heddleston JM, Nirschl JJ, Obara CJ, Guedes-Dias P, Boecker CA, et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature. 2021;591:659–664. doi: 10.1038/s41586-021-03309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau HD, Lennon-Duménil A-M, Pierobon P. “If you please… draw me a cell”. Insights from immune cells. J Cell Sci. 2020;133:jcs244806. doi: 10.1242/jcs.244806. [DOI] [PubMed] [Google Scholar]
- Moriarty RA, Mili S, Stroka KM. RNA localization in confined cells depends on cellular mechanical activity and contributes to confined migration. iScience. 2022;25:103845. doi: 10.1016/j.isci.2022.103845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier JA, Fabiano ED, Ludolph CM, White AE, Reinhart-King CA. Confinement primes cells for faster migration by polarizing active mitochondria. Nanoscale Adv. 2023;6:209–220. doi: 10.1039/D3NA00478C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastały P, Purushothaman D, Marchesi S, Poli A, Lendenmann T, Kidiyoor GR, Beznoussenko GV, Lavore S, Romano OM, Poulikakos D, et al. Role of the nuclear membrane protein Emerin in front-rear polarity of the nucleus. Nat Commun. 2020;11:2122. doi: 10.1038/s41467-020-15910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, Sánchez-Díaz M, Díaz-García E, Santiago DJ, Rubio-Ponce A, Li JL, Balachander A, Quintana JA, et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell. 2020;183:94–109.e23. doi: 10.1016/j.cell.2020.08.031. [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354:aaf3928–aaf3928. doi: 10.1126/science.aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourshargh S, Hordijk PL, Sixt M. Breaching multiple barriers: leukocyte motility through venular walls and the interstitium. Nat Rev Mol Cell Bio. 2010;11:366–378. doi: 10.1038/nrm2889. [DOI] [PubMed] [Google Scholar]
- Olins AL, Olins AL, Hoang TV, Hoang TV, Zwerger M, Herrmann H, Herrmann H, Zentgraf H, Zentgraf H, Noegel AA, et al. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–214. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan Y-N. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons . Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch EK, Aspalter IM, Sixt M. Focal adhesion–independent cell migration. Ann Rev Cell Dev Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- Paterson N, Lämmermann T. Macrophage network dynamics depend on haptokinesis for optimal local surveillance. eLife. 2021;11:e75354. doi: 10.7554/eLife.75354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Montell DJ (2022) Nuclei function as wedges to pry open spaces and promote collective, confined cell migration in vivo. Preprint at bioRxiv: 2021.12.16.473064
- Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Yamada KM. Fibroblasts lead the way: a unified view of 3D cell motility. Trends Cell Biol. 2015;25:666–674. doi: 10.1016/j.tcb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau J, Moreau H, Duménil A-ML, Pierobon P. Cell polarity in development and disease. Curr Top Dev Biol. 2023;154:197–222. doi: 10.1016/bs.ctdb.2023.02.011. [DOI] [PubMed] [Google Scholar]
- Pouthas F, Girard P, Lecaudey V, Ly TBN, Gilmour D, Boulin C, Pepperkok R, Reynaud EG. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 2008;121:2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- Raab M, Gentili M, Belly H, de, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER–endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Kopf A, Stopp J, Vries I, de, Driscoll MK, Merrin J, Hauschild R, Welf ES, Danuser G, Fiolka R, et al. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature. 2019;568:546–550. doi: 10.1038/s41586-019-1087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz J, Schumann K, Weber M, Lämmermann T, Pflicke H, Piel M, Polleux J, Spatz JP, Sixt M. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. 2009;11:1438–1443. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Sixt M. Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 2010;11:744–750. doi: 10.1038/embor.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Gomes ER. Nuclear positioning in skeletal muscle. Semin Cell Dev Biol. 2018;82:51–56. doi: 10.1016/j.semcdb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Roman W, Martins JP, Carvalho FA, Voituriez R, Abella JVG, Santos NC, Cadot B, Way M, Gomes ER. Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat Cell Biol. 2017;19:1189–1201. doi: 10.1038/ncb3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Pinheiro H, Pimentel MR, Segalés J, Oliveira LM, García-Domínguez E, Gómez-Cabrera MC, Serrano AL, Gomes ER, Muñoz-Cánoves P. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science. 2021;374:355–359. doi: 10.1126/science.abe5620. [DOI] [PubMed] [Google Scholar]
- Schmidt CJ, Stehbens SJ. Microtubule control of migration: coordination in confinement. Curr Opin Cell Biol. 2024;86:102289. doi: 10.1016/j.ceb.2023.102289. [DOI] [PubMed] [Google Scholar]
- Schmied C, Ebner M, Ferré PS, Haucke V, Lehmann M (2023) OrgaMapper: a robust and easy-to-use workflow for analyzing organelle positioning. Preprint at bioRxiv: 2023.07.10.548452
- Silva CG, Peyre E, Adhikari MH, Tielens S, Tanco S, Damme PV, Magno L, Krusy N, Agirman G, Magiera MM, et al. Cell-intrinsic control of interneuron migration drives cortical morphogenesis. Cell. 2018;172:1063–1078.e19. doi: 10.1016/j.cell.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starborg T, Lu Y, Kadler KE, Holmes DF. Electron microscopy of collagen fibril structure in vitro and in vivo including three-dimensional reconstruction. Methods Cell Biol. 2008;88:319–345. doi: 10.1016/S0091-679X(08)00417-2. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–465. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- Stoitzner P, Pfaller K, Stössel H, Romani N. A close-up view of migrating Langerhans cells in the skin. J Invest Dermatol. 2002;118:117–125. doi: 10.1046/j.0022-202x.2001.01631.x. [DOI] [PubMed] [Google Scholar]
