Summary
Background
Gallbladder cancer (GBC) is highly aggressive. Diagnosis of GBC is challenging as benign gallbladder lesions can have similar imaging features. We aim to develop and validate a deep learning (DL) model for the automatic detection of GBC at abdominal ultrasound (US) and compare its diagnostic performance with that of radiologists.
Methods
In this prospective study, a multiscale, second-order pooling-based DL classifier model was trained (training and validation cohorts) using the US data of patients with gallbladder lesions acquired between August 2019 and June 2021 at the Postgraduate Institute of Medical Education and research, a tertiary care hospital in North India. The performance of the DL model to detect GBC was evaluated in a temporally independent test cohort (July 2021–September 2022) and was compared with that of two radiologists.
Findings
The study included 233 patients in the training set (mean age, 48 ± (2SD) 23 years; 142 women), 59 patients in the validation set (mean age, 51.4 ± 19.2 years; 38 women), and 273 patients in the test set (mean age, 50.4 ± 22.1 years; 177 women). In the test set, the DL model had sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) of 92.3% (95% CI, 88.1–95.6), 74.4% (95% CI, 65.3–79.9), and 0.887 (95% CI, 0.844–0.930), respectively for detecting GBC which was comparable to both the radiologists. The DL-based approach showed high sensitivity (89.8–93%) and AUC (0.810–0.890) for detecting GBC in the presence of stones, contracted gallbladders, lesion size <10 mm, and neck lesions, which was comparable to both the radiologists (p = 0.052–0.738 for sensitivity and p = 0.061–0.745 for AUC). The sensitivity for DL-based detection of mural thickening type of GBC was significantly greater than one of the radiologists (87.8% vs. 72.8%, p = 0.012), despite a reduced specificity.
Interpretation
The DL-based approach demonstrated diagnostic performance comparable to experienced radiologists in detecting GBC using US. However, multicentre studies are warranted to explore the potential of DL-based diagnosis of GBC fully.
Funding
None.
Keywords: Gallbladder cancer, Deep learning, Ultrasound
Research in context.
Evidence before this study
We searched PubMed, Embase, and Google databases on July 1, 2019, for original studies of deep learning-based detection of gallbladder cancer from imaging studies and updated the search on Jan 1, 2023. We used the search terms “deep learning” OR “convolutional neural network” AND “gallbladder cancer” OR “gallbladder malignancies” AND “diagnosis” without date or language restrictions. The references of identified studies were also reviewed. We found five non-overlapping studies that used deep learning to diagnose gallbladder cancer on ultrasound (four studies, three on trans-abdominal and one on endoscopic ultrasound) or computed tomography (one study). The existing studies were limited by the small number of patients in the test set (or the need for a held-out test set). Importantly, these studies did not evaluate the performance of deep learning models to detect gallbladder cancer in diverse real-world scenarios, including different morphological types (polyps, mural thickening, masses), gallbladders with stones, contracted gallbladders, lesions of different sizes and at various sites within the gallbladder. Hence, the potential for deep learning-enabled ultrasound-based detection of gallbladder cancer was unclear. As ultrasound is the most practical and universally available non-invasive initial diagnostic test for gallbladder cancer, the voids in the existing literature necessitated this study.
Added value of this study
We trained, validated, and tested a deep learning algorithm to detect gallbladder cancer based on ultrasound using data from 565 prospective patients. The test cohort was temporally independent. To our knowledge, we report the largest sample size and the most comprehensive metadata of gallbladder lesions. We used advanced deep learning techniques (visual acuity-based learning and multiscale second-order pooling) described previously to address the challenges associated with developing deep learning models from ultrasound images. We found that the deep learning model was non-inferior to the radiologists to detect gallbladder cancer in the overall test cohort and subgroups covering all common scenarios in the real world. In some subgroups, the deep learning model had better sensitivity than the radiologists.
Implications of all the available evidence
Our study shows the promising diagnostic performance of the deep learning-based model for detecting gallbladder cancer. With multicentre validation, our model can be implemented for timely gallbladder cancer diagnosis in large community hospitals with a limited number of expert radiologists and may lead to an improved prognosis for this deadly cancer.
Introduction
Gallbladder cancer (GBC) is a lethal biliary tract malignancy with a grave prognosis. According to GLOBOCAN 2020 data, GBC incidence is 115,949 per year, and it accounts for 84,695 deaths every year worldwide.1 The highest incidence is in Asia and among the Asian countries, the highest incidence and mortality have been recorded in East Asia, followed by South-Central Asia. India accounts for 10% of the global GBC burden and GBC is the leading cause of cancer-related deaths among Indian women.2 There has been a steady rise in the incidence of GBC in men and women.2
GBC is often detected at an advanced stage in most patients, impeding curative resection and resulting in a dismal prognosis.3 The overall mean survival rate for patients with advanced GBC is six months, with a 5-year survival rate of <5%.4 Early diagnosis is critical for improving the survival rates of patients with GBC. Due to the absence of ionising radiation exposure, low cost, portability, and accessibility, ultrasound (US) is the initial diagnostic modality for evaluating patients with suspected gallbladder diseases. Although identifying gallstones and abnormalities such as gallbladder wall thickening at routine US is easy, accurate characterisation of early signs of GBC is challenging.5 If malignancy is not suspected, usually no further testing is performed, due to which early GBC could silently progress. Also, accurate analysis of US images requires a high degree of expertise and several years of training.
