Summary
Broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin (HA) stem of influenza A viruses (IAV) tend to be effective against either group 1 or group 2 viral diversity. In rarer cases, intergroup protective bnAbs can be generated by human antibody paratopes that accommodate the conserved glycan differences between the group 1 and group 2 stems. We applied germline-engaging nanoparticle immunogens to elicit a class of cross-group bnAbs from physiological precursor frequency within a humanized mouse model. Cross-group protection depended on the presence of the human bnAb precursors within the B cell repertoire, and the vaccine-expanded antibodies enriched for a N55T substitution in the CDRH2 loop, a hallmark of the bnAb class. Structurally, this single mutation introduced a flexible fulcrum to accommodate glycosylation differences and could alone enable cross-group protection. Thus broad IAV immunity can be expanded from the germline repertoire via minimal antigenic input and an exceptionally simple antibody development pathway.
Graphical Abstract
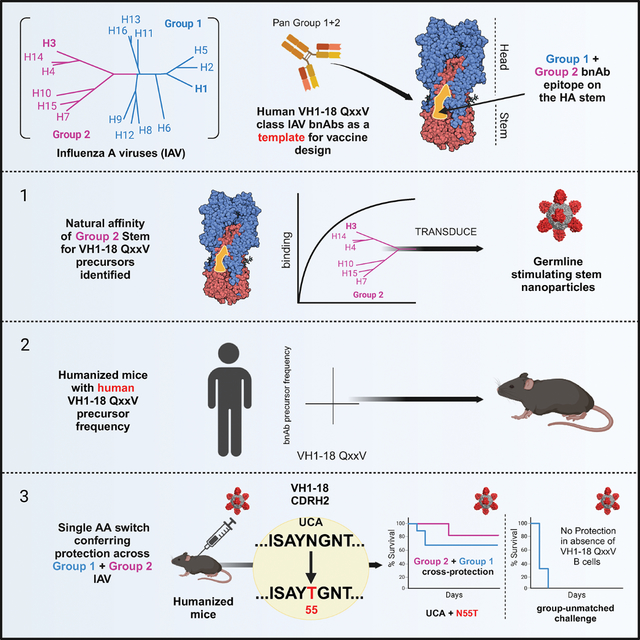
eTOC:
Differences in glycosylation prevent antibodies from widely engaging the otherwise conserved hemagglutinin (HA) stem of influenza A viruses (IAV). Ray et al. deploy a humanized mouse to elicit a broadly neutralizing antibody (bnAb) response that results in group 1 and group 2 IAV protection via a single amino acid mutation that tilts the immunoglobulin to accommodate glycan diversity.
Introduction
Influenza virus is a highly mutable pathogen that presents an ongoing diversity challenge to the immune system. A core issue is that hemagglutinin (HA), the major viral spike protein and seasonal vaccine immunogen, preferentially elicits antibodies against its hypervariable regions, limiting coverage and compromising pandemic preparedness1–4. Of the four influenza virus types (A, B, C, D), influenza A viruses (IAV) are responsible for a majority of infections in humans and are the source of influenza pandemics5–7. IAVs are phylogenetically divided into group 1 and group 2 viruses, and intragroup diversity is further subdivided into subtypes categorized by 18 distinct IAV HAs8,9. Broadly neutralizing antibodies (bnAbs) covering different levels of IAV diversity can be generated by targeting invariant features of HA, however these responses are immune-recessive/subdominant and will require rationally designed immune-focusing concepts to elicit higher titers1,2,10–13.
The majority of HA sequence diversity resides in its globular head domain, and ‘universal’ vaccine concepts currently under clinical evaluation include antibody-focusing to the relatively conserved stem or stalk domain of this protein14–16. These vaccines involve structure-based presentation of stem-only nanoparticles or sequential immunization with chimeric/head domain varying HAs to boost serum antibodies against conserved stem epitopes. Monoclonal human bnAbs can engage these sites to provide broad levels of coverage: heterosubtypic and the more exceptional cases of cross-group protection2,17–25. Within preclinical vaccine models, protective activity likely involves infection-blocking and Fc-mediated effector functions that are maximized when the bnAb sites are engaged26–29.
A feature of vaccine expanded stem responses in preclinical models is that coverage remains heterosubtypic and is not cross-group28,30–33. Differences between the glycosylation positions on the group 1 vs group 2 stem also impose steric constraints that hinder intergroup-reactivity to bnAb supersites32,34,35. There is no single stem immunogen that has been shown capable of eliciting intergroup IAV bnAbs. Rather, intergroup coverage has required co-formulation of group 1 + group 2 stem immunogens36.
In this study, we sought to elicit intergroup protective antibodies using a single vaccine immunogen by expanding germline BCR precursors that give rise to these bnAbs in humans. Functionally convergent human intergroup bnAbs are generated through public/shared antibody classes marked by usage of specific VH genes, DH genes and/or CDRH3 signatures20,22,24,25 and they have the potential to be triggered and expanded in vivo by different stem-focusing vaccine modalities37,38. We applied our one-step CRISPR/Cas9-induced homology-directed repair (HDR) platform39,40 to produce a humanized mouse vaccine model containing physiologically relevant recombination frequency of germline BCRs giving rise to intergroup bnAbs of the VH1–18 QxxV class24,37. We then immunized this mouse with recombinant nanoparticle displays of the group 2 HA stem37, which we found harbor natural binding affinity to VH1–18 QxxV bnAb precursors. The particles induced protection against group-unmatched IAV, and this was dependent on the bnAb precursors within our humanized mice. We found that after a single immunization, bnAb precursors were recruited to B cell germinal centers (GC) and underwent somatic hypermutation (SHM). Within 28 days post vaccination, potent and cross-protective bnAbs were elicited, and with only a fraction of mutations seen in the mature antibody from humans. Notably the immunogens enriched for N55T, a CDRH2 mutation present in all human VH1–18 QxxV bnAbs. We show that this mutation alone enables cross-group protection by providing a flexible fulcrum to pivot the antibody and accommodate the conserved glycosylation differences between the group 1 and 2 HA stems. Our findings thus reveal a simple molecular switch that can be triggered with minimal antigenic input to elicit exceptionally broad humoral immunity against IAV.
Results
VH1–18 QxxV bnAb precursors harbor natural affinity for HA stems from group 2 IAV
We began by examining the structural biology underscoring a prototypic pathway for development of VH1–18 QxxV class bnAbs: the elicitation of 09–1B12, a cross-group protective bnAb expanded in humans 24,37 (Figures 1A–C, S1, Tables S1, S2). Notably, the germline-inferred configuration of this antibody, 09–1B12-UCA, has reactivity to group 2 HA stems37 (see also Figure 1D) and we found that natural affinity for group 2 HA (H3 trimer from A/Perth/16/2009 and H7 trimer from A/Shanghai/02/2013) is also a generic feature of inferred VH1–18 QxxV bnAb precursors (10−6-10−8M) (Table S3). We leveraged this property to generate a 2.8 Å high resolution cryo-EM structure of 09–1B12-UCA in complex with the H3 trimer (A/Perth/16/2009) (Figure 1B,C). Comparison with the crystal structure of the mature 09–1B12, along with 16.g.07 (5KAN), another VH1–18 QxxV class bnAb20 indicates that germline affinity to the group 2 H3 stem is supplied by hallmark contacts of the VH1–18 QxxV class20,24,38: engagement via the “QxxV” positions in the CDRH3 along with VH-gene encoded contact by CDRH2 (Y54 and N/T55) (Figures 1B,C, S1). In addition, 16.g.07-like contacts in the antibody light chain CDRL1 (S/R at amino acid position 32) and FRW3 (S/R at amino acid positions 64, 66) were observed for both mature and UCA forms of 09–1B12 (Figure 1B,C, S1). The UCA also shares a 16.g.07-like CDRL1 (Q27) contact with the neighboring HA protomer (Q49), while in 09–1B12, the CDRL3 loop (D93 and T94) shows closer proximity to HA Q49 (Figure 1B,C). Collectively, these data point to the hallmark structural features of VH1–18 QxxV bnAbs as also responsible for the natural affinity between the bnAb precursor forms of this class and group 2 HA stems.
Figure 1. Germline VH1–18 QxxV bnAb shows natural affinity for HA from group 2 IAV.
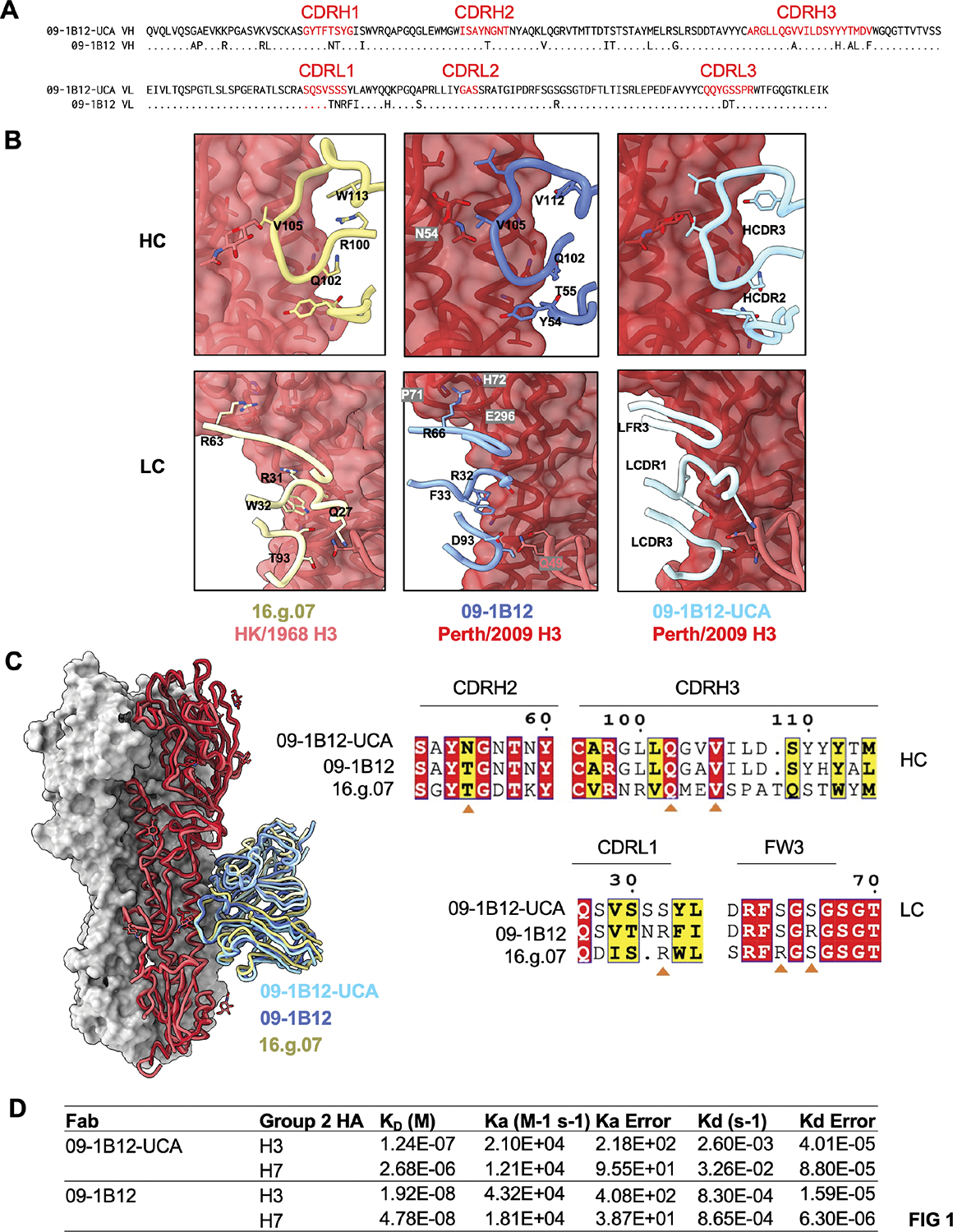
(A) Alignment of VH and VL amino acid sequences of 09–1B12-UCA vs mature 09–1B12. (B) CryoEM structure of 09–1B12-UCA and crystal structure of 09–1B12 in complex with H3 trimer bearing the H3ssF stem sequence (A/Perth/16/2009= red). Additional comparison is made with 16.g.07 (5KAN), another VH1–18 QxxV class bnAb20 (with H3 trimer A/Hong Kong/1/1968 = pink). (C) Overlay of the three antibodies along with conservation of antibody contacts sites in their HC and LC sequences (orange triangle). (D) Affinities of the germline versus mature forms of 09–1B12 for group 2 HA trimers (H3 from A/Perth/16/2009 and H7 from A/Shanghai/02/2013) [global fitting from four dilution curves for each antibody type (0.625 mM, 1.25 mM, 2.5 mM, and 5 mM), one experiment]. The HC and LC nucleotide sequences of 09–1B12-UCA was derived previously37. Natural gHgL affinities for H7 and H3 trimers are also seen for multiple VH1–18 QxxV bnAb class members (10−6-10−8 M) (Table S3). See also Figure S1 and Tables S1, S2 (in relation to B,C).
Generating a humanized mouse model that contains a physiologically relevant frequency of VH1–18 QxxV bnAb precursors within the B cell repertoire
Within pre-clinical vaccine models, generating cross-group protective IAV coverage has required co-formulation of group 1 + group 2 stem immunogens36. We developed a humanized mouse model to define whether individual stem immunogens could now elicit cross-group protective antibodies when VH1–18 QxxV class precursors were present, and at physiologically relevant frequency. In the first step, a 09–1B12-UCA KI mouse (H09−1B12-UCA/WT, κ09−1B12-UCA/WT) was generated using our CRISPR/Cas9-induced homologous directed recombination method 39,40 (Figures 2A–D, S2, S3). This mouse contained ~11% of paired HC + LC 09–1B12-UCA BCRs within the B220+ B cell repertoire (Figure 2A, S2A,B) and showed WT C57Bl/6-like B cell development in the bone marrow and periphery (Figure S3A–D). Importantly, we found that B220+ B cell cross-reactivity to H3ssF and H7ssF, two structure-guided group 2 stem-only ferritin nanoparticle immunogens derived from A/Perth/16/2009 and A/Shanghai/02/201337, was markedly elevated in the H09−1B12-UCA/WT, κ09−1B12-UCA/WT genotype relative to WT C57Bl/6 (Figure 2B,C). This cross-reactivity was specific to the central stem, as it did not include reactivity to epitope KO probes (H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO−, where KO = N-linked glycan at 45HA2) (Figure S2A). H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO− B cells were also >80% enriched for the UCA sequence, demonstrating the capacity of the H3ssF and H7ssF immunogens to select for this germline BCR from a diverse B cell repertoire (Figure 2C,D, S2A,C).
Figure 2. A group 2 HA stem immunogen elicits vaccine protection from group-unmatched influenza virus, but only when VH1–18 QxxV bnAb precursors are present at physiological frequency.
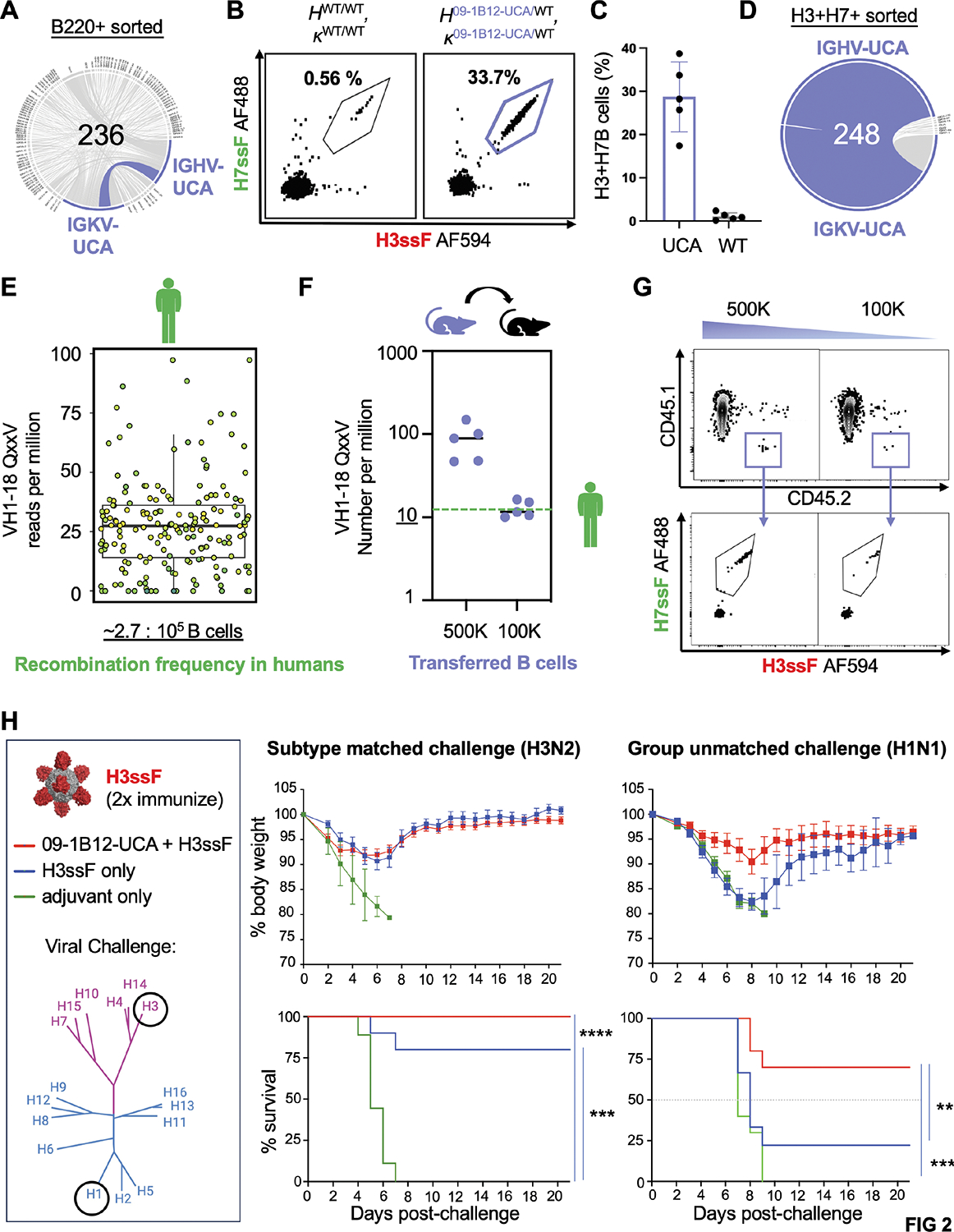
(A) HC and LC sequencing of B220+ cells from CD45.2 09–1B12-UCA donor mice (H09–1B12-UCA/WT, κ09–1B12-UCA/WT) (one mouse, one experiment). Of these cells 11.02% were paired IGHV/IGKV for the VH1–18 QxxV 09–1B12-UCA sequence. (B) B220+ B cell cross-reactivity to H3ssF-AF594 and H7ssF-AF488 in WT C57Bl/6 versus 09–1B12-UCA donor mice. These cross-reactive B cells were also not specific for the ferritin scaffold as they did not bind H3ssF-KO or H7ssF-KO (central stem epitope KO = N-linked glycan at 45HA2). (C) Proportion of the H3+/H7+ cross-reactive B cells in WT C57Bl/6 versus 09–1B12-UCA donor mice (H3ssF+/H7ssF+/ H3ssF-KO−/H7ssF-KO−) (n=5, mean ± SD, one experiment). (D) Enrichment of the VH1–18 QxxV UCA in cross-reactive B cells was observed at 83.5% (one mouse, one experiment). (E) Box plot of frequencies of the VH1–18 QxxV bnAb precursors among IgM antibody repertoires of n=10 human subjects41. A box plot containing the biological and technical replicates is presented; a separate breakdown of these values in each individual human subject is presented in Figure S4A. From this data we obtain a median bnAb precursor frequency of 2.7 per 100,000 B cells. (F,G) To recapitulate this value in the B cell repertoires of CD45.1 mice, we transferred CD45.2 09–1B12-UCA B cells (H09–1B12-UCA/WT, κ09–1B12-UCA/WT) at increasing amounts and then measured the VH1–18 QxxV bnAb precursor frequency (H3ssF+/H7ssF+/ H3ssF-KO−/H7ssF-KO−) in the spleen of the recipient mice. Transfer of 100,000 09–1B12-UCA B cells reproducibly gave a value of ~1 in 100,000 B cells (n=5 recipient mice per transfer amount, one experiment), consequently this was the VH1–18 QxxV bnAb precursor frequency used in all subsequent immunization experiments. (H) 09–1B12-UCA B cells were transferred (or not transferred) to recipient mice that were 2x sequentially immunized with H3ssF (day 0 prime + day 42 boost) and then lethally challenged (day 56) with subtype matched H3N2 influenza virus [108 TCID50/ml X-31] or group unmatched H1N1 influenza virus [104 TCID50/ml mouse-adapted A/California/2009 (maA/Cal/09)75,76]. The genetic distance of these influenza virus subtypes is also shown by the accompanying phylogenetic tree of HA IAV diversity (group 2 = purple, group 1 = blue). Weight loss and survival were monitored over 21 days. For subtype-matched H3N2 challenge: ****P<0.0001 between 09–1B12-UCA cells + H3ssF versus Sigma adjuvant; ***P<0.001 between H3ssF alone vs Sigma adjuvant (n=10 mice per group, Mantel-Cox test of survivorship, one experiment). For group-unmatched H1N1: **P<0.025 between 09–1B12-UCA cells + H3ssF vs H3ssF alone; ***P<0.001 between 09–1B12-UCA cells + H3ssF vs Sigma adjuvant alone, where the n=2 surviving from H3ssF only group (absence of 09–1B12-UCA cells) was not statistically significant (P>0.05) (n=10 mice per group, Mantel-Cox test of survivorship, one experiment). See also Figure S2 and S3 (in relation to A–D) and Figure S4 (in relation to E).
