Abstract
Plant family 1 glycosyltransferases (UGTs) represent a formidable tool to produce valuable natural and novel glycosides. Their regio- and stereo-specific one-step glycosylation mechanism along with their inherent wide acceptor scope are desirable traits in biotechnology. However, their donor scope and specificity are not well understood. Since different sugars have different properties in vivo and in vitro, the ability to easily glycodiversify target acceptors is desired, and this depends on our improved understanding of the donor binding site. In the aim to unlock the full potential of UGTs, studies have attempted to elucidate the structure-function relationship governing their donor specificity. These efforts have revealed a complex phenomenon, and general principles valid for multiple enzymes are elusive. Here, we review the studies of UGT donor specificity, and attempt to group the information into key concepts which can help shape future research. We zoom in on the family-defining PSPG motif, on two loop residues reported to interact with the C6 position of the sugar, and on the role of active site arginines in donor specificity. We continue to discuss attempts to alter and expand the donor specificity by enzyme engineering, and finally discuss future research directions.
Keywords: glycosyltransferase, donor specificity, enzyme engineering, glycodiversification, structure-function
1 Introduction
Glycosides are abundant in the biosphere and are represented by numerous high-value industrial compounds. This includes natural sweeteners such as steviol glycosides (Libik-Konieczny et al., 2021) and mogrosides (Li et al., 2022), therapeutic compounds such as amygdalin (Thodberg et al., 2018) and glycyrrhizin (Li et al., 2010), and natural colourants such as betanin (Zhang L. et al., 2023). The current or potential high demand for these compounds and other glycosides requires their efficient production (Nidetzky, 2021). Since glycosylation using conventional chemistry is notoriously challenging and usually inefficient, attention has been turned to biocatalysts for the glycosylation of organic compounds (De Roode et al., 2003). Here, family 1 glycosyltransferases (Drula et al., 2022) are in focus due to their ability to regio- and stereoselectively transfer a sugar molecule from a uridine diphosphate (UDP)-activated donor to a wide variety of acceptors in one step (Yao et al., 2022). These enzymes are also commonly referred to as UDP-dependent glycosyltransferases (UGTs). The catalytic cavity of UGTs is known to accommodate a varied panel of acceptors while still retaining catalytic activity, making them suitable for the synthesis of new-to-nature glycosides (De Bruyn et al., 2015).
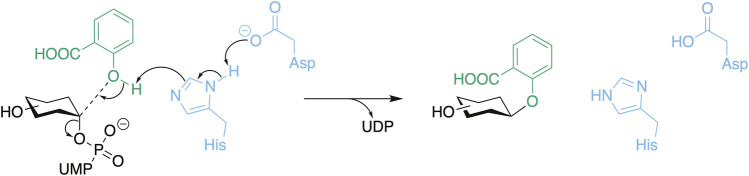
UGTs have been extensively studied, which has led to a broad understanding of their catalytic mechanism and acceptor specificity (Franconetti et al., 2021; Teze et al., 2021). Their mechanism involves a highly conserved catalytic dyad, typically comprising His and Asp. In O-glycosylation, this dyad deprotonates the acceptor, facilitating nucleophilic attack (Scheme 1) (Brazier-Hicks et al., 2007; Teze et al., 2021). However, O-/N-/S-glycosylation all occur via an SN2-like reaction and inversion of the anomeric configuration (Scheme 1) (Teze et al., 2021). This knowledge has yielded several successful engineering efforts leading to enhanced catalytic efficiency (e.g., Li et al., 2020), altered regiospecificity (e.g., Li et al., 2021; Zhang M. et al., 2022), and altered acceptor substrate scope (e.g., Wetterhorn et al., 2017). However, the structure-function relationship determining the donor specificity of the enzyme family is still poorly understood. UGTs adopt a GT-B fold consisting of two Rossmann-type fold domains where the acceptor predominantly interacts with the N-terminal domain while the donor substrate predominantly interacts with the C-terminal domain (Wang M. et al., 2023). A highly conserved motif resides within the C-terminal domain, denoted as the Plant Secondary Product Glycosyltransferase (PSPG) motif (Hughes and Hughes, 1994; Paquette et al., 2003). This motif is recognised as a critical part of donor binding (Li et al., 2007) and has laid grounds for several mutational studies with mixed results (vide infra). However, residues outside of the PSPG motif and from the N-terminal domain have also been demonstrated to be important for the donor specificity, illustrating the complexity of this structure-function relationship (Osmani et al., 2009).
SCHEME 1.
The general reaction mechanism of UGTs with the catalytic residues presented in blue, the donor substrate in black, and the acceptor substrate (salicylic acid) in green. Here, O-glycosylation is used as an example which includes a deprotonation step.
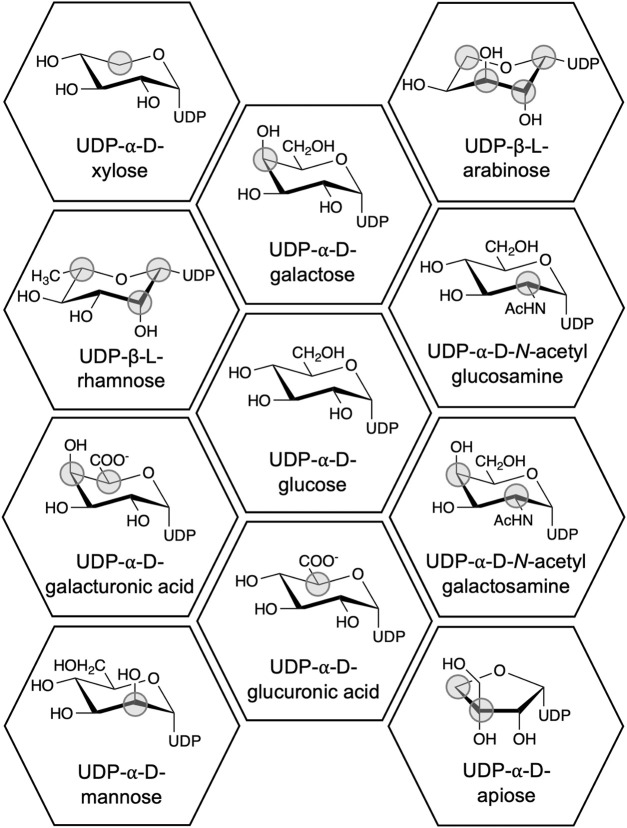
UGTs from plants most often use UDP-glucose (UDP-Glc) as donor (Louveau and Osbourn, 2019). However, UDP-galactose (UDP-Gal), -glucuronic acid (-Glu), and -rhamnose (-Rha) are also known plant UGT substrates (Figure 1). These sugars can potentially provide specific properties to an organic molecule upon conjugation (Goel et al., 2021; Zhang L. J. et al., 2022), but understanding these properties is hindered by the narrow donor substrate scope of UGTs, which usually includes only one UDP-sugar (Osmani et al., 2008). Hence, understanding the structure-function relationship of UDP-sugar specificity is a coveted goal in the research field. Despite numerous successful alterations of donor specificity (e.g., Liu S. et al., 2021; Chen et al., 2021; Huang et al., 2022), the structure-function relationship is still not well-defined (Rahimi et al., 2019). Instead, we now have an extensive knowledge base of the effects of mutating specific residues in specific UGTs. In addition, several high-resolution structures of UGTs in complex with UDP-sugars have been solved (Table 1).
FIGURE 1.
Chemical structures of UDP-sugars which have been discovered to be donor substrates for UGTs. The differences between the UDP-sugars have been marked with a grey circle using UDP-Glc as the reference structure. UDP-apiose was compared to the Haworth projection of UDP-α-d-glucofuranose.