- Stowers L, Yelon D, Berg LJ, Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc Natl Acad Sci USA. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzyz PJ, Lee HO, Sidhaye J, Weber IP, Leung LC, Norden C. Interkinetic nuclear migration is centrosome independent and ensures apical cell division to maintain tissue integrity. Dev Cell. 2015;32:203–219. doi: 10.1016/j.devcel.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Takakuwa H, Yamazaki T, Souquere S, Adachi S, Yoshino H, Fujiwara N, Yamamoto T, Natsume T, Nakagawa S, Pierron G, et al. Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles. Nat Cell Biol. 2023;25:1664–1675. doi: 10.1038/s41556-023-01254-1. [DOI] [PubMed] [Google Scholar]
- Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, Kapitein LC. Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron. 2017;96:1264–1271.e5. doi: 10.1016/j.neuron.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Huttner WB. Neural progenitor nuclei in motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Taylor RB, Duffus WPH, Raff MC, de Petris S. Redistribution and pinocytosis of lymphocyte surface immunoglobulin molecules induced by anti-immunoglobulin antibody. Nat New Biol. 1971;233:225–229. doi: 10.1038/newbio233225a0. [DOI] [PubMed] [Google Scholar]
- Théry M, Racine V, Piel M, Pépin A, Dimitrov A, Chen Y, Sibarita J-B, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam H-R, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–412. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F-C, Meyer T. Ca2+ pulses control local cycles of lamellipodia retraction and adhesion along the front of migrating cells. Curr Biol. 2012;22:837–842. doi: 10.1016/j.cub.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol. 2017;18:229–245. doi: 10.1038/nrm.2016.153. [DOI] [PubMed] [Google Scholar]
- Valenzuela A, Meservey L, Nguyen H, Fu M. Golgi outposts nucleate microtubules in cells with specialized shapes. Trends Cell Biol. 2020;30:792–804. doi: 10.1016/j.tcb.2020.07.004. [DOI] [PubMed] [Google Scholar]
- Vargas P, Maiuri P, Bretou M, Sáez PJ, Pierobon P, Maurin M, Chabaud M, Lankar D, Obino D, Terriac E, et al. Innate control of actin nucleation determines two distinct migration behaviours in dendritic cells. Nat Cell Biol. 2016;18:43–53. doi: 10.1038/ncb3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini V, Pezzano F, Castro FC, Häkkinen H-M, Jiménez-Delgado S, Colomer-Rosell M, Marro M, Tolosa-Ramon Q, Paz-López S, Valverde MA, et al. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science. 2020;370:eaba2644. doi: 10.1126/science.aba2644. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Gregory J, Parsons SF, Salmon ED. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulovesicular networks in interphase Xenopus egg extracts. J Cell Biol. 1995;130:1161–1169. doi: 10.1083/jcb.130.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier A-K, Homrich M, Ebbinghaus S, Juda P, Miková E, Hauschild R, Zhang L, Quast T, Mass E, Schlitzer A, et al. Multiple centrosomes enhance migration and immune cell effector functions of mature dendritic cells. J Cell Biol. 2022;221:e202107134. doi: 10.1083/jcb.202107134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. Intravital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem. 2015;84:1–21. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Dema A, Haren Jvan. Lights, cytoskeleton, action: optogenetic control of cell dynamics. Curr Opin Cell Biol. 2020;66:1–10. doi: 10.1016/j.ceb.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Alexander S, Alexander S, Schacht V, Schacht V, Coussens LM, Coussens LM, Andrian UH, von, Rheenen J, van, Rheenen Jvan, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Lindert M, te, Krause M, Alexander S, Riet J, te, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Linstedt AD. Golgi positioning. Cold Spring Harb Perspect Biol. 2011;3:a005322. doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Sixt M. Mechanisms of 3D cell migration. Nat Rev Mol Cell Biol. 2019;20:738–752. doi: 10.1038/s41580-019-0172-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Gaillard J, Vianay B, Guerin C, Orhant‐Prioux M, Blanchoin L, Théry M. Actin network architecture can ensure robust centering or sensitive decentering of the centrosome. EMBO J. 2022;41:e111631. doi: 10.15252/embj.2022111631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AL, Goldman YE, Holzbaur ELF, Ostap EM. Local cytoskeletal and organelle interactions impact molecular-motor-driven early endosomal trafficking. Curr Biol. 2013;23:1173–1180. doi: 10.1016/j.cub.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Obara CJ, Szczesna E, Nixon-Abell J, Mahalingan KK, Roll-Mecak A, Lippincott-Schwartz J, Blackstone C. ER proteins decipher the tubulin code to regulate organelle distribution. Nature. 2022;601:132–138. doi: 10.1038/s41586-021-04204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Ho CY, Lammerding J. Nuclear mechanics in disease. Ann Rev Biomed Eng. 2011;13:397–428. doi: 10.1146/annurev-bioeng-071910-124736. [DOI] [PMC free article] [PubMed] [Google Scholar]