Abstract
Background
Clinical outcomes in bacterial bloodstream infections (BSIs) are influenced by bacterial species, host immunity, and antibiotic therapy. The mechanisms by which such factors influence outcomes are poorly understood. We aimed to identify bacterial- and antibiotic-specific host transcriptional signatures in patients with bacterial BSI.
Methods
RNA sequencing was performed on blood samples from patients with BSI due to gram-negative (GN) versus gram-positive (GP) pathogens: Escherichia coli (n = 30) or Klebsiella pneumoniae (n = 28) versus methicillin-susceptible Staphylococcus aureus (MSSA) (n = 24) or methicillin-resistant S. aureus (MRSA) (n = 58). Patients were matched by age, sex, and race.
Results
No significant host transcriptome differences were detected in patients with E. coli versus K. pneumoniae BSI, so these were considered together as GN BSI. Relative to S. aureus BSI, patients with GN BSI had increased activation of the classic complement system. However, the most significant signal was a reduction in host transcriptional signatures involving mitochondrial energy transduction and oxidative burst in MRSA versus MSSA. This attenuated host transcriptional signature remained after controlling for antibiotic therapy.
Conclusions
Given the importance of immune cellular energetics and reactive oxygen species in eliminating hematogenous or intracellular MRSA, these findings may offer insights into its persistence relative to other bacterial BSIs.
Keywords: Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, transcriptome, bacteremia
RNA sequencing performed with blood samples from patients with gram-positive or gram-negative bacteremia demonstrated reduced host transcriptional signatures involving mitochondrial energy transduction in methicillin-resistant versus methicillin-susceptible Staphylococcus aureus. Patients with gram-negative infection had increased activation of the classic complement system.
Bacterial bloodstream infections (BSIs) are common and associated with high morbidity and mortality rates, healthcare costs, and antibiotic resistance [1–3]. One of the challenges in managing BSI is that patient clinical outcomes are dependent on a complex set of interconnected variables. For example, BSI outcomes have been associated with differences in clinical variables such as age, race, medical comorbid conditions, infecting bacterial species, choice of antibiotic, and timing of antibiotic therapy [3–6]. These clinical variables likely influence patient outcomes at least in part through changes in the host transcriptional response, though the links between clinical variables and the host transcriptional response are not well understood. In addition, biomarkers such as host or pathogen omics signatures that may explain or predict outcomes of BSIs have not been well established.
To better understand how clinical and bacterial variables influence patient response to infection, we aimed to identify the host transcriptional signatures associated with the infecting bacterial species and choice of antibiotic in patients with bacterial BSI. Previous approaches to address these questions have used animal models or cultured human cells. In these systems, infection with Staphylococcus aureus, for example, has been shown to influence transcription of genes involved in processes such as innate immunity and cytokine/chemokine signaling [7–10]. Antibiotic use has been shown to influence mitochondrial [11–13] and immune cell function [11, 14–16].
To further extend this work into humans, we enrolled a matched cohort of patients with methicillin-resistant S. aureus (MRSA), methicillin-susceptible S. aureus (MSSA), Escherichia coli, and Klebsiella pneumoniae BSIs. We captured detailed clinical information from these patients, as well as the infecting bacterial bloodstream isolate and patient whole-blood samples for RNA sequencing (RNA-Seq) analysis. Using these data, we identified host transcriptional signals specific to bacterial groups and antibacterial therapies. While RNA-Seq of whole-blood samples in infected patients has been used to address sepsis diagnostics [17–21], to our knowledge no prior studies have used such patient samples to define host transcriptional signals associated with BSIs from particular bacterial species or antibiotic therapies. Transcriptional profiling via RNA-Seq was used, as this technique allows for measurement of all expressed human genes in the blood with high accuracy and reproducibility and with a modest amount of biological material [22, 23].
Other omics approaches, such as unbiased proteomics or metabolomics. are promising but were not pursued because these techniques are less well established for the purposes intended in the current study [24, 25]. Identifying the ways that bacterial species and antibiotic therapies affect the patient transcriptional response will improve our understanding of both bacterial pathogenesis and the host feedback loop and may lead to improved diagnostics and antibacterial therapeutics.
METHODS
Patient Selection
Patients were enrolled into the Duke Bloodstream Infection Biorepository (BSIB), an ongoing cohort of prospectively enrolled adult inpatients with monomicrobial S. aureus or gram-negative (GN) bacterial BSI at Duke University Hospital or Duke Regional Hospital. Detailed clinical information, bloodstream bacterial isolate, and patient whole-blood samples are collected and stored through an institutional review board–approved protocol. A prior study identified a cohort of patients from the BSIB with MRSA BSI.
In the current study, we used the BSIB to match the patients with MRSA BSI to patients with MSSA, E. coli, or Klebsiella pneumoniae BSI. Those in each bacterial group (MSSA, E. coli, and K. pneumoniae) were independently matched to patients with MRSA BSI by age, sex, and race. In the current study, patients were enrolled between 2007 and 2014. Only patients with a full complement of clinical data and biospecimens were considered for matching. Of these patients, we ultimately matched 37% of patients with MRSA BSI (58 of 155), 15% of those with MSSA BSI (24 of 165), 4% of those with E. coli BSI (30 of 669), and 7% of those with K. pneumoniae BSI (28 of 379). For patients with MSSA BSI, we preferentially selected those treated with vancomycin to better match patients with MRSA BSI. However, given the low number of patients with MSSA treated with vancomycin (generally just those with severe β-lactam allergies), we also included some patients with MSSA BSI who were treated with a β-lactam antibiotic. For patients with E. coli, or K. pneumoniae BSI, we preferentially selected those treated with a β-lactam to provide consistency between the 2 groups.