TABLE 1.
Reported crystal structures of UGT:UDP-sugar complexes.
| UGT isoform | Plant species | UDP-sugar | Resolution (Å) | Crystallisation method | PDB | Reference |
|---|---|---|---|---|---|---|
| UGT71G1 | Medicago truncatula | UDP-Glc | 2.60 | Soaking | 2ACW | Shao et al. (2005) |
| UGT708C1 | Fagopyrum esculentum | UDP-Glc | 2.01 | Co-crystallisation | 6LLZ | Liu et al. (2020a) |
| UGT74AC1 | Siraitia grosvenorii | UDP-Glc | 2.10 | Soaking | 6L8Z | Li et al. (2020) |
| SbCGTb | Scutellaria baicalensis | UDP-Glc | 2.87 | Co-crystallisation | 6LFZ | Wang et al. (2020) |
| PtUGT1 | Polygonum tinctorium | UDP-Glc | 2.40 | Soaking | 6SU6 | Teze et al. (2021) |
| MiCGT | Mangifera indica | UDP-Glc | 2.85 | Co-crystallisation | 7VA8 | Wen et al. (2021) |
| UGT74AN2 | Calotropis gigantea | UDP-Glc | 2.15 | Soaking | 7W1H | Huang et al. (2022) |
| GgCGT | Glycyrrhiza glabra | UDP-Glc | 2.60 | Co-crystallisation | 6L5P | Zhang et al. (2020) |
| GgCGT | Glycyrrhiza glabra | UDP-Gal | 2.89 | Co-crystallisation | 6L5Q | Zhang et al. (2020) |
| UGT89C1 | Arabidopsis thaliana | UDP-Rha | 3.21 | Co-crystallisation | 6IJA | Zong et al. (2019) |
| UGT76G1 | Stevia rebaudiana | UDP-Xyl | 2.50 | Co-crystallisation | 6KVJ | Liu et al. (2020b) |
To foster additional research into UGT donor specificity, this review aims to delineate and establish connections within the existing knowledge base. Furthermore, the research direction will be discussed and related to future biocatalytic research.
2 The role of the PSPG motif in the donor specificity of UGTs
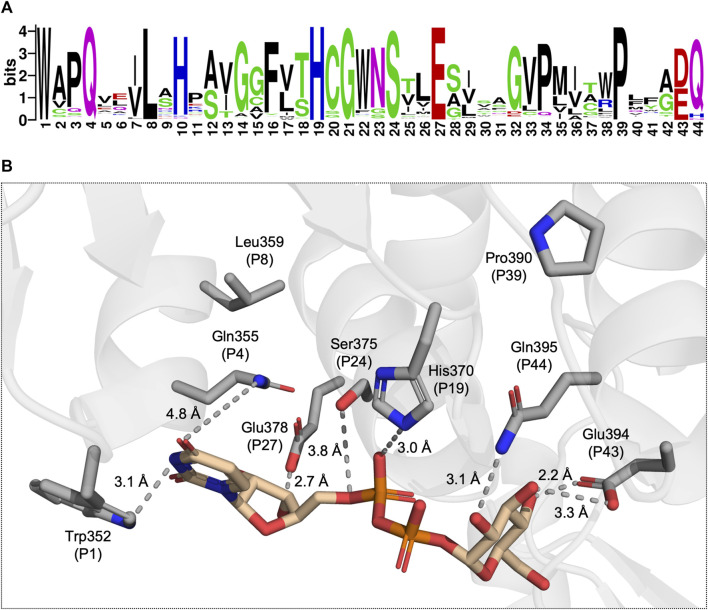
The PSPG motif is a key characteristic of plant UGTs and is commonly used in genome mining to identify UGTs (Fan et al., 2017; Rehman et al., 2018). The highly conserved motif is constituted by 44 residues with Trp as the initial residue and an acidic residue followed by Gln/His/Asn as the terminal residue pair. The PSPG motif contains several highly conserved positions, namely, positions 4, 8, 19, 21, 24, 27, and 39 (Figure 2A). Observing these residues in a crystal structure reveals that all, except Leu P8 and Pro P39, can participate in intermolecular interactions with the UDP moiety of the UDP-sugar (Figure 2B). Meanwhile, among the conserved residues within the PSPG motif, only P43 and P44 can interact with the sugar moiety (Figure 2B). Given conservation of the PSPG motif along with its role in UDP-sugar recognition, studies have targeted the motif to alter the donor specificity of UGTs.
FIGURE 2.
(A) A sequence logo generated from a multiple sequence alignment of 224 UGTs, each with determined donor specificity. (B) A view of the donor substrate binding site with the conserved residues of the PSPG motif displayed as grey sticks and UDP-Glc displayed in tan (PDB: 6SU6). The dotted lines represent potential intermolecular interactions between the UGT and UDP-Glc.
Due to its direct interaction with the sugar moiety and high conservation, P44 of the PSPG motif has often been the target of site-directed mutagenesis (e.g., Zong et al., 2019; Yamashita et al., 2023). Noticeably, the bifunctional glucosyl-/galactosyltransferase VvGT6 from Vitis vinifera was converted into a monofunctional galactosyltransferase by a single substitution of Gln373 with His (Ono et al., 2010a). However, the substitution of this Gln with His in the glucosyltransferases, UBGT, VvGT1, MrUFGT, and MrUGT72B67, did not confer UDP-Gal activity (Kubo et al., 2004; Offen et al., 2006; Ren et al., 2022a). Furthermore, wildtype galactosyltransferases with Gln or Asn at P44 are known (Xie et al., 2020; 2022; Ren et al., 2022b). Even though it is not possible to directly correlate the residue at P44 with UDP-Gal activity, many studies have demonstrated the importance of this residue in UDP-sugar specificity. Particularly mutating P44 into Gln has led to interesting discoveries. In the galactosyltransferase, AcGaT from Aralia cordata, His to Gln substitution at P44 improved UDP-Glc activity (Kubo et al., 2004). Similarly, changing His to Gln in the UDP-Rha-specific UGT89C1 led to an increase in the UDP-Rha activity while also enabling activity with UDP-Glc (Zong et al., 2019). In the arabinosyltransferases AsAAT1 from Avena strigose and UGT78D3 from A. thaliana, swapping P44 from His to Gln improved or enabled activity with UDP-Glc and UDP-Xyl while it significantly reduced UDP-Ara activity in the case of AsAAT1 (not tested for UGT78D3) (Han et al., 2014; Louveau et al., 2018).
PSPG positions other than P44 have also been mutated to understand their function. The glucosyltransferase UGT76E2 from A. thaliana was converted into a bifunctional UGT accepting both UDP-Glc and UDP-Gal (Akere et al., 2020). This was facilitated by mutating Asp374 (P43) to Glu, where the extra carbon atom enabled hydrogen bonding between Glu and the C4-OH of galactose. Meanwhile, mutating P43 to Ala has in several instances been shown to abolish glycosyltransferase activity, suggesting the importance of this residue, the acidity of which might be important for UDP-sugar recognition (Shao et al., 2005; Offen et al., 2006; Zong et al., 2019; Zhang S. et al., 2023). Another study expanded the donor specificity of UGT78H2 by mutating P3 and P23 (Chen et al., 2021). Wildtype UGT78H2 accepted UDP-Glu and -Gal while the introduction of mutations Asn340Pro (P3) and Lys360Asn (P23) expanded the donor scope to include UDP-Glc, and in addition improved the catalytic efficiency with UDP-Glu and -Gal by 7% and 30%, respectively. Molecular docking with UDP-Gal and -Glu showed that Asn in P23 produced an additional intermolecular interaction with the uridine moiety and a direct interaction with galactose as compared with Lys. Meanwhile, Liu S. et al., 2021 were able to increase the UDP-Glc activity in the galactosyltransferase, TcOGT4, by swapping P22 of the PSPG from Pro to Trp. This residue was identified through multiple sequence alignment with other glucosyltransferases. In addition to these examples, other studies have successfully altered the UDP-sugar donor specificity of UGTs by mutating residues in the PSPG motif. These are related to topics which are discussed in sections 4 and 5 and are accordingly referenced within those sections.
