Abstract
Mitochondria are dynamic organelles that respond to cellular stress through changes in global mass, interconnection, and subcellular location. As mitochondria play an important role in tumor development and progression, alterations in energy metabolism allow tumor cells to survive and spread even in challenging conditions. Alterations in mitochondrial bioenergetics have been recently proposed as a hallmark of cancer, and positive regulation of lipid metabolism constitutes one of the most common metabolic changes observed in tumor cells. Acyl-CoA synthetase 4 (ACSL4) is an enzyme catalyzing the activation of long chain polyunsaturated fatty acids with a strong substrate preference for arachidonic acid (AA). High ACSL4 expression has been related to aggressive cancer phenotypes, including breast cancer, and its overexpression has been shown to positively regulate the mammalian Target of Rapamycin (mTOR) pathway, involved in the regulation of mitochondrial metabolism genes. However, little is known about the role of ACSL4 in the regulation of mitochondrial function and metabolism in cancer cells. In this context, our objective was to study whether mitochondrial function and metabolism, processes usually altered in tumors, are modulated by ACSL4 in breast cancer cells. Using ACSL4 overexpression in MCF-7 cells, we demonstrate that this enzyme can increase the mRNA and protein levels of essential mitochondrial regulatory proteins such as nuclear respiratory factor 1 (NRF-1), voltage-dependent anion channel 1 (VDAC1) and respiratory chain Complex III. Furthermore, respiratory parameters analysis revealed an increase in oxygen consumption rate (OCR) and in spare respiratory capacity (SRC), among others. ACSL4 knockdown in MDA-MB-231 cells led to the decrease in OCR and in SCR, supporting the role of ACSL4 in the regulation of mitochondrial bioenergetics. Moreover, ACSL4 overexpression induced an increase in glycolytic function, in keeping with an increase in mitochondrial respiratory activity. Finally, there was a decrease in mitochondrial mass detected in cells that overexpressed ACSL4, while the knockdown of ACSL4 expression in MDA-MB-231 cells showed the opposite effect. Altogether, these results unveil the role of ACSL4 in mitochondrial function and metabolism and expand the knowledge of ACSL4 participation in pathological processes such as breast cancer.
Keywords: ACSL4, Acyl-CoA synthetase 4, Mitochondria, Glycolysis, Bioenergetics, Oxygen consumption rate, Breast cancer
1. Introduction
ACSL enzymes catalyze the production of acyl-CoA from long chain fatty acids. Five isoforms of ACSLs have been identified in humans and rodents; where acyl-CoA synthetase 4 (ACSL4) has a strong preference for polyunsaturated fatty acids, particularly arachidonic acid (AA). This substrate preference assumes that ACSL4 is an important enzyme for the control of free AA levels. ACSL4 is located in endoplasmic reticulum (ER), mitochondria-associated membranes and peroxisomes [1]. In addition to fulfilling physiological functions such as embryonic and nervous system development, cell death and steroidogenesis [[2], [3], [4]]. ACSL4 expression increases in cancer cells and thus promotes a more aggressive phenotype [[5], [6], [7], [8]]. Indeed, ACSL4 expression has been shown to promote elevated breast cancer cell proliferation, invasion and migration both in vitro and in vivo [[9], [10], [11]].
Our group, using a stable line of MCF-7 cells that overexpress ACSL4 under the control of tetracycline or doxycycline (MCF-7 tet-off/ACSL4), has previously demonstrated that ACSL4 overexpression can single-handedly produce tumorigenic MCF-7 cells. We have further shown that downregulation of ACSL4 by tetracycline treatment reduces MCF-7 cell aggressiveness and ability to progress to mammary tumors in vivo [9]. These results support our previous in vitro observations in which tetracycline treatment resulted in the inhibition of MCF-7 tet-off/ACSL4 cell proliferation and migration [5]. Moreover, it is known that ACSL4 negatively regulates estrogen receptor alpha (ERα) expression, leading to greater tumor progression and aggressiveness, thus generating insensitivity to hormonal therapy [10]. It has been shown that ACSL4 is involved in resistance to chemotherapy treatment in breast cancer, demonstrating that its expression increases the levels of ATP Binding Cassette transporters and consequently, the efflux of drugs, causing the ineffectiveness of chemotherapy in breast tumor lines that overexpress ACSL4 [7]. Through RNA-seq and functional proteomic analysis, we reported that ACSL4 regulates a broad spectrum of signaling pathways [10,12]. Among them, mTOR pathway activation is one of the main specific signatures of ACSL4 expression [10]. In this context, several proteins involved in mTOR cascade have been considered as putative targets for treatment of different types of tumors [13]. It is known that mTOR acts as a multichannel processor in a cellular nutrient-sensing network that constantly receives multiple inputs from distinct environmental signals and derives different outputs to appropriate downstream effectors. Furthermore, several studies demonstrated that mTOR modulates mitochondrial functions such as activity and biogenesis [14].
In regard of tumorigenesis and chemotherapy resistance, mitochondria play an important role in tumor development and progression and alterations in energy metabolism allow tumor cells to survive and spread even in challenging conditions [15,16].
Alterations in cellular bioenergetics have been recently proposed as a hallmark of cancer [17]. Through electron transport chain (ETC), normal cells obtain energy through cytosolic glycolysis followed by mitochondrial oxidative phosphorylation (OXPHOS) in aerobic conditions. Instead, cancer cells use glycolysis in the cytosol even in the presence of oxygen (aerobic glycolysis) with lactate production, a phenomenon first observed by Otto Warburg [18] and now known as the Warburg effect. Since the discovery of this effect and for many years, the switch to aerobic glycolysis in cancer cells was thought to correlate with a complete shutdown of mitochondrial OXPHOS leading to an increase in mitochondrial dysfunction [[19], [20], [21]]. However, recent studies have shown that mitochondrial OXPHOS is unaltered in most types of cancer [[22], [23], [24], [25]]. Aerobic glycolysis in many types of cancer results from changes in the oncogenes/tumor suppressor genes ratio, the tumor microenvironment, alterations in DNA and mitochondrial mass, and the tissue of origin, among other factors [26,27]. Even though the efficiency of ATP production per molecule of glucose is much lower through glycolysis, yield rates are much faster than those of OXPHOS and thus meet the demand for rapid cancer cell growth and proliferation. The reprogramming of glucose metabolism has been validated in many tumors, while an increase in glycolysis facilitates biomass biosynthesis (e.g., nucleotides, amino acids, and lipids) by providing glycolytic intermediates as feedstock [17,27,28].
Therefore, considering the role of ACSL4 in tumor aggressiveness and the signal transduction pathways that this enzyme regulates, the aim of this work was to establish whether mitochondrial function and metabolism, usually altered in tumors, are modulated by ACSL4 in breast cancer cells, and thus expand the knowledge of the role of this enzyme in pathological processes.
2. Materials and methods
2.1. Materials
Penicillin-streptomycin, trypsin solution ethylene diamine tetra-acetic acid (EDTA), and Dulbecco's modified Eagle's medium (DMEM) were provided by Gibco-Life Technologies Inc. (Gaithersburg, MD, USA). Fetal bovine serum (FBS) was from PAA laboratories GmbH (Pasching, Austria). Bovine serum albumin (BSA), molecular weight standards for electrophoresis, EDTA, hydroxymethyl aminomethane (Tris), ethylene glycol-bis (2 aminoethyl ether) –N, N, N ′, N′ tetraacetic acid (EGTA), leupeptin, pepstatin A, sodium dodecyl sulfate (SDS), glycine, Tetramethylethylenediamine (TEMED), and poly-l-lysine were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Non-protic detergent NP-40 (Tergitol type NP40) was obtained from Sigma Aldrich-Merck (Darmstadt, Germany). PCR primers were purchased from ThermoFisher Scientific (Waltham, MA, USA), Macrogen Inc. (Seoul, South Korea) and Integrated DNA Technologies (Coralville, IA, USA). MitoTracker Red 580 and MitoTracker Green were obtained from Invitrogen (Molecular Probes, Eugene, OR, USA). The monoclonal anti-β-tubulin antibody was purchased from Upstate Biotechnology (Lake Placid, NY, USA). Monoclonal antibodies anti-UQCRC2 OxPhos Complex III core 2 subunit (Complex III) (RRID:AB_2213640) was obtained from Abcam (Cambridge, UK), anti-succinate dehydrogenase (SDHA) was obtained from Cell Signaling Technologies (Boston, MA, USA) (RRID:AB_2750900) while anti-voltage-dependent anion-selective channel 1 (VDAC1) (RRID:AB_2750920), anti-nuclear respiratory factor 1 (NRF-1) (RRID:AB_1126766), anti-proliferating cell nuclear antigen (PCNA) (RRID:AB_628109), anti-translocator of the outer mitochondrial membrane (TOM20) (RRID:AB_628381) and anti-ACSL4 (RRID:AB_10843105) were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Peroxidase-conjugated goat-raised mouse anti-IgG secondary antibody (RRID:AB_11125547) was purchased from BioRad Laboratories (Hercules, CA, USA). PVDF membranes, reagents (acrylamide, bisacrylamide, Bradford and others) and electrophoresis equipment (Mini-Protean II and transfer kits) were provided by BioRad Laboratories. Enhanced chemiluminescence was from GE Healthcare (Buckinghamshire, UK). Sterile and plastic material for tissue culture was from Orange Scientific (Brainel'Alleud, Belgium). All other reagents were of the highest grade available.
2.2. Cell culture and treatments
Human breast cancer cell lines MDA-MB-231 and MCF-7 were generously provided by Dr. Vasilios Papadopoulos (School of Pharmacy, University of Southern California, Los Angeles, CA, USA) or obtained from the Lombardi Comprehensive Cancer Center (Georgetown University Medical Center, Washington D.C., USA) and validated by ATCC Cell Line Authentication Service. Both cell lines were cultured in DMEM with glucose (4500 mg/L) supplemented with 10 % heat-inactivated FBS plus 100 U/ml penicillin and 10 μg/ml streptomycin (complete DMEM) and maintained in a 5 % CO2 humidified atmosphere at 37 °C. The absence of mycoplasma contamination was tested routinely by polymerase chain reaction (PCR) assay.
2.3. Modified human breast cancer cell lines
2.3.1. MCF-7 tet-off control and MCF-7 tet-off/ACSL4
The tetracycline-repressible MCF-7 cell line, named MCF-7 tet-off, was used for the stable transfection of ACSL4 cDNA under the control of the tetracycline response element using the tet-off gene expression system (Clontech Laboratories, Inc, Mountain View, CA, USA). This line was developed in our laboratory prior to this work [9]. Tetracycline repression was not used in this work because this drug could affect certain mitochondrial parameters [[29], [30], [31]]. Western blot was carried out to confirm ACSL4 overexpression (Supplementary Fig. 1A).
2.3.2. MDA-MB-231 shControl and MDA-MB-231 shACSL4
The MDA-MB-231 cell line was used for stable transfection with pSUPER.retro plasmid (OligoEngine, Seattle, WA, USA) containing shRNA ACSL4 (5′AAGATTATTCTGTGGATGA-3′) or the empty vectors as control in Opti-MEM medium and Lipofectamine 2000 reagent (Invitrogen) as previously described by for our group [5]. ACSL4 knockdown was evaluated through Western blot (Supplementary Fig. 1B).
