Abstract
The carboxy-terminal region of the rabbit hemorrhagic disease virus p37 polyprotein cleavage product has been expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein. The recombinant GST-Δ2C protein showed in vitro ATP-binding and ATPase activities. Site-directed mutagenesis studies of the conserved residues G522 and T529 in motif A, D566 and E567 in motif B, and K600 in motif C were also performed. These results provide the first experimental characterization of a 2C-like ATPase activity in a member of the Caliciviridae.
Rabbit hemorrhagic disease virus (RHDV), the etiologic agent of a lethal pathology causing severe losses of farmed and wild rabbit populations (11), has been characterized as a member of the Caliciviridae family (16, 18) and, more recently, designated the type species of the genus Lagovirus (21). Purified virions contain two positive-polarity polyadenylated single-stranded RNA species, one of about 7.5 kb and the other of about 2.2 kb, bearing a virus-encoded VPg protein covalently attached to its 5′ end (13, 31). The 2.2-kb subgenomic RNA is colinear for its complete length to the 3′-terminal region of the genomic RNA. The data obtained from in vitro translation (30), Escherichia coli expression studies (12, 31), and the detection of viral protein products after RHDV infection of cultured hepatocytes (6) revealed that the viral RNA is translated into a polyprotein, which is subsequently cleaved to give rise to at least nine mature structural and nonstructural proteins. Nevertheless, only four of the eight necessary cleavage sites have been experimentally demonstrated.
Amino acid sequence motifs conserved among proteins with known biological activities have been used to predict the functions of uncharacterized polypeptides. Comparative sequence studies between RHDV and picornaviruses have predicted similarities between the RHDV mature cleavage products p37, p15, and p58 and picornaviral 2C nucleoside triphosphatase (NTPase), 3C protease, and 3D RNA-dependent RNA polymerase, respectively. Experimental data have proved that the prediction was correct for the p15 protease (12) and the p58 polymerase (10), but so far functional studies have not been published for p37, a putative 2C NTPase.
A detailed sequence analysis of the carboxy-terminal region of RHDV cleavage product p37 revealed the presence of two amino acid sequences, 522GAPGIGKT529 and 566DE567, which correspond to the conserved motifs A (GxxGxGKS/T) and B (DD/E) found in NTP-binding proteins, which have been described to have a broad range of functions (27). The A site is required for NTP binding, whereas the B site chelates Mg2+, which is then complexed to the β and γ phosphates of the NTP molecule bound at the A motif (1, 24). The presence of an additional conserved motif C (600KxxxFxSxxxxxS/TTN614), which is also involved in ATP hydrolysis (20), indicated that this RHDV protein might be a member of helicase superfamily III, which includes the picornavirus-like (2C-like) proteins (5).
In the picornavirus-like supergroup, the poliovirus 2C protein exhibits NTPase activity (14, 20, 22), but so far a helicase activity has not been demonstrated (22).
