Abstract
PURPOSE
Family history (FH) and pathogenic variants (PVs) are used for guiding risk surveillance in selected high-risk women but little is known about their impact for breast cancer screening on population level. In addition, polygenic risk scores (PRSs) have been shown to efficiently stratify breast cancer risk through combining information about common genetic factors into one measure.
METHODS
In longitudinal real-life data, we evaluate PRS, FH, and PVs for stratified screening. Using FinnGen (N = 117,252), linked to the Mass Screening Registry for breast cancer (1992-2019; nationwide organized biennial screening for age 50-69 years), we assessed the screening performance of a breast cancer PRS and compared its performance with FH of breast cancer and PVs in moderate- (CHEK2)- to high-risk (PALB2) susceptibility genes.
RESULTS
Effect sizes for FH, PVs, and high PRS (>90th percentile) were comparable in screening-aged women, with similar implications for shifting age at screening onset. A high PRS identified women more likely to be diagnosed with breast cancer after a positive screening finding (positive predictive value [PPV], 39.5% [95% CI, 37.6 to 41.5]). Combinations of risk factors increased the PPVs up to 45% to 50%. A high PRS conferred an elevated risk of interval breast cancer (hazard ratio [HR], 2.78 [95% CI, 2.00 to 3.86] at age 50 years; HR, 2.48 [95% CI, 1.67 to 3.70] at age 60 years), and women with a low PRS (<10th percentile) had a low risk for both interval- and screen-detected breast cancers.
CONCLUSION
Using real-life screening data, this study demonstrates the effectiveness of a breast cancer PRS for risk stratification, alone and combined with FH and PVs. Further research is required to evaluate their impact in a prospective risk-stratified screening program, including cost-effectiveness.
Polygenic risk provides a tool for risk-stratified breast cancer screening.
INTRODUCTION
Organized population-based screening has a prominent role in early detection of breast cancer in many countries, and reduction of breast cancer mortality has followed adoption of such screening programmes.1,2 Yet, the programs have also generated much controversy around the balance of benefits and harms, particularly regarding the age of initiation and screening interval. Instead of the one-size-fits-all regimen, increasing evidence suggests that cost-efficiency and the benefit-harm balance could be improved by risk-tailored screening, giving the opportunity to personalize the start and stop ages, and the screening interval.3 Such risk-tailored surveillance has long been used for specific subgroups, such as carriers of pathogenic variants (PVs) in moderate- or high-risk breast cancer susceptibility genes (eg, BRCA1 and PALB2), and with accumulation of early-onset breast cancer in the family.4,5
CONTEXT
Key Objective
The current approach to population breast cancer screening uses a one-size-fits-all regimen, yet studies on inherited risk factors, including polygenic risk scores (PRS) for breast cancer, family history, and pathogenic variants (PVs) in susceptibility genes, suggest the potential for personalized screening on the basis of individual risk profiles. How do such inherited risk factors, particularly the PRS, which is a more recently identified risk factor, perform in real-life screening data?
Knowledge Generated
The breast cancer PRS served for risk stratification of breast cancer screening both alone and combined with FH and PVs. A high PRS correlated with a high positive predictive value for breast cancer screening and conferred an elevated risk for interval breast cancer.
Relevance (G. Fleming)
-
Future trials attempting to personalize type and schedule of breast cancer screening should strongly consider incorporation of a PRS along with other risk factors.*
*Relevance section written by JCO Associate Editor Gini Fleming, MD.
In addition to moderate- and high-risk variants in susceptibility genes, breast cancer has a polygenic inheritance where many common variants across the genome jointly contribute to disease risk. Polygenic risk scores (PRS) combine such information into a single metric of inherited disease susceptibility.6 Compared with individuals with an average breast cancer PRS, a high PRS confers an up to three- to five-fold risk increase to breast cancer, with a lifetime risk of over 30%.7,8 The breast cancer PRS also considerably modifies the risk conferred by moderate- and high-risk variants.9,10 Previous studies show that breast cancer PRS offers opportunities for risk-tailored surveillance,7,8 but there is limited evidence on impact of PRSs for identifying high-risk women for stratified screening on the basis of population-based screening data.
In Finland, biennial screening is offered free for all women between age 50 and 69 years through a nationwide screening program. We estimate the impact of PRS in this screening setting, comparing the effect of PRS in risk stratification to family history (FH) and known moderate- to high-risk PVs in breast cancer susceptibility genes. FinnGen combines genomewide genotyping to nationwide health registries, including the Mass Screening Registry, allowing assessment of PRS in the breast cancer screening context starting from the initiation of a nationwide breast cancer screening program in 1992.11
METHODS
Patients and Outcomes
The FinnGen study, a collection of Finnish prospective epidemiologic cohorts, disease-based cohorts, and hospital biobank collections linked to nationwide health registries, has been previously described.12 A detailed ethics statement is provided in the Data Supplement (online only). In Finland, the population-based breast cancer screening program is managed by local authorities, and all screenings are monitored through the Mass Screening Registry for breast cancer, maintained by the Finnish Cancer Registry (FCR). We used FinnGen Data Freeze 9, studying women free from breast cancer in the beginning of 1992 with ≥1 screening invitation between 1992 and 2019 within the Mass Screening Registry. We identified the breast cancer cases from the FCR (available since 1953; nationwide completeness of breast cancer at 99.5%13) with International Classification of Diseases for Oncology (ICD-O-3 C50*), and from the nationwide death registry with ICD-10 C50* (available since 1969).
In Finland, biennial mammographic screening is offered to all women between age 50 and 69 years. Breast cancer cases were classified into the following three categories, following previous classification11: (1) nonattendees—nonparticipation in the previous screening, (2) screen-detected—a positive screening result (malignant histologic finding assigned to follow-up examinations or surgery) and a breast cancer diagnosis within 6 months after screening, and (3) interval—negative result in the previous screening or diagnosis later than 6 months from the screening. Our category frequencies are in line with age-matched proportions from nationwide data from the Mass Screening Register (Data Supplement, Table S1). Positive predictive value (PPV) was defined as the proportion of women diagnosed with breast cancer out of women with a positive screening finding, defined as women called back for a complementary follow-up examination (ultrasound with or without biopsy or additional imaging; most being biopsies) or surgery because of an abnormal screening mammogram.
PRS, PVs, and FH
We used a previously published genomewide breast cancer PRS,10 which has shown similar performance in another study.14 In short, the PRS was built with the software PRS-CS which applies continuous shrinkage (CS) priors, using summary statistics from a large genomewide association study independent of FinnGen. The PRS consists of 1,079,089 variants (PGS Catalog ID PGS000335). The PRS was studied either as a (1) continuous variable (per standard deviation [SD] increment), (2) by deciles, or (3) divided into three categories, with high PRS defined as the top decile of the PRS distribution (>90th percentile), low PRS as the bottom decile (<10th), with average risk (10th-90th) as reference to contrast the effect sizes to the average population.
The PV carriers were identified from the genotypes, and we studied three variants: CHEK2 c.1100delC, CHEK2 c.319+2T>A, and PALB2 c.1592delT, which are, respectively, 3.7-, 19.7-, and 242-fold enriched in the Finnish population compared with Non-Finnish-Swedish-Estonian Europeans.15 The CHEK2 variants are considered moderate-risk variants in Finland and the PALB2 a high-risk variant. These represent the most frequent breast cancer susceptibility genes within Finland, and the variants are identified with high quality on the genotyping array used in FinnGen. Variants were analyzed jointly to increase power. We used a registry-based composite end point for FH, identified through (1) parental causes of death from the death registry, (2) study participants' ICD-10 diagnoses denoting FH, and (3) by identifying breast cancers of the study participants' first-degree relatives included in FinnGen. Further information on genotyping, imputation, PRS, PVs, and FH is provided in the Data Supplement.
Statistical Analysis
For the general performance of PRS across the data, the start of the follow-up was set to birth, with follow-up ending at diagnosis of invasive breast cancer on in situ lesions, death, or on December 31, 2019. Hazard ratios (HRs) and their 95% CIs were estimated with Cox proportional hazards model implemented in survival package in R. Regression models were adjusted with birth year, genotyping array, subcohort, and the first 10 principal components of genetic ancestry. Statistical analyses (Data Supplement) were performed with R 4.1.2.
RESULTS
The current target age of the national screening program in Finland is biennial screening between age 50 and 69 years. Among all 117,252 women invited for a breast cancer screen, we first studied their overall effects on breast cancer risk before, during, and after screening age, for evaluating the performance of PRS compared with PVs and FH in each age group (Fig 1). We then evaluated the impact of the risk factors on screening events and metrics. Among the 117,252 women, we observed 11,556 breast cancer cases, of which 10,570 (91.6%) were invasive and 974 (8.4%) were in situ breast cancers (12 with missing information). One thousand four hundred fifty-three with prevalent breast cancer before the screening start were excluded from analyses on women of screening age.
FIG 1.
Study overview. aMost frequent susceptibility genes in Finland (CHEK2 and PALB2).
The three variants were analyzed jointly (2,437 mutation carriers, 2.1%; CHEK2 c.1100delC 1.6%, CHEK2 c.319+2T>A 0.2%, and PALB2 c.1592delT 0.3%). FH of breast cancer was defined through health care registries on the basis of parental causes of death, first-degree relatives in FinnGen diagnosed with breast cancer, or an ICD-10 diagnosis for FH (Methods).
Stratifying the three risk factors into three age groups (Table 1), the HRs of PRS, PVs, and FH decreased with increasing age, and the highest effect sizes were observed in the group before screening age. Similar patterns and effect sizes were observed for all three risk factors, with high PRS (>90th percentile of the PRS distribution) being the most common risk factor. Effect sizes by PRS decile are provided in the Data Supplement (Table S2).
TABLE 1.
