Abstract
We explored the utility of brief Mycobacterium tuberculosis whole-genome sequencing (WGS) “snapshots” at a sentinel site within Lima, Peru, for evaluating local transmission dynamics over time. Within a 17-km2 area, 15 of 70 (21%) isolates with WGS collected during 2011–2012 and 22 of 81 (27%) collected during 2020–2021 were clustered (P = .414), and additional isolates clustered with those from outside the area. Isolates from the later period were disproportionately related to large historic clusters in Lima from the earlier period. WGS snapshots at a sentinel site may not be useful for monitoring transmission, but monitoring the persistence of large transmission clusters might be.
Keywords: disease transmission, cluster analysis, molecular epidemiology
We explore the potential utility and limitations of Mycobacterium tuberculosis whole-genome sequencing at a sentinel site in a high-incidence setting for assessing changes in transmission dynamics over time.
Despite curative treatment, declines in global tuberculosis (TB) incidence have been slow [1]. Many public health interventions in settings with high TB incidence focus on early detection and treatment to reduce transmission [2]. However, in these settings, transmission is rarely monitored or assessed directly. In high-income settings with low TB incidence, genotyping, and later whole-genome sequencing (WGS), of Mycobacterium tuberculosis (Mtb) isolates has been part of TB surveillance for decades, allowing detection of transmission networks based on the relatedness of the strains involved [3].
In low- and middle-income countries (LMICs), health systems often lack the resources for universal mycobacterial culture for people with TB, let alone WGS (which requires a cultured Mtb isolate). A few LMIC studies have performed WGS on comprehensive population-based samples, including two that collected samples for over a decade [4–6]. In a district of Malawi, the proportion of transmission-linked cases decreased over time as TB incidence decreased [4], whereas in 2 counties of China, the proportion of clustered cases increased over time, which was attributed to more complete case finding [5]. Thus, it is possible to detect changes in evidence of transmission over time in high-incidence settings with longitudinal comprehensive WGS studies, but most LMICs lack resources to do so.
Given the comparative feasibility of performing WGS for limited samples of TB cases, we sought to explore the utility of brief WGS “snapshots” within a sentinel site to assess changes in transmission over time in a high-incidence setting. While such snapshots might underestimate the actual proportion of cases attributable to recent transmission due to a short sampling window, the proportion of clustered cases within the sample might still be a useful indicator for comparing snapshots over time. In this study, we assessed the utility of comparing WGS results for 2 sets of isolates collected by 2 studies implemented almost 10 years apart, both of which attempted to comprehensively sample all TB cases within a concentrated geographic area in Lima, Peru.
METHODS
Setting and Sample Collection
Lima, Peru is a megacity comprising 43 districts and a population of 11 million. In 2 studies implemented at different times, we attempted to obtain Mtb isolates from all people newly diagnosed with TB who lived in a 17-km2 area of the Carabayllo district (hereafter referred to as the study area) of Lima. The study area population was 178 000 in the 2017 census.
Samples and data were collected with written informed consent from adults and parental written informed consent with assent from minors <18 years old, in accordance to ethical standards of the Helsinki Declaration. The earlier study collected samples from individuals ≥16 years old with pulmonary TB. Samples were collected from the study area during June 2011–July 2012, and from 106 health centers in 10 of the 43 districts of Lima during 2009–2012 [7, 8]. The later study collected samples from individuals ≥8 years old with pulmonary TB in the study area during October 2020–June 2021. Of note, pandemic-related delays and logistical considerations resulted in a shorter sampling window than originally planned (9 rather than 12 months). For the earlier study, WGS was performed on all available isolates from the study area and a subset of the isolates from other regions. For the later study, WGS was performed on all available isolates. Sequencing details are provided in the Supplementary Methods.
Clustering Analyses
We conducted 3 different clustering analyses, each considering alternative cutoffs of ≤5 single-nucleotide polymorphisms' (SNPs) difference and ≤10 SNPs difference to define related Mtb isolates. Isolates that were related to at least 1 other isolate were considered to be clustered. Details of the analyses are shown in the Supplementary Methods. In brief, the first analysis used a Pearson χ2 test to compare the proportion of isolates during each period that was attributable to recent local transmission within the study area, assuming clustered isolates to reflect recent local transmission. The second analysis assessed the geographic scale of transmission to the study area. We identified all instances in which an isolate collected from the study area in the later period was related to any clustered case from the earlier period. We calculated the genomic distance between these 2 cases as well as the Euclidean geographic distance between where the patients resided. We used linear regression to assess whether there was an association between genomic distance and geographic distance, and conducted a sensitivity analysis adjusting for estimated local TB burden of the area where the patient from the earlier period resided [8]. The third analysis evaluated whether large transmission clusters present during the earlier period disproportionately contributed to the Mtb strains present in the study area during the later period. We categorized all of the isolates collected during 2009–2012 as being either unclustered, part of a small cluster (2–5 related isolates), or part of a large cluster (≥6 related isolates). We then identified whether each isolate from the later period was related to a large cluster, small cluster, or unclustered isolate from the earlier period and evaluated whether large clusters from the earlier period contributed disproportionately to cases in the later period. Statistical analyses were conducted in SAS version 9.4 (SAS Institute, Cary, North Carolina) and R version 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria) software.