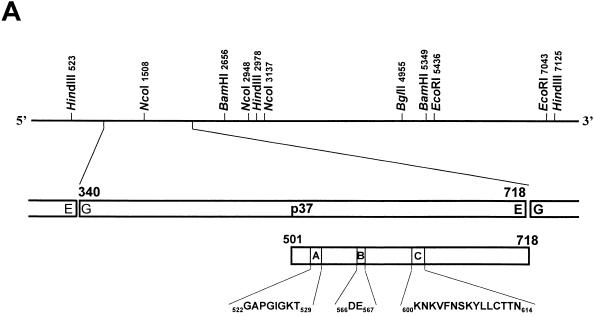
To investigate the putative NTPase activity of the p37 RHDV cleavage product, its corresponding coding region, encompassing the codons for amino acid residues 340 to 718 of the product of RHDV open reading frame 1, was amplified by PCR from plasmid pRC49 (12) using a sense primer, 5′-GGATCCAAGAGTTTCTGGGACAAGG-3′, in which a BamHI recognition sequence (underlined) has been added preceding the RHDV sequence, and an antisense primer, 5′-GAATTCtcaCTCAAATGAGGCCACGTC-3′, which incorporated an EcoRI restriction site (underlined) and a translation stop codon (lowercase). A 1,146-bp DNA fragment corresponding to nucleotides (nt) 1027 to 2163 of the Spanish AST/89 RHDV isolate (EMBL accession number Z49271) was amplified and cloned into the pGEM-T vector (Promega). This region of the RHDV genome, which encodes the conserved A, B, and C motifs (Fig. 1A) of the helicase superfamily III members, was selected, taking into account the location of the segment encoding the p37 carboxyl terminus, which was originated by cleavage at the 718EG719 peptide bond (12), and also considering the coding capacity needed for a 37-kDa polypeptide. Nevertheless, it should be mentioned that the p37 NH2-terminal residue has not been experimentally mapped. For this reason, and taking into account the known cleavage specificity of the RHDV 3C protease, the dipeptide 339EG340 could be hypothesized to be the amino-terminal cleavage site giving rise to a 2C-like protein with a calculated molecular mass of 42 kDa.
FIG. 1.
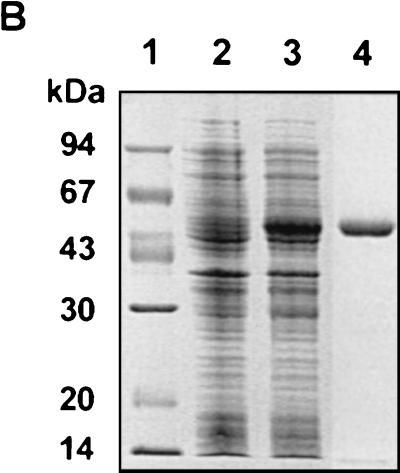
Genomic localization of the RHDV p37 cleavage product and SDS-PAGE analysis of the recombinant GST-Δ2C protein. (A) Schematic representation of the full-length RHDV cDNA indicating relevant restriction sites. Numbers indicate nucleotide residues of the RHDV genome or amino acid residues of the RHDV polyprotein. Amino acid residues in conserved motifs A, B, and C are indicated. (B) SDS-PAGE analysis of recombinant RHDV GST-Δ2C preparations. Lane 1, molecular mass markers; lane 2, cell extract from pGEX-Δ2C-transformed E. coli; lane 3, cell extract from pGEX-Δ2C-transformed, isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli; lane 4, purified GST-Δ2C fusion protein run on a separate gel.
All attempts to express the amplified cDNA (nt 1027 to 2163) coding for the putative full-length 2C protein in E. coli by using pGEX or pQE vector systems were unsuccessful. No transformants were obtained using these constructs, probably due to the toxic effects of basal levels of the recombinant proteins. Similar cytotoxic effects have been described for the recombinant 2C protein from hepatitis A virus (7). Expression of the hepatitis A virus 2C protein was inhibitory to the growth and protein synthesis of bacteria, but deletion of the 2C N-terminal amphipathic helix (21 amino acid residues) abrogated both this effect and the ability of 2C to associate with eukaryotic membranes. For this reason, and taking into account that a carboxy-terminal portion of the Flaviviridae NS3 protein showed NTPase (25, 28, 29) and helicase (9) activities, an amino-terminally truncated version of this polypeptide (Δ2C) encompassing amino acid residues 501 to 718 of the RHDV polyprotein (Fig. 1A), which contained the putative helicase consensus motifs A, B, and C (5), was expressed in E. coli as a glutathione S-transferase (GST) fusion protein. For this purpose, the amplified p37 cDNA (nt 1027 to 2163) cloned into the pGEM-T vector was digested with NcoI at a unique target sequence found within the cloned cDNA (nt 1508 to 1513 of the RHDV genome) (Fig. 1A), and the cohesive ends were filled using the Klenow fragment. The plasmid was further digested with EcoRI, and the 655-bp fragment was purified and inserted into a blunt-ended BamHI- and EcoRI-digested pGEX-2T expression vector (Amersham Pharmacia Biotech). The resulting pGEX-Δ2C expression vector included the coding region for the carboxy terminus of the putative 2C protein (Fig. 1A) fused in frame to the 3′ end of the Schistosoma mansoni GST gene.