Study Characteristics and Associations Between Breast Cancer and PRS, PVs, and FH
| Category | Before Screening Age | During Screening Age | After Screening Age |
|---|---|---|---|
| Any breast cancer, No. | 1,453 | 7,905 | 2,198 |
| Invasive breast cancer, No. | 1,377 | 7,145 | 2,058 |
| In situ breast cancer, No. | 74 | 760 | 140 |
| Bilateral breast cancer, No. | 20 | 96 | 36 |
| Age at disease onset, years, median (IQR) | 45.9 (42.9-47.9) | 59.1 (54.2-64.1) | 73.5 (70.1-76.7) |
| PRS >90% in cases, No. (%) | 341 (23.5) | 1,663 (21.0) | 404 (18.4) |
| PRS >90% in controls, No. (%) | 11,210 (9.9) | 9,547 (8.8) | 3,652 (8.5) |
| PV carriers in cases, No. (%) | 94 (6.5) | 345 (4.4) | 73 (3.3) |
| PV carriers in controls, No. (%) | 2,343 (2.0) | 1,998 (1.9) | 740 (1.7) |
| Positive FH in cases, No. (%) | 107 (7.4) | 489 (6.2) | 53 (2.4) |
| Positive FH in controls, No. (%) | 3,605 (31) | 3,116 (2.9) | 865 (2.0) |
| HR (95% CI) for PRS, continuous | |||
| Any breast cancer | 1.78 (1.69 to 1.87) | 1.66 (1.63 to 1.70) | 1.63 (1.56 to 1.70) |
| Invasive breast cancer | 1.75 (1.66 to 1.84) | 1.67 (1.63 to 1.71) | 1.64 (1.57 to 1.71) |
| In situ breast cancer | 2.39 (1.91 to 3.00) | 1.73 (1.61 to 1.86) | 1.58 (1.33 to 1.87) |
| Bilateral breast cancer | — | 2.49 (2.03 to 3.05) | 2.75 (1.96 to 3.85) |
| HR (95% CI) for PRS, PRS >90% v 10%-90% | |||
| Any breast cancer | 2.50 (2.21 to 2.83) | 2.38 (2.25 to 2.51) | 2.11 (1.90 to 2.36) |
| Invasive breast cancer | 2.45 (2.16 to 2.79) | 2.40 (2.26 to 2.54) | 2.07 (1.85 to 2.31) |
| In situ breast cancer | 3.62 (2.20 to 5.94) | 2.51 (2.10 to 2.99) | 3.14 (2.10 to 4.68) |
| Bilateral breast cancer | — | 4.71 (3.07 to 7.23) | — |
| HR (95% CI) for PVs | |||
| Any breast cancer | 3.13 (2.53 to 3.86) | 2.30 (2.06 to 2.56) | 1.95 (1.54 to 2.47) |
| Invasive breast cancer | 3.12 (2.51 to 3.87) | 2.31 (2.06 to 2.59) | 1.93 (1.51 to 2.47) |
| In situ breast cancer | — | 2.43 (1.71 to 3.45) | — |
| Bilateral breast cancer | — | 4.37 (2.01 to 9.51) | — |
| HR (95% CI) for FH | |||
| Any breast cancer | 1.97 (1.62 to 2.40) | 1.96 (1.79 to 2.15) | 1.68 (1.28 to 2.21) |
| Invasive breast cancer | 2.00 (1.64 to 2.46) | 2.02 (1.83 to 2.22) | 1.72 (1.30 to 2.27) |
| In situ breast cancer | — | 1.62 (1.17 to 2.25) | — |
| Bilateral breast cancer | — | 5.04 (2.74 to 9.29) | — |
NOTE. Cells containing — were not assessed because of small case counts. PVs: CHEK2 c.1100delC, CHEK2 c.319+2T>A, PALB2 c.1592delT, analyzed jointly for power, and heterozygotes were considered jointly with homozygotes. Of the 152 bilateral breast cancers, 144 were invasive breast cancers and eight were in situ cancers (on the basis of the most severe lesion). Twelve individuals had missing data on information about in situ versus invasive breast cancer. For before screening age (age <49 years), women diagnosed during or after screening age were considered as controls. For during screening age, women diagnosed after screening age were considered as controls, and cases diagnosed before screening age were excluded. For after screening age (age >71 years), women diagnosed before or during screening age were excluded. Analyses for in situ breast cancer excluded invasive breast cancer cases and vice versa. Similarly, analyses on bilateral breast cancer excluded nonbilateral breast cancer cases.
Abbreviations: FH, family history; HR, hazard ratio; PRS, polygenic risk score; PV, pathogenic variant.
Figure 2 and the Data Supplement (Fig S1) show lifetime risks of breast cancer for the risk factors and their combinations, and indicate when each group reaches a 2% cumulative incidence, aligning with the population's average at the start of organized screening at age 50 years. This age provides a guideline for when to begin screening on the basis of individual risk. Groups of high PRS, PV carriers, or positive FH reached this 2% prevalence at age 42 years, while women with high PRS and FH or PV reached it at age 38 years. By contrast, women without PVs or FH who have a low PRS reached a similar risk level two decades later, at age 62 years. The estimates are calibrated to the general population (see the Data Supplement for details).
FIG 2.
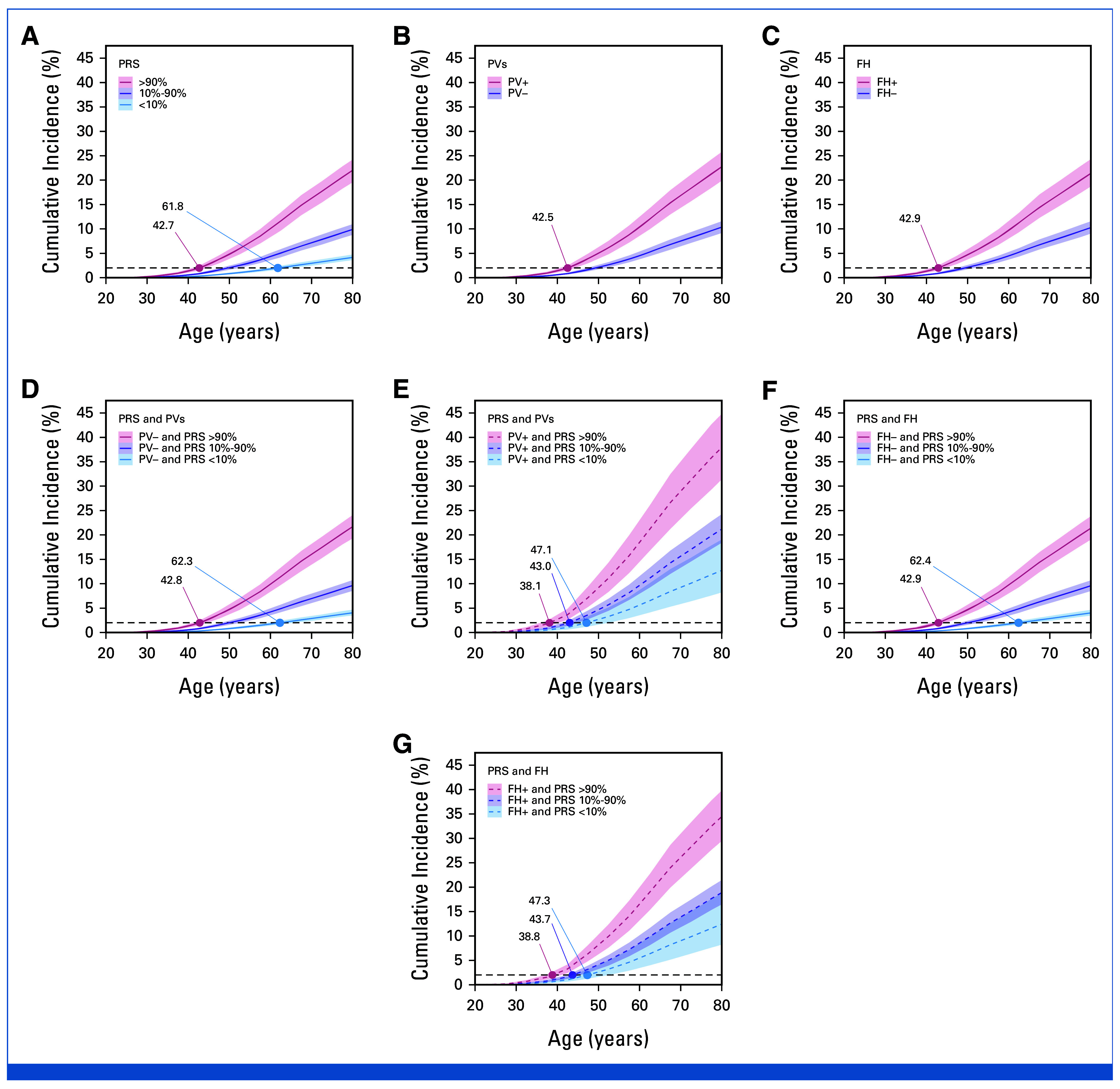
Lifetime risk of breast cancer for categories of (A) PRS, by carrier status for (B) PV, (C) FH status, (D and E) the combination of PRS and PVs, and (F and G) the combination of PRS and FH. The numbers indicate when each category reaches a 2% cumulative incidence, aligning with the population's average at the start of organized screening at age 50 years. The cumulative incidences by PRS decile are provided in the Data Supplement (Fig S1). The estimates are calibrated to the general population, with the estimates accounting for competing risks of death from other causes than breast cancer (see the Data Supplement for details). FH, family history; PRS, polygenic risk score; PV, pathogenic variant.
The screening events and metrics were studied in 115,799 individuals (excluding 1,453 women with breast cancer before first screening invitation). Of the 7,905 breast cancers diagnosed during the screening age window, 4,691 (59.3% of all) were screen-detected breast cancers (of which 11.8% in situs), 1,880 (23.8% of all) were interval breast cancers (7.0% in situs), and 1,334 (16.9% of all) were nonattendee breast cancers (nonparticipation in previous screening, 5.6% in situs). The overall screening participation rate for the 693,730 screening invitations was 88.5%. One hundred thirteen thousand nine hundred sixty-nine (97.0%) women had participated at least once in a screening (mean 5.4 screenings, SD 2.8, mean age at individual screenings 57.3 years, SD 4.9).
PPV
For each of the three risk factors, we assessed the PPV (proportion of women diagnosed with breast cancer out of women with a positive screening finding, defined as women called back for a complementary follow-up examination for an abnormal screening mammogram; see Methods for details). Of all screening events, 3.1% of women were referred for follow-up examinations, of whom 25.0% were diagnosed with breast cancer (0.8% of all screenings).
The PPVs increased considerably as a function of the PRS (Fig 3A), ranging from 12.7% (95% CI, 11.0 to 14.6) in the lowest PRS decile to 39.5% (95% CI, 37.6 to 41.5) in the highest decile. Similarly, being a PV carrier or having positive FH both increased the PPV (Figs 3B and 3C), with the PPVs slightly lower than what we observed for the highest PRS decile (35.9% [95% CI, 31.7 to 40.2%] for PV carriers; 35.5% [95% CI, 32.1 to 38.9] for positive FH). Combinations of PRS with PVs or FH risk led to the highest PPVs (Figs 3G and 3H). Women with positive FH and high PRS (>90%) had a PPV of 44.6% (95% CI, 37.2 to 52.3) and PV carriers with high PRS had an even higher PPV at 50.6% (95% CI, 39.6 to 61.5). In individuals with low PRS, FH and PVs had a particularly strong impact, although with wide CIs. As baseline risk increases with age, we also assessed the risk factors by screening age and observed clear PPV trends by age (Fig 3D-3F). Detailed PPVs, and PPVs by PRS decile for all ages are provided in the Data Supplement (Table S3 and Fig S2, respectively).
FIG 3.
