Abstract
Thogoto virus (THOV) is a tick-transmitted orthomyxovirus with a segmented, negative-stranded RNA genome. In this study, we investigated the coding strategy of RNA segment 6 and found that it contains 956 nucleotides and codes for the matrix (M) protein. The full-length cDNA contains a single, long reading frame that lacks a stop codon but has coding capacity for a putative 35-kDa protein. In contrast, the M protein of THOV has an apparent molecular mass of 29 kDa as assessed by polyacrylamide gel electrophoresis. Therefore, we investigated the possibility of posttranscriptional processing of segment 6 transcripts by reverse transcription-PCR and identified a spliced mRNA that contains a stop codon and is translated into the 29-kDa M protein. Interestingly, the nontemplated UGA stop codon is generated by the splicing event itself. Thus, the unusual M coding strategy of THOV resembles that of Influenza C virus.
Thogoto virus (THOV) is a tick-borne orthomyxovirus (24) and contains a genome of six single-stranded RNA segments of negative polarity. These genomic RNA segments are encapsidated by nucleoprotein (NP) and associate with the viral polymerase complex to form viral ribonucleoprotein complexes (vRNPs) (7, 30) in a manner similar to that of influenza viruses (5). The three largest segments code for the subunits of the viral RNA polymerase complex (PB2, PB1, and PA [19, 34]), segment 4 encodes the viral surface glycoprotein (21), and segment 5 encodes NP (37). The smallest RNA segment, segment 6, has not previously been characterized but presumably codes for the matrix (M) protein. In influenza viruses, it forms the major component of the virus particle and is expressed at high levels late in infection (16). Similarly, the M protein of THOV is also produced in abundance in virus-infected cells (31). This protein has a molecular weight of approximately 29,000 (30), comparable to that of the M protein of Dhori virus (DHOV), another tick-borne orthomyxovirus (7). DHOV M protein shows only weak sequence similarity to the matrix proteins of influenza viruses, and to date no functional studies have been undertaken (6).
The role of M protein in virus multiplication has been studied most intensively for Influenza A virus (FLUAV) (41). Upon infection, incoming vRNPs are transported into the nucleus, where viral transcription and replication take place. Newly assembled vRNPs are then coated by matrix protein (M1) (43) and exported from the nucleus to the cytoplasm (4, 20). To accomplish this export, M1 directly interacts with the FLUAV-encoded nuclear export protein (NEP; previously called NS2 [25]). Moreover, cytoplasmic M1 prevents vRNPs from relocating to the nucleus (40), allowing the vRNPs to be directed to the plasma membrane, where they are packaged into new virus particles (16).
To address the question of nuclear export and virus assembly in THOV-infected cells, we have cloned the genomic RNA of segment 6 that encodes the THOV M protein. Here we report that THOV M is produced from a spliced mRNA, using a coding strategy similar to that of Influenza C virus (FLUCV) (42).
Cloning of THOV segment 6.
