Abstract
Background
Atherosclerotic cardiovascular disease (ASCVD) is a leading cause of death globally, and early detection of high-risk individuals is essential for initiating timely interventions. The authors aimed to develop and validate a deep learning (DL) model to predict an individual’s elevated 10-year ASCVD risk score based on retinal images and limited demographic data.
Methods
The study used 89,894 retinal fundus images from 44,176 UK Biobank participants (96% non-Hispanic White, 5% diabetic) to train and test the DL model. The DL model was developed using retinal images plus age, race/ethnicity, and sex at birth to predict an individual’s 10-year ASCVD risk score using the pooled cohort equation (PCE) as the ground truth. This model was then tested on the US EyePACS 10K dataset (5.8% non-Hispanic White, 99.9% diabetic), composed of 18,900 images from 8969 diabetic individuals. Elevated ASCVD risk was defined as a PCE score of ≥7.5%.
Results
In the UK Biobank internal validation dataset, the DL model achieved an area under the receiver operating characteristic curve of 0.89, sensitivity 84%, and specificity 90%, for detecting individuals with elevated ASCVD risk scores. In the EyePACS 10K and with the addition of a regression-derived diabetes modifier, it achieved sensitivity 94%, specificity 72%, mean error -0.2%, and mean absolute error 3.1%.
Conclusion
This study demonstrates that DL models using retinal images can provide an additional approach to estimating ASCVD risk, as well as the value of applying DL models to different external datasets and opportunities about ASCVD risk assessment in patients living with diabetes.
Keywords: Cardiovascular disease risk, Pooled cohort equation, Retinal imaging, Artificial intelligence
Introduction
Atherosclerotic cardiovascular disease (ASCVD) is the most common cause of hospitalization and premature death in the United States.1 The risk of an individual experiencing an ASCVD event includes both nonmodifiable variables (age, sex, and race/ethnicity) and modifiable variables such as diabetes,2 hypertension,3 dyslipidemia,4 and smoking.5 Across a population, the risk of experiencing an ASCVD event varies greatly. Risk-based equations have therefore been developed to identify those who are at greatest risk of ASCVD so that preventive treatments can be initiated appropriate to the individual’s risk.6 The landmark Framingham Heart Study was the first to demonstrate that multivariable equations could identify an individual’s ASCVD risk with far greater accuracy than the existing metrics based solely on blood pressure and cholesterol.7 Since the Framingham-based equations were first published, other equations have been developed to serve different and more diverse populations with refined accuracy.8,9
The retina is unique in being the only part of the human vasculature where the microvascular system is visible at micron-level resolution by noninvasive means. The automated detection of components within retinal images to predict ASCVD risk has been used with moderate degrees of success previously,10,11 but in recent years there has been an exponential increase in the number of studies that have used artificial intelligence (AI), and deep learning (DL) in particular, to extract data from retinal images.12,13 There is now growing interest in using the retinal image data generated by DL models to augment the traditional means of estimating ASCVD risk.14 In this study we used retinal photographs and limited demographic data from the UK Biobank to develop and validate a DL model designed to predict an individual’s elevated 10-year ASCVD risk, based on the US-derived pooled cohort equation (PCE).15
The primary aim of this study was to develop and test a DL model to predict an individual’s 10-year ASCVD risk based on their retinal photographs plus age, race/ethnicity, and sex at birth and then further validate the findings in an external database. This model was built upon our previous work on detecting and grading retinopathy, maculopathy, macular degeneration, and effects of smoking in retinal images.16, 17, 18 Although the Framingham risk score is recommended to perform cardiovascular risk assessment in some countries,19 the 2018 American Heart Association (AHA) Cholesterol Clinical Practice Guidelines recommend using the US-derived PCE to estimate the 10-year risk for hard ASCVD events (coronary heart disease death, nonfatal myocardial infarction, fatal or nonfatal stroke).15 Our DL model was trained on and validated against the 10-year ASCVD risk as calculated by the PCE from individuals in the UK Biobank dataset (Level 3 in Figure 1; Supplemental Appendix 2). Further external validation was provided by testing on a US-based dataset, EyePACS 10K, with substantially greater racial diversity and predominantly from patients living with diabetes.
Figure 1.
The process of training and validation of the deep learning prediction model using the UK Biobank dataset. ASCVD = atherosclerotic cardiovascular disease; CNN =convolutional neural network; PCE = pooled cohort equation.
Methods
Datasets
The composition of datasets used in this study is shown in Table 1. The UK Biobank (IRB UOA-86299) was used for training and internal validation. The validation subset represented 20% of the data, selected randomly prior to development. The data from the UK Biobank can be accessed via a direct request to the UK Biobank, and was obtained using approved data management and data transfer protocols. A total of 89,894 fundus images from 44,176 unique participants from the UK Biobank were used in this study. Participants in the UK Biobank were recruited from a UK general population with only approximately 5% of the UK Biobank population self-identified as having diabetes “diagnosed by doctor.”
Table 1.
