The tetravalent PD-L1×4-1BB bispecific antibody ATG-101 activates 4-1BB+ T cells in a PD-L1 cross-linking–dependent manner, minimizing the hepatotoxicity of existing 4-1BB agonists and suppressing growth of ICI-resistant tumors.
Abstract
Immune checkpoint inhibitors (ICI) have transformed cancer treatment. However, only a minority of patients achieve a profound response. Many patients are innately resistant while others acquire resistance to ICIs. Furthermore, hepatotoxicity and suboptimal efficacy have hampered the clinical development of agonists of 4-1BB, a promising immune-stimulating target. To effectively target 4-1BB and treat diseases resistant to ICIs, we engineered ATG-101, a tetravalent "2+2″ PD-L1×4-1BB bispecific antibody. ATG-101 bound PD-L1 and 4-1BB concurrently, with a greater affinity for PD-L1, and potently activated 4-1BB+ T cells when cross-linked with PD-L1–positive cells. ATG-101 activated exhausted T cells upon PD-L1 binding, indicating a possible role in reversing T-cell dysfunction. ATG-101 displayed potent antitumor activity in numerous in vivo tumor models, including those resistant or refractory to ICIs. ATG-101 greatly increased the proliferation of CD8+ T cells, the infiltration of effector memory T cells, and the ratio of CD8+ T/regulatory T cells in the tumor microenvironment (TME), rendering an immunologically “cold” tumor “hot.” Comprehensive characterization of the TME after ATG-101 treatment using single-cell RNA sequencing further revealed an altered immune landscape that reflected increased antitumor immunity. ATG-101 was well tolerated and did not induce hepatotoxicity in non-human primates. According to computational semimechanistic pharmacology modeling, 4-1BB/ATG-101/PD-L1 trimer formation and PD-L1 receptor occupancy were both maximized at around 2 mg/kg of ATG-101, providing guidance regarding the optimal biological dose for clinical trials. In summary, by localizing to PD-L1–rich microenvironments and activating 4-1BB+ immune cells in a PD-L1 cross-linking–dependent manner, ATG-101 safely inhibits growth of ICI resistant and refractory tumors.
Significance:
The tetravalent PD-L1×4-1BB bispecific antibody ATG-101 activates 4-1BB+ T cells in a PD-L1 cross-linking–dependent manner, minimizing the hepatotoxicity of existing 4-1BB agonists and suppressing growth of ICI-resistant tumors.
Introduction
Programmed death-ligand 1 (PD-L1) and programmed cell death protein 1 (PD-1) blockade therapy, or immune checkpoint inhibitors (ICI), have revolutionized the treatment landscape of multiple malignancies. However, only a minority of patients demonstrate deep and durable responses, with many patients showing innate resistance or subsequently developing resistance to ICI therapy. Tumors with little T-cell infiltration or lack of pre-existing antitumor immune responses are defined as immunologically “cold.” Patients with these tumors have a poor prognosis following PD-(L)1 blocking therapy (1). On the other hand, malignancies that initially respond well to ICIs may develop acquired drug resistance, frequently as a consequence of T-cell exhaustion (2–4).
4-1BB (CD137, TNFRSF9) is a costimulatory receptor of the TNF receptor superfamily (TNFRSF) and accumulates on the T-cell surface upon activation. Receptor stimulation by endogenous 4-1BB ligand (4-1BBL) or agonistic 4-1BB antibodies can markedly augment T-cell activation and upregulate inflammatory cytokine response. 4-1BB agonism averts tumor-infiltrating lymphocyte (TIL) exhaustion and enhances antitumor activity of ICIs (5, 6), providing a rationale for the exploration of 4-1BB targeting immunotherapies in ICI-resistant patients.
Agonistic 4-1BB antibodies have exhibited potent anticancer efficacy in a variety of syngeneic mouse models (7). However, the on-target-off-tumor liver toxicity has hampered the successful clinical development of therapeutic 4-1BB agonists, such as urelumab, a fully human IgG4 mAb (8). The activation of 4-1BB–positive liver-infiltrating T cells by the agonistic antibodies has been implicated as the underlying mechanism of liver toxicity (9). Unlike urelumab, utomilumab, a humanized IgG2 mAb, requires Fc-mediated cross-linking to activate the 4-1BB signaling pathway (10, 11). Utomilumab was better tolerated in patients with a MTD > 10 mg/kg, albeit with a relatively modest antitumor activity (12).
One of the most promising next-generation 4-1BB targeting strategies is the use of bispecific antibodies (BsAb) or bispecific fusion proteins, which are designed to be enriched in the tumor microenvironment (TME) enabling tumor cell–mediated 4-1BB activation, thereby minimizing on-target-off-tumor toxicity. Typically, such bispecific proteins are composed of a tumor-associated antigen (TAA)-recognizing arm and a 4-1BB–agonistic arm. Several TAA/4-1BB targeting bifunctional proteins, such as HER2×4-1BB (13), CD30×4-1BB (14), B7H3×4-1BB (15), and 5T4×4-1BB (16, 17), have been evaluated in preclinical models, showing potent antitumor efficacy with reduced liver toxicity.
The combination of PD-L1 blockade with 4-1BB agonism has shown enhanced antitumor responses in preclinical cancer models (5, 6, 18). Therefore, dual targeting of both PD-L1 and 4-1BB by a BsAb might not only permit tumor cell–mediated 4-1BB activation but may also enable the optimal engagement of antitumor immunity. Multiple PD-L1×4-1BB BsAbs have been developed and are being evaluated in clinical trials. These molecules bind to an even number of 4-1BB and PD-L1 with a ratio of 1+1 or 2+2, such as GEN1046 (19) and MCLA-145 (10), or INBRX-105 (20), FS222 (21), ABL503 (22), and PRS-344 (23). It has been shown that a high-affinity differential between the cross-linking target (e.g., TAA or PD-L1) and 4-1BB results in possible functional benefits of the bispecific molecules (24). Nevertheless, the majority of clinical PD-L1×4-1BB bispecific molecules demonstrated less than a 10-fold difference in the binding affinities of PD-L1 and 4-1BB (10, 20–23, 25). Whether a greater affinity difference may result in improved clinical performance of the bispecific molecules remains to be explored.
In this study, we developed a “2+2″ tetravalent PD-L1×4-1BB BsAb, ATG-101. ATG-101 binds to PD-L1 and 4-1BB simultaneously, with a greater affinity for PD-L1, and potently activates 4-1BB–positive T cells when cross-linked with PD-L1–positive (PD-L1+) cells. ATG-101 demonstrated strong antitumor efficacy in a variety of tumor models, including those that were resistant to or had progressed on anti-PD-(L)1 therapy, rendering a “cold” tumor “hot.” The landscape of TME at the single-cell, transcriptome-level after ATG-101 treatment was investigated utilizing single-cell RNA sequencing (scRNA-seq) technology. ATG-101 was also well tolerated with no evidence of hepatotoxicity and cytokine release syndrome (CRS) in non-human primates (NHP).
Materials and Methods
Cell line
The HEK293T- or CHO-overexpressing cell lines were generated by the transduction of 293T or CHO (ATCC) cells with lentivirus from a PD-L1 or 4-1BB full-length sequence encoding vector. The HEK293T-overexpressing cell lines were maintained in DMEM (GIBCO) supplemented with 10% FBS (GIBCO), 50 U/mL penicillin, and 50 mg/mL streptomycin in a humidified atmosphere at 37°C with 5% CO2. The CHO-overexpressing cell lines were maintained in F12K (GIBCO) supplemented with 10% FBS (GIBCO), 50 U/mL penicillin, and 50 mg/mL streptomycin in a humidified atmosphere at 37°C with 5% CO2. NK92MI-FcγRIIIA were generated by the transduction of NK92MI cells (Procell) with lentivirus from an FcγRIIIA-GFP-encoding vector and maintained in NK92MI special medium (Procell, CM-0533). MCF7, RKO, NCI-H358, MC38, Pan02, B16F10, and EL-4 were purchased from ATCC and cultured according to manufacturer's guidelines. All cells were cultured for less than 20 passages. Human peripheral blood mononuclear cells (PBMC) were obtained as cryopreserved vials of cells from individual donors from Sailybio Corporation.
Antibody
The sequences of the ATG-101, analogs of atezolizumab (IgG1 with N297A mutation), urelumab (IgG4), and utomilumab (IgG2) were obtained from patents WO2019196309A1, WO 2010/077634 A1, US8137667B2, and US2012/0237498A1, respectively. Heavy and light chain genes were cloned into mammalian expression vector pcDNA3.1, expressed using the ExpiCHO system (Invitrogen) and protein was purified by protein A chromatography (GE). IgG1 isotype control with wild-type Fc was used in most experiments except that isotype control with LALA mutation at Fc domain was employed for single-cell sequencing.
Binding capability of ATG-101 to hPD-L1/h4-1BB by ForteBio
ATG-101 was biotin labeled (Thermo Fisher Scientific, A39257) and purified with desalting column (Thermo Fisher Scientific, 89890) as manufacturer's instructions. The concentration of biotinylated protein was measured using a bicinchoninic acid kit (Thermo Fisher Scientific, Pierce, 23225). Biotinylated ATG-101 at 100 nmol/L was captured on Streptavidin (SA) biosensor, followed by loading first protein hPD-L1-Fc (Sino Biology, 10377-H05H) or h4-1BB-Fc (ACROBiosystems, 41B-H5258)) at 100 nmol/L for 5 minutes, then loading the second protein h4-1BB-Fc (or hPD-L1-Fc) at 100 nmol/L for 5 minutes, IgG-Fc was used as control. First, ATG-101 was labeled with biotin and immobilized on the SA biosensors. Injection of human 4-1BB-Fc fusion protein resulted in the detection of a binding signal, indicating the formation of ATG-101–4-1BB complexes. The addition of human PD-L1-Fc fusion protein elicited a second signal. Injection of hPD-L1 followed by h4-1BB protein had a comparable effect.
Binding affinity of ATG-101 to hPD-L1/h4-1BB protein by Biacore
Biacore running buffer was 1×HBS-EP+ Buffer prepared from 10×HBS-EP+ Buffer (GE Healthcare, BR-1006-69). An anti-human IgG chip was prepared by immobilizing antihuman IgG Fc antibody via amine coupling (GE Healthcare, BR-1003-50) on 1–8 flow cells a CM5 chip (GE Healthcare, 29-1496-03). The sensor chip surfaces were then flushed with HBS-EP+ buffer to stabilize the baseline. Then 25 nmol/L monomeric hPD-L1 (ACROBiosystems, PD1-H5229) solution was 2-fold serially diluted and 50 nmol/L monomeric h41-BB (ACROBiosystems, 41B-H5227) solution was 2-fold serially diluted. The serially diluted hPD-L1 and h4-1BB were injected into flow cells 3 and 4 sequentially. The association and dissociation time were 180 and 400 seconds, respectively. The surface was regenerated with 10 mmol/L glycine pH 1.5 (GE Healthcare, BR-1003-54) by injecting into flow cells 3 and 4 at 30 μL/minute for 30 seconds followed by a stabilization period of 60 seconds. Binding kinetics was calculated using Biacore Insight Evaluation Software (Version 2.0.15.12933) and a 1:1 binding model for curve fitting.
Binding affinity of ATG-101 to FcγRs and FcRn by ForteBio
Biotinylated hFcγRIIIA (ACROBiosystems, CD8-H52H4) was loaded to the SA sensor (PALL Life Sciences ForteBio, 18-5019) for 2 minutes. Then 2-fold serially diluted ATG-101 down from 750 nmol/L was loaded to bind for 2 minutes then dissociating for 2 minutes. Ni-NTA sensor (ForteBio18-5101) was used for the other Fc receptors. Loading 15 nmol/L FcγRs [hFcγRI (ACROBiosystems, FCA-H52H2), hFcγRIIA (ACROBiosystems, CDA-H5221), hFcγRIIB (ACROBiosystems, CDB-H5228), FcRn (ACROBiosystems, FCM-h5286)] for 6 minutes, then loading 2-fold serial diluted ATG-101 from 1,000 nmol/L for 3 minutes. The dissociation was performed with Ni-NTA sensor for 10 minutes. Data were analyzed using Data Analysis 9.0.
Binding affinity of ATG-101 to monocytes and NK92MI-FcγRIIIa cells by flow cytometry
Human monocytes and NK92MI cell overexpressing human FcγRIIIA (NK92MI-FcγRIIIA) cells were incubated with Human Fc Receptor Blocking Solution (BioLegend, 422302) at room temperature for 10 minute or not. Then the cells were incubated with 100, 30, 10, 3 nmol/L ATG-101 and hIgG1 isotype at 4°C for 1 hour, respectively, followed by fluorophore (PE) anti-human IgG secondary antibody (Abcam, ab98596) for 30 minutes at 4°C. Mean fluorescence intensity (MFI) was measured with high-throughput flow cytometry.
Binding affinity of ATG-101 to hPD-L1/h4-1BB by flow cytometry
HEK293T cell overexpressing human PD-L1 (293T-hPDL1) and HEK293T cells overexpressing h4-1BB (293T-h41BB) cells were incubated with serial dilutions of ATG-101 at 4°C for 1 hour, respectively, followed by fluorophore (PE) anti-human IgG secondary antibody (Abcam, ab98596) and APC anti-human IgG secondary antibody (BioLegend, 410712) for 30 minutes at 4°C, respectively. MFI was measured with high-throughput flow cytometry.