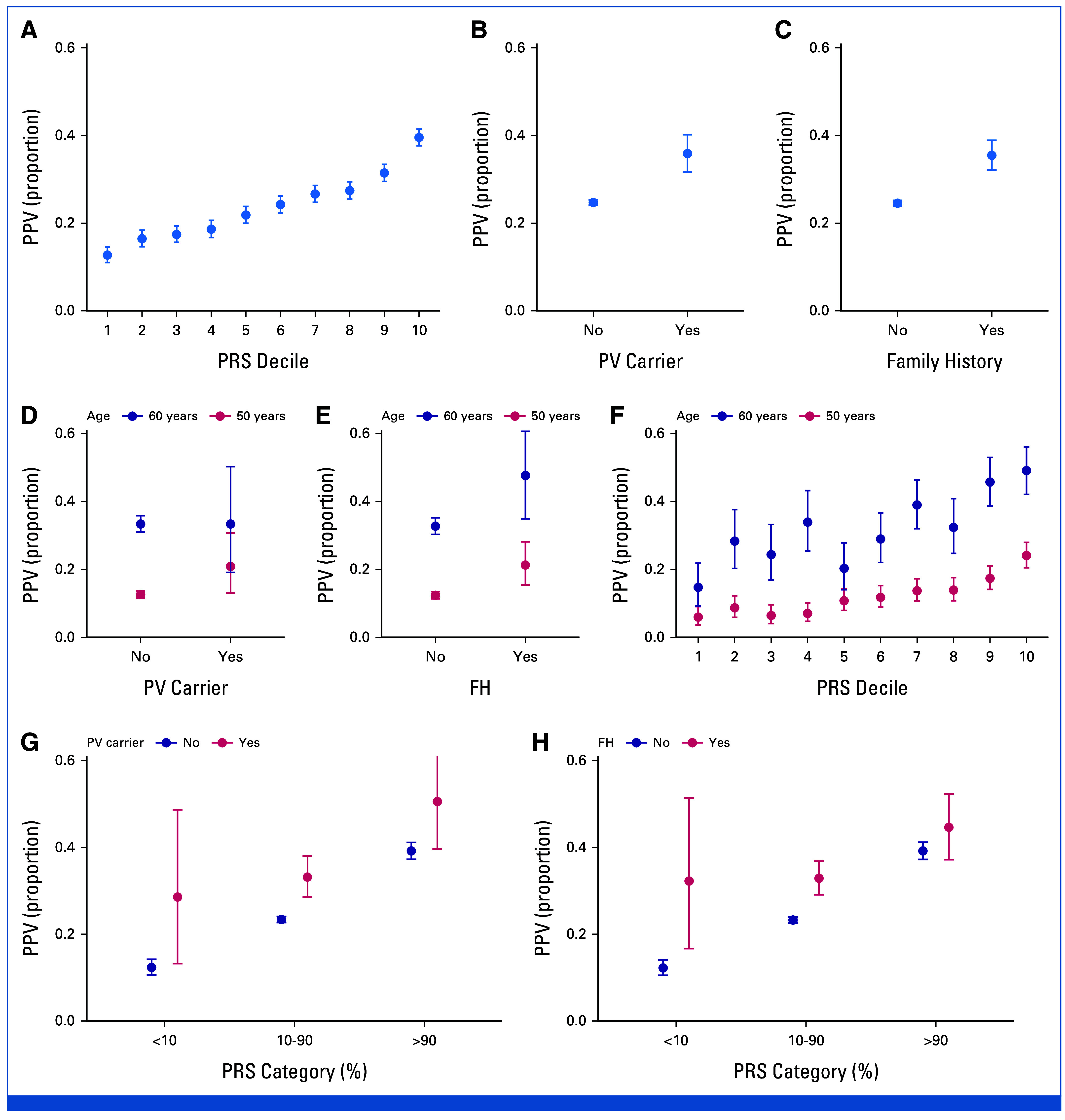
Observed PPV (proportion of screen-detected cancers among women with a screening finding defined as women called back for complementary follow-up examination for an abnormal screening mammogram; see Methods for details) with different risk factors. The PPV (A) by PRS decile, (B) by PV carrier status, and (C) by FH status, with corresponding PPVs for two screening ages, (D-F) 50 and 60 years. (D-F) Also highlight the impact of age on PPV, because of differences in baseline risk of breast cancer, which increases with age. (G and H) The PPV by combinations of (G) PRS and PV carrier and (H) PRS and FH status, with PRS divided into three categories. PPVs at all screening ages (biennially from age 50 years to 69 years) are provided in the Data Supplement (Fig S2), and the detailed numbers for the Figure are provided in the Data Supplement (Table S1). Error bars show the 95% CIs. PVs: CHEK2 c.1100delC, CHEK2 c.319+2T>A, PALB2 c.1592delT, analyzed jointly for power, and heterozygotes were considered jointly with homozygotes. FH, family history; PPV, positive predictive value; PRS, polygenic risk score; PV, pathogenic variant.
Mammography Screening Findings
We first assessed the proportion of cases in each category by PRS decile (Fig 4A). By definition, each PRS decile contains 10% of individuals, whereby proportions over 10% indicate enrichment of a screening finding. The proportion of cases was evenly distributed across PRS deciles for no findings and for benign breast lesions, whereas both in situ lesions and invasive breast cancers were in a similar manner enriched for high PRS. The highest enrichment was observed for high PRS for bilateral breast cancer, where 35.2% (95% CI, 27.4 to 43.5) of the cases had a high PRS (>90th percentile).
FIG 4.

Proportion of individuals with risk factors by type of most severe screening finding. Women with no finding have never had a positive screening finding. (A) Prevalence of women in different PRS deciles by screening finding. Each PRS decile contains by definition 10% of individuals (dotted line), whereby proportions over 10% indicate enrichment of the histologic finding. For instance, of women with bilateral breast cancer, 35.2% (95% CI, 27.4 to 43.5) had a PRS in the top decile. (B) Prevalence of each risk factor (PRS category, PV carrier, and family history) in women by screening finding. The PRS >90% contains the same information as the top deciles for each screening finding in (A). In addition to the final breast cancer diagnosis available in the Finnish Cancer Registry, information on histology from screening biopsies is available within the Mass Screening Registry, on the basis of ICD-O-3 morphology codes. The screening findings were divided into the following categories, with each woman classified on the basis of her most severe screening finding: (1) no finding (ie, no suspicion of malignancy, N = 110,185), (2) benign (nonmalignant) breast lesion (N = 758), (3) in situ lesion (N = 613), (4) unilateral invasive breast cancer (N = 4,098), and (5) bilateral breast cancer (N = 145; in situ or invasive lesions simultaneously in both breasts). Because of a small case count, the prevalence of PRS deciles one to three for bilateral breast cancer was calculated by pooling their prevalence and dividing it by three. Error bars show the 95% CIs. The P value for trend was calculated by randomly sampling 1,000 individuals in screening finding categories that had over 1,000 individuals (the categories were no finding and unilateral invasive breast cancer). PVs: CHEK2 c.1100delC, CHEK2 c.319+2T>A, PALB2 c.1592delT, analyzed jointly for power, and heterozygotes were considered jointly with homozygotes. ICD-O, International Classification of Diseases for Oncology; PRS, polygenic risk score; PV, pathogenic variant.
Next, we assessed the prevalence of high PRS, PVs, and FH by the most severe screening finding (Fig 4B). Being a PV carrier or having positive FH was less common than a high PRS, and similar to PRS, the risk factors were not enriched in women with benign breast lesions, and they did not distinguish women with situ lesions and invasive breast cancer. Similar to PRS, positive FH was enriched in women with bilateral breast cancer, with 10.3% (95% CI, 5.9 to 16.5) of the cases having positive FH.
Impact of Polygenic Risk on Screen- and Interval-Detected Breast Cancers
In women with high PRS, the effect size for screen-detected and interval breast cancers was similar: at the screening at age 50 years, the HR was 3.09 (95% CI, 2.38 to 4.03) for screen-detected and HR 2.78 (95% CI, 2.00 to 3.86) for interval breast cancer, with no clear trends for effect size by age or systematic differential effects by detection type (Data Supplement, Table S4). Women with a high PRS with a negative screen had an elevated risk for interval breast cancer, and a screen-detected cancer in the next screen (Fig 5, Data Supplement, Fig S3). Cumulative incidences for interval breast cancer by 1 year and 2 years after the negative screen, and for screen-detected breast cancer are provided in the Data Supplement (Table S5). For instance, at age 50 years, the cumulative incidence of interval breast cancer for PRS >90% was at 1 year 0.3% (95% CI, 0.2 to 0.5) and at 2 years 0.7% (95% CI, 0.5 to 0.9). For PRS 10%-90%, the corresponding cumulative incidences are much lower, at 1 year 0.1% (95% CI, 0.1 to 0.1) and at 2 years 0.2% (95% CI, 0.2 to 0.3).
FIG 5.