Written informed consent was obtained from patients or their legal representatives. If a patient died before notification of blood culture results, the patient was enrolled using an institutional review board–approved notification of decedent research. In patients with multiple hospitalizations with over the study period, only the first such hospitalization was included. Study definitions are provided in the Supplementary Methods.
RNA-Seq
Patient whole-blood samples were captured within 48–72 hours after the initial positive blood culture during the index hospitalization. Total RNA was isolated with the Qiagen RNA Blood kit, and quality control of the RNA was performed with the Nanodrop 8000 and Agilent Bioanalyzer 2100. Globin RNA was removed using a Life Technologies GLOBINCLEAR (human) kit. Libraries for RNA-Seq were prepared with a KAPA Stranded mRNA-Seq Kit, and 50–base pair length single-end reads were generated on an Illumina HiSeq 3000 sequencing system. Quality-control of raw FASTQ files was performed using FastQC software (Version 0.11.9; Babraham Bioinformatics) [26]. Picard software (Version 3.1.1; Broad Institute) [27] was used to mark duplicate reads. Alignment of reads to a reference genome was performed using STAR software (Version 2.7.11a; Cold Spring Harbor Laboratory) [28] with the GRCh38 human reference genome. Samtools software (Version 1.18; Samtools) [29] was used to perform the sorting and indexing of aligned reads. Count table generation was performed using featureCounts software (Version 2.0.2; Subread) [30].
Statistical Analysis
Differential expression analysis was performed using the DESeq2 package in R software (Version 1.42.0) [31]. Gene transcript level changes were considered significant if the following 2 conditions were met: (1) the absolute value of the log2 fold-change between 2 groups was ≥2 and (2) the false discovery rate (FDR) was ≤0.10. Biological processes associated with sets of differential gene transcript levels were identified using g:Profiler software (Ensembl 109, Ensembl genomes 55) [32], which performs functional enrichment analysis (also known as overrepresentation analysis or gene set enrichment analysis) on input gene lists. It maps genes to known functional information sources and detects statistically significantly enriched terms. Genes with altered transcript levels (eg, GN vs S. aureus BSI) were input as ordered lists by magnitude of transcript level between the 2 groups of interest.
RESULTS
Patient Cohorts
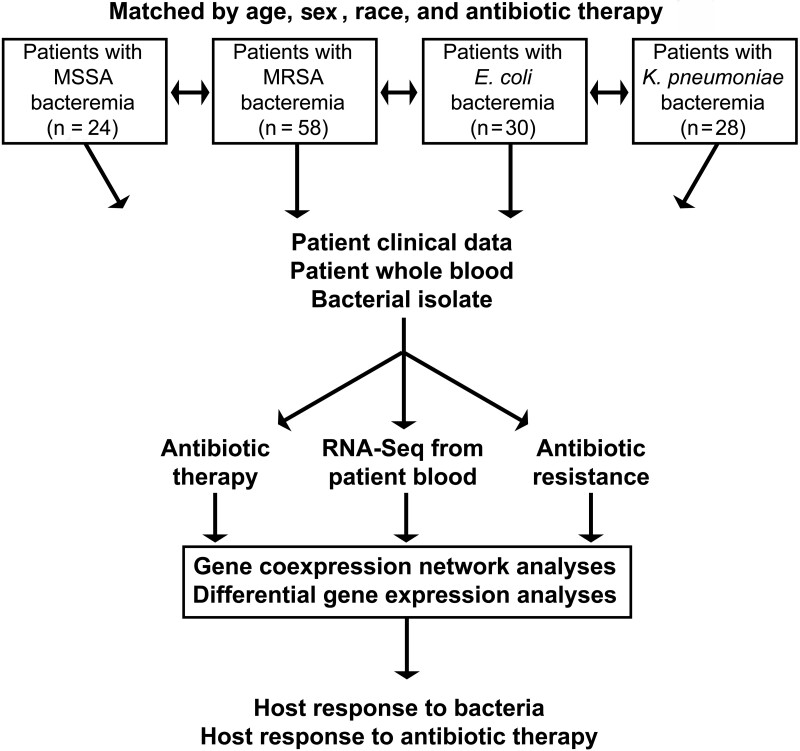
The overall schematic of the study design is shown in Figure 1. In total, 140 patients with bacterial BSI were enrolled in this study. This includes patients with MSSA (n = 24), MRSA (n = 58), E. coli (n = 30), and K. pneumoniae (n = 28) BSI (Table 1). Patients in the 4 groups were similar with respect to age, sex, and race as they were matched on these variables. Patients with gram-positive BSI (GP; ie, MSSA and MRSA) were more commonly hemodialysis dependent than those with GN BSI (ie, E. coli and K. pneumoniae) (24 of 82 [29%] vs 2 of 58 [3%], respectively; P < .001) and more commonly experienced complications of metastatic infection (46 of 82 [56%] vs 2 of 58 [3%]; P < .001). The higher rate of septic shock in patients with GP BSI (10 of 82 [12%]), relative to those with GN BSI (2 of 58 [3%]) did not reach statistical significance (P = .12).
Figure 1.
Overview of study design. Abbreviations: E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible S. aureus; RNA-Seq, RNA sequencing.
Table 1.