Blockade of PD-1–PD-L1 interaction with ATG-101
Biotinylated human or mouse PD-1 at a final concentration of 40 μg/mL and serially diluted ATG-101 or its PD-L1 parental antibody was cocultured with 293T-hPDL1 or 293T-mPDL1 cells. Then incubated for 1 hour at 4°C, followed by SA and PE (BioLegend, 405204) for 30 minutes at 4°C at 1:500 ratio. The high-throughput flow cytometer was used to determine MFI.
Exhausted T-cell induction and reactivation
Human CD3+ T cells were isolated from fresh PBMC with magnetic microbeads (Stemcell, 17951). Purified T cells were seeded in T25 flask and induced with prewashed and resuspended CD3/CD28 Dynabeads (Gibco, 11161D) with a bead-to-cell ratio of 1:1. Every 2 days or 3 days, cells were counted, washed, and restimulated with a fresh batch of Dynabeads. T-cell exhaustion markers were detected by flow cytometry using PD-1 (BioLegend, 367412), LAG3 (BioLegend, 369312), Tim3 (BioLegend, 345052), and TOX (eBioscience, 12-6502-82) antibody and cytokines (IL2, IFNγ, and TNFα) were analyzed with ELISA after 2 or 3 days of T cell stimulated with Dynabeads. Upon stimulation, the expression of the exhaustion markers TIM3, LAG3, PD-1, and TOX were increased, with a steady decline in the production of IL2, INFγ, and TNFα by the T cells. After six stimulations, the Dynabeads were removed, and the T cells were collected and mixed in a 10:1 ratio with 293T-PDL1 cells and then treated with ATG-101 (5 and 50 nmol/L), parental PD-L1 antibody (5 and 50 nmol/L), parental 4-1BB antibody (5 and 50 nmol/L) or isotype control (50 nmol/L) for 1 day and 3 days. Supernatants were collected, human IL2 (BD Pharmingen, 555190), IFNγ (BD Pharmingen, 555142), and TNFα (BD Pharmingen, 555212) were analyzed by ELISA according to manufacturer's instructions.
In vivo efficacy study on murine syngeneic tumor models
The protocol and any amendment(s) or procedures involving the care and use of animals in the mice studies presented in the article were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of CrownBio prior to execution. During the study, the care and use of animals were conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All studies were conducted following an approved IACUC protocol. h4-1BB knock-in (KI) mice [hCD137 (4-1BB) mice, C57BL/6-Tnfrsf9], whose 4-1BB gene is replaced by the h4-1BB gene, were supplied by Beijing Biocytogen, Inc. or Shanghai Model Organisms Center, Inc. These mice were housed in an SPF barrier. Six to 8 weeks old female h4-1BB KI mice were used for animal experiments. For all groups, the route of drug administration was intraperitoneal injection. The first treatment was done in the grouping day, which was defined as day 0. Body weight was recorded, and the tumors were measured with digital calipers two to three times per week. Tumor volumes (TV) were calculated by the formula: length × width2 × 0.5. The animals were euthanized at the end of the study. Phenotype of tumor cell lines used in vivo was shown in Supplementary Table S1. ATG-101 was administered at dosages that were equivalent in molar terms to anti-PD-L1 mAb in the tumor models. Specifically, a dosage of 10 mg/kg of anti-PDL1 is molar equivalent to 13 mg/kg of ATG-101. Detailed methods for each mice study are as follows.
MC38 colon cancer xenograft model (starting TV = 60 mm3)
MC38 cells were inoculated subcutaneously into the right side of h4-1BB KI mice (supplied by Beijing Biocytogen, Inc) at a concentration of 3 × 106 cells/0.2 mL. When the TV reached a size of about 60 mm3, they were separated into six groups based on TV, 5 mice for PBS group and 6 mice for the other groups. The mice were intraperitoneally injected with PBS (vehicle control), IgG (isotype control, 7.5 mg/kg), atezolizumab (7.5 mg/kg), avelumab (7.5 mg/kg), parental PD-L1 antibody (7.5 mg/kg), and parental 4-1BB antibody (3 mg/kg), ATG-101 (7.5 mg/kg) and parental PD-L1 antibody (7.5 mg/kg) + parental 4-1BB antibody (3 mg/kg) twice a week for 2 weeks.
MC38 colon cancer xenograft model (starting TV = 239 mm3)
The MC38 cells were inoculated subcutaneously on the right side of h4-1BB KI mice (supplied by Beijing Biocytogen, Inc.) at a concentration of 5 × 105 cells/0.1 mL. When the TV grew to about 239 mm3, mice were randomly divided into each group, 6 mice per group. The mice were intraperitoneally injected with IgG (10 mg/kg) or ATG-101 (3.25 and 13 mg/kg). A total of 13 mg/kg ATG-101 and 10 mg/kg IgG (or mAb) gave a comparable molar concentration. Treatment was provided six times continuously on days 0, 3, 6, 9, 12, and 15. Mice with a completely eradicated tumor in the efficacy study were rechallenge with MC38 on day 30, whereas 3 control mice were newly injected with the tumor cells at the same time. The efficacy project study terminated on day 69 for all groups.
B16F10 melanoma xenograft model
The h4-1BB KI mice (supplied by Shanghai Model Organisms Center, Inc) were implanted with 5 × 105 B16F10 cells. Tumor-bearing mice were randomly divided into seven groups based on TV (average TV≈48 mm3). The mice were intraperitoneally injected with PBS, IgG (10 mg/kg), atezolizumab (10 mg/kg), utomilumab (4 mg/kg), ATG-101 (3.25 and 13 mg/kg), and atezolizumab (10 mg/kg) + utomilumab (4 mg/kg).
Pan02 pancreatic cancer xenograft model
Each female C57BL/6 mouse (supplied by Beijing Biocytogen, Inc) was inoculated subcutaneously in the right front flank region with Pan02 tumor cells (3 × 106) in 0.1 mL of PBS for tumor development. The randomization was started when the mean tumor size reached approximately 149 mm3. A total of 24 tumor-bearing mice were enrolled in these groups and randomly assigned to three study groups. All of mice were injected intraperitoneally, IgG (7.5 mg/kg), aterzolizumab (7.5 mg/kg) or ATG-101 (10 mg/kg) once every 3 days.
Drug pharmacodynamic effect in the MC38-hPD-L1 xenograft model
The baseline PD-L1 expression of the wild-type MC38 is relatively low until INFγ stimulation, to ensure sufficient tumor-expressing PD-L1 for drug binding and cross-linking, we utilized an hPDL1-overexpressing MC38 cell line that consistently exhibited elevated levels of PD-L1 instead of MC38. MC38-hPD-L1 cells were inoculated subcutaneously into the right flank of h-41BB KI mice (supplied by Beijing Biocytogen, Inc) at a concentration of 5 × 105 cells/0.1 mL. When the TV reached about 281 mm3, the mice were randomly divided into two groups based on tumor size (4 mice per group). Both groups received intraperitoneal injections with IgG (7.5 mg/kg) or ATG-101 (9.9 mg/kg) on day 0, every 2 days for two times. The experiment was terminated 48 hours after the last administration, and the animals were euthanized at the conclusion. The body weight and TV of the mice were measured, and the tumor, spleen, and liver were excised and weighed. Blood was collected, digested, and processed into a single-cell suspension for flow cytometry analysis. CD8+ T cells and regulatory T cells (Treg; CD25+ and Foxp3+) were analyzed with antibodies against CD45 (BioLegend 103188), CD3 (BioLegend 100210), CD4 (BioLegend, 100438), CD8 (BioLegend 100759), CD25 (BioLegend 102008), and Foxp3 (eBioscience, 25-5773-82).
In vivo efficacy and PD evaluation in EL-4 xenograft model
The mice were subcutaneously inoculated with 5 × 105 EL-4 cells on the right side of h4-1BB KI mice (supplied by Shanghai Model Organisms Center, Inc). When the average size of the tumor reached 71 mm3, randomization was performed on the basis of the tumor size. All of mice were injected intraperitoneally every 2 days. The first treatment was done in the grouping day, which was defined as day 0. On day 10, all groups (8 mice per group) were treated with PBS (vehicle control), IgG (isotype control, 10 mg/kg), and ATG-101 (1, 3.25, 13 mg/kg), and mouse tumors were collected for flow cytometry analysis. All samples were processed into single-cell suspensions and stained cells followed by flow cytometry data collecting. Tumor cells were analyzed using viability dye for live/dead cells and fluorochrome-conjugated antibodies against human CD45 (BioLegend, 103138) as a leukocyte marker. Expression of T-cell and natural killer (NK) cell markers was analyzed using fluorochrome-conjugated antibodies against CD3 (BD Biosciences, 740268), CD8 (BioLegend, 100705) and CD4 (BioLegend, 100438), CD25 (BioLegend, 102012), CD69 (BioLegend, 104536), CD44 (BioLegend, 103032), CD62 L (BioLegend, 104440), Foxp3 (eBiosciences, 12-5773-82), Ki67 (eBiosciences, 56-5698-82), and CD335 (BioLegend, 137619). The markers for different immune cell population were shown in Supplementary Table S2.
In vivo efficacy of ATG-101 in atezolizumab-progressed MC-38 syngeneic mouse model
The h4-1BB KI mice (supplied by Beijing Biocytogen, Inc) were inoculated subcutaneously in the right rear flank region with 1 × 106 MC38 tumor cells in 0.1 mL of PBS for tumor development. Mice were randomized when the mean tumor size reached approximately 51 mm3. The date of randomization was designated as day 0, and treatments were commenced began on that day. After randomization, mice were treated with PBS (vehicle control, 6 mice per group) and atezolizumab (10 mg/kg, 14 mice per group). Atezolizumab was dosed to the disease progression group initially (14 mice per group), and when an individual mouse reached the predefined onset of disease progression, ATG-101 (13 mg/kg) would be dosed as the replacement medication for the mouse; for progression-free animals, atezolizumab monotherapy would be maintained. Progression of disease was defined when all the following conditions were met: during three consecutive measurements of TV from an individual mouse, (i) TV > 200 mm3 at the first measurement; (ii) the tumor size keeps growing and the growth rate accelerates between measurements; and (iii) the increase% in tumor size (ΔTV) >25% between two measurements. At the end of the study, tumors were collected, formalin-fixed, and paraffin-embedded blocks were produced, and sections were sectioned with a manual rotary microtome, 4 m thickness/section. CD8 (Cell Signaling Technology, 98941), CD4 (Cell Signaling Technology, 25229), F4/80 (Cell Signaling Technology, 70076), MHCII (Invitrogen, 14-5321-82), FoxP3 (Cell Signaling Technology, 12653), PD-L1 (Abcam, ab174838), and DAPI (Sigma, 28718-90-3) staining were all performed by multiplex IHC. Goat anti Rb IgG (Leica, DS9800 and Vector, MP-7444) were used as secondary antibodies. Vectra Polaris automated image system (Akoyabio) and inForm image analysis software was used to capture the pictures.
scRNA-seq data analysis
The h4-1BB KI mice were inoculated subcutaneously in the right rear flank region with MC38 tumor cells. Mice were randomized when the mean tumor size reached approximately 216 mm3. The date of randomization was designated as day 0, and hIgG and ATG-101 were dosed at day 0 and day 3. Tumors were collected at day 4. Tumor tissues were cut into small pieces of 2–4 mm and transferred into the gentleMACS C Tube containing the enzyme mix (Miltenyi Biotec, catalog no. 130-096-730). Tissues were dissociated at 37 °C. Dead cells were removed by Dead Cell Removal Kit (Miltenyi Biotec, catalog no. 130-090-101). CD45+ immune cells were isolated by CD45 (TIL) MicroBeads (Miltenyi Biotec, catalog no. 130-110-618) according to the manufacture's protocol. Raw scRNA-seq fastq data were processed by Cell Ranger 5.0.1, 10x Genomics (26) and aligned to mouse reference genome mm10 to obtain cell-gene count matrix for six samples. Data were collected from a total of 20,038 QC-positive TILs from 3 mice treated with ATG-101 and 20171 TILs from 3 mice treated with IgG control. The processed data matrix was further analyzed by Seurat as well as Scanpy (27–31). Cells are filtered by Unique Molecular Identifier (UMI) per cell (≥ 500), number of genes per cell (≥ 300), novelty score (log10 genes per UMI > 0.8), and mitochondrial gene ratio (< 0.2). Uniform Manifold Approximation and Projection (UMAP) algorithm was applied for expression matrix of each cell transcriptome to visualize cell populations in two-dimensional space. Biologically similar cells from different sample groups for were integrated by Harmony algorithm (32). Cell types were annotated to each cell by using ImmGen (https://www.immgen.org/) reference dataset (33). Specific immune lineages, such as T cells and dendritic cells (DC), are collected for deeper analysis, separately. Clustering of cells was performed by Leiden graph-clustering method and visualized in UMAP. Differential expression analysis is performed by DESeq2 (34) and Wilcoxon rank-sum (Mann–Whitney U) test (35). Upregulated and downregulated genes between treated and control group were identified by fold change > 1.5 and P value < 0.05, and functional analysis was performed on upregulated and downregulated genes by Gene Ontology enrichment analysis using ClusterProfiler (36). Gene set variation analysis (37) was performed for different cell types using 1,500+ canonical pathway gene sets derived from MSigDB database (38). Cell-cell communications analysis was performed using CellPhoneDB (39).