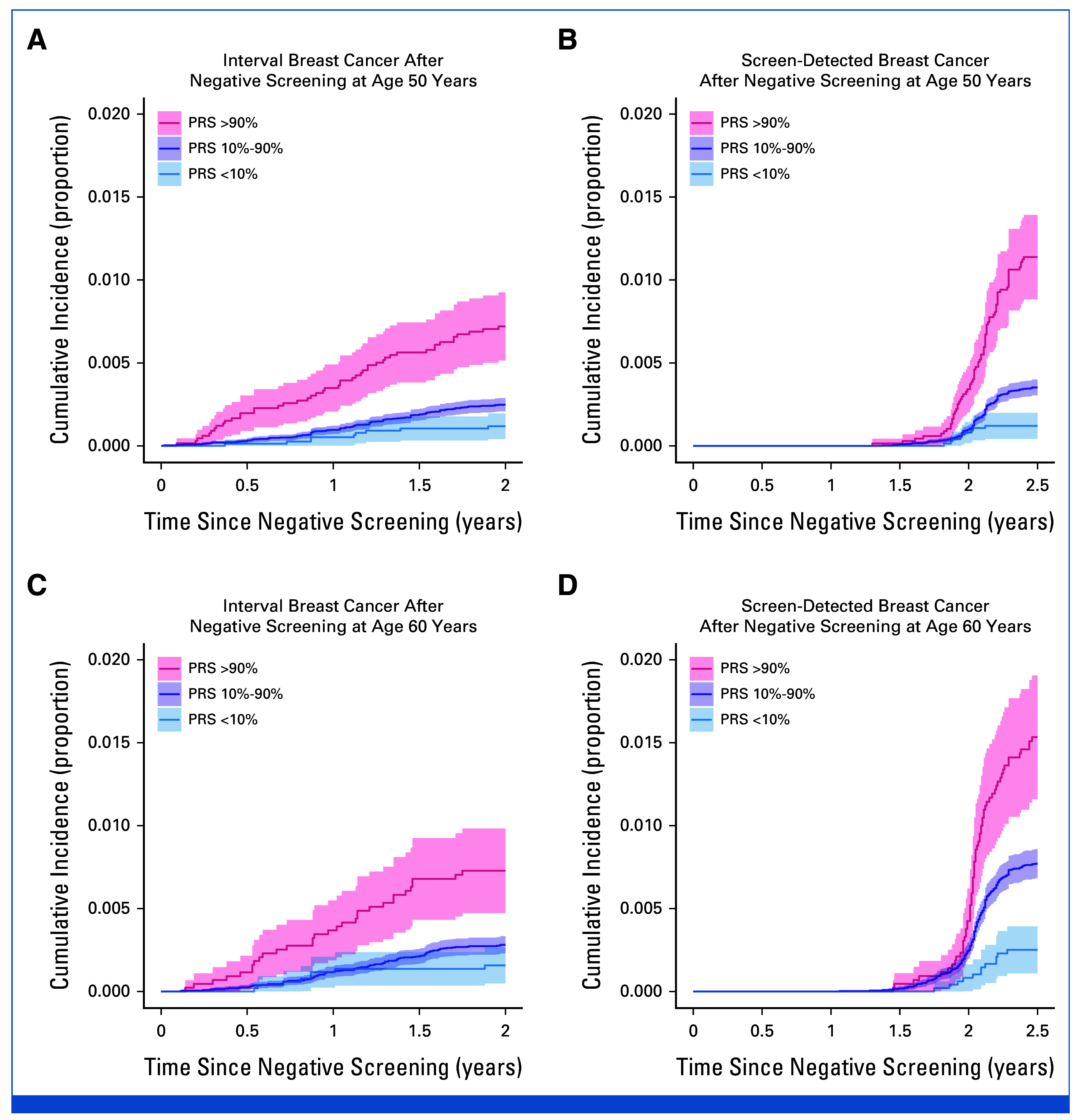
Survival curves showing cumulative incidence of breast cancer for interval and screen-detected cancers after a negative screen at age (A and B) 50 years and (C and D) 60 years by PRS category. Survival curves for a broader set of screening ages are shown in the Data Supplement (Fig S3). Cumulative incidence represents the proportion of individuals diagnosed by each time point shown on the x-axis. PRS, polygenic risk score.
For interval breast cancer, the risk sharply increased with high PRS 2 months after the negative screen at age 50 years. The cumulative incidence of interval breast cancer was low for women with average or low PRS. We did not observe distinct age-related patterns for risk of interval cancer. After a negative screen, the PRS also stratified women according to risk of screen-detected breast cancer in the next screen at all ages, with the cumulative incidence increasing consistently with age, because of baseline risk increases (Data Supplement, Fig S3).
DISCUSSION
PRS studies have recently provided us a broader understanding about the genetic risk factors underlying breast cancer and how it stratifies women on the basis of their future risk of breast cancer, thereby bringing opportunities for risk-tailored screening. We showed the impact of breast cancer PRS on detection of breast cancers in real-life screening data on a large number of women of screening age, comparing the performance of the PRS with PVs and FH, both of which can currently be used for tailoring screening on the basis of individual risk, but which are also not used on a population level. The findings for women with high PRS were similar to PV carriers and women with positive FH, and the risk factors showed complementary performance. The PRS showed clear patterns for a high PRS identifying women more likely to be diagnosed with breast cancer after a positive screening finding, without increasing the detection rate of benign breast lesions. Women with a high PRS with a negative screening finding had an elevated risk of interval breast cancer, and a screen-detected cancer in the next screen, indicating that women with high PRS could benefit from more frequent screenings. Evaluations of the impact of PRS before screening age also indicated that women with a high PRS could benefit from earlier screening initiation, which in many countries, including Finland, is at age 50 years. On the contrary, women with a low PRS had a very low risk of both interval- and screen-detected cancers, suggesting opportunities for less frequent screens for this low-risk group. The ages at which each risk group reaches a 2% cumulative incidence, aligning with the population at the start of organized screening, provides a guideline for when to begin screening on the basis of individual risk. Cumulative incidences of interval cancer at 1 and 2 years after a negative screen can be used to evaluate the potential impact of incorporating PRS into an annual screening program, depending on the accepted risk levels.
The potential role of breast cancer PRS for stratified screening has previously been evaluated through prospective and retrospective observational studies,8,16-19 with supporting evidence from cost-effectiveness modeling.20,21 However, none of these studies have used real-life screening data for screen-detected and interval breast cancers, which is necessary for understanding the impact of PRS in a screening context. Only a few small studies have used such screening data, with most of the studies evaluating PRSs that contain much fewer genetic variants than our genome-wide PRS.22,23 We linked polygenic risk information to observational population-based data on 693,730 screening events in a country with nationwide biennial mammography screening between age 50 and 69 years and high attendance to the screenings. In addition to a lack of large real-life breast cancer screening studies on breast cancer PRSs, population-scale studies are sparse also for the impact of PVs and FH, but ongoing clinical trials are underway evaluating the performance of personalized breast cancer screening.24,25
Risk-tailored surveillance is currently implemented in many countries for women with strong FH of early-onset breast cancer and in many women who have been identified to be carriers of PVs in breast cancer susceptibility genes such as BRCA1, PALB2, and CHEK2. Our results support adding PRS assessment to guide these decisions, as PRS complemented PV and FH information, which we and others have also shown previously.7,10 In particular, we show that women in the top 10% of the PRS have an elevated risk for interval cancers, showing that these women may benefit for shorter time intervals between screening visits. These women also had an elevated risk for bilateral breast cancer. Moreover, to evaluate the impact of PRSs, PVs, and FH outside of the screening program, we extended the general effect size evaluations to timelines before and after the screening age. We observed similar patterns for all the three risk factors with respect to age, which could support earlier initiation of breast cancer screening in women with high PRS. Second, this study shows that women with low PRS (<10%) had a very low risk of both interval- and screen-detected breast cancers even at age 50 years. At the very least, this argues against benefits of screening women with low PRS and no other risk factors before age 50 years, which is common practice in many countries.26,27 Further research is needed on the impact of delaying the onset of screening with low PRS.
The large FinnGen study links genotypes to nationwide registries containing 27 years of data on breast cancer screenings and up to 66 years of follow-up within the FCR. The screening attendance and other screening parameters are in line with nationwide data (Data Supplement, Table S1)11 and the parameters are comparable with reports from other European countries.27 However, our results are generalizable primarily to countries with biennial screening recommendations. The PVs evaluated are enriched in the Finnish population, allowing reliable identification of carriers. We used a contemporary genomewide PRS outperforming previously published PRSs with a small number of variants.14
Despite our substantial sample size, we lacked power to systematically assess risk factor combinations throughout the study or less common BRCA1 and BRCA2 PVs, which are rarer in Finland compared with many European countries. The dynamics of PRS and PVs are, however, similar across different susceptibility genes.9,28 Moreover, the high risk associated with BRCA1/2 mutations would often necessitate specialized screening protocols beyond traditional organized breast cancer screening.5 First-degree FH was identified through health care registries, with an effect size comparable with published estimates,29 and the dynamic between FH and PRS is similar regardless of the source of FH information.7,8,30,31 Within this registry-based study, variant carriers were not informed of being carriers. The study contains only individuals of European ancestry, but risk- and cost-effectiveness evaluations for breast cancer screening in Asian populations have reached similar conclusions for potential utility for PRSs.20,32
Using a large biobank study of women with data from real-life screening events, we show that a breast cancer PRS predicted the outcome of an initial positive screening finding. Moreover, a high PRS was associated with the risk of interval cancer among women with a negative screening result, and with an elevated risk for bilateral breast cancer. The findings support the use of a breast cancer PRS for risk stratification, with optimal stratification reached through combining PRS information with FH of breast cancer and PVs in breast cancer susceptibility genes. Further studies are needed to assess how to optimally integrate these factors into clinical care33 in addition to assessments of their impact incorporated into a prospective risk-stratified screening program, with cost-effectiveness evaluations.
ACKNOWLEDGMENT
The authors thank Sari Kivikko, Huei-Yi Shen, and Ulla Tuomainen for management assistance, and Bradley Jermy for methodologic support. The authors want to acknowledge the participants and investigators of FinnGen study. The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc, AstraZeneca UK Ltd, Biogen MA Inc, Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc, Merck Sharp & Dohme LCC, Pfizer Inc, GlaxoSmithKline Intellectual Property Development Ltd, Sanofi US Services Inc, Maze Therapeutics Inc, Janssen Biotech Inc, Novartis AG, and Boehringer Ingelheim International GmbH. The following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta), and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). Finnish Biobank Cooperative-FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland.
A list of FinnGen contributors is listed in Appendix Table A1 (online only).
APPENDIX
TABLE A1.
