Abstract
Point mutations and inserts in the β3-β4 region of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) are associated with resistance to nucleoside analog inhibitors. This report describes HIV-1 strains from seven patients that were found to have a 3-bp deletion in the β3-β4 region of the RT gene. These patient strains also had a mean of 6.2 drug resistance-associated mutations in their RT genes (range, 3 to 10 mutations). The deletion was most frequently found in strains with the Q151M mutation. Nonnucleoside RT inhibitor mutations were found in six of seven strains. Culture-based drug sensitivity assays showed that deletion-containing isolates had reduced susceptibility to four to eight RT inhibitors. Site-directed mutagenesis experiments showed that the deletion alone conferred reduced susceptibility to nucleoside analogs. Changes in the three-dimensional models of the RT deletion mutants were consistently observed at the β3-β4 loop and at helices C and E in both the presence and the absence of dTTP. Loss of hydrogen bonds between the RT and dTTP were also observed in the RT deletion mutant. These results suggest that the deletion in the RT gene contributes to resistance to several nucleoside analogs through a complex interaction with other mutations in the RT gene.
Treatment of human immunodeficiency virus type 1 (HIV-1)-infected individuals with combinations of protease and reverse transcriptase (RT) inhibitors has been highly effective in increasing both their duration and quality of life (3). Strong adherence to these treatment regimens often reduces the plasma virus concentrations to below the limits of detection by currently available assays. Treatment failure, typically defined as a significant rise from previously suppressed levels of circulating virus, is often associated with the emergence of virus strains resistant to antiretroviral drugs (7).
Mutations in the protease and RT genes of HIV-1 have been shown to confer resistance to antiretroviral drugs (compiled in reference 23). For protease inhibitors and nonnucleoside RT inhibitors (nnRTI), a relatively limited number of mutations provide resistance to all members of each respective class. Resistance mutations selected by nucleoside RT inhibitors (nRTI) are generally drug specific and provide limited cross-resistance to other nRTI. However, patients may have virus strains resistant to many nRTI through either the accumulation of many drug-specific mutations or the acquisition of unique multidrug resistance mutations.
A substantial number of mutations conferring resistance to nRTI have been demonstrated to appear in the β3-β4 region (codons 62 to 78) of HIV-1 RT (18, 23, 29). Point mutations associated with reduced drug susceptibility have been demonstrated at codons 62, 65, 67, 69, 70, 74, 75, and 77 (2, 6, 14, 27). In addition, an insert between codons 69 and 70 has recently been shown to participate in resistance to multiple nucleoside analogs (5, 16, 32). This insert pattern, along with the Q151M complex of mutations (24, 26), comprises two multinucleoside resistance patterns seen in the RT gene. These multinucleoside resistance patterns confer resistance to all nRTI but do not affect susceptibility to nnRTI or protease inhibitors. Recently, the occurrence of a single-amino-acid deletion in the RT gene of an HIV-1-infected individual was reported and associated with high-level zidovudine (AZT) resistance (9). We report the appearance of a similar 1-amino-acid deletion in the β3-β4 region of the HIV-1 RT gene of seven patients and describe its frequency and impact on drug susceptibility.
(This work was presented in part at the 3rd International Workshop on HIV Drug Resistance and Treatment Strategies, San Diego, California, in June 1999.)
MATERIALS AND METHODS
Sequences.
Patient-derived HIV-1 sequences were obtained from a population of patients failing their current antiretroviral therapy who had submitted blood samples for genotyping to either Stanford Hospital or Quest Diagnostics. Sequences were obtained by RT-PCR of plasma-derived HIV-1 RNA followed by dye-labeled dideoxyterminator sequencing using methods and quality-control procedures that have been previously described (32).
Drug susceptibility.
Recombinant isolates were prepared by homologous recombination and tested for susceptibility to RT inhibitors in a peripheral blood mononuclear cell (PBMC)-based cell culture assay as previously described (11, 32). The performance characteristics of this method have been presented (11), and evaluation of replicate results indicated that changes in susceptibility of fourfold or greater were significant. A fourfold change is also accepted as a clinically significant level of resistance (7).
Homology protein structure modeling.