Toxicity study of ATG-101 in cynomolgus monkeys with a 4-week recovery period
The protocol and any amendment(s) or procedures involving the care and use of animals in the NHP studies presented in the article were reviewed and approved by the IACUC of InnoStar prior to execution. During the study, the care and use of animals were conducted in accordance with the regulations of the AAALAC. All studies were conducted following an approved IACUC protocol. ATG-101 was given to four groups of cynomolgus monkeys, each with 5 males and 5 females, once a week at doses of 0 (0.9% sodium chloride injection), 10, 50, and 100 mg/kg. After treatment was ended, 3 animals/sex/group were necropsied at the end of the dosing period, and the surviving animals proceeded to recover for 28 days, and 2 animals/sex/group were necropsied at the end of the recovery period.
Alanine transaminase/aspartate transaminase test
Blood was collected before feeding. Samples were collected into the tubes with separation gel and coagulant and centrifuged to obtain serum. The Hitachi-7180 Automatic Clinical Analyzer was used to evaluate serum samples.
Cytokine Detection
Blood samples were collected and electrochemiluminescence was used to test cytokine release [The information of assay kit: Name:U-PLEX TH17 Combo 1(NHP) SECTOR(25PL)].
Receptor occupancy
AF647-labeled ATG-101 parental anti-PD-L1 mAb YN035 (AF647-YN035) was employed to assist in the measurement of free receptor occupancy (RO) rate of CD3+ T cells. Each test sample was divided into two wells (well A and well B). The well A was treated with saturating concentration of ATG-101 while the well B was only sample as nonspecific fluorescence adsorption control (namely background fluorescence signal). The fluorescence signal of test sample was the recorded fluorescence signal of well A subtracted well B. The RO rate (RO percent) of the same individual calculated with a zero-concentration sample. The calculation formula was RO% = [1 − (MFIB − MFIA)/(MFIpre-dosing B − MFIpre-dosing A)]×100%.
Computational semimechanistic pharmacology modeling
Model structure
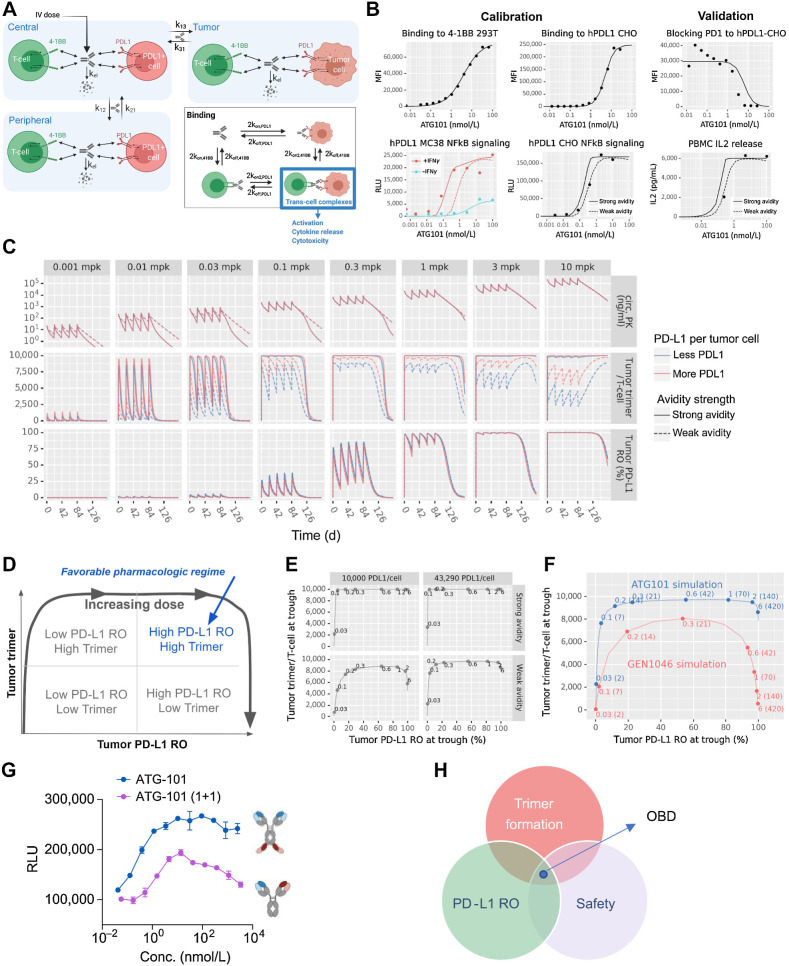
A schematic of the in vivo model is shown in Fig. 7A. The model has three compartments: a central compartment, representing the circulation; a tumor compartment; and a peripheral compartment, representing other tissues into which the drug distributes.
Figure 7.
ATG-101 demonstrated good safety profile. A and B, Mean serum concentrations of AST (A) and ALT (B) of cynomolgus monkeys over the study period at the indicated doses. C, Representative images of hematoxylin and eosin staining of liver tissue collected from the animals in the study. D, PD-L1 RO on CD3+ T cells over the study period at the indicated doses. AF647-labeled parental anti-PD-L1 antibody was employed to assist in the measurement of the ratio of free receptor. Median is shown in the figure (n = 10 per group, 5 female mice and 5 male mice.). E, Concentrations of IL2 and IL6 of cynomolgus monkeys in the study over the study period at the indicated doses. Means ± SEM are shown.
Pharmacokinetics and transport
Drug is dosed into the central compartment as an instantaneous bolus and distributes into and out of the tumor and peripheral compartments by first-order transport processes. Nonspecific elimination is modeled as a first-order process, with the same elimination rate is assumed for all compartments.
Drug binding
The drug can bind to either 4-1BB or PD-L1 initially and can then cross-link to the other receptor. To model the 2+2 format of the ATG-101 antibody, ATG-101 simultaneously binds two 4-1BB receptors or PD-L1 receptors. The microscopic binding rate constants are doubled to account for the two binding sites on ATG-101. GEN1046a is a 1+1 format antibody and binds only one receptor at a time. Binding interactions are modeled via mass-action kinetics, with the exception of ATG-101 binding to PD-L1, which is modeled using a Hill equation inspired on-rate to better match the on-cell binding data presented in the Results section.
Receptors
Each compartment contains a single 4-1BB+ cell population and a single PD-L1+ cell population. In the tumor, these cell populations are interpreted as tumor-infiltrating T cells and tumor cells, respectively. In the central and peripheral compartments, these cell populations represent all cells expressing the receptor. Drug bound receptors internalize at the same rate as free receptors in all compartments. In vitro cell binding was used to calibrate the monovalent binding affinities, and NFκB signaling assays were used to calibrate the cross-linking rate of ATG-101. Serial dilutions of ATG-101 were incubated with 293T-h41BB-NFκB-Luc or CHO-hPDL1. NFκB signaling assays was detected in the presence or absence IFNγ stimulation on hPD-L1–expressing MC38 cells.
Human model parameterization
Typical pharmacokinetic parameters were used for both ATG-101. The total amount and turnover of targets in the nominal patient was established by estimating the number of cells, the percent of receptor positive cells, the number of receptors per cell in each of the model compartments, and the internalization rate of the receptor. The effector–target (E:T) ratios in the tumor compartment were established by reviewing the density of CD3+ T cells in a select panel of solid tumors in indications of interest. To determine the PD-L1 target burden, the model was benchmarked against clinical atezolizumab pharmacokinetic data. Atezolizumab pharmacokinetic was modeled by disabling 4-1BB binding and setting the PD-L1 binding affinity to literature-derived estimates and the half-life and transport parameters to typical parameters for IgG antibodies. A PD-L1 burden was chosen to strike a good balance between consistency with the pharmacokinetic data and prior literature-based estimates of the burden. A body weight of 70 kg was assumed. Avidity values are chosen to be conservative for the comparison (i.e., the best-fit avidity for 1+1 format and the less favorable avidity for ATG-101).
Statistical analysis
Statistical analyses were performed using GraphPad Prism Software, version 9.0. For in vitro cell binding and luciferase assay, nonlinear regression was performed and EC50 values were determined. A one-way ANOVA was performed for cytokines release study with more than two groups. A two-way ANOVA was performed to compare data with more than two groups treated with different conditions including cytokines release studies, tumor growth curves, and lymphocyte change analyses. For mouse survival studies, the log-rank test (Mantel–Cox) was used. Differences with P < 0.05 were defined as statistically significant. Data are shown as means ± SEM (n = 2 or more).
Data and materials availability
All data are available in the main text or the Supplementary Materials and Methods, and upon request from the corresponding author. The raw sequencing data generated in this study are publicly available in the Sequence Read Archive database with BioProject ID PRJNA1083734, at http://www.ncbi.nlm.nih.gov/bioproject/1083734.
Code availability
Experimental protocols and the data analysis pipelines used in our work followed the 10X Genomics, Seurat official websites, and Scanpy official websites. The analysis steps, functions, and parameters used are described in detail in Materials and Methods.
Results
ATG-101 binds to PD‐L1 and 4-1BB simultaneously, and blocks PD-1–PD-L1 interaction
ATG-101 is a fully human IgG1-like PD-L1×4-1BB BsAb capable of concurrently binding to human PD-L1 and 4-1BB (hPD-L1 and h4-1BB). The ATG-101 molecule is structurally comprised of two identical IgG1 heavy chains, two identical light chains, with a short peptide (linker 1) linking a single-chain variable fragment (scFv) to the C terminal of each heavy chain. The scFv consists of a heavy chain variable domain (VH) and a light chain variable domain (VL) joined by a linker (linker 2; Fig. 1A).
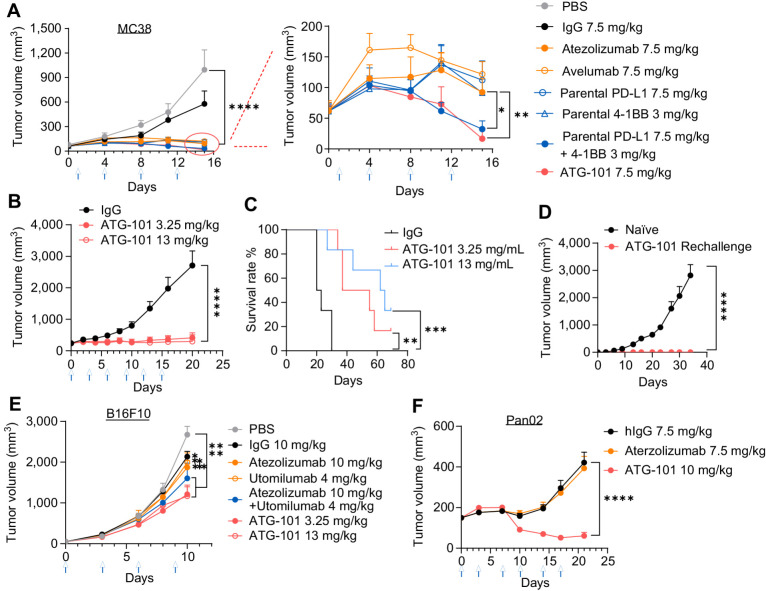
Figure 1.
ATG-101 binds to hPD‐L1 and h4-1BB simultaneously. A, Structure of ATG-101 BsAb. ATG-101 employs the IgG(H)-scFv structure, with the Fab arm targeting PD-L1 and the scFv targeting 4-1BB linked to the C-terminus of the FC domain. DS, disulphide bond. B, Binding capability of ATG-101 to hPD-L1/h4-1BB as determined by ForteBio. ATG-101 was immobilized on the biosensor, and the h4-1BB Fc (left) or hPD-L1 Fc (right) protein was injected first to bind ATG-101, whereafter hPD-L1 Fc (left) or h4-1BB Fc (right) protein was injected to bind. IgG-Fc was used as negative control. C, Kinetics parameter of binding affinity of ATG-101 to hPD-L1/h4-1BB. Kon, association rate constant; Koff, dissociation rate constant; Kd, dissociation constant, Kd = Koff/Kon. D, Binding of ATG-101 to 293T-hPDL1 (left) or 293T-h41BB (right). E, Blockade of the binding of biotinylated PD-1 protein to PD-L1 over expressed HEK293T cells by ATG-101 or parental PDL1 Ab. In D and E, n = 3, data representative of three independent experiments. F, Summary of binding affinity of ATG-101 to Fcγ receptor and FcRn as determined by ForteBio. ND, not detected. G, Binding of ATG-101 and human IgG1 control antibody to human monocytes and NK92MI-FcγRIIIA cells detected by flow cytometry. Representative of n = 3.
The fragment antigen-binding (Fab) arm of ATG-101 binds to PD-L1 while the scFv recognizes 4-1BB. ATG-101 could concurrently bind to human PD-L1 and human 4-1BB to create a trimer at the protein level (Fig. 1B). The binding affinity of ATG-101 to hPD-L1-His and h4-1BB-His protein was assessed by surface plasmon resonance. The binding affinity [dissociation constant (Kd)] of ATG-101 with human PD-L1 (hPD-L1-His) and human 4-1BB (h4-1BB-His) were 0.177 nmol/L and 11.4 nmol/L, respectively (Fig. 1C), reflecting a 64-fold difference in binding affinity. ATG-101 cross-reacted with cynomolgus monkey PD-L1 (Kd = 0.043 nmol/L), mouse PD-L1 (Kd = 1.58 nmol/L) and cynomolgus monkey 4-1BB (Kd = 3.57 nmol/L; Supplementary Fig. S1A). In a competitive binding assay with ATG-101 and analogs of clinical benchmark 4-1BB antibodies, urelumab very effectively blocks binding capability to human 4-1BB of biotinylated ATG-101 while, ATG-101 did not compete with biotinylated utomilumab or biotinylated urelumab for the binding to human 4-1BB in flow cytometry analysis, showing that they bind to distinct epitopes (Supplementary Fig. S1B).