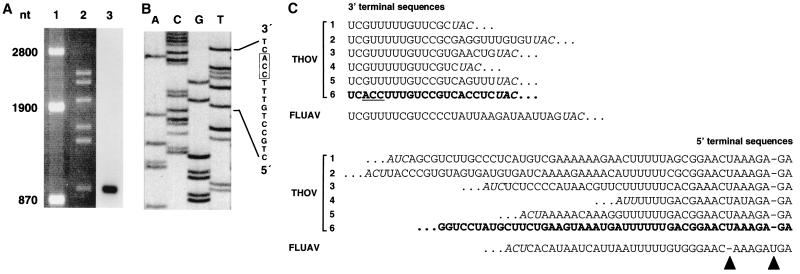
Genomic viral RNA was extracted from purified THOV particles (strain SiAr 126) (1), and a cDNA phage library was generated (34, 37). A segment 6-specific cDNA clone (pBK-M15) was isolated from the library by selection for recombinant phage plaques that were not recognized by cDNA probes coding for THOV segments 1 to 5. Northern blot analysis with virion RNA demonstrated that the insert of pBK-M15 hybridized to segment 6 (Fig. 1A). The insert contained 805 nucleotides (nt) but lacked the conserved terminal sequences that are characteristic for the segments of THOV. The sequences of the extreme 5′ and 3′ ends of segment 6 were determined by rapid amplification of 5′ cDNA ends (5′-RACE) and reverse transcription-PCR (RT-PCR) with genomic RNA circularized by intramolecular ligation using M-specific internal primers as described elsewhere (37). The sequence of the 3′ end was further confirmed by 5′-RACE of the viral transcripts of segment 6 by using poly(A)+-selected RNA from THOV-infected cells (Fig. 1B). Figure 1C shows a comparison of the 5′- and 3′-terminal sequences of segment 6 with the other five segments of THOV. Interestingly, the 3′ ends are highly conserved among all six segments and similar to those of FLUAV and other influenza viruses. The 3′ end of segment 6 is exceptional in that the three nucleotides at positions 3 to 5 are different from the sequences of the other segments. This sequence variation is not found in the genomic RNA of DHOV segment 6 (6) or any other orthomyxovirus (7). The 5′ end of segment 6 shows the characteristic nucleotide sequence of THOV, which differs from that of FLUAV at two positions. The U at position 3 of FLUAV vRNA is absent in the THOV sequence, whereas THOV has an additional U at position 8 (Fig. 1C). The THOV-specific 5′-end sequence allows intrastrand base pairing between positions 2 and 9 and positions 3 and 8 and thus favors the formation of a hook-like promoter structure (18, 35). Intrastrand base pairing has also been postulated for the 3′ end of FLUAV genomic RNAs (8, 9). With regard to this secondary structure, it is proposed that the unique 3′-end nucleotide sequence of THOV segment 6 may increase intrastrand base pairing at the 3′ terminus, since it allows an A-U base pair between positions 3 and 8 instead of the noncanonical G-U base pair seen for the other gene segments. Such a stabilized hook-like conformation may affect segment 6 gene expression.
FIG. 1.
Identification of vRNA segment 6 of THOV. (A) The cloned cDNA corresponds to the smallest vRNA segment of THOV. Genomic RNA from purified THOV virions was separated on a 1.2% agarose gel containing 3.7% formaldehyde and ethidium bromide. RNA markers (lane 1) and the six vRNA segments of THOV (lane 2) were visualized under UV light. Blotted RNA was probed with radiolabeled full-length M cDNA (lane 3). (B) Determination of the 5′ end of the viral mRNA encoded by segment 6 by 5′-RACE (37). Poly(A)+-selected RNA isolated from virus-infected cells was reverse transcribed using an oligonucleotide complementary to nt 438 to 419 of genomic segment 6. After poly(dC) tailing, the first-strand cDNA was PCR amplified using a poly(dG) anchor primer and an oligonucleotide corresponding to nt 379 to 356 of segment 6. The sequence of the resulting product was then determined using an oligonucleotide corresponding to nt 128 to 109 of segment 6. (C) Multiple sequence alignment of the noncoding 3′- and 5′-end sequences of the genomic segments of THOV and FLUAV. The vRNA terminal sequences of THOV segment 1 (34), segments 2 and 3 (19), segment 4 (21), segment 5 (37), and segment 6 (bold letters) are compared with a representative sequence of FLUAV segment 6 (44). The three differing nucleotides at the 3′-end of THOV segment 6 are underlined. A dash indicates a gap introduced into the sequence for optimal alignment. Start and stop codons, which are in antisense orientation, are shown in italic letters; the arrowheads indicate the U residues conserved at position 3 of the FLUAV and position 8 of the THOV 5′ noncoding regions.
RNA segment 6 of THOV encodes the M gene.
