Abstract
Background
Chronic pulmonary conditions such as asthma and chronic obstructive pulmonary disease increase the risk of morbidity and mortality during infection with the Middle East respiratory syndrome coronavirus (MERS-CoV). We hypothesized that individuals with such comorbidities are more susceptible to MERS-CoV infection due to increased expression of its receptor, dipeptidyl peptidase 4 (DPP4).
Methods
We modeled chronic airway disease by treating primary human airway epithelia with the Th2 cytokine interleukin 13 (IL-13), examining how this affected DPP4 protein levels with MERS-CoV entry and replication.
Results
IL-13 exposure for 3 days led to greater DPP4 protein abundance, while a 21-day treatment raised DPP4 levels and caused goblet cell metaplasia. Surprisingly, despite this increase in receptor availability, MERS-CoV entry and replication were not significantly affected by IL-13 treatment.
Conclusions
Our results suggest that greater DPP4 abundance is likely not the primary mechanism leading to increased MERS severity in the setting of Th2 inflammation. Transcriptional profiling analysis highlighted the complexity of IL-13–induced changes in airway epithelia, including altered expression of genes involved in innate immunity, antiviral responses, and maintenance of the extracellular mucus barrier. These data suggest that additional factors likely interact with DPP4 abundance to determine MERS-CoV infection outcomes.
Keywords: airway epithelia, DPP4, IL-13, MERS-CoV, th2 inflammation
Stimulating airway epithelia with interleukin 13 increased DPP4, the receptor for Middle East respiratory syndrome coronavirus (MERS-CoV). However, this failed to increase MERS-CoV infection, suggesting that elevated airway epithelial DPP4 cannot explain increased risk of severe MERS in chronic airway disease.
Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in Saudi Arabia in 2012 [1]. Since its emergence, the virus has spread to 27 countries and caused 936 deaths out of 2604 confirmed cases (as of May 2023; World Health Organization [https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html]). Its high case fatality rate (∼35%) is reminiscent of the severe acute respiratory syndrome epidemic and has caused worldwide concern. The MERS clinical spectrum ranges from asymptomatic infection to mild upper respiratory disease and life-threatening acute pulmonary disease, which can progress to respiratory failure, shock, and multiorgan failure [2, 3]. Epidemiologic studies indicate that mortality is significantly higher in persons with underlying comorbidities, including diabetes, obesity, cardiovascular diseases, renal disease, immunodeficiency, and chronic lung disease [4–7].
MERS-CoV uses dipeptidyl peptidase 4 (DPP4/CD26) as its cellular receptor [8]. DPP4 is a serine exopeptidase that cleaves X-proline or X-alanine dipeptides from the N-terminus of many bioactive proteins, including chemokines, growth factors, neuropeptides, and vasoactive peptides [9–11]. DPP4 tissue expression is altered in association with disease states, such as inflammation, cancer, obesity, and diabetes [12]. One study reported that DPP4 is highly expressed in bronchial epithelial cells of patients with untreated asthma [13]. Notably, asthma is one of the most frequent comorbidities in patients with MERS [4–7]. Additional studies report that the DPP4 protein is increased in alveolar epithelial cells from smokers and patients with chronic obstructive pulmonary disease (COPD): 2 high-risk comorbidities associated with severe MERS-CoV infection outcomes [14, 15]. Thus, greater DPP4 expression in respiratory epithelia is a possible mechanism underlying MERS-CoV susceptibility and outcomes.
Here, we test the hypothesis that during MERS-CoV infection, higher morbidity and mortality associated with chronic conditions such as asthma and COPD arise from inflammation-mediated increases in DPP4. Using interleukin 13 (IL-13) exposure in well-differentiated primary human airway epithelia (HAE) cultures as an in vitro model of type 2 inflammation, we found that short-term IL-13 exposure (3 days) significantly increased DPP4 abundance and the number of DPP4-expressing cells. Longer-term IL-13 exposure (21 days) also increased DPP4 expression and induced goblet cell metaplasia. Interestingly, despite the greater abundance of DPP4 in both settings, we observed no significant enhancement of MERS-CoV entry and/or replication. Transcriptional profiling demonstrated that IL-13–induced inflammation has complex effects on airway epithelia, including increased DPP4 abundance and changes in genes influencing virus binding and entry, innate immunity, and mucin production. The predicted effects of these changes include pro- and antiviral functions, with the overall outcome that IL-13 has no significant effect on MERS-CoV output. Overall, our findings suggest that greater DPP4 protein abundance associated with Th2 inflammation is not a significant factor in determining MERS-CoV infection and spread.
METHODS
Primary Cultures of Airway Epithelia
Human primary airway epithelia derived from human donor lungs were obtained from the University of Iowa Cells and Tissue Core. To establish airway epithelial cultures, cells were isolated from bronchi by enzymatic dispersion, seeded onto collagen-coated semipermeable membranes with a 0.4-μm pore size (0.6 cm2; Costar), and grown at an air-liquid interface in 1:1 DMEM/Ham's F-12 medium containing Ultroser G at 37 °C with 5% CO2 as described [16]. Only well-differentiated cultures were used in this study (>3 weeks postseeding), as assessed by the presence of tight junctions (transepithelial resistance > 1000 Ω × cm2). The study was approved by the Institutional Review Board at the University of Iowa.
Cytokine Stimulation of Human Airway Epithelia and Measurement of DPP4 Protein Abundance
Cytokines were added to the basolateral medium at the indicated concentrations and changed every 2 days. DPP4 abundance was measured by enzyme-linked immunosorbent assay (ELISA), and DPP4-positive cells were quantified by flow cytometry. To minimize the effects of HAE donor variability, experiments were designed so that the IL-13 and phosphate-buffered saline (PBS) treatment conditions were compared within donors. Detailed methods are provided in the supplementary material.
Viruses
The EMC/2012 strain of MERS-CoV (passage 8, designated MERS-CoV) was provided by Dr Bart Haagmans and Ron Fouchier (Erasmus Medical Center). See the supplementary material for detailed methods regarding MERS-CoV infection in HAE and pseudoparticle generation and transduction.