The binding capacity of ATG-101 to cell surface PD-L1 and 4-1BB were determined using flow cytometry. ATG-101 binds to hPD-L1–overexpressed HEK293T cells with a EC50 of 2.35 nmol/L (1 μg/mL translates to 5.1 nmol/L for ATG-101). It also binds with an EC50 of 7.55 nmol/L to HEK293T cells overexpressing human 4-1BB (Fig. 1D). No specific binding signal of ATG-101 has been detected on parental 293T and CHOS cells (Supplementary Fig. S1C). ATG-S101 blocked PD-1 protein binding to cell surface expressed PD-L1, with IC50 of 1.55 and 3.47 nmol/L for human and mouse PD-1–PD-L1 interactions, respectively (Fig. 1E).
To minimize undesired 4-1BB activation through Fc–FcγR interaction–mediated cross-linking of ATG-101 and FcγR-positive immune cells, the Asparagine 297 on CH2 domain of the Fc region of ATG-101 was mutated to alanine to eliminate its binding to the majority of FcγRs (40). ATG-101 exhibited weak binding to FcγRI, IIa/b, and IIIa; however, it maintained binding to FcRn with a Kd of 18.3 nmol/L (Fig. 1F). The binding capacity of ATG-101 to FcRn may prevent the degradation of IgG, and extend its half-life in circulation (41). In addition, wild-type human IgG1 control antibody (hIgG1) positively bound to cells that express FcγRs such as primary human monocytes and NK92MI overexpressing FcγRIIIa (NK92MI-FcγRIIIa), and FcR blockage reduced the binding signal detected by flow cytometry. While ATG-101 demonstrated low to no binding to these cells and FcR blockage did not affect the binding signal (Fig. 1G).
ATG-101 potently activates 4-1BB signaling in immunocytes upon PD-L1 cross-linking
One of the primary causes of liver damage generated by 4-1BB agonists is the stimulation of 4-1BB–positive liver-infiltrating T cells (9). PD-L1 cross-linking–dependent activation of 4-1BB may localize the T-cell activation in the TME and reduce off-tumor toxicity. Multiple assays were conducted to establish the agonistic characteristics of ATG-101 toward 4-1BB. ATG-101 activated HEK293T cells expressing h4-1BB that were integrated with NFκB fluorescence reporter (293T-h41BB-NFκB-Luc) in the presence of CHO cells overexpressing human (CHO-hPDL1) or mouse PD-L1 (CHO-mPDL1), with EC50 of 0.17 nmol/L and 1.23 nmol/L, respectively, while the addition of mock transfected CHO cells did not induce activation of the 4-1BB signaling pathway (Fig. 2A). Furthermore, as a tetravalent “2+2” bsAb, ATG-101 induced significantly higher 4-1BB activation compared with a bivalent PD-L1×4-1BB bsAb, which incorporates Fabs from parental mAbs of ATG-101 [ATG-101(1+1)], suggesting a superior activity of ATG-101 over bivalent PD-L1×4-1BB bsAb (Fig. 2B). In the murine colon carcinoma cell line MC38, elevated cell surface localization of PD-L1 was observed upon stimulation with mouse IFN gamma (mIFNγ; Supplementary Fig. S2A). ATG-S101 activated 4-1BB signaling in the presence of mIFNγ-stimulated MC38 cells. However, neither the combined treatment of parental anti-4-1BB and anti-PD-L1 mAbs nor ATG-101 with nonstimulated MC38 induced significant activation of the 4-1BB signaling (Supplementary Fig. S2B and S2C).
Figure 2.
ATG-101 activates 4-1BB signaling upon PD-L1 cross-linking. A and B, The 4-1BB agonistic activity of ATG-101 in NFκB luciferase assay. A, 293T-h41BB-NFκB-Luc cells were incubated with ATG-101 and CHO-hPDL1 (left) or CHO-mPDL1 (right) for 24 hours. Wild-type CHO cells as negative control. B, 4-1BB activation by ATG-101, bivalent PD-L1×4-1BB bsAb [ATG-101(1+1)], or isotype control antibodies in 293T-h41BB-NFκB-Luc cells with the presence of 293T-hPDL1. Tetravalent and bivalent ATG-101 activated 4-1BB signal pathway with EC50 of 5.25 nmol/L and 15.44 nmol/L, respectively. C, Effect of ATG-101 on IL2 release by CD8+ T cells with the presence of PD-L1+ cell lines. PD-L1 densities (total antibody binding sites) on MCF7, RKO, NCI-H358, and 293T-hPDL1 cell surfaces were determined using QIFIKIT (left). All the indicated cells cocultured with CD8+ T cells and ATG-101 for 3 days, and supernatants were harvested for IL2 ELISA to assess T-cell activation (right). D, ATG-101–induced IFNγ release by human primary CD8+ T cells coculturing with varying proportions of PD-L1+ cells. HEK293-PDL1 cells were mixed with parental HEK293 at different proportions. The IFNγ release was detected by ELISA. E, The individual and mean expression pattern of surface markers and cytokine release of CD3+ T cells from three healthy donors through six rounds of CD3/CD28 activation. F, Individual and mean IL2, IFNγ, and TNFα production by T cells after six rounds of activation (from E), in response to ATG-101, parental PD-L1 antibody, parental 4-1BB antibody, or isotype control. In A, C, and D, data representative of three independent experiments. In A, B, C, and F, data are presented as means ± SEM. Statistical analysis used two-way ANOVA in C and one-way ANOVA in F. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To evaluate the in vitro costimulatory activity of ATG-101 on primary human T cells, CD8+ T cells were prestimulated with anti-CD3 and cocultured with cell lines expressing different levels of PD-L1. ATG-101 activated CD8+ T lymphocytes to produce IL2 at concentrations of 0.1 and 1 nmol/L, with the quantity of IL2 production positively correlating with the cell surface expression or total antibody binding sites of PD-L1 (Fig. 2C). In addition, when HEK293 cells expressing PD-L1 (HEK293-PDL1) were cocultured with parental HEK293 cells in various proportions, ATG-101–induced IFNγ release increased with the proportion of PD-L1+ cells. ATG-101 did not activate CD8+ T cells in the absence of PD-L1 cross-linking (HEK293-PDL1 0%; Fig. 2D). The agonistic activity of ATG-101 was also detected with human PBMCs (hPBMC) stimulated by the superantigen Staphylococcus aureus enterotoxin A. ATG-101 significantly boosted IL2 production by stimulated-hPBMCs in the presence of CHO-PDL1 cells. In the absence of CHO-PDL1 cells or ATG-101, little IL2 production was observed (Supplementary Fig. S2D). ATG-101 caused hPBMCs to produce minimal proinflammatory cytokines when cocultured with human umbilical vein endothelial cells expressing moderate levels of PD-L1, but significantly higher levels when cocultured with CHO-PDL1, which expresses much more PD-L1 (Supplementary Fig. S2E and S2F).
Exhaustion of T cells has been linked to the acquired resistance of ICIs (2–4). To assess the potential effect of ATG-101 on exhausted-like T cells, human T cells were isolated, and the cell exhaustion was induced through consistent, strong activation with anti-CD3/CD28 beads for six cycles (42). The sixth stimulation by beads barely caused IL2/TNFα production, indicating some features of exhaustion (Fig. 2E). In the presence of PD-L1+ cells, ATG-101 could stimulates the IL2, INFγ, and TNFα production from these exhausted-like T cells, while IgG control or parental mAbs did not demonstrate such effect (Fig. 2F).
Taken together, ATG-101 strongly activates 4-1BB in immunocytes in a PD-L1 cross-linking–dependent manner. Agonistic potency correlates positively with PD-L1 expression and the proportion of PD-L1+ cells.
ATG-101 shows potent antitumor effect in vivo
The in vivo antitumor efficacy of ATG-101 was evaluated in h4-1BB KI mice (C57BL/6) bearing murine MC38 colon tumors, Hepa1–6 liver cancer, B16F10 melanoma, or Pan02 pancreatic tumors (Fig. 3; Supplementary Fig. S3).
Figure 3.
ATG-101 shows antitumor efficacy in vivo. A–D, MC38 colon cancer xenograft model on h4-1BB KI mice (C57BL/6). A, When the TV reached about 60 mm3 (day 0), the mice were treated with indicated antibody twice a week for 2 weeks (n = 5 for PBS; n = 6 for the others). B and C, In another MC38 xenograft model, when the TV reached about 239 mm3, the mice were grouped and treated with indicated antibody twice a week for 2 weeks (n = 5 for PBS; n = 6 for the other groups). Growth curves of MC38 tumor (B) and Kaplan–Meier survival plot of mice (C). D, Tumor-free mice from B were rechallenged with MC38. ATG-101 cured (n = 2) or naïve mice (n = 3). E, and F, B16F10 melanoma model or Pan02 pancreatic tumor model on h4-1BB KI mice. The mice were intraperitoneally injected with tested articles once every 3 days (n = 8). In A, B, D, E, and F, means ± SEM are shown. Statistical analysis used two-way ANOVA for tumor growth comparison and log-rank test (Mantel–Cox) for survival analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. In the tumor growth curves, only P values on the last day were shown except for B16F10 whose P value on day 10 is shown.
In the MC38 model, mice with an average TV of 60 mm3 were administered PBS control, human IgG Fc, ATG-101, atezolizumab (analog), avelumab (analog), parental 4-1BB mAbs, parental PD-L1 mAbs, or a combination of 4-1BB and PD-L1 parental mAbs twice weekly. ATG-101 at 7.5 mg/kg exhibited a tumor growth inhibition (TGI) of 109% on day 15 after treatment. TGI values for PD-L1 mAbs atezolizumab and avelumab were 94.28% and 88.35%, respectively. The efficacy of the combination of 4-1BB and PD-L1 mAbs was comparable to that of ATG-101 (Fig. 3A). No body weight loss was observed in any of the treatment groups (Supplementary Fig. S3A). ATG-101 treatment also demonstrated antitumor efficacy and survival benefit in a Hepa1-6 syngeneic model (Supplementary Fig. S3B).
To further evaluate the antitumor efficacy of ATG-101 on large, established tumors, IgG-Fc (10 mg/kg, control), and ATG-101 at 3.25 and 13 mg/kg (once every 3 days for a total of six doses, Q3D×6) were delivered to mice carrying MC38 tumor with an average TV of 239 mm3. Compared with the IgG-Fc control, ATG-101 treatment significantly suppressed tumor growth and prolonged the survival of tumor-bearing mice, without reducing body weight (Fig. 3B and C; Supplementary Fig. S3C and S3D). The average TGI on day 20 of 3.25 and 13 mg/kg ATG-101 doses were 93.0%, and 97.7%, respectively. Particularly, 2 out of 6 mice treated with 13 mg/kg ATG-101 rejected the tumor completely (Supplementary Fig. S3D). The two tumor-free animals were rechallenged with the MC38 cells, and these mice did not acquire tumors (Fig. 3D).
In addition, ATG-101 has been evaluated in h4-1BB KI mice bearing immunologically “cold” B16F10 syngeneic melanoma tumors. Mice received PBS, IgG, ATG-101, utomilumab analog, atezolizumab analog, or the combination of analogs (Q3D×5). ATG-101 was well tolerated, with no substantial weight loss or serious side effects (Supplementary Fig. S3E). On day 10, ATG-101 at 3.25 and 13 mg/kg exhibited TGIs of 55.74% and 57.42%, respectively, while utomilumab, atezolizumab, or the combination demonstrated TGIs of 26.03%, 30.52%, and 40.75%, respectively (Fig. 3E). In addition, ATG-101 at a dose of 13 mg/kg demonstrated a survival advantage over the control groups (Supplementary Fig. S3F).
Finally, in h4-1BB KI mice (C57BL/6J) bearing Pan02 murine pancreatic tumors, atezolizumab did not show antitumor efficacy at 7.5 mg/kg, while ATG-101 at a molar equivalent dosage of 10 mg/kg was well tolerated and significantly inhibited tumor growth, resulting in 78.48% TGI on day 24 (Fig. 3F; Supplementary Fig. S3G and S3H). These data suggest that ATG-101 has the potential to treat cancers with primary resistance to anti-PD-(L)1 therapies.
Taken together, ATG-101 demonstrated potent in vivo antitumor activity in mouse tumor models, including those with large established tumors, immunologically “cold” tumors, and tumors with primary resistance to ICIs.