Demographics and Outcomes in Patients With Bacterial Bloodstream Infections Included in the Study
| Characteristic or and Outcome | Patients, No. (%)a | |||
|---|---|---|---|---|
| MSSA BSI (n = 24) | MRSA BSI (n = 58) | Escherichia coli BSI (n = 30) | Klebsiella pneumoniae BSI (n = 28) | |
| Age, mean (SD), y | 63.7 (11.8) | 63.0 (11.5) | 65.0 (11.5) | 63.2 (14.5) |
| Female sex | 5 (21) | 19 (33) | 10 (33) | 8 (29) |
| Race | ||||
| White | 14 (58) | 30 (52) | 18 (60) | 16 (57) |
| Black | 10 (42) | 27 (47) | 11 (37) | 11 (39) |
| Other/unknown | 0 | 1 (2) | 1 (3) | 1 (4) |
| Hemodialysis dependent | 3 (13) | 21 (36) | 1 (3) | 1 (4) |
| Diabetes mellitus | 12 (50) | 30 (52) | 12 (40) | 6 (21) |
| Transplant recipient | 4 (17) | 6 (10) | 4 (13) | 5 (18) |
| HIV positive | 0 | 1 (2) | 3 (10) | 0 |
| Recent corticosteroid use | 4 (17) | 15 (26) | 6 (20) | 7 (25) |
| Antibiotic used | ||||
| Vancomycin | 13 (54) | 58 (100) | 0 | 0 |
| β-Lactam | 11 (46) | 0 | 30 (100) | 26 (93) |
| Fluoroquinolone | 0 | 0 | 0 | 2 (7) |
| Length of hospital stay, mean (SD), d | 18.1 (15.6) | 17.6 (12.3) | 9.1 (5.4) | 13.6 (20.2) |
| APACHE II acute physiology score, mean (SD) | 9.9 (9.4) | 8.4 (5.9) | 6.1 (3.3) | 5.6 (3.4) |
| Hospital-acquired BSI | 6 (25) | 5 (9) | 4 (13) | 7 (25) |
| Source of infection | ||||
| Endovascular | 4 (17) | 19 (33) | 1 (3) | 5 (18) |
| Respiratory tract | 0 (0) | 2 (3) | 0 (0) | 0 (0) |
| Skin/soft tissue | 4 (17) | 16 (28) | 1 (3) | 1 (4) |
| Genitourinary tract | 0 (0) | 1 (2) | 16 (53) | 7 (25) |
| Intra-abdominal | 0 (0) | 2 (3) | 6 (20) | 5 (18) |
| Other | 3 (12) | 6 (10) | 5 (17) | 3 (11) |
| None/unknown | 13 (54) | 12 (21) | 1 (3) | 7 (25) |
| Duration of symptoms, median (IQR), db | 4 (2–5) | 3 (1–5) | 2 (1–5) | 2 (1–7) |
| Complications of BSI | ||||
| Septic shock | 3 (13) | 7 (12) | 1 (3) | 1 (4) |
| Acute kidney injury | 8 (33) | 15 (26) | 16 (53) | 9 (32) |
| ALI/ARDS | 1 (4) | 4 (7) | 0 | 1 (4) |
| Metastatic infection | 12 (50) | 34 (59) | 0 | 2 (7) |
| Death in hospital | 0 | 3 (5) | 0 | 0 |
Abbreviations: ALI, acute lung injury; APACHE, Acute Physiology and Chronic Health Evaluation; ARDS, acute respiratory distress syndrome; BSI, bloodstream infection; HIV, human immunodeficiency virus; IQR, interquartile range; MRSA and MSSA, methicillin-resistant and methicillin-susceptible Staphylococcus aureus, respectively; SD, standard deviation.
aData represent no. (%) of patients unless otherwise specified.
bDays from symptom onset to obtaining of initial blood culture.
Patients with S. aureus BSI had higher Acute Physiology and Chronic Health Evaluation (APACHE) II acute physiology scores than those patients with GN BSI (mean 8.9 vs 5.9; P = .001). The APACHE II acute physiology score did not differ significantly between patients with MRSA versus MSSA BSI (mean 8.4 vs 9.9; P = .49). In the GP cases relative to the GN cases, the source of BSI was more commonly skin/soft-tissue infection (20 of 82 [24%] vs 2 of 58 [3%], respectively; P = .001) or endovascular (23 of 82 [28%] vs 6 of 58 [10%]; P = .01) and less commonly genitourinary (1 of 82 [1%] vs 23 of 58 [40%]; P < .001). Patients with MSSA BSI were treated with either vancomycin (13 of 24 [54%]) or a β-lactam antibiotic (11 of 24 [46%]), while those with MRSA BSI were treated with vancomycin. Most patients with E. coli or K. pneumoniae BSI were treated with a β-lactam antibiotic (30 of 30 [100%] and 26 of 28 [93%], respectively). No statistically significant difference was detected in the duration of symptoms before initial blood culture in patients with GP versus GN BSI (P = .28).