Artificial intelligence (AI) techniques have the potential to reduce human effort to a great extent. Unlike traditional image-dependent “semantic” feature evaluation by human experts, deep learning (DL) can automatically learn feature representations from sample images with convolutional neural networks (CNNs).6 These neural networks have been shown to match or surpass human performance in applying specific tasks and may even discover additional differential features not yet identified in current radiological practice.6 Machine learning has made transformational advancements in radiology and medical diagnosis for oncological diseases such as breast cancer, lung cancer, pancreatic cancer, and ovarian cancer.7, 8, 9, 10, 11, 12
We summarize the DL-based literature in gallbladder in Supplementary Table S1. The published literature has focused mainly on the detection of gallstones and classification of polyps on US. A few recent papers have reported DL-based GBC detection on US. A CNN architecture (GBCNet) recently showed state-of-the-art diagnostic performance in categorizing gallbladder lesions on US.13 The existing studies are limited by the small number of patients in the test set (or the need for a held-out test set). Additionally, the performance of these DL models in actual world practice is not known.14 For widespread implementation of DL-based diagnosis, it is critical to evaluate DL model performance in situations that are expected to affect radiologists' performance (e.g., polyps and mural thickening, contacted gallbladder, gallbladder neck lesions.15, 16, 17 Thus, we performed a large prospective study to develop and validate a DL model for the automatic detection of GBC at abdominal US and compare its diagnostic performance with that of radiologists. We also perform extensive subgroup analysis to demonstrate the robustness of DL-model.
Methods
Study design and participants
Consecutive patients with gallbladder diseases (based on prior US, CT, or MRI) underwent systematic US of the gallbladder at the Postgraduate Institute of Medical Education and Research, Chandigarh, a tertiary care hospital in Northern India, between August 2019 and July 2022. Patients whose final diagnosis could be established were included in the study. The diagnosis of GBC was based on histopathology of cholecystectomy specimen or percutaneous US or endoscopic US-guided biopsy or fine needle aspiration cytology. The diagnosis of benign gallbladder diseases was based on the histopathology of cholecystectomy specimens or clinical follow-up of at least three months, demonstrating the stability of lesions. Patients with polyps ≤ 5 mm, acute cholecystitis, or gallbladder abnormalities secondary to extracholecystitis causes (e.g., pancreatitis, hepatitis) or systematic illnesses (e.g., viral infections, fluid overload states) were excluded. This prospective study was approved by the Institutional Ethics Committee (approval number IEC-11/2019-1403), and informed written consent was obtained from all the patients.
Procedures
US acquisition and interpretation
Radiologists (with 1–8 years of post-training experience in abdominal US) performed gallbladder US on the Logiq S8 US scanner (GE Healthcare, US) using a convex transducer with a frequency range of 1–5 MHz after at least 6 h of fasting. For patients with gallbladder polyps, high-resolution US was also performed using a transducer with a frequency range of 2–8 MHz. US assessment was done in supine and lateral decubitus positions to visualise the entire gallbladder wall and lumen.5 All the US images were stored on the local hard disk. Colour or spectral Doppler images were not recorded. The US images were later reviewed independently by two radiologists with two and eight years of post-training experience in the abdominal US. The radiologists performing and reviewing the US images were aware that the patients had gallbladder diseases but were blinded to the findings of the previous imaging tests and the final diagnosis. The patients' records were handled by a data entry operator who archived the clinical and imaging data.
The US diagnosis of GBC was based on the presence of gallbladder masses infiltrating adjacent liver; diffuse or focal, symmetric, or asymmetric gallbladder wall thickening (GWT) showing indistinct interface with the liver or directly involving biliary or vascular structures; and hypoechoic polyp >10 mm with lobular surface and internal hypoechoic foci.18 The US diagnosis of benign diseases was based on the presence of well-defined masses <10 mm with a lack of invasion into adjacent structures or intraluminal polyps without the features described above. The presence of diffuse or focal, symmetric, or asymmetric GWT with mural layering, intramural echogenic foci or intramural cysts, and distinct interface with liver also led to a diagnosis of benign GWT.5
Data annotation
The US images were anonymised and saved in.jpeg format before feeding them to the CNN model. The input to the CNN model was the entire image. Before inputting the images for classification by the CNN model, no other data labelling or region of interest was drawn. The training cohort's data preprocessing of US images were performed as described below. Patient-level labels (benign and malignant) based on the reference standard described above were used for training.
Neural network implementation
We employed the multiscale, second-order pooling-based (MS-SoP) classifier proposed by Basu and colleagues for detecting GBC from US images.13 The codebase for the MS-SoP classifier is publicly available at https://github.com/sbasu276/GBCNet. The details of neural network implementation are given in supplementary data. Fig. 1 and Supplementary Figure S1 show the proposed architecture.
Fig. 1.
Schematic overview of the training and testing phases of the deep convolutional neural network (CNN) model. (a). We use the Gaussian Blurring-based curriculum proposed by Basu and colleagues13 during the training phase. During the initial epochs, blurred input images are used to train the network. Gradually the blur is lowered and towards the later phases, original high-resolution images are used for training. (b). For predicting the diagnosis of a patient using the CNN model, image-level predictions are generated for all images corresponding to the patient. The majority predicted class at the image-level predictions is chosen as the predicted class for the patient.