ATG-101 bolsters antitumor immunity in the TME
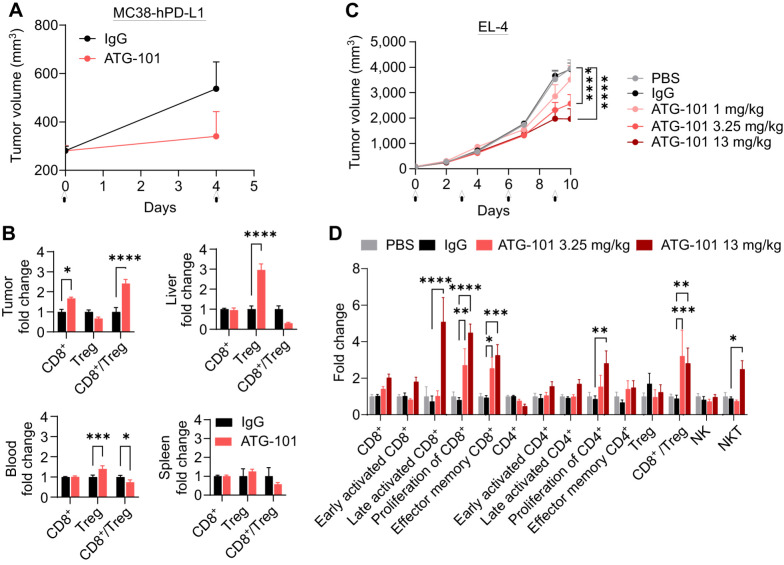
To determine the pharmacodynamic effects of ATG-101, h4-1BB KI mice bearing MC38-hPD-L1 tumors were treated with IgG control or ATG-101 (9.9 mg/kg) on day 0 and day 4 after grouping. Treatment with ATG-101 resulted in TGI (Fig. 4A). Forty-eight hours after the last dose administration, T cells were isolated from tumor, liver, spleen, and blood, and analyzed by flow cytometry (Fig. 4B). ATG-101 increased the infiltration of cytotoxic CD8+ T cells and the ratio of CD8+ T cells/Tregs in the tumor, compared with the IgG control. Interestingly, increased Treg levels and a decreased CD8+ T/Treg ratio were detected in blood and liver in response to ATG-101, possibly due to a compensatory effect (Fig. 4B; Supplementary Fig. S4A and S4B). The decreased CD8+ T/Treg ratio in liver suggested an anti-inflammatory state and decreased risk of ATG-101–induced liver damage.
Figure 4.
ATG-101 enhances the antitumor immunity in the TME. A, Growth curves of MC38-hPD-L1 colon tumors treated with IgG or ATG-101 on day 0 and day 4, as indicated by arrow. B, Normalized CD8+ T-cell and Treg number in tumor, liver, blood, and spleen from A. Forty-eight hours after the last administration, the samples were collected. Fold change indicates ATG-101 treatment group versus mean value of IgG control group. C, Growth curves of EL4 T-cell lymphoma. When the tumor size reached about 71 mm3, the mice (n = 8) were grouped and treated with PBS, IgG, or ATG-101. D, Normalized TIL analysis results from tumors from C. The makers used to classify the cell subsets in flow cytometry analysis are summarized in Supplementary Table S2. Fold change indicates IgG or ATG-101 treatment group versus mean valve of PBS group. Means ± SEM are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA. In the tumor growth curves, only P values on the last day are shown.
In lymphoma EL-4 model, ATG-101 at 3.25 and 13 mg/kg (once every 3 days) exhibited significant TGI compared with PBS or IgG group, with TGIs of 35% and 50%, respectively (Fig. 4C). Compared with PBS and IgG controls, ATG-101 at 3.25 and 13 mg/kg boosted the proliferation of CD8+ T cells, infiltration of effector memory T cells, and CD8+ T/Treg ratio in the TME. Specifically, 13 mg/kg ATG-101 increased the infiltration of late activated T cells and NK T cells (NKT) by 5 and 2.5 folds, respectively (Fig. 4D; Supplementary Fig. S4C). These findings suggest that ATG-101 may enhance antitumor immunity in the TME.
ATG-101 demonstrates in vivo efficacy in a tumor model progressing on anti-PD-L1 treatment
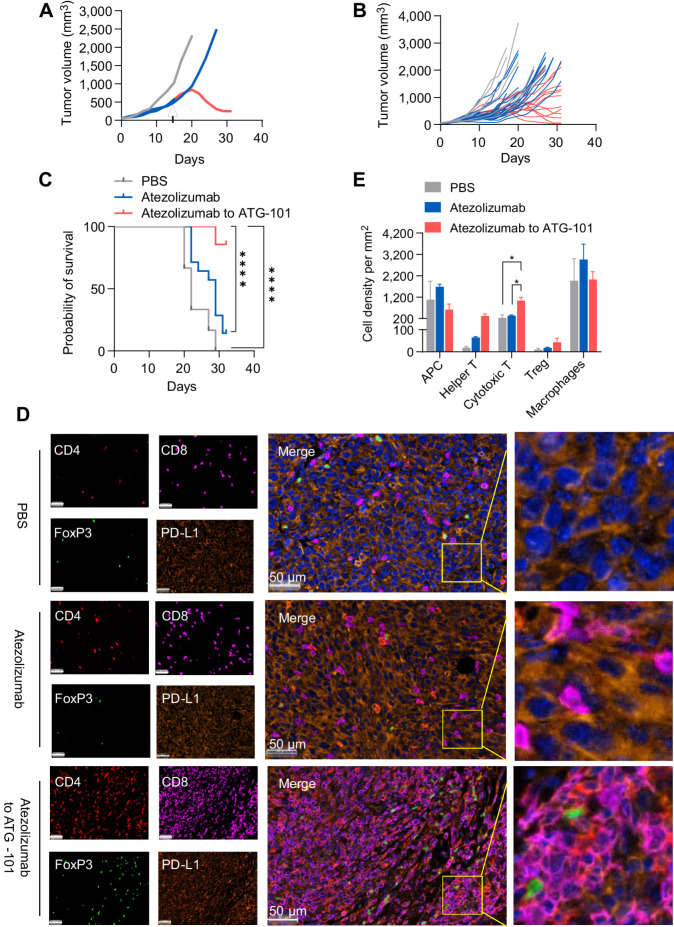
To evaluate the potential of ATG-101 in treating diseases with acquired resistance to ICI, antitumor activity of ATG-101 in a MC38 tumor model progressing on anti-PD-L1 treatment was tested. Mice bearing MC38 tumors were treated with anti-PD-L1 initially to achieve TGI and upon disease progression, half of the mice were switched to ATG-101 (“atezolizumab to ATG-101”), while the other half remained on anti-PD-L1 treatment (atezolizumab; Fig. 5A and B). Both atezolizumab and “atezolizumab to ATG-101” inhibited tumor growth compared with the PBS control group, whereas ATG-101 induced growth inhibition or regression in tumors that had progressed on atezolizumab (Fig. 5A and B). In addition, “atezolizumab to ATG-101” resulted in a significant survival advantage over atezolizumab group and the PBS control group (Fig. 5C). Furthermore, tumor specimens were collected at the end of the study, and the cell types in the TME were analyzed using multiplex immunofluorescence staining. ATG-101 significantly augmented the number of CD8+ T cells (cytotoxic T cells) and tended to upregulate CD4+ T cells (Th cells) in the TME, in comparison with the PBS control or atezolizumab groups (Fig. 5D and E; Supplementary Fig. S5).
Figure 5.
ATG-101 demonstrates in vivo efficacy in MC38 tumor model progressing on anti-PD-L1 treatment, enhancing the antitumor immunity in the TME. A, Representative individual MC38 tumor growth curve. The mice were treated with PBS (black), 10 mg/kg atezolizumab only (red), or mice initially treated with 10 mg/kg atezolizumab and switched to 13 mg/kg ATG-101 upon disease progression (red to blue). Arrow, the day switching atezolizumab to ATG-101. B, Individual tumor growth spaghetti plots of mice treated with PBS (black; n = 6), atezolizumab only (red; n = 14), and atezolizumab to ATG-101 (red-blue; n = 14). C, Survival data of mouse shown in B. ****, P < 0.0001 by log-rank (Mantel–Cox) test. D, Representative images of multiplex IHC staining of tumor samples collected from mouse from B. The tumor slices were stained for CD4 (Th cell; red), CD8 (effector T cell; purple), Foxp3 (Treg; green), and PD-L1 (cancer cells; dark orange). Nucleus was labeled with DAPI (blue). Insets in the merged images are enlarged. Scale bars, 50 μm. E, Quantitative analysis of cell density of infiltrated immune cell in D. Statistical analysis used one-way ANOVA. *, P < 0.05. Means ± SEM are shown. n = 3 in PBS and atezolizumab group; n = 6 in ATG-101 group.
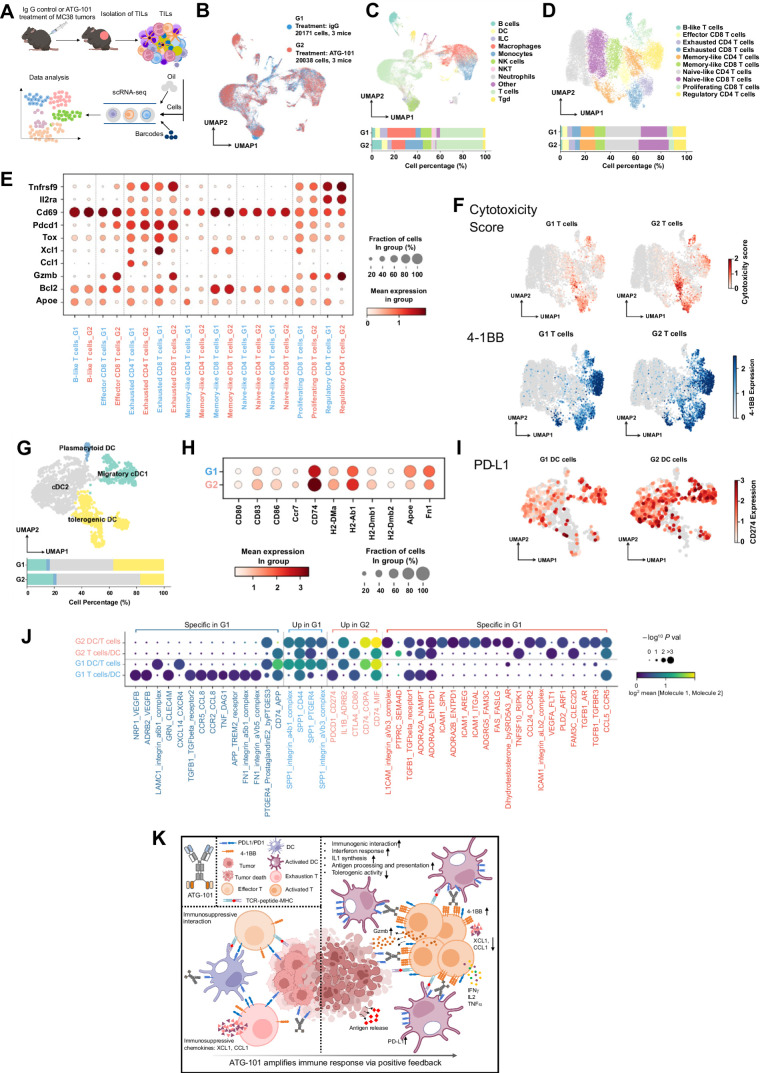
A scRNA-seq study revealed the landscape of TME at the single-cell, transcriptome level after ATG-101 treatment
scRNA-seq has been utilized to profile TMEs (43). To better understand the biology of immune responses elicited by ATG-101 in the TME, scRNA-seq was applied to TILs isolated from MC38 tumors treated with IgG control (Group 1, G1) or ATG-101 (Group 2, G2; Fig. 6A). G2 had a higher proportion of T cells, DCs, B cells, and monocytes, and a lower proportion of NK cells and macrophages (Fig. 6B and C). In accordance with the dual-binding ability of ATG-101 to 4-1BB expressed on T cells and PD-L1 on DCs, the gene expression profiles of T cells and DCs between G1 and G2 varied substantially (Supplementary Table S3). To study the dynamics of gene expression in different T-cell subpopulations, each cluster of T cells was annotated with canonical T-cell markers (Fig. 6D; Supplementary Table S4). G2 T cells exhibited a decrease in the proportion of exhausted CD8+ T cells and Tregs (Fig. 6D). There was a significant increase in cytotoxicity-related granzyme B (Gzmb) expression in effector CD8+ T cells, exhausted CD8+ T cells, proliferative CD8+ T cells, and Tregs in G2 compared with G1 (Fig. 6E and F). Decreased expression of Tox, a T-cell exhaustion marker was noticed in G2, together with a reduction of immunosuppressive chemokines Xcl1 and Ccl1 (Fig. 6E). Expression of 4-1BB was elevated in most of the T-cell subsets in G2 (Fig. 6E; Supplementary Fig. S6A). In addition, consistent with previously described downstream signaling of 4-1BB activation (44), most T-cell subsets in G2, especially exhausted T cells, exhibited increased expression of Traf (Supplementary Fig. S6A). ATG-101 treatment also upregulated IL2 and INFγ-related genes in the T-cell subpopulations (Supplementary Fig. S6B and S6C). These changes suggested that ATG-101 enhanced the antitumor immunity of T cells.
Figure 6.