Overall, the PSPG motif naturally serves as an integral part of donor substrate recognition. However, it is limited how much information can be extracted from the constituents of the motif and related to the donor specificity of a UGT. Many efforts have targeted the terminal residue with mixed results. Meanwhile, targeting other parts of the PSPG motif has proven an interesting path to altering donor specificity even though they, most of the time, do not interact with the sugar moiety, indicating a role in the orientation of the UDP-sugar.
3 A loop in the N-terminal domain can significantly alter activity with UDP-Rha, -Glc, and -Xyl
Rhamnose differs from the previously mentioned sugars by not containing a hydroxy group on C6. In addition, UDP-Rha naturally occurs in the β-l configuration, where the other substrates are in the α-d configuration (Figure 1), which impacts the overall geometry of UDP-Rha. These characteristics need to be accommodated to enable activity with UDP-Rha.
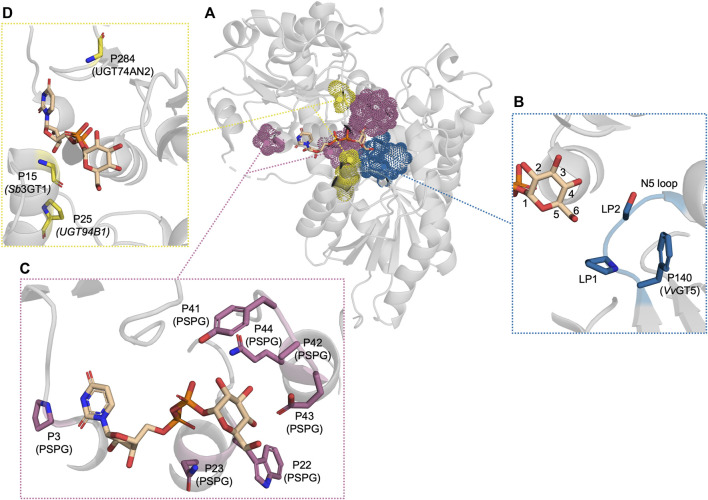
Several studies have focused on residues in proximity to C6 to understand how the activity with UDP-Rha can be enabled and disabled. In the UDP-Rha-bound crystal structure of the rhamnosyltransferase, UGT89C1 (Table 1), Pro147 (loop position 1 (LP1)) and Ile148 (LP2) were found to be in the vicinity of C6 (Figure 3B) (Zong et al., 2019). These two residues reside in a flexible loop, which was first described as an important loop for donor specificity by Osmani et al., 2009 and dubbed the N5 loop (Figure 3B). Swapping LP1 in UGT89C1 for the corresponding residue in the UDP-Glc-specific UGT71G1 (Thr) abolished rhamnosyltransferase activity. Meanwhile, swapping LP2 for the corresponding residue in UGT71G1 (Ser) reduced the activity to 20% relative to WT. Similar observations were made in a study of two UGTs from Viola tricolor, VtCGTa and VtCGTc, which displayed a preference for UDP-Glc and UDP-Rha, respectively (Han et al., 2023). Aligning VtCGTc and other rhamnosyltransferases with VtCGTa and other glucosyltransferases showed that a Thr occupied LP1 in the glucosyltransferases while Val, Pro, or Ile occupied this position in the rhamnosyltransferases. Mutating this position in VtCGTa its homologs to Val or Pro improved UDP-Rha activity in all instances while mutating the position into Ile only enabled UDP-Rha activity in some of the tested CGTa enzymes. Mutating LP1 in VtCGTc from Val to Thr significantly reduced the UDP-Rha activity and established activity with UDP-Glc. These results indicate that a hydrophobic residue in LP1 could be an important factor for UDP-Rha activity. However, these effects were not observed in the rhamnosyltransferase, HmF3RT, which also has activity with UDP-Xyl, and UDP-Glc (Zhang S. et al., 2023). Mutating Val129 (LP1) to Ala or Thr, improved activity with UDP-Rha and significantly improved the UDP-Glc activity to a level comparable to the UDP-Rha activity, while it decreased the activity with UDP-Xyl.
FIGURE 3.
The residues which have been reported to be important for UDP-sugar donor specificity, displayed in PDB: 6SU6. (A) An overview of the residues displayed as dot spheres representing the van der Waals surface area. Each color corresponds to a specific area: blue indicates the N5 loop, magenta indicates the PSPG motif, and yellow indicates residues near the cavity entrance. (B) A close-up view of the N5 loop area. (C) A close-up view of the PSPG motif area. (D) A close-up view of the residues close to the cavity entrance. “P” refers to the position in the corresponding UGT or PSPG motif mentioned in parentheses.
UDP-Glc and -Xyl activity has been demonstrated to be susceptible to changes when altering LP1 and LP2. One study identified two UGT89A2 homologues from 2 A. thaliana accessions which had distinct donor specificities (Chen and Li, 2017). UGT89A2 from Col-0 accepted UDP-Xyl and harboured an Ile at LP2 while UGT89A2 from C24 accepted UDP-Glc and UDP-Xyl and harboured a Ser at LP2. Swapping LP2 residues also led to a swap in donor specificity. A similar method was employed with Ib3GGT from Ipomoea batatas, which accepted UDP-Glc and UDP-Xyl as donor substrate, and its homologue from A. thaliana, At3GGT, which solely accepted UDP-Xyl (Wang et al., 2018). Scanning the donor binding pocket led to the identification of LP1 as the only variable residue, where Ib3GGT has a Thr while At3GGT has an Ile. Swapping the residues in the two UGTs also swapped their respective donor specificities. Other examples are described in Table 2.
TABLE 2.
Site-directed mutagenesis of LP1/LP2 and the change in donor specificity compared to WT *VtCGTa T145P was only tested with UDP-Rha and VtCGTc was only tested with UDP-Glc and UDP-Rha.