2.4. Bioinformatics analysis
2.4.1. Ingenuity pathway analysis
To identify biological functions and signaling pathways affected by differentially expressed genes in MCF-7 cells, further analysis was carried out on previously reported RNA-seq data using Ingenuity Pathways Analysis (IPA; Ingenuity Systems, Inc, Little Elm, TX, USA) [10,12]. IPA is the largest raw database and analysis system available for the study of metabolic and signaling pathways, molecular networks, and biological processes which are most significantly modified in a dataset of interest. Using this continuously updated database, the ranking and importance of biofunctions and canonical pathways were tested using p-values (<0.001). Furthermore, the canonical pathways were ordered by the ratio: number of genes from the input dataset that map to the pathway divided by the total number of molecules that exist in the canonical pathway.
2.4.2. Pathway, process and protein-protein interaction enrichment analysis
Metascape software [32] was used to analyze and visualize the canonical pathways and protein-protein interaction in which 299 mitochondrial genes affected by ACSL4 expression participate. Through this analysis, statistically enriched terms (GO/KEGG terms, canonical pathways, hallmark gene sets, etc.) were identified. Terms with a p-value <0.01, a minimum count of 3, and an enrichment factor >1.5 (the enrichment factor is the ratio between the observed counts and the counts expected by chance) were collected and grouped into clusters based on their membership similarities. Cumulative hypergeometric p-values and enrichment factors were calculated and used for analysis. Significant terms were grouped hierarchically in a tree based on statistical kappa similarities across genetic members. A threshold score of 0.3 kappa was then applied to convert the tree into groups of terms. Next, a representative set of terms was selected from this group and a network design was constructed. Protein-protein interaction enrichment analysis was carried out with the following databases: STRING, BioGrid, OmniPath, InWeb_IM. Only physical interactions in STRING (physical score >0.132) and BioGrid were used. The resultant network contained the subset of proteins that form physical interactions with at least one other member in the list. The Molecular Complex Detection (MCODE) algorithm10 was applied to identify densely connected network components if networks contained between 3 and 500 proteins. Pathway and process enrichment analysis was applied to each MCODE component independently, and the three most interesting terms were retained as the functional description of the components, shown in corresponding network plots (Supplementary Fig. 2). The network was visualized with Cytoscape v3.9.11.2 software.
2.5. Isolation of the mitochondrial fraction
Mitochondrial protein analysis was carried out using mitochondria isolation through differential centrifugation. Cells were washed with PBS and harvested in a buffer solution containing 10 mM Tris-HCl pH 7.4, 250 mM sucrose, 0.1 mM EDTA (TSE). Cells were then homogenized with a manual homogenizer (Kontes Pellet Pestle Motor Homogenizer) and centrifuged at 1,000×g for 15 min to remove nuclei and cell debris. The supernatant obtained was centrifuged at 16,000×g for 15 min to obtain the mitochondrial fraction (pellet). The mitochondrial fraction was washed once with TSE buffer, centrifuged at 16,000×g for 15 min, and then resuspended in the same buffer [33].
2.6. Isolation of nuclear extracts
Cells were grown in 6-well plates and cultured at 37 °C and 5 % CO2 prior to nuclear protein extraction. Cells were scraped from wells into 500 μl ice-cold PBS and recovered by centrifugation for 5 min at 13,000×g. The supernatant was discarded before cells were resuspended in 400 μl low salt buffer [10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 μg/ml leupeptin, 0.5 μg/ml aprotinin pH 7.9] and incubated on ice for 15 min. Twenty-five microliters 10 % NP-40 (v/v) was then added followed by brief vortexing. Cytosolic extracts were collected after centrifugation. The nuclear pellet was then extracted in 50 μl high salt buffer [20 mM HEPES, 25 % glycerol (v/v), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 0.5 mg/ml leupeptin, 0.5 mg/ml aprotinin pH 7.9] at 4 °C with shaking for 15 min, followed by disruption of nuclei by resuspension in a tuberculin syringe for 30 s. Non-soluble fractions were removed by centrifugation at 13,000×g for 15 min at 4 °C [34].
2.7. Western blot
Mitochondrial and nuclear proteins were separated on 12 % or 10 % SDS-PAGE and electrotransferred to PVDF membranes (Bio-Rad Laboratories) as previously described [5]. The proteins transferred were visualized using a Ponceau S staining solution containing 0.2 % Ponceau S in 1 % acetic acid. Membranes were then incubated with 1 % BSA in 500 mM NaCl, 20 mM Tris-HCl pH 7.5, and 0.5 % Tween 20 (T-TBS) for 2 h at room temperature, with gentle shaking. The membranes were then rinsed twice with T-TBS and incubated overnight at 4 °C with 1:2000 dilutions of primary antibodies mouse monoclonal anti-ACSL4, anti-PCNA, anti- SDHA, anti-NRF-1, anti-VDAC1 and anti-Complex III. Bound antibodies were visualized by incubation with 1:5000 horseradish peroxidase-conjugated secondary goat anti-mouse or anti-rabbit antibodies and detected by chemiluminescence (BioLumina, Kalium Tech, Buenos Aires, Argentina). The immunoblots were then quantified using Gel Pro Analyzer software.
2.8. RNA extraction and real-time PCR
For real-time qPCR total RNA was isolated using Tri Reagent following the manufacturer's instructions (Molecular Research Center Inc., Cincinnati, OH, USA). Extracted RNA was deoxyribonuclease-treated using RNAse-free DNase RQ1 (Promega, Madison, WI, USA). Reverse transcription was performed using total RNA (2 μg) and M-MLV Reverse Transcriptase (Promega). The expression of NRF-1 (forward primer: 5′-GTACAAGAGCATGATCCTGGA-3′, reverse primer: 5′-GCTCTTCTGTGCGGACATC-3′), NRF-2 (forward primer: 5′-CAGCGACGGAAAGAGTATGA-3′, reverse primer: 5′-TGGGCAACCTGGGAGTAG-3′), uncoupling protein 2 (UCP2) (forward primer: 5′-TGGTCGGAGATACCAAAGCAC-3′, reverse primer: 5′-GCTCAGCACAGTTGACAATGGC-3′) and adenine nucleotide translocase (ANT1) (forward primer: 5′-TCAACGTCTCTGTCCAAGGC-3′, reverse primer: 5′-GTCAACTGTCCCCGTGTACA-3′) were assessed by real time qPCR (Mic PCR instrument, Molecular Biosystems, San Diego, CA, USA) using the SYBR Green Master Mix reagent kit (Applied Biosystems, Carlsbad, CA, USA). The reaction conditions were: 5-min in one cycle at 95 °C, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Gene mRNA expression levels were normalized to human cyclophilin (forward primer: 5′-TGGCAAGTCCATCTATGGGGA-3′, reverse primer: 5′-ACTTATTCGAGTTGTCCAACAGTCAGCA-3′). Real-time PCR data were analyzed by calculating the 2−ΔΔCt value (comparative Ct method) for each experimental sample (relative gene expression).
2.9. MitoTracker red staining
MitoTracker Red 580 (MTR) dye (Invitrogen) was used to assess mitochondrial activity through fluorescence microscopy. This compound accumulates in mitochondria with active respiration depending on mitochondrial membrane potential (ΔΨm). MCF-7 tet-off control, MCF-7 tet-off/ACSL4, MDA-MB-231 shControl and MDA-MB-231 shACSL4 cells were seeded on 24-well plates on coverslips previously treated with poly-l-lysine for 24 h. Cells were then incubated in serum-free medium with MTR at a final concentration of 150 nM for 45 min at 37 °C in the dark. After incubation, MTR was removed from each well, and wells were washed with PBS. Cells were then fixed with a 4 % paraformaldehyde (PFA) and 5 % sucrose solution for 10 min at room temperature and processed for immunofluorescence. Samples were analyzed with a Nikon E200 epifluorescence microscope. Images were obtained at equal exposure time and subsequently analyzed with FIJI software. MTR signal was measured as integrated fluorescence density per number of cells.
2.10. MitoTracker green staining
Mitochondrial mass was determined using MitoTracker Green FM (MTG) staining (Invitrogen), which localizes to mitochondria regardless of ΔΨm. MTG was used in suspension, as it is not well retained after aldehyde fixation. MCF-7 tet-off control, MCF-7 tet-off/ACSL4, MDA-MB-231 shControl and MDA-MB-231 shACSL4 cells were seeded in T25 flasks until confluence. After trypsinization, cells were resuspended in serum-free medium. An aliquot was removed for cell counting using a Countes II FL automated counter (Life Technologies). Cells in suspension were incubated with MTG (150 nM) for 30 min at 37 °C in the dark, centrifuged at 5,000×g for 5 min, and washed with PBS. Then, 1 × 106 cells were resuspended in PBS and analyzed by flow cytometry using a FACS Aria II (INIGEM-Hospital de Clínicas “José de San Martín'', Facultad de Medicina, Universidad de Buenos Aires) to measure the level of MTG incorporated. Propidium iodide treatment was used to discard the dead cell population from the MTG staining analysis. The results obtained were analyzed using Cyflogic software v.1.2.1.
2.11. Immunofluorescence analysis
TOM20 expression was analyzed by immunofluorescence. Cells were grown to approximately 80 % confluence on 24-well plates on poly-l-lysine-pre-treated coverslips. Coverslips were then removed, and cells were washed with PBS buffer. Cells were fixed with a 4 % PFA and 5 % sucrose solution, pH 7.3, for 10 min at room temperature and then washed three times with PBS for 10 min each. Samples were permeabilized for 10 min at room temperature with a 0.1 % Triton X-100, 1 % BSA in PBS solution. To block nonspecific antibody binding, coverslips were incubated with blocking solution (1 % BSA and 0.05 % Tween 20 in PBS) for 2 h in a humid chamber at room temperature. Cells were then incubated overnight with anti-TOM20 (1:200) monoclonal primary antibody in a humid chamber at 4 °C. After several washes with 0.05 % Tween 20-PBS, cells were incubated for 1 h at room temperature with an anti-mouse antibody (1:500) conjugated to cy2 (green) or cy3 (red). Before mounting, cells were incubated with DAPI (1:7000) for 10 min at room temperature. Finally, coverslips were placed on the slides with DAKO Fluorescence mounting medium (DAKO, Carpinteria, CA, USA) and samples were analyzed with a Nikon E200 epifluorescence microscope. Controls omitting the primary antibody were included in all trials. Images were obtained at equal exposure time and subsequently analyzed with FIJI software.
2.12. Bioenergetics analysis
A Seahorse XFe8 analyzer (Agilent Technologies, Santa Clara, CA, USA) was used to assess the bioenergetic phenotypes of MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells and MDA-MB-231 shControl and shACSL4 cells. The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured through Mito Stress and Glycolysis Stress Tests, respectively. Briefly, in both tests, cells were seeded at a density of 3 × 104 cells per well in a XFe8 plate for 24 h before measurement in a XFe8 V7 cell culture microplate (Seahorse Bioscience, North Billerica, MA, USA) in DMEM complete medium. After 24 h incubation at 37 °C in a 5 % CO2 atmosphere, the medium was replaced with 600 μL of XF assay media (3.5 mM KCl, 120 mM NaCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1.2 mM Na2SO4, 2 mM MgSO4, 15 mM d-glucose with or without 10 mM pyruvate, 4 mg/ml BSA, pH 7.4). Plates were kept at 37 °C for 1 h in a non-CO2 incubator and loaded onto the Seahorse XFe8 extracellular flow analyzer following the manufacturer's instructions. All experiments were carried out at 37 °C.
