Abstract
Until now, the analysis of the genetic diversity of bovine respiratory syncytial virus (BRSV) has been based on small numbers of field isolates. In this report, we determined the nucleotide and deduced amino acid sequences of regions of the nucleoprotein (N protein), fusion protein (F protein), and glycoprotein (G protein) of 54 European and North American isolates and compared them with the sequences of 33 isolates of BRSV obtained from the databases, together with those of 2 human respiratory syncytial viruses and 1 ovine respiratory syncytial virus. A clustering of BRSV sequences according to geographical origin was observed. We also set out to show that a continuous evolution of the sequences of the N, G, and F proteins of BRSV has been occurring in isolates since 1967 in countries where vaccination was widely used. The exertion of a strong positive selective pressure on the mucin-like region of the G protein and on particular sites of the N and F proteins is also demonstrated. Furthermore, mutations which are located in the conserved central hydrophobic part of the ectodomain of the G protein and which result in the loss of four Cys residues and in the suppression of two disulfide bridges and an α helix critical to the three-dimensional structure of the G protein have been detected in some recent French BRSV isolates. This conserved central region, which is immunodominant in BRSV G protein, thus has been modified in recent isolates. This work demonstrates that the evolution of BRSV should be taken into account in the rational development of future vaccines.
Bovine respiratory syncytial virus (BRSV) and human respiratory syncytial virus (HRSV) are members of the genus Pneumovirus, subfamily Pneumovirinae, and family Paramyxoviridae (38, 39, 40, 49). These two related viruses share common epidemiological, clinical, and pathological characteristics. The respiratory syncytial viruses (RSV) are the most common and important cause of lower respiratory tract illness in cattle and young infants (71). More than 70% of calves exhibit a positive serological response against BRSV by the age of 12 months.
Neutralization tests (9) and reaction patterns with specific monoclonal antibodies (2, 48) have revealed that HRSV contains two major groups, A and B. The main differences between these two groups are located in the glycoprotein (G protein), while others are located in the fusion protein (F protein) and nucleoprotein (N protein) (48, 51). Subgroups within the G and F proteins of BRSV have been characterized more recently by serological analysis of a limited number of isolates and confirmed by phylogenetic analysis (20, 54, 60). Further studies of BRSV variability have focused on the G protein, which was shown to be the most variable protein and, with the F protein, one of the targets for neutralizing antibodies. A low percentage of sequence divergence between and within the G proteins of BRSV subgroups has been reported, suggesting that BRSV has the same extent of diversity as HRSV (15, 21, 36, 44, 66).
The G protein is responsible for virus binding to the cell surface receptor (41). It is a type II G protein with a signal/anchor domain between residues 38 and 66. It is synthesized as a 32-kDa polypeptide precursor which is extensively modified by the addition of both N- and O-linked oligosaccharides to achieve the mature form of 80 to 90 kDa (10, 26, 32, 77). This protein is structurally different from its counterparts (hemagglutinin-neuraminidase and hemagglutinin) in other paramyxoviruses. It has been proposed that the BRSV G protein contains several independently folding regions, in which the ectodomain consists of a conserved central hydrophobic region located between two polymeric mucin-like regions (33, 34). This conserved central hydrophobic region contains four conserved cysteines which form two disulfide bridges. Peptides based on this region were found to be immunodominant in the G proteins of both HRSV (1) and BRSV (34). They also conferred protection against HRSV challenge in mice (69). Recently, the three-dimensional structure of a segment of this conserved central region was elucidated by nuclear magnetic resonance spectroscopy (13). The major epitope of this region was described as being located at the tip of a loop, overlapping a relatively flat surface formed by the double disulfide-bonded cysteine noose and lined by highly conserved residues (33, 34).
Attempts to develop a vaccine against HRSV in humans have been unsuccessful (28). However, inactivated and modified live vaccines against BRSV have been developed and used for more than 15 years in France and Belgium and for 2 or 3 years in other countries, such as Denmark or Sweden. Three types of vaccines are mostly used in Europe. Rispoval RS (strain RB-94 attenuated vaccine) is the vaccine most commonly used in bovines. It has been commercialized in Belgium and The Netherlands since 1978 and in France since 1983. Bayovac BRSV (strain Lehmkuhl 375 attenuated vaccine) has been commercialized in The Netherlands, France, and Belgium since 1994, 1996, and 1997, respectively. The third vaccine is Vacores (strain 220/69 inactivated with beta-propiolactone and associated with the adjuvants aluminum hydroxide and saponin). It has been commercialized in France and Belgium from 1996 to 1999. Such vaccination prevents the disease and its economic consequences in herds. It does not suppress the circulation of BRSV, however. In fact, some of the characteristics of RSV infection are that the virus can replicate in spite of detectable levels of specific antibodies (3, 72) and that previous immunization of animals through infection or vaccination does not completely eliminate the circulation of the virus (53, 61). In addition, reinfection occurs in human and bovine adults (14, 25, 27, 72).
Given the high rates of mutation of RNA viruses (12), it would not be surprising if previously stimulated immunity (i.e., former infection or vaccination) could select new variants, at least in the G protein, which is known to favor the accommodation of sequence changes. Studying the sequence of BRSV isolates in Europe therefore could provide an indication of how the evolution of RSV might be modified in nature by immune escape mechanisms. We therefore examined the changes that have occurred in the N, F, and G protein genes of 87 BRSV isolates over the past 32 years. Some of these isolates originated from vaccinated or nonvaccinated calves in geographical areas where vaccination had been widely used for many years, while others came from nonvaccinated calves in countries where vaccination had been absent or very limited. In this study, we demonstrate that a continuous evolution of the sequences of the N, G, and F proteins of BRSV has been occurring in isolates since 1967. The exertion of a strong positive selective pressure on the G protein and on particular sites of the N and F proteins is also demonstrated.
MATERIALS AND METHODS
Viruses and field samples. (i) BRSV with cell culture passages.
The five BRSV strains isolated from Belgium (220/69, RB-94, FV 160, MVR553, and 5761) between 1969 and 1990 were kindly provided by Wellemans and Leunen (76) (Table 1). The Lelystad strain isolated in 1974 from The Netherlands and strain 4642 isolated in 1976 from the United Kingdom were kindly provided by T. J. Kimman. The Lelystad strain has been described by Kimman et al. (29). Strain V347 isolated in 1990 from The Netherlands was kindly provided by R. S. Schrijver. D80 was isolated by C. Delacourt in 1986 in the Somme Veterinary Laboratory (Dury, France). B35 and 90504, which were kindly provided by E. Le Drean, were isolated in 1986 and 1990, respectively, in the Ille-et-Vilaine Veterinary Laboratory (Rennes, France). W6 was isolated in our laboratory in 1993 from a calf from a herd involved in a respiratory tract infection outbreak. Isolates A1 and A2Gelfi were obtained from the same herd in 1995. Vaccine strains were obtained from vaccine bottles for RB-94 (Rispoval RS; batch 31863L) and Lehmkuhl 375 (Bayovac; batch 2071); the strain 220/69 vaccine (Vacores) was kindly provided by Merial.
TABLE 1.