To construct a humanized mouse model containing physiological bnAb precursor frequency, we measured recombination frequency of VH1–18 QxxV bnAb precursors within a publicly available IgM database of heavy-chain repertoire-sequenced human subjects 41 (Figure 2E, S4). In these subjects, VH1–18 QxxV bnAb precursors were found with a median recombination frequency of ~2.7 in 100 000 B cells (Figure 2E, S4), similar to the value reported for VH6–1 class influenza bnAb precursors 42. We then performed adoptive transfer of B cells from H09−1B12-UCA/WT, κ09−1B12-UCA/WT to CD45.1 C57Bl/6 mice and measured the resultant H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO−/CD45.2+/CD45.1− B cells (= bnAb precursor) in the recipient animals (Figure 2F,G). There was a dose-response relationship between the number of B cells transferred and the VH1–18 QxxV bnAb precursor frequency established within the recipient spleen, akin to other humanized mouse systems deploying adoptive B cell transfer39,40,43–45. To match humans, we set the VH1–18 QxxV bnAb precursor frequency to 1 in 100 000 B cells in all downstream immunization / immune challenge experiments. Collectively, this established a humanized mouse model containing VH1–18 QxxV bnAb class precursors at a B cell repertoire frequency that matched VH1–18 QxxV recombination frequency seen in humans.
Physiological B cell repertoire frequency of VH1–18 QxxV bnAb precursors enables vaccine-protection from group-unmatched IAV
H3ssF elicits heterosubtypic immunity within group 2 IAV, but this does not extend to group 1 IAV37; coformulation of group 1 and group 2 immunogens is needed for cross group protection36. To initially define whether the presence of VH1–18 QxxV bnAb precursors (at human-like recombination frequency, see also Figure 2E–F) could enable cross group protection, we sequentially (2x) immunized animals containing (or not containing) 09–1B12-UCA B cells (at human precursor frequency) with H3ssF (+ Sigma adjuvant) and then lethally challenged with subtype-matched H3N2 or group-unmatched H1N1 influenza viruses (Figure 2H). We found that H3ssF protected against subtype-matched H3N2 virus in the absence of UCA cells, but required their presence for protection against the group-unmatched H1N1 virus. Hence, the inclusion of VH1–18 QxxV bnAb precursors into a diverse host B cell repertoire at human-like recombination frequency enabled the elicitation of cross-group protective immunity, all using a single recombinant vaccine immunogen.
Individual stem nanoparticles selectively expand and mature the bnAb precursors within B cell germinal centers in the humanized mice
To investigate the B cell pathways underscoring cross-group protection, we tracked CD45.2 VH1–18 QxxV UCA lineage expansion after a single immunization. In these experiments, we deployed H3ssF and H7ssF37 as higher and lower affinity germline stimulating immunogens (Figure 3A); monomeric UCA BCR affinities for the H3 and H7 stems from these antigens is separated by a logfold difference (1.24e-07 vs 2.68e-06 M) (see also Figure 1D, Table S3). H3ssF and H7ssF (+Sigma adjuvant) triggered comparable recruitment of CD45.2 B cells to germinal centers (GCs) within the CD45.1 hosts after one immunization step (Figure 3B–D, Figure S5A,B). Naked ferritin particles were also injected with Sigma Adjuvant (Figure 3A) and while this elevated GC reactions in the CD45.1 host (Figure 3B,C), it was not accompanied by recruitment of CD45.2 UCA B cells (Figure 3D). Thus, expansion of the CD45.2 UCA B cells was dependent on the stem-antigens. We further confirmed that GC-recruited CD45.2 B cells were targeting the central stem epitope (H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO−, epitope KO = insertion of N-linked glycan at 45HA2) (Figure 3E, S5A,B). If we exchanged our nanoparticle flow cytometry probes for H7 and H3 trimers, we recapitulated our key findings: expansion of H3+/H7+ cross-reactive CD45.2 B cells into GCs at 8, 15 and 28 days after immunizing with H3ssF or H7ssF, and failure to trigger this response if ferritin alone is deployed as the immunogen (Figure S5C–G).
Figure 3. Stem nanoparticles selectively expand VH1–18 QxxV bnAb precursors from physiological frequency in the antibody repertoire and induce diversification through somatic hypermutation in GCs.
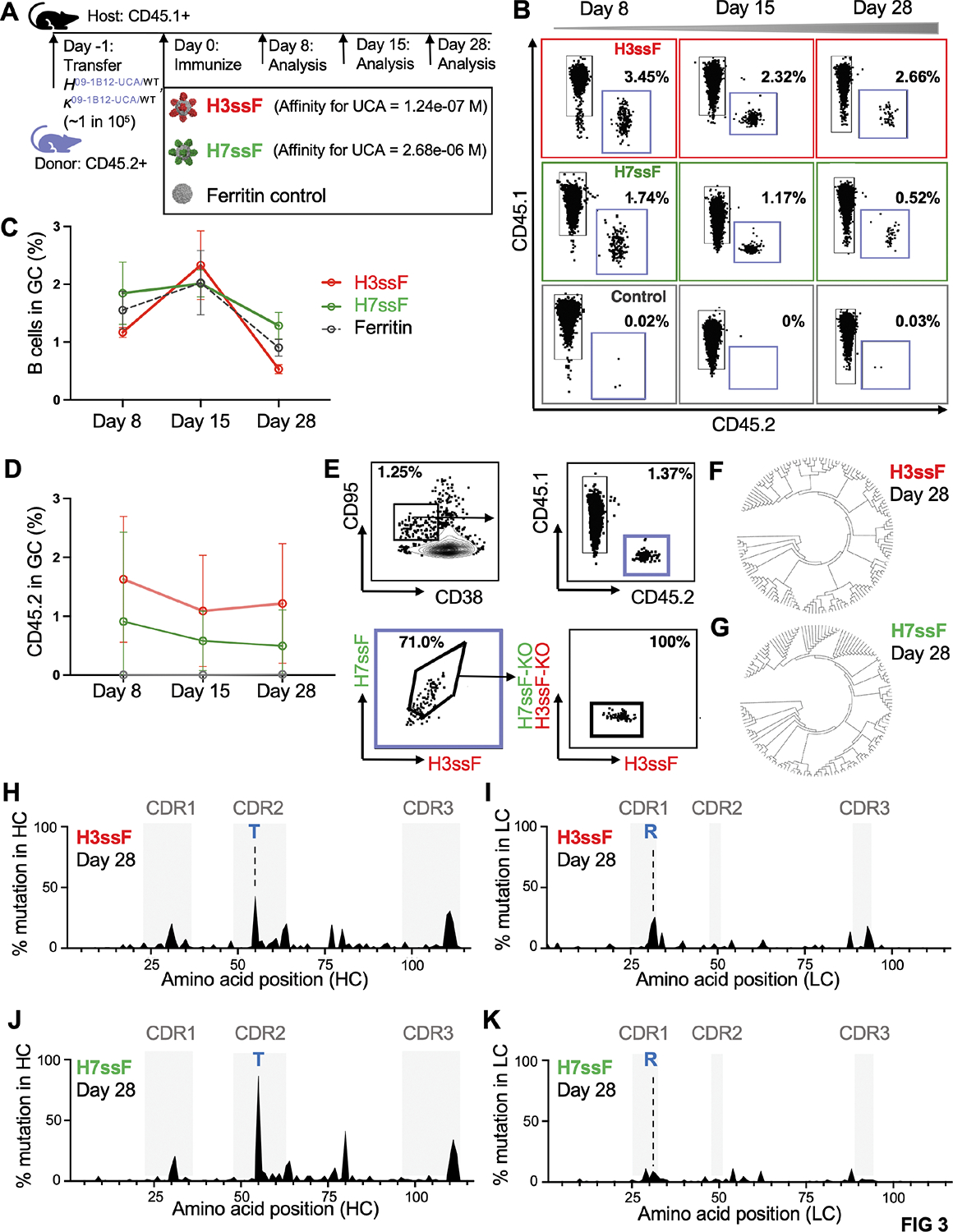
(A) Schematic of adoptive transfer performed at precursor frequencies of ~1 per 105 09–1B12-UCA B cells into WT mice at day −1 and subsequent single immunization of higher affinity H3ssF or lower affinity H7ssF. Naked ferritin particles were also given as a control. All vaccines were adjuvanted by the Sigma Adjuvant System. Spleens were sampled at the time points indicated. (B) Representative flow plots of CD45.2 B cells being recruited to GCs at days 8, 15, and 28 post-vaccination. (C) The percentage of GC B cells in the CD45.1 host was quantified at each time point (n=5 mice per immunogen, mean ± SD, one experiment). (D) The percentage of CD45.2 B cells within the host GCs each time point (n=5 mice per immunogen, mean ± SD, one experiment). (E) The GC CD45.2 B cells were also marked by epitope specificity to the central stem site [H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO− (central stem epitope KO =N-linked glycan at 45HA2)] and single GC CD45.2 B cells in this gate were sorted by FACS and subjected to BCR sequencing. Results presented in B-E were recapitulated if H7 and H3 trimers were used (instead of nanoparticles) as the antigen B cell probes (Figure S5C–G). (F, G) HC nucleotide diversification of H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO− B cell clones at 28 days after immunization with H3ssF or H7ssF. (H) Mutation frequency in the HC amino acid sequence at 28 days post immunization with H3ssF. (I) Mutation frequency in the paired LC amino sequence at 28 days post immunization with H3ssF. (J) Mutation frequency in the HC amino sequence at 28 days post immunization with H7ssF. (K) Mutation frequency in the paired LC amino sequence at 28 days post immunization with H7ssF. (H-K) Blue letters mark enrichment of amino acid mutations also present in mature 09–1B12 and 16.g.07. Data from (F-K) is from 125 GC BCRs for H3ssF (n=3 mice), 111 GC BCRs for H7ssF (n=3 mice); one experiment. See also Figure S5 (in relation to A–E) and Table S4 (in relation to F–K).
Single cell BCR sequencing of the CD45.2+/CD45.1−/H3ssF+/H7ssF+/H3ssF-KO−/ H7ssF-KO− GC B cells revealed diversification through SHM in response to both H3ssF and H7ssF (Figure 3F–K; Table S4). SHM was concentrated in the CDR1–3 regions of the HC and LC (Figure 3H–K), and mutations also accumulated in FW3 of the HC and LC (Figure 3H–K). Notably, the mutations N55T (CDRH2) and S32R (CDRL1) enriched in the vaccine-expanded BCRs (Figure 3H–K) are also present in the mature 09–1B12 and 16.g.07 (Figure 1B,C). In the mature antibodies, contact to the group 1 stem is greatly strengthened by S32R in CDRL1 (Figure 1). The importance of N55T, a hallmark of the VH1–18 QxxV bnAb class in humans24, is detailed in the following sections. Collectively, these data indicate that our immunogens: (A) selectively trigger and expand the VH1–18 QxxV bnAb lineage from a diverse B cell repertoire bearing bnAb precursors at human-like recombination frequency; and (B) guide SHM and affinity selection to enrich for some hallmark mutations seen for this bnAb class in humans.
The stem nanoparticles elicit cross-group protective bnAbs with minimal SHM after a single immunization.
We expressed two example mAbs (O1 and O2) that were expanded by H3ssF or H7ssF, respectively (Figure 4A, Table S4). These antibodies harbored some of the amino acid mutations present in mature 09–1B12, however they also showed far lower SHM (Figure 4A). Despite this, these vaccine elicited antibodies showed 09–1B12-equivalent activities: (1) comparable neutralization activity across group 2 IAV, along with more limited neutralization of group 1 IAV, as expected for the VH1–18 QxxV bnAb class24,46 (Figure 4B); (2) comparable protection against H3N2 viral challenge following passive transfer to C57Bl/6 WT mice (Figure 4C); and (3) comparable protection against H1N1 viral challenge following passive transfer to C57Bl/6 WT mice (Figure 4D). These results indicate that cross-group protective antibodies were elicited after a single immunization, and achieved fully functional somatic activity with minimal SHM.
Figure 4. Vaccine expansion of cross-group protective IAV bnAbs that use minimal SHM.
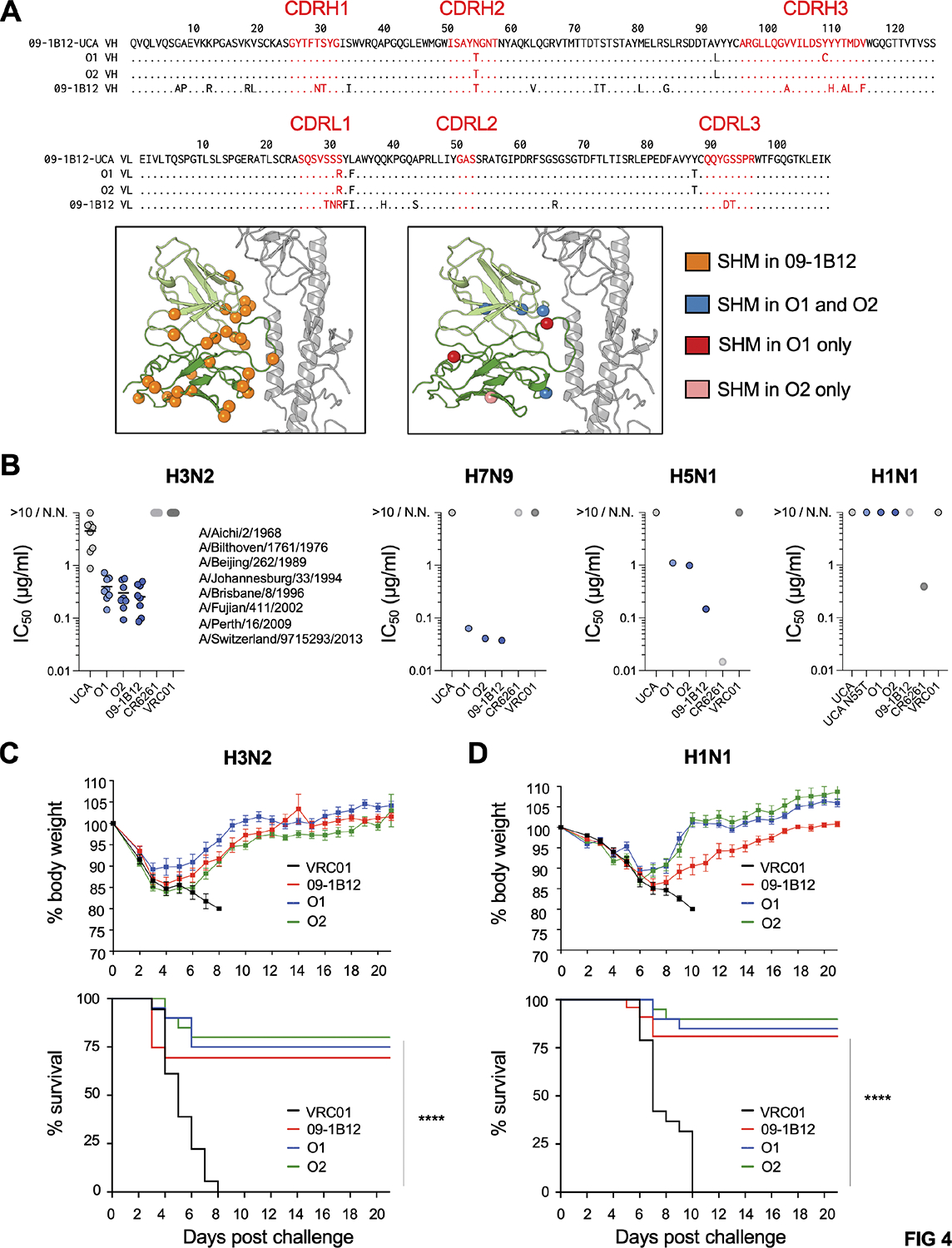
(A) O1 and O2 were example mAbs expanded after a single immunization with H3ssF or H7ssF, respectively. O1 and O2 sequences contain some of the mutations present in mature 09–1B12, but have far lower SHM, as noted in the amino acid alignments and antibody structure. (B) Neutralization activities of 09–1B12-UCA, O1, O2 and 09–1B12 across: H3N2 (eight diverse viral strains covering >50 years of diversification: A/Aichi/2/1968, A/Bilthoven/1761/1976 A/Beijing/353/1989, A/Johannesburg/33/1994, A/Brisbane/8/1996, A/Fujian/411/2002, A/Perth/16/2009, A/Switzerland/9715293/2013); H7N9 (A/Shanghai/02/2013); H5N1 (A/Vietnam/1203/2004); and H1N1 (A/California/07/2009). Cross-group protecting VH1–18 QxxV bnAbs neutralize group 2 viruses but show more limited neutralizing activity against group 1 IAV24,46. VRC01 served as an isotype control, 09–1B12 was a positive control for group 2 IAV neutralization and CR6261 was a positive control for group 1 IAV neutralization. N.N. = non-neutralizing (each antibody was run in triplicate, one experiment). (C) Weight loss and survival from lethal H3N2 virus challenge (108 TCID50/ml X-31) following passive transfer of 5 mg/kg O1, O2, 09–1B12 or VRC01 as isotype control [n=20 mice per group, ****P<0.001 (Mantel-Cox test of survivorship), one experiment]. (D) Weight loss and survival from lethal H1N1 challenge [104 TCID50/ml maA/Cal/0975,76] following passive transfer of 5 mg/kg O1, O2, 09–1B12 or VRC01 as isotype control [n=20 mice per group, ****P<0.0001 (Mantel-Cox test of survivorship), one experiment]. See also Table S4 (in relation to A).
Enrichment of N55T within the CDRH2 of germinal center BCRs and serum antibodies reveals a simple molecular switch conferring group 1 + group 2 IAV protection
The H3+/H7+ cross-reactive 09–1B12-UCA BCRs expanded within GCs were enriched for the mutation N55T within the CDRH2 loop (Figure 3H,J, Table S4). Skewed use of this mutation was seen at 28 days post-immunization with H3ssF or H7ssF, but it also enriched earlier at 15 days when the higher affinity H3ssF was used as the immunogen (Figure 5A,B, Table S4). This enrichment was also observed in the serum antibodies elicited following sequential immunization with either H3ssF or H7ssF (day 0 prime + day 42 boost), as evaluated by cryoEMPEM 47,48 at 15 and 28 days post-boost (Figure 5C–H, S1, Table S1). At these timepoints, the serum antibodies had specificity to the central stem epitope, as judged by differential reactivity to H3ssF/H7ssF ± stem epitope KO (N-linked glycan at 45HA2) (Figure 5C,F). For cryoEMPEM, Fabs purified from the immune sera were complexed with HA trimers matched to H3ssF or H7ssF (A/Perth/16/2009 and A/Shanghai/02/2013, respectively), confirming antibody targeting to the central stem (Figure 5D,G, I). The significant differences of the local map resolutions at CDRH2 loop as well as the volumes around residue 55 between H3 and H7 complexes suggest H7 preferably binds to Fabs that harboring T55 but not N55, which is consistent with the binding affinity data (KDs of H7 binding to 09–1B12-UCA vs 09–1B12-UCA + N55T, Table S3). Notably, when the same H3ssF or H7ssF immunization regimens were applied to WT C57Bl/6 mice (not bearing 09–1B12-UCA cells), we failed to resolve Fab density for the full-length H3 or H7 trimer, further underscoring importance of B cell repertoire in eliciting antibodies against the central stem epitope (Figure 5E,H). When the bnAb precursors are in place, our results highlight N55T as a vaccine-selected mutation, both in the GC and serum antibody response.
Figure 5. Stem focused VH1–18 QxxV responses enrich for the public mutation N55T within germinal centers and serum antibodies elicited by stem nanoparticles.