The demographic and risk factor makeup of the UK Biobank–derived training and internal test datasets and the EyePACS 10K external validation dataset used in this study
| UK Biobank: training N = 35,570 |
UK Biobank: test N = 8606 |
EyePACS 10K N = 8969 |
||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 56 | 8.3 | 57 | 8.3 | 56 | 10 |
| Systolic blood pressure (mm Hg) | 134 | 18.1 | 134∗ | 17 | 131∗∗ | 12 |
| Diastolic blood pressure (mm Hg) | 81.4 | 10.1 | 81∗∗ | 9.8 | 71∗∗ | 8.9 |
| HbA1c (%) | 5.4% | 0.6% | 5.4%∗∗ | 0.5% | 8.1%∗ | 1.7% |
| Total cholesterol (mg/dL) | 220 | 44.1 | 219∗∗ | 43.2 | 179∗∗ | 43.7 |
| HDL cholesterol (mg/dL) | 58 | 15.1 | 58∗∗ | 15.1 | 47∗∗ | 12.3 |
| BMI | 27.2 | 4.7 | 27.1 | 4.6 | Not known | Not known |
| Sex at birth | Male | Female | Male | Female | Male | Female |
| 16,313 (46%) | 19,257 (54%) | 3991 (46%) | 4615 (54%) | 3923 (46%) | 5046 (54%) | |
| Current smoker | True | False | True | False | True | False |
| 4526 (13%) | 29,535 (87%) | 1100 (13%)† | 7122 (87%) | 507 (6%)† | 8414 (94%) | |
| Diabetes (%) | 5.0% | 4.8% | 99.9% | |||
| Race/ethnicity | Non-Hispanic White | 93.2% | 93.0% | 5.8% | ||
| South Asian | 2.1% | 2.1% | 6.5% | |||
| East Asian | 0.36% | 0.27% | 0.40% | |||
| Black/African American | 2.14% | 1.93% | 6.80% | |||
| Hispanic | N.A. | N.A. | 65.7% | |||
| Multiracial | 0.71% | 0.80% | 0.05% | |||
| Native American | N.A. | N.A. | ?? | |||
| Other | 1.16% | 1.16% | 1.80% | |||
| Prefer not to answer | 0.33% | 0.35% | 0.05% | |||
| Declined/do not know | 0.04% | 0.03% | 12.9% | |||
BMI = body mass index; N.A. = not available.
Significance test was performed between UK Biobank training and test, as well as Biobank training and EyePACS 10K external validation (∗P < .01 z test; ∗∗P < .001 z test; †P < .01 χ2). Refer to Supplemental Appendix 2 for aligning race/ethnicity terminology between the UK Biobank and EyePACS 10K datasets. The UK Biobank did not include Hispanic ethnicity as an option and the White participants were predominantly of British and Irish origin. EyePACS 10K included choices of Hispanic, Black, or White, so separating Hispanic Black participants from Hispanic White participants was not possible.
As described below, a dataset from the US-based EyePACS study (IRB UCB 2017-09-10340), with multiple differences from UK Biobank, was used for external validation. The dataset used for this analysis (EyePACS 10K) consisted of a subset of 9947 individuals who had sufficient clinical data to calculate a traditional PCE risk score. Of these, 978 were excluded because they had established ASCVD prior to the date of retinal imaging. The external validation dataset thus comprised 18,900 images from 8969 individuals. The research presented in this study, and the datasets used in it, adhered to the principles outlined in the Declaration of Helsinki. The mean age of individuals in the EyePACS 10K dataset was 56 ± 10 years (compared to 57 ± 8.3 years in the UK Biobank). They predominantly self-identified as Hispanic, whereas the UK Biobank population was predominantly non-Hispanic White. As the EyePACS 10K population consisted almost exclusively of people living with diabetes presenting for diabetic retinopathy screening, the mean hemoglobin A1c (HbA1c) level of this population was high (8.1% ± 1.7%). Additional demographics from both datasets are summarized in Table 1.
Inclusion/exclusion criteria
Individuals in both datasets who had established ASCVD (Supplemental Appendix 1) prior to the acquisition of the retinal images were excluded. A previously trained DL-based image quality model was used to screen all retinal images for acceptable image quality.17 Only the earliest acceptable images and the individual’s accompanying biometrics obtained on that same visit were used from the UK Biobank data. For the EyePACS 10K dataset, the earliest acceptable images which had accompanying biodata, taken no earlier than 1 year before image acquisition, were used.
Model assessment
Receiver operating characteristic curve, sensitivity, and specificity metrics
To assess the DL model’s ability to predict an elevated 10-year ASCVD risk score, we compared the score the DL model issued with that generated by the PCE equation. The DL model’s performance was based on a PCE score of ≥7.5% being the ground truth for elevated risk. The receiver operating characteristic (ROC) curve, the precision recall curve, and the overall performance of the DL model on the UK Biobank and EyePACS 10K datasets were determined. The sensitivity and specificity of the model to detect individuals with an elevated ASCVD risk in the UK Biobank and EyePACS 10K datasets were also calculated. To investigate the performance of the model in different demographic groups in the UK Biobank, the performance of the model across sex, age, and race/ethnicity were also calculated. To investigate the impact the retinal image had on the DL model’s performance, and to ascertain whether the retinal data contributed positively to the output, we reran the DL model withholding the retinal image input data and recalculated the area under the receiver operating characteristic curve (AUROC), sensitivity, and specificity.
Comparison of the performance of the DL model and the PCE equation to predict 10-year ASCVD risk and events
The performance of the DL model vs PCE was also assessed by way of a data binning technique.20 Patients in the UK Biobank and EyePACS 10K datasets were first arranged in ascending order by their DL model predicted scores and then by their PCE-calculated 10-year ASCVD risk scores. Both datasets were then divided into 20 bins with equal population numbers by an ascending order of their ASCVD risk scores, ie, from the lowest 5%, every 5% to the top 5%. The mean 10-year ASCVD risk scores generated by the DL model and the PCE equation were then plotted for each bin in both the UK Biobank and EyePACS 10K datasets. The magnitude of the deviation between the 2 sets of results generated for each dataset was then assessed by measuring the mean error and the mean absolute error per case.