In the Mito Stress Test, OCR was analyzed as a measure of mitochondrial respiratory function. Before the test, a dose response curve (0.25, 0.5 and 1 μM) was constructed to establish the concentration of carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) needed to achieve maximal OCR. OCR and baseline were measured at the beginning of the assay, followed by the sequential addition of oligomycin, FCCP, and Rotenone/Antimycin A (Rot/AA) combination, the concentrations used of these compounds differs between cell lines, being of 2.5 μM oligomycin, 0.5 μM FCCP, and a 0.5 μM Rot/AA for MCF-7 tet-off cells and of 1.5 μM oligomycin, 1 μM FCCP, and a 0.5 μM Rot/AA for MDA-MB-231 silenced cell line. OCR was measured as picomole O2 per minute (pmol O2/min). Respiratory parameters like basal, maximal and ATP-linked respiration, proton leak, efficiency coupling, and spare respiratory capacity were calculated on the basis of OCR data obtained in the Mito Stress Tests using Seahorse Wave software for XF analyzers. In the Glycolysis Stress Test, the ECAR was evaluated as a measure of glycolytic function. For ECAR measurements, the base assay medium was supplemented with 2 mM l-glutamine and the pH was adjusted to 7.4. Cells were pre-incubated for 1 h without CO2 before the assay, and sequential injections were performed with 10 mM glucose, 1.5 μM oligomycin, and 50 mM 2-deoxy-d-glucose (2-DG). Glycolytic parameters such as glycolysis, glycolytic capacity, and non-glycolytic acidification were calculated as indicated by the manufacturer using Seahorse Wave software. All Seahorse measurements were normalized to seeded cells or total protein. The experiments were performed at least three times independently.
2.13. Statistical analysis
Statistical significance was determined using 2-way student's t-test, unless otherwise indicated. In all statistical analyses, significance was set at p < 0.05, with * p < 0.05, **p < 0.01 and ***p < 0.001 vs control. Data analysis was performed using GraphPad Prism 8 software.
3. Results
3.1. Bioinformatics analysis of gene expression profiles in MCF-7 breast cancer cells
In order to analyze the effect of ACSL4 expression on mitochondrial function, we conducted bioinformatics analysis of gene expression profiles previously obtained from MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells [10,12]. IPA analysis was carried out using RNA-seq data previously obtained. This analysis revealed several canonical pathways which may be regulated by ACSL4 in connection with mitochondrial processes, such as OXPHOS, mitochondrial dysfunction, and mTOR (Table 1).
Table 1.
Canonical signaling pathways affected by ACSL4 overexpression in MCF-7 breast cancer cells.
| Ingenuity Canonical Pathways |
-log (p value) | Affected genes |
|---|---|---|
| OXPHOS | 2.31E01 | ATP5F1A,ATP5F1B,ATP5F1C,ATP5F1D,ATP5F1E,ATP5MC1,ATP5MC2,ATP5MC3,ATP5MF,ATP5PB,ATP5PD,ATP5PF,ATPAF1,COX11,COX15,COX17,COX4I1,COX5A,COX5B,COX6A1,COX7A2L,COX7B,COX7B2,COX7C,CYC1,MT-CO1,MT-CO2,MT-CYB,MT-ND1,MT-ND2,MT-ND3,MT-ND4,MTND5,NDUFA1,NDUFA2,NDUFA4,NDUFA5,NDUFA6,NDUFA7,NDUFA8,NDUFB11,NDUFB3,NDUFB4,NDUFB6,NDUFB7,NDUFB8,NDUFB9,NDUFS2,NDUFS5,NDUFS6,NDUFS,NDUFV2,SDHA,SDHB,SURF1,UQCR10,UQCR11,UQCRB,UQCRC1,UQCRH,UQCRQ |
| Mitochondrial dysfunction | 2.28E01 | ACO2,AIFM1,APH1A,ATP5F1A,ATP5F1B,ATP5F1C,ATP5F1D,ATP5F1E,ATP5MC1,ATP5MC2,ATP5MC3,ATP5MF,ATP5PB,ATP5PD,ATP5PF,ATPAF1,COX11,COX15,COX17,COX4I1,COX5A,COX5B,COX6A1,COX7A2L,COX7B,COX7B2,COX7C,CYB5R3,CYC1,FURIN,GLRX2,GPX4,HSD17B10,HTRA2,MAPK8,MT-CO1,MT-CO2,MT-CYB,MT-ND1,MT-ND2,MT-ND3,MT-ND4,MT-ND5,NCSTN,NDUFA1,NDUFA2,NDUFA4,NDUFA5,NDUFA6,NDUFA7,NDUFA8,NDUFB11,NDUFB3,NDUFB4,NDUFB6,NDUFB7,NDUFB8,NDUFB9,NDUFS2,NDUFS5,NDUFS6,NDUFS7,NDUFV2,PARK7,PDHA1,PRDX3,PSEN1,SDHA,SDHB,SDHC,SURF1,TXN2,UCP2,UQCR10,UQCR11,UQCRB,UQCRC1,UQCRH,UQCRQ,VDAC1,VDAC3 |
| mTOR | 1.56E01 | AKT1,AKT1S1,AKT2,ARHGAP8/PRR5,ARHGAP8,ATG13,CDC42,DDIT4,DGKZ,EIF3A,EIF3D,EIF3E,EIF3F,EIF3G,EIF3H,EIF3I,EIF3J,EIF3L,EIF3M,EIF4A1,EIF4A2,EIF4A3,EIF4B,EIF4EBP1,EIF4G3,FKBP1A,HRAS,IRS1,MAPK1,MAPKAP1,PIK3C2A,PIK3R2,PPP2R2A,PRKAA1,PRKAB1,PRKCD,PTPA,RAC3,RALB,RAP1B,RASD1,RHEB,RHOA,RHOB,RHOF,RHOQ,RND1,RPS10,RPS11,RPS12,RPS14,RPS15,RPS16,RPS17,RPS18,RPS19,RPS2,RPS20,RPS21,RPS23,RPS24,RPS25,RPS26,RPS27,RPS27A,RPS28,RPS3,RPS3A,RPS4X,RPS5,RPS6,RPS6KA1,RPS6KB1,RPS7,RPS8,RPS9,RPSA,STK11,TSC2,ULK1,VEGF |
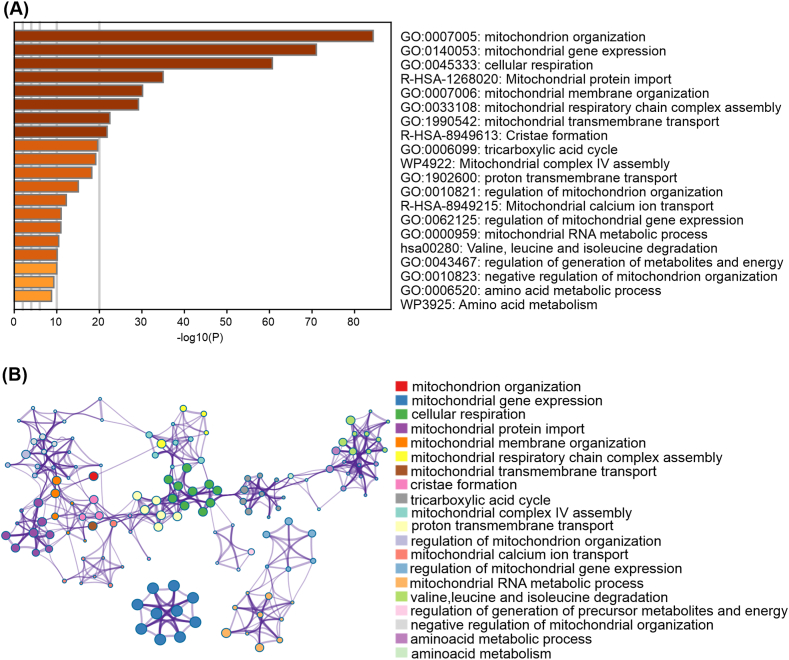
Fig. 1A shows pathways mostly affected by ACSL4 including the respiratory transport chain, mitochondrial organization and proton transmembrane transport. Nodes representing enriched clusters of genes affected by ACSL4 overexpression in MCF-7 cells after Metascape analysis (Fig. 1B). An extended analysis of protein-protein interactions enrichment in mitochondrial processes is depicted in Supplementary material (Supplementary Fig. 2). These findings support a role for ACSL4 in mitochondrial function and metabolism.
Fig. 1.
Enriched ontology clusters. (A) Bar graph of enriched terms in the mitochondrial RNA-seq gene list, colored according to p-values. (B) Visualization of results in the form of a network. Each term is represented by a circular node whose size is proportional to the number of input genes and whose color represents group membership (nodes of the same color belong to the same group). Terms with a similarity score >0.3 are linked by a border (border thickness represents the similarity score). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. ACSL4 overexpression up-regulates mitochondrial proteins involved in mitochondrial metabolism and function in MCF-7 breast cancer cells
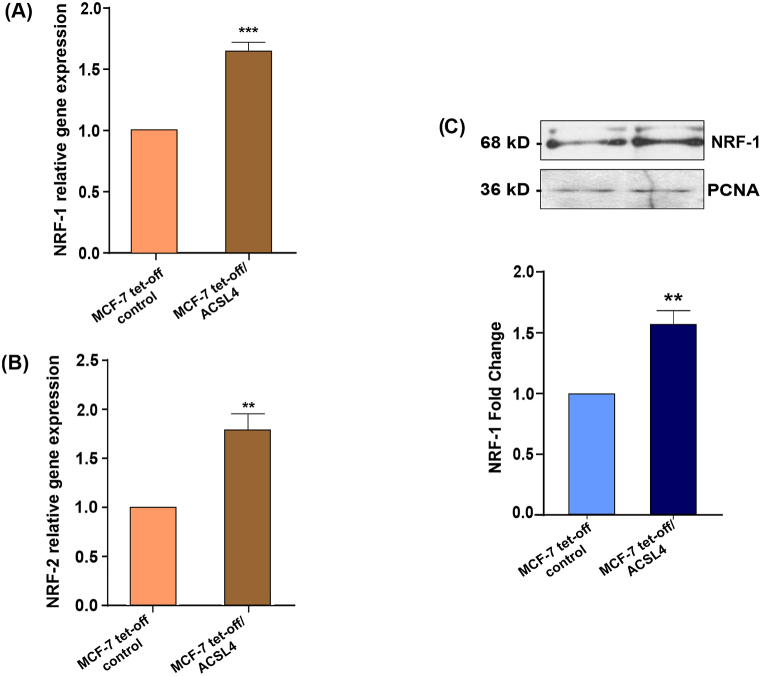
The upstream regulators analysis (that predict whether a transcription factor is activated or inhibited, given the observed gene expression changes in our experimental dataset) predicted that NRF-1 was activated (z score 3.074, p value 0.00388), suggesting that it could be positively modulated by ACSL4 overexpression. NRF-1 and NRF-2 are transcription factors widely known to take part in the regulation of proteins associated with mitochondrial biogenesis and antioxidant defenses [35,36]. For these reasons, we next studied NRF-1 and NRF-2 mRNA expression by real-time qPCR. Results showed that ACSL4 overexpression significantly increased NRF-1 and NRF-2 mRNA expression in MCF-7 cells (Fig. 2A and B). Furthermore, we analyzed NRF-1 protein levels from nuclear extracts by Western blot. Our results demonstrated that NRF-1 levels were significantly higher in MCF-7 tet-off/ACSL4 than in MCF-7 tet-off control cells (Fig. 2C).
