Abstract
Biomechanical forces, and their molecular transducers, including key mechanosensitive transcription factor genes, such as KLF2, are required for cardiac valve morphogenesis. However, klf2 mutants fail to completely recapitulate the valveless phenotype observed under no-flow conditions. Here, we identify the transcription factor EGR3 as a conserved biomechanical force transducer critical for cardiac valve formation. We first show that egr3 null zebrafish display a complete and highly penetrant loss of valve leaflets, leading to severe blood regurgitation. Using tissue-specific loss- and gain-of-function tools, we find that during cardiac valve formation, Egr3 functions cell-autonomously in endothelial cells, and identify one of its effectors, the nuclear receptor Nr4a2b. We further find that mechanical forces up-regulate egr3/EGR3 expression in the developing zebrafish heart and in porcine valvular endothelial cells, as well as during human aortic valve remodeling. Altogether, these findings reveal that EGR3 is necessary to transduce the biomechanical cues required for zebrafish cardiac valve morphogenesis, and potentially for pathological aortic valve remodeling in humans.
EGR3, a mechanosensitive transcription factor gene in zebrafish and mammals, is necessary for cardiac valve morphogenesis.
INTRODUCTION
Tissue morphogenesis is intrinsically linked to the mechanical forces that are required for the cellular and molecular processes defining cell fate (1–4). In this context, endothelial cells, which line the blood vessels throughout the body, are specialized according to the function and mechanical properties of the tissues they reside in as well as blood flow patterns (5–8). During heart development, the specialized endothelial cells lining the cardiac wall, known as endocardial cells (EdCs), invade the adjacent extracellular matrix (ECM) in the atrioventricular (AV) canal and outflow tract (OFT) in response to oscillatory shear stress (9–11). These migrating EdCs acquire mesenchymal characteristics, differentiate into valve interstitial cells (VICs), and proliferate to give rise to the valve leaflets (11–21), which are essential for cardiac function as they guarantee unidirectional blood flow.
Extensive work in vertebrates has shown that perturbation of mechanical forces in the heart severely affects valve formation (10, 22–25). In particular, the zebrafish model allows for the observation and manipulation of intracardiac mechanical forces while imaging valve morphogenesis at single-cell resolution. For example, stopping cardiac contraction, and consequently blood flow, results in a valveless phenotype (26). Molecularly, the expression of several genes has been shown to be flow sensitive (11, 27), and the classical endothelial flow-responsive transcription factor Klf2 has been described as the main mechanosensitive transcription factor required for cardiac valve formation (9, 10, 12, 16, 18, 28–31). However, zebrafish klf2a/b double mutants only display a partially penetrant valveless phenotype (12, 29, 32), indicating that other transcription factors play critical roles in transducing the mechanical signals that promote valvulogenesis.
Here, we show that the transcription factor Early growth response 3 (Egr3) is a critical transducer of mechanical signaling during cardiac valve morphogenesis in zebrafish. We first show that loss of egr3 causes a complete and highly penetrant lack of cardiac valves. Data from loss- and gain-of-function experiments indicate that mechanical forces activate the expression of egr3 in a subset of EdCs, thereby inducing their migration toward the adjacent ECM. Using transcriptomic analyses followed by functional investigation, we also show that the nuclear receptor Nr4a2b is a target and effector of Egr3 during cardiac valve formation. Furthermore, we show that mechanical forces up-regulate EGR3 expression in porcine valvular endothelial cells (VECs) and that EGR3 and its target/effector NR4A2 may be involved in human cardiac valve remodeling upon ectopic biomechanical overload.
RESULTS
Egr3 is a critical regulator of cardiac valve formation
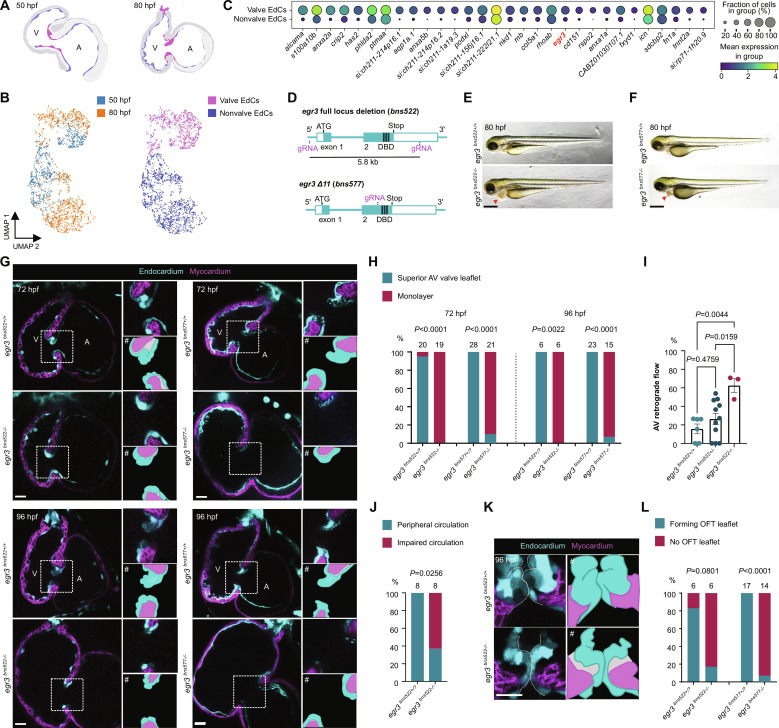
Seeking to identify additional transcription factors required for cardiac valve formation in zebrafish, we first determined and explored the transcriptional landscape of EdCs at the time when key morphogenetic events underlying valve development take place. Thus, we isolated wild-type zebrafish hearts at 50 hours postfertilization (hpf) when valve identity has just been established, and at 80 hpf when forming valves are first observed (Fig. 1A) (12–14, 33). We carried out single-cell RNA sequencing (scRNA-seq) of whole hearts and obtained 1310 and 2194 EdCs at 50 and 80 hpf, respectively (Fig. 1B). The valve population was clearly identified at both time points by the expression of valve endocardium specific genes such as has2 (34) and alcama (33) (fig. S1A). Differential expression analysis between valve and nonvalve EdCs revealed early growth response 3 (egr3) in the top 30 most enriched genes in the valve cluster and the only one encoding a DNA binding protein with transcription factor activity (Fig. 1C). Notably, egr3 expression appears more enriched in the valve EdCs when compared with other established valve transcription factor genes (16, 18, 23, 35), including klf2a/b (29) and nfatc1 (14) (fig. S1, B and C). We further analyzed egr3 expression by in situ hybridization and observed a similarly restricted mRNA localization in the AV canal (fig. S1D). Two other Egr family members have been implicated in cardiac valve development. In mouse, loss of Egr2 (Krox20) results in aortic valve defects and regurgitation (36, 37), while in zebrafish and humans, egr1/EGR1 has been shown to be expressed in cardiac valves (38, 39) and this expression requires mechanical forces (38, 40). We observed egr1 expression mostly at 80 hpf and in most EdCs (fig. S1, B and C), and neither egr2a/b nor egr4 was detectable in our dataset (fig. S1, B and C). Although the expression pattern and potential role of Egr3 in heart development have not been investigated yet, analyses of human and mouse heart scRNA-seq datasets (41, 42) reveal that EGR3/Egr3 is expressed in cardiac valve endocardium and AV mesenchyme (fig. S2, A and B). Sequence alignment shows that the Egr1 to Egr4 DNA binding domains are highly conserved (fig. S2, C and D), indicating the potential existence of common targets.
Fig. 1. Egr3 is required for cardiac valve formation.
(A) Schematic of hearts isolated for scRNA-seq; valve EdCs (magenta), nonvalve EdCs (blue). (B) scRNA-seq of EdCs from 50 and 80 hpf dissected zebrafish hearts. (C) Dot plot of 30 most differentially expressed genes in valve compared with nonvalve EdCs. (D) Schematic of generation of egr3 full locus deletion (bns522) and Δ11 (bns577) alleles. (E and F) Brightfield images of egr3+/+ and egr3−/− sibling larvae at 80 hpf; red arrowheads point to pericardial edema. (G) Confocal images of representative hearts from 72 and 96 hpf egr3+/+ and egr3−/− sibling larvae. (H) Percentage of egr3+/+ and egr3−/− sibling larvae with a superior valve leaflet and without a superior valve leaflet (i.e., endocardial monolayer) at 72 and 96 hpf; seven and four independent experiments, respectively. (I) AV retrograde blood flow shown as a fraction of three cardiac cycles; n = 6, 11, and 3; one experiment; mean ± SEM, one-way ANOVA followed by Tukey’s post hoc test. (J) Peripheral circulation in the caudal vein plexus at 80 hpf, one experiment. (K) Confocal images of representative OFT valves from 96 hpf egr3+/+ and egr3−/− sibling larvae. (L) Percentage of egr3+/+ and egr3−/− sibling larvae with and without forming OFT leaflets at 96 hpf; four independent experiments. (G and K) EdCs are marked by Tg(kdrl:eGFP) expression (cyan), and myocardial cells by Tg(myl7:BFP-CAAX) expression (magenta). # indicates illustrative vectorized cartoons of the valves. (H), (J), and (L) Fisher’s exact test. AV, atrioventricular; V, ventricle; A, atrium; OFT, OFT; DBD, DNA binding domain; EdC, EdC. Scale bars, 400 μm [(E) and (F)] and 20 μm [(G) and (K)].
Given the strong and early enrichment of egr3 in the valve endocardium, we investigated its role in valvulogenesis by generating two mutant alleles using the CRISPR-Cas9 system, a 5.8-kb full locus deletion (bns522) (i.e., a null allele), and an 11–base pair (bp) deletion (Δ11) (bns577), resulting in a premature termination codon that is predicted to disrupt the DNA binding domain (Fig. 1D and fig. S1, E and F). During early development, the valveless embryonic heart achieves unidirectional blood flow in part because of localized contraction waves and a simple vascular network (43) whose capacitance provides pressure storage and diminishes cardiac efforts (44). By 72 hpf, the AV EdCs have folded into a functional prevalvular structure capable of reducing retrograde blood flow and therefore supporting cardiac output (10, 14, 15). egr3 mutants, of both alleles, are indistinguishable from their wild-type siblings until 72 hpf when they start to exhibit noticeable pericardial edema (Fig. 1, E and F). In contrast to their wild-type siblings, egr3 mutants fail to form AV valve leaflets as their EdCs remain as a monolayer in a fully penetrant fashion in the null allele (19/19) and a nearly fully penetrant fashion in the Δ11 allele (19/21) (Fig. 1, G and H, and fig. S1, G and H). This complete lack of AV valves results in severe retrograde blood flow (Fig. 1I and movies S1 and S2) and leads to heart failure and impaired peripheral circulation (Fig. 1J and movies S3 and S4); egr3 mutants no longer display cardiac output by 96 hpf (movies S1 to S4). Despite this impaired circulation, cardiac contraction and heart rate are not obviously affected in egr3 mutants (movies S1 and S2). In 96 hpf wild-type larvae, the AV valves are more mature and elongated and the forming OFT valve leaflets are also present (14, 16, 45). At this stage, the AV EdCs in egr3 mutants still remain as a monolayer (Fig. 1, G and H, and fig. S1, G and H) and, similarly, the forming OFT valves also fail to develop (Fig. 1, K and L, and fig. S1I). Together, these data show that Egr3 is essential for cardiac valve formation in zebrafish.