For the UK Biobank the above exercise was then repeated using actual observed ASCVD events, categorized by risk as determined by the DL model and the PCE equation. This exercise was not possible for the EyePACS 10K dataset, as there were no future ASCVD events recorded after the retinal images were acquired. In accordance with ACC/AHA Work Group guidelines, an ASCVD event for the purposes of outcomes was defined as the first nonfatal acute myocardial infarction, fatal or nonfatal stroke, or fatal coronary artery disease.21 The ICD codes used to define a hard ASCVD event are provided in Supplemental Appendix 1.
Finally, to assess the probabilistic performance of the 2 methods for predicting 10-year ASCVD risk, the Brier score loss was calculated for both the DL model and the PCE equation.
Model development
The DL model pipeline encompasses approximately 50 distinct DL models, classified into 2 primary categories: image-based models and non-image-based models. The former uses image data as input, while the latter relies on vectorized interpretations of participant information, including their biometrics. This dichotomy results in a diversity of input data and target formats within the pipeline. For instance, the systolic blood pressure (SBP) model uses a retinal image as input, with the corresponding SBP value serving as the target. This model’s function is independent of additional factors, thereby enabling it to be trained on samples missing the HbA1c value, provided the SBP data are accessible. Another model of note is the PCE ASCVD prediction model. It uses outputs from preceding image-based models, with the calculated PCE results serving as the target. However, the PCE equation requires multiple variables, including SBP, HbA1c, total cholesterol, and HDL (high-density lipoprotein) cholesterol. Only participants with all of the above-mentioned components available were used in this study.
Model ensemble
The DL prediction model ensemble used is demonstrated in Figure 2 and comprises 3 different levels.
Figure 2.
The structure of the deep learning prediction model developed in this work. ASCVD = atherosclerotic cardiovascular disease; PCE = pooled cohort equation.
The first level (Level 1) includes an image quality check convolutional neural network (CNN) (described above), a laterality (left eye / right eye) detector CNN, and an image location (fovea / non-fovea) detector CNN. The input of this layer is retinal images only. This process ensures that only foveal-centered images that are of sufficient quality are accepted into the model. Identifying the laterality of the image ensures that only a single fovea-centered image for each eye for each individual is used during the analysis. The final available dataset after requiring high- or medium-quality, fovea-centered images, 1 per eye, consisted of 89,894 images, representing 44,176 patients. This was divided into an 80/20 split for training and validation, respectively. The training dataset thus consisted of 76,321 images (representing 35,570 patients). Meanwhile, the test dataset comprised 19,080 images representing 8606 patients who had valid biometrics. The demographics of the final dataset did not significantly differ from those that included images of low quality. The patient demographics of the test and training datasets were statistically compared with a 2-sample Kolmogorov-Smirnov test to ensure that the demographic distribution of the training and test group were similar. There was no statistically meaningful difference between the 2 datasets, comparing the age, sex, and race/ethnicity makeup of the 2 groups (Supplementary material). The training of these models is explained elsewhere, and these models were not tuned or retrained for this study.17
The second level (Level 2) includes 9 ensembles of AIs, each consisting of 5 CNNs (45 in total). The retinopathy, maculopathy, drusen, pigmentary abnormality, advanced age-related macular degeneration, and smoking CNNs were previously trained on other datasets.16,22,23 The rest of the CNNs were trained using the unique UK Biobank labels in the fundus images: “hba1c_result,” “tchdl_result,” “systolic_bp,” “systolic_bp2,” “smoking_status.” “Systolic_bp” and “systolic_bp2” are 2 consecutive blood pressure measurements in the UK Biobank and in this study we used the mean of the two.
These CNNs follow modified versions of the Inception-Resnet-V2 or ResNet50 structures. Taking the single retinopathy CNN model as an example, the model has a deep structure, consisting of 164 layers, and uses a combination of inception and residual blocks. The inception blocks use a combination of convolutional layers with different filter sizes, while the residual blocks use skip connections to enable the model to learn from previous layers. We also employed batch normalization and bottleneck layers to improve training efficiency. Overall, the model architecture is designed to extract features at multiple scales and capture fine-grained details in images, making it well suited to detect the level of retinopathy or other biomarkers. For each CNN in Level 2, the image plus biomarkers dataset was split for training, validation, and testing: 70%, 15%, 15%, respectively. The excessive background of the fundus images was cropped, and the resulting image was resized to 800 × 800 pixels. A batch size of 8 was chosen to optimize GPU memory during training. Adam optimizer was adopted with a learning rate 1 × 10e-3 to update parameters toward the minimization of the loss. Dropout was enabled with a rate p = 0.2, and the model was trained for at least 100 EPOCHs. All codes related to this work were implemented using Python 3.7.
Additionally, we developed a complex jury system to arrive at the ultimate prediction for each biomarker. To elaborate, using the retinopathy model as an example, there exist 6 distinct levels of retinopathy (R0–R5). Five jury models were employed to assess each eye, resulting in 30 probability values per eye. These probabilities were merged and consolidated for both eyes, thereby yielding a final value for each patient.
The third level (Level 3) is a multilayer perceptron, which uses the output of the second-level CNNs, plus the patient’s chronological age, sex, and race/ethnicity to estimate their PCE ASCVD risk score. This PCE-derived ASCVD risk score is the ground-truth label, calculated from the relevant 9 fields in the UK Biobank dataset for each participant (age, sex, race/ethnicity, smoking status, blood pressure, diabetes, serum total cholesterol, HDL cholesterol, blood pressure–lowering medication; https://tools.acc.org/asASCVD-risk-estimator-plus/#!/calculate/estimate/). The architecture of the model comprises an input layer, followed by 5 dense layers that exhibit a gradual decrease in neuron counts, namely 1024, 512, 256, 128, and 32. These layers are interspersed with batch normalization and LeakyReLU activation functions with a leaky rate of 0.1. To address overfitting concerns, dropout layers with a rate of 0.3 were incorporated after the third, fourth, and fifth dense layers. The ultimate layer, encompassing a single neuron and a linear activation function, predicts the target value. For optimization purposes, an Adam optimizer is used with an exponentially decaying learning rate schedule, initialized at 3e-3 and decaying by a factor of 0.95 every 1000 steps. The Huber loss function was employed to guide the model parameters’ updating. To curb overfitting and ensure efficient training, early stopping was implemented.