Host Transcriptional Responses to GN Versus GP Bacteria
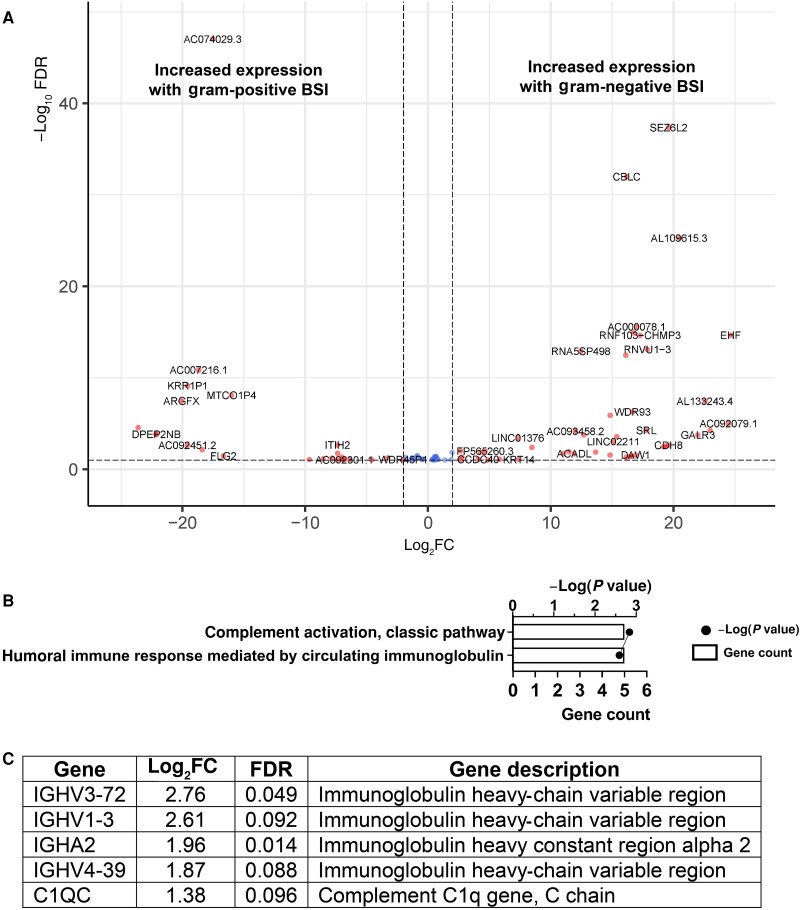
We began by investigating host transcriptional responses to prototypic BSI pathogens. Patients with E. coli BSI, relative to those with K. pneumoniae BSI, differed in the transcript level of a single gene—ADGRG7 (adhesion G–protein-coupled receptor G7)—with a log2 fold change of 21.8 (FDR P = 6.31 × 10−5). Given the similarity in transcript level profiles between patients with E. coli and those with K. pneumoniae BSI, the 2 groups were combined into a GN BSI group for all further analyses. When we compared patients with GN and those with S. aureus BSI, we identified 77 host genes with differential transcript levels. Of these, 48 were more highly present in patients with GN relative to S. aureus BSI, and 29 were more highly present in those with S. aureus BSI. The genes with altered transcript levels are listed in Supplementary Table 1, and a volcano plot of these genes is shown in Figure 2A. Biological process analysis of the genes with differential transcript levels revealed activation of the classic complement pathway (adjusted P = .001) and the related pathway of the humoral immune system mediated by circulating immunoglobulin (adjusted P = .003) (Figure 2B) in patients with GN BSI. The identification of these biological processes was due to increased gene transcript levels in patients with GN BSI of 5 genes involved in immune function—the C1q protein subunit C and 4 immunoglobulin heavy-chain genes (Figure 2C).
Figure 2.
Whole-blood gene transcript level changes in patients with gram-negative bloodstream infection (BSI) relative to gram-positive (ie, Staphylococcus aureus) BSI. A, Volcano plot showing gene transcript level differences in patients with gram-negative (GN) versus S. aureus BSI. Genes that had a log2-transformed fold change of >1 or less than −1 and a false discovery rate (FDR) of <0.10 are shown in red and genes meeting only the FDR criteria are shown in blue. B, g:Profiler biological processes associated with the gene transcript differences in patients with GN versus S. aureus BSI. For each biological process, the number of genes associated with the process (gene count) and the P value associated with the process are shown. C, Genes associated with the processes shown in B.
Host Transcriptional Responses to MSSA Versus MRSA
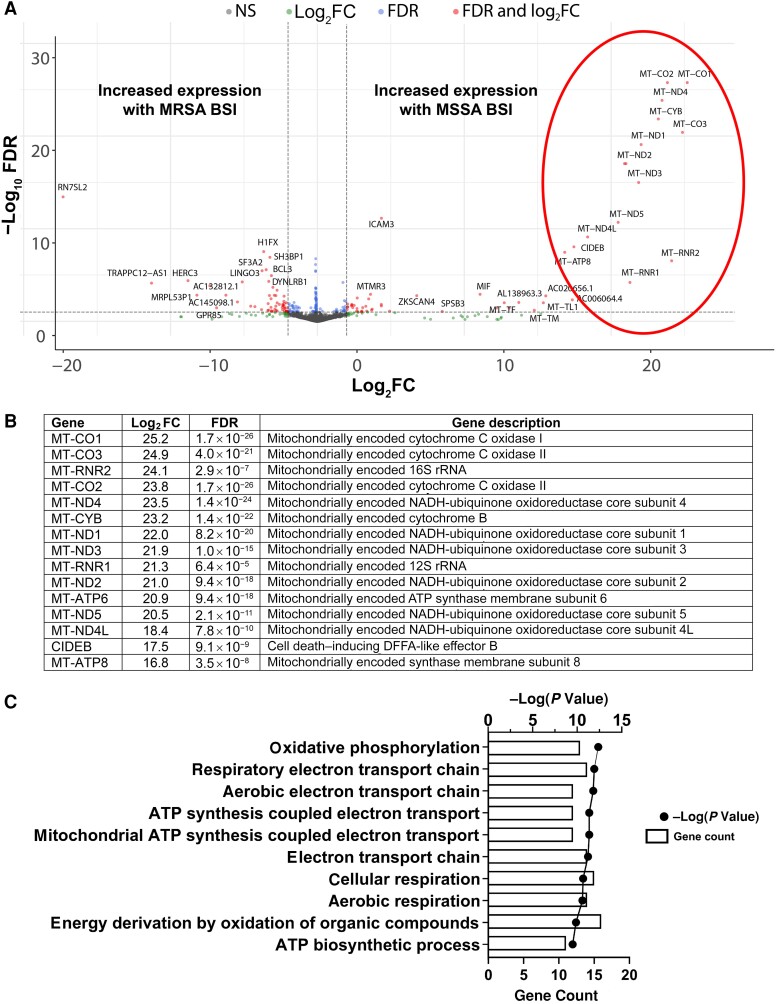
In our exploratory analyses, we unexpectedly noted significant differences in the serum transcriptional profiles of patients with MSSA versus MRSA BSI; these were explored further. In total, we identified 157 human genes that were differentially expressed in patients with MSSA versus MRSA BSI. Of these transcripts, 73 were more highly expressed in MSSA BSI and 84 were more highly present with MRSA BSI. Illustrated in Figure 3A, there was a large set of mitochondrial-encoded genes that were highly up-regulated in patients with MSSA BSI relative to MRSA BSI. These included almost all the protein-coding genes in the mitochondrial genome: adenosine triphosphate (ATP) synthase (MT-ATP8 and MT-ATP6), cytochrome C oxidase (MT-CO1, MT-CO2, and MT-CO3), cytochrome B (MT-CYB), and nicotinamide adenine dinucleotide, reduced (NADH) dehydrogenase (MT-ND1, MT-ND2, MT-ND3, MT-ND4L, MT-ND4, and MT-ND5) (Figure 3B). The proteins encoded by these genes, along with some nuclear gene-encoded proteins, form the enzyme complexes that drive ATP synthesis through oxidative phosphorylation. A g:Profiler biological processes analysis was performed to identify processes that are differentially regulated in patients with MSSA versus MRSA BSI (Figure 3C). Energy transduction pathways, such as oxidative phosphorylation and mitochondrial ATP synthesis–coupled electron transport, were driven by differences in transcript levels of the mitochondrial genes described above. A full list of genes differentially present in patients with MSSA versus MRSA BSI is found in Supplementary Table 2.
Figure 3.