Endothelial cell–specific deletion of egr3 results in a lack of AV valves
To more precisely assess egr3 expression in the developing heart, we generated a Gal4 knock-in reporter line [Pt(egr3:Gal4-VP16)], hereafter referred to as egr3:Gal4 (bns576), by targeting exon 2 using the CRISPR-Cas9 system, thereby disrupting the zinc-finger DNA binding domain (Fig. 2A). Consequently, the egr3:Gal4 line has endogenous egr3 reporter activity and is also an egr3 mutant allele (fig. S3B). egr3 reporter+ embryos display a strong expression in the olfactory bulb as well as in specific structures of the central and peripheral nervous systems (fig. S3A). egr3 reporter expression is also evident in the presumptive AV canal starting at 34 hpf (fig. S3A), mostly in the AV endocardium, but also in the AV myocardium (Fig. 2B). Moreover, egr3 reporter expression is observed in the OFT endocardium starting at 72 hpf (Fig. 2B), coinciding with the onset of OFT valve morphogenesis (16). Notably, most egr3 reporter–expressing EdCs were observed in, or near, the AV canal or OFT.
Fig. 2. Endothelial-specific deletion of egr3 recapitulates the global mutant phenotype.
(A) Schematic of the knock-in reporter [Pt(egr3:Gal4-VP16)] generated by inserting Gal4-VP16 in exon 2 of egr3. (B) Pt(egr3:Gal4-VP16); Tg(UAS:eGFP) expression (gray) at 72 hpf; EdCs are marked by Tg(kdrl:Hsa.HRAS-mCherry) expression (cyan) and myocardial cells by Tg(myl7:BFP-CAAX) expression (magenta); yellow arrowheads point to AV canal egr3 reporter+ cardiomyocytes. (B′) Maximum projection of Pt(egr3:Gal4-VP16); Tg(UAS:eGFP) expression with image segmentation of egr3 reporter+ AV canal EdCs (cyan), egr3 reporter+ AV canal myocardial cells (magenta), and egr3 reporter+ OFT cells (yellow). (C) Schematic of egr3 floxed allele Pt(egr3:loxP-egr3-loxP). (D) Confocal images of representative hearts from 72 hpf egr3+/+, endothelial cell–specific egr3−/−, and myocardial cell–specific egr3−/− larvae using the egr3 floxed allele line. (E and F) Percentage of 72 hpf egr3+/?, endothelial cell–specific egr3−/− (E), and myocardial cell–specific egr3−/− (F) larvae with a superior valve leaflet and without a superior valve leaflet (i.e., endocardial monolayer); one and two independent experiments, respectively; Fisher’s exact test. Scale bars, 20 μm.
Considering that both endocardial and myocardial cells in the AV canal express egr3 based on the egr3:Gal4 reporter expression, we sought to determine whether egr3 deletion in one of these cell types was sufficient to recapitulate the egr3 mutant phenotype. Thus, we used the CRISPR-Cas9 system to generate a sequential loxP site knock-in in the egr3 locus, specifically 440 bp upstream of the transcription start site and 1.9 kb downstream of the stop codon (Fig. 2C). Injection of Cre mRNA into embryos from egr3 flox/+ zebrafish crossed with egr3+/− zebrafish resulted in 72 hpf larvae displaying pericardial edema and no valve leaflets, correlating with the recombination of the egr3 floxed allele (fig. S3, D to F). No phenotype was observed in the uninjected larvae, indicating that the egr3 floxed allele is functional. When crossing egr3 flox/+ zebrafish with egr3+/− zebrafish carrying an endothelial Cre driver (kdrl:Cre), we observed that deleting egr3 in endothelial cells was sufficient to recapitulate the global egr3 mutant phenotype, thereby resulting in the complete lack of AV valve leaflets (fig. S3G and Fig. 2, D and E). Moreover, when crossing egr3 flox/+ zebrafish with egr3+/− zebrafish carrying a myocardial Cre driver (myl7:Cre), we observed that deleting egr3 in myocardial cells led to no noticeable differences with their control siblings including in their valve leaflets (fig. S3H and Fig. 2, D to F). Collectively, these data show that egr3 functions in the endothelium to drive AV valve formation.
egr3 directs AV canal EdC migration through its target Nr4a2b
To gain more insight into the cellular and molecular causes leading to the absence of the AV valve in egr3 mutants, we analyzed egr3+/? and egr3−/− sibling embryos at 48 hpf, a time when they cannot be visually distinguished from each other. At this stage, the AV canal is already morphologically and molecularly distinct from the atrial and ventricular chambers (33). Tissue convergence and a decrease in AV canal EdC size are important morphogenetic steps that precede AV valve formation (46, 47). Thus, to evaluate possible morphological differences in the endocardium leading to the egr3 mutant phenotype, we analyzed EdC volume in each cardiac region (fig. S4, A and B). Consistent with published data (47), AV canal EdCs had a smaller volume when compared with ventricular and atrial EdCs (fig. S4C). Notably, there were no differences in EdC cell number or volume in any cardiac region between egr3 mutants and their egr3+/? siblings (fig. S4, C and D) such that egr3 mutant EdCs were observed in expected numbers and underwent the cell shape changes typically observed during early AV valve morphogenesis.
To identify the molecular causes of the egr3 mutant phenotype, we performed bulk RNA-seq analysis of 48 hpf egr3+/+ and egr3−/− dissected hearts (Fig. 3A). One hundred fifty-four genes were differentially expressed between egr3 wild-type and mutant sibling embryos. Notably, important valve regulators such as klf2a/b, notch1b, dll4, nfatc1, or wnt9a/b did not appear to be differentially expressed in egr3 mutants (fig. S5A). Using in situ hybridization for egr1, has2, hey2, klf2a, klf4, piezo2a, and immunostaining for Alcama, we observed that the patterning of the AV canal was obviously not affected in egr3 mutants (fig. S5, B and C), altogether suggesting that lack of egr3 leads to the absence of the AV valve despite apparently unaffected expression of established cardiac valve markers and regulators. We further screened our dataset for differentially expressed genes with cardiac valve expression and found that nr4a2b, spp1, and nrg1 were significantly down-regulated in egr3 mutants (Fig. 3A). Nr4a2/Nurr1 is a nuclear receptor with transcription factor activity (48), and nr4a2b exhibits cardiac valve expression according to our scRNA-seq data (Fig. 3B), while spp1/osteopontin and nrg1 are expressed by valve EdCs (Fig. 3, B and C) (49, 50). Our observations that nr4a2b and spp1 are down-regulated in egr3 mutants agree with published reports in mammals showing that EGR3 is upstream of Nr4a2 (51) and that NR4A2 activates SPP1 expression by binding to its promoter (52). Moreover, EGR3 (53, 54), NR4A2 (55), and SPP1 (56, 57) promote endothelial cell migration downstream of vascular endothelial growth factor (VEGF), supporting the model that Egr3 is necessary for EdC migration during valvulogenesis.
Fig. 3. egr3 is necessary for EdC migration, which is required for cardiac valve formation.
(A) RNA-seq heatmap analysis of differentially expressed genes; fold change ± 1.5, Padj < 0.05, mean > 5. (B) scRNA-seq endocardial expression pattern of Egr3 targets nr4a2b and spp1. (C) In situ hybridization for Egr3 targets nr4a2b and spp1 in egr3+/+ and egr3−/− siblings at 48 hpf. (D) Confocal images of representative hearts from 48 hpf egr3+/? and egr3−/− sibling embryos displaying, respectively, AV EdC protrusions toward the ECM and no protrusions; EdCs are marked by Tg(kdrl:eGFP) expression (cyan). (E) Percentage of hearts from 48 hpf egr3+/? and egr3−/− sibling embryos displaying AV EdC protrusions toward the ECM; three independent experiments. (F) Confocal images of representative hearts from 55 hpf egr3+/− and egr3−/− sibling embryos displaying, respectively, collective AV EdC migration and no migration; EdCs are marked by Tg(kdrl:NLS-mCherry) expression (gray) and AV EdCs by Pt(egr3:Gal4-VP16);Tg(UAS:eGFP) expression (green). (G) Percentage of hearts from 55 hpf egr3+/− and egr3−/− sibling embryos displaying AV EdC migration; three independent experiments. (H) Number of migrating AV EdCs per heart in 55 hpf egr3+/− and egr3−/− sibling embryos; n = 18 and 17 embryos; three independent experiments. Student’s t test, mean ± SEM. (E) and (G) Fisher’s exact test. Scale bars, 20 μm.
To test whether deficient cellular migration could be the cause of the egr3 mutant phenotype, we evaluated EdC migration in the initial steps of AV valve morphogenesis. From 48 hpf onward, the AV canal EdCs start to extend protrusions into the adjacent ECM and, within a few hours, two to four AV canal EdCs collectively migrate in a ventricle to atrium direction (Fig. 3D) (13, 14). egr3 mutants exhibit a twofold reduction in the number of AV canal EdC protrusions toward the ECM (Fig. 3E), although this reduction was not significant. Strikingly, the subsequent collective EdC migration is severely affected, as only 6% (n = 1/17) of the mutant embryos engaged in EdC migration compared with 72% (n = 13/18) of their heterozygous siblings at 55 hpf (Fig. 3, F and G). Between two to four migrating EdCs were observed in these heterozygous hearts, while zero to two migrating EdC was generally observed in these egr3 mutant hearts (Fig. 3H).