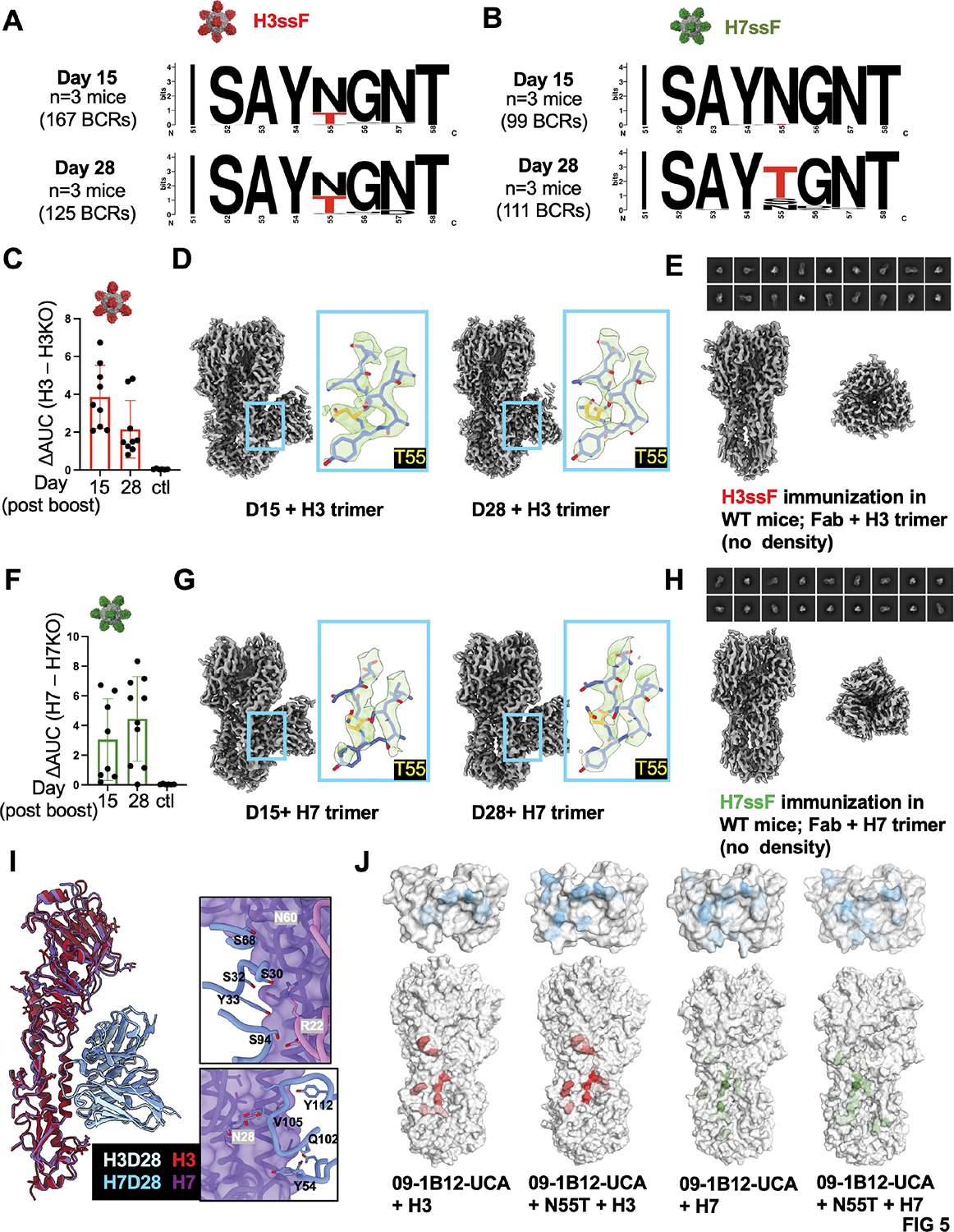
(A, B) Logo plots of the CDRH2 domain of the VH1–18 QxxV class precursors enriched in the GCs at 15 and 28 days post-immunizations with H3ssF or H7ssF (see also Figure 3F–K; 167 GC BCRs for H3ssF at Day 15 (n=3 mice), 125 GC BCRs for H3ssF at Day 28 (n=3 mice), 99 GC BCRs for H7ssF at Day 15, 111 GC BCRs for H7ssF at Day 28 (n=3 mice); one experiment). (C-I) CryoEMPEM was performed on VH1–18 QxxV antibodies elicited in the serum after sequential immunization (day 0 prime + day 42 boost) with either H3ssF or H7ssF. In all cases immune sera were evaluated at 15 and 28 days post-boost. (C) H3ssF elicited IgG showing differential reactivity to H3 ± epitope KO (central stem epitope KO= N-linked glycan at 45HA2); adjuvant only is the control (n=9 at Day 15 and n=9 mice at Day 28; n=6 control mice received adjuvant only; one experiment). (D) CryoEMPEM of H3ssF immune sera at 15 and 28 days post-boost, with antibodies in complex with H3 trimer (A/Perth/16/2009) [immune sera pooled from all mice at Day 15 (one experiment); immune sera pooled from all mice at Day 28 (one experiment)]. The 09–1B12-UCA Fab was docked into the maps and the HC N55 residues are boxed in red. (E) No density of 09–1B12-like Fab in complex with H3 trimer was found in the immune sera from WT C57Bl/6 mice (lacking VH1–18 QxxV bnAb precursors) subjected to the same H3ssF immunization regimen (28 days post-boost is shown) (immune sera pooled from n= 10 mice, one experiment). (F) H7ssF-elicited IgG showing differential reactivity to H7 ± epitope KO (central stem epitope KO= N-linked glycan at 45HA2); adjuvant only is the control (n=8 mice at Day 15 and n=9 mice at Day 28; n=6 control mice received adjuvant only; one experiment). (G) CryoEMPEM of H7ssF immune sera at 15 and 28 days post-boost, with antibodies in complex with H7 trimer (A/Shanghai/02/2013) [immune sera pooled from all mice at Day 15 (one experiment); immune sera pooled from all mice at Day 28 (one experiment)]. The 09–1B12-UCA Fab was docked into the maps and the HC T55 residues are yellow. (H) No density of 09–1B12-like Fab in complex with H7 trimer was found in the immune sera from WT C57Bl/6 mice (lacking VH1–18 QxxV bnAb precursors) subjected to the same H7ssF immunization regimen (28 days post-boost is shown) (immune sera pooled from n= 10 mice, one experiment). (I) Overlapping cryo-EMPEM structure of H7ssF and H3ssF immune sera at 28 days post-boost [09–1B12-UCA Fab docked in; with LC contacts (upper panel) and HC contacts (lower panel) shown]. (J) MD simulations of 09–1B12-UCA in contact with H3 (A/Perth/16/2009) or H7 (A/Shanghai/02/2013) trimers ± N55T. The color gradient on the surface indicates the interaction time for hydrogen bonds and charged interactions (darker colors means longer interaction time). See also Table S4 (in relation to A,B) and Figure S1, Table S1 (in relation to D,G,I).
Given that N55T is a marker of the VH1–18 QxxV bnAb class in humans24, we initially assessed the consequences of this mutation by performing molecular dynamics (MD) simulations of the 09–1B12-UCA in complex with H3 or H7 trimers (Figure 5J). In both scenarios T55 increased the interaction time and interaction areas of the QxxV motif, and also increased LC interactions with HA (Figure 5J). Accordingly, we biochemically evaluated the contribution of N55T to recognition of both group 2 and group 1 HAs for UCAs inferred from six different human VH1–18 QxxV class bnAbs (reversion of sHsL to gHgL) (Figure 6A). We found that for all antibodies, N55T strengthened germline-encoded antibody affinity to group 2 HA and also enabled binding to group 1 HAs, which was otherwise undetectable with the UCA (Figure 6A, Table S3). Alone, N55T also provided broad neutralizing activity against group 2 IAV (Figure 6B), a hallmark activity of mature VH1–18 QxxV bnAbs24,46. To define whether this single mutation alone enabled broad cross-group protection, we performed passive antibody transfer of 09–1B12-UCA mAb ± N55T into C57Bl/6 WT mice and then challenged with lethal doses of H3N2 or H1N1 viruses (Figure 6C,D). We observed a similar result after challenging from both viruses: the UCA form of the antibody failed to protect, however addition of N55T provided cross-group protection for >50% of recipient animals, approaching the activity of the mature 09–1B12 (Figure 6C,D). Hence, this single vaccine-selected mutation could alone enable cross-group protection from IAV.
Figure 6. N55T alone enables cross-group recognition and protection.
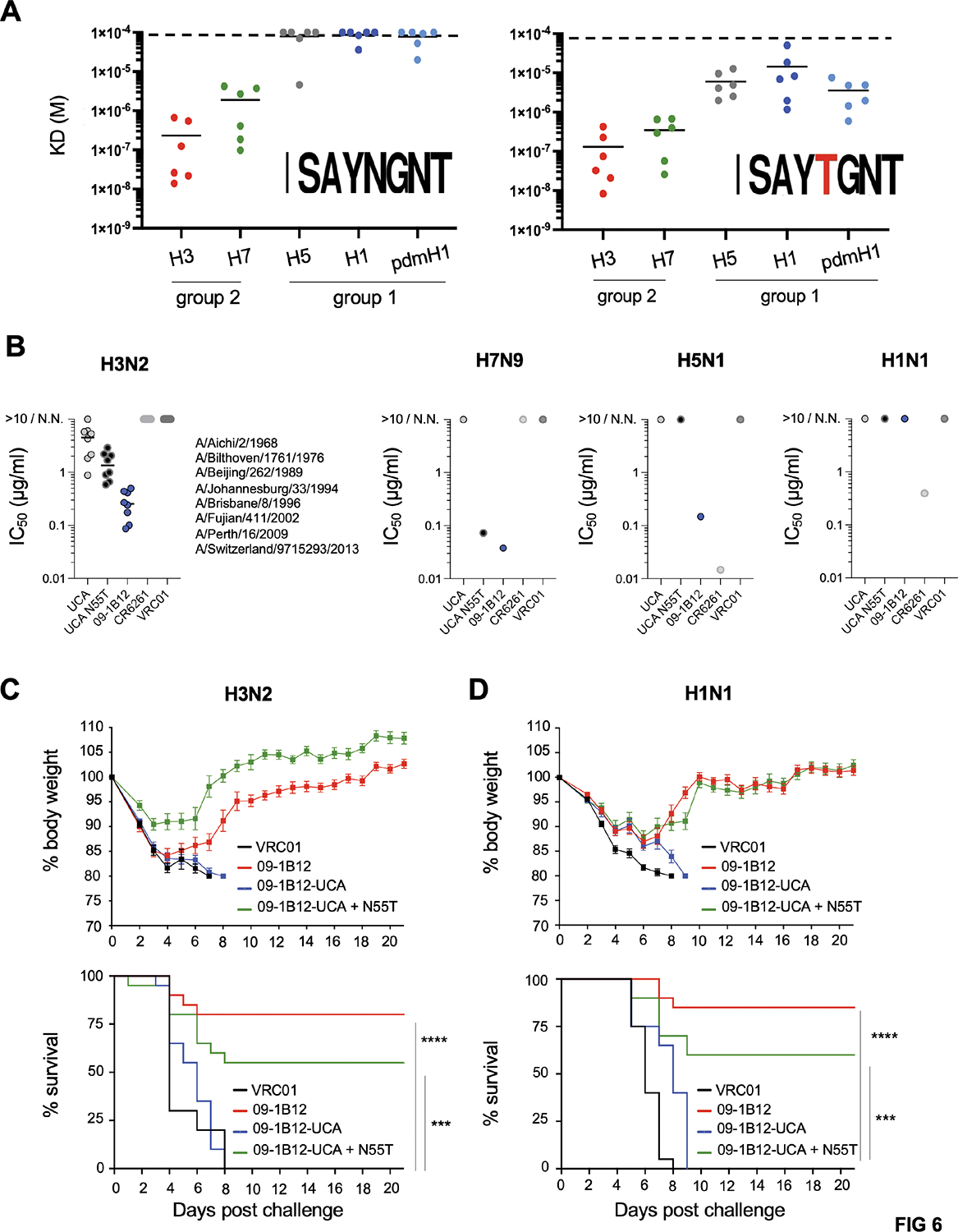
(A) Binding affinities for UCA-inferred Fabs from six VH1–18 QxxV bnAb class members ± N55T [09–1B12, 05–2A09, 05–2D04, 27–1D08, 21–1A01, 06–1F04] to group 1 and group 2 IAV HAs (H3 = A/Perth/16/2009; H7 = A/Shanghai/02/2013; H5 = A/Indonesia/05/2005; H1 = A/Michigan/45/2015; pdmH1 = A/California/07/2009) as measured by biolayer interferometry [global fitting from four dilution curves for each antibody type (0.625 mM, 1.25 mM, 2.5 mM, and 5 mM), one experiment]. Values above 100 uM are undetectable with our instrument77. (B) Neutralization activities of 09–1B12-UCA, 09–1B12-UCA + N55T, and 09–1B12 across: H3N2 (eight diverse viral strains covering >50 years of diversification: A/Aichi/2/1968, A/Bilthoven/1761/1976 A/Beijing/353/1989, A/Johannesburg/33/1994, A/Brisbane/8/1996, A/Fujian/411/2002, A/Perth/16/2009, A/Switzerland/9715293/2013); H7N9 (A/Shanghai/02/2013); H5N1 (A/Vietnam/1203/2004); and H1N1 (A/California/07/2009). Cross-group protecting VH1–18 QxxV bnAbs neutralize group 2 viruses but show more limited neutralizing activity against group 1 IAV24,46. N.N. = non-neutralizing. VRC01 served as an isotype control, 09–1B12 was a positive control for group 2 IAV neutralization and CR6261 was a positive control for group 1 IAV neutralization. The neutralization values obtained were generated within the same experiment shown in Figure 4B and therefore contain the same positive and negative control values (each antibody was run in triplicate, one experiment). (C) Weight loss and survival from lethal H3N2 virus challenge (108 TCID50/ml X-31) following passive transfer of 5 mg/kg 09–1B12-UCA, 09–1B12-UCA + N55T, 09–1B12, or VRC01 as the isotype control [n=20 mice per group, ****P<0.0001, ***P<0.001 (Mantel-Cox test of survivorship)]. (D) Weight loss and survival from lethal H1N1 virus challenge [104 TCID50/ml maA/Cal/0975,76] following passive transfer of 5 mg/kg 09–1B12-UCA, 09–1B12-UCA + N55T, 09–1B12 or VRC01 as isotype control [n=20 mice per group, ****P<0.0001, ***P<0.001 (Mantel-Cox test of survivorship)]. See also Table S3 (in relation to A) and Figure S6 (in relation to A–D).
N55T provides a ‘fulcrum release’ to accommodate conserved group 1 N-glycans, enabling dual recognition of group 1 and group 2 stems.
By overlaying the complexes of H3 + UCA or 09–1B12, a shift in Fab binding angles was observed, driven by the movements of HC N55T and LC S66R mutations (Figure 7A, S1). Notably the CDRH3 loop (QxxV) is at the fulcrum of this rotation (Figure 7A). This flexibility around the QxxV fulcrum is underscored by our finding that N55 forms more hydrogen bonds with the other residues within CDRH2/3 loops (S52, N57, and Q102), while T55 showed fewer interactions with those residues (Figure 7B,C). Collectively, this indicates that the CDRH2 loop with T55 is more flexible to accommodate the changes in Fab binding angles. To determine the structural contribution of N55T for group 1 HA, we resolved a co-complex of H1 trimer (A/Michigan/45/2015) with 09–1B12-UCA + N55T (3.5 Å) and mature 09–1B12 (3.3 Å) (Figure 7D, S1). Notably, group 2 bnAbs can be prevented from recognizing the group 1 stem by clashing with conserved HA1 N-linked glycans32,34,35. We find that N55T driven flexibility enables dual recognition of group 1 HA through a mechanism we term ‘fulcrum release’. Here the group 1 IAV N-glycans at positions N289, N278, and N33 from the neighboring protomer interact with the antibody LC, tilting the antibody contact angle backward (Figure 7D). Fulcrum release enables UCA + N55T (Figure 7E) and 09–1B12 (Figure 7F) to accommodate this tilt and approach the central stem epitope and to avoid clashing with the group 1 N-glycans. Hence, we provide the structural basis for a single amino acid mutation in the germline encoded CDRH2 loop that enables VH1–18 QxxV antibodies to engage group 1 and group 2 HA stems.
Figure 7. N55T provides a ‘fulcrum release’ to enable recognition of group 1 and group 2 HA stems.
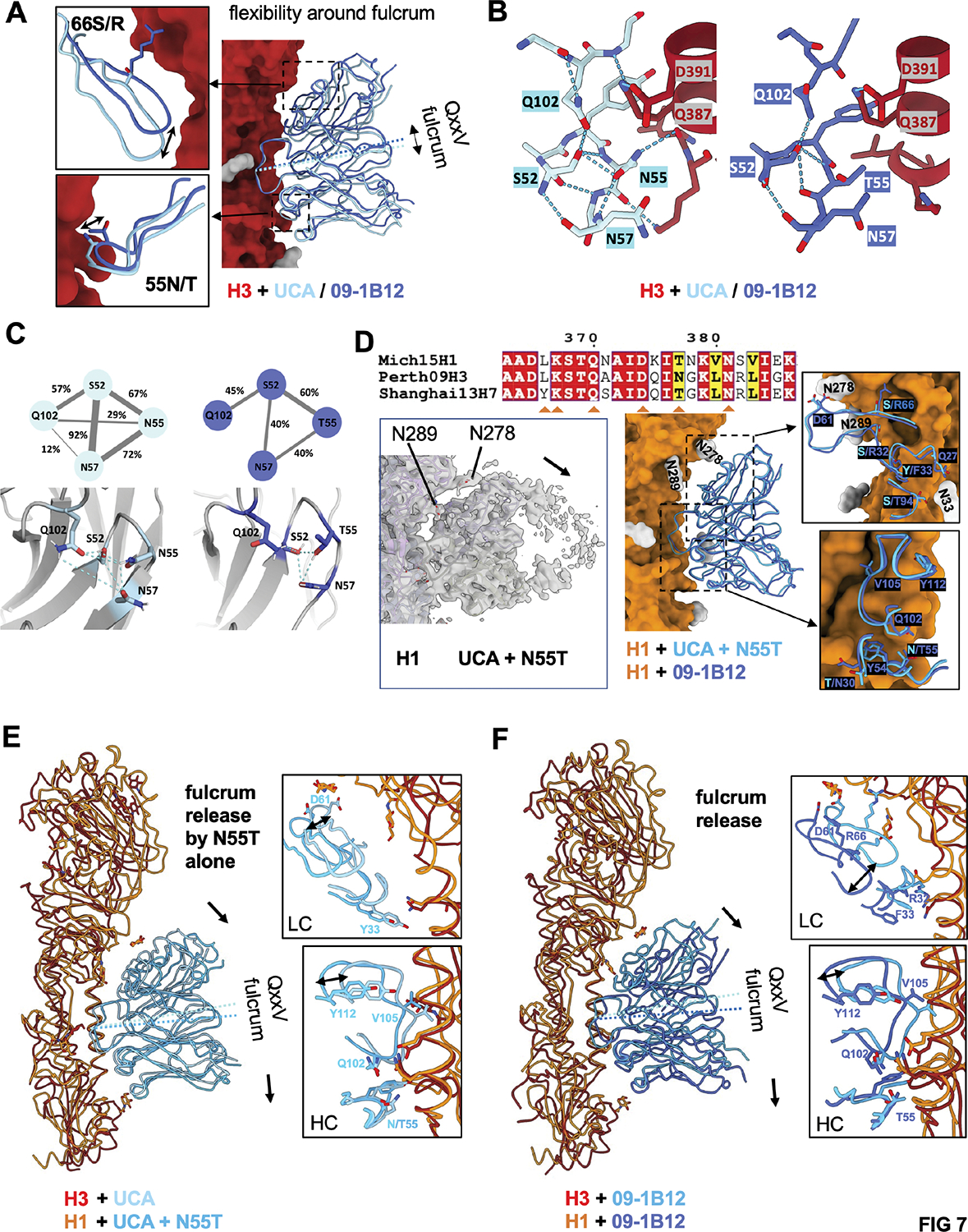
(A) Overlay of the 09–1B12-UCA and mature 09–1B12 in complex with H3 (A/Perth/16/2009; see also Figures 1B,C, S1, Tables S1, S2) illustrates the angular change where the HC and LC pivot around the QxxV fulcrum (=flexibility around the fulcrum). (B) N55 forms more hydrogen bonds with S52, N57, and Q102 in the CDRH2 and CDRH3 loops (visualized using ChimeraX). By contrast, T55 shows substantially fewer interactions to better accommodate changes in the binding angles. (C) MD simulations on the Fabs alone (without HA) show the percentages of interactions between paired residues (S52, N/T55, N57, and Q102) during simulations. (D) The cryoEM map of 09–1B12-UCA + N55T and overlayed structures of 09–1B12-UCA + N55T/09–1B12 in complex with H1 (A/Michigan/45/2015) (3.5 and 3.3 Å, respectively). A rotational tilt is imposed by group 1 IAV N-glycans at positions N289, N278, and N33 from the neighboring protomer interacting with the antibody LC. (E) N55T alone enables this tilting around the QxxV fulcrum (which we term fulcrum release) as visualized by overlay of H3 + 09–1B12-UCA co-complex with H1 + 09–1B12-UCA-N55T co-complex. (F) Fulcrum release as seen in the mature antibody as visualized by overlay of H3 + 09–1B12 co-complex with H1 + 09–1B12 co-complex. Arrows indicate the flexible antibody tilting enabled by fulcrum release to accommodate the conserved group 1 IAV glycan positions. See also Figure S1, Tables S1, S2.