| UGT isoform | Plant species | LP1/LP2 (WT) | Donor specificity (WT) | LP1/LP2 (mutant) | Donor specificity (mutant) | Reference |
|---|---|---|---|---|---|---|
| UGT89A2 | Arabidopsis thaliana (Col-0) | Ser/Ile | UDP-Xyl | Ser/Ser | UDP-Glc/UDP-Xyl | Chen and Li (2017) |
| UGT89A2 | Arabidopsis thaliana (C24) | Ser/Ser | UDP-Glc/UDP-Xyl | Ser/Ile | UDP-Xyl | Chen and Li (2017) |
| Ib3GGT | Ipomoea batatas | Thr/Ile | UDP-Glc/UDP-Xyl | Ile/Ile | UDP-Xyl | Wang et al. (2018) |
| At3GGT | Arabidopsis thaliana | Ile/Val | UDP-Xyl | Thr/Val | UDP-Glc/UDP-Xyl | Wang et al. (2018) |
| CsGT2 | Camellia sinensis | Ile/Thr | UDP-Glc/UDP-Xyl | Ser/Thr | UDP-Glc | Ohgami et al. (2015) |
| UGT74AN3 | Catharanthus roseus | Thr/Asn | UDP-Glc | Val/Asn | UDP-Xyl | Huang et al. (2023) |
| UGT73F4 | Glycine max | Ser/Tyr | UDP-Xyl | Gly/Tyr | UDP-Glc/UDP-Xyl/UDP-Gal | Sayama et al. (2012) |
| UGT73F2 | Glycine max | Gly/Phe | UDP-Glc | Ser/Tyr | UDP-Xyl | Sayama et al. (2012) |
| UGT78W1 | Morella rubra | Val/Ser | UDP-Gal | Val/Ala | UDP-Glc/UDP-Gal | Ren et al. (2022b) |
| UGT76D1 | Arabidopsis thaliana | Pro/Ser | UDP-Glc | Thr/Ser | UDP-Glc/UDP-Gal | Akere et al. (2020) |
| AsAAT1 | Avena strigosa | Pro/Met | UDP-Ara/UDP-Xyl | Ser/Met | UDP-Ara/UDP-Gal | Louveau et al. (2018) |
| VtCGTa | Viola tricolor | Thr/Ser | UDP-Glc/UDP-Xyl/UDP-Gal | Pro/Ser | UDP-Rha* | Han et al. (2023) |
| VtCGTc | Viola tricolor | Val/Ser | UDP-Glc/UDP-Xyl/UDP-Rha/UDP-Ara | Thr/Ser | UDP-Glc/UDP-Rha* | Han et al. (2023) |
Recently, a study identified a donor substrate-promiscuous UGT, UGT74AN3, with a preference for UDP-Glc (Huang et al., 2023). This preference was changed to UDP-Xyl and the UDP-Glc activity was significantly reduced upon mutating LP1 from Thr to Val. A molecular dynamics simulation showed that Val could form a hydrophobic interaction with C5 of xylose. Along the same lines, other studies which alter the preference between UDP-Glc and -Xyl make the same argument based on structural modelling and docking, that a hydrophobic residue in LP1/LP2 would clash with C6-OH of UDP-Glc and potentially interact with C5 of xylose, while Ser or Thr would enable the possibility of hydrogen bonding with the hydroxyl group of glucose (Ohgami et al., 2015; Wang et al., 2018). However, when looking at the gathered examples of mutating LP1/LP2 and the alteration in donor specificity (Table 2), it is difficult to identify a simple relationship between the residues of LP1/LP2 and donor specificity. Additionally, reports of enabled UDP-Gal activity upon mutating LP1 have also been reported (Table 2) (Louveau et al., 2018; Akere et al., 2020). In both cases, the WT harbours a Pro at LP1 while mutating LP1 to a hydrophilic residue, Thr or Ser, enabled UDP-Gal activity. This could indicate that the flexibility of the loop plays a significant role, since Pro would rigidify the loop while Thr or Ser would both enhance flexibility and enable potential hydrogen bonding. Finally, newly discovered apiosyltransferases were also found to be specific to UDP-apiose partly via bulky hydrophobic residue on LP1 and LP2, which shows the importance of this loop in the binding of furanoses as well (Wang H. T. et al., 2023; Yamashita et al., 2023).
Altogether, LP1 and LP2 in this N-terminal loop can be considered a hot spot for governing the donor specificity of UGTs. The flexibility along with the hydrophilicity/hydrophobicity of the loop seem to be the most important factors for the donor specificity of UGTs in this region. Lastly, it is worth noting that a loop very similar to the UGT N5 loop is observed in prokaryotic GT-B fold GTs, where structural and mutational studies indicate similar importance in the donor specificity arguing for evolutionary conservation and potentially functional importance (Liu B. et al., 2021; Vuksanovic et al., 2024).
4 The role of an active site-arginine in UDP-glucuronic acid activity
Glucuronosyltransferases play a vital role in the phase II metabolism of exogenous compounds in mammals (Mazerska et al., 2016). In addition, natural glucuronides are found in plants where some isolated glucuronides such as scutallerin-7-O-glucuronide (Guo et al., 2014) and apigenin-7-O-glucuronide (Ma et al., 2017; Yue et al., 2019) display pharmacological activity. Even though there is a wide variety of glucuronides identified in plants (Yue et al., 2019), only a few plant glucuronosyltransferases have been discovered and characterised. One of these is UGT94B1 from Bellis perennis. (Sawada et al., 2005; Osmani et al., 2008). Mutating Arg25 (Figure 4) to either Ser, Gly, Pro, or even another basic residue, Lys, significantly decreased the activity with UDP-Glu, while activity with UDP-Glc was improved in all mutants except Arg25Pro (Osmani et al., 2008). Low UDP-Gal activity was also observed, compared to none with WT. The corresponding Arg in UGT73P12 (Arg32) from Glycyrrhiza uralensis was also demonstrated to be important for glucuronosyltransferase activity (Nomura et al., 2019). Mutating residue 32 from Arg to Ser led to a significant decrease in the activity with UDP-Glu while UDP-Glc and -Gal activity was increased 7-fold and 73-fold, respectively.
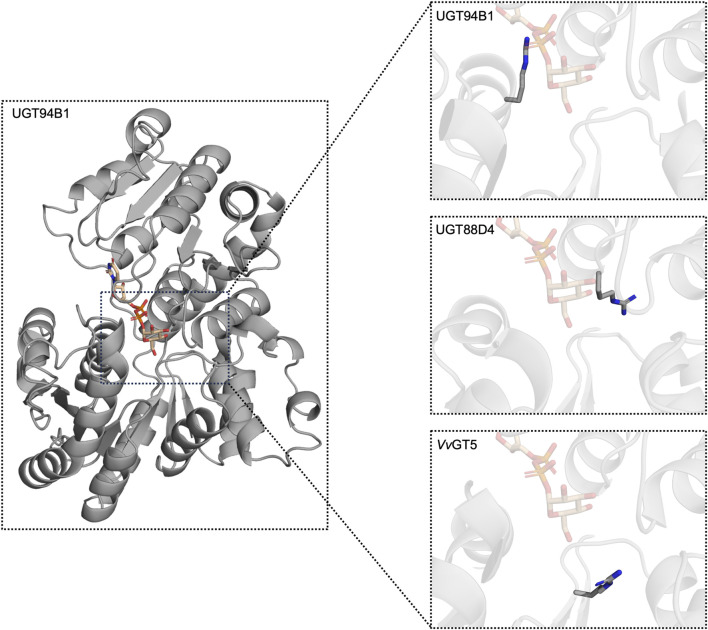
FIGURE 4.
A view of the three different Arg positions described in the text. Alphafold (Jumper et al., 2021) models of UGT94B1, UGT88D4, and VvGT5 were used and had UDP-Glc from PDB 6SU6, displayed in tan, superimposed into the models.
Even though these studies allude to an Arg being important for UDP-Glu activity, studies have found that the specific position of the Arg is not conserved amongst glucuronosyltransferases. Noguchi et al. discovered another conserved Arg in P22 of the PSPG motif across Lamiales flavonoid 7-O-glucuronosyltransferases (Figure 4) (Noguchi et al., 2009). A multiple sequence alignment revealed that flavonoid 7-O-glucosyltransferases had a conserved Trp in P22 of the PSPG motif instead. Swapping Arg for Trp in flavonoid 7-O-glucuronosyltransferases UGT88D4, D6, and D7 changed their specificity from UDP-Glu to UDP-Glc. This Arg is also conserved in other glucuronosyltransferases such as UGT88D1 (Nagashima et al., 2000), UGT88D8 (Ono et al., 2010b), and UBGAT (Nagashima et al., 2000). Meanwhile, in VvGT5, an Arg in another position was discovered to be important for UDP-Glu activity (Ono et al., 2010a). Through structural modelling and docking, the authors hypothesised Arg140 (Figure 4) to be interacting with the carboxylic acid of the glucuronic acid moiety. When mutating this residue into a Trp, which is the corresponding residue in glucosyltransferase VvGT1 and galactosyltransferase VvGT6 (Ono et al., 2010a), UDP-Glu activity was abolished while activity with UDP-Glc and -Gal was gained. Interestingly, this Arg is placed in the intersection between a β-sheet and the N5 loop (Section 3) further arguing for the importance of this region.