Fig. 2.
ACSL4 overexpression increases the expression of NRF-1 and NRF-2 in MCF-7 breast cancer cells. NRF-1 and NRF-2 expression was assessed through RT-qPCR and Western blot in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. (A)NRF-1 and (B) NRF-2 mRNA expression levels. mRNA relative expression was calculated as the 2−ΔΔCt value (comparative Ct method) for each experimental sample (relative gene expression) (**p < 0.01, ***p < 0.001). (C) Representative Western blot and quantification of NRF-1 protein levels. NRF-1 levels were normalized to proliferating cell nuclear antigen (PCNA) levels. Optical density for each band was obtained, and control sample intensity was arbitrarily defined as 1. Proteins levels were relativized to respective controls. Results are expressed as the mean arbitrary units ± SEM of at least three independent experiments (**p <0.01). The full, non-adjusted blot image is included in Supplementary Material (Supplementary Fig. 3).
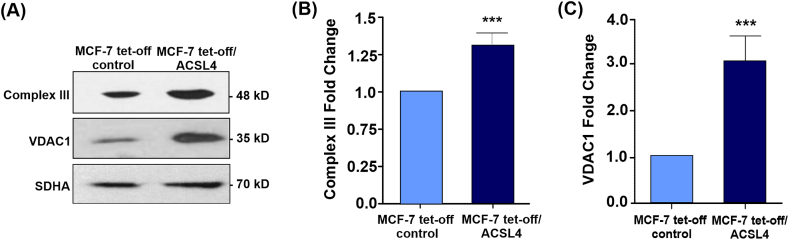
Next, Western blot analyses from mitochondrial extracts were carried out on the protein levels of some genes involved in mitochondrial function such as the UQCRQ2 subunit of complex III (Complex III) of the ETC and VDAC1 (Fig. 3A). Mitochondrial Complex III accepts electrons both from complex I and complex II through ubiquinone and increased activity of this complex has been reported in cancer [[37], [38], [39]]. In turn, VDAC1 is the most extensively characterized and abundant isoform of the VDAC protein family, and it has been implicated in pathologies such as cancer or neurodegenerative diseases [[40], [41], [42]]. Our results showed a significant increase in Complex III (Fig. 3B) and VDAC1 protein levels (Fig. 3C) in ACSL4-overexpressing cells. In agreement with bioinformatics analysis, these findings indicate that ACSL4 may modulate genes related to mitochondrial functions.
Fig. 3.
ACSL4 overexpression up-regulates mitochondrial proteins involved in mitochondrial metabolism and function in MCF-7 breast cancer cells. The mitochondrial fraction of MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells was analyzed through Western blot. (A) Representative Western blot of Complex III and VDAC1 protein levels. (B) Quantitative representation of three independent blots of Complex III. (C) Quantitative representation of three independent blots of VDAC1. Protein levels were normalized to SDHA levels. Optical density for each band was obtained, and control sample intensity was arbitrarily defined as 1. Protein levels were relativized to respective controls. Results are expressed as the mean arbitrary units ± SEM of at least three independent experiments (***p <0.001). The full, non-adjusted blots images are included in Supplementary Material (Supplementary Fig. 3).
3.3. ACSL4 overexpression stimulates mitochondrial active respiration in MCF-7 breast cancer cells
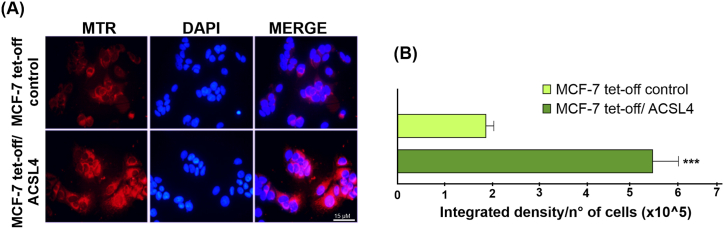
ΔΨm is essential for the maintenance of cellular health and viability and the driving force for ATP production. Although optimal ΔΨm values cannot be conclusively determined for cells and mitochondria [43] they are known to correlate with active mitochondrial respiration. Since MTR is used to study the presence of active mitochondria depending on ΔΨm [[44], [45], [46]] and based on our obtained data showing that ACSL4 promotes an increase in relevant mitochondrial proteins, we next used MTR staining to elucidate whether ACSL4 favors the presence of healthier and more active mitochondria in MCF-7 cells. Results showed a higher MTR signal intensity in MCF-7 tet-off/ACSL4 than in MCF-7 tet-off control cells (Fig. 4A), which is also reflected in the quantification of MTR signal intensity per number of cells (Fig. 4B). This result suggests healthier and more viable mitochondria in ACSL4-overexpressing cells.
Fig. 4.
ACSL4 overexpression favors healthier mitochondria with active respiration in MCF-7 breast cancer cells. Mitochondrial respiratory activity and health evaluated in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. (A) Fluorescence microscopy images of mitochondria stained with MTR (red) and nuclei stained with DAPI (blue). Images were obtained at equal exposure time and intensity. (B) Quantification of integrated MTR signal as reference of mitochondrial activity. Results are expressed as the mean integrated signal relative to number of cells ± SEM of at least three independent experiments (***p < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
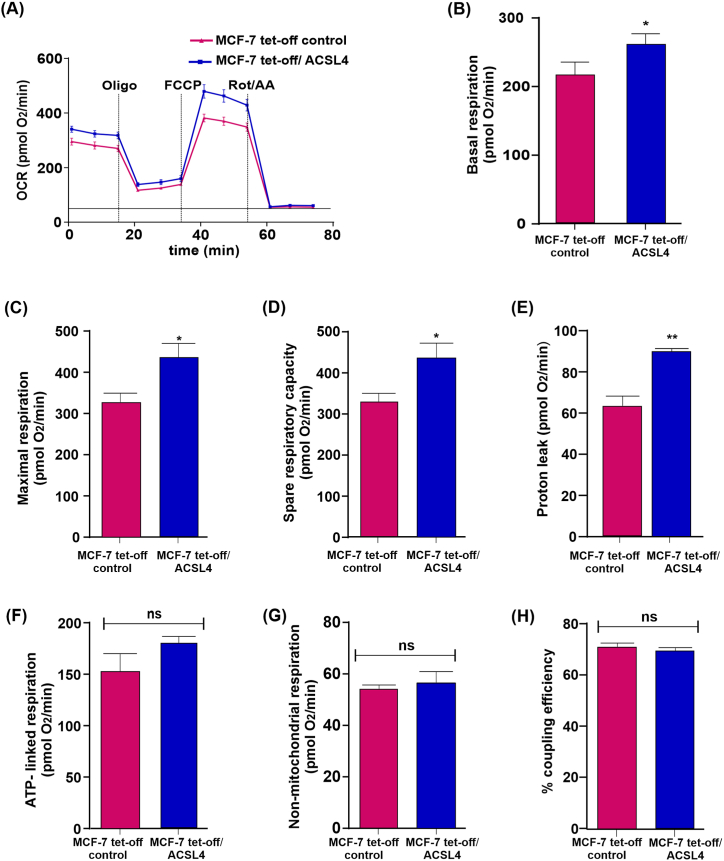
3.4. ACSL4 overexpression positively modulates OCR and respiratory parameters inMCF-7 breast cancer cells
To evaluate OXPHOS and respiratory status, we further studied the role of ACSL4 overexpression in mitochondrial bioenergetics. To this end, OCR and respiratory parameters were measured in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells through the Mito Stress Test. As shown in Fig. 5A, ACSL4 induced an increase in OCR. The analyses of respiratory parameters using data obtained from OCR measurements rendered significantly higher basal respiration, maximal respiration, spare respiratory capacity, and proton leak (Fig. 5B, C, 5D and 5E) in MCF-7 tet-off/ACSL4 than in MCF-7 tet-off control cells. Nevertheless, other mitochondrial respiratory parameters such as ATP-linked respiration, non-mitochondrial respiration, and percentage of coupling efficiency were unaffected by ACSL4 overexpression (Fig. 5F, G and 5H).
Fig. 5.
ACSL4 overexpression induces an increase in OCR and respiratory parameters in MCF-7 breast cancer cells. The mitochondrial stress test was used to obtain OCR and respiratory parameters of mitochondrial bioenergetic function in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. ATP synthase inhibitor Oligomycin A (2.5 μM) was added to derive ATP-linked OCR, FCCP (0.5 μM) to uncouple the mitochondria for maximal OCR, and Rotenone/Antimycin A (0.5 μM) to inhibit electron transport chain. (A) OCR representative trace in MCF-7 tet-off/ACSL4 (blue square) and MCF-7 tet-off control cells (pink triangle) over time. Using OCR data, respiratory parameters were determined and represented in bar graphs: (B) Basal respiration, (C) maximal respiration,(D) spare respiratory capacity, (E) proton leak, (F) ATP-linked respiration, (G) non-mitochondrial respiration, and (H) percentage of coupling efficiency. Results are expressed as the mean of pmol O2/min ±SEM of three independent measurements (ns: non-significant, *p < 0.05, **p < 0.01). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
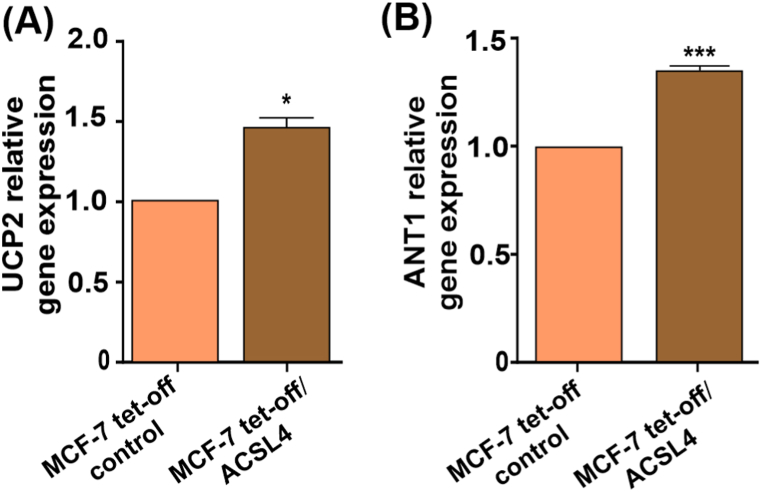
3.5. UCP2 and ANT1 mRNA levels are stimulated by ACSL4 overexpression in MCF-7 breast cancer cells
UCPs and ANTs are known to regulate processes such as proton leak, electron leak, and proton slip [[47], [48], [49]]. Given the increase reported above in proton leak, further assays were conducted on UCP2 and ANT1 mRNA expression levels in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. Results showed ACSL4 ability to induce a significant increase in the expression levels of UCP2 and ANT1 mRNA (Fig. 6A and B), which hints at ACSL4 involvement in the regulation of mitochondrial uncoupling in MCF-7 breast cancer cells.