Homology protein structure modeling by satisfaction of spatial restraints was performed by the method of Sali (20). Homology protein structure modeling is founded on building a structural model of a protein on the basis of close similarity to a template protein of known structure. In the first stage of protein structure modeling, the alignment between the unknown sequence and related template structures was obtained. Second, restraints on various distances, angles, and dihedral angles in the sequence were derived from its alignment with the template structures. Finally, the three-dimensional models were obtained by minimizing violations of homology-derived and energy restraints, using conjugate gradients and molecular dynamics procedures.
An important step in homology protein modeling experiments is the evaluation of the model quality. To ensure the quality, we used internal consistency checks as implemented in the software package MODELLER 4 (21) to test the three-dimensional profile. This software package has been tested extensively in structural genomics projects (22) and has been reported to have accuracy approaching that of low-resolution X-ray structures (2.8 Å) or medium-resolution nuclear magnetic resonance structures (10 distance restraints per residue) when there is a high degree of homology between the template structure and the target sequence (17). We have tested the accuracy of MODELLER extensively for HIV protease enzyme-inhibitor complexes, for which a large number of crystal structures are available. At sequence identity values of higher than 85% (as is the case for HIV protease and RT mutants), the accuracy of the modeled structures is better than that obtained by low-resolution X-ray structures (unpublished observations).
Two crystal structure RT templates (open and closed complexes) were used in our homology modeling experiments to examine structural changes both in the presence and in the absence of deoxynucleoside triphosphates (dNTP). One of the templates for the modeling experiments is the open structure of the HIV-1 RT-DNA complex (10). The second template is the closed structure of the HIV-1 RT-DNA-dTTP complex (8). Since in vitro susceptibility assays were carried out with HIV constructs based on wild-type NL4-3, conversions from BH10 and HXB2 to pNL4-3 were performed by using the open (BH10) or closed (HXB2) crystal structure as a template and the NL4-3 sequence as a target in the modeling calculations. No significant changes were observed between the open HIV RT NL4-3 model and the open RT structure from BH10 or the closed HIV RT NL4-3 model and the closed RT structure from HXB2 (unpublished data). The homology models of the HIV RT mutant and wild-type enzymes were obtained by optimizing an objective function (combined spatial restraints and CHARMM energy terms enforcing proper stereochemistry) in Cartesian space. The optimization was carried out using the variable-target function method employing methods of conjugate gradients and molecular dynamics with simulated annealing. Default settings in MODELLER 4.0 (number of iterations in optimization, 200; nonbonded restraint type, dynamic soft-sphere repulsion terms) were used. The modeled structures of the HIV RT variants were least-squares superimposed using the program suite O (12), and the figures were prepared using the program RIBBONS, written by M. Carson (4). The network of interactions between the RT and the dTTP ligand was examined with the program LIGPLOT (31). Files for the homology models of the RT deletion mutants (in Protein Data Bank [PDB] format) are available upon request and include complete main chain and side chain information for both the p66 and p51 components.
RESULTS
RT sequences.
Examination of the HIV protease and RT gene sequences obtained from 8,396 patients showed that seven patients (0.083%) had strains which had a 1-amino-acid deletion in the RT gene. Alignment of these sequences with the clade B consensus sequence (13) indicated that this deletion occurred in the codon 67 to 69 region of the RT gene (Table 1). All patient-derived deletion strains had at least two resistance-associated RT gene mutations, with an average of six mutations per RT gene (range, 3 to 9 mutations). There was a significant association of the deletion with the Q151M mutation (four deletion strains of 158 strains with Q151M versus three deletion strains of 8,241 strains without Q151M; P < 0.0001, chi-square analysis). The four deletion strains with the Q151M mutation had all or part of the Q151M-associated mutation complex (A62V, V75I, F77L, and F116Y). Six of the seven deletion strains possessed at least 1 nnRTI mutation (mean, 2.0 mutations; range, 0 to 3), and all patients had an average of three major protease inhibitor resistance-associated mutations (data not shown).
TABLE 1.