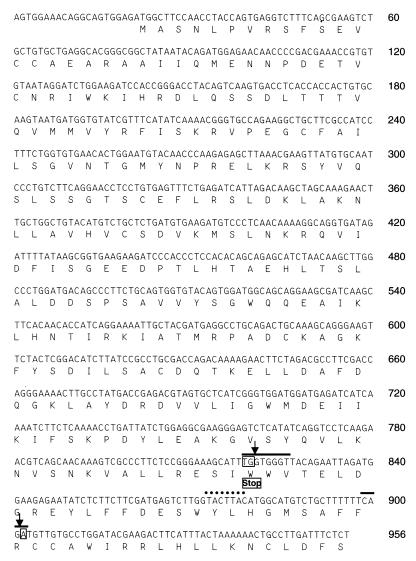
A full-length cDNA clone of segment 6 was amplified by RT-PCR using a primer pair that anneals to the terminal regions of segment 6. The PCR product was inserted into the pBSK(+) vector and sequenced. The complete segment 6 RNA contains 956 nt (Fig. 2), a length consistent with that determined by agarose gel electrophoresis and similar to that of the smallest (962-nt) segment of DHOV (6). THOV segment 6 contains a single, long reading frame of 936 nt that starts with an AUG at positions 21 to 23. Surprisingly, a stop codon could not be identified. The nonterminated reading frame has a coding capacity for 312 amino acids, corresponding to a polypeptide with a calculated molecular weight of 35,679. No additional reading frames longer than 42 amino acids were found.
FIG. 2.
Complete cDNA nucleotide sequence of THOV segment 6 (in antigenomic orientation) and deduced amino acid sequence. Arrows indicate the splice site positions for the M protein-specific mRNA. Black bars indicate the splice donor and acceptor sites; the dotted line indicates the putative splice branch site. The boxed nucleotides TG and A represent the termination codon generated in the spliced product.
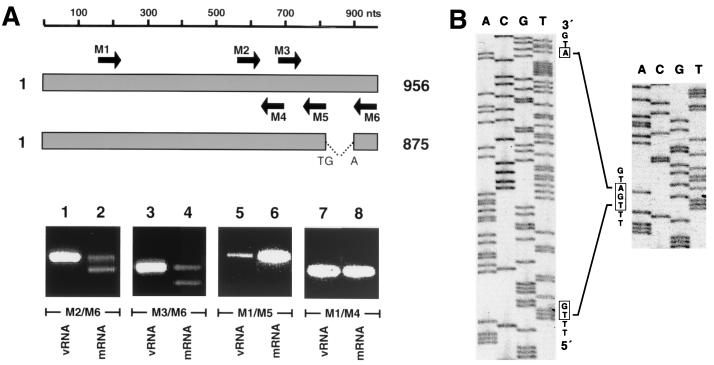
As the longest reading frame lacks a stop codon, we analyzed virus transcripts for posttranscriptional modifications using RT-PCR. Genomic RNA isolated from THOV particles and poly(A)+-selected RNA from THOV-infected cells were amplified in parallel and with different primer combinations. The downstream primer M6 is complementary to the 3′ end of THOV cRNA, and upstream primers M1 and M2 anneal to central sequences (Fig. 3A). With these primers, RT-PCR amplification of vRNA template resulted in products of expected lengths (Fig. 3A, lanes 1 and 3). An additional, smaller band was apparent when poly(A)+-selected RNA was used as a template (lanes 2 and 4). In contrast, both vRNA and mRNA templates gave rise to a single product when the downstream primers M4 and M5 were used (lanes 5 to 8). These results suggest posttranscriptional modification of segment 6 transcripts, most likely a splicing event which affects the sequences between nt 779 and 906. To further investigate this, the two RT-PCR products amplified from poly(A)+ RNA using primers M2 and M6 (lane 2) were isolated, sequenced, and compared to the genomic vRNA sequence.
FIG. 3.