To investigate whether Egr3 is sufficient to induce EdC migration into the ECM, we generated a stable egr3 overexpression line, Tg(5XUAS:egr3-p2a-dTomato), hereafter referred to as “egr3 OE.” To induce ectopic egr3 overexpression in the endothelium, we crossed egr3 OE zebrafish with the endothelial fli1a:Gal4 driver, Tg(fli1a:Gal4FF). egr3-overexpressing cells were clearly distinguished by dTomato fluorescence (Fig. 4A). In control hearts, VICs were present in the ECM of the AV canal, but not in the ECM of the ventricle or atrium (Fig. 4, B and C). However, in egr3 OE hearts, not only was there more than a 2.5-fold increase in VIC number in the ECM of the AV canal (Fig. 4, B and C), but dTomato+ EdCs were also present in the ventricular and atrial ECM, adjacent to the endocardial monolayer (Fig. 4, A to C). To better characterize the increased ECM invasion by EdCs in egr3 OE hearts, we assessed the expression of the endocardial valve marker Alcama (33). Notably, egr3 overexpression in the endothelium was sufficient to induce ectopic Alcama expression in ventricular and atrial EdCs (Fig. 4, D and E), as well as a significant increase of Alcama-expressing EdCs in the AV canal (Fig. 4E). Next, we tested whether overexpressing the Egr3 target, Nr4a2b, would be sufficient to rescue AV canal EdC migration in egr3 mutants. Thus, we generated a stable UAS-driven nr4a2b overexpression line, Tg(5XUAS:nr4a2b-p2a-dTomato), hereafter referred to as “nr4a2b OE,” and used it in the background of the egr3 bns577 mutant allele and egr3:Gal4+/−. The resulting transheterozygous progeny fail to produce a functional Egr3 protein and overexpress nr4a2b in egr3:Gal4+ cells. Transheterozygous larvae displayed the expected egr3 mutant phenotype at 72 hpf (Fig. 4F and fig. S3B). egr3 transheterozygous larvae overexpressing nr4a2b in their AV canal endocardium displayed a significant increase in the number of VICs in the AV canal ECM, indicating a partial rescue of EdC migration (Fig. 4, F and G). Not all VICs in nr4a2b OE hearts were dTomato+, indicating that Nr4a2b may also act in a cell nonautonomous fashion in this process. As previously reported for klf2a overexpression in the AV canal, using an nfatc:Gal4 line (15), we found that nr4a2b has an inhibitory effect on valve formation in egr3+/− larvae (fig. S5, D and E), suggesting that nr4a2b expression needs to be tightly regulated during valvulogenesis. Collectively, these data indicate that the absence of cardiac valves in egr3 mutants is a result of failed EdC migration into the adjacent ECM and that Egr3 is sufficient to induce endocardial Alcama expression and migration; they also suggest that Egr3 promotes AV canal EdC migration through its target Nr4a2b.
Fig. 4. egr3 triggers EdC migration and Alcama expression.
(A) 3D reconstruction of representative hearts from control [Tg(fli1a:Gal4);Tg(UAS:Kaede)] and endothelial-specific egr3-overexpressing [Tg(fli1a:Gal4);Tg(UAS:Kaede);Tg(UAS:egr3-p2a-dTomato)] 72 hpf larvae; magnified regions show single z planes of the atrium from control and endothelial-specific egr3-overexpressing 72 hpf larvae; yellow arrowheads point to Tg(UAS:egr3-p2a-dTomato)+ atrial EdCs in the adjacent ECM. (B) Confocal images of representative AV VIC quantification through the spot render function in Imaris; VICs (magenta spots). (C) Quantification of EdCs present in the ECM per cardiac region in 72 hpf endothelial-specific egr3-overexpressing larvae; n = 9 control and 22 egr3 OE larvae, n = 0 and 0.14 EdCs (ventricle), n = 5 and 13.36 EdCs (AV canal), n = 0 and 0.59 EdCs (atrium). (D) Confocal images of representative hearts from control [Tg(fli1a:Gal4);Tg(UAS:Kaede)] and endothelial-specific egr3-overexpressing [Tg(fli1a:Gal4);Tg(UAS:Kaede);Tg(UAS:egr3-p2a-dTomato)] 54 hpf embryos immunostained for Alcama; magnified regions show Alcama+ cells in the AV canal of the control heart and in the atrium of the Tg(UAS:egr3-p2a-dTomato) heart. (E) Quantification of Alcama+ EdCs per cardiac region of control and endothelial-specific egr3-overexpressing embryos at 54 hpf; n = 13 control and 14 egr3 OE embryos, n = 0.07 and 3.28 EdCs (ventricle), n = 31.77 and 42.86 EdCs (AV canal), n = 0.38 and 4.71 EdCs (Atrium); three independent experiments. (F) Confocal images of representative hearts from control Pt(egr3:Gal4-VP16) and egr3-driven nr4a2b-overexpressing [Pt(egr3:Gal4-VP16); Tg(UAS:nr4a2b-p2a-dTomato)] egr3−/− larvae at 72 hpf. (G) Quantification of EdCs present in the AV canal ECM of control and nr4a2b-overexpressing egr3−/− larvae at 72 hpf; n = 16 and 15 larvae; two independent experiments. (C), (E), and (G) Student’s t test, mean ± SEM. Scale bars, 20 μm.
egr3 is a mechanosensitive transcription factor gene in cardiac valve development and disease
Considering that EGR3 is involved in mediating VEGF-induced endothelial migration (53) and that it is a VEGF target via extracellular signal–regulated kinase (ERK) activation (58), we decided to test whether egr3 AV canal expression was dependent on VEGF signaling. We found that egr3 AV canal expression appeared to be unaltered after treatment with VEGF receptor inhibitors or a mitogen-activated protein kinase kinase (MEK) inhibitor (fig. S6A), suggesting that it is independent of VEGF/ERK signaling. Given the fundamental role of mechanical signaling in cardiac valve formation (10, 23, 26), we then decided to test whether egr3 expression was downstream of mechanical forces. We simulated a no-flow/no-contraction condition in vivo by taking advantage of the zebrafish embryo’s ability to withstand severe cardiac dysfunction (59). We used silent heart zebrafish mutants, which lack the sarcomeric protein Tnnt2a and, therefore, do not display cardiac contraction (59). Notably, egr3 AV canal expression was completely abrogated in tnnt2a mutant hearts (Fig. 5A), and we also observed the loss of egr3 expression when treating zebrafish embryos with the myosin inhibitor BDM (fig. S6A). To test whether egr3 expression responds to ectopic mechanical stimulation, we performed microsurgical insertion of a bead inside the embryonic heart through the inflow tract (22, 60) at 48 hpf (Fig. 5B). The bead remained inside the heart for 24 hours and moved rhythmically across the ventricular chamber following the heartbeats and without stopping the blood flow. Upon this ectopic mechanical stimulation, egr3 expression was expanded in the hearts of the bead-inserted larvae when compared with sham controls, as shown by the expression of the egr3:Gal4 reporter (Fig. 5, C and D). We did not find ectopic EdCs in the ECM or an increased number of VICs in the bead inserted hearts when compared with sham controls (fig. S6B). As mechanical forces are required for valve formation, we next tested whether overexpressing egr3 in endothelial cells in no-flow conditions could rescue some of the early steps of valve development. We injected tnnt2a morpholinos (MOs) into one-cell stage embryos of egr3 OE zebrafish crossed with the endothelial fli1a:Gal4 driver (Fig. 5E). Valvular cell identity is absent in no-flow condition hearts as the endocardium remains as a monolayer and the expression of valve markers is absent (Fig. 5E) (26, 61). Notably, some egr3-overexpressing endothelial cells in no-flow condition hearts expressed Alcama, displayed a partly cuboidal valvular cell shape, and were often found in the ECM (Fig. 5E). These Alcama+ egr3-overexpressing cells were present in the endocardium of both the ventricle and atrium as well as in the AV canal of no-flow condition hearts (Fig. 5F and fig. S6C), suggesting a lack of spatial specification of valve identity in no-flow hearts and/or the ability of Egr3 to drive this specification on its own.
Fig. 5. egr3 expression in the heart is regulated by biomechanical forces.
(A) In situ hybridization for egr3 expression in 48 hpf tnnt2a+/+ and tnnt2a−/− sibling embryos. (B) Illustration of bead insertion into a 48 hpf heart. (C) Pt(egr3:Gal4-VP16); Tg(UAS:eGFP) expression (gray) at 72 hpf in bead-inserted larva versus sham; myocardial cells are marked by Tg(myl7:BFP-CAAX) expression (magenta) and EdCs by Tg(kdrl:Hsa.HRAS-mCherry) expression (cyan). Red dotted circle highlights inserted bead; yellow arrowhead points to egr3+ cells near the bead. (D) Quantification of total volume of egr3 reporter+ cells in bead-inserted larvae and sham; n = 3 sham and 5 experimental larvae; one experiment. (E) Confocal images of representative hearts from control [Tg(fli1a:Gal4);Tg(UAS:Kaede)] and endothelial-specific egr3-overexpressing [Tg(fli1a:Gal4);Tg(UAS:Kaede);Tg(UAS:egr3-p2a-dTomato)] tnnt2a MO–injected embryos immunostained for Alcama at 54 hpf; magnified regions show the AV canal of tnnt2a MO–injected embryos with rescued Alcama expression in the Tg(UAS:egr3-p2a-dTomato) embryo. (F) Quantification of Alcama+ EdCs per cardiac region of tnnt2a MO–injected embryos at 54 hpf; n = 6 control and 6 egr3 OE embryos, n = 0.00 and 8.00 EdCs (ventricle), n = 0.00 and 13.00 EdCs (AV canal), n = 0.00 and 4.83 EdCs (Atrium); two independent experiments. (G) Oscillatory shear stress induces the expression of the mechanosensitive transcription factor gene egr3 in valve EdCs, where it orchestrates valvulogenesis by promoting Alcama expression and their migration via Nr4a2b. (H) Schematic of aortic valve collection from LVAD patients and control donors. (I) Relative mRNA levels of EGR3, KLF2, NR4A2, and SPP1 from aortic valves of LVAD patients and control donors; n = 4 and 4 to 6; one experiment. Ct values can be found in data S1. (D), (F), and (I) Student’s t test, mean ± SEM. (B) and (H) Illustrations were generated with Biorender.com. Scale bars, 20 μm.