RT gene mutations in HIV-1 strains possessing a 1-amino-acid deletion in the RT gene
| Straina | Codons in β3-β4 regionb
|
Drug resistance mutations (other changes)c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 64 | 65 | 66 | 67–69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | ||
| Wild type | K | K | K | D S T | K | W | R | K | L | V | D | |
| q712 | – | – | – | S G | R | – | – | – | – | – | – | Y181C, G190G/A, T215F, K219E (K20R, K102E, I135T, Q197K, I202I/V, R211K) |
| v1966 | – | – | – | S G | R | – | – | – | I | – | – | M41L, L100I, K103N, T215F, K219Q (E6D, K20R, K102R, D123E, K166R, D177E, V189I, G196K, T200I, Q207E) |
| q489 | – | R | – | D G | – | – | – | – | – | – | – | Y181C (V35L, S48E, D123E, A158S, Q197E, R211K, F214L, K219R) |
| q759 | – | – | – | N G | – | – | – | – | – | I | – | F77L, Y115F, F116Y, Q151M, Y181C, M184V, G190A, K219E (K101E, K122E, I135T, R172K, E203E/Q, R211K) |
| q514 | – | – | – | D G | – | – | – | – | – | T | – | A62V, Q151M (T200A, Q207E, R211K, F214L) |
| q020 | – | – | – | D G | S | – | – | – | – | I | – | A62V, F77L, K101Q, Y115F, F116Y, Q151M, M184V (K122E, S162C) |
| v620 | – | – | – | D G | – | – | – | – | – | T | – | A62V, A98G, V108I, Y115F, Q151M, Y181C, M184V (K20R, K22R, I31R, K122E, F214L) |
Strains identified with Q or V were identified from patient plasma by population-based sequencing. The wild-type HIV-1 sequence is based on the consensus clade B sequence.
Dashes indicate no difference from the HIV-1 consensus B sequence.
Mutations listed are outside the β3-β4 region. Changes in brackets have not been associated previously with drug resistance.
Drug susceptibility.
Recombinant isolates from three patients were tested for susceptibility to nRTI in a PBMC-based cell culture assay. The results (Table 2) indicated that changes in susceptibility to nRTI were, in general, consistent with the known drug resistance mutations in the RT gene. For example, zidovudine resistance was consistent with the presence of the T215F and K70R mutations (with or without M41L) or the Q151M complex, and nevirapine and efavirenz resistance was consistent with the presence of the Y181C or K103N mutation. However, reduced susceptibility of strain v1966 to lamividine, adefovir, and stavudine was not explained by known drug resistance mutations, nor was the reduced susceptibility of strain q712 to lamivudine (3TC).
TABLE 2.
In vitro susceptibility of deletion-containing HIV-1 isolates to RT inhibitors
| Strain | Genotypic changes | Fold change in IC90 compared to NL4-3a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AZT | ddI | 3TC | D4T | PMEA | ABC | NVP | EFV | ||
| v1966 | del(SG)b, M41L, L74I, L100I, K103N, T215F, K219Q | >100 | 4.6 | 10.7 | 9.6 | 10.6 | 8.8 | >150 | >500 |
| q712 | del(SG), Y181C, G190G/A, T215F, K219E | 40 | <2 | 6.2 | 2.1 | 3.3 | <2 | >150 | 42.5 |
| q759 | del(NG), V75I, F77L, F116Y, Q151M, Y181C, M184V, G190A, K219E | >100 | >100 | >100 | 21.9 | <2 | 27.6 | >150 | >500 |
| Construct A | del(ST) | 2.5 | 2.7 | 16.7 | 2.4 | 2.9 | 3.4 | <2 | <2 |
| Construct B | del(ST) + T215Y | 16.2 | 5.2 | 16.4 | 10.0 | <2 | 8.8 | <2 | <2 |
| Construct C | T215Y | 15.1 | 2.9 | 3.7 | 2.9 | 2.0 | nt | <2 | nt |
| Construct D | del(SG) | <2 | 6.4 | 9.3 | 3.0 | 2.8 | 3.7 | nt | nt |
| Construct E | del(SG) + Q151M | 4.2 | 4.0 | 5.7 | 12.3 | <2 | 4.6 | nt | nt |
| Construct F | Q151M | 4.7 | 5.1 | 3.7 | 9.2 | <2 | 3.5 | nt | nt |
| Construct K | V75I, F77L, Y116F, Q151M | >100 | >100 | >100 | >100 | 2.5 | >50 | <2 | <2 |
The 90% inhibitory concentration (IC90) values for NL4-3 are zidovudine (AZT), 0.08 μM; didanosine (ddI), 0.083 μM; lamivudine (3TC), 0.09 μM; stavudine (d4T), 0.35 μM; adefovir (PMEA), 6.4 μM; abacavir (ABC), 1.0 μM; nevirapine (NVP), 0.07 μM; and efavirenz (EFV), 0.004 μM. nt, not tested.
del(SG) indicates a deletion between codons 67 and 69; the letters in parentheses are the single-letter codes for the two amino acids remaining between codons 67 and 69.