Transcripts of THOV segment 6 are modified by splicing. (A) Segment 6-specific RT-PCR products of vRNA prepared from virus particles and mRNA isolated from THOV-infected cells. The bars at the top schematically represent unspliced and spliced transcripts of segment 6. Arrows indicate the positions and orientations of the PCR primers. The cDNAs were synthesized using random hexanucleotides and amplified by PCR using segment 6-specific primers (M1, nt 151 to 170, 5′ TACAGTCAAGTGACCTCACC 3′; M2, nt 550 to 569, 5′CCATCAGGAAAATTGCTACG 3′; M3, nt 664 to 684, 5′ GAAAACTTGCCTATGACCGAG 3′; M4, nt 634 to 617, 5′ GTCTGGTCGCAGGCGGAT 3′; M5, nt 779 to 760, 5′ CTTGAGGACCTGATATGAGA 3′; M6, nt 927 to 906, 5′ GAAGTCTTCGTATCCAGGCACA 3′). The PCR products were analyzed by electrophoresis on an ethidium bromide-stained agarose gel. (B) Sequence determination of the RT-PCR products. The cDNA fragments of the slower-migrating upper (left panel) and faster-migrating lower (right panel) bands shown in lane 2 of panel A were isolated from the agarose gel and sequenced using primer M3.
The nucleotide sequence of the larger RT-PCR product proved to be identical to the sequence of segment 6 (Fig. 3B, left) that was determined from genomic RNA. In contrast, the sequence of the smaller product contained a deletion of 81 nt between positions 820 and 902 (Fig. 3B, right). Visual examination of the nucleotide sequences flanking these positions revealed similarities to consensus donor and acceptor splice sites (Fig. 2 and reference 22). These splice recognition sites were identical to those of FLUCV segment 6 (42) and share minor sequence similarities to FLUAV segment 7 coding for the M1 and the M2 proteins (17). In addition, a sequence motif of 7 nt located 22 nt upstream of the potential 3′ splice site of THOV segment 6, shares similarity to the branch site of cellular introns (Fig. 2), a motif required for the splicing process (23). The splicing of THOV segment 6 transcripts results in the formation of an UGA stop codon which terminates the open reading frame (ORF) at nt 819, whereby UG originates from the 5′ splice site and A originates from the 3′ splice site. This ORF encodes a polypeptide of 266 amino acids with a calculated molecular weight of 29,851, a size consistent with that of the THOV M protein found in virus particles (see below). We concluded that M of THOV is translated from a spliced mRNA and that the larger RT-PCR product represents unspliced mRNA transcripts or full-length genomic RNA copurifying with the mRNA preparation.
The deduced amino acid sequence was compared with published M sequences of DHOV and FLUAV, FLUBV, and FLUCV, using the Jotun-Hein algorithm (11). M protein of THOV has amino acid sequence similarities of 25% with M protein of DHOV and 10 to 15% with M protein of the influenza viruses. No conserved regions were detectable between these sequences. In contrast to the findings for THOV, no evidence for a splicing event has been reported for the DHOV segment 6 transcripts. The M protein of DHOV is encoded by a full-length unspliced mRNA (6), as are the M1 proteins of FLUAV and FLUBV (16). The M gene encoded on segment 6 of FLUCV, however, codes for a single long ORF processed by posttranscriptional splicing, creating a new UGA stop codon at the splice junction (42). Thus, THOV appears to use the same coding strategy as FLUCV to express its M protein.
THOV M protein is encoded by a spliced mRNA.