The observations of these studies suggest the presence of an Arg in the binding site to be vital for UDP-Glu activity. The studies propose the positively charged guanidinium group of Arg to coordinate the negatively charged carboxylic acid of glucuronic acid (pKa of α-d-Glu = 2.93 (Wang et al., 1991)) to stabilise its charge (Osmani et al., 2008; Louveau and Osbourn, 2019). To support this hypothesis, studies have attempted to enable glucuronosyltransferase activity in UGTs with no UDP-Glu specificity by adding an Arg into one of the conserved Arg positions. However, no studies have been able to facilitate UDP-Glu specificity by employing this strategy. UGT88A7 mutant Thr139Ser/Trp367Arg (Noguchi et al., 2009), UGT88E3 mutant Thr150Ser/Trp371Arg (Noguchi et al., 2009), and UGT85B1 mutant Pro26Arg (Osmani et al., 2008) did not confer glucuronosyltransferase activity. These observations could be caused by the fact that for each specific UGT, the exact position of the Arg plays a significant role in whether UDP-Glu activity is enabled. Otherwise, other residues could play a part in correctly coordinating the anomeric carbon with the acceptor substrate or coordinating the carboxylic acid with the Arg. Finally, it is important to mention that glucuronosyltransferases where no Arg can be located close to the sugar moiety of UDP-Glu have also been reported. This includes UGT78H2 (Chen et al., 2021), and GuUGAT (Xu et al., 2016). This demonstrates that there are still unresolved aspects of the structure-function relationship of glucuronosyltransferases. Currently, no crystal structure of a glucuronosyltransferase in a complex with UDP-Glu has been published (Table 1). The elucidation of such a structure would provide valuable insights into the structure-function relationship of glucuronosyltransferases.
5 Expanding the donor specificityof UGTs
UGTs that can accept several UDP-sugars serve as the epitome in glycodiversification (Wen et al., 2023; Wu et al., 2023). Hence, it is important to understand which traits lead to UDP-sugar promiscuity. Several natural UGTs accepting several UDP-sugars have been discovered. These UGTs have then been utilised in attempts to either restrict their broad donor substrate scope or use either their structure or sequence information to widen the donor substrate scope of other UGTs. Sb3GT1 from S. baicalensis was discovered to accept all five tested donors (UDP-Glc, UDP-Gal, UDP-GlcNAc, UDP-Ara, and UDP-Xyl) (Wang et al., 2019). It was hypothesised that the donor substrate promiscuity was caused by the large active site. To test this hypothesis, the volume of Sb3GT1’s active site was decreased by site-directed mutagenesis using the tight active site of VvGT1 as a template. Gly15Ser, Gly15Phe, and Gly15Val all significantly decreased UDP-Gal and UDP-GlcNAc activity which supported the hypothesis of the cavity volume playing a significant role in the donor promiscuity of Sb3GT1 (Figure 3D). Similarly, Wen et al. tried to alter the donor substrate scope of the promiscuous UGT72AS1 from Helleborus thibetanus through site-directed mutagenesis by comparing the sequence information with sequences of both donor promiscuous and donor-specific UGTs (Wen et al., 2023). However, constructed mutants either exhibited conservation of function with all tested donors or a significant decrease with all donor substrates. Only Tyr377Ala (PSPG motif P41) improved activity with UDP-GlcNAc without impacting the function on other donor substrates which could be accredited to reduced steric hindrance.
The same approach has been employed to expand the donor specificity of a UGT with a narrow donor substrate scope (Zhang et al., 2020). This study used the donor substrate-promiscuous GgCGT, which accepts UDP-Glc, -Gal, -Xyl, -Ara, and -GlcNAc, as a template to widen the promiscuity of UGT74AN2 from Calotropis gigantea, which could efficiently utilise UDP-Glc and weakly utilise UDP-Gal and -GlcNAc (Huang et al., 2022). This led to the construction of a UGT74AN2 triple mutant with mutations Ile284Arg, Trp390His (PSPG motif P41), and Val391Gly (PSPG motif P42). It was argued that Arg284 would enable a hydrogen bond with C2-OH while His390 and Gly391 would reduce steric hindrance (Figure 3C). Indeed, the triple mutant had improved activity with UDP-Glc, UDP-GlcNAc, and UDP-Gal and gained UDP-Rha activity. The sequence alignment of UGT74AN2 with GgCGT and other donor-promiscuous UGTs showed that a common trait between the donor promiscuous UGTs, which was not observed in the more restricted UGT74AN2, was a non-bulky residue at PSPG motif P42. However, the authors did not test mutating only this position to Ala or Gly. Overall, these studies indicate that active site volume plays an important role in allowing a wide donor substrate scope in UGTs.
6 Current state and future directions for elucidating the structure-function relationship governing UGT sugar donor specificity
Based on the described studies, key concepts of the structure-function relationship of UGT sugar donor specificity can be made. First, the PSPG motif plays an integral part in the binding of UDP donors. However, several mutational studies have focused on the terminal residue of the motif leaving the rest of the motif mostly unexplored. While a Gln as the terminal residue seems to be important for UDP-Glc activity, no results have consistently shown that this residue position can be used as a lever to activate and de-activate activity with other UDP-sugars. As demonstrated by several studies (Noguchi et al., 2009; Zhang et al., 2020; Liu S. et al., 2021; Chen et al., 2021), the rest of the motif contains hotspots which are responsible for determining the donor specificity of UGTs and should be further explored. As suggested by Kurze et al., 2022, there could be an interplay between specific positions in the PSPG motif which could be important in the donor specificity of UGTs.
Second, LP1 and LP2 in the N5 loop play a significant role in the donor substrate scope. This is the case for UDP-sugars which differ in the C5/C6 position such as UDP-Glc, -Xyl, and -Rha. Additionally, effects on UDP-Gal, which differs in the C4 position, and on the furanose UDP-apiose, have also been observed. A simple relationship between LP1 and LP2 along with the resulting donor substrate scope could not be described. However, the important factors seem to be the overall polarity along with the flexibility of the loop.
Third, even though discrepancies are observed, an Arg in the binding site of glucuronosyltransferases seems to play an important role in UDP-Glu activity. The Arg potentially serves as a neutraliser of the negatively charged carboxylic acid of UDP-Glu through its positively charged guanidinium group. However, the position of the Arg is not conserved and has, to the best of our knowledge, been described in three separate positions. Solving the structure of a glucuronosyltransferase in complex with UDP-Glu could also help further understand the structure-function relationship and shed light on the binding mode.
Fourth, UGTs with a wide donor substrate scope have mainly been found to possess this trait through a spacious binding site. Mutational studies have supported this theory, but no other theories have currently been tested.
Altogether, the donor substrate scope of UGTs is a complex concept in which several factors seem to play a significant role. The results of these studies can be used as a guiding map to determine which areas and residues to target for altering the donor specificity of UGTs. However, the lack of an in-depth understanding of the structure-function of UGT donor specificity hinders the efficient discovery of UGTs with a specific donor specificity from, e.g., multi-omics data. Studies that have attempted to address and understand this structure-function relationship have mainly resorted to mutational studies adopting a rational engineering mindset Figure 3A. However, studies have shown that residues outside the active site can impact the donor substrate scope of UGTs (Akere et al., 2018). Since no true high-throughput method for assaying UGT activity is currently available, random mutagenesis-based methods, which could help elucidate important areas outside the binding cavity, are currently a time- and material-consuming option. In addition, previous studies have found that the donor substrate scope is also dependent on the acceptor substrate (Wang et al., 2019; Huang et al., 2023), further complicating the situation. Hence, future research efforts and data set curations should also consider the acceptor substrate.