Origins of virus strains and classification according to nucleotide variability in segments of the N, G, and F protein genesa
| Virus | Subgroup | Strain | Stock farming | Yr | Countryb |
|---|---|---|---|---|---|
| HRSV | |||||
| Group A | A2 | ||||
| Group B | 18537 | ||||
| ORSV | ORSV | ||||
| BRSV | I | 4642 | 1976 | United Kingdom | |
| Maryland | 1975 | US (Maryland) | |||
| BovX | 1967 | Switzerland | |||
| II | Lelystad | 1974 | The Netherlands | ||
| RB-94 | 1969 | Belgium | |||
| RB-94 (Rispoval RS) | 1969 | Belgium | |||
| MRV553 | 1983 | Belgium | |||
| W2 | 1969 | Belgium | |||
| 220/69 (Vacores) | 1969 | Belgium | |||
| FV160 | 1969 | Belgium | |||
| NMK7 | 1968 | Japan | |||
| B35-1 | 1986 | France | |||
| D80 | 1986 | France | |||
| 9511581DK | 1995 | Denmark | |||
| 9510237DK | 1995 | Denmark | |||
| 9510236DK | 1995 | Denmark | |||
| 9402020DK | 1994 | Denmark | |||
| 4556SW | 1995 | Sweden | |||
| 271SW | 1995 | Sweden | |||
| 8307027DK | 1983 | Denmark | |||
| 9616348DK | 1996 | Denmark | |||
| 87LU195 | 1987 | Denmark | |||
| 9510597DK | 1995 | Denmark | |||
| 9304899DK | 1993 | Denmark | |||
| 9510796DK | 1995 | Denmark | |||
| 37NL | 1974 | The Netherlands | |||
| III | 1344R | ? | US (Kansas) | ||
| 1540R | ? | US (Kansas) | |||
| 88-294 | ? | US (South Dakota) | |||
| 236-652 | ? | US (Nebraska) | |||
| 85-1330 | 1985 | US | |||
| Lehmkuhl 375 (Bayovac) | ? | US | |||
| FS-1 | 1975 | US (Iowa) | |||
| 375.1 | 197? | US (Iowa) | |||
| 375.2 | ? | US (Iowa) | |||
| 234-489 | ? | US (New York) | |||
| 87-14594 | ? | US (South Dakota) | |||
| NDKS-7 | ? | US (North Dakota) | |||
| 391.2 | 1985 | US (North Carolina) | |||
| IV | VC464 | 1974 | US (Missouri) | ||
| Dorset | 1971 | United Kingdom | |||
| Snook | 1976 | United Kingdom | |||
| 5761 | 1990 | Belgium | |||
| 90504 | 1990 | France | |||
| V347 | 1990 | The Netherlands | |||
| WBH | 1986 | The Netherlands | |||
| V | W6 | A | 1993 | France | |
| A1 | A | 1995 | France | ||
| A2Gelfi | A | 1995 | France | ||
| B1 | B | 1996 | France | ||
| B2 | B | 1996 | France | ||
| C1 | C | 1996 | France | ||
| C2 | C | 1996 | France | ||
| D1 | D | 1996 | France | ||
| D2 | D | 1996 | France | ||
| 394 | E | 1995 | France | ||
| 673 | E | 1995 | France | ||
| Bioch | E | 1995 | France | ||
| F1 | F | 1997 | France | ||
| F2 | F | 1997 | France | ||
| I1 | I | 1997 | France | ||
| I2 | I | 1997 | France | ||
| I3 | I | 1997 | France | ||
| J1 | J | 1997 | France | ||
| J2 | J | 1997 | France | ||
| L1 | L | 1997 | France | ||
| L2 | L | 1997 | France | ||
| 8352c | M | 1998 | France | ||
| 8356c | M | 1998 | France | ||
| 8346c | M | 1998 | France | ||
| 7313c | M | 1998 | France | ||
| G2 | 1997 | France | |||
| 58Pd | 1998 | France | |||
| 88P | 1998 | France | |||
| P1 | 1998 | Belgium | |||
| P2d | 1998 | Belgium | |||
| P3d | 1998 | Belgium | |||
| P4 | 1998 | Belgium | |||
| P5c | N | 1998 | Belgium | ||
| P7c | N | 1998 | Belgium | ||
| P6c | 1998 | Belgium | |||
| P8c | 1998 | Belgium | |||
| P9 | 1998 | Belgium | |||
| P10 | 1998 | Belgium | |||
| VI | K1 | K | 1997 | France | |
| K2 | K | 1997 | France | ||
| 75Pd | 1998 | France |
GenBank accession numbers are AF188554 to AF188604, M11486, D00390, M17213, D00334, U07233, L08470, Y08718, M35076, A51908, M82816, U57823, U33539, L27840, L27802, D00953, U24714, U24713, U92112, U92108, U92107, U92103, U55247, U55246, U92098, U92114, U92100, U92109, U92101, U92110, U55254, L08415, L08416, L08411, L08414, U24716, L10925, L08410, L08413, L08412, L08417, M58307, U24715, Y08719, Y08717, S40504, and M58350.
US, United States.
Calf was vaccinated with Vacores.
Calf was vaccinated with Rispoval RS.
(ii) BRSV without culture passage.
Twenty-three calves from 11 herds exhibiting an acute respiratory tract infection outbreak compatible with BRSV were sampled between 1995 and 1997 by bronchoalveolar lavage (A1, A2Gelfi, B1, B2, C1, C2, D1, D2, 394, 673, Bioch, F1, F2, I1, I2, I3, J1, J2, L1, L2, G2, K1, and K2). In 1998, nasal secretions (8346, 8352, and 8356) and a necropsy sample of lung tissue (7313) from four calves in the same herd with BRSV-associated respiratory tract infection signs were sampled. During the same year, lungs from calves which had died of BRSV infection were collected from three French herds (58P, 75P, and 88P) and nine Belgian herds (P1, P2, P3, P4, P5, P6, P7, P8, P9, and P10). All the samples were stored at −80°C until analysis. These isolates were never passaged in cell cultures before RNA extraction.
Extraction of RNA and PCR. (i) RNA extraction.
Tissue samples were homogenized directly in their storage tubes with a Diax 600 homogenizer (Heidolph, Bioblock, Illkirch, France). The homogenizer arbor was replaced between each tissue sample and sterilized (NaOH for 10 min and then 30 min at 120°C) before further use. The RNA used as a template for cDNA synthesis was extracted from cells and tissues as previously described (70). Briefly, RNA was extracted with a solution of guanidine isothiocyanate and phenol (TRIzol reagent; Gibco BRL), purified with chloroform, and precipitated with isopropanol overnight at 4°C. After being washed with 70% ethanol, the RNA pellet was dried and then resuspended in 50 ml of diethyl pyrocarbonate (DEPC)-treated water.
(ii) Nested RT-PCR.
Different nested reverse transcription (RT)-PCRs were used to amplify the BRSV RNA of the N protein gene, the G protein gene, and the F protein gene (Table 2). The nested RT-PCR for the N protein gene was performed as previously described (70). Primers used for the G and F protein gene nested RT-PCRs were based on the published sequences of the BRSV G protein (GenBank accession numbers M58307, L10925, L08410, L08411, L08412, L08413, L08414, L08415, L08416, and L08417) and the BRSV F protein (GenBank accession numbers D00953, M58350, and M82816). Primer sequences were selected using the program PRIMER, version 0.5 (43). The different nested RT-PCRs were performed with a GeneAmp PCR system 480 (Perkin-Elmer, Courtaboeuf, France).
TABLE 2.
Sequences of primers and characteristics of each nested RT-PCR
| Protein gene | Primers | Positions | Sequence (5′→3′) | Cycle profilea | Length of amplified product (bp) |
|---|---|---|---|---|---|
| N | N2.1 | 1–19 | ATG GCT CTC AGC AAG GTC A | 94°C, 60 s; 58°C, 60 s; 72°C, 90 s | 1,034 |
| N2.2 | 1034–1013 | TCT TGG TTT CTT GGT GTA CCT C | |||
| N2.3 | 127–146 | CAT CTC AAT AAG TTG TGT GG | 94°C, 45 s; 49°C, 60 s; 72°C, 60 s | 731 | |
| N2.4 | 857–838 | TCT ACA ACC TGT TCC ATT TC | |||
| G | G2.5 | 11–31 | AGA CAT TAA AGA GGG CTT GGA | 94°C, 60 s; 55°C, 60 s; 72°C, 90 s | 1,030 |
| F2.7 | 118–99 | CTG CAC TGC ATG TTG ATT GA | |||
| VG1 | 131–155 | CCA CCC TAG CAA TGA TAA CCT TGA C | 94°C, 45 s; 58°C, 60 s; 72°C, 60 s | 541 | |
| VG4 | 672–648 | GCT AGT TCT GTG GTG GAT TGT TGT C | |||
| F | F2.1 | 136–157 | AGT GCA TTA AGA ACT GGA TGG T | 94°C, 60 s; 58°C, 60 s; 72°C, 90 s | 1,335 |
| F2.2 | 1470–1450 | TGC ATC AAA CTC ATC AGA AGG | |||
| F2.3 | 433–452 | GGA TCT GCT ATT GCA AGT GG | 94°C, 45 s; 62°C, 60 s; 72°C, 60 s | 833 | |
| F2.4 | 1265–1244 | CAT TTT GTC TTC CCA TAG CAT G |
There were 35 cycles. They were preceded by a denaturation step (94°C, 12 min) and followed by an elongation step (72°C, 10 min).
RT of viral RNA into specific cDNAs of the N, G, and F proteins was done using primers N2.1, G2.5, and F2.1, respectively. One microliter of each primer (2 pmol) was mixed with 9 μl of DEPC-treated water containing RNA, incubated at 68°C for 10 min, and finally chilled on ice. Each tube received 4 μl of a solution containing each nucleoside triphosphate (10 mmol), 1 μl of RNasin (40 U) (Promega, Charbonnières, France), 2 μl of dithiothreitol (Gibco BRL), 4 μl of Superscript TM II buffer (250 mM Tris HCl, 375 mM KCl, 15 mM MgCl2) (Gibco BRL), and 1 μl of SuperScript TM II (200 U) (Gibco BRL) and was incubated at 42°C for 50 min. The reverse transcriptase was inactivated at 70°C for 15 min. The RNA-cDNA hybrids were diluted 10 times in DEPC-treated water.