To investigate whether EGR3 is activated in mammalian VECs in response to shear stress, we used a unique fluid activation device specifically designed to expose cells to physiologically relevant pulsatile wall shear stress (WSS) conditions (62). We observed an up-regulation of EGR3 expression in porcine VECs exposed to pulsatile WSS compared with static conditions (fig. S6D), indicating that EGR3 responds to pulsatile shear stress in VECs. In addition to playing an important role in valvulogenesis, mechanical forces are also pivotal during pathological valve remodeling. For instance, the use of a left ventricular assist device (LVAD) has become an established treatment option as well as a bridge for end-stage heart failure patients awaiting a heart transplant (63). Despite improving survival and heart function, LVAD use is correlated with aortic insufficiency, and 15 to 52% of patients may present worsening of previous aortic valve pathology or develop de novo aortic valve insufficiency (64). The LVAD increases cardiac output by diverting the blood from the left ventricle straight into the aorta, thereby decreasing the strain in the mitral valve and causing biomechanical overload in the aortic root and valves (65). This increased biomechanical overload in the aortic valves induces valve remodeling (i.e., aortic valve fusion) (66, 67). However, how the aortic valves transduce the biomechanical overload into a molecular response remains elusive. As zebrafish egr3 expression responds to lack of, as well as increase in, mechanical forces during valve development (Fig. 5, A to D) and EGR3 responds to pulsatile WSS in VECs (fig. S6D), we sought to investigate whether EGR3 expression was altered during human aortic valve remodeling following LVAD placement. We collected samples of aortic valves from control donors and LVAD patients and performed quantitative reverse transcription polymerase chain reaction (RT-qPCR) (Fig. 5H). EGR3 expression was increased by almost sixfold in LVAD patient valves compared with donor control valves (Fig. 5I). Expression of the classical mechanosensitive transcription factor gene KLF2 was increased by nearly fourfold (Fig. 5I). We also tested EGR3 targets and observed that in LVAD patient valves, NR4A2 expression was increased by more than 13-fold and SPP1 expression by more than 20-fold, although the latter was not significant (Fig. 5I).
DISCUSSION
Here, we identify the transcription factor Egr3 as a master transducer of the mechanical forces that guide valvulogenesis in zebrafish, and show that Egr3 is required for EdC migration into the adjacent ECM by activating the expression of the migration regulator nr4a2b (55). Klf2 has long been considered to be the main mechanosensitive transcription factor gene in cardiac valve formation. However, the broad endocardial expression of klf2a/b compared with the restricted expression of egr3 in valve EdCs suggests that Egr3 has a more specific role in transducing mechanical forces during valvulogenesis. klf2 mutations in zebrafish result in a 47 to 56% penetrance of the cardiac valveless phenotype observed in no-flow conditions (29, 32). Moreover, klf2 mutants also display cardiomyocyte extrusion (32) and it is not clear how this phenotype affects cardiac valve formation. In contrast, the egr3 mutant phenotype is highly penetrant and it is specific to the valves, with no obvious defects in other cardiac regions.
EGR3 is expressed in AV canal EdCs and fibroblasts in mammalian hearts (41, 68), and it responds to pulsatile WSS in VECs, consistent with a conserved function in valvulogenesis. Egr3 mutant mice display an increased frequency of perinatal mortality (69), and it would be worth investigating them for any underlying cardiac defects. In humans, EGR3 single-nucleotide variants resulting in loss-of-function alleles are highly underrepresented with a loss-of-function observed/expected upper bound fraction of 0.396 (gnomAD v4.0.0), possibly suggesting that EGR3 is haploinsufficient. Haploinsufficiency is also typically observed in genes associated with bicuspic aortic valve disease, such as GATA6 (70) and NOTCH (71, 72). Of note, case reports have shown that chromosomal duplications of the region where EGR3 is located (8p21.3) are associated with congenital heart disease (i.e., valve and septal defects) (73, 74), while a case-control study has reported an association between an EGR3 locus polymorphism and coronary artery disease (CAD) (75), though with no mention of possible valve defects.
In parallel to egr3 up-regulation following mechanical stimulation in the developing zebrafish heart, EGR3 is up-regulated in porcine VECs and in LVAD aortic valves upon biomechanical overload. In addition, EGR3 expression does not seem to respond to fluid shear stress in human umbilical vein endothelial cells (HUVECs) (76), suggesting a cell- and/or flow-specific response. In addition, EGR3 is necessary and sufficient to partially mediate the transforming growth factor–β (TGF-β) fibrotic response in fibroblasts (77). Thus, we speculate that the biomechanical stimuli that promote EGR3 expression during cardiac development also promote it in LVAD aortic valves, thereby contributing to pathological aortic valve remodeling and insufficiency (65–67). It will be important to investigate the role of EGR3, KLF2, and other mechanosensitive transcription factor genes in LVAD aortic valve pathogenesis.
Transduction of mechanical forces, which is mediated in part by VEGF signaling (78), also plays a role in lymphatic endothelial cell (LEC) development and physiology. VEGF is a known regulator of EGR3 and NR4A2 expression in endothelial cells (53, 58, 79). Human LECs up-regulate EGR3 and NR4A2 expression following VEGF stimulation (80, 81), and a recent report on the transcriptional coactivator Zmiz1 suggests that Egr3 is also involved in lymphatic valve formation (82).
In summary, our study reveals a previously unknown signaling axis in which EGR3 is necessary to transduce the mechanical signals required for cardiac valve formation and potentially also for LVAD valve remodeling. We anticipate that perturbations in the function or expression of EGR3, or its targets, also lead to cardiac valve defects in humans.
MATERIALS AND METHODS
Zebrafish handling and lines
All zebrafish husbandry was performed under standard conditions in accordance with institutional (Max Planck Gesellschaft) and national (German) ethical and animal welfare regulations. All procedures performed on animals conform to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes and were approved by the Animal Protection Committee (Tierschutzkommission) of the Regierungspräsidium Darmstadt (reference: B2/1218). The following mutant, knock-in reporter, and transgenic lines were generated for this study: egr3bns522, egr3bns577, Pt(egr3:Gal4-VP16)bns576 abbreviated as egr3:Gal4, Pt(egr3:loxP-egr3-loxP)bns661, Tg(5xUAS:egr3-p2a-dTomato)bns607 abbreviated as egr3:OE, and Tg(5xUAS:nr4a2b-p2a-dTomato)bns719 abbreviated as nr4a2b:OE. The following transgenic and mutant lines were used in this study: tnnt2amn0031Gt or Gt(GBT-R14) (83), Tg(kdrl:EGFP)s843 (33), Tg(kdrl:Hsa.HRAS-mCherry)s896 (84), Tg(kdrl:NLS-mCherry)is4 (85), Tg(fli1a:Gal4FF)ubs4 (86), Tg(myl7:BFP-CAAX)bns193 (87), Tg(myl7:mCherry-CAAX)bns7 (88), Tg(myl7:EGFP)twu26 (89), Tg(5xUAS:EGFP)nkuasgfp1a (90), Tg(UAS:Kaede)rk8 (91), Tg(myl7:Cre)sd55 (92), Tg(kdrl:Cre)s898 (93), and Tg(Mmu.Hhex-E1B:GFP)bns321 (14).
Generation of zebrafish lines
For CRISPR-Cas9 mutagenesis, in vitro synthesis of Cas9 mRNA and design of guide RNAs (gRNAs) were performed as previously described (94, 95). gRNA efficiency was evaluated in single AB wild-type injected embryos by high-resolution melting analysis (HRMA) or T7 endonuclease assay. Generation of egr3 full locus deletion (bns522) was achieved by using one gRNA in the immediate 5′ intergenic region and another in the 3′ UTR, and F1 adults were screened by PCR followed by sequencing of PCR products. The egr3 Δ11 allele (bns577) was generated with commercially available egr3 gRNA [Integrated DNA Technologies (IDT)], and F1 adults were screened by HRMA followed by sequencing of PCR products. The egr3:Gal4 reporter line (bns576) was generated according to the GeneWeld method (96) with 48-bp homology arms flanking the p2a-Gal4-VP16-beta-actin sequence, and using the egr3 gRNA (IDT) as for the egr3 Δ11 allele (bns577). We used the previously published sequential loxP knock-in method (97) to generate the egr3 floxed allele (bns661) with single-stranded donor oligonucleotides containing loxp sites flanked by asymmetric 21- and 49-bp homology arms. For the 5′ loxP knock-in, we used the same 5′ gRNA as for the full locus deletion allele (bns522). Adult F1 zebrafish were screened for the 5′ loxP integration using a loxP-specific PCR primer and sequenced after performing a loxP-flanking PCR. The 5′ loxP founder zebrafish was then crossed with AB wild type, and the resulting one-cell stage embryos were injected with a 3′ gRNA designed to cut the neighboring intergenic region. Adult F1 egr3 floxed allele zebrafish were screened by loxP-specific and flanking PCRs followed by sequencing of PCR products. Despite observing a complete and adequate integration of the 5′ loxP site, we have not been able to sequence the full extent of the 3′ loxP site. Genetic polymorphisms and/or integration of multiple copies after mutagenesis of the 3′ region in the zebrafish used for 3′ loxP knock-in might explain why we could not sequence the loxP integration. However, the floxed egr3 allele was confirmed by the loxP-specific and flanking PCRs, as well as by functional analysis after Cre mRNA injection, in which a recombined band of 400 bp was identified by PCR and the egr3 phenotype was observed at the expected Mendelian ratios. Conditional knockouts were obtained by crossing egr3flox/+ zebrafish with egr3bns577/+ zebrafish carrying Tg(kdrl:Cre) or Tg(myl7:Cre), for endothelial or myocardial cell–specific knockout, respectively. The low number of myocardial cell–specific knockout larvae (3 of 46) (Fig. 2, D and F) is under the expected ratio of 1:8 for a cross of egr3bns577/+, egr3flox/+, and myl7:Cre+/− zebrafish and without selecting the progeny for a phenotype. We used tol2 transgenesis to generate the egr3 (bns607) and nr4a2b (bns719) overexpression lines, respectively egr3:OE and nr4a2b:OE. Adult egr3:OE and nr4a2b:OE F1 zebrafish were screened by crossing with Tg(fli1a:Gal4FF) zebrafish and using the dTomato+ expression in the progeny as a proxy. dTomato+ embryos were used to confirm transgenesis by PCR amplification and sequencing of the PCR products. Of note, the data presented in this work were generated using progeny from F1 to F3 animals (obtained from sequential outcrosses to different transgenic lines or nontransgenic AB zebrafish); while doing the revisions, we observed a reduction in the penetrance of the cardiac valve phenotype in the progeny of F4 animals. Genotyping PCRs were performed with KAPA2G Fast Ready Mix (Sigma-Aldrich 2GFRMKB) and HRMAs with Maxima SYBR Green/Fluorescein qPCR Master Mix (2X) (Thermo Fisher Scientific K0241). The sequences of all genotyping primers, CRISPR sites, and donor oligonucleotides are available in data S3.