Molecular constructs were created to assess the impact of the deletion alone and in the presence of other RT gene mutations. HIV-1 constructs possessing only the deletion showed reduced susceptibility to lamivudine in cell culture assays (Table 2). However, when combined with mutations at either codon 215 or codon 151, reduced susceptibility to a greater number of nRTI was found compared to constructs with only the codon 215 or 151 mutation. Susceptibility to nnRTI was unaffected.
RT homology modeling.
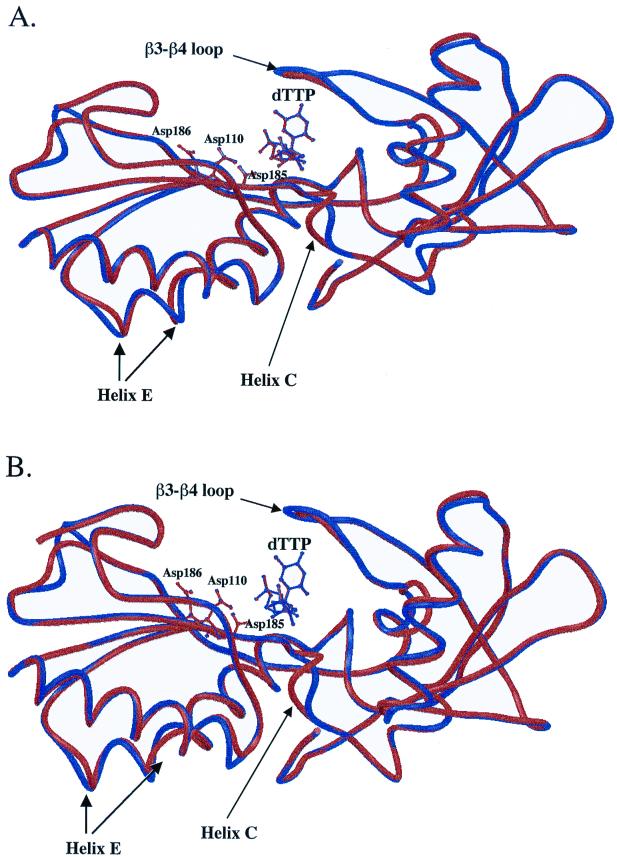
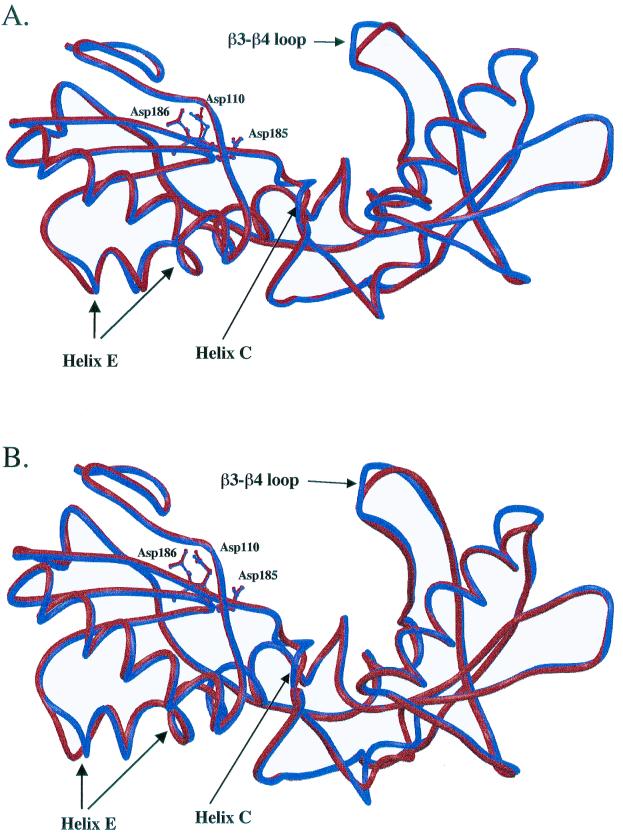
Protein homology modeling experiments of HIV-1 RT deletion mutants were carried out with two templates, the RT-DNA open (10) and RT-DNA-dTTP closed structure (8), to examine changes in the absence and in the presence of dNTP. Both the closed and open models have consistent changes at the β3-β4 helix C (codons 114 to 118) and helix E (codons 167 to 178) regions (Fig. 1 and 2). These results indicate that the long-range effects observed in the deletion mutants are not mediated by contacts through the bound dNTP. Differences in the β3-β4 loop between the wild type and the deletion mutant q759 are more pronounced for the open complex than in the closed complex (Fig. 1A and 2A). In contrast, differences in helix C are more pronounced for the closed complex (Fig. 1A and 2A). In the closed complex, the position of the dNTP is unchanged. The position of the nucleic acid is also unchanged for both the closed and the open complexes. For the closed RT complex model, the conformation of the three active-site residues (D110, D185, and D186) is identical between the RT deletion strains q759 and 67delSG and the wild type (Fig. 1A and B). For the open RT complex model, the conformation of the side chain for D110 is different from the wild type (Fig. 2A and B). Contacts between RT and the ligand for the wild type and the deletion mutant are listed in Table 3. The analysis of interactions between the RT and dTTP shows that four hydrogen bonds with residues R72, D185, Q151, and V111 were lost in the patient isolate q759 (Table 3). Other changes were also found in the patient-derived RT enzymes, but the position of the changes varied among the different strains.
FIG. 1.
Superimposition of the closed structural models of wild-type (NL4-3) HIV-1 RT (shown in blue) with the RT from (A) patient strain q759 and (B) a molecular construct containing only the 67delSG deletion (shown in red). Only the polypeptide backbone of the fingers and palm domains (residues 1 to 235) is shown. The orientations of the three active-site residue side chains are also illustrated.