RESULTS
Short-term IL-13 Exposure Increases DPP4 in Primary Human Airway Epithelia
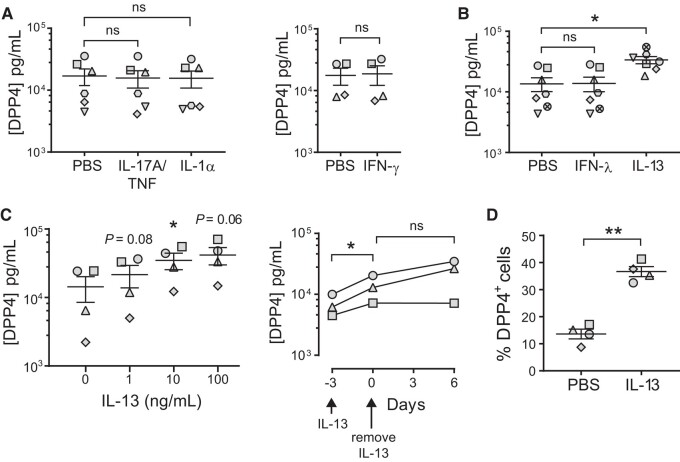
We sought to better understand how DPP4 protein abundance is regulated by proinflammatory cytokines that drive chronic airway inflammation. Interleukin 17, tumor necrosis factor α (TNF-α), interleukin 1, interferon γ (IFN-γ), and other cytokines contribute to the development of the Th2-low asthma endotype and regulate DPP4 protein in a cell type–specific manner [17–19]. We investigated whether these cytokines influence DPP4 protein abundance in human airways, using primary HAE derived from donor lungs and differentiated at an air-liquid interface. We treated HAE with interleukin 17A/tumor necrosis factor α, interleukin 1α, or IFN-γ for 3 days and quantified the cellular abundance of DPP4 by ELISA. Consistent with our previous work, we found that baseline levels of DPP4 protein vary widely among HAE donors [20]; however, we observed no significant increase in DPP4 relative to baseline levels in any of the HAE donors in response to these cytokines (Figure 1A). Similarly, we tested the effects of interferon λ (IFN-λ), a cytokine that increases in HAE during MERS-CoV infection [20]. The HAE donors exhibited no significant changes in DPP4 levels after 3-day stimulation with IFN-λ (Figure 1B).
Figure 1.
IL-13 treatment increases the abundance of DPP4 in human airway epithelia. A (left), Well-differentiated air-liquid interface cultures of primary HAE were maintained for 3 days in medium containing IL-17A/TNF-α (20 ng/mL; 10 ng/mL), IL-1α (20 ng/mL), or PBS. After 3 days, ELISA was used to determine DPP4 protein abundance in cell lysates (n = 6 HAE donors). Data are presented as mean ± SE. A (right), HAE were stimulated for 3 days with PBS or IFN-γ (100 ng/mL), and DPP4 protein in cell lysates was quantified by ELISA (n = 4). B, HAE were stimulated for 3 days with PBS, IFN-λ (1 ng/mL), or IL-13 (10 ng/mL), and DPP4 protein was measured in cell lysates (n = 7). A and B, A ratio paired t test was used to test for statistically significant differences in cytokine-treated epithelia relative to the PBS control. *P < .05. ns = not significant. C (left), HAE (n = 4) were grown in medium containing IL-13 at the indicated concentrations for 3 days; then, ELISA was used to determine DPP4 protein abundance in cell lysates. Repeated measures 1-way analysis of variance, followed by a Dunnett multiple-comparison test, was used to test for significant differences between IL-13 treatment at each concentration and baseline (0 ng/mL). *P < .05. C (right), HAE (n = 3) were stimulated with IL-13 (20 ng/mL) for 3 days; then, IL-13 was removed from the growth medium and the epithelia maintained for an additional 6 days. DPP4 abundance in cell lysates was measured by ELISA. A ratio paired t test was used to test for significant differences between time points. *P < .05. D, HAE (n = 4) were treated for 3 days with IL-13 (20 ng/mL) or PBS, followed by flow cytometry to quantify DPP4+ cells. Percentages of DPP4+ cells are plotted for each HAE donor. A ratio paired t test was used to test for a statistically significant difference between the PBS and IL-13 conditions. **P < .01. DPP4, dipeptidyl peptidase 4; ELISA, enzyme-linked immunosorbent assay; HAE, human airway epithelia; IFN-γ, interferon γ; IFN-λ, interferon λ; IL-1α, interleukin 1α; IL-13, interleukin 13; IL-17A, interleukin 17A; ns, not significant; PBS, phosphate-buffered saline; TNF-α, tumor necrosis factor α.
We next asked whether DPP4 abundance is altered under conditions that model the Th2-high asthma endotype. Th2-high asthma is driven by type 2 cytokines (interleukin 4 [IL-4], interleukin 5, and IL-13) [21, 22]. Among these Th2 cytokines, IL-13 plays the most important role in inducing phenotypic changes associated with asthma in cell and animal models [23–26]. Interestingly, IL-4 and IL-13 raise DPP4 expression in cultured renal carcinoma cells and renal tubular epithelial cells [27, 28], and IL-13 treatment increases DPP4 mRNA expression in human bronchial epithelial cells [13]. To test whether treatment with IL-13 increases DPP4, we exposed epithelia to IL-13 for 3 days and measured cellular DPP4 abundance by ELISA. We found that IL-13 treatment significantly increased the DPP4 protein in epithelia from all HAE donors (Figure 1B). This IL-13–induced increase in DPP4 expression was dose dependent, with some variability in the magnitude of the increase among donors (Figure 1C). The higher DPP4 protein levels persisted for at least 6 days following removal of IL-13 from the media (Figure 1C) and were accompanied by an increase in the number of DPP4-expressing cells as measured by flow cytometry (Figure 1D).