Training the neural network
The weights of the classification network were initialised from a model pre-trained on the publicly available GBCU dataset, which contains 1255 US images of GB from 218 patients.13 Since we modified the classification head, we initialised the weights of the classification head (the last layer) using the Xavier initialization method.19,20 We fine-tuned the neural network on our dataset.
We used the binary cross-entropy loss (BCE) as the objective function. We trained the network end-to-end with a stochastic gradient descent optimizer with an initial learning rate of 0.003, a momentum of 0.9, a weight decay of 0.0005, and mini-batches of size 32 on images of 233 patients (training cohort). The learning rate decays by a factor of 0.9 after every 5th epoch. We used the resize, random crop, random horizontal flip with probability 0.1, and normalisation for data augmentations during the training to mitigate overfitting issues. The input image size to the network was 224 × 224. We trained the model for 60 epochs using a Gaussian blurring-based training curriculum introduced by Basu and colleagues.13 For the first 10 epochs, the training images are blurred by convoluting with a Gaussian kernel with . Then, after every 5th epoch, the of the Gaussian kernel is halved, thus reducing the blur. After 30 epochs, the model is trained with original resolution images. Such a training curriculum mitigates the tendency of the neural network to learn from spurious echogenic textures and enhances the network's ability to learn from the low-frequency features as well.13 We used a validation set of 59 patients to optimize the network weights and the hyperparameters. The best-performing model on the validation set in terms of accuracy was selected for evaluation on a held-out test set of 273 patients (test set). We also evaluated the performance of two popular pre-trained CNN-models (ResNet-50, DenseNet-121) on the held-out test cohort. Both the models were fine-tuned using the training data of 233 patients. The models were trained and evaluated in a system with Intel(R) Xeon(R) Gold 5218 processor and four Nvidia Tesla V100 32 GB GPUs.
Statistical analysis
We evaluated the performance of the CNN and both the radiologists [Radiologist 1 (with two years post-training experience) and Radiologist 2 (with eight years of post-training experience) for the entire cohort and multiple subgroups. Subgroup analysis was performed for gallbladder diseases with stones, contracted gallbladder, different morphological types of gallbladder cancer (including masses, GWT, mass with GWT, and polyps), size ≤10 mm and >10 mm, different sites (body, fundus, and neck), and focal vs. diffuse disease.
To evaluate the classification results of CNN and radiologists, we used sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and area under the receiver operating characteristic (ROC) curve (AUC). Diagnostic performance measures were calculated using the mean and 95% CI. CIs for sensitivity, specificity and accuracy are the “exact” Clopper-Pearson CIs.21 CIs for the PPV and NPV are the standard logit CIs.21 CIs for AUC were calculated using pROC package. CNN and radiologists' sensitivity, specificity, and accuracy were compared using the Mc Nemar test. The AUCs were compared using the DeLong method. The statistical analyses were performed using IBM® SPSS® version 22 (IBM India Pvt Ltd, India), MedCal® (MedCalc Software Ltd, Belgium), SciPy 1.1.0 (Austin, TX, USA), Analyse-it® version 6.15 (Analyse-it Software Ltd. UK), and R.22, 23, 24, 25 A p-value of <0.05 was considered statistically significant.
Role of the funding source
Not applicable.
Results
Over the study period, we recruited 565 patients (Supplementary Figure S2). The mean age (±SD) was 50.8 ± 22.6 years. There were 208 (36.8%) males and 357 (63.2%) females. There were 116 (20.5%), 333 (58.9%), 75 (13.3%), and 41 (7.3%) patients with the mass-replacing gallbladder, GWT, mass with GWT, and polyp, respectively, in the overall group. The mean size of the masses and polypoidal lesions was 4.6 ± 2.2 cm and 1.32 ± 0.54 cm, respectively. The mean GWT was 1.1 ± 0.7 cm. There were 189 (33.5%) patients with benign and 376 (66.5%) patients with malignant diagnoses. The characteristics of the overall group and the training, validation, and test sets are given in Table 1.
Table 1.
Clinical characteristics of the patients in the overall group, training, validation, and testing cohorts.
| All | Training set | Validation set | Test set | |
|---|---|---|---|---|
| Number of individuals | 565 | 233 | 59 | 273 |
| Diagnosis | ||||
| Benign | 189 (33.5) | 75 (32.2) | 24 (40.7) | 90 (33) |
| Malignant | 376 (66.5) | 158 (67.8) | 35 (59.3) | 183 (67) |
| Age (years) | 50.8 ± 22.6 | 48 ± 23 | 51.4 ± 19.2 | 50.4 ± 22.1 |
| Sex | ||||
| Male | 208 (36.8) | 91 (39.1) | 21 (35.6) | 96 (31.2) |
| Female | 357 (63.2) | 142 (60.9) | 38 (64.4) | 177 (64.8) |
| Morphological type | ||||
| Masses | 116 (20.5) | 45 (19.3) | 14 (23.7) | 57 (20.9) |
| Thickening | 333 (58.9) | 139 (59.6) | 31 (52.5) | 163 (59.7) |
| Mass with thickening | 75 (13.3) | 30 (12.9) | 8 (13.6) | 37 (13.5) |
| Polyp | 41 (7.3) | 19 (8.2) | 6 (10.2) | 16 (5.9) |
| Cholelithiasis | 334 (59.1) | 150 (64.4) | 28 (47.4) | 156 (57.1) |
| Gallbladder statusa | ||||
| Distended | 285 (50.4) | 115 (49.4) | 25 (42.4) | 145 (54.9) |
| Contracted | 164 (29) | 73 (31.3) | 20 (33.9) | 71 (26) |
| Size (cm) | ||||
| Mass | 4.69 ± 2.21 | 4.77 ± 2.46 | 4.61 ± 2.21 | 4.29 ± 2.33 |
| Wall thickness | 1.12 ± 0.71 | 1.16 ± 0.73 | 1.0 ± 0.71 | 0.95 ± 0.67 |
| Polyp | 1.32 ± 0.54 | 1.36 ± 0.67 | 1.41 ± 0.59 | 1.29 ± 0.55 |
| Sitea, b | ||||
| Neck | 72 (12.7) | 28 (12) | 2 (3.4) | 42 (15.4) |
| Body | 51 (9) | 24 (10.3) | 11 (18.6) | 16 (5.9) |
| Fundus | 49 (8.7) | 18 (7.7) | 10 (16.9) | 21 (7.7) |
| Diffuse | 249 (44.1) | 100 (42.9) | 24 (40.7) | 125 (45.8) |
Masses replacing gallbladder were excluded.