The recombinant GST-Δ2C protein (51 kDa) could be efficiently purified from E. coli BL21 cultures harboring the pGEX-Δ2C plasmid by affinity chromatography (Fig. 1B) using a bulk GST purification module (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions. The protein pattern of the eluted fractions was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8), and the protein concentrations were calculated using the Bio-Rad protein assay. The purified recombinant GST-Δ2C protein was stored at −70°C in 50 mM Tris-HCl (pH 8.0) in the presence of 25% glycerol. All attempts to release the Δ2C moiety from the GST carrier protein by in situ thrombin proteolysis gave rise to low-molecular-weight degraded products (data not shown), and consequently the intact recombinant GST-Δ2C fusion was used for the subsequent functional assays.
In previous works, thin-layer chromatography (TLC) was used successfully for the separation of ribonucleoside triphosphates and Pi (22). Using a similar experimental approach, we have been able to demonstrate that the purified recombinant GST-Δ2C protein is able to hydrolyze [γ-32P]ATP, resulting in the production of 32Pi (Fig. 2A). Similarly to RHDV 2C-like protein, the poliovirus 2C protein conserved its NTPase activity as a fusion protein with either maltose binding protein (22) or GST (20).
FIG. 2.
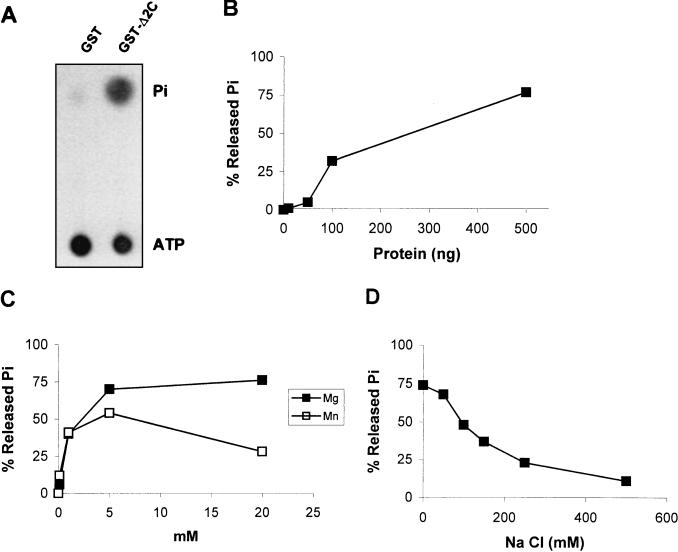
Biochemical characterization of GST-Δ2C ATPase activity. (A) Autoradiograph of a TLC plate from a standard assay showing [γ-32P]ATP and 32Pi mobility. (B) Effect of GST-Δ2C concentration on ATPase activity. (C) Effects of Mg2+ and Mn2+ on GST-Δ2C ATPase activity. (D) Effect of NaCl concentration on GST-Δ2C ATPase activity.
The standard ATPase assay was performed in a 20-μl reaction mixture containing 20 mM HEPES-KOH (pH 7.5), 5 mM magnesium acetate, 1 mM dithiothreitol, 0.4 μCi of [γ-32P]ATP (7,000 Ci mmol−1) (ICN Biomedicals, Inc.), and 0.1 μg of recombinant wild-type or mutant GST-Δ2C protein. The reaction was carried out for 5 min at 37°C and stopped by adding 0.1 M EDTA and placing the mixture in ice. One-microliter aliquots were then transferred onto plastic polyethyleneimine cellulose F sheets (Merck), which were then developed with 0.15 M formic acid–0.15 M LiCl in a TLC chamber and exposed to X-ray film. The amount of radioactivity in each spot was measured with an Instant-Imager (Packard Instrument Company).