N55T is a generic feature that enables cross-group protection by VH1–18 QxxV bnAb class members.
Given that N55T is present in all VH1–18 QxxV bnAbs, we evaluated its contribution to cross-group protection by two other members of this bnAb class: 21–1A01; and 05–2A0924 (Figure S6). We passively transferred the mature and inferred UCA forms of these antibodies (± N55T) into C57Bl/6 WT mice and then challenged with H3N2 or H1N1 viruses (Figures S6A,B). Akin to our previous findings with 09–1B12 ± N55T (Figure 6C,D), the UCA forms were comparable to the isotype control, and failed to enable statistically significant protection. However, addition of N55T provided significant cross-group protection for >50% of the mice, approaching the activity of mature forms (Figures S6A,B). Hence, we conclude that N55T, identified by selection within the antibody responses of our humanized mouse system, provides a simple and general molecular ‘switch’ that enables cross-group protection by VH1–18 QxxV bnAbs.
Discussion
Enhancing coverage of influenza virus diversity traditionally involves inclusion of more or different HA antigens in the seasonal vaccine49,50. This principle of additivity has been greatly extended by the recent development of an mRNA vaccine that encodes for HA representatives from all influenza A and B virus categories that elicits broad protection in mice51. By contrast, a central challenge for antibody-focusing concepts is to elicit broad coverage through a minimal set of rationally designed antigens1,2. Indeed, the compression of multiple protein functionalities within a single molecule is an important theme in the design of therapeutics and scalable biologics52,53. Our results highlight a simple molecular ‘switch’ that can be triggered by a single (and simple) recombinant HA immunogen to deliver exceptionally broad coverage across IAV.
In the absence of human bnAb precursors, HA nanoparticle immunogens (including H3ssF and H7ssF) can elicit heterosubtypic immunity against IAV subtypes, however these responses are not cross-group protective28,30–32,37. Under these conditions, elicitation of cross-group IAV antibodies relies on addition: co-formulation of group 1 + group 2 stem immunogens36. By contrast, our results demonstrate that vaccine-elicitation of cross-group protective immunity by a single stem immunogen can be enabled by specific germline-encoded features within the human antibody repertoire. We previously reported a human-repertoire prerequisite for vaccine-expanding group 1 bnAb classes within mice28,54,55 and there are analogous repertoire-requirements for expanding these bnAbs within non-human primates33. A low level of H3+/H7+cross-reactive B cells were initially present in the C57Bl/6 repertoire, however these cells likely lacked sufficient antigen affinity and/or frequencies to enable either vaccine protection against group-unmatched IAV or elicitation of high serum titers of central stem bnAbs that could be detected by cryoEMPEM. While there may be limits on our capacity to detect expansion of these host lineages, our results identify a decisive repertoire effect when VH1–18 QxxV bnAb precursors are present at physiological frequency, namely vaccine elicitation of cross-group protecting IAV bnAbs.
Both precursor frequency and BCR affinity for cognate antigen modulate recruitment to B cell GCs after immunization43,45,56. In our system, precursors could be expanded into cross-group protective bnAbs over a logfold range of germline affinities for the group 2 stem (10−6 - 10−7 M). Vaccine expansion of protective bnAbs across this affinity range was likely supported by fundamental avidity factors, such as stem nanoparticle valency and cell surface arrayed BCR57–64, and ultimately by the exceptionally low SHM requirement that we find is needed for full protective activity by VH1–18 QxxV class bnAbs. This minimal SHM greatly contrasts the complex affinity maturation pathways seen for human bnAbs against other hypervariable pathogens such as HIV65,66 and is exemplified by the N55T molecular switch enabling protection against group 1 and 2 IAV. The correlates of protection we observe are also consistent with VH1–18 QxxV bnAb class: broad neutralizing activity against group 2 viruses with non-neutralizing protection from group 1 IAV24.
The N55T substitution is not catalyzed by conventional mutation hotspots67,68, but it is nevertheless a marker of the human VH1–18 QxxV bnAb class20,24. Enrichment of N55T in the GCs expanded within our humanized mouse system pointed to a previously unrecognized functional importance for this signature that we then verified in other VH1–18 QxxV class bnAbs from humans. Our data indicates that this public amino acid substitution provides a ‘fulcrum release’ action that pivots the antibody to accommodate a conserved group 1 N-glycan and extend ‘hardcoded’ germline recognition of group 2-only to group 1 + 2 stems. Achieving this clash relief through a quite minor shift in the organization of the antibody paratope further underscores the low threshold for expanding this bnAb class.
It is uncertain how pre-existing immunity to HA antigens from prior infection and/or vaccination will modulate this simple pathway for bnAb development. Obligate recall of strain specific memory B cells through ‘primary addiction’ or related feedback effects has the potential to hamper expansion of the pathway69–72. However, the structurally similar group 1 stem nanoparticle, H1ssF, does successfully expand germline-encoded group 1 IAV bnAbs in humans14,15, suggesting that absence of an ‘immunodistractive’ HA head domain may be important for avoiding off-target memory recall when pre-existing immunity is present.
Collectively, our studies demonstrate proof-of-concept for vaccine expansion of unusually broad, cross-group protecting IAV bnAbs using a single recombinant immunogen. This is underscored by a simple molecular signature / switch that can be triggered from the human germline antibody repertoire to enable coverage through minimal immunological complexity.
Limitations of study
There are additional contacts that support cross-group-protection by VH1–18 QxxV class bnAbs20,24 and we have not assigned their hierarchy in relation to fulcrum release. There are also orthogonal/additional, non-VH1–18 QxxV bnAb classes in humans that enable cross-group protection20,22,24,25,38 and it is not yet clear if single immunogens will be capable of collectively expanding these pathways and if these bnAbs are also enabled by minimal SHM. We have also used germline inferred BCRs as opposed to bona fide bnAb precursors, although we would note that rationally designed germline stimulating immunogens originally based on inferred UCA have thus far all succeeded in selectively expanding their ‘authentic’ counterparts in human clinical trials14,15,31,73,74. Lastly, and as mentioned earlier, have not accounted for prior exposure to influenza virus or seasonal vaccines where different individual immune-histories may skew ‘intended’ humoral immunity through ‘unintended’ memory recall69–72.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Lingwood (dlingwood@mgh.harvard.edu).
Materials Availability
There are no restrictions on the availability on the materials used in this study.
Data and Code Availability
3D maps and models from the EM analysis have been deposited to the Protein Data Bank (http://www.rcsb.org/) and Electron Microscopy Databank (http://www.emdatabank.org/), respectively, and are publicly available from the date of publication. Accession numbers are listed in the Key Resources Table. Paired HC and LC sequences from the GCs expanded by H3ssF and H7ssF are deposited in Genbank. Accession numbers are provided in the Key Resources Table and in Table S4.
The complete code used to compute the frequencies of the VH1–18 QxxV class in human IgM repertoires has been deposited in Zenodo and is publicly available from the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PerCP/Cy5.5 anti-mouse CD45.1 Antibody | Biolegend | Cat#110728; RRID:AB_893348 |
| PE-Cy™7 Hamster Anti-Mouse CD95 | BD Biosciences | Cat#557653; RRID:AB_396768 |
| Brillant Violet 785™ anti-mouse CD45.2 | Biolegend | Cat#109839; RRID:AB_2562604 |
| Anti-mouse CD38, BV510 | BD Biosciences | Cat#740129; RRID:AB_2739886 |
| BV711 Rat Anti-Mouse Ig, λ1, λ2 & λ3 Light Chain | BD Biosciences | Cat#744527; RRID:AB_2742301 |
| BUV395 Rat Anti-Mouse Ig, κ light chain | BD Biosciences | Cat#742839; RRID:AB_2741090 |
| BV421 Rat Anti-Mouse IgM | BD Biosciences | Cat#743323; RRID:AB_2741424 |
| PE/Cy7 anti-mouse IgD Antibody | Biolegend | Cat#405720; RRID:AB_2561875 |
| PE anti-mouse CD45.2 Antibody | Biolegend | Cat#109808; RRID:AB_313445 |
| Anti-mouse CD38 Alexa700 | Invitrogen | Cat#56-0381-82 |
| BV786 Rat Anti-Mouse IgD | BD Biosciences | Cat#563618; RRID:AB_2738322 |
| Alexa Fluor® 700 anti-mouse IgD Antibody | BD Biosciences | Cat#405730 |
| BV605 Rat anti-mouse CD45R/B220 | BD Biosciences | Cat#563708; RRID:AB_2738383 |
| PE anti-mouse CD138 (Syndecan-1) Antibody | Biolegend | Cat#142504; RRID:AB_10915989 |
| BV510 Rat Anti-Mouse IgD | BD Biosciences | Cat#563110; RRID:AB_2737003 |
| Brilliant Violet 421™ anti-mouse IgD Antibody | Biolegend | Cat#405725; RRID:AB_2562743 |
| BV510 Rat Anti-Mouse B220/CD45R Clone RA3-6B2 | BD Biosciences | Cat#563103; RRID:AB_2738007 |
| BV421 Rat Anti-Mouse IgG1 | BD Biosciences | Cat#562580; RRID:AB_2737664 |
| BUV395 Rat Anti-Mouse IgM | BD Biosciences | Cat#743329; RRID:AB_2741430 |
| CD4 Monoclonal Antibody (GK1.5), APC-eFluor 780 | Invitrogen | Cat#47-0042-80 |
| CD8a Monoclonal Antibody (53-6.7), APC-eFluor 780 | Invitrogen | Cat#47-0081-80 |
| F4/80 Monoclonal Antibody (BM8), APC-eFluor 780 | Invitrogen | Cat#47-4801-80 |
| Ly-6G Monoclonal Antibody (1A8-Ly6g), APC-eFluor 780 | Invitrogen | Cat#47-5931-80 |
| FITC anti-mouse/human CD45R/B220 Antibody | Biolegend | Cat#103206; RRID:AB_312991 |
| APC anti-mouse CD43 Antibody | Biolegend | Cat#143208; RRID:AB_11149685 |
| APC/Cy7 anti-mouse CD21/CD35 (CR2/CR1) Antibody | Biolegend | Cat#123418; RRID:AB_1953275 |
| BV421 Rat Anti-Mouse CD24 | BD Biosciences | Cat#562563; RRID:AB_2737002 |
| Anti-mouse CD249 (BP1) FITC | Thermo | Cat#11-5891-82; RRID:AB_465295 |
| Alexa Fluor 700 anti-mouse TCRb chain | BioLegend | Cat#109224; RRID:AB_1027654 |
| FITC anti-mouse CD21/CD35 (CR2/CR1) Antibody | BioLegend | Cat#123408; RRID:AB_940403 |
| PerCP/Cy5.5 anti-mouse/human CD45R/B220 Antibody | BioLegend | Cat#103236; RRID:AB_893354 |
| APC anti-mouse CD23 Antibody | BioLegend | Cat#101620; RRID:AB_2563438 |
| PE anti-mouse CD24 Antibody | BioLegend | Cat#101808; RRID:AB_312840 |
| Anti Mouse CD4 BV510 | BioLegend | Cat#100559; RRID:AB_2562608 |
| BV421 Rat Anti-Mouse IgM | BD Biosciences | Cat#743323; RRID:AB_2741424 |
| Alkaline Phosphatase AffiniPure Goat Anti-Mouse IgG, Fcγ Fragment Specific | Jackson ImmunoResearch | Cat#115-055-071 |
| Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block™) | BD Biosciences | Cat#553142; RRID:AB_394657 |
| Alexa Fluor® 488 anti-mouse CD45.2 Antibody | Biolegend | Cat#109816; RRID:AB_492868 |
| Alexa Fluor® 594 anti-mouse CD3 Antibody | Biolegend | Cat#100240; RRID:AB_2563427 |
| TotalSeq™-C0301 anti-mouse Hashtag 1 Antibody | Biolegend | Cat#155861; RRID:AB_2800693 |
| TotalSeq™-C0302 anti-mouse Hashtag 2 Antibody | Biolegend | Cat#155863; RRID:AB_2800694 |
| TotalSeq™-C0303 anti-mouse Hashtag 3 Antibody | Biolegend | Cat#155865; RRID:AB_2800695 |
| TotalSeq™-C0304 anti-mouse Hashtag 4 Antibody | Biolegend | Cat#155867; RRID:AB_2800696 |
| TotalSeq™-C0305 anti-mouse Hashtag 5 Antibody | Biolegend | Cat#155869; RRID:AB_2800697 |
| TotalSeq™-C0306 anti-mouse Hashtag 6 Antibody | Biolegend | Cat#155871; RRID:AB_2819910 |
| TotalSeq™-C0307 anti-mouse Hashtag 7 Antibody | Biolegend | Cat#155873; RRID:AB_2819911 |
| TotalSeq™-C0308 anti-mouse Hashtag 8 Antibody | Biolegend | Cat#155875; RRID:AB_2819912 |
| TotalSeq™-C0309 anti-mouse Hashtag 9 Antibody | Biolegend | Cat#155877; RRID:AB_2819913 |
| TotalSeq™-C0310 anti-mouse Hashtag 10 Antibody | Biolegend | Cat#155879; RRID:AB_2819914 |
| 09-1B12-UCA | Produced in house (Corbett et al. 2019) | N/A |
| 09-1B12-UCA + N55T | Produced in house | N/A |
| 09-1B12 | Produced in house (Andrews et al. 2017) | N/A |
| 05-2A09-UCA | Produced in house | N/A |
| 05-2A09-UCA + N55T | Produced in house | N/A |
| 05-2A09 | Produced in house (Andrews et al. 2017) | N/A |
| 21-1A01-UCA | Produced in house | N/A |
| 21-1A01-UCA- + N55T | Produced in house | N/A |
| 21-1A01 | Produced in house (Andrews et al. 2017) | N/A |
| 05-2D04-UCA | Produced in house | N/A |
| 05-2D04-UCA + N55T | Produced in house | N/A |
| 05-2D04 | Produced in house (Andrews et al. 2017) | N/A |
| 27-1D08-UCA | Produced in house | N/A |
| 27-1D08-UCA + N55T | Produced in house | N/A |
| 27-1D08 | Produced in house (Andrews et al. 2017) | N/A |
| O1 | Produced in house | N/A |
| O2 | Produced in house | N/A |
| VRC01 | Produced in house | N/A |
| CR6261 | Produced in house | N/A |
| Biological samples | ||
| H3N2 X-31 | BEI Resources | Cat#NR-3483 |
| Mouse adapted H1N1 A/California/07/2009 (maA/Cal/09) | Sabra Klein and Andrew Pekosz John Hopkins University (Fink et al. 2018) | N/A |
| Influenza reporter viruses | Masaru Kanekiyo, NIH (Creanga et al. 2021) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Pan B Cell Isolation Kit II, mouse | Miltenyi Biotec | Cat#130-104-443 |
| LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit, for UV excitation | Thermo Fisher Scientific | Cat#L34962 |
| CountBright™ Absolute Counting Beads, for flow cytometry | Thermo Fisher Scientific | Cat#C36950 |
| SIGMAFAST™ p-Nitrophenyl phosphate Tablets | Sigma | Cat#N2770-50SET |
| NP40 | Millipore | Cat#492016-100ML |
| UltraComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Cat#01-2222-42 |
| H3ssF | Produced in house (Corbett et al. 2019) | N/A |
| H7ssF | Produced in house (Corbett et al. 2019) | N/A |
| Invitrogen™ Molecular Probes™ DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat#D1306 |
| Chromium Next GEM Single Cell 5’ Kit v2 | 10x Genomics | Cat#PN-1000263 |
| Library Construction Kit | 10x Genomics | Cat#PN-1000190 |
| Chromium Single Cell Mouse BCR Amplification Kit | 10x Genomics | Cat#PN-1000255 |
| Chromium Next GEM Chip K Single Cell Kit | 10x Genomics | Cat#PN-1000286 |
| Dual Index Kit TT Set A | 10x Genomics | Cat#PN-1000215 |
| Dual Index Kit TN Set A | 10x Genomics | Cat#PN-1000250 |
| Penicillin/streptomycin | Gibco | Cat#15140163 |
| Puromycin | Gibco | Cat#A1113803 |
| Opti-MEM | Gibco | Cat#31985088 |
| Bovine Serum Albumin | Sigma | Cat#A9418 |
| TransIT-LT1 | MirusBio | Cat#2306 |
| Gibson assembly kit | NEB | Cat#E5510S |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher Scientific | Cat#A14525 |
| Ni Sepharose resin | GE Healthcare | N/A |
| A/Perth/16/2009 H3 trimer | Produced in house (Joyce et al. 2016) | N/A |
| A/Shanghai/02/2013 H7 trimer | Produced in house (Joyce et al. 2016) | N/A |
| A/Michigan/45/2015 H1 trimer | This paper | N/A |
| A/California/07/2009 H1 trimer | Produced in house (Sangesland et al. 2019) | N/A |
| A/Japan/305/1957 H2 trimer | This paper | |
| A/Indonesia/05/2005 H5 trimer | Produced in house (Whittle et al. 2014) | N/A |
| A/Perth/16/2009 H3 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Shanghai/02/2013 H7 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Michigan/45/2015 H1 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/California/07/2009 H1 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Indonesia/05/2005 H5 trimerDstem (KO, 45HA2) | This paper | N/A |
| H3ssF-KO (45HA2) | This paper | N/A |
| H7ssF-KO (45HA2) | This paper | N/A |
| Sigma Adjuvant System | Sigma-Aldrich | Cat#S6322 |
| Protein G Sepharose | GE Healthcare | Cat#17061802 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#KK2602 |
| Maxima H Minus Reverse Transcriptase | Thermo Fisher | Cat#EP0751 |
| HotStarTaq Plus DNA Polymerase | QIAGEN | Cat#203603 |
| Buffer RLT | QIAGEN | Cat#79216 |
| BirA Biotin-Protein Ligase Bulk Reaction Kit | Avidity | Cat#Bulk BirA |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher | Cat#A20173 |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher | Cat#A10235 |
| Alexa Fluor 594 Protein Labeling Kit | Thermo Fisher | Cat#A10239 |
| Alexa Fluor 546 Protein Labeling Kit | Thermo Fisher | Cat#A20183 |
| MiSeq Reagent Kit, V2 500 cycles | Illumina | Cat#MS-102-2003 |
| 10% PEG 10k | Hampton Research | Cat#HR2-084 |
| Lys-C endoproteinase | NEB | Cat#P8109S |
| Protease inhibitor cocktail (PIC) | Roche | Cat#11697498001 |
| Deposited data | ||
| VH1-18 heavy chain (HC) and light chain (LC) sequences (see also Data S1) | This paper | GenBank IDs: PP507169 - PP508172 |
| X-ray structure of 09-1B12 bound to A/Perth/14/2009 H3N2 hemagglutinin | This paper | PDB ID: 8UWA |
| CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6 | This paper | PDB ID: 8UT3, EMDB ID: EMD-42528 |
| CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12 | This paper | PDB ID: 8UT4, EMDB ID: EMD-42529 |
| CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T | This paper | PDB ID: 8UT5, EMDB ID: EMD-42530 |
| CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | This paper | PDB ID: 8UT6, EMDB ID: EMD-42531 |
| CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | This paper | PDB ID: 8UT7, EMDB ID: EMD-42532 |
| CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-15 days post-immunization | This paper | PDB ID: 8UT8, EMDB ID: EMD-42533 |
| CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization | This paper | PDB ID: 8UT9, EMDB ID: EMD-42534 |
| CryoEM structure of A/Perth/16/2009 H3 | This paper | EMDB ID: EMD-42535 |
| CryoEM map of A/Shanghai/1/2013 H7 HA | This paper | EMDB ID: EMD-42536 |
| Code used to compute frequencies of the VH1-18 QxxV class in human IgM repertoires | This paper | https://www.doi.org/10.5281/zenodo.10800716 |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID:CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID:CVCL_D615 |
| Canine: MDCK | ATCC | Cat#CCL-34; RRID:CVCL_0422 |
| Human: HEK293F | Gibco | Cat#R79007; RRID:CVCL_6642 |
| MDCK-SIAT1-PB1 | Produced in house (Creanga et al. 2021) | N/A |
| MDCK-SIAT1-H5 | Produced in house (Creanga et al. 2021) | N/A |
| MDCK-SIAT1-H7 | Produced in house (Creanga et al. 2021) | N/A |
| Expi293F GnTI− | Produced in house | N/A |
| HEK293T-PB1 | Produced in house (Creanga et al. 2021) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: B6.SJL-Ptprcapepcb/BoyJ mice | The Jackson Laboratory | JAX: 002014; RRID:IMSR_JAX:002014 |
| Mouse: C57BL/6 | The Jackson Laboratory | JAX: 000664; RRID:IMSR_JAX:000664 |
| Mouse: 09-1B12-UCA KI mouse (H09-1B12-UCA/WT, k09-1B12-UCA/WT) | This paper | N/A |
| Oligonucleotides | ||
| TaqMan probes for genotyping | TransnetYX | N/A |
| Cocktails of IgG- and IgK-specific primers and thermocycling conditions described previously | von Boehmer et al. 2016 | N/A |