FinnGen
| Full Name | Affiliation |
|---|---|
| Aarno Palotie | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Mark Daly | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Bridget Riley-Gills | AbbVie, Chicago, IL |
| Howard Jacob | AbbVie, Chicago, IL |
| Dirk Paul | Astra Zeneca, Cambridge, United Kingdom |
| Athena Matakidou | Astra Zeneca, Cambridge, United Kingdom |
| Adam Platt | Astra Zeneca, Cambridge, United Kingdom |
| Heiko Runz | Biogen, Cambridge, MA |
| Sally John | Biogen, Cambridge, MA |
| George Okafo | Boehringer Ingelheim, Ingelheim am Rhein, Germany |
| Nathan Lawless | Boehringer Ingelheim, Ingelheim am Rhein, Germany |
| Robert Plenge | Bristol Myers Squibb, New York, NY |
| Joseph Maranville | Bristol Myers Squibb, New York, NY |
| Mark McCarthy | Genentech, San Francisco, CA |
| Julie Hunkapiller | Genentech, San Francisco, CA |
| Margaret G. Ehm | GlaxoSmithKline, Collegeville, PA |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Simonne Longerich | Merck, Kenilworth, NJ |
| Caroline Fox | Merck, Kenilworth, NJ |
| Anders Mälarstig | Pfizer, New York, NY |
| Katherine Klinger | Translational Sciences, Sanofi R&D, Framingham, MA |
| Deepak Raipal | Translational Sciences, Sanofi R&D, Framingham, MA |
| Eric Green | Maze Therapeutics, San Francisco, CA |
| Robert Graham | Maze Therapeutics, San Francisco, CA |
| Robert Yang | Janssen Biotech, Beerse, Belgium |
| Chris O'Donnell | Novartis Institutes for BioMedical Research, Cambridge, MA |
| Tomi P. Mäkelä | HiLIFE, University of Helsinki, Finland, Finland |
| Jaakko Kaprio | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Petri Virolainen | Auria Biobank/University of Turku/Hospital District of Southwest Finland, Turku, Finland |
| Antti Hakanen | Auria Biobank/University of Turku/Hospital District of Southwest Finland, Turku, Finland |
| Terhi Kilpi | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Markus Perola | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Jukka Partanen | Finnish Red Cross Blood Service/Finnish Hematology Registry and Clinical Biobank, Helsinki, Finland |
| Anne Pitkäranta | Helsinki Biobank/Helsinki University and Hospital District of Helsinki and Uusimaa, Helsinki |
| Juhani Junttila | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Raisa Serpi | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Tarja Laitinen | Finnish Clinical Biobank Tampere/University of Tampere/Pirkanmaa Hospital District, Tampere, Finland |
| Veli-Matti Kosma | Biobank of Eastern Finland/University of Eastern Finland/Northern Savo Hospital District, Kuopio, Finland |
| Jari Laukkanen | Central Finland Biobank/University of Jyväskylä/Central Finland Health Care District, Jyväskylä, Finland |
| Marco Hautalahti | FINBB—Finnish Biobank Cooperative |
| Outi Tuovila | Business Finland, Helsinki, Finland |
| Raimo Pakkanen | Business Finland, Helsinki, Finland |
| Jeffrey Waring | AbbVie, Chicago, IL |
| Bridget Riley-Gillis | AbbVie, Chicago, IL |
| Fedik Rahimov | AbbVie, Chicago, IL |
| Ioanna Tachmazidou | Astra Zeneca, Cambridge, United Kingdom |
| Chia-Yen Chen | Biogen, Cambridge, MA |
| Heiko Runz | Biogen, Cambridge, MA |
| Zhihao Ding | Boehringer Ingelheim, Ingelheim am Rhein, Germany |
| Marc Jung | Boehringer Ingelheim, Ingelheim am Rhein, Germany |
| Shameek Biswas | Bristol Myers Squibb, New York, NY |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Julie Hunkapiller | Genentech, San Francisco, CA |
| Margaret G. Ehm | GlaxoSmithKline, Collegeville, PA |
| David Pulford | GlaxoSmithKline, Stevenage, United Kingdom |
| Neha Raghavan | Merck, Kenilworth, NJ |
| Adriana Huertas-Vazquez | Merck, Kenilworth, NJ |
| Jae-Hoon Sul | Merck, Kenilworth, NJ |
| Anders Mälarstig | Pfizer, New York, NY |
| Xinli Hu | Pfizer, New York, NY |
| Katherine Klinger | Translational Sciences, Sanofi R&D, Framingham, MA |
| Robert Graham | Maze Therapeutics, San Francisco, CA |
| Eric Green | Maze Therapeutics, San Francisco, CA |
| Sahar Mozaffari | Maze Therapeutics, San Francisco, CA |
| Dawn Waterworth | Janssen Research & Development, LLC, Spring House, PA |
| Nicole Renaud | Novartis Institutes for BioMedical Research, Cambridge, MA |
| Ma'en Obeidat | Novartis Institutes for BioMedical Research, Cambridge, MA |
| Samuli Ripatti | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Johanna Schleutker | Auria Biobank/University of Turku/Hospital District of Southwest Finland, Turku, Finland |
| Markus Perola | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Mikko Arvas | Finnish Red Cross Blood Service/Finnish Hematology Registry and Clinical Biobank, Helsinki, Finland |
| Olli Carpén | Helsinki Biobank/Helsinki University and Hospital District of Helsinki and Uusimaa, Helsinki |
| Reetta Hinttala | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Johannes Kettunen | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Arto Mannermaa | Biobank of Eastern Finland/University of Eastern Finland/Northern Savo Hospital District, Kuopio, Finland |
| Katriina Aalto-Setälä | Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland |
| Mika Kähönen | Finnish Clinical Biobank Tampere/University of Tampere/Pirkanmaa Hospital District, Tampere, Finland |
| Jari Laukkanen | Central Finland Biobank/University of Jyväskylä/Central Finland Health Care District, Jyväskylä, Finland |
| Johanna Mäkelä | FINBB—Finnish Biobank Cooperative |
| Reetta Kälviäinen | Northern Savo Hospital District, Kuopio, Finland |
| Valtteri Julkunen | Northern Savo Hospital District, Kuopio, Finland |
| Hilkka Soininen | Northern Savo Hospital District, Kuopio, Finland |
| Anne Remes | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Mikko Hiltunen | University of Eastern Finland, Kuopio, Finland |
| Jukka Peltola | Pirkanmaa Hospital District, Tampere, Finland |
| Minna Raivio | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Pentti Tienari | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Juha Rinne | Hospital District of Southwest Finland, Turku, Finland |
| Roosa Kallionpää | Hospital District of Southwest Finland, Turku, Finland |
| Juulia Partanen | Institute for Molecular Medicine Finland, HiLIFE, University of Helsinki, Finland |
| Ali Abbasi | AbbVie, Chicago, IL |
| Adam Ziemann | AbbVie, Chicago, IL |
| Nizar Smaoui | AbbVie, Chicago, IL |
| Anne Lehtonen | AbbVie, Chicago, IL |
| Susan Eaton | Biogen, Cambridge, MA |
| Heiko Runz | Biogen, Cambridge, MA |
| Sanni Lahdenperä | Biogen, Cambridge, MA |
| Shameek Biswas | Bristol Myers Squibb, New York, NY |
| Julie Hunkapiller | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Edmond Teng | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Fanli Xu | GlaxoSmithKline, Brentford, United Kingdom |
| David Pulford | GlaxoSmithKline, Stevenage, United Kingdom |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Laura Addis | GlaxoSmithKline, Brentford, United Kingdom |
| John Eicher | GlaxoSmithKline, Brentford, United Kingdom |
| Qingqin S Li | Janssen Research & Development, LLC, Titusville, NJ 08560 |
| Karen He | Janssen Research & Development, LLC, Spring House, PA |
| Ekaterina Khramtsova | Janssen Research & Development, LLC, Spring House, PA |
| Neha Raghavan | Merck, Kenilworth, NJ |
| Martti Färkkilä | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Jukka Koskela | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Sampsa Pikkarainen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Airi Jussila | Pirkanmaa Hospital District, Tampere, Finland |
| Katri Kaukinen | Pirkanmaa Hospital District, Tampere, Finland |
| Timo Blomster | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Mikko Kiviniemi | Northern Savo Hospital District, Kuopio, Finland |
| Markku Voutilainen | Hospital District of Southwest Finland, Turku, Finland |
| Mark Daly | Institute for Molecular Medicine, Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Ali Abbasi | AbbVie, Chicago, IL |
| Jeffrey Waring | AbbVie, Chicago, IL |
| Nizar Smaoui | AbbVie, Chicago, IL |
| Fedik Rahimov | AbbVie, Chicago, IL |
| Anne Lehtonen | AbbVie, Chicago, IL |
| Tim Lu | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Linda McCarthy | GlaxoSmithKline, Brentford, United Kingdom |
| Amy Hart | Janssen Research & Development, LLC, Spring House, PA |
| Meijian Guan | Janssen Research & Development, LLC, Spring House, PA |
| Jason Miller | Merck, Kenilworth, NJ |
| Kirsi Kalpala | Pfizer, New York, NY |
| Melissa Miller | Pfizer, New York, NY |
| Xinli Hu | Pfizer, New York, NY |
| Kari Eklund | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Antti Palomäki | Hospital District of Southwest Finland, Turku, Finland |
| Pia Isomäki | Pirkanmaa Hospital District, Tampere, Finland |
| Laura Pirilä | Hospital District of Southwest Finland, Turku, Finland |
| Oili Kaipiainen-Seppänen | Northern Savo Hospital District, Kuopio, Finland |
| Johanna Huhtakangas | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Nina Mars | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Ali Abbasi | AbbVie, Chicago, IL |
| Jeffrey Waring | AbbVie, Chicago, IL |
| Fedik Rahimov | AbbVie, Chicago, IL |
| Apinya Lertratanakul | AbbVie, Chicago, IL |
| Nizar Smaoui | AbbVie, Chicago, IL |
| Anne Lehtonen | AbbVie, Chicago, IL |
| David Close | Astra Zeneca, Cambridge, United Kingdom |
| Marla Hochfeld | Bristol Myers Squibb, New York, NY |
| Natalie Bowers | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Jorge Esparza Gordillo | GlaxoSmithKline, Brentford, United Kingdom |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Dawn Waterworth | Janssen Research & Development, LLC, Spring House, PA |
| Fabiana Farias | Merck, Kenilworth, NJ |
| Kirsi Kalpala | Pfizer, New York, NY |
| Nan Bing | Pfizer, New York, NY |
| Xinli Hu | Pfizer, New York, NY |
| Tarja Laitinen | Pirkanmaa Hospital District, Tampere, Finland |
| Margit Pelkonen | Northern Savo Hospital District, Kuopio, Finland |
| Paula Kauppi | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Hannu Kankaanranta | University of Gothenburg, Gothenburg, Sweden/Seinäjoki Central Hospital, Seinäjoki, Finland/Tampere University, Tampere, Finland |
| Terttu Harju | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Riitta Lahesmaa | Hospital District of Southwest Finland, Turku, Finland |
| Nizar Smaoui | AbbVie, Chicago, IL |
| Alex Mackay | Astra Zeneca, Cambridge, United Kingdom |
| Glenda Lassi | Astra Zeneca, Cambridge, United Kingdom |
| Susan Eaton | Biogen, Cambridge, MA |
| Hubert Chen | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Joanna Betts | GlaxoSmithKline, Brentford, United Kingdom |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Rajashree Mishra | GlaxoSmithKline, Brentford, United Kingdom |
| Majd Mouded | Novartis, Basel, Switzerland |
| Debby Ngo | Novartis, Basel, Switzerland |
| Teemu Niiranen | Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Felix Vaura | Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Veikko Salomaa | Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Kaj Metsärinne | Hospital District of Southwest Finland, Turku, Finland |
| Jenni Aittokallio | Hospital District of Southwest Finland, Turku, Finland |
| Mika Kähönen | Pirkanmaa Hospital District, Tampere, Finland |
| Jussi Hernesniemi | Pirkanmaa Hospital District, Tampere, Finland |
| Daniel Gordin | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Juha Sinisalo | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Marja-Riitta Taskinen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Tiinamaija Tuomi | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Timo Hiltunen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Jari Laukkanen | Central Finland Health Care District, Jyväskylä, Finland |
| Amanda Elliott | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA; and Massachusetts General Hospital, Boston, MA |
| Mary Pat Reeve | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Sanni Ruotsalainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Benjamin Challis | Astra Zeneca, Cambridge, United Kingdom |
| Dirk Paul | Astra Zeneca, Cambridge, United Kingdom |
| Julie Hunkapiller | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Audrey Chu | GlaxoSmithKline, Brentford, United Kingdom |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Dermot Reilly | Janssen Research & Development, LLC, Boston, MA |
| Mike Mendelson | Novartis, Boston, MA |
| Jaakko Parkkinen | Pfizer, New York, NY |
| Melissa Miller | Pfizer, New York, NY |
| Tuomo Meretoja | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Heikki Joensuu | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Olli Carpén | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Johanna Mattson | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Eveliina Salminen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Annika Auranen | Pirkanmaa Hospital District, Tampere, Finland |
| Peeter Karihtala | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Päivi Auvinen | Northern Savo Hospital District, Kuopio, Finland |
| Klaus Elenius | Hospital District of Southwest Finland, Turku, Finland |
| Johanna Schleutker | Hospital District of Southwest Finland, Turku, Finland |
| Esa Pitkänen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Nina Mars | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mark Daly | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Relja Popovic | AbbVie, Chicago, IL |
| Jeffrey Waring | AbbVie, Chicago, IL |
| Bridget Riley-Gillis | AbbVie, Chicago, IL |
| Anne Lehtonen | AbbVie, Chicago, IL |
| Jennifer Schutzman | Genentech, San Francisco, CA |
| Julie Hunkapiller | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Diptee Kulkarni | GlaxoSmithKline, Brentford, United Kingdom |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Alessandro Porello | Janssen Research & Development, LLC, Spring House, PA |
| Andrey Loboda | Merck, Kenilworth, NJ |
| Heli Lehtonen | Pfizer, New York, NY |
| Stefan McDonough | Pfizer, New York, NY |
| Sauli Vuoti | Janssen-Cilag Oy, Espoo, Finland |
| Kai Kaarniranta | Northern Savo Hospital District, Kuopio, Finland |
| Joni A Turunen | Helsinki University Hospital and University of Helsinki, Helsinki, Finland; Eye Genetics Group, Folkhälsan Research Center, Helsinki, Finland |
| Terhi Ollila | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Hannu Uusitalo | Pirkanmaa Hospital District, Tampere, Finland |
| Juha Karjalainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Esa Pitkänen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mengzhen Liu | AbbVie, Chicago, IL |
| Heiko Runz | Biogen, Cambridge, MA |
| Stephanie Loomis | Biogen, Cambridge, MA |
| Erich Strauss | Genentech, San Francisco, CA |
| Natalie Bowers | Genentech, San Francisco, CA |