Fig. 6.
ACSL4 overexpression stimulates UCP2 and ANT1 mRNA levels in MCF-7 breast cancer cells. UCP2 and ANT1 mRNA expression was evaluated in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells.(A) UCP2 and (B) ANT1 relative mRNA expression measured by real-time qPCR after RNA extraction and calculated as the 2−ΔΔCt value (comparative Ct method) for each experimental sample (relative gene expression). Results are expressed as the mean arbitrary units ± SEM of at least three independent experiments (*p < 0.05, ***p < 0.001).
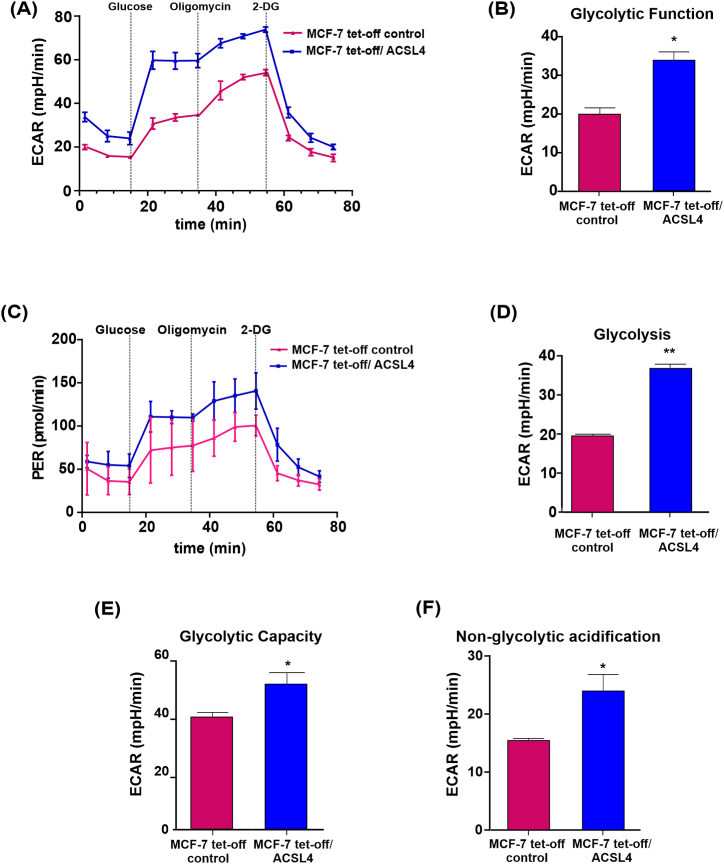
3.6. ACSL4 overexpression stimulates glycolytic function in MCF-7 breast cancer cells
As glycolytic activity has been extensively associated with cancer cells, we next characterized glycolytic metabolism in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells using the Glycolysis Stress Test. Before glucose injection, MCF-7 tet-off/ACSL4 cells showed higher ECAR than MCF-7 tet-off control cells (Fig. 7A), which translated into a significant increase in glycolytic function (Fig. 7B). After the injection of a saturating concentration of glucose, MCF-7 tet-off/ACSL4 cells showed a significant increase in glycolysis (Fig. 7D), in keeping with the increase observed in glycolytic function and ECAR (Fig. 7A and B). The injection of oligomycin, which blocks ATP synthase, significantly increased ECAR in MCF-7 tet-off/ACSL4, which correlated with higher glycolytic capacity (Fig. 7E) and a higher proton efflux rate (PER) (Fig. 7C). Results also showed significantly higher non-glycolytic acidification in MCF-7 tet-off/ACSL4 cells than control cells (Fig. 7F), which suggests the contribution of other sources of extracellular acidification that are not attributed to glycolysis.
Fig. 7.
ACSL4 overexpression modulates mitochondrial glycolytic function in MCF-7 cells. Analysis of glycolytic metabolism in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells by sequential application of glucose, pharmacological inhibitors of OXPHOS (oligo) and glycolysis (2-DG). Glycolytic parameters were calculated. (A) ECAR representative trace over time in MCF-7 tet-off/ACSL4 (blue square) and MCF-7 tet-off control cells (pink triangle), (B) glycolytic function, (C) PER representative trace over time in MCF-7 tet-off/ACSL4 (blue square) and MCF-7 tet-off control cells (pink triangle), (D) glycolysis, (E) glycolytic capacity, and (F) non-glycolytic acidification are represented. Results are expressed as the mean of mpH/min ±SEM of at least three independent experiments (*p < 0.05, **p < 0.01). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
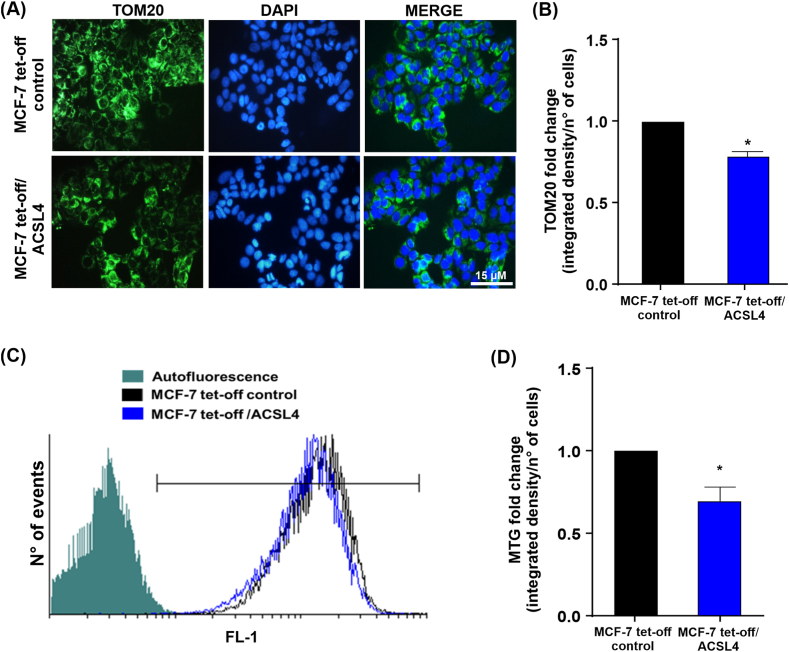
3.7. ACSL4 overexpression reduces mitochondrial mass in MCF-7 breast cancer cells
Mitochondrial mass reflects a balance between biogenesis and mitophagy [50], and its dysregulation has been demonstrated in different pathologies, including cancer [15,51,52]. TOM20 is often used as a marker of mitochondrial mass [53] due to its constitutive expression in the outer mitochondrial membrane. In addition, the disappearance of fluorescent signal for individual proteins such as TOM20, cytochrome c, HtrA2/Omi, or mitochondrial-targeting fluorescent proteins is frequently used to assess mitophagy mechanisms [54]. Consequently, TOM20 expression was evaluated through immunofluorescence as a marker of mitochondrial mass and a possible indicator of mitophagy in MCF-7 cells. Results showed lower TOM20 fluorescence intensity in MCF-7 tet-off/ACSL4 cells (Fig. 8A and B), which indicates that ACSL4 induced a significant decrease in mitochondrial mass (Fig. 8B).
Fig. 8.
ACSL4 overexpression reduces mitochondrial mass in MCF-7 breast cancer cells. Mitochondrial mass analysis in MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. (A) Fluorescence microscopy images of cells stained with anti-TOM20 (green), and nuclei stained with DAPI (blue). Images were obtained at equal exposure time and intensity. (B) Quantification of TOM20 immunofluorescence. (C) Representative histogram of MTG staining as a measure of mitochondrial mass. Fluorescence intensity was measured at 516 nm emission (FL-1). (D) Quantification of MTG signal is expressed as mean fold change of integrated signal density per number of cells ± SEM of at least three independent experiments (*p < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In order to support our immuno-microscopy results on mitochondrial mass, MTG staining was used to assess fluorescence intensity by flow cytometry in MCF-7 cells. Results revealed that ACSL4 overexpression reduced mitochondrial mass, which correlated with a lower MTG fluorescence intensity (Fig. 8C and D). These data confirm the results of measurement of TOM20 expression.
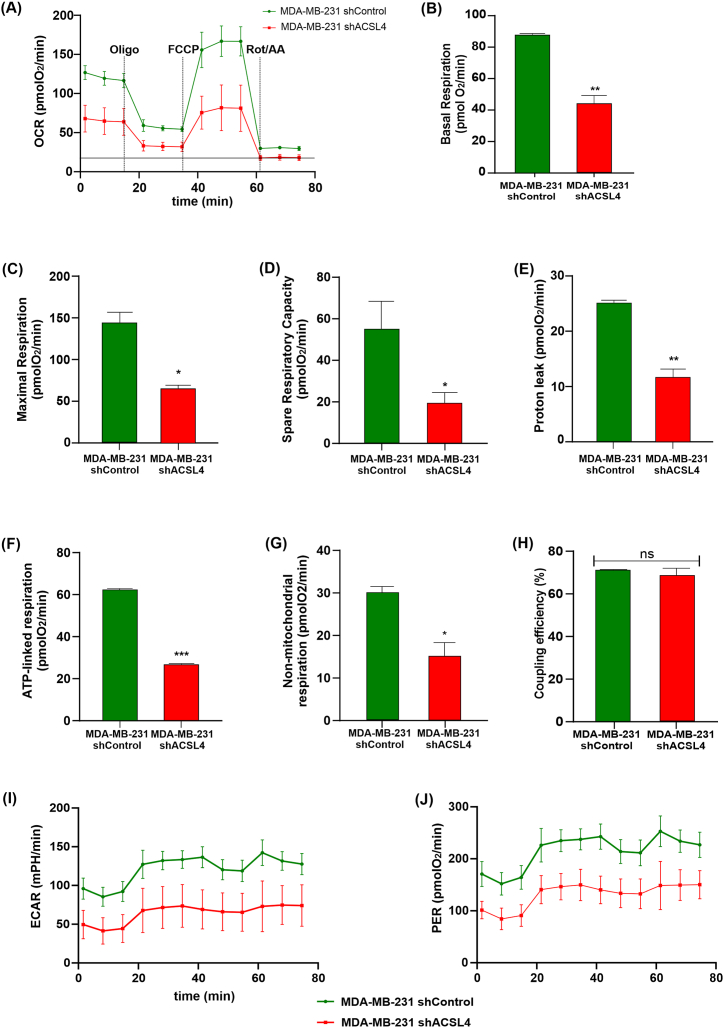
3.8. OCR and ECAR are decreased by ACSL4 silencing in MDA-MB-231 cells
We further investigated the role of ACSL4 in mitochondrial function by using small hairpin RNA (shRNA) to disrupt endogenous expression of the enzyme in the more aggressive MDA-MB-231 cell line (Supplementary Fig. 1B). To enhance our studies on bioenergetics and mitochondrial metabolism, we assessed OCR and ECAR together with respiratory parameters using the Mito Stress Test in MDA-MB-231 shACSL4 and shControl cells. Consistent with the results previously obtained in the ACSL4-overexpressing MCF-7 cell line, OCR was significantly reduced in MDA-MB-231 shACSL4 cells compared to control cells (Fig. 9A). Furthermore, prior to glucose injection, the basal respiration was significantly higher in control cells compared to ACSL4-silenced cells (Fig. 9B). The analysis of respiratory parameters demonstrated that ACSL4 silencing in the MDA-MB-231 cell line significantly reduced the maximal respiration, proton leak, SRC, ATP-linked respiration, and non-mitochondrial respiration (Fig. 9C, D, 9E, 9F and 9G). However, there was no observed effect on the percentage of coupling efficiency (Fig. 9H). Additionally, knockdown of ACSL4 in this particular cell line resulted in a significant decrease of ECAR and PER in comparison to control cells (Fig. 9I and J).