The first set of primers, N2.1 and N2.2, was used for the first round of PCR for the N protein gene, and the internal set of primers, N2.3 and N2.4, was used for the second round of PCR. The first set of primers used to amplify a segment of the G protein gene consisted of G2.5 and F2.7, and the second set consisted of VG1 and VG4 (73). The primer sets F2.1-F2.2 and F2.3-F2.4 were successively used to amplify a segment of the F protein gene. The dilutions at each different step and the compositions of the PCR mixtures were identical for all three nested RT-PCRs. The first round of PCR was performed with 10 μl of diluted cDNA which was mixed in a final volume of 100 μl with 5 μl of each primer (50 pmol) from the first set, 200 mM each nucleoside triphosphate, 10 mM Tris HCl (pH 8.3), 50 mM KCl, 1 mM MgCl2, and 2.5 IU of AmpliTaq Gold polymerase (Perkin-Elmer). The products of the first PCR were diluted 10 times. Ten milliliters was taken to perform the second round of PCR using the same mixture except for the substitution of a set of internal primers.
Sequence analysis.
Sequencing of amplified products was performed using primers of the second set for each corresponding nested RT-PCR according to the Applied Biosystems (Foster City, Calif.) protocol. The samples were loaded on an Applied Biosystems 373A sequencer. The nucleotide sequences of BRSV isolates were analyzed using the program TRANSLATE of the University of Wisconsin GCG 9.1 package (11). They were compared with the sequences of other BRSV isolates, when available, obtained from GenBank (the accession numbers of the sequences are reported in Table 1). Multiple sequence alignments were generated with the CLUSTAL W 1.60 program (68). Representative isolates of HRSV subgroups A and B and ovine RSV (ORSV), when available, were also included in this analysis.
Phylogenetic trees of nucleotide sequences were constructed using the maximum-likelihood (ML) method available in version 1.2 of the FastDNAml program (52) derived from version 3.3 of the DNA Maximum Likelihood program (17). Trees were computed using empirical base composition and transition/transversion ratios obtained from the data sets. Localized rearrangements were made to improve tree search efficiency. To reduce computing time, all calculations were performed with starting trees obtained by the neighbor-joining (NJ) method (57). The robustness of the topologies was assessed with 100 bootstrap replicates (18) using the same parameters and program options. The NJ method implemented in CLUSTAL W (68) was also used for comparisons, in which case 1,000 bootstrap replicates were used.
Finally, the numbers of nucleotide substitutions per synonymous site (ds) and nonsynonymous site (dn) were estimated using the method of Nei and Gojobori (50) as implemented in the MEGA sequence analysis package (version 1.01) (30) for all possible pairs of BRSV sequences from each coding region. They were also plotted for individual codons using the SNAP program (available at http://hiv-web.lanl.gov/SNAP/WEBSNAP/SNAP.html). For both types of substitution, only non-zero values were retained for the analysis so that the nonsynonymous/synonymous substitution rate ratio could be calculated for each pair of fragments, together with the average. The rates of nonsynonymous and synonymous substitutions were calculated using the method of Li et al. (42), in which the rate is calculated for pairs of sequences with a third sequence as an outgroup (46).
Modeling of secondary and tertiary structures.
Hydrophilicity was calculated using PEPTIDESTRUCTURE in the GCG package according to the algorithm of Kyte and Doolittle (31) with a window set to seven residues. The prediction of solvent accessibility was performed using PHDacc (55) (available at http://dodo.cpmc.columbia.edu/predictprotein).
Secondary structures for the conserved central region of the G protein of BRSV were obtained from the Brookhaven Protein Data Bank (http://www.pdb.bnl.gov/index.html). The structures were aligned with the computer program SWISSPDBVIEWER (http://www.expasy.ch/spdbv/mainpage.html). Amino acid replacements were performed on the alignments, and the new structure was modelled using version 4 of the MODELLER program. This program models the three-dimensional structure of proteins by satisfaction of spatial restraints (58, 59). The output of MODELLER was then graphically visualized using SWISSPDBVIEWER.
RESULTS
Nucleotide sequences of BRSV isolates are closely related.
The nucleotide sequences of fragments of the N protein gene (690 nucleotides corresponding to codon positions 50 to 279), of the G protein gene (489 nucleotides corresponding to codon positions 53 to 215), and of the F protein gene (741 nucleotides corresponding to codon positions 167 to 413) from 54 BRSV isolates were determined and compared with those of 33 isolates taken from GenBank. These included the sequences of the three vaccine strains (RB-94 [Rispoval RS], 220/69 [Vacores], and Lehmkuhl 375 [Bayovac]) used mostly in Europe. Alignment of the nucleotide sequences determined in this study did not reveal any gaps or insertions. After removal of the nucleotide sequences which were identical, 25 partial N protein gene sequences (690 bases), 63 partial G protein gene sequences (437 to 489 bases), and 25 partial F protein gene sequences (741 bases) were available for analysis. The vaccine strains RB-94 and 220/69 were identical in all three coding regions. The average percentage of pairwise divergences between fragments of BRSV was lowest for the N and F protein genes (2%) and highest for the G protein gene (8%). The average percentage of pairwise divergences between fragments of BRSV and HRSV was lowest for the N protein gene (20%), intermediate for the F protein gene (22%), and highest for the G protein gene (48%).