| Hao Chen | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Kaisa Tasanen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Laura Huilaja | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Katariina Hannula-Jouppi | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Teea Salmi | Pirkanmaa Hospital District, Tampere, Finland |
| Sirkku Peltonen | Hospital District of Southwest Finland, Turku, Finland |
| Leena Koulu | Hospital District of Southwest Finland, Turku, Finland |
| Nizar Smaoui | AbbVie, Chicago, IL |
| Fedik Rahimov | AbbVie, Chicago, IL |
| Anne Lehtonen | AbbVie, Chicago, IL |
| David Choy | Genentech, San Francisco, CA |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Dawn Waterworth | Janssen Research & Development, LLC, Spring House, PA |
| Kirsi Kalpala | Pfizer, New York, NY |
| Ying Wu | Pfizer, New York, NY |
| Pirkko Pussinen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Aino Salminen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Tuula Salo | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| David Rice | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Pekka Nieminen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Ulla Palotie | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Maria Siponen | Northern Savo Hospital District, Kuopio, Finland |
| Liisa Suominen | Northern Savo Hospital District, Kuopio, Finland |
| Päivi Mäntylä | Northern Savo Hospital District, Kuopio, Finland |
| Ulvi Gursoy | Hospital District of Southwest Finland, Turku, Finland |
| Vuokko Anttonen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Kirsi Sipilä | Research Unit of Oral Health Sciences Faculty of Medicine, University of Oulu, Oulu, Finland; Medical Research Center, Oulu, Oulu University Hospital and University of Oulu, Oulu, Finland |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Hannele Laivuori | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Venla Kurra | Pirkanmaa Hospital District, Tampere, Finland |
| Laura Kotaniemi-Talonen | Pirkanmaa Hospital District, Tampere, Finland |
| Oskari Heikinheimo | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Ilkka Kalliala | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Lauri Aaltonen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Varpu Jokimaa | Hospital District of Southwest Finland, Turku, Finland |
| Johannes Kettunen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Marja Vääräsmäki | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Outi Uimari | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Laure Morin-Papunen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Maarit Niinimäki | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Terhi Piltonen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Katja Kivinen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Elisabeth Widen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Taru Tukiainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mary Pat Reeve | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mark Daly | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Niko Välimäki | University of Helsinki, Helsinki, Finland |
| Eija Laakkonen | University of Jyväskylä, Jyväskylä, Finland |
| Jaakko Tyrmi | University of Oulu, Oulu, Finland/University of Tampere, Tampere, Finland |
| Heidi Silven | University of Oulu, Oulu, Finland |
| Eeva Sliz | University of Oulu, Oulu, Finland |
| Riikka Arffman | University of Oulu, Oulu, Finland |
| Susanna Savukoski | University of Oulu, Oulu, Finland |
| Triin Laisk | Estonian biobank, Tartu, Estonia |
| Natalia Pujol | Estonian biobank, Tartu, Estonia |
| Mengzhen Liu | AbbVie, Chicago, IL |
| Bridget Riley-Gillis | AbbVie, Chicago, IL |
| Rion Pendergrass | Genentech, San Francisco, CA |
| Janet Kumar | GlaxoSmithKline, Collegeville, PA |
| Kirsi Auro | GlaxoSmithKline, Espoo, Finland |
| Iiris Hovatta | University of Helsinki, Finland |
| Chia-Yen Chen | Biogen, Cambridge, MA |
| Erkki Isometsä | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Kumar Veerapen | Broad Institute, Cambridge, MA |
| Hanna Ollila | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Jaana Suvisaari | Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Thomas Damm Als | Aarhus University, Denmark |
| Antti Mäkitie | Department of Otorhinolaryngology—Head and Neck Surgery, University of Helsinki and Helsinki University Hospital, Helsinki, Finland |
| Argyro Bizaki-Vallaskangas | Pirkanmaa Hospital District, Tampere, Finland |
| Sanna Toppila-Salmi | University of Helsinki, Finland |
| Tytti Willberg | Hospital District of Southwest Finland, Turku, Finland |
| Elmo Saarentaus | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Antti Aarnisalo | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Eveliina Salminen | Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Elisa Rahikkala | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Johannes Kettunen | Northern Ostrobothnia Hospital District, Oulu, Finland |
| Kristiina Aittomäki | Department of Medical Genetics, Helsinki University Central Hospital, Helsinki, Finland |
| Fredrik Åberg | Transplantation and Liver Surgery Clinic, Helsinki University Hospital, Helsinki University, Helsinki, Finland |
| Mitja Kurki | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA |
| Samuli Ripatti | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mark Daly | Institute for Molecular Medicine, Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute of MIT and Harvard; Massachusetts General Hospital |
| Juha Karjalainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Aki Havulinna | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Juha Mehtonen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Priit Palta | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Shabbeer Hassan | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Pietro Della Briotta Parolo | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Wei Zhou | Broad Institute, Cambridge, MA |
| Mutaamba Maasha | Broad Institute, Cambridge, MA |
| Kumar Veerapen | Broad Institute, Cambridge, MA |
| Shabbeer Hassan | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Susanna Lemmelä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Manuel Rivas | University of Stanford, Stanford, CA |
| Mari E. Niemi | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Aarno Palotie | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Aoxing Liu | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Arto Lehisto | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Andrea Ganna | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Vincent Llorens | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Hannele Laivuori | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Taru Tukiainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mary Pat Reeve | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Henrike Heyne | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Nina Mars | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Joel Rämö | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Elmo Saarentaus | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Hanna Ollila | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Rodos Rodosthenous | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Satu Strausz | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Tuula Palotie | University of Helsinki and Hospital District of Helsinki and Uusimaa, Helsinki, Finland |
| Kimmo Palin | University of Helsinki, Helsinki, Finland |
| Javier Garcia-Tabuenca | University of Tampere, Tampere, Finland |
| Harri Siirtola | University of Tampere, Tampere, Finland |
| Tuomo Kiiskinen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Jiwoo Lee | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA |
| Kristin Tsuo | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA |
| Amanda Elliott | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA and Massachusetts General Hospital, Boston, MA |
| Kati Kristiansson | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Mikko Arvas | Finnish Red Cross Blood Service/Finnish Hematology Registry and Clinical Biobank, Helsinki, Finland |
| Kati Hyvärinen | Finnish Red Cross Blood Service, Helsinki, Finland |
| Jarmo Ritari | Finnish Red Cross Blood Service, Helsinki, Finland |
| Olli Carpén | Helsinki Biobank/Helsinki University and Hospital District of Helsinki and Uusimaa, Helsinki |
| Johannes Kettunen | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Katri Pylkäs | University of Oulu, Oulu, Finland |
| Eeva Sliz | University of Oulu, Oulu, Finland |
| Minna Karjalainen | University of Oulu, Oulu, Finland |
| Tuomo Mantere | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Eeva Kangasniemi | Finnish Clinical Biobank Tampere/University of Tampere/Pirkanmaa Hospital District, Tampere, Finland |
| Sami Heikkinen | University of Eastern Finland, Kuopio, Finland |
| Arto Mannermaa | Biobank of Eastern Finland/University of Eastern Finland/Northern Savo Hospital District, Kuopio, Finland |
| Eija Laakkonen | University of Jyväskylä, Jyväskylä, Finland |
| Nina Pitkänen | Auria Biobank/University of Turku/Hospital District of Southwest Finland, Turku, Finland |
| Samuel Lessard | Translational Sciences, Sanofi R&D, Framingham, MA |
| Clément Chatelain | Translational Sciences, Sanofi R&D, Framingham, MA |
| Perttu Terho | Auria Biobank/University of Turku/Hospital District of Southwest Finland, Turku, Finland |
| Sirpa Soini | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Jukka Partanen | Finnish Red Cross Blood Service/Finnish Hematology Registry and Clinical Biobank, Helsinki, Finland |
| Eero Punkka | Helsinki Biobank/Helsinki University and Hospital District of Helsinki and Uusimaa, Helsinki |
| Raisa Serpi | Northern Finland Biobank Borealis/University of Oulu/Northern Ostrobothnia Hospital District, Oulu, Finland |
| Sanna Siltanen | Finnish Clinical Biobank Tampere/University of Tampere/Pirkanmaa Hospital District, Tampere, Finland |
| Veli-Matti Kosma | Biobank of Eastern Finland/University of Eastern Finland/Northern Savo Hospital District, Kuopio, Finland |
| Teijo Kuopio | Central Finland Biobank/University of Jyväskylä/Central Finland Health Care District, Jyväskylä, Finland |
| Anu Jalanko | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Huei-Yi Shen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Risto Kajanne | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mervi Aavikko | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mitja Kurki | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Broad Institute, Cambridge, MA |
| Juha Karjalainen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Pietro Della Briotta Parolo | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Arto Lehisto | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Juha Mehtonen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Wei Zhou | Broad Institute, Cambridge, MA |
| Masahiro Kanai | Broad Institute, Cambridge, MA |
| Mutaamba Maasha | Broad Institute, Cambridge, MA |
| Kumar Veerapen | Broad Institute, Cambridge, MA |
| Hannele Laivuori | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Aki Havulinna | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland; Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Susanna Lemmelä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Tuomo Kiiskinen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| L. Elisa Lahtela | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mari Kaunisto | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Elina Kilpeläinen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Timo P. Sipilä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Oluwaseun Alexander Dada | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Awaisa Ghazal | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Anastasia Kytölä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Rigbe Weldatsadik | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Kati Donner | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Timo P. Sipilä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Anu Loukola | Helsinki Biobank/Helsinki University and Hospital District of Helsinki and Uusimaa, Helsinki |
| Päivi Laiho | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Tuuli Sistonen | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Essi Kaiharju | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Markku Laukkanen | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Elina Järvensivu | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Sini Lähteenmäki | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Lotta Männikkö | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Regis Wong | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Auli Toivola | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Minna Brunfeldt | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Hannele Mattsson | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Kati Kristiansson | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Susanna Lemmelä | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Sami Koskelainen | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Tero Hiekkalinna | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Teemu Paajanen | THL Biobank/Finnish Institute for Health and Welfare (THL), Helsinki, Finland |
| Priit Palta | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Kalle Pärn | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Mart Kals | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Shuang Luo | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Vishal Sinha | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Tarja Laitinen | Pirkanmaa Hospital District, Tampere, Finland |
| Mary Pat Reeve | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Marianna Niemi | University of Tampere, Tampere, Finland |
| Kumar Veerapen | Broad Institute, Cambridge, MA |
| Harri Siirtola | University of Tampere, Tampere, Finland |
| Javier Gracia-Tabuenca | University of Tampere, Tampere, Finland |
| Mika Helminen | University of Tampere, Tampere, Finland |
| Tiina Luukkaala | University of Tampere, Tampere, Finland |
| Iida Vähätalo | University of Tampere, Tampere, Finland |
| Jyrki Pitkänen | Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland |
| Marco Hautalahti | Finnish Biobank Cooperative—FINBB |
| Johanna Mäkelä | Finnish Biobank Cooperative—FINBB |
| Sarah Smith | Finnish Biobank Cooperative—FINBB |
| Tom Southerington | Finnish Biobank Cooperative—FINBB |
Sini Kerminen
Employment: Nightingale Health PLC
Stock and Other Ownership Interests: Nightingale Health PLC
Research Funding: Nightingale Health PLC
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at the American Society of Human Genetics Annual Meeting, Los Angeles, CA, October 25-29, 2022.