Post hoc regression–based diabetes modifier
We anticipated that our DL model, initially trained on the UK Biobank dataset, may require calibration to accurately predict the 10-year ASCVD risk scores for individuals with diabetes as represented in the EyePACS 10K dataset. To address this, we derived a “diabetes modifier,” an adjustment factor derived from a linear regression model, which would correct the output of the DL model trained on the general population of the UK Biobank to predict ASCVD risk more accurately in people living with diabetes. This modifier considers individual patient factors such as age and sex, as well as the initial prediction of ASCVD risk (pce_pred), to tailor the risk assessment for diabetic individuals. The regression equation that we subsequently developed for this adjustment is as follows:
Final risk score = DL model risk score + max(diabetes_adjustment, 0) where the diabetes_adjustment is calculated as: diabetes_adjustment = -10.4156 + (0.3560 ∗ age) − (4.5422 ∗ sex) − (0.5410 ∗ pce_pred) with sex encoded as 0 for male and 1 for female.
Results
The ability of the DL model to identify elevated-risk individuals (PCE-generated ASCVD score ≥7.5%), compared to the PCE equation, for both the UK Biobank and EyePACS 10K datasets are shown in Table 2. In UK Biobank, the DL model achieved an AUROC of 0.89, a sensitivity of 83%, and a specificity of 90%. Withholding the retinal image data from the DL model resulted in lower performance, with AUROC 0.84, sensitivity 70%, and specificity 88%. Assessment of the performance of DL model across different subgroups revealed there was a significant difference between age groups, with the model performing better for individuals over 60 compared to those under 60. There was no significant difference in DL model performance between male/female or different race/ethnic groups (Table 3).
Table 2.
Confusion matrices comparing the deep learning model–predicted atherosclerotic cardiovascular disease score vs pooled cohort equation–calculated scores
| UK BioBank |
EyePACS 10K |
EyePACS 10K + diabetes modifier |
||||
|---|---|---|---|---|---|---|
| PCE-generated ASCVD score <7.5% | PCE-generated ASCVD score ≥7.5% | PCE-generated ASCVD score <7.5% | PCE-generated ASCVD score ≥7.5% | PCE-generated ASCVD score <7.5% | PCE-generated ASCVD score ≥7.5% | |
| DL model–generated ASCVD score <7.5% | 5923 (69%) | 331 (4%) | 4126 (46%) | 2238 (25%) | 3015 (34%) | 279 (3%) |
| DL model–generated ASCVD score ≥7.5% | 642 (7%) | 1710 (20%) | 217 (2%) | 2388 (27%) | 1328 (15%) | 4347 (48%) |
ASCVD = atherosclerotic cardiovascular disease; DL = deep learning; PCE = pooled cohort equation.
Results are n (%) of people.
Table 3.
Performance of deep learning model (area under receiver operating characteristic curve score) across different races/ethnicities and demographics in the UK Biobank test dataset
| Demographic segment | AUROC‡ | Group size | N where PCE ≥7.5% | |
|---|---|---|---|---|
| Sex assigned at birth | Female | 0.88 | 4615 | 217 |
| Male | 0.89 | 3991 | 1824 | |
| Race/ethnicity† | Non-Hispanic White | 0.87 | 8023 | 1945 |
| South Asian | 0.83 | 206 | 44 | |
| East Asian | ||||
| Black/African American | 0.89 | 166 | 24 | |
| Other/multiracial | 0.93 | 202 | 25 | |
| Age bracket | Age ≤60 | 0.72 | 5112 | 417 |
| Age >60 | 0.84 | 3494 | 1624 | |
AUROC = area under the receiver operating characteristic curve; PCE = pooled cohort equation.
Participants self-identified as belonging to a particular race/ethnicity group (Supplemental Appendix 2).
Sex subgroups comparison (female to male), Delong test statistic: -0.6114, P value: .54. Age subgroups comparison, Delong test: -14.3876, P value: .000. Race/ethnicity subgroup comparisons: White vs Asian, Delong test statistic: 1.513, P value: 0.13; White vs Black, Delong test statistic: -0.814, P value: .416; Asian vs Black, Delong test statistic: -1.681, P value: .09.
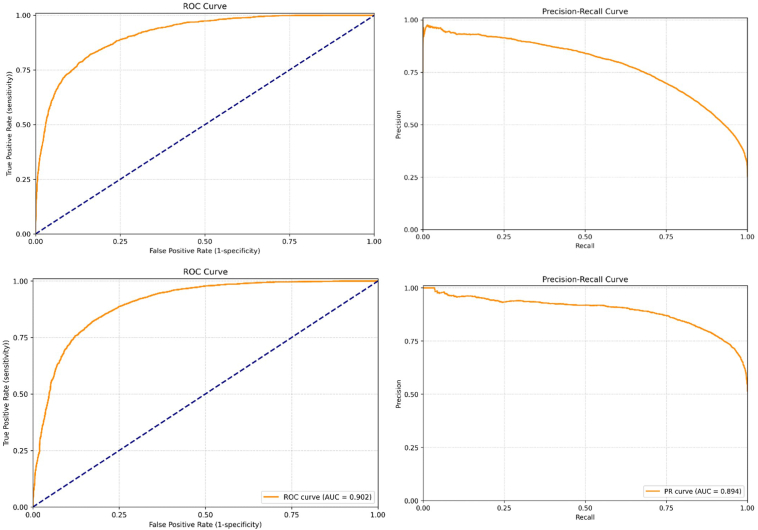
In EyePACS 10K, the DL model achieved a similar AUROC: 0.90, with a lower sensitivity of 52% and a higher specificity of 95%. The ROC and the accompanying precision-recall curve plots are shown in Figure 2. Assessment of the performance of DL model across different subgroups revealed there was a small but significant difference between those individuals of Black/African-American race compared to all other groups. The model also performed better for individuals over 60 years old compared to those under 60. There was no significant difference in DL model performance between the other race/ethnic groups (Table 4).