Whole-blood gene transcript level changes in patients with methicillin-susceptible Staphylococcus aureus (MSSA) bloodstream infection (BSI) relative to methicillin-resistant S. aureus (MRSA) BSI. A, Volcano plot showing gene transcript level differences in patients with MSSA versus MRSA BSI. Genes that had log2-transformed fold change (log2FC) >1 or less than −1 and had a false discovery rate (FDR) <0.10 are shown in red; genes meeting only the log2FC or FDR criteria are shown in green or blue, respectively; and genes meeting neither criterion (NS) are shown in gray. Circled in red is a group of genes that particularly differed between the 2 groups and are further described in B. B, Log2-transformed fold changes (log2FC). C, g:Profiler biological processes analysis of gene transcript level differences in patients with MSSA versus MRSA BSI. For each pathway, the number of genes associated with the pathway (count) and the P value associated with the pathway are shown. Abbreviations: ATP, adenosine triphosphate; DFFA, DNA fragmentation factor-α; NADH, nicotinamide adenine dinucleotide, reduced; rRNA, ribosomal RNA.
Given the observed differences in gene transcript levels among those with MSSA versus MRSA BSI, we next sought to determine the extent to which these changes correlated with antibiotic therapy (eg, vancomycin or β-lactam) versus bacteria (eg, MSSA vs MRSA). To examine how variation in antibiotic therapy may be associated with host transcriptional response, we identified differences in gene transcript levels among patients with MSSA BSI treated with vancomycin (n = 13) relative to a β-lactam antibiotic (n = 11). We found only 2 genes that were significantly differentially expressed, both higher in patients treated with vancomycin: KCNQ1OT1 and MTCO3P12. KCNQ101T1 is a non–protein-encoding antisense RNA gene that appears to be involved in gene silencing through chromatin remodeling interactions [33]. KCNQ101T1 has been shown to regulate multiple processes, including cell proliferation, cell cycle, migration and invasion, and immune evasion, among others [34]. MTCO3P12 is a pseudogene of mitochondria-encoded cytochrome oxidase III, and low gene expression levels have been associated with atopic dermatitis in children [35].
The fact that only 2 genes were differentially present in patients with MSSA BSI treated with vancomycin versus β-lactams suggested to us that the effect of different antibiotics on host transcriptional response is negligible in patients with MSSA BSI. To examine how bacterial etiology and antibiotic therapy influenced host gene expression, we identified transcript level changes in patients with MSSA BSI treated with vancomycin (n = 13) relative to those with MRSA BSI (n = 58). In this subgroup analysis, we identified 216 genes that were differentially present in patients with MSSA versus MRSA BSI, of which 96 were more highly present with MSSA BSI and 120 were more highly present with MRSA BSI (Supplementary Table 3).
Biological processes—including proteasomal protein catabolic process (adjusted P = .002) and proteolysis involved in protein catabolic process (adjusted P = .003) were identified owing to transcript level differences in genes of the ubiquitin-proteasome pathway. Such transcriptional profiles included ubiquitin conjugating enzymes E2 H and D2 (UBE2H and UBE2D2), ubiquitin C (UBC), Pellino E3 ubiquitin protein ligase 1 (PEL1), deubiquitinase (YOD1), proteasome activator subunit 4 (PSME4), and proteasome 26S subunit (PSMD7) (Supplementary Figure 1). These gene transcript levels are higher in patients with MSSA BSI than in those with MRSA BSI. In addition to the proteosome-related pathways, biological processes associated with the autophagosome were identified, such as maintenance of protein localization in an organelle (adjusted P = .001), regulation of autophagosome assembly (adjusted P = .002), and regulation of vacuole organization (adjusted P = .003). These processes were identified based on transcript level differences in the Ras oncogene member RAB1B, phosphatidylinositol-5-phosphate 4-kinase type 2 alpha gene PIP4K2A, and the myotubalarin-related protein 3, among others.