Effects of Short-term IL-13 Treatment on MERS-CoV Entry and Infection
We hypothesized that an IL-13–induced increase in DPP4 would raise MERS-CoV binding and entry in airway epithelia. To test for effects on viral entry, we treated HAE with IFN-λ or IL-13 for 3 days and then transduced cells with vesicular stomatitis virus (VSV)–based particles pseudotyped with the MERS S protein and bearing a luciferase reporter (Figure 2A). At 24 hours posttransduction, pseudoparticle-driven luciferase activity in the IFN-λ–treated cells was significantly reduced relative to the PBS control condition, while pseudoparticle transduction was not significantly different with IL-13 treatment (Figure 2B). We speculate that pseudoparticle transduction was reduced in the IFN-λ–treated epithelia due to interferon (IFN)–mediated upregulation of host innate immune factors such as the IFITM proteins, which are known to block VSV and VSV-derived pseudoparticle entry by inhibiting viral fusion with host membranes (as reviewed Majdoul and Compton [29]). To assess the impact of the IL-13 or IFN-λ treatment on infection with authentic virus, we applied MERS-CoV to epithelia and measured apically released viral progeny by plaque assay at 1 and 2 days postinfection (Figure 2A). As shown in Figure 2C, IFN-λ treatment produced the expected inhibition of viral replication at 1 and 2 days postinfection; however, viral titers were unaffected by IL-13 treatment. Together, these data suggest that a short-term IL-13 exposure (3 days) and the concomitant increase in DPP4 do not significantly raise MERS-CoV entry or infection.
Figure 2.
Short-term exposure to IL-13 has little effect on MERS-CoV entry and infection. A, Experimental design for testing MERS-CoV entry and replication in HAE. Briefly, HAE were maintained for 3 days in medium containing IFN-λ (1 ng/mL) or IL-13 (20 ng/mL) and then transduced apically with MERS-S pps or infected with MERS-CoV (MOI, 0.5). B, MERS-S pps entry was quantified by measuring luciferase activity in cell lysates at 24 hours posttransduction. Data points represent individual HAE donors, and data are plotted as mean ± SE (n = 5). Log-transformed data were tested for significant differences by repeated measures 1-way ANOVA, followed by a Tukey multiple-comparison test. C, Following apical infection with MERS-CoV, a plaque assay was used to measure apically released viral progeny at 1 and 2 days postinfection. Data represent mean ± SE (n = 4). Log-transformed data were tested for statistically significant differences relative to the PBS control at the indicated time points by repeated measures 2-way ANOVA, followed by a Sidak multiple-comparison test. *P < .05. **P < .01. ANOVA, analysis of variance; HAE, human airway epithelia; IFN-λ, interferon λ; IL-13, interleukin 13; MERS-CoV, Middle East respiratory syndrome coronavirus; MERS-S pps, MERS S protein pseudoparticles; MOI, multiplicity of infection; ns, not significant; PBS, phosphate-buffered saline; PFU, plaque-forming units; RLU, relative light units.
Long-term Exposure to IL-13 Increases DPP4 Abundance and Induces Goblet Cell Metaplasia in Human Airway Epithelia
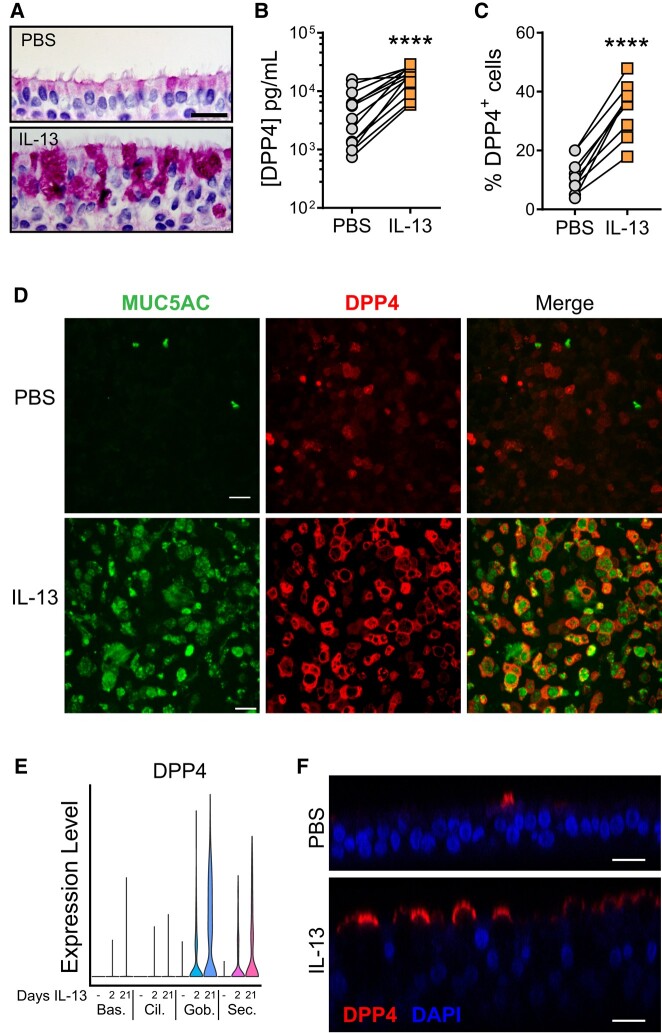
We next examined whether DPP4-mediated MERS-CoV entry and infection are altered in the setting of goblet cell metaplasia. Goblet cell metaplasia and mucus hypersecretion are hallmarks of many chronic airway diseases, including asthma, cystic fibrosis, and COPD. In asthma, goblet cell metaplasia is associated with chronic exposure to Th2 cytokine–driven inflammation. We previously showed that 21-day exposure to IL-13 induced goblet cell metaplasia in HAE [30]. Using this IL-13 treatment protocol, we observed the expected dramatic increase in goblet cells and epithelium remodeling through dPAS staining (diastase and periodic acid–Schiff; Figure 3A), which was accompanied by increases in cellular DPP4 and the number of DPP4-expressing cells (Figure 3B and 3C). In keeping with this, immunofluorescent staining indicated significantly higher DPP4 expression, as well as greater goblet cell abundance as assessed by immunostaining for MUC5AC (Figure 3D). While most DPP4-positive cells were MUC5AC positive, not every goblet cell expressed DPP4. We further investigated the distribution of DPP4-expressing cells in IL-13–treated epithelia using a publicly available single-cell RNA sequencing (RNAseq) data set (GEO accession GSE229202). This study confirmed that DPP4 mRNA increases after 2 or 21 days of exposure to IL-13, primarily in secretory cell types such as goblet cells (Figure 3E). Immunofluorescence staining revealed that much of the DPP4 protein in IL-13–treated cultures localized to the apical surface (Figure 3F), where it is likely available to interact with the spike protein on the MERS-CoV envelope.