Several isolates were collected within the same herd. Twenty-nine isolates were collected from different calves exhibiting respiratory tract illness in 11 different herds located in France and 1 herd located in Belgium. The isolates of a definite herd were obtained on the same day, except for those from herd A, which was sampled over an interval of 2 years (Table 1). The nucleotide sequences of the N, F, and G protein gene regions obtained from five of these herds (C, I, J, M, and K) were identical. Differences were found in the F protein gene sequence between samples from four of the other herds. Only a very limited number of differences were apparent, within the N and G protein gene sequences, in the three remaining herds. The sequences of the N, F, and G protein genes of the viruses (W6, A1, and A2Gelfi) isolated at a 2-year interval from the same herd (A) were different. Thus, a single virus or a group of very closely related viruses would seem to predominantly infect a given herd at a given time.
Temporal and geographical clustering of BRSV sequences.
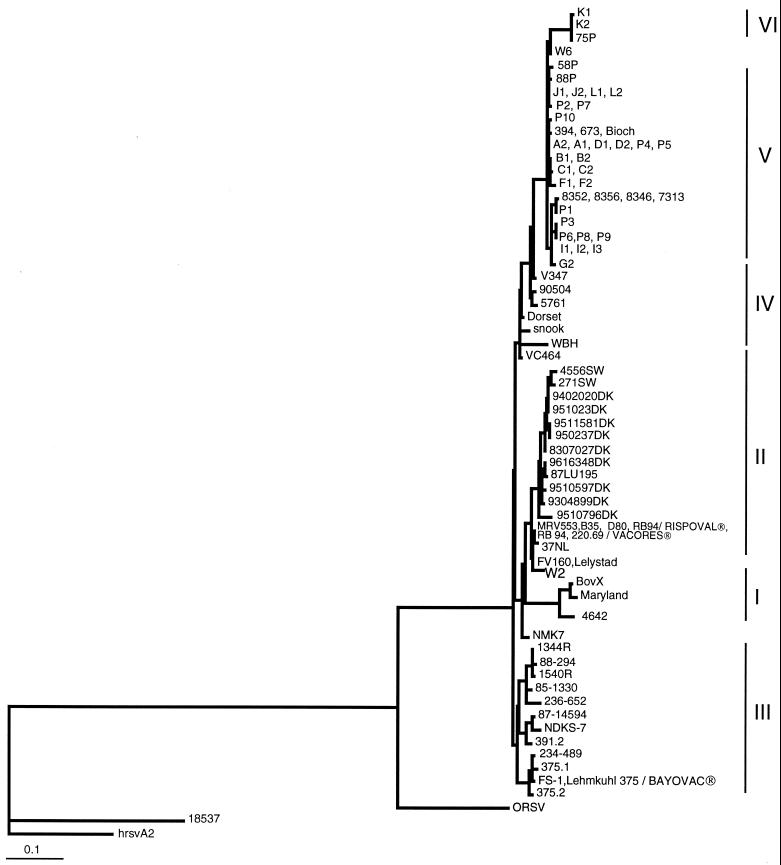
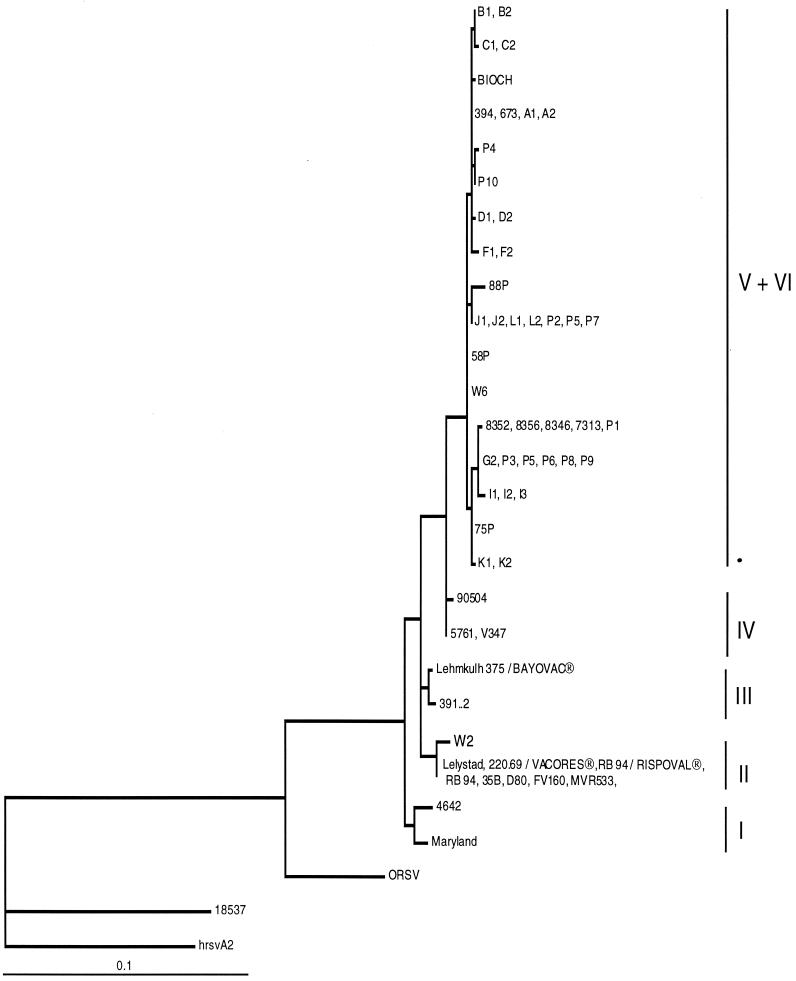
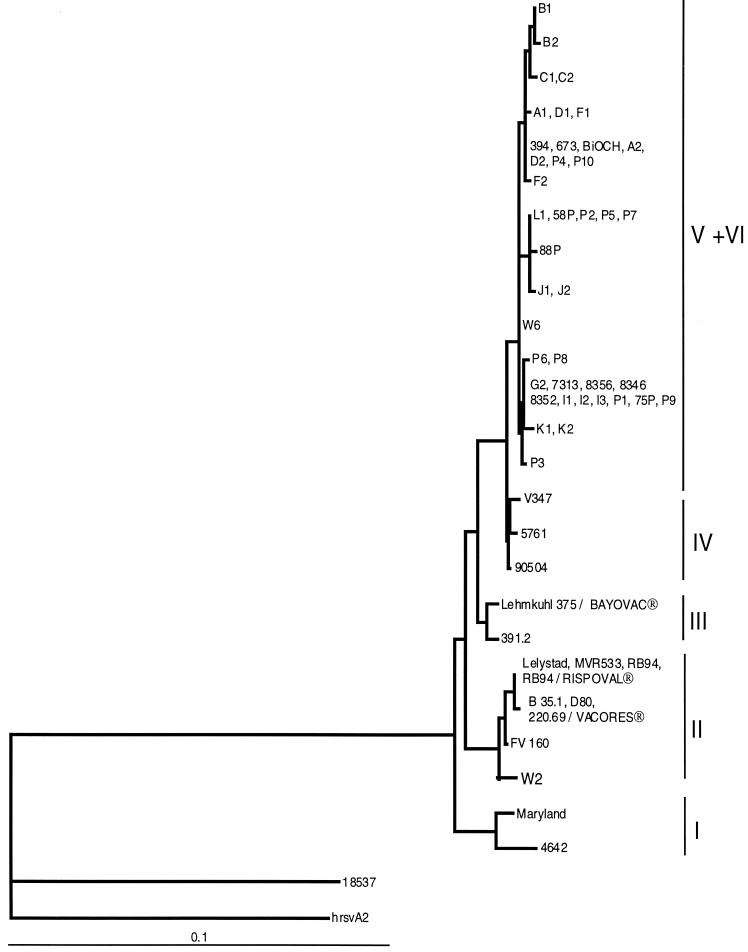
The ML method was used to compute phylogenetic trees, based on the alignment of the different gene fragments. For clarity, it was possible, from the phylogenetic tree obtained with the G protein fragment, to arbitrarily divide the data into six subgroups (I to VI) (Fig. 1 and Table 1). The topology and significance of the different nodes giving rise to these six subgroups were verified by bootstrap analysis using both the ML and the NJ methods (data not shown). However, the node giving rise to the isolates of subgroup IV was not statistically corroborated by bootstrap analysis, but the subgroup was retained because of its intermediate location between the isolates of subgroups V and VI and the others. Isolate NMK7 from Japan was not related to any of these subgroups and was arbitrarily included in group II. Trees were also constructed using the alignments of the nucleotide sequences of the N and F protein genes (Fig. 2 and 3). In this case, as for the G protein gene, the topology and significance of the important nodes of the trees were verified by bootstrap analysis using both the ML and the NJ methods (data not shown). No sequence from subgroup III isolates was available to be included in these trees, except for the Lehmkuhl 375 and 391.2 strains. These trees showed the same general clustering of isolates as that already described for the G protein gene. In both cases, the subgroup IV isolates were still in an intermediate position between those of subgroups V and VI and the others. A striking difference between these trees and the tree obtained for the G protein gene is that the isolates belonging to subgroup VI (K1, K2, and 75P) could no longer be distinguished from those of subgroup V. The vaccine strains RB-94 (and 220/69) and Lehmkuhl 375 were localized in subgroups II and III, respectively.
FIG. 1.
ML phylogenetic tree showing the relationships among the G protein genes of 87 BRSV isolates, 1 ovine RSV (ORSV) isolate, and 2 human RSV isolates (18537 and hrsvA2). Horizontal branches are drawn to scale, and the tree is rooted with isolate hrsvA2. Designations at the ends of the branches refer to the identifying code of the isolate (Table 1).
FIG. 2.
ML phylogenetic tree showing the relationships among the N protein genes of 58 BRSV isolates, 1 ovine RSV (ORSV) isolate, and two human RSV isolates (18537 and hrsvA2). Horizontal branches are drawn to scale, and the tree is rooted with isolate hrsvA2. Designations at the ends of the branches refer to the identifying code of the isolate (Table 1).
FIG. 3.
ML phylogenetic tree showing the relationships among the F protein genes of 56 BRSV isolates and 2 human RSV isolates (18537 and hrsvA2). Horizontal branches are drawn to scale, and the tree is rooted with isolate hrsvA2. Designations at the ends of the branches refer to the identifying code of the isolate (Table 1).
Subgroup I, which consisted of strains isolated before 1976, no longer seems to exist in Europe, as has previously been suggested (15). The isolates of subgroup III came exclusively from the United States. Those belonging to the remaining subgroups, II, IV, V, and VI, came exclusively from Europe, except for the VC464 isolate, which originated from the United States. A correlation therefore exists between the continent of origin of the isolates and their clustering in the phylogenetic tree. In Europe, the isolates from northern Europe, Denmark, and Sweden clustered in subgroup II, whatever their date of isolation (from 1983 to 1995). Those from The Netherlands, Belgium, and France were found in subgroups II, IV, V, and VI, depending on their date of isolation. In general, the close clustering of European isolates is related to their date of isolation, although several isolates could be distant from each other for a particular year. For example, when the strains originating from The Netherlands, Belgium, the United Kingdom, and France are considered, the dates of isolation of the subgroup II, IV, V, and VI isolates are 1969 to 1986, 1971 to 1990, 1993 to 1998, and 1997 to 1998, respectively (Fig. 1 and Table 1). All the recent isolates from these countries are located at the top of the tree in subgroups V and VI. This result could indicate the presence of a temporal clustering in addition to the geographical clustering already described. The recent isolates of subgroups V and VI are more distantly related to the vaccine strains than to the other isolates.
During 1998, field samples were obtained from 11 calves that had been vaccinated approximately 2 to 3 months before the onset of symptoms. These calves were less than 6 months old and exhibited respiratory tract illness. Seven of them had previously been vaccinated with strain 220/69 (Vacores), and four of them had been vaccinated with strain RB-94 (Rispoval RS). Analysis of the N, F, and G protein gene sequences indicated that 10 of these isolates belonged to subgroup V and 1 belonged to subgroup VI, in accordance with their dates of isolation (Fig. 1, 2, 3, and 4). These results suggest also that vaccination sometimes does not prevent infection of calves with BRSV of subgroups V and VI, indicating that vaccinated calves may be poorly protected against infection by recent BRSV isolates of subgroups V and VI.
FIG. 4.
Sequences of amino acids 53 to 215 of the G protein of BRSV isolates. Designations to the left of the sequences indicate the isolate code (Table 1). Subgroup designations are shown to the left of the isolate designations.
Four highly conserved Cys residues have been replaced by Arg in the sequence of the G protein of recent French isolates.