These gene transcript levels were higher in patients with MSSA BSI relative to MRSA BSI. Of note, the mitochondrial gene transcript levels that differed in the overall cohort of patients with MSSA versus MRSA BSI (Figure 3) again had higher transcript levels in patients with vancomycin-treated MSSA relative to MRSA BSI, though the changes did not meet the FDR threshold of <0.10 in this subgroup analysis. In total, we interpret the observed transcriptional profile differences between patients with MSSA and MRSA BSI as being due to a fundamental difference in the host response to the bacteria (ie, MSSA vs MRSA), rather than a treatment-specific effect (ie, effect of vancomycin vs β-lactam antibiotic).
Bacteria-Specific Host Transcriptional Response Signatures
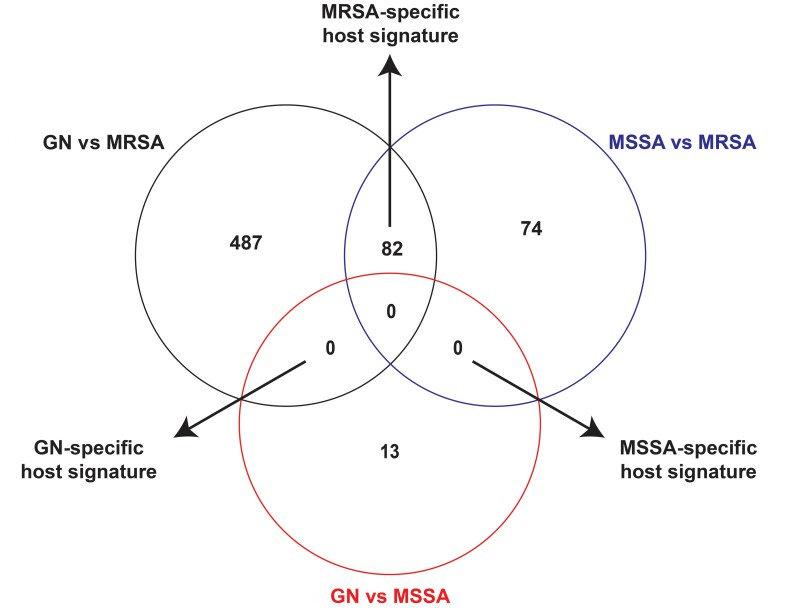
In the above analyses we identified host transcriptional genes that had differential transcript levels in patients with GN versus S. aureus BSI and MSSA versus MRSA BSI. We next sought to identify the host transcriptional signatures that are specific to individual pathogen groups (ie, GN, MRSA, or MSSA). As in Figure 4, pairwise comparisons of the host serum transcriptional changes in patients with GN versus MSSA BSI, GN versus MRSA BSI, and MSSA versus MRSA BSI were used to identify pathogen-specific host transcriptional signatures. Interestingly, only MRSA-specific host transcriptional changes were identified. The MRSA-specific host transcriptional response consisted of 82 genes, of which 38 were more highly present in MRSA and 44 more highly present in MSSA and GN BSI. Genes in the MRSA-specific host transcriptional response are listed in Supplementary Table 4. Biological processes associated with the MRSA-specific host transcriptional signature are primarily related to energy transduction (eg, immune cell oxidative phosphorylation, respiratory energetics, and respiratory electron transport chain) (Supplementary Figure 2) and essentially identical to the processes that were differentially regulated in patients with MRSA relative to MSSA BSI (as detailed in Figure 3). The biological processes associated with the MRSA-specific transcriptional signature were driven almost entirely by decreased transcript levels of mitochondrial genes (eg, MT-ATP8 and MT-ATP6) listed in Figure 3B.
Figure 4.
Identification of bacteria-specific host transcriptional signatures. Within each circle is the number of genes with transcript levels that differ in each comparison (eg, patients with gram-negative [GN] vs methicillin-susceptible Staphylococcus aureus [MSSA] bloodstream infection [BSI]). Gene transcript levels shared between 2 different comparisons are determined to be part of bacteria-specific response. For example, if a gene is up-regulated in patients with methicillin-resistant S. aureus (MRSA) BSI relative to both MSSA BSI and GN BSI, then the gene is defined as part of the MRSA-specific host signature. See Supplementary Table 4 for specific identities of the MRSA-specific host signature genes.
DISCUSSION
In this work we sought to understand how the host transcriptional response differs in GN BSI relative to S. aureus BSI. To address this issue, we performed RNA-Seq on whole-blood samples of patients with E. coli or K. pneumoniae BSI, compared to MSSA or MRSA BSI. There were 2 primary findings from this study. First, we found that, relative to S. aureus BSI, the host transcriptional signature in GN BSI was enriched in the activation of immunoglobulin and classic complement system transcription. Interestingly, this signature suggests that antibody-dependent cell cytotoxicity may play a more important role in defending against GN compared with GP pathogens. Second, we identified a MRSA-specific host transcriptional signature that reflected reduced mitochondrial responses involved in immune cell respiratory burst and energy transduction. Each of these outcomes is discussed in detail below.
We found that the host transcription signature in patients with GN BSI, relative to S. aureus BSI, was activation of the immunoglobulin and classic complement pathway responses. Complement is a component of the innate immune system that functions, in part, to eliminate pathogens such as bacteria from the bloodstream. The classic pathway of complement fixation is antibody dependent; it is triggered by binding of immunoglobulin M or G immunoglobulins to the bacterial surface, followed by C1q binding to the bacterial/immunoglobulin complex. Binding of C1q triggers a well-defined cascade of events that ultimately leads to formation of the membrane attack complex in the bacterial membrane. In GN bacteria, formation of the membrane attack complex leads to loss of membrane integrity and bacterial death. In GP bacteria like S. aureus, however, the thick peptidoglycan layer prevents cell death [36]. In these respects, increased expression of the classic complement pathway in response to GN infection, as opposed to S. aureus infection, is biologically plausible given that the complement proteins can directly lead to loss of GN outer membrane integrity and ensuing bacterial death. By comparison, many strains of S. aureus are serum resistant, corresponding to refractivity to complement-mediated killing.