Under the conditions used, the extent of ATP hydrolysis was dependent on the recombinant protein concentration (Fig. 2B). No ATPase activity was observed when the fusion protein was replaced by purified GST (Fig. 2A), supporting the idea that the hydrolytic activity was due to the Δ2C moiety.
Various concentrations of magnesium and manganese salts were used to investigate the divalent ion requirements of the Δ2C ATPase activity (Fig. 2C). The recombinant enzyme showed a strict requirement of Mg2+ or Mn2+ under these circumstances, and therefore an appropriate concentration of magnesium acetate (5 mM) was added to all subsequent ATPase assays.
The Δ2C enzyme activity was also found to be negatively affected by the ionic strength of the incubation medium, as demonstrated by the inhibitory effects observed after the addition of increasing amounts of NaCl (Fig. 2D).
Stimulation of NTPase activity by polynucleotides appears to be a general feature of helicase proteins (2, 3, 23). Nevertheless, the addition of 2 μg of poly(U) to the RHDV Δ2C ATPase incubation mixture using the optimized reaction conditions gave rise to an unexpected inhibition of the enzyme activity (data not shown). Similar inhibitory effects were also described for the ATPase activity of the poliovirus 2C protein (20).
To show that the presence of unlabeled UTP in the poly(U) preparation was not responsible for the observed inhibition, we also used 0.5 μg of gel-purified (10) oligo(U) (approximately 50 residues long). A similar (fourfold) inhibition of the GST-Δ2C protein ATPase activity was found (data not shown), supporting the notion that the negative effects of poly(U) were specifically due to the polynucleotide.
Experiments were also conducted to directly address the putative helicase activity of the GST-Δ2C protein, by following previously described procedures (22). Nevertheless, after repeated attempts we could not detect any unwinding activity (data not shown). This result could be a consequence of the use of an amino-terminally truncated protein or could indicate that RHDV 2C is not, in fact, a helicase, an activity that also could not be demonstrated for the poliovirus 2C protein (20, 22).
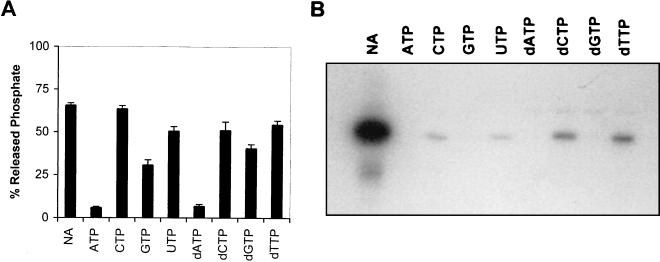
In order to investigate GST-Δ2C nucleotide preferences, we used an indirect approach by studying the inhibitory effects of unlabeled NTPs and deoxy-NTPs (dNTPs) on the observed GST-Δ2C ATPase activity. For this purpose, the standard assay was carried out in the presence of a molar excess (1 mM) of each individually assayed NTP or dNTP. The resulting residual ATPase activity was measured after 20 min of incubation. The results, summarized in Fig. 3A, showed that 1 mM ATP or dATP strongly inhibited Pi release (91 or 89%, respectively), indicating that these are the preferred nucleotides for the recombinant enzyme. In contrast, the presence of CTP hardly inhibited (3%) the release of labeled Pi. Intermediate inhibitory effects could be found in the presence of GTP (53%) and dGTP (38%), whereas the remaining compounds, UTP (23%), dCTP (23%), and dTTP (17%), gave rise to even lower inhibition levels. These results indicate a preference of the enzyme for purine substrates, as was also described for the poliovirus 2C protein (20, 22).