The mutations observed in the amino acid sequences of the different European subgroups are shown in Fig. 4. The sequences can be divided into three domains: two highly variable domains corresponding to the mucin-like regions and one central conserved domain. The two mucin-like regions of the BRSV G protein, the first of which was located between the transmembrane domain and the central conserved domain and the second of which was located from this central conserved domain to the COOH terminus of the molecule, had very high Ser and Thr contents—31% for the first region (amino acid positions 67 to 153) and 30.4% for the second region (amino acid positions 193 to 215)—thereby confirming the results of previous studies (34).
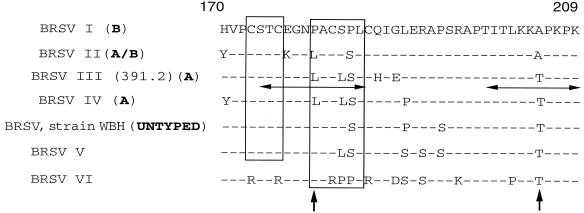
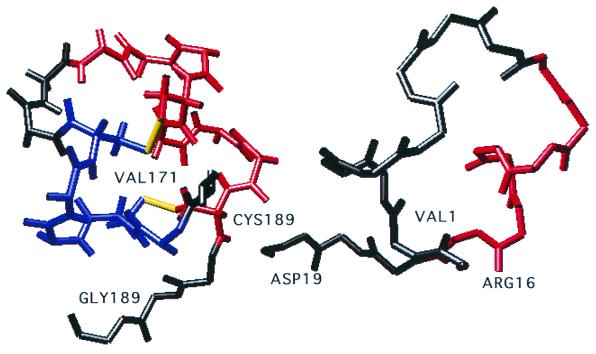
The central conserved domain also exhibited important amino acid changes (Fig. 4 and 5). Based on the availability of the structural coordinates of and the experimental restraints on the central conserved region of the G protein deposited in the Protein Data Bank of Brookhaven National Laboratory, we were able to examine the modifications induced in the three-dimensional structure by the mutations observed in this domain in field isolates. This domain is particularly important, as it was shown to be immunodominant in the G protein (34). The initial resolution of the structure was performed on strain 391.2 (13), which belongs to subgroup III. As most of the important residues in subgroups I to VI were conserved, the characteristic structure of the central conserved region of the G protein, as indicated by MODELLER, was probably also conserved. This structure consists of two disulfide bridges: Cys173-Cys186 (outer bridge) and Cys176-Cys182 (inner bridge). It begins with 1 turn of the α helix (Cys173-Cys176) which runs antiparallel to the 1.5 α-helical turn formed by Leu180-Leu185 (13) (Fig. 6). The two helices are linked by a type I turn (Cys176-Asn179). Asparagine 179 is involved in three hydrogen bonds.
FIG. 5.
Consensus sequence of BRSV subgroups from residues 170 to 209. Horizontal arrows indicate linear epitopes from the study of Langedijk et al. (33). Vertical arrows indicate mutations determining antigen subgrouping (33). Boxes indicate α helices.
FIG. 6.
Representation of the superimposition of the cysteine noose of BRSV strain 391.2 (left) and of the consensus sequence of isolates from subgroup VI (right). α Helices are indicated in blue and red. Disulfide bridges are drawn in yellow. Residues 171 to 189 in the representation of strain 391.2 correspond to residues 1 to 19 in that of subgroup VI.
The most striking mutations are located in the isolates of subgroup VI. In these isolates, the two disulfide bridges no longer exist due to replacement of the four Cys residues by four Arg residues. Resolution of the structure indicates that the first α helix is disrupted (Fig. 6). As a consequence, Asp179 is involved in only one hydrogen bond with Arg176 (data not shown). Near the central conserved region, the Ala205→Thr mutation distinguishing the G protein sequences of subgroup II (which includes most of the currently used vaccine strains) from those of subgroups III, IV, V, and VI is sufficient to cause escape from antibody binding. This mutation results in a potential additional O-glycosylation site which could modify the antigenicity of the linear epitope described for positions 199 to 209 (33).
Nonsynonymous changes are localized in specific regions.
The following analysis is restricted to the European BRSV isolates and to the vaccine strains Lehmkuhl 375 (subgroup III), RB-94 (subgroup II), and 220/69 (subgroup II). This analysis includes 38 different nucleotide sequences of the G protein gene which were available for all 489-base sequences, 24 different N protein gene nucleotide sequences (690 bases), and 24 different F protein gene nucleotide sequences (741 bases). The percentages of amino acid divergence between the European BRSV sequences were 0.7, 0.8, and 16.8 for the N, F, and G protein gene regions, respectively.
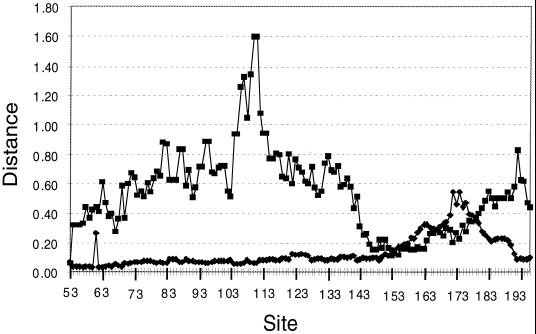
ds and dn as well as the dn/ds ratios for these three coding regions were assessed. ds, dn, and the dn/ds ratios were calculated as the means of the ratios determined in the pairwise sequence comparisons. Overall differences in the dn/ds ratios between the N, F, and G protein gene regions were significant (Pearson chi-square analysis). As expected, ds, dn, and the dn/ds ratios of the G protein fragment were significantly higher than those of the N and F protein fragments (Table 3). However, the dn/ds ratio was not identical over the entire G protein gene sequence (Fig. 7) and rose to values exceeding 0.8 in the two different coding parts of the genes corresponding to the mucin-like regions described above. The first region that exhibited a major increase in the dn/ds ratio was located between codons 82 and 153, in particular, between codons 104 and 132, where two dn/ds ratios reached 1.587. Another, minor region was located between codons 192 and 202. These ratios are considered evidence of positive selection (16) in the G protein. Our analysis shows a slight increase in ds between these two regions, starting from codon 149 and reaching a maximum of 0.545 between codons 174 and 183. ds then decreases slowly toward codons 184 to 203, where ds is 0.206 (Fig. 7). The region between codons 154 and 192 seems to be particularly well conserved.
TABLE 3.
ds, dn, and dn/ds ratiosa
| Protein gene (no. of isolates analyzed) | Avg pairwise divergence (SD) for:
|
dn/ds ratio | |
|---|---|---|---|
| ds | dn | ||
| N (24) | 0.0595 (0.0110) | 0.0029 (0.0015) | 0.05 |
| F (24) | 0.0692 (0.0124) | 0.0029 (0.0012) | 0.05 |
| G (38) | 0.1131 (0.0166) | 0.0490 (0.0061) | 0.58 |
| Gb (11) | 0.0802 (0.0165) | 0.0493 (0.0069) | 0.48 |
| Gc (32) | 0.0718 (0.0145) | 0.0446 (0.0066) | 0.62 |
The analysis of the N, F, and G protein genes was performed with 690, 741, and 489 nucleotides, respectively.
Analysis was performed with virus isolated in the absence of selective pressure due to vaccination.
Analysis was performed with virus isolated in countries where vaccination was widely used.
FIG. 7.
Genetic changes in the G protein gene according to codon number. ds (diamonds) and the dn/ds ratio (squares) are represented along the G protein length from codons 53 to 215, with a window of 20.
Similarly, to evaluate the effects of natural selection on individual amino acids that large-scale pairwise comparisons are unlikely to reveal, we calculated the mean dn for each codon in the N and F protein gene sequences and identified the codons with rates higher than the mean ds across all codons (4). In the N protein gene, two regions that exhibited a major increase in the average dn/ds ratio (the average dn/ds ratio rose to values exceeding 0.62) were located between codons 56 and 67 (region N1) and codons 147 and 153 (region N2). The mean dn for codon 98 in the N protein gene sequence was also above the mean ds. Three such regions in the F protein gene sequence (the average dn/ds ratio rose to values exceeding 0.66) were identified between codons 202 and 218 (region F1), codons 294 and 305 (region F2), and codons 389 and 401 (region F3). These results indicate that these regions concentrate most of the positive selection in the N and F proteins. Regions N2, F1, and F3, which exhibit high values for hydrophilicity and relative solvent accessibility (the accessibility of F3 being the lowest of the three regions), are potentially exposed at the surface of the proteins (data not shown). In contrast, region F2 is hydrophobic and has a very low relative accessibility. Region N1 has values for hydrophilicity and relative solvent accessibility that decrease from its amino terminus to its carboxy terminus (data not shown).
Rates of evolution of the synonymous and nonsynonymous substitutions in the G protein gene.