FIG. 2.
Superimposition of the open structural models of wild-type (NL4-3) HIV-1 RT (shown in blue) with the RT from (A) patient strain q759 and (B) a molecular construct containing only the 67delSG deletion (shown in red). Only the polypeptide backbone of the fingers and palm domains (residues 1 to 235) is shown. The orientations of the three active-site aspartic acid residue side chains (110, 185, and 186) are also indicated.
TABLE 3.
Changes in the network of interactions between the HIV-1 RT and dTTP for the isolate q759a
| RT structural model | Changes
|
|
|---|---|---|
| Hydrogen bonds | Nonbonded contacts | |
| Wild type | Y115, A114, D113, R72, D185, Q151, V111 | D185, Q151, Y115, A114 |
| Isolate q759 | F115, A114, D113 | D185, M151, F115, A114 |
Wild-type RT contacts were calculated based on the structure reported in reference 8. Boldface indicates mutations compared to the wild type.
DISCUSSION
Genotypic analysis of patients failing their current antiretroviral therapy revealed that seven patients had HIV-1 strains with a 1-amino-acid deletion in their RT gene. These RT deletions occurred in the β3-β4 region of the RT gene, where many nucleoside inhibitor resistance mutations occur (23) and where resistance-conferring insertions have been recently reported (5, 16, 28, 32). Mutation analysis of the HIV-1 strains suggested that nearly all patients had used (and failed) at least one drug from all three classes of antiretroviral drugs. In addition to the deletion, the RT genes also possessed up to nine drug resistance-associated mutations, with six of seven strains having evidence of resistance to both nRTI and nnRTI and all patients having protease inhibitor resistance mutations. Whether the deletion appeared as an early event in developing drug resistance or appeared later in association with other mutations is unclear, as reliable drug histories and earlier genotypes were not available for these patients. However, a recent report showed the emergence of a deletion-containing strain in one patient during a course of zidovudine-didanosine therapy following zidovudine monotherapy (9), suggesting that deletion-containing strains may appear without extensive courses of treatment. This early appearance of the nonconventional genotypic change is similar to that seen in patients who developed the T69S+XX insert-containing strains (32).