FIG. 3.
Effects of NTPs and dNTPs on GST-Δ2C ATP hydrolysis and ATP-binding activity. (A) Hydrolysis of [γ-32P]ATP by GST-Δ2C protein in the presence of a 1 mM concentration of each individually assayed unlabeled NTP or dNTP. (B) Autoradiograph of an SDS-PAGE gel showing the effects of an excess of unlabeled NTPs or dNTPs on the ATP-binding activity of GST-Δ2C protein. NA, no unlabeled nucleotide added.
To further substantiate the nucleotide preferences of recombinant GST-Δ2C, we investigated the effect of an excess of unlabeled nucleotides on the ATP-binding activity of the RHDV GST-Δ2C protein. For this purpose, the purified recombinant protein was incubated with [α-32P]ATP in the presence or absence of unlabeled competing nucleotides. The cross-linking reaction mixtures (20 μl), containing 1 μg of wild-type or mutant GST-Δ2C protein in 25 mM Tris-HCl (pH 8)–5 mM magnesium acetate–1 μCi of [α-32P]ATP (400 Ci mmol−1) (Amersham Pharmacia Biotech)–12.5% glycerol–1.5 mM dithiothreitol, were incubated on ice for 15 min before irradiation for about 8 min at 8 cm from the light source (0.78 J cm−2) using a Stratalinker (Stratagene). After UV cross-linking, the samples were boiled for 5 min in the presence of electrophoresis sample buffer and analyzed by SDS–12% PAGE (8). The gel was stained with Coomassie blue, dried, and autoradiographed. The radioactivity in each band was measured using an Instant-Imager (Packard Instrument Company). After correcting for the amount of protein in each band, as measured by densitometry, the percentage of ATP binding to the mutant proteins was calculated relative to the amount of ATP bound to the wild-type protein.
As expected from its ATPase activity, the RHDV Δ2C protein was also able to efficiently bind ATP, as indicated by the major labeled band observed (Fig. 3B) corresponding to the mobility of recombinant GST-Δ2C protein. The radioactive label was efficiently competed by the presence of 1 mM unlabeled ATP, GTP, dATP, or dGTP in the incubation medium. In the presence of 1 mM CTP, UTP, dCTP, or dTTP, only small amounts of ADP-protein complexes could still be observed, indicating a lower binding capacity for the RHDV Δ2C protein.
The ATPase and ATP-binding studies, in the presence of a molar excess of unlabeled competing NTPs or dNTPs both confirmed a preference order of RHDV Δ2C for the various nucleotides used, with the purines being the preferred compounds.
In order to investigate the contribution of the A, B, and C conserved motifs to ATP binding and hydrolysis by the Δ2C polypeptide, we performed a limited number of site-directed mutagenesis experiments. Highly conserved amino acid residues were chosen as targets for mutagenesis, and the residues that were introduced (replacing those naturally occurring in RHDV) could be classified into two groups. The first group included changes to residues that were never found at the corresponding position in other NTP-binding proteins (4), such as mutations G522I and T529A in domain A, D566L in motif B, and K600Q in the conserved C sequence. The second type of mutants included amino acid changes to residues similar to those found at the equivalent positions of 2C-like proteins of picornaviruses, such as T529S in region A and E567D in the B motif.
For mutant construction, the BamHI-EcoRI fragment from the pGEX-Δ2C plasmid was ligated into the BamHI and EcoRI sites of pBluescript SK(+), originating the recombinant plasmid pBS-Δ2C, which was used to perform in vitro site-directed mutagenesis using the Chameleon double-stranded kit (Stratagene) and following the manufacturer's instructions. Mutagenic oligonucleotide primers, ranging from 27 to 33 nt (Table 1), were designed to produce point mutations in the conserved sequence motifs A, B, and C. The resulting mutant BamHI-EcoRI DNA fragments were then inserted into the BamHI-EcoRI-digested pGEX-2T expression plasmid. Each of the mutated expression vectors was transformed into E. coli BL21 cells, and the GST-Δ2C mutant proteins were produced and purified as described above for the wild-type protein. To assess the overall purity and molecular size of the resulting recombinant polypeptides, equivalent amounts of each purified protein preparation were analyzed by SDS-PAGE, and the gel was stained with Coomassie blue. A major 51-kDa protein band of comparable intensity was observed for the wild-type and all mutant GST-Δ2C polypeptides (data not shown).