The relative selective pressures exerted on the G protein gene of BRSV isolates in the presence or in the absence of vaccination were investigated. First, this analysis was carried out with the three vaccine strains and 29 subgroup I, II, IV, V, and VI isolates originating from different herds in the United Kingdom, The Netherlands, Belgium, and France between 1969 and 1998 (29 years). This time frame includes the period when vaccination was widely practiced, at least in Belgium, France, and The Netherlands. Second, the same analysis was performed with 11 isolates of subgroups I and II, including the three vaccine strains, beginning with the oldest Belgian isolate (1969) and ending with a Danish isolate with the most recent complete sequence (1995). After 1977, the date of first use of vaccination against BRSV in Europe, this analysis was restricted to isolates from Denmark and Sweden. This time frame represents 26 years of BRSV evolution in the absence of selective pressure due to cattle vaccination. ds and dn as well as the dn/ds ratios were calculated for these two groups of isolates. The dn/ds ratio for the 32 sequences potentially subjected to vaccine-induced selection (dn/ds ratio, 0.62) was slightly higher than that obtained for the 11 sequences not subjected to any vaccine selective pressure (dn/ds ratio, 0.47) (Table 3).
To more precisely evaluate the effects of selection in these two epidemiological situations, the rates of evolution of the synonymous and nonsynonymous substitutions were determined. The vaccine strain Lehmkuhl 375 (subgroup III) was chosen as the outgroup. For the isolates potentially subjected to vaccine selective pressure, the correlations obtained between time of isolation and synonymous (syn) or nonsynonymous (nsy) substitutions were highly significant (n = 703, Rsyn = 0.189, Rnsy = 0.458, P < 0.001). The rates were 5.2 × 10−4 and 6.4 × 10−4 nucleotide changes per year per synonymous site and per nonsynonymous site, respectively. These results are indicative of a continuous evolution of BRSV G protein gene sequences from subgroups I and II to subgroup VI. For the isolates collected in the absence of vaccination, no significant correlation was obtained between the intervals of isolation and synonymous or nonsynonymous changes.
Rates of evolution of the synonymous and nonsynonymous substitutions in the N and F protein genes.
The same analysis concerning the rate of evolution was performed with 31 N and 40 F protein gene sequences belonging to subgroups II, IV, V, and VI (all the European sequences were included in this analysis, except for the identical sequences from the same herds). This sample includes viruses isolated when and where vaccination was widely practiced in Europe. This analysis could not be performed in the absence of selective pressure due to cattle vaccination, as the N and F protein gene sequences of isolates from Denmark and Sweden were not available from GenBank. The correlations obtained were highly significant for the N protein gene (n = 465, Rsyn = 0.731, Rnsy = 0.396, P < 0.001) and for the F protein gene (n = 780, Rsyn = 0.574, Rnsy = 0.751, P < 0.001). For the N protein gene, the rates were 1.6 × 10−3 and 5.4 × 10−5 nucleotide changes per year per synonymous site and per nonsynonymous site, respectively. For the F protein gene, the rates were 6.3 × 10−4 and 1.9 × 10−4 nucleotide changes per year per synonymous site and per nonsynonymous site, respectively. These values are also indicative of a continuous evolution of BRSV N and F protein gene sequences in countries where vaccination was widely used.
DISCUSSION
The genetic variability of the two antigenically distinct HRSV subgroups has been well characterized (8). However, this type of analysis has, in the case of BRSV, been limited until now to a small number of isolates and mostly to the G protein (15, 20, 21, 36, 44, 54, 60, 66). In the work reported here, we examined the divergence within nucleotide and amino acid sequences of the G protein in 87 BRSV isolates. The coding sequences of the N and F proteins in at least 58 of these isolates were also compared.
The rate of evolution of the BRSV sequences varies according to the gene, with the G protein gene evolving more rapidly and being highly tolerant to the fixation of mutations, whereas the N and F protein genes appear to be under stronger selective structural constraints which limit their evolution. However, a progressive accumulation of nucleotide changes was noted for the three fragments of these genes, as previously described for HRSV (8).
This work confirms the primary structure of the G protein determined for HRSV (for a review, see reference 47). It can be divided into three different parts, with one region located between amino acids 67 and 153 and another, starting with amino acid 193, constituting two mucin-like regions, with their characteristic high Ser and Thr contents. The end of this second region could not be determined in our study due to partial sequencing of the G protein gene. As with the G protein gene of HRSV (7, 22), nonsynonymous mutations are not randomly distributed along the G protein gene of BRSV but are localized predominantly in two areas of the gene (codons 82 to 153 and codons 192 to 202) coding for the mucin-like regions; presumably, these areas represent antigenic regions in the BRSV G protein. Both regions have already been shown to be of high antigenic importance in both BRSV and HRSV (8, 23, 34, 56, 66).
In contrast, the central hydrophobic region of the G protein, located between the two mucin-like regions and comprising amino acid positions 154 to 192, has remained highly stable during evolution. This region has been shown to accept predominantly synonymous mutations. It has been postulated to contain the region interacting with the putative RSV cell receptor. However, a very limited number of changes still occurred in the sequence; these changes modified the three-dimensional structure of the internal loop in the central hydrophobic region and an O-glycosylation site at codon position 205. The cysteine noose motif connecting the strands of the loop was disrupted due to mutation of Cys to Arg. Cysteine noose motifs are known to induce a high surface accessibility for the residues contained in the loop (35).
Some of these mutations presented by field isolates were similar to some of those already described for in vitro HRSV antibody escape mutants (45, 74). However, this work constitutes the first evidence that natural in vivo infections can occur with mutants lacking the four cysteines involved in the two disulfide bridges. Since at least one calf died of its infection, there is nothing about the clinical illness due to these viruses to suggest that they are attenuated. These mutations probably lead to important modifications in the antigenic sites located in this region. Some of these mutations had been tested by Pepscan analysis and had been shown to severely modify the recognition of antibodies (33). This finding is of concern with regard to recent BRSV isolates from France, where vaccination is widely used. Whether or not these mutations are linked to the positive selection exerted on other parts of the molecule remains to be proved. However, it has been shown that this central part of the G protein molecule constitutes a major domain involved in protection against HRSV infection (63, 67). Furthermore, this region, particularly amino acids 193 to 203, is involved in the enhancement of illness and lung eosinophilia in mice (65). The consequences of the modification of the conformational structure within the G protein in recent isolates from France should be investigated to determine if this modification could have an effect on illness enhancement and on protection. Indeed, strains from vaccination failures (subgroups V and VI) described in this paper are the most divergent from the vaccine strains and also the most recent.
The rate of evolution per nonsynonymous sites indicated 6.4 × 10−4 nucleotide changes per year for isolates originating from countries where vaccination was widely used (France, The Netherlands, and Belgium) during the period from 1969 to 1998. This high value cannot be related to Taq polymerase-induced errors (5, 64). Conversely, the same evaluation performed on isolates originating from countries where calf vaccination was limited and very recent (Denmark and Sweden) or from countries before the start of extensive vaccination (France, The Netherlands, and Belgium before 1977) did not indicate any significant evolution of the sequence. This result also favors the existence of a stronger positive selection pressure on the BRSV G protein in countries where BRSV vaccination had been more widely used. In some ways, this finding is equivalent to the sequence evolution due to immunological pressure on the hemagglutinin of influenza A viruses (for a review, see reference 24). The rates of evolution per synonymous and nonsynonymous sites determined for the N, G, and F protein fragments also indicate continuous evolution of BRSV sequences in the presence of vaccination. The values obtained for the evolution of BRSV are not surprising compared to those obtained for other RNA viruses (for a review, see reference 12). They are higher than those obtained for stable G protein genes, such as those of lyssaviruses (4), but lower than that of the hemagglutinin gene of influenza A virus, which is known to show rapid and temporal rather than geographical variations (19). In this respect, the molecular epidemiology of BRSV is intermediate between those of influenza B virus (6, 78) and influenza A virus (75). The clustering of BRSV isolates according to their continent of origin has not been observed for HRSV (8). This difference might reflect different circulation patterns for the human populations and calf herds between the American and European continents.
Some localized regions in the N protein (regions N1 [positions 56 to 67] and N2 [positions 143 to 153]) and in the F protein (regions F1 [positions 202 to 218], F2 [positions 294 to 305], and F3 [positions 389 to 401]) contained most of the amino acid variability of these proteins. No data are available concerning the localization of antigenic sites in the BRSV N protein. However, region N2, which is predicted to be exposed at the surface of the molecule, overlaps with a peptide of the HRSV N protein (positions 131 to 150) which was shown to be a linear antigenic domain (37). There is complete identity in this region between the two N proteins. The regions defined in the F protein do not correspond to any of the antigenic sites described for BRSV and HRSV. Further work will be needed to study the relationships between these relatively hypervariable regions in the N and F proteins and the immunological response against BRSV. An alternative explanation is that some of the variable sites are of high flexibility and accumulate accompanying mutations that are not directly implicated in positive immune selection.