Table 4.
Performance of deep learning model (area under receiver operating characteristic curve score) across different races/ethnicities and demographics in EyePACS 10K dataset
| Demographic segment | AUROC‡ | Segment size | N where PCE ≥7.5% | |
|---|---|---|---|---|
| Sex assigned at birth | Female | 0.89 | 5352 | 1975 |
| Male | 0.86 | 3612 | 2648 | |
| Race/ethnicity† | Hispanic | 0.91 | 5893 | 2817 |
| Black/African American | 0.85 | 609 | 426 | |
| Non-Hispanic White | 0.91 | 517 | 323 | |
| East Asian | 0.9 | 586 | 361 | |
| Declined/do not know | 0.92 | 1158 | 575 | |
| South Asian | 0.95 | 37 | 18 | |
| Other | 0.89 | 169 | 106 | |
| Age bracket | Age ≤60 | 0.88 | 5572 | 1751 |
| Age >60 | 0.84 | 3392 | 2872 | |
AUROC = area under the receiver operating characteristic curve; PCE = pooled cohort equation.
Race/ethnicity was self-reported by study participants (Supplemental Appendix 2).
Age subgroups comparison, Delong test statistic: 5.9290, P value: <.001. Race/ethnicity subgroup comparisons: White vs African American, Delong test statistic: 3.290, P value: .001; White vs Asian, Delong test statistic: 0.624, P value: .53; Asian vs African American, Delong test statistic: 2.730, P value: .006; Latin American vs White, Delong test statistic: -0.057, P value: .95; Latin American vs African American, Delong test statistic: 4.157, P value: <.001.
In post hoc analysis, applying a regression-based diabetes modifier to the DL model output, the sensitivity and specificity to detect individuals with elevated risk against PCE in the EyePACS 10K dataset were 94% and 72%, respectively. Assessment of the performance of DL model across different subgroups revealed there was a small but significant difference between those individuals of Black/African American race compared to all other groups. The model also performed better for individuals over 60 years old compared to those under 60. There was no significant difference in DL model performance between the other race/ethnic groups (Table 5).
Table 5.
Performance of deep learning model (area under receiver operating characteristic curve score) across different races/ethnicities and demographics in EyePACS 10K dataset when diabetes modifier is applied
| Demographic segment | AUROC‡ | Segment size | N where PCE ≥7.5% | |
|---|---|---|---|---|
| Sex assigned at birth | Female | 0.92 | 5352 | 1975 |
| Male | 0.91 | 3612 | 2648 | |
| Race/ethnicity† | Hispanic | 0.94 | 5893 | 2817 |
| Black/African American | 0.87 | 609 | 426 | |
| Non-Hispanic White | 0.92 | 517 | 323 | |
| East Asian | 0.92 | 586 | 361 | |
| Declined/do not know | 0.94 | 1158 | 575 | |
| South Asian | 0.97 | 37 | 18 | |
| Other | 0.92 | 169 | 106 | |
| Age bracket | Age ≤60 | 0.9 | 5572 | 1751 |
| Age >60 | 0.86 | 3392 | 2872 | |
AUROC = area under the receiver operating characteristic curve; PCE = pooled cohort equation.
Race/ethnicity was self-reported by study participants (Appendix 2).
Sex subgroups comparison, Delong test: 0.7657, P value: .44. Age subgroups comparison, Delong test: 5.6842, P value: .00. Race/ethnicity subgroups comparison: White vs African American: Delong test statistic: 2.778, P value: .005; White vs Asian, Delong test statistic: -0.339, P value: .74; Asian vs African American, Delong test statistic: 3.203, P value: .001; Latin American vs White, Delong test statistic: 1.635, P value: .10; Latin American vs African American, Delong test statistic: 5.054, P value: <.001.
The AUROC values were then calculated for each demographic segment of both the UK Biobank and EyePACS 10K datasets (Table 3, Table 4, Table 5). The DL model performed well across both sexes and all races/ethnicities, but its performance in younger individuals (≤ 60 years) in UK Biobank was lower than in older individuals (AUROC 0.72 vs 0.84).
Relationship between ASCVD risk scores generated and observed ASCVD event rates using the traditional PCE score and the DL model predicted score
UK Biobank
The predicted 10-year ASCVD risk scores generated by the PCE equation and the DL model, plotted for each bin in the UK Biobank, are shown in Figure 3A. The predicted 10-year ASCVD risk scores rose equally and proportionally for all risk categories in both cohorts. Across the UK Biobank, the predicted 10-year ASCVD risk scores from PCE and the DL model were very similar (mean error 0.3%, mean absolute error 2.4%). The actual ASCVD event rates observed in both the PCE and DL model, when categorized by ascending order of predicted risk, are also shown in Figure 3A. The actual ASCVD event rate rose steadily from the nonelevated to elevated risk categories in both the PCE and DL models across all individuals in the UK Biobank. The magnitude of the actual ASCVD event rates was again very similar to the predicted 10-year ASCVD risk score produced by the PCE and DL models for all risk categories. The actual ASCVD event rates observed when the results generated by the PCE and DL models were subdivided into the binary classification (predicted risk score <7.5% or ≥7.5%) are shown in Table 6. The ASCVD event rate observed in those individuals from the UK Biobank allocated a “nonelevated” score by the traditional PCE method was the same as those allocated a “nonelevated” score by the DL prediction model (2.2% vs 2.0%). The same finding was observed in the “elevated” risk groups (ASCVD event rate: traditional PCE 7.5%, DL model 7.5%). Analysis with the point-biserial correlation coefficient revealed that the 10-year ASCVD risk score produced by the PCE and the DL model were both significantly correlated with actual ASCVD events: calculated PCE vs ASCVD events: 0.144 (P < .01); DL model predicted risk score vs ASCVD events: 0.152 (P < .01). The accuracy of the PCE equation to correctly predict an ASCVD event was identical to that of the DL model, with the Brier score loss for both 0.067.