TABLE 1.
DNA oligomers used for site-directed mutagenesis
| Domain | Point mutanta | Amino acid sequence | Mutagenic oligonucleotide sequenceb |
|---|---|---|---|
| A | WTc | 522GAPGIGKT529 | |
| G522I | I------- | 5′ ATGCCTGGGGCTATTTTGAAAATGACAGCCACG 3′ | |
| T529A | -------A | 5′ CTGTGGACTAAGTAGGCTTTACCAATGCCTGG 3′ | |
| T529S | -------S | 5′ GTGGACTAAGTAGGATTTACCAATGCCTGG 3′ | |
| B | WT | 566DE567 | |
| D566L | L- | 5′ GTGTTGAACTCGAGCGCAATAGCAACC 3′ | |
| E567D | -D | 5′ CCACAAGTGTTGAAGTCGTCCGCAATAGC 3′ | |
| C | WT | 600KNKVFNSKYLLCTTN614 | |
| K600Q | Q-------------- | 5′ GAAAACTTTGTTCTGGTTCTCAGCCTTGTC 3′ |
The number refers to the amino acid position in the RHDV polyprotein.
Mutated nucleotide residues are underlined.
WT, wild type.
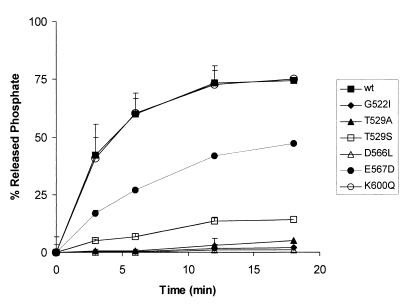
To test the ability of each mutant protein to hydrolyze [γ-32P]ATP, a time course experiment was performed using the standard ATPase assay. Aliquots (1 μl) from the reaction mixture were withdrawn at 3, 6, 12, and 18 min and spotted onto polyethyleneimine cellulose sheets. After the TLC separation and plate autoradiography, the released Pi was measured using an Instant-Imager (Packard Instrument Company) and expressed as a percentage of the total phosphate (Fig. 4).
FIG. 4.
ATPase activity of wild-type (wt) and mutant GST-Δ2C proteins. Each data point represents the average of three independent determinations.
The data indicated that the nonconserved amino acid substitutions directed to domains A (G522I and T529A) and B (D566L) completely abolished ATPase activity. In contrast, the nonconserved mutation in domain C (K600Q) showed wild-type activity. The conserved Glu-to-Asp change in domain B (E567D) resulted in the loss of about 50% of the wild-type NTPase activity. Surprisingly, the conservative threonine-to-serine substitution in domain A (T529S) resulted in a severe loss of the ATPase activity (12 to 15% of the wild-type levels).
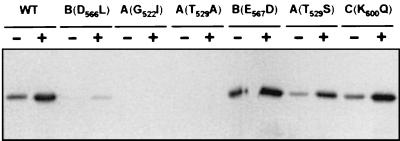
It has been previously shown that the A and B sites play important roles in NTP binding. X-ray crystallography data have indicated that the invariant K residue of site A was in direct contact with the β and γ phosphates of the bound nucleotide, while the first Asp residue of the B site interacted, via a water molecule, with a magnesium ion complexed with the β and γ phosphates (1, 17, 24). To investigate the ATP-binding properties of the Δ2C point mutants, we used a UV cross-linking assay, in which ATP is photolyzed by UV light in the presence or absence of Mg2+, resulting in a covalent bond between the protein and the [α-32P]ATP.