| sgRNA | Lin et al. 2018; Wang et al. 2021 | N/A |
| Primers for Single Cell BCR Amplification, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Flowjo 10.9.0 | Treestar | https://www.flowjo.com/; RRID:SCR_008520 |
| Prism 10.1.0 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| IMGT/V-quest | Brochet et al. 2008; Giudicelli et al. 2011 | http://www.imgt.org/IMGTindex/V-QUEST.php; RRID:SCR_010749 |
| Geneious Prime | Biomatters | https://www.geneious.com/; RRID:SCR_010519 |
| Cell Ranger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger; RRID:SCR_017344 |
| WebLogo | Crooks et al., 2004 | https://weblogo.berkeley.edu; RRID:SCR_010236 |
| Seurat R package | Standard R Package | https://satijalab.org/seurat/; RRID:SCR_016341 |
| R circlize 0.4.15 | R package | https://cran.r-project.org/web/packages/circlize/index.html; RRID:SCR_002141 |
| Bash 5.1.16 | GNU Project shell | https://www.gnu.org/software/bash/; RRID:SCR_021268 |
| Python 3.10 | Python Programming Language | https://www.python.org/: RRID:SCR_008394 |
| R 4.1.2 | The R project for statistical computing | https://www.r-project.org/; RRID:SCR_001905 |
| Tidyverse (dplyr 1.1.4, readr 2.1.4, ggplot2 3.4.4) | Wickham et al. 2019 | https://www.tidyverse.org/packages/; RRID:SCR_019186 |
| Pandas 2.1.1 | Pandas development team, 2020 | https://pandas.pydata.org; RRID:SCR_018214 |
| Pandaseq | Masella et al. 2012 | https://github.com/neufeld/pandaseq; RRID:SCR_002705 |
| MigMAP | Sangesland et al. 2019 | https://github.com/mikessh/migmap |
| MEGA11 software | Tamura et al. 2011, 2021 | https://www.megasoftware.net; RRID:SCR_023017 |
| UCSF ChimeraX | Pettersen et al.2021 | https://www.cgl.ucsf.edu/chimerax/; RRID:SCR_015872 |
| Coot 0.9.8 | Casanal et al. 2020 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID:SCR_014222 |
| Phenix | Afonine et al. 2018 | https://phenix-online.org; RRID:SCR_014224 |
| Rosetta | Conway et al. 2014 | https://www.rosettacommons.org/home; RRID:SCR_015701 |
| XDSGUI | XDS program package | https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/XDSGUI; RRID:SCR_015652 |
| Protonate3D tool | Molecular Operating Environment, 2020; Labute, 2009 | RRID:SCR_014882 |
| Amber Tools20 package | Case et al. 2020 | https://ambermd.org/index.php; RRID:SCR_014230 |
| NpT ensemble | Salomon-Ferrer et al. 2013 | N/A |
| SHAKE algorithm | Miyamoto, 1992 | N/A |
| Berendsen algorithm | Berendsen, 1984 | N/A |
| GetContacts software | This paper | https://getcontacts.github.io/ |
| Other | ||
| Mastercycler PCR machine | Eppendorf | N/A |
| ELISA readers | BioTek | N/A |
| 5 laser ARIA II | BD Biosciences | RRID:SCR_018091 |
| 5 laser LSR Fortessa | BD Biosciences | RRID:SCR_019601 |
| Qubit | Thermo Fisher | RRID:SCR_018095 |
| MiSeq | Illumina | RRID:SCR_016379 |
| Nextseq2000 | Illumina | RRID:SCR_023614 |
| Tapestation | Agilent | RRID:SCR_019394 |
| 5 Laser Aria SORP | BD Biosciences | N/A |
| Superdex 200 10/300 Column | GE Healthcare | Cat#17517501 |
| Superose 6 10/300 Column | GE Healthcare | Cat#17517201 |
| 96 Well glass bottom plate with high performance #1.5 cover glass | CellVis | Cat#P96-1.5P |
| CellDiscoverer7 | Zeiss | N/A |
| Tangential flow filtration system | Cytiva | N/A |
| ViewDrop II seals | SPT LabTech | Cat#4150-05600 |
| Erythrina cristagalli Gel-ECA-Immobilized Lectin | EY Laboratories | N/A |
| AKTA size exclusion chromatography (SEC) | Cytiva | RRID:SCR_023461 |
| IgG Elution Buffer | Pierce | Cat#21009 |
| Cu grids | EMS | Cat#CF300-Cu-UL |
| Vitrobot | Thermo Fisher | N/A |
| Glacios-2 cryoEM | Thermo Fisher | N/A |
| Beam line 24-ID-E | Advanced Photon Source | N/A |
| BLItz System (Octet N1) | Fortebio | RRID:SCR_023267 |
| Streptavidin biosensors (SA) | Sartorious | Cat#18-5019 |
| Prolab Isopro RMH 3000, 5P75-5P76 mice diet | LabDiet | https://www.labdiet.com/product/detail/5p76-prolab-isopro-rmh-3000 |
EXPERIMENTAL MODELS AND PARTICIPANT DETAILS
Generation of 09–1B12-UCA knock-in (KI) mice
09–1B12-UCA KI mice were generated following published protocols39,40. In brief, the targeting vector 4E1078 was modified by the incorporation of human rearranged 09–1B12-UCA KI VDJ (heavy chain construct) or VJ (light chain construct) sequences downstream of the promoter region and by elongation of the 5’ and 3’ homology regions using the Gibson assembly method (NEB). The targeting vector DNA was confirmed by Sanger sequencing (Eton Bioscience Inc.). Next, fertilized mouse oocytes were microinjected with a donor plasmid containing the pre-rearranged 09–1B12-UCA IGH with the mouse VHJ558 promoter, or the pre-rearranged 09–1B12-UCA IGK with the mouse Vk4–53 promoter (200 ng/mL); two pair of single-guide RNAs (sgRNAs, 25 ng/mL) targeting either the H or the k locus; and AltR-Cas9 protein (50 ng/mL) and injection buffer 39. The nucleotide sequence for 09–1B12-UCA IGH and IGK was from Corbett et al. 37. Following culture, resulting zygotes were implanted into the uteri of pseudopregnant surrogate C57Bl/6J mothers. F0-mice from the 09–1B12-UCA KI mouse (CD45.2+/+) colony were bred at the animal facility of the Gene Modification Facility (Harvard University) and breeding for colony expansion and experimental procedures was subsequently performed at the Ragon Institute of Mass General, MIT, and Harvard. Ear or tail snips from 09–1B12UCA KI mice were genotyped by TaqMan assay under a fee for service agreement (TransnetYX). TaqMan probes for the genotyping assay were developed by TransnetYX.
All experiments were performed under the approval by the Institutional Animal Care and Use Committee (IACUC) of Harvard University and the Massachusetts General Hospital (MGH) (Animal Study Protocols 2016N000022 and 2016N000286, 2014N000252) and conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Both male and female animals were used at 8–12 weeks of age. The light cycles in the animal room were set on a 12 hour light cycle [7AM-7PM (ON) 7PM-7AM (OFF)]. The temperature range for the room was 68 – 73 degrees Fahrenheit and the humidity index was from 30% – 70%. The feed was replaced every two weeks with fresh pelleted ration (Prolab Isopro RMH 3000), concomitant with changing fresh bedding in the cage. The cages were also inspected daily and additional pellets were added if food was low or empty.
METHOD DETAILS
Adoptive transfer
For experiments male B6.SJL-Ptprcapepcb/BoyJ mice (CD45.1+/+) 8–12 weeks of age were purchased from The Jackson Laboratory (Bar Harbor, ME). CD45.2+ B cells from male or female 09–1B12-UCA KI mice (H09–1B12-UCA/WT, κ09−1B12-UCA/WT) were enriched using the Pan B Cell Isolation Kit II (Miltenyi Biotec), counted, diluted to desired cell numbers in PBS and adoptively transferred into CD45.1+ recipient mice as reported previously43. Ages of the recipient animals ranged from 8–10 weeks.
Recombinant HA antigens and B cell probes
Recombinant H3 and H7 trimer ectodomains from A/Perth/16/2009 and A/Shanghai/02/2013, along with ferritin nanoparticle display of their trimeric stem domains (H3ssF and H7ssF) were affinity purified following expression in Expi293 cells according to established methodology37,55,60,79. The Expi293 cells were transfected with expression vector using the ExpiFectamine™ 293 Transfection Kit (Thermofisher), which supplies the transfection reagent and enhancer solution. Five days after transfection, the culture supernatants were harvested, filtered (VacuCap 8/0.2 μm filters, Pall Corporation) and buffer exchanged into PBS using a tangential flow filtration system [Pall Corporation; T-Series Centramate cassettes with Omega PES membrane 10 kDa (Cytiva, OS010T12)]. For HA trimers, the buffer exchanged supernatant was equilibrated with Ni Sepharose resin (GE Healthcare), whereas Erythrina cristagalli Gel-ECA-Immobilized Lectin (EY Laboratories) was mixed with the buffer exchanged supernatant containing H3ssF or H7ssF. Ni Sepharose was washed with 20 mM imidazole and the HA trimers subsequently eluted with 500 mM imidazole. For H3ssF and H7ssF, the resin was washed with PBS and the HA nanoparticles were eluted with 0.2 M lactose. All proteins were further purified by size exclusion chromatography (SEC) (AKTA pure protein purification system, Cytiva): HA trimers were resolved on a Superdex increase 200 10/300 column (Cytiva) and the stem nanoparticles were separated using a Superose 6 10/300 column (Cytiva). These same expression and purification procedures were applied to central stem epitope KO versions of the HA trimers and HA nanoparticles (containing N-linked glycan at 45HA2) and also for the recombinant H1 and H5 trimers used in this study (A/Michigan/45/2015, A/California/07/2009, A/Indonesia/05/2005). The HA trimers used in this study also contained the Y98F mutation in the RBS to prevent binding to sialyl oligosaccharide and foldon avi his sequence for trimerization/site specific biotinylation/affinity purification28,55,60,80,81.
B cell flow cytometry probes for H3ssF, H3ssF-KO, H7ssF, H7ssF-KO (central stem epitope KO = insertion of N-linked glycan at 45HA2) were generated by fluorescently labelled using amine reactive labeling kits (H3ssF-AF594, H7ssF-AF488, H7ssF-KO -AF647 + H3ssF-KO-AF647), as per our established procedure for ferritin nanoparticle-based B cell probes28. In some applications, HA trimers were avi-tagged and biotinylated at this site using the enzyme BirA and then flow probes (H3-APC and H7-AF488) were generated by adding fluorescent SA conjugates in five sequential increments (final molar ratio of HA to streptavidin label was 4:1) so as to saturate the site60.
For structural studies, HA trimers were produced following transfection into Expi293F GnTI− (Gibco, #A39240) using ExpiFectamine (Gibco, #A14524). The proteins were purified over TALON metal affinity resin (Takara, #635653) and Superdex increase 200 10/300 column (Cytiva) on an AKTA Pure (Cytiva). C-terminal tags were cleaved with HRV 3C protease (Thermo Scientific, #88946) which was removed by TALON resin and purification over the S200 column.
B cell flow cytometry, FACS and sequencing to characterize 09–1B12-UCA KI mice (H09–1B12-UCA/WT, κ09–1B12-UCA/WT).
Cells from bone marrows (femurs and tibia) were isolated, filtered and resuspended in ACK lysis buffer to remove erythrocytes. Whole spleens were mechanically dissociated to generate single-cell suspensions and resuspended in ACK lysis buffer. Bone marrow cells and splenocytes were then separately resuspended in FACS buffer (2% FBS/PBS), Fc-blocked (clone 2.4G2, BD Biosciences) and stained for viability with Live/Dead Blue (Thermo Fisher Scientific) for 20 min at 4°C. For flow analysis and FACS, we applied H3ssF-AF594, H7ssF-AF488, H7ssF-KO-AF647, and H3ssF-KO-AF647 (0.25μg of each probe / 100 μl staining mix) along with the following fluorescent antibodies (1:200 final dilution): CD4-APCeF780, CD8-APC-eF780, Gr-1-APC-eF780, F4/80-APC-eF780, B220-PerCP Cy5.5, IgD-BV421 and IgM-BUV395. To measure B cell development, different fluorescent antibody panels were applied to splenocytes [TCRβ-AF700, B220-PerCP Cy5.5, CD24-PE, CD21-FITC, CD23-APC, CD8a-APC-e780, CD4-BV510, IgM-BV421 and IgD-PE Cy7] and bone marrow cells [B220-FITC, TCRb-AF700, CD43-APC, CD21-APC Cy7, BP-1-PE, CD24-BV421, IgD-PE Cy7, IgM-BUV395] at 1:200 final dilution. Flow cytometry was performed using a BD LSRFortessa (BD Biosciences) and the data were analyzed using FlowJo software (Tree Star). For single cell FACS, mature B cells gated as lymphocytes+/singlets+/Live-Dead−/dump− (anti-mouse CD4, CD8, Ly-6G, F4/80)/ B220+/IgD+/IgM+ and antigen-specific CD45.2 B cells gated as lymphocytes+/ singlets+/ Live-Dead−/dump− (anti-mouse CD4, CD8, Ly-6G, F4/80) /B220+/IgD+/IgM+/H3ssF+/H7ssF+/H3ssF-KO−/H7ssF-KO− were sorted (BD FACS Aria II instrument with 70 μm nozzle) into 96-well PCR plates, and then rapidly frozen on dry ice and stored at −80°C.
Sorted single-cell suspensions were encapsulated and converted into several DNA libraries following the 10x Next GEM Single cell 5’ protocol (10x Genomics). Briefly, single cells were isolated with Gel Bead-In-EMulsions (GEMs) using the Chromium controller provided by 10x Genomics, resulting in uniquely barcoded transcriptome for each individual cell. After initial cDNA amplification and conversion to dsDNA, individual sequencing libraries were generated for gene expression, VDJ repertoires and hashtag oligos82. Library quality was analyzed using a Tapestation 4200 (Agilent). Libraries were pooled at a ratio based on depth requirements established by 10x Genomics and subsequently sequenced using a Nextseq2000 sequencer (llumina). Raw base call files generated by sequencing were demultiplexed, aligned and aggregated using the pipeline offered as part of Cell Ranger (10x Genomics). For VDJ repertoire analysis, immunoglobulin v genes were determined by Cell Ranger. Heavy and Light chains were subsequently paired based on 10x barcodes using a custom R script after doublet determination and removal using hashtag antibody sequences and the Seurat R package (https://satijalab.org/seurat/). Chord plots were produced in R circlize version 0.4.15 (https://cran.r-project.org/web/packages/circlize/index.html) package.
VH1–18-QxxV bnAb precursor frequency in humans
To compare VH1–18 QxxV bnAb precursor frequency within our adoptive transfer model to the natural value in humans, we measured this value in publicly available IgM BCR repertoires from n=10 human subjects sequenced to high depth41. From this dataset, we considered only those antibody sequences annotated as encoding productive IgM heavy chains. The complete code used to compute the frequencies of the VH1–18 QxxV class in these repertoires is available at https://www.doi.org/10.5281/zenodo.10800716. The Bash, R, and Python languages were used, as well as tidyverse 83 and pandas software packages84.
Sequential (2x) immunization of humanized mice and viral challenge
One day after adoptive transfer of CD45.2 B cells from 09–1B12-UCA KI mice, the recipient CD45.1 mice were given an intraperitoneal injection of 50 μg of H3ssF within 100 ml of inoculum containing 50% w/v Sigma adjuvant (Sigma, Cat# S6322; also known as Ribi), or Sigma adjuvant-only. We also included a CD45.1 mouse group that received H3ssF, but no UCA cells were present. Mice were boosted 42 days after the initial immunization (H3ssF + Sigma adjuvant for the mice primed with H3ssF; or Sigma adjuvant-only for the non-immunogen group). Fourteen days after the boost (day 56), the mice were intranasally infected with 100% lethal doses of either: subtype-matched H3N2 X-31 (BEI Resources cat# NR-3483) (108 TCID50/ml); or group-unmatched, mouse-adapted H1N1 A/California/07/2009 (maA/Cal/09)75,76 (104 TCID50/ml). Mice were monitored each day for 21 days for survival and body weight loss. The humane endpoint was set at 20% weight loss. Both viruses were cultured in MDCK cells and quantified by TCID50 in MDCK cells85. X-31 was obtained from BEI Resources and maA/Cal/09 was kindly provided by Sabra Klein and Andrew Pekosz, John Hopkins University.
Single shot immunization and lineage tracking within B cell germinal centers
One day after adoptive transfer of CD45.2 B cells from 09–1B12-UCA KI mice, the recipient CD45.1 animals were immunized intraperitoneally with either: 50 μg H3ssF; 50 μg H7ssF; or 50 μg of ferritin-only nanoparticle. In all cases, a 200 μl inoculum containing 50% w/v Sigma adjuvant was deployed. On days 8, 15, and 28, whole spleens were mechanically dissociated to generate single-cell suspensions. ACK lysis buffer was used to remove red blood cells and splenocytes were then resuspended in FACS buffer (2% FBS/PBS), Fc-blocked (clone 2.4G2, BD Biosciences) and stained for viability with Live/Dead Blue (Thermo Fisher Scientific) for 20 min at 4°C. For flow staining we applied H3ssF-A594, H7ssF-A488, H7ssF-KO-A647, H3ssF-KO-A647 (or trimer probes: H3-APC and H7-AF488) at 0.25μg of probe/100 μl staining mix along with following fluorescent antibodies (1:200 final dilution): CD4-APCeF780, CD8-APC-eF780, Gr-1-APC-eF780, F4/80-APC-eF780, B220-BV605, CD95-PE-Cy7, CD38-BV510, CD45.1-PerCPCy5.5, CD45.2-BV785. Cells were acquired by a BD LSR Fortessa (BD Biosciences) and the data were analyzed using FlowJo software (Tree Star). Single GC CD45.2 B cells with specificity to the central stem [lymphocytes+/singlets+/Live-Dead−/dump− (anti-mouse CD4, CD8, Ly-6G, F4/80) /B220+/CD95+/CD38−/CD45.1−/CD45.2+/Ag+(H3ssF-AF594+/H7-AF488+/H3ssF-KO−/H7ssF-KO−)] were also were sorted using a BD FACS Aria II instrument (BD Biosciences) with 70 μm nozzle into 96-well plates containing RLT lysis buffer supplemented with 1% beta-mercaptoethanol (BME). The sorted cells were promptly frozen and stored at −80°C for future analysis.
BCR sequencing of GC B cells
Single cell BCR libraries were generated from products of whole transcriptome amplification (WTA) using the Smart-Seq2 protocol86. The WTA product from each single cell reaction were first subjected to two 0.8x (v/v) SPRI bead-based cleanups followed by cDNA quantification/normalization. BCR sequences from heavy and light chains were enriched from single B cells using a V region and J region specific primer set in which the primers were also attached to Illumina P7 (V region) and P5 sequences (J region) (final concentration: 0.5 μM each) (Table S5). After amplification, a 0.8x (v/v) SPRI cleanup was performed, and we quantified and normalized the amplicons to 0.2–0.5 ng/μL. Within a subsequent step-out PCR [Kapa HiFi HotStart ReadyMix; Kapa Biosystems], we added cellular barcodes and Illumina sequencing adapters (based on Nextera XT Index Adapters, Illumina Inc.) to each single cell-amplified heavy and light chain, as we have performed previously28,55,61. After a 0.8x (v/v) SPRI cleanup, the heavy chain and light chain products were pooled and sequenced using paired end 250×250 reads and 8×8 index reads on an Illumina MiSeq System [MiSeq Reagent Kit v2 (500-cycle)]. The BCR heavy and light chains were paired and reconstructed using PandaSeq87 and aligned against the human IMGT database88,89 with PCR/sequencing error correction using MigMAP (a wrapper for IgBlast: https://github.com/mikessh/migmap). Consensus V-chain and L/k-chain sequences for each single cell was performed by collapsing all reads with the same CDR3 sequence and then identifying the top heavy and light chain sequences based on frequency. Any heavy or light chain sequence with fewer than 15 reads or a frequency less than twice that of the next sequence of the same chain was considered without consensus. Phylogenetic trees to visualize relatedness were generated from the heavy chain nucleotide sequences using the maximum likelihood method with the Tamura-Nei model in MEGA11 software90,91. Heavy chain and light chain sequences from 09–1B12-UCA were used as the baseline for tree construction. Sequence logos were generated using WebLogo (https://weblogo.berkeley.edu)92.