Figure 3.
Receiver operating characteristic (ROC) curves (left-hand graphs) and precision-recall curves (right-hand graphs) for UK Biobank (top) and EyePACS 10K (bottom) datasets.
Table 6.
UK Biobank atherosclerotic cardiovascular disease (ASCVD) event rates in those with and without elevated ASCVD risk scores by pooled cohort equation–based ASCVD risk score compared to dep learning model–predicted ASCVD risk score.
| 10-year ASCVD risk score: PCE | Individuals (ASCVD events) | 10-year ASCVD risk score: DL model | Individuals (ASCVD events) |
|---|---|---|---|
| <7.5% | 6565 (146) 2.2% |
<7.5% | 6254 (123) 2.0% |
| ≥7.5% | 2041 (153) 7.5% |
≥7.5% | 2352 (176) 7.5% |
ASCVD = atherosclerotic cardiovascular disease; DL = deep learning; PCE = pooled cohort equation.
Values in each cell represent #people (#ASCVD events) and % events/cases.
EyePACS 10K dataset
The ASCVD risk scores generated by the PCE equation and the DL model, plotted for each bin in the EyePACS 10K dataset, are shown in Figure 3B. For all risk categories the predicted 10-year ASCVD risk score, as measured by the PCE equation, was substantially higher than that produced by the DL model (mean error 3.6%, mean absolute error 4.4%). Comparison between Figure 3A and 3B reveals that the performance of the DL model was very consistent across both the UK Biobank and EyePACS 10K. However, the PCE equation predicted consistently higher scores across all risk profiles in the EyePACS dataset, unlike in UK Biobank.
As the ROC curves indicated that the performance of the DL model was very similar in both the internal (AUROC 0.89) and external (AUROC 0.90) validation datasets, but with differing sensitivities and specificities, we hypothesized that there was a systematic difference between the 10-year ASCVD risk score produced by the PCE and our DL model when it was applied to people living with diabetes. We therefore derived a correction factor: a “diabetes modifier,” which would translate the 10-year ASCVD risk score produced by our DL model to that produced by the PCE, across all risk profiles.24 The ASCVD risk score generated by the DL model with the diabetes modifier applied, plotted for each bin in the EyePACS 10K dataset, is shown in Figure 3B. The results produced by the DL model with the diabetes modifier applied were now very similar to PCE (mean error -0.2%, mean absolute error 3.1%). The sensitivity and specificity of the new diabetes-modified DL model to detect individuals with elevated risk against PCE in the EyePACS 10K dataset were 94% and 72%, respectively.
Discussion
In this study we developed and validated a novel DL model to calculate a 10-year ASCVD risk score using more than 90,000 retinal images from the UK Biobank dataset and then externally validated it on more than 18,000 retinal images from the EyePACS 10K dataset. Our DL model reliably detected those individuals with elevated ASCVD risk scores (≥7.5%) in both datasets using only the retinal image and the individuals’ age, race/ethnicity, and sex with AUROCs of 0.89–0.90.
The actual observed ASCVD event rate in UK Biobank was very similar between the DL model and PCE. There was also a significant correlation between the risk scores produced by both models and actual ASCVD events and the probabilistic accuracy of the DL model to correctly predict an ASCVD event was identical to that of the PCE.
There was a clear difference between the PCE and the DL model–predicted 10-year ASCVD risk scores in the EyePACS 10K dataset. When assessed by the PCE, 48% of individuals in the EyePACS 10K dataset were deemed to be “nonelevated” and 52% were deemed “elevated” risk. The DL model apportioned the risk differently: 70% “nonelevated” risk, 30% “elevated” risk. Unlike what was observed in the UK Biobank, when the 10-year ASCVD risk scores generated by the PCE and the DL model from the EyePACS 10K dataset were grouped into bins of ascending risk scores, the magnitude and distribution of the predicted 10-year ASCVD risk scores generated by the PCE and the DL model were quite different; mean error was 3.6%, mean absolute error as measured by each index case was 4.4% (Figure 3B). As the performance of the DL model assessed by the AUROC was very similar in both the UK Biobank and EyePACS 10K datasets, and the principal difference in the 2 datasets was low vs high rate of diabetes, we hypothesized that it should be possible to regress a correction factor—a “diabetes modifier”—to transform the ASCVD risk score produced by our DL model to that produced by the PCE. Applied to the EyePACS 10K dataset, this post hoc analysis showed that 10-year ASCVD risk scores generated by the DL model were substantially better aligned to the PCE after the diabetes modifier was applied (Figure 3B). The subsequent sensitivity and specificity of the “diabetes-modified” DL model to detect individuals with “elevated” risk in the EyePACS 10K dataset were also improved, at 94% and 72%, respectively (Figure 4).
Figure 4.