Single-cell transcriptomic analysis of TILs of IgG or ATG-101–treated MC38 tumors. A, Scheme of the study. B, The two-dimensional UMAP embedding plot of all TILs labeled by treatment groups. C, The two-dimensional UMAP embedding plot of 11 identified main cell types in TILs. The relative proportions of each cell type across two different treatment groups are calculated and shown below. D, The two-dimensional UMAP embedding plot of T cells colored by T-cell subtypes. The relative proportions of each subtype across two different treatment groups are calculated and shown below. E, Dot plot of key genes in T-cell subtypes across two treatment groups. F, Scatter plots showing cytotoxicity score and 4-1BB expression of T cells, projecting on the UMAP plot. Cytotoxicity score was calculated on the basis of the expression of Gzma, Gzmb, Gzmk, Prf1, Nkg7, Ifng, and Il2. G, The two-dimensional UMAP embedding plot of DCs colored by DC subtypes. The relative proportions of each subtype across two different treatment groups are calculated and shown below. H, Dot plot of key genes in DC across two treatment groups. I, Scatter plots showing expression of PD-L1 in DCs, projecting on the UMAP plot. J, Dot plots of significant interactions between DC and T cells detected by Cellphone DB. Dark blue, significant interactions specifically detected in G1; light blue, significant interactions detected in both G1 and G2, while expression of interaction pairs decreased in G2; light red, significant interactions detected in both G1 and G2, while expression of interaction pairs increased in G2; dark red, significant interactions specifically detected in G2. K, Predicted mechanism of ATG-101.
We also discovered substantial differences between G1 and G2 in DC subpopulations and gene expression profiles (Fig. 6G and H; Supplementary Fig. S6D; Supplementary Table S3). Each cluster of DCs was annotated with signature genes (Supplementary Table S4). G2 DCs demonstrated an increase in the proportion of cDC2 and migratory cDC1 subsets and a decrease in the proportion of tolerogenic DCs (Fig. 6G). Upregulated expression of the DC costimulatory molecules, such as Cd80 and Cd86, and DC activation marker Cd83 were observed cDCs in G2 (Fig. 6H; Supplementary Fig. S6E). In addition, Ccr7, an important molecule for DC migration, and a group of genes involved in antigen processing and presentation, including Cd74, H2-DMa, H2-Ab1, H2-DMb1, and H2-DMb2, were upregulated in G2 DCs. G2 DCs also demonstrated downregulation of genes associated with the tolerogenic or immunosuppressive activity of DCs, such as Apoe (45) and Fn1 (Fig. 6H; ref. 46). Gene ontology enrichment analysis of G2-upregulated genes revealed that interferon response, IL1 synthesis, and antigen processing and presentation signaling pathways were activated in G2 (Supplementary Fig. S6F). Within the macrophage subpopulation, the administration of ATG-101 resulted in an increase in the expression of genes associated with M1 macrophages, such as Fcgr1, Cd86, and Stat1, while simultaneously decreasing the expression of genes associated with M2 macrophages, such as Csf1r and Mrc1 (Supplementary Fig. S6G). The expression pattern of some other immune-related genes across different cell subpopulations were analyzed and listed in Supplementary Fig. S6H and Supplementary Table S3. Because of the increased gene expression of PD-L1 in DCs (Fig. 6I) and 4-1BB in T cells following ATG-101 treatment, it is anticipated that ATG-101 will promote greater T cell-dendritic cell (T-DC) cross-linking over time, further activating 4-1BB signaling, strengthening T cell–DC interaction, and generating a positive feedback loop. To test the hypothesis, we analyzed T-DC communication and interaction using CellPhoneDB (39). In G1 and G2, 133 and 135 interactions were detected, respectively, of which, 119 interactions were detected in both groups (Supplementary Table S5). There were 14 G1-unique interactions between DCs and T cells, including tumor-promoting interactions such as CCR5/CCL8 (47, 48), FN1/integrin (49), and TREM2/APP (Fig. 6J; refs. 50, 51). While 21 G2-unique interactions were identified, including four ICAM1-involved interactions, which have been reported to be important for CD8+ T-cell memory and T-cell activation (Fig. 6J; refs. 52, 53). These findings provide support for the concept that ATG-101 altered the interaction between DCs and T cells, hence enhancing antitumor immunity in the TME via positive feedback (Fig. 6K).
ATG-101 demonstrated good safety profile in vitro and in vivo
The potential toxicity profile of ATG-101 was examined in a GLP-compliant 4-week repeated dose toxicity study in cynomolgus monkeys via intravenous infusion, followed by a 4-week recovery period. During the study, there were no significant changes in serum alanine transaminase (ALT) and aspartate transaminase (AST) concentrations between ATG-101 groups and vehicle groups at any time point, indicating that ATG-101 exhibited no hepatotoxicity in cynomolgus monkeys even at the highest 100 mg/kg dose level (Fig. 7A and B). No ATG-101–related macroscopic findings in liver were observed in all treatment groups (Fig. 7C). The highest non-severely toxic dose of ATG-101 was determined to be 100 mg/kg.
A RO study was also performed to measure the changing conditions of PD-L1 RO rate on CD3+ T cells during the GLP toxicology study. On day 1, 2 hours following the initial dose, the RO of each ATG-101 dosing group was close to 100% (97.04%–103.86%). Throughout the dosing and recovery period (day 14, day 28, day 43, and day 57), the RO at 10, 50, and 100 mg/kg maintained at or above 83.28% (83.28%–110.13%), indicating adequate exposure to the ATG-101–related targets during the experiment (Fig. 7D).
CRS is a common adverse event induced by therapeutic immune agonists (54). ATG-101 had no significant impact on monkey plasma cytokine levels (Fig. 7E; Supplementary Table S6). Besides, the effects of ATG-101 on cytokine release were also evaluated utilizing hPBMCs from 6 distinct donors in both soluble and plate-bound forms of the antibody. The strength of cytokine release induced by ATG-101 at concentrations of 0.3 or 300 μg/mL was significantly less than the positive controls, and no significantly increased cytokine levels were detected in both soluble and plate-bound formats of the cytokine release assay (Supplementary Fig. S7). In addition, serum proinflammatory cytokines, such as IL6, TNFα, and IL1ß, were not significantly altered by ATG-101 in MC38 mice model from Fig. 6A (Supplementary Fig. S8). These data suggest a low risk of ATG-101–induced CRS both in vitro and in vivo.
A computational semimechanistic pharmacology model of ATG-101 provides a rational basis for clinical dose selection and prediction of optimal biological dose
A quantitative systems pharmacology (QSP) model for ATG-101 was developed to predict clinical starting and efficacious doses for first-in-human studies using the available preclinical data (Fig. 8A). The three-compartment model predicts the formation of the tetrameric complex between the ATG-101, 4-1BB, and PD-L1, referred to as the “trimer,” as well as the RO to individual targets. This allows the model to quantify 4-1BB–driven activation as well as checkpoint blockade along the PD-1: PD-L1 axis. The binding parameters (e.g., individual target affinities, the avid trans-cell complex formation rates) were calibrated to in vitro pharmacodynamic readouts (on-cell binding, NFκB signaling, etc.) and validated against PD1-blocking assays and IL2 release (Fig. 8B).
Figure 8.
Semimechanistic pharmacology model of ATG-101. A,In vitro model diagram. The model has three compartments: a central compartment, representing the circulation; a tumor compartment; and a peripheral compartment, representing other tissues into which the drug distributes. Drug binds to either receptor initially, then cross-link to the other. Trans-cell complexes (i.e., trimers) are assumed to drive the pharmacologic activity of ATG-101. B,In vitro model calibration and validation. Two cross-linking rates (“strong avidity” and “weak avidity” lines) were calibrated to capture the uncertainty in the parameter (calibration; left). The model was validated with PD-1/PD-L1 blocking assays (validation; right). Lines show model predictions and points show data values for CHO cells expressing hPD-L1 and mPD-L1. C,In vivo human solid tumor model predictions. The in vivo human solid tumor patient model was used to simulate five doses administered every 3 weeks at levels from 0.001 to 10 mg/kg and predict circulating free drug levels, tumor trimer formation, and tumor PD-L1 RO over time. The cross-linking rate and PD-L1 per tumor cell were varied to include the effects of parameter uncertainty and variability in the predictions. D–F, Human model simulated trimer versus PD-L1 RO. D, Schematic representing the pharmacologic regimes between trimer formation and RO of PD-L1. E, Simulations of trimer formation versus PD-L1 RO in high versus low PD-L1 density and high versus low cross-linking avidity for ATG-101 at once every 3 weeks. F, As in the previous panel, the x-axis indicates the tumor cell PD-L1 RO at trough. The y-axis indicates the number of trimers per tumor-infiltrating T cell at trough. Points indicate select doses, with text indicating the dose level in mg/kg outside the parentheses and mg inside the parentheses. Panels indicate the dosing frequency. G, NFκB signaling assays of tetravalent ATG-101 and bivalent ATG-101 in 4-1BB activation. H, Schematic outlining the features of the optimal dose of ATG-101. There is a “sweet spot” between trimer formulation, PD-L1 RO, and BsAb safety. OBD, optimal biological dose.
Human predictions were performed by translating the calibrated binding parameters with human-specific parameters for receptor concentrations, receptor turnover, drug transport, and elimination. Predicted time series for the pharmacokinetics in blood as well as trimer formation and PD-L1 RO in the tumor compartment are shown in Fig. 8C for once every 3 weeks dosing. Given the uncertainty in the avidity parameters of the trans-cell binding and the clinical variability of PD-L1 expression, high and low bookend values were carried forward in the analyses.
The model was then used to evaluate an appropriate efficacious dose that gives the overall best efficacy on both the 41BB-activation and checkpoint inhibition axes. Note that at high doses, the formation of trimer is inhibited because the potential cross-arm binding receptors are occupied by single-bound drug molecules, a “hook effect” phenomenon common to all bispecific molecules (55). This presents a potential tradeoff between complete coverage of PD-L1 with the optimal formation of trans-cell complexes (Fig. 8D). The most favorable regimen lay at a high PD-L1 RO with maximal trimer formation at a single dose level. The ability to achieve this balance depends on the overall avidity of the molecule, especially the trans-cell binding. The model-predicted landscape of trimer versus PD-L1 RO for ATG-101 is visualized in Fig. 8E. Even in a range of parameter regimes given the uncertainty in the model, trimer and PD-L1 RO are both maximized at approximately 2 mg/kg, with less likely hood of inducing hook effect compared with a monovalent format (Fig. 8F). To validate these results, in vitro 4-1BB activation experiments were performed with the full version of ATG-101 and a monovalent format similar to the GEN1046 structure (Fig. 8G). The binding results indicate a similar trend as predicted by the systems pharmacology model. This demonstrates the differences in format can affect the overall avidity of the molecule in ways that are consistent with the model predictions between the two molecules.
Overall, these simulations predict that tetravalent ATG-101 is less susceptible than bivalent molecules to loss of trimer formation at higher doses due to its stronger cross-target binding. This may allow the drug to be dosed such that the benefits of 4-1BB activation and checkpoint inhibition are maximized simultaneously, while presenting a safe therapeutic for patients (Fig. 8H).
Discussion
The clinical development of agonistic 4-1BB antibodies has been hampered by hepatotoxicity (urelumab) or suboptimal efficacy (utomilumab; ref. 9). Bifunctional proteins targeting both a TAA and 4-1BB have been investigated in clinical trials and preclinical models, and they have been shown to circumvent these difficulties by inducing tumor cell–mediated 4-1BB agonism (13–16). As a tetravalent PD-L1×4-1BB BsAb, ATG-101 potently activates 4-1BB–positive T cells in a PD-L1 cross-linking–dependent manner. The higher affinity of PD-L1 binding versus 4-1BB binding ensures TME distribution of ATG-101, and the deficiency of FcγRs binding ensures that ATG-101 shows 4-1BB agonist activity only when cross-linked by PD‐L1+ cells. As a result, ATG-101 strongly inhibits the growth of anti-PD-(L)1–resistant tumors in vivo, without causing liver damage in cynomolgus monkeys at doses levels up to 100 mg/kg.
It has been reported that a high affinity difference between cross-linking target (e.g., TAA or PD-L1) and 4-1BB leads to potential functional advantages of the bispecific molecules. However, most clinical PD-L1×4-1BB bispecific molecules demonstrated less than a 10-fold difference between PD-L1 and 4-1BB binding affinities (4-1BB Kd/PD-L1 Kd; refs. 10, 20–23, 25). The 4-1BB Kd/PD-L1 Kd for GEN1046, MCLA-145, ABL503, FS222, and PRS-344 was 0.15 nmol/L/0.16 nmol/L, 1.9 nmol/L/0.51 nmol/L, 13.8 nmol/L/3.07 nmol/L, 0.66 nmol/L/0.19 nmol/L, and 4.84 nmol/L/0.68 nmol/L, respectively (10, 20–23, 25, 56). ATG-101, however, demonstrated a significantly greater affinity difference, with a 4-1BB Kd/PD-L1 Kd ratio of 64.4 (11.4 nmol/L/0.177 nmol/L). ATG-101 and ABL503 (22) have lower binding affinities to 4-1BB (Kd > 10 nmol/L) compared with other PD-L1×4-1BB bispecific molecules. Yet, a recent study revealed that low-affinity agonistic antibodies displayed enhanced agonism compared with higher affinity parental molecules (56). Although preclinical advantages have been revealed, it must be determined to what extent the reduced 4-1BB arm binding affinity and greater 4-1BB Kd/PD-L1 Kd ratio of ATG-101 translate into clinical benefit.