Plasmid construction
The open reading frames (ORFs) for egr3, nr4a2b, spp1, nrg1, hey2, and has2 were amplified from cDNA generated from a pool of 78 hpf larvae, and cloned into a pCS2+ vector. To generate the egr3:Gal4 line, the pGTag-Gal4-VP16-beta-actin plasmid was a gift from J. Essner (Addgene # 117817) and homology arms were added by vector ligation (98). To generate the egr3:OE line (bns607) and nr4a2b:OE line (bns719), egr3 and nr4a2b were amplified from pCS2+ and cloned into a 5XUAS:p2a-dTomato vector. Vector ligation was performed by PCR amplification followed by in vivo cloning (99). All primers are listed in data S3.
Microscopy
Live imaging of stopped zebrafish hearts was performed by mounting N-Phenylthiourea (PTU)-treated embryos and larvae in 1% agarose containing 0.2% tricaine. We used the following confocal microscopes: LSM700 Axio Imager 2 and LSM880 Axio Examiner with a W Plan-Apochromat 40×/1.0 dipping lens for image acquisition. We used Fiji (ImageJ 2.1.0/1.53o) to adjust the brightness and contrast of representative images. For three-dimensional (3D) rendering and analysis of confocal images, we used Imaris x64 (Bitplane, 10.0.1). We used the Imaris cell identification function to automatically segment EdCs based on their nucleus, marked by Tg(kdrl:NLS-mCherry) expression, and their cytoplasm, marked by Tg(kdrl:EGFP) expression. The EdCs were further segregated into ventricular, AV, and atrial with the help of the x/y position object filtering followed by manual curation. Illustrative cartoons were created using Inkscape vector graphics editor unless otherwise indicated in the figure legends.
In situ hybridization and immunostaining
Whole-mount RNA in situ hybridization was performed as previously described (100). In situ probes for egr3, spp1, nrg1, hey2, and has2 were synthesized from previously cloned ORFs in pCS2+ vectors using PCR products with an incorporated T7 promoter. An in situ probe for egr1 was provided by J. Vermot in a pBSK vector. In situ probes for klf2a (15), klf4, nr4a2b, and piezo2a (15) were synthesized directly from a PCR product of 78 hpf wild-type AB cDNA, using primers with an incorporated T7 promoter. To visualize the cardiac outline, we used the primary mouse anti-MHC (myosin heavy chain) antibody [1:500, MF-20, Developmental Studies Hybridoma Bank (DSHB) AB_2147781], followed by the secondary goat anti-mouse immunoglobulin G (IgG) Alexa Fluor 488 antibody (1:500, Invitrogen A11029). To assess AV valve identity, we performed immunostaining for Alcama using mouse anti-Alcama antibody (1:50, ZN-8, DSHB AB_531904) followed by goat anti-mouse Alexa Fluor 647 antibody (1:500, Invitrogen A21236). In situ hybridizations were imaged using an SMZ25 stereomicroscope (Nikon) with a 2×/0.3 objective. The sequence of primers used to synthesize the in situ probes can be found in data S3.
AV blood flow fraction
Functional analysis of blood regurgitation was performed in 72 hpf zebrafish larvae mounted in 1% agarose without tricaine. The zebrafish hearts were imaged live using an inverted Cell Observer Spinning Disk microscope with a 25×/0.8 water-immersion objective at 240 frames per second (fps). Quantification of AV blood flow fraction was performed using a custom-made ImageJ script as previously described (15). For each imaged larva, AV retrograde blood flow is shown as a fraction of three averaged cardiac cycles.
Microsurgical insertion of a bead
To assess the influence of ectopic mechanical stimulation on egr3 expression in vivo, a bead was kept inside of the beating zebrafish heart for 24 hours (22, 60). For that manipulation, 48 hpf zebrafish embryos were mounted ventrally in 1% agarose and a microsurgical incision was performed in the yolk sac using thin forceps. A bead (Cube Biotech 32201) was inserted with the forceps into the yolk cavity and gently pushed into the inflow tract, eventually reaching the heart through suction. Embryos were released from the agarose and kept in egg water at 28.5°C for 24 hours, when egr3:Gal4; UAS:eGFP expression was assessed by confocal microscopy. Only larvae that had the bead inside of the ventricle and still displayed blood flow were considered for the analysis. Sham embryos underwent the same procedures of mounting and yolk piercing, but without bead insertion.
Chemical treatments
To evaluate the influence of cardiac contraction on egr3 expression, we treated wild-type embryos with 15 mM of the myosin inhibitor BDM (Sigma-Aldrich B0753). Possible regulation of AV egr3 expression by VEGF signaling was tested by treating embryos with 1 and 2.5 μM SKLB1002 (Selleck Chemicals S7258) (101) and 0.5 μM SU5416 (Sigma-Aldrich S8442). Possible regulation of AV egr3 expression by ERK signaling was tested by treating embryos with 1 μM of the MEK inhibitor PD0325901 (Sigma-Aldrich PZ0162) (2). Concentrations were adjusted to diminish side effects. Dimethyl sulfoxide (DMSO; 0.1%) was administered as vehicle, and all treatments were performed from 36 to 48 hpf, when embryos were collected and fixed for in situ hybridization.
Porcine aortic valve endothelial cell isolation and culture
We used a fluid activation device that applies physiologically relevant pulsatile WSS on the surface of porcine aortic VECs as previously described (62). VECs were isolated from porcine aortic valve leaflets. Briefly, aortic valve leaflets were isolated and incubated in collagenase type II/Dulbecco’s modified Eagle medium (DMEM) (Life Technologies) for 3 × 7 min. After incubation, cells were gently scraped to isolate VECs from the two sides of the leaflets. After isolation, VECs were seeded and cultured on flask coated with rat collagen type I (50 μl/ml) (10 mg/ml concentration, BD Biosciences). VECs were cultured at 37°C and 5% CO2 in Endothelial Cell Growth Medium (Promocell) supplemented with manufacturer’s adjuvants and 5% fetal bovine serum (FBS). After 1 hour of culture, VECs were first removed from the collagen gels and lysed in TRIzol (Thermo Fisher Scientific 15596026). RNAs were extracted using RNeasy Mini Kit (Qiagen 74104).
Research ethics for donated aortic valve samples
The study population consisted of LVAD (n = 6) patients referred for heart transplantation at La Timone Hospital (102). Patients were not included if they had connective tissue disease and previous aortic valve dysfunction. The patients provided their written informed consent to participate in this study (approved by the Marseille ethics committee n°13.061). Aortic regurgitation (AR) was graded through an integrative approach (103, 104). Mechanism of AR was separated into prolapse of the fused leaflet, cusp restriction, or both. Aortic valves were recognized by their anatomical landmarks under the microscope, and leaflets were isolated with minimal aortic wall contamination. Immediately following surgical removal, a portion of the aortic valve leaflet was placed in RNAlater solution (Sigma-Aldrich) and stored at −80°C until processing. Control valve samples were obtained at autopsy of individuals without cardiac problems who suffered a traumatic death (n = 8). The control group was composed of individuals with an average age of 59 ± 4 years, and a male to female ratio (M/F) of 85%. The LVAD group was composed of individuals with an average age of 54 ± 12 years, and M/F ratio of 71%. This study was performed according to the principles of the Declaration of Helsinki and in accordance with institutional guidelines.
Quantitative RT-qPCR
Total RNA from human aortic valve samples was purified using TRIzol (Life Technologies) and RNeasy Mini Kit. RNA was reverse-transcribed using the AffinityScript Multiple Temperature cDNA synthesis kit (Agilent). Quantitative PCR was performed using SYBR Green (Roche)–based qPCR on a LightCycler 480 (Roche). Graphs show the mean ± SD for six to eight biological replicates. Samples were normalized to TBP or GAPDH as endogenous housekeeping genes. Relative expression levels were calculated by the comparative cycle threshold (ΔΔCt) method. All Ct and ΔCt values are listed in data S1. The sequence of primers used for RT-qPCR can be found in data S3.
scRNA-seq sample preparation and data analysis
To obtain a longitudinal transcriptional signature of single EdCs, we isolated hearts from Tg(myl7:EGFP) wild-type zebrafish at 50 and 80 hpf. Up to 150 hearts were isolated per condition as previously described (105). Briefly, zebrafish embryos and larvae were fragmented using a needle and syringe in an Eppendorf tube with 1 ml of DMEM (Thermo Fisher Scientific 88281) with 10% FBS (Sigma-Aldrich F2442). The homogenate was filtered through a 100-μm mesh filter. An additional wash with DMEM with 10% FBS was performed to remove the remaining hearts in the Eppendorf tube, followed by filtering. The isolated hearts were manually collected from the flow-through under a fluorescence microscope. The hearts were further dissociated using the Pierce Primary Cardiomyocyte Isolation Kit (Thermo Fisher Scientific 88281) and incubated for 25 min at 30°C (shaker 300 rpm) with pipetting every 5 to 10 min. Dead cells were removed from the final cell isolate by fluorescence-activated cell sorting in phosphate-buffered saline (PBS) without calcium or magnesium (Lonza, 17-516F) and with 0.04% bovine serum albumin (BSA). The cell suspensions were counted with a Moxi cell counter and diluted according to manufacturer’s protocol to obtain 10,000 single-cell data points per sample. Each sample was run separately on a lane in a Chromium controller with Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 (10x Genomics). scRNA-seq library preparation was done using a standard protocol, and sequencing was done on Nextseq2000. scRNA-seq analysis was performed as previously described (106), and final data visualization was done using a CellxGene package (doi:10.5281/zenodo.3235020). EdCs were identified by the expression of endothelial markers (e.g., cdh5, fli1a, kdrl, and tie1) and represented 3504 from the 6476 sequenced heart cells. Valve EdCs were identified by the expression of valve markers (e.g., alcama and has2) and represented 1343 cells. Data analysis was performed using only the EdCs, and the other cell types are not displayed for visualization purposes. The classification for transcription factor used was based on the Gene Ontology (GO) terms containing direct DNA binding domain, such as “DNA-binding transcription factor activity, RNA polymerase II-specific” and “DNA binding.” Thus, crip2 did not appear in our gene enrichment analysis as the encoded protein seems to lack a direct DNA binding domain (107).