Fig. 9.
ACSL4 knockdown in MDA-MB-231 cells decreases OCR, respiratory parameters and ECAR. The mitochondrial stress test was used to obtain OCR and respiratory parameters and ECAR of mitochondrial bioenergetics function in MDA-MB-231 cells knocked down for ACSL4 expression (MDA-MB-231 shACSL4) compared to control cells (MDA-MB-231 shControl). The ATP synthase inhibitor oligomycin A (1.5 μM) was added to obtain ATP-linked OCR, FCCP (1 μM) to uncouple mitochondria for maximal OCR, and rotenone/Antimycin A (0.5 μM) to inhibit the electron transport chain. (A) Representative OCR curve in MDA-MB-231 shControl cells (green circle) and MDA-MB-231 shACSL4 cells (red square) over time. Respiratory parameters were determined from the OCR data and plotted as bar graphs: (B) basal respiration, (C) maximal respiration, (D) spare respiratory capacity, (E) proton leak, (F) ATP-linked respiration, (G) non-mitochondrial respiration, and (H) percentage of coupling efficiency. Results are expressed as the mean of pmol O2/min ±SD relative to total protein of three independent measurements (*p < 0.05, **p < 0.01, ***p < 0.001). In addition, ECAR and PER were obtained; (I) ECAR representative curve over time in MDA-MB-231 shACSL4 (red square) and MDA-MB-231 shControl cells (green circle), (J) PER representative curve over time in MDA-MB-231 shACSL4 (red square) and MDA-MB-231 shControl cells (green circle). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
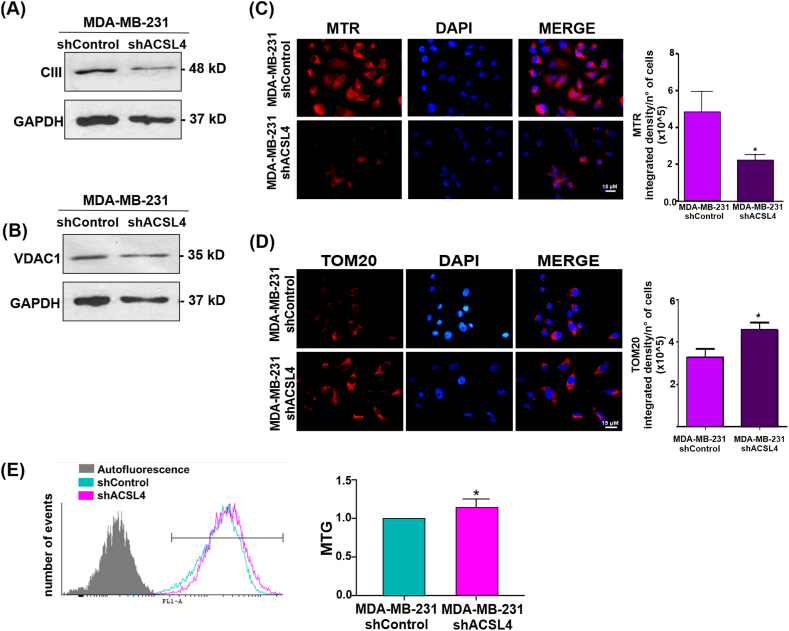
3.9. ACSL4 knockdown reduces mitochondrial markers, discourages mitochondrial active respiration and increases mitochondrial mass in MDA-MB-231 breast cancer cells
To further study the role of ACSL4 in mitochondrial metabolism and function, Complex III and VDAC1 expression levels were evaluated in whole lysates of MDA-MB-231 shACSL4 and MDA-MB-231 shControl cells (Fig. 10). In addition, analyses were conducted on mitochondrial activity using MTR, mitochondrial mass through TOM20 expression and MTG staining through flow cytometry. As expected, and supporting our previous results, all the parameters analyzed revealed values opposite to those obtained in MCF-7 cells. Indeed, ACSL4 silencing reduced the levels of mitochondrial markers Complex III (Fig. 10A) and VDAC1 (Fig. 10B). Additionally, ACSL4 knockdown affected mitochondrial health and discouraged active mitochondrial respiration (Fig. 10C). Furthermore, ACSL4 silencing in MDA-MB-231 cells induced a significant increase in mitochondrial mass, observed by TOM20 immunofluorescence (Fig. 10D) and flow cytometry analysis (Fig. 10E).
Fig. 10.
ACSL4 knockdown in MDA-MB-231 cell line induces opposite effects in mitochondrial metabolism and function than those observed in MCF-7 tet-off/ACSL4 cells. Western blot analyses of (A) Complex III and (B) VDAC1 levels on protein extracts of MDA-MB-231 shASCL4 and MDA-MB-231 shControl triple negative breast cancer cells. GAPDH signal was used as loading control protein. The full, non-adjusted blots images are included in Supplementary Material (Supplementary Fig. 3). (C) MTR was used to evaluate mitochondria active respiration. Picture shows fluorescence microscopy images of mitochondria stained with MTR (red) and nuclei stained with DAPI (blue). Images were obtained at equal exposure time and intensity (left). Quantification of the signal is expressed as the mean integrated density per number of cells ± SEM of at least three independent experiments (right). Mitochondrial mass was measured by (D) TOM20 immunofluorescence and (E) MTG staining followed by flow cytometry. (D) Fluorescence microscopy images of mitochondria stained with TOM20 (red) and nuclei stained with DAPI (blue). Images were obtained at equal exposure time and intensity (left). Quantification of fluorescence signal is expressed as the mean integrated density per number of cells ± SEM of at least three independent experiments (*p < 0.05, **p˂ 0.01). (E) Representative histogram of MTG staining as a measure of mitochondrial mass. Fluorescence intensity was measured at 516 nm emission (FL-1) (left). Quantification of MTG signal is expressed as mean fold change of integrated signal density per number of cells ± SEM of at least three independent experiments (*p < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
This study was undertaken to clarify the role of ACSL4 in mitochondrial function and cellular bioenergetics in breast cancer cells. Our results provide evidence of the involvement of ACSL4 in mitochondrial function and metabolism. These results may also explain the increased aggressiveness and tumorigenic capacity of cells overexpressing ACSL4.
Bioinformatics analysis of RNA-seq data from MCF-7 cells stably overexpressing ACSL4 unveiled cellular pathways which could be affected by ACSL4 expression, including cancer-related processes such as cell proliferation, migration, and invasion [10,12]. In addition, analyses revealed that ACSL4 may not only modulate canonical pathways related to mitochondrial processes such as OXPHOS, mitochondrial dysfunction, and the mTOR pathway but also affect the expression of genes associated with mitochondrial function and metabolism, including Complex III of the ETC, NRF-1/2, VDAC1, and uncoupling proteins. Taken together, these results raised the question whether ACSL4 expression could also affect energy metabolism and mitochondrial function in breast cancer cells.
Interestingly, the analysis of mitochondrial protein levels showed that the stable overexpression of ACSL4 in MCF-7 cells induced an increase in the protein expression of NRF-1, VDAC1 and Complex III, all genes related to mitochondrial metabolism and function [36,37,55]. Furthermore, ACSL4 also up-regulated NRF-1 and NRF-2 mRNA. NRF-1 and NRF-2 are widely known to play an important role in the nuclear-mitochondrial interactions involved in mitochondrial biogenesis [35,56]. NRF-1 is a key transcription factor regulating the expression of genes involved in mitochondrial biogenesis and the ETC [57], as it is known to bind to GC-rich DNA elements in the promoters of these genes [36]. Previous studies indicate that NRF-1 acts as an oncoprotein driving breast carcinogenesis induced by estrogens [58]. Worth pointing out, higher NRF-1 expression has been detected in breast cancer cells with an aggressive phenotype resistant to tamoxifen [59], which is in line with our results in MCF-7 cells overexpressing ACSL4. In turn, NRF-2 is not only involved in mitochondrial biogenesis but also implicated in breast carcinogenesis, its expression correlates with poor prognosis in breast cancer patients. Moreover, elevated NRF-2 levels have been found in drug-resistant tumor cells. In addition, NRF-2 takes part in cellular antioxidant defense and regulates the expression of various protective genes [60,61]. In this sense, the significant increase observed in NRF-2 levels in MCF-7 tet-off/ACSL4 cells correlates not only with ACSL4-induced aggressiveness but also with previously reported ACSL4-induced resistance to antitumor drugs [7].
Regarding mitochondrial proteins, a recent study has shown numerous NRF-1 binding sites in the promoter of the VDAC1. Situations compatible with a tumoral environment, e.g., nutrient depletion or hypoxia, induce an increase in VDAC1 protein expression gene [62]. The increase observed in Complex III could also be induced by the increase in NRF-1 expression, as this transcription factor can directly or indirectly stimulate the transcription of ETC complexes [63,64]. Moreover, the silencing of ACSL4 expression in the MDA-MB-231 cell line produced a significant decrease in Complex III and VDAC1 expression. Altogether, these results show an association between ACSL4 and the expression of key mitochondrial proteins in breast cancer cells and suggest alterations in mitochondrial function and bioenergetics.
As revealed by our mitochondrial bioenergetics’ studies, ACSL4 overexpression induced an increase in OCR which correlated with higher basal respiration than that of MCF-7 tet-off control cells. Analysis of respiratory parameters based on total OCR data showed that non-mitochondrial respiration, ATP-linked respiration, and coupling efficiency remained unaltered by ACSL4 overexpression, whereas parameters such as maximal respiration, SRC, and proton leak significantly differed between MCF-7 tet-off/ACSL4 and MCF-7 tet-off control cells. The increase observed in maximal respiration in ACSL4-overexpressing cells may be associated with an increase in the expression of mitochondrial complexes, among them, the Complex III. These data suggest that, in the context of growing energy demand, cells with higher ACSL4 levels may be better prepared to use and oxidize more substrates. This behavior correlates with energy requirements observed in most aggressive tumor cells to maintain high rates of proliferation, invasion, and migration [23,26,50].
Likewise, the increase observed in SRC in MCF-7 tet-off/ACSL4 cells suggests that ACSL4 overexpression gives cells greater capacity to respond to energy demand in stressful conditions where ATP requirements increase, and the utilization of oxidizable substrates becomes a limiting factor. Of note, malignant tumoral cells with high SRC are supposed to bypass this limitation and further proliferate, migrate, and expand over non-malignant cells [27,65]. The increase in SRC may be explained by an ACSL4-induced increase in the expression of NRF-1, VDAC1 and Complex III, which facilitates the oxidation of ETC substrates. SRC is regulated by various mechanisms, particularly the PI3K/AKT/mTOR pathway [65]. Our laboratory has previously shown that ACSL4 produces an increase in the mTOR pathway in breast cancer cells [10], which indicates that the increase in SRC observed in MCF-7 tet-off/ACSL4 cells is mediated, at least in part, by mTOR activation.