Following these observations, future research efforts could include the development of a true high-throughput UGT activity assay, which would greatly accelerate data generation and unlock a treasure-trove of powerful techniques such as directed evolution and machine learning-based prediction. This would enable this research topic to enter a new paradigm. Moreover, it could be advantageous to establish and strengthen connections between information from diverse organisms and even between various types of carbohydrate-active enzymes (CAZymes) (Quirke and Crich, 2021; Franceus et al., 2022). This could potentially contribute to a more comprehensive understanding of the structure-function relationship, particularly in terms of what determines the specificity of enzymes that share similar substrates. Other research directions could include in silico studies such as molecular dynamics simulations with the N5 loop being a potential target of these studies or ancestral sequence reconstruction to uncover a potential evolutionary element of the structure-function relationship (Franceus et al., 2024). Lastly, the emergence of machine learning and explainable artificial intelligence as tools to uncover enzyme mechanisms and structure-function relationships could also prove useful (Taujale et al., 2020; Kouba et al., 2023). Machine learning could allow the exploration of enzyme features outside the binding site and epistatic effects, the combination of donor- and acceptor-substrate features, and the inclusion of UGTs or other CAZymes across organisms.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We thank The Novo Nordisk Foundation for supporting this work through grant NNF20CC0035580.
Author contributions
HG: Conceptualization, Writing–original draft, Writing–review and editing. DW: Conceptualization, Supervision, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Akere A., Chen S. H., Liu X., Chen Y., Dantu S. C., Pandini A., et al. (2020). Structure-based enzyme engineering improves donor-substrate recognition of Arabidopsis thaliana glycosyltransferases. Biochem. J. 477, 2791–2805. 10.1042/BCJ20200477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akere A., Liu Q., Wu S., Hou B., Yang M. (2018). High throughput mass spectrometry-based characterisation of Arabidopsis thaliana group H glycosyltransferases. RSC Adv. 8, 30080–30086. 10.1039/C8RA03947J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M., Offen W. A., Gershater M. C., Revett T. J., Lim E. K., Bowles D. J., et al. (2007). Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. U. S. A. 104, 20238–20243. 10.1073/PNAS.0706421104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Li X. (2017). Identification of a residue responsible for UDP-sugar donor selectivity of a dihydroxybenzoic acid glycosyltransferase from Arabidopsis natural accessions. Plant J. 89, 195–203. 10.1111/TPJ.13271 [DOI] [PubMed] [Google Scholar]
- Chen Q., Liu X., Hu Y., Wang Y., Sun B., Chen T., et al. (2021). Broaden the sugar donor selectivity of blackberry glycosyltransferase UGT78H2 through residual substitutions. Int. J. Biol. Macromol. 166, 277–287. 10.1016/J.IJBIOMAC.2020.10.184 [DOI] [PubMed] [Google Scholar]
- De Bruyn F., Maertens J., Beauprez J., Soetaert W., De Mey M. (2015). Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 33, 288–302. 10.1016/J.BIOTECHADV.2015.02.005 [DOI] [PubMed] [Google Scholar]
- De Roode B. M., Franssen M. C. R., Van Der Padt A., Boom R. M. (2003). Perspectives for the industrial enzymatic production of glycosides. Biotechnol. Prog. 19, 1391–1402. 10.1021/BP030038Q [DOI] [PubMed] [Google Scholar]
- Drula E., Garron M. L., Dogan S., Lombard V., Henrissat B., Terrapon N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. 10.1093/NAR/GKAB1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Chen T., Zhang S., Wu B., He B. (2017). Mining of efficient microbial UDP-glycosyltransferases by motif evolution cross plant kingdom for application in biosynthesis of salidroside. Sci. Rep. 7 (1), 463–469. 10.1038/s41598-017-00568-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceus J., Lormans J., Desmet T. (2022). Building mutational bridges between carbohydrate-active enzymes. Curr. Opin. Biotechnol. 78, 102804. 10.1016/J.COPBIO.2022.102804 [DOI] [PubMed] [Google Scholar]
- Franceus J., Rivas-Fernández J. P., Lormans J., Rovira C., Desmet T. (2024). Evolution of phosphorylase activity in an ancestral glycosyltransferase. ACS Catal. 14, 3103–3114. 10.1021/ACSCATAL.3C05819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franconetti A., Ardá A., Asensio J. L., Blériot Y., Thibaudeau S., Jiménez-Barbero J. (2021). Glycosyl oxocarbenium ions: structure, conformation, reactivity, and interactions. Acc. Chem. Res. 54, 2552–2564. 10.1021/ACS.ACCOUNTS.1C00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel B., Tripathi N., Mukherjee D., Jain S. K. (2021). Glycorandomization: a promising diversification strategy for the drug development. Eur. J. Med. Chem. 213, 113156. 10.1016/J.EJMECH.2021.113156 [DOI] [PubMed] [Google Scholar]
- Guo C., Zhu Y., Weng Y., Wang S., Guan Y., Wei G., et al. (2014). Therapeutic time window and underlying therapeutic mechanism of breviscapine injection against cerebral ischemia/reperfusion injury in rats. J. Ethnopharmacol. 151, 660–666. 10.1016/J.JEP.2013.11.026 [DOI] [PubMed] [Google Scholar]
- Han B. Y., Wang Z. L., Li J., Jin Q., Wang H. T., Chen K., et al. (2023). A highly selective C-rhamnosyltransferase from Viola tricolor and insights into its mechanisms. Acta Pharm. Sin. B 13, 3535–3544. 10.1016/J.APSB.2023.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. H., Kim B. G., Yoon J. A., Chong Y., Ahn J. H. (2014). Synthesis of flavonoid O-pentosides by Escherichia coli through engineering of nucleotide sugar pathways and glycosyltransferase. Appl. Environ. Microbiol. 80, 2754–2762. 10.1128/aem.03797-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., He Y., Jiang R., Deng Z., Long F. (2022). Functional and structural dissection of a plant steroid 3-O-glycosyltransferase facilitated the engineering enhancement of sugar donor promiscuity. ACS Catal. 12, 2927–2937. 10.1021/ACSCATAL.1C05729 [DOI] [Google Scholar]
- Huang W., Zhang X., Li J., Lv J., Wang Y., He Y., et al. (2023). Substrate promiscuity, crystal structure, and application of a plant UDP-glycosyltransferase UGT74AN3. ACS Catal. 14, 475–488. 10.1021/ACSCATAL.3C05309 [DOI] [Google Scholar]
- Hughes J., Hughes M. A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (manihot esculenta crantz) cotyledons. Mitochondrial DNA 5, 41–49. 10.3109/10425179409039703 [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba P., Kohout P., Haddadi F., Bushuiev A., Samusevich R., Sedlar J., et al. (2023). Machine learning-guided protein engineering. ACS Catal. 13, 13863–13895. 10.1021/ACSCATAL.3C02743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Arai Y., Nagashima S., Yoshikawa T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429, 198–203. 10.1016/J.ABB.2004.06.021 [DOI] [PubMed] [Google Scholar]
- Kurze E., Wüst M., Liao J., McGraphery K., Hoffmann T., Song C., et al. (2022). Structure–function relationship of terpenoid glycosyltransferases from plants. Nat. Prod. Rep. 39, 389–409. 10.1039/D1NP00038A [DOI] [PubMed] [Google Scholar]