Multiple sites of involvement were excluded.
The diagnostic performance of the CNN and the radiologists in the training and validation cohorts are shown in the Supplementary Table S2 and Supplementary Table S3. The diagnostic performances of ResNet-50 and DenseNet-121 are reported in Supplementary Table S4.
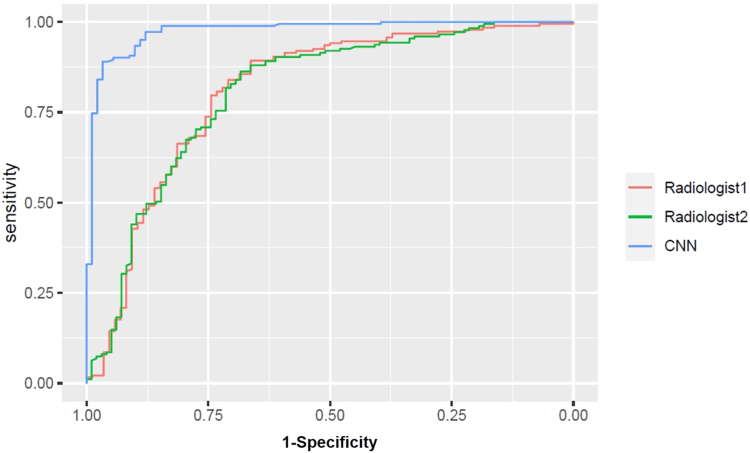
The sensitivity, specificity, accuracy, and AUC of CNN in the test cohort were 92.3% (95% CI, 88.1–95.6), 74.4% (95% CI, 65.3–79.9), 86.4% (95% CI, 82.2–90.5), and 0.887 (95% CI, 0.844–0.930), respectively compared to 86.8% (95% CI, 81.1–91.4), 67% (95% CI, 56.3–76.5), 80.2% (95% CI, 75–84.8), and 0.826 (95% CI, 0.767–0.884), respectively for Radiologist 1 and 87.9% (95% CI, 82.3–92.3), 80% (95% CI, 70.2–87.7), 75.2% (95% CI, 65.4–83.4), and 0.837 (95% CI, 0.781–0.892), respectively for Radiologist 2. There were no significant differences in the sensitivities, specificities, and AUCs of CNN and the radiologists (Fig. 2).
Fig. 2.
Performance of the deep convolutional neural network (CNN). Area under receiver operating characteristic curve of CNN and radiologists in the test cohort for detecting gallbladder cancer in the overall group.
In the patients with gallstones, CNN had diagnostic performance comparable to the radiologists for detecting GBC. There was no difference in the diagnostic performance of the CNN and the radiologists in detecting GBC on mass, mass with thickening, and polyp morphological subtypes. However, the CNN had better sensitivity than the radiologist 2 in detecting wall thickening type of GBC (87.8% vs. 72.8%, p = 0.012). This higher sensitivity was achieved at the cost of lower specificity. In the contracted gallbladder, CNN had a greater sensitivity than Radiologist 2. However, the difference was not statistically significant (93% vs. 77.3%, p = 0.050). Additionally, the specificity of CNN was lower than radiologists. The sensitivity and AUC of CNN for lesions <10 mm was greater than the radiologists. However, the differences were not statistically significant. The diagnostic performance of the CNN and the radiologists for lesions >10 mm was comparable. There were no significant differences between CNN and radiologists based on the thickening site and whether the thickening was focal or diffuse. Table 2 shows the diagnostic performance of CNN and the radiologists in the test cohort. Supplementary Tables S5–S7 show the p-values of differences in the diagnostic performance of CNN and the two radiologists.
Table 2.