Figure 3.
Long-term exposure to IL-13 increases DPP4 abundance and induces goblet cell metaplasia in human airway epithelia. A, Representative images from HAE treated for 21 days with PBS or IL-13. dPAS staining was performed to confirm increased mucin production and goblet cell frequency. Scale bar: 20 µm. B, DPP4 protein abundance was measured by ELISA in cell lysates from HAE treated with PBS or IL-13 (20 ng/mL) for 21 days (n = 13). Data points represent individual HAE donors. C, Flow cytometry was used to quantitate total percentages of DPP4-expressing cells after a 21-day treatment with PBS or IL-13 (n = 9). B and C, A ratio paired t test was used to test for statistically significant differences between IL-13– and PBS-treated epithelia. ****P < .0001. D, Representative images from HAE treated for 21 days with vehicle (PBS) or IL-13. Immunofluorescent staining for the goblet cell marker MUC5AC was used to confirm goblet cell metaplasia in the IL-13–treated cells, with increased DPP4. Scale bars: 20 µm. E, Single-cell RNA-sequencing data show normalized gene expression levels for DPP4 in basal, ciliated, goblet, and secretory cell populations in HAE treated with IL-13 for 2 or 21 days. Cell type identifications were made with canonical cell–type markers. Bas, basal cells; Cil, ciliated cells; Gob, goblet cells; Sec, secretory cells. F, Orthogonal z-stack images from HAE treated for 21 days with PBS or IL-13. Epithelia were stained for DPP4 and nuclei with DAPI. Scale bars: 20 µm. dPAS, diastase and periodic acid–Schiff; DPP4, dipeptidyl peptidase 4; ELISA, enzyme-linked immunosorbent assay; HAE, human airway epithelia; IL-13, interleukin 13; PBS, phosphate-buffered saline.
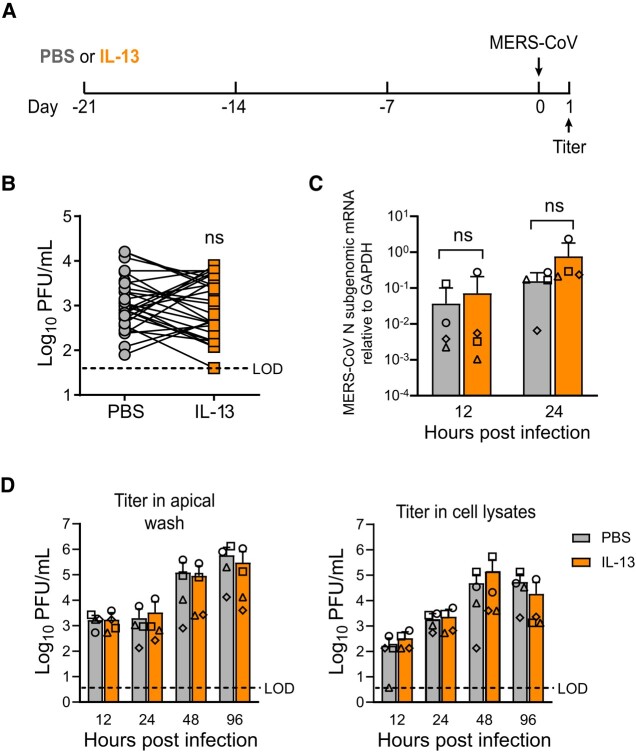
MERS-CoV Entry and Infection in the Setting of IL-13–Induced Goblet Cell Metaplasia
HAE were treated with PBS or IL-13 for 21 days and then infected with MERS-CoV (Figure 4A). We found that infection of IL-13–treated cells with MERS-CoV produced variable results: in some donors, IL-13 treatment was associated with increased virus titers in apical secretions at 1 day postinfection, while the opposite was observed in other donors. Overall, there was no significant increase in released progeny virus in IL-13–treated cells, despite their higher DPP4 expression (Figure 4B). Similarly, viral mRNA levels were not significantly different in the PBS- and IL-13–treated cells at 12 or 24 hours postinfection (Figure 4C). In a subsequent time course experiment, peak titers and virus growth kinetics were similar in paired IL-13– and PBS-treated HAE cultures infected with MERS-CoV, with respect to virus shed into the apical compartment as well as intracellular viral load (Figure 4D). Overall, these data suggest that greater receptor abundance in IL-13–treated cells has little overall impact on the progression of MERS-CoV infection.
Figure 4.
MERS-CoV entry and infection in the setting of IL-13–induced goblet cell metaplasia. A, Experimental design for testing MERS-CoV entry and replication in HAE. HAE were treated with PBS or IL-13 (20 ng/mL) for 21 days and then infected with MERS-CoV (MOI, 0.5). B, Following apical infection with MERS-CoV, plaque assay was used to measure apically released viral progeny at 1 day postinfection. Data points represent individual HAE donors (n = 30). Log-transformed data were tested for statistically significant differences relative to the PBS control through paired t tests. C, HAE (n = 4) were treated for 21 days with PBS or IL-13 and then infected with MERS-CoV (MOI, 0.5). Subgenomic mRNAs for the MERS-CoV N gene were quantified by reverse transcriptase–qualitative polymerase chain reaction. Data are plotted as mean ± SD. D, HAE (n = 4) were treated for 21 days with PBS or IL-13 and then infected with MERS-CoV (MOI, 0.5). Viral growth kinetics were assessed by quantifying apically released viral progeny in the apical wash fluid (left) as well as intracellular titers from cell lysates (right). Data are plotted as mean ± SD. Log-transformed data were tested for statistically significant differences between PBS- and IL-13–treated epithelia at each time point via a repeated measures 2-way analysis of variance, followed by a Sidak multiple-comparison test. HAE, human airway epithelia; IL-13, interleukin 13; MERS-CoV, Middle East respiratory syndrome coronavirus; MOI, multiplicity of infection; ns, not significant; PBS, phosphate-buffered saline; PFU, plaque-forming units.