RNA-seq sample preparation and data analysis
To further investigate the egr3 mutant phenotype, we conducted a bulk RNA-seq analysis of dissected zebrafish hearts at 48 hpf. As egr3 mutants cannot be distinguished from their siblings at this stage, embryos were live genotyped at 36 hpf as previously described (108). We dissected 20 hearts for each biological duplicate of egr3bns577−/− and egr3bns577+/+ sibling samples in cold DMEM with 10% FBS. Total RNA was isolated using the miRNeasy Micro Kit (Qiagen 217084), followed by on-column deoxyribonuclease (DNase) digestion (DNase-Free DNase Set, Qiagen 79254). Final elution was performed in 12 μl of ribonuclease (RNase)–free water, and subsequent RNA quality control, cDNA preparation, sequencing, and analyses were performed as previously described (101). A total of 154 genes were significantly dysregulated based on Padj < 0.05. Heatmaps were obtained using the Webbased Interactive Omics visualizatioN—Applications (WIlsON) (109).
Statistical analysis
GraphPad Prism (v.9) was used to perform all statistical analyses. We used two-tailed Student’s t test for comparing two samples (Figs. 3H, 4, C, E, and G, and 5, D, F, and I, and figs. S4D and S6D), and one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test for multiple comparisons (Fig. 1I and fig. S4C); data are presented as mean ± SEM, and actual P values are shown. Two-sided Fisher’s exact test was used to compare categorical data between two groups (Figs. 1, H, J, and L, 2, E and F, and 3, E and G, and figs. S3C and S5E), and data were represented as percentages after statistical analysis to improve visualization. To allow for statistical analysis, the classifications “Superior valve leaflet” and “Superior s-shape leaflet” were collapsed into “Superior valve leaflet,” and the “Monolayer / one VIC” and “Monolayer” were collapsed into “Monolayer” (Fig. 1H); all classifications and proportions are in figs. S1 (G to I) and S3I. All genetic experiments with zebrafish were performed independently at least twice, using different batches of embryos and on different days unless otherwise indicated. Exact sample sizes and P values are described in the figures and figure legends. P values <0.05 were considered significant.
Acknowledgments
We thank Y. Xu for discussion and comments on the manuscript. We also thank A. Atzberger, K. Khrievono, N. Fukuda, S. Howard, S. Perathoner, and H.-M. Maischein for their technical support and J. Vermot for providing the egr1 in situ probe. We thank S. Abdelilah-Seyfried and V. Lecaudey for their comments and suggestions as thesis advisory committee members. We also thank A. Gentile, C. Dooley, J. Martinez-Meyer, and P. Panza for discussions and important input during the development of this work and the CPI hub for Systems Biology and Medicine, run by M. Looso (EXC 2026, DFG), for providing computational resources, cloud infrastructure, and permanent web services.
Funding: This research was supported by funds from the Max Planck Society and awards from the European Research Council (ERC) under the European Union’s research and innovation programmes (AdG 694455-ZMOD and AdG 101021349-TAaGC) to D.Y.R.S. Work in S.Z.’s laboratory was supported by the “Fédération Française de Cardiologie” and the “Fondation Lefoulon-Delalande.”
Author contributions: Conceptualization: A.R.d.S., F.G., S.Z., T.J., and D.Y.R.S. Methodology: A.R.d.S., F.G., E.F., A.T., J.-F.A., R.R., S.G., M.L., and T.J. Investigation: A.R.d.S., F.G., G.L.M.B., E.F., A.T., J.-F.A., S.L., S.G.J., and T.J. Visualization: A.R.d.S., R.R., and T.J. Supervision: F.G., S.Z., T.J., and D.Y.R.S. Writing—original draft: A.R.d.S. Writing—review and editing: F.G., G.L.M.B., S.Z., T.J., and D.Y.R.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: scRNA-seq data of 50 and 80 hpf whole isolated hearts have been deposited in NCBI’s Gene Expression Omnibus under accession number GSE242483, and they can be interactively accessed at: https://bioinformatics.mpi-bn.mpg.de/ribeiro-da-silva-et-al-2024. Analysis of bulk RNA-seq data of 48 hpf dissected hearts is provided in data S2. Raw bulk RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under accession number GSE241935, and they can be interactively accessed at: https://bioinformatics.mpi-bn.mpg.de/dst179-ribeiro-da-silva-et-al-2024. Previously published data are available at: Human snRNA-seq data are available online at the Broad Single Cell Portal under study number SCP1852 (41). Mouse scRNA-seq data are available online at the Broad Single Cell Portal (42). Uniprot was used to align zebrafish, human, and mouse EGR1 to EGR4: P26632, P18146, P08046, E7F266, Q05159, P11161, P08152, Q6DG27, Q06889, P43300, A9JTG0, Q05215, and Q9WUF2. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for movies S1 to S4
Legends for data S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S4
Data S1 to S3
REFERENCES AND NOTES
- 1.Vignes H., Vagena-Pantoula C., Vermot J., Mechanical control of tissue shape: Cell-extrinsic and -intrinsic mechanisms join forces to regulate morphogenesis. Semin. Cell Dev. Biol. 130, 45–55 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Priya R., Allanki S., Gentile A., Mansingh S., Uribe V., Maischein H. M., Stainier D. Y. R., Tension heterogeneity directs form and fate to pattern the myocardial wall. Nature 588, 130–134 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Daems M., Peacock H. M., Jones E. A. V., Fluid flow as a driver of embryonic morphogenesis. Development 147, dev185579 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Sidhwani P., Yelon D., Fluid forces shape the embryonic heart: Insights from zebrafish. Curr. Top. Dev. Biol. 132, 395–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trimm E., Red-Horse K., Vascular endothelial cell development and diversity. Nat. Rev. Cardiol. 20, 197–210 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aitken C., Mehta V., Schwartz M. A., Tzima E., Mechanisms of endothelial flow sensing. Nat. Cardiovasc. Res. 2, 517–529 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Nakajima H., Chiba A., Fukumoto M., Morooka N., Mochizuki N., Zebrafish vascular development: General and tissue-specific regulation. J. Lipid Atheroscler. 10, 145–159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das R. N., Tevet Y., Safriel S., Han Y., Moshe N., Lambiase G., Bassi I., Nicenboim J., Bruckner M., Hirsch D., Eilam-Altstadter R., Herzog W., Avraham R., Poss K. D., Yaniv K., Generation of specialized blood vessels via lymphatic transdifferentiation. Nature 606, 570–575 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heckel E., Boselli F., Roth S., Krudewig A., Belting H. G., Charvin G., Vermot J., Oscillatory flow modulates mechanosensitive klf2a expression through trpv4 and trpp2 during heart valve development. Curr. Biol. 25, 1354–1361 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Vermot J., Forouhar A. S., Liebling M., Wu D., Plummer D., Gharib M., Fraser S. E., Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLOS Biol. 7, e1000246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Donnell A., Yutzey K. E., Mechanisms of heart valve development and disease. Development 147, dev183020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steed E., Faggianelli N., Roth S., Ramspacher C., Concordet J. P., Vermot J., klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat. Commun. 7, 11646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunawan F., Gentile A., Fukuda R., Tsedeke A. T., Jimenez-Amilburu V., Ramadass R., Iida A., Sehara-Fujisawa A., Stainier D. Y. R., Focal adhesions are essential to drive zebrafish heart valve morphogenesis. J. Cell Biol. 218, 1039–1054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunawan F., Gentile A., Gauvrit S., Stainier D. Y. R., Bensimon-Brito A., Nfatc1 promotes interstitial cell formation during cardiac valve development in Zebrafish. Circ. Res. 126, 968–984 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Juan T., Ribeiro da Silva A., Cardoso B., Lim S., Charteau V., Stainier D. Y. R., Multiple pkd and piezo gene family members are required for atrioventricular valve formation. Nat. Commun. 14, 214 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duchemin A. L., Vignes H., Vermot J., Mechanically activated piezo channels modulate outflow tract valve development through the Yap1 and Klf2-Notch signaling axis. eLife 8, e44706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff J., Endothelial-to-mesenchymal transition. Circ. Res. 124, 1163–1165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paolini A., Fontana F., Pham V. C., Rodel C. J., Abdelilah-Seyfried S., Mechanosensitive Notch-Dll4 and Klf2-Wnt9 signaling pathways intersect in guiding valvulogenesis in zebrafish. Cell Rep. 37, 109782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lincoln J., Garg V., Etiology of valvular heart disease-genetic and developmental origins. Circ. J. 78, 1801–1807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestel J., Ramadass R., Gauvrit S., Helker C., Herzog W., Stainier D. Y., Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development 143, 2217–2227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherz P. J., Huisken J., Sahai-Hernandez P., Stainier D. Y., High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development 135, 1179–1187 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Hove J. R., Koster R. W., Forouhar A. S., Acevedo-Bolton G., Fraser S. E., Gharib M., Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature 421, 172–177 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wang M., Lin B. Y., Sun S., Dai C., Long F., Butcher J. T., Shear and hydrostatic stress regulate fetal heart valve remodeling through YAP-mediated mechanotransduction. eLife 12, e83209 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang K. L., Parnall M., Loughna S., Effect of altered haemodynamics on the developing mitral valve in chick embryonic heart. J. Mol. Cell. Cardiol. 108, 114–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalogirou S., Malissovas N., Moro E., Argenton F., Stainier D. Y., Beis D., Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc. Res. 104, 49–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartman T., Walsh E. C., Wen K. K., McKane M., Ren J., Alexander J., Rubenstein P. A., Stainier D. Y., Early myocardial function affects endocardial cushion development in zebrafish. PLOS Biol. 2, E129 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haack T., Abdelilah-Seyfried S., The force within: Endocardial development, mechanotransduction and signalling during cardiac morphogenesis. Development 143, 373–386 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Goddard L. M., Duchemin A. L., Ramalingan H., Wu B., Chen M., Bamezai S., Yang J., Li L., Morley M. P., Wang T., Scherrer-Crosbie M., Frank D. B., Engleka K. A., Jameson S. C., Morrisey E. E., Carroll T. J., Zhou B., Vermot J., Kahn M. L., Hemodynamic forces sculpt developing heart valves through a KLF2-WNT9B paracrine signaling axis. Dev. Cell 43, 274–289.