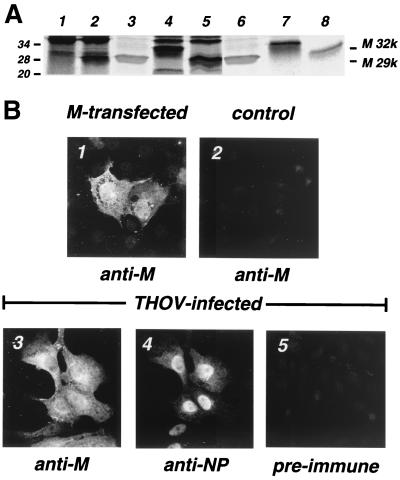
To confirm that the M protein is translated from a spliced mRNA, full-length and spliced transcripts were translated and compared to the M protein of virus particles. To this end, poly(A)+-selected transcripts were amplified, using primers complementary to nt 1 to 24 and 908 to 936 of THOV segment 6 cRNA. Two PCR products that corresponded in size to the expected full-length or spliced transcripts were obtained. The PCR products were both cloned into the pBSK(+) vector under the control of a bacteriophage T7 RNA polymerase promoter and into a mammalian expression vector, pSUPERcatch (pSC) (10). In pSC, the ORF of M protein is N-terminally tagged with a Flag epitope (15) and under the control of a cytomegalovirus promoter. In vitro translation of the two cDNAs resulted in the formation of protein products that were recognized by immunoprecipitation using a rabbit serum raised against purified Escherichia coli-produced recombinant M protein. The immunoprecipitate from the full-length transcript had an apparent molecular weight of about 32,000, whereas that from the spliced transcript had an apparent molecular weight of 29,000 (Fig. 4A, lanes 7 and 8). The smaller product corresponded in size to the authentic M protein extracted from purified THOV particles (lanes 3 and 6). Similar results were obtained when the two cDNAs were expressed in vivo under the control of the T7 RNA polymerase promoter (lanes 4 and 5). In this in vivo system, transcripts are synthesized in the cytoplasm by the T7 RNA polymerase provided by a recombinant vaccinia virus and are not modified posttranscriptionally. Synthesis of M protein in THOV-infected cells was analyzed by immunoprecipitation of 35S-labeled proteins from cell extracts using M-specific antibodies. Only the smaller gene product was detected in virus-infected cells (lane 2). Two bands which migrated above the 29,000-molecular-weight protein were also precipitated from uninfected cell extracts (lane 1) and probably represent cellular proteins. These data support the assumption that the M protein is translated from a spliced mRNA and that the unspliced transcript does not lead to a readily detectable viral protein.
FIG. 4.
Comparison of recombinant THOV M protein with the authentic viral protein. (A) 35S-labeled proteins immunoprecipitated from cell lysates using a rabbit polyclonal antiserum raised against recombinant M expressed in E. coli. The proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. Lane 1, mock-infected cells; lane 2, cells infected with THOV for 16 h; lanes 4 and 5, cells transfected with the cDNA encoding the full-length (M 32k) and spliced (M 29k), respectively, transcripts of THOV segment 6; lanes 7 and 8, in vitro translation products of the full-length (M 32k) and spliced (M 29k), respectively, transcripts of THOV segment 6. Lanes 3 and 6 show authentic M protein isolated from purified THOV particles and stained with Coomassie blue. Molecular mass markers (in kilodaltons) are indicated on the left. (B) Subcellular localization of THOV M protein in mammalian cells. Cells were transfected with an M expression construct coding for the spliced variant of the segment 6 transcript (panel 1), infected with THOV for 16 h (panels 3 to 5), or left untreated (panel 2). The recombinant M protein was detected by immunofluorescence using the M-specific polyclonal rabbit antiserum (panel 1). Panels 3 and 4 show double-immunofluorescence pictures of THOV-infected cells stained with the anti-M antiserum (panel 3) or a monoclonal antibody directed against the viral NP (panel 4) (30). To demonstrate the specificity of the staining, untreated control cells were incubated with the M-specific antiserum (panel 2) and THOV-infected cells were stained with the preimmune serum (panel 5).