IL-13 Treatment Alters Expression of Genes Involved in MERS-CoV Infection and Antiviral Defense
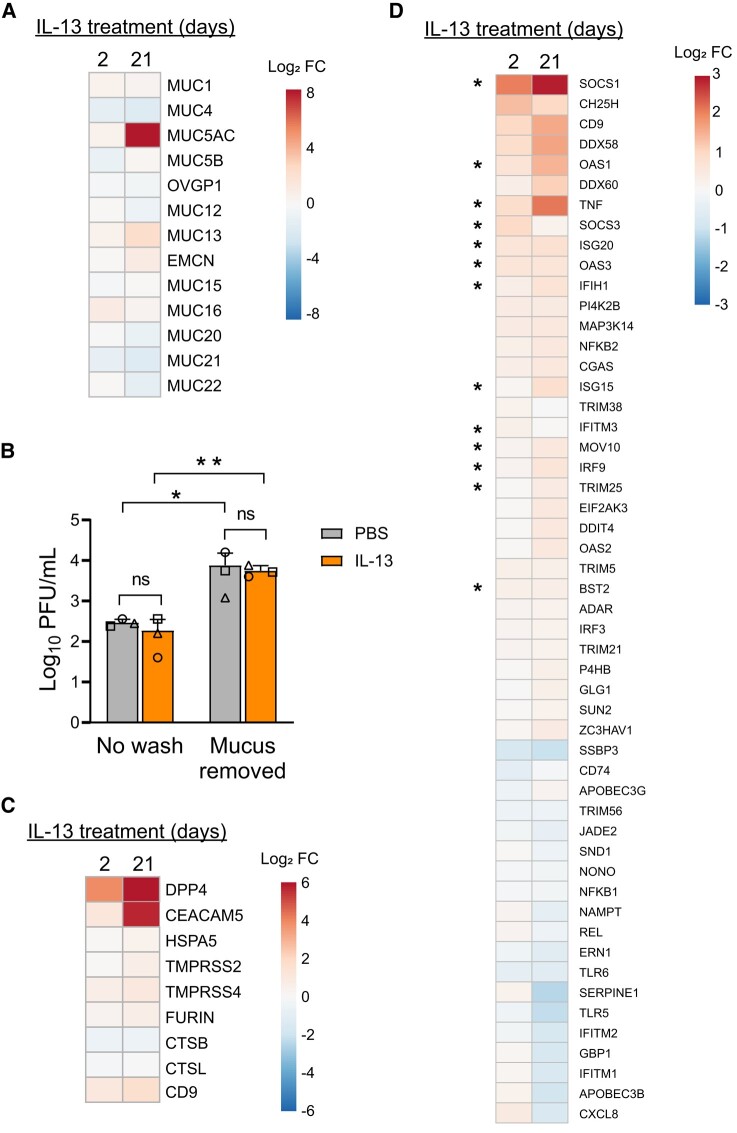
We were surprised that greater DPP4 abundance did not consistently lead to increased MERS-CoV infection. Previous MERS-CoV infection experiments in the HAE model suggest that it is unlikely that the DPP4 concentrations measured in the PBS- or IL-13–treated cultures are sufficient to saturate the virus under these infection conditions [20]. We thus considered the possibility that IL-13 treatment may produce additional changes that moderate the impact of raising receptor abundance. We interrogated a data set from a previous study in which HAE were treated with IL-13 for 2 or 21 days and transcriptional profiling was carried out by bulk RNAseq (GEO accession GSE240741). One prominent gene signature in the IL-13–treated epithelia was the up- and downregulation of mucin genes, particularly following the 21-day treatment. Most striking was the elevation in MUC5AC after 21 days of IL-13 exposure, accompanied by increased transcripts for MUC1, MUC13, MUC15, and MUC16 (summarized in the heat map in Figure 5A). These transcriptional changes are consistent with the increased dPAS staining and epithelial remodeling observed in cultured cells after 21 days of IL-13 stimulation (Figure 3A) and with earlier studies of Th2 cytokine–induced goblet cell metaplasia [30–32].
Figure 5.
IL-13 treatment alters expression of genes that influence MERS-CoV entry and infection, antiviral defense, and the mucin barrier. A, HAE (n = 8) were treated with PBS or IL-13 (20 ng/mL) for 2 or 21 days, and transcriptional profiling was carried out by bulk RNAseq. Displayed on the heat map are expression changes in all detected mucin genes after 2 or 21 days of IL-13 treatment. Expression levels are represented as log2 fold change. B, HAE (n = 3) were stimulated with PBS or IL-13 (20 ng/mL) for 21 days. Accumulated mucus on the apical surface was removed by rinsing with 10mM DTT (“Mucus removed”), while control cells were not rinsed (“No wash”). Epithelia were then infected with MERS-CoV (MOI, 0.5), and apical surface liquid was collected at 1 day postinfection for titering. Data are presented as mean ± SD. Log-transformed data were tested for statistically significant differences between conditions by 2-way analysis of variance, followed by a Sidak multiple-comparison test. *P < .05. **P < .01. C, Heat map depicts genes with known roles in MERS-CoV attachment/entry that were differentially expressed in IL-13–treated epithelia relative to PBS-treated cells (P < .01 for at least 1 treatment condition). D, Heat map summarizes interferon-stimulated genes that were up- or downregulated by IL-13 treatment in the RNAseq data set (P < .01 in at least 1 treatment condition). Asterisks denote genes that have been reported to be targets of STAT6 signaling. All heat maps were generated through ClustVis (https://biit.cs.ut.ee/clustvis/). DTT, dithiothreitol; FC, fold change; HAE, human airway epithelia; IL-13, interleukin 13; MOI, multiplicity of infection; ns, not significant; PBS, phosphate-buffered saline; PFU, plaque-forming units; RNAseq, RNA sequencing.
We hypothesized that this mucus layer might provide protection from MERS-CoV infection in the IL-13–treated epithelia by impeding access to DPP4 at the cell surface. To test this, epithelia were stimulated for 21 days with PBS or IL-13; then, cells were left unwashed or rinsed apically with dithiothreitol to remove accumulated mucus. After apical mucus removal, epithelia were infected with MERS-CoV, and apically shed virus was quantified at 1 day postinfection. As shown in Figure 5B, for the PBS- and IL-13–treated epithelia, titers were significantly higher at 1 day postinfection when the mucus layer was removed prior to infection, suggesting that airway mucus does provide some protection against MERS-CoV infection. However, as infection levels were similarly affected by mucus removal in the PBS- and IL-13–treated cells, it does not appear that the enhanced mucus layer in the IL-13–stimulated condition provides any additional benefit beyond this baseline level.