Recombinant Monoclonal Antibodies and Fabs
The variable sequences from BCR heavy and light chains were synthesized as gene blocks by GenScript. The sequences were cloned into human IgG1 heavy chain and light chain expression plasmids28,55,61. The antibodies were transfected into Expi293 cells, as described for the recombinant HA trimers and HA nanoparticles, except that we harvested the supernatant after six days of culture. Following filtration of the supernatant (VacuCap 8/0.2 μm filters, Pall Corporation) the material was equilibrated with Protein G Sepharose resin. After washing with PBS, the IgG was eluted with IgG elution buffer (Pierce, Cat# 21028) into 1M Tris, pH 8. The eluate was further purified and buffer exchanged to PBS using SEC with a Superdex 200 10/300 column (GE Healthcare). Additional monoclonal antibodies were expressed and purified in the same way: 09–1B12-UCA, 09–1B12, 21–1A01-UCA, 21–1A01; 05–2A09-UCA, 05–2A09, 05–2D04-UCA, 27–1D08-UCA, 06–1F04-UCA24,37; 09–1B12-UCA + N55T, 21–1A01-UCA + N55T, 05–2A09-UCA + N55T, 05–2D04-UCA + N55T, 27–1D08-UCA + N55T, 06–1F04-UCA + N55T; CR626193,94; VRC0195. The HC and LC nucleotide sequences of 09–1B12-UCA was from Corbett et al.37, the other UCAs were inferred by gHgL reversion of the mature antibody sequence. For structural studies, antibody Fab fragments were produced by transfection into HEK293F (Gibco, #R79007) using PEI at a 3:1 ratio, followed by expression and his tag-based affinity purification and cleavage of the tag with HRV 3C protease, as described earlier for HA trimers.
Neutralization Assay
Microneutralization assays were performed according to Creanga et al.46. HEK293T, HEK293T-PB1 (constitutively expressing A/WSN/1933 PB1), MDCK-SIAT1-PB1, MDCK-SIAT1-H5 (constitutively expressing A/Vietnam/1203/2005 HA), and MDCK-SIAT1-H7 (constitutively expressing A/Shanghai/02/2013 HA) were maintained in DMEM (Gibco, #11965084) supplemented with 10% heat-inactivated FBS (Peak Serum) and 100 U of penicillin/streptomycin (Gibco, #15140163)46. Media for MDCK-SIAT1-PB1, -H5, and -H7 cells was further supplemented with 1 mg/ml geneticin (Gibco, #10131035) and 0.25 μg/ml puromycin (Gibco, #A1113803). Prior to infection, cells were seeded in “flu media” composed of Opti-MEM (Gibco, #31985088) supplemented with 0.3% BSA (Sigma, #A9418), 100 U of penicillin/streptomycin (Gibco), 0.1 mg/ml of CaCl2, and 0.01% heat-inactivated FBS (Peak Serum).
Influenza reporter viruses were generated as described previously46. Briefly, H1N1 and H3N2 viruses were generated using plasmids encoding bidirectional cassettes for PB2, PA, NP, M, and NS from A/WSN/193396, tdKatushka2 containing PB1 packing sequences, a plasmid encoding human TMPRSS2, and HA and NA segments flanked with non-coding regions from A/WSN/1933 (for H1N1 viruses) or A/Netherlands/009/2010 (for H3N2 viruses). Plasmids were transfected into HEK293T-PB1 cells in a 6-well plate using TransIT-LT1 (MirusBio, #2306). After three days, the supernatant was clarified by centrifugation at 800 ×g for 5 minutes and added to a monolayer of MDCK-SIAT1-PB1 cells in a 6-well plate. Two to three days later when cytopathic effect was evident, the supernatant was clarified and added to a monolayer of MDCK-SIAT1-PB1 cells in T75 or T150 flasks. Two to three days later, the supernatant was clarified, and aliquots of viral stocks were frozen at −80 °C. To generate H5N1 and H7N9 viruses, the internal genes (PB1, PB2, PA, NP, M, NS) from A/Puerto Rico/8/1934 were used with tdKatushka2 bearing the HA packing sequences from A/Puerto Rico/8/1934. For H5N1 and H7N9 viruses, a plasmid expressing H5 HA (A/Vietnam/1203/2004) or H7 HA (A/Shanghai/02/2013) were additionally transfected into HEK293T cells. H5N1 and H7N9 viruses were passaged and propagated as above in MDCK-SIAT1-HA cells constitutively expressing H5 or H7 HA.
One day prior to titering, MDCK-SIAT-PB1 (or -HA) cells were seeded in 96-well plates at 20,000–30,000 cells per well (CellVis, #P96–1.5P). The next day, viral stocks were thawed and diluted 2-fold 23 times in quadruplicate. The virus dilutions were then mixed with an equal volume of flu media additionally supplemented with 2 μg/ml of TPCK-treated trypsin (Sigma, #T1426). Virus was incubated at 37 °C / 5% CO2 for 1 hour before removing flu media from MDCK-SIAT1-PB1 (or -HA) cells and adding 100 μl of the virus. Approximately 18–20 hours later, the cells in each well were imaged on a CellDiscoverer7 (Zeiss) instrument and fluorescent cells were counted. The cell counts were plotted, and a sigmoidal curve was fitted in GraphPad Prism. The dilution that corresponded to the half maximal positive cell count was used for subsequent neutralization assays.
For microneutralization measurements, the viruses were diluted in flu media with 2 μg/ml of TPCK-treated trypsin and antibodies were diluted in a four-fold series in flu media. The virus and antibody dilutions were mixed in equal volumes in a 96-well plate and incubated at 37 °C / 5% CO2 for 1 hour. The flu media was then removed from the MDCK-SIAT1-PB1 (or -HA) cells and the antibody/virus mixture was added. Approximately 18–20 hours later, the cells were counted as above. Each plate contained ten virus only control wells and two cell only controls. Each value had the cell only control background subtracted and was normalized to the average of the virus only controls. The percent neutralization was plotted and fitted with a sigmoidal curve (GraphPad Prism) to determine the half maximal inhibitory concentration (IC50). Each neutralization assay was run in duplicate.
ELISA
To assess the IgG titers of immunized mice, 200 ng per well of either H3ssF, H7ssF, or their respective epitope KO versions, were pre-coated in 96-well plates at 4°C overnight. After incubation with blocking buffer (3% BSA in PBS + 0.01% tween 20) for 2 h at RT, 3-fold serial diluted sera (7 dilutions) with starting dilution 1:100 from priming or boosting mice were incubated with pre-coated protein for 2 h at RT. Wells were washed and incubated with Alkaline Phosphatase AffiniPure Goat Anti-Mouse IgG (Jackson ImmunoResearch, #115–055-071) at 1:1,000 in PBS with 0.5% BSA for 1 h at RT. P-Nitrophenyl phosphate (Sigma, # N2770) dissolved in ddH2O (50 μL/well, RT, 25 min) was used for detection. Absorbance at 405 nm was determined with a plate reader (Synergy Neo2, BioTek). ELISA curves were calculated and analyzed using GraphPad Prism 9.5.1 (GraphPad).
Immunization of humanized mice and serum processing for CryoEMPEM
One day after adoptive transfer of CD45.2 B cells from 09–1B12-UCA KI mice, the recipient CD45.1 mice were immunized intraperitoneally with either 50 μg H3ssF or 50 μg H7ssF; each delivered in a 100 ml inoculum containing 50% w/v Sigma adjuvant. The mice were boosted again at 42 days with these same inoculums and immune sera were obtained at 15 and 28 days post-boost. These sera were then processed for cryoEMPEM analysis48. Briefly, Serum IgG was purified from pooled sera (in the same group) with Protein G resin and subsequently digested with papain. Digested polyclonal Fabs were purified over Superdex 200 Increase column (Cytiva).
CryoEM
For cryoEM, HA (30 μg) was incubated with the purified monoclonal Fab (40 ug) or polyclonal Fabs (0.5–1 mg) overnight at 4 °C. The complex was then purified over Superdex 200 Increase column (GE Healthcare) and concentrated to ~0.7 mg/ml. Next, 3 μL of the complex was mixed with 0.5 μL 0.7% (w/v) Octyl-beta-Glucoside (OBG) before deposition onto glow-discharged 1.2/1.3 Cu 300 grids (EMS), directly preceding the deposition in a Vitrobot (Thermo Fisher Scientific) with following settings: 4°C, 100% humidity, 0 s wait time, 4.5–6 s blot time, blot force 1. Once sample was deposited, the grids were blotted and plunged into liquid to immobilize the particles in vitreous ice. The micrographs were collected with EPU image acquisition software at a nominal magnification of ×190,000 with a TFS Falcon 4 detector mounted on a Glacios set to 200 kV set to counting mode, with a total exposure dose of ~50 e−/Å2. Micrographs were collected in CryoSPARC Live and followed by gain reference correction, motion correction, defocus estimation, and micrograph curation (Table S1). Template Picker was used to pick particles, which were then extracted and 2D-classified in cryoSPARC. The particles in selected 2D classes were further cleaned up by Heterogenous refinement using C1 symmetry and 3D Variability with symmetry expanded particles. Final maps were made by homogeneous refinements with tight masks on HA and Fab on one protomer.
For model building, the initial models were generated from PDBID: 3LZG (H1), 4KVN (H3), 4N5J (H7), and 5KAN (V1–18 Fab), and docked into cryoEM map using UCSF ChimeraX97. Coot 0.9.898, Phenix99, and Rosetta100 were used for model building and refinement (Table S1). The final models and cryoEM maps have been deposited in PDB (8UT3, 8UT4, 8UT5, 8UT6, 8UT7, 8UT8, 8UT9) and EMDB (EMD-42528, EMD-42529, EMD-42530, EMD-42531, EMD-42532, EMD-42533, EMD-42534, EMD-42535, EMD-42536).
X-ray crystallography
09–1B12 Fab and H3 Perth/2009 HA were complexed at a 4:1 molar ratio for 20 minutes at room temperature prior to concentration and purification over an S200 column (Cytiva). Crystals were generated by the hanging drop method over 0.1 M sodium citrate tribasic dihydrate (pH 5.0), 10% v/v 2-propanol, and 10% PEG 10k (Hampton Research, #HR2–084) for ~7 days at 18 °C using 96-well plates (Greiner, #655101) with ViewDrop II seals (sptlabtech, #4150–05600). Crystals were harvested after adding 1 μl of 12% (+/−)-2-methyl-2,4-pentanediol (MPD) in the crystallization solution and plunged into liquid nitrogen to freeze.
X-ray diffraction data was collected at beam line 24-ID-E (Advanced Photon Source) and processed with XDSGUI (https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/XDSGUI). The work used NE-CAT beamlines (GM124165), a Pilatus detector (RR029205), and an Eiger detector (OD021527) at the APS (DE-AC02–06CH11357). The structure was initially solved by molecular replacement using the HA protomer from PDB 4KVN, 16.a.26 variable heavy chain with the CDRH3 removed (from PDB 5K9Q), and a model of the variable light chain with the CDRL3 removed101. PHENIX102 was used to initially refine the coordinates and B factors prior to manual model building in COOT103. Final refinement additionally included Translation Libration Screw refinements and Ramachandran restraints (Table S2). The final model was deposited to the Protein Data Bank (8UWA).
Structure Modelling and MD simulations
To characterize the interaction profiles of the UCA/09–1B12 antibodies in complex with H1, H3 and H7 we performed molecular dynamics simulations of these antibody complexes, respectively. As starting structures for our simulations, we used the available cryo-EM structures, presented in this study. For all these antibody-antigen complexes we performed each three replicas of 1μs classical molecular dynamics simulations. Furthermore, we also performed simulations of the free variable domains (Fvs), namely 09–1B12-UCA, 09–1B12-UCA + N55T and 09–1B12, to characterize the effect of the mutations on the dynamic properties of the Fvs.
The starting structures for our simulations were prepared in MOE using the Protonate3D tool104,105. With the help tool of the Amber Tools20 package106, we explicitly bonded all existing disulfide bridges and placed the antibody-antigen complexes into cubic water boxes of TIP3P water molecules with a minimum wall distance to the protein of 12 Å107. Parameters for all antibody-antigen and free Fv simulations were derived from the CHARMM36m 108. To neutralize the charges, we used uniform background charges106,109. Each system was equilibrated using a multistep equilibration protocol110. Molecular dynamics simulations were performed using pmemd.cuda in an NpT ensemble to be as close to the experimental conditions as possible and to obtain the correct density distributions of both protein and water111. Bonds involving hydrogen atoms were restrained by applying the SHAKE algorithm112, allowing a timestep of 2.0 fs. Atmospheric pressure of the system was preserved by weak coupling to an external bath using the Berendsen algorithm113. The Langevin thermostat was used to maintain the temperature at 300K during simulations114,115.
To calculate contacts of the antibody-antigen complexes, we used the GetContacts software (https://getcontacts.github.io/).This tool can compute interactions within one protein structure, but also between different protein interfaces and allows to monitor the evolution of contacts during the simulation. The contacts are defined based on the default geometrical criteria provided by GetContacts.
Biolayer interferometry (BLI)
For BLI, Fabs were produced by digestion of recombinant IgGs with Lys-C endoproteinase (New England Biolabs Cat. # P8109S)28,55,61. The reaction was in PBS containing 1 mM EDTA, with a ratio of 5 mg of Lys-C per milligram of IgG. After incubating for 12 hours at room temperature, the reaction was stopped by addition of 1x complete protease inhibitor cocktail (Roche, Cat # 11697498001) and Protein A/G-agarose was applied to remove any uncleaved IgG. The resin was washed with PBS, and the supernatant was concentrated using Amicon Ultra concentrators with a 10 kDa cutoff. The Fabs were further purified by SEC, using a Superdex 200 10/300 column (GE Healthcare). BLI was performed using the Personal Assay BLItz System (Fortebio). Following avi-tag biotinylation (see earlier), the biotinylated forms of HA trimers spanning group 1 and group 2 diversity (A/Perth/16/2009, A/Shanghai/02/2013, A/Michigan/45/2015, A/California/07/2009, A/Indonesia/05/2005) were immobilized onto streptavidin biosensors (Sartorius, Cat#18–5019). After establishing a baseline in kinetic buffer (PBS containing 0.02% Tween20, 0.1% BSA), the Fabs were introduced at 0.625 mM, 1.25 mM, 2.5 mM, and 5 mM, with 120 seconds of association and 120 seconds of dissociation. The equilibrium dissociation constant (KD) values were calculated by fitting a 1:1 binding isotherm using software provided by the vendor77.
Passive transfer of monoclonal antibodies and viral challenge
Monoclonal antibodies were infused at 5 mg/kg intraperitoneally into WT C57Bl/6J mice24. Two hours later, the mice were intranasally infected with 100% lethal doses of either: subtype-matched H3N2 X-31 (BEI Resources cat# NR-3483) (108 TCID50/ml); or group-unmatched, mouse-adapted H1N1 A/California/07/2009 (maA/Cal/09)75,76 (104 TCID50/ml). The mice were monitored each day for 21 days for survival and body weight loss. The humane endpoint was set at 20% weight loss, as described above.
QUANTIFICATION AND STATISTICAL ANALYSES
All statistical analysis were conducted using Prism 9.01 (GraphPad). Sample sizes and statistical tests are indicated in the figure legends. Data were considered statistically significant at P<0.05.
Supplementary Material
Highlights.
Natural HA affinity for human bnAb precursors is transduced into vaccine nanoparticles
A humanized mouse is engineered to contain human bnAb precursor frequency
Vaccine protection against Group 1 + Group 2 IAV is generated by stem bnAb elicitation
The antibodies undergo a single amino acid ‘switch’ to accommodate IAV glycan diversity
Acknowledgements
This work was supported by funding to D.L. and F.D.B. (R01AI153098, and flexible funding from the Ragon Institute of Mass General, MIT and Harvard). D.L. was also supported by NIH grants (R01AI12437, R01AI155447, U19AI057229, P30AI060354). A.G.S. was supported by NIH grants (R01A146779, P01AI89618). A.B.W. was supported by a grant from the Bill and Melinda Gates Foundation (INV-004923). B.A.A. was the recipient of an NSF Graduate Research Fellowship (DGE-210743). M.F-Q. was supported by the Austrian Academy of sciences APART-MINT postdoctoral fellowship. We acknowledge CHRONOS for granting access to Piz Daint at CSCS, Switzerland. We further recognize EuroHPC Joint Undertaking for enabling access to Karolina at IT4Innovations, Czech Republic. The computational results presented here were also achieved (in part) using the LEO HPC infrastructure of the University of Innsbruck. We thank Thomas Dupic for feedback on our analysis of human bnAb precursor frequency and Jesse Bloom for providing reagents used to generate the influenza reporter viruses. We thank Sabra Klein and Andrew Pekosz for providing maA/Cal/09. We are indebted to the beamline staff at NE-CAT for help with x-ray data collection. We thank the members of Schmidt - Batista - Lingwood joint lab meetings for critical feedback on this project.
Footnotes
Declaration of Interests
D.L. reports SAB membership for Metaphore Bio (a Flagship company), and consultancy relationships with Tendel Therapies, and Lattice Therapeutics Inc. F.D.B. has consultancy relationships with Adimab and Third Rock Ventures, and is Chief Editor or The EMBO Journal. M.K. is listed as inventor of patents on vaccine immunogens used in this study filed by the U.S. Department of Health and Human Services. The remaining authors declare no competing interests.