In conclusion, the evolution of BRSV combines a high rate of sequence evolution (providing local genetic differentiation, which explains some of the geographical clustering observed in the phylogenetic trees) with a greatly elevated rate of amino acid changes in some regions of the G protein. Since these regions of the G protein are believed to be antigenically important, these changes provide an opportunity for the virus to potentially escape from previously established immunity, as determined in vitro with HRSV (67). As a consequence, recent isolates originating from countries where vaccination is widely used exhibit greater changes in the amino acid sequences in these critical regions. Further studies are now needed to reevaluate the level of protection provided by vaccine strains, taking into account this rapid evolution and, in particular, some of the mutations in the G protein described here. In addition, the ability of RSV to evade immune responses in humans should continue to be investigated. We are suggesting that particular attention should be given to the development of candidate HRSV subunit vaccines including only the central region of the G protein (62). Considering the mutations in this region described here, this subunit-vaccine approach should be used with caution.
ACKNOWLEDGMENTS
We are greatly indebted to veterinarians, practitioners, and farmers who allowed us to collect BRSV isolates; to Chantal Delacourt for strain D80 isolated in the Veterinary Laboratory of the Somme; to Eric Le Dréan for viruses B35-1 and 90504 isolated in the Veterinary Laboratory of Ille et Vilaine; and to D. Desmecht for lung samples from Belgium (P1 to P10). We also thank Suzanne Bonhoure, Martine Deplanche, Frédéric Lasserre, Martine Moulignier, and Jean-Luc Pingret for expert technical assistance and Edward Holmes, Nicolas Le Novere, and Wayne Sullender for helpful comments and critical reading of the manuscript.
This study was supported in part by grants from ENVT and from Institut Pasteur (Air, Environnement, et Santé grant).
REFERENCES
- 1.Åkerlind-Stopner B, Utter G, Mufson M A, Örvell C, Lerner R A, Norrby E. A subgroup-specific antigenic site in the G protein of respiratory syncytial virus forms a disulfide-bonded loop. J Virol. 1990;64:5143–5148. doi: 10.1128/jvi.64.10.5143-5148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson L J, Heirholzer J C, Tson C, Hendry R M, Fernie B N, Stone Y, McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 3.Beem M. Repeated infections with respiratory syncytial virus. J Immunol. 1987;98:1115–1122. [PubMed] [Google Scholar]
- 4.Bourhy H, Kissi B, Audry L, Smreczak M, Sadkowska-Todys M, Kulonen K, Tordo N, Zmudzinski J F, Holmes E C. Ecology and evolution of rabies virus in Europe. J Gen Virol. 1999;99:2545–2557. doi: 10.1099/0022-1317-80-10-2545. [DOI] [PubMed] [Google Scholar]
- 5.Bracho M A, Moya A, Barrio E. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J Gen Virol. 1998;79:2921–2928. doi: 10.1099/0022-1317-79-12-2921. [DOI] [PubMed] [Google Scholar]
- 6.Buonagurio D A, Nakada S, Desselberger U, Krystal M, Palese P. Noncumulative sequence changes in the hemagglutinin genes of influenza C virus isolates. Virology. 1985;146:221–232. doi: 10.1016/0042-6822(85)90006-6. [DOI] [PubMed] [Google Scholar]
- 7.Cane P A, Matthews D A, Pringle C R. Identification of variable domains of the attachment (G) protein of subgroup A respiratory syncytial viruses. J Gen Virol. 1991;72:2091–2096. doi: 10.1099/0022-1317-72-9-2091. [DOI] [PubMed] [Google Scholar]
- 8.Cane P A, Pringle C R. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol. 1995;69:2918–2925. doi: 10.1128/jvi.69.5.2918-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coates H V, Alling D W, Chanock R M. An antigenic analysis of respiratory syncytial virus isolates by plaque reduction neutralization assay. Am J Epidemiol. 1966;83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 10.Collins P L, Mottet G. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus. Altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 1992;73:849–863. doi: 10.1099/0022-1317-73-4-849. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the Vax. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 13.Doreleijers J F, Langedijk J P M, Hard K, Boelens R, Rullmann J A C, Schaaper W M, van Oirschot J T, Kaptein R. Solution structure of immunodominant region of protein G of bovine respiratory syncytial virus. Biochemistry. 1996;35:14684–14688. doi: 10.1021/bi9621627. [DOI] [PubMed] [Google Scholar]
- 14.Dowell S F, Anderson L J, Gary H E, Erdman D D, Plouffe J F, File T M, Martson B J, Breiman R F. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J Infect Dis. 1996;174:456–462. doi: 10.1093/infdis/174.3.456. [DOI] [PubMed] [Google Scholar]
- 15.Elvander M, Vilcek S, Baule C, Uttenthal A, Ballagi-Pordany A, Belak S. Genetic and antigenic analysis of the G attachment protein of bovine respiratory syncytial virus strains. J Gen Virol. 1998;79:2939–2946. doi: 10.1099/0022-1317-79-12-2939. [DOI] [PubMed] [Google Scholar]
- 16.Endo T, Ikeo K, Gojobori T. Large-scale search for genes on which positive selection may operate. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Fitch W M, Bush R M, Bender C A, Cox N J. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc Natl Acad Sci USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furze J, Wertz G, Lerch R A, Taylor G. Antigenic heterogeneity of the attachment protein of bovine respiratory syncytial virus. J Gen Virol. 1994;75:363–370. doi: 10.1099/0022-1317-75-2-363. [DOI] [PubMed] [Google Scholar]
- 21.Furze J M, Roberts S R, Wertz G W, Taylor G. Antigenically distinct G glycoproteins of BRSV strains share a high degree of genetic homogeneity. Virology. 1997;231:48–58. doi: 10.1006/viro.1997.8490. [DOI] [PubMed] [Google Scholar]
- 22.Garcia O, Martin M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Brena P, Martinez I, Garcia-Barreno B, Melero J A. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the glycoprotein. J Virol. 1994;68:5448–5459. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Barreno B, Portela A, Delgado T, Lopez J A, Melero J A. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO J. 1990;9:4181–4187. doi: 10.1002/j.1460-2075.1990.tb07642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnett G P, Antia R. Population biology of virus-host interactions. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 51–73. [Google Scholar]
- 25.Glezen W P, Paredes A, Allison J E, Taber L H, Frank A L. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J Pediatr. 1981;98:708–715. doi: 10.1016/s0022-3476(81)80829-3. [DOI] [PubMed] [Google Scholar]
- 26.Gruber C, Levine S. Respiratory syncytial virus polypeptides. IV. The oligosaccharides of the glycoproteins. J Gen Virol. 1985;66:417–432. doi: 10.1099/0022-1317-66-3-417. [DOI] [PubMed] [Google Scholar]
- 27.Hall C B, Douglas R G, Geiman J M. Respiratory syncytial virus infections in infants: quantification and duration of shedding. J Pediatr. 1976;89:11–15. doi: 10.1016/s0022-3476(76)80918-3. [DOI] [PubMed] [Google Scholar]
- 28.Kim H W, Canchola J G, Brandt C D, Pyles G, Chanock R M, Jensen K, Parrott R H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 29.Kimman T G, Zimmer G M, Westenbrink F, Mars J, de Leeuw P W. Diagnosis of bovine respiratory syncytial virus infections improved by virus detection in lung lavage samples. Am J Vet Res. 1986;47:143–147. [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetic Analysis, version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 31.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 32.Lambert D M. Role of oligosaccharides in structure and in function of respiratory syncytial virus glycoproteins. Virology. 1988;164:458–466. doi: 10.1016/0042-6822(88)90560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langedijk J P M, Meloen R H, Taylor G, Furze J M, van Oirschot J T. Antigenic structure of the central conserved region of protein G of bovine respiratory syncytial virus. J Virol. 1997;71:4055–4061. doi: 10.1128/jvi.71.5.4055-4061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langedijk J P M, Schaaper V M M, Meloen R H, van Oirschot J T. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J Gen Virol. 1996;77:1249–1257. doi: 10.1099/0022-1317-77-6-1249. [DOI] [PubMed] [Google Scholar]
- 35.Lapthorn A J, Janes R W, Isaacs N W, Wallace B A. Cystine nooses and protein specificity. Nat Struct Biol. 1995;2:266–268. doi: 10.1038/nsb0495-266. [DOI] [PubMed] [Google Scholar]
- 36.Larsen L E, Uttenthal A, Arctander P, Tjornehoj K, Viuff B, Rontved C, Ronsholtved L, Alexandersen S, Blixenkrone-Moller M. Serological and genetic characterisation of bovine respiratory syncytial virus (BRSV) indicates that Danish isolates belong to the intermediate subgroup: no evidence of a selective effect on the variability of G protein nucleotide sequence by prior cell culture adaptation and passages in cell culture or calves. Vet Microbiol. 1998;62:265–279. doi: 10.1016/s0378-1135(98)00226-0. [DOI] [PubMed] [Google Scholar]
