To the editor:
The identification, curation, and interpretation of genomic variants plays a key role in the diagnosis and clinical care of individuals with genetic diseases and related research. Variant databases such as ClinVar1, Human Gene Mutation Database (HGMD®)2, and Leiden Open Variant Database (LOVD)3 provide interpretation of genetic variants, but the information they contain is often incomplete due to the sheer volume and rapid growth of medical literature and the high cost of expert curation.
Finding and keeping up with the latest variant related information from relevant genomic literature is thus of critical importance for genomic research and precision medicine. Unfortunately, traditional keyword-based search systems such as PubMed often yield suboptimal results for variant searches because of two major challenges: (I) the same genetic variant is commonly described with different names (e.g., ‘A146T’ vs. ‘c.436G>A’ vs. ‘rs121913527’) in the literature. As a result, PubMed fails to return the same results for these queries. (II) Many variant results are missed in PubMed output because its search is restricted to the article title and abstract data only. There also exist a few other literature resources on genomic variants such as Mastermind‡ but their full content is not freely available nor is access to their proprietary variant search software.
To help researchers, clinical laboratory and healthcare professionals, and database curators stay up to date with published research on genomic variants, we previously developed LitVar4, a semantic search system that makes use of advanced text and data mining techniques to identify and normalize variant information in full-length articles. Since its launch in 2018, LitVar has been widely accessed millions of times through the web interface and/or programmatic API calls for various use cases5–9 such as assisting variant identification/interpretation/curation (see Table 1); it is also cross-linked by several major variant information resources such as dbSNP10, ClinVar1 and SNPedia11.
Table 1. LitVar has been widely accessed millions of times by various users from the research community and the clinical testing laboratories.
Based on what has been reported in the cited articles and direct user input, we have summarized some of the main use cases of the LitVar system, ranging from assisting variant identification/interpretation/curation5,6, to facilitating in-silico variant annotation in genomics/bioinformatics research7,8, to supporting precision medicine in clinical research/patient care9.
| Use Cases | Description/Example |
|---|---|
| Variant Identification | Locating all previously reported variants of a target gene. |
| Variant Interpretation | Searching for published information about a variant of interest (e.g., finding variant-related disease information). |
| Variant Curation | Assisting manual variant curation and classification (e.g., speeding up variant curation in ClinGen). |
| Bioinformatics and Genomics | Facilitating in-silico variant annotation for genetic disease analysis (e.g., LitVar data is integrated by other computational methods for in-depth variant analysis). |
| Clinical Research and Care | Supporting precision medicine research and patient care (e.g., helping variant evaluation within the context of molecular tumor boards). |
Here we present LitVar 2.0 (https://www.ncbi.nlm.nih.gov/research/litvar2/), a significantly improved system that features several major expansions over its predecessor, including but not limited to (1) improved variant recognition accuracy; (2) the inclusion of variant information from article supplementary data; (3) more powerful search capabilities; and (4) a redesigned user interface for more convenient results navigation.
In addition to the entire set of PubMed abstracts and open-access PMC full-text articles, we included article-associated supplementary materials in LitVar 2.0 as they were found to be enriched with variant information12,13. All input text is then processed by tmVar 3.014, our newly improved text-mining tool for variant extraction and normalization that addresses some previously difficult edge cases as well as offers multiple options to normalize a variant (see more technical details and its benchmarking performance in Table S1). Additionally, LitVar 2.0 also makes use of PubTator15, a state-of-the-art tool for tagging other pertinent bio-entities such as genes and diseases. In total, approximately 14 million unique variants (~70 million variant mentions in total) are found among the entire set of PubMed abstracts, PMC full texts, and associated supplementary materials (see more detailed statistics in Table 2). A significant fraction (~85%) of the unique variants appear only in the article supplementary data, thus not available previously (see detailed statistics in Table 3). Through its monthly update, new content is continuously added to LitVar 2.0.
Table 2. Total number of variants (unique) in LitVar 2.0 vs. its original system.
As can be seen, the new system contains significantly more unique variants compared to the original system. This is largely due to the inclusion of supplementary materials in LitVar 2.0 (see Table S3 also). It is also because new published articles on variants have been included since the original system was first published in 2018.
| System | Total variants (unique) | Date of Access |
|---|---|---|
| LitVar | 1,968,872 | March 2018 |
| LitVar 2.0 | 13,829,591 | December 2022 |
Table 3. Supplementary materials of full-length articles are highly rich for variant information.