SUPPORT
Supported by Academy of Finland (grant numbers 331671 and 355567 to N.M., 285380 to S.R., 338507 to M.P.); University of Helsinki HiLIFE Fellows Grant 2023-2025 to N.M.; Finska Läkaresällskapet (to N.M.); Academy of Finland Center of Excellence in Complex Disease Genetics (grant number 312062 to S.R., 336825 to M.P.); European Union's Horizon 2020 research and innovation program under grant agreement No 101016775; the Sigrid Jusélius Foundation (to S.R.); The Finnish Innovation Fund Tekes (grant number 2273/31/2017 to E.W.); Cancer Foundation Finland (to T.M.); and The Doctoral Programme in Population Health, University of Helsinki (to M.T.).
See accompanying Editorial, p. 1462
Contributor Information
Collaborators: Aarno Palotie, Mark Daly, Bridget Riley-Gills, Howard Jacob, Dirk Paul, Athena Matakidou, Adam Platt, Heiko Runz, Sally John, George Okafo, Nathan Lawless, Robert Plenge, Joseph Maranville, Mark McCarthy, Julie Hunkapiller, Margaret G. Ehm, Kirsi Auro, Simonne Longerich, Caroline Fox, Anders Mälarstig, Katherine Klinger, Deepak Raipal, Eric Green, Robert Graham, Robert Yang, Chris O'Donnell, Tomi P. Mäkelä, Jaakko Kaprio, Petri Virolainen, Antti Hakanen, Terhi Kilpi, Markus Perola, Jukka Partanen, Anne Pitkäranta, Juhani Junttila, Raisa Serpi, Tarja Laitinen, Veli-Matti Kosma, Jari Laukkanen, Marco Hautalahti, Outi Tuovila, Raimo Pakkanen, Jeffrey Waring, Bridget Riley-Gillis, Fedik Rahimov, Ioanna Tachmazidou, Chia-Yen Chen, Heiko Runz, Zhihao Ding, Marc Jung, Shameek Biswas, Rion Pendergrass, Julie Hunkapiller, Margaret G. Ehm, David Pulford, Neha Raghavan, Adriana Huertas-Vazquez, Jae-Hoon Sul, Anders Mälarstig, Xinli Hu, Katherine Klinger, Robert Graham, Eric Green, Sahar Mozaffari, Dawn Waterworth, Nicole Renaud, Ma'en Obeidat, Samuli Ripatti, Johanna Schleutker, Markus Perola, Mikko Arvas, Olli Carpén, Reetta Hinttala, Johannes Kettunen, Arto Mannermaa, Katriina Aalto-Setälä, Mika Kähönen, Jari Laukkanen, Johanna Mäkelä, Reetta Kälviäinen, Valtteri Julkunen, Hilkka Soininen, Anne Remes, Mikko Hiltunen, Jukka Peltola, Minna Raivio, Pentti Tienari, Juha Rinne, Roosa Kallionpää, Juulia Partanen, Ali Abbasi, Adam Ziemann, Nizar Smaoui, Anne Lehtonen, Susan Eaton, Heiko Runz, Sanni Lahdenperä, Shameek Biswas, Julie Hunkapiller, Natalie Bowers, Edmond Teng, Rion Pendergrass, Fanli Xu, David Pulford, Kirsi Auro, Laura Addis, John Eicher, Qingqin S Li, Karen He, Ekaterina Khramtsova, Neha Raghavan, Martti Färkkilä, Jukka Koskela, Sampsa Pikkarainen, Airi Jussila, Katri Kaukinen, Timo Blomster, Mikko Kiviniemi, Markku Voutilainen, Mark Daly, Ali Abbasi, Jeffrey Waring, Nizar Smaoui, Fedik Rahimov, Anne Lehtonen, Tim Lu, Natalie Bowers, Rion Pendergrass, Linda McCarthy, Amy Hart, Meijian Guan, Jason Miller, Kirsi Kalpala, Melissa Miller, Xinli Hu, Kari Eklund, Antti Palomäki, Pia Isomäki, Laura Pirilä, Oili Kaipiainen-Seppänen, Johanna Huhtakangas, Nina Mars, Ali Abbasi, Jeffrey Waring, Fedik Rahimov, Apinya Lertratanakul, Nizar Smaoui, Anne Lehtonen, David Close, Marla Hochfeld, Natalie Bowers, Rion Pendergrass, Jorge Esparza Gordillo, Kirsi Auro, Dawn Waterworth, Fabiana Farias, Kirsi Kalpala, Nan Bing, Xinli Hu, Tarja Laitinen, Margit Pelkonen, Paula Kauppi, Hannu Kankaanranta, Terttu Harju, Riitta Lahesmaa, Nizar Smaoui, Alex Mackay, Glenda Lassi, Susan Eaton, Hubert Chen, Rion Pendergrass, Natalie Bowers, Joanna Betts, Kirsi Auro, Rajashree Mishra, Majd Mouded, Debby Ngo, Teemu Niiranen, Felix Vaura, Veikko Salomaa, Kaj Metsärinne, Jenni Aittokallio, Mika Kähönen, Jussi Hernesniemi, Daniel Gordin, Juha Sinisalo, Marja-Riitta Taskinen, Tiinamaija Tuomi, Timo Hiltunen, Jari Laukkanen, Amanda Elliott, Mary Pat Reeve, Sanni Ruotsalainen, Benjamin Challis, Dirk Paul, Julie Hunkapiller, Natalie Bowers, Rion Pendergrass, Audrey Chu, Kirsi Auro, Dermot Reilly, Mike Mendelson, Jaakko Parkkinen, Melissa Miller, Tuomo Meretoja, Heikki Joensuu, Olli Carpén, Johanna Mattson, Eveliina Salminen, Annika Auranen, Peeter Karihtala, Päivi Auvinen, Klaus Elenius, Johanna Schleutker, Esa Pitkänen, Nina Mars, Mark Daly, Relja Popovic, Jeffrey Waring, Bridget Riley-Gillis, Anne Lehtonen, Jennifer Schutzman, Julie Hunkapiller, Natalie Bowers, Rion Pendergrass, Diptee Kulkarni, Kirsi Auro, Alessandro Porello, Andrey Loboda, Heli Lehtonen, Stefan McDonough, Sauli Vuoti, Kai Kaarniranta, Joni A Turunen, Terhi Ollila, Hannu Uusitalo, Juha Karjalainen, Esa Pitkänen, Mengzhen Liu, Heiko Runz, Stephanie Loomis, Erich Strauss, Natalie Bowers, Hao Chen, Rion Pendergrass, Kaisa Tasanen, Laura Huilaja, Katariina Hannula-Jouppi, Teea Salmi, Sirkku Peltonen, Leena Koulu, Nizar Smaoui, Fedik Rahimov, Anne Lehtonen, David Choy, Rion Pendergrass, Dawn Waterworth, Kirsi Kalpala, Ying Wu, Pirkko Pussinen, Aino Salminen, Tuula Salo, David Rice, Pekka Nieminen, Ulla Palotie, Maria Siponen, Liisa Suominen, Päivi Mäntylä, Ulvi Gursoy, Vuokko Anttonen, Kirsi Sipilä, Rion Pendergrass, Hannele Laivuori, Venla Kurra, Laura Kotaniemi-Talonen, Oskari Heikinheimo, Ilkka Kalliala, Lauri Aaltonen, Varpu Jokimaa, Johannes Kettunen, Marja Vääräsmäki, Outi Uimari, Laure Morin-Papunen, Maarit Niinimäki, Terhi Piltonen, Katja Kivinen, Elisabeth Widen, Taru Tukiainen, Mary Pat Reeve, Mark Daly, Niko Välimäki, Eija Laakkonen, Jaakko Tyrmi, Heidi Silven, Eeva Sliz, Riikka Arffman, Susanna Savukoski, Triin Laisk, Natalia Pujol, Mengzhen Liu, Bridget Riley-Gillis, Rion Pendergrass, Janet Kumar, Kirsi Auro, Iiris Hovatta, Chia-Yen Chen, Erkki Isometsä, Kumar Veerapen, Hanna Ollila, Jaana Suvisaari, Thomas Damm Als, Antti Mäkitie, Argyro Bizaki-Vallaskangas, Sanna Toppila-Salmi, Tytti Willberg, Elmo Saarentaus, Antti Aarnisalo, Eveliina Salminen, Elisa Rahikkala, Johannes Kettunen, Kristiina Aittomäki, Fredrik Åberg, Mitja Kurki, Samuli Ripatti, Mark Daly, Juha Karjalainen, Aki Havulinna, Juha Mehtonen, Priit Palta, Shabbeer Hassan, Pietro Della Briotta Parolo, Wei Zhou, Mutaamba Maasha, Kumar Veerapen, Shabbeer Hassan, Susanna Lemmelä, Manuel Rivas, Mari E. Niemi, Aarno Palotie, Aoxing Liu, Arto Lehisto, Andrea Ganna, Vincent Llorens, Hannele Laivuori, Taru Tukiainen, Mary Pat Reeve, Henrike Heyne, Nina Mars, Joel Rämö, Elmo Saarentaus, Hanna Ollila, Rodos Rodosthenous, Satu Strausz, Tuula Palotie, Kimmo Palin, Javier Garcia-Tabuenca, Harri Siirtola, Tuomo Kiiskinen, Jiwoo Lee, Kristin Tsuo, Amanda Elliott, Kati Kristiansson, Mikko Arvas, Kati Hyvärinen, Jarmo Ritari, Olli Carpén, Johannes Kettunen, Katri Pylkäs, Eeva Sliz, Minna Karjalainen, Tuomo Mantere, Eeva Kangasniemi, Sami Heikkinen, Arto Mannermaa, Eija Laakkonen, Nina Pitkänen, Samuel Lessard, Clément Chatelain, Perttu Terho, Sirpa Soini, Jukka Partanen, Eero Punkka, Raisa Serpi, Sanna Siltanen, Veli-Matti Kosma, Teijo Kuopio, Anu Jalanko, Huei-Yi Shen, Risto Kajanne, Mervi Aavikko, Mitja Kurki, Juha Karjalainen, Pietro Della Briotta Parolo, Arto Lehisto, Juha Mehtonen, Wei Zhou, Masahiro Kanai, Mutaamba Maasha, Kumar Veerapen, Hannele Laivuori, Aki Havulinna, Susanna Lemmelä, Tuomo Kiiskinen, L. Elisa Lahtela, Mari Kaunisto, Elina Kilpeläinen, Timo P. Sipilä, Oluwaseun Alexander Dada, Awaisa Ghazal, Anastasia Kytölä, Rigbe Weldatsadik, Kati Donner, Timo P. Sipilä, Anu Loukola, Päivi Laiho, Tuuli Sistonen, Essi Kaiharju, Markku Laukkanen, Elina Järvensivu, Sini Lähteenmäki, Lotta Männikkö, Regis Wong, Auli Toivola, Minna Brunfeldt, Hannele Mattsson, Kati Kristiansson, Susanna Lemmelä, Sami Koskelainen, Tero Hiekkalinna, Teemu Paajanen, Priit Palta, Kalle Pärn, Mart Kals, Shuang Luo, Vishal Sinha, Tarja Laitinen, Mary Pat Reeve, Marianna Niemi, Kumar Veerapen, Harri Siirtola, Javier Gracia-Tabuenca, Mika Helminen, Tiina Luukkaala, Iida Vähätalo, Jyrki Pitkänen, Marco Hautalahti, Johanna Mäkelä, Sarah Smith, and Tom Southerington
DATA SHARING STATEMENT