Next, we compared the subcellular localization of recombinant M expressed from plasmids with that of authentic M expressed during viral infection. Mouse 3T3 cells were transfected with the mammalian expression vector (pSC-M) containing the spliced M cDNA. In parallel, 3T3 cells were infected with THOV. The cells were analyzed 24 h later by immunofluorescence, using M-specific antibodies. The recombinant protein showed cytoplasmic as well as nuclear localization (Fig. 4B, panel 1). In THOV-infected cells, M protein was also found in both compartments (panel 3), while, as expected, NP localized predominantly in the nucleus (panel 4 and reference 39). To confirm the specificity of the M protein staining, we treated control cells with the same M-specific antibodies. No cross-reaction with cellular proteins was detectable (panel 2). In addition, a preimmune serum of the rabbit that was immunized with the recombinant M protein showed only weak signals in THOV-infected cells (panel 5), indicating the specificity of the antiserum used.
Our results demonstrate that RNA segment 6 of THOV encodes the M protein. The transcripts coding for this protein are modified by splicing, a process which creates a termination codon at the splice junction. A nuclear phase of replication has previously been shown for THOV (30, 31). The nucleus provides an environment for the cap-stealing mechanism involved in THOV mRNA synthesis (2, 37). The present data demonstrate that THOV mRNA synthesis requires an additional nuclear function, namely, the cellular splicing machinery.
Unspliced M protein mRNA should be capable of encoding a longer polypeptide which contains an additional 46 amino acids of the C terminus. However, a protein product translated from unspliced mRNA has not been identified in virus-infected cells, indicating that the unspliced mRNA may not be translated. If this larger product does exist, it may do so at undetectable levels or be subject to posttranslational modifications. Usually splicing is a means by which viruses expand the coding capacity of the genome. In the present case, splicing may serve another function and contribute to regulated expression of the M gene during the virus life cycle.
The role of the orthomyxovirus M gene is best studied in FLUAV (41). M1 of FLUAV is expressed late in the viral life cycle and is involved in diverse steps, such as export of newly synthesized vRNPs out of the nucleus (3, 20), retention of these vRNPs in the cytoplasm (40), and assembly of vRNPs into virus particles at the plasma membrane (16). It is possible that THOV M serves similar and, possibly, additional functions. In FLUAV-infected cells, nuclear export of vRNPs requires an additional protein called NS2 or NEP. NEP is encoded by RNA segment 8 and binds to M1 which is associated to newly synthesized vRNPs in the cell nucleus (25). In contrast to FLUAV, THOV has only six genomic RNA segments and seems to lack an NEP gene. It will be of interest to see whether THOV M protein itself has NEP function or whether the putative larger product of the unspliced transcripts could provide this function.
In addition to M1, RNA segment 6 of FLUAV codes for M2, a transmembrane protein which forms an ion channel in the virus envelope allowing acidification of the virion interior upon virus entry into host cells (29, 32). A corresponding protein has not been detected in THOV-infected cells. In FLUCV, however, a protein with similar properties is produced in an unusual way. An unspliced mRNA species transcribed from FLUCV segment 6 is expressed in virus-infected cells at a low level. It codes for a 374-amino-acid protein that can be detected in cell lysates (12). This protein, P42, is an integral membrane protein containing two hydrophobic regions. Recently, it has been reported that the P42 protein is cleaved by a signal peptidase, yielding a membrane-integrated C-terminal fragment, CM2, with a molecular weight of 18,000 (14, 27). CM2 has biochemical properties similar to those of the M2 protein of FLUAV (13, 26). By analogy, the long unspliced reading frame of THOV segment 6 might code for a THOV protein with ion channel activity. However, structure prediction analyses of the peptide sequence of the unspliced reading frame did not reveal features characteristic for transmembrane domains.
M1 of FLUAV has been shown to inhibit the activity of the viral polymerase complex (28, 33). Preliminary results indicate that THOV M has a similar negative effect on the activity of the THOV polymerase complex (G. Kochs, unpublished data). In the early stage of infection, inhibition of the polymerase by M must be avoided. Therefore, a strong restriction of M expression is necessary to guarantee a successful infection cycle. Splicing of segment 6 could contribute to a tight control of M expression. Furthermore, the unusual promoter sequence of RNA segment 6 correlates with low transcriptional activity (Kochs, unpublished). We have recently established a system to reconstitute the viral polymerase complex of THOV in transfected cells (36, 38). This system will allow us to further analyze the role of the segment 6 promoter structure and the splicing event for M gene expression.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the GenBank data bank with accession number AF236794.