RESULTS
During 2009–2012, a total of 4500 people with TB were enrolled from different regions of Lima, of whom 3851 (86%) had a positive mycobacterial culture. A total of 2748 participants with WGS isolates from this period were included in the analysis. Within the study area, 70 monoisolates with WGS represented 56% of the 124 individuals ≥16 years old who were treated for pulmonary TB during June 2011–July 2012, and 81 monoisolates with WGS represented 59% of the 138 individuals ≥8 years old who were treated for pulmonary TB during October 2020–June 2021 (Supplementary Table 1).
Recent Transmission Within the Study Area
Using either SNPs difference cutoff, the percentage of clustered cases was not significantly different between the 2 periods (Table 1). Using a cutoff of ≤10 SNPs, 27% of isolates were clustered in the later period compared to 21% in the earlier period (P = .414); using a cutoff of ≤5 SNPs, 24% of isolates were clustered in the later period compared to 14% in the earlier period (P = .110). Limiting isolates in the later period to people ≥16 years old yielded similar results. In both periods, related isolates within households represented ≤10% of all isolates, and household clusters represented 36%–60% of clustered isolates.
Table 1.
Clustering Among Isolates Collected Within the Study Area During 2 Time Periods
| Clustering Type (SNP Cutoff Used to Define Clustering) | 2011–2012 (n = 70) | 2020–2021 (n = 81) | 2020–2021, Age ≥16 Years (n = 79) | P Value for 2011–2012 vs 2020–2021 | P Value Limited to Age ≥16 Years in Both Periods |
|---|---|---|---|---|---|
| Clustered (≤10 SNPs) | 15 (21%) | 22 (27%) | 20 (25%) | .414 | .576 |
| Clustered (≤5 SNPs) | 10 (14%) | 20 (24%) | 18 (23%) | .110 | .185 |
| Household clustered (≤10 SNPs) | 6 (9%) | 8 (10%) | 6 (8%) | .783 | .827 |
| Household clustered (≤5 SNPs) | 6 (9%) | 8 (10%) | 6 (8%) | .783 | .827 |
Abbreviation: SNP, single-nucleotide polymorphism.
Geographic Distance and Relatedness of Strains Between Periods
Using a cutoff of ≤10 SNPs, 45 (56%) isolates from the study area in the later period were related to at least 1 isolate collected from throughout Lima during the earlier period. However, only 4 (5%) were related to an isolate collected from the study area during the earlier period. Using a cutoff of ≤5 SNPs, 23 (28%) isolates from the study area in the later period were related to at least 1 isolate collected from throughout Lima during the earlier period, and only 2 (2%) were related to an isolate collected from the study area during the earlier period. Genomic distance between isolates from the 2 periods was inversely associated with geographic distance when using a cutoff of ≤10 SNPs to define clusters (slope: −0.17 [95% confidence interval {CI}, −.31 to −.019]) and was not associated when using a cutoff of ≤5 SNPs (slope: 0.01 [95% CI, −.07 to .09]). Adjusting for local TB burden yielded similar results. Thus, we found no evidence that Mtb strains in the study area during the later period were more related to strains that had been circulating in nearby regions in the earlier period compared to strains from other parts of the city.