The biological activity of a PD-L1×4-1BB bispecific is primarily mediated by two mechanisms: (i) PD-L1 cross-linking–dependent 4-1BB activation, which occurs as a result of PD-L1-antibody-4-1BB trimer formation; and (ii) blockade of PD-1–PD-L1 interaction, the rate of which can be estimated by the RO of PD-L1. Therefore, the saturation biological activity is supplied by maximal trimer formation in addition to 100% PD-L1 RO. GEN1046, a "1+1″ bivalent BsAb, is the most advanced PD-L1×4-1BB BsAb. It has demonstrated encouraging early clinical activity in a heavily pretreated patient population with advanced solid tumors, including those resistant to prior anti-PD-(L)1 treatments (19, 57). In addition, GEN1046 appears to have a manageable safety profile with flat dosages up to 1,200 mg (19). However, a much lower 100 mg flat dose of GEN1046, which was predicted to yield only 70% PD-L1 RO (58), was chosen for the dose expansion phase and the combination with anti-PD-1 is being evaluated in a clinical trial (59). This was owing to the fact that greater dosages resulted in decreased trimer formation or the “hook effect” (58). These findings suggest that although being safely administered, GEN1046 could not reach a dose level at which it could deliver saturated biological activity.
The formation of this trimeric complex is subject to various factors such as binding affinity to each target, avidity effects between the two targets, the density of target receptors, and the E:T ratios. While these trimeric complexes are difficult to detect directly, they can be predicted via mechanistic modeling. By our QSP modeling, ATG-101 dosed at 2 mg/kg (140 mg) once every 3 weeks is projected to achieve greater than 90% trimer formation and greater than 90% PD-L1 RO in humans, suggesting the potential of ATG-101 to hit the “sweet spot” in clinic. In addition, the simulation implies that tetravalent binding of the "2+2″ BsAb may result in greater trimer formation and reduced hook effect than the "1+1″ format.
Anti-PD1/PD-L1 therapy has shown great success in the treatment of malignancies. However, only a small subset of patients exhibit deep and durable clinical responses, and many of those who initially respond are likely to develop acquired resistance (60). A “cold” TME is one of the major causes for the primary resistance to ICI treatment (61). Tumors with little antitumor immune cell infiltration in the TME or lack of pre-existing antitumor immune responses due to immunosuppressive TME have been defined as “cold,” and have characteristically poor response to ICIs (1, 61). B16F10 is a “cold,” syngeneic murine melanoma model with poor T-cell infiltration (62). mAbs targeting PD-L1 or 4-1BB shows little antitumor efficacy in the B16F10 model, while ATG-101 demonstrated potent antitumor activity. EL-4 is a syngeneic murine T-cell lymphoma model that is resistant to anti-PD-L1 treatment (63). ATG-101 inhibited EL-4 growth in a dose-dependent manner. Notably, ATG-101 significantly increased the infiltration, proliferation, and activation of CD8+ T cells, the infiltration of NKT cells, and the CD8+/Treg ratio in the TME, implying a role in turning “cold” tumors “hot” and increasing antitumor immunity. This may be the underlying mechanism of ATG-101’s activity in treating ICI-resistant tumors.
Although the downstream signaling of 4-1BB has been extensively studied, the dynamics of TME at the single-cell, transcriptome level in response to a PD-L1×4-1BB BsAb treatment has not been reported. Utilizing scRNA-seq, we observed increased expression of cytotoxic marker Gzmb and decreased expression of exhaustion marker Tox in CD8+ T cells in response to ATG-101, indicating that ATG-101 may play a role in reversing T-cell exhaustion and enhancing cytotoxicity. In addition, ATG-101 activated IFN response, IL1 synthesis, and antigen processing and presentation signaling pathways in DCs, downregulating the proportion of tolerogenic DCs and the expression of genes associated with the tolerogenic or immunosuppressive activity of DCs. Notably, ATG-101 treatment elevated the gene expression of PD-L1 in DCs and 4-1BB in T cells, which suggested that ATG-101 may promote greater T-DC cross-linking over time, further activating 4-1BB signaling, strengthening T cell–DC interaction, and generating a positive feedback loop. Consistent with the hypothesis, we found ATG-101 altered the interaction between DCs and T cells. Twenty-one DC–T interactions were only observed in the ATG-101–treated group, four of which are ICAM-1 related and reported to be essential for CD8+ T-cell memory and T-cell activation (52, 53). These results deepened the understanding of the mechanism underlying PD-L1×4-1BB BsAb regulation of TME at the transcriptome level. However, this study still has some limitations. In this study, for instance, TILs were only analyzed at a single time point, obscuring the evolution of the immune landscape after drug treatment. In addition, the biological function and downstream mechanism of ATG-101–related T-DC interactions discovered in this study require additional experimental validation and investigations.
Recent studies have demonstrated that a portion of the efficacy of anti-PD(L)1 antibodies is derived from nontumor tissue, such as lymph nodes, and nontumor cell types, such as T cells, macrophages, B cells, DCs, neutrophils, and fibroblasts. By interacting with tumor cells or other cells types, PD-L1 expressed on nontumor cells regulates antitumor immunity and influences the efficacy of anti-PD-1/PD-L1 therapy in patients (64) In addition, PD(L)1 blockade–induced DC activation in lymph nodes can promote T-cell recruitment to the tumor in both early and advanced stages of cancer (65). In the current study, scRNA-seq analysis also revealed altered T-DC cross-talk in response to ATG-101. Therefore, it is also possible that a part of the antitumor efficacy of PD-L1×41BB BsAbs is due to a mechanism unrelated to tumor cells. To validate this hypothesis and determine the importance of the nontumor mechanism to the drug's efficacy, however, further research is required.
In summary, the preclinical results illustrated in this study are consistent with the contention that ATG-101 will provide safe and effective combined PD-L1 antagonism and tumor-specific 4-1BB agonism. A phase I, multicenter, dose-escalating clinical trial evaluating ATG-101 in patients with solid tumors and hematologic malignancies is ongoing (66).
Supplementary Material
Supplementary Methods, Supplementary Figures and Supplementary Table S1-2
ScRNAseq data reavealed the cell types and differential expressed genes of TILs from IgG and ATG-101 groups
Genetic marker or signature genes for cell subtypes described in scRNAseq analysis
Significant interactions between DC and T cells detected by Cellphone DB
ATG-101 did not affecct cytokine release in monkey
Acknowledgments
We thank Innostar, Crown Bioscience, and Biocytogen for the support of several animal studies. We thank Tamara Etto and Nirmal Lorensuhewa for proofreading the article.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
Y.-k. Zhang reports personal fees from Antengene during the conduct of the study and personal fees from Antengene outside the submitted work. P. Chen reports grants from Shanghai Antengene Corporation Limited outside the submitted work. A. Sun reports personal fees from Shanghai Antengene Corporation Limited outside the submitted work. G. Bian reports personal fees from Shanghai Antengene Corporation Limited outside the submitted work. D. Flowers reports personal fees from Antengene Corporation during the conduct of the study. K. Subramanian reports other support from Antengene during the conduct of the study. K. Lynch reports personal fees and other support from Antengene during the conduct of the study; personal fees and other support from Antengene outside the submitted work; and K. Lynch holds the role of Chief Medical Officer with Antengene between 2021 and 2023, during which time some of this work was completed. J. Mei reports personal fees from Antengene Biotech LLC and Antengene Corporation Co., Ltd. outside the submitted work. B. Shan reports personal fees from Antengene Corporation Co., Ltd. outside the submitted work. B. Hou reports personal fees from Antengene Corporation Co., Ltd. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
H. Yuwen: Investigation, writing–original draft, project administration, writing–review and editing. H. Wang: Investigation. T. Li: Investigation, writing–review and editing. Y. Ren: Investigation, writing–original draft, writing–review and editing. Y.-k. Zhang: Formal analysis, investigation. P. Chen: Investigation, writing–review and editing. A. Sun: Formal analysis, writing–review and editing. G. Bian: Writing-review and editing. B. Li: Conceptualization, investigation. D. Flowers: Investigation, writing–original draft, writing–review and editing. M. Presler: Investigation, writing–original draft, writing–review and editing. K. Subramanian: Investigation, writing–original draft, writing–review and editing. J. Xue: Data curation, formal analysis. J. Wang: Investigation. K. Lynch: Investigation. J. Mei: Project administration. X. He: Investigation. B. Shan: Investigation, project administration. B. Hou: Conceptualization, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. [DOI] [PubMed] [Google Scholar]
- 2. Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 2016;354:1160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sarmento-Ribeiro AB, Scorilas A, Goncalves AC, Efferth T, Trougakos IP. The emergence of drug resistance to targeted cancer therapies: clinical evidence. Drug Resist Updat 2019;47:100646. [DOI] [PubMed] [Google Scholar]
- 4. Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T cell exhaustion. Science 2016;354:1165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim HD, Park S, Jeong S, Lee YJ, Lee H, Kim CG, et al. 4–1BB delineates distinct activation status of exhausted tumor-infiltrating CD8(+) T cells in hepatocellular carcinoma. Hepatology 2020;71:955–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woroniecka KI, Rhodin KE, Dechant C, Cui X, Chongsathidkiet P, Wilkinson D, et al. 4–1BB agonism averts TIL exhaustion and licenses PD-1 blockade in glioblastoma and other intracranial cancers. Clin Cancer Res 2020;26:1349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho SK, Xu Z, Thakur A, Fox M, Tan SS, DiGiammarino E, et al. Epitope and Fc-mediated cross-linking, but not high affinity, are critical for antitumor activity of CD137 agonist antibody with reduced liver toxicity. Mol Cancer Ther 2020;19:1040–51. [DOI] [PubMed] [Google Scholar]
- 8. Segal NH, Logan TF, Hodi FS, McDermott D, Melero I, Hamid O, et al. Results from an integrated safety analysis of urelumab, an agonist anti-CD137 monoclonal antibody. Clin Cancer Res 2017;23:1929–36. [DOI] [PubMed] [Google Scholar]
- 9. Chester C, Sanmamed MF, Wang J, Melero I. Immunotherapy targeting 4–1BB: mechanistic rationale, clinical results, and future strategies. Blood 2018;131:49–57. [DOI] [PubMed] [Google Scholar]
- 10. Geuijen C, Tacken P, Wang LC, Klooster R, van Loo PF, Zhou J, et al. A human CD137xPD-L1 bispecific antibody promotes anti-tumor immunity via context-dependent T cell costimulation and checkpoint blockade. Nat Commun 2021;12:4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Tan S, Zhang C, Chai Y, He M, Zhang CW, et al. Limited cross-linking of 4–1BB by 4–1BB ligand and the agonist monoclonal antibody utomilumab. Cell Rep 2018;25:909–20. [DOI] [PubMed] [Google Scholar]
- 12. Segal NH, Gopal AK, Bhatia S, Kohrt HE, Levy R, Pishvaian MJ, et al. A phase 1 study of PF-05082566 (anti-4–1BB) in patients with advanced cancer. J Clin Oncol 32:15s, 2014. (suppl; abstr 3007). [Google Scholar]
- 13. Hinner MJ, Aiba RSB, Jaquin TJ, Berger S, Durr MC, Schlosser C, et al. Tumor-localized costimulatory T-cell engagement by the 4–1BB/HER2 bispecific antibody-anticalin fusion PRS-343. Clin Cancer Res 2019;25:5878–89. [DOI] [PubMed] [Google Scholar]