Acknowledgments
We thank Patricia A. Nuttall for the generous gift of the THOV NP-specific antibody and Christian Janzen for cloning the spliced and unspliced M expression plasmids.
This work was supported by grant Ko 1579/3-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Albanese M, Bruno-Smiraglia C, Di Cuonzo G, Lavagnino A, Srihongse S. Isolation of Thogoto virus from Rhipicephalus bursa ticks in western Sicily. Acta Virol. 1972;16:267. [PubMed] [Google Scholar]
- 2.Albo C, Martin J, Portela A. The 5′ ends of Thogoto virus (Orthomyxoviridae) mRNAs are homogeneous in both length and sequence. J Virol. 1996;70:9013–9017. doi: 10.1128/jvi.70.12.9013-9017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui M, Wills E G, Helenius A, Whittaker G R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choppin P W, Compans R W. The influenza viruses and influenza. New York, N.Y: Academic Press; 1975. [Google Scholar]
- 6.Clay W C, Fuller F J. Nucleotide sequence of the tick-borne orthomyxo-like Dhori/India/1313/61 virus membrane protein gene. J Gen Virol. 1992;73:2609–2616. doi: 10.1099/0022-1317-73-10-2609. [DOI] [PubMed] [Google Scholar]
- 7.Clerx J P M, Fuller F, Bishop D H. Tick-borne viruses structurally similar to orthomyxoviruses. Virology. 1983;127:205–219. doi: 10.1016/0042-6822(83)90384-7. [DOI] [PubMed] [Google Scholar]
- 8.Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the corkscrew conformation. J Gen Virol. 1999;80:2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- 9.Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 10.Georgiev O, Bourquin J P, Gstaiger M, Knoepfel L, Schaffner W, Hovens C. Two versatile eukaryotic vectors permitting epitope tagging, radiolabelling and nuclear localisation of expressed proteins. Gene. 1996;168:165–167. doi: 10.1016/0378-1119(95)00764-4. [DOI] [PubMed] [Google Scholar]
- 11.Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- 12.Hongo S, Gao P, Sugawara K, Muraki Y, Matsuzaki Y, Tada Y, Kitame F, Nakamura K. Identification of a 374 amino acid protein encoded by RNA segment 6 of influenza C virus. J Gen Virol. 1998;79:2207–2213. doi: 10.1099/0022-1317-79-9-2207. [DOI] [PubMed] [Google Scholar]
- 13.Hongo S, Sugawara K, Muraki Y, Kitame F, Nakamura K. Characterization of a second protein (CM2) encoded by RNA segment 6 of influenza C virus. J Virol. 1997;71:2786–2792. doi: 10.1128/jvi.71.4.2786-2792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hongo S, Sugawara K, Muraki Y, Matsuzaki Y, Takashita E, Kitame F, Nakamura K. Influenza C virus CM2 protein is produced from a 374-amino-acid protein (P42) by signal peptidase cleavage. J Virol. 1999;73:46–50. doi: 10.1128/jvi.73.1.46-50.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopp T P. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 16.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–88. [Google Scholar]
- 17.Lamb R A, Horvarth C M. Diversity of coding strategies in influenza viruses. Trends Genet. 1991;7:261–266. doi: 10.1016/0168-9525(91)90326-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leahy M, Dessens J, Nuttall P. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J Virol. 1997;71:8352–8356. doi: 10.1128/jvi.71.11.8352-8356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leahy M B, Nuttall P A, Weber F, Kochs G, Dessens J T. The fourth genus in the Orthomyxoviridae: sequence analysis of two Thogoto virus polymerase proteins and comparison to influenza viruses. Virus Res. 1997;50:215–224. doi: 10.1016/s0168-1702(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 20.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 21.Morse M A, Marriott A C, Nuttall P A. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein gp64. Virology. 1992;186:640–646. doi: 10.1016/0042-6822(92)90030-s. [DOI] [PubMed] [Google Scholar]