The influence of the deletion on drug susceptibility was somewhat limited by the number and nature of other drug resistance mutations present in the patient-derived viral strains. Resistance to lamivudine is typically conferred by a mutation at RT codon 184 (30). However, two strains showed reduced susceptibility to lamivudine in the absence of the typical lamivudine resistance mutation M184V or the Q151M complex. Data from susceptibility tests on deletion-containing molecular constructs indicate a role for the deletion in influencing lamivudine susceptibility. Thus, in some patients, acquisition of the deletion may provide a sufficient change in susceptibility to overcome the level of lamivudine exposure. The addition of the M184V mutation in some deletion-containing strains may be necessary to overcome a higher level of pharmacologic exposure compared to other patients. Studies have shown that the M184V mutation suppresses the degree of resistance conferred by the T215Y mutation (15) but not by the Q151M mutation (25). The deletion-containing viral constructs with either T215Y or Q151M do not appear to differ in susceptibility to AZT despite having reduced susceptibility to 3TC. These results suggest that the deletion may provide the viral strain with a moderate level of 3TC resistance without compromising AZT resistance. Recent studies have suggested that other mutation patterns may similarly confer 3TC resistance in the absence of M184V (1). Changes in susceptibility to other nRTI in the deletion-alone constructs were consistently small. However, the interaction of the deletion with other mutations appears to have a greater impact on drug susceptibility than the deletion alone. This has also been shown for strains possessing the T69S+XX insert, which is also found in the β3-β4 region (16, 32).
The deletion in the β3-β4 region of the RT gene appears to have both local and long-range effects on RT enzyme structure. Local changes in the models of the β3-β4 region are believed to alter the orientation of the primer-template complex passing through the enzyme, which is believed to alter nucleotide selectivity (29). In the protein homology models, changes were consistently found in the alpha helix C region (codons 114 to 118), which is involved in both coordinating the triphosphate moiety and interacting with the 3′OH of the incoming dNTP (8). These changes were observed in both the deletion-only construct and all deletion-containing patient strains. The reduced susceptibility of the deletion-containing strains is likely to be at least partially mediated through changes in the dNTP binding pocket and through changes in the β3-β4 loop, which likely make the enzymes less susceptible to nRTI by being selective against the modified dNTP.
There are significant changes in the network of interactions between the dTTP and the deletion-containing RT relative to the wild type. Four of the seven hydrogen bonds were lost in patient isolate q759. These changes are likely to be the combined result of the one-residue deletion and point mutations present in this isolate. One of the hydrogen bonds lost in the deletion mutant involves the active-site residue D185 (Table 3). A second hydrogen bond is lost at position 151. The loss of hydrogen bonds is expected to result in significantly weaker binding of the dNTP and/or inhibitor. The Q151M mutation present in isolate q759 is associated with the multinucleoside resistance phenotype. As shown in Table 3, hydrogen bonds were more significantly affected than nonbonded contacts, as the cumulative result of the deletion and point mutations. Since hydrogen bonds are stronger than nonbonding contacts, the loss of hydrogen bonds affects dNTP binding more significantly than the loss of nonbonding contacts.
There were also consistent changes in the alpha helix E region (codons 167 to 178) of the deletion-containing strains, in both the open and closed structures. This region is involved in nnRTI binding (29). However, the deletion-only molecular construct showed no change in susceptibility of nnRTIs, and six of seven patient strains had significant nnRTI mutations. These results suggest that it is unlikely that the deletion contributes directly to nnRTI resistance in spite of structural differences detected by the modeling. Additional structural changes were observed in the different patient-derived RT models, but these changes were not consistent between strains. This is likely due to the impact of other drug-selected and/or polymorphic differences from wild-type enzyme.
Long-distance structural changes have already been reported for another flexible molecule, the HIV protease (19). One of the HIV protease drug resistance hypotheses suggests that HIV protease subdomain flexibility may contribute to drug resistance. The support for this hypothesis is based on the work of Stroud and coworkers (19), who identified five subdomains in retroviral proteases by differential distance matrix analysis: one terminal subdomain encompassing the N- and C-terminal β sheet of the dimer, two core subdomains containing the catalytic aspartic acids, and two flap subdomains. Rigid body rotation of the five subdomains and movement within their interfacial joints may provide a rational context for understanding HIV protease drug resistance (19). The HIV RT enzyme is also a plastic protein defined by the major structural changes observed between the open and closed complexes. Our models suggest that the flexibility of the HIV RT is responsible for structural changes in the finger subdomain (due to the 67delSG deletion) being transmitted to trigger additional changes in the palm subdomain (in particular, movement of helices C and E). X-ray crystallographic studies will be necessary to more thoroughly investigate the relationship between these local and distant structural changes, drug susceptibility, and enzyme function.