In our analysis of the RNAseq data set, we investigated transcriptional changes in genes and pathways known to be involved in viral infection and antiviral host defense. The heat map in Figure 5C displays genes with identified roles in MERS-CoV binding and entry that were differentially expressed in IL-13–treated epithelia relative to PBS-treated cells. In addition to DPP4, IL-13 treatment raised the transcript abundances for CEACAM5, HSPA5, TMPRSS2, TMPRSS4, FURIN, and CD9. While these changes suggest that IL-13 treatment should heighten susceptibility to MERS-CoV, we identified altered transcript levels for several IFN-stimulated genes (ISGs) and genes related to innate immune sensing and signaling that might be expected to affect viral permissivity (see heat map in Figure 5D). Reverse transcriptase–qualitative polymerase chain reaction was used to confirm the altered transcript abundances for a subset of these genes, as presented in Supplementary Figure 1. In the presence of IL-13, many ISGs increased: SOCS1, OAS1-3, ISG15, DDX58 (RIG-I), DDX60, IFIH1 (MDA5), IRF3, IRF9, CGAS, and NFKB2. Of note, IL-4 and IL-13 are known to exert their effects through a signal transduction pathway that utilizes STAT6 [33, 34]. Many of the genes that increased in response to IL-13 are known targets of STAT6 signaling (Figure 5D, asterisks), suggesting that IL-13 treatment influences the antiviral state by upregulating baseline levels of many STAT6-regulated antiviral molecules. These transcriptional changes were particularly pronounced with longer-term IL-13 exposure (21 days).
DISCUSSION
Underlying comorbidities such as chronic lung disease influence DPP4 expression in the respiratory tract and may affect individual MERS-CoV susceptibility. We used an in vitro model of well-differentiated HAE to investigate MERS-CoV infection in the setting of IL-13–induced inflammation. We found that DPP4 protein abundance and the number of DPP4-positive cells were reliably higher following IL-13 treatment on short (3 days) and longer (21 days) time scales. However, MERS-CoV replication was not universally increased in IL-13–treated cells. Instead, we observed that IL-13 treatment had variable and donor-dependent effects on viral replication early in the infection, and these differences did not translate to measurably different infection outcomes. These results indicate that increasing DPP4 abundance in airway epithelia is insufficient to enhance MERS-CoV infection in the setting of IL-13–induced inflammation.
We explored an RNAseq data set to ask whether global transcriptional changes induced by IL-13 exposure might contribute to these findings. This analysis suggested that IL-13 exposure changes the expression of MUC5AC and other mucin genes in ways that have the potential to affect mucus barrier function. There is debate about whether the increased presence of mucins is helpful or harmful in airway host defense, and it may be that the consequences of these changes are context and microbe dependent. For example, there is evidence that dysregulated mucin production contributes to impaired innate immunity in individuals with Th2 inflammation by reducing ciliary beat frequency and airway mucociliary transport [31, 32, 35]. However, during respiratory viral infections, an intact mucin barrier can help protect against infection by restricting viral access to cell surface receptors. A protective effect of airway mucus has been observed for another coronavirus, SARS-CoV-2 [36, 37], as well as for influenza [38]. Our results suggest that increased mucins at the apical surface in IL-13–treated epithelia is relatively neutral with respect to their effects on MERS-CoV. The extra mucus was not a barrier to MERS-CoV infection (Figure 5B), and we saw no evidence that the mucus layer prevented shedding of viral progeny in infected HAE cultures (Figure 4D). Thus, it seems unlikely that the relatively similar levels of MERS-CoV infection observed in PBS- and IL-13–treated epithelia can be explained by a protective effect from this enhanced mucus layer. It remains possible that Th2 cytokine–induced mucus production might influence MERS-CoV infection in an in vivo setting through effects on mucociliary clearance.
We noted that the expression of many ISGs and innate host defense factors was altered by IL-13–induced inflammation (Figure 5D). This observation is consistent with the concept established in the literature that chronic IL-13 exposure affects the “antiviral state” of airway epithelia. For instance, Jakiela et al also noted that baseline levels of ISGs were broadly higher in airway cells after IL-13 stimulation [39]. Jackson et al [31] similarly reported changes to the host defense repertoire of HAE in the setting of IL-13–induced inflammation and goblet cell metaplasia. The authors found that while chronic IL-13 exposure reduced the proportion of defensive secretory cells and caused a general downregulation of innate immune genes across most cell subsets, there was a subset with greater abundance of ISGs: IFI27, IFITM2, IFITM3, IFI6, IFIH1, IRF6, and DDX58. These findings suggest that chronic IL-13 exposure has complex and variable effects on innate immune gene expression that operate at the level of individual cells. In our study, it is unknown whether higher DPP4 expression and altered expression of antiviral genes occurred in distinct or overlapping cell populations and how these changes related to a given cell's infection status. It is difficult to predict whether the observed IL-13–induced transcriptional changes help or hinder the innate immune response to MERS-CoV; however, our results imply that the net effect of these changes may be to blunt the impact of greater DPP4 abundance by fortifying intracellular antiviral defenses.
Our results suggest that in people with airway disease characterized by Th2 inflammation, increased MERS-CoV risk cannot be attributed solely to upregulation of DPP4 and other factors required for viral attachment and entry on airway epithelia. Several alternative explanations have been proposed. One possibility is that drugs used to prevent or treat asthma exacerbations or COPD, which suppress inflammation and innate immunity, may compromise host immune responses to MERS-CoV infection. Additionally, there are reports that chronic Th2 cytokine exposure dampens IFN responses in airway epithelia. Reduced production of type I and type III IFNs was observed in response to viral infection in cells derived from people with asthma and COPD [40–42] and in airway epithelia from smokers [43]. There are also reports that IL-4/IL-13 treatment inhibited TLR3 expression and IRF3 signaling and blunted ISG expression in airway epithelia infected with rhinovirus [39, 44]. Similarly, when goblet cell metaplasia was modeled by FOXA3 overexpression in a human airway cell line, type I and type III IFN production in response to rhinovirus infection was suppressed [45]. We found that IL-13 exposure altered the baseline transcript abundances of several ISGs (Figure 5); however, we did not investigate whether IL-13 affects the epithelia's responsiveness to MERS-CoV infection—that is, whether IFNs and ISGs are more or less robustly induced upon infection with MERS-CoV in IL-13–treated cells.