Contribution of Historic Large Transmission Clusters to Study Area Strains
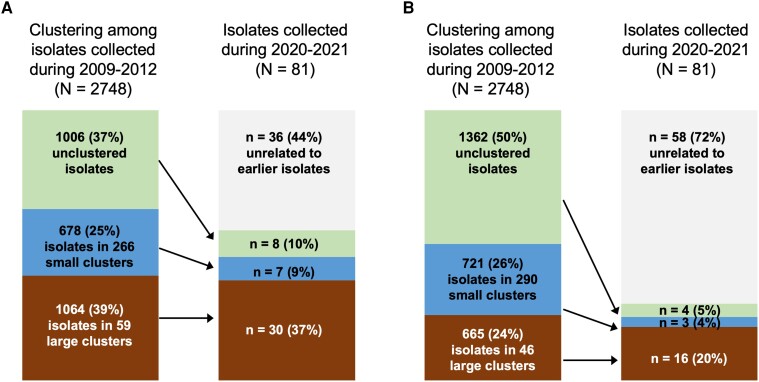
Using a cutoff of ≤10 SNPs, 39% of isolates during 2009–2012 were part of large clusters, 25% were part of small clusters, and 36% were unclustered (Figure 1A). Among the 2020–2021 isolates, 37% and 9% were related to large and small clusters from the earlier period, respectively, 10% were related to unclustered isolates from the earlier period, and 44% were not related to any isolates from the earlier period. Isolates from the later period had >3 times the odds of being related to a large cluster versus an unclustered isolate from the earlier period compared to what would be expected if all isolates from the earlier period had the same likelihood of being related to a later isolate (odds ratio [OR], 3.55 [95% CI, 1.62–7.77]). Isolates from the later period were not more likely to be related to a small cluster from the earlier period compared to what would be expected (OR, 1.30 [95% CI, .47–3.60]). The same trend of large but not small historic clusters being disproportionately related to later isolates was observed when a cutoff of ≤5 SNPs was used (Figure 1B; OR for being related to a large cluster, 8.20 [95% CI, 2.73–24.62]; OR for being related to a small cluster, 1.42 [95% CI, .32–6.35]).
Figure 1.
Clustering among 2009–2012 isolates and relatedness of 2020–2021 isolates to historic clusters. Cutoffs of ≤10 single-nucleotide polymorphisms (SNPs) difference (A) and ≤5 SNPs difference (B) were used to define related isolates. Small clusters were defined as comprising 2–5 related isolates, and large clusters as comprising ≥6 related isolates. Arrows indicate relatedness of isolates between the 2 time periods.
DISCUSSION
We found that WGS “snapshots,” which attempt to comprehensively sequence isolates over brief periods of time from a sentinel site within a megacity, may be insufficient to draw conclusions about the total contribution of recent transmission to current cases. Our geographic analysis suggests that persistent transmission clusters may stretch over long distances, so clustering of isolates within a small sentinel site may not be a meaningful indicator of transmission. However, we found evidence that large transmission networks from a decade ago continue to contribute to current cases, suggesting a potential alternative strategy of monitoring the persistence of these networks through periodic WGS surveys encompassing a broad geographic area with lower coverage.
While we found evidence of local recent transmission even within the geographically compact study area, our results confirm that transmission networks stretch across the city [8]. We believe that this phenomenon reflects the daily mobility of residents of the study area, given that many work in areas of the city far from where they live [9], a phenomenon that has been tied to transmission in other high-incidence settings as well [10]. Monitoring clustering within a limited geographic area may be of limited public health utility if the majority of cases in an area are due to transmission that occurred somewhere else.
The observation that recent cases are disproportionately related to large transmission clusters from a decade ago reflects the individual-level heterogeneity in Mtb transmission that is well documented but poorly understood. A recent modeling study estimated that 2%–31% of cases were responsible for 80% of transmission across diverse settings [11]. The persistence of large transmission clusters suggests a potential surveillance strategy of periodic WGS surveys to track the proportion of cases in these established transmission clusters among representative samples of isolates from throughout the city. As has been shown in Rwanda with drug-resistant TB, interventions that reduce transmission can result in a decreased percentage of cases of a dominant transmissible clone [12].
Our study was subject to several limitations. First, the coronavirus disease 2019 pandemic shortened the planned 2020–2021 isolate collection period, reduced detection of TB in Peru by 25% [1], and increased delays in diagnosis [13]. As a result, the proportion of clustered cases in the later time period is likely to be more of an underestimate than in the earlier time period, but we do not know by how much. Second, in the later time period we collected limited data about participants, preventing a rigorous assessment of epidemiologic linkages. Third, we used SNP thresholds for defining clusters that were consistent with previous literature [14] but not based on assessment of epidemiological linkages in this particular cohort.
In conclusion, in urban settings where people travel long distances for work, WGS “snapshots” of Mtb isolates from residents of a sentinel site are of limited value for monitoring the impact of TB programs on the general population. Periodic WGS surveys comprising sparser sampling of a broader area to monitor the proportion of cases in persistent transmission clusters could be useful.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Courtney M Yuen, Division of Global Health Equity, Brigham and Women's Hospital, Boston, Massachusetts, USA; Department of Global Health and Social Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Chuan-Chin Huang, Division of Global Health Equity, Brigham and Women's Hospital, Boston, Massachusetts, USA.