As shown below, over 11 million unique variants are found only in the article supplementary materials.
| Data Type | Data Source | Size |
|---|---|---|
| Total variants (unique) | Abstract + Full text | 2,116,802 |
| Abstract + Full text + Supplementary materials | 13,829,591 |
Due to variant name normalization and standardization, LitVar 2.0 supports variant searches with a variety of different formats, including DNA (e.g., c.2612C>T or g.13843A>G), protein (e.g., P871L or Pro871Leu or BRCA1 p.P871L), or multiple standardized nomenclature (e.g., HGVS, RSID, SPDI, and ClinGen Allele Registry IDs). Real-time variant disambiguation and query autocomplete are available so that users can conveniently and precisely select their variant of interest from a drop-down list. Moreover, LitVar 2.0 allows users to combine a variant/gene search with any free-text keywords, to find articles matching both the variant and the text query.
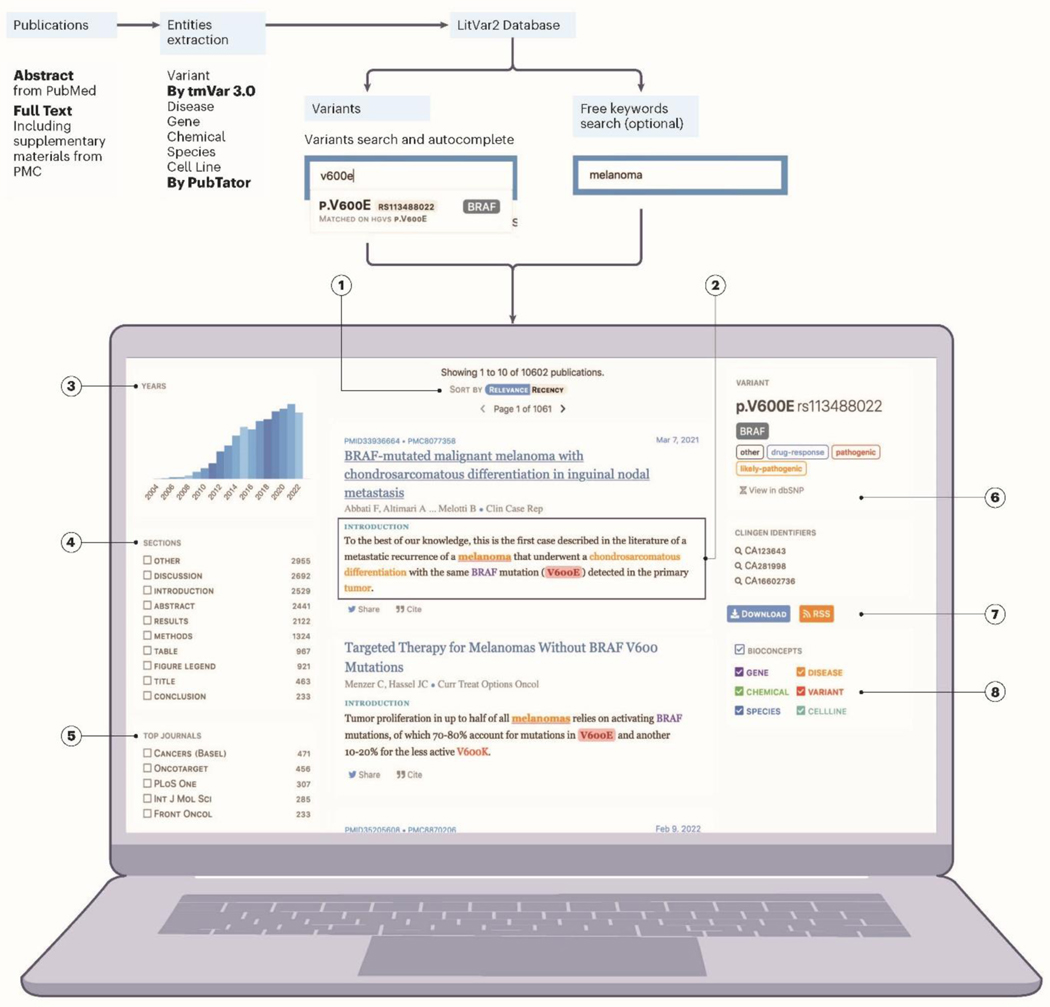
When examining retrieved publications, search results can be sorted either by date or relevance (Fig. 1.1). The former uses reverse time order while the latter ranks results based on query-document relatedness via Solr, an open-source enterprise-search platform (https://solr.apache.org). Each search result is accompanied by a brief snippet containing the variant and pre-annotated biological concepts when available (Fig. 1.2). Search results can be further narrowed using three filters: year (Fig. 1.3), journal (Fig. 1.5), or article section (e.g., show results only found in “Methods”; Fig. 1.4).
Figure 1: The overall workflow of the LitVar system and its search results page.