One limitation of this study is that the HAE donors were not stratified by measures of health status, such as smoking or chronic lung disease; therefore, it is difficult to know whether and to what extent such underlying conditions might have influenced the cells’ responses to IL-13 stimulation or MERS-CoV infection. It will be important to perform studies in HAE derived from donors with asthma and/or COPD to further explore how the complex changes in these disease states affect DPP4 levels and responses to MERS-CoV infection. This study also does not address the effects of increasing DPP4 abundance in alveolar and other epithelia. In addition to its expression in secretory cells of the conducting airways, DPP4 is expressed in alveolar epithelial cells (type I and II cells) and endothelial cells [14, 46], and alveolar epithelial cells are an important site of MERS-CoV infection. Alveolar epithelia may respond to Th2 inflammation in ways that are distinct from airway-derived epithelia, perhaps resulting in fewer compensatory changes in innate immunity to overcome the effects of higher viral receptor availability. We further acknowledge that cultured epithelia do not fully capture the in vivo complexity of the Th2 phenotype, which involves interactions between epithelia and immune cells (many of which express DPP4) in multiple organs and tissue types. In related work with the coronavirus SARS-CoV-2, human clinical studies and experiments in mice suggest that systemic effects arising from IL-13–mediated inflammation are important determinants of disease severity [47–50].
Our findings indicate that the increased risk for severe MERS outcomes in the setting of IL-13–induced inflammation is more complex than simply greater DPP4 abundance. The potential effects of increased DPP4 abundance appear to be counteracted by other changes arising from chronic Th2 cytokine exposure that may limit viral permissivity. Importantly, our findings point to factors not present in the in vitro HAE cell culture model—including immune and other cell types, as well as systemic effects—as potentially more significant contributors to the increased MERS severity in people with underlying comorbidities. Future studies involving animal models will be helpful in exploring these complex virus-host interactions during MERS-CoV infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Kun Li, Department of Pediatrics, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Jennifer A Bartlett, Department of Pediatrics, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Christine L Wohlford-Lenane, Department of Pediatrics, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Biyun Xue, Department of Internal Medicine, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Andrew L Thurman, Department of Internal Medicine, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Thomas M Gallagher, Department of Microbiology and Immunology, Loyola University Chicago, Maywood, IL.
Alejandro A Pezzulo, Department of Internal Medicine, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Paul B McCray, Jr, Department of Pediatrics, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA; Department of Microbiology, Pappajohn Biomedical Institute, Carver College of Medicine, University of Iowa, Iowa City, IA.
Notes
Acknowledgments. We thank Patrick Ten Eyck in the Biostatistics, Epidemiology, and Research Design Core in the Institute for Clinical and Translational Science at the University of Iowa for assistance with statistical analysis for this study. We also acknowledge valuable technical support from the University of Iowa Cells and Tissue Core, part of the Center for Gene Therapy of Cystic Fibrosis.
Financial support. This work was supported by the National Institutes of Health (P01 AI060699 to T. M. G. and P. B. M., R01 HL163024 and K01 HL140261 to A. A. P.). The Center for Gene Therapy of Cystic Fibrosis is supported by the National Institutes of Health (P30 DK054759) and the Cystic Fibrosis Foundation. Support was also provided by the Cystic Fibrosis Foundation (PEZZUL20A1-KB to A. A. P.). P. B. M. is supported by the Roy J. Carver Charitable Trust.
References
- 1. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–20. [DOI] [PubMed] [Google Scholar]
- 2. Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med 2014; 160:389–97. [DOI] [PubMed] [Google Scholar]
- 3. Assiri A, McGeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med 2013; 369:407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alqahtani FY, Aleanizy FS, Ali El Hadi Mohamed R, et al. Prevalence of comorbidities in cases of Middle East respiratory syndrome coronavirus: a retrospective study. Epidemiol Infect 2018; 147:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hui DS, Azhar EI, Kim YJ, Memish ZA, Oh MD, Zumla A. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis 2018; 18:e217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mobaraki K, Ahmadzadeh J. Current epidemiological status of Middle East respiratory syndrome coronavirus in the world from 1.1.2017 to 17.1.2018: a cross-sectional study. BMC Infect Dis 2019; 19:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang YM, Hsu CY, Lai CC, et al. Impact of comorbidity on fatality rate of patients with Middle East respiratory syndrome. Sci Rep 2017; 7:11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus–EMC. Nature 2013; 495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen X. Biochemical properties of recombinant prolyl dipeptidases DPP-IV and DPP8. Adv Exp Med Biol 2006; 575:27–32. [DOI] [PubMed] [Google Scholar]
- 10. Lambeir AM, Durinx C, Scharpe S, De Meester I. Dipeptidyl-peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit Rev Clin Lab Sci 2003; 40:209–94. [DOI] [PubMed] [Google Scholar]
- 11. Mentlein R. Dipeptidyl-peptidase IV (CD26)—role in the inactivation of regulatory peptides. Regul Pept 1999; 85:9–24. [DOI] [PubMed] [Google Scholar]
- 12. Rohrborn D, Wronkowitz N, Eckel J. DPP4 in diabetes. Front Immunol 2015; 6:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiobara T, Chibana K, Watanabe T, et al. Dipeptidyl peptidase-4 is highly expressed in bronchial epithelial cells of untreated asthma and it increases cell proliferation along with fibronectin production in airway constitutive cells. Respir Res 2016; 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meyerholz DK, Lambertz AM, McCray PB Jr. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol 2016; 186:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seys LJM, Widagdo W, Verhamme FM, et al. DPP4, the Middle East respiratory syndrome coronavirus receptor, is upregulated in lungs of smokers and chronic obstructive pulmonary disease patients. Clin Infect Dis 2018; 66:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karp PH, Moninger TO, Weber SP, et al. An in vitro model of differentiated human airway epithelia: methods for establishing primary cultures. In: Wise C, ed. Epithelial cell culture protocols. Totowa: Humana Press, 2002:115–37. [DOI] [PubMed] [Google Scholar]