Additional Supplemental Items
Table S4. Paired heavy and light chain antibody sequences from the H3/H7 cross-reactive germinal center B cells expanded after 1x immunization with H3ssF or H7ssF and sorted at days 15 and 28. Related to Figure 3–5.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Caradonna TM, and Schmidt AG (2021). Protein engineering strategies for rational immunogen design. npj Vaccines 6, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sangesland M, and Lingwood D (2021). Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, McDermot AB, and Yewdell JW (2017). Defining B cell immunodominance to viruses. Nat. Immunol. 18, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouma S, Anderson EM, Hensley SE. (2020). Challenges of Making Effective Influenza Vaccines. Ann. Rev. Virol. 7, 495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrie JG, and Gordon A (2018). Epidemiological Studies to Support the Development of Next Generation Influenza Vaccines. Vaccines 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paules C, and Subbarao K Influenza. (2017). Lancet 390, 697–708. [DOI] [PubMed] [Google Scholar]
- 7.Petrova VN, and Russell CA (2018). The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 16, 47–60. [DOI] [PubMed] [Google Scholar]
- 8.Chen JM, Sun YX, Chen JW, Liu S, Yu JM, Shen CJ, Sun XD, and Peng D (2009). Panorama phylogenetic diversity and distribution of type A influenza viruses based on their six internal gene sequences. Virol. J. 6, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang Q, Wang S, S. Liu, Hou G, Li J, Jiang W, Wang K, Peng C, Liu D, Guo A, and Chen J (2019). Diversity and distribution of type A influenza viruses: an updated panorama analysis based on protein sequences. Virol. J.16, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei CJ, Crank MC, Shiver J, Graham BS, Mascola JR, and Nabel GJ. (2020). Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 19, 239–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krammer F (2019). The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 19, 383–397. [DOI] [PubMed] [Google Scholar]
- 12.Guthmiller JJ, and Wilson PC (2018). Harnessing immune history to combat influenza viruses. Curr. Opin. Immunol. 53,187–195. [DOI] [PubMed] [Google Scholar]
- 13.Erbelding EJ, Post D, Stemmy E, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, and Fauci AS (2018). A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 218, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews SF, Cominsky LY, Shimberg GD, Gillespie RA, Gorman J, Raab JE, Brand J, Creanga A, Gajjala SR, Narpala S, Cheung CSF, Harris DR, Zhou T, Gordon I, Holman L, Mendoza F, Houser KV, Chen GL, Mascola JR, Graham BS, Kwong PD, Widge A, Dropulic LK, Ledgerwood JE Kanekiyo M, and McDermott AB (2023). An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Sci. Transl. Med. 15, eade4976. [DOI] [PubMed] [Google Scholar]
- 15.Widge AT, Hofstetter AR, Houser KV, Awan SF, Chen GL, Burgos Florez MC, Berkowitz NM, Mendoza F, Hendel CS, Holman LA, Gordon IJ, Apte P, Lian CJ, Gaudinski MR, Coates EE, Strom L, Wycuff D, Vazquez S, Stein JA, Gall JG, Adams WC, Carlton K, Gillespie RA, Creanga A, Crank MC, Andrews SF, Castro M, Serebryannyy LA, Narpala SR, Hatcher C, Lin BC, O’Connell S, Freyn AW, Rosado VC, Nachbagauer R, Palese P, Kenekiyo M, McDermott AB, Koup RA, Dropulic LK, Graham BS, Mascola JR, Ledgerwood JE, and VRC 321 study team. (2023). An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 15, eade4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachbagauer R, Feser J, Naficy A, Bernstein DI, Guptill J, Walter EB, Berlanda-Scorza F, Stadlbauer D, Wilson PC, Aydillo T, Behazadi MA, Bhavsar D, Bliss C, Capuano C, Carreno JM, Chromilkova V, Claeys C, Coughlan L, Freyn AW, Gast C, Javier A, Jiang K, Mariottini C, McMahon M, McNeal M, Solorzano A, Strohmeier S, Sun W, Van der Wielen M, Innis BL, Garcia-Sastre A, Palese P, Krammer F (2021). A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 27, 106–114. [DOI] [PubMed] [Google Scholar]
- 17.Nabel GJ, and Fauci AS (2010). Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat. Med. 16, 1389–1391. [DOI] [PubMed] [Google Scholar]
- 18.Cho A, and Wrammert J (2016). Implications of broadly neutralizing antibodies in the development of a universal influenza vaccine. Curr. Opin. Virol. 17, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guthmiller JJ, Han J, Utset HA, Li L, Lan LY, Henry C, Stamper CT, McMahon M, O’Dell G, Fernandez-Quintero ML, Freyn AW, Amanat F, Stovicek O, Gentles L, Richey ST, de la Pena AT, Rosado VC, Dugan HL, Zheng N-Y, Tepora ME, Bitar DJ, Changrob S, Strohmeier S, Huang M, Garcia-Sastre A, Liedl KR, Bloom JD, Nachbagauer R, Palese P, Krammer F, Coughlan L, Ward AB, and Wilson PC (2022). Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature 602, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, Kong WP, Leung K, Narpala SN, Prabhakaran MS, Yang ES, Zhang B, Zhang Y, Asokan M, Boyington JC, Bylund T, Darko S, Lees CR, Ransier A, Shen CH, Wang L, Whittle JR, Wu X, Yassine HM, Santos C, Matsuoka Y, Tsybovsky Y, Baxa U, NISC Comparative Sequencing Program, Mullikin. JC, Subbarao K, Douek DC, Graham BS, Koup RA, Ledgerwood JE, Roederer M, Shapiro L, Kwong PD, Mascola JR, and McDermott AB (2016). Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Frenandez-Rodrigues BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JPM, Skehel JJ, and Lanzavecchia A (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856. [DOI] [PubMed] [Google Scholar]
- 22.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, Vorlaender MK, Bianchi S, Guarino B, De Marco A, Vanzetta F, Agatic G, Foglierini M, Pinna D, Fernandez-Rodriguez B, Fruehwirth A, Silacci C, Ogrodowicz RW, Martin SR, Sallusto F, Suzich JA, Lanzavecchia A, Zhu Q, Gamblin SJ, and Skehel JJ (2016). Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, and Friesen RH (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, Boyington JC, Prabhakaran MS, Narpala SR, Chen X, Bailer RT, Chen G, Coates E, Kwong PD, Koup RA, Mascola JR, Graham BS, Ledgerwood JE, and McDermott AB. (2017). Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci. Immunol 2, eaan2676. [DOI] [PubMed] [Google Scholar]
- 25.Chuang GY, Shen CH, Cheung CS, Gorman J, Creanga A, Joyce MG, Leung K, Rawi R, Wang L, Yang ES, Yang Y, Zhang B, Zhang Y, Kanekiyo M, Zhou T, DeKosky BJ, Graham BS, Mascola JR, and Kwong PD (2021). Sequence-Signature Optimization Enables Improved Identification of Human HV6–1-Derived Class Antibodies That Neutralize Diverse Influenza A Viruses. Front. Immunol. 12, 662909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderven HA, Jegaskanda S, Wheatley AK, and Kent SJ (2017). Anti-bodydependent cellular cytotoxicity and influenza virus. Curr. Opin. Virol. 22, 89–96. [DOI] [PubMed] [Google Scholar]
- 27.He W, Tan GS, Mullarkey CE, Lee AJ, Lam MM, Krammer F, Henry C, Wilson PC, Ashkar AA, Palese P, and Miller MS (2016). Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Nat Acad. Sci. USA. 2016; 113, 11931–11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangesland M, Ronsard L, Kazer SW, Bals J, Boyoglu-Barnum S, Yousif AS, Barnes R, Feldman J, Quirindongo-Crespo M, McTamney PM, Rohrer D, Lonberg N, Chackerian B, Graham BS, Kanekiyo M, Shalek A, and Lingwood D (2019). Germline-Encoded Affinity for Cognate Antigen Enables Vaccine Amplification of a Human Broadly Neutralizing Response against Influenza Virus. Immunity 51, 735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiLillo DJ, Palese P, Wilson PC, and Ravetch JV (2016). Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 126, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kükrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, and Radošević K (2015). A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306. [DOI] [PubMed] [Google Scholar]
- 31.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, and Graham BS (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070. [DOI] [PubMed] [Google Scholar]
- 32.Boyoglu-Barnum S, Hutchinson GB, Boyington JC, Moin SM, Gillespie RA, Tsybovsky Y, Stephens T, Vaile JR, Lederhofer J, Corbett KS, Fisher BE, Yassine HM, Andrews SF, Crank MC, McDermott AB, Mascola JR, Graham BS, and Kanekiyo M (2020). Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Comm. 11, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darricarrere N, Qiu Y, Kanekiyo M, Creanga A, Gillespie RA, Moin SM, Saleh J, Sancho J, Chou T-H, Zhou Y, Zhang R, Dai S, Moody A, Saunders KO, Crank MC, Mascola JR, Graham BS, Wei C-J, and Nabel GJ (2021). Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 13, eabe5449. [DOI] [PubMed] [Google Scholar]
- 34.Liu WC, Jan JT, Huang YJ, Chen TH, and Wu SC (2016). Unmasking Stem-Specific Neutralizing Epitopes by Abolishing N-Linked Glycosylation Sites of Influenza Virus Hemagglutinin Proteins for Vaccine Design. J. Virol. 90, 8496–8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Chen S, Yang D, Wang X, Zhu J, Peng D, and Liu X (2015). Role of stem glycans attached to haemagglutinin in the biological characteristics of H5N1 avian influenza virus. J. Gen. Virol. 96,1248–1257. [DOI] [PubMed] [Google Scholar]
- 36.Moin SM, Boyington JC, Boyoglu-Barnum S, Gillespie RA, Cerutti G, Cheung CS, Cagigi A, Gallagher JR, Brand J, Prabhakaran M, Tsybovsky Y, Stephens T, Fisher BE, Creanga A, Ataca S, Rawi R, Corbett KS, Crank MC, Karlsson Hedestam GB, Gorman J, McDermott AB, Harris AK, Zhou T, Kwong PD, Shapiro L, Mascola JR, Graham BS, and Kanekiyo M (2022). Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses. Immunity. 2022; 55, 2405–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, Gillespie RA, Shi W, Wang L, Zhang Y, Andrews SF, Joyce MG, Crank MC, Douek DC, McDermott AB, Mascola JR, Graham BS, and Boyington JC (2019). Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 10, e02810–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Han J, Sun W, Puente-Massaguer E, Yu W, Palese P, Krammer F, Ward AB, Wilson IA (2022). Influenza chimeric hemagglutinin structures in complex with broadly protective antibodies to the stem and trimer interface. Proc. Natl. Acad. Sci. USA 119, e2200821119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Ray R, Kratochvil S, Melzi E, Lin YC, Giguere S, Xu L, Warner J, Cheon D, Liguori A, Groschel B, Phelps N, Adachi Y, Tingle R, Wu L, Crotty S, Kirsch KH, Nair U, Schief WR, and Batista FD (2021). Multiplexed CRISPR/CAS9-mediated engineering of pre-clinical mouse models bearing native human B cell receptors. EMBO J. 40, e105926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin YC, Pecetta S, Steichen JM, Kratochvil S, Melzi E, Arnold J, Dougan SK, Wu L, Kirsch KH, Nair U, Schief WR, and Batista FD One-step CRISPR/Cas9 method for the rapid generation of human antibody heavy chain knock-in mice. EMBO J. 37, e99243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briney B, Inderbitzin A, Joyce C, Burton DR (2019). Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung CS, Fruehwirthm A, Paparoditis PCG, Shen CH, Foglierini M, Joyce MG, Leung K, Piccoli L, Rawi R, Silacci-Fregni C, Tsybovsky Y, Verardi R, Wang L, Wang S, Yang ES, Zhang B, Zhang Y, Chuang GY, Corti D, Mascola JR, Shapiro L, Kwong PD, Lanzavecchia A, and Zhou T (2020). Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation. Cell Rep. 2020; 32,108088. [DOI] [PubMed] [Google Scholar]
- 43.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, Schief WR, Nemazee D, and Crotty S (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity. 48, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tas JMJ, Koo JH, Lin YC, Xie Z, Steichen JM, Jackson AM, Hauser BM, Wang X, Cottrell CA, Torres JL, Warner JE, Kirsch KH, Weldon SR, Groschel B, Nogal B, Ozorowski G, Bangaru S, Phelps N, Adachi Y, Eskandarzadeh S, Kubitz M, Burton DR, Lingwood D, Schmidt AG, Nair U, Ward AB, Schief WR, and Batista FD Antibodies from primary humoral responses modulate the recruitment of naive B cells during secondary responses. Immunity 55, 1856–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dosenovic P, Kara EE, Pettersson AK, McGuire AT, Gray M, Hartweger H, Thientosapol ES, Stamatatos L, and Nuzzenzweig MC (2018). Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. USA 115, 4743–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, Ledgerwood JE, McDermott AB, Mascola JR, Graham BS, and Kanekiyo M A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat. Comm. 12, 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antanasijevic A, Sewall LM, Cottrell CA, Carnathan DG, Jimenez LE, Ngo JT, Silverman JB, Groschel B, Georgeson E, Bhiman J, Bastidas R, LaBranche C, Allen JD, Copps J, Perrett HR, Rantalainen K, Cannac F, Yang YR, de la Peña AT, Rocha RF, Berndsen ZT, Baker D, King NP, Sanders RW, Moore JP, Crotty S, Crispin M, Montefiori DC, Burton DR, Schief WR, Silvestri G, and Ward AB (2021) Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Nat. Comm. 12, 4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han J, Schmitzm AJ, Richey ST, Dai YN, Turner HL, Mohammed BM, Fremont DH, Ellebedy AH, and Ward AB (2021). Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep. 34, 108682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandor AM, Sturdivant MS, and Ting JPY (2021). Influenza Virus and SARS-CoV-2 Vaccines. J. Immunol. 206, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiore AE, Bridgesm CB, and Cox NJ (2009). Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 2009; 333, 43–82. [DOI] [PubMed] [Google Scholar]
- 51.Arevalo CP, Bolton MJ, Le Sage V, Ye N, Furey C, Muramatsu H, Alameh M-G, Pardi N, Drapeau EM, Parkhouse K, Garretson T, Morris JS, Moncla LH, Tam YK, Fan SHY, Lakdawala SS, Weissman DW, and Hensley SE (2022). A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science 378, 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu L, Pegu A, Rao E, Doria-Rose N, Beninga J, McKee K, Lord DM, Wei RR, Deng G, Louder M, Schmidt SD, Mankoff Z, Wu L, Asokan M, Beil C, Lange C, Leuschner WD, Kruip J, Sendak R, Kwon YD, Zhou T, Chen X, Bailer RT, Wang K, Choe M, Tartaglia LJ, Barouch DH, O’Dell S, Todd JP, Burton DR, Roederer M, Connors M, Koup RA, Kwong PD, Yang ZY, Mascola JR, and Nabel GJ (2017) Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seung E, Xing Z, Wu L, Rao E, Cortez-Retamozo V, Ospina B, Chen L, Beil C, Song Z, Zhang B, Levit M, Deng G, Hebert A, Kirby P, Li A, Poulton EJ, Vicente R, Garrigou A, Piepenhagen P, Ulinski G, Sanicola-Nadel M, Bangari DS, Qiu H, Pao L, Wiederschain D, Wei R, Yang ZY, and Nabel GJ (2022). A trispecific antibody targeting HER2 and T cells inhibits breast cancer growth via CD4 cells. Nature 603, 328–334. [DOI] [PubMed] [Google Scholar]
- 54.Amitai A, Sangesland M, Barnes RM, Rohrer D, Lonberg N, Lingwood D, Chakraborty AK. Defining and Manipulating B Cell Immunodominance Hierarchies to Elicit Broadly Neutralizing Antibody Responses against Influenza Virus. Cell Systems. 2020; 11, 573–88.e9. 10.1016/j.cels.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sangesland M, de la Peña AT, Boyoglu-Barnum S, Ronsard L, Nait Mohamed FA, Barcamonte-Moreno T, Barnes RM, Rohrer D, Lonberg N, Ghebremichael M, Kanekiyo M, Ward A, and Lingwood D. (2022). Allelic polymorphism controls autoreactivity and vaccine elicitation of human broadly neutralizing antibodies against influenza virus. Immunity 55, 1693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbott RK, and Crotty S (2020). Factors in B cell competition and immunodominance. Immunol. Rev. 296, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batista FD, Iber D, and Neuberger MS (2001). B cells acquire antigen from target cells after synapse formation. Nature 411, 489–94. [DOI] [PubMed] [Google Scholar]
- 58.Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, and Nabel GJ. (2012). Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kato Y, Abbott RK, Freeman BL, Haupt S, Groschel B, Silva M, Menis S, Irvine DJ, Schief WR, and Crotty S Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 53, 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver GC, Villar RF, Kanekiyo M, Nabel GJ, Mascola JR, and Lingwood D (2016). In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat. Protoc. 11, 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ronsard L, Yousif AS, Nait Mohamed FA, Feldman J, Okonkwo V, McCarthy C, Schnabel J, Caradonna T, Barnes RM, Rohrerm D, Lonbergm N, Schmidt A, and Lingwood D (2023). Engaging an HIV vaccine target through the acquisition of low B cell affinity. Nat. Comm. 14, 5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fleire SJ, Goldman JP, Carrasco YR, Weber M, Bray D, and Batista FD (2006) B cell ligand discrimination through a spreading and contraction response. Science 312, 738–741. [DOI] [PubMed] [Google Scholar]
- 63.Batista FD, and Neuberger MS (1998). Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8, 751–759. [DOI] [PubMed] [Google Scholar]
- 64.Batista FD, and Neuberger MS (2000). B cells extract and present immobilized antigen: implications for affinity discrimination. EMBO J. 19, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrabi R, Bhiman JN, and Burton DR (2018). Strategies for a multi-stage neutralizing antibody-based HIV vaccine. Curr. Opin. Immunol. 53, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams WB, Wiehe K, Saunders KO, and Haynes BF (2021). Strategies for induction of HIV-1 envelope-reactive broadly neutralizing antibodies. J. Int. AIDS Soc. 7, e25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang C, Bagnara D, Chiorazzi N, Scharff MD, and MacCarthy T (2020). AID Overlapping and Poleta Hotspots Are Key Features of Evolutionary Variation Within the Human Antibody Heavy Chain (IGHV) Genes. Front. Immunol. 11, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Noia JM, and Neuberger MS (2007). Molecular mechanisms of antibody somatic hypermutation. Ann. Rev. Biochem. 76, 1–22. [DOI] [PubMed] [Google Scholar]
- 69.Schiepers A, van ‘t Wout MFL, Greaney AJ, Zang T, Muramatsu H, Lin PJC, Tam YK, Mesin L, Starr TN, Bieniasz PD, Pardi N, Bloom JD, and Victora GD (2023). Molecular fate-mapping of serum antibody responses to repeat immunization. Nature 615, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand GJ, Taylor WM, Lim S, Huang M, Qu X, Lee J-H, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, and Wilson PC (2015). Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7, 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen YQ, Neu KE, Henry C, Zhang N-Y, Yu-Ling LL, Tepora ME, Stovicek O, Bitar D, Palm AE, Stamper CT, Changrob S, Utset HA, Coughlan L, Krammer F, Cobey S, and Wilson PC (2020). Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med. 12, eabd3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abreu RB, Kirchenbaum GA, Clutter EF, Sautto GA, and Ross TM (2020). Preexisting subtype immunodominance shapes memory B cell recall response to influenza vaccination. JCI Insight 5, e132155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, and Schief WR (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leggat DJ, Cohen KW, Willis JR, Fulp WJ, deCamp AC, Kalyuzhniy O, Cottrell CA, Menis S, Finak G, Ballweber-Fleming L, Srikanth A, Plyler JR, Schiffner T, Liguori A, Rahaman F, Lombardo A, Philiponis V, Whaley RE, Seese A, Brand J, Ruppel AM, Hoyland W, Yates NL, Williams LD, Greene K, Gao H, Mahoney CR, Corcoran MM, Cagigi A, Taylor A, Brown DM, Ambrozak DR, Sincomb T, Hu X, Tingle R, Georgeson E, Eskandarzadeh S, Alavi N, Lu D, Mullen TM, Kubitz M, Groschel B, Maenza J, Kolokythas O, Khati N, Bethony J, Crotty S, Roederer M, Karlsson Hedestam GB, Tomaras GD, Montefiori D, Diemert D, Koup RA, Laufer DS, McElrath MJ, McDermott AB, and Schief WR (2022). Vaccination induces HIV broadly neutralizing antibody precursors in humans. Science 378, eadd6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ursin RL, Dhakal S, Liu H, Jayaraman S, Park HS, Powell HR, Sherer ML, Littlefield KE, Fink AL, Ma Z, Mueller AL, Chen AP, Seddu K, Woldetsadik YA, Gearhart PJ, Larman HB, Maul RW, Pekosz A, and Klein SL (2022). Greater Breadth of Vaccine-Induced Immunity in Females than Males Is Mediated by Increased Antibody Diversity in Germinal Center B Cells. mBio 13, e0183922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fink AL, Engle K, Ursin RL, Tang WY, Klein SL (2028). Biological sex affects vaccine efficacy and protection against influenza in mice. Proc. Natl. Acad. Sci. USA 115, 12477–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015). Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell Rep. 13, 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ota T, Doyle-Cooper C, Cooper AB, Doores KJ, Aoki-Ota M, Le K, Schief WR, Wyatt RT, Burton DR, and Nemazee D (2013). B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J. Immunol. 191, 3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong W-P, Wang L, and Nabel GJ (2013). Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei C-J, McDermott AB, Graham BS, Koup RA, and Nabel GJ (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wheatley AK, Whittle JR, Lingwood D, Kanekiyo M, Yassine HM, Ma SS, Narpala SR, Prabhakaran MS, Matus-Nicodemos RA, Bailer RT, Nabel GJ, Graham BS, Ledgerwood JE, Koup RA, McDermott AB (2015). H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J. Immunol. 195, 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, and Nussenzeig MC (2016). Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 11, 1908–1923. [DOI] [PubMed] [Google Scholar]
- 83.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, and Yutani H (2019). Welcome to the Tidyverse. JOSS 4, 1686. [Google Scholar]
- 84.Panda Development Team. (2020). pandas-dev/pandas: Pandas. Zenodo, latest. 10.5281/zenodo.3509134. [DOI] [Google Scholar]
- 85.Balish AL, Katz JM, and Klimov AI (2013). Influenza: propagation, quantification, and storage. Curr. Protoc. Microbiol. Chapter 15. 10.1002/9780471729259. [DOI] [PubMed] [Google Scholar]
- 86.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A (2014). Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–4.22.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masella AP, Bartram AK, Truszkowskim JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brochet X, Lefranc MP, and Giudicelli V (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giudicelli V, Brochet X, and Lefranc MP (2011). IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011, 695–715. [DOI] [PubMed] [Google Scholar]
- 90.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura K, Stecher G, and Kumar S (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38, 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res.14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinati J, Meulen JT, Carsetti R, Peiris M, de Kruif K, and Goudsmit J (2008). Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 3, e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, and Wilson IA (2009). Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, Mckee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, and Mascola JR. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, and Webster RG (2001) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97, 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Casanal A, Lohkamp B, and Emsley P (2020). Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 29, 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conway P, Tyka MD, DiMaio F, Konerding DE, and Baker D (2014). Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, and Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Struct. Biol 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- 103.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D. Struct. Biol 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Molecular Operating Environment (MOE). (2020). Chemical Computing Group ULC. 10–1010 Sherbrooke St W, Montreal, QC H3A 2R7, Canada. [Google Scholar]
- 105.Labute P (2009). Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 75, 187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Case DA, Aktulga HM, Belfon K, Ben-Shalom IY, Berryman JT, Brozell SR, Cerutti DS, Cheatham III TE, Cisneros GA, Cruzeiro VWD, Darden TA, Forouzesh N, Giambaşu G, Giese T, Gilson MK, Gohlke H, Goetz AW, Harris J, Izadi S, Izmailov SA, Kasavajhala K, Kaymak MC King E, Kovalenko A, Kurtzman T Lee TS, Li P, Lin C Liu J, Luchko T Luo R Machado M, Man V, Manathunga M, Merz KM, Miao Y Mikhailovskii O, Monard G,Nguyen H, O’Hearn KA, Onufriev A, Pan F Pantano S, Qi R, Rahnamoun A, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shajan A, Shen J, Simmerling CL, Skrynnikov NR, Smith J, Swails J, Walker RC, Wang J, Wang J, Wei H, Wu X, Wu Y, Xiong Y, Xue Y, York DM, Zhao S, Zhu Q and Kollman PA (2020) Amber 2020. University of California, San Francisco. [Google Scholar]
- 107.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- 108.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, and Mackerell AD Jr (2017). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hub JS, de Groot BL, Grubmuller H, and Groenhof G (2014). Quantifying Artifacts in Ewald Simulations of Inhomogeneous Systems with a Net Charge. J. Chem. Theory Comput. 10, 381–390. [DOI] [PubMed] [Google Scholar]
- 110.Wallnoefer HG, Liedl KR, and Fox T (2011) A challenging system: free energy prediction for factor Xa. J. Comp. Chem. 32, 1743–1752. [DOI] [PubMed] [Google Scholar]
- 111.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, and Walker RC (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comp. 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- 112.Miyamoto SK, and Settle PA (1992). An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comp. Chem. 8, 952–62. [Google Scholar]
- 113.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, and Haak JR (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. [Google Scholar]
- 114.Doll DM, Meyers LE and Adelman SA (1975). Generalized Langevin equation approach for atom/solid-surface scattering: inelastic studies. J. Chem. Phys. 63, 4908–4914. [Google Scholar]
- 115.Adelman SA, and Doll JD (1976). Generalized Langevin Equation Approach for Atom/Solid-surface Scattering: General Formulation for Classical Scattering off Harmonic Solids. J. Chem. Phys. 64, 2375–2388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
3D maps and models from the EM analysis have been deposited to the Protein Data Bank (http://www.rcsb.org/) and Electron Microscopy Databank (http://www.emdatabank.org/), respectively, and are publicly available from the date of publication. Accession numbers are listed in the Key Resources Table. Paired HC and LC sequences from the GCs expanded by H3ssF and H7ssF are deposited in Genbank. Accession numbers are provided in the Key Resources Table and in Table S4.