In the absence of Mg2+, the ATP-binding capacities of the wild-type and mutant proteins were reduced about fivefold (Fig. 5), supporting the relevance of this ion for nucleotide binding. In addition, nonconserved substitutions in either domain A (G522I and T529A) or domain B (D566L) resulted in a dramatic decrease in ATP cross-linking (Fig. 5). On the other hand, the conservative mutations T529S (domain A) and E567D (domain B) had moderate effects (Fig. 5) on the ATP-binding capacities of the mutant proteins.
FIG. 5.
ATP-binding assay of wild-type (WT) and mutant GST-Δ2C proteins in the presence (+) or absence (−) of Mg2+. The amino acid substitutions in domains A, B, and C are indicated in parentheses.
In contrast with the results found for sites A and B, a nonconservative substitution (K600Q) in domain C did not alter the ATP-binding capacity of the mutant protein (Fig. 5).
From the site-directed mutagenesis studies it could be concluded that the replacement of the motif A conserved Gly and Thr residues completely abolished the ATP-binding capacity and ATPase activity of the RHDV Δ2C protein. Similar changes in the poliovirus 2C protein (Gly129Ile [14] and Ser136Ala [26]) produced nonviable viruses after transfection of cultured cells. It was also found that the replacement of the conserved Asp566 residue with Leu in motif B severely impaired its ATP-binding and ATPase activities. Similarly, this type of mutation was lethal in poliovirus 2C, as no viral RNA replication could be detected in cells transfected with mutant transcripts, suggesting a functional role for the NTP-binding motif of 2C in the RNA replication and proliferation of poliovirus (14). The lack of ATP binding obtained with the RHDV Δ2C site B mutant (D566L) does not agree with the results of an earlier mutational study involving the B motif (DEAD) of the mammalian translation initiation factor eIF-4A, which is included in the superfamily II helicases (5). In this case, ATP binding was not affected by mutations in the B motif, although ATP hydrolysis was abolished (19). This discrepancy may reflect differences in the ATP-binding capacities of the members of superfamilies II and III of RNA helicases. In addition, mutations in other conserved amino acid residues, such as Lys135 (domain A) and Asp177 (motif B), also abolished the ATPase activity of the poliovirus 2C protein (15, 20).
Conservative-replacement studies, with amino acid residues from RHDV Δ2C motifs A and B replaced by residues observed at the equivalent picornavirus 2C protein positions, showed that this type of changes more severely affected ATP hydrolysis than ATP-binding capacity, suggesting the biological relevance of these residues, particularly that of Thr529. Nevertheless, in the poliovirus system, where in vivo studies can be performed, no viable mutants could be isolated after transfection of cultured cells using mutated transcripts at the conserved domain A Ser (S136T) or domain B Asp (D177E) from poliovirus 2C (26).
The analysis of the purified RHDV mutant protein carrying the K600Q change (motif C) showed wild-type ATPase and ATP-binding activities. In contrast to this observation, it has been previously reported that replacement of the conserved Asn residue in poliovirus 2C motif C inhibited the ATPase activity to undetectable levels, suggesting the requirement of motif C for the hydrolytic activity (20). Our results might indicate that not all the conserved residues in motif C play similar roles or that, despite the high similarities observed, some significant functional differences could be found between the 2C polypeptides from poliovirus and caliciviruses.
The in vivo significance of these replacements could not be assessed in the RHDV system due to the lack of both a permissive cell culture and the possibility of producing infective viral transcripts. Nevertheless, the data reported in this work provide the first experimental evidence of the existence of a 2C-like ATPase activity in a member of the Caliciviridae, thus supporting the previous predictions of a 2C NTPase protein based on amino acid sequence analysis.
Acknowledgments
This work was supported by grant PB96-0552-CO2-O1 from Dirección General Enseñanza Superior, Spain.