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontana F., Haack T., Reichenbach M., Knaus P., Puceat M., Abdelilah-Seyfried S., Antagonistic activities of Vegfr3/Flt4 and Notch1b fine-tune mechanosensitive signaling during zebrafish cardiac valvulogenesis. Cell Rep. 32, 107883 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wunnemann F., Ta-Shma A., Preuss C., Leclerc S., van Vliet P. P., Oneglia A., Thibeault M., Nordquist E., Lincoln J., Scharfenberg F., Becker-Pauly C., Hofmann P., Hoff K., Audain E., Kramer H. H., Makalowski W., Nir A., Gerety S. S., Hurles M., Comes J., Fournier A., Osinska H., Robins J., Puceat M., MIBAVA Leducq Consortium principal investigators, Andelfinger G., Dietz H. C., Mc Callion A. S., Andelfinger G., Loeys B. L., Van Laer L., Eriksson P., Mohamed S. A., Mertens L., Franco-Cereceda A., Mital S., Loss of ADAMTS19 causes progressive non-syndromic heart valve disease. Nat. Genet. 52, 40–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiplunkar A. R., Lung T. K., Alhashem Y., Koppenhaver B. A., Salloum F. N., Kukreja R. C., Haar J. L., Lloyd J. A., Krüppel-Like factor 2 is required for normal mouse cardiac development. PLOS ONE 8, e54891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasouli S. J., El-Brolosy M., Tsedeke A. T., Bensimon-Brito A., Ghanbari P., Maischein H. M., Kuenne C., Stainier D. Y., The flow responsive transcription factor Klf2 is required for myocardial wall integrity by modulating Fgf signaling. eLife 7, e38889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beis D., Bartman T., Jin S. W., Scott I. C., D’Amico L. A., Ober E. A., Verkade H., Frantsve J., Field H. A., Wehman A., Baier H., Tallafuss A., Bally-Cuif L., Chen J. N., Stainier D. Y., Jungblut B., Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 132, 4193–4204 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Smith K. A., Lagendijk A. K., Courtney A. D., Chen H., Paterson S., Hogan B. M., Wicking C., Bakkers J., Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development 138, 4193–4198 (2011). [DOI] [PubMed] [Google Scholar]
- 35.MacGrogan D., Munch J., de la Pompa J. L., Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 15, 685–704 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Odelin G., Faure E., Kober F., Maurel-Zaffran C., Theron A., Coulpier F., Guillet B., Bernard M., Avierinos J. F., Charnay P., Topilko P., Zaffran S., Loss of Krox20 results in aortic valve regurgitation and impaired transcriptional activation of fibrillar collagen genes. Cardiovasc. Res. 104, 443–455 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Odelin G., Faure E., Coulpier F., Di Bonito M., Bajolle F., Studer M., Avierinos J. F., Charnay P., Topilko P., Zaffran S., Krox20 defines a subpopulation of cardiac neural crest cells contributing to arterial valves and bicuspid aortic valve. Development 145, dev151944 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Banjo T., Grajcarek J., Yoshino D., Osada H., Miyasaka K. Y., Kida Y. S., Ueki Y., Nagayama K., Kawakami K., Matsumoto T., Sato M., Ogura T., Haemodynamically dependent valvulogenesis of zebrafish heart is mediated by flow-dependent expression of miR-21. Nat. Commun. 4, 1978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghazvini-Boroujerdi M., Clark J., Narula N., Palmatory E., Connolly J. M., DeFelice S., Xu J., Jian B., Hazelwood S., Levy R. J., Transcription factor Egr-1 in calcific aortic valve disease. J. Heart Valve Dis. 13, 894–903 (2004). [PubMed] [Google Scholar]
- 40.Gaste A., Bertrand E., Avierinos J.-F., Deplano V., Ait-Oufella H., Zaffran S., Wall shear stress activation of EGR1 and KLF2 transcription in valvular cells is mediated by the ERK1/2 mitogen-activated protein kinase pathway. Arch. Cardiovasc. Dis. Suppl. 15, 220 (2023). [Google Scholar]
- 41.Hill M. C., Kadow Z. A., Long H., Morikawa Y., Martin T. J., Birks E. J., Campbell K. S., Nerbonne J., Lavine K., Wadhwa L., Wang J., Turaga D., Adachi I., Martin J. F., Integrated multi-omic characterization of congenital heart disease. Nature 608, 181–191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotto J., Cullum R., Drissler S., Arostegui M., Garside V. C., Fuglerud B. M., Clement-Ranney M., Thakur A., Underhill T. M., Hoodless P. A., Cell diversity and plasticity during atrioventricular heart valve EMTs. Nat. Commun. 14, 5567 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forouhar A. S., Liebling M., Hickerson A., Nasiraei-Moghaddam A., Tsai H. J., Hove J. R., Fraser S. E., Dickinson M. E., Gharib M., The embryonic vertebrate heart tube is a dynamic suction pump. Science 312, 751–753 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Anton H., Harlepp S., Ramspacher C., Wu D., Monduc F., Bhat S., Liebling M., Paoletti C., Charvin G., Freund J. B., Vermot J., Pulse propagation by a capacitive mechanism drives embryonic blood flow. Development 140, 4426–4434 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Faucherre A., Maati H. M. O., Nasr N., Pinard A., Theron A., Odelin G., Desvignes J. P., Salgado D., Collod-Beroud G., Avierinos J. F., Lebon G., Zaffran S., Jopling C., Piezo1 is required for outflow tract and aortic valve development. J. Mol. Cell. Cardiol. 143, 51–62 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Boselli F., Steed E., Freund J. B., Vermot J., Anisotropic shear stress patterns predict the orientation of convergent tissue movements in the embryonic heart. Development 144, 4322–4327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignes H., Vagena-Pantoula C., Prakash M., Fukui H., Norden C., Mochizuki N., Jug F., Vermot J., Extracellular mechanical forces drive endocardial cell volume decrease during zebrafish cardiac valve morphogenesis. Dev. Cell 57, 598–609.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Amoasii L., Sanchez-Ortiz E., Fujikawa T., Elmquist J. K., Bassel-Duby R., Olson E. N., NURR1 activation in skeletal muscle controls systemic energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 116, 11299–11308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peal D. S., Burns C. G., Macrae C. A., Milan D., Chondroitin sulfate expression is required for cardiac atrioventricular canal formation. Dev. Dyn. 238, 3103–3110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai D., Liu X., Forrai A., Wolstein O., Michalicek J., Ahmed I., Garratt A. N., Birchmeier C., Zhou M., Hartley L., Robb L., Feneley M. P., Fatkin D., Harvey R. P., Neuregulin 1 sustains the gene regulatory network in both trabecular and nontrabecular myocardium. Circ. Res. 107, 715–727 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Marballi K. K., Alganem K., Brunwasser S. J., Barkatullah A., Meyers K. T., Campbell J. M., Ozols A. B., McCullumsmith R. E., Gallitano A. L., Identification of activity-induced Egr3-dependent genes reveals genes associated with DNA damage response and schizophrenia. Transl. Psychiatry 12, 320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lammi J., Huppunen J., Aarnisalo P., Regulation of the osteopontin gene by the orphan nuclear receptor NURR1 in osteoblasts. Mol. Endocrinol. 18, 1546–1557 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Liu D., Evans I., Britton G., Zachary I., The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene 27, 2989–2998 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Suehiro J., Hamakubo T., Kodama T., Aird W. C., Minami T., Vascular endothelial growth factor activation of endothelial cells is mediated by early growth response-3. Blood 115, 2520–2532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao D., Desai S., Zeng H., VEGF stimulates PKD-mediated CREB-dependent orphan nuclear receptor Nurr1 expression: Role in VEGF-induced angiogenesis. Int. J. Cancer 128, 2602–2612 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Liaw L., Lindner V., Schwartz S. M., Chambers A. F., Giachelli C. M., Osteopontin and beta 3 integrin are coordinately expressed in regenerating endothelium in vivo and stimulate Arg-Gly-Asp-dependent endothelial migration in vitro. Circ. Res. 77, 665–672 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Senger D. R., Ledbetter S. R., Claffey K. P., Papadopoulos-Sergiou A., Peruzzi C. A., Detmar M., Stimulation of endothelial cell migration by vascular permeability factor/vascular endothelial growth factor through cooperative mechanisms involving the alphavbeta3 integrin, osteopontin, and thrombin. Am. J. Pathol. 149, 293–305 (1996). [PMC free article] [PubMed] [Google Scholar]
- 58.Liu D., Jia H., Holmes D. I., Stannard A., Zachary I., Vascular endothelial growth factor-regulated gene expression in endothelial cells: KDR-mediated induction of Egr3 and the related nuclear receptors Nur77, Nurr1, and Nor1. Arterioscler. Thromb. Vasc. Biol. 23, 2002–2007 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Sehnert A. J., Huq A., Weinstein B. M., Walker C., Fishman M., Stainier D. Y., Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 31, 106–110 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Fukui H., Chow R. W., Xie J., Foo Y. Y., Yap C. H., Minc N., Mochizuki N., Vermot J., Bioelectric signaling and the control of cardiac cell identity in response to mechanical forces. Science 374, 351–354 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Juan T., Bellec M., Cardoso B., Athea H., Fukuda N., Albu M., Gunther S., Looso M., Stainier D. Y. R., Control of cardiac contractions using Cre-lox and degron strategies in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 121, e2309842121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faure E., Bertrand E., Gaste A., Plaindoux E., Deplano V., Zaffran S., Side-dependent effect in the response of valve endothelial cells to bidirectional shear stress. Int. J. Cardiol. 323, 220–228 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Miller L., Birks E., Guglin M., Lamba H., Frazier O. H., Use of ventricular assist devices and heart transplantation for advanced heart failure. Circ. Res. 124, 1658–1678 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Truby L. K., Garan A. R., Givens R. C., Wayda B., Takeda K., Yuzefpolskaya M., Colombo P. C., Naka Y., Takayama H., Topkara V. K., Aortic insufficiency during contemporary left ventricular assist device support: Analysis of the INTERMACS registry. JACC Heart Fail. 6, 951–960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.John R., Mantz K., Eckman P., Rose A., May-Newman K., Aortic valve pathophysiology during left ventricular assist device support. J. Heart Lung Transplant. 29, 1321–1329 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Rose A. G., Park S. J., Bank A. J., Miller L. W., Partial aortic valve fusion induced by left ventricular assist device. Ann. Thorac. Surg. 70, 1270–1274 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Hata H., Fujita T., Ishibashi-Ueda H., Nakatani T., Kobayashi J., Pathological analysis of the aortic valve after long-term left ventricular assist device support. Eur. J. Cardiothorac. Surg. 46, 193–197 (2014). [DOI] [PubMed] [Google Scholar]