- 37.Leonov S V, Waris M, Norrby E. Linear antigenic and immunogenic regions of human respiratory syncytial virus N protein. J Gen Virol. 1995;76:357–364. doi: 10.1099/0022-1317-76-2-357. [DOI] [PubMed] [Google Scholar]
- 38.Lerch R A, Anderson K, Wertz G W. Nucleotide sequence analysis and expression from recombinant vectors demonstrate that attachment protein G of bovine respiratory syncytial virus is distinct from that of human respiratory syncytial virus. J Virol. 1990;64:5559–5569. doi: 10.1128/jvi.64.11.5559-5569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerch R A, Anderson K, Wertz G W. Nucleotide sequence analysis of the bovine respiratory syncytial virus fusion protein mRNA and expression from a recombinant vaccinia virus. Virology. 1991;181:118–131. doi: 10.1016/0042-6822(91)90476-r. [DOI] [PubMed] [Google Scholar]
- 40.Lerch R A, Stott E J, Wertz G W. Characterization of bovine respiratory syncytial virus proteins and mRNAs and generation of cDNA clones to the viral mRNAs. J Virol. 1989;63:833–840. doi: 10.1128/jvi.63.2.833-840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 42.Li W-H, Tanimura M, Sharp P M. Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol. 1988;5:313–330. doi: 10.1093/oxfordjournals.molbev.a040503. [DOI] [PubMed] [Google Scholar]
- 43.Lincoln S E, Daly M J, Lander E S. PRIMER: a computer program for automatically selecting PCR primers, Version 0.5. Cambridge, Mass: MIT Center for Genome Research and Whitehead Institute for Biomedical Research; 1991. [Google Scholar]
- 44.Mallipeddi S K, Samal S K. Sequence variability of the glycoprotein gene of bovine respiratory syncytial virus. J Gen Virol. 1993;74:2001–2004. doi: 10.1099/0022-1317-74-9-2001. [DOI] [PubMed] [Google Scholar]
- 45.Martinez I, Dopazo J, Melero J A. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol. 1997;78:2419–2429. doi: 10.1099/0022-1317-78-10-2419. [DOI] [PubMed] [Google Scholar]
- 46.McGuire K, Holmes E C, Gao G F, Reid H W, Gould E A. Tracing the origins of louping ill virus by molecular phylogenetic analysis. J Gen Virol. 1998;79:981–988. doi: 10.1099/0022-1317-79-5-981. [DOI] [PubMed] [Google Scholar]
- 47.Melero J A, Garcia-Barreno B, Martinez I, Pringle C R, Cane P A. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol. 1997;78:2411–2418. doi: 10.1099/0022-1317-78-10-2411. [DOI] [PubMed] [Google Scholar]
- 48.Mufson M A, Örvell C, Rafner B, Norrby E. Two distinct subgroups of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 49.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. [Google Scholar]
- 50.Nei M, Gojobori T. Simple methods for estimating the number of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 51.Norrby E, Mufson M A, Sheshberadaran H. Structural differences between subtype A and B strains of respiratory syncytial virus. J Gen Virol. 1986;67:2721–2729. doi: 10.1099/0022-1317-67-12-2721. [DOI] [PubMed] [Google Scholar]
- 52.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comp Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 53.Piazza F M, Johnson S A, Darnell M E R, Porter D D, Hemming V G, Prince G A. Bovine respiratory syncytial virus protects cotton rats against human respiratory syncytial virus infection. J Virol. 1993;67:1503–1510. doi: 10.1128/jvi.67.3.1503-1510.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prozzi D, Walravens K, Langedijk J P M, Daus F, Kramps J A, Letesson J J. Antigenic and molecular analyses of the variability of bovine respiratory syncytial virus G glycoprotein. J Gen Virol. 1997;78:359–366. doi: 10.1099/0022-1317-78-2-359. [DOI] [PubMed] [Google Scholar]
- 55.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 56.Rueda P, Delgado T, Portela A, Melero J A, García-Barreno B. Premature stop codons in the G glycoprotein of human respiratory syncytial viruses resistant to neutralization by monoclonal antibodies. J Virol. 1991;65:3374–3378. doi: 10.1128/jvi.65.6.3374-3378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitou N, Nei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 58.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 59.Sanchez R, Sali A. Evaluation of comparative protein structure modeling by MODELER-3. Proteins. 1997;1(Suppl.):50–58. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 60.Schrijver R S, Daus F, Kramps J A, Langedijk J P M, Buijs R, Middel W G J, Taylor G, Furze J, Huyben M W C, van Oirschot J T. Subgrouping of bovine respiratory syncytial virus strains detected in lung tissue. Vet Microbiol. 1996;53:253–260. doi: 10.1016/s0378-1135(96)01223-0. [DOI] [PubMed] [Google Scholar]
- 61.Schrijver R S, Langedijk J P M, Middel W G J, Kramps J A, Rijsewijk F A M, van Oirschot J T. A bovine respiratory syncytial virus strain with mutations in subgroup-specific antigenic domains of the G protein induces partial heterologous protection in cattle. Vet Microbiol. 1998;63:159–175. doi: 10.1016/s0378-1135(98)00244-2. [DOI] [PubMed] [Google Scholar]
- 62.Siegrist C A, Plotnicky-Gilquin H, Cordova M, Berney M, Bonnefoy J Y, Nguyen T N, Lambert P H, Power U F. Protective efficacy against respiratory syncytial virus following murine neonatal immunization with BBG2Na vaccine: influence of adjuvants and maternal antibodies. J Infect Dis. 1999;179:1326–1333. doi: 10.1086/314778. [DOI] [PubMed] [Google Scholar]
- 63.Simard C, Nadon F, Seguin C, Trudel M. Evidence that amino acid region 124–203 of glycoprotein G from the respiratory syncytial virus (RSV) constitutes a major part of the polypeptide domain that is involved in the protection against RSV infection. Antiviral Res. 1995;28:303–315. doi: 10.1016/0166-3542(95)00053-4. [DOI] [PubMed] [Google Scholar]
- 64.Smith D, McAllister J, Casino C, Simmonds P. Virus “quasispecies”: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 65.Sparer T E, Matthews S, Hussel T, Rae A J, Garcia-Barreno B, Melero J A, Openshaw J M. Eliminating a region of respiratory syncytial virus attachment protein allows induction of protective immunity without vaccine-enhanced lung eosinophilia. J Exp Med. 1998;187:1921–1926. doi: 10.1084/jem.187.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stine L C, Hoppe D K, Kelling C L. Sequence conservation in the attachment glycoprotein and antigenic diversity among bovine respiratory syncytial virus isolates. Vet Microbiol. 1997;54:201–221. doi: 10.1016/s0378-1135(96)01288-6. [DOI] [PubMed] [Google Scholar]
- 67.Sullender W M, Edwards K G. Mutations of respiratory syncytial virus attachment glycoprotein G associated with resistance to neutralization by primate polyclonal antibodies. Virology. 1999;264:230–236. doi: 10.1006/viro.1999.9987. [DOI] [PubMed] [Google Scholar]
- 68.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trudel M, Stott E J, Taylor G, Oth D, Mercier G, Nadon F, Seguin C, Simard C, Lacroix M. Synthetic peptides corresponding to the F protein of RSV stimulate murine B and T cells but fail to confer protection. Arch Virol. 1991;117:59–71. doi: 10.1007/BF01310492. [DOI] [PubMed] [Google Scholar]
- 70.Valarcher J-F, Bourhy H, Gelfi J, Schelcher F. Evaluation of a nested reverse transcription-PCR assay based on the nucleoprotein gene for diagnosis of spontaneous and experimental bovine respiratory syncytial virus infections. J Clin Microbiol. 1999;37:1858–1862. doi: 10.1128/jcm.37.6.1858-1862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Poel W H M, Brand A, Kramps J A, van Oirschot J T. Respiratory syncytial virus infections in human beings and in cattle. J Infect. 1994;29:215–228. doi: 10.1016/s0163-4453(94)90866-4. [DOI] [PubMed] [Google Scholar]
- 72.Van der Poel W H M, Kramps J A, Middel W G J, van Oirschot J T, Brand A. Dynamics of bovine respiratory syncytial virus infections, a longitudinal epidemiological study in dairy herds. Arch Virol. 1993;133:309–321. doi: 10.1007/BF01313771. [DOI] [PubMed] [Google Scholar]
- 73.Vilcek S, Elvander M, Ballagi-Pordany A, Belak S. Development of nested PCR assays for detection of bovine respiratory syncytial virus in clinical samples. J Clin Microbiol. 1994;32:2225–2231. doi: 10.1128/jcm.32.9.2225-2231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walsh E E, Falsey A R, Sullender W M. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J Gen Virol. 1998;79:479–487. doi: 10.1099/0022-1317-79-3-479. [DOI] [PubMed] [Google Scholar]
- 75.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wellemans G, Leunen J. Le virus respiratoire syncytial et les troubles respiratoires des bovins. Ann Med Vet. 1975;119:359–369. [Google Scholar]
- 77.Wertz G W, Krieger M, Ball L A. Structure and cell face maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J Virol. 1989;63:4767–4776. doi: 10.1128/jvi.63.11.4767-4776.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamashita M, Krystal M, Fitch W M, Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988;163:112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]