Diagnostic performance of the convolutional neural networks (CNN) and radiologists in test cohort.
| Groups | % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | % Accuracy (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|---|
| Overall | ||||||
| CNN | 92.3 (88.1–95.6) | 74.4 (65.3–79.9) | 90.1 (84.9–94.1) | 80 (70.2–87.6) | 86.4 (82.2–90.5) | 0.887 (0.844–0.930) |
| Radiologist 1 | 86.8 (81.1–91.4) | 67 (56.3–76.5) | 87 (81.31–91.5) | 76.1 (65.8–84.5) | 80.2 (75–84.8) | 0.826 (0.767–0.884) |
| Radiologist 2 | 87.9 (82.3–92.3) | 80 (70.2–87.7) | 89.7 (84.32–93.8) | 75.2 (65.4–83.4) | 85.3 (80.5–89.3) | 0.837 (0.781–0.892) |
| Stones | ||||||
| CNN | 92.2 (87–95.2) | 79.6 (71.9–93.1) | 90.1 (82.5–95.1) | 80.0 (67.0–89.5) | 87.8 (82.3–93) | 0.890 (0.836–0.945) |
| Radiologist 1 | 90.2 (82.7–95.2) | 72.2 (58.4–83.5) | 85.5 (77.3–91.7) | 76.9 (63.1–87.4) | 83.9 (77.3–89.4) | 0.812 (0.733–0.891) |
| Radiologist 2 | 90.1 (82.5–95.2) | 77.8 (64.4–87.9) | 88.24 (80.3–93.7) | 81.1 (68–90.5) | 85.8 (79.3–90.9) | 0.835 (0.761–0.909) |
| Mass | ||||||
| CNN | 98.2 (90.4–99.9) | 100 (2.5–100%) | 99.1 (95.1–99.9) | 20 (0.5–71.6) | 98.2 (90.6–99.6) | 1 |
| Radiologist 1 | 96.4 (87.6–99.5) | 100 (2.5–100) | 100 (93.4–100) | 25 (0.6–80.6) | 96.5 (87.9–99.6) | 1 |
| Radiologist 2 | 100 (93.6–100) | 100 (2.5–100) | 100 (93.6–100) | 100 (2.5–100) | 100 (93.7–100) | 1 |
| Thickening | ||||||
| CNN | 87.8 (78.7–93.9) | 74.1 (64.4–84.2) | 84.1 (74.7–91) | 86.6 (76.8–93.4) | 81 (74.7–87.2) | 0.859 (0.802–0.917) |
| Radiologist 1 | 81.7 (71.6–89.3) | 72.8 (61.8–82.1) | 76.1 (65.8–84.5) | 80 (69.1–88.3) | 77.3 (70.1–83.4) | 0.733 (0.698–0.847) |
| Radiologist 2 | 72.8 (61.8–82.1) | 79 (68.5–87.3) | 77.6 (66.6–86.4) | 74.7 (64.2–83.4) | 75.9 (68.6–82.2) | 0.755 (0.687–0.831) |
| Mass + Thickening | ||||||
| CNN | 94.6 (81.8–99.3) | – | 96.9 (84.2–99.9) | – | 94.6 (81.8–99.3) | – |
| Radiologist 1 | 94.4 (81.3–99.3) | – | 97.1 (84.6–99.9) | – | 94.4 (81.3–99.3) | – |
| Radiologist 2 | 97.1 (85.1–99.9) | – | 100 (90.5–100) | – | 97.1 (85.1–99.9) | |
| Polyp | ||||||
| CNN | 87.5 (47.3–99.6) | 75 (34.9–96.8) | 77.7 (39.9–97.1) | 85.7 (42.1–99.6) | 81.2 (54.3–95.9) | 0.779 (0.529–0.994) |
| Radiologist 1 | 85.7 (42.1–99.6) | 62.5 (24.5–91.5) | 80 (44.3–97.4) | 85.7 (42.1–99.6) | 73.3 (44.9–92.2) | 0.759 (0.497–0.994) |
| Radiologist 2 | 75 (34.9–96.8) | 75 (34.9–96.8) | 85.7 (42.1–99.6) | 77.7 (39.9–97.2) | 75 (47.6–92.7) | 0.753 (0.497–0.994) |
| Contracted | ||||||
| CNN | 93 (80.9–98.5) | 71.4 (55.1–89.3) | 78.7 (64.3–89.3) | 57.5 (39.2–74.5) | 84.5 (75.6–93) | 0.860 (0.768–0.952) |
| Radiologist 1 | 81.4 (66.6–91.6) | 75 (55.1–89.3) | 83.3 (68.6–93.0) | 72.4 (52.7–87.2) | 78.9 (67.5–87.6) | 0.794 (0.680–0.907) |
| Radiologist 2 | 77.3 (62.2–88.5) | 77.8 (57.7–91.3) | 82.5 (67.2–92.6) | 67.7 (48.6–83.3) | 77.5 (66–86.5) | 0.759 (0.640–0.877) |
| Size (<10 mm) | ||||||
| CNN | 89.8 (78.8–96.1) | 76.1 (68.6–87.9) | 79.6 (67.7–88.7) | 88.7 (79–95) | 81.7 (76.4–89.5) | 0.875 (0.832–0.949) |
| Radiologist 1 | 81 (68.5–90.1) | 76.6 (65.5–85.5) | 73.4 (60.9–83.7) | 84.5 (73.9–92) | 78.5 (70.6–85.1) | 0.782 (0.696–0.883) |
| Radiologist 2 | 77.1 (64.2–87.2) | 80.7 (70.2–88.8) | 75.8 (62.8–86.1) | 83.1 (72.8–90.1) | 79.3 (71.4–85.7) | 0.788 (0.707–0.870) |
| Size (>10 mm) | ||||||
| CNN | 84.8 (75.1–94.4) | 60 (39.1–80.7) | 95.8 (90.4–98.6) | 47.3 (24.4–71.1) | 79.1 (73.4–84.3) | 0.769 (0.661–0.916) |
| Radiologist 1 | 91.4 (85.7–96.1) | 42.9 (17.6–71.1) | 94.2 (88.4–97.6) | 41.1 (18.4–67.1) | 86.9 (80.2–92.1) | 0.678 (0.506–0.850) |