A: Ten-year atherosclerotic cardiovascular disease (ASCVD) risk scores (crosses) and ASCVD event rates (dots) when categorized according to the traditional pooled cohort equation (PCE)–calculated risk score (red) or deep learning (DL) model–predicted risk score (green) in the UK Biobank. B: Ten-year ASCVD risk when categorized according to the traditional PCE-calculated risk score (red) or DL model–predicted risk score (green) or DL model–predicted score after application of the diabetes modifier (blue) in the EyePACS 10K dataset.
Regardless of diabetes type and status, the PCE equation currently treats all people living with diabetes as a uniform group and it effectively adds a modifier to the regression equation, which serves to elevate their risk score compared to nondiabetic individuals. Recently it has been suggested that the traditional regression-based equations, like the PCE, may overestimate ASCVD risk in many people living with diabetes.25,26 The application of DL algorithms to predict ASCVD risk-related outcomes from retinal images has been comprehensively reviewed by Hu and colleagues.27 This review revealed that, to date, a heterogeneous array of models have been developed using a variety of different inputs: retinal images only, retinal images + various biodata, and reporting against different cardiac-related outcomes. To date, only 2 other groups have reported the results of a DL model trained and then externally validated to predict the ASCVD 10-year risk from retinal photographs.24, 25, 26 Our results support the accumulating evidence that indicates DL algorithms can use retinal images to accurately predict ASCVD risk and they compare favorably with the 1 other DL model that has been validated on the UK Biobank, which had sensitivity and specificity of 83% and 88%, respectively.28, 29, 30
Strengths and limitations
The primary strengths of this study include the following: we have trained and validated a DL model designed to predict an individual’s ASCVD 10-year risk based on nothing more than a retinal photograph and limited demographic data; the DL model was not only able to reliably match the PCE scores, but also showed similar prediction to PCE of actual ASCVD events; and the DL model was externally validated on a very different dataset. Our DL model performed similarly (by AUROC) on the external EyePACS 10K dataset as it did on the UK Biobank, and we investigated the application of a “diabetes modifier” to the output of our DL model to better match the sensitivity and specificity of the PCE when applied to individuals living with diabetes. However, as there were no future hard ASCVD events in the EyePACS 10K dataset, we could not validate the DL model on this key metric. Although the DL model performed well on all races/ethnicities, (AUROC scores ≥0.87 in all groups), there was a small but significant difference in its performance on individuals of Black/African American race compared to all other race/ethnic groups. This highlights the need to ensure that AI tools are trained and thoroughly tested in the target population groups to avoid exacerbating existing health inequalities. Finally, it is highly probable that the ground truth for the smoking DL model, namely self-reported smoking status, lacks robustness, as it is widely accepted that individuals tend to under-report their smoking habit.31
Conclusion
In conclusion, our results show that it is possible to train a DL model that can assess ASCVD risk as well as the traditional PCE method, using nothing more than a retinal photograph and limited demographic data. We have also shown that the application of a “diabetes modifier” to our DL model is a promising approach to matching the PCE in people living with diabetes. If these results can be replicated in other large population-based datasets, DL models like ours may offer the potential to significantly improve access to ASCVD risk detection strategies, as the risk predictions these models produce do not require multiple clinical and laboratory assessments to generate an individual’s ASCVD risk score.
Acknowledgments
Funding Sources
Toku Eyes has funded this study.
Disclosures
Ehsan Vaghefi, David Squirrell, Song Yang, Songyang An, and Li Xie are staff members of Toku, which funded this study. Jacqueline Shreibati, John Marshall, Michael V. McConnell, and Matthew Budoff are consultants of Toku, which has funded this study. Toku holds the patent for the technology introduced here (US11766223B1).
Patient Consent and Ethics
The data used here were obtained using ethical approvals from the original data guardians, IRB UOA-86299 and IRB UCB 2017-09-10340.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2023.12.004.
Appendix. Supplementary data
References
- 1.Centers for Disease Control and Prevention. About Underlying Cause of Death. 2020:1999–2020. https://wonder.cdc.gov/ucd-icd10.html [Google Scholar]
- 2.Almourani R., Chinnakotla B., Patel R., Kurukulasuriya L.R., Sowers J. Diabetes and cardiovascular disease: an update. Curr Diab Rep. 2019;19:1–13. doi: 10.1007/s11892-019-1239-x. [DOI] [PubMed] [Google Scholar]
- 3.Kjeldsen S.E. Hypertension and cardiovascular risk: general aspects. Pharmacol Res. 2018;129:95–99. doi: 10.1016/j.phrs.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Alloubani A., Nimer R., Samara R. Relationship between hyperlipidemia, cardiovascular disease and stroke: a systematic review. Curr Cardiol Rev. 2021;17:52–66. doi: 10.2174/1573403X16999201210200342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo T., Nakano Y., Adachi S., Murohara T. Effects of tobacco smoking on cardiovascular disease. Circ J. 2019;83:1980–1985. doi: 10.1253/circj.CJ-19-0323. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd-Jones D.M., Braun L.T., Ndumele C.E., et al. Use of risk assessment tools to guide decision-making in the primary prevention of atherosclerotic cardiovascular disease: a special report from the American Heart Association and American College of Cardiology. Circulation. 2019;139:e1162–e1177. doi: 10.1161/CIR.0000000000000638. [DOI] [PubMed] [Google Scholar]