We also observed unforeseen and extensive differences in host transcriptomes in patients with MRSA BSI, as compared with MSSA BSI. These changes were durable even when controlling for differences in antibiotic therapy. Patients with MRSA BSI had significantly lower transcript levels of mitochondrial genes that are critical components of the electron transport chain and hence energy transduction. To our knowledge, this is the first study to identify a difference in host mitochondrial gene transcript levels in MRSA versus MSSA infection. Differences in mitochondrial function are likely to significantly influence disease progression as multiple studies have shown the organelle to be critical for generation of antimicrobial reactive oxygen species necessary for killing intracellular S. aureus [37, 38]. The underlying cause of this compelling difference in host transcriptional profiles in response to MSSA versus MRSA phenotypes of S. aureus is under investigation.
The current study had several limitations. Patients were enrolled from a single hospital system, which may have introduced bias owing to lack of variability in the patient population, clinical presentation, and treatment practices. We have attempted to mitigate this bias by enrolling a large cohort of patients that are diverse with respect to race, sex, and clinical presentation. Furthermore, patients were matched with respect to age, sex, race, and antibiotic therapy to limit variability between subgroups of interest. We measured host transcript level data from a single time point in the first few days following the first positive blood culture, and we did not capture temporal changes within and between patient subgroups. While our matching scheme did account for potential confounding variables—including age, sex, race, and antibiotic therapy—we note that other variables that were not accounted for (eg, source of BSI, level of acute illness, etc) could have biased the results. In comparisons involving groups that differed in severity of illness (eg, patients with S. aureus vs GN BSI), we cannot parse whether the observed host transcriptomic differences stemmed from variability in severity of illness or some other pathogen-specific response. Finally, we did not measure protein levels, so we cannot address the extent to which the observed changes in the global transcriptome are correlated with alterations in the proteome.
In conclusion, the current findings may shed new light on host transcriptional responses in specific contexts of pathogen-induced and antibiotic-induced immunity. Interestingly, the importance of immunoglobulin responses in clearing GN bacteria from the bloodstream appears to differ considerably from their importance in clearing GP bacteria. This finding is of interest given that no vaccines aimed at preventing invasive S. aureus infections through generation of high titers of opsonic antibodies against the bacterium have demonstrated efficacy in clinical trials [39–41]. The optimal immune response to invasive S. aureus infection is an area of ongoing research [42]. Ultimately, we uncovered an MRSA-specific host transcriptional signature that was driven in large part by reduced mitochondrial responses necessary for essential immune functions in oxidative killing of S. aureus. Given the importance of such responses in clearance of S. aureus from intracellular reservoirs, this finding may offer insights into the propensity for MRSA to persist in BSI relative to other bacteria. Moreover, these results suggest that transcriptional or other omics biomarkers may enable molecular differentiation of host responses that predict successful versus poor outcomes of BSIs.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Joshua T Thaden, Division of Infectious Diseases, Duke University School of Medicine, Durham, North Carolina, USA.
Richard Ahn, Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, Los Angeles, California, USA; Institute for Quantitative and Computational Biosciences, University of California, Los Angeles, Los Angeles, California, USA.
Felicia Ruffin, Division of Infectious Diseases, Duke University School of Medicine, Durham, North Carolina, USA.
David W Gjertson, Department of Biostatistics, University of California, Los Angeles, Los Angeles, California, USA; Department of Pathology and Laboratory Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Alexander Hoffmann, Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, Los Angeles, California, USA; Institute for Quantitative and Computational Biosciences, University of California, Los Angeles, Los Angeles, California, USA.
Vance G Fowler, Jr, Division of Infectious Diseases, Duke University School of Medicine, Durham, North Carolina, USA.
Michael R Yeaman, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California, USA; Department of Medicine, Divisions of Molecular Medicine and Infectious Diseases, Harbor-UCLA Medical Center, Torrance, California, USA; Institute for Infection & Immunity, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, California, USA.
Notes
Author contributions. Conception: V. G. F. and M. R. Y. Data acquisition: F. R. Data analysis: J. T. T., R. A., D. W. G., and A. H. Data interpretation: J. T. T., R. A., D. W. G., A. H., V. G. F., and M. R. Y. Supervision: V. G. F. and M. R. Y.
Financial support. This work was the National Institute of Allergy and Infectious Diseases (systems biology grants NIAID U01-AI124319 and NIAID U19-AI172713-01 to M. R. Y., grant K08-AI171183-01 to J. T. T., and grant 1R01-AI165671-01 to V. G. F.).