The FinnGen data may be accessed through Finnish Biobanks' FinBB portal (www.finbb.fi; email: info.fingenious@finbb.fi). The weights for our polygenic risk scores are available at PGS Catalog (https://www.pgscatalog.org/), PGS ID PGS000335.
AUTHOR CONTRIBUTIONS
Conception and design: Nina Mars, Sini Kerminen, Samuli Ripatti
Collection and assembly of data: Nina Mars, Sirpa Heinävaara, Samuli Ripatti
Data analysis and interpretation: Nina Mars, Sini Kerminen, Kirsimari Aaltonen, Max Tamlander, Matti Pirinen, Eveliina Jakkula, Tuomo Meretoja, Elisabeth Widén, Samuli Ripatti
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Comprehensive Inherited Risk Estimation for Risk-Based Breast Cancer Screening in Women
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Sini Kerminen
Employment: Nightingale Health PLC
Stock and Other Ownership Interests: Nightingale Health PLC
Research Funding: Nightingale Health PLC
No other potential conflicts of interest were reported.
REFERENCES
- 1. Iwamoto Y, Kaucher S, Lorenz E, et al. Development of breast cancer mortality considering the implementation of mammography screening programs—A comparison of western European countries. BMC Public Health. 2019;19:823. doi: 10.1186/s12889-019-7166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zielonke N, Gini A, Jansen EEL, et al. Evidence for reducing cancer-specific mortality due to screening for breast cancer in Europe: A systematic review. Eur J Cancer. 2020;127:191–206. doi: 10.1016/j.ejca.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 3. Clift AK, Dodwell D, Lord S, et al. The current status of risk-stratified breast screening. Br J Cancer. 2022;126:533–550. doi: 10.1038/s41416-021-01550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly MB, Pal T, Berry MP, et al. Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 19:77–102, 2021 [DOI] [PubMed] [Google Scholar]
- 5. Sessa C, Balmana J, Bober SL, et al. Risk reduction and screening of cancer in hereditary breast-ovarian cancer syndromes: ESMO clinical practice guideline. Ann Oncol. 2023;34:33–47. doi: 10.1016/j.annonc.2022.10.004. [DOI] [PubMed] [Google Scholar]
- 6. Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mars N, Koskela JT, Ripatti P, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med. 2020;26:549–557. doi: 10.1038/s41591-020-0800-0. [DOI] [PubMed] [Google Scholar]
- 9. Fahed AC, Wang M, Homburger JR, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mars N, Widén E, Kerminen S, et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat Commun. 2020;11:6383. doi: 10.1038/s41467-020-19966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarkeala T, Luostarinen T, Dyba T, et al. Breast carcinoma detection modes and death in a female population in relation to population-based mammography screening. Springerplus. 2014;3:348. doi: 10.1186/2193-1801-3-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leinonen MK, Miettinen J, Heikkinen S, et al. Quality measures of the population-based Finnish Cancer Registry indicate sound data quality for solid malignant tumours. Eur J Cancer. 2017;77:31–39. doi: 10.1016/j.ejca.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 14. Mars N, Kerminen S, Feng Y-CA, et al. Genome-wide risk prediction of common diseases across ancestries in one million people. Cell Genom. 2022;2:100118. doi: 10.1016/j.xgen.2022.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans DGR, Harkness EF, Brentnall AR, et al. Breast cancer pathology and stage are better predicted by risk stratification models that include mammographic density and common genetic variants. Breast Cancer Res Treat. 2019;176:141–148. doi: 10.1007/s10549-019-05210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lall K, Lepamets M, Palover M, et al. Polygenic prediction of breast cancer: Comparison of genetic predictors and implications for risk stratification. BMC Cancer. 2019;19:557. doi: 10.1186/s12885-019-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van den Broek JJ, Schechter CB, van Ravesteyn NT, et al. Personalizing breast cancer screening based on polygenic risk and family history. J Natl Cancer Inst. 2021;113:434–442. doi: 10.1093/jnci/djaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong JZY, Chai JH, Yeoh YS, et al. Cost effectiveness analysis of a polygenic risk tailored breast cancer screening programme in Singapore. BMC Health Serv Res. 2021;21:379. doi: 10.1186/s12913-021-06396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pashayan N, Morris S, Gilbert FJ, et al. Cost-effectiveness and benefit-to-harm ratio of risk-stratified screening for breast cancer: A life-table model. JAMA Oncol. 2018;4:1504–1510. doi: 10.1001/jamaoncol.2018.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J, Holm J, Bergh J, et al. Breast cancer genetic risk profile is differentially associated with interval and screen-detected breast cancers. Ann Oncol. 2015;26:517–522. doi: 10.1093/annonc/mdu565. [DOI] [PubMed] [Google Scholar]
- 23. Grassmann F, He W, Eriksson M, et al. Interval breast cancer is associated with other types of tumors. Nat Commun. 2019;10:4648. doi: 10.1038/s41467-019-12652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shieh Y, Eklund M, Madlensky L, et al. Breast cancer screening in the precision medicine era: Risk-based screening in a population-based trial. J Natl Cancer Inst. 2017;109:djw290. doi: 10.1093/jnci/djw290. [DOI] [PubMed] [Google Scholar]
- 25. Roux A, Cholerton R, Sicsic J, et al. Study protocol comparing the ethical, psychological and socio-economic impact of personalised breast cancer screening to that of standard screening in the “My Personal Breast Screening” (MyPeBS) randomised clinical trial. BMC Cancer. 2022;22:507. doi: 10.1186/s12885-022-09484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ren W, Chen M, Qiao Y, et al. Global guidelines for breast cancer screening: A systematic review. Breast. 2022;64:85–99. doi: 10.1016/j.breast.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponti A, Anttila A, Ronco G, et al. Cancer Screening in the European Union (2017). Report on the implementation of the Council Recommendation on cancer screening (second report). https://screening.iarc.fr/EUreport.php
- 28. Gallagher S, Hughes E, Wagner S, et al. Association of a polygenic risk score with breast cancer among women carriers of high- and moderate-risk breast cancer genes. JAMA Netw Open. 2020;3:e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brewer HR, Jones ME, Schoemaker MJ, et al. Family history and risk of breast cancer: An analysis accounting for family structure. Breast Cancer Res Treat. 2017;165:193–200. doi: 10.1007/s10549-017-4325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mars N, Lindbohm JV, Della Briotta Parolo P, et al. Systematic comparison of family history and polygenic risk across 24 common diseases. Am J Hum Genet. 2022;109:2152–2162. doi: 10.1016/j.ajhg.2022.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Feng B, Miron A, et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: A prospective study from the Breast Cancer Family Registry and kConFab. Genet Med. 2017;19:30–35. doi: 10.1038/gim.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho WK, Tan MM, Mavaddat N, et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat Commun. 2020;11:3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Menko FH, Monkhorst K, Hogervorst FBL, et al. Challenges in breast cancer genetic testing. A call for novel forms of multidisciplinary care and long-term evaluation. Crit Rev Oncol Hematol. 2022;176:103642. doi: 10.1016/j.critrevonc.2022.103642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The FinnGen data may be accessed through Finnish Biobanks' FinBB portal (www.finbb.fi; email: info.fingenious@finbb.fi). The weights for our polygenic risk scores are available at PGS Catalog (https://www.pgscatalog.org/), PGS ID PGS000335.