| Radiologist 2 | 91.2 (84.8–95.5) | 61.5 (31.6–86.1) | 96.6 (91.5–99.1) | 45 (23.1–68.4) | 88.4 (81.9–93.2) | 0.741 (0.579–0.904) |
| Site—body | ||||||
| CNN | 88.9 (51.7–99.7) | 71.4 (29.1–96.3) | 87.8 (71.8–96.6) | 72.7 (39.–93.9) | 81.2 (54.3–95.9) | 0.891 (0.690–1) |
| Radiologist 1 | 88.9 (51.7–99.7) | 85.7 (42.1–99.6) | 90.6 (74.9–98.0) | 66.6 (34.8–90.1) | 87.5 (61.6–98.4) | 0.810 (0.557–1) |
| Radiologist 2 | 77.8 (39.9–97.2) | 100 (59–100) | 100 (87.6–100) | 62.5 (35.4–84.8) | 87.5 (61.6–98.4) | 0.857 (0.631–1) |
| Site—fundus | ||||||
| CNN | 86.7 (57.2–98.2) | 100 (69.5–100) | 94.4 (72.7–99.8) | 60 (26.2–87.8) | 90.5 (79.1–93.3) | 0.981 (0.937–1) |
| Radiologist 1 | 80 (51.9–95.6) | 50 (11.8–88.2) | 86.3 (65.1–97.1) | 50 (11.8–88.2) | 71.4 (47.8–88.7) | 0.656 (0.373–0.921) |
| Radiologist 2 | 86.7 (59.5–98.3) | 66.7 (22.3–95.7) | 90 (68.3–98.7) | 50 (15.7–84.3) | 80.9 (58.1–94.5) | 0.922 (0.804–1) |
| Site—neck | ||||||
| CNN | 86.1 (78.5–94.2) | 66.7 (22.2–95.6) | 94.3 (84.3–98.8) | 53.8 (25.1–80.7) | 83.3 (74.5–91.7) | 0.810 (0.641–0.995) |
| Radiologist 1 | 91.6 (77.5–98.2) | 50 (11.8–88.2) | 90.7 (79.7–96.9) | 45.4 (16.7–76.6) | 85.7 (71.5–94.6) | 0.708 (0.447–0.969) |
| Radiologist 2 | 85.7 (69.7–95.2) | 57.1 (18.4–90.1) | 94.1 (83.7–98.7) | 50 (23–76.9) | 80.9 (65.9–91.4) | 0.680 (0.421–0.891) |
| Focal | ||||||
| CNN | 92.8 (86.7–96.6) | 72.7 (49.7–89.2) | 93.4 (86.9–97.3) | 60 (40.6–77.3) | 89.8 (83.7–94.2) | 0.844 (0.805–0.946) |
| Radiologist 1 | 91.4 (85.7–96.1) | 60.8 (38.5–80.3) | 92.6 (86.5–96.6) | 58.3 (36.6–77.8) | 87.1 (80.5–92.1) | 0.764 (0.639–0.889) |
| Radiologist 2 | 90.3 (83.7–94.9) | 78.3 (56.3–92.5) | 95.7 (90.3–98.6) | 60 (40.6–77.3) | 88.4 (82.1–93.1) | 0.839 (0.735–0.942) |
| Diffuse | ||||||
| CNN | 91.2 (80.4–97) | 75 (62.6–85.9) | 83.1 (71.7–91.2) | 90 (79.4–96.2) | 82.4 (75.3–89.2) | 0.889 (0.830–0.946) |
| Radiologist 1 | 80.7 (68.1–89.9) | 77.9 (66.2–87.1) | 75.4 (62.7–85.5) | 82.8 (71.3–91.1) | 79.2 (71–85.9) | 0.811 (0.731–0.890) |
| Radiologist 2 | 78.9 (66.1–88.6) | 80.9 (69.5–89.4) | 77.5 (64.7–87.4) | 82.1 (70.8–90.3) | 80 (71.9–86.6) | 0.799 (0.717–0.881) |
CNN: convolutional neural network, AUC: area under receiver operating characteristic curve, PPV: positive predictive value, NPV: negative predictive.
The AUCs of all the overall cohort are shown in the Supplementary Figure S3. The confusion matrix of CNN diagnosis is shown in Supplementary Figure S4. Class activation maps in Fig. 3 and Supplementary Figure S5 and Supplementary Figure S6.
Fig. 3.
Class activation maps for deep convolutional neural network (CNN) detection of gallbladder cancer. Arrows in each image point to the abnormality.
Discussion
In this prospective study evaluating DL-based GBC detection on US, we found that the performance of the DL-based approach was comparable to the experienced radiologists. Overall, the DL-based approach achieved high sensitivity, specificity, and AUC for detecting GBC. In different subgroups also, the DL-based approach achieved high sensitivity and AUC. These results suggest the potential of DL-assisted GBC detection in improving the diagnostic performance of non-experienced radiologists and impacting the prognosis of GBC.
GBC is prevalent in certain geographical regions of the world.2 Accurate GBC detection in imaging studies is challenging and delayed diagnosis of GBC is associated with poor prognosis.4,5 The DL-based approach is associated with improved diagnostic performance in several cancers.7, 8, 9, 10, 11, 12 However, there is limited literature on DL-based GBC detection in imaging.