- 17. Bauvois B, Djavaheri-Mergny M, Rouillard D, Dumont J, Wietzerbin J. Regulation of CD26/DPPIV gene expression by interferons and retinoic acid in tumor B cells. Oncogene 2000; 19:265–72. [DOI] [PubMed] [Google Scholar]
- 18. Nemoto E, Sugawara S, Takada H, Shoji S, Horiuch H. Increase of CD26/dipeptidyl peptidase IV expression on human gingival fibroblasts upon stimulation with cytokines and bacterial components. Infect Immun 1999; 67:6225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silva AP, Cavadas C, Baisse-Agushi B, Spertini O, Brunner HR, Grouzmann E. NPY, NPY receptors, and DPP IV activity are modulated by LPS, TNF-alpha and IFN-gamma in HUVEC. Regul Pept 2003; 116:71–9. [DOI] [PubMed] [Google Scholar]
- 20. Li K, Wohlford-Lenane C, Bartlett JA, McCray PB Jr. Inter-individual variation in receptor expression influences MERS-CoV infection and immune responses in airway epithelia. Front Public Health 2021; 9:756049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caminati M, Pham DL, Bagnasco D, Canonica GW. Type 2 immunity in asthma. World Allergy Organ J 2018; 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fahy JV. Type 2 inflammation in asthma—present in most, absent in many. Nat Rev Immunol 2015; 15:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grunig G, Warnock M, Wakil AE, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science 1998; 282:2261–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hershey GK. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol 2003; 111:677–90. [DOI] [PubMed] [Google Scholar]
- 25. Kuperman DA, Huang X, Koth LL, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med 2002; 8:885–9. [DOI] [PubMed] [Google Scholar]
- 26. Wills-Karp M, Luyimbazi J, Xu X, et al. Interleukin-13: central mediator of allergic asthma. Science 1998; 282:2258–61. [DOI] [PubMed] [Google Scholar]
- 27. Kehlen A, Gohring B, Langner J, Riemann D. Regulation of the expression of aminopeptidase A, aminopeptidase N/CD13 and dipeptidylpeptidase IV/CD26 in renal carcinoma cells and renal tubular epithelial cells by cytokines and cAMP-increasing mediators. Clin Exp Immunol 1998; 111:435–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riemann D, Kehlen A, Langner J. Stimulation of the expression and the enzyme activity of aminopeptidase N/CD13 and dipeptidylpeptidase IV/CD26 on human renal cell carcinoma cells and renal tubular epithelial cells by T cell-derived cytokines, such as IL-4 and IL-13. Clin Exp Immunol 1995; 100:277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Majdoul S, Compton AA. Lessons in self-defence: inhibition of virus entry by intrinsic immunity. Nat Rev Immunol 2022; 22:339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pezzulo AA, Tudas RA, Stewart CG, et al. HSP90 inhibitor geldanamycin reverts IL-13- and IL-17-induced airway goblet cell metaplasia. J Clin Invest 2019; 129:744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jackson ND, Everman JL, Chioccioli M, et al. Single-cell and population transcriptomics reveal pan-epithelial remodeling in type 2-high asthma. Cell Rep 2020; 32:107872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrison CB, Edwards CE, Shaffer KM, et al. SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc Natl Acad Sci U S A 2022; 119:e2119680119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 1996; 4:313–9. [DOI] [PubMed] [Google Scholar]
- 34. Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol 1996; 157:3220–2. [PubMed] [Google Scholar]
- 35. Laoukili J, Perret E, Willems T, et al. IL-13 alters mucociliary differentiation and ciliary beating of human respiratory epithelial cells. J Clin Invest 2001; 108:1817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonser LR, Eckalbar WL, Rodriguez L, et al. The type 2 asthma mediator IL-13 inhibits severe acute respiratory syndrome coronavirus 2 infection of bronchial epithelium. Am J Respir Cell Mol Biol 2022; 66:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chatterjee M, Huang LZX, Mykytyn AZ, et al. Glycosylated extracellular mucin domains protect against SARS-CoV-2 infection at the respiratory surface. PLoS Pathog 2023; 19:e1011571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cohen M, Zhang XQ, Senaati HP, et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 2013; 10:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jakiela B, Rebane A, Soja J, et al. Remodeling of bronchial epithelium caused by asthmatic inflammation affects its response to rhinovirus infection. Sci Rep 2021; 11:12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Contoli M, Message SD, Laza-Stanca V, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med 2006; 12:1023–6. [DOI] [PubMed] [Google Scholar]
- 41. Mallia P, Message SD, Gielen V, et al. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am J Respir Crit Care Med 2011; 183:734–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wark PA, Johnston SL, Bucchieri F, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005; 201:937–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu W, Zhang W, Booth JL, et al. Human primary airway epithelial cells isolated from active smokers have epigenetically impaired antiviral responses. Respir Res 2016; 17:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Contoli M, Ito K, Padovani A, et al. Th2 cytokines impair innate immune responses to rhinovirus in respiratory epithelial cells. Allergy 2015; 70:910–20. [DOI] [PubMed] [Google Scholar]
- 45. Chen G, Korfhagen TR, Karp CL, et al. Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am J Respir Crit Care Med 2014; 189:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Widagdo W, Raj VS, Schipper D, et al. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J Virol 2016; 90:4838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chhiba KD, Patel GB, Vu THT, et al. Prevalence and characterization of asthma in hospitalized and nonhospitalized patients with COVID-19. J Allergy Clin Immunol 2020; 146:307–14.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Donlan AN, Sutherland TE, Marie C, et al. IL-13 is a driver of COVID-19 severity. JCI Insight 2021; 6:e150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramakrishnan RK, Al Heialy S, Hamid Q. Implications of preexisting asthma on COVID-19 pathogenesis. Am J Physiol Lung Cell Mol Physiol 2021; 320:L880–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu Z, Hasegawa K, Ma B, Fujiogi M, Camargo CA Jr, Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J Allergy Clin Immunol 2020; 146:327–9.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.