- Li J., Mu S., Yang J., Liu C., Zhang Y., Chen P., et al. (2022). Glycosyltransferase engineering and multi-glycosylation routes development facilitating synthesis of high-intensity sweetener mogrosides. iScience 25, 105222. 10.1016/J.ISCI.2022.105222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Qu G., Shang N., Chen P., Men Y., Liu W., et al. (2021). Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases. Green Synthesis Catal. 2, 45–53. 10.1016/J.GRESC.2021.01.005 [DOI] [Google Scholar]
- Li J., Yang J., Mu S., Shang N., Liu C., Zhu Y., et al. (2020). Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 10, 3629–3639. 10.1021/ACSCATAL.9B05232 [DOI] [Google Scholar]
- Li L., Modolo L. V., Escamilla-Trevino L. L., Achnine L., Dixon R. A., Wang X. (2007). Crystal structure of medicago truncatula UGT85H2 – insights into the structural basis of a multifunctional (Iso)flavonoid glycosyltransferase. J. Mol. Biol. 370, 951–963. 10.1016/J.JMB.2007.05.036 [DOI] [PubMed] [Google Scholar]
- Li Y., Luo H. M., Sun C., Song J. Y., Sun Y. Z., Wu Q., et al. (2010). EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genomics 11, 268. 10.1186/1471-2164-11-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libik-Konieczny M., Capecka E., Tuleja M., Konieczny R. (2021). Synthesis and production of steviol glycosides: recent research trends and perspectives. Appl. Microbiol. Biotechnol. 105, 3883–3900. 10.1007/S00253-021-11306-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhao C., Xiang Q., Zhao N., Luo Y., Bao R. (2021a). Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis. Int. J. Biol. Macromol. 166, 806–817. 10.1016/J.IJBIOMAC.2020.10.238 [DOI] [PubMed] [Google Scholar]
- Liu M., Wang D., Li Y., Li X., Zong G., Fei S., et al. (2020a). Crystal structures of the C-glycosyltransferase UGT708C1 from buckwheat provide insights into the mechanism of C-glycosylation. Plant Cell 32, 2917–2931. 10.1105/TPC.20.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang M., Bao Y., Chen K., Xu L., Su H., et al. (2021b). Characterization of a highly selective 2″- O-galactosyltransferase from trollius chinensis and structure-guided engineering for improving UDP-glucose selectivity. Org. Lett. 23, 9020–9024. 10.1021/ACS.ORGLETT.1C02581 [DOI] [PubMed] [Google Scholar]
- Liu Z., Li J., Sun Y., Zhang P., Wang Y. (2020b). Structural insights into the catalytic mechanism of a plant diterpene glycosyltransferase SrUGT76G1. Plant Commun. 1, 100004. 10.1016/J.XPLC.2019.100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau T., Orme A., Pfalzgraf H., Stephenson M. J., Melton R., Saalbach G., et al. (2018). Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase Family1 provides insights into disease resistance and sugar donor specificity. Plant Cell 30, 3038–3057. 10.1105/TPC.18.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau T., Osbourn A. (2019). The sweet side of plant-specialized metabolism. Cold Spring Harb. Perspect. Biol. 11, a034744. 10.1101/CSHPERSPECT.A034744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Zhang X. M., Jiang J. G., Zhu W. (2017). Apigenin-7-O-β-D-glucuronide inhibits modified low-density lipoprotein uptake and foam cell formation in macrophages. J. Funct. Foods 35, 615–621. 10.1016/J.JFF.2017.06.026 [DOI] [Google Scholar]
- Mazerska Z., Mróz A., Pawłowska M., Augustin E. (2016). The role of glucuronidation in drug resistance. Pharmacol. Ther. 159, 35–55. 10.1016/J.PHARMTHERA.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Nagashima S., Hirotani M., Yoshikawa T. (2000). Purification and characterization of UDP-glucuronate: baicalein 7-O-glucuronosyltransferase from Scutellaria baicalensis Georgi. cell suspension cultures. Phytochemistry 53, 533–538. 10.1016/S0031-9422(99)00593-2 [DOI] [PubMed] [Google Scholar]
- Nidetzky B. (2021). Glycosyltransferase cascades made fit for the biocatalytic production of natural product glycosides. Biocatal. Pract. Tech. React. Appl., 225–243. 10.1002/9783527824465.CH8 [DOI] [Google Scholar]
- Noguchi A., Horikawa M., Fukui Y., Fukuchi-Mizutani M., Iuchi-Okada A., Ishiguro M., et al. (2009). Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21, 1556–1572. 10.1105/TPC.108.063826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Seki H., Suzuki T., Ohyama K., Mizutani M., Kaku T., et al. (2019). Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 99, 1127–1143. 10.1111/TPJ.14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405. 10.1038/SJ.EMBOJ.7600970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami S., Ono E., Horikawa M., Murata J., Totsuka K., Toyonaga H., et al. (2015). Volatile glycosylation in tea plants: sequential glycosylations for the biosynthesis of aroma β-primeverosides are catalyzed by two Camellia sinensis glycosyltransferases. Plant Physiol. 168, 464–477. 10.1104/PP.15.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Homma Y., Horikawa M., Kunikane-Doi S., Imai H., Takahashi S., et al. (2010a). Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22, 2856–2871. 10.1105/TPC.110.074625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Ruike M., Iwashita T., Nomoto K., Fukui Y. (2010b). Co-pigmentation and flavonoid glycosyltransferases in blue Veronica persica flowers. Phytochemistry 71, 726–735. 10.1016/J.PHYTOCHEM.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Osmani S. A., Bak S., Imberty A., Olsen C. E., Møller B. L. (2008). Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 148, 1295–1308. 10.1104/PP.108.128256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S. A., Bak S., Møller B. L. (2009). Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70, 325–347. 10.1016/J.PHYTOCHEM.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Paquette S., Møller B. L., Bak S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62, 399–413. 10.1016/S0031-9422(02)00558-7 [DOI] [PubMed] [Google Scholar]
- Quirke J. C. K., Crich D. (2021). Side chain conformation restriction in the catalysis of glycosidic bond formation by leloir glycosyltransferases, glycoside phosphorylases, and transglycosidases. ACS Catal. 11, 5069–5078. 10.1021/ACSCATAL.1C00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi S., Kim J., Mijakovic I., Jung K. H., Choi G., Kim S. C., et al. (2019). Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 37, 107394. 10.1016/J.BIOTECHADV.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Rehman H. M., Nawaz M. A., Shah Z. H., Ludwig-Müller J., Chung G., Ahmad M. Q., et al. (2018). Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 8 (1), 1875. 10.1038/s41598-018-19535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Cao Y., Xing M., Guo Y., Li J., Xue L., et al. (2022a). Genome-wide analysis of UDP-glycosyltransferase gene family and identification of members involved in flavonoid glucosylation in Chinese bayberry (Morella rubra). Front. Plant Sci. 13, 998985. 10.3389/FPLS.2022.998985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Guo Y., Xie L., Zhao Z., Xing M., Cao Y., et al. (2022b). Identification of UDP-rhamnosyltransferases and UDP-galactosyltransferase involved in flavonol glycosylation in Morella rubra. Hortic. Res. 9, uhac138. 10.1093/HR/UHAC138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Suzuki H., Ichimaida F., Yamaguchi M. A., Iwashita T., Fukui Y., et al. (2005). UDP-Glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. J. Biol. Chem. 280, 899–906. 10.1074/JBC.M410537200 [DOI] [PubMed] [Google Scholar]