- 68.J. Lotto, R. Cullum, S. Drissler, M. Arostegui, V. Garside, B. Fuglerud, A. Thakur, T. Underhill, P. Hoodless, Single-cell transcriptomics reveals diversity during heart valve epithelial-to-mesenchymal transitions (2022). 10.21203/rs.3.rs-1647065/v1. [DOI]
- 69.Tourtellotte W. G., Milbrandt J., Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat. Genet. 20, 87–91 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Gharibeh L., Komati H., Bosse Y., Boodhwani M., Heydarpour M., Fortier M., Hassanzadeh R., Ngu J., Mathieu P., Body S., Nemer M., C., GATA6 regulates aortic valve remodeling, and its haploinsufficiency leads to right-left type bicuspid aortic valve. Circulation 138, 1025–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garg V., Muth A. N., Ransom J. F., Schluterman M. K., Barnes R., King I. N., Grossfeld P. D., Srivastava D., Mutations in NOTCH1 cause aortic valve disease. Nature 437, 270–274 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Luna-Zurita L., Flores-Garza B. G., Grivas D., Siguero-Alvarez M., de la Pompa J. L., Cooperative response to endocardial Notch reveals interaction with Hippo pathway. Circ. Res. 133, 1022–1039 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fan Y. S., Siu V. M., Molecular cytogenetic characterization of a derivative chromosome 8 with an inverted duplication of 8p21.3→p23.3 and a rearranged duplication of 8q24.13→qter. Am. J. Med. Genet. 102, 266–271 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Gug C., Stoicanescu D., Mozos I., Nussbaum L., Cevei M., Stambouli D., Pavel A. G., Doros G., De novo 8p21.3→ p23.3 duplication with t(4;8)(q35;p21.3) translocation associated with mental retardation, autism spectrum disorder, and congenital heart defects: Case report with literature review. Front. Pediatr. 8, 375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X., Ma Y. T., Xie X., Yang Y. N., Ma X., Zheng Y. Y., Pan S., Liu F., Chen B. D., Association of Egr3 genetic polymorphisms and coronary artery disease in the Uygur and Han of China. Lipids Health Dis. 13, 84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsaryk R., Yucel N., Leonard E. V., Diaz N., Bondareva O., Odenthal-Schnittler M., Arany Z., Vaquerizas J. M., Schnittler H., Siekmann A. F., Shear stress switches the association of endothelial enhancers from ETV/ETS to KLF transcription factor binding sites. Sci. Rep. 12, 4795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang F., Shangguan A. J., Kelly K., Wei J., Gruner K., Ye B., Wang W., Bhattacharyya S., Hinchcliff M. E., Tourtellotte W. G., Varga J., Early growth response 3 (Egr-3) is induced by transforming growth Factor-β and regulates fibrogenic responses. Am. J. Pathol. 183, 1197–1208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angeli V., Lim H. Y., Biomechanical control of lymphatic vessel physiology and functions. Cell. Mol. Immunol. 20, 1051–1062 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen J., Liang X., Zhang S., Wang S., Garcia S. P., Yan P., Yu H., Li Z., Liu L., Zhang F., Wei W., Le H., Zhang Y., Yuan G. C., Chen S., Chen Y., Sun K., Pu W. T., Zhang B., Two faces of bivalent domain regulate VEGFA responsiveness and angiogenesis. Cell Death Dis. 11, 75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dieterich L. C., Ducoli L., Shin J. W., Detmar M., Distinct transcriptional responses of lymphatic endothelial cells to VEGFR-3 and VEGFR-2 stimulation. Sci. Data 4, 170106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shin J. W., Huggenberger R., Detmar M., Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 112, 2318–2326 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.K. C. Rajan, N. R. Patel, A. Shenoy, J. P. Scallan, M. Y. Chiang, M. J. Galazo, S. M. Meadows, Zmiz1 is a novel regulator of lymphatic endothelial cell gene expression and function. bioRxiv 2023.07.22.550165 [Preprint] (2023). 10.1101/2023.07.22.550165. [DOI] [PMC free article] [PubMed]
- 83.Clark K. J., Balciunas D., Pogoda H. M., Ding Y., Westcot S. E., Bedell V. M., Greenwood T. M., Urban M. D., Skuster K. J., Petzold A. M., Ni J., Nielsen A. L., Patowary A., Scaria V., Sivasubbu S., Xu X., Hammerschmidt M., Ekker S. C., In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Methods 8, 506–512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi N. C., Shaw R. M., De Val S., Kang G., Jan L. Y., Black B. L., Stainier D. Y., Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734–739 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y., Kaiser M. S., Larson J. D., Nasevicius A., Clark K. J., Wadman S. A., Roberg-Perez S. E., Ekker S. C., Hackett P. B., McGrail M., Essner J. J., Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development 137, 3119–3128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herwig L., Blum Y., Krudewig A., Ellertsdottir E., Lenard A., Belting H. G., Affolter M., Distinct cellular mechanisms of blood vessel fusion in the zebrafish embryo. Curr. Biol. 21, 1942–1948 (2011). [DOI] [PubMed] [Google Scholar]
- 87.Guerra A., Germano R. F., Stone O., Arnaout R., Guenther S., Ahuja S., Uribe V., Vanhollebeke B., Stainier D. Y., Reischauer S., Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. eLife 7, e32833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uribe V., Ramadass R., Dogra D., Rasouli S. J., Gunawan F., Nakajima H., Chiba A., Reischauer S., Mochizuki N., Stainier D. Y. R., In vivo analysis of cardiomyocyte proliferation during trabeculation. Development 145, dev164194 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Ho Y. L., Lin Y. H., Tsai I. J., Hsieh F. J., Tsai H. J., In vivo assessment of cardiac morphology and function in heart-specific green fluorescent zebrafish. J. Formos. Med. Assoc. 106, 181–186 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K., Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 105, 1255–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hatta K., Tsujii H., Omura T., Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 1, 960–967 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Han P., Bloomekatz J., Ren J., Zhang R., Grinstein J. D., Zhao L., Burns C. G., Burns C. E., Anderson R. M., Chi N. C., Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature 534, 700–704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertrand J. Y., Chi N. C., Santoso B., Teng S., Stainier D. Y., Traver D., Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jao L. E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B., Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982–4987 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Welker J. M., Wierson W. A., Almeida M. P., Mann C. M., Torrie M. E., Ming Z., Ekker S. C., Clark K. J., Dobbs D. L., Essner J. J., McGrail M., GeneWeld: Efficient targeted integration directed by short homology in Zebrafish. Bio Protoc. 11, e4100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burg L., Palmer N., Kikhi K., Miroshnik E. S., Rueckert H., Gaddy E., MacPherson Cunningham C., Mattonet K., Lai S. L., Marin-Juez R., Waring R. B., Stainier D. Y. R., Balciunas D., Conditional mutagenesis by oligonucleotide-mediated integration of loxP sites in zebrafish. PLOS Genet. 14, e1007754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wierson W. A., Welker J. M., Almeida M. P., Mann C. M., Webster D. A., Torrie M. E., Weiss T. J., Kambakam S., Vollbrecht M. K., Lan M., McKeighan K. C., Levey J., Ming Z., Wehmeier A., Mikelson C. S., Haltom J. A., Kwan K. M., Chien C. B., Balciunas D., Ekker S. C., Clark K. J., Webber B. R., Moriarity B. S., Solin S. L., Carlson D. F., Dobbs D. L., McGrail M., Essner J., Efficient targeted integration directed by short homology in zebrafish and mammalian cells. eLife 9, e53968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watson J. F., Garcia-Nafria J., In vivo DNA assembly using common laboratory bacteria: A re-emerging tool to simplify molecular cloning. J. Biol. Chem. 294, 15271–15281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Boezio G. L. M., Zhao S., Gollin J., Priya R., Mansingh S., Guenther S., Fukuda N., Gunawan F., Stainier D. Y. R., The developing epicardium regulates cardiac chamber morphogenesis by promoting cardiomyocyte growth. Dis. Model. Mech. 16, dmm049571 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Theron A., Touil A., Resseguier N., Collod-Beroud G., Norscini G., Simoni A. S., Odelin G., Habib G., Collart F., Zaffran S., Avierinos J. F., Clinical insights into a tertiary care center cohort of patients with bicuspid aortic valve. Int. J. Cardiovasc. Imaging 38, 51–59 (2022). [DOI] [PubMed] [Google Scholar]
- 103.Baumgartner H., Falk V., Bax J. J., De Bonis M., Hamm C., Holm P. J., Iung B., Lancellotti P., Lansac E., Rodriguez Munoz D., Rosenhek R., Sjogren J., Tornos Mas P., Vahanian A., Walther T., Wendler O., Windecker S., Zamorano J. L., ESC Scientific Document Group , 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur Heart J 38, 2739–2791 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Nishimura R. A., Otto C. M., Bonow R. O., Carabello B. A., Erwin J. P. III, Fleisher L. A., Jneid H., Mack M. J., McLeod C. J., O’Gara P. T., Rigolin V. H., Sundt T. M. III, Thompson A., 2017 AHA/ACC focused update of the 2014 AHA/ACC guideline for the management of patients with valvular heart disease: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 70, 252–289 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Burns C. G., MacRae C. A., Purification of hearts from zebrafish embryos. Biotechniques 40, 278–282 (2006). [PubMed] [Google Scholar]
- 106.Mattonet K., Riemslagh F. W., Guenther S., Prummel K. D., Kesavan G., Hans S., Ebersberger I., Brand M., Burger A., Reischauer S., Mosimann C., Stainier D. Y. R., Endothelial versus pronephron fate decision is modulated by the transcription factors Cloche/Npas4l, Tal1, and Lmo2. Sci. Adv. 8, eabn2082 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheung A. K., Ko J. M., Lung H. L., Chan K. W., Stanbridge E. J., Zabarovsky E., Tokino T., Kashima L., Suzuki T., Kwong D. L., Chua D., Tsao S. W., Lung M. L., Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 8390–8395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang X., Zhang Z., Zhao Q., Lou X., Rapid and efficient live zebrafish embryo genotyping. Zebrafish 17, 56–58 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Schultheis H., Kuenne C., Preussner J., Wiegandt R., Fust A., Bentsen M., Looso M., WIlsON: Web-based interactive omics visualization. Bioinformatics 35, 1055–1057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for movies S1 to S4
Legends for data S1 to S3
Movies S1 to S4
Data S1 to S3