Two recent studies evaluated the performance of DL-based characterisation of gallbladder polyps.26 In a transabdominal US-based study, 535 patients were divided into the development dataset (n = 437) and test dataset (n = 98). The polyps were classified into neoplastic and non-neoplastic by a CNN model, and the performance was compared with three radiologists. The DL-based approach showed sensitivity of 74.3% (95% CI, 56.7–87.5), specificity of 92.1% (95% CI, 82.4–97.4), accuracy of 85.7% (95% CI, 73.2–93.9), and AUC of 0.92 (95% CI, 0.87–0.95) for detection of neoplastic polyps. The AUC of the three reviewers was 0.94, 0.78, and 0.87. With the DL-assisted approach (combining CNN and radiologists' evaluation), the specificity (65.1–85.7 to 71.4–93.7), AUC (0.78–0.94 to 0.91–0.95), and intraclass correlation coefficient (0.87–0.93) for detection of neoplastic polyps improved. In another study, the DL-based classification model was developed on 1039 endoscopic US images.27 The model performance was tested in an external cohort of 83 patients. For the differential diagnosis of neoplastic and non-neoplastic GB polyps, the sensitivity and specificity of the DL model were 60.3% and 77.4%, respectively, compared to 74.2% and 44.9%, respectively, for the endoscopists. The accuracy of the DL model is 77.4% vs. 65.3% for endoscopists.
DL models' potential to detect GBC on US images has also been explored in a few recent studies. Basu et al. proposed state-of-the-art CNN model based on multiscale, second-order pooling architecture (GBCNet).13 The dataset comprised 1255 US images, of which 1133 were used for training and 122 for testing. The image level performance of the GBCNet was better than the two radiologists and other state-of-the-art image classification models.13 The sensitivity, specificity, and accuracy of the GBCNet to classify images as normal, benign, and malignant were 92.9%, 90%, and 87.7%, respectively, compared to 70.7–73.2%, 81.1–87.3%, and 68.3–70%, respectively for the radiologists. The accuracy of GBCNet for binary classification of images into malignant and non-malignant was 91% compared to 78.4–81.6% for radiologists. Recently, a transformer-based DL model (RadFormer) showed performance comparable to GBCNet with a faster test time inference.28 RadFormer also allowed human-readable explanations of the model decisions.28 The pretrained CNN-based models for detecting GBC in US images have been reported to be suboptimal in literature on public datasets.13,28 Our findings are consistent with that reported in the literature. The pretrained CNN models perform well in detecting non-malignant control gallbladder with high accuracy due to its regular shape and appearance. However, these models struggle to identify malignancy due to the high variability of appearance in malignant gallbladder lesions. Thus, we see a high specificity, but a low sensitivity for pretrained ResNet-50 and DenseNet-121 models.
Despite these recent promising technical works on DL-based GBC detection on US images, there is no published work demonstrating the performance of DL models in GBC detection in diverse clinical scenarios. We evaluated the DL-based patient-level GBC detection [unlike previous works where image level predictions were performed13,28] on US images in a large cohort recruited prospectively. We also assessed the performance of CNN in various clinically relevant subgroups, including distinct morphological subtypes, gallbladder lesions with stones, contracted gallbladders, lesions <10 mm, and distinct gallbladder sites. We found that CNN performed well in all the subgroups. In some subgroups, including mural thickening and polyp subtypes, contracted gallbladders, and gallbladder neck lesions, which pose a challenge for the radiologists, CNN performed better than the experienced radiologists (although the difference was not statistically significant in all these subgroups).
Our study had few limitations. First, although we tested the performance on a temporally independent held-out dataset, single-centre data was used. Second, the number of patients in some subgroups, e.g., polyps, was small. The relative paucity of polyps in our cohort is due to the geographical variations in the presentation of gallbladder lesions and the exclusion of polyps ≤5 mm. Third, we must assert that the US images were read by two academic radiologists with expertise in abdominal US. We expect these models to outperform the non-expert radiologists in detecting GBC. However, this needs to be confirmed in future studies. Finally, we did not explicitly evaluate the impact of CNN on early diagnosis and prognosis of GBC.
In conclusion, the DL-based approach demonstrated comparable or better diagnostic performance than expert radiologists in detecting GBC at US. However, multicentre studies are warranted to explore the potential of DL-based diagnosis of GBC fully.
Contributors
Conceptualisation: PanG.
Writing-original draft and project administration: PanG, SB.
Methodology, data curation, investigation, formal analysis, and writing-review and editing: PanG, SB, PR, UD, RS, DK, MC, SS, TDY, VG, LK, CD, ParG, UNS, RS, MSS, CA.
Visualisation, software, validation, resources, and supervision: PanG, SB, CA.
Data sharing statement
Restrictions apply to the availability of these data due to the patient privacy. Hence, the data are not publicly available. Deidentified data can be made available upon reasonable request to the corresponding author.
Declaration of interests
None.