- 7.Mahmood S.S., Levy D., Vasan R.S., Wang T.J. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008. doi: 10.1016/S0140-6736(13)61752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kavousi M., Leening M.J., Nanchen D., et al. Comparison of application of the ACC/AHA guidelines, Adult Treatment Panel III guidelines, and European Society of Cardiology guidelines for cardiovascular disease prevention in a European cohort. JAMA. 2014;311:1416–1423. doi: 10.1001/jama.2014.2632. [DOI] [PubMed] [Google Scholar]
- 9.Grundy S.M., Stone N.J., Bailey A.L., et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e1082–e1143. doi: 10.1161/CIR.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Yin Y., Cao G., Wei B., Zheng Y., Yang G. Hierarchical retinal blood vessel segmentation based on feature and ensemble learning. Neurocomputing. 2015;149:708–717. [Google Scholar]
- 11.Mishra V., Samuel C., Sharma S. Use of machine learning to predict the onset of diabetes. International Journal of Recent Advances in Mechanical Engineering (IJMECH) 2015;4:9–14. [Google Scholar]
- 12.Poplin R., Varadarajan A.V., Blumer K., et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2:158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 13.Rim T.H., Lee C.J., Tham Y.-C., et al. Deep-learning-based cardiovascular risk stratification using coronary artery calcium scores predicted from retinal photographs. Lancet Digit Health. 2021;3:e306–e316. doi: 10.1016/S2589-7500(21)00043-1. [DOI] [PubMed] [Google Scholar]
- 14.Miyazawa A.A. Artificial intelligence: the future for cardiology. Heart. 2019;105:1214. doi: 10.1136/heartjnl-2018-314464. [DOI] [PubMed] [Google Scholar]
- 15.Grundy S.M., Stone N.J. 2018 American Heart Association/American College of Cardiology/Multisociety Guideline on the Management of Blood Cholesterol–Secondary Prevention. JAMA Cardiol. 2019;4:589–591. doi: 10.1001/jamacardio.2019.0911. [DOI] [PubMed] [Google Scholar]
- 16.Vaghefi E., Yang S., Xie L., et al. A multi-centre prospective evaluation of THEIA™ to detect diabetic retinopathy (DR) and diabetic macular oedema (DMO) in the New Zealand screening program. Eye. 2023;37:1683–1689. doi: 10.1038/s41433-022-02217-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaghefi E., Yang S., Xie L., et al. THEIA™ development, and testing of artificial intelligence-based primary triage of diabetic retinopathy screening images in New Zealand. Diabet Med. 2021;38 doi: 10.1111/dme.14386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie L., Yang S., Squirrell D., Vaghefi E. Towards implementation of AI in New Zealand national diabetic screening program: cloud-based, robust, and bespoke. PLoS One. 2020;15 doi: 10.1371/journal.pone.0225015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson G.J., Thanassoulis G., Anderson T.J., et al. 2021 Canadian Cardiovascular Society guidelines for the management of dyslipidemia for the prevention of cardiovascular disease in adults. Can J Cardiol. 2021;37:1129–1150. doi: 10.1016/j.cjca.2021.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Java T Point What is Binning in Data Mining? 2010. https://www.javatpoint.com/what-is-binning-in-data-mining#:∼:text=Binning%2C%20also%20called%20discretization%2C%20is,the%20number%20of%20distinct%20values
- 21.Goff D.C., Jr., Lloyd-Jones D.M., Bennett G., et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S49–S73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 22.Xie L., Vaghefi E., Yang S., Han D., Marshall J., Squirrell D. Automation of macular degeneration classification in the AREDS dataset, using a novel neural network design. Clin Ophthalmol. 2023;17:455–469. doi: 10.2147/OPTH.S396537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaghefi E., Yang S., Hill S., Humphrey G., Walker N., Squirrell D. Detection of smoking status from retinal images; a Convolutional Neural Network study. Sci Rep. 2019;9:7180. doi: 10.1038/s41598-019-43670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsur A., Batsry L., Toussia-Cohen S., et al. Development and validation of a machine-learning model for prediction of shoulder dystocia. Ultrasound Obstet Gynecol. 2020;56:588–596. doi: 10.1002/uog.21878. [DOI] [PubMed] [Google Scholar]
- 25.Vaghefi E., Squirrell D., Yang S., et al. Development and validation of a deep-learning model to predict 10-year ASCVD risk from retinal images using the UK Biobank and EyePACS 10K datasets. medRxiv. 2023 doi: 10.1016/j.cvdhj.2023.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnett D.K. Widespread diabetes screening for cardiovascular disease risk estimation. Lancet. 2021;397:2228–2230. doi: 10.1016/S0140-6736(21)00764-9. [DOI] [PubMed] [Google Scholar]
- 27.Hu W., Yii F.S., Chen R., et al. A systematic review and meta-analysis of applying deep learning in the prediction of the risk of cardiovascular diseases from retinal images. Transl Vis Sci Technol. 2023;12:14. doi: 10.1167/tvst.12.7.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rim T.H., Lee G., Kim Y., et al. Prediction of systemic biomarkers from retinal photographs: development and validation of deep-learning algorithms. Lancet Digit Health. 2020;2:e526–e536. doi: 10.1016/S2589-7500(20)30216-8. [DOI] [PubMed] [Google Scholar]
- 29.Yi J.K., Rim T.H., Park S., et al. Cardiovascular disease risk assessment using a deep-learning-based retinal biomarker: a comparison with existing risk scores. Eur Heart J Digit Health. 2023;4:236–244. doi: 10.1093/ehjdh/ztad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y., Xiong J., Zhu Y., et al. Deep learning algorithm using fundus photographs for 10-year risk assessment of ischemic cardiovascular diseases in China. Sci Bull (Beijing) 2022;67:17–20. doi: 10.1016/j.scib.2021.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Curry L.E., Richardson A., Xiao H., Niaura R.S. Nondisclosure of smoking status to health care providers among current and former smokers in the United States. Health Educ Behav. 2013;40:266–273. doi: 10.1177/1090198112454284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.