The results of the bioenergetic analysis performed in the silenced ACSL4 MDA-MB-231 cells reinforced the role of ACSL4 in the regulation of these mitochondrial parameters in breast cancer cells. In addition, ATP-linked respiration and non-mitochondrial respiration were significantly reduced in MDA-MB-231 shACSL4 cells compared to control, suggesting that these processes may be highly dependent on ACSL4 expression and evident in the more aggressive breast cancer cells. Regarding this, non-mitochondrial respiration is due to the activity of non-mitochondrial oxidases, desaturases and detoxification enzymes, being quite different among cell types [66].
Interestingly, it has been described in breast cancer cells of aggressive phenotype, that proton leak is increased with respect to the less aggressive tumor cells, in agreement with our results. In this context, it has been demonstrated that both UCP2 and ANT1 are related to the regulation of proton leak in different cell models [47,49,67]. Our work showed an increase in UCP2 and ANT1 mRNA levels in MCF-7 cells that overexpress ACSL4 and a significant decrease in MDA-MB-231 shACSL4 cells compared to control cells, however the percentage of coupling efficiency was not altered in both models. These results suggest that, although ACSL4 induces the expression of uncoupling genes, mitochondria are not uncoupled and OXPHOS is active, favoring the aggressive properties of these cells. In this way, cells that overexpress ACSL4 are able to develop and establish a tumor in in vivo models, as has already been described by our group [9].
We further demonstrate that ACSL4 overexpression promotes an increase in MTR staining, indicative of an increase in ΔΨm triggered by a rise in mitochondrial activity. Indeed, higher ΔΨm values have been shown in different types of tumors as compared to normal cells [68]. Of note, our results were corroborated using ACSL4 silencing in MDA-MB-231 cells.
Metabolic reprogramming is known to play a key role in cancer development and progression [17,28,69]. Studies in colon cancer cells have shown ACSL4 ability to induce a switch to glycolytic metabolism, which gives cells an energetic advantage and thus favors tumor migration and proliferation [70]. In line with this evidence, our glycolytic analysis revealed an increase in ECAR, PER and glycolytic capacity in MCF-7 tet-off/ACSL4 cells, which suggests that ACSL4 is capable of increasing glycolytic metabolism in breast cancer cells as well as in colon cancer cells. To support these results, a significant decrease in both, ECAR and PER, was observed in MDA-MB-231 shACSL4 cells respect to control. This metabolic reprogramming possesses an energetic advantage for breast cancer cells in tumor establishment, development, and aggressiveness. Worth highlighting, both our results and existing background show that tumor establishment and progression do not always rely on a switch to glycolytic metabolism at the expense of a complete OXPHOS shutdown but may rather involve both OXPHOS and glycolysis [[22], [23], [24], [25]].
It is well established that the PI3K/AKT/mTOR signaling pathway is critical for the regulation of mitochondrial homeostasis and plays an important role in the development and proliferation of a wide range of cancers [71]. We propose that the role of ACSL4 in mitochondrial function is mediated, at least in part, through this pathway. As mentioned above, our laboratory showed that overexpression of ACSL4 in MCF-7 cells results in mTORC1/2 and AKT activation, which was confirmed in MDA-MB-231 shACSL4 cells compared to their controls [10]. Moreover, activation of mTORC1/2 by angiotensin II is mediated by ACSL4 in adrenocortical human H295R cells [72]. In particular, mTORC1 is known to control cellular metabolism mainly by regulating the translation and transcription of metabolic genes, including peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1α), an upstream regulator of NRF-1/2 [73]. Furthermore, it has been shown that mTORC1 modulates glycolysis through 4E-BP1-dependent translational activation of hypoxia-inducible factor 1 α (HIF1α) and glutaminolysis by inhibiting SIRT4, and that hyperactivation of the mTOR/4E-BP/eIF4E pathway coordinates mitochondrial functions, nucleotide and lipid synthesis, and translational programs to drive neoplasic growth [71].
In addition, we have previously shown that ACSL4 promotes cell growth and tumor progression through the release of AA and its metabolism to lipoxygenase products [9]. In this context, the lipoxygenase pathway has been implicated in the activation of mTOR in breast cancer cells. AA levels have been shown to correlate strongly with the signaling activity of mTORC1 and mTORC2 in human breast tumor tissue. AA is effective in activating both mTOR complexes and its effect is mediated by lipoxygenase but not cycloxygenase metabolites [10,74].
Certain pathologies, including cancer, involve modifications in the mitochondrial mass [75]. Our studies show that ACSL4 overexpression in MCF-7 cells induced a decrease in mitochondrial mass, while ACSL4 silencing in MDA-MB-231 cells promoted an increase in this parameter. These findings suggest that ACSL4 has a role in the maintenance of mitochondrial mass in the breast cancer cells under study and that, in turn, this role varies with cell aggressiveness. Changes in mitochondrial mass are associated with an imbalance between mitochondrial biogenesis and mitophagy [15,75]. In this context, our studies revealed a decrease in TOM20 expression levels in MCF-7 tet-off/ACSL4 cells respect to control while opposite results were observed in MDA-MB-231 shACSL4 cells. TOM20 has been previously reported as a marker of early mitophagy whose expression decreases at the onset of mitophagy in correlation with a reduction in mitochondrial mass [53,76].
Therefore, our results suggest that ACSL4 may stimulate mitophagic mechanisms and play a role in modulating mitochondrial mass and membrane potential, although further studies are needed to determine whether these mechanisms underlie the decrease in the number of mitochondria.
5. Conclusion
In conclusion, our findings on mitochondrial bioenergetics and glycolytic metabolism show that ACSL4 overexpression stimulates mitochondrial turnover, which may favor tumor aggressiveness and metabolic response to stressful situations in breast cancer cells. These results open a novel field of study on the action of ACSL4 in the mitochondria of tumor cells and/or pathological situations.
Data availability statement
Data included in article/supp. material/referenced in article.
Funding
This work was supported by National Scientific and Technical Research Council (CONICET) – Argentina, P.I.P.: (2021–2023, 11220200102166CO, to P.M. Maloberti and 11220200101892CO to C. Poderoso), University of Buenos Aires – Argentina (UBACYT 2020: 20020190100040BA, to P.M. Maloberti), National Agency for Scientific and Technological Promotion, MinCyT, (PICT-2018-02254 to C. Poderoso, PICT-2020-SERIEA-01204, to P.M. Maloberti). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
CRediT authorship contribution statement
Yanina Benzo: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Jesica G. Prada: Methodology, Investigation. Melina A. Dattilo: Methodology, Investigation. María Mercedes Bigi: Methodology, Data curation. Ana F. Castillo: Methodology, Investigation. María Mercedes Mori Sequeiros Garcia: Methodology, Investigation. Cecilia Poderoso: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Paula M. Maloberti: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Mayra Ríos Medrano, Lucía M. Herrera and Nadin Calvente for technical support. We thank María M. Rancez for providing language help and writing assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30639.
Contributor Information
Cecilia Poderoso, Email: cpoderoso@fmed.uba.ar.
Paula M. Maloberti, Email: pmaloberti@fmed.uba.ar.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Radif Y., et al. The endogenous subcellular localisations of the long chain fatty acid-activating enzymes ACSL3 and ACSL4 in sarcoma and breast cancer cells. Mol. Cell. Biochem. 2018 doi: 10.1007/s11010-018-3332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faergeman N.J., Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang M.J., et al. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc. Natl. Acad. Sci. U. S. A. 1997;94(7):2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuwata H., Hara S. Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism. Prostag. Other Lipid Mediat. 2019;144:1–27. doi: 10.1016/j.prostaglandins.2019.106363. [DOI] [PubMed] [Google Scholar]
- 5.Maloberti P.M., et al. Functional interaction between acyl-coa synthetase 4, lipooxygenases and cyclooxygenase-2 in the aggressive phenotype of breast cancer cells. PLoS One. 2010;5(11):1–12. doi: 10.1371/journal.pone.0015540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dattilo M.A., et al. Regulatory mechanisms leading to differential Acyl-CoA synthetase 4 expression in breast cancer cells. Sci. Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-46776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlando U.D., Castillo A.F., Medrano M.A.R., Solano A.R., Maloberti P.M., Podesta E.J. Acyl-CoA synthetase-4 is implicated in drug resistance in breast cancer cell lines involving the regulation of energy-dependent transporter expression. Biochem. Pharmacol. 2019;159:52–63. doi: 10.1016/J.BCP.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Chen J., et al. ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis. 2020;9(4):1–18. doi: 10.1038/s41389-020-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlando U.D., et al. The functional interaction between acyl-coa synthetase 4, 5-lipooxygenase and cyclooxygenase-2 controls tumor growth: a novel therapeutic target. PLoS One. 2012;7(7):1–14. doi: 10.1371/journal.pone.0040794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlando U.D., Castillo A.F., Dattilo M.A., Solano A.R., Maloberti P.M., Podesta E.J. Acyl-CoA synthetase-4, a new regulator of mTOR and a potential therapeutic target for enhanced estrogen receptor function in receptor-positive and -negative breast cancer. Oncotarget. 2015;6(40):42632–42650. doi: 10.18632/oncotarget.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X., et al. Long chain fatty Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone resistance in human breast cancer. PLoS One. 2013;8(10):1–20. doi: 10.1371/journal.pone.0077060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo A.F., Orlando U.D., Lopez P., Solano A.R., Maloberti P.M., Podesta E.J. Gene expression profile and signaling pathways in MCF-7 breast cancer cells mediated by acyl-coa synthetase 4 overexpression. Transcr Open Access. 2016;3(2):1–9. doi: 10.4172/2329-8936.1000120. [DOI] [Google Scholar]
- 13.Doghman M., Lalli E. Efficacy of the novel dual PI3-kinase/mTOR inhibitor NVP-BEZ235 in a preclinical model of adrenocortical carcinoma. Mol. Cell. Endocrinol. Nov. 2012;364(1–2):101–104. doi: 10.1016/J.MCE.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Ramanathan A., Schreiber S.L. Direct control of mitochondrial function by mTOR. Proc. Natl. Acad. Sci. U. S. A. Dec. 2009;106(52):22229–22232. doi: 10.1073/PNAS.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. Jul. 2016;166(3):555–566. doi: 10.1016/J.CELL.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porporato P.E., Filigheddu N., Pedro J.M.B.S., Kroemer G., Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. Mar. 2018;28(3):265–280. doi: 10.1038/CR.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lunetti P., et al. Metabolic reprogramming in breast cancer results in distinct mitochondrial bioenergetics between luminal and basal subtypes. FEBS J. Feb. 2019;286(4):688–709. doi: 10.1111/FEBS.14756. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O. On the origin of cancer cells. Science (1979) Feb. 1956;123(3191):309–314. doi: 10.1126/SCIENCE.123.3191.309/ASSET/A8D38B53-799F-4009-AAD3-E77CEF33D301/ASSETS/SCIENCE.123.3191.309.FP.PNG. [DOI] [PubMed] [Google Scholar]