REFERENCES
- 1.Cho H S, Ha N C, Kang L W, Chung K M, Back S H, Jang S K, Oh B H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 2.Eagles R M, Balmori-Melian E, Beck D L, Gardner R C, Forster R L. Characterization of NTPase, RNA-binding and RNA-helicase activities of the cytoplasmic inclusion protein of tamarillo mosaic potyvirus. Eur J Biochem. 1994;224:677–684. doi: 10.1111/j.1432-1033.1994.t01-1-00677.x. [DOI] [PubMed] [Google Scholar]
- 3.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkühler C, De Francesco R. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 5.Kadaré G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.König M, Thiel H-J, Meyers G. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J Virol. 1998;72:4492–4497. doi: 10.1128/jvi.72.5.4492-4497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusov Y Y, Probst C, Jecht M, Jost P D, Gauss-Muller V. Membrane association and RNA binding of recombinant hepatitis A virus protein 2C. Arch Virol. 1998;143:931–944. doi: 10.1007/s007050050343. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Clum S, You S, Ebner K E, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López Vázquez A, Martín Alonso J M, Casais R, Boga J A, Parra F. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J Virol. 1998;72:2999–3004. doi: 10.1128/jvi.72.4.2999-3004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcato P S, Benazzi C, Vecchi G, Galeotti M, Della S L, Sarli G, Lucidi P. Clinical and pathological features of viral haemorrhagic disease of rabbits and the European brown hare syndrome. Rev Sci Tech Off Int Epizoot. 1991;10:371–392. doi: 10.20506/rst.10.2.560. [DOI] [PubMed] [Google Scholar]
- 12.Martín Alonso J M, Casais R, Boga J A, Parra F. Processing of rabbit hemorrhagic disease virus polyprotein. J Virol. 1996;70:1261–1265. doi: 10.1128/jvi.70.2.1261-1265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyers G, Wirblich C, Thiel H-J. Rabbit hemorrhagic disease virus: molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991;184:664–676. doi: 10.1016/0042-6822(91)90436-f. [DOI] [PubMed] [Google Scholar]
- 14.Mirzayan C, Wimmer E. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology. 1992;189:547–555. doi: 10.1016/0042-6822(92)90578-d. [DOI] [PubMed] [Google Scholar]
- 15.Mirzayan C, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 16.Ohlinger V F, Haas B, Meyers G, Weiland F, Thiel H-J. Identification and characterization of the virus causing rabbit hemorrhagic disease. J Virol. 1990;64:3331–3336. doi: 10.1128/jvi.64.7.3331-3336.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra F, Prieto M. Purification and characterization of a calicivirus as the causative agent of a lethal hemorrhagic disease in rabbits. J Virol. 1990;64:4013–4015. doi: 10.1128/jvi.64.8.4013-4015.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 21.Pringle C R. Virus taxonomy—San Diego 1998. Arch Virol. 1998;143:1449–1459. doi: 10.1007/s007050050389. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 23.Staüber N, Martinez-Costas J, Sutton G, Monastyrskaya K, Roy P. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J Virol. 1997;71:7220–7226. doi: 10.1128/jvi.71.10.7220-7226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Story R M, Steitz T A. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 25.Suzich J A, Tamura J K, Palmer-Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teterina N L, Kean K M, Gorbalenya A E, Agol V I, Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992;73:1977–1986. doi: 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- 27.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α and β subunits of ATP synthetase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrener P, Tamura J K, Collett M S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wengler G, Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 30.Wirblich C, Sibilia M, Boniotti M B, Rossi C, Thiel H-J, Meyers G. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J Virol. 1995;69:7159–7168. doi: 10.1128/jvi.69.11.7159-7168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirblich C, Thiel H-J, Meyers G. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J Virol. 1996;70:7974–7983. doi: 10.1128/jvi.70.11.7974-7983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]