The complete code used to compute the frequencies of the VH1–18 QxxV class in human IgM repertoires has been deposited in Zenodo and is publicly available from the date of publication. DOI is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PerCP/Cy5.5 anti-mouse CD45.1 Antibody | Biolegend | Cat#110728; RRID:AB_893348 |
| PE-Cy™7 Hamster Anti-Mouse CD95 | BD Biosciences | Cat#557653; RRID:AB_396768 |
| Brillant Violet 785™ anti-mouse CD45.2 | Biolegend | Cat#109839; RRID:AB_2562604 |
| Anti-mouse CD38, BV510 | BD Biosciences | Cat#740129; RRID:AB_2739886 |
| BV711 Rat Anti-Mouse Ig, λ1, λ2 & λ3 Light Chain | BD Biosciences | Cat#744527; RRID:AB_2742301 |
| BUV395 Rat Anti-Mouse Ig, κ light chain | BD Biosciences | Cat#742839; RRID:AB_2741090 |
| BV421 Rat Anti-Mouse IgM | BD Biosciences | Cat#743323; RRID:AB_2741424 |
| PE/Cy7 anti-mouse IgD Antibody | Biolegend | Cat#405720; RRID:AB_2561875 |
| PE anti-mouse CD45.2 Antibody | Biolegend | Cat#109808; RRID:AB_313445 |
| Anti-mouse CD38 Alexa700 | Invitrogen | Cat#56-0381-82 |
| BV786 Rat Anti-Mouse IgD | BD Biosciences | Cat#563618; RRID:AB_2738322 |
| Alexa Fluor® 700 anti-mouse IgD Antibody | BD Biosciences | Cat#405730 |
| BV605 Rat anti-mouse CD45R/B220 | BD Biosciences | Cat#563708; RRID:AB_2738383 |
| PE anti-mouse CD138 (Syndecan-1) Antibody | Biolegend | Cat#142504; RRID:AB_10915989 |
| BV510 Rat Anti-Mouse IgD | BD Biosciences | Cat#563110; RRID:AB_2737003 |
| Brilliant Violet 421™ anti-mouse IgD Antibody | Biolegend | Cat#405725; RRID:AB_2562743 |
| BV510 Rat Anti-Mouse B220/CD45R Clone RA3-6B2 | BD Biosciences | Cat#563103; RRID:AB_2738007 |
| BV421 Rat Anti-Mouse IgG1 | BD Biosciences | Cat#562580; RRID:AB_2737664 |
| BUV395 Rat Anti-Mouse IgM | BD Biosciences | Cat#743329; RRID:AB_2741430 |
| CD4 Monoclonal Antibody (GK1.5), APC-eFluor 780 | Invitrogen | Cat#47-0042-80 |
| CD8a Monoclonal Antibody (53-6.7), APC-eFluor 780 | Invitrogen | Cat#47-0081-80 |
| F4/80 Monoclonal Antibody (BM8), APC-eFluor 780 | Invitrogen | Cat#47-4801-80 |
| Ly-6G Monoclonal Antibody (1A8-Ly6g), APC-eFluor 780 | Invitrogen | Cat#47-5931-80 |
| FITC anti-mouse/human CD45R/B220 Antibody | Biolegend | Cat#103206; RRID:AB_312991 |
| APC anti-mouse CD43 Antibody | Biolegend | Cat#143208; RRID:AB_11149685 |
| APC/Cy7 anti-mouse CD21/CD35 (CR2/CR1) Antibody | Biolegend | Cat#123418; RRID:AB_1953275 |
| BV421 Rat Anti-Mouse CD24 | BD Biosciences | Cat#562563; RRID:AB_2737002 |
| Anti-mouse CD249 (BP1) FITC | Thermo | Cat#11-5891-82; RRID:AB_465295 |
| Alexa Fluor 700 anti-mouse TCRb chain | BioLegend | Cat#109224; RRID:AB_1027654 |
| FITC anti-mouse CD21/CD35 (CR2/CR1) Antibody | BioLegend | Cat#123408; RRID:AB_940403 |
| PerCP/Cy5.5 anti-mouse/human CD45R/B220 Antibody | BioLegend | Cat#103236; RRID:AB_893354 |
| APC anti-mouse CD23 Antibody | BioLegend | Cat#101620; RRID:AB_2563438 |
| PE anti-mouse CD24 Antibody | BioLegend | Cat#101808; RRID:AB_312840 |
| Anti Mouse CD4 BV510 | BioLegend | Cat#100559; RRID:AB_2562608 |
| BV421 Rat Anti-Mouse IgM | BD Biosciences | Cat#743323; RRID:AB_2741424 |
| Alkaline Phosphatase AffiniPure Goat Anti-Mouse IgG, Fcγ Fragment Specific | Jackson ImmunoResearch | Cat#115-055-071 |
| Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block™) | BD Biosciences | Cat#553142; RRID:AB_394657 |
| Alexa Fluor® 488 anti-mouse CD45.2 Antibody | Biolegend | Cat#109816; RRID:AB_492868 |
| Alexa Fluor® 594 anti-mouse CD3 Antibody | Biolegend | Cat#100240; RRID:AB_2563427 |
| TotalSeq™-C0301 anti-mouse Hashtag 1 Antibody | Biolegend | Cat#155861; RRID:AB_2800693 |
| TotalSeq™-C0302 anti-mouse Hashtag 2 Antibody | Biolegend | Cat#155863; RRID:AB_2800694 |
| TotalSeq™-C0303 anti-mouse Hashtag 3 Antibody | Biolegend | Cat#155865; RRID:AB_2800695 |
| TotalSeq™-C0304 anti-mouse Hashtag 4 Antibody | Biolegend | Cat#155867; RRID:AB_2800696 |
| TotalSeq™-C0305 anti-mouse Hashtag 5 Antibody | Biolegend | Cat#155869; RRID:AB_2800697 |
| TotalSeq™-C0306 anti-mouse Hashtag 6 Antibody | Biolegend | Cat#155871; RRID:AB_2819910 |
| TotalSeq™-C0307 anti-mouse Hashtag 7 Antibody | Biolegend | Cat#155873; RRID:AB_2819911 |
| TotalSeq™-C0308 anti-mouse Hashtag 8 Antibody | Biolegend | Cat#155875; RRID:AB_2819912 |
| TotalSeq™-C0309 anti-mouse Hashtag 9 Antibody | Biolegend | Cat#155877; RRID:AB_2819913 |
| TotalSeq™-C0310 anti-mouse Hashtag 10 Antibody | Biolegend | Cat#155879; RRID:AB_2819914 |
| 09-1B12-UCA | Produced in house (Corbett et al. 2019) | N/A |
| 09-1B12-UCA + N55T | Produced in house | N/A |
| 09-1B12 | Produced in house (Andrews et al. 2017) | N/A |
| 05-2A09-UCA | Produced in house | N/A |
| 05-2A09-UCA + N55T | Produced in house | N/A |
| 05-2A09 | Produced in house (Andrews et al. 2017) | N/A |
| 21-1A01-UCA | Produced in house | N/A |
| 21-1A01-UCA- + N55T | Produced in house | N/A |
| 21-1A01 | Produced in house (Andrews et al. 2017) | N/A |
| 05-2D04-UCA | Produced in house | N/A |
| 05-2D04-UCA + N55T | Produced in house | N/A |
| 05-2D04 | Produced in house (Andrews et al. 2017) | N/A |
| 27-1D08-UCA | Produced in house | N/A |
| 27-1D08-UCA + N55T | Produced in house | N/A |
| 27-1D08 | Produced in house (Andrews et al. 2017) | N/A |
| O1 | Produced in house | N/A |
| O2 | Produced in house | N/A |
| VRC01 | Produced in house | N/A |
| CR6261 | Produced in house | N/A |
| Biological samples | ||
| H3N2 X-31 | BEI Resources | Cat#NR-3483 |
| Mouse adapted H1N1 A/California/07/2009 (maA/Cal/09) | Sabra Klein and Andrew Pekosz John Hopkins University (Fink et al. 2018) | N/A |
| Influenza reporter viruses | Masaru Kanekiyo, NIH (Creanga et al. 2021) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Pan B Cell Isolation Kit II, mouse | Miltenyi Biotec | Cat#130-104-443 |
| LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit, for UV excitation | Thermo Fisher Scientific | Cat#L34962 |
| CountBright™ Absolute Counting Beads, for flow cytometry | Thermo Fisher Scientific | Cat#C36950 |
| SIGMAFAST™ p-Nitrophenyl phosphate Tablets | Sigma | Cat#N2770-50SET |
| NP40 | Millipore | Cat#492016-100ML |
| UltraComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Cat#01-2222-42 |
| H3ssF | Produced in house (Corbett et al. 2019) | N/A |
| H7ssF | Produced in house (Corbett et al. 2019) | N/A |
| Invitrogen™ Molecular Probes™ DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat#D1306 |
| Chromium Next GEM Single Cell 5’ Kit v2 | 10x Genomics | Cat#PN-1000263 |
| Library Construction Kit | 10x Genomics | Cat#PN-1000190 |
| Chromium Single Cell Mouse BCR Amplification Kit | 10x Genomics | Cat#PN-1000255 |
| Chromium Next GEM Chip K Single Cell Kit | 10x Genomics | Cat#PN-1000286 |
| Dual Index Kit TT Set A | 10x Genomics | Cat#PN-1000215 |
| Dual Index Kit TN Set A | 10x Genomics | Cat#PN-1000250 |
| Penicillin/streptomycin | Gibco | Cat#15140163 |
| Puromycin | Gibco | Cat#A1113803 |
| Opti-MEM | Gibco | Cat#31985088 |
| Bovine Serum Albumin | Sigma | Cat#A9418 |
| TransIT-LT1 | MirusBio | Cat#2306 |
| Gibson assembly kit | NEB | Cat#E5510S |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher Scientific | Cat#A14525 |
| Ni Sepharose resin | GE Healthcare | N/A |
| A/Perth/16/2009 H3 trimer | Produced in house (Joyce et al. 2016) | N/A |
| A/Shanghai/02/2013 H7 trimer | Produced in house (Joyce et al. 2016) | N/A |
| A/Michigan/45/2015 H1 trimer | This paper | N/A |
| A/California/07/2009 H1 trimer | Produced in house (Sangesland et al. 2019) | N/A |
| A/Japan/305/1957 H2 trimer | This paper | |
| A/Indonesia/05/2005 H5 trimer | Produced in house (Whittle et al. 2014) | N/A |
| A/Perth/16/2009 H3 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Shanghai/02/2013 H7 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Michigan/45/2015 H1 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/California/07/2009 H1 trimerDstem (KO, 45HA2) | This paper | N/A |
| A/Indonesia/05/2005 H5 trimerDstem (KO, 45HA2) | This paper | N/A |
| H3ssF-KO (45HA2) | This paper | N/A |
| H7ssF-KO (45HA2) | This paper | N/A |
| Sigma Adjuvant System | Sigma-Aldrich | Cat#S6322 |
| Protein G Sepharose | GE Healthcare | Cat#17061802 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#KK2602 |
| Maxima H Minus Reverse Transcriptase | Thermo Fisher | Cat#EP0751 |
| HotStarTaq Plus DNA Polymerase | QIAGEN | Cat#203603 |
| Buffer RLT | QIAGEN | Cat#79216 |
| BirA Biotin-Protein Ligase Bulk Reaction Kit | Avidity | Cat#Bulk BirA |
| Alexa Fluor 647 Protein Labeling Kit | Thermo Fisher | Cat#A20173 |
| Alexa Fluor 488 Protein Labeling Kit | Thermo Fisher | Cat#A10235 |
| Alexa Fluor 594 Protein Labeling Kit | Thermo Fisher | Cat#A10239 |
| Alexa Fluor 546 Protein Labeling Kit | Thermo Fisher | Cat#A20183 |
| MiSeq Reagent Kit, V2 500 cycles | Illumina | Cat#MS-102-2003 |
| 10% PEG 10k | Hampton Research | Cat#HR2-084 |
| Lys-C endoproteinase | NEB | Cat#P8109S |
| Protease inhibitor cocktail (PIC) | Roche | Cat#11697498001 |
| Deposited data | ||
| VH1-18 heavy chain (HC) and light chain (LC) sequences (see also Data S1) | This paper | GenBank IDs: PP507169 - PP508172 |
| X-ray structure of 09-1B12 bound to A/Perth/14/2009 H3N2 hemagglutinin | This paper | PDB ID: 8UWA |
| CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6 | This paper | PDB ID: 8UT3, EMDB ID: EMD-42528 |
| CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12 | This paper | PDB ID: 8UT4, EMDB ID: EMD-42529 |
| CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T | This paper | PDB ID: 8UT5, EMDB ID: EMD-42530 |
| CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | This paper | PDB ID: 8UT6, EMDB ID: EMD-42531 |
| CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | This paper | PDB ID: 8UT7, EMDB ID: EMD-42532 |
| CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-15 days post-immunization | This paper | PDB ID: 8UT8, EMDB ID: EMD-42533 |
| CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization | This paper | PDB ID: 8UT9, EMDB ID: EMD-42534 |
| CryoEM structure of A/Perth/16/2009 H3 | This paper | EMDB ID: EMD-42535 |
| CryoEM map of A/Shanghai/1/2013 H7 HA | This paper | EMDB ID: EMD-42536 |
| Code used to compute frequencies of the VH1-18 QxxV class in human IgM repertoires | This paper | https://www.doi.org/10.5281/zenodo.10800716 |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID:CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID:CVCL_D615 |
| Canine: MDCK | ATCC | Cat#CCL-34; RRID:CVCL_0422 |
| Human: HEK293F | Gibco | Cat#R79007; RRID:CVCL_6642 |
| MDCK-SIAT1-PB1 | Produced in house (Creanga et al. 2021) | N/A |
| MDCK-SIAT1-H5 | Produced in house (Creanga et al. 2021) | N/A |
| MDCK-SIAT1-H7 | Produced in house (Creanga et al. 2021) | N/A |
| Expi293F GnTI− | Produced in house | N/A |
| HEK293T-PB1 | Produced in house (Creanga et al. 2021) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: B6.SJL-Ptprcapepcb/BoyJ mice | The Jackson Laboratory | JAX: 002014; RRID:IMSR_JAX:002014 |
| Mouse: C57BL/6 | The Jackson Laboratory | JAX: 000664; RRID:IMSR_JAX:000664 |
| Mouse: 09-1B12-UCA KI mouse (H09-1B12-UCA/WT, k09-1B12-UCA/WT) | This paper | N/A |
| Oligonucleotides | ||
| TaqMan probes for genotyping | TransnetYX | N/A |
| Cocktails of IgG- and IgK-specific primers and thermocycling conditions described previously | von Boehmer et al. 2016 | N/A |
| sgRNA | Lin et al. 2018; Wang et al. 2021 | N/A |
| Primers for Single Cell BCR Amplification, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Flowjo 10.9.0 | Treestar | https://www.flowjo.com/; RRID:SCR_008520 |
| Prism 10.1.0 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| IMGT/V-quest | Brochet et al. 2008; Giudicelli et al. 2011 | http://www.imgt.org/IMGTindex/V-QUEST.php; RRID:SCR_010749 |
| Geneious Prime | Biomatters | https://www.geneious.com/; RRID:SCR_010519 |
| Cell Ranger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger; RRID:SCR_017344 |
| WebLogo | Crooks et al., 2004 | https://weblogo.berkeley.edu; RRID:SCR_010236 |
| Seurat R package | Standard R Package | https://satijalab.org/seurat/; RRID:SCR_016341 |
| R circlize 0.4.15 | R package | https://cran.r-project.org/web/packages/circlize/index.html; RRID:SCR_002141 |
| Bash 5.1.16 | GNU Project shell | https://www.gnu.org/software/bash/; RRID:SCR_021268 |
| Python 3.10 | Python Programming Language | https://www.python.org/: RRID:SCR_008394 |
| R 4.1.2 | The R project for statistical computing | https://www.r-project.org/; RRID:SCR_001905 |
| Tidyverse (dplyr 1.1.4, readr 2.1.4, ggplot2 3.4.4) | Wickham et al. 2019 | https://www.tidyverse.org/packages/; RRID:SCR_019186 |
| Pandas 2.1.1 | Pandas development team, 2020 | https://pandas.pydata.org; RRID:SCR_018214 |
| Pandaseq | Masella et al. 2012 | https://github.com/neufeld/pandaseq; RRID:SCR_002705 |
| MigMAP | Sangesland et al. 2019 | https://github.com/mikessh/migmap |
| MEGA11 software | Tamura et al. 2011, 2021 | https://www.megasoftware.net; RRID:SCR_023017 |
| UCSF ChimeraX | Pettersen et al.2021 | https://www.cgl.ucsf.edu/chimerax/; RRID:SCR_015872 |
| Coot 0.9.8 | Casanal et al. 2020 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID:SCR_014222 |
| Phenix | Afonine et al. 2018 | https://phenix-online.org; RRID:SCR_014224 |
| Rosetta | Conway et al. 2014 | https://www.rosettacommons.org/home; RRID:SCR_015701 |
| XDSGUI | XDS program package | https://strucbio.biologie.uni-konstanz.de/xdswiki/index.php/XDSGUI; RRID:SCR_015652 |
| Protonate3D tool | Molecular Operating Environment, 2020; Labute, 2009 | RRID:SCR_014882 |
| Amber Tools20 package | Case et al. 2020 | https://ambermd.org/index.php; RRID:SCR_014230 |
| NpT ensemble | Salomon-Ferrer et al. 2013 | N/A |
| SHAKE algorithm | Miyamoto, 1992 | N/A |
| Berendsen algorithm | Berendsen, 1984 | N/A |
| GetContacts software | This paper | https://getcontacts.github.io/ |
| Other | ||
| Mastercycler PCR machine | Eppendorf | N/A |
| ELISA readers | BioTek | N/A |
| 5 laser ARIA II | BD Biosciences | RRID:SCR_018091 |
| 5 laser LSR Fortessa | BD Biosciences | RRID:SCR_019601 |
| Qubit | Thermo Fisher | RRID:SCR_018095 |
| MiSeq | Illumina | RRID:SCR_016379 |
| Nextseq2000 | Illumina | RRID:SCR_023614 |
| Tapestation | Agilent | RRID:SCR_019394 |
| 5 Laser Aria SORP | BD Biosciences | N/A |
| Superdex 200 10/300 Column | GE Healthcare | Cat#17517501 |
| Superose 6 10/300 Column | GE Healthcare | Cat#17517201 |
| 96 Well glass bottom plate with high performance #1.5 cover glass | CellVis | Cat#P96-1.5P |
| CellDiscoverer7 | Zeiss | N/A |
| Tangential flow filtration system | Cytiva | N/A |
| ViewDrop II seals | SPT LabTech | Cat#4150-05600 |
| Erythrina cristagalli Gel-ECA-Immobilized Lectin | EY Laboratories | N/A |
| AKTA size exclusion chromatography (SEC) | Cytiva | RRID:SCR_023461 |
| IgG Elution Buffer | Pierce | Cat#21009 |
| Cu grids | EMS | Cat#CF300-Cu-UL |
| Vitrobot | Thermo Fisher | N/A |
| Glacios-2 cryoEM | Thermo Fisher | N/A |
| Beam line 24-ID-E | Advanced Photon Source | N/A |
| BLItz System (Octet N1) | Fortebio | RRID:SCR_023267 |
| Streptavidin biosensors (SA) | Sartorious | Cat#18-5019 |
| Prolab Isopro RMH 3000, 5P75-5P76 mice diet | LabDiet | https://www.labdiet.com/product/detail/5p76-prolab-isopro-rmh-3000 |