- 14. Rajendran S, Li Y, Ngoh E, Wong HY, Cheng MS, Wang CI, et al. Development of a bispecific antibody targeting CD30 and CD137 on Hodgkin and reed-sternberg cells. Front Oncol 2019;9:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. You G, Lee Y, Kang YW, Park HW, Park K, Kim H, et al. B7-H3×4–1BB bispecific antibody augments antitumor immunity by enhancing terminally differentiated CD8(+) tumor-infiltrating lymphocytes. Sci Adv 2021;7:eaax3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dahlman A, Nelson M, Bannink J, Johnson S, Werchau D, Nilsson A, et al. Preclinical safety and efficacy of a tumor-directed T cell activating 4–1BB x 5T4 ADAPTIR™ bispecific antibody [abstract]. In:Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019Mar 29–Apr 3; Atlanta, GA. Philadelphia (PA): AACR; Cancer Res 2019;79(13 Suppl):Abstract nr 2380. [Google Scholar]
- 17. Liu L, Huang L, Long V, Yang Y, Burns R, Li J, et al. Tumor-antigen 5T4-dependent activation of the CD137 costimulatory pathway by bispecific 5T4 x CD137 x CD137 TRIDENT™ molecules [abstract]. In:Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019Mar 29–Apr 3; Atlanta, GA. Philadelphia (PA): AACR; Cancer Res 2019;79(13 Suppl):Abstract nr 554. [Google Scholar]
- 18. Vezys V, Penaloza-MacMaster P, Barber DL, Ha SJ, Konieczny B, Freeman GJ, et al. 4–1BB signaling synergizes with programmed death ligand 1 blockade to augment CD8 T cell responses during chronic viral infection. J Immunol 2011;187:1634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muik A, Garralda E, Altintas I, Gieseke F, Geva R, Ben-Ami E, et al. Preclinical characterization and phase I trial results of a bispecific antibody targeting PD-L1 and 4–1BB (GEN1046) in patients with advanced refractory solid tumors. Cancer Discov 2022;12:1248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elshiaty M, Schindler H, Christopoulos P. Principles and current clinical landscape of multispecific antibodies against cancer. Int J Mol Sci 2021;22:5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lakins MA, Koers A, Giambalvo R, Munoz-Olaya J, Hughes R, Goodman E, et al. FS222, a CD137/PD-L1 tetravalent bispecific antibody, exhibits low toxicity and antitumor activity in colorectal cancer models. Clin Cancer Res 2020;26:4154–67. [DOI] [PubMed] [Google Scholar]
- 22. Jeong S, Park E, Kim HD, Sung E, Kim H, Jeon J, et al. Novel anti-4–1BBxPD-L1 bispecific antibody augments anti-tumor immunity through tumor-directed T-cell activation and checkpoint blockade. J Immunother Cancer 2021;9:e002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peper-Gabriel JK, Pavlidou M, Pattarini L, Morales-Kastresana A, Jaquin TJ, Gallou C, et al. The PD-L1/4–1BB bispecific antibody-anticalin fusion protein PRS-344/S095012 elicits strong T-cell stimulation in a tumor-localized manner. Clin Cancer Res 2022;28:3387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warmuth S, Gunde T, Snell D, Brock M, Weinert C, Simonin A, et al. Engineering of a trispecific tumor-targeted immunotherapy incorporating 4–1BB co-stimulation and PD-L1 blockade. Oncoimmunology 2021;10:2004661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Claus C, Ferrara-Koller C, Klein C. The emerging landscape of novel 4–1BB (CD137) agonistic drugs for cancer immunotherapy. MAbs 2023;15:2167189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heng TS, Painter MW; Immunological Genome Project Consortium. The immunological genome project: networks of gene expression in immune cells. Nat Immunol 2008;9:1091–4. [DOI] [PubMed] [Google Scholar]
- 34. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soneson C, Robinson MD. Bias, robustness and scalability in single-cell differential expression analysis. Nat Methods 2018;15:255–61. [DOI] [PubMed] [Google Scholar]
- 36. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 40. Wang X, Mathieu M, Brezski RJ. IgG Fc engineering to modulate antibody effector functions. Protein Cell 2018;9:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu R, Oldham RJ, Teal E, Beers SA, Cragg MS. Fc-engineering for modulated effector functions-improving antibodies for cancer treatment. Antibodies 2020;9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dunsford LS, Thoirs RH, Rathbone E, Patakas A. A human in vitro T cell exhaustion model for assessing immuno-oncology therapies. In: Tan S-L, editor. Immuno-oncology: cellular and translational approaches. New York (NY): Springer US; 2020. p. 89–101. [Google Scholar]
- 43. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018;18:35–45. [DOI] [PubMed] [Google Scholar]
- 44. Kim AMJ, Nemeth MR, Lim SO. 4–1BB: a promising target for cancer immunotherapy. Front Oncol 2022;12:968360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kemp SB, Carpenter ES, Steele NG, Donahue KL, Nwosu ZC, Pacheco A, et al. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-kappaB-mediated production of CXCL1. Cancer Res 2021;81:4305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paavola KJ, Roda JM, Lin VY, Chen P, O'Hollaren KP, Ventura R, et al. The fibronectin-ILT3 interaction functions as a stromal checkpoint that suppresses myeloid cells. Cancer Immunol Res 2021;9:1283–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ward ST, Li KK, Hepburn E, Weston CJ, Curbishley SM, Reynolds GM, et al. The effects of CCR5 inhibition on regulatory T-cell recruitment to colorectal cancer. Br J Cancer 2015;112:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hemmatazad H, Berger MD. CCR5 is a potential therapeutic target for cancer. Expert Opin Ther Targets 2021;25:311–27. [DOI] [PubMed] [Google Scholar]
- 49. Wang JP, Hielscher A. Fibronectin: how its aberrant expression in tumors may improve therapeutic targeting. J Cancer 2017;8:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolf EM, Fingleton B, Hasty AH. The therapeutic potential of TREM2 in cancer. Front Oncol 2022;12:984193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lim S, Yoo BK, Kim HS, Gilmore HL, Lee Y, Lee HP, et al. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer 2014;14:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scholer A, Hugues S, Boissonnas A, Fetler L, Amigorena S. Intercellular adhesion molecule-1-dependent stable interactions between T cells and dendritic cells determine CD8+ T cell memory. Immunity 2008;28:258–70. [DOI] [PubMed] [Google Scholar]
- 53. Comrie WA, Li S, Boyle S, Burkhardt JK. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J Cell Biol 2015;208:457–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shimabukuro-Vornhagen A, Godel P, Subklewe M, Stemmler HJ, Schlosser HA, Schlaak M, et al. Cytokine release syndrome. J Immunother Cancer 2018;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Betts A, van der Graaf PH. Mechanistic quantitative pharmacology strategies for the early clinical development of bispecific antibodies in oncology. Clin Pharmacol Ther 2020;108:528–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu X, Orr CM, Chan HTC, James S, Penfold CA, Kim J, et al. Reducing affinity as a strategy to boost immunomodulatory antibody agonism. Nature 2023;614:539–47. [DOI] [PubMed] [Google Scholar]
- 57. Aix SP, Calvo E, Moreno V, Garralda E, Cervantes A, Ramalingam S, et al. 516 Peripheral and tumoral immune activity in the expansion part of the first-in-human DuoBody®-PD-L1×4–1BB (GEN1046) trial. J Immunother Cancer 2021;9:A546. [Google Scholar]
- 58. Bajaj G, Nazari F, Presler M, Thalhauser C, Forssmann U, Jure-Kunkel M, et al. 786 Dose selection for DuoBody®-PD-L1×4–1BB (GEN1046) using a semimechanistic pharmacokinetics/pharmacodynamics model that leverages preclinical and clinical data. J Immunother Cancer 2021;9:A821. [Google Scholar]
- 59. Genmab. Safety and efficacy study of GEN1046 as a single agent or in combination with another anti-cancer therapy for treatment of recurrent (non-small cell) lung cancer. NCT05117242; 2021.
- 60. Lei Q, Wang D, Sun K, Wang L, Zhang Y. Resistance mechanisms of anti-PD1/PDL1 therapy in solid tumors. Front Cell Dev Biol 2020;8:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Petroni G, Buque A, Coussens LM, Galluzzi L. Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat Rev Drug Discov 2022;21:440–62. [DOI] [PubMed] [Google Scholar]
- 62. Yu JW, Bhattacharya S, Yanamandra N, Kilian D, Shi H, Yadavilli S, et al. Tumor-immune profiling of murine syngeneic tumor models as a framework to guide mechanistic studies and predict therapy response in distinct tumor microenvironments. PLoS One 2018;13:e0206223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen P, Liu Y, Deng M, Wang J, Hoenemann D, Lynch K, et al. 608 Synergistic effect of the combination of ATG-017, an ERK1/2 inhibitor, and immune checkpoint inhibitor in preclinical cancer models. J Immunother Cancer 2021;9:A638. [Google Scholar]
- 64. Zhao X, Bao Y, Meng B, Xu Z, Li S, Wang X, et al. From rough to precise: PD-L1 evaluation for predicting the efficacy of PD-1/PD-L1 blockades. Front Immunol 2022;13:920021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Pul KM, Fransen MF, van de Ven R, de Gruijl TD. Immunotherapy goes local: the central role of lymph nodes in driving tumor infiltration and efficacy. Front Immunol 2021;12:643291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Antengene Corporation Limited. A study of evaluating the safety and efficacy of ATG-101 in patients with metastatic/advanced solid tumors and mature B-cell Non-Hodgkin lymphomas (PROBE). NCT04986865; 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Figures and Supplementary Table S1-2
ScRNAseq data reavealed the cell types and differential expressed genes of TILs from IgG and ATG-101 groups
Genetic marker or signature genes for cell subtypes described in scRNAseq analysis
Significant interactions between DC and T cells detected by Cellphone DB
ATG-101 did not affecct cytokine release in monkey



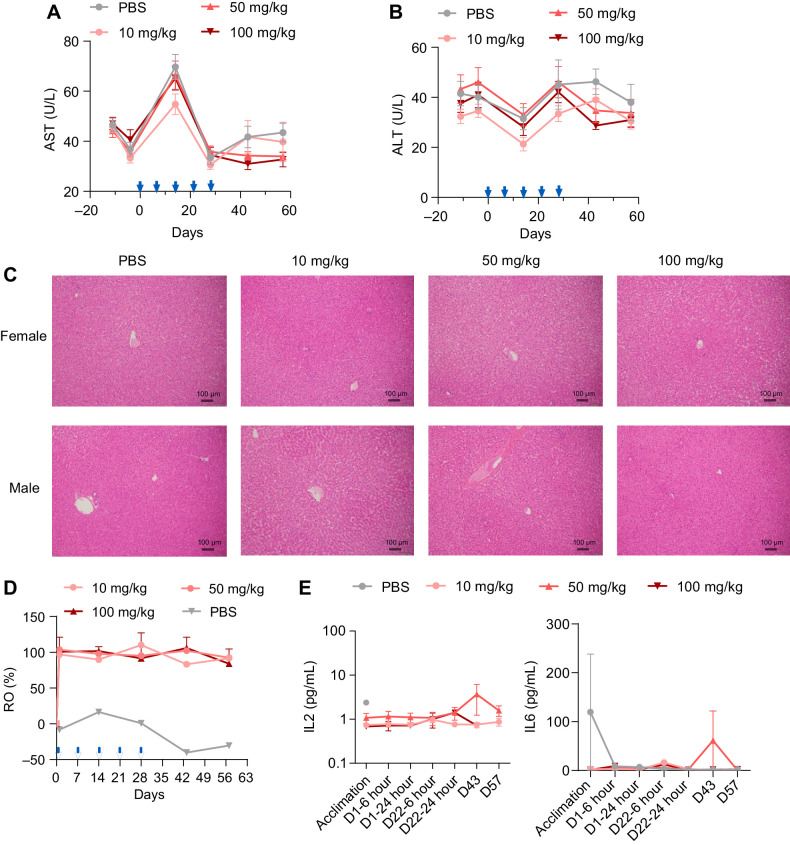
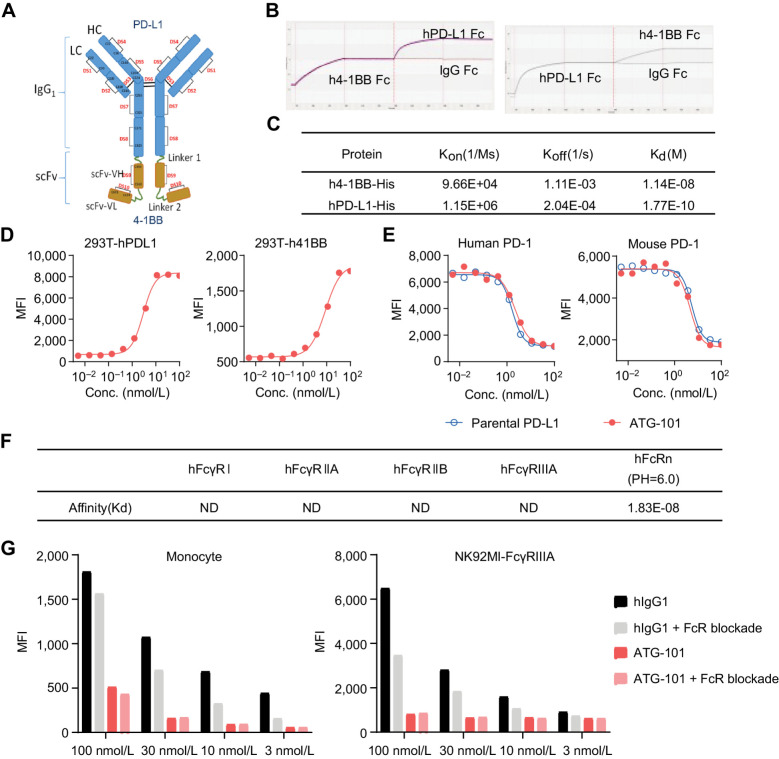
![Figure 2. ATG-101 activates 4-1BB signaling upon PD-L1 cross-linking. A and B, The 4-1BB agonistic activity of ATG-101 in NFκB luciferase assay. A, 293T-h41BB-NFκB-Luc cells were incubated with ATG-101 and CHO-hPDL1 (left) or CHO-mPDL1 (right) for 24 hours. Wild-type CHO cells as negative control. B, 4-1BB activation by ATG-101, bivalent PD-L1×4-1BB bsAb [ATG-101(1+1)], or isotype control antibodies in 293T-h41BB-NFκB-Luc cells with the presence of 293T-hPDL1. Tetravalent and bivalent ATG-101 activated 4-1BB signal pathway with EC50 of 5.25 nmol/L and 15.44 nmol/L, respectively. C, Effect of ATG-101 on IL2 release by CD8+ T cells with the presence of PD-L1+ cell lines. PD-L1 densities (total antibody binding sites) on MCF7, RKO, NCI-H358, and 293T-hPDL1 cell surfaces were determined using QIFIKIT (left). All the indicated cells cocultured with CD8+ T cells and ATG-101 for 3 days, and supernatants were harvested for IL2 ELISA to assess T-cell activation (right). D, ATG-101–induced IFNγ release by human primary CD8+ T cells coculturing with varying proportions of PD-L1+ cells. HEK293-PDL1 cells were mixed with parental HEK293 at different proportions. The IFNγ release was detected by ELISA. E, The individual and mean expression pattern of surface markers and cytokine release of CD3+ T cells from three healthy donors through six rounds of CD3/CD28 activation. F, Individual and mean IL2, IFNγ, and TNFα production by T cells after six rounds of activation (from E), in response to ATG-101, parental PD-L1 antibody, parental 4-1BB antibody, or isotype control. In A, C, and D, data representative of three independent experiments. In A, B, C, and F, data are presented as means ± SEM. Statistical analysis used two-way ANOVA in C and one-way ANOVA in F. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/453f/11094422/2fc62d920828/1680fig2.jpg)