Acknowledgements
None.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lansea.2023.100279.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Roa J.C., García P., Kapoor V.K., Maithel S.K., Javle M., Koshiol J. Gallbladder cancer. Nat Rev Dis Primers. 2022;8:69. doi: 10.1038/s41572-022-00398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misra S., Chaturvedi A., Misra N.C., Sharma I.D. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–176. doi: 10.1016/s1470-2045(03)01021-0. [DOI] [PubMed] [Google Scholar]
- 4.Hundal R., Shaffer E.A. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99–109. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta P., Dutta U., Rana P., et al. Gallbladder reporting and data system (GB-RADS) for risk stratification of gallbladder wall thickening on ultrasonography: an international expert consensus. Abdom Radiol (NY) 2022;47:554–565. doi: 10.1007/s00261-021-03360-w. [DOI] [PubMed] [Google Scholar]
- 6.Litjens G., Kooi T., Bejnordi B.E., et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Balkenende L., Teuwen J., Mann R.M. Application of deep learning in breast cancer imaging. Semin Nucl Med. 2022;52:584–596. doi: 10.1053/j.semnuclmed.2022.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Park C.M., Lee J.H. Deep learning for lung cancer nodal staging and real-world clinical practice. Radiology. 2022;302:212–213. doi: 10.1148/radiol.2021211981. [DOI] [PubMed] [Google Scholar]
- 9.Armato S.G., 3rd Deep learning demonstrates potential for lung cancer detection in chest radiography. Radiology. 2020;297:697–698. doi: 10.1148/radiol.2020203538. [DOI] [PubMed] [Google Scholar]
- 10.Le Berre C., Sandborn W.J., Aridhi S., et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020;158:76–94.e2. doi: 10.1053/j.gastro.2019.08.058. [DOI] [PubMed] [Google Scholar]
- 11.Liu K.L., Wu T., Chen P.T., et al. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: a retrospective study with cross-racial external validation. Lancet Digit Health. 2020;2:e303–e313. doi: 10.1016/S2589-7500(20)30078-9. [DOI] [PubMed] [Google Scholar]
- 12.Van Calster B., Timmerman S., Geysels A., Verbakel J.Y., Froyman W. A deep-learning-enabled diagnosis of ovarian cancer. Lancet Digit Health. 2022;4 doi: 10.1016/S2589-7500(22)00130-3. [DOI] [PubMed] [Google Scholar]
- 13.Basu S., Gupta M., Rana P., et al. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (CVPR) 2022. Surpassing the human accuracy: detecting gallbladder cancer from USG images with curriculum learning; pp. 20886–20896. [DOI] [Google Scholar]
- 14.Aggarwal R., Sounderajah V., Martin G., et al. Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. NPJ Digit Med. 2021;4:65. doi: 10.1038/s41746-021-00438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta P., Kumar M., Sharma V., et al. Evaluation of gallbladder wall thickening: a multimodality imaging approach. Expert Rev Gastroenterol Hepatol. 2020;14:463–473. doi: 10.1080/17474124.2020.1760840. [DOI] [PubMed] [Google Scholar]
- 16.Lopes Vendrami C., Magnetta M.J., Mittal P.K., et al. Gallbladder carcinoma and its differential diagnosis at MRI: what radiologists should know. Radiographics. 2021;41:78–95. doi: 10.1148/rg.2021200087. [DOI] [PubMed] [Google Scholar]
- 17.Kamaya A., Fung C., Szpakowski J.L., et al. Management of incidentally detected gallbladder polyps: society of radiologists in ultrasound consensus conference recommendations. Radiology. 2022;305:277–289. doi: 10.1148/radiol.213079. [DOI] [PubMed] [Google Scholar]
- 18.Kim J.H., Lee J.Y., Baek J.H., et al. High-resolution sonography for distinguishing neoplastic gallbladder polyps and staging gallbladder cancer. AJR Am J Roentgenol. 2015;204:W150–W159. doi: 10.2214/AJR.13.11992. [DOI] [PubMed] [Google Scholar]
- 19.Paszke A., Gross S., Massa F., et al. Proceedings of the 33rd international conference on neural information processing systems. 2019. PyTorch: an imperative style, high-performance deep learning library; pp. 8026–8037. [Google Scholar]
- 20.Glorot X., Bengio Y. Proceedings of the thirteenth international conference on artificial intelligence and statistics. JMLR; 2010. Understanding the difficulty of training deep feedforward neural networks; pp. 249–256. [Google Scholar]
- 21.MedCalc Software Ltd Diagnostic test evaluation calculator. https://www.medcalc.org/calc/diagnostic_test.php Version 20.210.
- 22.IBM Corp . IBM SPSS statistics for windows, version 22.0. IBM Corp.; Armonk, NY: 2013. [Google Scholar]
- 23.Virtanen P., Gommers R., Oliphant T.E., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Analyse-it software, Ltd. 2012. http://analyse-it.com/ [Google Scholar]
- 25.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong Y., Kim J.H., Chae H.D., et al. Deep learning-based decision support system for the diagnosis of neoplastic gallbladder polyps on ultrasonography: preliminary results. Sci Rep. 2020;10:7700. doi: 10.1038/s41598-020-64205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang S.I., Kim Y.J., Kim E.J., et al. Diagnostic performance of endoscopic ultrasound-artificial intelligence using deep learning analysis of gallbladder polypoid lesions. J Gastroenterol Hepatol. 2021;36(12):3548–3555. doi: 10.1111/jgh.15673. [DOI] [PubMed] [Google Scholar]
- 28.Basu S., Gupta M., Rana P., Gupta P., Arora C. RadFormer: transformers with global-local attention for interpretable and accurate Gallbladder Cancer detection. Med Image Anal. 2023;83 doi: 10.1016/j.media.2022.102676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.