After a search is performed, search results are displayed and sorted either by date or relevance (1). Search results consist of publication title with a snippet (2) containing highlighted entities, explaining why the publication matched the search query. A yearly histogram displays the evolution of the popularity of the search topic over time, while facet filters – year (3), section (4), and journal (5) – allow users to further narrow down the search results. In addition to search results, detailed information about the variant of interest is displayed as well as options to restrict the search to a specific ClinGen identifier (6), download the results, subscribe to the RSS field (7), or toggle which biological entities should be highlighted in text (8).
In addition to matching publications, the search results page displays detailed information about the variant of interest (Fig. 1.6), obtained from its cross-linked dbSNP and ClinVar databases. Search results can be downloaded or users can subscribe to the corresponding RSS feed to immediately be notified when new publications mentioning that variant become available in LitVar (Fig. 1.7).
LitVar is freely available as a web tool for all users and supports recent versions of all major browsers. Its entire content can also be accessed programmatically via its APIs (https://www.ncbi.nlm.nih.gov/research/litvar2/api) or downloaded in bulk via our ftp site (https://ftp.ncbi.nlm.nih.gov/pub/lu/LitVar/).
The development of LitVar has been greatly benefitted from user feedback. We encourage our users and the biomedical research community to continue using LitVar and help us jointly improve LitVar. In the future, we plan to further enhance variant disambiguation accuracy, to include copy number variations, and to categorize articles by various evidence types (e.g., Genome-wide association studies; functional studies; segregation data; etc).
Supplementary Material
Acknowledgment:
This research is supported by the NIH Intramural Research Program, National Library of Medicine. HLR was supported by NIH/NHGRI grant U24HG006834. We thank Dr. Qingyu Chen for his help on the early draft of this manuscript.
Footnotes
Competing Interests: The authors declare no competing interests.
Keeping up with the latest variant related publications is of critical importance for genomic research and precision medicine. Here we present LitVar 2.0 (https://www.ncbi.nlm.nih.gov/research/litvar2/), a significantly improved web-based system to accurately search for genetic variants and related information in the unstructured literature. Unlike traditional literature search engines, LitVar 2.0 provides a unified search over abstracts, full text, and supplementary data, as well as precise variant recognition through AI-based disambiguation.
Data Availability:
LitVar 2.0 and its data are made freely available to the scientific community at https://www.ncbi.nlm.nih.gov/research/litvar2/.
References:
- 1.Landrum MJ et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Research 46, D1062–D1067, doi: 10.1093/nar/gkx1153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenson PD et al. The human gene mutation database: 2008 update. Genome Medicine 1, 1–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fokkema IFAC et al. LOVD v. 2.0: the next generation in gene variant databases. Human Mutation 32, 557–563 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Allot A. et al. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic Acids Research 46, W530–W536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Setlere S, Jurcenko M, Gailite L, Rots D. & Kenina V. Alanyl-tRNA Synthetase 1 Gene Variants in Hereditary Neuropathy: Genotype and Phenotype Overview. Neurol Genet 8, e200019, doi: 10.1212/NXG.0000000000200019 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadler KV et al. Re-evaluation of missense variant classifications in NF2. Hum Mutat 43, 643–654, doi: 10.1002/humu.24370 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M. et al. SNPMap-An integrated visual SNP interpretation tool. Front Genet 13, 985500, doi: 10.3389/fgene.2022.985500 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baux D. et al. MobiDetails: online DNA variants interpretation. Eur J Hum Genet 29, 356–360, doi: 10.1038/s41431-020-00755-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papageorgiou L. et al. Epione application: An integrated web‑toolkit of clinical genomics and personalized medicine in systemic lupus erythematosus. Int J Mol Med 49, doi: 10.3892/ijmm.2021.5063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherry ST, Ward M. & Sirotkin K. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Research 9, 677–679 (1999). [PubMed] [Google Scholar]
- 11.Cariaso M. & Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Research 40, D1308–D1312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimeno Yepes A. & Verspoor K. Literature mining of genetic variants for curation: quantifying the importance of supplementary material. Database 2014, bau003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naderi N, Mottaz A, Teodoro D. & Ruch P. Analyzing the Information Content of Text-Based Files in Supplementary Materials of Biomedical Literature. Studies in Health Technology Informatics 294, 876–877 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Wei C-H, Allot A, Riehle K, Milosavljevic A. & Lu Z. tmVar 3.0: an improved variant concept recognition and normalization tool. Bioinformatics, submitted (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei C-H, Allot A, Leaman R. & Lu Z. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Research 47, W587–W593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LitVar 2.0 and its data are made freely available to the scientific community at https://www.ncbi.nlm.nih.gov/research/litvar2/.