- Sayama T., Ono E., Takagi K., Takada Y., Horikawa M., Nakamoto Y., et al. (2012). The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 24, 2123–2138. 10.1105/TPC.111.095174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., He X., Achnine L., Blount J. W., Dixon R. A., Wang X. (2005). Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from medicago truncatula. Plant Cell 17, 3141–3154. 10.1105/TPC.105.035055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taujale R., Venkat A., Huang L. C., Zhou Z., Yeung W., Rasheed K. M., et al. (2020). Deep evolutionary analysis reveals the design principles of fold a glycosyltransferases. Elife 9, e54532. 10.7554/ELIFE.54532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teze D., Coines J., Fredslund F., Dubey K. D., Bidart G. N., Adams P. D., et al. (2021). O-/N-/S-specificity in glycosyltransferase catalysis: from mechanistic understanding to engineering. ACS Catal. 11, 1810–1815. 10.1021/ACSCATAL.0C04171 [DOI] [Google Scholar]
- Thodberg S., Del Cueto J., Mazzeo R., Pavan S., Lotti C., Dicenta F., et al. (2018). Elucidation of the amygdalin pathway reveals the metabolic basis of bitter and sweet almonds (prunus dulcis). Plant Physiol. 178, 1096–1111. 10.1104/PP.18.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuksanovic N., Clasman J. R., Imperiali B., Allen K. N. (2024). Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA. Protein Sci. 33, e4848. 10.1002/PRO.4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang C., Fan W., Yang J., Appelhagen I., Wu Y., et al. (2018). A novel glycosyltransferase catalyses the transfer of glucose to glucosylated anthocyanins in purple sweet potato. J. Exp. Bot. 69, 5444–5459. 10.1093/JXB/ERY305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. M., Loganathan D., Linhardt R. J. (1991). Determination of the pKa of glucuronic acid and the carboxy groups of heparin by 13C-nuclear-magnetic-resonance spectroscopy. Biochem. J. 278, 689–695. 10.1042/BJ2780689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. T., Wang Z. L., Chen K., Yao M. J., Zhang M., Wang R. S., et al. (2023a). Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants. Nat. Commun. 14 (1), 6658. 10.1038/s41467-023-42393-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ji Q., Lai B., Liu Y., Mei K. (2023b). Structure-function and engineering of plant UDP-glycosyltransferase. Comput. Struct. Biotechnol. J. 21, 5358–5371. 10.1016/J.CSBJ.2023.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang S., Xu Z., Li M., Chen K., Zhang Y., et al. (2019). Highly promiscuous flavonoid 3- O-glycosyltransferase from scutellaria baicalensis. Org. Lett. 21, 2241–2245. 10.1021/ACS.ORGLETT.9B00524 [DOI] [PubMed] [Google Scholar]
- Wang Z. L., Gao H. M., Wang S., Zhang M., Chen K., Zhang Y. Q., et al. (2020). Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc. Natl. Acad. Sci. U. S. A. 117, 30816–30823. 10.1073/PNAS.2012745117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Wu H.-C., Ouyang W.-H., Nie J.-X., Guo Y.-P., Wang F., et al. (2023). Exploring the catalytic flexibility and reversibility of plant glycosyltransferase HtUGT72AS1 for glycodiversification of phenolic compounds. J. Agric. Food Chem. 71, 8998–9008. 10.1021/ACS.JAFC.3C01459 [DOI] [PubMed] [Google Scholar]
- Wen Z., Zhang Z. M., Zhong L., Fan J., Li M., Ma Y., et al. (2021). Directed evolution of a plant glycosyltransferase for chemo- and regioselective glycosylation of pharmaceutically significant flavonoids. ACS Catal. 11, 14781–14790. 10.1021/ACSCATAL.1C04191 [DOI] [Google Scholar]
- Wetterhorn K. M., Gabardi K., Michlmayr H., Malachova A., Busman M., McCormick S. P., et al. (2017). Determinants and expansion of specificity in a trichothecene UDP-glucosyltransferase from oryza sativa. Biochemistry 56, 6585–6596. 10.1021/ACS.BIOCHEM.7B01007 [DOI] [PubMed] [Google Scholar]
- Wu Y., Yang Y., Du L., Zhuang Y., Liu T. (2023). Identification of a highly promiscuous glucosyltransferase from Penstemon barbatus for natural product glycodiversification. Org. Biomol. Chem. 21, 4445–4454. 10.1039/D3OB00370A [DOI] [PubMed] [Google Scholar]
- Xie L., Cao Y., Zhao Z., Ren C., Xing M., Wu B., et al. (2020). Involvement of MdUGT75B1 and MdUGT71B1 in flavonol galactoside/glucoside biosynthesis in apple fruit. Food Chem. 312, 126124. 10.1016/J.FOODCHEM.2019.126124 [DOI] [PubMed] [Google Scholar]
- Xie L., Guo Y., Ren C., Cao Y., Li J., Lin J., et al. (2022). Unravelling the consecutive glycosylation and methylation of flavonols in peach in response to UV-B irradiation. Plant Cell Environ. 45, 2158–2175. 10.1111/PCE.14323 [DOI] [PubMed] [Google Scholar]
- Xu G., Cai W., Gao W., Liu C. (2016). A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol. 212, 123–135. 10.1111/NPH.14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Fujimori T., An S., Iguchi S., Takenaka Y., Kajiura H., et al. (2023). The apiosyltransferase celery UGT94AX1 catalyzes the biosynthesis of the flavone glycoside apiin. Plant Physiol. 193, 1758–1771. 10.1093/PLPHYS/KIAD402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Xing X., Yu L., Wang Y., Zhang X., Zhang L. (2022). Structurefunction relationships in plant UDP-glycosyltransferases. Ind. Crops Prod. 189, 115784. 10.1016/J.INDCROP.2022.115784 [DOI] [Google Scholar]
- Yue T., Chen R., Chen D., Liu J., Xie K., Dai J. (2019). Enzymatic synthesis of bioactive O-glucuronides using plant glucuronosyltransferases. J. Agric. Food Chem. 67, 6275–6284. 10.1021/ACS.JAFC.9B01769 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu X., Li J., Meng Y., Zhao G. R. (2023a). Improvement of betanin biosynthesis in Saccharomyces cerevisiae by metabolic engineering. Synth. Syst. Biotechnol. 8, 54–60. 10.1016/J.SYNBIO.2022.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. J., Wang D. G., Zhang P., Wu C., Li Y. Z. (2022a). Promiscuity characteristics of versatile plant glycosyltransferases for natural product glycodiversification. ACS Synth. Biol. 11, 812–819. 10.1021/ACSSYNBIO.1C00489 [DOI] [PubMed] [Google Scholar]
- Zhang M., Li F. D., Li K., Wang Z. L., Wang Y. X., He J. B., et al. (2020). Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J. Am. Chem. Soc. 142, 3506–3512. 10.1021/JACS.9B12211 [DOI] [PubMed] [Google Scholar]
- Zhang M., Yi Y., Gao B. H., Su H. F., Bao Y. O., Shi X. M., et al. (2022b). Functional characterization and protein engineering of a triterpene 3-/6-/2′-O-glycosyltransferase reveal a conserved residue critical for the regiospecificity. Angew. Chem. Int. Ed. 61, e202113587. 10.1002/ANIE.202113587 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wang Y., Cui Z., Li Q., Kong L., Luo J. (2023b). Functional characterization of a Flavonol 3-O-rhamnosyltransferase and two UDP-rhamnose synthases from Hypericum monogynum. Plant Physiology Biochem. 197, 107643. 10.1016/J.PLAPHY.2023.107643 [DOI] [PubMed] [Google Scholar]
- Zong G., Fei S., Liu X., Li J., Gao Y., Yang X., et al. (2019). Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-l-rhamnose and rhamnosylation mechanism. Plant J. 99, 257–269. 10.1111/TPJ.14321 [DOI] [PubMed] [Google Scholar]