References
- 1. Kern WV, Rieg S. Burden of bacterial bloodstream infection—a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin Microbiol Infect 2020; 26:151–7. [DOI] [PubMed] [Google Scholar]
- 2. Thaden JT, Li Y, Ruffin F, et al. Increased costs associated with bloodstream infections caused by multidrug-resistant gram-negative bacteria are due primarily to patients with hospital-acquired infections. Antimicrob Agents Chemother 2017; 61:10–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG Jr, van Duin D. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by Pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother 2017; 61:10–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi SH, Dagher M, Ruffin F, et al. Risk factors for recurrent Staphylococcus aureus bacteremia. Clin Infect Dis 2020; 72:1891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee CC, Lee CH, Hong MY, Tang HJ, Ko WC. Timing of appropriate empirical antimicrobial administration and outcome of adults with community-onset bacteremia. Crit Care 2017; 21:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzalez C, Rubio M, Romero-Vivas J, Gonzalez M, Picazo JJ. Bacteremic pneumonia due to Staphylococcus aureus: a comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis 1999; 29:1171–7. [DOI] [PubMed] [Google Scholar]
- 7. Zwack EE, Chen Z, Devlin JC, et al. Staphylococcus aureus induces a muted host response in human blood that blunts the recruitment of neutrophils. Proc Natl Acad Sci U S A 2022; 119:e2123017119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gronnemose RB, Garde C, Wassmann CS, et al. Bacteria-host transcriptional response during endothelial invasion by Staphylococcus aureus. Sci Rep 2021; 11:6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brady RA, Bruno VM, Burns DL. RNA-Seq Analysis of the host response to Staphylococcus aureus skin and soft tissue infection in a mouse model. PLoS One 2015; 10:e0124877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thanert R, Goldmann O, Beineke A, Medina E. Host-inherent variability influences the transcriptional response of Staphylococcus aureus during in vivo infection. Nat Commun 2017; 8:14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang JH, Bhargava P, McCloskey D, Mao N, Palsson BO, Collins JJ. Antibiotic-induced changes to the host metabolic environment inhibit drug efficacy and alter immune function. Cell Host Microbe 2017; 22:757–65.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chatzispyrou IA, Held NM, Mouchiroud L, Auwerx J, Houtkooper RH. Tetracycline antibiotics impair mitochondrial function and its experimental use confounds research. Cancer Res 2015; 75:4446–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leach KL, Swaney SM, Colca JR, et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell 2007; 26:393–402. [DOI] [PubMed] [Google Scholar]
- 14. Asberg SE, Mediaas SD, Marstad A, et al. Frontline science: antibiotic treatment routes Mycobacterium avium to phagolysosomes without triggering proinflammatory cytokine production in human Mϕs. J Leukoc Biol 2021; 109:23–33. [DOI] [PubMed] [Google Scholar]
- 15. Yoon GS, Sud S, Keswani RK, et al. Phagocytosed clofazimine biocrystals can modulate innate immune signaling by inhibiting TNFα and boosting IL-1RA secretion. Mol Pharm 2015; 12:2517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye C, Li W, Yang Y, et al. Inappropriate use of antibiotics exacerbates inflammation through OMV-induced pyroptosis in MDR Klebsiella pneumoniae infection. Cell Rep 2021; 36:109750. [DOI] [PubMed] [Google Scholar]
- 17. Kalantar KL, Neyton L, Abdelghany M, et al. Integrated host-microbe plasma metagenomics for sepsis diagnosis in a prospective cohort of critically ill adults. Nat Microbiol 2022; 7:1805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li D, Gai W, Zhang J, Cheng W, Cui N, Wang H. Metagenomic next-generation sequencing for the microbiological diagnosis of abdominal sepsis patients. Front Microbiol 2022; 13:816631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci U S A 2018; 115:E12353–E62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McHugh L, Seldon TA, Brandon RA, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med 2015; 12:e1001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ko ER, Henao R, Frankey K, et al. Prospective validation of a rapid host gene expression test to discriminate bacterial from viral respiratory infection. JAMA Netw Open 2022; 5:e227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang C, Gong B, Bushel PR, et al. The concordance between RNA-Seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 2014; 32:926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timp W, Timp G. Beyond mass spectrometry, the next step in proteomics. Sci Adv 2020; 6:eaax8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang XW, Li QH, Xu ZD, Dou JJ. Mass spectrometry-based metabolomics in health and medical science: a systematic review. RSC Adv 2020; 10:3092–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrews S. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 18 October 2023.
- 27.Picard. http://broadinstitute.github.io/picard/. Accessed 18 October 2023.
- 28. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30:923–30. [DOI] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019; 47:W191–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32:232–46. [DOI] [PubMed] [Google Scholar]
- 34. Cagle P, Qi Q, Niture S, Kumar D. KCNQ1OT1: an oncogenic long noncoding RNA. Biomolecules 2021; 11:1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nousbeck J, McAleer MA, Irvine AD. Peripheral blood gene expression profile of infants with atopic dermatitis. JID Innov 2023; 3:100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muller-Eberhard HJ. The membrane attack complex of complement. Annu Rev Immunol 1986; 4:503–28. [DOI] [PubMed] [Google Scholar]
- 37. Abuaita BH, Schultz TL, O'Riordan MX. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 2018; 24:625–36.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dunham-Snary KJ, Surewaard BG, Mewburn JD, et al. Mitochondria in human neutrophils mediate killing of Staphylococcus aureus. Redox Biol 2022; 49:102225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fattom A, Matalon A, Buerkert J, Taylor K, Damaso S, Boutriau D. Efficacy profile of a bivalent Staphylococcus aureus glycoconjugated vaccine in adults on hemodialysis: phase III randomized study. Hum Vaccin Immunother 2015; 11:632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fattom AI, Horwith G, Fuller S, Propst M, Naso R. Development of StaphVAX, a polysaccharide conjugate vaccine against S. aureus infection: from the lab bench to phase III clinical trials. Vaccine 2004; 22:880–7. [DOI] [PubMed] [Google Scholar]
- 41. Fowler VG, Allen KB, Moreira ED, et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 2013; 309:1368–78. [DOI] [PubMed] [Google Scholar]
- 42. Miller LS, Fowler VG, Shukla SK, Rose WE, Proctor RA. Development of a vaccine against Staphylococcus aureus invasive infections: evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol Rev 2020; 44:123–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.