- 19.Hume D.A., Weidemann M.J. Role and regulation of glucose metabolism in proliferating cells. J. Natl. Cancer Inst. 1979;62(1):3–8. doi: 10.1093/jnci/62.1.3. [DOI] [PubMed] [Google Scholar]
- 20.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. Mar. 2016;41(3):211. doi: 10.1016/J.TIBS.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Luis C., et al. Warburg Effect Inversion: adiposity shifts central primary metabolism in MCF-7 breast cancer cells. Life Sci. Apr. 2019;223:38–46. doi: 10.1016/J.LFS.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Viale A., Corti D., Draetta G.F. Tumors and mitochondrial respiration: a neglected connection. Cancer Res. Sep. 2015;75(18):3685–3686. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- 23.Nayak A.P., Kapur A., Barroilhet L., Patankar M.S. Oxidative phosphorylation: a target for novel therapeutic strategies against Ovarian cancer. Cancers. Sep. 2018;10(9) doi: 10.3390/CANCERS10090337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G., Luo Q., Li H., Liu Q., Ju Y., Song G. Increased oxidative phosphorylation is required for Stemness maintenance in liver cancer stem cells from hepatocellular carcinoma cell line HCCLM3 cells. Int. J. Mol. Sci. Jul. 2020;21(15):5276. doi: 10.3390/IJMS21155276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greene J., Segaran A., Lord S. Targeting OXPHOS and the electron transport chain in cancer; Molecular and therapeutic implications. Semin. Cancer Biol. 2022;86:851–859. doi: 10.1016/J.SEMCANCER.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Solaini G., Sgarbi G., Baracca A. Oxidative phosphorylation in cancer cells. Biochim. Biophys. Acta Bioenerg. Jun. 2011;1807(6):534–542. doi: 10.1016/J.BBABIO.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J. ‘Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (Review)’. Oncol. Lett. Dec. 2012;4(6):1151. doi: 10.3892/OL.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiliro C., Firestein B.L. Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells. Apr. 2021;10(5):1056. doi: 10.3390/CELLS10051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.I. A. Chatzispyrou, N. M. Held, L. Mouchiroud, J. Auwerx, and R. H. Houtkooper, ‘Tetracycline Antibiotics Impair Mitochondrial Function and its Experimental Use Confounds Research’, doi: 10.1158/0008-5472.CAN-15-1626. [DOI] [PMC free article] [PubMed]
- 30.Dijk S.N., Protasoni M., Elpidorou M., Kroon A.M., Taanman J.-W. Mitochondria as target to inhibit proliferation and induce apoptosis of cancer cells: the effects of doxycycline and gemcitabine. Sci. Rep. 2020;10:4363. doi: 10.1038/s41598-020-61381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozicz T., Rahman S., Morava E. The doxycycline paradox in primary mitochondrial diseases. J. Inherit. Metab. Dis. Jul. 2022;45(4):659–660. doi: 10.1002/JIMD.12531. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duarte A., et al. Mitochondrial fusion is essential for steroid biosynthesis. PLoS One. 2012;7(9):1–12. doi: 10.1371/journal.pone.0045829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sloss C.M., Cadalbert L., Finn S.G., Fuller S.J., Plevin R. Disruption of two putative nuclear localization sequences is required for cytosolic localization of mitogen-activated protein kinase phosphatase-2. Cell. Signal. Jun. 2005;17(6):709–716. doi: 10.1016/J.CELLSIG.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta Mol. Cell Res. Jul. 2011;1813(7):1269–1278. doi: 10.1016/J.BBAMCR.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiyama T., et al. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol. Neurodegener. Oct. 2018;13(1) doi: 10.1186/S13024-018-0287-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens K.M., Kulawiec M., Desouki M.M., Vanniarajan A., Singh K.K. Impaired OXPHOS complex III in breast cancer. PLoS One. Aug. 2011;6(8) doi: 10.1371/JOURNAL.PONE.0023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiorillo M., et al. Repurposing atovaquone: targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. Apr. 2016;7(23):34084–34099. doi: 10.18632/ONCOTARGET.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Reyes I., et al. Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature. Sep. 2020;585(7824):288–292. doi: 10.1038/S41586-020-2475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoshan-Barmatz V., Ben-Hail D. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion. 2012;12(1):24–34. doi: 10.1016/j.mito.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Magrì A., Reina S., De Pinto V. VDAC1 as pharmacological target in cancer and neurodegeneration: focus on its role in apoptosis. Front. Chem. Apr. 2018;6:108. doi: 10.3389/FCHEM.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y., Wang W., Yang T., Thomas E.R., Dai R., Li X. The potential role of voltage-dependent anion channel in the treatment of Parkinson's disease. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/4665530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zorova L.D., et al. Mitochondrial membrane potential. Anal. Biochem. Jul. 2018;552:50–59. doi: 10.1016/J.AB.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jayaraman S. Flow cytometric determination of mitochondrial membrane potential changes during apoptosis of T lymphocytic and pancreatic beta cell lines: comparison of tetramethylrhodamineethylester (TMRE), chloromethyl-X-rosamine (H2-CMX-Ros) and MitoTracker Red 580 (MTR580) J. Immunol. Methods. Nov. 2005;306(1–2):68–79. doi: 10.1016/J.JIM.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Imamura K., Takeshima T., Kashiwaya Y., Nakaso K., Nakashima K. D-β-hydroxybutyrate protects dopaminergic SH-SY5Y cells in a rotenone model of Parkinson's disease. J. Neurosci. Res. Nov. 2006;84(6):1376–1384. doi: 10.1002/JNR.21021. [DOI] [PubMed] [Google Scholar]
- 46.Sargiacomo C., Stonehouse S., Moftakhar Z., Sotgia F., Lisanti M.P. MitoTracker deep red (MTDR) is a metabolic inhibitor for targeting mitochondria and eradicating cancer stem cells (CSCs), with anti-tumor and anti-metastatic activity in vivo. Front. Oncol. 2021;11:2569. doi: 10.3389/FONC.2021.678343/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fink B.D., Hong Y.S., Mathahs M.M., Scholz T.D., Dillon J.S., Sivitz W.I. UCP2-dependent proton leak in isolated mammalian mitochondria. J. Biol. Chem. Feb. 2002;277(6):3918–3925. doi: 10.1074/JBC.M107955200. [DOI] [PubMed] [Google Scholar]
- 48.Demine S., Renard P., Arnould T. Mitochondrial uncoupling: a key controller of biological processes in physiology and diseases. Cells. 2019;8(8) doi: 10.3390/cells8080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreiter J., et al. ANT1 activation and inhibition patterns support the fatty acid cycling mechanism for proton transport. Int. J. Mol. Sci. Mar. 2021;22(5):1–14. doi: 10.3390/IJMS22052490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ksiȩzakowska-Łakoma K., Zyła M., Wilczyński J.R. Mitochondrial dysfunction in cancer. Przeglad Menopauzalny. 2014;18(2):136–144. doi: 10.5114/pm.2014.42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fulda S., Galluzzi L., Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 52.Wallace D.C. Mitochondrial and cancer. Nat. Rev. Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Contino S., et al. Presenilin 2-dependent maintenance of mitochondrial oxidative capacity and morphology. Front. Physiol. Oct. 2017;8(OCT) doi: 10.3389/FPHYS.2017.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu J., Dagda R.K., Chu C.T. Monitoring mitophagy in neuronal cell cultures. Methods Mol. Biol. 2011;793:325–339. doi: 10.1007/978-1-61779-328-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shoshan-Barmatz V., De Pinto V., Zweckstetter M., Raviv Z., Keinan N., Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Aspect. Med. 2010;31(3):277–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways. Front. Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarpulla R.C. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci. 2008;1147:321. doi: 10.1196/ANNALS.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das J., Felty Q., Poppiti R., Jackson R., Roy D. Nuclear respiratory factor 1 acting as an oncoprotein drives estrogen-induced breast carcinogenesis. Cells. 2018;7(12) doi: 10.3390/cells7120234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radde B.N., et al. Nuclear respiratory factor-1 and bioenergetics in tamoxifen-resistant breast cancer cells. Exp. Cell Res. Sep. 2016;347(1):222–231. doi: 10.1016/j.yexcr.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C., et al. NRF2 promotes breast cancer cell proliferation and metastasis by increasing RhoA/ROCK pathway signal transduction. Oncotarget. Oct. 2016;7(45):73593–73606. doi: 10.18632/ONCOTARGET.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng X., et al. The Nrf2/PGC1α pathway regulates antioxidant and proteasomal activity to alter cisplatin sensitivity in Ovarian cancer. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/4830418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guarino F., et al. NRF-1 and HIF-1α contribute to modulation of human VDAC1 gene promoter during starvation and hypoxia in HeLa cells. Biochim. Biophys. Acta Bioenerg. 2020;1861(12) doi: 10.1016/j.bbabio.2020.148289. [DOI] [PubMed] [Google Scholar]
- 63.Herzig R.P., Scacco S., Scarpulla R.C. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J. Biol. Chem. 2000;275(17):13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 64.Satoh J.I., Kawana N., Yamamoto Y. Pathway analysis of ChIP-seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul. Syst. Biol. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marchetti P., Fovez Q., Germain N., Khamari R., Kluza J. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB (Fed. Am. Soc. Exp. Biol.) J. 2020;34(10):13106–13124. doi: 10.1096/fj.202000767R. [DOI] [PubMed] [Google Scholar]
- 66.Hill B.G., et al. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012 doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valle A., Oliver J., Roca P. Role of uncoupling proteins in cancer. Cancers. Jun. 2010;2(2):567. doi: 10.3390/CANCERS2020567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulikov A.V., Luchkina E.A., Gogvadze V., Zhivotovsky B. Mitophagy: link to cancer development and therapy. BBRC (Biochem. Biophys. Res. Commun.) 2017;482(3):432–439. doi: 10.1016/j.bbrc.2016.10.088. [DOI] [PubMed] [Google Scholar]
- 69.Phan L.M., Yeung S.C.J., Lee M.H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11(1):1. doi: 10.7497/J.ISSN.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sánchez-Martínez R., Cruz-Gil S., García-Álvarez M.S., Reglero G., De Molina A.R. Complementary ACSL isoforms contribute to a non-Warburg advantageous energetic status characterizing invasive colon cancer cells. Sci. Rep. 2017;7(1):1–15. doi: 10.1038/s41598-017-11612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita M., et al. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14(4):473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helfenberger K.E., Argentino G.F., Benzo Y., Herrera L.M., Finocchietto P., Poderoso C. Angiotensin II regulates mitochondrial mTOR pathway activity dependent on acyl-CoA synthetase 4 in adrenocortical cells. Endocrinology. Oct. 2022;163(12) doi: 10.1210/ENDOCR/BQAC170. [DOI] [PubMed] [Google Scholar]
- 73.de la Cruz López K.G., Toledo Guzmán M.E., Sánchez E.O., García Carrancá A. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front. Oncol. Dec. 2019;9 doi: 10.3389/FONC.2019.01373/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen Z.H., et al. Critical role of arachidonic acid-activated mTOR signaling in breast carcinogenesis and angiogenesis. Oncogene. 2013;32(2):160–170. doi: 10.1038/onc.2012.47. [DOI] [PubMed] [Google Scholar]
- 75.Chourasia A.H., Boland M.L., Macleod K.F. Mitophagy and cancer. Cancer Metabol. 2015;3(4) doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Villa E., et al. Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20(12):2846–2859. doi: 10.1016/j.celrep.2017.08.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.