The deletion was significantly associated with the presence of Q151M. In contrast, no strains containing the Q151M mutation have been reported to also have a β3-β4 insert. The Q151M mutation is typically associated with mutations at codons 62, 75, 77, and 116, and the presence of these additional mutations affects the magnitude and breadth of drug resistance (24, 26). Several of the deletion-containing strains with the Q151M mutation had some but not all of these associated mutations. Thus, it is possible that the deletion compensates for one or more of these Q151M-associated mutations in producing high-level resistance to many nRTI.
The results presented here further indicate the importance of the β3-β4 region in modulating the effectiveness of nRTI. In addition to point mutations, insertions and deletions in this region have now been described in patients failing nRTI therapy. New antiretroviral drugs need to be designed and developed with the impact of changes in this region in mind. Drugs that will be effective against strains with the common resistance mutation patterns are necessary to effectively treat patients who have failed on other antiretroviral drugs.
ACKNOWLEDGMENTS
We thank Andrej Sali and Andras Fiser for advice on the use of the protein homology modeling package MODELLER.
REFERENCES
- 1.Bloor S, Hertogs K, DeVroey V, Miller V, Sturmer M, Larder B. Lamivudine-resistant HIV-1 clinical isolates lacking the Met184Val mutation have novel polymorphisms in RT. Antivir Ther. 1999;4(Suppl. 1):19. [Google Scholar]
- 2.Boyer P L, Lisziewicz J, Lori F, Hughes S H. Analysis of amino insertion mutations in the fingers subdomain of HIV-1 reverse transcriptase. J Mol Biol. 1999;285:995–1008. doi: 10.1006/jmbi.1998.2508. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter C C J, Cooper D A, Fischl M A, Gatell H M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the international AIDS society-USA panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 4.Carson M, Bugg C E. Algorithm for ribbon models of proteins. J Mol Graphics. 1986;4:121–122. [Google Scholar]
- 5.De Antoni A, Foli A, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:889–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 6.Gu Z, Gao Q, Fang H, et al. A novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7228–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 9.Imamichi T, Sinha T, Imamichi H, Zhang Y-M, Metcalf J A, Falloon J, Lane H C. High-level resistance to 3′-azido-3′-deoxythymidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J Virol. 1999;74:1023–1028. doi: 10.1128/jvi.74.2.1023-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S the RV-43 Study Group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 13.Korber B, Foley B, Leitner T, McCutchan F E, Hahn B H, Menendez-Arias L, Myers G, Kuiken C L. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 14.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 15.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;369:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 16.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martí-Renom M A, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 18.Mayers D. Prevalence and incidence of resistance to zidovudine and other antiretroviral drugs. Am J Med. 1997;102:70–75. doi: 10.1016/s0002-9343(97)00067-3. [DOI] [PubMed] [Google Scholar]
- 19.Rose R B, Craik C S, Stroud R M. Domain flexibility in retroviral proteases: structural implications for drug resistant mutations. Biochemistry. 1998;37:2607–2621. doi: 10.1021/bi9716074. [DOI] [PubMed] [Google Scholar]
- 20.Sali A, Potteron L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 21.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez R, Sali A. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc Natl Acad Sci USA. 1998;95:13597–13602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:2–14. http://hiv-web.lanl.gov . [Online.] http://hiv-web.lanl.gov. . [Google Scholar]
- 24.Shafer R W, Iversen A K N, Winters M A, Aguiniga E, Katzenstein D A, Merigan T C the AIDS Clinical Trials Group 143 Virology Team. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J Infect Dis. 1995;172:70–78. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- 25.Shafer R W, Winters M A, Iverson A K N, Merigan T C. Genotypic and phenotypic changes during culture of a multinucleoside-resistant human immunodeficiency virus type 1 strain in the presence and absence of additional reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1996;40:2887–2890. doi: 10.1128/aac.40.12.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kaojima E, Alcaide M L, Chockekuchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 28.Tamalet C, Izopet J, Koch N, Fantini J, Yahi N. Stable rearrangements of the β3-β4 hairpin loop of HIV-1 reverse transcriptase in plasma viruses from patients receiving combination therapy. AIDS. 1998;12:F161–F166. doi: 10.1097/00002030-199814000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three dimensional structure of HIV-1 RT. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 30.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistance to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace A C, Laskowski R A, Thornton J M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 32.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair deletion in the reverse transcriptase gene of HIV-1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]