- 22.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–470. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsen T W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 24.Nuttall P A, Morse M A, Jones L D, Portela A. Orthoacariviruses. In: Gibbs A J, Calisher C H, editors. Molecular evolution of viruses. Cambridge, England: Cambridge University Press; 1995. pp. 416–425. [Google Scholar]
- 25.O'Neill R, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pekosz A, Lamb R A. The CM2 protein of influenza C virus is an oligomeric integral membrane glycoprotein structurally analogous to influenza A virus M2 and influenza B virus NB proteins. Virology. 1997;237:439–451. doi: 10.1006/viro.1997.8788. [DOI] [PubMed] [Google Scholar]
- 27.Pekosz A, Lamb R A. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc Natl Acad Sci USA. 1998;95:13233–13238. doi: 10.1073/pnas.95.22.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez D R, Donis R O. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology. 1998;249:52–61. doi: 10.1006/viro.1998.9318. [DOI] [PubMed] [Google Scholar]
- 29.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 30.Portela A, Jones L D, Nuttall P. Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. J Gen Virol. 1992;73:2823–2830. doi: 10.1099/0022-1317-73-11-2823. [DOI] [PubMed] [Google Scholar]
- 31.Siebler J, Haller O, Kochs G. Thogoto and Dhori virus replication is blocked by inhibitors of cellular polymerase II activity but does not cause shutoff of host cell protein synthesis. Arch Virol. 1996;141:1587–1594. doi: 10.1007/BF01718257. [DOI] [PubMed] [Google Scholar]
- 32.Sugrue R J, Hay A J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber F, Gruber S, Haller O, Kochs G. PB2 polymerase subunit of Thogoto virus (Orthomyxoviridae family) Arch Virol. 1999;144:1601–1609. doi: 10.1007/s007050050613. [DOI] [PubMed] [Google Scholar]
- 35.Weber F, Haller O, Kochs G. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggest a new panhandle structure. Arch Virol. 1997;142:1029–1033. doi: 10.1007/s007050050138. [DOI] [PubMed] [Google Scholar]
- 36.Weber F, Haller O, Kochs G. MxA GTPase blocks reporter gene expression of reconstituted Thogoto virus ribonucleoprotein complexes. J Virol. 2000;74:560–563. doi: 10.1128/jvi.74.1.560-563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber F, Haller O, Kochs G. Nucleoprotein vRNA and mRNA of Thogotovirus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J Virol. 1996;70:8361–8367. doi: 10.1128/jvi.70.12.8361-8367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber F, Jambrina E, Gonzalez S, Dessens H, Leahy M, Kochs G, Portela A, Nuttall P, Haller O, Ortin J, Zürcher T. In vivo reconstitution of active Thogoto virus polymerase: assays for the compatibility with other orthomyxovirus core proteins and template RNAs. Virus Res. 1998;58:13–20. doi: 10.1016/s0168-1702(98)00096-3. [DOI] [PubMed] [Google Scholar]
- 39.Weber F, Kochs G, Gruber S, Haller O. A classical bipartite nuclear localization signal on the Thogoto and influenza A virus nucleoproteins. Virology. 1998;250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 40.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittaker G, Bui M, Helenius A. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 1996;6:67–71. doi: 10.1016/0962-8924(96)81017-8. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita M, Krystal M, Palese P. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J Virol. 1988;62:3348–3355. doi: 10.1128/jvi.62.9.3348-3355.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye Z, Liu T, Offringa D P, McInnis J, Levandowski R A. Association of influenza virus matrix protein with ribonucleoproteins. J Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng H, Palese P, Garcia-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]