Ana Karina Millones, Socios En Salud Sucursal Peru, Lima, Peru.
Roger I Calderon, Socios En Salud Sucursal Peru, Lima, Peru.
Abigail L Manson, Infectious Disease and Microbiome Program, Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, Massachusetts, USA.
Judith Jimenez, Socios En Salud Sucursal Peru, Lima, Peru.
Carmen Contreras, Socios En Salud Sucursal Peru, Lima, Peru.
Ashlee M Earl, Infectious Disease and Microbiome Program, Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, Massachusetts, USA.
Mercedes C Becerra, Department of Global Health and Social Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Leonid Lecca, Department of Global Health and Social Medicine, Harvard Medical School, Boston, Massachusetts, USA; Socios En Salud Sucursal Peru, Lima, Peru.
Megan B Murray, Department of Global Health and Social Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Notes
Author contributions. C. M. Y. conceptualized the current study and wrote the first draft of the manuscript. M. C. B. and M. B. M. conceptualized and led the earlier study. A. K. M., J. J., C. C., and L. L. oversaw the implementation of sample collection in Peru. R. I. C., A. L. M., and A. M. E. oversaw mycobacterial culture and WGS procedures. C.-C. H., C. M. Y., and A. L. M. conducted the analyses.
Data availability. Sequencing data from the 2020–2021 study are available at the National Center for Biotechnology Information Sequence Read Archive under Bioproject PRJNA778463. Sequencing data from the 2009–2012 study will be available at the Harvard Dataverse repository (https://dataverse.harvard.edu).
Disclaimer. The funder had no role in the design, analysis, or writing of this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funder.
Financial support. This work was supported by the National Institutes of Health (grant numbers DP2MD015102 to C. M. Y.; U01AI057786 to M. C. B.; U19AI076217, U19AI109755, and U19AI111224 to M. B. M.; and U19AI110818 to A. M. E.).
References
- 1. World Health Organization . Global tuberculosis report 2022. Geneva, Switzerland: World Health Organization, 2022. [Google Scholar]
- 2. Yuen CM, Amanullah F, Dharmadhikari A, et al. Turning off the tap: stopping tuberculosis transmission through active case-finding and prompt effective treatment. Lancet 2015; 386:2334–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nikolayevskyy V, Niemann S, Anthony R, et al. Role and value of whole genome sequencing in studying tuberculosis transmission. Clin Microbiol Infect 2019; 25:1377–82. [DOI] [PubMed] [Google Scholar]
- 4. Guerra-Assuncao JA, Crampin AC, Houben RM, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife 2015; 4:e05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li M, Guo M, Peng Y, et al. High proportion of tuberculosis transmission among social contacts in rural China: a 12-year prospective population-based genomic epidemiological study. Emerg Microbes Infect 2022; 11:2102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saavedra Cervera B, Lopez MG, Chiner-Oms A, et al. Fine-grain population structure and transmission patterns of Mycobacterium tuberculosis in southern Mozambique, a high TB/HIV burden area. Microb Genom 2022; 8:mgen000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becerra MC, Huang CC, Lecca L, et al. Transmissibility and potential for disease progression of drug resistant Mycobacterium tuberculosis: prospective cohort study. BMJ 2019; 367:l5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang CC, Trevisi L, Becerra MC, et al. Spatial scale of tuberculosis transmission in Lima, Peru. Proc Natl Acad Sci U S A 2022; 119:e2207022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuen CM, Brooks MB, Millones AK, et al. Geospatial analysis of reported activity locations to identify sites for tuberculosis screening. Sci Rep 2022; 12:14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson KN, Shah NS, Mathema B, et al. Spatial patterns of extensively drug-resistant tuberculosis transmission in KwaZulu-Natal, South Africa. J Infect Dis 2018; 218:1964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith JP, Cohen T, Dowdy D, Shrestha S, Gandhi NR, Hill AN. Quantifying Mycobacterium tuberculosis transmission dynamics across global settings: a systematic analysis. Am J Epidemiol 2023; 192:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ngabonziza JCS, Rigouts L, Torrea G, et al. Multidrug-resistant tuberculosis control in Rwanda overcomes a successful clone that causes most disease over a quarter century. J Clin Tuberc Other Mycobact Dis 2022; 27:100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millones AK, Lecca L, Acosta D, et al. The impact of the COVID-19 pandemic on patients’ experiences obtaining a tuberculosis diagnosis in Peru: a mixed-methods study. BMC Infect Dis 2022; 22:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatherell HA, Colijn C, Stagg HR, Jackson C, Winter JR, Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med 2016; 14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.